- 1Department of Medical Research, Taipei Veterans General Hospital, Taipei, Taiwan
- 2Department of Life Sciences and Institute of Genomic Sciences, National Yang-Ming University, Taipei, Taiwan
- 3Institute of Food Safety and Health Risk Assessment, National Yang-Ming University, Taipei, Taiwan
- 4School of Medicine, National Yang-Ming Medical University, Taipei, Taiwan
- 5Institute of Pharmacology, National Yang-Ming University, Taipei, Taiwan
- 6School of Pharmaceutical Sciences, National Yang-Ming University, Taipei, Taiwan
- 7Department of Chest Medicine, Taipei Veterans General Hospital, Taipei, Taiwan
- 8Division of Infectious Diseases, Department of Medicine, Taipei Veterans General Hospital, Taipei, Taiwan
- 9Institute of Clinical Medicine, National Yang-Ming University, Taipei, Taiwan
- 10Department of Pediatrics, Renai Branch, Taipei City Hospital, Taipei, Taiwan
- 11Institute of Microbiology and Immunology, National Yang-Ming University, Taipei, Taiwan
- 12Genomic Research Center, Academia Sinica, Taipei, Taiwan
A sudden outbreak of COVID-19 caused by a novel coronavirus, SARS-CoV-2, in Wuhan, China in December 2019 quickly grew into a global pandemic, putting at risk not only the global healthcare system, but also the world economy. As the disease continues to spread rapidly, the development of prophylactic and therapeutic approaches is urgently required. Although some progress has been made in understanding the viral structure and invasion mechanism of coronaviruses that may cause severe cases of the syndrome, due to the limited understanding of the immune effects caused by SARS-CoV-2, it is difficult for us to prevent patients from developing acute respiratory distress syndrome (ARDS) and pulmonary fibrosis (PF), the major complications of coronavirus infection. Therefore, any potential treatments should focus not only on direct killing of coronaviruses and prevention strategies by vaccine development, but also on keeping in check the acute immune/inflammatory responses, resulting in ARDS and PF. In addition, potential treatments currently under clinical trials focusing on killing coronaviruses or on developing vaccines preventing coronavirus infection largely ignore the host immune response. However, taking care of SARS-CoV-2 infected patients with ARDS and PF is considered to be the major difficulty. Therefore, further understanding of the host immune response to SARS-CoV-2 is extremely important for clinical resolution and saving medication cost. In addition to a breif overview of the structure, infection mechanism, and possible therapeutic approaches, we summarized and compared the hematopathologic effect and immune responses to SARS-CoV, MERS-CoV, and SARS-CoV-2. We also discussed the indirect immune response caused by SARS and direct infection, replication, and destroying of immune cells by MERS-CoV. The molecular mechanisms of SARS-CoV and MERS-CoV infection-induced lymphopenia or cytokine storm may provide some hint toward fight against SARS-CoV-2, the novel coronavirus. This may provide guidance over using immune therapy as a combined treatment to prevent patients developing severe respiratory syndrome and largely reduce complications.
Introduction
Coronaviruses belong to the Coronaviridae family of the subfamily Coronavirinae. The viruses of this family have a broad range of animal hosts, and zoonotic transfer between species is common. Within the Coronavirinae subfamily, there are four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (1, 2). Coronaviruses are non-segmented positive-sense RNA viruses, whose RNA is covered by the solar corona-shaped envelope, from which they acquired their name. They are characterized by having the largest genome among all RNA viruses with an average size of 30 kb (3). Two-thirds of the coronaviral genome encodes non-structural proteins responsible for the virus replication, including RNA-dependent RNA polymerase, proteases, and helicase. The 3′ end of the genome encodes four main structural proteins of the coronavirus particles, which are the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins (4).
Coronaviruses have a long history of infecting humans. HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1 are the prevalent human coronaviruses, which are estimated to have been circulating in the human population for centuries (4). These viruses cause mild upper respiratory infection, or in other words, common cold symptoms (5). On the other hand, three members of the Betacoronavirus genus were zoonotically transferred to humans from other mammalian species in the past two decades and caused major epidemics with high mortality rates. Severe Acute Respiratory Syndrome (SARS), caused by SARS-CoV, started in Guangdong province of China in 2002 and affected 8,096 people worldwide, resulting in 774 deaths (10% mortality rate) (https://www.cdc.gov/sars/about/faq.html). Middle East respiratory syndrome (MERS) caused by MERS-CoV started in Saudi Arabia in 2012 and affected 2,506 people, causing 862 deaths worldwide with a 35% mortality rate (https://www.who.int/csr/don/31-january-2020-mers-united-arab-emirates/en/). In December 2019, a novel coronavirus, SARS-CoV-2, caused an outbreak of Coronavirus Disease 2019 (COVID-19) in Wuhan city in China, which quickly spread throughout the world and grew into a global pandemic affecting hundreds of thousands of people as of March 2020. Notably, although SARS-CoV-2 is characterized by higher contagiousness in comparison with SARS-CoV and MERS-CoV, it causes a much lower mortality rate (2.3% from the epidemic in China in Jan.-Feb, 2020) (6). All three viruses can cause acute respiratory distress syndrome (ARDS), the most acute and fatal stage of the disease, characterized by wide-spread inflammation in the lungs resulting from the aberrant immune response to the viral infection (7–9).
Therefore, in this review, we discuss three coronaviruses, SARS-CoV, MERS-CoV, and SARS CoV-2, from an immunological point of view. We describe their structure and protein composition, mechanisms of entering host cells, and mechanisms to evade innate immune responses. Comparing their hosts, invading mechanisms, and inflammatory responses will help us understand more about coronaviruses, aid in solving the global SARS-CoV-2 epidemic happening now, and find out possible effective treatments to deal with the public health crises caused by coronaviruses in the future.
Virus Structure
As was demonstrated by cryoelectron tomography and cryoelectron microscopy, coronavirus virions are of spherical shape with diameters of approximately 65–125 nm (10). The club-shaped spikes on the surface of the virion are the most prominent feature of coronaviruses. These spikes confer them a solar corona-like appearance from which the name “coronavirus” is derived. The nucleocapsids are helically symmetrical and are packed by the envelope of the virion (5). Coronavirus particles contain four main structural proteins, namely the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins.
S Protein
Coronavirus S protein is a large multifunctional class I viral transmembrane protein, whose size varies from 1,160 amino acids in Infectious Bronchitis Virus (IBV) in poultry to 1,400 amino acids in Feline Coronavirus (FCoV) (11). It is a trimer located on the virion surface, giving the virion a crown-like appearance. As for its function, it mediates the entry of the infectious virion particles into the cells by making attachments between virion particles and host cell membranes through interaction with various host cellular receptors (12). Furthermore, it plays an important role in tissue tropism and the determination of host range (13). In addition, S protein is capable of inducing host immune response (13). S proteins in all coronaviruses can be divided into two domains, S1 and S2 (11). S1 functions as the receptor-binding domain (RBD) while S2 acts as a membrane fusion subunit. The S1 domain can be further divided into two subdomains, named the N-terminal domain (NTD) and the C-terminal domain (CTD). Both of these subdomains act as the receptor-binding domains, interacting efficiently with various host receptors (13). The S1 CTD contains the receptor-binding motif (RBM).
M Protein
The M protein is the most abundant structural protein of the coronavirus virion. It is a small (~25–30 kDa) protein with three transmembrane domains that is responsible for maintaining the shape of the virion (14). The amino acid sequences of the M protein are diverse in different coronaviruses, however, the structural similarity is maintained overall (15). It has a short N-terminal glycosylated domain outside the virion and a much larger C-terminal domain inside the virion that extends 6–8 nm into the viral particle (16). Most M proteins are co-translationally inserted into the ER membrane without a signal sequence. The viral scaffold is maintained by interactions between M proteins. Recent studies suggest that the M protein exists as a dimer in the virion, and may adopt two different conformations allowing it to promote membrane curvature, as well as bind to the nucleocapsid (14).
E Protein
The E protein is the smallest structural protein (~8–12 kDa) within the virion. It plays a multifunctional role in the pathogenesis, assembly, and release of the virus. The virulence of the virus is also related to the E protein (17). The E proteins from different coronaviruses are highly diverse in their amino acid sequences but are characterized by a common structure (18). There are three domains in the E protein: short hydrophilic amino-terminal domain, large hydrophobic transmembrane domain, and C terminal domain (19). The deletion of the E protein-encoding gene results in slower amplification of the virus, but the protein does not seem to be essential for the replication of SARS-CoV (20). Besides its role in assembly and release of the virus, the E protein still has other functions, for instance, the ion channel activity. Compared to SARS-CoV, the SARS-CoV-2 (2019-nCoV) E protein reveals a similar amino acid constitution without any substitution (21).
N Protein
The N protein is the only structural protein present in the nucleocapsid. It is composed of three highly conserved and separate domains: an N-terminal domain (NTD), RNA-binding domain or a linker region (LKR), and a C-terminal domain (CTD) (22). The NTD binds to the 3′ end of the viral RNA and is highly divergent from virus to virus (23). The LKR region [also called SR (Serine and Arginine) domain] is charged because of its serine and arginine-rich sequence (24). It has been reported to interact directly with RNA in vitro and play a part in cell signaling (25, 26). The N protein has two RNA substrates that have already been identified, the transcriptional regulatory sequence (TRS) (25) and the genomic packaging signal (27). In addition, it can also act as a viral suppressor of RNA silencing in mammalian cells (28). N protein is also heavily phosphorylated (29), so that it can change its conformation to enhance the affinity for viral vs. non-viral RNA. N protein also binds nsp3 (24, 30) and the M protein (31). These proteins may interact to help tether the viral genome packaging.
HE Protein
The hemagglutinin-esterase (HE) is a structural protein present in a subset of Betacoronavirus. The protein acts as a hemagglutinin, which binds sialic acids of surface glycoproteins. It also contains acetylesterase activity (32). These activities are thought to enhance the cell entry mediated by the S protein and virus spread through the mucosa (33).
Structure OF SARS-CoV
SARS-CoV virus particles are spherical with an average diameter of 78 nm. The virus contains a helical nucleocapsid, surrounded by an envelope (34), covered with rod-shaped long envelope particles of about 20 nm in length, with typical coronal features. The structure of SARS-CoV is similar to that of other coronaviruses. The gene sequence is 5′ end, replicase [rep], spike [S], envelope [E], membrane [M], nucleocapsid [N], 3′ end. There are short untranslated regions at both ends. The sequences of the other five non-structural proteins may be distributed between ORF S and N (35).
The SARS-CoV genome contains a total of 11 ORFs and encodes 23 mature proteins (36). Among them, two major ORFs (ORF1a and ORF1b) account for about two-thirds of the genome size and encode two important polyproteins, pp1a and pp1ab. Polyproteins are proteolytically cleaved to produce non-structural proteins, the most important of which are RNA-dependent RNA polymerase and ATPase helicase. Only several nucleotides are different among different viruses (37).
Structure OF MERS-CoV
The genome of MERS-CoV consists of genes encoding the replicase and structural proteins (spike-envelope-membrane-nucleocapsid)-poly (A)−3′, similar to other coronaviruses. The virus has 10 ORFs and encodes 16 putative non-structural proteins involved in the viral transcription and replication process (38, 39).
Structure OF SARS-CoV-2
Basically, the structure of SARS-CoV-2 shares all the typical characteristics with other coronaviruses. Several recent studies considering the structure of SARS-CoV-2 were all focused on the S protein. Wrapp et al. (40) reported a structure at 3.5 Å resolution of SARS-CoV-2 S protein. Yan et al. (41) reported the complex structure of B0AT1, an amino acid transporter protein, with human host cell binding receptor angiotensin-converting enzyme 2 (ACE2), which provided important insights into the molecular basis of coronavirus infection. Lan et al. (42) reported a crystal structure of SARS-CoV-2 S protein's receptor binding domain (RBD) region bound to ACE2. The viral architecture of SARS-CoV-2 with post-fusion spike was observed by Cyro-EM, which showed the image of disassociated spikes (43).
Infection (Entering Host Cells)
SARS-CoV
Similar to other coronaviruses, SARS-CoV enters cells through endocytosis and membrane fusion, and its host receptor is ACE2 (35, 44). SARS-CoV enters into target cells and can be inhibited by polyanionic compounds, suggesting that the SARS-CoV envelope protein may be positively charged. At the same time, SARS-CoV needs to be in the acidified endosome to produce effective infection, indicating that its effect is pH-dependent (45). Viral RNA is replicated in the unique bottle-shaped bilayer membrane compartments (46). Several studies have found that SARS-CoV infection can cause ultrastructural changes in vivo and in cultured cells, including the formation of double-membrane vesicles and nucleocapsid inclusions and particles in the cytoplasm (34).
MERS-CoV
MERS-CoV has been reported as being able to infect and kill not only alveolar epithelial cells but also T cells (47). MERS-CoV enters host cells by binding to a DPP4 receptor expressed in the kidney and other organs (48), and uses proteases of the host to enter lung cells. Furin activates the S protein on the viral envelope, mediating the membrane fusion and virus entry into host cells (49). Like SARS-CoV, MERS-CoV can overcome the host's natural immune response, produce high virus titers, and induce cytokine imbalance (38, 50).
SARS-CoV-2
SARS-CoV-2 is mainly considered to infect respiratory epithelial cells, but a recent study confirmed that it can also infect T lymphocytes (51), spleens, and lymph nodes (52). There are already some solid studies that confirm that ACE2 serves as the receptor for the entry of SARS-CoV-2. Analysis of the receptor binding motif (RBM), a portion of the receptor binding domain (RBD) that makes contact with ACE2 (53), revealed that most amino acid residues essential for ACE2 binding by SARS-CoV were conserved in SARS-CoV-2. Hoffmann et al. blocked ACE2 in Vero cells and found that both SARS-CoV and SARS-CoV-2 infection was dramatically inhibited, and that serine protease TMPRSS2 played an important role in SARS-CoV-2's infection (54). The difference of the host cells among SARSCoV, MERS-CoV and SARS-CoV-2 is summarized in Figure 1.
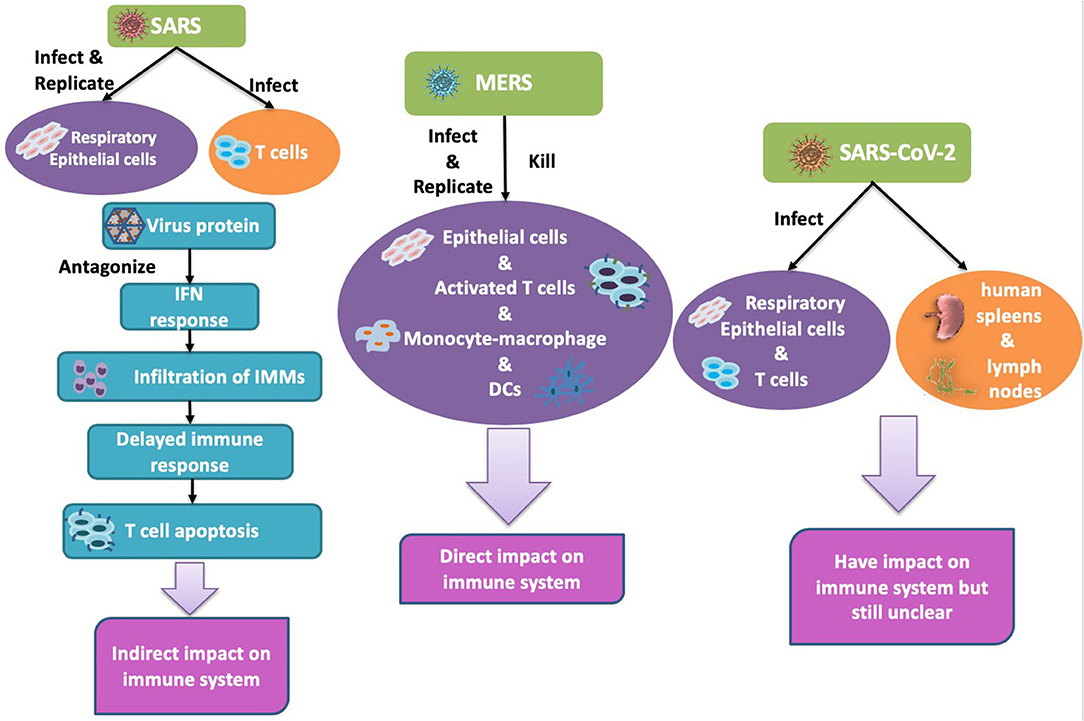
Figure 1. Human coronavirus infects different types of cells. Left: SARS-CoV can infect alveolar epithelial cells and immune cells but can only replicate in epithelial cells. Middle: MERS-CoV infected and replicated in both alveolar epithelial cells and immune cells. Right: SARS-CoV2: infected lung and damaged lung and immune system.
ACE2 Receptor
Angiotensin-converting enzyme 2 (ACE2) is an essential component of the renin-angiotensin system (55). It was shown to bind to the S protein of SARS-CoV in 2003 by mass spectrometry (56) and was also confirmed to be a receptor of SARS-CoV-2 required to enter human cells (57). Xu et al. (58) drafted the currently available world's largest human kidney cell atlas with 42,589 cells and identified 19 clusters through unsupervised hierarchical clustering analysis. ACE2 and TMPRSS genes were significantly co-expressed in podocytes and proximal convoluted tubules as potential host cells targeted by SARS-CoV-2. Comparative analysis showed that ACE2 expression in kidney cells was no less than that in the lung, esophagus, small intestine, and colon, suggesting that the kidney may be an important target organ for SARS-CoV-2.
As for the susceptibility of different population groups to SARS-CoV-2, Chen et al. (59) showed that the expression of ACE2 in Asians was similar to that in other races, and was also not related to sex. Surprisingly, ACE2 was shown to be significantly upregulated after virus infection, including SARS-CoV and SARS-CoV-2 (60). According to the public data analysis, the level of ACE2 expression in adipose tissue was higher than that in lung tissue, which was indicative of the possibility that adipose tissue was also a potential target of SARS-CoV-2 (61).
Inflammatory Response to Coronavisuses
Human coronaviruses can be divided into two groups by their pathogenicity. Whereas, low pathogenic coronaviruses (HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1) cause mild cold-like respiratory illness, the highly pathogenic SARS-CoV, MERS-CoV and SARS-CoV-2 cause immunopathological events that result in fatal pneumonia. The invasion of such coronaviruses is associated with severe immune responses, which may eventually lead to acute respiratory distress syndrome (ARDS). The innate immune system constitutes the primary line of defense against the invading viruses. The pathogen-associated molecular patterns (PAMPs), represented by the viral RNA or dsRNA formed during viral replication, are recognized by intracellular sensors such as RIG-I and MDA5. After recognition, the downstream signaling cascade results in activation of NF-κB and IRF3 transcriptional activity (62). This leads to the expression of type I interferon (IFN) and pro-inflammatory cytokines, which constitute the defense line against the virus infection at an early stage (63).
SARS-CoV and MERS-CoV have evolved a number of strategies to suppress type I IFN response during their invasion. SARS-CoV can interfere with the downstream signaling of the RNA sensors, including MAVS and TRAF3/6, directly or indirectly (64). As for MERS-CoV, it can downregulate interferon-stimulated genes (ISG) by activating repressive histone modification as its strategy (64).
As part of the adaptive immunity, T cells also play important roles in the primary defense line against coronaviruses. There are many T cell epitopes identified to induce an IFN-γ-specific T cell response or cytotoxic T lymphocyte (CTL) response. Studies have found that epitopes in the S protein (64, 65) and the N protein of coronaviruses (66, 67) can induce antibody responses in both mice models or patients. IgM and IgG, produced by B lymphocytes, are formed after the infection of coronaviruses (68, 69). The induction of IgM is an early and transient response to neoantigens, which is later replaced by the induction of IgG to play the role as the predominant and long-term antibody. IgG is characterized by a longer half-life and lower molecular weight, which gives it the ability to provide long-lasting protection and effective tissue penetration (68).
The Immune Response to SARS-CoV
The combined induction of antibodies and virus-specific T cells provides optimal protective immunity. Following the infection, a strong humoral immune response with a high titer of neutralization antibodies targeting the SARS-CoV S protein that show a protective effect are found in the serum of most patients. In addition, CD4+ T cells targeting N protein and HLA-A2 restricted CD8+ T cells targeting S protein were observed in SARS patients (70–72). However, the dramatic loss of CD4+ T cells (in ~90–100% of patients) and CD8+ T cells (in ~80–90% of patients) was observed in the acute phase of SARS patients (73). The delayed adaptive immune response resulted in prolonged virus clearance and correlated with the severity of the SARS disease (74). One possible reason for the decreased number of T cells is that after infecting alveolar epithelial cells, SARS-CoV encodes multiple structural and non-structural proteins that antagonize innate IFN response (75–78). The delayed IFN response orchestrates infiltration of pathogenic inflammatory monocyte-macrophages (IMMs) and elevation of pro-inflammatory cytokines (79). IMM-derived pro-inflammatory cytokines, such as type I INF (80), may sensitize T cells to undergo apoptosis through Bim (81) or Bcl-xL (82)-mediated intrinsic pathway by E protein, thus consequently impeding the viral clearance (79). Depletion of IMMs or neutralization of pro-inflammatory cytokines was shown to protect mice from lethal SARS-CoV infection (79). Another possible explanation of the reduction of virus-specific T cells is the alteration in antigen presenting cell (APC) function and impaired dendritic cell (DC) migration, resulting in the reduced priming of T Cells (83, 84). This mechanism was supported by animal studies using SARS-CoV-MA15, the mouse-adapted strain of SARS-CoV. Inefficient activation of respiratory DCs by SARS-CoV-MA15 attributed to poor virus-specific CD4+ and CD8+ T cells responses (84). Moreover, the age-dependent reduction in the magnitude of T cell response may also explain the higher susceptibility to SARS-CoV with advanced age (85). Consistently, depletion of CD4+ T cells delayed SARS-CoV (Urbani strain) clearance and enhanced pneumonitis (86). In contrast, transfer of SARS-CoV-specific CD4+ and CD8+ T cells resulted in rapid virus clearance and amelioration of the disease (87). Mechanistically, the pattern recognition receptors such as MyD88 (88) and TRIF (89) are required for protection against SARS-CoV infection.
In addition to the humoral response, a 3-year follow-up study of 176 SARS patients showed that the level of IgM peaked at ~1 month after symptoms onset, and IgG peaked at 2–4 months (90). Patients with a longer illness period showed a lower neutralizing antibody response compared to patients with a shorter illness duration (91). It was reported that vaccine-elicited, neutralizing monoclonal antibody (MAb) targeting the S protein of SARS-CoV facilitates viral entry into host cells and enhances viral infectivity (92). This phenomenon is the so called antibody-dependent enhancement (ADE) (69), which is regarded as a great burden for vaccine development.
The Immune Response to MERS-CoV
The immune response mechanism triggered by MERS-CoV has still not been fully studied. It is known that the S protein of MERS-CoV can upregulate the levels of the repressors of the TLR signaling pathways, such as of IL-1R-associated kinase (IRAK-M) and peroxisome proliferator-activated receptor-gamma (PPARY). IRAK-M and PPARY negatively regulate IRF7, which normally induces the expression of IFN-alpha and IFN-beta (93). If these negative regulators can maintain their persistence in the long-term, the clearance of MERS-CoV infections will be impaired.
Comparatively less is known about the fate of T cells in MERS-CoV infection and little information is known about the recognized epitopes (72). Similar to SARS-CoV, MERS-CoV-specific CD8+ T cells are also important for clearing the virus (94). Though both SARS-CoV and MERS-CoV infects monocyte-macrophages, DCs, and activated T cells, only MERS-CoV was able to replicate in the infected immune cells, which consequently resulted in aberrant induction of inflammatory cytokines in macrophages and DCs (95, 96) and of both extrinsic and intrinsic apoptosis pathway in T cells (47). Such active replication of MERS-CoV in these immune cells may underlie the comparatively higher fatality rate of MERS disease.
As for humoral immunity, antibody response to MERS-CoV is typically detected on the second and third week after the onset of infection. But the longevity of the antibodies seemed to be correlated to the severity of disease. In patients who had pneumonia caused by MERS-CoV, the antibodies were still detectable 13 months after infection (97). However, in patients after mild or subclinical infection of MERS-CoV, MERS antibodies were detected at low levels (98). Similar to SARS-CoV, MAb that has a strong binding affinity to the spike protein of MERS-CoV also facilitates ADE viral entry into host cells (99).
The Immune Response to SARS-CoV-2
According to case reports, the pathogenesis of SARS-CoV-2 includes immunological responses of both innate and adaptive immunity systems.
Compared to normal patients, patients requiring ICU admission had higher concentrations of GCSF, IP10, MCP1, MIP1A, and TNFα. These cytokines may help to judge the condition of patients (100).
Secondary hemophagocytic lymphohistiocytosis (sHLH), which is mostly triggered by virus infection in adults, is a condition in which the body makes too many activated immune cells (macrophages and lymphocytes) (https://primaryimmune.org/disease/hemophagocytic-lymphohistiocytosis-hlh). The cytokine profile of sHLH is associated with the severity of COVID-19, which is characterized by increased interleukin (IL)-2, IL-7, GCSF, IP10 (CXCL10), MCF1 (CCL2), MIP1A (CCL3), and TNF-α (100, 101). Therefore, it is possible that this phenomenon happens in COVID-19 patients. The current explanation for the sHLH phenomenon is that the body has experienced a cytokine storm caused by excessive immunity. However, the details of immune and inflammatory response to SARS-CoV-2 infection are still under scrutiny.
Similar to SARS-CoV, SARS-CoV-2 also targeted pneumocytes (both types I and II) and alveolar macrophages (102). Consistently, pathological examination of patients who were infected by SARS-CoV-2 revealed the infiltration of plasma cells and macrophages and a high density of macrophages and foam cells in the alveolar cavities (103). However, compared with SARS-CoV, SARS-CoV-2 did not significantly induce types I, II, or III interferons in the infected human lung tissues (102). As cytokine storm may be the main cause for the severity of the coronavirus infection, these findings support the relevant severity of SARS-CoV and SARS-CoV-2.
As for adaptive immunity, it is known that the low levels of CD4+ and CD8+ T cells are related to the mortality of SARS-CoV-2 patients (104, 105). The up-regulation of apoptosis and autophagy in PBMC of SARS-CoV-2 patients (106) suggested that, similar to how MERS can directly infect T cells and induce apoptosis (47), SARS-CoV-2 may cause lymphocytopenia through inducing T-cell apoptosis or autophagic cell death. It was supported by a recent report showing that SARS-CoV-2 could infect T cells through receptor-dependent or S protein-mediated membrane fusion (51).
As for the antibody response, it was reported that in 23 patients with COVID-19, the viral load peaked during the first week and then began to fall. Both IgG and IgM antibodies which targeted the nucleoprotein and the surface spike receptor began to rise around 10 days after symptom onset, and the seroconversion of most patients happened within the first 3 weeks (107). Another study among 173 patients reported that the seroconversion rate of Ab, IgM, and IgG was 93.1, 82.7, and 64.7%, respectively. And the median seroconversion time for Ab, IgM, and IgG were day 11, day 12, and day 14 after onset, respectively (108). Whether ADE can happen in SARS-CoV-2 infection is still not confirmed, but as humans have already experienced a SARS-CoV epidemic and several other coronavirus infection such as 229E (109), according to former studies of SARS-CoV (92), it is possible that ADE can also happen in the infection of SARS-CoV-2 (110).
Discussion and Summary
The recent outbreak of the SARS-CoV-2 infection has caused a worldwide crisis in the epidemiology and medical systems. Since SARS-CoV-2 was confirmed to share the same host receptor, ACE2, with SARS-CoV, the strategies used to tackle SARS-CoV are under investigation for treating SARS-CoV-2 infection. However, despite both attacking lungs and using the same host receptor to enter target cells, the three coronaviruses causing three serious pneumonia epidemics are different in the range of infected cell types and their effects on infected cells.
SARS-CoV is mainly replicated in respiratory epithelial cells, though it can also infect a variety of immune cells such as monocytes, macrophages, dendritic cells, and activated T cells (111–114). MERS-CoV, in contrast, not only infects the immune cells and epithelial cells, but is also able to replicate in the former cells and lyse them, which may be one of the reasons for the high mortality of MERS (47, 96, 115). The details of the infection and lytic replication mechanisms of SARS-CoV-2 in host cells are currently unclear (Table 1). However, diarrhea, liver and kidney damage, and hemophagocytic lymphohistiocytosis have been reported in patients with SARS-CoV-2, indicating that the host cell range of the virus may be wider than currently recognized (100).
Following the invasion of a pathogen, the host triggers a serious response from the immune system. SARS-CoV does not directly lyse and kill T cells, but indirectly induces T cell apoptosis (82). MERS-CoV, which is a more severe and aggressive virus, directly targets T cells and undergoes lytic replication, thus directly causing their death (47). Therefore, MERS-CoV has a direct impact on the immune system. SARS-CoV-2 is reported to infect T lymphocytes (51), but we still don't know how this affects the immune system in particular (Figure 2). A severe reduction of immune cells was observed in patients infected with SARS-CoV-2, but whether this phenomenon is directly caused by the virus or indirectly caused by dysregulated cytokine production by immune cells has yet to be determined (100). Direct viral effects require treatment strategies that target viral replication. Indirect viral effects through dysregulated cytokine production by residential macrophages/dendritic cells or antigen presentation by APCs can now be treated by immune therapy approaches. Understanding the behavior of SARS-CoV-2 in the host cells of the human body and its effect on the immune system may provide important tips for combating the disease.
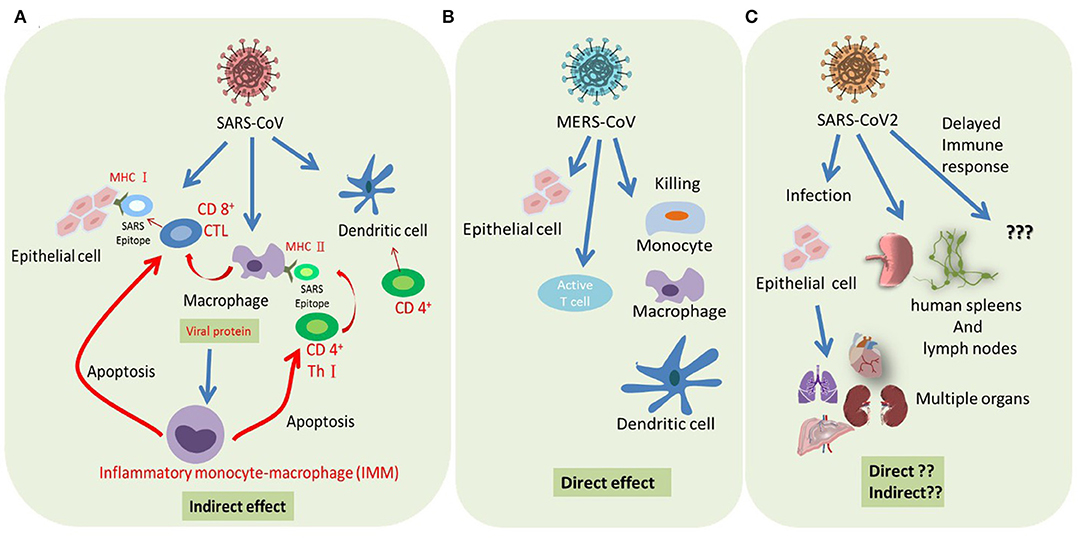
Figure 2. Summary of host immune response modulated by severe coronaviruses. (A) SARS-CoV infected epithelial cells represents SARS epitope by MHC I to recruit CD8+ cytotoxic T cells (CTL). Macrophage and dendritic cells (DCs) are infected by SARS-CoV and represent SARS epitope by MHC II to recruit CD4+ helper T cells (Th1). Abortive replication of SARS in macrophage impaired its cytokine production, resulting in a delayed IFN response, infiltration of inflammatory monocyte-macrophages (IMMs), and T cells apoptosis. In addition, SARS-CoV infection impaired dendritic cell (DC) function, resulting in reduced T cell activation. (B) Successful replication of MERS-CoV in both alveolar epithelial cells and immune cells resulted in the direct killing of these infected cells. (C) SARS-CoV-2 can probably infect both lung epithelial cells and immune cells and damage the tissue through a direct or cytokine-mediated indirect effect.
There have already been some studies showing that other tissues and organs may also be the target of SARS-CoV-2, further reminding us to focus on other organs besides lungs, such as the kidneys, spleen, and lymph nodes (52, 116). This may give us some hints for the multiple organ dysfunction syndrome in some severe COVID-19 cases (117). Moreover, research on the expression pattern of ACE2 in different population groups and races indicates that there is no sex or race bias in susceptibility to SARS-CoV-2 (118).
It is still unclear whether reinfection with SARS-CoV-2 can occur in recovered patients. There has been some news about “reoccurring COVID-19 cases” (https://www.scmp.com/news/china/society/article/3065091/coronavirus-recovered-patient-dies-china-reports-139-new-cases), but as they are not formal case reports, it is not certain whether these patients had fully recovered from COVID-19 before the symptoms relapse. In a study on rhesus macaques re-exposed to SARS-CoV-2 after disappearance of symptoms and positive antibody response to primary infection, no evidence of reinfection was found (119). Also, how long the antibodies will remain in recovered patients is still unclear. Since the vaccine against COVID-19 has still not been developed, the recommendations of the CDC, which advises people to wear cloth face masks and keep a 6-foot distance from others, should be the best way to prevent reinfection (https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/diy-cloth-face-coverings.html).
To deepen the research on coronavirus, humans must learn enough lessons from this pandemic. In the wave of globalization and scientific and technological progress, infectious diseases have become more prone to spreading, which has made it harder for humans to deal with them. How to understand the infectious biomolecular mechanism and immune pathological environment in the future will be a more important proposition for us than ever before. According to China's response to the epidemic, it did not take long to identify what the pathogen was, but the imperfect public health emergency system is the main reason for the spread of the epidemic. Therefore, in addition to developing drugs, vaccines, and updating treatment plans, scholars should also call on governments to strengthen the construction and improvement of social public health emergency systems to prevent similar pandemics from happening again.
Author Contributions
S-HC and P-CC designed, constructed, and organized this work, and reviewed article. YL, M-LW, C-SC, Y-PY, and AY collected all references and wrote up the manuscript. W-YL, Y-HL, Y-TL and Y-JC prepared the study materials and organized figures and table.
Funding
This work was funded by Ministry of Science and Technology (MOST 108-2319-B-001-004, MOST 108-2119-M-010-001, MOST 108-2320-B-010-006, MOST 108-2320-B-010 -019 -MY3, MOST 107-2923-B-010 -002 -MY2, MOST 108-2321-B-006-032, and MOST 108-2321-B-010-006) and Taipei Veterans General Hospital (V107E-002-2, V108D46-004-MY2-1, V108E-006-4, 108E-006-5 and 109VACS-003), Taiwan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. New Engl J Med. (2020) 382:727–33. doi: 10.1056/NEJMoa2001017
2. Chen Y, Liu Q, Guo D. Coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. (2020) 92:418–23. doi: 10.1002/jmv.25681
3. Chan JF-W, Kok K-H, Zhu Z, Chu H, To KK-W, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. (2020) 9:221–36. doi: 10.1080/22221751.2020.1719902
4. Pyrc K, Dijkman R, Deng L, Jebbink MF, Ross HA, Berkhout B, et al. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J Mol Biol. (2006) 364:964–73. doi: 10.1016/j.jmb.2006.09.074
5. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biology Clifton N J. (2015) 1282:1–23. doi: 10.1007/978-1-4939-2438-7_1
6. Wu Z, McGoogan JM. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. (2020) 323:1239–42. doi: 10.1001/jama.2020.2648
7. Cleri DJ, Ricketti AJ, Vernaleo JR. Severe acute respiratory syndrome (SARS). Infect Dis Clin N Am. (2010) 24:175–202. doi: 10.1016/j.idc.2009.10.005
8. Naeem Z. Middle East Respiratory Syndrome (MERS) : an update. Int J Heal Sci. (2013) 7:5–6. doi: 10.12816/0006053
9. Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. (2020) doi: 10.1016/S2213-2600(20)30079-5
10. Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. (2020) 24:91–8. doi: 10.1016/j.jare.2020.03.005
11. Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. (2012) 4:1011–33. doi: 10.3390/v4061011
12. Beniac DR, Andonov A, Grudeski E, Booth TF. Architecture of the SARS coronavirus prefusion spike. Nat Struct Mol Biol. (2006) 13:751–2. doi: 10.1038/nsmb1123
13. Li F. Structure, function, and evolution of Coronavirus spike proteins. Ann Rev Virol. (2015) 3:1–25. doi: 10.1146/annurev-virology-110615-042301
14. Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. (2010) 174:11–22. doi: 10.1016/j.jsb.2010.11.021
15. Arndt AL, Larson BJ, Hogue BG. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J Virol. (2010) 84:11418–28. doi: 10.1128/JVI.01131-10
16. Nal B, Chan C, Kien F, Siu L, Tse J, Chu K, et al. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J Gen Virol. (2005) 86:1423–34. doi: 10.1099/vir.0.80671-0
17. Nieto-Torres JL, DeDiego ML, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PloS Pathog. (2014) 10:e1004077. doi: 10.1371/journal.ppat.1004077
18. Godet M, L'Haridon R, Vautherot J-F, Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. (1992) 188:666–75. doi: 10.1016/0042-6822(92)90521-P
19. Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. (2019) 16:69. doi: 10.1186/s12985-019-1182-0
20. DeDiego ML, Alvarez E, Almazan F, Rejas MT, Lamirande E, Roberts A, et al. A severe acute respiratory syndrome Coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol. (2006) 81:1701–13. doi: 10.1128/JVI.01467-06
21. Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, et al. Genome composition and divergence of the novel Coronavirus (2019-nCoV) originating in China. Cell Host Microbe. (2020) 27:325–8. doi: 10.1016/j.chom.2020.02.001
22. McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. (2014) 6:2991–3018. doi: 10.3390/v6082991
23. Fan H, Ooi A, Tan YW, Wang S, Fang S, Liu DX, et al. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. (2005) 13:1859–68. doi: 10.1016/j.str.2005.08.021
24. Hurst KR, Koetzner CA, Masters PS. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J Virol. (2009) 83:7221–34. doi: 10.1128/JVI.00440-09
25. Stohlman SA, Baric RS, Nelson GN, Soe LH, Welter LM, Deans RJ. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol. (1988) 62:4288–95. doi: 10.1128/JVI.62.11.4288-4295.1988
26. You J, Dove BK, Enjuanes L, DeDiego ML, Alvarez E, Howell G, et al. Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J Gen Virol. (2005) 86:3303–10. doi: 10.1099/vir.0.81076-0
27. Molenkamp R, Spaan WJM. Identification of a specific interaction between the Coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. (1997) 239:78–86. doi: 10.1006/viro.1997.8867
28. Cui L, Wang H, Ji Y, Yang J, Xu S, Huang X, et al. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J Virol. (2015) 89:9029–43. doi: 10.1128/JVI.01331-15
29. Stohlman SA, Lai MM. Phosphoproteins of murine hepatitis viruses. J Virol. (1979) 32:672–75. doi: 10.1128/JVI.32.2.672-675.1979
30. Hurst KR, Koetzner CA, Masters PS. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol. (2013) 87:9159–72. doi: 10.1128/JVI.01275-13
31. Sturman LS, Holmes KV, Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. (1980) 33:449–62. doi: 10.1128/JVI.33.1.449-462.1980
32. Klausegger A, Strobl B, Regl G, Kaser A, Luytjes W, Vlasak R. Identification of a Coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J Virol. (1999) 73:3737–43. doi: 10.1128/JVI.73.5.3737-3743.1999
33. Cornelissen LA, Wierda CM, van der Meer FJ, Herrewegh AA, Horzinek MC, Egberink HF, et al. Hemagglutinin-esterase, a novel structural protein of torovirus. J Virol. (1997) 71:5277–86. doi: 10.1128/JVI.71.7.5277-5286.1997
34. Goldsmith CS, Tatti KM, Ksiazek TG, Rollin PE, Comer JA, Lee WW, et al. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. (2004) 10:320–6. doi: 10.3201/eid1002.030913
35. Du L, He Y, Zhou Y, Liu S, Zheng B-J, Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat Rev Microbiol. (2009) 7:226–36. doi: 10.1038/nrmicro2090
36. Ruan Y, Wei CL, Ling AE, Vega VB, Thoreau H, Thoe SYS, et al. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. (2003) 361:1779–85. doi: 10.1016/S0140-6736(03)13414-9
37. Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. (2003) 300:1394–9. doi: 10.1126/science.1085952
38. Skariyachan S, Challapilli SB, Packirisamy S, Kumargowda ST, Sridhar VS. Recent aspects on the pathogenesis mechanism, animal models and novel therapeutic interventions for middle east respiratory syndrome Coronavirus infections. Front Microbiol. (2019) 10:569. doi: 10.3389/fmicb.2019.00569
39. Chan JF-W, Yao Y, Yeung M-L, Deng W, Bao L, Jia L, et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis. (2015) 212:1904–13. doi: 10.1093/infdis/jiv392
40. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Sci New York N Y. (2020) 367:1260–3. doi: 10.1126/science.abb2507
41. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Sci New York N Y. (2020) 367:1444–8. doi: 10.1126/science.abb2762
42. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Crystal structure of the 2019-nCoV spike receptor-binding domain bound with the ACE2 receptor. Biorxiv. (2020). doi: 10.1101/2020.02.19.956235
43. Liu C, Yang Y, Gao Y, Shen C, Ju B, Liu C, et al. Viral architecture of SARS-CoV-2 with post-fusion spike revealed by Cryo-EM. Biorxiv. (2020). doi: 10.1101/2020.03.02.972927
44. Kuba K, Imai Y, Ohto-Nakanishi T, Penninger JM. Trilogy of ACE2: a peptidase in the renin-angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol Ther. (2010) 128:119–28. doi: 10.1016/j.pharmthera.2010.06.003
45. Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci USA. (2004) 101:4240–5. doi: 10.1073/pnas.0306446101
46. Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol. (2002) 76:3697–708. doi: 10.1128/JVI.76.8.3697-3708.2002
47. Chu H, Zhou J, Wong BH-Y, Li C, Chan JF-W, Cheng Z-S, et al. Middle east respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis. (2015) 213:904–14. doi: 10.1093/infdis/jiv380
48. Abdel-Moneim AS. Middle East respiratory syndrome coronavirus (MERS-CoV): evidence and speculations. Arch Virol. (2014) 159:1575–84. doi: 10.1007/s00705-014-1995-5
49. Banik GR, Khandaker G, Rashid H. Middle East respiratory syndrome coronavirus “MERS-CoV”: current knowledge gaps. Paediatr Respir Rev. (2015) 16:197–202. doi: 10.1016/j.prrv.2015.04.002
50. Gralinski LE, Baric RS. Molecular pathology of emerging coronavirus infections. J Pathol. (2015) 235:185–95. doi: 10.1002/path.4454
51. Wang X, Xu W, Hu G, Xia S, Sun Z, Liu Z, et al. SARS-CoV-2 infects T lymphocytes through its spike protein-mediated membrane fusion. Cell Mol Immunol. (2020) 1–3. doi: 10.1038/s41423-020-0424-9
52. Yongwen C, Feng Z, Diao B, Wang R, Wang G, Wang C, et al. The novel severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. Medrxiv. (2020). doi: 10.1101/2020.03.27.20045427
53. Li W, Sui J, Huang I-C, Kuhn JH, Radoshitzky SR, Marasco WA, et al. The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2. Virology. (2007) 367:367–74. doi: 10.1016/j.virol.2007.04.035
54. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. (2020) 181:271–80.e8. doi: 10.1016/j.cell.2020.02.052
55. Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, et al. A novel Angiotensin-Converting Enzyme–related carboxypeptidase (ACE2) converts angiotensin i to angiotensin 1-9. Circ Res. (2000) 87:E1–9. doi: 10.1161/01.RES.87.5.e1
56. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. (2003) 426:450–4. doi: 10.1038/nature02145
57. Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. (2020) 579:270–3. doi: 10.1038/s41586-020-2012-7
58. Xu D, Zhang H, Gong H, Chen J, Ye J, Meng T, et al. Identification of a potential mechanism of acute kidney injury during the Covid-19 outbreak: a study based on single-cell transcriptome analysis. Intens Care Med. (2020) 2020020331. doi: 10.1007/s00134-020-06026-1
59. Chen Y, Shan K, Qian W. Asians and other races express similar levels of and share the same genetic polymorphisms of the SARS-CoV-2 cell-entry receptor. Preprints. (2020) 2020020258. doi: 10.20944/preprints202002.0258.v1
60. Wang P-H, Cheng Y. Increasing host cellular receptor—Angiotensin-Converting Enzyme 2 (ACE2) expression by Coronavirus may facilitate 2019-nCoV infection. Biorxiv. (2020). doi: 10.1101/2020.02.24.963348
61. Jia X, Yin C, Lu S, Chen Y, Liu Q, Bai J, et al. Two things about COVID-19 might need attention. Preprints. (2020) 2020020315. doi: 10.20944/preprints202002.0315.v
62. Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy. (2020) 38:1–9. doi: 10.12932/AP-200220-077
63. Wit E de, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. (2016) 14:523–34. doi: 10.1038/nrmicro.2016.81
64. Lu L, Manopo I, Leung BP, Chng HH, Ling AE, Chee LL, et al. Immunological characterization of the spike protein of the severe acute respiratory syndrome Coronavirus. J Clin Microbiol. (2004) 42:1570–6. doi: 10.1128/JCM.42.4.1570-1576.2004
65. Buchholz UJ, Bukreyev A, Yang L, Lamirande EW, Murphy BR, Subbarao K, et al. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc National Acad Sci USA. (2004) 101:9804–9. doi: 10.1073/pnas.0403492101
66. Peng H, Yang L, Wang L, Li J, Huang J, Lu Z, et al. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology. (2006) 351:466–75. doi: 10.1016/j.virol.2006.03.036
67. Leung DTM, Tam FCH, Ma CH, Chan PKS, Cheung JLK, Niu H, et al. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J Infect Dis. (2004) 190:379–86. doi: 10.1086/422040
68. Watson JG, Bird AG. (eds). “Chapter 1 - Basic immunology,' in Handbook of Immunological Investigations in Children (Butterworth-Heinemann) (1990).
69. Tirado S, Yoon K. Antibody-dependent enhancement of virus infection and disease. Viral Immunol. (2003) 16:69–86. doi: 10.1089/088282403763635465
70. Wang Y-D, Sin W-YF, Xu G-B, Yang H-H, Wong T, Pang X-W, et al. T-cell epitopes in Severe Acute Respiratory Syndrome (SARS) Coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J Virol. (2004) 78:5612–18. doi: 10.1128/JVI.78.11.5612-5618.2004
71. Oh H-LJ, Gan SK-E, Bertoletti A, Tan Y-J. Understanding the T cell immune response in SARS coronavirus infection. Emerg Microbes Infec. (2012) 1:e23. doi: 10.1038/emi.2012.26
72. Liu WJ, Zhao M, Liu K, Xu K, Wong G, Tan W, et al. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antivir Res. (2017) 137:82–92. doi: 10.1016/j.antiviral.2016.11.006
73. Li T, Qiu Z, Zhang L, Han Y, He W, Liu Z, et al. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. (2004) 189:648–51. doi: 10.1086/381535
74. Li T, Qiu Z, Han Y, Wang Z, Fan H, Lu W, et al. Rapid loss of both CD4+ and CD8+ T lymphocyte subsets during the acute phase of severe acute respiratory syndrome. Chin Med J. (2003) 116:985–7.
75. Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. Severe acute respiratory syndrome Coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/golgi membrane. J Virol. (2007) 81:9812–24. doi: 10.1128/JVI.01012-07
76. Kindler E, Thiel V, Weber F. Advances in virus research. Adv Virus Res. (2016) 96:219–43. doi: 10.1016/bs.aivir.2016.08.006
77. Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng C-TK, et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol. (2008) 82:4471–9. doi: 10.1128/JVI.02472-07
78. Totura AL, Baric RS. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol. (2012) 2:264–75. doi: 10.1016/j.coviro.2012.04.004
79. Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, et al. Dysregulated Type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. (2016) 19:181–93. doi: 10.1016/j.chom.2016.01.007
80. Welsh RM, Bahl K, Marshall HD, Urban SL. Type 1 interferons and antiviral CD8 T-cell responses. Plos Pathog. (2012) 8:e1002352. doi: 10.1371/journal.ppat.1002352
81. Bahl K, Hüebner A, Davis RJ, Welsh RM. Analysis of apoptosis of memory T cells and dendritic cells during the early stages of viral infection or exposure to toll-like receptor agonists. J Virol. (2010) 84:4866–77. doi: 10.1128/JVI.02571-09
82. Yang Y, Xiong Z, Zhang S, Yan Y, Nguyen J, Ng B, et al. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem J. (2005) 392:135–43. doi: 10.1042/BJ20050698
83. Yoshikawa T, Hill T, Li K, Peters CJ, Tseng C-TK. Severe acute respiratory syndrome (SARS) coronavirus-induced lung epithelial cytokines exacerbate SARS pathogenesis by modulating intrinsic functions of monocyte-derived macrophages and dendritic cells. J Virol. (2008) 83:3039–48. doi: 10.1128/JVI.01792-08
84. Zhao J, Zhao J, Rooijen NV, Perlman S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. Plos Pathog. (2009) 5:e1000636. doi: 10.1371/journal.ppat.1000636
85. Zhao J, Zhao J, Legge K, Perlman S. Age-related increases in PGD(2) expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Investig. (2011) 121:4921–30. doi: 10.1172/JCI59777
86. Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, Zaki SR, et al. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol. (2009) 84:1289–301. doi: 10.1128/JVI.01281-09
87. Zhao J, Zhao J, Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol. (2010) 84:9318–25. doi: 10.1128/JVI.01049-10
88. Sheahan T, Morrison TE, Funkhouser W, Uematsu S, Akira S, Baric RS, et al. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. Plos Pathog. (2008) 4:e1000240. doi: 10.1371/journal.ppat.1000240
89. Totura AL, Whitmore A, Agnihothram S, Schäfer A, Katze MG, Heise MT, et al. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome Coronavirus infection. Mbio. (2015) 6:e00638–15. doi: 10.1128/mBio.00638-15
90. Wu L, Wang N, Chang Y, Tian X, Na D, Zhang L, et al. Duration of Antibody responses after severe acute respiratory syndrome. Emerg Infect Dis. (2007) 13:1562–4. doi: 10.3201/eid1310.070576
91. Ho M, Chen W, Chen H, Lin S, Wang M, Di J, et al. Neutralizing antibody response and SARS severity. Emerg Infect Dis. (2005) 11:1730–7. doi: 10.3201/eid1111.040659
92. Jaume M, Yip M, Cheung C, Leung H, Li P, Kien F, et al. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J Virol. (2011) 85:10582–97. doi: 10.1128/JVI.00671-11
93. Al-Qahtani A, Lyroni K, Aznaourova M, Tseliou M, Al-Anazi M, Al-Ahdal M, et al. Middle east respiratory syndrome corona virus spike glycoprotein suppresses macrophage responses via DPP4-mediated induction of IRAK-M and PPAR3B3. Oncotarget. (2017) 8:9053–66. doi: 10.18632/oncotarget.14754
94. Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, Gale MJ, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA. (2014) 111:4970–5. doi: 10.1073/pnas.1323279111
95. Tynell J, Westenius V, Rönkkö E, Munster VJ, Melén K, Österlund P, et al. Middle East respiratory syndrome coronavirus shows poor replication but significant induction of antiviral responses in human monocyte-derived macrophages and dendritic cells. J Gen Virol. (2015) 97:344–55. doi: 10.1099/jgv.0.000351
96. Zhou J, Chu H, Li C, Wong BH-Y, Cheng Z-S, Poon VK-M, et al. Active replication of middle East respiratory syndrome Coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis. (2013) 209:1331–42. doi: 10.1093/infdis/jit504
97. Al-Abdallat M, Payne D, Alqasrawi S, Rha B, Tohme R, Abedi G, et al. Hospital-associated outbreak of middle east respiratory syndrome Coronavirus: a serologic, epidemiologic, and clinical description. Clin Infect Dis. (2014) 59:1225–33. doi: 10.1093/cid/ciu359
98. Drosten C, Meyer B, Müller M, Corman V, Al-Masri M, Hossain R, et al. Transmission of MERS-Coronavirus in household contacts. New Engl J Med. (2014) 371:828–35. doi: 10.1056/NEJMoa1405858
99. Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, et al. Molecular mechanism for antibody-dependent enhancement of Coronavirus entry. J Virol. (2019) 94. doi: 10.1128/JVI.02015-19
100. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet Lond Engl. (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
101. Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet Lond Engl. (2020) doi: 10.1016/S0140-6736(20)30628-0
102. Chu H, Chan JF-W, Wang Y, Yuen TT-T, Chai Y, Hou Y, et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis. (2020) pi:ciaa410. doi: 10.1093/cid/ciaa410
103. Cai Y, Hao Z, Gao Y, Ping W, Wang Q, Peng S, et al. COVID-19 in the perioperative period of lung resection: a brief report from a single thoracic surgery department in Wuhan, China. J Thorac Oncol. (2020) doi: 10.1016/j.jtho.2020.04.003
104. Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. (2020) 17:533–5. doi: 10.1038/s41423-020-0402-2
105. Zeng Q, Li Y, Huang G, Wu W, Dong S, Xu Y. Mortality of COVID-19 is associated with cellular immune function compared to immune function in Chinese Han population. Medrxiv. (2020). doi: 10.1101/2020.03.08.20031229
106. Xiong Y, Liu Y, Cao L, Wang D, Guo M, Guo D, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Ssrn Electron J. (2020) 9:761–770. doi: 10.2139/ssrn.3549993
107. To K, Tsang O, Leung W, Tam A, Wu T, Lung D, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. (2020) doi: 10.1016/S1473-3099(20)30196-1
108. Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. (2020) pi:ciaa344. doi: 10.1101/2020.03.02.20030189
109. Warnes S, Little Z, Keevil C. Human Coronavirus 229E remains infectious on common touch surface materials. Mbio. (2015) 6:e01697–15. doi: 10.1128/mBio.01697-15
110. Tetro J. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. (2020) 22:72–3. doi: 10.1016/j.micinf.2020.02.006
111. Groneberg DA, Hilgenfeld R, Zabel P. Molecular mechanisms of severe acute respiratory syndrome (SARS). Respir Res. (2005) 6:8. doi: 10.1186/1465-9921-6-8
112. Hamming I, Timens W, Bulthuis M, Lely A, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. (2004) 203:631–7. doi: 10.1002/path.1570
113. Law H, Cheung C, Ng H, Sia S, Chan Y, Luk W, et al. Chemokine up-regulation in SARS-coronavirus–infected, monocyte-derived human dendritic cells. Blood. (2005) 106:2366–74. doi: 10.1182/blood-2004-10-4166
114. Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med. (2005) 202:415–24. doi: 10.1084/jem.20050828
115. Chu H, Zhou J, Wong B, Li C, Cheng Z, Lin X, et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology. (2014) 454–455:197–205. doi: 10.1016/j.virol.2014.02.018
116. Diao B, Wang C, Wang R, Feng Z, Tan Y, Wang H, et al. Human kidney is a target for novel severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) infection. Medrxiv. (2020). doi: 10.1101/2020.03.04.20031120
117. Liu Y, Chen H, Tang K, Guo Y. Clinical manifestations and outcome of SARS-CoV-2 infection during pregnancy. J Infect. (2020) doi: 10.1016/j.jinf.2020.02.028
118. Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Biorxiv. (2020). doi: 10.1101/2020.01.26.919985
Keywords: SARS-CoV, MERS-CoV, SARS-CoV-2, hematopathologic effect, immune responses, immune therapy
Citation: Liang Y, Wang M-L, Chien C-S, Yarmishyn AA, Yang Y-P, Lai W-Y, Luo Y-H, Lin Y-T, Chen Y-J, Chang P-C and Chiou S-H (2020) Highlight of Immune Pathogenic Response and Hematopathologic Effect in SARS-CoV, MERS-CoV, and SARS-Cov-2 Infection. Front. Immunol. 11:1022. doi: 10.3389/fimmu.2020.01022
Received: 30 March 2020; Accepted: 28 April 2020;
Published: 12 May 2020.
Edited by:
Lucia Lopalco, San Raffaele Hospital (IRCCS), ItalyReviewed by:
Kartika Padhan, National Institutes of Health (NIH), United StatesElena Martinelli, Population Council, United States
Copyright © 2020 Liang, Wang, Chien, Yarmishyn, Yang, Lai, Luo, Lin, Chen, Chang and Chiou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei-Ching Chang, cGNjaGFuZ0B5bS5lZHUudHc=; Shih-Hwa Chiou, c2hjaGlvdUB2Z2h0cGUuZ292LnR3
†These authors have contributed equally to this work and share first authorship
 Yanwen Liang
Yanwen Liang Mong-Lien Wang
Mong-Lien Wang Chian-Shiu Chien
Chian-Shiu Chien Aliaksandr A. Yarmishyn
Aliaksandr A. Yarmishyn Yi-Ping Yang
Yi-Ping Yang Wei-Yi Lai
Wei-Yi Lai Yung-Hung Luo
Yung-Hung Luo Yi-Tsung Lin
Yi-Tsung Lin Yann-Jang Chen2,9,10
Yann-Jang Chen2,9,10 Pei-Ching Chang
Pei-Ching Chang Shih-Hwa Chiou
Shih-Hwa Chiou