- 1Department of Natural Sciences, Yuzhang Normal University, Nanchang, China
- 2State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 3Department of Bioscience, College of Life Sciences, Nanchang University, Nanchang, China
PKZ is a novel and unique eIF2α protein kinase identified in fish. Although PKZ is most homologous to PKR, particularly in the C-terminal catalytic domain, it contains two N-terminal Z-DNA-binding domains (Zα1 and Zα2) instead of the dsRNA binding domains (dsRBDs) in PKR. As a novel member of eIF2α kinase family, the available data suggest that PKZ has some distinct mechanisms for recognition, binding, and B-Z DNA transition. Functionally, PKZ seems to be activated by the binding of Zα to Z-DNA and participates in innate immune responses. In this review, we summarize the recent progress on fish PKZ.
Introduction
In eukaryotes, translation initiation is an extremely complicated process, and regulation of mRNA translation is very important for a cell to adapt to various stress conditions. As an important mechanism for control of protein synthesis, phosphorylation, or dephosphorylation of eukaryotic translation initiation factor 2 α subunit (eIF2α) represents a core molecular switch for stress adaptation and rapid metabolic regulation (1). In mammals, eIF2α is phosphorylated at Ser51 by a family of four kinases, HRI (heme-regulated inhibitor), PERK (PKR-like endoplasmic reticulum kinase), GCN2 (general control non-derepressible-2), and PKR (double-stranded RNA-dependent protein kinase). HRI is a sensor of heme deprivation and arsenite exposure. PERK is primarily activated by endoplasmic reticulum stress. GCN2 is activated under conditions of amino acid and glucose deprivation. PKR is a well-known dsRNA-activated and interferon (IFN)-stimulated protein, and plays significant roles in antiviral immune response (2–4).
The past few decades have witnessed significant progress in understanding of fish innate immune, and four eIF2α kinases analogous to those in mammals have been identified in fish (5–13). Intriguingly, fish have some unique antiviral-relevant or immune-relevant genes such as PKZ, the fifth member of eIF2α kinase family. PKZ has a closer evolutionary relationship with PKR (14). Functionally, it protects fish cells against viral infection through phosphorylation of eIF2α and may act as a cytosolic DNA sensor to initiate innate immune response (15). In this review, we summarize the recent progress on fish PKZ and its role in innate immune response.
Discovery of PKZ as a PKR-Like Protein Kinase
When fish cells are infected with the virus, many fish IFN-stimulated genes (ISGs) are induced, most of which are homologous to the known mammalian ISGs, and some of which are novel. Crucian carp PKZ (CaPKZ) is such a fish-specific gene, and was named CaPKR-like at that time when it was first identified from IFN-producing CAB cells after treatment with UV-inactivated GCRV in 2004 (16). The full-length cDNA of CaPKZ is 2192 bp with an ORF encoding a polypeptide of 513 amino acid residues. Although the protein size and C-terminal 11 catalytic sub-domains of CaPKZ are most similar to that of mammalian PKR proteins, CaPKZ catalytic domain possesses a large and variant insert (≈85 residues) between sub-domains IV and V, in contrast to a short insert (≈10–34 residues) in PKR. Specially, CaPKZ has two Z-DNA binding domains (Zα1 and Zα2) within its N-terminus instead of typical dsRNA binding domains (dsRBDs) in PKR.
Since then, the homologous genes of CaPKZ have been cloned from Zebrafish (Danio rerio) (17), Atlantic salmon (Salmo salar) (18), Rare minnow (Gobiocypris rarus) (19) and Grass carp (Ctenopharyngodon idellus) (20), indicating there exists a novel eIF2α kinase exclusively in fish. In Zebrafish, PKZ gene is made up of 11 exons and transcribes four alternative splicing variants (A–D, named in order of decreasing abundance) (17, 21). Variant A codes the entire protein. Variant B codes a protein that lacks the kinase insert domain of 78 aa. Variants C and D encode truncated proteins because they retain an intron providing a premature stop codon (17).
Similarities and Differences of PKZ and PKR
Structurally, PKZ resembles PKR, containing an N-terminal regulatory domain and a C-terminal eIF2α kinase domain. Also, the kinase domain of fish PKZ is around 65.42% identical to that of fish PKR (Figure 1A). The striking difference between PKZ and PKR lies in the kinase insert (KI) linking kinase subdomains IV and V, which is necessary for phosphorylation of eIF2α (21). As for the N-terminal regulatory region, it is highly divergent. PKZ has two Zα domains, while PKR has dsRBDs in its N-terminus. It is noted that unlike mammalian and amphibian PKRs possessing two dsRBDs, fish PKRs own one, two or three dsRBDs (21).
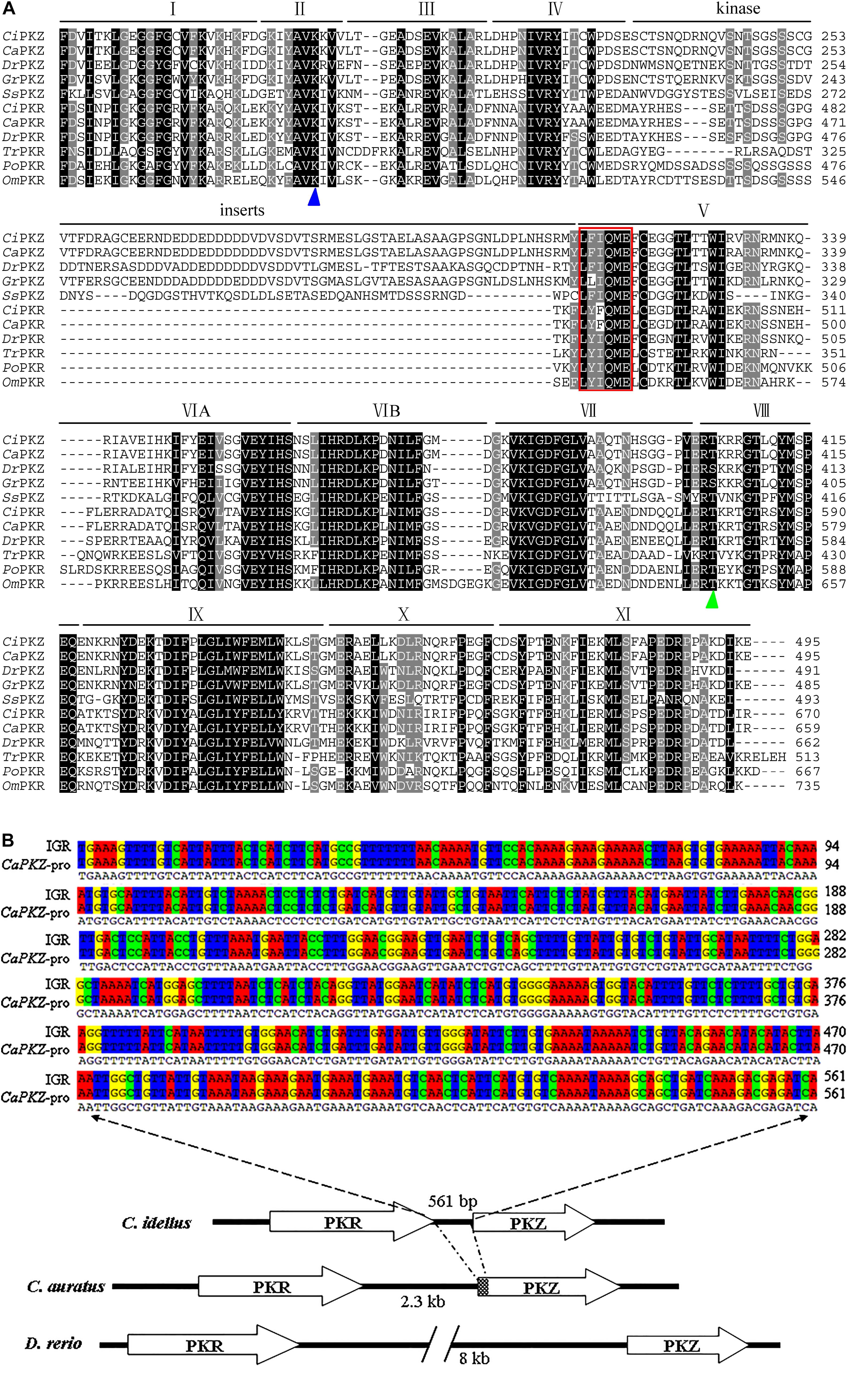
Figure 1. Multiple sequence alignment and genomic arrangement of fish PKR and PKZ genes. (A) Multiple sequence alignment of the kinase domains of fish PKR and PKZ by Clustal X 2.0 program. Residues Lysine (K) for PKR/PKZ catalytic activity (blue triangle) and Serine/Threonine for autophosphorylation (green triangle) are marked under the sequences. The conserved sequence of LFIQME(Y/F)C(D/E) in subdomain V is surrounded by a red box. Identical (shaded in black) and similar (shaded in gray and light gray) residues are indicated. The following abbreviations were used: Ci, Ctenopharyngodon idellus; Ca, Carassius auratus; Dr, Danio rerio; Ss, Salmo salar; Gr, Gobiocypris rarus; Tr, Takifugu rubripes; Po, Paralichthys olivaceus; Om, Oncorhynchus mykiss. (B) Genomic arrangement of tandemly arranged known fish PKR and PKZ genes. The genomic arrangement and relative orientation of PKR and PKZ genes in C. idellus, C. auratus, and D. rerio are shown. Arrows indicate the 5’ to 3’ orientation of the genes. The approximate sizes of intergenic regions are indicated below. Two sequence alignment of the intergenic region (IGR) between CiPKR and CiPKZ and the partial sequence of CaPKZ promoter are displayed above.
Similar to the genomic arrangement of the three X. tropicalis PKR genes, fish PKR, and PKZ genes are tandemly arranged in head-to-tail (parallel) orientation in the genome of zebrafish, crucian carp or grass carp (21, 22). However, searching the Grass Carp Genome Database (GCGD) shows that the length of intergenic region (IGR) between PKZ and PKR in grass carp genome is very short, only 561 bp, which is different from that of zebrafish or crucian carp. Interestingly, the nucleotide sequence of the IGR and the partial sequence of CaPKZ promoter show 99.8% (560/561) sequence identity (Figure 1B). Whether the size divergence of IGR is related to the intensity and rapidity of expression and regulation of PKZ gene in different species is not known. Phylogenetic analysis shows that the kinase domains of fish PKRs are more closely related to those of fish PKZs than to non-fish vertebrate PKRs. It is speculated that a gene duplication event generating fish PKR and PKZ genes occurred early during teleost fish evolution after the divergence of the tetrapod lineage, and the Zα domains replaced the dsRBDs in fish (21).
Since PKZ and PKR coexist in fish as neighboring genes, their functions in cells may be overlapping and distinct. In CAB cells, both PKZ and PKR are activated by IFN or some IFN stimuli resulting in phosphorylation of eIF2α and inhibition of virus replication. The effect of both kinases together is much more significant than either of them, and PKZ seems to exhibit a weaker antiviral ability than PKR, correlating with its lower ability to phosphorylate eIF2α than PKR. Moreover, the activation patterns and functions of PKZ and PKR in cells vary greatly (22).
Function Domains of PKZ
Zα Domains and Its Interaction With Z-DNA
Zα domains belong to a subfamily of wHTH domains with the unique property of specific binding to Z-DNA (23). The highly conserved Zα domain is observed in human ADAR1 (24), DAI (25), vaccinia virus E3L protein (26), and PKZ. Typically, the Zα domain contains three α-helices and three antiparallel β-strands arranged in α1β1α2α3β2β3 topology. Helix 3 and the β-loop between strands β2 and β3 have been found to interact with DNA (27). The β-loop participates in Z-DNA binding in conjunction with the recognition helix 3 and forms the “wing” structure. The conserved proline residues in the wing make hydrophobic contacts with the phosphate backbone of Z-DNA (23, 28).
Current structural analyses imply that the Zα domain of PKZ (ZαPKZ) has distinguished features from other Zα domains. There is an extended loop between β2 and β3 strands of DrZαPKZ, forming the largest β-wing in the known Zα domains (29). The Arg62 residue of the extended wing of DrZαPKZ leads to interact with the primary DNA strand and lost for binding at the edge of the DNA duplex (30). As for CaPKZ Zα (CaZαPKZ), it recognizes P0–P4 phosphates of Z-DNA, while other Zα domains interact with P1–P5 phosphates (31).
As a novel protein containing the Zα domain, fish PKZ Zα exhibits a high binding potential to Z-DNA (17, 18, 32–34), and the sub-domains Zα1 and Zα2 undertake diverse functions, showing that Zα1 is more efficient than Zα2 in B–Z conformational transition (32, 33). ZαPKZ can recognize and convert B-DNA or B-RNA to Z-conformation, which serves as a “flippase” similar to ZαADAR1 (33, 35, 36). Several conserved residues in Zα are essential for Z-DNA recognition and binding. In ZαADAR1, nine residues (K169, K170, N173, R174, Y177, T191, P192, P193, and W195) are important for Z-DNA recognition and binding (25, 27). Similarly, the subdomain Zα1 of PKZ contains nine residues as ZαADAR1. These conserved residues in ZαPKZ play a critical role in the B–Z transition of DNA (33).
Intriguingly, the B-Z transition activity of ZαPKZ is strongly salt concentration-dependent, unlike other Zα proteins such as hZαADAR1 and YabZαE3L. With increasing of [NaCl] from 10 to 250 mM, the B-Z transition activity of CaZαPKZ is impaired severely (37). Similarly, DrZαPKZ has the ability to convert poly(dG-dC) into the Z-DNA in the presence of low amounts of cobalt hexamine (17). In addition, DrZαPKZ has two positively charged residues (Lys61 and Arg62) in the extended β-wing involved in B–Z DNA transition, whereas CaZαPKZ has only one positively charged residue (Lys56). It may be a reason why the B–Z transition induced by DrZαPKZ is faster than by CaZαPKZ (29).
Kinase Domains and eIF2α Kinase Activity of PKZ
Gene organization of CiPKZ and DrPKZ reveals that the kinase domain is encoded by seven exons, and the kinase insert is encoded by four exons. A distinguishing sequence of LFIQME(Y/F)C(D/E) in subdomain V of PKZ is proposed to be important for eIF2α kinase activity (16, 17) (Figure 1A). Sequence alignment and site-directed mutagenesis studies indicate that a conserved Lys is important for catalytic activity, located at position 199 in DrPKZ, 198 in CaPKZ or CiPKZ. At position 402 in DrPKZ is a Ser (corresponding to Thr-404 in CaPKZ or CiPKZ), allowing for autophosphorylation (17, 20) (Figure 1A).
In mammals, PKR functions as an eIF2α kinase. EIF2α can be phosphorylated at Ser51 by PKR, leading to inhibition of general protein synthesis and initiation of apoptosis (38, 39). Given a highly homologous kinase region, fish PKZ has similar function features as PKR. In vitro, recombinant PKZ autophosphorylates and phosphorylates eIF2α in the absence of any activators. The dephosphorylated PKZ is activated again by poly(dG-dC) but not poly(I:C). PKZ shows an ability to phosphorylate recombinant wild-type eIF2α but not the recombinant non-phosphorylatable variant eIF2α (S51A). Furthermore, the wild-type PKZ, but not the kinase defective variant (K198R in CaPKZ/CiPKZ, K199R in DrPKZ, and K217R in SsPKZ), exhibits a direct inhibitory effect on reporter gene expression (17, 18, 20, 22). In addition, both of two DrPKZ isoforms (DrPKZ-A and DrPKZ-B) functionally interact with eIF2α and inhibit protein synthesis in vivo. Deletion of the insert domain of DrPKZ-A or DrPKZ-B results in abrogating the kinase activity completely. It indicates that the insert domain is required for DrPKZ kinase activity. Kinase activity appears to be independent of the insert length, while it depends on the presence of specific amino acids within the insert domain (40).
Roles of PKZ in Fish Innate Immune Responses
Being Induced as a Typical ISG
It is well known that IFN exerts antiviral effects through induction of hundreds of ISGs, including PKR, Mx1, ADAR1, ISG15, viperin, and so on (14). Coexistence of PKR and PKZ in a head-to-tail orientation in fish genomes is believed to be very important for similar transcriptional activation after immunostimulation. Actually, PKZ is up-regulated by virus infection and lots of IFN stimuli, including IFN, poly(I:C), poly(dA-dT), poly(dG-dC), genomic DNA, and even Aeromonas hydrophila (16–22, 40). poly(I:C)-induced of PKZ requires novel protein synthesis and IFN stimulates PKZ expression through Stat1 pathway, indicating that PKZ is a novel IFN stimulated gene. On the contrary, the suppressor of cytokine signaling 1 (SOCS-1), an inhibitor of the IFN signaling pathways, can significantly suppress the expression of PKZ (41).
Consistently, PKZ and PKR promoters contain IFN-stimulated response element (ISRE) that is required for their expression induced by IFN (42). There is at least one typical ISRE within PKZ promoter (one ISRE in CiPKZ, two ISREs in CaPKZ) (22, 43), which is significantly induced by poly(I:C) and IFN (22). Also, IRF3 and IRF7 can significantly activate CiPKZ promoter, but cannot effectively activate the truncated mutant CiPKZ-nISRE-pro that lacked ISRE (43), highlighting the relevance of PKZ during IFN-mediated antiviral response.
Function as an Antiviral Effector
As a typical ISG encoding a novel eIF2α kinase, PKZ might inhibit viral replication, like PKR, through phosphorylating eIF2α, and thus resulting in inhibition of the synthesis of viral proteins. In vitro assays have shown that overexpression of PKZ significantly inhibit GCRV replication, while knockdown of this protein makes fish cells more vulnerable to virus infection. It is no doubt that PKZ can bind and phosphorylate eIF2α, and upon GCRV infection, the expression level of PKZ is consistent with the relatively level of eIF2α phosphorylation and the virus titer (22). Despite that PKR and PKZ are simultaneously induced during viral infection, fish PKR and PKZ form homodimers, but not heterodimers, to phosphorylate eIF2α independently, indicating fish PKZ and PKR play a non-redundant but cooperative role in IFN antiviral response.
In addition, PKZ might function as an antiviral effector by facilitating cell apoptosis. Overexpression of wild-type CiPKZ (PKZ-wt) in CIK cells results in a striking decrease of cell viability rate. When PKZ-deficient cells were transfected with PKZ-wt rather than mutant PKZ-K198R, a significant increase of apoptotic cell number is observed. Also, this apoptosis is related to the eIF2α phosphorylation level (44). Due to host cells undergo apoptosis in response to virus infection and the induction of apoptosis is an antiviral innate immune mechanism (45, 46), it can be deduced that fish PKZ is indeed an antiviral effector. A question is how PKZ is activated during virus infection. It is noted that Z-DNA binding is indispensable to the regulation of PKZ activity. The enzyme missing the Zα domain (PKZΔN) is less effective than wild-type PKZ at inhibiting luciferase activity (20). As for DrPKZ, deletion of the N-terminus leads to weakening the kinase activity compared with the wild-type (17).
Function as a Cytosolic Sensor
It is interesting that two Z-DNA binding domain-containing proteins identified in mammals, DAI (ZBP1/DLM-1) and ADAR1, have been confirmed to be involved in IFN signaling. As a cytosolic sensor, DAI binds to cytosolic dsDNA initiating innate immune signal by activation of NF-κB and upregulation of type I IFN (47). In mammals, in addition to being an antiviral effector, PKR also acts as a dsRNA sensor detecting virus infection in the cytoplasm. Since fish PKZ can bind to Z-DNA or Z-RNA with high affinity, it is reasonable to infer that PKZ is a new sensor for Z-DNA/RNA in the cytoplasm (22). A recent study has shown that PKZ can trigger immune responses via IRF3- or ISGF3-like mediated pathways in fish (15). On account of the fact that DAI has not been found in the fish genome, perhaps PKZ is a kind of compensator for the lack of DAI, functioning as a cytosolic DNA sensor to trigger innate antiviral immune response. Nevertheless, a lot of questions need to be addressed. What’s the manner of PKZ recognizes DNA? What’s the relationship between PKZ and the candidate substrates? What’re the biological significances for PKZ-triggered innate immune response?
Conclusion
In mammals, there are four members of eIF2α kinase family, and PKZ is the fifth member found in this family. Like PKR, PKZ inhibits viral replication through a similar mechanism to phosphorylate eIF2α. PKR has N-terminal dsRBDs, which enables PKR as a cytosolic receptor to recognize dsRNA generated during virus infection. Similarly, fish PKZ might function as a Z-DNA/Z-RNA sensor, to capture nucleic acid molecules and subsequently activate innate immune response. Despite of these findings, what is the source of Z-DNA/Z-RNA in the cytoplasm? What is the source of negative supercoiling in the cytoplasm? Given that Z-DNA is a transcription-dependent structure, it can be stabilized by the negative supercoiling generated by a moving RNA polymerase as it plows through the DNA double helix in vivo. We also do not known why PKZ is exclusively in fish genome. It is likely that the coexistence of PKR and PKZ is especially important to taxonomically lower fish species that live in a complex water environment, since harboring distinct N-terminal domains enable fish PKR and PKZ to recognize different viral nucleic acids, which makes the fish innate immune system more effective by broadening their ability to sense viruses and to defense against viral infection cooperatively.
Author Contributions
CW created the figure and drafted the manuscript under the guidance of YZ and CH. YZ and CH edited and revised the drafts, respectively. All authors approved the final version.
Funding
This work was supported by Major projects of the National Natural Science Foundation of Jiangxi Province (20171ACB20004), the National Natural Science Foundation of China (Grant 31472304), and the Science and Technology Project of Education Department of Jiangxi Province (GJJ191210).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ADAR, double-stranded RNA adenosine deaminase; Ca, Carassius auratus; Ci, Ctenopharyngodon idellus; DAI, DNA-dependent activator of interferon regulatory factors; Dr, Danio rerio; dsRBD, dsRNA binding domain; eIF2 α, the α subunit of eukaryotic translation initiation factor 2; IFN, interferon; IRF3, interferon transcription factor 3; ISGF3, interferon-stimulated gene factor 3; PKR, double-stranded RNA-dependent protein kinase; PKZ, protein kinase containing Z-DNA binding domains; Ss, Salmo salar; Z α, Z-DNA binding domain.
References
1. Yong J, Grankvist N, Han J, Kaufman RJ. Eukaryotic translation initiation factor 2 alpha phosphorylation as a therapeutic target in diabetes. Expert Rev Endocrinol Metab. (2014) 9:345–56. doi: 10.1586/17446651.2014.927309
2. Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci. (2013) 70:3493–511. doi: 10.1007/s00018-012-1252-6
3. Masson GR. Towards a model of GCN2 activation. Biochem Soc Trans. (2019) 47:1481–8. doi: 10.1042/BST20190331
4. Clemens MJ, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. (1997) 17:503–24. doi: 10.1089/jir.1997.17.503
5. del Castillo CS, Hikima J, Ohtani M, Jung TS, Aoki T. Characterization and functional analysis of two PKR genes in fugu (Takifugu rubripes). Fish Shellfish Immunol. (2012) 32:79–88. doi: 10.1016/j.fsi.2011.10.022
6. Hu YS, Li W, Li DM, Liu Y, Fan LH, Rao ZC, et al.Cloning, expression and functional analysis of PKR from grass carp (Ctenopharyngodon idellus). Fish Shellfish Immunol. (2013) 35:1874–81. doi: 10.1016/j.fsi.2013.09.024
7. Komoike Y, Matsuoka M. Exposure to tributyltin induces endoplasmic reticulum stress and the unfolded protein response in zebrafish. Aquat Toxicol. (2013) 142–3:221–9. doi: 10.1016/j.aquatox.2013.08.017
8. Wang Z, Mai KS, Xu W, Zhang YJ, Liu YL, Ai QH. Dietary methionine level influences growth and lipid metabolism via GCN2 pathway in cobia (Rachycentron canadum). Aquaculture. (2016) 454:148–56. doi: 10.1016/j.aquaculture.2015.12.019
9. Zang SQ, Zhang X, Li C, Wang LQ, Wei JG, Qin QW. HRI of Epinephelus coioides is a critical factor in the grouper immune response to RGNNV infection. Fish Shellfish Immunol. (2019) 87:659–68. doi: 10.1016/j.fsi.2019.02.011
10. Zenke K, Nam YK, Kim KH. Molecular cloning and expression analysis of double-stranded RNA-dependent protein kinase (PKR) in rock bream (Oplegnathus fasciatus). Vet Immunol Immunopathol. (2010) 133:290–5. doi: 10.1016/j.vetimm.2009.08.009
11. Zhang T, Li DM, Wan LJ, Chen X, Wang XQ, Zhong B, et al.Ctenopharyngodon idella PERK (EIF2AK3) decreases cell viability by phosphorylating eIF2alpha under ER stress. Fish Shellfish Immunol. (2017) 70:568–74. doi: 10.1016/j.fsi.2017.09.044
12. Zhu R, Zhang YB, Chen YD, Dong CW, Zhang FT, Zhang QY, et al. Molecular cloning and stress-induced expression of Paralichthys olivaceus heme-regulated initiation factor 2alpha kinase. Dev Comp Immunol. (2006) 30:1047–59. doi: 10.1016/j.dci.2006.02.001
13. Zhu R, Zhang YB, Zhang QY, Gui JF. Functional domains and the antiviral effect of the double-stranded RNA-dependent protein kinase PKR from Paralichthys olivaceus. J Virol. (2008) 82:6889–901. doi: 10.1128/JVI.02385-07
14. Zhang YB, Gui JF. Molecular regulation of interferon antiviral response in fish. Dev Comp Immunol. (2012) 38:193–202. doi: 10.1016/j.dci.2012.06.003
15. Xu XW, Li MF, Wu CX, Li DM, Jiang ZY, Liu CX, et al. The fish-specific protein kinase (PKZ) initiates innate immune responses via IRF3- and ISGF3-like mediated pathways. Front Immunol. (2019) 10:582. doi: 10.3389/fimmu.2019.00582
16. Hu CY, Zhang YB, Huang GP, Zhang QY, Gui JF. Molecular cloning and characterisation of a fish PKR-like gene from cultured CAB cells induced by UV-inactivated virus. Fish Shellfish Immunol. (2004) 17:353–66. doi: 10.1016/j.fsi.2004.04.009
17. Rothenburg S, Deigendesch N, Dittmar K, Koch-Nolte F, Haag F, Lowenhaupt K, et al. A PKR-like eukaryotic initiation factor 2alpha kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc Natl Acad Sci USA. (2005) 102:1602–7. doi: 10.1073/pnas.0408714102
18. Bergan V, Jagus R, Lauksund S, Kileng O, Robertsen B. The Atlantic salmon Z-DNA binding protein kinase phosphorylates translation initiation factor 2 alpha and constitutes a unique orthologue to the mammalian dsRNA-activated protein kinase R. FEBS J. (2008) 275:184–97. doi: 10.1111/j.1742-4658.2007.06188.x
19. Su JG, Zhu ZY, Wang YP. Molecular cloning, characterization and expression analysis of the PKZ gene in rare minnow Gobiocypris rarus. Fish Shellfish Immunol. (2008) 25:106–13. doi: 10.1016/j.fsi.2008.03.006
20. Yang PJ, Wu CX, Li W, Fan LH, Lin G, Hu CY. Cloning and functional analysis of PKZ (PKR-like) from grass carp (Ctenopharyngodon idellus). Fish Shellfish Immunol. (2011) 31:1173–8. doi: 10.1016/j.fsi.2011.10.012
21. Rothenburg S, Deigendesch N, Dey M, Dever TE, Tazi L. Double-stranded RNA-activated protein kinase PKR of fishes and amphibians: varying the number of double-stranded RNA binding domains and lineage-specific duplications. BMC Biol. (2008) 6:12. doi: 10.1186/1741-7007-6-12
22. Liu TK, Zhang YB, Liu Y, Sun F, Gui JF. Cooperative roles of fish protein kinase containing Z-DNA binding domains and double-stranded RNA-dependent protein kinase in interferon-mediated antiviral response. J Virol. (2011) 85:12769–80. doi: 10.1128/JVI.05849-11
23. Tome AR, Kus K, Correia S, Paulo LM, Zacarias S, de Rosa M, et al. Crystal structure of a poxvirus-like zalpha domain from cyprinid herpesvirus 3. J Virol. (2013) 87:3998–4004. doi: 10.1128/JVI.03116-12
24. Herbert A, Alfken J, Kim YG, Mian IS, Nishikura K, Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc Natl Acad Sci USA. (1997) 94:8421–6. doi: 10.1073/pnas.94.16.8421
25. Schwartz T, Behlke J, Lowenhaupt K, Heinemann U, Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat Struct Biol. (2001) 8:761–5. doi: 10.1038/nsb0901-761
26. Kim YG, Muralinath M, Brandt T, Pearcy M, Hauns K, Lowenhaupt K, et al. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc Natl Acad Sci USA. (2003) 100:6974–9. doi: 10.1073/pnas.0431131100
27. Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. (1999) 284:1841–5. doi: 10.1126/science.284.5421.1841
28. Kim K, Khayrutdinov BI, Lee CK, Cheong HK, Kang SW, Park H, et al. Solution structure of the Zbeta domain of human DNA-dependent activator of IFN-regulatory factors and its binding modes to B- and Z-DNAs. Proc Natl Acad Sci USA. (2011) 108:6921–6. doi: 10.1073/pnas.1014898107
29. Subramani VK, Kim D, Yun K, Kim KK. Structural and functional studies of a large winged Z-DNA-binding domain of Danio rerio protein kinase PKZ. FEBS Lett. (2016) 590:2275–85. doi: 10.1002/1873-3468.12238
30. de Rosa M, Zacarias S, Athanasiadis A. Structural basis for Z-DNA binding and stabilization by the zebrafish Z-DNA dependent protein kinase PKZ. Nucleic Acids Res. (2013) 41:9924–33. doi: 10.1093/nar/gkt743
31. Kim D, Hur J, Park K, Bae S, Shin D, Ha SC, et al. Distinct Z-DNA binding mode of a PKR-like protein kinase containing a Z-DNA binding domain (PKZ). Nucleic Acids Res. (2014) 42:5937–48. doi: 10.1093/nar/gku189
32. Wu CX, Wang SJ, Lin G, Hu CY. The Zalpha domain of PKZ from Carassius auratus can bind to d(GC)(n) in negative supercoils. Fish Shellfish Immunol. (2010) 28:783–8. doi: 10.1016/j.fsi.2010.01.021
33. Lu PD, Deng SL, Zhu YJ, Yan YB, Liu Y, Hu CY. The Zalpha domain of fish PKZ facilitates the B-Z conformational transition of oligonucleotide DNAs with d(GC)(n) inserts. Acta Biochim Biophys Sin (Shanghai). (2012) 44:957–63. doi: 10.1093/abbs/gms081
34. Lu PD, Deng SL, Wu CX, Zhu YJ, Liu Y, Lin G, et al. The Zalpha domain of fish PKZ converts DNA hairpin with d(GC)(n) inserts to Z-conformation. Acta Biochim Biophys Sin (Shanghai). (2013) 45:1062–8. doi: 10.1093/abbs/gmt114
35. Kim YG, Lowenhaupt K, Maas S, Herbert A, Schwartz T, Rich A. The zab domain of the human RNA editing enzyme ADAR1 recognizes Z-DNA when surrounded by B-DNA. J Biol Chem. (2000) 275:26828–33. doi: 10.1074/jbc.M003477200
36. Kang YM, Bang J, Lee EH, Ahn HC, Seo YJ, Kim KK, et al. NMR spectroscopic elucidation of the B-Z transition of a DNA double helix induced by the Z alpha domain of human ADAR1. J Am Chem Soc. (2009) 131:11485–91. doi: 10.1021/ja902654u
37. Lee AR, Seo YJ, Choi SR, Ryu KS, Cheong HK, Lee SS, et al. NMR elucidation of reduced B-Z transition activity of PKZ protein kinase at high NaCl concentration. Biochem Biophys Res Commun. (2017) 482:335–40. doi: 10.1016/j.bbrc.2016.11.064
38. Jeffrey IW, Bushell M, Tilleray VJ, Morley S, Clemens MJ. Inhibition of protein synthesis in apoptosis: differential requirements by the tumor necrosis factor alpha family and a DNA-damaging agent for caspases and the double-stranded RNA-dependent protein kinase. Cancer Res. (2002) 62:2272–80. doi: 10.1016/S0165-4608(01)00623-9
39. Sonenberg N, Hinnebusch AG. Regulation of translation initiation ineukaryotes: mechanisms and biological targets. Cell. (2009) 136:731–45. doi: 10.1016/j.cell.2009.01.042
40. Liu, ZY Jia KT, Li C, Weng SP, Guo CJ, He JG. A truncated Danio rerio PKZ isoform functionally interacts with eIF2alpha and inhibits protein synthesis. Gene. (2013) 527:292–300. doi: 10.1016/j.gene.2013.05.043
41. Nie L, Xiong R, Zhang YS, Zhu LY, Shao JZ, Xiang LX. Conserved inhibitory role of teleost SOCS-1s in IFN signaling pathways. Dev Comp Immunol. (2014) 43:23–9. doi: 10.1016/j.dci.2013.10.007
42. Sun F, Zhang YB, Liu TK, Gan L, Yu FF, Liu Y, et al. Characterization of fish IRF3 as an IFN-inducible protein reveals evolving regulation of IFN response in vertebrates. J Immunol. (2010) 185:7573–82. doi: 10.4049/jimmunol.1002401
43. Liu D, Mao HL, Gu MH, Xu XW, Sun ZC, Lin G, et al. The transcription regulation analysis of Ctenopharyngodon idellus PKR and PKZ genes. Gene. (2016) 576:512–9. doi: 10.1016/j.gene.2015.10.070
44. Wu CX, Hu YS, Fan LH, Wang HZ, Sun ZC, Deng SL, et al.Ctenopharyngodon idella PKZ facilitates cell apoptosis through phosphorylating eIF2alpha. Mol Immunol. (2016) 69:13–23. doi: 10.1016/j.molimm.2015.11.006
45. Barber GN. Host defense, viruses and apoptosis. Cell Death Differ. (2001) 8:113–26. doi: 10.1038/sj.cdd.4400823
46. Nainu F, Shiratsuchi A, Nakanishi Y. Induction of apoptosis and subsequent phagocytosis of virus-infected cells as an antiviral mechanism. Front Immunol. (2017) 8:1220. doi: 10.3389/fimmu.2017.01220
Keywords: PKZ, Zα domain, Z-DNA, kinase activity, fish
Citation: Wu C, Zhang Y and Hu C (2020) PKZ, a Fish-Unique eIF2α Kinase Involved in Innate Immune Response. Front. Immunol. 11:585. doi: 10.3389/fimmu.2020.00585
Received: 03 December 2019; Accepted: 13 March 2020;
Published: 31 March 2020.
Edited by:
Philip John Griebel, University of Saskatchewan, CanadaReviewed by:
Francesco Buonocore, University of Tuscia, ItalyHao-Ching Wang, Taipei Medical University, Taiwan
Copyright © 2020 Wu, Zhang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yibing Zhang, eWJ6aGFuZ0BpaGIuYWMuY24=; Chengyu Hu, aHVjeTIwMDhAMTYzLmNvbQ==
 Chuxin Wu
Chuxin Wu Yibing Zhang
Yibing Zhang Chengyu Hu
Chengyu Hu