- 1Herman B Wells Center for Pediatric Research, IAPUI, Indianapolis, IN, United States
- 2Department of Pediatrics, University of Florida, Gainesville, FL, United States
- 3Department of Genetics, Cell Biology and Anatomy, University of Nebraska Medical Center, Omaha, NE, United States
- 4Division of Immunology, Beth Israel Deaconess Medical Center (BIDMC), Harvard Medical School, Boston, MA, United States
- 5Department of Basic and Translational Sciences, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA, United States
Fusion proteins, which consist of factor VIII or factor IX and the transmucosal carrier cholera toxin subunit B, expressed in chloroplasts and bioencapsulated within plant cells, initiate tolerogenic immune responses in the intestine when administered orally. This approach induces regulatory T cells (Treg), which suppress inhibitory antibody formation directed at hemophilia proteins induced by intravenous replacement therapy in hemophilia A and B mice. Further analyses of Treg CD4+ lymphocyte sub-populations in hemophilia B mice reveal a marked increase in the frequency of CD4+CD25−FoxP3−LAP+ T cells (but not of CD4+CD25+FoxP3+ T cells) in the lamina propria of the small but not large intestine. The adoptive transfer of very small numbers of CD4+CD25−LAP+ Treg isolated from the spleen of tolerized mice was superior in suppression of antibodies directed against FIX when compared to CD4+CD25+ T cells. Thus, tolerance induction by oral delivery of antigens bioencapsulated in plant cells occurs via the unique immune system of the small intestine, and suppression of antibody formation is primarily carried out by induced latency-associated peptide (LAP) expressing Treg that likely migrate to the spleen. Tolerogenic antigen presentation in the small intestine requires partial enzymatic degradation of plant cell wall by commensal bacteria in order to release the antigen. Microbiome analysis of hemophilia B mice showed marked differences between small and large intestine. Remarkably, bacterial species known to produce a broad spectrum of enzymes involved in degradation of plant cell wall components were found in the small intestine, in particular in the duodenum. These were highly distinct from populations of cell wall degrading bacteria found in the large intestine. Therefore, FIX antigen presentation and Treg induction by the immune system of the small intestine relies on activity of a distinct microbiome that can potentially be augmented to further enhance this approach.
Introduction
The incidence of hemophilia is ~1 in 5,000 male births worldwide. Mutations in either the serine protease factor IX (FIX) or its co-factor, factor VIII (FVIII) that reduce coagulation activity to <1% of normal typically result in severe disease, which is characterized by frequent and potentially life-threatening bleeds. Deficiency of FVIII is referred to as hemophilia A, while FIX deficiency is called hemophilia B. Bleeding can be prevented by frequent intravenous injections of recombinant or plasma-derived factor product (2–3 times per week). However, 20–30% of severe hemophilia A patients develop neutralizing antibodies that inhibit coagulation activity and are therefore referred to as “inhibitors.”
Antibody formation directed at the newly-introduced proteins represents a serious complication in protein replacement therapy for the X-linked bleeding disorder hemophilia (1–5). One partial remedy is ITI (“immune tolerance induction,” consisting of daily high-dose intravenous infusion of FVIII), which however may take months to years and can cost >$1M (1). Incidence of inhibitor formation against FIX is lower in treatment of hemophilia B (estimated ~5% of patients). However, up to 50% of patients with FIX inhibitors experience anaphylactic reactions and/or nephrotic syndrome upon further exposure to FIX (4).
Bypassing agents are available to restore hemostasis, but these are very expensive and have to be more carefully dosed to avoid thrombosis. More recently, a bispecific antibody has been developed that mimics FVIII and promotes coagulation in hemophilia A patients even in the presence of an inhibitor (6, 7). No such therapy is available for hemophilia B, although progress has been made in development of an RNAi therapy that down-regulates expression of the anti-coagulant protein anti-thrombin III, thereby promoting coagulation in hemophilia A or B patients (8). However, none of these bypassing therapies completely restore hemostasis nor do they induce tolerance.
Hence, we and others have been developing diverse novel protocols aimed at reversal of inhibitor formation by immune tolerance induction (ITI), which have primarily been tested in hemophilic mice in which either the F8 or F9 gene had been deleted (9–14). These studies employ a range of strategies, including lymphocyte-based therapies and administration of small molecule/protein/antibody drugs, which modulate distinct immune responses (5). However, methodologies that allow for a prediction of inhibitor formation by individual patients need to be improved and a better understanding of risk factors will be requisite.
We are currently evaluating an alternative approach, which employs introduction of the coagulation factor antigen through a tolerogenic route without the use of immune suppressive drugs or genetic engineering. To this end, we have developed a plant cell-based oral tolerance approach (15–21). FVIII and FIX antigens have been expressed in chloroplast transgenic (transplastomic) crop plants for high levels of antigen production in green leaves. Initially developed in tobacco, this platform has now been optimized in the edible crop plant lettuce, thereby moving closer to clinical application (16, 18, 20, 22). While early studies expressed the native human genes, subsequence studies employed codon optimization to increase antigen expression 10–50-fold in chloroplasts (18). Plants can be grown under soil-free conditions, and leaves harvested and freeze-dried and ultimately converted to a dry powder. This cost-effective production system does not require extraction and purification of the antigen. In fact, antigens are stable in lyophilized plant cells for 2–3 years when stored at ambient temperature (16, 20, 23). Commercial scale production in cGMP hydroponic facility has been demonstrated for several human blood proteins (16, 20, 24). Most importantly, methods have been developed to remove antibiotic resistance genes from chloroplast genomes of edible plant cells producing enzymes or biopharmaceuticals (20, 24, 25). Plant cell wall protects antigens from acid and enzymes in the stomach because they do not cleave Beta1–4, 1–6 linkages in plant cell wall polymers (17, 26). However, commensal bacteria release plant cell wall degrading enzymes thereby releasing antigens in the gut lumen (17, 24). Moreover, antigens are expressed as fusion proteins between the coagulation factor and a transmucosal carrier. N-terminal fusion of CTB (cholera toxin B subunit, an FDA approved antigen), results in pentamer formation and, upon release in the intestine, binding to GM receptor on gut epithelial cells and transmucosal delivery to the immune system (13, 19, 27–29). A furin cleavage site has been engineered between CTB and the antigen so the antigen is released, while CTB is retained in cells that have taken up the fusion protein (30). A major advantage of targeted delivery is efficacy at low antigen doses (18, 20, 21).
Repeated oral delivery of plant cells expressing CTB-fused antigen has been effective in suppression of inhibitor formation against FVIII in hemophilia A mice and against FIX in hemophilia B mice and dogs that were subsequently treated with intravenous FVIII or FIX therapy (18–21). Moreover, IgE formation and thus anaphylaxis against FIX was prevented in hemophilia B mice and dogs (13, 16, 20, 21). Studies in hemophilia B mice revealed a complex mechanism of tolerance induction that involves changes in subsets of dendritic cell (DCs) and regulatory T cell (Treg) populations (13, 15, 19). Here, we demonstrate induction of CD4+CD25−FoxP3−LAP+ Treg (“LAP+ Treg”) in the small but not large intestine of hemophilia B mice and further support their role in suppressing antibody formation.
Materials and Methods
Animal Experiments and Adoptive T Cell Transfer
Hemophilia B mice with targeted F9 gene deletion on C3H/HeJ background were as published (20, 21, 31). Male mice 6–8 weeks of age were used for the experiments and housed under special pathogen-free conditions. For oral tolerance induction, freeze-dried powder of lettuce (Lactuca sativa) cv. Simpson Elite with CTB-FIX transgene integrated into chloroplast genome (homoplasmic transplastomic plants) were prepared as published (16, 20). The transgene expresses cholera toxin B subunit (CTB) fused to the N terminus of the mature form of human coagulation factor IX (FIX). Plant material (containing 5 μg CTB-FIX antigen per dose suspended in 200 μl of sterile PBS) was orally delivered via gavage twice per week for 2 months using a 20-G bulb-tipped gastric gavage needle. For intravenous challenge with human FIX antigen, mice were administrated 1 IU FIX (Benefix, Pfizer, New York, NY) into the tail vein once per week for 2 months, starting 4 weeks after initiation of the oral tolerance regimen. Control groups received only intravenous FIX but no gavages. Subsequently, splenocyte preparations were pooled for each experimental group (n = 10 per group), and CD4+ T cells were isolated by magnetic sorting using the CD4+ T cell isolation kits from Miltenyi Biotech (Auburn, CA). Isolated CD4+ T cells were stained with anti-mouse CD4-eflour 450 (clone RM4–5, eBioscience, San Diego, CA), LAP-APC (clone TW7–16B4, BioLegend, San Diego, CA) and CD25-PE (clone PC61, BD Biosciences, San Jose, CA) antibodies. Flow sorting was employed to purify CD4+CD25+ T cells and CD4+CD25−LAP+ T cells using FACS Aria II cell-sorter (BD Biosciences, San Jose, CA). Post-sort analysis of cells confirmed more than 95% purity. Purified live cells (as determined by trypan blue staining) were adoptively transferred into naive strain-matched mice via tail vein injection (300,000 cells/mouse). After 24 h, recipient mice were immunized by subcutaneous injection of 1 IU FIX formulated in Sigma Adjuvant System. Blood samples were collected 3 weeks later, and plasma levels of FIX-specific immunoglobulins were measured by ELISA. Statistical analysis was performed using unpaired student t-test.
Isolation of Gut Lymphocytes
Mice were tolerized as explained above, and small intestinal (ileum and distal jejunum) and large intestinal (colon) tissues were harvested, homogenized, and treated with RBC lysis buffer (Sigma, St. Louis, MO). Intestine was placed in and flushed with ice-cold PBS. Attached tissues such as fat tissue or Peyer's patches were removed, and the intestine cut longitudinally. Tissue was further cut into small pieces (<0.5 cm) and washed multiple times with ice-cold PBS. For pre-digestion, tissue was transferred into 50 ml tube, containing 20 ml pre-digestion solution (pre-heated to 37°C): 1 × HBSS (w/o Ca2+, or Mg2+, or phenol red), 10 mM HEPES, 5 mM EDTA, 1 mM DTT, and 5% FCS (fetal calf serum). Tissue was then incubated for 20 min at 37°C while slowly rotating horizontally. After brief vortex (10 s), tissue was passed through a 100-μm cell strainer (without application of pressure). Incubation and cell straining were repeated once. All flow throughs were collected and subsequently combined to isolate intraepithelial lymphocytes (IELs), while the cells retained by the strainer were combined to isolate lamina propria lymphocytes (LPLs).
For LPLs isolation, cells were placed into pre-warmed buffer of the following composition: 1 × HBSS (w/o Ca2+, or Mg2+, or phenol red), 10 mM HEPES, 0.5 mg/ml collagenase D, 0.5 mg/ml DNase, and 5% FCS. Incubation was again performed at 37°C with horizontal tube rotation, this time for 30 min. Cells were passed through 100-μm cell strainer, followed by addition of excess FACS buffer. Cells were collected by centrifugation for 10 min at 300 g, followed by resuspension in FACS buffer and storage on ice. Finally, LPL or IEL fractions were each purified by Percoll gradient centrifugation (40:80%, 20 min, 1,000 g, room temperature). Cells were recovered, washed in FACS buffer, and ultimately resuspended in FACS buffer for fluorescent antibody staining and flow cytometry.
Flow Cytometry
Analysis by flow cytometry was performed as published (13). Surface staining with antibodies was performed at 4°C for 30 min in PBS, followed by addition of viability dye eFluor 506 (or APC-Cy7) at 4°C for 30 min in PBS. Fixation and Foxp3 Alexa Fluor 647 stain was performed with the transcriptional factor staining buffer set from eBiosciences (San Diego, CA). Isotype control, single positive, and unstained cells served as controls. Flow cytometry was performed using the LSR II system (BD Bioscience, San Jose, CA), and data were analyzed with FlowJo (BD Life Sciences, Franklin Lake, NJ) or FCSExpress software (De Novo Software, Los Angeles, CA). Antibodies against murine antigens were obtained from eBiosciences and included anti-CD4 (eFluor450 conjugated), -CD25 (PE), -LAP (PerCP-eFluor710), and -FoxP3 (Alexa Fluor 647).
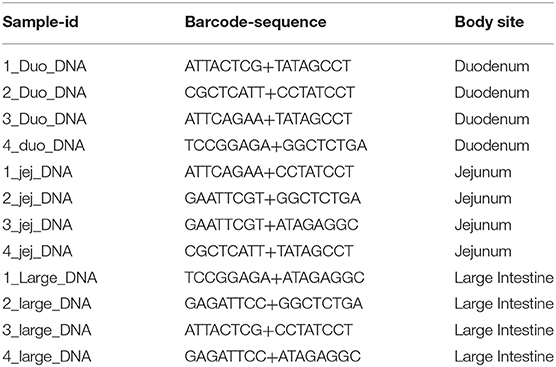
Microbiome Analysis
Gut content from duodenum, jejunum, and ileum segments of small intestine as well as large intestine were collected, and DNA was extracted using QIAamp PowerFecal Pro DNA Kit from Qiagen (Hilden, Germany) according to the manufacturer's protocols. To assess the microbial diversity of duodenum, jejunum/ileum and large intestine, the QIAseq 16S/ITS libraries were developed for nine variable regions of 16S rRNA (six amplicons covering v1v2, v2v3, v3v4, v4v5, v5v7, and v7v9 regions) and eukaryote ITS (internal transcribed spacer) using QIAseq 16S/ITS screening panel from the extracted DNA. Sequencing libraries were labeled with different multiplex indexing barcodes (Table 1) for each sample. The indexed libraries were quantified and paired-end (2 × 251 bp) MiSeq sequencing was performed at the Center for Medical Genomics at Indiana University School of Medicine, using reagents from Qiagen. Raw sequencing reads were demultiplexed into separate FASTQ files for each sample with reads from each variable region. Sequence quality of the demultiplexed FASTQ files were assessed using FASTQC tool and imported into QIIME2 environment. Sequencing data were analyzed using QIIME2 2019.10 pipeline to obtain the microbial diversity and abundance in the duodenum, jejunum, and large intestine regions (32). The reads from all the samples were separated based on amplified variable regions (v1v2, v2v3, v3v4, v4v5, v5v7, v7v9, and ITS) using a QIIME2 cutadapt plugin. QIIME2 with DADA2 denoising method was used for quality control and to identify amplicon sequence variants (ASVs) based on amplified variable regions. The parameters “–p-trunc-len” and “–p-trim-left” in QIIME2 DADA2 denoise-paired plugin were altered based on read quality distribution. The QIIME2 Naïve Bayes classifiers were built from SILVA 132 99% OTUs specifically for primer sets used for amplification of variable regions (33). Then the QIIME2 q2-feature-classifier plugin was used for taxonomic assignments of ASVs using the default settings. For ITS-based classification of eukaryotic species, QIIME2 classifier based on UNITE database was used (34), however as we found a small number of taxa only from Protista, the eukaryotic analysis was not advanced to the next level. The identified taxonomy tables were filtered for rare and unclassified ASVs. The taxonomic composition based on each variable region was compared and viewed using QIIME2 taxa barplot plugin at each taxonomic level from kingdom down to species. Alpha diversity analysis was performed using QIIME2 q2-diversity plugin. The enzyme producing potentials of microbes in duodenum, jejunum, and large intestine regions were identified using PICRUSt2 v2.2 algorithm based on the sequences and abundance profiles of the ASVs identified with QIIME2 (35). Predicted microbial enzymes were mapped to microbes at various taxonomic levels. The abundance of enzymes involved in the plant cell wall degradation were compared among duodenum, jejunum, and large intestine regions using STAMP 2.1.3 software (36). Because different amplicon regions show different microbial compositions and consequently different potentials for producing the enzymes of interest, the highest abundance of each enzyme in any amplicon region was used for plotting the enzyme box plots across all three regions.
Results
LAP+CD25− but Not CD4+CD25+ T Splenocytes Suppress Anti-FIX Formation After Adoptive Transfer of Low Cell Numbers
During oral tolerance induction, total CD4+CD25+FoxP3+ Treg frequencies do not significantly change in various organs examined, which is in contrast to the increase in frequencies of LAP+ Treg. Nonetheless, we found suppression of antibody formation against FIX to be equally potent as with CD4+CD25−LAP+ following adoptive transfer (while non-CD4+ cells or CD4+CD25−LAP− failed to suppress) (13). In these studies, we had transferred 1 × 106 cells of either subset from tolerized to recipient mice. Here, we transferred more limited cell numbers (300,000 per recipient mouse) from hemophilia B mice that had been orally tolerized to FIX to naive mice of the same strain (Figure 1A). Recipient mice were challenged with FIX in adjuvant, followed by measurement of antibody titers 3 weeks later. Antibody titers were similar for mice that received CD4+CD25+ T cells isolated from the spleens of orally tolerized or untreated control mice (n = 5/experimental recipient group, Figure 1B). In contrast, CD4+CD25−LAP+ T cells from orally tolerized animals suppressed antibody formation, with titers significantly lower as compared to after CD4+CD25+ T cells transfer (Figure 1B). Therefore, LAP+ Treg are the main source of systemic suppression of antibody formation. These cells are induced by antigen administration on mucosal interphases, and hence we were unable to obtain sufficient numbers of CD4+CD25−LAP+ T cells in unfed control mice as illustrated in Figure 1A.
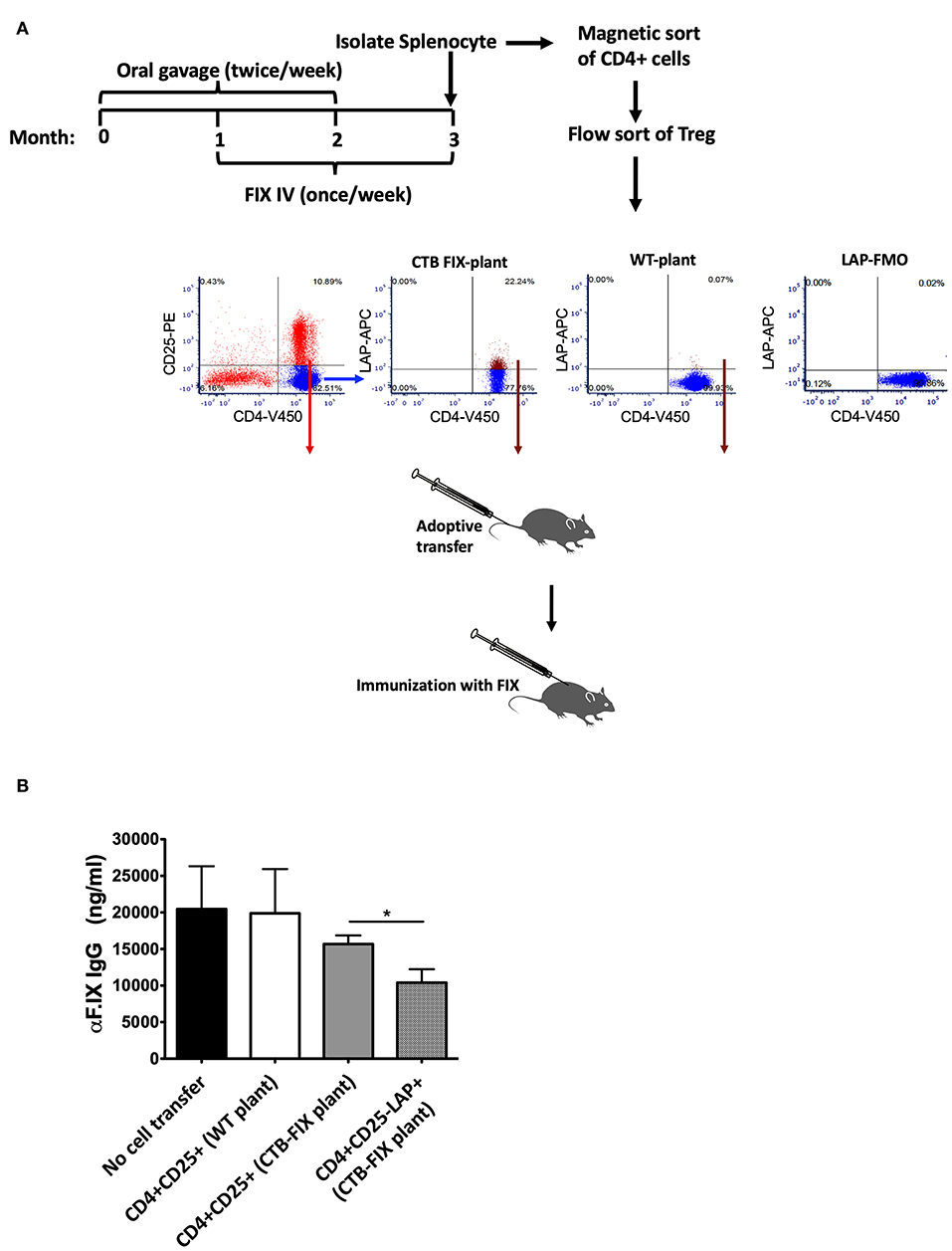
Figure 1. Suppression of antibody formation against FIX after adoptive transfer of Treg. (A) Experimental outline of oral tolerance with bioencapsulated CTB-FIX/intravenous treatment with FIX, T cell isolation, adoptive transfer, and immunization. Donor hemophilia B mice had been treated in this manner or were naïve control mice. Flow cytometry plots show results from purification of splenic CD4+CD25+ T cells and CD4+CD25−LAP+ T cells by flow sorting. (B) FIX-specific IgG titers in mice that received either CD4+CD25+ T cells or CD4+CD25−LAP+ T cells after immunization with FIX. Results are average ±SD (n = 5/group). * indicates P < 0.05.
LAP+ Treg Are Expanded in the Lamina Propria of the Small but Not the Large Intestine
Next, we examined the relative frequencies of FoxP3+ and LAP+ Treg in the immune system lining the small and large intestines in hemophilia B mice upon completion of our oral tolerance regimen (Figure 2 and Supplementary Figure S1). Consistent with our prior findings (13, 19), no increases in total frequencies of FoxP3+ Treg were found in intraepithelial lymphocytes (IEL) or lamina propria lymphocytes (LPL) of either organ (Figure 3A). Compared to untreated mice, frequencies of LAP+ Treg also did not change in the large intestine (n = 5 per group). However, LAP+ Treg frequencies were significantly increased (by 4-fold, Figure 3B) among LPLs of the small intestine (a more modest 2-fold, non-significant increase was seen in IPLs of the small intestine, Figure 3B). Therefore, our results further support the prevailing model that oral tolerance induction primarily occurs in the immune system of the small intestine.
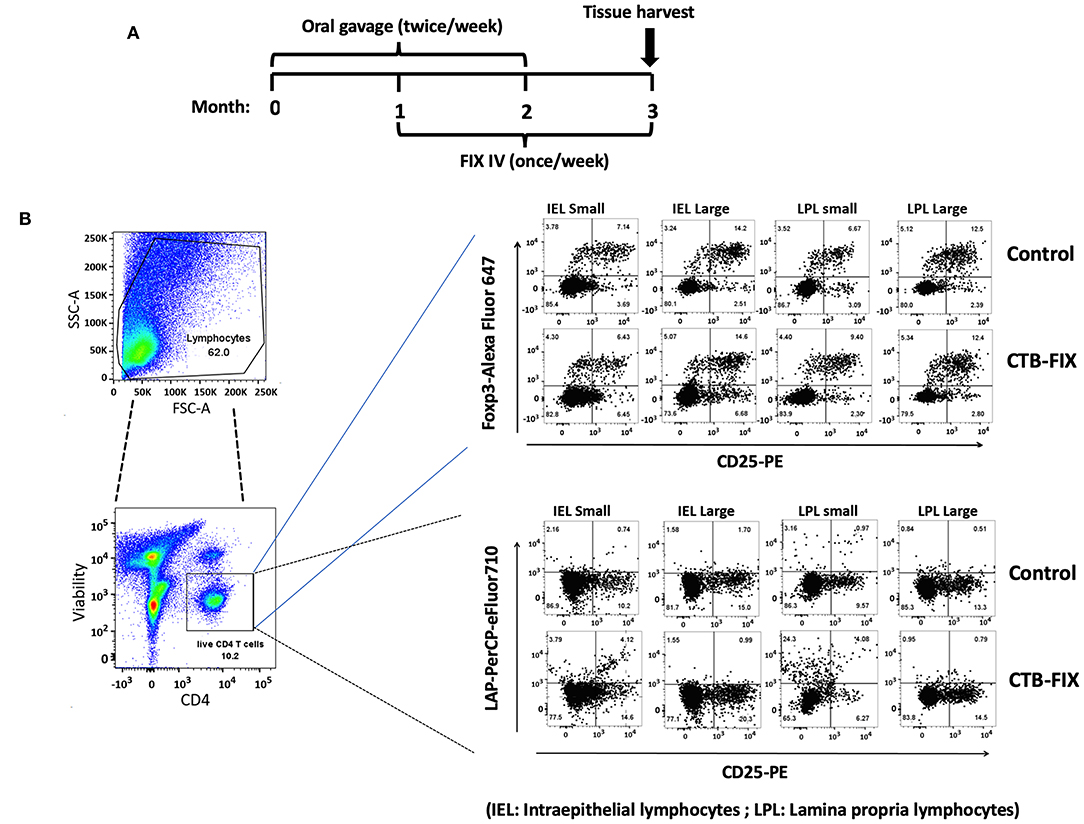
Figure 2. Analysis of Treg subsets in intraepithelial lymphocyte (IEL) and lamina propria lymphocyte (LPL) populations of small and large intestine of hemophilia B mice using flow cytometry. (A) Time course of oral tolerance regimen with CTB-FIX expressing plant cells and treatment by intravenous injection of recombinant FIX protein. (B) Gating scheme and examples of flow cytometry results. Examples of small intestine (“small”) and large intestine (“large”) results are shown for CD4+CD25+FoxP3+ T cells and CD4+CD25−LAP+ T cells. Control mice had not received any treatment.
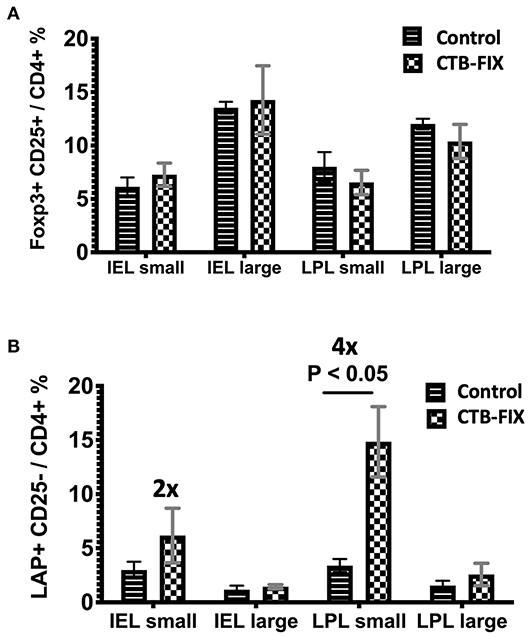
Figure 3. Frequencies of (A) CD4+CD25+FoxP3+ T cells and (B) CD4+CD25−LAP+ T cells among IEL and LPL populations of small and large intestines of orally tolerized and naïve control hemophilia B mice. Results are average ±SD (n = 5/group).
Microbiome Analysis Reveals Presence of Bacteria in Duodenum That Are Capable of Producing Plant Cell Wall Degrading Enzymes
To study the microbiome of the hemophilia B mice, intestinal contents of four C3H/HeJ F9−/− mice of the same colony used in the oral tolerance experiments were collected for DNA extraction. Contents of duodenum and jejunum/ileum portions of the small intestine were collected separately, in addition to contents from the large intestine (colon). All the nine variable regions of the 16S ribosomal RNA gene covered by six amplicons were analyzed by next-generation sequencing (NGS). Consistent with findings by others (37), the microbiomes of the duodenum and jejunum/ileum portion of the small intestine were dominated by bacteria of the Lactobacillales order (Figure 4, Supplementary Figures S2A–E, and Table 1). The microbiome of the large intestine was distinct from the small intestine with more alpha diversity (Supplementary Figures S3A,B), and consisted predominantly of bacteria of the Bacteroidales, Clostridiales, Campylobacterales, and Deferribacterales orders. Although both locations contain considerable populations of Firmicutes, these are predominantly Lactobacillales in the small intestine and Clostridiales in the large intestine. Also, bacteria from phylum, Bacteriodetes are significantly higher in the large intestine compared to the small intestine.
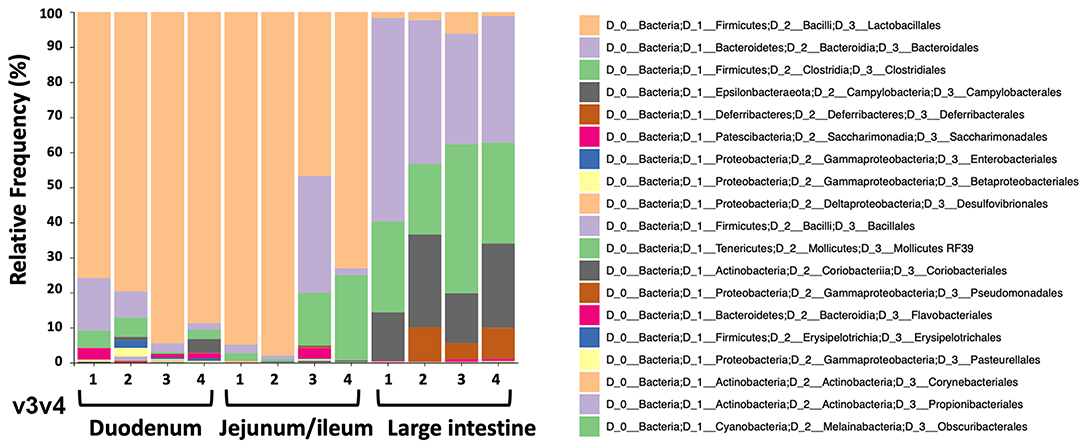
Figure 4. Relative frequencies of bacterial orders in duodenum, jejunum/ileum, and small intestine of hemophilia B mice (n = 4) as determined by bioinformatic analysis of NGS data. The example shown here is for data obtained from amplification of variable region v3v4.
Next, we mined the data for frequencies of species that are known to produce enzymes involved in degradation of plant cell wall components such as cellulose, xylan, mannan, and pectin, or in degradation of lipids or generation of glucose (Table 2 and Supplementary Figure S4). As expected, the large intestine provided higher frequencies of bacteria producing β-N-acetylhexosaminidase [EC 3.2.1.52], cellulase (β-1,4-endoglucanase) [EC 3.2.1.4], amino-acid N-acetyltransferase [EC 2.3.1.1], β-glucosidase [EC 3.2.1.21], mannan endo-1,4-β-mannosidase [EC 3.2.1.78], and pectinesterase [EC 3.1.1.11] (Figures 5D–G,I–K) compared to duodenum and jejunum/ileum because of its higher microbial diversity. Producers of endo-1,4-β-xylanase [EC 3.2.1.8] were only detected in the large intestine (Figure 5K). The jejunum/ileum section only contained higher levels of bacteria producing 6-phospho-β-glucosidase [EC 3.2.1.86], while producers of carboxylesterase [EC 3.1.1.1] were similarly abundant as for large intestine (Figure 5C). Especially, the duodenum population was dominated by producers of 6-phospho-β-glucosidase [EC 3.2.1.86] and higher levels of triacylglycerol lipase [EC 3.1.1.3] than other locations (Figures 5A,C). Interestingly, there was also strong evidence for cellulase, β-N-acetylhexosaminidase, amino-acid N-acetyltransferase, β-glucosidase, xylan 1,4-β-xylosidase, and pectinesterase producers in the small intestine region (Figures 5B,D–H,J). Although differences between duodenum and ileum/jenunum did not reach statistical significance, the duodenum of the hemophilia B mice trended to show higher frequencies of bacteria producing enzymes for breakdown of multiple cell wall components (Figures 5A,C,E,H,J and Supplementary Table S1). Comparison of the relative contribution of different families of bacteria to the production of these particular enzymes in different segments of the gut revealed that these populations are highly distinct between the duodenum, jejunum/ileum, and the large intestine (Figures 6A–H). For example, carboxylesterase [EC 3.1.1.1] producers are dominated by Defferibacteracea in the large intestine and Lactobacillaceae in the jejunum/ileum, while three different families (Burkholderiaceae, Lactobacillaceae, and Staphylococcaceae) contribute similarly in the duodenum (Figure 6A). Another example is the high contribution of Flavobacteriaceae to cellulase production in the duodenum, while lacking contribution of Clostridiales that is seen in the colon (Figure 5H). The exception was 6-phospho-β-glucosidase [EC 3.2.1.86], which appears to be predominantly produced by Lactobacillaceae in all segments (Figure 6I).
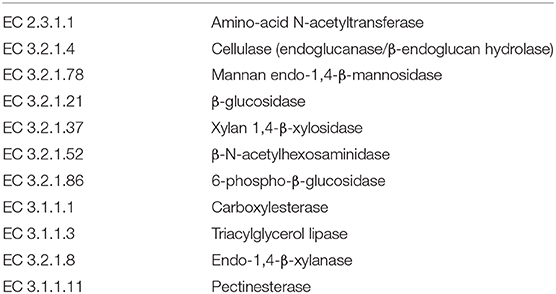
Table 2. Plant cell wall degrading bacterial enzymes produced by species identified in microbiome of hemophilia B mice.
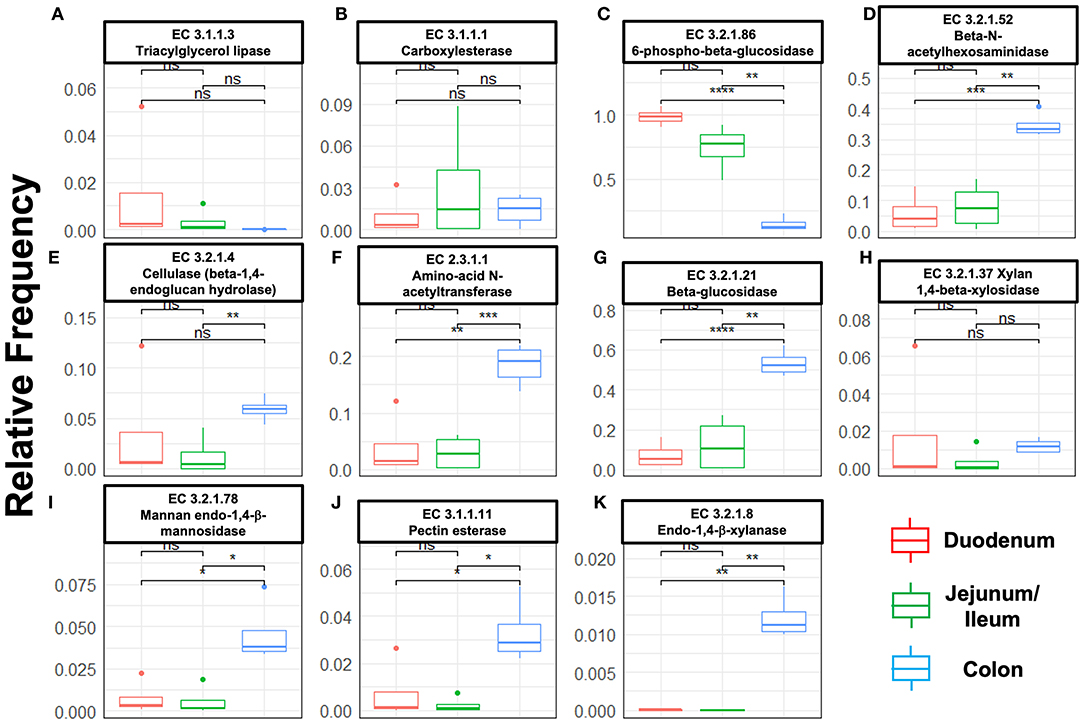
Figure 5. Species relative frequencies of bacteria producing the following enzymes in duodenum, jejunum/ileum, and large intestine of hemophilia B mice as predicted by PICRUSt2 on different 16S rRNA variable region taxonomic profiles. Results are shown for six variable regions analyzed by NGS (Statistical method t-test was used, asterisks represent significance level of a pairwise t-test with P-values i.e., ‘****' ≤ 1e-04, ‘***' ≤ 0.001, ‘**' ≤ 0.01, ‘*' ≤ 0.05, ns > 0.05 (not significant). (A) Triacylglycerol lipase. (B) Carboxylesterase. (C) 6-phospho-β-glucosidase. (D) β-N-acetylhexosaminidase. (E) Cellulase (β-1,4-endoglucan hydrolase). (F) Amino-acid N-acetyltransferase. (G) β-glucosidase. (H) Xylan 1,4-β-xylosidase. (I) Mannan endo-1,4-β-mannosidase. (J) Pectinesterase. (K) Endo-1,4-β-xylanase.
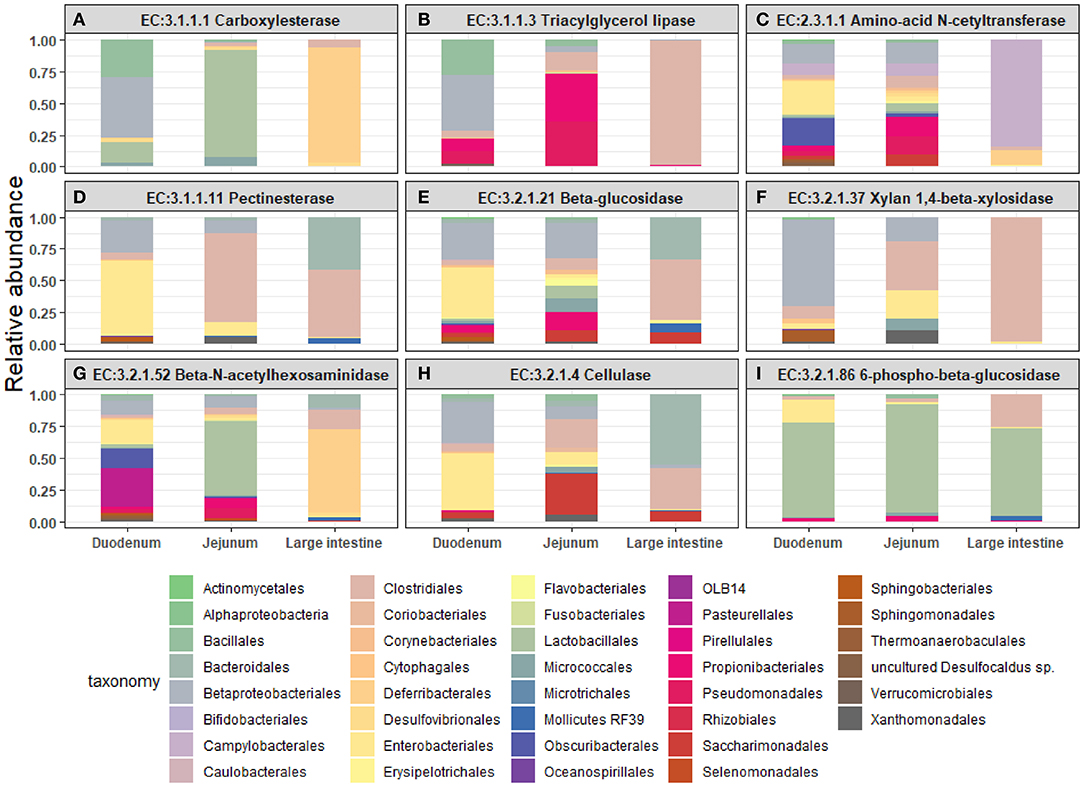
Figure 6. Relative abundance of bacterial orders producing enzymes that degrade plant cell wall components were identified in the duodenum, jejunum, and large intestine of hemophilia B mice. Results are shown as highest abundance of each enzyme from the tested amplicon regions in duodenum, jejunum/ileum, and large intestine of the hemophilia B mice. (A) Carboxylesterase. (B) Triacylglycerol lipase. (C) Amino-acid N-acetyltransferase. (D) Pectinesterase. (E) β-glucosidase. (F) Xylan 1,4-β-xylosidase. (G) β-N-acetylhexosaminidase. (H) Cellulase (β-1,4-endoglucan hydrolase). (I) 6-phospho-β-glucosidase.
Discussion
In our prior studies, up-take of FIX antigen by epithelial cells and DCs (including CD103+ DCs, which are critical for Treg induction) was observed in all portions of the small intestine (13). Although the immune system of the colon contains large numbers of CD4+CD25+FoxP3+ Treg (“FoxP3+ Treg”; in part to prevent inflammatory responses to the large bacterial population), oral tolerance induction is believed to be a function of the small intestine's immune system (38–41). Among the various subsets of Treg, our protocol most robustly induces CD4+CD25−LAP+ Treg, whose frequencies are substantially increased in Peyer's patches and mesenteric lymph nodes (MLNs), while still showing significant increases in spleens (13, 19), which is further supported by our new data. Additionally, we now demonstrate that plant cell-based oral tolerance induces LAP+ Treg in the mucosal immune system of the small intestine only, thus further supporting its crucial role in this approach. Detection of LAP (latency-associated peptide of TGF-β) on the surface of T cells reflects high expression of TGF-β, a cytokine that is expressed by CD103+ DCs and that is required for induction of FoxP3+ Treg and LAP+ Treg (42–44). The latter suppress in a TGF-β dependent manner and may serve as a biomarker for oral tolerance induction (18).
Antigen-specific Treg induction in the small intestine requires antigen release from the plant cells. Bioencapsulated antigens are protected from acid hydrolysis and digestive enzymes in the stomach through the β-1,4 and β-1,6 linkages of plant cell wall components that mammalian enzymes cannot hydrolyze. Therefore, enzymatic activities provided by the microbiome of the small intestine are critical to release FVIII or FIX antigens for delivery to the immune system and Treg induction. The microbiome of the small intestine is distinct from that of the large intestine and dominated by Firmicutes that are of the order Lactobacillales. This finding is consistent with the greater frequency of producers of 6-phospho-β-glucosidase as compared to the large intestine, highlighting the role of the small intestine in nutrient absorption. The large intestine has overall greater microbial diversity but also contains a large proportion of Firmicutes. These are however mostly Clostridiales, which are known to induce FoxP3+ Treg in the colon (45). Less is known about the role of the small intestine's microbiome in Treg induction, a function that is critical for oral tolerance.
In order to begin to define the role of the small intestine's microbiome in Treg induction in plant cell-based oral tolerance, we reasoned that we needed to first address the question of antigen release. Plant cell wall degrading microbes are mostly studied for the colon, addressing end digestion rather than nutrient absorption and oral immune tolerance, which are functions of the small intestine. Here, we find that the microbiome of the small intestine, while not as capable as the large intestine, does provide diversity of enzyme producers that degrade various components of the plant cell wall and greater levels of triacylglycerol lipase and 6-phospho-β-glucosidase producers. Within the small intestine, the duodenum tended to have a greater capacity and diversity of such enzyme producers compared to the jejunum/ileum. It should also be noted that not all enzymes required for complete cell wall degradation are needed for antigen release which merely relies on sufficient disruption of the cell wall integrity. The duodenum likely functions as an initial place for breakdown of plant cells, so that antigens can be released for tolerance induction. The jejunum/ileum location contains Peyer's patches and has important functions in antigen uptake and processing (39, 41).
Interestingly, composition of the bacterial population that produces the diverse cell wall producing enzymes is highly distinct between the duodenum and the large intestine. This feature could potentially be exploited to design probiotics for duodenum-specific delivery of enzyme producers. For example, with the exception of 6-phospho-β-glucosidase, the enzymatic activities that we investigated here are provided by bacteria that do not belong to the order of Lactobacillales, which are by far the most abundant order in the small intestine. Instead, the enzymatic activities are contributed by other types of bacteria that are present but infrequent in the small intestine, such as Burkholderiales or Flavobacteriales. Therefore, Daniell et al. have recently generated plants expressing cell wall degrading enzymes (24, 25). Importantly, oral delivery of pectinase, endoglucanase, exoglucanase, and mannanase before feeding plants cells expressing therapeutic proteins almost doubled drug levels in plasma (46). This method would provide more precise dosing of the required enzymatic activity than a probiotic approach.
While not further investigated here, we previously also found evidence for induction of Tr1 (type 1 regulatory T) cells in the lamina propria of orally tolerized mice (13). These cells are known to produce large amounts of IL-10. FoxP3+ Treg on mucosal interphases and LAP+ Treg also produce IL-10, and our oral tolerance protocol was unsuccessful in hemophilia B mice deficient in IL-10 (13). Even though our studies on other tolerance induction protocols showed that IL-10 is not generally required for Treg induction or their ability to suppress antibody formation, IL-10 is critical for plant cell-based oral tolerance induction (13, 47, 48). Moreover, we found that IL-10 and TGF-β is mainly expressed by LAP+ Treg upon antigen re-stimulation of splenocytes from orally tolerized mice. Although our previous adoptive transfer studies do not entirely rule out that contamination with small numbers of LAP+ Treg contributed to suppression by CD4+CD25+ T cells, the bulk of the literature on oral tolerance to food antigens supports our interpretation that FoxP3+ Treg contribute to oral tolerance induction to FVIII and FIX in our studies (38, 40, 49). It has been proposed that FoxP3+ Treg induced in the MLN by CD103+ DCs (following antigen uptake in the LP and migration to the MLN) subsequently migrate to the LP, where additional stimulation with antigen results in their further expansion (49). Through expression of IL-10 and other molecules, FoxP3+ Treg and Tr1 cells may help shape a local environment that supports induction of LAP+ Treg, which are most robustly induced and thus constitute the main type of Treg that suppresses systemic antibody/inhibitor formation against FVIII or FIX in plant-based oral tolerance. Certain species of Clostridium and Bacteriodes have been shown to induce IL-10 producing FoxP3+ Treg, e.g., via certain polysaccharides (50). Such effects have not yet been documented for other subsets of Treg such as LAP+ Treg or Tr1 cells. Clostridium species are controlling FoxP3+ Treg and Th17 differentiation and expansion, e.g., through production of short chain fatty acids (SCFAs) (50–52), and elimination of FoxP3+ Treg can lead to increased populations of Firmicutes (53). Similar to FoxP3+ Treg and Th17 cells, induction of LAP+ Treg depends on TGF-β, and LAP+ Treg are major contributors of IL-10 production in our plant cell-based induced immune regulation of responses against FVIII or FIX (13). One would therefore expect similar effects of the microbiome on LAP+ Treg induction. However, the findings summarized above are based on observations on the colonic microbiome and colonic Treg, while LAP+ Treg are induced in the mucosa of the small intestine. Analogous mechanisms in the small intestine that may impact oral tolerance remain to be discovered.
In conclusion, suppression of antibody formation by oral tolerance by administration of CTB-FIX bioencapsulated in plant cells is primarily performed by LAP+ Treg, which are expanded in the immune system of the small but not the large intestine. Therefore, optimal release of antigen from the plant cells in the small intestine and delivery to the associated immune system is key for success of this strategy. Bacteria capable of providing the required enzymatic activities are present in the small intestine (in particular in the duodenum). The composition of this population is distinct from that of the large intestine, and their augmentation may further enhance plant cell-based oral tolerance induction.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
All animals are maintained under specific pathogen-free conditions at the University of Florida animal facility. Experiments were performed according to the guidelines of the Institutional Animal Care and Use Committee (IACUC) at the University of Florida.
Author Contributions
SK, XW, and TB performed experiments. SK, XW, NA, CT, CG, HD, and RH designed, analyzed, and interpreted experiments. SK, NA, TB, CT, CG, HD, and RH wrote the manuscript. RH supervised the study.
Funding
This work was supported by NIH grants R01 HL133191 to RH and HD, R01 HL107904 to HD, R01 HL131093 to RH and CT, U54 HL142012 to RH and 5P20GM103427, 5P30CA036727 to CG, as well as National Science Foundation, United States EPSCoR Award (grant OIA-1557417). RH is supported by the Riley Children's Foundation and the Lilly Foundation.
Conflict of Interest
HD and RH have received funds from Takeda Therapeutics to further develop oral tolerance to factor VIII. RH has received funds from Luye R&D (Boston) to develop gene therapy for hemophilia.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer HW declared a shared affiliation, with no collaboration, with one of the authors, CT, to the handling editor at the time of review.
Acknowledgments
The authors thank the Center for Medical Genomics at Indiana University School of Medicine for microbiome sequencing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.00844/full#supplementary-material
References
1. Ljung R, Auerswald G, Benson G, Dolan G, Duffy A, Hermans C, et al. Inhibitors in haemophilia A and B: management of bleeds, inhibitor eradication and strategies for difficult-to-treat patients. Eur J Haematol. (2019) 102:111–22. doi: 10.1111/ejh.13193
2. Pipe SW, Sabatino DE, Nugent DJ, Hooper WC, Soucie JM, Keith Hoots W, et al. Executive summary of the NHLBI State of the Science (SOS) workshop: overview and next steps in generating a national blueprint for future research on factor VIII inhibitors. Haemophilia. (2019) 25:610–5. doi: 10.1111/hae.13713
3. Pratt KP. Anti-drug antibodies: emerging approaches to predict, reduce or reverse biotherapeutic immunogenicity. Antibodies (Basel). (2018) 7:E19. doi: 10.3390/antib7020019
4. Santoro C, Quintavalle G, Castaman G, Baldacci E, Ferretti A, Riccardi F, et al. Inhibitors in hemophilia B. Semin Thromb Hemost. (2018) 44:578–89. doi: 10.1055/s-0038-1660817
5. Sherman A, Biswas M, Herzog RW. Tolerance induction in hemophilia: innovation and accomplishments. Curr Opin Hematol. (2018) 25:365–72. doi: 10.1097/MOH.0000000000000446
6. Mahlangu JN. Bispecific antibody emicizumab for haemophilia A: a breakthrough for patients with inhibitors. BioDrugs. (2018) 32:561–70. doi: 10.1007/s40259-018-0315-0
7. Young G, Callaghan M, Dunn A, Kruse-Jarres R, Pipe S. Emicizumab for hemophilia A with factor VIII inhibitors. Expert Rev Hematol. (2018) 11:835–46. doi: 10.1080/17474086.2018.1531701
8. Nogami K, Shima M. New therapies using nonfactor products for patients with hemophilia and inhibitors. Blood. (2019) 133:399–406. doi: 10.1182/blood-2018-07-820712
9. Biswas M, Sarkar D, Kumar SR, Nayak S, Rogers GL, Markusic DM, et al. Synergy between rapamycin and FLT3 ligand enhances plasmacytoid dendritic cell-dependent induction of CD4+CD25+FoxP3+ Treg. Blood. (2015) 125:2937–47. doi: 10.1182/blood-2014-09-599266
10. Kim YC, Zhang AH, Su Y, Rieder SA, Rossi RJ, Ettinger RA, et al. Engineered antigen-specific human regulatory T cells: immunosuppression of FVIII-specific T- and B-cell responses. Blood. (2015) 125:1107–15. doi: 10.1182/blood-2014-04-566786
11. Moghimi B, Sack BK, Nayak S, Markusic DM, Mah CS, Herzog RW. Induction of tolerance to factor VIII by transient co-administration with rapamycin. J Thromb Haemost. (2011) 9:1524–33. doi: 10.1111/j.1538-7836.2011.04351.x
12. Reipert BM, Sasgary M, Ahmad RU, Auer W, Turecek PL, Schwarz HP. Blockade of CD40/CD40 ligand interactions prevents induction of factor VIII inhibitors in hemophilic mice but does not induce lasting immune tolerance. Thromb Haemost. (2001) 86:1345–52. doi: 10.1055/s-0037-1616733
13. Wang X, Su J, Sherman A, Rogers GL, Liao G, Hoffman BE, et al. Plant-based oral tolerance to hemophilia therapy employs a complex immune regulatory response including LAP+CD4+ T cells. Blood. (2015) 125:2418–27. doi: 10.1182/blood-2014-08-597070
14. Waters B, Qadura M, Burnett E, Chegeni R, Labelle A, Thompson P, et al. Anti-CD3 prevents factor VIII inhibitor development in hemophilia A mice by a regulatory CD4+CD25+-dependent mechanism and by shifting cytokine production to favor a Th1 response. Blood. (2009) 113:193–203. doi: 10.1182/blood-2008-04-151597
15. Daniell H, Kulis M, Herzog RW. Plant cell-made protein antigens for induction of Oral tolerance. Biotechnol Adv. (2019) 37:107413. doi: 10.1016/j.biotechadv.2019.06.012
16. Herzog RW, Nichols TC, Su J, Zhang B, Sherman A, Merricks EP, et al. Oral tolerance induction in hemophilia B dogs fed with transplastomic lettuce. Mol Ther. (2017) 25:512–22. doi: 10.1016/j.ymthe.2016.11.009
17. Kwon KC, Daniell H. Oral delivery of protein drugs bioencapsulated in plant cells. Mol Ther. (2016) 24:1342–50. doi: 10.1038/mt.2016.115
18. Kwon KC, Sherman A, Chang WJ, Kamesh A, Biswas M, Herzog RW, et al. Expression and assembly of largest foreign protein in chloroplasts: oral delivery of human FVIII made in lettuce chloroplasts robustly suppresses inhibitor formation in haemophilia A mice. Plant Biotechnol J. (2018) 16:1148–60. doi: 10.1111/pbi.12859
19. Sherman A, Su J, Lin S, Wang X, Herzog RW, Daniell H. Suppression of inhibitor formation against FVIII in a murine model of hemophilia A by oral delivery of antigens bioencapsulated in plant cells. Blood. (2014) 124:1659–68. doi: 10.1182/blood-2013-10-528737
20. Su J, Zhu L, Sherman A, Wang X, Lin S, Kamesh A, et al. Low cost industrial production of coagulation factor IX bioencapsulated in lettuce cells for oral tolerance induction in hemophilia B. Biomaterials. (2015) 70:84–93. doi: 10.1016/j.biomaterials.2015.08.004
21. Verma D, Moghimi B, Loduca PA, Singh HD, Hoffman BE, Herzog RW, et al. Oral delivery of bioencapsulated coagulation factor IX prevents inhibitor formation and fatal anaphylaxis in hemophilia B mice. Proc Natl Acad Sci U S A. (2010) 107:7101–6. doi: 10.1073/pnas.0912181107
22. Kwon KC, Chan HT, León IR, Williams-Carrier R, Barkan A, Daniell H. Codon-optimization to enhance expression yields insights into chloroplast translation. Plant Physiol. (2016) 172:62–77. doi: 10.1104/pp.16.00981
23. Park J, Yan G, Kwon KC, Liub M, Gonnellaa PA, Yangb S, et al. Oral delivery of novel human IGF-1 bioencapsulated in lettuce cells promotes musculoskeletal cell proliferation, differentiation and diabetic fracture healing. Biomaterials. (2020) 233:119591. doi: 10.1016/j.biomaterials.2019.119591
24. Daniell H, Ribeiro T, Lin S, Saha P, Mcmichael C, Chowdhary R, et al. Validation of leaf and microbial pectinases: commercial launching of a new platform technology. Plant Biotechnol J. (2019) 17:1154–66. doi: 10.1111/pbi.13119
25. Kumari U, Singh R, Ray T, Rana S, Saha P, Malhotra K, et al. Validation of leaf enzymes in the detergent and textile industries: launching of a new platform technology. Plant Biotechnol J. (2019) 17:1167–82. doi: 10.1111/pbi.13122
26. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. (2012) 3:289–306. doi: 10.4161/gmic.19897
27. Daniell H, Lee SB, Panchal T, Wiebe PO. Expression of the native cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J Mol Biol. (2001) 311:1001–9. doi: 10.1006/jmbi.2001.4921
28. Ruhlman T, Ahangari R, Devine A, Samsam M, Daniell H. Expression of cholera toxin B-proinsulin fusion protein in lettuce and tobacco chloroplasts – oral administration protects against development of insulitis in non-obese diabetic mice. Plant Biotechnol J. (2007) 5:495–510. doi: 10.1111/j.1467-7652.2007.00259.x
29. Xiao Y, Kwon KC, Hoffman BE, Kamesh A, Jones NT, Herzog RW, et al. Low cost delivery of proteins bioencapsulated in plant cells to human non-immune or immune modulatory cells. Biomaterials. (2016) 80:68–79. doi: 10.1016/j.biomaterials.2015.11.051
30. Limaye A, Koya V, Samsam M, Daniell H. Receptor-mediated oral delivery of a bioencapsulated green fluorescent protein expressed in transgenic chloroplasts into the mouse circulatory system. FASEB J. (2006) 20:959–61. doi: 10.1096/fj.05-5134fje
31. Cao O, Hoffman BE, Moghimi B, Nayak S, Cooper M, Zhou S, et al. Impact of the underlying mutation and the route of vector administration on immune responses to factor IX in gene therapy for hemophilia B. Mol Ther. (2009) 17:1733–42. doi: 10.1038/mt.2009.159
32. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. (2019) 37:852–7. doi: 10.1038/s41587-019-0209-9
33. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. (2013) 41:D590–6. doi: 10.1093/nar/gks1219
34. Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. (2019) 47:D259–D64. doi: 10.1093/nar/gky1022
35. Douglas GM, Maffei VJ, Zaneveld J, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2: an improved and extensible approach for metagenome inference. bioRxiv. (2019) 672295. doi: 10.1101/672295
36. Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. (2014) 30:3123–4. doi: 10.1093/bioinformatics/btu494
37. Gu S, Chen D, Zhang JN, Lv X, Wang K, Duan LP, et al. Bacterial community mapping of the mouse gastrointestinal tract. PLoS One. (2013) 8:e74957. doi: 10.1371/journal.pone.0074957
38. Kuhn C, Weiner HL. Immunology. How does the immune system tolerate food? Science. (2016) 351:810–1. doi: 10.1126/science.aaf2167
39. Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol. (2012) 5:232–9. doi: 10.1038/mi.2012.4
40. Rezende RM, Weiner HL. History and mechanisms of oral tolerance. Semin Immunol. (2017) 30:3–11. doi: 10.1016/j.smim.2017.07.004
41. Wang X, Sherman A, Liao G, Leong KW, Daniell H, Terhorst C, et al. Mechanism of oral tolerance induction to therapeutic proteins. Adv Drug Deliv Rev. (2013) 65:759–73. doi: 10.1016/j.addr.2012.10.013
42. Gandhi R, Farez MF, Wang Y, Kozoriz D, Quintana FJ, Weiner HL. Cutting edge: human latency-associated peptide+ T cells: a novel regulatory T cell subset. J Immunol. (2010) 184:4620–4. doi: 10.4049/jimmunol.0903329
43. Ochi H, Abraham M, Ishikawa H, Frenkel D, Yang K, Basso AS, et al. Oral CD3-specific antibody suppresses autoimmune encephalomyelitis by inducing CD4+CD25−LAP+ T cells. Nat Med. (2006) 12:627–35. doi: 10.1038/nm1408
44. Oida T, Zhang X, Goto M, Hachimura S, Totsuka M, Kaminogawa S, et al. CD4+CD25− T cells that express latency-associated peptide on the surface suppress CD4+CD45RBhigh-induced colitis by a TGF-β-dependent mechanism. J Immunol. (2003) 170:2516–22. doi: 10.4049/jimmunol.170.5.2516
45. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. (2011) 331:337–41. doi: 10.1126/science.1198469
46. Daniell H, Mangu V, Yakubov B, Park J, Habibi P, Shi Y, et al. Investigational new drug enabling angiotensin oral-delivery studies to attenuate pulmonary hypertension. Biomaterials. (2020) 233:119750. doi: 10.1016/j.biomaterials.2019.119750
47. Hoffman BE, Martino AT, Sack BK, Cao O, Liao G, Terhorst C, et al. Nonredundant roles of IL-10 and TGF-β in suppression of immune responses to hepatic AAV-factor IX gene transfer. Mol Ther. (2011) 19:1263–72. doi: 10.1038/mt.2011.33
48. Wang X, Terhorst C, Herzog RW. In vivo induction of regulatory T cells for immune tolerance in hemophilia. Cell Immunol. (2016) 301:18–29. doi: 10.1016/j.cellimm.2015.10.001
49. Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. (2011) 34:237–46. doi: 10.1016/j.immuni.2011.01.016
50. Pandiyan P, Bhaskaran N, Zou M, Schneider E, Jayaraman S, Huehn J. Microbiome dependent regulation of Tregs and Th17 cells in mucosa. Front Immunol. (2019) 10:426. doi: 10.3389/fimmu.2019.00426
51. Duscha A, Gisevius B, Hirschberg S, Yissachar N, Stangl GI, Eilers E, et al. Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell. (2020) 180:1067–80.e16. doi: 10.1016/j.cell.2020.02.035
52. Omenetti S, Pizarro TT. The Treg/Th17 axis: a dynamic balance regulated by the gut microbiome. Front Immunol. (2015) 6:639. doi: 10.3389/fimmu.2015.00639
Keywords: hemophilia, oral tolerance, transgenic plant, regulatory T (Treg) cell, factor IX
Citation: Kumar SRP, Wang X, Avuthu N, Bertolini TB, Terhorst C, Guda C, Daniell H and Herzog RW (2020) Role of Small Intestine and Gut Microbiome in Plant-Based Oral Tolerance for Hemophilia. Front. Immunol. 11:844. doi: 10.3389/fimmu.2020.00844
Received: 04 December 2019; Accepted: 14 April 2020;
Published: 20 May 2020.
Edited by:
Kathleen P. Pratt, Uniformed Services University of the Health Sciences, United StatesReviewed by:
David William Scott, Uniformed Services University of the Health Sciences, United StatesHoward L. Weiner, Harvard Medical School, Brigham and Women's Hospital, United States
Copyright © 2020 Kumar, Wang, Avuthu, Bertolini, Terhorst, Guda, Daniell and Herzog. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Henry Daniell, aGRhbmllbGxAdXBlbm4uZWR1; Roland W. Herzog, cndoZXJ6b2dAaXUuZWR1
†These authors have contributed equally to this work
 Sandeep R. P. Kumar
Sandeep R. P. Kumar Xiaomei Wang2†
Xiaomei Wang2† Cox Terhorst
Cox Terhorst Roland W. Herzog
Roland W. Herzog