- 1Department of Biosciences, Manipal University Jaipur, Jaipur, India
- 2Department of Microbiology, School of Life Sciences, Central University of Rajasthan, Ajmer, India
- 3Department of Biology, College of Sciences, University of Ha'il, Ha'il, Saudi Arabia
- 4Department of Biochemistry, College of Medicines, University of Ha'il, Ha'il, Saudi Arabia
- 5Department of Botany, Munshi Singh College, Babasaheb Bhimrao Ambedkar Bihar University, Muzaffarpur, India
- 6Amity Institute of Biotechnology, Amity University Rajasthan, Jaipur, India
Human milk is a complex liquid that contains multifaceted compounds which provide nutrition to infants and helps to develop their immune system. The presence of secretory immunoglobulins (IgA), leucocytes, lysozyme, lactoferrin, etc., in breast milk and their role in imparting passive immunity to infants as well as modulating development of an infant's immune system is well-established. Breast milk miRNAs (microRNAs) have been found to be differentially expressed in diverse tissues and biological processes during various molecular functions. Lactation is reported to assist mothers and their offspring to adapt to an ever-changing food supply. It has been observed that certain subtypes of miRNAs exist that are codified by non-human genomes but are still present in circulation. They have been termed as xeno-miRNA (XenomiRs). XenomiRs in humans have been found from various exogenous sources. Route of entry in human systems have been mainly dietary. The possibility of miRNAs taken up into mammalian circulation through diet, and thereby effecting gene expression, is a distinct possibility. This mechanism suggests an interesting possibility that dietary foods may modulate the immune strength of infants via highly specific post-transcriptional regulatory information present in mother's milk. This serves as a major breakthrough in understanding the fundamentals of nutrition and cross-organism communication. In this review, we elaborate and understand the complex crosstalk of XenomiRs present in mother's milk and their plausible role in modulating the infant immune system against infectious and inflammatory diseases.
Introduction
Human milk is a complex liquid, containing multifaceted compounds that provide nutrition and help to develop infant's immune systems (1). The presence of secretory immunoglobulins (IgA), leucocytes, lysozyme, lactoferrin, etc., in breast milk and their role in imparting passive immunity to infants as well as modulating the of development of an infant's immune system is well-established. Furthermore, parental dietary signals can be passed to tens of succeeding generations. miRNA in milk appeared to be endogenous for mammary glands and employed as biomarkers to assess the performance and healthiness of glands during lactation (2). The abundance of variants in miRNAs in mammalian milk has led to curiosity over their influence on developing infant gastrointestinal and immune system, along with tissue-specific developmental functions. Over 600 miRNAs have been identified from human breast milk; a range of these possess immunological roles viz. regulation of T-cells and B-cells differentiation, while others have regulatory effects on various physiological and metabolic responses (3, 4).
Breast milk miRNAs exert differential expression in diverse tissues and modulate diverse molecular functions. Lactation has been considered as the first step in postnatal defense and immune-modulation, thus enabling adaptation in spite of an ever-changing food supply (5). The research illustrated that maternally-secreted miRNA plays regulatory roles, i.e., macromolecule biosynthesis and macromolecule metabolism in infants, and are also considered as contributors to the nutritional richness of breast milk. Human breast milk may be fractionated into cells, skim milk, and lipids. Each fraction contains varying concentrations of microRNA, with the highest in the cell and lipid fractions, and the lowest in skim milk. Transcriptome profiling revealed the inclusive presence of known and novel miRNA species in fat globules of human breast milk (6). An electropherogram profiling generated for miRNA using miRCURY-Biofluids kit indicate the following concentrations: 12.74 ± 24.44 ng/mL (lipid fraction), (2.17 ± 5.66 ng/mL) (skim milk), and 1,443 ± 3,448 ng/mL (cells) (3, 4).
Thus, we aim to confirm the significance of the correlative deviation of the relative composition of breast milk miRNA with maternal fat diet. Dietary comparisons viz. high fat-high carbohydrate and relative glucose-galactose resulted in notably increased detection of miR-67 and miR-27 in high fat intake as opposed to high carbohydrate intake. Deciphering human breast milk miRNomics may pave new avenues in infant disease alleviation and health augmentation.
The Long and Short of miRNA
miRNAs are approximately 22-nucleotide-long non-coding small RNAs that mediate post-transcriptional gene silencing by binding to their mRNA targets. Discovered in 2004, miRNAs were involved in a wide range of biological processes including cell proliferation, cell differentiation, cell migration, disease initiation, and disease progression. The circulating miRNA are actively secreted by living cells or passively released during cell death. The miRNA profiles vary based on the type of cell/tissue or organ and are being suggested to serve as biomarkers for indication of different diseases.
The advent of next generation sequencing and genome-wide analysis has highlighted the existence of exogenous miRNAs. As a result of their stability and presence in bodily fluids, studies have hinted at an inter-individual epigenetic communication, whereby by having contact with their body fluids miRNA's released by one individual could have actions in another (7).
XenomiRs and Their Horizontal Transfer in Humans
It has been observed that certain subtypes of miRNAs exist that are codified by non-human genomes but are present in the circulation. They have been termed as xeno-miRNA (XenomiRs). One of the earliest mentions of XenomiRs was reported by Zhang et al. (8), where they discovered exogenous plant miRNAs (miR168a in rice) existing in the sera and tissues of animals (8). XenomiRs in humans have been found from plant (9, 10), animal (11), and viral sources. Approximately 872 circulating miRNAs, belonging to 42 plant families, have been identified by genomic analysis screening and RNA sequencing, with 325 miRNAs fully annotated from the plant model Arabidopsis thaliana (12).
The route of entry of miRNAs in human systems has been mainly via the diet. Multiple studies based on cloning, RNA sequencing, dietary miRNA transfer experiments and analysis, as well as experimentation relating to the strong iodine-containing oxidizing agent, suggest that approximately 5% of the different types of miRNAs detected so far in the human serum are plant miRNAs containing a 2′ -O-methyl modified 3′ end making them resistant to oxidative degeneration (13). The prevalence of exogenous miRNAs in food has been evident irrespective of the intense amount of processing techniques. On comparison, food derived from animals demonstrate a higher incidence of the miRNA. The elusive question remains whether the XenomiRs are good or bad for humans? Evidence has cropped over the years that circulating XenomiRs may regulate the progression or alleviation of chronic maladies. A freely circulating candidate, miR2911 from honeysuckle, was found to have a singular effect of targeting Influenza A viruses (IAVs) and demonstrated anti-viral activity (14).
Extracellular miRNAs (EcmiRNA) when naked are rapidly degraded by extracellular RNases, and hence have been found to be ingested as either membrane vesicles or as stable macromolecular complexes. AGO-bound miRNAs (Argonaute-bound) have shown the capability of crossing gap junctions, thereby enabling modulation of gene expression in the host organism. Studies confirmed that dietary miRNA remains stable despite processing and a dedicated vegetarian-diet was found to be associated with an increase of plant miR-168 in GI (gastro-intestinal) mucosa, feces, and in fecal samples of colorectal cancer (15). Conversely, no changes in miR-21 were reported after 1-week meat-rich diet in feces. On the contrary, another study demonstrated red meat upregulates multiple tumor-associated miRNAs (miR-17-92 cluster and miR-21) (16).
Validation of xenomiR in human milk generated an unexpected differential profile in human subjects with diminutive food habit fluctuations. A distinct demarcation could be observed in the miRNA profile of candidates with a vegetarian and non-vegetarian diet. Five plant food-derived miRNAs (miR166a, miR156a, miR157a, miR172a, and miR168a) were found to be significantly present in human breast milk. In contrast, quantification of the miRNA indicated a relatively higher concentration of miR166a and miR168a [whole milk] and miR156a and miR168a [exosomal fraction] in candidates with a non-vegetarian diet. miR168a was significantly lower in vegetarian candidates whereas miR172a was found to be prevalent in non-vegetarian ones (17). Milk-derived XenomiRs may have a different modus operandi in inter-species epigenetic communication. Surprisingly, homology has been found between animal and human miRNAs. Mir-155, present in the milk derived from bovine and human sources, was sequentially compared and observed to have a high percentage of sequence similarity (Figure 1). Hence, presence of such types of homologous miRNAs may generate evident physiological responses. Similar sequence similarities have also been cited in miR-21-5p and miR-30a-5p sequences in human and bovine samples (18). In addition to a varied diet, certain amounts of conservation of xenomiR present in the milk exosomes from different organisms has been observed. let-7 family members like let-7a-5p, let-7b-5p, and let-7f, as well as miR-148a, were found to be conserved in human, cow, pig, and panda species (19). Its uptake by human cell line [THP-1] was substantiated, thus suggesting that bovine milk exosomes containing miRNA can enter the circulatory system of humans (20). Time course analysis in a murine model indicated that there was distribution of exosomal miRNA in the liver, spleen, heart, and lungs even 3 h after intake (21). Galactose and sialo-galactose glycan modifications of the exosomal proteins have been implicated in the uptake of bovine miRNA in non-bovine species (22).
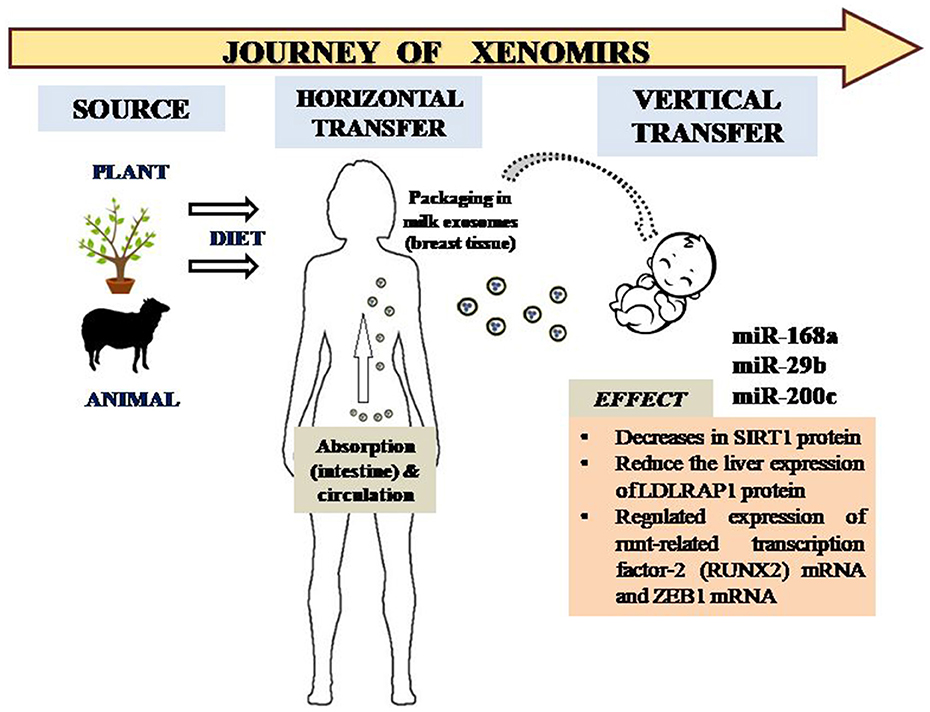
Figure 1. Transfer of XenomiRs from exogenous sources to humans and the subsequent journey into infants via packaged exosomes in milk. The XenomiRs affect specific targets in the human infant, thus effecting particular pathways corresponding to genome expression and regulation.
A study based on C. elegans indicated that uptake of miRNA from the human gut is mediated by two transport proteins, Sid-1 and Sid2. Sid-1, a multipass trans-membrane protein in stomach lumen, and Sid-2, an intestinal luminal transporter, channelizes dsRNA across the membrane. Sid-2 binds long dsRNA first for endocytosis, then the dsRNA is presented to Sid-1. An efficient uptake of dsRNA to the intestinal cell cytoplasm is facilitated by the activation of Sid-1 by Sid-2. Sid-1 is able to transfer small interfering RNA (siRNA) and miR-21 (23).
Vertical Transfer of XenomiRs in Maternal Fetal Axis
Breast milk contains a high level of nutritional content and confers the first line of immunological security to the infant. miRNA have been detected in whey fractions of human breast milk, bovine milk, and rat milk. Profiling of 681 mature miRNAs present in human milk and fat and a comparative analysis revealed higher miRNA concentration in human milk and maternal peripheral blood mononuclear cells. Furthermore, the presence of epithelial cells in mature milk justified the predominant origin of human milk miRNAs from the mammary epithelium (4). Similarly, bovine milk miRNA was persistent in supernatant as well as in exosomes.
Exosomes, a subtype of extracellular vesicle of 30–120 nm diameter, constitute the highly bioactive fraction enabling cell-to-cell communication (24–27). Colostrum has a comparatively higher prevalence of range of miRNA as compared to mature milk (28). Tolerance of exosomes to acidic pH and RNases ensures their persistence in humans as well as in the infant gastro-intestinal (GI) tract, thus aiding functional effects.
Mother's milk is rich in miRNAs, whose expression is variable and dependent upon the developmental stage and nutritional requirements of the fetus. Although initial studies have indicated the miRNA transferred via milk was prone to digestion in the stomach (29), progressive studies have confirmed the uptake and their subsequent physiological post-transcriptional regulatory effects (30, 31). This serves as a major breakthrough in understanding the fundamentals of nutrition and cross-organism communication.
When monitored, the compositional variations of miRNA content indicated variation after 2 months of lactation, especially in breast milk for preterm babies (32). miR-16 and miR-21 have been observed to be stably expressed during a study period in raw pooled milk samples, whereas miR-146b showed a higher stability than miR-16 and miR-21 in lipids and skim milk fractions. Clock-time dependent variability in miR-16-5p emphasizes dynamic fluctuations variations in breast milk with the nourishment requirements of infants. miR-16 was found to be involved in the regulation of gastrointestinal rhythmicity of the lactating infant along with development of its coordination with maternal rhythms.
In a report by Zhao et al. (12), breast milk samples were analyzed and found to contain plant-derived miRNA with an 100% prevalence, whereas analysis of the sera indicated the presence of circulating exosomes with a 42.9% prevalence rate. Another group (33), has shown the consistent presence of plant XenomiRs in human plasma and exosomal fractions. Diet derived miRNA tend to have 2′-O-methylation and, hence, were easily distinguishable. Plant-derived miRNA are packed into microvescicles [MV] and released into the sera. These MVs with AGO2-associated exogenous miRNA are transported to other organs (8). Another prominent mechanism, especially in the case of miR2911, is association of the miRNA with proteinase K-resistant complex, enabling stable passage through the serum (34). Two plant-derived miRNAs, namely miR156a and miR168a, have been consistently found in human breast milk, thus confirming the uptake of plant XenomiRs by mammary glands. So as per the partial evidences generated, the passage of foreign miRNA is via the GI tract followed by encapsulation in exosomes, uptake of exosomes by the mammary gland, and subsequent transfer of the exosomes into the fetus via breast milk. The route of transfer of serum exosomes in breast milk, however, remains unclear. These human exosomes are resistant to proteinase K, pepsin, and pancreatin digestion but have the capability of being uptaken by macrophages (3, 34–36).
Studies have confirmed that transfer of human milk microRNA into infants enables alleviation of the immune system in infants, especially in the first 6 months of life. Milk miR-155 have been found to regulate T and B cells in the innate immune response of infants. As far as XenomiRs uptake, studies have been conducted on transgenic animals. However, there is definite gap in knowledge confirming the effect milk-derived XenomiRs have in infants. The presence of XenomiRs in soluble fractions of human breast milk ensures the uptake by the infant. Studies have already indicated that there is a vertical transfer of miRNAs across the fetal placental axis (37).
Milk Nutrimiromics in Infant Immunity
Emerging research on mammary gland biology and bioactive molecules stated that miRNAs are one of the most important immunological agents. Studies have confirmed that transfer of human milk microRNA into infants enables alleviation of the immune system in infants, especially in the first 6 months of life. Milk miR-155 have been found to regulate T and B cells in the innate immune response of infants. As far as uptake of XenomiRs, validated studies have been conducted on transgenic animals. Analysis of lactation-related miRNA expression profiles in porcine milk exosomes during the lactation period (from newborn to 28 days after birth) revealed the presence of high numbers of immune-related miRNAs in colostrum (28). Thirteen unique immune-related miRNAs have observed to be present in breast milk exosomal vesicles (Table 1). miR-148a-3p was observed to be overexpressed during lactation period in porcine milk and observed to down-regulate the expression of DNA methyltransferase 3b (28, 55). However, the correlation between these and the subsequent impact on infant's growth and development is still unclear, and there is definite lacunae confirming milk-derived XenomiRs exercising their effect in infants. The presence of XenomiRs in soluble fractions of human breast milk ensures the uptake by the infant.
Studies have already indicated the presence of XenomiRs in the umbilical cord and cord blood, and the possibility of vertical transfer of miRNAs across the fetal placental axis (56). Transport of miRNA from mother to fetus in vesicles, mainly lipid encapsulated, provides them enhanced stability to cross through various barriers, particularly gastrointestinal ones (57). miR155 is the first miRNA associated with the immune system (58). It is transferred to the fetus via placental axis to neonates, as well as via milk to infants, and plays a key role in the maturation of macrophages and dendritic cells into active phenotype through toll-like receptors (59). miR223 was reported to act as a differentiation factor for monocytes and is responsible for proliferation and activation of granulocytes (60, 61). LPS (lipopolysaccharide) and pro-inflammatory cytokines (TNF-α, IL-1β) appeared as stimulants of miR146a and 146b, which in turn negatively regulate an acute innate immune response. miR146a is mainly responsible for NF-kB a mediated inflammatory response and type 1 interferon induction and signaling (62–64). However, miR125b is transcriptionally down-regulated via LPS stimulation and plays a key role in maintaining the levels of TNF-α (65).
Another set of miRNA involved in natal adipogenesis are miRNA-30b, miRNA-378, and let-7a. These have been found both in colostrum and mature milk. Higher incidence of let-7a and miRNA-378 has been quite evident in colostrum as compared to mature milk (66). Furthermore, let-7a/b/f-5p and miR-148-3p in vivo regulates transcription factor NF-κB, resulting in a suppression of the immune response (19). miR-148a-3p, along with miR-30b-5p, miR-182-5p, and miR-200a-3p, has also been designated as major immune-related pre-miRNAs (3), modulating the expression of TGIF2, PXR, and DNMT3B. Furthermore, miR-30b-5p stimulates cellular invasion and immunosuppression, miR-182-5p facilitates T-cell mediated immune responses, and miR-200a-3p has been linked with Hodgkin lymphoma and oral cancers. Other miRNAs associated with pathological and immune responses include miR-29a-3p (target interferon-γ, suppresses immune response to intracellular pathogens), miR-141-3p (biomarker for colon cancer), miR-378-3p (molecular switch involved in breast cancer cell metabolism), and miR-146b-5p (NF-kB signaling in innate immune responses). Colostrum contain miR-181a and hsa-miR-223, whose targets tend to be T and granulocytes cell populations, and hence affect the developmental stage of adaptive immune response in infants (67). Necrotizing enterocolitis afflicts 1–5% of premature neonates and there is a marked oxidative stress associated in the intestinal epithelial compartment. miRNA-125b present in breast milk-derived exosomes exhibits a prospective role in decreasing cell toxicity by targeting TLR4-dependent regulation of p53 (68). A distinct correlation has been found between Hsa-miR-195-5p and HsamiR-191-5p and CD4+ T-cell counts.
XenomiRs of Breast Milk and Their Effect
To date, few XenomiRs have been functionally annotated. Plant-specific miR-168a, obtained from Magnifera indica, is acquired by humans via ingestion, and propelled into circulation via the GI tract. Magnifera Oleifera miR-168a is a functional homolog of human miR-579. Transfection of miR-168a in hepatic cancer cell lines effects the expression of NAD-dependent deacetylase sirtuin-1(SIRT1) protein, a valid target of miR-579, further supporting this cross-kingdom regulation (69, 70). Sirt1 catalyzes the formation of unique metabolite O-acetyl-ADP ribose, which regulates the functioning of several transcription factors. Another instance of the regulatory action of miR-168a was found in the case of low-density lipoprotein receptor adapter protein 1 (LDLRAP1), involved in vesicular endocytosis, whereby expression of hepatic LDLRAP1 was down-regulated upon exposure to miR-168a. LDLRAP1 is one of the major transport proteins involved in cholesterol uptake by the cells (8, 54).
miR-29b and miR-200c found in bovine milk tends to target the runt-related transcription factor-2 (RUNX2) mRNA and zinc finger E-box-binding homeobox 1 (ZEB1) mRNA and, furthermore, no homeostatic maintenance of the depleted levels of these miRNAs were compensated by intracellular synthesis. RUNX2 is involved in osteoblast differentiation and bone mineralization, whereas ZEB1 modulates the expression of IL2 in vivo (Figure 1).
Conclusions
The potential implication of XenomiRs affecting and inducing epigenetic modification in humans is vast. There is a marked absence of validated data regarding the vertical transfer of xenomiR to infants. With the advent of state of the art technology, procedures have emerged which can distinguish the dietary miRNAs and endogenous miRNAs even at minimal concentrations and their progressive packaging in the milk exosomes. Enrichment of the functionally annotated XenomiRs has taken a major leap via in silico method. Techniques like RNase H-dependent PCR (rhPCR), combined with exhaustive high throughput sequencing, may further add to the plethora of knowledge pertaining to XenomiRs (71).
Author Contributions
MD and SR conceived the idea. BS, MD, and NP contributed to writing of the manuscript. MD and BS prepared figures and/or tables. SR, MS, and MK reviewed drafts of the paper and approved the final draft.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wholeheartedly thank all the authors whose research work has made this review possible.
References
1. Sharp JA, Modepalli V, Enjapoori AK, Bisana S, Abud HE, Lefevre C, et al. Bioactive functions of milk proteins: a comparative genomics approach. J Mammary Gland Biol Neoplasia. (2014) 19:289–302. doi: 10.1007/s10911-015-9331-6
2. Silveri L, Tilly G, Vilotte JL, Le Provost F. MicroRNA involvement in mammary gland development and breast cancer. Reprod Nutr Dev. (2006) 46:549–56. doi: 10.1051/rnd:2006026
3. Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, et al. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. (2012) 8:118–23. doi: 10.7150/ijbs.8.118
4. Alsaweed M, Lai CT, Hartmann PE, Geddes DT, Kakulas F. Human milk cells and lipids conserve numerous known and novel miRNAs, some of which are differentially expressed during lactation. PLoS ONE. (2016) 11:e0152610. doi: 10.1371/journal.pone.0152610
5. Dall SR, Boyd IL. Evolution of mammals: lactation helps mothers to cope with unreliable food supplies. Proc Biol Sci. (2004) 271:2049–57. doi: 10.1098/rspb.2004.2830
6. Munch EM, Harris RA, Mohammad M, Benham AL, Pejerrey SM, Showalter L, et al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS ONE. (2013) 8:e50564. doi: 10.1371/journal.pone.0050564
7. Melnik BC, Schmitz G. DNA methyltransferase 1-targeting miRNA-148a of dairy milk: a potential bioactive modifier of the human epigenome. Funct Foods Health and Dis. (2017) 7:671–87. doi: 10.31989/ffhd.v7i9.379
8. Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. (2012) 22:107–26. doi: 10.1038/cr.2011.158
9. Fritz JV, Heintz-Buschart A, Ghosal A, Wampach L, Etheridge A, Galas D, et al. Sources and functions of extracellular small RNAs in human circulation. Annu Rev Nutr. (2016) 36:301–36. doi: 10.1146/annurev-nutr-071715-050711
10. Liu YC, Chen WL, Kung WH, Huang HD. Plant miRNAs found in human circulating system provide evidences of cross kingdom RNAi. BMC Genom. (2017) 18:112. doi: 10.1186/s12864-017-3502-3
11. Li Z, Xu R, Li N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr Metab. (2018) 15:68. doi: 10.1186/s12986-018-0311-x
12. Zhao Y, Cong L, Lukiw WJ. Plant and animal microRNAs (miRNAs) and Their Potential for Inter-kingdom Communication. Cell Mol Neurobiol. (2018) 38:133–40. doi: 10.1007/s10571-017-0547-4
13. Luo Y, Wang P, Wang X, Wang Y, Mu Z, Li Q, et al. Detection of dietetically absorbed maize-derived microRNAs in pigs. Sci Rep. (2017) 7:645. doi: 10.1038/s41598-017-00488-y
14. Zhou Z, Li X, Liu J, Dong L, Chen Q, Kong H, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. (2015) 25:39–49. doi: 10.1038/cr.2014.130
15. Link J, Thon C, Schanze D, Steponaitiene R, Kupcinskas J, Zenker M, et al. Food-derived xeno-microRNAs: influence of diet and detectability in gastrointestinal tract-proof-of-principle study. Mol Nutr Food Res. (2019) 63:e1800076. doi: 10.1002/mnfr.201800076
16. Gavrilas LI, Ionescu C, Tudoran O, Lisencu C, Balacescu O, Miere D. The role of bioactive dietary components in modulating miRNA expression in colorectal cancer. Nutrients. (2016) 8:590. doi: 10.3390/nu8100590
17. Lukasik A, Brzozowska I, Zielenkiewicz U, Zielenkiewicz P. Detection of plant miRNAs abundance in human breast milk. Int J Mol Sci. (2017) 19:37. doi: 10.3390/ijms19010037
18. Fromm B, Tosar JP, Lu Y, Halushka MK, Witwer KW. Human and cow have identical miR-21–5p and miR-30a-5p sequences, which are likely unsuited to study dietary uptake from cow milk. J Nutr. (2018) 148:1506–7. doi: 10.1093/jn/nxy144
19. Van Herwijnen MJC, Driedonks TAP, Snoek BL, Kroon AMT, Kleinjan M, et al. Abundantly present miRNAs in milk-derived extracellular vesicles are conserved between mammals. Front Nutr. (2018) 5:81. doi: 10.3389/fnut.2018.00081
20. Izumi H, Tsuda M, Sato Y, Kosaka N, Ochiya T, Iwamoto H, et al. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J Dairy Sci. (2015) 98:2920–33. doi: 10.3168/jds.2014-9076
21. Manca S, Upadhyaya B, Mutai E, Desaulniers AT, Cederberg RA, White BR, et al. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci Rep. (2018) 8:11321. doi: 10.1038/s41598-018-29780-1
22. Sukreet S, Silva BVRE, Adamec J, Cui J, Zempleni J. Galactose and sialo-galactose modifications in glycoproteins on the surface of bovine milk exosome are essential for exosome uptake in non-bovine species (OR34–07-19). Curr Dev Nutr. (2019) 3(Suppl. 1):pp.nzz031-OR34. doi: 10.1093/cdn/nzz031.OR34-07-19
23. Mcewan DL, Weisman AS, Hunter CP. Uptake of extracellular double-stranded RNA by SID-2. Mol Cell. (2012) 47:746–54. doi: 10.1016/j.molcel.2012.07.014
24. Zamanian M, Fraser LM, Agbedanu PN, Harischandra H, Moorhead AR, Day TA, et al. Release of small RNA-containing exosome-like vesicles from the human filarial parasite brugia malayi. PLoS Negl Trop Dis. (2015) 9:e0004069. doi: 10.1371/journal.pntd.0004069
25. Jan AT, Azam M, Rahman S, Almigeiti AMS, Choi DH, Lee EJ, et al. Perspective insights into disease progression, diagnostics, and therapeutic approaches in alzheimer's disease: a judicious update. Front Aging Neurosci. (2017) 9:356. doi: 10.3389/fnagi.2017.00356
26. Jan AT, Malik MA, Rahman S, Yeo HR, Lee EJ, Abdullah TS, et al. Perspective insights of exosomes in neurodegenerative diseases: a critical appraisal. Front Aging Neurosci. (2017) 9:317. doi: 10.3389/fnagi.2017.00317
27. Jan AT, Rahman S, Khan S, Tasduq SA, Choi I. Biology, pathophysiological role, and clinical implications of exosomes: a critical appraisal. Cells. (2019) 8:99. doi: 10.3390/cells8020099
28. Gu Y, Li M, Wang T, Liang Y, Zhong Z, Wang X, et al. Lactation-related microRNA expression profiles of porcine breast milk exosomes. PLoS ONE. (2012) 7:e43691. doi: 10.1371/journal.pone.0043691
29. Title AC, Denzler R, Stoffel M. Uptake and function studies of maternal milk-derived microRNAs. J Biol Chem. (2015) 290:23680–91. doi: 10.1074/jbc.M115.676734
30. Benmoussa A, Lee CH, Laffont B, Savard P, Laugier J, Boilard E, et al. Commercial dairy cow milk microRNAs resist digestion under simulated gastrointestinal tract conditions. J Nutr. (2016) 146:2206–15. doi: 10.3945/jn.116.237651
31. Liao Y, Du X, Li J, Lonnerdal B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol Nutr Food Res. (2017) 61:1700082. doi: 10.1002/mnfr.201700082
32. Floris I, Billard H, Boquien CY, Joram-Gauvard E, Simon L, Legrand A, et al. MiRNA analysis by quantitative PCR in preterm human breast milk reveals daily fluctuations of hsa-miR-16–5p. PLoS ONE. (2015) 10:e0140488. doi: 10.1371/journal.pone.0140488
33. Beatty M, Guduric-Fuchs J, Brown E, Bridgett S, Chakravarthy U, Hogg RE, et al. Small RNAs from plants, bacteria and fungi within the order Hypocreales are ubiquitous in human plasma. BMC Genomics. (2014) 15:933. doi: 10.1186/1471-2164-15-933
34. Yang J, Hirschi KD, Farmer LM. Dietary RNAs: new stories regarding oral delivery. Nutrients. (2015) 7:3184–99. doi: 10.3390/nu7053184
35. Lonnerdal B, Du X, Liao Y, Li J. Human milk exosomes resist digestion in vitro and are internalized by human intestinal cells. FASEB J. (2015) 29:121.123.
36. Hock A, Miyake H, Li B, Lee C, Ermini L, Koike Y, et al. Breast milk-derived exosomes promote intestinal epithelial cell growth. J Pediatr Surg. (2017) 52:755–9. doi: 10.1016/j.jpedsurg.2017.01.032
37. Simpson MR, Brede G, Johansen J, Johnsen R, Storrø O, Sætrom P, et al. Human breast milk miRNA, maternal probiotic supplementation and atopic dermatitis in offspring. PLoS ONE. (2015) 10:e0143496. doi: 10.1371/journal.pone.0143496
38. Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. (2008) 105:13556–61. doi: 10.1073/pnas.0803055105
39. Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem. (2008) 283:9674–80. doi: 10.1074/jbc.M709382200
40. Chen X, Gao C, Li H, Huang L, Sun Q, Dong Y, et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. (2010) 20:1128–37. doi: 10.1038/cr.2010.80
41. Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. MicroRNA expression in human airway smooth muscle cells: role of miR-25 in regulation of airway smooth muscle phenotype. Am J Respir Cell Mol Biol. (2010) 42:506–13. doi: 10.1165/rcmb.2009-0123OC
42. Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, Vega-Saenz De Miera E, et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. (2011) 20:104–18. doi: 10.1016/j.ccr.2011.05.027
43. Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. (2010) 11:1057–62. doi: 10.1038/ni.1945
44. Brabletz T, Jung A, Hlubek F, Lohberg C, Meiler J, Suchy U, et al. Negative regulation of CD4 expression in T cells by the transcriptional repressor ZEB. Int Immunol. (1999) 11:1701–8. doi: 10.1093/intimm/11.10.1701
45. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. (2008) 10:593–601. doi: 10.1038/ncb1722
46. Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, et al. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. (2009) 113:3754–64. doi: 10.1182/blood-2008-10-184077
47. Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, et al. Prognostic values of microRNAs in colorectal cancer. Biomark Insights. (2006) 2:113–21. doi: 10.1177/117727190600100009
48. O'hara AJ, Vahrson W, Dittmer DP. Gene alteration and precursor and mature microRNA transcription changes contribute to the miRNA signature of primary effusion lymphoma. Blood. (2008) 111:2347–53. doi: 10.1182/blood-2007-08-104463
49. Elyakim E, Sitbon E, Faerman A, Tabak S, Montia E, Belanis L, et al. hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res. (2010) 70:8077–87. doi: 10.1158/0008-5472.CAN-10-1313
50. Biton M, Levin A, Slyper M, Alkalay I, Horwitz E, Mor H, et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat Immunol. (2011) 12:239–46. doi: 10.1038/ni.1994
51. Sheedy FJ, Palsson-Mcdermott E, Hennessy EJ, Martin C, O'leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. (2010) 11:141–7. doi: 10.1038/ni.1828
52. Jennewein C, Von Knethen A, Schmid T, Brune B. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor gamma. (PPARgamma) mRNA destabilization. J Biol Chem. (2010) 285:11846–53. doi: 10.1074/jbc.M109.066399
53. Meng F, Henson R, Wehbe-Janek H, Smith H, Ueno Y, Patel T. The MicroRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. J Biol Chem. (2007) 282:8256–64. doi: 10.1074/jbc.M607712200
54. Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr. (2014) 144:1495–500. doi: 10.3945/jn.114.196436
55. Duursma AM, Kedde M, Schrier M, Le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. (2008) 14:872–7. doi: 10.1261/rna.972008
56. Li J, Zhang Y, Li D, Liu Y, Chu D, Jiang X, et al. Small non-coding RNAs transfer through mammalian placenta and directly regulate fetal gene expression. Protein Cell. (2015) 6:391–6. doi: 10.1007/s13238-015-0156-2
57. Tome-Carneiro J, Fernandez-Alonso N, Tomas-Zapico C, Visioli F, Iglesias-Gutierrez E, Davalos A. Breast milk microRNAs harsh journey towards potential effects in infant development and maturation. Lipid encapsulation can help. Pharmacol Res. (2018) 132:21–32. doi: 10.1016/j.phrs.2018.04.003
58. Cione E, Lucente M, Gallelli L, De Sarro G, Luciani F, Caroleo MC. Innate immunity and human milk MicroRNAs content: a new perspective for premature newborns. J Compr Ped. (2017) 8:e43359. doi: 10.5812/compreped.43359
59. O'connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. (2007) 104:1604–9. doi: 10.1073/pnas.0610731104
60. Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. (2008) 451:1125–9. doi: 10.1038/nature06607
61. Haneklaus M, Gerlic M, O'neill LA, Masters SL. miR-223: infection, inflammation and cancer. J Intern Med. (2013) 274:215–26. doi: 10.1111/joim.12099
62. Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. (2008) 10:R101. doi: 10.1186/ar2493
63. Williams AE, Perry MM, Moschos SA, Larner-Svensson HM, Lindsay MA. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem Soc Trans. (2008) 36:1211–5. doi: 10.1042/BST0361211
64. Nahid MA, Pauley KM, Satoh M, Chan EK. miR-146a is critical for endotoxin-induced tolerance: IMPLICATION IN INNATE IMMUNITY. J Biol Chem. (2009) 284:34590–9. doi: 10.1074/jbc.M109.056317
65. Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. (2007) 179:5082–9. doi: 10.4049/jimmunol.179.8.5082
66. Xi Y, Jiang X, Li R, Chen M, Song W, Li X. The levels of human milk microRNAs and their association with maternal weight characteristics. Eur J Clin Nutr. (2016) 70:445–9. doi: 10.1038/ejcn.2015.168
67. Perri M, Lucente M, Cannataro R, De Luca IF, Gallelli L, Moro G, et al. Variation in immune-related microRNAs profile in human milk amongst lactating women. Microrna. (2018) 7:107–14. doi: 10.2174/2211536607666180206150503
68. Martin C, Patel M, Williams S, Arora H, Brawner K, Sims B. Human breast milk-derived exosomes attenuate cell death in intestinal epithelial cells. Innate Immun. (2018) 24:278–84. doi: 10.1177/1753425918785715
69. Pirro S, Zanella L, Kenzo M, Montesano C, Minutolo A, Potesta M, et al. MicroRNA from Moringa oleifera: identification by high throughput sequencing and their potential contribution to plant medicinal value. PLoS ONE. (2016) 11:e0149495. doi: 10.1371/journal.pone.0149495
70. Navarro E, Funtikova AN, Fito M, Schroder H. Prenatal nutrition and the risk of adult obesity: long-term effects of nutrition on epigenetic mechanisms regulating gene expression. J Nutr Biochem. (2017) 39:1–14. doi: 10.1016/j.jnutbio.2016.03.012
Keywords: breast milk, exosomes, xeno-miRNA, vertical transfer, micro RNA
Citation: Stephen BJ, Pareek N, Saeed M, Kausar MA, Rahman S and Datta M (2020) Xeno-miRNA in Maternal-Infant Immune Crosstalk: An Aid to Disease Alleviation. Front. Immunol. 11:404. doi: 10.3389/fimmu.2020.00404
Received: 31 October 2019; Accepted: 20 February 2020;
Published: 24 March 2020.
Edited by:
Afsar Raza Naqvi, University of Illinois at Chicago, United StatesReviewed by:
Tomohide Takaya, Shinshu University, JapanDileep Kumar, The University of Utah, United States
Copyright © 2020 Stephen, Pareek, Saeed, Kausar, Rahman and Datta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Safikur Rahman, c2hhZmlxdWUyQGdtYWlsLmNvbQ==; Manali Datta, bWRhdHRhQGpwci5hbWl0eS5lZHU=
†These authors have contributed equally to this work
 Bjorn John Stephen
Bjorn John Stephen Nidhi Pareek
Nidhi Pareek Mohd Saeed3
Mohd Saeed3 Safikur Rahman
Safikur Rahman Manali Datta
Manali Datta