- 1Department of Animal Science, Iowa State University, Ames, IA, United States
- 2Veterinary Diagnostic Laboratory, Kansas State University, Manhattan, KS, United States
- 3Food Safety and Enteric Pathogen Unit, National Animal Disease Center, Agricultural Research Service, U.S. Department of Agriculture, Ames, IA, United States
- 4Immunobiology Graduate Program, College of Veterinary Medicine, Iowa State University, Ames, IA, United States
- 5Agricultural Research Service Participation Program, Oak Ridge Institute for Science and Education, Oak Ridge, TN, United States
- 6Laboratory Animal Research, Iowa State University, Ames, IA, United States
- 7Diagnostic Medicine and Pathobiology Department, Kansas State University, Manhattan, KS, United States
Severe combined immunodeficiency (SCID) is described as the lack of functional T and B cells. In some cases, mutant genes encoding proteins involved in the process of VDJ recombination retain partial activity and are classified as hypomorphs. Hypomorphic activity in the products from these genes can function in the development of T and B cells and is referred to as a leaky phenotype in patients and animals diagnosed with SCID. We previously described two natural, single nucleotide variants in ARTEMIS (DCLR1EC) in a line of Yorkshire pigs that resulted in SCID. One allele contains a splice site mutation within intron 8 of the ARTEMIS gene (ART16), while the other mutation is within exon 10 that results in a premature stop codon (ART12). While initially characterized as SCID and lacking normal levels of circulating lymphoid cells, low levels of CD3ε+ cells can be detected in most SCID animals. Upon further assessment, we found that ART16/16, and ART12/12 SCID pigs had abnormally small populations of CD3ε+ cells, but not CD79α+ cells, in circulation and lymph nodes. Newborn pigs (0 days of age) had CD3ε+ cells within lymph nodes prior to any environmental exposure. CD3ε+ cells in SCID pigs appeared to have a skewed CD4α+CD8α+CD8β− T helper memory phenotype. Additionally, in some pigs, rearranged VDJ joints were detected in lymph node cells as probed by PCR amplification of TCRδ V5 and J1 genomic loci, as well as TCRβ V20 and J1.1, providing molecular evidence of residual Artemis activity. We additionally confirmed that TCRα and TCRδ constant region transcripts were expressed in the thymic and lymph node tissues of SCID pigs; although the expression pattern was abnormal compared to carrier animals. The leaky phenotype is important to characterize, as SCID pigs are an important tool for biomedical research and this additional phenotype may need to be considered. The pig model also provides a relevant model for hypomorphic human SCID patients.
Introduction
Artemis is involved in DNA repair and is critical for proper T and B cell receptor gene arrangement and subsequent development of naïve lymphocytes. Mutations in Artemis results in severe combined immunodeficiency (SCID), characterized by the lack of circulating T and B cells (1). We previously described two natural mutations within the ARTEMIS gene in two separate alleles (termed ART16 and ART12) in a line of Yorkshire pigs, which leads to SCID in a homozygous or heterozygous state (2). The ART16 allele contains a splice site mutation within intron 8 of ARTEMIS and all transcripts in homozygous ART16/16 fibroblasts are missing exon 8. The ART12 allele contains a nonsense mutation in exon 10 that leads to a premature stop codon and many of the transcripts in ART12/12 fibroblasts are missing large portions of all exons (2). Upon the discovery of the ART SCID pigs, bone marrow transplantation (BMT) was performed to reconstitute the animal immune system and to further study the animals. Among the SCID pigs that underwent BMT, two ART16/16 pigs developed host derived CD3ε+ T-cell lymphoma (3). This led us to hypothesize that the ART16 allele potentially produced Artemis protein with residual activity, resulting in development of T cells. Hypomorphic mutations are reported in RAG (4–6) and ARTEMIS (7–10) in human patients, and can lead to complications such as cancer (8, 9) and Omenn's syndrome (11). Mice have also been genetically modified with hypomorphic ARTEMIS mutations (12, 13). Thus, it is important to understand and characterize the CD3ε+ cells that developed in ARTEMIS mutant SCID pigs, a phenotype referred to as leaky.
We describe here a leaky CD3ε+ cellular phenotype in pigs with SCID due to mutated ART. Tissues and blood were collected from pigs across the three different SCID genotypes (ART12/12, ART12/16, and ART16/16) at various ages, and pigs from all genotypes had some level of CD3ε+ cells. We detected CD79α+ cells in the lymph node in only one ART16/16 pig out of 25 total SCID pigs tested. We performed PCR on SCID pig lymph node DNA and found evidence of VDJ recombination in both the TCRδ and TCRβ loci. We additionally confirmed the expression of TCRα and TCRδ constant region transcripts utilizing a two-color RNA in-situ hybridization technique to observe αβ and γδ T cells, respectively. Taken together, occurrence of VDJ recombination within the TCR loci and expression of TCR transcripts in lymphoid tissues show that CD3ε+ cells are capable of developing in SCID pigs with mutations in ART. Documentation of this leaky CD3ε+ cellular phenotype is important as this animal model is further developed for biomedical research.
Materials and Methods
Ethics Statement
All animal procedures and protocols were approved by Iowa State University's Institutional Animal Care and Use Committee.
Generation of Piglets and Rearing
SCID pigs were derived as previously described (2) and housed in either clean conventional rooms or in biocontainment facilities (14). All piglets were either handfed 250 mL colostrum or naturally suckled on the sow after birth. All pigs, genotypes, housing conditions, age, CD3ε result summaries, analysis methods, and figure references are in Table 1. There was no overlap in pigs between this study and those described in Waide et al. (2).
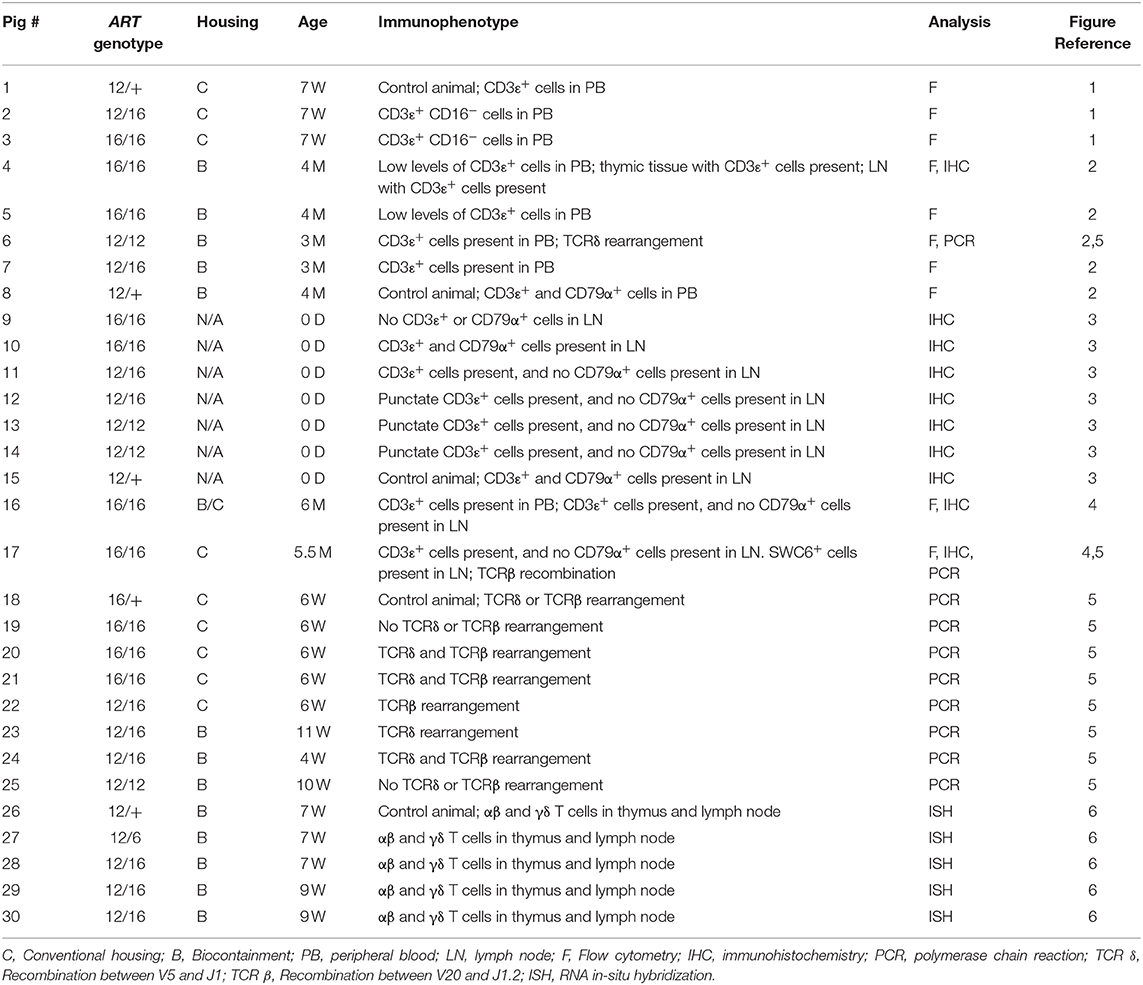
Table 1. Overview of SCID pigs, genotypes, and description of CD3ε+ and CD79α+ cells in blood or lymph nodes.
Flow Cytometry of PBMCs and MNCs
PBMCs were isolated as previously described (15). To collect lymph node mononuclear cells (MNCs), lymph node tissue was dissected and placed in HBSS with 10 μg/mL gentamicin. In the lab, tissues were minced in a digestion solution of HBSS with 300 mg/L of collagenase, 3% FBS, and 2 mM HEPES (referred to herein as d-HBSS). The tissue and d-HBSS mixture was incubated for 1 h at 37°C with vortexing every 15 min. The suspension was passed through a 100 μm cell strainer to isolate MNCs and was washed once in d-HBSS. Isolated PBMCs and MNCs were counted by flow cytometry using a BD cell viability counting kit.
PBMCs and MNCs were stained with the following antibodies: anti-pig CD16 (G7, Bio-Rad) with a goat anti-mouse IgG1 secondary antibody, anti-pig CD3ε (BB23-8E6-8C8), anti-human CD79α (HM7), anti-pig CD8β (PPT23), anti-pig CD8α (76-12-4), anti-pig CD4α (74-12-4), and anti-pig SWC6 (MAC320). SWC6 is an uncharacterized membrane protein and the antibody (MAC320) identifies a population of γδ TCR+ cells in the blood (16, 17). Cells were fixed in 2% PBS formaldehyde and data was acquired with a FACS Canto II and subsequently analyzed using FlowJo (Tree Star).
Immunohistochemistry of Collected Lymphoid Tissues
Pigs were euthanized via intravenous injection of pentobarbital sodium (Fatal Plus). Lymph node and thymic tissues were collected and placed into 10% buffered formalin for 24 hours. Tissues were then moved to 70% ethanol until processing. Immunohistochemical staining for T and B lymphocyte markers was performed in paraffin-embedded tissue thin sections at Kansas State Veterinary Diagnostic Laboratory (KSVDL). Briefly, deparaffinized slide-mounted thin sections were pre-treated for 5 min with a peroxide block, followed by incubation with primary antibody. Primary antibodies included mouse anti-CD3ε (clone LN10, Leica Biosystems) and mouse anti-CD79α (clone HM57, Abcam). Primary antibodies were incubated for 25 min with PowerVision Poly-HRP anti-mouse IgG at room temperature, with DAB chromagen, and counterstained with hematoxylin.
PCR of Lymph Node DNA for TCRβ and TCRγ V and J Rearrangement
PCR analysis was performed on DNA isolated from various lymph node tissues (thoracic, inguinal, and subscapular) collected from SCID and carrier pigs. We used a protocol similar to Suzuki et al. (18) to assess TCR rearrangements, with the addition of an in-house designed TCRβ J1.2 reverse primer (see below)(19). Zymo Research Genomic-DNA™ Tissue Miniprep Kit was used to extract genomic DNA from the tissue. A 40 μl PCR reaction volume was used with 160 ng of gDNA was used as a template, and amplification was performed with Promega GoTaq® Green Master Mix. TCRβ recombination was assessed using primers designed for TCRβV20 (5′ GATGTCATGGACATCATTTGCCATC 3′) (18) and TCRβJ1.2 (5′ GGGCCGAAGTTATAGTCATA 3′). TCRδ recombination was assessed using primers designed for TCRδV5 (5′ TTCAGACACGTGACCTTCAG 3′) (18) and TCRδJ1 (5′ GTTCCACAACCAGCTGAGTC 3′) (18). Germline sequence of TCRβV20 was amplified as a positive control using the TCRβV20 primer listed above with a reverse primer (5′ GCTGAGATTCTGGGATTCAC 3′). All samples used the same thermal cycling parameters of 94°C for 10 min, followed by 39 cycles of 95°C for 30 s, 62°C for 30 s, 72°C for 1 min 30 s, followed by a final step of 72°C for 10 min.
In-situ Hybridization of TRDC and TRAC Constant Region Transcripts in Lymphoid Tissues
Fixed tissues were prepared as described above. Chromogenic 2-color RNA in-situ hybridization was performed using the RNAscope 2.5 HD Duplex kit (Advanced Cell Diagnostics). Custom probes complementary to porcine TRDC (ACD catalog no. 553141) and TRAC (ACD catalog no. 565291) mRNA were created to detect γδ and αβ T cells, respectively. RNAscope staining was carried out according to manufacturer's instructions with the following adjustments: (1) target retrieval was carried out by incubating slides in 1X Target Retrieval Solution (ACD) at 95°C for 15 min in a pressurized Decloaking Chamber NxGen (BioCare) and (2) Protease Plus (ACD) was applied to each tissue section for 15 min at 40°C.
Results
Peripheral CD16− CD3ε+ Cells Detected in SCID Pigs Raised in Conventional Housing
The initial detection of circulating CD3ε+ cells in SCID pigs occurred during a routine engraftment check on an ART−/− SCID pig that had previously received a bone marrow transplantation. During the engraftment check, peripheral blood mononuclear cells (PBMCs) were assessed for CD3ε expression from the putative BMT, as well as a carrier (ART+/−) and SCID (ART−/−) pigs as control cells for the assay. Flow cytometry analysis revealed the SCID pig tested had a small population of CD3ε+ cells in circulation (data not shown). Blood was tested again from a carrier (Pig 1; p1), ART12/16 (p2), and ART16/16 (p3) littermate pigs, all of which were 7 weeks of age and raised in conventional (non-biocontainment) rooms (Figure 1). NK cells develop in ART−/− SCID pigs and retain function in vitro(15); thus PBMCs from the indicated animals were labeled for CD3ε and CD16 (20) to evaluate for NKT cells. CD3ε+ cells in SCID pigs were detected again, confirming the earlier observation of “leaky” SCID phenotype (13). All SCID pig CD3ε+ populations were CD16−, suggesting that they were not NKT cells (Figure 1).
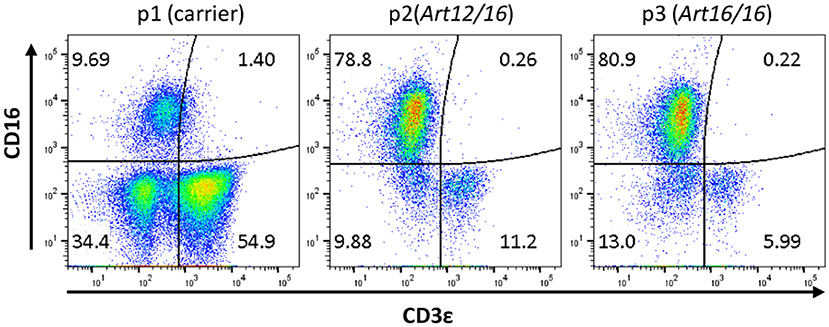
Figure 1. SCID pigs have CD3ε+ CD16− cells in circulation. PBMCs from a carrier (p1) and SCID pigs (p2 [ART12/16] and p3 [ART16/16]) housed in conventional clean rooms were stained to determine CD16 and CD3ε expression.
SCID Pigs in Biocontainment Facilities Develop CD3ε+, But Not CD79α+ Cells
We next investigated when CD3ε+ cells developed in ART−/− SCID pigs and if raising the pigs in clean biocontainment facilities (14) would reduce the prevalence of CD3ε+ cellular expansion, as there would be a reduced pathogen exposure compared to standard conventional housing settings (14). PBMCs from four pigs born and raised in positive pressure biocontainment facilities (14) over a 3- to 4-month period were isolated and labeled for CD3ε and CD79α expression to assess T and B cell development. PBMC from carrier animals (ART+/−) were stained alongside SCID pig PBMC samples, and carriers consistently had 70–90% CD3ε+ cells and 5–15% CD79α+ cells within the lymphocyte population (data not shown). PBMCs from SCID pigs were first gated on total mononuclear cells (lymphocytes and myeloid cells) as opposed to lymphocytes only due to inability to distinguish between the two populations by forward scatter (FSC) and side scatter (SSC) in the blood samples. ART16/16 animals (p4 and p5) had low to no circulating CD3ε+ cells, while the ART12/12 (p6) and ART12/16 (p7) pigs had variable frequencies of CD3ε+ cells (Figure 2A). Interestingly, upon necropsy at 4 months of age, p4 had thymic tissue (Supplemental Figure 1) which immunohistochemistry indicated as predominantly CD3ε+ cells. CD79α+ cells were not detected in the blood during the 3–4 month period tested (data not shown).
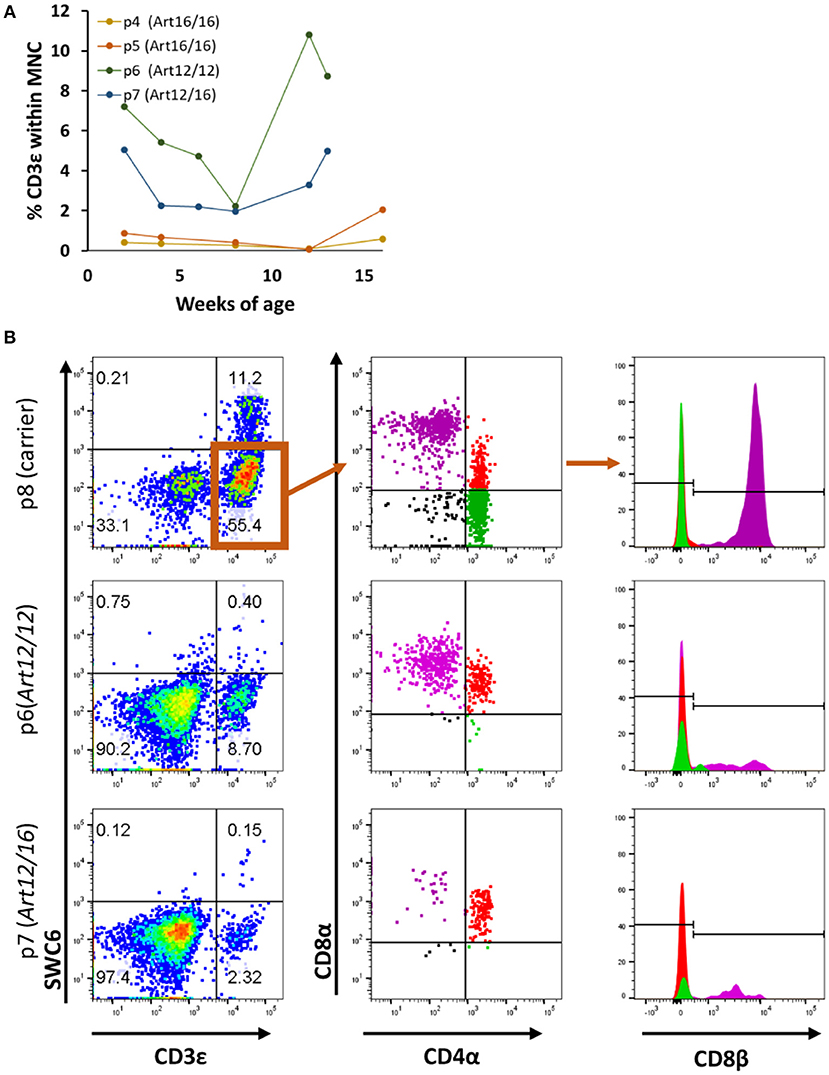
Figure 2. SCID pig CD3ε+ cells are primarily cytotoxic T and memory T cell in circulation. (A) Four SCID pigs (p4 [ART16/16], p5 [ART16/16], p6 [ART12/16], and p7 [ART12/12]) were monitored over a three to four-month period for circulating CD3ε+ cells. P6 and p7 had variable levels of CD3ε+ cells throughout this period. (B) CD3ε+ cells from p6 and p7 were assed for expression of SWC6, CD4α, CD8α, and CD8β. CD3ε+ cells within the SCID pigs were primarily CD8α+ CD8β+ CD4α− and CD8α+ CD8β− CD4α+.
We next sought to determine the cellular phenotypes of the CD3ε+ cells in the SCID pigs. Normal swine have T helper cells (CD4+CD8α−CD8β−), memory T helper cells (CD4+CD8α+CDβ−) and cytotoxic T cells (CD4−CD8α+CD8β+) (20). To assess the cellular phenotypes, PBMCs were stained for SWC6, CD4α, CD8α, and CD8β. SWC6 is expressed on a majority of γδ T cells found within the blood (16, 17). PBMCs from p6 (ART12/12), p7 (ART12/16), and p8 (carrier), as a control, were analyzed for these T cell markers at 3 months of age. Both SCID animals appeared to have a very small population of CD3ε+ SWC6+ cells (Figure 2B). The CD3ε+ SWC6− population in both p6 and p7 were primarily CD8α+ CD8β+ CD4α− and CD8α+ CD8β− CD4α+. T helper memory cells in swine have a CD8α+ CD8β− CD4α+ cellular phenotype (21).
CD3ε+ Cells in the Lymph Nodes of Neonatal SCID Pigs
Since we found that CD3ε+ cells circulate in older animals, we next asked if these cells were present in neonatal SCID pigs. Lymph nodes were collected from 0 day old (0-6 hours old and given no colostrum) SCID pigs with ART16/16 (p9 and p10), ART12/16 (p11 and p12), ART12/12 (p13 and p14), and carrier (p15) genotypes and assessed for the presence of CD3ε+ and CD79α+ cells by immunohistochemistry. Two animals for each genotype were assessed; representative histology images are shown in Figure 3. Lymph nodes from SCID animals had abnormal architecture with poorly defined medulla and cortex structures. All SCID genotypes had some level of CD3ε+ cells present. The p13 and p14 (ART12/12) animal's lymph nodes had fewer CD3ε+ cells, which were punctate within the lymph node. CD79α+ cells were only found in one pig (p10, ART16/16), out of 25 that were tested. These results indicate that CD3ε+ cells, and in rare cases CD79α+ cells, initiate development in utero in ART−/− SCID pigs.
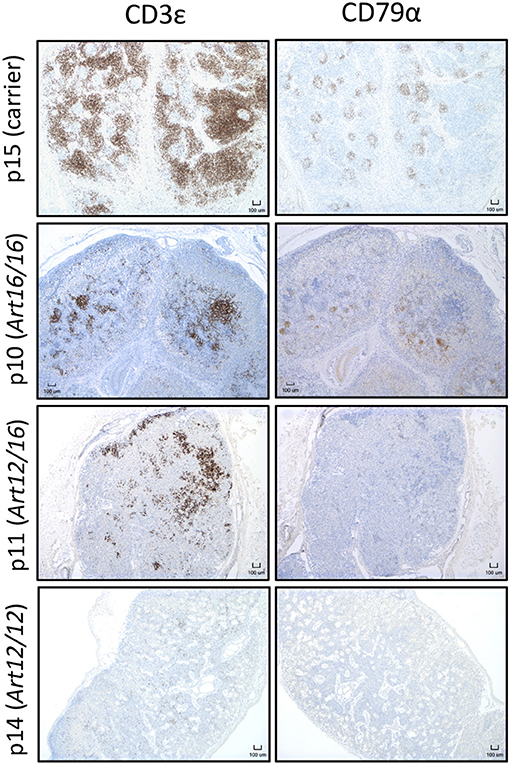
Figure 3. CD3ε+ cells are found in neonatal SCID pig lymph nodes. Lymph nodes were collected from 0 day old SCID pigs (p15 [carrier], p10 [ART16/16], p11 [ART12/16], p14 [ART12/12]) and were stained for CD3ε and CD79α. Indicated cells are labeled as brown.
CD3ε+ Cells in 5- and 6-Month-Old SCID Pigs
We additionally collected lymph nodes from ART16/16 SCID pigs that were 5.5 and 6 months of age. Pig p16 was originally raised in biocontainment facilities and was later moved to conventional housing at approximately 3 months of age. At 6 months of age the pig developed severe skin lesions and was euthanized due to failure to thrive. At the time of euthanasia, a blood sample was collected from this animal, and 33% of lymphocytes were found to be CD3ε+ (Figure 4A). Other T cell markers were not assessed at this time. Lymph nodes were also collected, and CD3ε+, but not CD79α+, cells were present (Figure 4B).
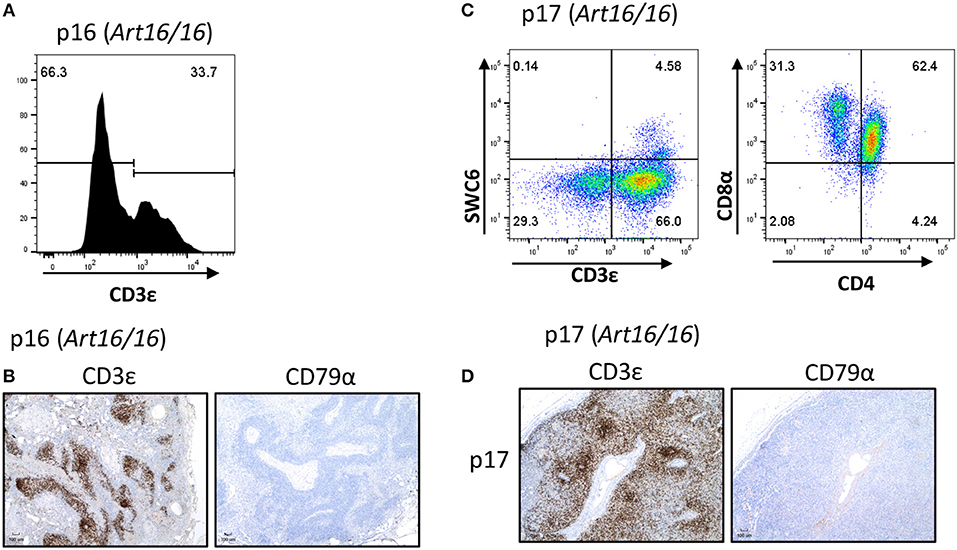
Figure 4. CD3ε+ cells in a 5- and 6-month-old SCID pigs. (A) PBMCs (gated on lymphocytes) were stained for CD3ε expression from a 6-month-old ART16/16 SCID (p16). (B) Lymph nodes were stained for CD3ε and CD79α expression from p16. (C) Mononuclear cells were isolated from a popliteal lymph node from p17 and stained for CD3ε, SWC6, CD4, and CD8α. A small population of CD3ε+ SWC6+ cells were present. A majority of the CD3+ SWC6− population were CD4α+ CD8α+. (D) Lymph nodes were stained for CD3 and CD79α expression from p17 [ART16/16].
Another older ART16/16 animal, p17, was also raised to 5 and a half months of age in biocontainment facilities. The pig was diagnosed with an E. coli bacterial infection and was euthanized due to failure to thrive. Lymph nodes were collected from this animal and were assessed by flow cytometry and histological analysis (Figures 4C,D). MNCs from a popliteal lymph node were isolated and analyzed by flow cytometry. Of gated MNCs, 66% of cells were CD3ε+. A small portion of the CD3ε+ cells were also SWC6+. CD3ε+ cells in p17 lymph nodes consisted of CD8α+ CD4α− and CD8α+ CD4α+. Expression of CD8β was not assessed at this time. Immunohistochemistry revealed high prevalence of CD3ε+ cells in the lymph node. CD79α+ cells were not present in lymph nodes.
Evidence of Limited TCR Rearrangement in ART–/– SCID Pigs
Artemis is involved in DNA repair and is a critical component of VDJ recombination. Since we observed that CD3ε+ cells develop in ART−/− SCID pigs, we assessed if there was sufficient residual Artemis function for productive somatic recombination by performing genomic PCR analysis on the TCRδ and TCRβ loci. PCR analysis was performed using primers that were specific for TCRδV5 and TCRδJ1, as well as TCRβV20 and TCRβJ1.2 in an assay originally described by Suzuki et al. (18) (Figure 5). We chose these primer pairs because they had previously been used to assess for VDJ recombination in a colony of RAG2 knock out pigs (18). Due to variation within V gene segments, amplification of specific recombined gene fragments can be difficult, however the previously mentioned primer sets were confirmed to amplify PCR fragments that specifically contained the designated V and J gene segments.
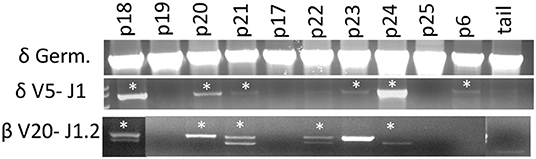
Figure 5. Cells with TCRδ V5-J1 and TCRβV20-J1.2 recombination are in SCID pig lymph nodes. DNA from lymph nodes were amplified with primers specific for TCRδV5 – TCRδJ1, as well as TCRβ V20- TCRβ J1.1 from a carrier and SCID pigs with all genotypes. Pigs shown include a carrier (p18), ART16/16 (p19, p20, p21, and p17), ART12/16 (p22, p23, p24), and ART12/12 (p25 and p6). Bands with asterisks were sequenced to confirm presence of V gene segment within the amplificon (Figures S2, S3).
SCID pigs of all three genotypes (p6, p16-25) were assessed. At least one pig from each SCID genotype had recombination between the V and J segments for either TCR complexes (TCRβ or TCRδ), although not every animal showed evidence of recombination. PCR was performed on a wild type, ART16/+ pig (p18) as a control and we detected rearrangements for both primer sets. PCR products (with asterisks) were sequenced to confirm amplicon contained V and J gene segments (Supplemental Figures 2, 3). In our control wildtype amplificons, we were unable to sequence through the VDJ joint, which we hypothesized was due to variable sequence joints using those specific V and J gene segments within wildtype animals. However, two SCID pigs (p20 and p24) had amplicons that sequenced through the TCRδ VDJ joint, indicating that the there was only a single clone for the V and J gene segments tested (Supplemental Figure 4). Together, we show VDJ recombination can occur in SCID pigs, although the repertoire may be limited.
TCR Transcripts Are Expressed in SCID Thymic and Lymph Node Tissue
Because we could detect productive somatic recombination products at the TCR loci, we wanted to determine if TCR transcripts were being expressed within SCID pig lymphoid tissues. Probes were created to target swine TRAC (pink) and TRDC (blue) transcripts and were used for the detection in thymic and lymph node tissue from one carrier (p26) and four (ART12/16) SCID pigs (p27-p30) (Figure 6). The carrier animal, p26, had a defined cortex and medullary areas within the thymus. ART12/16 SCID pigs lacked this architecture, although some structure was present within the thymus from SCID p30. Additionally, in the carrier lymph node there are defined areas of αβ and γδ T cells, with γδ localized near the connective tissue and αβ cells in the interior of the lymph node nodules. SCID pig lymph nodes had TRAC and TRAD expressing T cells near the connective tissue, however the localization of these cells was less organized compared to the carrier pig. All four ART12/16 SCID pig thymi and lymph node tissues expressed TCRα and TCRδ transcripts. Additionally, there appears to be greater abundance of TRDC transcripts in the SCID pigs compared to the carrier animal.
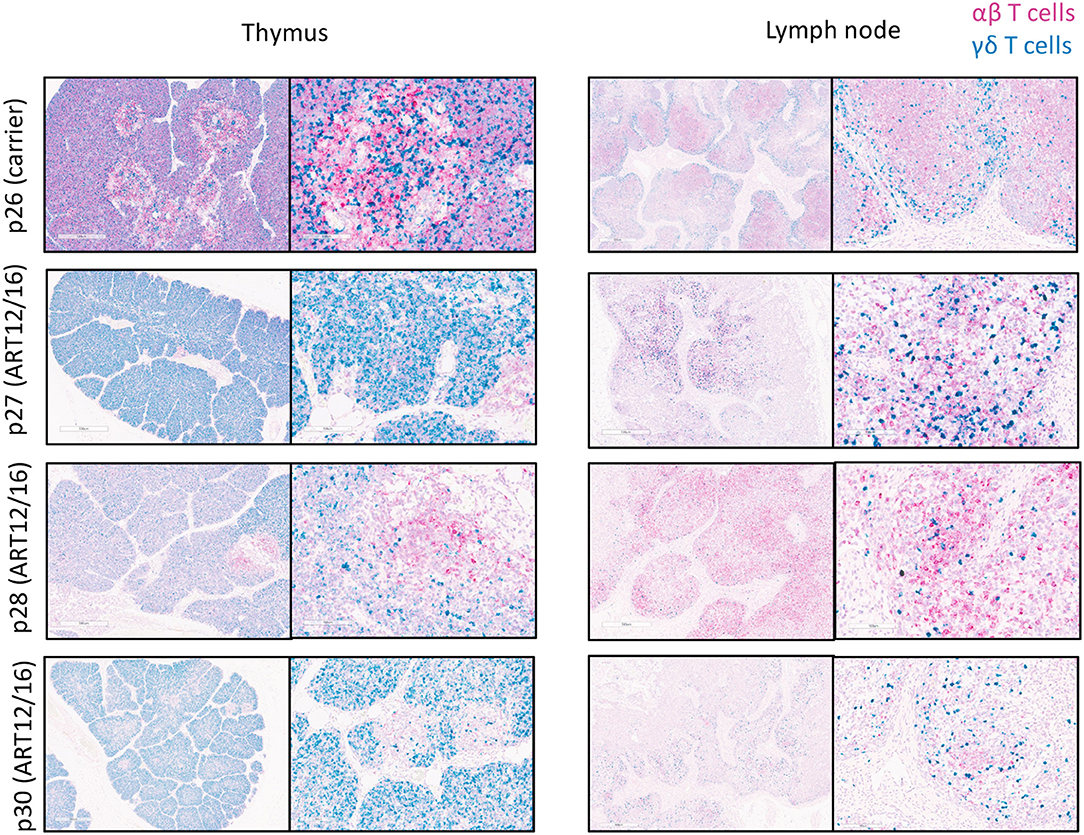
Figure 6. αβ and γδ T cells are in SCID pig thymic tissue and lymph nodes. Lymph nodes and remnant thymic tissue were collected from a carrier (p26) and ART12/16 SCID pigs (p27, p28, and p30). Probes were created to target the constant region of TCRα (red) and TCRδ (green) transcripts, to detect αβ and γδ T cells, respectively. All three ART12/16 SCID pig have both αβ and γδ T cells within the thymic and lymph node tissue, however the distribution of the cells and lymphoid organization differs from carrier animals.
Discussion
Here we report the description of T cells in SCID pigs with naturally occurring mutations in ART, a phenotype referred to as leaky SCID. We first observed that CD3ε+ cells were capable of developing in these SCID pigs when two ART16/16 bone marrow transplanted pigs developed host-derived T cell lymphoma (3). We followed these findings by asking if CD3ε+ cells were found in circulation of SCID pigs housed in conventional settings and biocontainment. We found that SCID pigs raised in either setting and across all three SCID genotypes (ART16/16, ART12/16, and ART12/12) had a leaky T cell phenotype. We next assessed if newborn SCID pigs had CD3ε+ cells in lymph nodes to gain insight into when these cells could be developing. We found CD3ε+ cells in lymph nodes of all SCID genotypes, although ART12/12 SCID pigs had fewer cells in pigs that we analyzed. We further assessed this phenotype and assayed for VDJ recombination by amplifying TCRβ and δ with V and J specific primers and found that all three SCID genotypes are capable of VDJ recombination. We suspect the TCR repertoire is limited in these SCIDs, as not all SCID pigs had recombination, and in some animals, there appears to be a clonal repertoire for the recombination events tested (Supplemental Figure 4). Lastly, we confirmed that TCRα and TCRδ transcripts are expressed in the thymic and lymph node tissues of SCID pigs.
Hypomorphic mutations are reported in human patients within RAG1 (5, 6), ARTEMIS (7–10), and IL2RG (22) genes. Such mutations can lead to Omenn's syndrome (11) or cancer development (7, 9). Omenn's syndrome is characterized by the development and expansion of a population of self-reactive T cells. Symptoms of Omenn's syndrome include diarrhea, eosinophilia, and lethargy (23). Additionally, T-cell lymphomas can develop due to recombination errors made during VDJ recombination (8). Identifying that SCID pigs with ART16 and ART12 alleles can produce CD3ε+ cells suggests that these types of complications could arise in SCID pigs. We have previously had SCID pigs with severe rashes, diarrhea, as well as masses of lymphoid tissue that may be explained as Omenn's syndrome or cancer (unpublished observations). The characterization of CD3ε+ cell development in our SCID pigs suggest that these ailments could occur in our ART SCID model. Additional investigation into the function of the ART16 and ART12 alleles and potential protein products in the development of CD3ε+ cells will also be important as this model is developed.
Further investigation into CD3ε+ cell function and cytokine production will be required for better understanding of these cells within the SCID pig. Additionally, assessment of the TCRα and TCRγ recombination events would help to better characterize these animals. RNA sequencing of the repertoire of these animals would be useful in gaining a better understanding about the development of these cells (24). We have recently generated an ART−/− IL2RG−/γ SCID pig model, in which we show that these “leaky” CD3ε+ cells are not present in piglets, as these animals do not have any CD3ε+ cells in blood or lymphoid organs (25). Researchers and veterinarians working with SCID pigs need to be aware of the potential of leaky T cell development. Presence of these cells could affect the health and potential use of these animals in different biomedical models.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The animal study was reviewed and approved by the Iowa State University Institutional Animal Care and Use Committee.
Author Contributions
AB designed experiments, ran flow cytometry, and wrote manuscript. AC-O and JO performed immunohistochemistry on tissues. YS and SAC performed TCR PCR analysis on lymph node DNA. JW performed RNA in-situ hybridization on lymphoid tissues. EP performed flow cytometry experiments. AA performed necropsies on animals and collected tissue and blood samples. CL and JC were involved in experimental planning and immunological data analysis. RR and JD were involved in experiment planning. SEC was involved with SCID pig care, genotyping, and tissue collection. CT was involved in all aspects of design and interpretation of the research. All authors read and edited the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Iowa State University's Laboratory Animal Resources staff for the care of our SCID pig colony. This project was funded by the Office of the Director, the Office of Research Infrastructure Programs, and the National Institutes of Health 1R24OD019813.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.00510/full#supplementary-material
Abbreviations
SCID, severe combined immunodeficient; TCR, T cell receptor; VDJ, variable; diversity; and joining; BMT, bone marrow transplant; PBMC, peripheral blood mononuclear cells
References
1. Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. (2001) 105:177–86. doi: 10.1016/S0092-8674(01)00309-9
2. Waide EH, Dekkers JCM, Ross JW, Rowland RRR, Wyatt CR, Ewen CL, et al. Not all SCID pigs are created equally: two independent mutations in the artemis gene cause SCID in pigs. J Immunol. (2015) 195:3171–9. doi: 10.4049/jimmunol.1501132
3. Powell EJ, Graham J, Ellinwood NM, Hostetter J, Yaeger M, Ho C-S, et al. T cell lymphoma and leukemia in severe combined immunodeficiency pigs following bone marrow transplantation: a case report. Front Immunol. (2017) 8:813. doi: 10.3389/fimmu.2017.00813
4. de Villartay J-P, Lim A, Al-Mousa H, Dupont S, Déchanet-Merville J, Coumau-Gatbois E, et al. A novel immunodeficiency associated with hypomorphic RAG1 mutations and CMV infection. J Clin Invest. (2005) 115:3291–9. doi: 10.1172/JCI25178
5. Ott de Bruin LM, Bosticardo M, Barbieri A, Lin SG, Rowe JH, Poliani PL, et al. Hypomorphic Rag1 mutations alter the preimmune repertoire at early stages of lymphoid development. Blood. (2018) 132:281–92. doi: 10.1182/blood-2017-12-820985
6. Giblin W, Chatterji M, Westfield G, Masud T, Theisen B, Cheng H-L, et al. Leaky severe combined immunodeficiency and aberrant DNA rearrangements due to a hypomorphic RAG1 mutation. Blood. (2009) 113:2965–75. doi: 10.1182/blood-2008-07-165167
7. Huang Y, Giblin W, Kubec M, Westfield G, St. Charles J, Chadde L, et al. Impact of a hypomorphic Artemis disease allele on lymphocyte development, DNA end processing, and genome stability. J Exp Med. (2009) 206:893–908. doi: 10.1084/jem.20082396
8. Jacobs C, Huang Y, Masud T, Lu W, Westfield G, Giblin W, et al. A hypomorphic Artemis human disease allele causes aberrant chromosomal rearrangements and tumorigenesis. Hum Mol Genet. (2011) 20:806–19. doi: 10.1093/hmg/ddq524
9. Moshous D, Pannetier C, de Chasseval R, Le Deist F, Cavazzana-Calvo M, Romana S, et al. Partial T and B lymphocyte immunodeficiency and predisposition to lymphoma in patients with hypomorphic mutations in Artemis. J Clin Invest. (2003) 111:381–7. doi: 10.1172/JCI16774
10. Felgentreff K, Lee YN, Frugoni F, Du L, van der Burg M, Giliani S, et al. Functional analysis of naturally occurring DCLRE1C mutations and correlation with the clinical phenotype of ARTEMIS deficiency. J Allergy Clin Immunol. (2015) 136:140–50.e7. doi: 10.1016/j.jaci.2015.03.005
11. Ege M, Ma Y, Manfras B, Kalwak K, Lu H, Lieber MR, et al. Omenn syndrome due to ARTEMIS mutations. Blood. (2005) 105:4179–86. doi: 10.1182/blood-2004-12-4861
12. Humblet-Baron S, Schonefeldt S, Garcia-Perez JE, Baron F, Pasciuto E, Liston A. Cytotoxic T-lymphocyte-associated protein 4-Ig effectively controls immune activation and inflammatory disease in a novel murine model of leaky severe combined immunodeficiency. J Allergy Clin Immunol. (2017) 140:1394–403.e8. doi: 10.1016/j.jaci.2016.12.968
13. Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell. (2002) 10:1379–90. doi: 10.1016/S1097-2765(02)00755-4
14. Powell EJ, Charley S, Boettcher AN, Varley L, Brown J, Schroyen M, et al. Creating effective biocontainment facilities and maintenance protocols for raising specific pathogen-free, severe combined immunodeficient (SCID) pigs. Lab Anim. (2018) 23:677217750691. doi: 10.1177/0023677217750691
15. Powell EJ, Cunnick JE, Knetter SM, Loving CL, Waide EH, Dekkers JCM, et al. NK cells are intrinsically functional in pigs with Severe Combined Immunodeficiency (SCID) caused by spontaneous mutations in the Artemis gene. Vet Immunol Immunopathol. (2016) 175:1–6. doi: 10.1016/j.vetimm.2016.04.008
16. Davis WC, Zuckermann FA, Hamilton MJ, Barbosa JI, Saalmuller A, Binns RM, et al. Analysis of monoclonal antibodies that recognize gamma delta T/null cells. Vet Immunol Immunopathol. (1998) 60:305–16. doi: 10.1016/S0165-2427(97)00107-4
17. Binns RM, Duncan IA, Powis SJ, Hutchings A, Butcher GW. Subsets of null and gamma delta T-cell receptor+ T lymphocytes in the blood of young pigs identified by specific monoclonal antibodies. Immunology. (1992) 77:219–27.
18. Suzuki S, Iwamoto M, Hashimoto M, Suzuki M, Nakai M, Fuchimoto D, et al. Generation and characterization of RAG2 knockout pigs as animal model for severe combined immunodeficiency. Vet Immunol Immunopathol. (2016) 178:37–49. doi: 10.1016/j.vetimm.2016.06.011
19. Uenishi H, Hiraiwa H, Yamamoto R, Yasue H, Takagaki Y, Shiina T, et al. Genomic structure around joining segments and constant regions of swine T-cell receptor alpha/delta (TRA/TRD) locus. Immunology. (2003) 109:515–26. doi: 10.1046/j.1365-2567.2003.01695.x
20. Gerner W, Kaser T, Saalmuller A. Porcine T lymphocytes and NK cells–an update. Dev Comp Immunol. (2009) 33:310–20. doi: 10.1016/j.dci.2008.06.003
21. Summerfield A, Rziha HJ, Saalmuller A. Functional characterization of porcine CD4+CD8+ extrathymic T lymphocytes. Cell Immunol. (1996) 168:291–6. doi: 10.1006/cimm.1996.0078
22. Kuijpers TW, van Leeuwen EMM, Barendregt BH, Klarenbeek P, aan de Kerk DJ, Baars PA, et al. A reversion of an IL2RG mutation in combined immunodeficiency providing competitive advantage to the majority of CD8(+) T cells. Haematologica. (2013) 98:1030–8. doi: 10.3324/haematol.2012.077511
23. Elnour IB, Ahmed S, Halim K, Nirmala V. Omenn's Syndrome: a rare primary immunodeficiency disorder. Sultan Qaboos Univ Med J. (2007) 7:133–8.
24. Hammer SE, Leopold M, Prawits L-M, Mair KH, Schwartz JC, Hammond JA, et al. Development of a RACE-based RNA-Seq approach to characterize the T-cell receptor repertoire of porcine gammadelta T cells. Dev Comp Immunol. (2020) 105:103575. doi: 10.1016/j.dci.2019.103575
Keywords: SCID, severe combined immunodeficiency, artemis, T cell, swine
Citation: Boettcher AN, Cino-Ozuna AG, Solanki Y, Wiarda JE, Putz E, Owens JL, Crane SA, Ahrens AP, Loving CL, Cunnick JE, Rowland RRR, Charley SE, Dekkers JCM and Tuggle CK (2020) CD3ε+ Cells in Pigs With Severe Combined Immunodeficiency Due to Defects in ARTEMIS. Front. Immunol. 11:510. doi: 10.3389/fimmu.2020.00510
Received: 23 August 2019; Accepted: 05 March 2020;
Published: 31 March 2020.
Edited by:
Jayne Hope, University of Edinburgh, United KingdomReviewed by:
Sabine Hammer, University of Veterinary Medicine Vienna, AustriaCaroline Fossum, Swedish University of Agricultural Sciences, Sweden
Copyright © 2020 Boettcher, Cino-Ozuna, Solanki, Wiarda, Putz, Owens, Crane, Ahrens, Loving, Cunnick, Rowland, Charley, Dekkers and Tuggle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher K. Tuggle, Y2t0dWdnbGUmI3gwMDA0MDtpYXN0YXRlLmVkdQ==
 Adeline N. Boettcher
Adeline N. Boettcher A. Giselle Cino-Ozuna2
A. Giselle Cino-Ozuna2 Yash Solanki
Yash Solanki Jayne E. Wiarda
Jayne E. Wiarda Ellie Putz
Ellie Putz Amanda P. Ahrens
Amanda P. Ahrens Crystal L. Loving
Crystal L. Loving Joan. E. Cunnick
Joan. E. Cunnick Raymond R. R. Rowland
Raymond R. R. Rowland Jack C. M. Dekkers
Jack C. M. Dekkers Christopher K. Tuggle
Christopher K. Tuggle