- 1Infectious Diseases Unit, I. Department of Medicine, University Medical Center Hamburg-Eppendorf, Hamburg, Germany
- 2German Center for Infection Research (DZIF), Partner Site Hamburg-Lübeck-Borstel-Riems, Hamburg, Germany
- 3Department of Transfusion Medicine, University Medical Center Hamburg-Eppendorf, Hamburg, Germany
- 4Division of Vaccine Discovery, La Jolla Institute for Immunology, La Jolla, CA, United States
- 5Division of Infectious Diseases, Department of Medicine, University of California, San Diego, La Jolla, CA, United States
- 6Protozoa Immunology, Bernhard-Nocht-Institute for Tropical Medicine, Hamburg, Germany
Background: T cells are thought to play a major role in conferring immunity against malaria. This study aimed to comprehensively define the breadth and specificity of the Plasmodium falciparum (P. falciparum)-specific CD4+ T cell response directed against the exported protein 1 (EXP1) in a cohort of patients diagnosed with acute malaria.
Methods: Peripheral blood mononuclear cells of 44 patients acutely infected with P. falciparum, and of one patient infected with P. vivax, were stimulated and cultured in vitro with an overlapping set of 31 P. falciparum-specific 13-17-mer peptides covering the entire EXP1 sequence. EXP1-specific T cell responses were analyzed by ELISPOT and intracellular cytokine staining for interferon-γ production after re-stimulation with individual peptides. For further characterization of the epitopes, in silico and in vitro human leukocyte antigen (HLA) binding studies and fine mapping assays were performed.
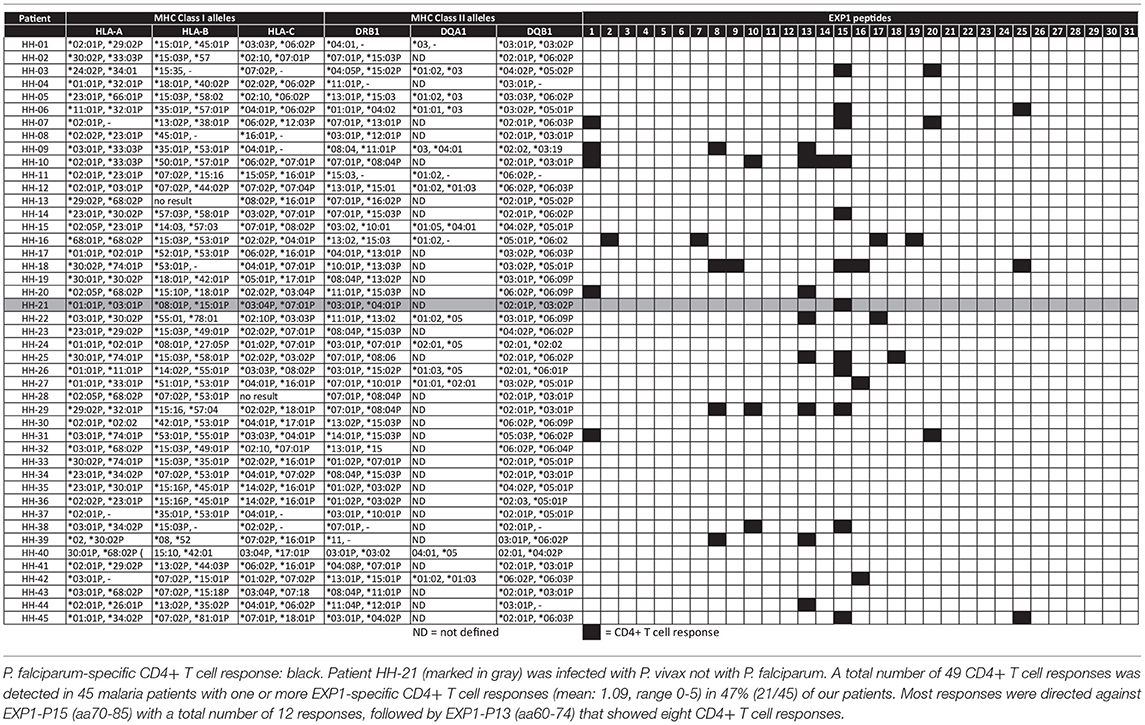
Results: We detected one or more EXP1-specific CD4+ T cell responses (mean: 1.09, range 0–5) in 47% (21/45) of our patients. Responses were directed against 15 of the 31 EXP1 peptides. Peptides EXP1-P13 (aa60-74) and P15 (aa70-85) were detected by 18% (n = 8) and 27% (n = 12) of the 45 patients screened. The optimal length, as well as the corresponding most likely HLA-restriction, of each of these two peptides was assessed. Interestingly, we also identified one CD4+ T cell response against peptide EXP1-P15 in a patient who was infected with P. vivax but not falciparum.
Conclusions: This first detailed characterization of novel EXP1-specific T cell epitopes provides important information for future analysis with major histocompatibility complex-multimer technology as well as for immunomonitoring and vaccine design.
Introduction
219 million cases of malaria and 435,000 deaths were recorded in 2017, mostly in Africa, and half of the world's population is at risk of the disease (1). Plasmodium falciparum (P. falciparum) is the species that causes the majority of deaths, and drug and insecticide resistance is increasing. A consistent solution would be the broad employment of a -thus far elusive- effective and durable protective malaria vaccine (2).
The exact mechanisms that lead to protection against malaria have not been fully defined, but both specific CD4+ and CD8+ T cells are thought to play a major role in conferring immunity against malaria (3–6). Obstacles in vaccine development are the large genome of P. falciparum, that encodes for more than 5,000 proteins (7). These malaria proteins exhibit great diversity and are partially only expressed at certain stages of the plasmodial life cycle, making them difficult targets for the immune system and vaccine design (8).
As a consequence, only relatively few P. falciparum-specific T cell epitopes located on a small number of malaria antigens have been identified and characterized in detail (9). Additionally, studies on the phenotype and function of P. falciparum-specific T cells are largely missing. Furthermore, detailed immunological studies that assess the T cell repertoire in correlation with patient characteristics [such as the origin and the human leukocyte antigen (HLA) alleles] and the clinical course of the disease [complicated vs. uncomplicated course, anemia, parasitaemia, or the C-reactive protein (CRP)] are lacking. Thus, fine mapping of the optimal length of T cell epitopes and investigation of their HLA-restriction will allow the synthesis of suitable major histocompatibility complex (MHC)-multimers for phenotypical analysis that are necessary to understand the function of T cells in malaria (10).
The 162 amino acid long exported protein 1 (EXP1) plays a pivotal role for the parasite at different stages of the life cycle. It is, for example, a component within the parasitophorous vacuole membrane (PVM) that separates and protects the parasite from the cytosol of the host cell (11). The sequence of EXP1 is well conserved across different strains of Plasmodium species (Supplementary Table 1). The expression of EXP1 during the liver and the blood stage of the plasmodial life cycle inside the human host potentially makes it an ideal target for P. falciparum-specific CD4+ and CD8+ T effector cells. CD8+ T cells are thought to play an important role during the liver stage because they are able to attack infected hepatocytes (that present peptides on MHC class I molecules) (3). CD4+ T cells are considered to mainly confer immunity during the blood stage in which erythrocytes (that do not carry MHC class I molecules) are being invaded by the parasite (12).
Former studies have shown that antibodies against EXP1 were able to inhibit parasite growth in vitro as well as in vivo (13, 14). Also, DNA vaccines containing the P. falciparum EXP1 molecule and synthetic peptides from the C-terminal region of EXP1 conferred protection in mice (15, 16).
EXP1 is expressed at two different life cycle stages where it is assumed to have important physiological functions (17), and it has shown encouraging results in previous vaccine studies. This makes EXP1 a promising target antigen for vaccine development (18–20). So far, only four CD4+ and six CD8+ EXP1-specific T cell epitopes have been described (Supplementary Figure 1) (21–25).
In this study we comprehensively defined 15 different P. falciparum-specific EXP1 CD4+ T cell epitopes using a previously established in vitro culture protocol for pathogen-specific T cells (26–29), and evaluated clinical parameters as well as patients' characteristics. We also performed fine mapping and HLA binding experiments that will allow the design of suitable MHC-multimers to characterize the phenotype of the T cell response in future studies (29).
Materials and Methods
Patient Cohort
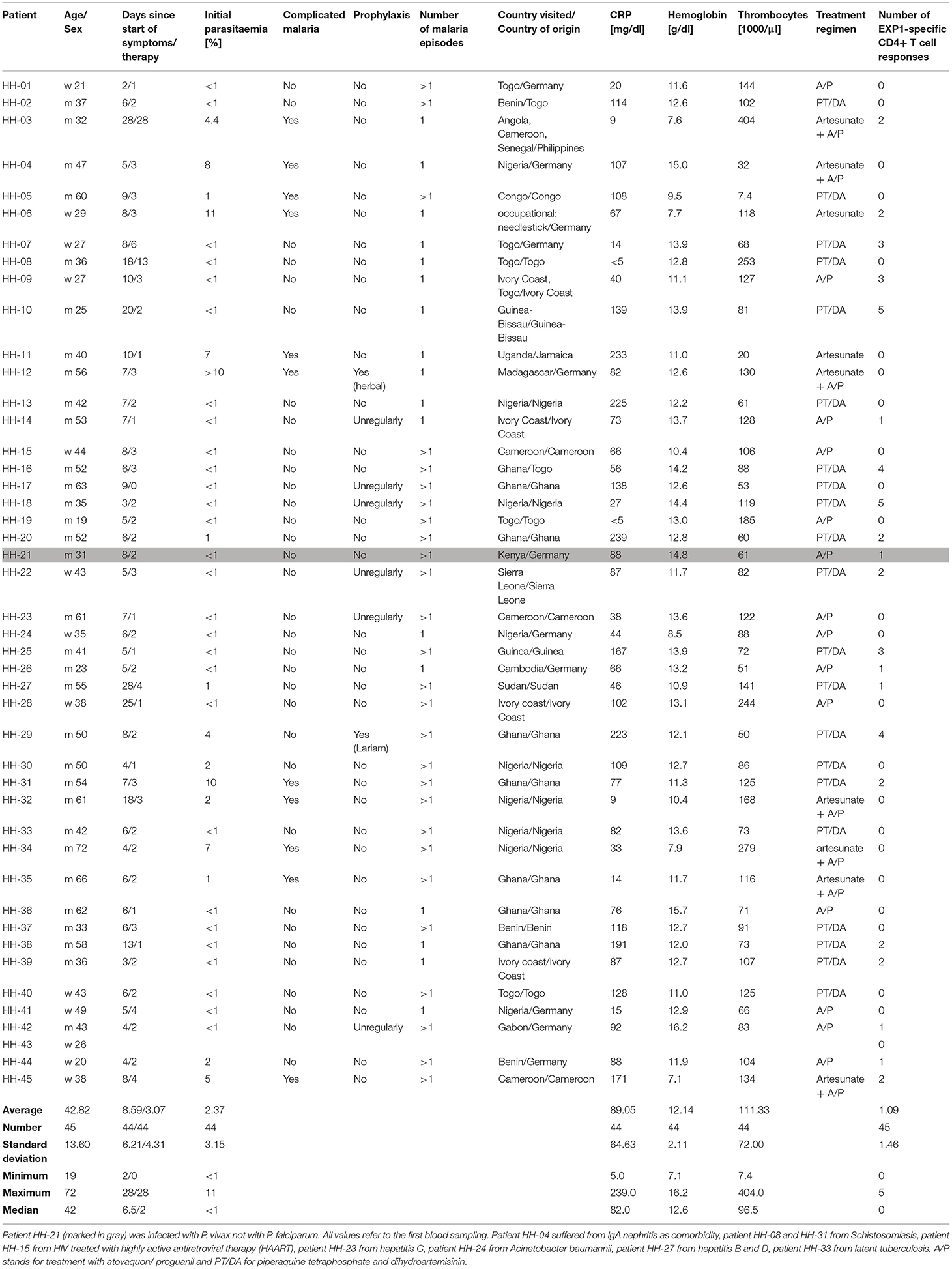
PBMCs (peripheral blood mononuclear cells) of P. falciparum infected patients (n = 44) and one patient infected with Plasmodium vivax, as well as uninfected healthy controls (n = 10) collected at University Medical Center Hamburg–Eppendorf, were either used freshly or stored in liquid nitrogen (−196°C). P. falciparum infection was verified microscopically by experienced lab technicians in the diagnostic department of the Bernhard-Nocht-Institute for Tropical Medicine, Hamburg, Germany. Thick and thin blood smears were stained with 4% Giemsa and examined under oil immersion (original magnification ×100). Patients were further sub-stratified into two groups for some of the analysis. Group 1 (n = 11) included patients originally from Germany and group 2 (n = 31) included patients who were born in Africa. Two patients came from the Philippines and Jamaica. We also compared patients with a complicated (30) (n = 11) and uncomplicated (n = 33) course of the disease and we distinguished between patients that had the first malaria infection (n = 17) and patients that have had prior malaria infections (n = 27). Other clinical parameters including parasitaemia, CRP, hemoglobin and thrombocytes were also assessed and are shown in Table 1 (clinical details for patient HH-43 are unknown).
EXP1 Peptides
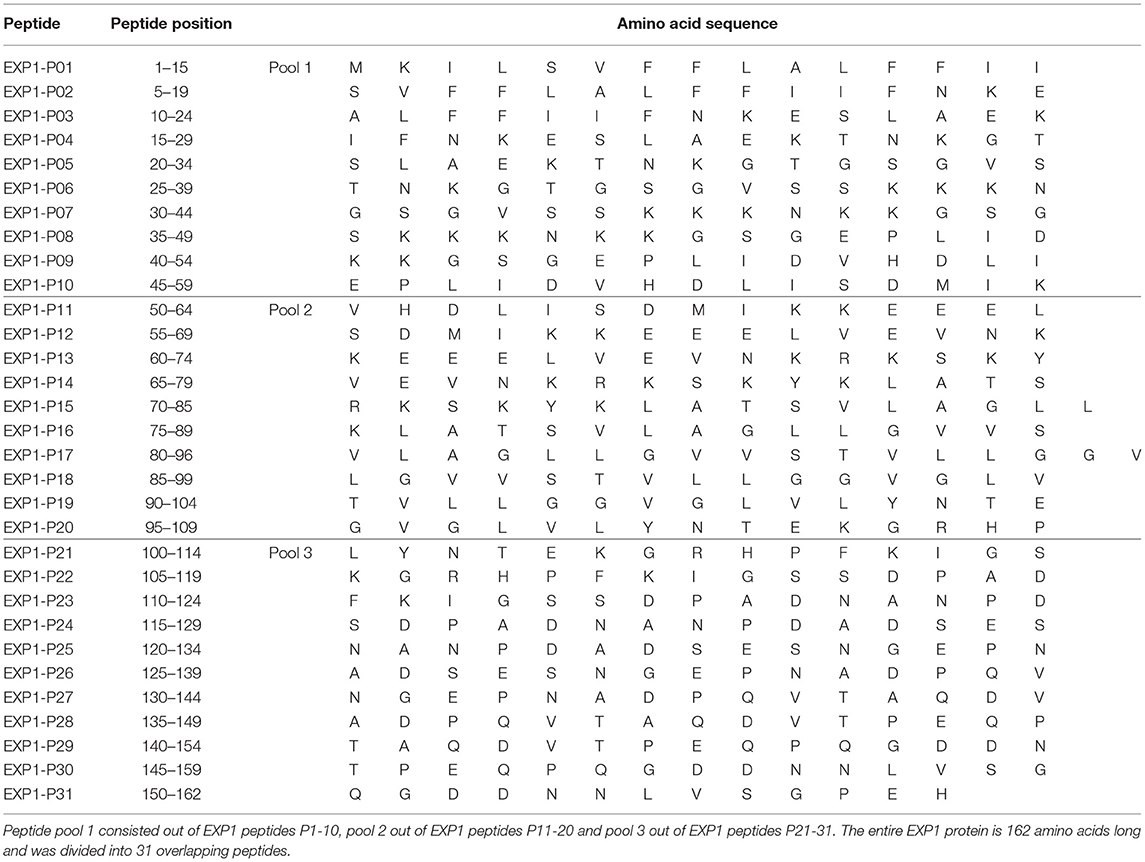
Thirty-one 13-17-mer peptides overlapping by ten amino acids corresponding to the complete amino acid sequence of the EXP1 protein [Uniprot.org; UniProt ID: Q9U590 (31)] were synthesized (peptides&elephants, Hennigsdorf, Germany), and pooled into 3 pools of 10, 10 and 11 peptides (Table 2). For in vitro culture peptide pools were used at a total concentration of 10 μg/ml [concentration of a single peptide within the pool was accordingly 1 μg/ml (1/10)]. For the enzyme linked immunospot assays (ELISPOT) the concentration of single peptides was 10 μg/ml. Further peptides for truncation experiments and variants from different strains were also synthesized. Various sequences were found on uniprot.org (Supplementary Table 1) (32). We used the terms [EXP1; exported protein 1; circumsporozoite-related protein 1; circumsporozoite-related antigen 1] for the protein name and P. falciparum [PLAFA] or P. vivax [PLAVI] as organism term.
HLA Typing
High definition molecular HLA class I and II typing was performed at the Institute of Transfusion Medicine at University Medical Center Hamburg-Eppendorf, by polymerase chain reaction-sequence specific oligonucleotide (PCR-SSO) using the commercial kit SSO LabType as previously described (One Lambda, Canoga Park, CA, USA) (33).
Bulk Stimulation of PBMCs
Cryopreserved PBMCs were thawed and cultivated at 1–5 × 106 cells/ml for optimal results in 500 μl of R10 medium (RPMI 1640 medium with 10% FCS (Sigma Aldrich), 1% HEPES buffer and 1% Penicillin-Streptomycin). PBMCs were stimulated with EXP1 peptide pool 1 [EXP1 peptide 1-10 (EXP1-P01-10)], pool 2 (EXP1-P11-20) or pool 3 (EXP1-P21-31) (Table 2) at a final concentration of 10 μg/ml, together with 1 μg/ml of anti-CD28 and anti-CD49d antibodies [BD FastImmune™, clone: L293 (CD28) and clone: L25 (CD49d)] for 14 days. Medium with recombinant IL-2 (50 U/ml) was added when necessary. After 14 days, cells were re-stimulated with single EXP1-peptides (final concentration 10 μg/ml) and then assayed for interferon-γ (IFNγ) production by ELISPOT and intracellular cytokine staining (ICS) as previously described (28).
ELISPOT Assay
ELISPOT assays were performed as previously described (26, 28, 34). We used 20,000 cells per well and responses were considered positive if the number of spots was at least three times the number of spots in the negative control. Single peptides were used at a concentration of 10 μg/ml. Anti-CD3-antibodies served as a positive control, DMSO and R10 as a negative control (35). All positive responses were confirmed by ICS assays following stimulation with the respective peptide.
Intracellular IFNγ Staining and Flow Cytometry
ICS was performed as previously described (26, 28). 5 × 105 PBMCs were stimulated with the corresponding EXP1 peptide at a final concentration of 10 μg/ml before blocking the secretion with 10 μg/ml Brefeldin A (Sigma Aldrich) 1 h after stimulation. Cells were then incubated at 37°C overnight and stained with the Zombie NIR Fixable Viability kit for live cells as well as surface antibodies anti-CD3 (clone: Okt3; AlexaFluor 700), anti-CD4 (clone: SK3; PerCP-Cy5.5) and anti-CD8 (clone: RPA-T8; Brilliant Violet 786) (all antibodies by BioLegend). After fixation and permeabilization (eBioscience™, Foxp3/Transcription Factor Staining Buffer Set), cells were stained with anti-IFNγ-antibodies (clone: 4S.B3; PE- Texas red; BioLegend). Cells were then analyzed on a BD LSRFortessa (BD Bioscience). An exemplary gating strategy is shown in Supplementary Figure 2. We defined a T cell response as positive when the percentage of CD4+ T cells within the gate for IFNγ was three times higher than the negative control and the population could be clearly separated from the negative control (26, 28, 36). DMSO and R10 were added to the negative control. Figure 1A shows an exemplary ICS result of an EXP1-specific CD4+ T cell response.
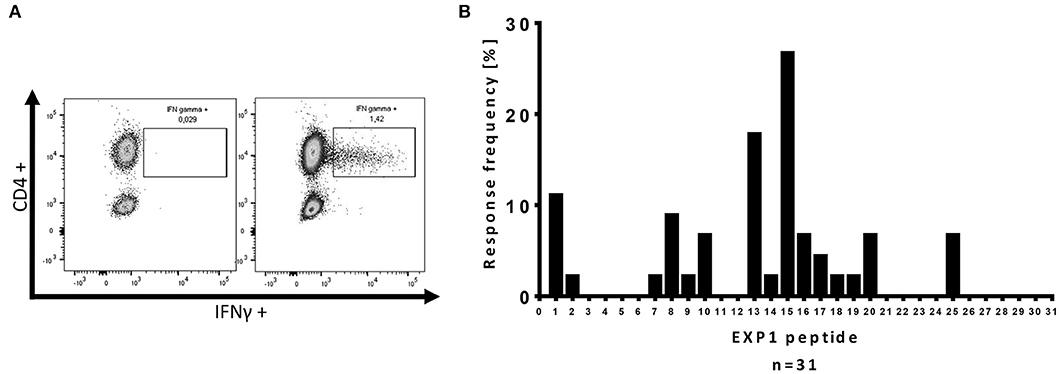
Figure 1. Intracellular cytokine staining (ICS) for IFNγ production of CD4+ T cells after re-stimulation with single EXP1 peptides. (A) Exemplary ICS dot blot of a P. falciparum-specific T cell response. Every positive response in the ELISPOT assay was confirmed by re-stimulation with a single peptide intracellular cytokine staining (ICS) assay for IFNγ production. CD4+ T cells were gated (an exemplary gating strategy is shown in Supplementary Figure 2), (left) negative control, (right) positive peptide response against EXP1-P15 by HH-03. DMSO and R10-medium were added to the negative control. (B) Overview of the relative location of the P. falciparum-specific T cell responses detected by one or more patients of the cohort against one or more of the 31 single EXP1 peptides. Response frequency (RF) is the number of patients that had a specific T cell response against a single peptide divided by the number of patients that were tested (n = 45). The center of EXP1, EXP1-P13 to EXP1-P20 (aa60-109), showed a high immunogenicity.
In vitro HLA Binding Assays and in silico Prediction
In vitro binding assays with 14 of the peptides that elicited a response were performed using purified HLA-DR molecules, as previously described (Table 3) (37). MHC class II in silico binding predictions were defined, using the IEDB analysis resource (38–40), combining predictions from the ANN (41, 42) and SMM (43) algorithms (Supplementary Table 2A).
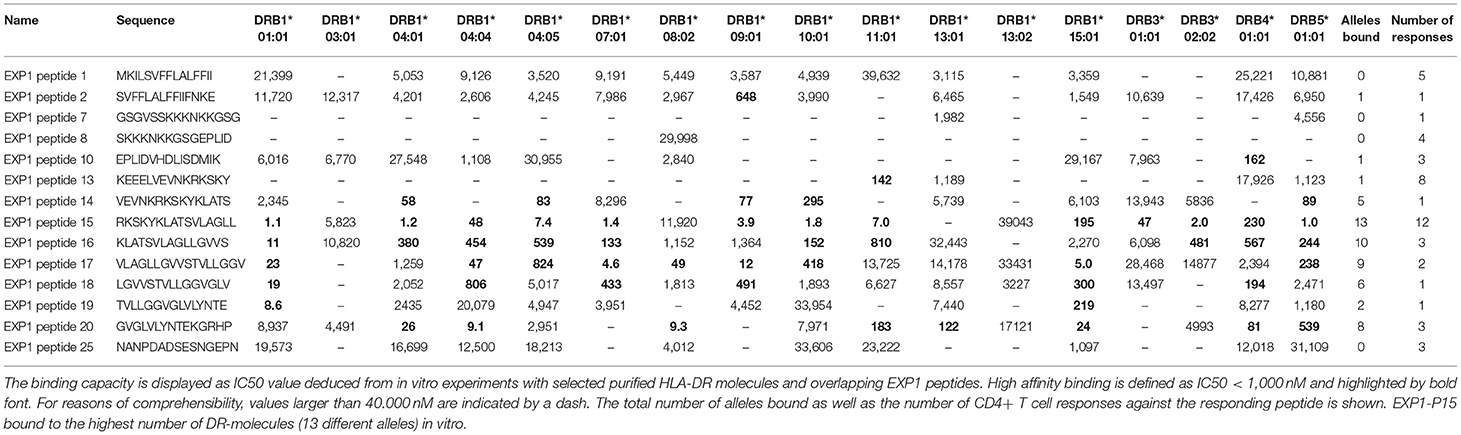
Table 3. In vitro binding predictions of 14 P. falciparum-specific peptides to 17 frequent DRB1, DRB3, DRB4, and DRB5 types.
Statistical Analysis
All flow cytometric data were analyzed using FlowJo 10.5.0 software (Treestar, Ashland, OR, USA). Statistical analyses were carried out using the Prism 7.0 software (GraphPad software, San Diego, CA). Mann Whitney test or the Kruskal–Wallis test with Dunn's post-test was performed throughout all samples for inter group comparisons. Spearman's correlation was performed for bivariate correlation analyses. Data are expressed as means with standard deviations (SD) or with standard error of mean (SEM). P-values less than or equal to 0.05 were considered significant.
Results
Clinical Features of the Study Cohort
The clinical data of all patients as well as other characteristics are summarized in Table 1. The clinical cohort consisted of 45 malaria patients of which 44 patients were infected with P. falciparum; one patient was infected with P. vivax (marked in gray). 13 patients were female (29%) and 32 were male (71%), and the average age was 42.8 years (range: 19–72 years). Eleven patients (24.4%) were born in Germany, 31 patients (68.9%) originated from the African continent, two patients were from the Philippines and Jamaica. The vast majority of patients who were treated for malaria at University Medical Center Hamburg-Eppendorf had traveled and returned from Western Africa: nine patients had traveled to Nigeria, eight patients to Ghana and five patients to Togo (Supplementary Figure 3). Patient HH-21 who was infected by P. vivax had traveled to Kenya, where growing evidence of P. vivax infection could be shown (44). P. falciparum infection was excluded by PCR and the patient had no medical record of a previous P. falciparum infection. Only one patient had traveled outside of Africa (to Cambodia). Eleven patients had a course of disease defined as complicated, while 34 courses of the disease were defined as uncomplicated. Further clinical parameters are shown in Table 1.
Initial parasitaemia was on average 2.37% (range: <1–11%), CRP 89 mg/dl (range: 5–239 mg/dl), hemoglobin 12.1 g/dl (range: 7.1–16.2 g/dl), and thrombocytes 111.300 /μl (range: 7,400–404,000 /μl). The average number of days between the start of symptoms and the blood sampling for our study was ~8.6 (range: 2–28 days). The number of days between the start of therapy and the blood sampling for our study was about 3.1 (range: 0–28 days). Patient HH-17 had a blood draw before therapy for malaria was initiated.
Detection of EXP1-Specific T Cell Responses After in vitro Cultivation With IL-2
The P. falciparum-specific T cell response has so far not been comprehensively assessed for neither the majority of P. falciparum-specific antigens in general, nor for EXP1 in particular (9). In order to evaluate the EXP1-specific T cell response on an epitope level, we first looked at the ex vivo IFNγ production of PBMCs of acutely infected malaria patients after stimulation with single peptides of an overlapping 13-17-mer peptide set covering the entire P. falciparum antigen EXP1. Most samples were drawn 1-4 days after admission to the hospital (average 3.07 days) shortly before, during or after initiation of anti-malaria therapy, only 3 samples were taken at later timepoints.
However, we detected only one T cell response with the ex vivo ELISPOT in a total of ten patients (data not shown). The fresh blood sample of patient HH-38 showed an ex vivo response against EXP1-P15. After the in vitro culture, the same patient responded again to EXP1-P15 and additionally to EXP1-P10. These results are in line with the results of previous studies that showed that the magnitude of the ex vivo P. falciparum-specific CD4+ and CD8+ T cell response is low (23, 45–47). No EXP1-specific T cell responses were detectable after 14 days of in vitro culture of 10 healthy controls that had never traveled to a malaria endemic area (data not shown).
Due to the presumably low ex vivo precursor frequencies of P. falciparum-specific T cells, we employed a previously used, ultra-sensitive, in vitro approach (26–28, 48). In short: our peptide set was divided into three different peptide pools (Table 2) and we started three parallel cell cultures per patient, stimulating them with each pool for 14 days (Pool 1: EXP1-P1-10, Pool 2: EXP1-P11-20, Pool 3: EXP1-P21-31). This was followed by a single peptide ELISPOT for IFNγ-production against re-stimulation with the individual peptides. The 31 overlapping single peptides covered the entire EXP1 protein, and PBMCs from 45 patients were tested. Every positive response in the ELISPOT assay was confirmed by re-stimulation with the according single peptide and subsequent intracellular cytokine staining (ICS) assay for IFNγ production. Figure 1A shows an exemplary ICS result of an EXP1-specific CD4+ T cell response.
With this approach, we were able to distinguish one or more responses directed against 15 of the 31 overlapping EXP1 peptide specificities (48.4%). We detected EXP1-specific CD4+ T cell responses in nearly half of our patients (21/45, 47%) but, surprisingly, only four patients showed a CD8+ T cell response (4/45, 8.9%) (Supplementary Figure 4B). Patients recognized an average of 1.09 CD4+ restricted peptides (range 0–5), with a total number of 49 CD4+ T cell responses in 45 patients. The average frequency of IFNγ+CD4+ T cells was 1.12% (range: 0.2–4.84%) (data not shown). Figure 1B gives an overview of the distribution of the different peptide-specific T cell responses. Most responses were directed against epitopes that are located in the center of the EXP1 protein. Eight patients (response frequency (RF): 17.8%; 8/45) showed a CD4+ T cell response against EXP1-P13 (aa60-74), and 12 patients responded to EXP1-P15 (aa70-85) (RF: 26.7%, 12/45). Interestingly, the patient who was infected with P. vivax also responded to EXP1-P15. The sequences of P. falciparum EXP1-P15 (RKSKYKLATSVLAGLL) and P. vivax EXP1 aa68-82 (KKSNYKLATTVLASAL) were then compared and show a high degree of sequence homology (Supplementary Table 1).
We also longitudinally analyzed, in selected patients, the EXP1-specific T cell responses in the long-term follow-up. The patient HH-06 showed a large response against EXP1-P15 (frequency: 1.86% IFNγ+CD4+) and EXP1-P25 (frequency: 1.88% IFNγ+CD4+) during acute infection. However, 12 months after the first sampling a specific T cell response could not be detected from a cell culture started directly with fresh blood (data not shown). The patient had stayed in Germany and no further malaria infections had occurred in the intervening 12 months. By contrast, in patient HH-27, who responded to EXP1-P16 (frequency: 3.91% IFNγ+CD4+), we could detect responses against EXP1-P16 (frequency: 9.73% IFNγ+CD4+) and EXP1-P10 (frequency: 0.25% IFNγ+CD4+) in a sample collected 7 years prior to the present assays. During these 7 years, several subsequent malaria infections had occurred as reported by the patient and patient's health provider.
Of the 45 patient samples utilized, seven samples were cultured from fresh samples, of which three patients showed a CD4+ T cell response (42.9%, 3/7). The other 38 samples were stored in liquid nitrogen before analysis, and of these 18 patients showed a T cell response (47.4%, 18/38). Thus, there were no significant differences in the breadth of the detected responses as a function of sample storage conditions (data not shown).
The Breadth of the EXP1-Specific CD4+ T Cell Response Does Not Correlate With the Clinical Course of Acute Malaria
Several clinical parameters, including parasitaemia, CRP, hemoglobin, and thrombocyte count, were examined, but no significant correlation between the number of T cell responses and any of the parameters could be identified (Figure 2). Similarly, no correlation was found between age or gender and the CD4+ T cell response (Supplementary Figure 5). The patients' origin, the duration of the malaria therapy and the clinical course of the disease (uncomplicated and complicated) did also not correlate with the breadth of the CD4+ T cell response (Supplementary Figures 6A–C). When comparing the number of in vitro responses between different patient groups, we could not see significant differences between patients with their first episode of diagnosed malaria infection (52.9%, 9/17) vs. patients who had a documented history of a past malaria infection (44.4%, 12/27) (Supplementary Figure 6D).
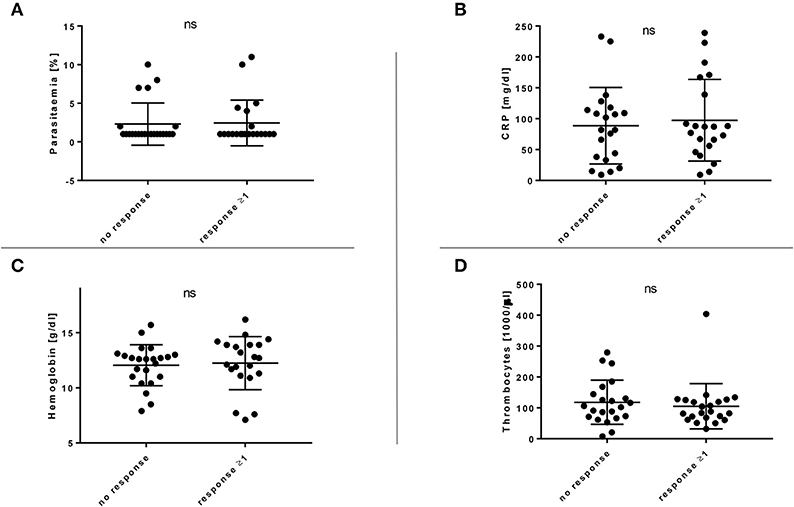
Figure 2. (A–D) Breadth of the EXP1-specific T cell response in comparison with relevant clinical parameters. Clinical parameters, including parasitaemia, CRP, hemoglobin and thrombocytes were examined but no significant (ns) correlation with the CD4+ T cell response could be shown. (A) Response or no response compared to parasitaemia [%]. (B) Response or no response compared to C-reactive protein (CRP) [mg/dl]. (C) Response or no response compared to hemoglobin [g/dl]. (D) Response or no response compared to thrombocytes [1,000/μl].
HLA Binding and Restriction Experiments
Table 4 lists the 45 donors examined in the present study along with their HLA-DRB1* typing data and summarizes the individual patient-specific EXP1-peptide response patterns. The finding that certain peptides were recognized by a large proportion of subjects with acute P. falciparum infection, despite the fact that they expressed different HLA class II molecules, let us hypothesize that these were comparatively promiscuous MHC class II binding EXP1-specific CD4+ T cell epitopes (i.e., peptides recognized in the context of multiple HLA class II molecules).
To test this hypothesis, we measured the in vitro capacity of the most frequently recognized EXP1 peptides to bind to a panel of 17 of the most prevalent human HLA-DR molecules, since the majority of HLA class II restricted CD4+ T cell responses are presented by these molecules (49, 50). Peptides with an IC50 of 1,000 nM or lower, a threshold previously found associated with the vast majority of HLA class II-restricted T cell epitopes, were considered binders to the respective DR molecule (51–53). As shown in Table 3, in general the peptides located in the middle of the protein (i.e., EXP1-P14-18; aa65-99) were associated with the most promiscuous binding (range: 5-13 of the 17 DR tested). These include EXP1-P14 which bound to five, EXP1-P15 to 13, EXP1-P16 to 10 and EXP1-P17 to nine different HLA molecules (Table 3).
The most frequently recognized peptides were promiscuous in binding to multiple HLA-DR molecules, with each of the peptides binding between zero and 13 different DR molecules. Furthermore, some of these peptides were able to bind to several HLA-DR molecules with extremely high affinities (IC50 <10 nM) (Table 3). EXP1-P15, for example, showed excellent binding to 13 HLA molecules: DRB1*01:01 (1.1 nM), DRB1*04:01 (1.2 nM), DRB1*04:04 (48 nM), DRB1*04:05 (7.4 nM), DRB1*07:01 (1.4 nM), DRB1*09:01 (3.9 nM), DRB1*10:01 (1.8 nM), DRB1*11:01 (7.0 nM), DRB1*15:01 (195 nM), DRB3*01:01 (47 nM), DRB3*02:02 (2.0 nM), DRB4*01:01 (230 nM) and DRB5*01:01 (1.0 nM). This binding pattern is in line with the HLA molecules of the patients that responded to EXP1-P15. Peptides that bound to zero alleles in our DRB in vitro assay may be restricted by DQ or DP molecules.
Supplementary Table 2B shows the most likely HLA-restriction considering the in vitro binding data as well as the responders' HLA molecules. Nine out of 12 patients who recognized EXP1-P15 expressed at least one of the HLA molecules mentioned above. The most common HLA molecule expressed in patients of our cohort who recognized EXP1-P15 was DRB1*07:01 (six patients). Interestingly, six patients from our cohort also expressed DRB1*07:01 but did not recognize EXP1-P15. EXP1-P13 bound to DRB1*11:01 in the in vitro assay (142 nM). This is consistent with the fact that out of eight patients who recognized EXP1-P13, five were carrying the HLA-DRB1*11 molecule.
These results indicate that certain EXP1 peptides, especially those that are located in the center of the protein like EXP1-P15 are potentially promiscuous T cell epitopes.
Fine Mapping of EXP1-P13 and P15
To narrow down the minimal length of the epitopes within EXP1-P13 and P15, we tested the magnitude of the frequency of responding CD4+ T cells by the use of truncations (Figure 3). We first cultivated PBMCs of patients who had shown a strong response against the corresponding peptide with the original peptide (concentration 10 μg/ml) and then tested the magnitude of the IFNγ responses by ICS. The highest CD4+ T cell frequency for EXP1-P13 was reached by patient HH-09 by EXP1-P13 truncation 1 (13-mer truncation: EELVEVNKRKSKY). For EXP1-P15 the 14-mer EXP1-P15 truncation 1 (SKYKLATSVLAGLL) triggered the biggest response by patient HH-03 and in vitro binding data as well as preliminary restriction experiments suggest that this epitope was likely restricted by DRB1*04 (Supplementary Figure 7) in this patient. However, restriction by other molecules like DP and DQ molecules were not tested and cannot be excluded as alternative but less likely restricting molecules.

Figure 3. (A) Fine-mapping of malaria epitope EXP1-P13 (HH-09). (B) Fine-mapping of EXP1-P15 (HH-03). (C) Testing of variants for EXP1-P20 (HH-03). Experiments were performed by ICS for IFNγ production after stimulation with peptides of different length and with varying sequences. (A) For EXP1-P13 we used 15-, 13-, and 11-mer N- as well as C- terminal truncations with PBMCs of patient HH-09 (DRB1*08:04, *11:01P). Truncation 1 showed the highest CD4+ T cell response. The core epitope of EXP1-P13 is marked in black. (B) 16-, 14-, and 12-mer N- as well as C- terminal truncations were tested for EXP1-P15. PBMCs of patient HH-03 (DRB1*04:05P,*15:02P) were used for this experiment. EXP1-P15 Truncation 1 showed the highest CD4+ T cell response as well as EXP1-P15 Variant 2 which contains Serine (S) instead of Alanine (A). For EXP1-P15 and P20 variants were tested. Variant 1 of EXP1-P20 contained the amino acid Arginine (R) instead of Histidine (H) and Variant 2 Arginine (R) instead of Lysine (K).
For both EXP1-P15 and EXP1-P20 we also tested variants found on UniProt (32). EXP1-P15 variant 2 contained serine (S) instead of alanine (A) at position 13 and showed the biggest response. EXP1-P20 variant 1 contained arginine (R) instead of histidine (H) at position 14 and EXP1-P20 variant 2 contained arginine (R) instead of lysine (K) at position 11. EXP1-P20 variant 2 triggered a much bigger response than variant 1. These results show that the change of a single amino acid can lead to an improvement of binding and therefore recognition and are in line with prior publications (54). No variants for EXP1-P13 could be found in online databases (Supplementary Table 1).
Discussion
Here, we present an immunological study of the P. falciparum EXP1-specific T cell response on a single epitope level using PBMCs of a large and well-characterized patient cohort. In concordance with the results of previous studies, frequencies of ex vivo EXP1-specific T cell responses were low. However, with our sensitive in vitro testing approach we detected T cell responses in 21 of the 45 patients, and discovered responses directed against 15 of 31 peptides (48.4%) spanning the entire EXP1 protein, making this the first study to comprehensively describe responses against a broad range of EXP1-specific epitopes. For most P. falciparum antigens previous studies only studied vaccine-primed T cell responses in malaria-naïve volunteers within human vaccine trials (25, 55) which cannot be directly compared with P. falciparum T cell responses generated during a natural malaria infection. Other studies used small and partly HLA-A2 pre-selected study populations (54). HLA-A2 is the most common expressed class I HLA in Caucasians, but not as commonly expressed in African individuals. As a result, these previous studies have limited applicability for vaccine design and immunomonitoring in malaria endemic countries. Other studies used in silico predicted epitopes or peptide pools instead of comprehensive, overlapping peptide sets (24, 56, 57). However, this approach does not allow the identification of single epitopes and immunogenic parts of antigens. Published P. falciparum specific CD4+ and CD8+ T cell epitopes as well as HLA-restriction (if known) and newly detected CD4+ T cell epitopes from this study are shown in Supplementary Figure 1. We also assessed important clinical parameters such as the HLA type, origin and parasitaemia in our study cohort, but no significant correlation with the CD4+ T cell response could be shown.
Of note, EXP1-P15 (aa70-85) and EXP1-P13 (aa60-74) elicited a CD4+ T cell response in a quarter of our study group that had a diverse HLA-background. The minimal length was experimentally narrowed down in two patients and the most likely restricting HLA-molecule of each epitope was evaluated using our in vitro HLA class II binding data (Table 3), in silico predictions (Supplementary Table 2A) and comparing it with the HLA molecules of responding patients (Supplementary Table 2B). EXP1-P15 is likely to be restricted by HLA-DRB1*01:01, DRB1*04:01, DRB1*04:02, DRB1*04:05, DRB1*07:01, DRB1*10:01, DRB1*13:03, DRB1*15:02, and DRB1*15:03. Interestingly, six patients in our cohort also expressed DRB1*07:01 but did not respond to EXP1-P15. Heterology of the protein sequence of the autologous infecting strain, or slightly differing immunogenic epitope cores between different patients with the same HLA molecules (29) could explain, why some patients were able to recognize a certain epitope unlike others even though they express the same HLA. In this study we have only tested the binding to DRB-molecules, since they restrict the majority of class II responses (~50%), but acknowledge that restrictions by HLA-DQ and DP molecules are also possible (58). One study examining the HLA-DP, DQ, and DRB3/4/5 restricted responses showed that Dengue virus-specific CD4+ T cell responses were of lower magnitude compared to HLA-DRB1 (58, 59). This is consistent with their lower levels of expression (58). Future studies should experimentally confirm the HLA-restriction of these EXP1 T cell epitopes and possibly evaluate restriction by HLA-DP or DQ molecules.
Further detailed in vitro HLA-restriction and peptide truncation experiments to establish optimal peptide length and HLA-restriction (i.e., with single HLA-molecule transfected cell lines or by blockade with anti-DR, and-DQ or anti-DP antibodies) of peptide specificities defined in this study need to be conducted in future follow-up studies.
We still might underestimate the breadth of the P. falciparum EXP1-specific T cell response. In this study design we aimed to expand and define dominant peptide-specific T cell responses producing IFNγ to define the breadth of the CD4+ T cell repertoire after short term cell cultures and using exogenous IL-2 which is known to create a type 1 T helper (Th1) cell cytokine shift (60) and no statement can be made about the original ex vivo phenotype or functionality of these cells. Future studies should therefore further characterize T cell responses by directly assessing ex vivo secretion (e.g., with ELISPOT) of different, additional cytokines (e.g., IL-4, IL-10) that potentially could have been missed using the current approach.
Further subdominant responses might be detectable using peptide sets of peptide variants matching autologous sequences or by applying more sensitive, but far more labor-intensive techniques, like single cell dilution cloning. Patient HH-21 was infected with P. vivax and not with P. falciparum but the EXP1-specific cultures nevertheless elicited a CD4+ T cell response against EXP1-P15 indicating that immunity against malaria can indeed be cross-species specific. Of note, the sequence of EXP1-P15 (aa70-85) is well-conserved (Supplementary Table 1), potentially making this epitope an interesting candidate for immunomonitoring by MHC-multimer technology or an epitope to be used for a suitable subunit vaccine.
Also, our analysis is limited to samples drawn directly after hospital admission during acute malaria and it is not known how long each patient had been infected. Of note, if we stratify our data according to patients with and without a response, surprisingly the time after malaria treatment did not significantly differ between either group at this early phase of infection (Supplementary Figure 6B). The single cases for which we were able to analyze the T cell response at later time points would let us hypothesize that the naturally primed EXP1 specific T cell response rapidly wanes over the weeks after successful treatment.
Although EXP1 is likely to be essential for the development and the survival of the parasite, its exact function in the P. falciparum malaria life cycle is far from clear and more investigation as well as linkage between important and conserved regions and immunopotent epitopes are necessary (61). Since EXP1 is expressed at the liver stage during a malaria infection we also expected to detect CD8+ T cell responses. However, all CD8+ T cell responses detected in this study were weak and coincided with a parallel CD4+ T cell response directed against the same peptide in the same patient and we did not follow-up on fine-mapping these subdominant T cell responses (Supplementary Figure 4). It has to be considered that the design of our overlapping peptide set, with an average length of 15 amino acids, is biased toward detection of CD4+ T cell responses and could therefore explain low CD8+ T cell responses. Furthermore, prior publications showed that the blood phase of a malaria infection suppresses immune cells of the liver stage (62, 63). This could also explain the lack of CD8+ T cell responses in our cohort since all patients had an extensive blood phase with strong symptoms and the need to be hospitalized. It has been discussed that CD4+ T cells might play an important role in priming other immune cells (i.e., NK cells) and in evoking a strong antibody response which is necessary for immune control (64, 65). If an efficient induction of immunity against plasmodial infection by CD8+ T cells depends on CD4+ T cells, is therefore an important question to address in the development of an effective vaccine.
EXP1 is also called circumsporozoite-related antigen due to a similarity between the amino acid sequence of EXP1 and the circumsporozoite protein (CSP). The CSP is by far the most investigated malaria antigen and it is also part of the RTS,S vaccine trial (66). The amino acid sequence NANPDADSESNGEPN of EXP1 is similar to the NANP-repeat region of the CSP which has so far only shown few T cell responses (67, 68). Interestingly, we detected T cell responses against EXP1-P25 which is located in this NANP-related region of EXP1 and future studies are necessary to evaluate if not only CD4+ T cell and antibody responses against the NANP-repeat region but also CD8+ T cell responses can be found. The cross reactivity of vaccine-induced CSP and EXP1-primed T cell responses should be tested in future studies.
The RTS,S vaccine only showed short lived efficacy and also naturally acquired immunity only lasts a few months (69). Epidemiological evidence described that antibodies to Plasmodium antigens are inefficiently generated and rapidly lost without continued parasite exposure (70). We also investigated longitudinal follow-up samples of a subset of patients and could show that the EXP1-specific T cell response primed by natural infection waned over time and was not detectable 12 months after the diagnosis and therapy of acute malaria (data not shown). Unfortunately, it was difficult in the present study to collect samples at later timepoints after the patients were discharged. And future, well-structured and prospective longitudinal studies that describe the dynamics of the priming and the breadth, magnitude and quality of the T cell response directed against different malaria antigens are necessary.
This current study aimed to primarily discover novel P. falciparum-specific epitopes with a frequency high enough to be detectable e.g., after enrichment by MHC class II multimer technology. This technology will allow the characterization of the phenotype of T cells which could help to understand the role but also the complications of T cells during malaria infections. In previous studies, EXP1 has shown to generate a strong antibody response in naturally exposed individuals and high antibody titers specific to EXP1 aa73-162 correlated with a high level of IL-6 production to the same peptide (20). This strong recognition by B and T cells suggest that the whole sequence of EXP1 may represent a suitable malaria antigen for a subunit vaccine construct (20).
In summary, a broad range of P. falciparum EXP1-specific CD4+ T cell responses can be detected after in vitro expansion in nearly half of our patients of different origin and within a diverse HLA molecule background. We did not find any correlation of the number of T cell responses and relevant clinical parameters in this cohort. These detailed data on P. falciparum EXP1-specific T cell epitopes will be helpful for the development of tools like MHC class II multimers or to monitor the immune response on an epitope level during future malaria vaccine trials.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
All subjects gave written informed consent in accordance with the Declaration of Helsinki and the study was approved by the local ethics board of the Ärztekammer Hamburg (PV 4238).
Author Contributions
JH, TJ, and JSzW: conception. JH and JSzW: first draft. MM performed the HLA-typing. JS: the in vitro binding restrictions. All authors: important contributions and proofreading.
Funding
This project has been funded by the Deutsche Forschungsgemeinschaft SFB841 (TJ und JSzW) und SFB1328 (JSzW), Deutsches Zentrum für Infektionsforschung DZIF (JH, TJ, and JSzW), and NIH R21Al134127 (AS).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all patients who participated in this study. We also thank Jonas Arand, Tom Diedrich, and Maria Sophia Mackroth for helping with the recruitment of the patients, Silke Kummer and Robin Woost for technical assistance, Sophia Schulte for performing further multi-cytokine ICS assays and Georg Lauer for his critical advice and discussion during the revision process.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.03037/full#supplementary-material
Supplementary Table 1. 13 different sequences of P. falciparum EXP1 and two sequences of P. vivax EXP1. We found 13 different sequences for P. falciparum EXP1 in the online database UniProt (31). The sequence of P. vivax EXP1 that resembles P. falciparum EXP1-P15 is framed. Some sequences are only fragments and therefore shorter than 162 amino acids.
Supplementary Table 2. In silico (A) and most likely (B) binding predictions for EXP1 peptides that elicited a CD4+ T cell response. (A) In silico binding predictions for HLA-DRB1 molecules. In silico binding predictions were performed as described for 15 EXP1 peptides that at least one patient's CD4+ T cells responded to. The threshold for the binding affinity was defined as 5,0; the most probable binders are displayed in bold. The MHC class II in silico binding predictions were made using the IEDB analysis resource consensus tool (38–40) combining predictions from ANN (41, 42) and SMM (43). (B) Most likely HLA-restrictions of the EXP1 peptides that elicited a response as determined by in silico and in vitro predictions matching the patients' HLA molecules.
Supplementary Figure 1. Epitope map of EXP1. EXP1 is a 162 amino acid long protein consisting of a signal sequence (aa1–23), an N-terminus (aa23–79), a transmembrane domain (aa79–101), and a C-terminus (aa101–162) (71). Already published CD4+ and CD8+ T cell epitopes as well as HLA-restriction (if known) and newly detected CD4+ T cell epitopes from this study are marked within this epitope map. Marked in light gray: Published CD8+ T cell epitopes: KILSVFFLA (23, 25), ALFFIIFNK (23), ATSVLAGL (21), VLAGLLGNV (23), GLLGNVSTV (23, 25), VLLGGVGLVL (9, 23). Marked in gray: Published CD4+ T cell epitopes: KSKYKLATSVLAGLL (22, 25), YKLATSVLAGLLGVVSTVLLGG (24, 25), AGLLG(V/N)VSTVLLGGV (25), GLVLYNTEKGRHPFKIGSSD (9, 24). Marked in dark gray: CD4+ T cell epitopes that elicited a CD4+ T cell response in the current study.
Supplementary Figure 2. Exemplary gating strategy. Dead cells, CD14+ cells and CD19+ cells were excluded. After gating on single cells and lymphocytes, CD3+ cells were selected. Subsequent IFNγ response was based on either CD8+ or CD4+ T cells.
Supplementary Figure 3. Countries that were visited by 41 malaria patients are marked by a black dot. Most patients who were treated for malaria at University Hospital of Hamburg had traveled and returned from Western Africa: nine patients had traveled to Nigeria, eight to Ghana, and five patients to Togo.
Supplementary Figure 4. Exemplary ICS dot blot of a P. falciparum-specific CD8+ T cell response of HH-45 against EXP1-P15. (A) CD8+ T cells are gated with a negative control (left) and a positive peptide response (right) against EXP1-P15 (aa70-85) by HH-45. DMSO and R10-medium were added to the negative control. (B) P. falciparum-specific CD4+ T cell response: black, P. falciparum-specific CD8+ T cell response: gray. Patient HH-16, HH-18, HH-20 and HH-25 showed a CD8+ T cell response that coincided with a CD4+ T cell response against EXP1-P02, P07, P09, P13, and P15. Interestingly, we did not detect a CD8+ response against a peptide without a CD4+ response against the same peptide.
Supplementary Figure 5. Breadth of the P. falciparum-specific T cell response in correlation with age [years] and sex. (A) No CD4+ or CD4+ T cell response compared to age. (B) No CD4+ or CD4+ T cell response compared to sex. No correlation with the CD4+ T cell response compared with age or sex could be shown.
Supplementary Figure 6. Breadth of the P. falciparum-specific T cell response in correlation with patient characteristics. (A) Number of P. falciparum-specific T cell responses detected in patients from Germany and in patients from the African continent. (B) Correlation of the P. falciparum-specific T cell response with the days since the start of the malaria therapy. (C) Correlation of the P. falciparum-specific T cell response compared to a complicated or uncomplicated course of the malaria infection. (D) Number of P. falciparum-specific T cell responses detected in patients with the first malaria infection and patients with prior malaria infection.
Supplementary Figure 7. Restriction assay for EXP1-P15 with PBMCs of patient HH-03 (DRB1*04:05P,*15:02P, DQB1*04:02P, *05:02P). This figure shows the results of the restriction assay for EXP1-P15. PBMCs of patient HH-03 (DRB1*04:05P,*15:02P, DQB1*04:02P, *05:02P) were cultivated for 14 days with EXP1-P15 (concentration 10 μg/ml). We then used PBMCs of matched donors as antigen presenting cells (APCs). These APCs were incubated with EXP1-P15 (concentration 10 μg/ml) for 20 minutes, then washed six times and used to stimulate the PBMCs of patient HH-03. Donor 1 (DRB1*15:02, *11:01; DQB1*03:01P, *06:01P) matched the HLA molecule HLA-DRB1*15:02 of patient HH-03 and Donor 2 (DRB1*04:01P,*04:04P; DQB1*03:02P) the HLA allele HLA-DRB1*04. The results show that HLA-DRB1*04 most likely presents EXP1-P15. As negative control PBMCs of Donor 1 and 2 that were not incubated with EXP1-P15 were used to stimulate PBMCs of patient HH-03. As positive control we used PBMCs of patient HH-03 incubated with EXP1-P15 for 20 min and washed for six times. Donor 1 and 2 expressed different HLA-DQB1 molecules than patient HH-03 and we could therefore exclude the possibility of presentation by DQB1. Unfortunately, HLA-DQA1 alleles were not assessed for Donor 1 and 2 and presentation by these molecules could therefore not be excluded and could explain, why the positive control (frequency: 9.51%) showed a higher frequency than the APCs of Donor 2 (frequency: 2.35%).
References
1. World Health Organization. World Malaria Report 2018. (2018). Available at: www.who.int/malaria
2. Cockburn IA, Seder RA. Malaria prevention: from immunological concepts to effective vaccines and protective antibodies. Nat Immunol. (2018) 19:1199–1211. doi: 10.1038/s41590-018-0228-6
3. Cockburn IA, Amino R, Kelemen RK, Kuo SC, Tse S-W, Radtke A, et al. In vivo imaging of CD8+ T cell-mediated elimination of malaria liver stages. Proc Natl Acad Sci USA. (2013) 110:9090–5. doi: 10.1073/pnas.1303858110
4. Seder RA, Chang L-J, Enama ME, Zephir KL, Sarwar UN, Gordon IJ, et al. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science. (2013) 341:1359–65. doi: 10.1126/science.1241800
5. Stevenson MM, Riley EM. Innate immunity to malaria. Nat Rev Immunol. (2004) 4:169–80. doi: 10.1038/nri1311
6. Dobaño C, Moncunill G. Naturally acquired immunity (NAI). In: Kremsner PG and Krishna S, editors. Encyclopedia of Malaria. New York, NY: Springer (2018). p. 1–15. doi: 10.1007/978-1-4614-8757-9_131-1
7. Nirmalan N, Sims PFG, Hyde JE. Quantitative proteomics of the human malaria parasite Plasmodium falciparum and its application to studies of development and inhibition. Mol Microbiol. (2004) 52:1187–1199. doi: 10.1111/j.1365-2958.2004.04049.x
8. Crompton PD, Moebius J, Waisberg M, Garver LS, Miller LH, Barillas C, et al. Malaria immunity in man and mosquito: insights into unsolved mysteries of a deadly infectious disease. Annu Rev Immunol. (2014) 32:157–87. doi: 10.1146/annurev-immunol-032713-120220
9. Heide J, Vaughan KC, Sette A, Jacobs T, Schulze zur Wiesch J. Comprehensive review of human Plasmodium falciparum-specific CD8+ T cell epitopes. Front Immunol. (2019) 10:397. doi: 10.3389/fimmu.2019.00397
10. Anthony DD, Lehmann PV. T-cell epitope mapping using the ELISPOT approach. Methods. (2003) 29:260–9. doi: 10.1016/S1046-2023(02)00348-1
11. Spielmann T, Gardiner DL, Beck H-P, Trenholme KR, Kemp DJ. Organization of ETRAMPs and EXP-1 at the parasite-host cell interface of malaria parasites. Mol Microbiol. (2006) 59:779–94. doi: 10.1111/j.1365-2958.2005.04983.x
12. Hill AVS, Elvin J, Willis AC, Aidoo M, Allsopp CEM, Gotch FM, et al. Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature. (1992) 360:434–9. doi: 10.1038/360434a0
13. Charoenvit Y, Mellouk S, Sedegah M, Toyoshima T, Leef MF, Delavega P, et al. Plasmodium yoelii: 17-kDa hepatic and erythrocytic stage protein is the target of an inhibitory monoclonal antibody. Exp Parasitol. (1995) 80:419–29. doi: 10.1006/expr.1995.1054
14. Kara UA, Stenzel DJ, Ingram LT, Bushell GR, Lopez JA, Kidson C. Inhibitory monoclonal antibody against a (myristylated) small-molecular-weight antigen from Plasmodium falciparum associated with the parasitophorous vacuole membrane. Infect Immun. (1988) 56:903–9.
15. Doolan DL, Hedstrom RC, Rogers WO, Charoenvit Y, Rogers M, de la Vega P, et al. Identification and characterization of the protective hepatocyte erythrocyte protein 17 kDa gene of Plasmodium yoelii, homolog of Plasmodium falciparum exported protein 1. J Biol Chem. (1996) 271:17861–8. doi: 10.1074/jbc.271.30.17861
16. Charoenvit Y, Majam VF, Corradin G, Sacci JB, Wang R, Doolan DL, et al. CD4(+) T-cell- and gamma interferon-dependent protection against murine malaria by immunization with linear synthetic peptides from a Plasmodium yoelii 17-kilodalton hepatocyte erythrocyte protein. Infect Immun. (1999) 67:5604–14.
17. Spielmann T, Montagna GN, Hecht L, Matuschewski K. Molecular make-up of the Plasmodium parasitophorous vacuolar membrane. Int J Med Microbiol. (2012) 302:179–86. doi: 10.1016/j.ijmm.2012.07.011
18. Coppel RL, Favaloro JM, Crewther PE, Burkot TR, Bianco AE, Stahl HD, et al. A blood stage antigen of Plasmodium falciparum shares determinants with the sporozoite coat protein. Proc Natl Acad Sci USA. (1985) 82:5121–5. doi: 10.1073/pnas.82.15.5121
19. Simmons D, Woollett G, Bergin-Cartwright M, Kay D, Scaife J. A malaria protein exported into a new compartment within the host erythrocyte. EMBO J. (1987) 6:485–91 doi: 10.1002/j.1460-2075.1987.tb04779.x
20. Meraldi V, Nebie I, Moret R, Cuzin-Ouattara N, Thiocone A, Doumbo O, et al. Recognition of synthetic polypeptides corresponding to the N- and C-terminal fragments of Plasmodium falciparum Exp-1 by T-cells and plasma from human donors from African endemic areas. Parasite Immunol. (2002) 24:141–50. doi: 10.1046/j.1365-3024.2002.00447.x
21. Aidoo M, Lalvani A, Gilbert SC, Hu JT, Daubersies P, Hurt N, et al. Cytotoxic T-lymphocyte epitopes for HLA-B53 and other HLA types in the malaria vaccine candidate liver-stage antigen 3. Infect Immun. (2000) 68:227–32. doi: 10.1128/IAI.68.1.227-232.2000
22. Doolan DL, Southwood S, Chesnut R, Appella E, Gomez E, Richards A, et al. HLA-DR-promiscuous T cell epitopes from Plasmodium falciparum pre-erythrocytic-stage antigens restricted by multiple HLA class II alleles. J Immunol. (2000) 165:1123–37. doi: 10.4049/jimmunol.165.2.1123
23. Doolan DL, Hoffman SL, Southwood S, Wentworth PA, Sidney J, Chesnut RW, et al. Degenerate cytotoxic T cell epitopes from P. falciparum restricted by multiple HLA-A and HLA-B supertype alleles. Immunity. (1997) 7:97–112. doi: 10.1016/S1074-7613(00)80513-0
24. Quakyi IA, Currier J, Fell A, Taylor DW, Roberts T, Houghten RA, et al. Analysis of human T cell clones specific for conserved peptide sequences within malaria proteins. Paucity of clones responsive to intact parasites. J Immunol. (1994) 153:2082–92.
25. Wang R, Richie TL, Baraceros MF, Rahardjo N, Gay T, Banania J, et al. Boosting of DNA vaccine-elicited gamma interferon responses in humans by exposure to malaria parasites boosting of DNA vaccine-elicited gamma interferon responses in humans by exposure to malaria parasites. Infect Immun. (2005) 73:2863–72. doi: 10.1128/IAI.73.5.2863-2872.2005
26. Lauer GM, Ouchi K, Chung RT, Nguyen TN, Day CL, Purkis DR, et al. Comprehensive analysis of CD8(+)-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J Virol. (2002) 76:6104–13. doi: 10.1128/JVI.76.12.6104-6113.2002
27. Day CL, Walker BD. Progress in defining CD4 helper cell responses in chronic viral infections. J Exp Med. (2003) 198:1773–7. doi: 10.1084/jem.20031947
28. Schulze zur Wiesch J, Lauer GM, Day CL, Kim AY, Ouchi K, Duncan JE, et al. Broad repertoire of the CD4+ Th cell response in spontaneously controlled hepatitis C virus infection includes dominant and highly promiscuous epitopes. J Immunol. (2005) 175:3603–13. doi: 10.4049/jimmunol.175.6.3603
29. Landahl J, Bockmann JH, Scheurich C, Ackermann C, Matzat V, Heide J, et al. Detection of a broad range of low level MHC class II-restricted Hepatitis Delta Virus (HDV)-specific T cell responses regardless of the clinical status. J Infect Dis. (2018) 219:568–77. doi: 10.1093/infdis/jiy549
30. DTG. Leitlinie: Diagnostik und Therapie der Malaria. Dtsch Gesellschaft für Tropenmedizin und Int Gesundh. (2016). Available online at: https://www.awmf.org/uploads/tx_szleitlinien/042-001l_S1_Malaria_Diagnostik_Therapie_2016-08_verlaengert.pdf (accessed April 25, 2018).
31. UniProt. Available at: https://www.uniprot.org/ (accessed July 30, 2019).
32. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. (2019) 47:D506–15. doi: 10.1093/nar/gky1049
33. da Costa Lima Caniatti MC, Borelli SD, Guilherme ALF, Tsuneto LT. Association between HLA genes and dust mite sensitivity in a Brazilian population. Hum Immunol. (2017) 78:88–94. doi: 10.1016/j.humimm.2016.10.014
34. Kim AY, Lauer GM, Ouchi K, Addo MM, Lucas M, Schulze Zur Wiesch J, et al. The magnitude and breadth of hepatitis C virus-specific CD8+ T cells depend on absolute CD4+ T-cell count in individuals coinfected with HIV-1. Blood. (2005) 105:1170–8. doi: 10.1182/blood-2004-06-2336
35. Posavad CM, Magaret AS, Zhao L, Mueller DE, Wald A, Corey L. Development of an interferon-gamma ELISPOT assay to detect human T cell responses to HSV-2. Vaccine. (2011) 29:7058–66. doi: 10.1016/j.vaccine.2011.07.028
36. Draenert R, Altfeld M, Brander C, Basgoz N, Corcoran C, Wurcel AG, et al. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J Immunol Methods. (2003) 275:19–29. doi: 10.1016/S0022-1759(02)00541-0
37. Sidney J, Southwood S, Moore C, Oseroff C, Pinilla C, Grey HM, et al. Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. Curr Protoc Immunol. (2013) 100:18.3.1–36. doi: 10.1002/0471142735.im1803s100
38. Kim Y, Ponomarenko J, Zhu Z, Tamang D, Wang P, Greenbaum J, et al. Immune epitope database analysis resource. Nucleic Acids Res. (2012) 40:W525–30. doi: 10.1093/nar/gks438
39. Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. (2008) 4:e1000048. doi: 10.1371/journal.pcbi.1000048
40. Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. (2010) 11:568. doi: 10.1186/1471-2105-11-568
41. Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, Buus S, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. (2003) 12:1007–17. doi: 10.1110/ps.0239403
42. Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. (2008) 36:W509–12. doi: 10.1093/nar/gkn202
43. Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. (2005) 6:132. doi: 10.1186/1471-2105-6-132
44. Twohig KA, Pfeffer DA, Baird JK, Price RN, Zimmerman PA, Hay SI, et al. Growing evidence of Plasmodium vivax across malaria-endemic Africa. PLoS Negl Trop Dis. (2019) 13:e0007140. doi: 10.1371/journal.pntd.0007140
45. Chakravarty S, Cockburn I, Kuk S, Medicine MO-N, 2007 U. CD8+ T lymphocytes protective against malaria liver stages are primed in skin-draining lymph nodes. Nat Med. (2007) 13:1035–41. doi: 10.1038/nm1628
46. Epstein JE, Tewari K, Lyke KE, Sim BKL, Billingsley PF, Laurens MB, et al. Live attenuated malaria vaccine designed to protect through hepatic CD8+ T cell immunity. Science. (2011) 334:475–80. doi: 10.1126/science.1211548
47. Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci USA. (2003) 100:9952–7. doi: 10.1073/pnas.1633254100
48. Schulze zur Wiesch J, Ciuffreda D, Lewis-Ximenez L, Kasprowicz V, Nolan BE, Streeck H, et al. Broadly directed virus-specific CD4 + T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J Exp Med. (2012) 209:61–75. doi: 10.1084/jem.20100388
49. Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, et al. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol. (2010) 185:943–55. doi: 10.4049/jimmunol.1000405
50. Lindestam Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, et al. Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset. PLoS Pathog. (2013) 9:e1003130. doi: 10.1371/journal.ppat.1003130
51. Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. (1998) 160:3363–73.
52. Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, et al. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J Immunol. (2010) 184:2492–503. doi: 10.4049/jimmunol.0903655
53. Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, et al. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J Immunol. (2010) 185:4189–98. doi: 10.4049/jimmunol.1001006
54. Gonzalez JM. HLA-A0201 restricted CD8 T-lymphocyte responses to malaria: identification of new Plasmodium falciparum epitopes by IFN-g ELISPOT. Parasite Immunol. (2000) 22:501–14. doi: 10.1046/j.1365-3024.2000.00331.x
55. Sedegah M, Kim Y, Peters B, McGrath S, Ganeshan H, Lejano J, et al. Identification and localization of minimal MHC-restricted CD8+ T cell epitopes within the Plasmodium falciparum AMA1 protein. Malar J. (2010) 9:241. doi: 10.1186/1475-2875-9-241
56. Sedegah M, Hollingdale MR, Farooq F, Ganeshan H, Belmonte M, Kim Y, et al. Sterile immunity to malaria after DNA prime/adenovirus boost immunization is associated with effector memory CD8+T cells targeting ama1 class I epitopes. PLoS ONE. (2014) 9:e106241. doi: 10.1371/journal.pone.0106241
57. Ganeshan H, Kusi KA, Anum D, Hollingdale MR, Peters B, Kim Y, et al. Measurement of ex vivo ELISpot interferon-gamma recall responses to Plasmodium falciparum AMA1 and CSP in Ghanaian adults with natural exposure to malaria. Malar J. (2016) 15:55. doi: 10.1186/s12936-016-1098-8
58. Grifoni A, Moore E, Voic H, Sidney J, Phillips E, Jadi R, et al. Characterization of magnitude and antigen specificity of HLA-DP, DQ, and DRB3/4/5 restricted DENV-specific CD4+ T cell responses. Front Immunol. (2019) 10:1568. doi: 10.3389/fimmu.2019.01568
59. Grifoni A, Angelo MA, Lopez B, O'Rourke PH, Sidney J, Cerpas C, et al. Global assessment of dengue virus-specific CD4+ T cell responses in dengue-endemic areas. Front Immunol. (2017) 8:1309. doi: 10.3389/fimmu.2017.01309
60. Rogers PR, Huston G, Swain SL. High antigen density and IL-2 are required for generation of CD4 effectors Secreting Th1 rather than Th0 cytokines. J Immunol. (1998) 161:3844–52.
61. Iriko H, Ishino T, Otsuki H, Ito D, Tachibana M, Torii M, et al. Plasmodium falciparum exported protein 1 is localized to dense granules in merozoites. Parasitol Int. (2018) 67:637–9. doi: 10.1016/j.parint.2018.06.001
62. Ocaña-Morgner C, Mota MM, Rodriguez A. Malaria blood stage suppression of liver stage immunity by dendritic cells. J Exp Med. (2003) 197:143–51. doi: 10.1084/jem.20021072
63. Keitany GJ, Kim KS, Krishnamurty AT, Hondowicz BD, Hahn WO, Dambrauskas N, et al. Blood stage malaria disrupts humoral immunity to the pre-erythrocytic stage circumsporozoite protein. Cell Rep. (2016) 17:3193–205. doi: 10.1016/j.celrep.2016.11.060
64. Wikenheiser DJ, Brown SL, Lee J, Stumhofer JS. NK1.1 expression defines a population of CD4+ effector T cells displaying Th1 and Tfh cell properties that support early antibody production during Plasmodium yoelii infection. Front Immunol. (2018) 9:2277. doi: 10.3389/fimmu.2018.02277
65. Illingworth JJ, Alanine DG, Brown R, Marshall JM, Bartlett HE, Silk SE, et al. Functional comparison of blood-stage Plasmodium falciparum malaria vaccine candidate antigens. Front Immunol. (2019) 10:1254. doi: 10.3389/fimmu.2019.01254
66. Zavala F, Tam J, Hollingdale M, Cochrane A, Quakyi I, Nussenzweig R, et al. Rationale for development of a synthetic vaccine against Plasmodium falciparum malaria. Science. (1985) 228:1436–40. doi: 10.1126/science.2409595
67. Kastenmüller K, Espinosa DA, Trager L, Stoyanov C, Salazar AM, Pokalwar S, et al. Full-length Plasmodium falciparum circumsporozoite protein administered with long-chain poly(I·C) or the toll-like receptor 4 agonist glucopyranosyl lipid adjuvant-stable emulsion elicits potent antibody and CD4 + T cell immunity and protection in mice. Infect Immun. (2013) 81:789–800. doi: 10.1128/IAI.01108-12
68. Bongfen SE, Ntsama PM, Offner S, Smith T, Felger I, Tanner M, et al. The N-terminal domain of Plasmodium falciparum circumsporozoite protein represents a target of protective immunity. Vaccine. (2009) 27:328–35. doi: 10.1016/j.vaccine.2008.09.097
69. Fowkes FJI, Boeuf B. Immunity to malaria in an era of declining malaria transmission. Parasitology. (2016) 143:139–53. doi: 10.1017/S0031182015001249
70. Ly A, Hansen DS. Development of B cell memory in malaria. Front Immunol. (2019) 10:559. doi: 10.3389/fimmu.2019.00559
Keywords: malaria, Plasmodium falciparum, Plasmodium vivax, CD4+, CD8+, T cell epitope, HLA binding, HLA class II
Citation: Heide J, Wildner NH, Ackermann C, Wittner M, Marget M, Sette A, Sidney J, Jacobs T and Schulze zur Wiesch J (2020) Detection of EXP1-Specific CD4+ T Cell Responses Directed Against a Broad Range of Epitopes Including Two Promiscuous MHC Class II Binders During Acute Plasmodium falciparum Malaria. Front. Immunol. 10:3037. doi: 10.3389/fimmu.2019.03037
Received: 12 September 2019; Accepted: 11 December 2019;
Published: 22 January 2020.
Edited by:
Moriya Tsuji, Aaron Diamond AIDS Research Center, United StatesReviewed by:
Adrian John Frederick Luty, Institut de recherche pour le développement (IRD), FranceKatsuyuki Yui, Nagasaki University, Japan
Copyright © 2020 Heide, Wildner, Ackermann, Wittner, Marget, Sette, Sidney, Jacobs and Schulze zur Wiesch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julian Schulze zur Wiesch, anVsaWFuc3p3QGdtYWlsLmNvbQ==
 Janna Heide
Janna Heide Nils H. Wildner
Nils H. Wildner Christin Ackermann1
Christin Ackermann1 Matthias Marget
Matthias Marget Alessandro Sette
Alessandro Sette John Sidney
John Sidney Thomas Jacobs
Thomas Jacobs Julian Schulze zur Wiesch
Julian Schulze zur Wiesch