- 1Center for Genetic Epidemiology and Genomics, School of Public Health, Medical College of Soochow University, Suzhou, China
- 2Department of Epidemiology and Biostatistics, School of Public Health, Anhui Medical University, Hefei, China
- 3Anhui Province Key Laboratory of Major Autoimmune Diseases, Hefei, China
- 4Department of Rheumatology and Immunology, Anhui Provincial Hospital, Hefei, China
In a previous study, we have reported an increased plasma midkine (MK) and pleiotrophin (PTN) concentrations in patients with systemic lupus erythematosus (SLE) and the increase in MK and PTN associated with inflammatory cytokines interleukin (IL)-17 level and some clinical manifestations, suggesting the underlying association of MK and PTN with SLE. This study was conducted to investigate the association between common single-nucleotide polymorphisms (SNPs) in the MK and PTN gene and SLE susceptibility. A total of 989 subjects (496 SLE patients and 493 healthy controls) were included and genotyped for three MK SNPs and seven PTN SNPs in using improved multiple ligase detection reaction (iMLDR). Results have demonstrated no significant differences for genotype and allele frequencies in all 10 SNPs between SLE patients and healthy controls. Case-only analysis in SLE revealed that, in MK gene, the genotype frequency of AA/AG (rs35324223) was significantly lower in patients with photosensitivity than those without; the allele frequency of A/G (rs20542) was significantly higher in patients without serositis. In PTN gene, the A/G allele frequency (rs322236), C/T allele frequency, and TT/CT genotype frequency (rs6970141) showed significantly increased results in patients with immunological disorder compared to those without. Furthermore, no significant differences in plasma MK and PTN concentrations with its SNPs genotypes were found. MK and PTN SNPs showed no associations with SLE genetic susceptibility, but it may be associated with the course of this disease; further studies are needed to focus on the mechanism of MK and PTN genes in the pathogenesis of SLE.
Introduction
Systemic lupus erythematosus (SLE) is a chronic and severe systemic autoimmune disease involving multiple organs/tissues, characterized by the production of antinuclear autoantibodies, complement and interferon activation, and tissue destruction (1–3). SLE could occur at any age, particularly during childbearing years and predominantly affecting women (at a 9:1 ratio) (2). Up to now, no single cause for SLE has been identified. Epidemiological evidence, together with recent linkage and association studies, have demonstrated that the interactions between predisposing genetic factors and environmental components are hypothesized to contribute to the pathogenesis of SLE (4–8).
Midkine (MK), representing as the founding member of heparin-binding growth factor family, was initially identified as the product of the MK gene, which is 1.5 kb in size and located on chromosome 11q11.2 (9, 10). MK has a similar structure and shares 50% homology in amino acid sequence with pleiotrophin (PTN) (11). MK and PTN are the only cytokines that constitute the PTN/MK developmental gene family and show a biological activity including the promotion of growth, cell migration, tissue morphogenesis, and chemokine expression in numerous target cell types (12, 13). During the past few years, increasing evidences suggested an essential role of MK and PTN in carcinomas and acute and chronic inflammatory diseases (14–16). Studies have demonstrated that MK and PTN are involved in several types of carcinomas; the overexpression of MK or PTN correlated with a poor prognosis for patients with neuroblastomas, urinary bladder carcinomas, and papillary thyroid cancer (17–20). Furthermore, studies have shown an association of intronic polymorphism of MK gene with human sporadic colorectal and gastric cancers (21, 22). In postmenopausal women, PTN gene promoter −1227C>T (rs321198) polymorphism contributed to the genetic background of osteoporosis (23).
In rheumatoid arthritis (RA), a study has found an increased serum MK level in RA patients and its correlation with disease activity score (DAS) 28-erythrocyte sedimentation rate (ESR) and rheumatoid factors (RFs) titer (24). Our previous study has revealed that, as compared to healthy controls, the levels of plasma MK and PTN are elevated in SLE and also associated with interleukin (IL)-17 levels and some clinical manifestations, including rash and anti-Sjögren's-syndrome-related antigen A (anti-SSA) (25). These findings suggest a potential role of MK and PTN involved in certain types of autoimmune diseases.
Although study has unveiled the genetic association of MK and PTN in human cancer, the association between common single-nucleotide polymorphisms (SNPs) in the MK and PTN genes and SLE susceptibility has not yet been elucidated. Therefore, the present study was undertaken to comprehensively evaluate the role of common genetic variations in MK and PTN gene with SLE susceptibility in a Chinese population.
Materials and Methods
Study Population
This case–control genotyping study consisted of 989 study subjects. Four hundred ninety-six SLE patients were recruited from the Department of Rheumatology and Immunology at Anhui Provincial Hospital, the First Affiliated Hospital of Anhui Medical University. All patients were diagnosed based on a strict medical history and immunological autoantibody screening results, as well as with at least four 1997 American College of Rheumatology (ACR) revised criteria for SLE; in addition, an attending rheumatologist (Xiao-Mei Li) was also invited to confirm the diagnosis of SLE. The patients were then classified based on the 1997 revised ACR classification criteria (26, 27). Individual disease activity was quantified using the SLE Disease Activity Index 2000 (SLEDAI-2K) score (28, 29). The disease activity of SLE was stratified according to SLEDAI-2K score, of which a SLEDAI-2K score of ≥10 or <10 was defined as more active or less active, respectively (30). Four hundred ninety-three geographically and ethnically matched healthy controls were enrolled in the current study. All of the healthy controls did not have any inflammatory/autoimmune diseases history. Demographics, clinical manifestations, and laboratory findings were collected from hospital medical records and then reviewed by experienced physicians.
The Ethical Committee of Anhui Medical University (Hefei, Anhui, China) approved this study. The present study was conducted in accordance with the Declaration of Helsinki. All subjects, both cases and controls, provided informed consent to participate in this study.
SNP Screening and Genotyping
We used Ensembl Gene Browser 37 (http://grch37.ensembl.org/index.html) to acquire the detailed genetic and location information of human MK and PTN genes (31), and downloaded the linkage pedigree file (PED) and marker information file in Chinese Han population (CHB) of Beijing. Then, candidate tag SNPs selection was applied by utilizing the Haploview 4.2 software (Broad Institute, Cambridge, MA, United States), with the linkage disequilibrium r2 ≥ 0.80 and minor allele frequency (MAF) ≥ 5%. In total, 41 candidate tag SNPs (2 MK tag SNPs, 39 PTN tag SNPs) were chosen based on prior criteria. The online bioinformatics tools (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html) were implemented to predict the function of 41 tag SNPs (32), including potentially deleterious functional impact at the splicing, transcriptional, translational, and post-translational level, as shown in Table S1.
In addition, relevant literatures reporting SNPs regarding MK and PTN gene polymorphisms were also reviewed. Finally, in our study cohort, we included three tag SNPs (rs116869512, rs20542, and rs35324223) in MK gene and seven tag SNPs (rs161335, rs321198, rs322236, rs3959914, rs6970141, rs919581, and rs322297) in PTN gene for further genotyping. The detailed information regarding the location of the SNPs within each gene locus is displayed in Table S2.
Plasma MK and PTN Detections
Intravenous blood samples (5 ml) of SLE patients were collected; the plasma sample was extracted and then frozen at −80°C in a refrigerator until assayed. Determinations of MK and PTN concentrations in plasma were performed using enzyme-linked immunosorbent assay (ELISA) kits (purchased from Anhui Xinle Biotechnology Co. LTD; expressed as pg/ml).
Statistical Analysis
Comparisons of genotype and allele frequencies of SNPs between cases and controls were undertaken using chi-square or Fisher's exact test. Differences in plasma MK and PTN levels between different genotypes were compared using non-parametric test. The unconditional logistic regression model was used to estimate the associations between genotypes and SLE susceptibility. Three models were considered for statistical analysis, including additive, dominant, and recessive models. Statistical analysis was performed with the use of the Statistical Package for the Social Sciences (SPSS) statistical software, version 23.0 (SPSS Inc., Chicago, IL, United States).
Hardy–Weinberg equilibrium (HWE) and haplotype analyses were implemented using the SHEsis software (http://analysis.bio-x.cn/myAnalysis.php) (33). All statistical tests with two-tailed P < 0.05 values were considered statistically significant. The Bonferroni correction was used for multiple testing to reduce the chances of obtaining false-positive results.
Results
Basic Characteristics of Study Subjects
This study recruited 496 SLE patients and 493 healthy controls. In SLE patients, there were 438 female and 58 male with an average age of 37.58 ± 11.44 years; the disease duration was 5.78 ± 5.59 years, and the average SLEDAI-2K scores were 11.40 ± 9.07. In addition, the body mass index (BMI) for patients was 21.83 ± 3.17. As for healthy controls, there were 434 female and 59 male with an average age of 38.45 ± 11.32 years. The age and gender distribution showed no significant differences between cases and controls. Demographic characteristics and clinical features of study subjects are summarized in Table 1. The major clinical features of SLE patients comprised immunological disorder (73.2%), hematological disorder (68.1%), malar rash (54.6%), arthritis (49.4%), photosensitivity (31.1%), and renal disorder (37.1%) (Table 1). In healthy controls, the presence of observed genotype frequency distributions of all included tag SNPs were not significantly different from the HWE at the 5% level.
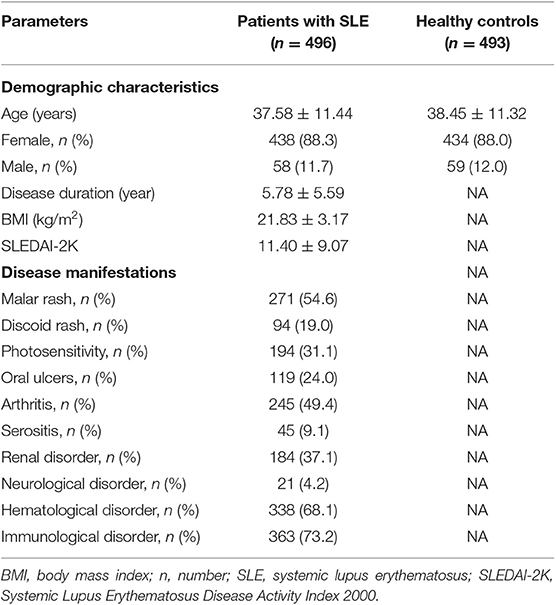
Table 1. Demographic characteristics and clinical features of patients with SLE and control subjects.
Association of MK and PTN Gene Polymorphisms With Susceptibility to SLE
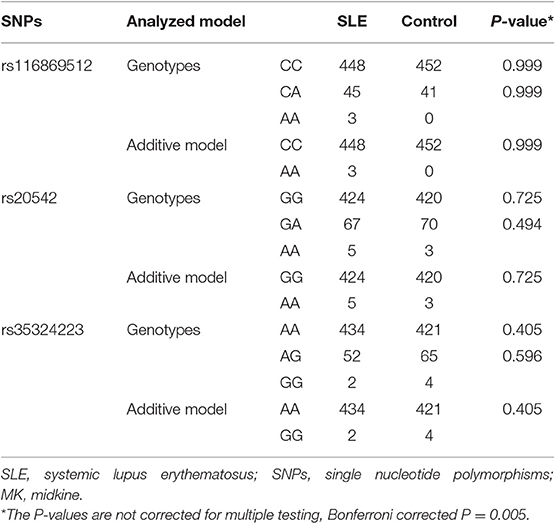
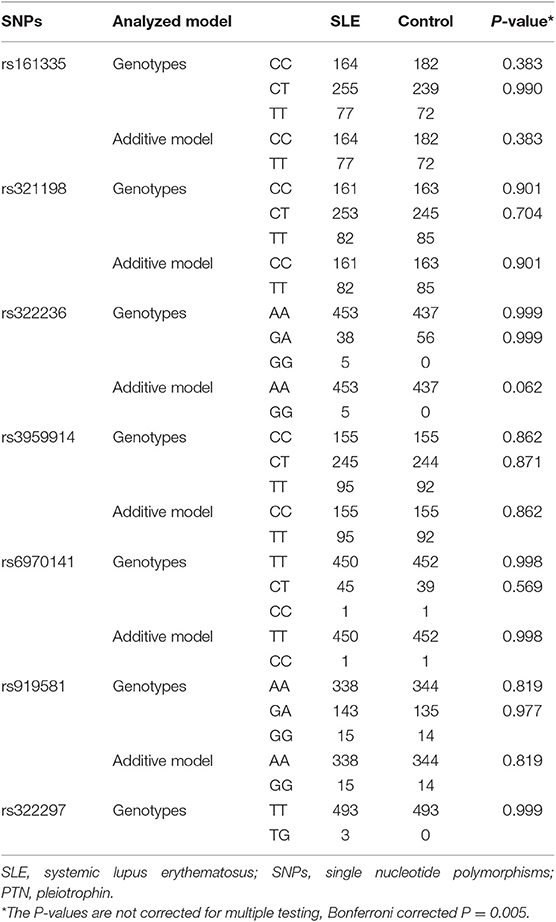
The genotype and allele frequencies of MK and PTN genes in SLE and healthy controls are shown in Tables 2, 3. In MK gene, there was no significant difference in genotype and allele distributions of three tag SNPs in SLE patients compared to healthy controls (Table 2). When analyzing seven tag SNPs in PTN gene, we did not find any significant differences in genotype and allele frequencies between SLE patients and healthy controls (Table 3).
Association of MK and PTN Gene Polymorphisms With Clinical Manifestations in SLE
To unveil the possible genetic associations in MK and PTN gene polymorphisms with clinical manifestations, case-only analysis was applied. In MK gene, as shown in Table 4, the frequency of AA/AG genotype (rs35324223) was significantly lower in patients with photosensitivity than those without (P = 0.012). The allele frequency of A/G (rs20542) was significantly higher in patients without serositis (P = 0.042). In PTN gene, the A/G allele frequency (rs322236), C/T allele frequency (rs6970141), and TT/CT genotype frequency appeared significantly increased risks in patients with immunological disorder compared to those without (P = 0.020, P = 0.027, P = 0.035, respectively) (Table 5). However, there were no significant associations for other tag SNPs in MK and PTN genes with clinical disease manifestations.
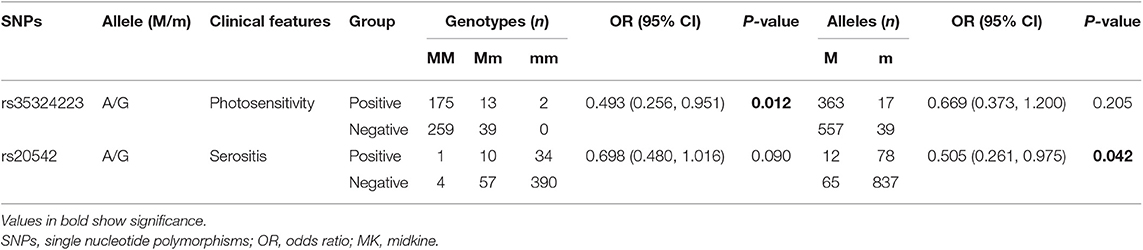
Table 4. The positive findings on association of clinical characteristics with genotype and allele frequencies in MK.
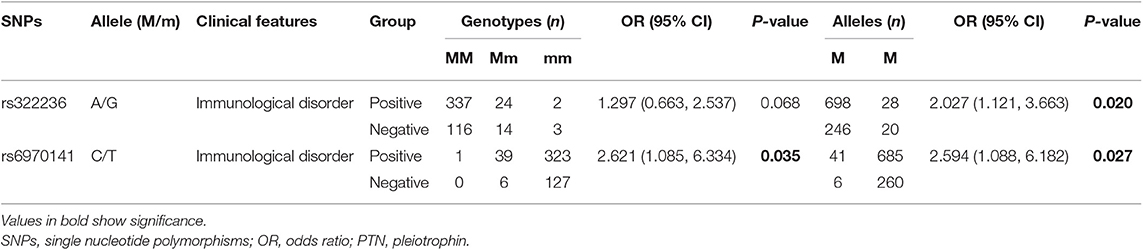
Table 5. The positive findings on association of clinical characteristics with genotype and allele frequencies in PTN.
Association of Plasma MK and PTN Levels With Its Genotypes
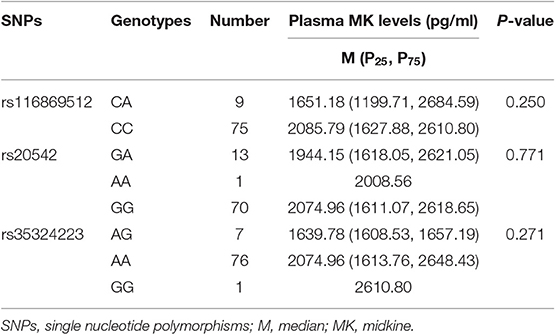
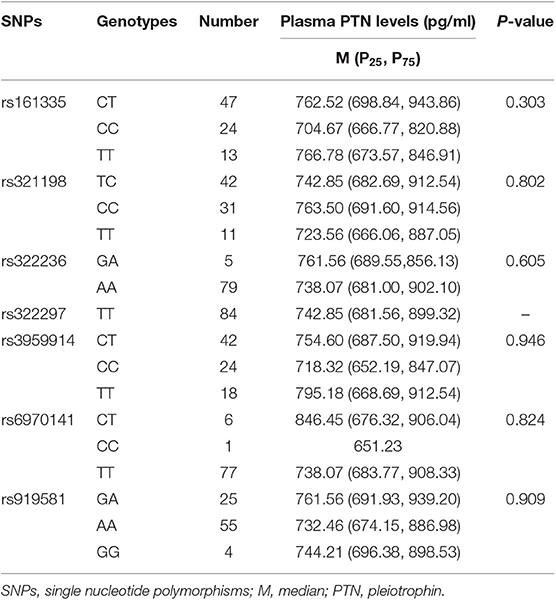
The results indicated that, in patients with SLE, there were no significant differences in plasma MK and PTN concentrations with its tag SNPs genotypes (Tables 6, 7).
Haplotype Analyses
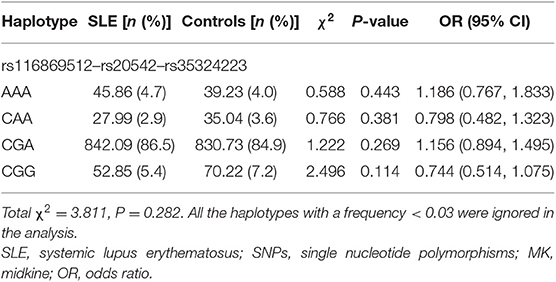
We have constructed four main haplotypes (AAA, CAA, CGA, and CGG) for MK gene and eight main haplotypes (CCATCTA, CCATTTG, CTATCTA, CTATTTA, CTATTTG, TCATCTA, TCATTTA, and TCATTTG) for PTN gene using SHEsis software. The results revealed that MK and PTN genes haplotypes were not associated with SLE susceptibility (Tables 8, 9).
Discussion
MK and PTN comprise a two-member family of heparin-binding cytokines. Previous studies have demonstrated that MK and PTN can be highly expressed in various human cancers and play a key role in the promotion of cancer cell survival, proliferation, and angiogenesis, contributing to tumor growth. MK gene promoter contains a putative nuclear factor-kappa B (NF-kB)-responsive element that can drive the induction of MK (34). In the early of 2000, Ahmed et al. has examined the entire coding region, four regions of the promoter and eight sets of intron-based and promoter region primers; they found that, in the MK promoter region, mainly a G/T polymorphism (G to T transition) at the 62 nd base of intron 3, there was a higher G/T genotype frequency in colorectal cancers, suggesting that this G/T genotype might be a risk factor contributing to the carcinoma in the colon and rectum (35, 36). However, Lai et al. reported that the genetic variation of MK gene (rs20542) was not associated with sporadic gastric cancers (21).
For PTN, interferon (IFN)-g could positively regulate and promote the expression of PTN via an IFN-g-responsive promoter element (37). Besides, hyaluronan (HA) has been shown to inhibit the toll-like receptor (TLR) 4 activity to downregulate PTN level in a Th1-type autoimmune disease model, implicating PTN as a TLR4-responsive gene (38). Mencej-Bedrac et al. has addressed the relationship of PTN gene with osteoporosis; they found that the PTN gene rs321198 polymorphisms and its CT haplotype were associated with genetic susceptibility of osteoporosis in postmenopausal women (23).
It has also been disclosed that MK plays critical roles in several types of inflammatory diseases. MK was expressed by macrophage- and fibroblast-like cells of the synovial membrane. Enhanced MK levels on both synovial fluid and sera were present in RA patients, and the increase in MK level positively correlated with RF (24). In RA patients with active inflammatory synovitis, the inflamed tissues showed an overexpression of MK, while a non-inflamed tissue had no MK expression at all (39).
We previously reported elevated plasma MK and PTN concentrations in SLE compared with healthy controls, and the increase in MK and PTN levels correlated with inflammatory cytokine IL-17 and some clinical features/parameters, including rash and anti-SSA antibody, suggesting that an aberrant expression of MK and PTN might be involved in the progress of inflammation and the course of SLE (25).
In the present study, we investigated the association of MK and PTN gene SNPs and its susceptibility to SLE in a Chinese population. A total of 10 tag SNPs of MK and PTN genes were genotyped and analyzed; however, we did not find any significant associations of 10 tag SNPs with SLE susceptibility. In addition, we also performed a case-only analysis to determine the relationship of 10 tag SNPs with the clinical features of SLE; our results showed that the SNPs in MK and PTN genes associated with some SLE clinical manifestations. In the MK gene, the frequency of AA/AG genotypes (rs35324223) was decreased in SLE patients with skin photosensitivity, suggesting that the lower AA/AG genotype of rs35324223 might correlate with decreased skin photosensitivity risk and play as a protective factor for the occurrence of skin photosensitivity. Moreover, we identified a decreased A/G (rs20542) allele frequency in patients with serositis; the decreased A/G allele frequency appeared to be protective against serositis in SLE. In the PTN gene, we found that two SNPs (rs322236 and rs6970141) associated with an increased risk for immunological disorder in SLE; the A allele of rs322236, T allele, and TT/CT genotype of rs6970141 were the risk alleles of immunological disorder, contributing to the disease clinical features. To observe the possible associations of MK and PTN SNPs with their plasma protein levels, we analyze the differences in plasma MK and PTN levels in different genotypes of each tag SNPs, but there were no correlations between plasma levels of MK and PTN levels and its tag SNPs. At last, the haplotype analysis in our study revealed no significant differences in MK and PTN gene haplotypes. Taken together, although there were no associations of the 10 tag SNPs in MK and PTN genes with SLE susceptibility, we have revealed that there are some SNPs that interact together in modulating the risk toward clinical manifestation of skin photosensitivity, serositis, and immunological disorder in SLE.
To the best of our knowledge, this is the first study to investigate the association of MK and PTN gene SNPs with SLE susceptibility. Nevertheless, several shortcomings of the present study should be acknowledged. First, only part of the patients had plasma MK and PTN measurements, which may cause the potential bias. Second, only common variants in MK and PTN genes were examined; it may reduce the effect of these SNPs in their association analyses. Furthermore, in the present study, only Chinese Han population was included; it may restrict the generalizability of our results.
Conclusions
Overall, results from the present study revealed that MK and PTN gene polymorphisms have no associations with SLE genetic susceptibility; however, we found associations of some tag SNPs with specific SLE clinical manifestations, suggesting that MK and PTN genes associated with the course of SLE. However, further studies with a larger study sample size and more ethnic lines covering the entire gene variability of both MK and PTN genes are still required to confirm our results.
Ethics Statement
This study was approved by the Ethical Committee of Anhui Medical University (Hefei, Anhui, China). All the study subjects provided informed consent to participate in this study.
Consent for Publication
We have obtained consent to publish from the participant (or legal parent or guardian for children) to report individual patient data.
Author Contributions
PW, H-FP, and D-QY participated in the design of this study, analyzed the results, and finalized the manuscript. J-BW and Y-MM carried out all the ELISA analyses. C-NZ and X-ML contributed sera of their SLE patients and assisted in analyzing the clinical data of these patients. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81573222).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the study participants as well as the staff involved in the collection of blood samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.00110/full#supplementary-material
References
1. Cervera R, Khamashta MA, Hughes GR. The Euro-lupus project: epidemiology of systemic lupus erythematosus in Europe. Lupus. (2009) 18:869–74. doi: 10.1177/0961203309106831
2. Tsokos GC. Systemic lupus erythematosus. N Engl J Med. (2011) 365:2110–21. doi: 10.1056/NEJMra1100359
3. Katsuyama T, Tsokos GC, Moulton VR. Aberrant T cell signaling and subsets in systemic lupus erythematosus. Front Immunol. (2018) 9:1088. doi: 10.3389/fimmu.2018.01088
4. Crispin JC, Hedrich CM, Tsokos GC. Gene-function studies in systemic lupus erythematosus. Nat Rev Rheumatol. (2013) 9:476–84. doi: 10.1038/nrrheum.2013.78
5. Olsson LM, Johansson AC, Gullstrand B, Jonsen A, Saevarsdottir S, Ronnblom L, et al. A single nucleotide polymorphism in the NCF1 gene leading to reduced oxidative burst is associated with systemic lupus erythematosus. Ann Rheum Dis. (2017) 76:1607–13. doi: 10.1136/annrheumdis-2017-211287
6. Wen L, Zhu C, Zhu Z, Yang C, Zheng X, Liu L, et al. Exome-wide association study identifies four novel loci for systemic lupus erythematosus in Han Chinese population. Ann Rheum Dis. (2017) 77:417. doi: 10.1136/annrheumdis-2017-211823.
7. Li J, Wu GC, Zhang TP, Yang XK, Chen SS, Li LJ, et al. Association of long non-coding RNAs expression levels and their gene polymorphisms with systemic lupus erythematosus. Sci Rep. (2017) 7:15119. doi: 10.1038/s41598-017-15156-4
8. Pacheco GV, Novelo Noh IB, Velasco Cardenas RM, Angulo Ramirez AV, Lopez Villanueva RF, Quintal Ortiz IG, et al. Expression of TLR-7, MyD88, NF-kB, and INF-α in B lymphocytes of mayan women with systemic lupus erythematosus in Mexico. Front Immunol. (2016) 7:22. doi: 10.3389/fimmu.2016.00022
9. Muramatsu T. Midkine and pleiotrophin: two related proteins involved in development, survival, inflammation, and tumorigenesis. J Biochem. (2002) 132:359–71. doi: 10.1093/oxfordjournals.jbchem.a003231
10. Muramatsu T. Midkine (MK), the product of a retinoic acid responsive gene, and pleiotrophin constitute a new protein family regulating growth and differentiation. Int J Dev Biol. (1993) 37:183–8.
11. Salaru DL, Arsenescu-Georgescu C, Chatzikyrkou C, Karagiannis J, Fischer A, Mertens PR. Midkine, a heparin-binding growth factor, and its roles in atherogenesis and inflammatory kidney diseases. Nephrol Dial Transplant. (2016) 31:1781–7. doi: 10.1093/ndt/gfw083
12. Xu C, Zhu S, Wu M, Han W, Yu Y. Functional receptors and intracellular signal pathways of midkine (MK) and pleiotrophin (PTN). Biol Pharm Bull. (2014) 37:511–20. doi: 10.1248/bpb.b13-00845
13. Kadomatsu K, Kishida S, Tsubota S. The heparin-binding growth factor midkine: the biological activities and candidate receptors. J Biochem. (2013) 153:511–21. doi: 10.1093/jb/mvt035
14. Weckbach LT, Muramatsu T, Walzog B. Midkine in inflammation. Sci World J. (2011) 11:2491–505. doi: 10.1100/2011/517152
15. Sorrelle N, Dominguez ATA, Brekken RA. From top to bottom: midkine and pleiotrophin as emerging players in immune regulation. J Leukoc Biol. (2017) 102:277–86. doi: 10.1189/jlb.3MR1116-475R
16. Hao H, Maeda Y, Fukazawa T, Yamatsuji T, Takaoka M, Bao XH, et al. Inhibition of the growth factor MDK/midkine by a novel small molecule compound to treat non-small cell lung cancer. PLoS ONE. (2013) 8:e71093. doi: 10.1371/journal.pone.0071093
17. Jee YH, Sadowski SM, Celi FS, Xi L, Raffeld M, Sacks DB, et al. Increased pleiotrophin concentrations in papillary thyroid cancer. PLoS ONE. (2016) 11:e0149383. doi: 10.1371/journal.pone.0149383
18. Meng Z, Tan J, Zhang G, Tian W, Fu Q, Li W, et al. Evaluation of serum midkine as a biomarker in differentiated thyroid cancer. Life Sci. (2015) 130:18–24. doi: 10.1016/j.lfs.2015.02.028
19. Kadomatsu K, Muramatsu T. Midkine and pleiotrophin in neural development and cancer. Cancer Lett. (2004) 204:127–43. doi: 10.1016/S0304-3835(03)00450-6
20. Ma J, Lang B, Wang X, Wang L, Dong Y, Hu H. Co-expression of midkine and pleiotrophin predicts poor survival in human glioma. J Clin Neurosci. (2014) 21:1885–90. doi: 10.1016/j.jocn.2014.02.020
21. Lai KC, Chen WC, Jeng LB, Li SY, Chou MC, Tsai FJ. Association of genetic polymorphisms of MK, IL-4, p16, p21, p53 genes, and human gastric cancer in Taiwan. Eur J Surg Oncol. (2005) 31:1135–40. doi: 10.1016/j.ejso.2005.07.005
22. Nobata S, Mogi H, Shinozawa T. Exon skipping of midkine pre-mRNA is enhanced by intronic polymorphism in a colon cancer cell line. Cancer Lett. (2004) 207:89–93. doi: 10.1016/j.canlet.2003.10.012
23. Mencej-Bedrac S, Prezelj J, Komadina R, Vindisar F, Marc J. −1227C>T polymorphism in the pleiotrophin gene promoter influences bone mineral density in postmenopausal women. Mol Genet Metab. (2011) 103:76–80. doi: 10.1016/j.ymgme.2011.01.017
24. Shindo E, Nanki T, Kusunoki N, Shikano K, Kawazoe M, Sato H, et al. The growth factor midkine may play a pathophysiological role in rheumatoid arthritis. Mod Rheumatol. (2017) 27:54–9. doi: 10.1080/14397595.2016.1179860
25. Wu GC, Yuan H, Pan HF, Ye DQ. Elevated plasma midkine and pleiotrophin levels in patients with systemic lupus erythematosus. Oncotarget. (2017) 8:40181–9. doi: 10.18632/oncotarget.13658
26. Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. (1997) 40:1725. doi: 10.1002/art.1780400928
27. Pan HF, Fang XH, Wu GC, Li WX, Zhao XF, Li XP, et al. Anti-neutrophil cytoplasmic antibodies in new-onset systemic lupus erythematosus and lupus nephritis. Inflammation. (2008) 31:260–5. doi: 10.1007/s10753-008-9073-3
28. Gladman DD, Ibanez D, Urowitz MB. Systemic lupus erythematosus disease activity index 2000. J Rheumatol. (2002) 29:288–91.
29. Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. (1992) 35:630–40. doi: 10.1002/art.1780350606
30. Pan HF, Wu GC, Fan YG, Leng RX, Peng H, Zhou M, et al. Decreased serum level of IL-21 in new-onset systemic lupus erythematosus patients. Rheumatol Int. (2013) 33:2337–42. doi: 10.1007/s00296-013-2724-1
31. Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, et al. Ensembl 2017. Nucleic Acids Res. (2017) 45:D635–42. doi: 10.1093/nar/gkw1104
32. Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. (2009) 37:W600–5. doi: 10.1093/nar/gkp290
33. Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. (2009) 19:519–23. doi: 10.1038/cr.2009.33
34. Rawnaq T, Dietrich L, Wolters-Eisfeld G, Uzunoglu FG, Vashist YK, Bachmann K, et al. The multifunctional growth factor midkine promotes proliferation and migration in pancreatic cancer. Mol Cancer Res. (2014) 12:670–80. doi: 10.1158/1541-7786.MCR-13-0467
35. Ahmed KM, Shitara Y, Kuwano H, Takenoshita S, Uchimuro K, Shinozawa T. Genetic variations of the midkine (MK) gene in human sporadic colorectal and gastric cancers. Int J Mol Med. (2000) 6:281–7. doi: 10.3892/ijmm.6.3.281
36. Ahmed KM, Shitara Y, Takenoshita S, Kuwano H, Saruhashi S, Shinozawa T. Association of an intronic polymorphism in the midkine (MK) gene with human sporadic colorectal cancer. Cancer Lett. (2002) 180:159–63. doi: 10.1016/S0304-3835(02)00040-X
37. Li F, Tian F, Wang L, Williamson IK, Sharifi BG, Shah PK. Pleiotrophin (PTN) is expressed in vascularized human atherosclerotic plaques: IFN-{gamma}/JAK/STAT1 signaling is critical for the expression of PTN in macrophages. FASEB J. (2010) 24:810–22. doi: 10.1096/fj.09-140780
38. Asari A, Kanemitsu T, Kurihara H. Oral administration of high molecular weight hyaluronan (900 kDa) controls immune system via Toll-like receptor 4 in the intestinal epithelium. J Biol Chem. (2010) 285:24751–8. doi: 10.1074/jbc.M110.104950
39. Takada T, Toriyama K, Muramatsu H, Song XJ, Torii S, Muramatsu T. Midkine, a retinoic acid-inducible heparin-binding cytokine in inflammatory responses: chemotactic activity to neutrophils and association with inflammatory synovitis. J Biochem. (1997) 122:453–8. doi: 10.1093/oxfordjournals.jbchem.a021773
Keywords: midkine, pleiotrophin, single nucleotide polymorphisms, systemic lupus erythematosus, autoimmune diseases
Citation: Wang P, Mao Y-M, Zhao C-N, Wang J-B, Li X-M, Ye D-Q and Pan H-F (2020) Association of Midkine and Pleiotrophin Gene Polymorphisms With Systemic Lupus Erythematosus Susceptibility in Chinese Han Population. Front. Immunol. 11:110. doi: 10.3389/fimmu.2020.00110
Received: 15 October 2018; Accepted: 15 January 2020;
Published: 21 February 2020.
Edited by:
Laurence Morel, University of Florida, United StatesReviewed by:
Mrinal K. Sarkar, University of Michigan, United StatesAlfred Hyoungju Kim, Washington University School of Medicine in St. Louis, United States
Copyright © 2020 Wang, Mao, Zhao, Wang, Li, Ye and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong-Qing Ye, eWRxQGFobXUuZWR1LmNu; Hai-Feng Pan, cGFuaGFpZmVuZzE5ODJAc2luYS5jb20=; cGFuaGFpZmVuZ0BhaG11LmVkdS5jbg==
†These authors have contributed equally to this work and share senior authorship
 Peng Wang1†
Peng Wang1† Hai-Feng Pan
Hai-Feng Pan