
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 30 April 2020
Sec. Comparative Immunology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.00711
This article is part of the Research Topic Evolution and Comparative Immunology of Immune Systems in Marine Organisms View all 14 articles
Crustacean cardioactive peptide (CCAP), a cyclic amidated non-apeptide, is widely found in arthropods. The functions of CCAP have been revealed to include regulation of heart rate, intestinal peristalsis, molting, and osmotic pressure. However, to date, there has not been any report on the possible involvement of CCAP in immunoregulation in crustaceans. In this study, a CCAP precursor (designated as Sp-CCAP) was identified in the commercially important mud crab Scylla paramamosain, which could be processed into four CCAP-associated peptides and one mature peptide (PFCNAFTGC-NH2). Bioinformatics analysis indicated that Sp-CCAP was highly conserved in crustaceans. RT-PCR results revealed that Sp-CCAP was expressed in nerve tissues and gonads, whereas the Sp-CCAP receptor gene (Sp-CCAPR) was expressed in 12 tissues of S. paramamosain, including hepatopancreas. In situ hybridization further showed that an Sp-CCAPR-positive signal is mainly localized in the F-cells of hepatopancreas. Moreover, the mRNA expression level of Sp-CCAPR in the hepatopancreas was significantly up-regulated after lipopolysaccharide (LPS) or polyriboinosinic polyribocytidylic acid [Poly (I:C)] challenge. Meanwhile, the mRNA expression level of Sp-CCAPR, nuclear transcription factor NF-κB homologs (Sp-Dorsal and Sp-Relish), member of mitogen-activated protein kinase (MAPK) signaling pathway (Sp-P38), pro-inflammatory cytokines factor (Sp-TNFSF and Sp-IL16), and antimicrobial peptide (Sp-Lysozyme, Sp-ALF, Sp-ALF4, and Sp-ALF5) in the hepatopancreas were all up-regulated after the administration of synthetic Sp-CCAP mature peptide both in vivo and in vitro. The addition of synthetic Sp-CCAP mature peptide in vitro also led to an increase in nitric oxide (NO) concentration and an improved bacterial clearance ability in the hepatopancreas culture medium. The present study suggested that Sp-CCAP signaling system might be involved in the immune responses of S. paramamosain by activating immune molecules on the hepatopancreas. Collectively, our findings shed new light on neuroendocrine-immune regulatory system in arthropods and could potentially provide a new strategy for disease prevention and control for mud crab aquaculture.
The neuroendocrine-immune (NEI) regulatory system refers to a complex network formed by the interaction of the nervous system, endocrine system, and immune system (1). The nervous and endocrine systems regulate various physiological processes by releasing neuropeptides, neurotransmitters, and hormones (2). The neuropeptides are usually synthesized and secreted by neurons or neuroendocrine cells and composed of 3–100 amino acid residues (3). As an extracellular chemical messenger, neuropeptides regulate a range of physiological functions, including immunity, growth, reproduction, metabolism, food intake, and circadian rhythm, by activating specific receptors (3). Neuropeptide receptors are mostly G protein-coupled receptors (GPCRs), which constitute the largest family of cell surface receptors. They play a vital role in physiological processes by promoting cellular communication via recognizing various ligands, including bioactive peptides, nucleosides, and amines (4).
A large number of studies have shown that neuropeptides also interact with the immune system by binding to receptors of immune cells (5–14). For example, it has been reported that in humans, through binding to their respective receptors, vasoactive intestinal peptide (VIP), pituitary adenylate cyclase-activating polypeptide (PACAP), urocortin 1 (UCN), and adrenomedullin (AM) reduced the production of pro-inflammatory factors (5–7). In rainbow trout Oncorhynchus mykiss, prolactin has been shown to increase mRNA expression of MyD88 and IL-1β during in vitro infection with the pathogen, Piscirickettsia salmonis (8), whereas in Japanese pufferfish Takifugu rubripes, neuromedin U elevated the mRNA expression of IL-6, IL-18, and TNF-α in peripheral blood leukocytes (9). Similarly, in invertebrates, FMRFamide reportedly regulated the expression of immune effectors and apoptosis-related genes via P38 mitogen-activated protein kinase (MAPK) signaling pathway in oyster Crassostrea gigas (10). In fruit fly Drosophila, allatostatin-C receptor 2 (ASTC-R2) played a crucial role in host survival when infected by the pathogenic bacterium Photorhabdus luminescens (11). In crustaceans, it has also been reported that crustacean hyperglycemic hormones (CHHs) promoted the elimination of the pathogen Vibrio harveyi in the hemolymph and significantly up-regulated the mRNA levels of antimicrobial peptides (AMPs) (PEN4 and crustin) in Pacific white shrimp Litopenaeus vannamei (12, 13). For the same species, the silencing of molt-inhibiting hormone (MIH) also led to significant increases in mortality of the shrimp infected by bacterium Vibrio parahaemolyticus and white spot syndrome virus (WSSV) (14).
In recent years, more and more evidence suggested that in addition to hemocytes, the hepatopancreas also plays an important role in the immunity of crustaceans (15–21). In addition to function as a digestive gland, the hepatopancreas is also a crucial organ for immunity in crustaceans (16). Indeed, crustacean hepatopancreas is a major source of immune response molecules, including lectins, nitric oxide (NO), stress proteins, antibacterial and antiviral proteins, enzymes, and apoptotic genes (15). Furthermore, many immune-related signal transduction pathways are also found in crustacean hepatopancreas, including MAPK, PPAR, Rap1, PI3K-Akts, cyclic adenosine monophosphate (cAMP), and NF-κB signaling pathway (15, 16, 22). However, more studies are needed to further clarify the immune mechanisms of the hepatopancreas in crustaceans.
Crustacean cardioactive peptide (CCAP) is a cyclic amidated non-apeptide first isolated from the pericardial organs in shore crab Carcinus maenas with a function of regulating heartbeat (23). It has since been derived mainly from the nervous system of various arthropods, with confirmed roles of neurohormone and neurotransmitter (24, 25). In recent studies, CCAP mRNA was found in the midgut of cockroach Periplaneta americana and Pacific white shrimp L. vannamei, as well as in the gills of oriental river prawn Macrobrachium nipponense (26–29). CCAP has been shown to be involved in various physiological processes in insects and crustaceans, such as modulation of heartbeat in fruit fly Drosophila melanogaster (30) and marine crabs C. maenas and Callinectes sapidus (23, 31), stimulation of American cockroach P. americana midgut contraction and stick insect Baculum extradentatum hindgut contraction (26, 32), regulation of ecdysis in prawn M. nipponense (29), modulation of oviduct and spermatheca contraction in grasshopper Locusta migratoria (33, 34), and increasing survival of shrimp L. vannamei subjected to freshwater stress (27).
Like other neuropeptide receptors, CCAP receptor is a GPCR. So far, CCAP receptor has been identified in various insects and crustaceans and has been shown to be involved with its ligand to regulate physiological processes. For instance, knockdown of CCAP receptor reportedly resulted in the loss of CCAP heartbeat regulation function in blood-suck bug Rhodnius prolixus (35), and interfering CCAP and its receptor reduced the success rate of ecdysis in red flour beetle Tribolium castaneum (36). In mud crab Scylla paramamosain, recently, CCAP partial transcript has been found from the cerebral transcriptome database, and its receptor is identified via ligand-receptor binding assay by our laboratory (37, 38).
Mud crab S. paramamosain is widely distributed in the Indo-Pacific region, and the species is an important mariculture species along the southeast coastal provinces of China (39). Mud crab in aquaculture is vulnerable to diverse bacterial, fungal, and viral pathogens, which could lead to severe economic losses to the industry. In order to prevent and control disease outbreaks in aquaculture, an increasing number of research has focused on the functions and enhancement of the immune system of cultured species (40–42). In this study, we first obtained and characterized the full-length cDNA of Sp-CCAP from the cerebral ganglia of S. paramamosain. The tissue distribution of Sp-CCAP and its receptor (Sp-CCAPR) were detected by semi-quantitative RT-PCR, and the locations of Sp-CCAPR in the hepatopancreas were further determined by in situ hybridization. Subsequently, we investigated Sp-CCAPR expression profiles following immune stimulation, and finally the immunomodulatory mechanisms of Sp-CCAP and its receptor were evaluated by in vivo and in vitro experiments. This is the first report on CCAP involvement in immunomodulation in an arthropod, and it potentially provides a new strategy for disease control based on neuroendocrine immunity for mud crab aquaculture.
The animal study protocol has been approved by the Animal Ethics Committee of Xiamen University.
Healthy mud crabs (36.36 ± 2.31 g) at the intermolt stage were purchased from a fish market in Haicang District, Xiamen City, Fujian Province, China. Prior to the experiments, the crabs were acclimated in small tanks (40 × 40 × 60 cm) filled with seawater with salinity 30 ppt and temperature 26 ± 0.5°C for 7 days. During the acclimation period, the crabs were fed fresh field snail Cipangopaludina chinensis Gray once daily, and half of tank water was renewed every day.
Total RNA from hemocytes and various tissues, that is, eyestalk ganglion, cerebral ganglion, thoracic ganglion, hepatopancreas, gill, stomach, midgut, heart, epidermis, gonad, and muscle, were extracted using TRIzol Reagent (Invitrogen, USA) according to the manufacturer's instructions. The concentrations and quality of RNAs were checked by a Q6000 spectrophotometer (Quawell), and the integrity was assessed by 1.5% (w/v) agarose gel electrophoresis. The first-strand cDNA was synthesized using PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa) for semi-quantitative RT-PCR and quantitative real-time PCR (qRT-PCR) analyses.
Partial cDNA sequence of Sp-CCAP was obtained from the transcriptome database of Scylla paramamosain (37). The full-length Sp-CCAP cDNA with 1 μg of total RNA extracted from cerebral ganglion was amplified with the SMART™ RACE cDNA Amplification Kit (BD Biosciences). The 3′-race and 5′-race PCR amplification was performed with universal primers [Universal Primer Mix (UPM)] and gene-specific primers for touchdown PCR and nested PCR amplification. PCR products were purified and cloned into the pMD19-T plasmids (TaKaRa). The positive colonies were screened and further confirmed by DNA sequencing. The primer sequences are listed in Table 1.
The ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi) was used to predict open reading frames (ORFs) and amino acid sequence of Sp-CCAP. The amino acid sequence was submitted to predict protein signal peptide with SignalP 5.0 Server (http://www.cbs.dtu.dk/services/SignalP/). The isoelectric point of Sp-CCAP was predicted by ExPASy software. The homology amino acid sequences of Sp-CCAP from other species in the National Center for Biotechnology Information (NCBI) database were obtained through the BlastX homology search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). These sequences were used to create the multiple sequence alignment by MEGA 7.0 software. Phylogenic trees were constructed via the neighbor-joining (NJ) method using MEGA 7.0 software. Bootstrap sampling was reiterated for 1,000 times.
Semi-quantitative RT-PCR was used to detect the distribution of Sp-CCAP and Sp-CCAPR mRNA in hemocytes and tissues from eyestalk ganglion, cerebral ganglion, thoracic ganglion, hepatopancreas, gill, stomach, midgut, heart, epidermis, gonad, and muscle using Sp-CCAP-qF, Sp-CCAP-qR, Sp-CCAPR-qF, and Sp-CCAPR-qR as primers (Table 1). The PCR was performed with the Ex-Taq® DNA polymerase (TaKaRa) under the following conditions: pre-denaturation at 94°C for 5 min and 40 cycles consisting of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s; and the final extension was carried out at 72°C for 10 min. PCR products were resolved on 1.5% agarose gel, and the results were observed and photographed by UV gel imager with water used as a template for negative control and β-actin as an internal control. The experiment was repeated three times.
The 217-bp fragment of Sp-CCAPR was amplified by PCR and cloned into PGEM-T EASY Vector (Promega) for the subsequent in situ hybridization experiment. The riboprobes were synthesized using the DIG RNA Labeling Kit (Roche Diagnostics, Germany) and transcribed by SP6 and T7 polymerases. The hepatopancreas was quickly removed from the crabs and immediately fixed in 4% paraformaldehyde solution for 12 h at 4°C. After being treated with serially diluted ethanol (75, 85, 95, and 100%) and xylene, the sample was embedded in paraffin and sectioned into 0.7-μm continuous sections. Hybridization was subsequently carried out according to the methods reported previously (46) and visualized by the BCIP/NBT Chromogen Kit (Solarbio).
Polyriboinosinic polyribocytidylic acid [Poly (I:C)] (Sigma, USA) was dissolved in crustacean physiological saline (1.13 × 10−2 mol/L of KCl, 1.33 × 10−2 mol/L of CaCl2, 0.44 mol/L of NaCl, 1.0 × 10−2 mol/L of Hepes, 2.3 × 10−2 mol/L of Na2SO4, and 2.6 × 10−2 mol/L of MgCl2, pH 7.4) at 1 mg/ml; and lipopolysaccharide (LPS) (Sigma, USA) was dissolved in crustacean physiological saline at 0.5 mg/ml. Seventy-five crabs were randomly divided into three groups and injected with 100 μl of Poly (I:C), LPS, or crustacean physiological saline (control). In addition, five untreated crabs were used for the initial measurements. Hepatopancreas tissues of five individuals from each treatment group were subsequently randomly sampled at 3, 6, 12, 24, and 48 h for RNA extraction and qRT-PCR analysis. The qPCR used a QuantStudio™ 6 Flex Real-Time PCR (Applied Biosystems) with SYBR® Select Master Mix (TaKaRa). The total reaction volume was 20 μl containing 10 μl of SYBR® Select Master Mix, 2 μl of the five-fold diluted cDNA, 0.5 μl (1.0 × 10−5 mol/L) each of the forward and reverse primers, and 7 μl of ultrapure water; and the procedure included 50°C for 2 min; 95°C for 2 min; followed by 40 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s; and followed by a melting curve analysis at 60–95°C. A relative transcript level was determined using the 2−ΔΔCt algorithm with β-actin from S. paramamosain as the internal control. The sequences of the primers used are listed in Table 1.
The predicted Sp-CCAP mature peptide (PFCNAFTGC-NH2) was synthesized by GL Biochem (Shanghai, China) with a purity of 98% for the subsequent experiments. Forty crabs were randomly divided into two groups: the CCAP treatment group was injected with 100 μl of CCAP mature peptide dissolved in crustacean physiological saline (final concentration in hemolymph about 5 × 10−6 mol/L), whereas the control group was injected with crustacean physiological saline of the same volume. Meanwhile, five untreated crabs were randomly selected for the initial measurement. The hepatopancreas tissues of five crabs from each group were sampled at 3, 6, 12, and 24 h after injection for RNA extraction and gene expression analysis (see Total RNA Extraction and First-Strand cDNA Synthesis and Immune Challenges With Lipopolysaccharide and Polyriboinosinic Polyribocytidylic Acid Injection).
S. paramamosain at the intermolt stage was anesthetized on ice for 10 min, followed by sterilization in 75% ethanol for 5 min. Hepatopancreas tissues were subsequently dissected and washed with crustacean physiological saline before being cut by a pair of scissors into fragments of ~20 mg. The fragments were then precultured at 26°C in a 24-well-plate with 200 μl of L15 medium, which contained penicillin (100 U/ml) and streptomycin (100 μg/ml, Sigma). After an hour, the culture was substituted with L15 medium containing Sp-CCAP peptide at one of three concentrations of 10−6, 10−7, and 10−8 mol/L or without adding any peptide (control), which were based on a previous study from our lab (47). Quadruple treatments were used. After 6 h of culture, tissue fragments were collected from each treatment for total RNA extraction and subsequent qRT-PCR analysis, whereas tissue culture medium was also collected for NO concentration measurement. The NO production was determined using the Total Nitric Oxide Assay Kit (Beyotime, China). Briefly, the absorbance of the nitrite was measured with the Griess reaction at OD540nm, and the nitrite concentration of each tissue culture medium was then calculated according to the standard curve constructed using NaNO2 for the calculation of total NO concentration.
The bacterial clearance assay was carried out based on the method described in a previous study (48), with some modifications. That is, after hepatopancreas tissue with Sp-CCAP mature peptide was added at three concentrations (10−8, 10−7, and 10−6 mol/L) plus a control without adding Sp-CCAP being cultured for 6 h, either Staphylococcus aureus or Vibrio parahaemolyticus suspension (were stored in our laboratory), both pathogenic to the mud crab, was added to culture wells at the final bacterial concentration of ~3 × 104 cfu/ml per well. After another 3 h of culture, each tissue culture medium was inoculated and cultured on either Luria–Bertani (LB) solid medium (for S. aureus) or 2216E solid medium (for V. parahaemolyticus) for 12 h at 37°C, and the number of colonies was then observed and recorded. The assay was performed in triplicates for each culture medium.
All data were presented as mean ± SEM. Statistical differences among treatments were analyzed using one-way ANOVA (followed by Duncan's test) or Student's t-test (SPSS 18.0). Differences were considered statistically significant at p < 0.05 and highly significant at p < 0.01.
The complete cDNA sequence of the Sp-CCAP precursor was obtained by using 3′/5′ RACE coupled to nested PCR. The full length of Sp-CCAP mRNA is 638 bp with a 64-bp 5′ untranslated region (UTR), a 142-bp 3′UTR, and a 432-bp ORF encoding a protein of 143 amino acids with a calculated molecular weight of 15.84 kDa and a theoretical isoelectric point of 9.43 (GenBank Accession MN923209). The deduced precursor peptide contained a signal peptide of 32 amino acids, four putative dibasic (37KR38 and 49KR50), tribasic (61KKR63), and tetrabasic (115RRKR118) cleavage sites, which could give rise to five peptides, including four precursor-related peptides (CCAP AP1: 33-36; CCAP AP2: 39-48; CCAP AP3: 64-114; and CCAP AP4: 115-143) and one mature peptide containing nine amino acids (PFCNAFTGC-NH2) (Figure 1).

Figure 1. Nucleotide and deduced-amino acid sequences of Sp-CCAP cDNA. The initiation codon, termination codon, signal peptide, crustacean cardioactive peptide (CCAP) mature peptide, CCAP-associated peptides, amidation site, cleavage sites, and cysteine residues are marked by different symbols.
Multiple alignment of the amino acid sequences of CCAP precursors from different crustaceans indicated that the CCAP mature peptides were fully identical among the crustacean species used for comparison (Figure 2). Phylogenetic analysis of the amino acid sequences of CCAP precursors among different arthropod species showed that Sp-CCAP and other crustacean CCAP clustered into one branch, whereas insect CCAP clustered into another branch (Figure 3).
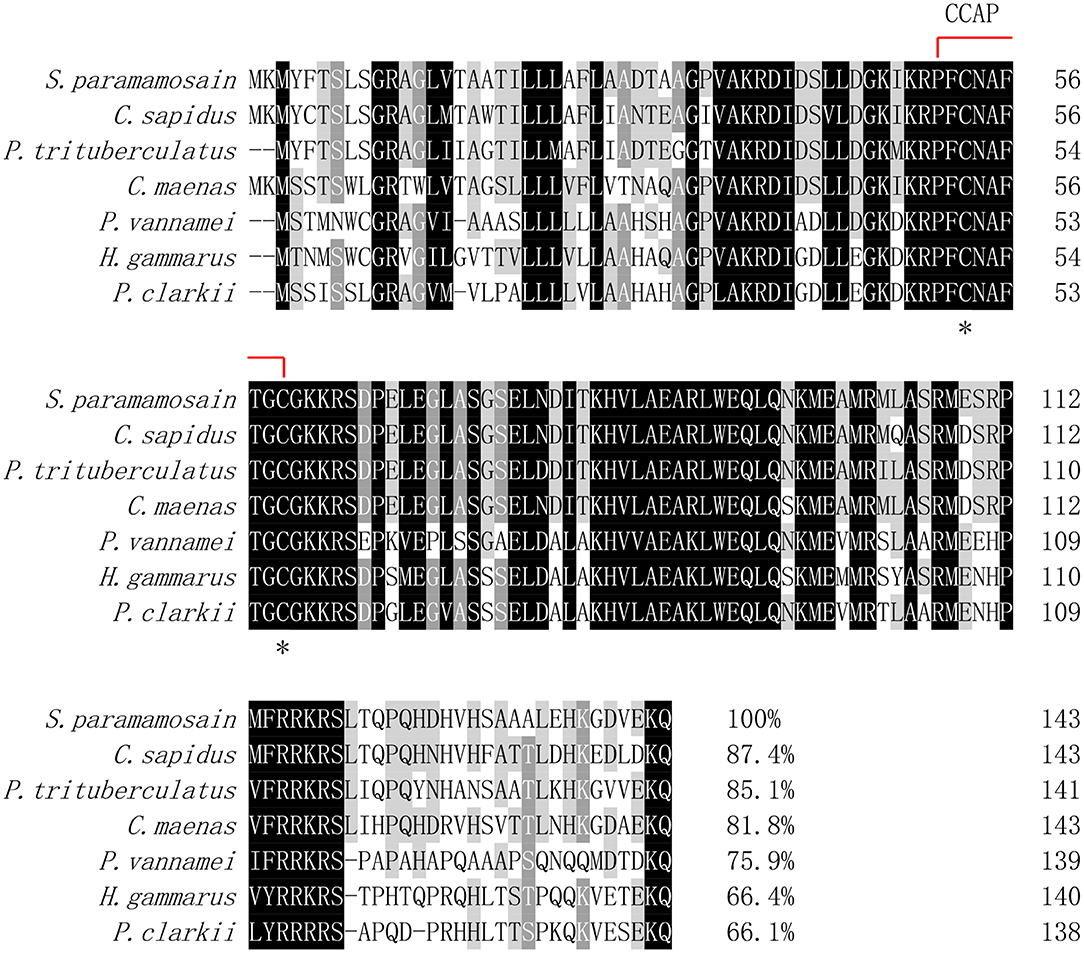
Figure 2. Multiple alignments of the deduced amino acid sequence of crustacean cardioactive peptides (CCAPs) among various crustacean species. The CCAP mature peptide is indicated by the red line; conserved amino acid residues are marked by asterisks. GenBank accession numbers of CCAPs are as follows: Callinectes sapidus (ABB46290.1); Carcinus maenas (ABB46291.1); Portunus trituberculatus (AVK43051.1); Penaeus vannamei (ALP06206.1); Homarus gammarus (ABB46292.1); and Procambarus clarkia (BAF34910.1).
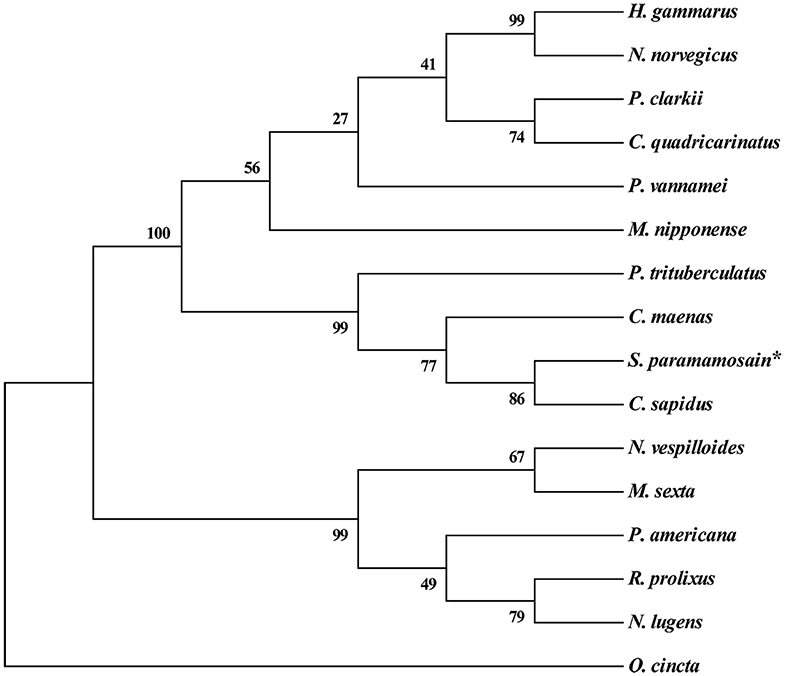
Figure 3. Phylogenetic analysis of crustacean cardioactive peptides (CCAPs) relative to various crustacean and insect species. The sequences used in evolutionary tree analysis include Callinectes sapidus (ABB46290.1); Carcinus maenas (ABB46291.1); Portunus trituberculatus (AVK43051.1); Homarus gammarus (ABB46292.1); Procambarus clarkia (BAF34910.1); Nephrops norvegicus (QBX89037.1); Cherax quadricarinatus (AWK57511.1); Penaeus vannamei (ALP06206.1); Macrobrachium nipponense (ASH96804.1); Periplaneta Americana (Q75UG5.1); Rhodnius prolixus (ACZ52615.1); Nilaparvata lugens (BAO00946.1); Nicrophorus vespilloides (XP_017778790.1); and Orchesella cincta (ODM98622.1).
The expression pattern of Sp-CCAP among various tissues was determined by semi-quantitative RT-PCR. The results showed that Sp-CCAP was expressed in nerve and gonad tissues (Figure 4). To identify the potential target sites of Sp-CCAP, the expression pattern of the Sp-CCAPR transcript was also determined, and Sp-CCAPR was found expressed in 12 tissues, including hemocytes and hepatopancreas (Figure 4).
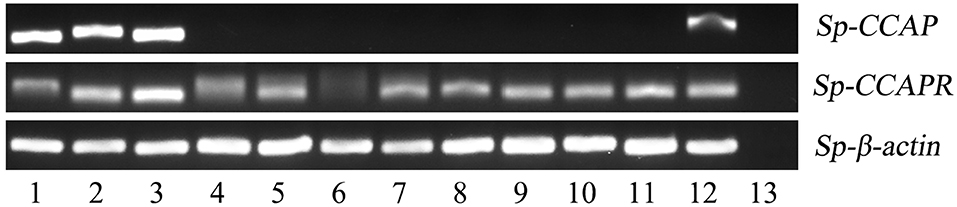
Figure 4. Tissue distribution of Sp-CCAP and Sp-CCAPR in S. paramamosain. 1, eyestalk ganglion; 2, cerebral ganglion; 3, thoracic ganglion; 4, gill; 5, hepatopancreas; 6, hemocytes; 7, stomach; 8, midgut; 9, heart; 10, epidermis; 11, muscle; 12, gonad; and 13, the negative control.
To precisely localize the Sp-CCAPR transcript in hepatopancreas, in situ hybridization was performed. Histological results showed that hepatopancreatic tubule epithelial cells of Scylla paramamosain include E-cells (embryonic), F-cells (fibrillar), B-cells (blisterlike), and R-cells (resorptive) (Figure 5C). In situ hybridization localized Sp-CCAPR-positive signal mainly in the F-cells of hepatopancreas (Figure 5A), whereas in the control group, no such positive signal was found (Figure 5B).
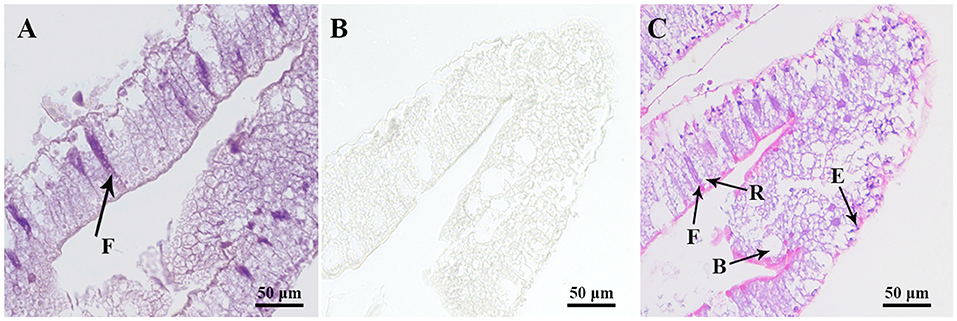
Figure 5. Localization of Sp-CCAPR mRNA in the hepatopancreas by in situ hybridization. (A) Positive signals with the antisense probes. (B) Sense probes of Sp-CCAPR served as the negative control. (C) Histological observation of hepatopancreatic tubule epithelial cells: E, E-cell (E: embryonic); F, F-cell (F: fibrillar); B, B-cell (B: blisterlike); and R-cell (R: resorptive).
Because involvement of neuropeptides in NEI regulation is mainly by binding to their receptors on immune cells, the temporal patterns of Sp-CCAPR mRNA expression in the hepatopancreas after LPS and Poly (I: C) injection were investigated. The results showed that following LPS challenge, a significantly up-regulated mRNA expression level of Sp-CCAPR was only observed at 12 h, which was 2.09-fold of that in the control (p < 0.05) (Figure 6A). However, after Poly (I:C) stimulation, the mRNA expression level of Sp-CCAPR was significantly up-regulated at both 3 and 24 h, with 4.16-fold and 2.28-fold increase, respectively, compared with that of the control at the same time point (p < 0.05) (Figure 6B).
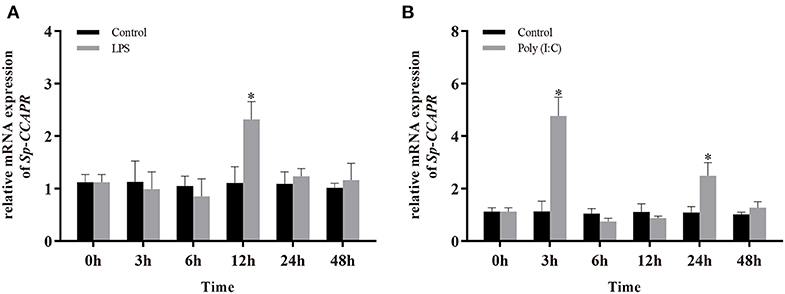
Figure 6. Changes in mRNA expression of Sp-CCAPR in the hepatopancreas after lipopolysaccharide (LPS) and polyriboinosinic polyribocytidylic acid [Poly (I:C)] injection. (A) After LPS stimulation. (B) After Poly (I:C) stimulation. All data are shown as mean ± SEM (n = 5); statistical analysis was performed using Student's t-test. *indicates significant difference (p < 0.05).
To evaluate the potential involvement of Sp-CCAP in immune regulation, after being injected with synthetic Sp-CCAP mature peptide, the changes in mRNA expression levels of immune-related genes, including Sp-CCAPR, nuclear transcription factor NF-κB homologs (Sp-Dorsal and Sp-Relish), member of MAPK signaling pathway (Sp-P38), pro-inflammatory cytokines factor (Sp-TNFSF and Sp-IL16), and AMP (Sp-Lysozyme, Sp-ALF, Sp-ALF4, and Sp-ALF5), in the hepatopancreas were quantified up to 24 h (Figure 7). The mRNA expression of Sp-CCAPR was shown to increase significantly at 6 and 12 h post-Sp-CCAP stimulation, which was 2.69-fold and 2.41-fold of that of the control, respectively (p < 0.05) (Figure 7A). Similarly, the mRNA expression of Sp-P38 was significantly up-regulated to 1.45-fold and 1.65-fold of that in the control at 6 and 12 h, respectively (p < 0.05) (Figure 7B). Likewise, the nuclear transcription factor Sp-Dorsal mRNA expression level was significantly up-regulated at 6, 12, and 24 h (Figure 7C), whereas the Sp-Relish mRNA expression level was significantly up-regulated at 6 and 24 h (p < 0.05) (Figure 7D). Moreover, pro-inflammatory factor Sp-TNFSF and Sp-IL16 mRNA expression levels were both up-regulated significantly at 6 and 12 h (p < 0.05) (Figures 7E,F). Additionally, the mRNA expression level of Sp-lysozyme and Sp-ALF4 increased significantly at 12 h (p < 0.05) but returned to normal at 24 h (p > 0.05) (Figures 7G,I). The mRNA expression level of Sp-ALF1 also increased significantly at 6, 12, and 24 h (p < 0.05) (Figure 7H). Finally, the Sp-ALF5 mRNA expression level was sharply up-regulated at 6 h (p < 0.05) but dropped back to similar levels to that of the control from 12 h onward (p > 0.05) (Figure 7J).
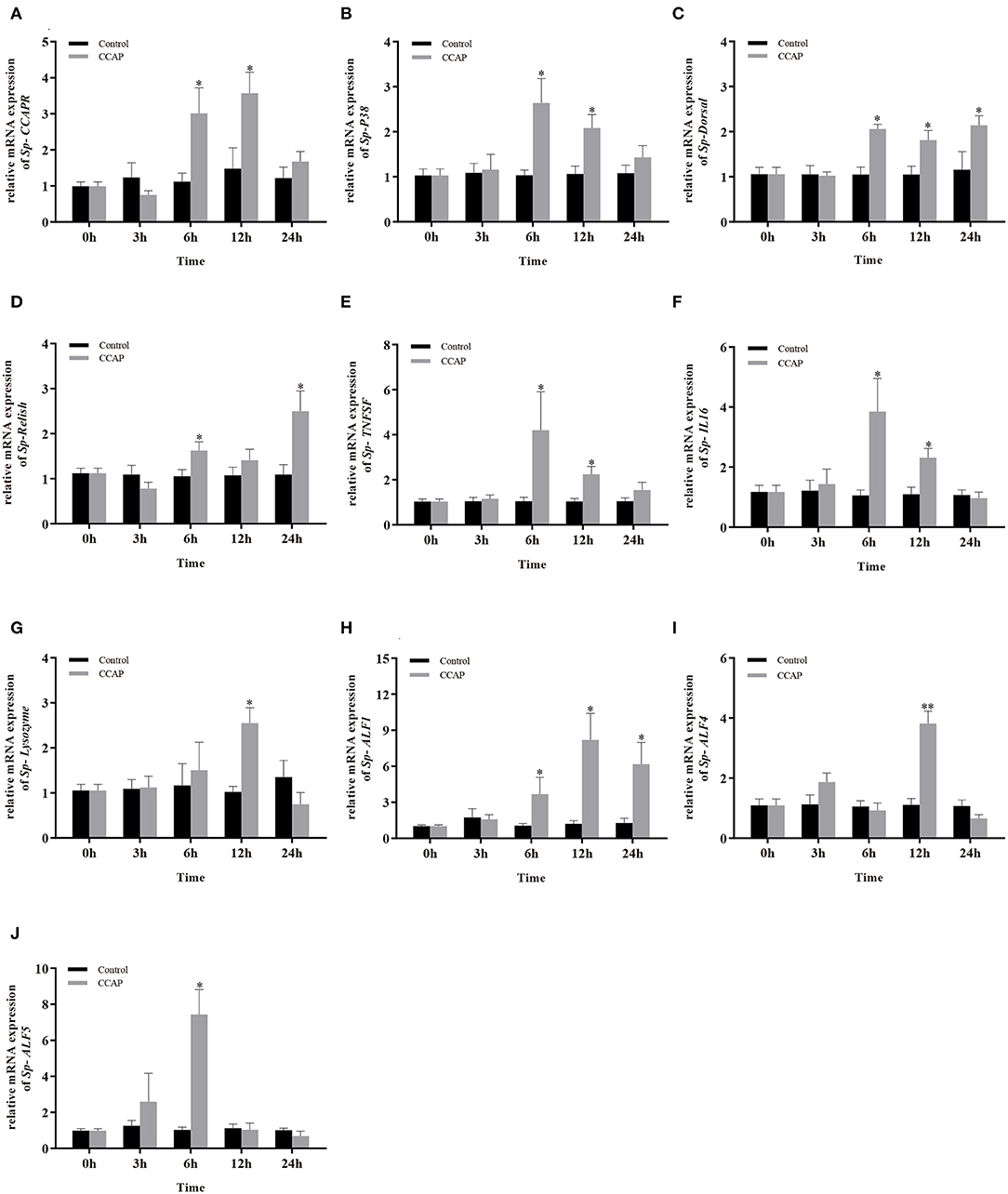
Figure 7. Effects of Sp-CCAP injection on the mRNA expressions of immune-related genes in the hepatopancreas. (A) Sp-CCAPR; (B) Sp-P38; (C) Sp-Dorsal; (D) Sp-Relish; (E) Sp-TNFSF; (F) Sp-IL16; (G) Sp-Lysozyme; (H) Sp-ALF1; (I) Sp-ALF4; (J) Sp-ALF5. All data are shown as mean ± SEM (n = 5); statistical analysis by Student's t-test. * and **on the top of bars indicate significant (p < 0.05) and highly significant differences (p < 0.01), respectively.
To further evaluate the immunoregulation function of Sp-CCAP, Sp-CCAP mature peptide was added to the hepatopancreatic explant cultures at three different concentrations (10−8, 10−7, and 10−6 mol/L), and the expression levels of immune-related genes were measured (Figure 8). The mRNA expression level of both Sp-CCAPR and Sp-P38 was both up-regulated following the addition of Sp-CCAP mature peptide at all three concentrations, and significant differences were detected at 10−8 and 10−7 mol/L as compared with those of the control (p < 0.05) (Figures 8A,B). Moreover, the Sp-Dorsal mRNA expression level was significantly up-regulated at all three concentrations (p < 0.05) (Figure 8C), and the Sp-Relish mRNA expression level was significantly up-regulated at 10−8 and 10−6 mol/L (p < 0.05) (Figure 8D). Similarly, the Sp-TNFSF mRNA expression level was significantly up-regulated at 10−8 and 10−7 mol/L (p < 0.05) (Figure 8E), whereas the Sp-IL16 mRNA expression level was significantly up-regulated at all three concentrations (p < 0.05) (Figure 8F). Finally, the mRNA expression levels of AMP genes Sp-lysozyme, Sp-ALF1, and Sp-ALF5 were significantly up-regulated at 10−8 mol/L (p < 0.05) (Figures 8G,H,J), whereas the mRNA expression level of Sp-ALF4 was not significantly different from that of the control at all concentrations (Figure 8I).
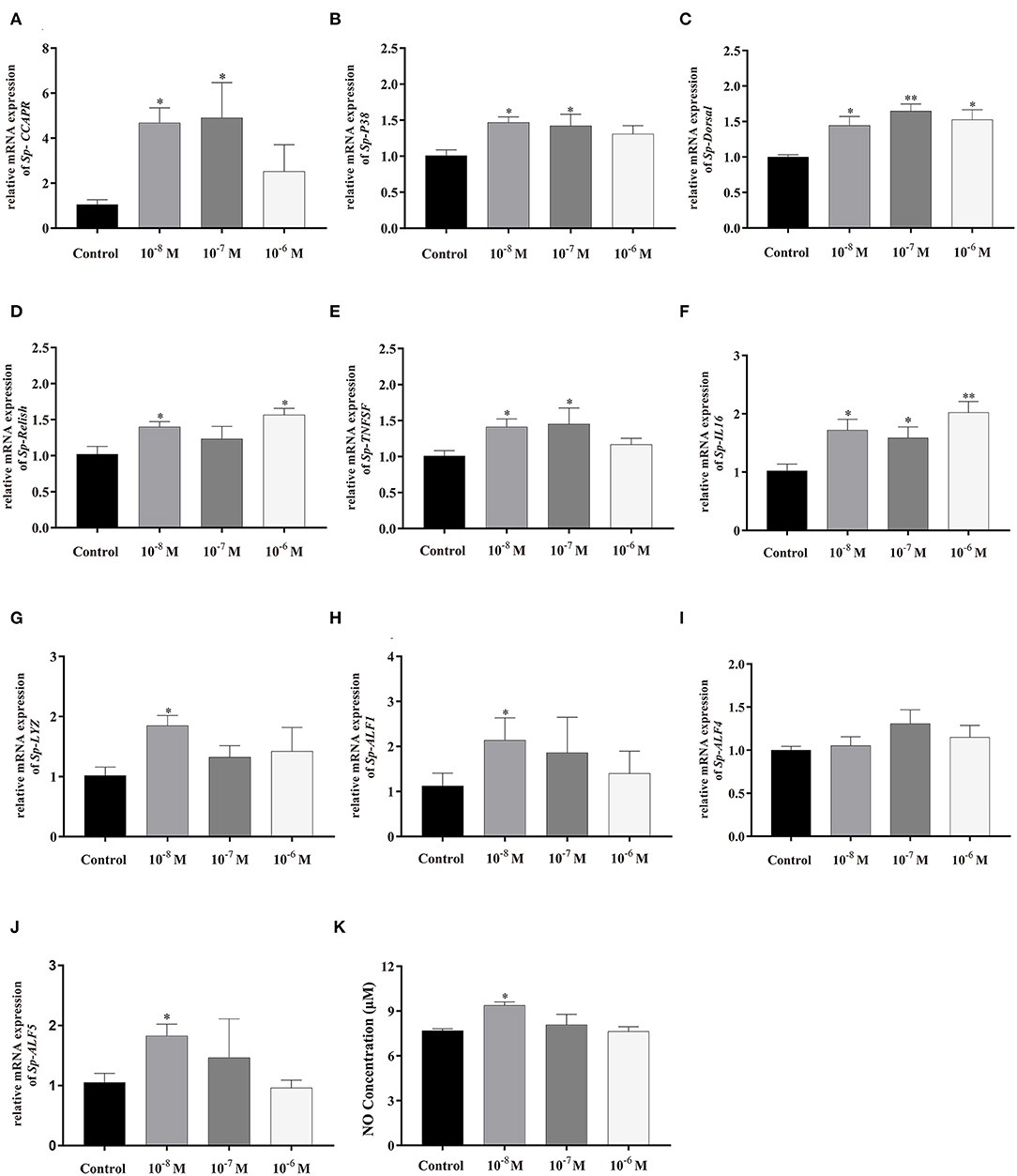
Figure 8. Effects of Sp-CCAP addition at different concentrations on mRNA expressions of immune-related genes in in vitro cultured hepatopancreas tissues and NO concentration in the culture media. (A) Sp-CCAPR; (B) Sp-P38; (C) Sp-Dorsal; (D) Sp-Relish; (E) Sp-TNFSF; (F) Sp-IL16; (G) Sp-Lysozyme; (H) Sp-ALF1; (I) Sp-ALF4; (J) Sp-ALF5; and (K) the concentration of NO. All data are shown as mean ± SEM (n = 4); statistical analysis performed by one-way ANOVA followed by Duncan's test. * and **on top of bars indicate significant (p < 0.05) and highly significant differences (p < 0.01), respectively.
NO is an important gaseous signaling molecule that plays a key role in the innate immune system; NO concentration changes in the hepatopancreas culture media were also determined after adding Sp-CCAP mature peptide at different concentrations (Figure 8J). It showed that when treated with Sp-CCAP at 10−8 M, NO concentration in the medium increased significantly (p < 0.05); however, NO content did not significantly vary from the control at the concentrations of 10−6 and 10−7 mol/L (Figure 8J).
The bacterial clearance capability of each hepatopancreas culture medium with Sp-CCAP mature peptide added at 10−8, 10−7, and 10−6 mol/L was evaluated against the control (no Sp-CCAP addition) to assess whether up-regulated immune molecules led to enhanced antibacterial capacity. The results showed that based on colony counts, in both cases of S. aureus and Vibrio parahaemolyticus, bacteria numbers in all tissue culture media with Sp-CCAP mature peptide addition decreased compared with those of the control; and the improvement in bacterial clearance capacity was significant when Sp-CCAP mature peptide was added at 10−8 and 10−7 mol/L (p < 0.05) (Figures 9A–D).
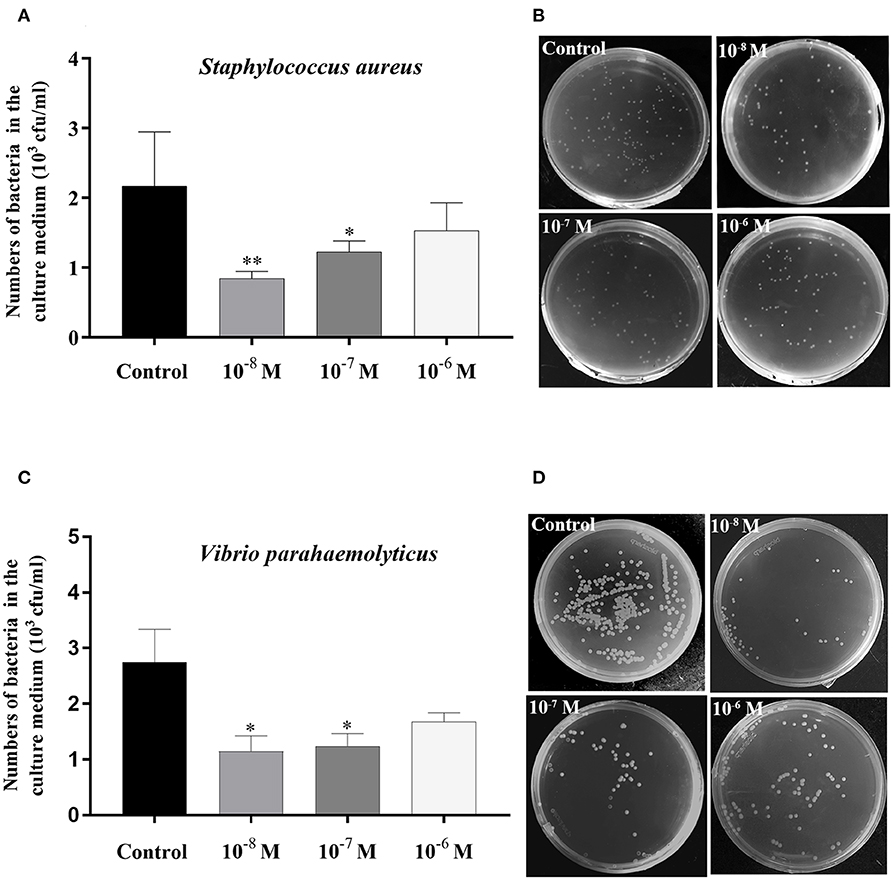
Figure 9. Bacterial clearance capacity of hepatopancreas culture medium treated with different concentrations of Sp-CCAP as compared with the no Sp-CCAP addition control. (A) Results with Staphylococcus aureus. (B) S. aureus colonies grown on Luria–Bertani (LB) plates. (C) Results with Vibrio parahaemolyticus. (D) S. aureus colonies grown on 2216E plates. All data are shown as mean ± SEM (n = 4); statistical analysis performed by one-way ANOVA followed by Duncan's test. * and **on top of bars indicate significant (p < 0.05) and highly significant differences (p < 0.01), respectively.
As a multifunctional peptide hormone, CCAP is known to play an important role in the regulation of various physiological processes in arthropods (23, 26–34). Its immunomodulatory function, however, has never been reported previously; the present study hence appears to be the first to report the involvement of CCAP in regulating hepatopancreas immunity in an arthropod.
In this study, a cDNA encoding CCAP precursor was identified from mud crab Scylla paramamosain. In the phylogenetic tree constructed, all CCAP precursor peptides could be divided into two branches of crustacean and insect. The Sp-CCAP precursor peptide fell into the branch of crustacean, indicating that Sp-CCAP shared high similarity with other crustaceans. By comparing precursor peptides of all sequences from crustaceans, four restriction sites were identified, suggesting that their translation processing modules are highly conserved. CCAP mature peptide sequences (PFCNAFTGC-NH2) are identical among different species, which indicates essential roles of CCAP to arthropods.
The distribution of genes in different tissues has a guiding role for exploring the physiological functions of neuropeptides (10). RT-PCR results showed that mRNA transcript of Sp-CCAP was mainly expressed in the nervous system and gonads of S. paramamosain, whereas the Sp-CCAPR mRNA was expressed in a wide range of different tissues. However, in American cockroach Periplaneta americana and Pacific white shrimp L. vannamei, CCAP mRNA was reportedly expressed in the midgut and the nervous system (26, 27), which suggests that likely there are different physiological regulation pathways of CCAP in arthropod species. In the present study, the detection of mRNA expression of both Sp-CCAP and Sp-CCAPR in gonads suggested that they may act as an autocrine/paracrine factor to regulate ovarian development, similar to that of short neuropeptide F (sNPF) identified in the same crab species (49). Neurohormones have been found to participate in immune regulation through receptors on hemocytes, the well-known immune-related cells that play crucial roles in host immune defense in crustaceans (50). For instance, in insects, 5-HT receptors were found expressed in the hemocytes, and 5-HT has been shown to modulate hemocyte phagocytosis through 5HT1B and 5-HT2B receptors (51). Similarly, Es-GPCR89 mRNA was expressed in the hemocytes of Chinese mitten crab Eriocheir sinensis and found to mediate cerebral antimicrobial activity (52). In this study, Sp-CCAPR was found expressed in the hemocytes, suggesting that Sp-CCAP may be involved in the immune regulation of hemocytes as a neurohormone in S. paramamosain. In crustaceans, the hepatopancreas is not only the major organ responsible for digestion but also an important immune organ (16). Indeed, in crayfish Procambarus clarkii, a putative GPCR gene, HP1R, was found expressed in the hepatopancreas and was suggested to play a role in protecting the host from bacterial invasion (53). In this study, in situ hybridization showed that Sp-CCAPR mRNA was mainly expressed in F-cells of hepatopancreas, indicating that Sp-CCAP may play an immunomodulatory role via its receptor on F-cells in S. paramamosain.
Increasingly studies have shown that neuropeptides play an important role in the NEI network; typically, they are activated by immune stimuli and bind to their receptors to participate in the innate immune responses (2, 54). In this study, Sp-CCAPR was found to distribute in the hepatopancreas of S. paramamosain, and its mRNA expression greatly increased after the stimulation of LPS and Poly (I:C), which suggested that both LPS and Poly (I:C) challenges might activate Sp-CCAP to bind to its receptor in the hepatopancreas to participate in immune responses. Interestingly, previous studies have reported similar results in other species. For example, HPR1 gene in the hepatopancreas of P. clarkii was significantly up-regulated by stimulation with Aeromonas hydrophila (53), the mRNA of LPSenhR-1 was significantly up-regulated after LPS stimulation in rainbow trout Oncorhynchus mykiss (55), and Poly (I:C) stimulation induced significantly higher neurokinin-2 receptor mRNA expression in human dendritic cell (56). It is well-known that the neuroendocrine system regulates the immune responses by releasing neuropeptide hormones, whereas the immune system activates the neuroendocrine system by secreting cytokines, thus forming a circular network of neuroendocrine and immune regulation (57). Sp-CCAPR showed two expression peaks at 3 and 24 h after Poly (I:C) stimulation; it is speculated that Sp-CCAP/Sp-CCAPR can respond quickly to Poly (I:C) stimulation and be indirectly affected by Poly (I:C)-induced immune factor activation.
In this study, the immunomodulatory effect of Sp-CCAP was further studied by in vivo injection and in vitro culture experiment. In the in vivo experiment, Sp-CCAP was found to significantly induce the expression of its receptor Sp-CCAPR, P38 MAPKs (Sp-P38), and nuclear transcription factor NF-κBs (Sp-Dorsal and Sp-Relish) in the hepatopancreas, which suggested that by stimulating the expression of Sp-CCAPR, Sp-CCAP likely induced increased amount of Sp-CCAPR on the cell membrane in the hepatopancreas, thus greatly enhancing the activity of signaling pathway mediated by Sp-CCAP. On the other hand, P38 MAPKs as a member of MAPK superfamily, can associate extracellular signals with intracellular mechanisms (58) and play a crucial role in the inflammatory response and the host defense against pathogen infections (59). The NF-κB pathway is an essential pathway for the innate immune response to pathogen invasion in both vertebrates and invertebrates (60). Therefore, in order to explore the possibility that immune molecules regulated by Sp-CCAP and its receptor Sp-CCAPR signaling pathway, the mRNA expression levels of several immune molecules with pro-inflammatory and antibacterial properties, that is, Sp-IL16, Sp-TNFSF, and AMPs, were detected. Of these immune molecules, AMPs are key effector molecules that induce innate immunity in various invertebrates (61). As one of the AMP family, the anti-LPS factor (ALF) is well-known to possess a wide range of antibacterial, antifungal, and antiviral properties (62). Another AMP, lysozyme, reportedly protects organisms by destroying the cell walls of infectious bacterial pathogens (63). Meanwhile, interleukin-16 (IL-16), a pleiotropic cytokine, plays essential roles in the regulation of various innate immune processes (64), and has been reported to play a strong positive role in antibacterial responses in L. vannamei (65). Relish and Dorsal are invertebrate NF-κB homologs, they function as essential transcription factors on mediating the activation of AMP genes in crustaceans (66), and the expression of IL-16 is dependent on the NF-κB pathway (67). The P38 MAPK plays a key role in a variety of immune responses by regulating the production of pro-inflammatory cytokines, including TNFs and IFNs (58, 59). In this study, the up-regulation of nuclear transcription factor NF-κBs, pro-inflammatory factor IL-16, and AMP genes suggests that Sp-CCAP might influence the expressions of Sp-IL16 and AMPs by mediating the NF-κB signaling pathway, whereas the up-regulation of Sp-P38 and Sp-TNFSF suggests that Sp-CCAP might affect the expression of Sp-TNFSF by mediating the P38 MAPK signaling pathway. Together, these results suggest that Sp-CCAP possibly activates and induces inflammatory and antimicrobial responses in S. paramamosain. Interestingly, the Sp-CCAPR expression level was not significantly elevated at 24 h post Sp-CCAP mature peptide injection, whereas Sp-Dorsal, Sp-Relish, and Sp-ALF1 expression levels were still up-regulated. This phenomenon may be explained by the possible accumulation of sufficient receptor proteins on the cell membrane, thus promoting these gene expressions.
The results of the in vitro experiment were similar to those of in vivo experiment; that is, adding Sp-CCAP mature peptide to the hepatopancreas cultures could promote the expressions of signal pathway-related genes and immune effector molecules. They provided further evidence that Sp-CCAP was involved in hepatopancreas immunity of S. paramamosain. Moreover, NO is an important gaseous signal molecule that plays anti-bacterial and inflammatory roles in invertebrates (68). In the present study, it was found that NO concentration in the hepatopancreas culture medium increased significantly when Sp-CCAP mature peptide was added at 10−8 mol/L. It provided additional evidence from the point of view of gaseous signal molecules that Sp-CCAP likely plays a significant role in the hepatopancreas immunity of S. paramamosain.
Finally, the in vitro bacterial clearance experiment showed that the up-regulation of immune effector molecules in the hepatopancreas mediated by Sp-CCAP signaling pathway could effectively resist bacterial infection. Indeed, the results demonstrated for the first time that neuropeptides play antibacterial roles in the hepatopancreas of a crustacean, likely via regulating immune-effector molecules.
In invertebrates, on the one hand, when neuropeptides act on immune cells, neuropeptide receptors on the membrane can activate the G-protein Gαs/Gαi subunit and react with adenylate cyclase to increase or decrease intracellular cAMP concentration (69). On the other hand, when neuropeptides bind to Go/Gq protein-coupled receptors on immune cells, they can activate the activity of phospholipase C, thereby altering intracellular Ca2+ concentration (69). The changes of these secondary messengers (cAMP and Ca2+) activate a series of signaling pathways, such as MAPK (JNK, ERK, and P38) and NF-κB signaling pathway, through cascade amplification. They synergistically promote the release of inflammatory factors and immune factors (70). CCAPR was first identified in Drosophila but has since been identified in many other insects; it is known that the involvement of CCAPR in various physiological processes, such as molt and heartbeat regulation, is mediated by CCAP (35, 36, 71). A previous study has shown that the binding of Sp-CCAP to Sp-CCAPR activated the cAMP level and Ca2+ signal response in the cytoplasm of S. paramamosain (38). Therefore, in this study, activating P38 MAPKs and NF-κB signaling pathways by the binding of Sp-CCAP to Sp-CCAPR were likely via cAMP and Ca2+ concentration changes in hepatopancreas cells, which promoted the expression of Sp-IL16, Sp-TNFSF, Sp-LYZ, Sp-ALF1, Sp-ALF4, and Sp-ALF5, hence enhancing the immune responses of S. paramamosain to pathogen infection.
In summary, the present study provides the first evidence that CCAP plays an immunomodulatory role in the hepatopancreas in a crustacean. It suggests that CCAP might activate immune effector molecules mediated by the P38 MAPK pathway and NF-κB pathway in the hepatopancreas to resist pathogen infection. This study also potentially provides a new strategy for disease control from the perspective of neuroendocrine immunity for the mud crab aquaculture.
The datasets generated for this study can be found in the National Center for Biotechnology Information GenBank Accession MN923209.
All the animals used in this study have been approved by the Animal Ethics Committee of Xiamen University.
YW and HY designed the experiments. YW and DL performed the experiments. YW and ZX analyzed the data. XG contributed reagents/materials tools. YW and HY contributed to the discussion. YW wrote the manuscript. HY and CZ revised the manuscript. All the authors read and approved the final manuscript.
This study was supported by the Natural Science Foundation of China (Nos. 31972765 and 31772827).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank all laboratory members for their constructive suggestions and discussions, as well as the reviewers for their valuable comments.
1. Besedovsky H, Sorkin E. Network of immune-neuroendocrine interactions. Clin Exp Immunol. (1977) 7:1–12.
2. Berczi I, Chalmers IM, Nagy E, Warrington RJ. The immune effects of neuropeptides. Baillières Clin Rheumatol. (1996) 10:227–57. doi: 10.1016/S0950-3579(96)80016-1
3. Merighi A. Neuropeptides and coexistence. In: Squire LR, editor. Encyclopedia of Neuroscience. Oxford: Academic Press (2009). p. 843–49. doi: 10.1016/B978-008045046-9.01467-4
4. Bockaert J, Claeysen S, Bécamel C, Pinloche S, Dumuis A. G protein-coupled receptors: dominant players in cell–cell communication. Int Rev Cytol. (2002) 212:63–132. doi: 10.1016/S0074-7696(01)12004-8
5. Delgado M, Martinez C, Pozo D, Calvo JR, Leceta J, Ganea D, et al. Vasoactive intestinal peptide (VIP) and pituitary adenylate cyclase-activation polypeptide (PACAP) protect mice from lethal endotoxemia through the inhibition of TNF-α and IL-6. J Immunol. (1999) 162:1200–05.
6. Martinez C, Abad C, Delgado M, Arranz A, Juarranz MG, Rodriguez-Henche N, et al. Anti-inflammatory role in septic shock of pituitary adenylate cyclase-activating polypeptide receptor. Proc Natl Acad Sci USA. (2002) 99:1053–58. doi: 10.1073/pnas.012367999
7. Gonzalez-Rey E, Chorny A, Varela N, Robledo G, Delgado M. Urocortin and adrenomedullin prevent lethal endotoxemia by down-regulating the inflammatory response. Am J Pathol. (2006) 168:1921–30. doi: 10.2353/ajpath.2006.051104
8. Peña B, Isla A, Haussmann D, Figueroa J. Immunostimulatory effect of salmon prolactin on expression of toll-like receptors in Oncorhynchus mykiss infected with Piscirickettsia salmonis. Fish Physiol Biochem. (2016) 42:509–16. doi: 10.1007/s10695-015-0155-5
9. Kono T, Ida T, Kawahara N, Watanabe F, Biswas J, Sato T, et al. Identification and immunoregulatory function of neuromedin U (Nmu) in the Japanese pufferfish, Takifugu rubripes. Dev Comp Immunol. (2017) 73:246–56. doi: 10.1016/j.dci.2017.03.007
10. Li M, Wang M, Wang W, Wang L, Liu Z, Sun J, et al. The immunomodulatory function of invertebrate specific neuropeptide FMRFamide in oyster Crassostrea gigas. Fish Shellfish Immunol. (2019) 88:480–88. doi: 10.1016/j.fsi.2019.03.023
11. Bachtel ND, Hovsepian GA, Nixon DF, Eleftherianos I. Allatostatin C modulates nociception and immunity in Drosophila. Sci Rep. (2018) 8:7501. doi: 10.1038/s41598-018-25855-1
12. Wanlem S, Supamattaya K, Tantikitti C, Prasertsan P, Graidist P. Expression and applications of recombinant crustacean hyperglycemic hormone from eyestalks of white shrimp (Litopenaeus vannamei) against bacterial infection. Fish Shellfish Immunol. (2011) 30:877–85. doi: 10.1016/j.fsi.2011.01.014
13. Wang L, Chen H, Xu J, Xu Q, Wang M, Zhao D, et al. Crustacean hyperglycemic hormones directly modulate the immune response of hemocytes in shrimp Litopenaeus vannamei. Fish Shellfish Immunol. (2017) 62:164–74. doi: 10.1016/j.fsi.2017.01.007
14. Zuo H, Yuan J, Niu S, Yang L, Weng S, He J, et al. A molting-inhibiting hormone-like protein from Pacific white shrimp Litopenaeus vannamei is involved in immune responses. Fish Shellfish Immunol. (2018) 72:544–51. doi: 10.1016/j.fsi.2017.11.031
15. Zhou M, Abbas MN, Kausar S, Jiang C, Dai L. Transcriptome profiling of red swamp crayfish (Procambarus clarkii) hepatopancreas in response to lipopolysaccharide (LPS) infection. Fish Shellfish Immunol. (2017) 71:423–33. doi: 10.1016/j.fsi.2017.10.030
16. Roszer T. The invertebrate midintestinal gland (“hepatopancreas”) is an evolutionary forerunner in the integration of immunity and metabolism. Cell Tissue Res. (2014) 358:685–95. doi: 10.1007/s00441-014-1985-7
17. Duan Y, Liu P, Li J, Li J, Chen P. Expression profiles of selenium dependent glutathione peroxidase and glutathione S-transferase from Exopalaemon carinicauda in response to Vibrio anguillarum and WSSV challenge. Fish Shellfish Immunol. (2013) 35:661–70. doi: 10.1016/j.fsi.2013.05.016
18. Cao J, Wang Z, Zhang Y, Qu F, Guo L, Zhong M, et al. Identification and characterization of the related immune-enhancing proteins in crab Scylla paramamosain stimulated with rhubarb polysaccharides. Mol Immunol. (2014) 57:263–73. doi: 10.1016/j.molimm.2013.10.003
19. Duan Y, Liu P, Li J, Wang Y, Li J, Chen P. Molecular responses of calreticulin gene to Vibrio anguillarum and WSSV challenge in the ridgetail white prawn Exopalaemon carinicauda. Fish Shellfish Immunol. (2014) 36:164–71. doi: 10.1016/j.fsi.2013.10.024
20. Zheng J, Mao Y, Su Y, Wang J. Effects of nitrite stress on mRNA expression of antioxidant enzymes, immune-related genes and apoptosis-related proteins in Marsupenaeus japonicus. Fish Shellfish Immunol. (2016) 58:239–52. doi: 10.1016/j.fsi.2016.08.058
21. Bao S, Zheng Z, Aweya JJ, Yao D, Li S, Sun C, et al. MicroRNA-589-5p modulates the expression of hemocyanin as part of the anti-WSSV immune response in Litopenaeus vannamei. Dev Comp Immunol. (2020) 107:103642. doi: 10.1016/j.dci.2020.103642
22. Ren X, Liu P, Li J. Comparative transcriptomic analysis of Marsupenaeus japonicus hepatopancreas in response to Vibrio parahaemolyticus and white spot syndrome virus. Fish Shellfish Immunol. (2019) 87:755–64. doi: 10.1016/j.fsi.2019.02.030
23. Stangier J, Hilbich C, Beyreuther K, Keller R. Unusual cardioactive peptide (CCAP) from pericardial organs of the shore crab Carcinus maenas. Proc Natl Acad Sci USA. (1987) 84:575–79. doi: 10.1073/pnas.84.2.575
24. Dircksen H, Homberg U. Crustacean cardioactive peptide-immunoreactive neurons innervating brain neuropils, retrocerebral complex and stomatogastric nervous system of the locust, Locusta migratoria. Cell Tissue Res. (1995) 279:495–515. doi: 10.1007/BF00318163
25. Dircksen H, Keller R. Immunocytochemical localization of CCAP, a novel crustacean cardioactive peptide, in the nervous system of the shore crab, Carcinus maenas L. Cell Tissue Res. (1988) 254:347–60. doi: 10.1007/BF00225807
26. Matsui T, Sakai T, Satake H, Takeda M. The pars intercerebralis affects digestive activities of the American cockroach, Periplaneta americana, via crustacean cardioactive peptide and allatostatin-A. J Insect Physiol. (2013) 59:33–37. doi: 10.1016/j.jinsphys.2012.06.010
27. Mikani A, Watari Y, Takeda M. Brain-midgut cross-talk and autocrine metabolastat via the sNPF/CCAP negative feed-back loop in the American cockroach, Periplaneta americana. Cell Tissue Res. (2015) 362:481–96. doi: 10.1007/s00441-015-2242-4
28. Chen T, Ren C, Wang Y, Gao Y, Wong NK, Zhang L. Crustacean cardioactive peptide (CCAP) of the Pacific white shrimp (Litopenaeus vannamei): molecular characterization and its potential roles in osmoregulation and freshwater tolerance. Aquaculture. (2016) 451:405–12. doi: 10.1016/j.aquaculture.2015.10.005
29. Shan D, Fu H, Qiao H, Sun S, Zhang W, Jin S. Molecular cloning, characterization, and expression of crustacean cardioactive peptide in oriental river prawn, Macrobrachium nipponense, during acute hypoxia and reoxygenation. J World Aquacult Soc. (2018) 49:356–65. doi: 10.1111/jwas.12502
30. Nichols R, Kaminski S, Walling E, Zornik E. Regulating the activity of a cardioacceleratory peptide. Peptides. (1999) 20:1153–58. doi: 10.1016/S0196-9781(99)00118-7
31. Fort TJ, Garcia-Crescioni K, Agricola HJ, Brezina V, Miller MW. Regulation of the crab heartbeat by crustacean cardioactive peptide (CCAP): central and peripheral actions. J Neurophysiol. (2007) 97:3407–20. doi: 10.1152/jn.00939.2006
32. Lange AB, Patel K. The presence and distribution of crustacean cardioactive peptide in the central and peripheral nervous system of the stick insect, Baculum extradentatum. Regul Pept. (2005) 129:191–201. doi: 10.1016/j.regpep.2005.02.011
33. Silva RD, Lange AB. The association of crustacean cardioactive peptide with the spermatheca of the African migratory locust, Locusta migratoria. J Insect Physiol. (2006) 52:399–409. doi: 10.1016/j.jinsphys.2006.01.006
34. Donini A, Lange AB. The effects of crustacean cardioactive peptide on locust oviducts are calcium-dependent. Peptides. (2002) 23:683–91. doi: 10.1016/S0196-9781(01)00662-3
35. Lee D, Broeck JV, Lange AB. Identification and expression of the CCAP receptor in the chagas' disease vector, Rhodnius prolixus, and its involvement in cardiac control. PLoS ONE. (2013) 8:e68897. doi: 10.1371/journal.pone.0068897
36. Arakane Y, Li B, Muthukrishnan S, Beeman RW, Kramer KJ, Park Y. Functional analysis of four neuropeptides, EH, ETH, CCAP and bursicon, and their receptors in adult ecdysis behavior of the red flour beetle, Tribolium castaneum. Mech Dev. (2008) 125:984–95. doi: 10.1016/j.mod.2008.09.002
37. Bao C, Yang Y, Huang H, Ye H. Neuropeptides in the cerebral ganglia of the mud crab, Scylla paramamosain: transcriptomic analysis and expression profiles during vitellogenesis. Sci Rep. (2015) 5:17055. doi: 10.1038/srep17055
38. Bao C, Yang Y, Zeng C, Huang H, Ye H. Identifying neuropeptide GPCRs in the mud crab, Scylla paramamosain, by combinatorial bioinformatics analysis. Gen Comp Endocrinol. (2018) 269:122–30. doi: 10.1016/j.ygcen.2018.09.002
39. Li Y, Xia X, Wu Q, Liu W, Lin Y. Infection with Hematodinium sp. in mud crabs Scylla serrata cultured in low salinity water in southern China. Dis Aquatic Organ. (2008) 82:145–50. doi: 10.3354/dao01988
40. Li S, Zhang Z, Li C, Zhou L, Liu W, Li Y. Molecular cloning and expression profiles of nitric oxide synthase (NOS) in mud crab Scylla paramamosain. Fish Shellfish Immunol. (2012) 32:503–12. doi: 10.1016/j.fsi.2011.12.002
41. Wei X, Wang L, Sun W, Zhang M, Ma H, Zhang Y. C-type lectin B (SpCTL-B) regulates the expression of antimicrobial peptides and promotes phagocytosis in mud crab Scylla paramamosain. Dev Comp Immunol. (2018) 84:213–29. doi: 10.1016/j.dci.2018.02.016
42. Gu WB, Liu ZP, Zhou YL, Li B, Wang LZ, Dong WR. The nuclear factor interleukin 3-regulated (NFIL3) transcription factor involved in innate immunity by activating NF-κB pathway in mud crab Scylla paramamosain. Dev Comp Immunol. (2019) 101:103452. doi: 10.1016/j.dci.2019.103452
43. Chen B, Fan DQ, Zhu KX, Shan ZG, Chen FY, Wang KJ. Mechanism study on a new antimicrobial peptide Sphistin derived from the N-terminus of crab histone H2A identified in haemolymphs of Scylla paramamosain. Fish Shellfish Immunol. (2015) 47, 833–846. doi: 10.1016/j.fsi.2015.10.010
44. Xie JW, Cheng CH, Ma HL, Feng J, Su YL, Guo ZX. Molecular characterization, expression and antimicrobial activities of a c-type lysozyme from the mud crab, Scylla paramamosain. Dev Comp Immunol. (2019) 98, 54–64. doi: 10.1016/j.dci.2019.04.002
45. Yu Z, Geng Y, Huang A, Wang K, Huang X, Wang J. Molecular characterization of a p38 mitogen-activated protein kinase gene from Scylla paramamosain and its expression profiles during pathogenic challenge. J Invertebr Pathol. (2017) 144, 32–36. doi: 10.1016/j.jip.2017.01.001
46. Wang Y, Chen Y, Han K, Zou Z, Zhang Z. A vasa gene from green mud crab Scylla paramamosain and its expression during gonadal development and gametogenesis. Mol Biol Rep. (2012) 39:4327–35. doi: 10.1007/s11033-011-1220-5
47. Liu J, Liu A, Liu F, Huang H, Ye H. Role of neuroparsin 1 in vitellogenesis in the mud crab, Scylla paramamosain. Gen Comp Endocrinol. (2020) 285:113248. doi: 10.1016/j.ygcen.2019.113248
48. An S, Dong S, Wang Q, Li S, Gilbert LI, Stanley D. Insect neuropeptide bursicon homodimers induce innate immune and stress genes during molting by activating the NF-κB transcription factor Relish. PLoS ONE. (2012) 7:e34510. doi: 10.1371/journal.pone.0034510
49. Bao C, Yang Y, Huang H, Ye H. Inhibitory role of the mud crab short neuropeptide F in vitellogenesis and oocyte maturation via autocrine/paracrine signaling. Front Endocrinol. (2018) 9:390. doi: 10.3389/fendo.2018.00390
50. Johansson MW, Keyser P, Sritunyalucksana K, Söderhäll K. Crustacean haemocytes and haematopoiesis. Aquaculture. (2000) 191:45–52. doi: 10.1016/S0044-8486(00)00418-X
51. Qi Y, Huang J, Li M, Wu Y, Xia R, Ye G. Serotonin modulates insect hemocyte phagocytosis via two different serotonin receptors. Elife. (2016) 5:e12241. doi: 10.7554/eLife.12241
52. Qin X, Jin X, Zhou K, Li H, Wang Q, Li W, et al. EsGPCR89 regulates cerebral antimicrobial peptides through hemocytes in Eriocheir sinensis. Fish Shellfish Immunol. (2019) 95:151–62. doi: 10.1016/j.fsi.2019.10.015
53. Dong C, Zhang P. A putative G protein-coupled receptor involved in innate immune defense of Procambarus clarkii against bacterial infection. Comp Biochem Physiol Mol Integr Physiol. (2012) 161:95–101. doi: 10.1016/j.cbpa.2011.09.006
54. Taghert PH, Nitabach MN. Peptide neuromodulation in invertebrate model systems. Neuron. (2012) 76:82–97. doi: 10.1016/j.neuron.2012.08.035
55. Kiryu I, Köllner B, Kuroda A, Ototake M, Dijkstra JM. A new putative G-protein coupled receptor gene associated with the immune system of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. (2003) 15:117–127. doi: 10.1016/S1050-4648(02)00143-2
56. Kitamura H, Kobayashi M, Wakita D, Nishimura T. Neuropeptide signaling activates dendritic cell-mediated type 1 immune responses through neurokinin-2 receptor. J Immunol. (2012) 188:4200–08. doi: 10.4049/jimmunol.1102521
57. Verburg-van Kemenade BML, Van der Aa LM, Chadzinska M. Neuroendocrine-immune interaction: regulation of inflammation via G-protein coupled receptors. Gen Comp Endocrinol. (2013) 188:94–101. doi: 10.1016/j.ygcen.2012.11.010
58. Ono K, Han J. The p38 signal transduction pathway activation and function. Cell Signal. (2000) 12:1–13. doi: 10.1016/S0898-6568(99)00071-6
59. Sun J, Wang L, Wu Z, Han S, Wang L, Li M. P38 is involved in immune response by regulating inflammatory cytokine expressions in the Pacific oyster Crassostrea gigas. Dev Comp Immunol. (2019) 91:108–114. doi: 10.1016/j.dci.2018.10.011
60. Jiang M, Tu D, Gu W, Zhou Y, Zhu Q, Guo X. Identification and functional analysis of inhibitor of NF-κB kinase (IKK) from Scylla paramamosain: the first evidence of three IKKs in crab species and their expression profiles under biotic and abiotic stresses. Dev Comp Immunol. (2018) 84:199–212. doi: 10.1016/j.dci.2018.02.014
61. Diamond G, Beckloff N, Weinberg A, Kisich KO. The roles of antimicrobial peptides in innate host defense. Curr Pharm Des. (2009) 15:2377–92. doi: 10.2174/138161209788682325
62. Li J, Zhang Y, Zhang Y, Mao F, Xiang Z, Xiao S. The first invertebrate NFIL3 transcription factor with role in immune defense identified from the Hong Kong oyster, Crassostrea hongkongensis. Dev Comp Immunol. (2017) 76:1–8. doi: 10.1016/j.dci.2017.05.011
63. Chen T, Ren C, Wang Y, Luo P, Jiang X, Huang W. Molecular cloning, inducible expression and antibacterial analysis of a novel i-type lysozyme (lyz-i2) in Pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. (2016) 54:197–203. doi: 10.1016/j.fsi.2016.04.008
64. Center DM, Cruikshank W. Modulation of lymphocyte migration by human lymphokines. I. Identification and characterization of chemoattractant activity for lymphocytes from mitogen-stimulated mononuclear cells. J Immunol. (1982) 128:2563–8.
65. Liang Q, Zheng J, Zuo H, Li C, Niu S, Yang L. Identification and characterization of an interleukin-16-like gene from Pacific white shrimp Litopenaeus vannamei. Dev Comp Immunol. (2017) 74:49–59. doi: 10.1016/j.dci.2017.04.011
66. Li F, Xiang J. Recent advances in researches on the innate immunity of shrimp in China. Dev Comp Immunol. (2013) 39:11–26. doi: 10.1016/j.dci.2012.03.016
67. Glas J, Török HP, Unterhuber H, Radlmayr M, Folwaczny C. The- 295T-to-C promoter polymorphism of the IL-16 gene is associated with crohn's disease. Clin Immunol. (2003) 106:197–200. doi: 10.1016/S1521-6616(03)00021-4
68. Chen T, Wong NK, Jiang X, Luo X, Zhang L, Yang D. Nitric oxide as an antimicrobial molecule against Vibrio harveyi infection in the hepatopancreas of Pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. (2015) 42:114–20. doi: 10.1016/j.fsi.2014.10.042
69. Xu G, Wu S, Wu Y, Gu G, Fang Q, Ye G. De novo assembly and characterization of central nervous system transcriptome reveals neurotransmitter signaling systems in the rice striped stem borer, Chilo suppressalis. BMC Genomics. (2015) 16:525. doi: 10.1186/s12864-015-1742-7
70. Gein SV, Baeva TA. Endogenous opioid peptides in regulation of innate immunity cell functions. Biochemistry. (2011) 76:309–19. doi: 10.1134/S0006297911030035
Keywords: neuropeptide, crustacean cardioactive peptide, hepatopancreas, immunoregulation, arthropod
Citation: Wei Y, Lin D, Xu Z, Gao X, Zeng C and Ye H (2020) A Possible Role of Crustacean Cardioactive Peptide in Regulating Immune Response in Hepatopancreas of Mud Crab. Front. Immunol. 11:711. doi: 10.3389/fimmu.2020.00711
Received: 15 January 2020; Accepted: 30 March 2020;
Published: 30 April 2020.
Edited by:
Gyri T. Haugland, University of Bergen, NorwayCopyright © 2020 Wei, Lin, Xu, Gao, Zeng and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haihui Ye, aGFpaHVpeWVAeG11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.