- Department of Medical Microbiology and Radboud Center for Infectious Diseases, Radboud University Medical Center, Nijmegen, Netherlands
Natural killer (NK) cells are known to be activated during malaria infection, exhibiting both cytokine production and cytotoxic functions. However, NK cells are heterogeneous in their expression of surface activatory and inhibitory receptors which may influence their response to malaria parasites. Here, we studied the surface marker profile and activation dynamics of NK cells during a Controlled Human Malaria Infection in 12 healthy volunteers. Although there was significant inter-patient variability in timing and magnitude of NK cell activation, we found a consistent and strong increase in expression of the activatory receptor NKp30. Moreover, high baseline NKp30 expression was associated with NK cell activation at lower parasite densities. Our data suggest that NKp30 expression may influence the NK cell response to P. falciparum, explaining inter-patient heterogeneity and suggesting a functional role for this receptor in malaria.
Introduction
Malaria infection in humans activates a broad cellular immune response involving monocytes, T cells, B cells, and NK cells. NK cells may play a functional role in protection against Plasmodium falciparum, as certain NK cell receptor genotypes are associated with decreases in malaria susceptibility and pathology [reviewed in (1)]. During the pathological blood stage of P. falciparum infection, circulating NK cells display a dual functional role, i.e., cytokine production (2–5) and killing of infected blood cells both via antibody-independent (6–8) and antibody-dependent cytotoxicity (9, 10). Their relative contribution to protection remains unknown.
NK cells are often considered a homogenous, unchanging population, but multicolored flow cytometry and mass cytometry have revealed that NK cells actually consist of many distinct populations, differing in their functionality against specific diseases (11–14). Artavanis-Tsakonas et al. previously demonstrated that in malaria naïve donors a specific subpopulation of NK cells expressing the lectin-type receptor NKG2A are the main IFN-γ producers in response to P. falciparum-infected RBC (15). Most studies determining the NK cell response against P. falciparum demonstrate that there is large inter-donor variability (16, 17). We hypothesized that this heterogeneity might at least in part be explained by differences in NK cell phenotype prior to infection.
To date most data on responsiveness of NK cells to P. falciparum has been obtained from ex vivo stimulation experiments or case-control studies in endemic areas. We took advantage of the Controlled Human Malaria Infection model to evaluate the activation and function of different NK cell subsets at multiple time points during a malaria infection. Our data show in vivo NK cell activation in all donors with an upregulation of IFN-γ and granzyme B production. There was indeed a significant variability both in the timing and magnitude of the NK cell response, and increased baseline receptor expression of NKp30 predicted a more rapid in vivo NK cell activation.
Materials and Methods
Clinical Trials
Study 1 was a single-center, open-label clinical trial in 12 malaria naïve individuals conducted at the Radboud university medical center (Nijmegen, The Netherlands) from May until June 2018. Study volunteers provided written informed consent and were screened as described previously (18). The trial was approved by the Central Committee on Research Involving Human Subjects (CCMO; NL63552.091.17) of the Netherlands, performed according to the Declaration of Helsinki and Good Clinical Practice and prospectively registered at ClinicalTrials.gov (NCT03454048). Volunteers were infected by the bites of five P. falciparum 3D7 strain-infected Anopheles mosquitoes, and followed up for parasitemia twice daily starting on day 6 post infection. Parasitemia was assessed by thick blood smear and qPCR. Volunteers were treated with a sub-optimal dose of piperaquine when parasitemia reached density detectable by thick blood smear or 5,000 parasites/milliliter by qPCR, and received curative treatment if recrudescent parasitemia occurred.
Study 2 was a single-center randomized placebo controlled malaria vaccine trial (CCMO NL39541.091.12; NCT01728701) published previously (19). Only study subjects that received placebo vaccination followed by CHMI were included in the current analysis. In short, volunteers received bites from five P. falciparum NF54 strain-infected Anopheles mosquitoes, and were followed up for parasitemia twice daily starting on day 5 post infection. Parasitemia was assessed by thick blood smear and/or qPCR, and volunteers received curative treatment with atovaquone/proguanil, either when parasitemia reached levels detectable by microscopy (n = 5) or after two consecutive qPCRs >500 parasites/milliliter (n = 4).
Whole Blood NK Cell Phenotyping
In study 1, 100 μL fresh EDTA blood was stained directly with a pre-prepared and antibody mixture containing: CD3-AlexaFluor700 (Biolegend; clone OKT3), pan-γδTCR-PE (Beckman Coulter; clone IMMU510), CD56-Brilliant Violet(BV)421 (Biolegend; clone HCD56), CD16-APC-eFluor780 (eBiosciences; clone CB16), CD69-PerCP-Cy5.5 (Biolegend; clone FN50), NKp30-APC (Biolegend; clone P30-15), NKG2D-Brilliant Violet(BV)510 (Biolegend; clone 1D11), NKG2A-PEVio770 (Miltenyi Biotec; clone REA110), and CD57-FITC (Biolegend; clone HCD57). A single mixture was prepared one day before the first time point, aliquotted per time point and stored in the dark until use. Samples were stained at 4°C in the dark for 30 min, followed by erythrocyte lysis with 1 mL FACS Lysis buffer (BD Biosciences) for exactly 5 min. Samples were centrifuged and then washed with 0.5% Bovine Serum Albumin (BSA) in PBS. Cell pellets were resuspended in 100 μL 1% paraformaldehyde (PFA) and analyzed on a Gallios flow cytometer (Beckman Coulter). At each time point, staining and fixation was completed within 4 h of blood draw and flow cytometry was performed the same day using identical acquisition settings and a standardized protocol. CD69 was used as a marker for lymphocyte activation after CHMI, as described earlier (20, 21).
PBMC Isolation and Cryopreservation
In study 2, blood samples for peripheral blood mononuclear cell (PBMC) isolation were taken pre-challenge, 3 days after antimalarial treatment and 35 days after challenge infection. Isolation and cryopreservation was performed as described previously (22). In short, PBMCs were isolated from citrate anti-coagulated blood using vacutainer cell preparation tubes (CPT; BD Diagnostics) by density gradient centrifugation. Cells were washed four times in ice-cold phosphate buffered saline (PBS), counted using 0·1% Trypan blue with 5% Zap-o-Globin II Lytic Reagent (Beckman Coulter), cryopreserved at a concentration of 10 × 106 cells/ml in ice-cold fetal calf serum (Gibco)/10% DMSO (Merck), and stored in vapor-phase nitrogen.
PBMC Thawing and Re-stimulation
Immediately prior to use, cells were thawed and washed twice in Dutch-modified RPMI 1640 (Gibco/Invitrogen). Cell viability was assessed by counting in 0·1% Trypan blue with 5% Zap-o-Globin II Lytic Reagent (Beckman Coulter) to assess cell viability. PBMCs were cultured at 2.5 × 106 cells/ml in RPMI 1640 (Dutch Modification; Gibco) with 5 mg/ml gentamycin (Centraform), 100 mM pyruvate (Gibco), 200 mM glutamax (Gibco), supplemented with 10% heat-inactivated pooled human A+ serum (obtained from Sanquin Bloodbank, Nijmegen, The Netherlands) at a final volume of 200 μL in 96-wells plates. Cells were stimulated with purified Plasmodium falciparum NF54 schizonts or uninfected red blood cells at a concentration of 5 × 106 RBC/ml. After 3 h, Brefeldin A (10 μg/mL; Sigma-Aldrich) and monansin (2 μM; eBioscience) were added to culture. After another 3 h (6 h total stimulation) cells from two stimulation replicates (1.0 × 106 cells total) were combined, washed and stained with Fixable Viability Stain 700 (BD Biosciences) for 30 min. After washing with PBS, cells were stained with extracellular antibodies, CD3-AlexaFluor700 (Biolegend; clone OKT3), CD56-PE (Biolegend; clone HCD56), CD16-APC-eFluor780 (eBiosciences; clone CB16), NKG2A-PEVio770 (Miltenyi Biotec; clone REA110), and CD57-APC (Biolegend; clone HCD57) for 30 min at 4°C in the dark. Cells were washed and fixed with Foxp3 fixation/permeabilization buffer (eBioscience). After washing with permeabilization buffer (eBioscience) cells were stained for intracellular cytokines with IFN-γ-PE-Dazzle (Biolegend; clone 4S.B3) and granzyme B-FITC (Biolegend; clone GB11). After another wash with permeabilization buffer, cells from two staining replicates (2.0 × 106 cells total) were taken up in 200 μL 1% paraformaldehyde (PFA) and analyzed on a Gallios flow cytometer (Beckman Coulter) the next day.
Data Analysis and Statistics
Flow cytometry data was analyzed using Flow Jo software (version 10.0.8 for Apple OS). Statistical analysis was performed using GraphPad Prism (version 5.03 for Windows). Gating strategy and representative plots are shown in Supplementary Figure 1 (whole blood) and Supplementary Figure 3 (PBMCs).
Results
Heterogeneity in NK Cell Activation After CHMI
After malaria infection, NK cell activation as defined by upregulated CD69 expression was determined daily from day 6 post-infection until 3 days after antimalarial treatment (Supplementary Figure 1). In study #1 the first activation of NK cells in a number of volunteers was observed 1 day after the first appearance of parasitemia detectable by qPCR (Figure 1).
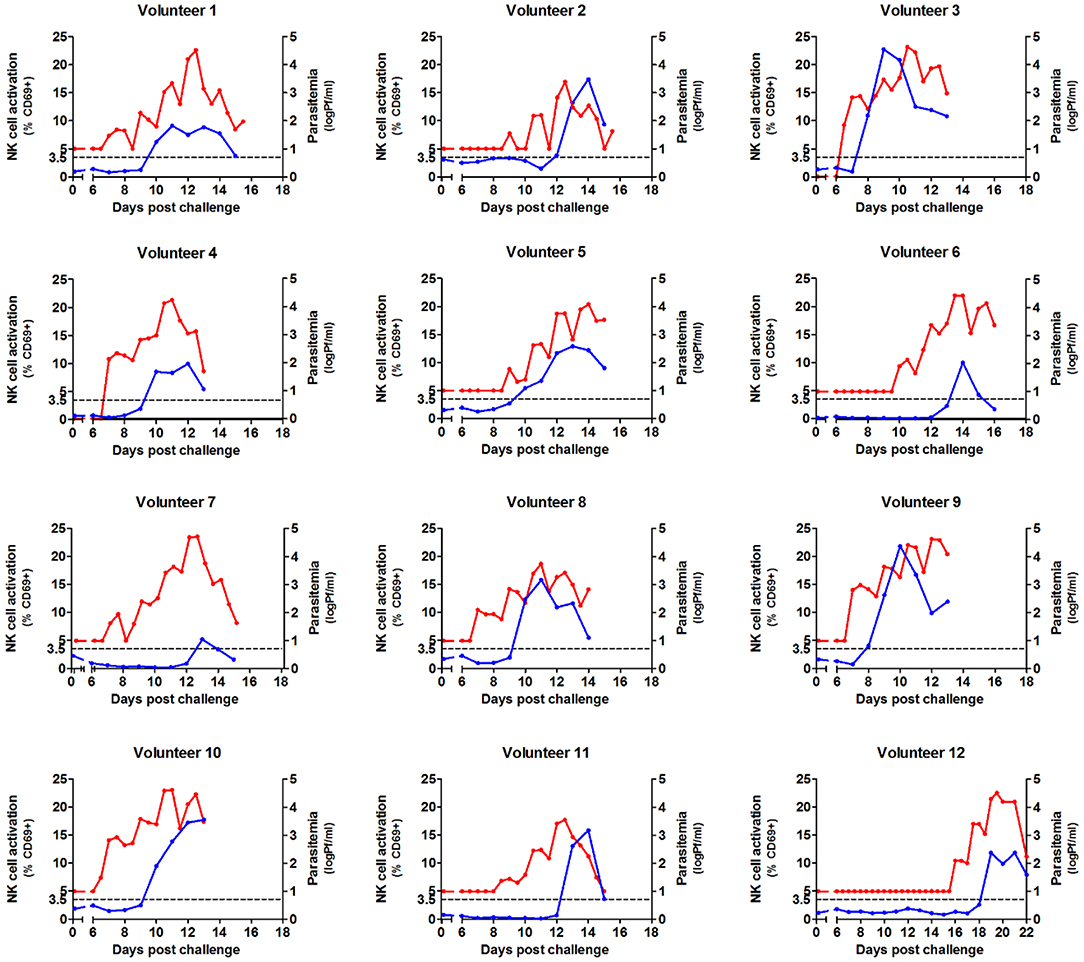
Figure 1. Kinetics of parasitemia and NK cell activation during Controlled Human Malaria Infection. NK cells were analyzed by flow cytometry daily in whole venous blood from 12 volunteers undergoing Controlled Human Malaria Infection. Antimalarial treatment was initiated when parasite densities reached levels detectable by microscopy. Each graph shows the activation of NK cells (defined by CD69 surface expression) from day 6 post infection until day 3 after antimalarial treatment (blue line, left axis). The same graph shows parasitemia measured by qPCR from day 6 after infection, until day 3 after antimalarial treatment (red line, right axis). NK cell activation is first seen 1–2 days after the first appearance of parasitemia. Each graph represents the data gathered for a single volunteer (n = 12).
In the absence of parasitemia, up to 3.5% of NK cells expressed CD69, therefore >3.5% CD69 expression was considered significant NK cell activation above background (Figure 2A). There was indeed a significant heterogeneity in the timing of first NK cell activation, ranging from 1 day after the first appearance of parasitemia (i.e., volunteer 5) to 5 days after parasitemia (volunteer 7). This may be partially explained by differences in starting parasite density. Parasitemia (prior to the initiation of antimalarial treatment) correlated strongly with the degree of NK cell activation (Spearman p = 0.0017; Figure 2A). However, this does not explain the diversity entirely, as some volunteers have significant NK cell activation (defined as CD69 expression >3.5%) at very low circulating parasitemia, such as volunteer 5, while others require very high parasitemia before NK cells become activated, such as volunteer 7. This circulating parasite density prior to NK cell activation was highly variable between volunteers (mean 4,798 Pf/ml, range 25–26,152 Pf/ml), suggestive for a host-dependent factor.
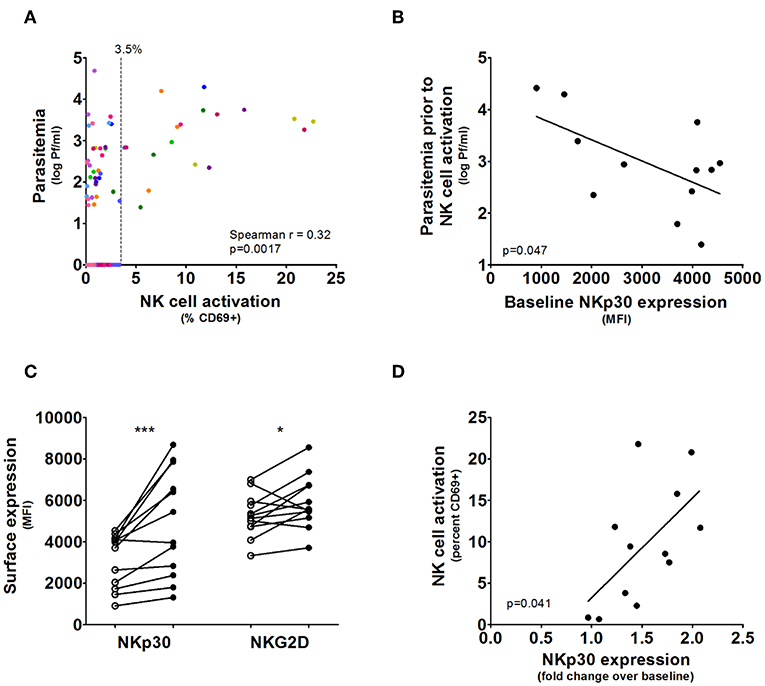
Figure 2. NKp30 predicts rapid NK cell activation during CHMI. (A) The graph shows NK cell activation correlated with parasitemia in all post-challenge but pre-treatment blood samples, where each color represents the samples from an individual volunteer. Blood samples without parasites showed up to 3.5% NK cell CD69 expression. (B) For each volunteer (n = 12) the maximum parasitemia measured by qPCR in prior to or at the moment of first NK cell activation measured in whole blood was determined. Increased baseline NK cell NKp30 surface expression determined by Mean Fluorescent Intensity correlated with NK cell activation at lower parasitemia. Line and p-value are the result of a linear regression analysis. (C) Surface expression of the activatory receptors NKp30 and NKG2D determined on total NK cells for each volunteer prior to malaria infection (open circles) and on the day of antimalarial treatment (closed circles). P-values are the result of Wilcoxon matched-pairs signed rank test; *p < 0.05; ***p < 0.001. (D) The fold change in NKp30 expression (determined by MFI) between measurements at baseline and antimalarial treatment for each volunteer were correlated to NK cell CD69 expression at on the day of antimalarial treatment. Line and p-value are the result of a linear regression analysis.
Baseline NKp30 Expression Predicts Activation After CHMI
NK cell activation is dependent on a delicate balance between activatory- and inhibitory receptors, and the expressed receptor profile may relate to the observed heterogeneity during CHMI. Therefore, we next determined whether the expression of activatory receptors NKp30 or NKG2D, the inhibitory receptor NKG2A or the differentiation marker CD57 predicted an individual's response to CHMI. Indeed, higher baseline NK cell NKp30 expression correlated with activation at lower parasitemia (linear regression p = 0.047; Figure 2B). NKp30 and NKG2D were expressed on nearly 100% of NK cells for all volunteers (Supplementary Figure 1).
NKp30 was strongly upregulated during CHMI (pre-challenge vs. day of antimalarial treatment: mean MFI 3,145 vs. 4,913, Wilcoxon matched-pairs signed rank test p = 0.0010; Figure 2C), while the upregulation of NKG2D was marginal (pre-challenge vs. day of antimalarial treatment: mean MFI 5,268 vs. 5,916, Wilcoxon matched-pairs signed rank test p = 0.043; Figure 2C). The increase in NKp30 expression was proportional to total NK cell activation at antimalarial treatment (linear regression p = 0.041; Figure 2D).
NK cells can be divided into distinct populations representing levels of differentiation based on their expression of CD56, CD16, NKG2A, and CD57 (11), and a previous study suggested NKG2A+ NK cells are more responsive to P. falciparum in vitro (15). We sought to determine whether this may result from differential expression of NKp30. However, while baseline expression of NKp30 varied between CD56dimNKG2A+ and CD56dimNKG2A– subsets (Figure 3A; Supplementary Figure 2), all NK cell subsets showed an upregulation of NKp30 (Figure 3A). Furthermore, we did not see any differences in activation as defined by CD69 upregulation between the CD56dim subsets, though there was significantly more activation of the CD56dim subset compared to the CD56bright subset (Figures 3B,C).
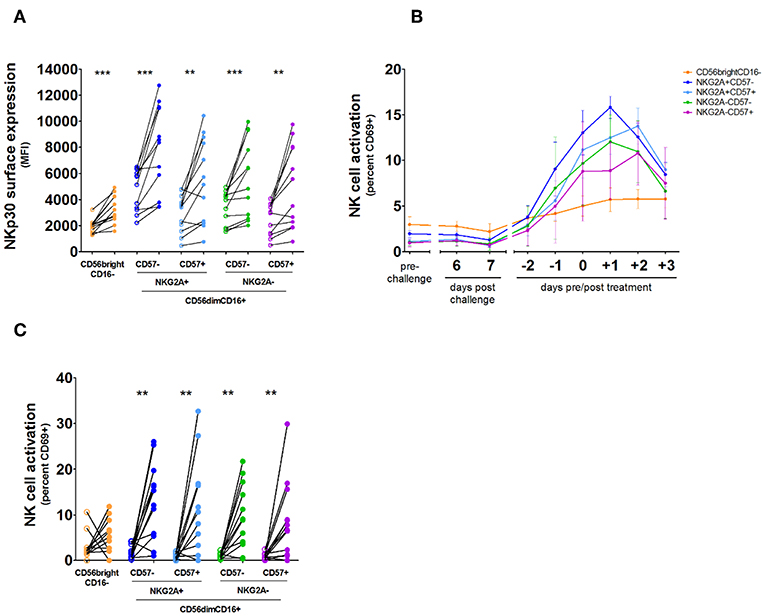
Figure 3. NKp30 expression and activation kinetics on NK cell subsets. NK cells were analyzed by daily flow cytometry in whole venous blood from 12 volunteers undergoing Controlled Human Malaria Infection. Antimalarial treatment was initiated when parasite densities reached levels detectable by microscopy. (A) Total NK cells were divided into five subpopulations based on their surface expression of CD56, CD16, NKG2A, and CD57: CD56brightCD16– (orange), CD56dimCD16+NKG2A+CD57– (dark blue), CD56dimCD16+NKG2A+CD57+ (light blue), CD56dimCD16+NKG2A-CD57– (green), and CD56dimCD16+NKG2A-CD57+ (purple). Surface expression of NKp30 for each NK cell subset prior to malaria infection (open circles) compared with NKp30 expression on each NK cell subset on the day of antimalarial treatment (closed circles). P-values are the result of Wilcoxon matched-pairs signed rank test; **p < 0.01; ***p < 0.001. (B) The graph shows the mean and error of NK cell CD69 surface expression on each subset per day in 12 volunteers. (C) Surface expression of CD69 was determined for each NK cell subset prior to malaria infection (open circles) and on the day of antimalarial treatment (closed circles). P-values are the result of Wilcoxon matched-pairs signed rank test; *p < 0.05; **p < 0.01; ***p < 0.001.
NK Cell Subsets Upregulate CD69, IFN-γ, and Granzyme B During CHMI
As there appears to be little activation of the CD56bright NK cell subset during the course of infection, we wanted to determine the ability of both the CD56brightCD16– and CD56dimCD16+ subsets to produce granzyme B and IFN-γ and degranulate during infection, using isolated and cryopreserved peripheral blood mononuclear cells (PBMCs) from study #2 (Supplementary Figure 3). We found that both subsets increase production of granzyme B and IFN-γ and show improved degranulation during infection (Figures 4A–C; Supplementary Figure 4).
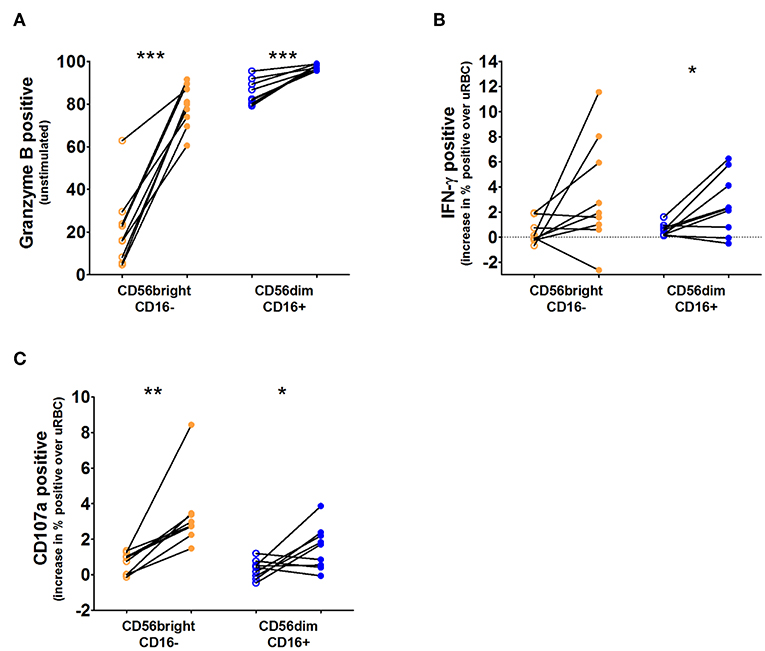
Figure 4. IFN-γ, granzyme B, and degranulation responses per NK cell subset. Cryopreserved PBMCs from nine volunteers taken before challenge (open circles) and 3 days after antimalarial treatment (closed circles) were thawed and stimulated for 6 h with Pf-infected red blood cells (PfRBC) or uninfected RBC (uRBC). Total NK cells were divided into two subpopulations based on their surface expression of CD56 and CD16. (A) Intracellular granzyme B content (% cells positive) determined by flow cytometry in unstimulated NK cells (those incubated with uninfected RBC cultures) at both time points. (B) Intracellular IFN-γ production (% cells positive) in response to PfRBC stimulation at both time points, IFN-γ production after PfRBC stimulation was corrected for production in response to uRBC. (C) CD107a staining (% cells positive) in response to PfRBC stimulation at both time points, CD107a staining after PfRBC stimulation was corrected for production in response to uRBC. For all three graphs open circles are pre-infection time points and closed circles are day 3 post antimalarial treatment. P-values are the result of paired samples t-test; *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
These data show that NKp30 is a marker for the NK cell response during a Controlled Human Malaria Infection, and suggests a possible functional role in the response to infected red blood cells. We demonstrate that the expression of this receptor at baseline relates to individual NK cell responses to P. falciparum in vivo. Furthermore, we show that NK cell activation during the course of infection is linked to an increase in NKp30 expression.
Both NKp30 and NKG2D have been shown to increase expression during NK cell activation (14, 23), however, during CHMI the magnitude of NKp30 upregulation of is particularly pronounced compared to NKG2D. It has previously been demonstrated in vitro that NKp30 binds to the P. falciparum protein Pf EMP1 leading to NK cell activation (24). This supports our finding that NK cells with higher resting NKp30 expression are more sensitive to activation at lower parasitemia. However, it is important to note that other in vitro studies suggest that Pf EMP1 may be dispensable (25) and that MDA5 signaling may be essential (26) for NK cell activation in response to Pf RBC. Therefore, multiple mechanisms may be involved in NK cell activation during malaria.
This is the first study with longitudinal daily samples from the initial phase of a malaria infection as parasites emerge from the liver that suggests an important role for NKp30. We thereby measured CD69 expression directly in patient blood samples, without re-stimulation, remaining close to the induced in vivo phenotype of an early natural infection. Furthermore, we show that baseline NKp30 expression is linked to a more rapid NK cell activation during subsequent infection.
Population based studies conducted in sub-Saharan Africa have identified a single nucleotide polymorphism (SNP) in the promoter for the NCR3 gene that encodes NKp30 that is associated with an increased number of clinical, uncomplicated malaria episodes in individuals over 5 years old (27–29). The combined data are highly suggestive for a potential functional role of NKp30-mediated NK cells in malaria. In our study we do not detect differences in time to parasitemia, maximum parasitemia, or parasite multiplication rate between those with high NKp30 expression and those with low NKp30 expression (data not shown). However, an important limitation of this study is that it was not designed to measure an effect on control of blood stage parasite replication. Even in volunteers with very rapid NK cell activation, this occurred only 2 days before the initiation of antimalarial treatment. This period between NK cell activation and drug treatment would be too short to measure an effect on parasite multiplication. Instead, cohort studies in endemic areas are better suited to answer this question. Future studies in endemic areas could determine NKp30 expression on NK cells at the beginning of a malaria season and during follow-up visits, and correlate this with number of clinical malaria episodes.
Broad inter-donor variability in the activation of NK cells in response to P. falciparum has been described in multiple studies (16, 17, 21). Our current finding suggest that baseline NK cell phenotype can play a role in this diversity. However, other immunological factors, including other activatory and inhibitory receptors not studied here, interactions with other immune cells and cytokine production likely also contribute to the NK cell response. Furthermore, parasitological factors, such as the initial starting parasitemia and parasite multiplication rate may also affect host response.
The phenotypic diversity of NK cells has been a topic of extensive study during the last decade (12, 14). Since the first discovery of NK cell memory in murine CMV infection (30), specific NK cell phenotypes have been identified as the main responders in human EBV (13), CMV (31), and HIV infection (14, 32) as well. Similarly, studies suggested that NKG2A+ NK cells, specifically respond to P. falciparum (15, 16). Interestingly, this does not appear to be the case during controlled human malaria infection in vivo.
Nevertheless, the finding that NKp30 expression predicts the response to CHMI, underscores the potential importance of NK cell phenotype in our susceptibility to disease. The diversity of the NK cell repertoire has been implicated in the risk of HIV acquisition (14, 33), and viral infections in turn have been shown to change its composition (34–37). Our study suggests that NK cell phenotype affects the response to a P. falciparum infection.
The current study was limited to analysis of CD56, CD16, NKG2A, CD57, NKp30, and NKG2D. In contrast, data from studies on other diseases using cytometry by time-of-flight (CyTOF) have suggested there may be more than 100,000 NK cell phenotypes, each characterized by a distinct combination of surface receptors (12). Furthermore, the expression of diverse killer cell immunoglobulin-like receptors (KIRs) plays an important role in NK differentiation and function (11). Therefore, it is likely that additional receptors, or combinations of inhibitory and activatory receptors, are also important for the interaction between NK cells and P. falciparum parasites. Future studies looking at a larger number of receptors and cytokines could unravel both these effects in more detail.
In conclusion, this study is the first to identify the expression the NK cell activatory receptor NKp30 as a marker that predicts a rapid NK cell response to parasitemia and suggest a potential role for this receptor in NK cell functionality against P. falciparum.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Central Committee on Research Involving Human Subjects, Netherlands. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JW and RS designed the study. JW performed the analysis and wrote the first draft of the manuscript which was supervised by RS.
Funding
JW and RS were supported by a grant from the Bill and Melinda Gates Foundation, OPP1091355.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank all the trial volunteers and the staff of the Radboud university medical center who were involved in the study, in particular Manon Alkema, Guido Bastiaens, Gerdie de Jong, Maurits van der Meer, and Isaie Reuling for conducting the clinical trials. We also thank Marga van de Vegte-Bolmer, Wouter Graumans, and Rianne Stoter for the generation and cryopreservation of the P. falciparum-infected red blood cells, and Anieck Lomans and Marleen Meij for their assistance in setting up the flow cytometry analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02864/full#supplementary-material
References
1. Wolf AS, Sherratt S, Riley EM. NK cells: uncertain allies against malaria. Front Immunol. (2017) 8:212. doi: 10.3389/fimmu.2017.00212
2. Artavanis-Tsakonas K, Riley EM. Innate immune response to malaria: rapid induction of IFN-gamma from human NK cells by live Plasmodium falciparum-infected erythrocytes. J Immunol. (2002) 169:2956–63. doi: 10.4049/jimmunol.169.6.2956
3. Mccall MB, Roestenberg M, Ploemen I, Teirlinck A, Hopman J, De Mast Q, et al. Memory-like IFN-gamma response by NK cells following malaria infection reveals the crucial role of T cells in NK cell activation by P. falciparum. Eur J Immunol. (2010) 40:3472–7. doi: 10.1002/eji.201040587
4. Agudelo O, Bueno J, Villa A, Maestre A. High IFN-gamma and TNF production by peripheral NK cells of Colombian patients with different clinical presentation of Plasmodium falciparum. Malar J. (2012) 11:38. doi: 10.1186/1475-2875-11-38
5. Coch C, Hommertgen B, Zillinger T, Dassler-Plenker J, Putschli B, Nastaly M, et al. Human TLR8 senses RNA from Plasmodium falciparum-infected red blood cells which is uniquely required for the IFN-gamma response in NK cells. Front Immunol. (2019) 10:371. doi: 10.3389/fimmu.2019.00371
6. Orago AS, Facer CA. Cytotoxicity of human natural killer (NK) cell subsets for Plasmodium falciparum erythrocytic schizonts: stimulation by cytokines and inhibition by neomycin. Clin Exp Immunol. (1991) 86:22–9. doi: 10.1111/j.1365-2249.1991.tb05768.x
7. Mavoungou E, Luty AJ, Kremsner PG. Natural killer (NK) cell-mediated cytolysis of Plasmodium falciparum-infected human red blood cells in vitro. Eur Cytokine Netw. (2003) 14:134–42.
8. Chen Q, Amaladoss A, Ye W, Liu M, Dummler S, Kong F, et al. Human natural killer cells control Plasmodium falciparum infection by eliminating infected red blood cells. Proc Natl Acad Sci USA. (2014) 111:1479–84. doi: 10.1073/pnas.1323318111
9. Arora G, Hart GT, Manzella-Lapeira J, Doritchamou JY, Narum DL, Thomas LM, et al. NK cells inhibit Plasmodium falciparum growth in red blood cells via antibody-dependent cellular cytotoxicity. Elife. (2018) 7:e36806. doi: 10.7554/eLife.36806.017
10. Hart GT, Tran TM, Theorell J, Schlums H, Arora G, Rajagopalan S, et al. Adaptive NK cells in people exposed to Plasmodium falciparum correlate with protection from malaria. J Exp Med. (2019) 216:1280–90. doi: 10.1084/jem.20181681
11. Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. (2010) 116:3853–64. doi: 10.1182/blood-2010-04-281675
12. Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. (2013) 5:208ra145. doi: 10.1126/scitranslmed.3006702
13. Azzi T, Lunemann A, Murer A, Ueda S, Beziat V, Malmberg KJ, et al. Role for early-differentiated natural killer cells in infectious mononucleosis. Blood. (2014) 124:2533–43. doi: 10.1182/blood-2014-01-553024
14. Strauss-Albee DM, Fukuyama J, Liang EC, Yao Y, Jarrell JA, Drake AL, et al. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci Transl Med. (2015) 7:297ra115. doi: 10.1126/scitranslmed.aac5722
15. Artavanis-Tsakonas K, Eleme K, Mcqueen KL, Cheng NW, Parham P, Davis DM, et al. Activation of a subset of human NK cells upon contact with Plasmodium falciparum-infected erythrocytes. J Immunol. (2003) 171:5396–405. doi: 10.4049/jimmunol.171.10.5396
16. Korbel DS, Newman KC, Almeida CR, Davis DM, Riley EM. Heterogeneous human NK cell responses to Plasmodium falciparum-infected erythrocytes. J Immunol. (2005) 175:7466–73. doi: 10.4049/jimmunol.175.11.7466
17. Korbel DS, Norman PJ, Newman KC, Horowitz A, Gendzekhadze K, Parham P, et al. Killer Ig-like receptor (KIR) genotype predicts the capacity of human KIR-positive CD56dim NK cells to respond to pathogen-associated signals. J Immunol. (2009) 182:6426–34. doi: 10.4049/jimmunol.0804224
18. Bijker EM, Bastiaens GJ, Teirlinck AC, Van Gemert GJ, Graumans W, Van De Vegte-Bolmer M, et al. Protection against malaria after immunization by chloroquine prophylaxis and sporozoites is mediated by preerythrocytic immunity. Proc Natl Acad Sci USA. (2013) 110:7862–7. doi: 10.1073/pnas.1220360110
19. Bastiaens GJ, Van Meer MP, Scholzen A, Obiero JM, Vatanshenassan M, Van Grinsven T, et al. Safety, immunogenicity, and protective efficacy of intradermal immunization with aseptic, purified, cryopreserved Plasmodium falciparum sporozoites in volunteers under chloroquine prophylaxis: a randomized controlled trial. Am J Trop Med Hyg. (2015) 94:663–73. doi: 10.4269/ajtmh.15-0621
20. Bijker EM, Schats R, Visser LG, Sauerwein RW, Scholzen A. Ex vivo lymphocyte phenotyping during Plasmodium falciparum sporozoite immunization in humans. Parasite Immunol. (2015) 37:590–8. doi: 10.1111/pim.12276
21. Walk J, De Bree LCJ, Graumans W, Stoter R, Van Gemert GJ, Van De Vegte-Bolmer M, et al. Outcomes of controlled human malaria infection after BCG vaccination. Nat Commun. (2019) 10:874. doi: 10.1038/s41467-019-08659-3
22. Walk J, Reuling IJ, Behet MC, Meerstein-Kessel L, Graumans W, Van Gemert GJ, et al. Modest heterologous protection after Plasmodium falciparum sporozoite immunization: a double-blind randomized controlled clinical trial. BMC Med. (2017) 15:168. doi: 10.1186/s12916-017-0923-4
23. Vendrame E, Fukuyama J, Strauss-Albee DM, Holmes S, Blish CA. Mass cytometry analytical approaches reveal cytokine-induced changes in natural killer cells. Cytometry B Clin Cytom. (2017) 92:57–67. doi: 10.1002/cyto.b.21500
24. Mavoungou E, Held J, Mewono L, Kremsner PG. A Duffy binding-like domain is involved in the NKp30-mediated recognition of Plasmodium falciparum-parasitized erythrocytes by natural killer cells. J Infect Dis. (2007) 195:1521–31. doi: 10.1086/515579
25. Baratin M, Roetynck S, Pouvelle B, Lemmers C, Viebig NK, Johansson S, et al. Dissection of the role of PfEMP1 and ICAM-1 in the sensing of Plasmodium-falciparum-infected erythrocytes by natural killer cells. PLoS ONE. (2007) 2:e228. doi: 10.1371/journal.pone.0000228
26. Ye W, Chew M, Hou J, Lai F, Leopold SJ, Loo HL, et al. Microvesicles from malaria-infected red blood cells activate natural killer cells via MDA5 pathway. PLoS Pathog. (2018) 14:e1007298. doi: 10.1371/journal.ppat.1007298
27. Delahaye NF, Barbier M, Fumoux F, Rihet P. Association analyses of NCR3 polymorphisms with P. falciparum mild malaria. Microbes Infect. (2007) 9:160–6. doi: 10.1016/j.micinf.2006.11.002
28. Brisebarre A, Kumulungui B, Sawadogo S, Atkinson A, Garnier S, Fumoux F, et al. A genome scan for Plasmodium falciparum malaria identifies quantitative trait loci on chromosomes 5q31, 6p21.3, 17p12, and 19p13. Malar J. (2014) 13:198. doi: 10.1186/1475-2875-13-198
29. Baaklini S, Afridi S, Nguyen TN, Koukouikila-Koussounda F, Ndounga M, Imbert J, et al. Beyond genome-wide scan: association of a cis-regulatory NCR3 variant with mild malaria in a population living in the Republic of Congo. PLoS ONE. (2017) 12:e0187818. doi: 10.1371/journal.pone.0187818
30. Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. (2009) 457:557–61. doi: 10.1038/nature07665
31. Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. (2012) 119:2665–74. doi: 10.1182/blood-2011-10-386995
32. Lisovsky I, Isitman G, Song R, Dafonseca S, Tremblay-Mclean A, Lebouche B, et al. A higher frequency of NKG2A+ than of NKG2A- NK cells responds to autologous HIV-infected CD4 cells irrespective of whether or not they coexpress KIR3DL1. J Virol. (2015) 89:9909–19. doi: 10.1128/JVI.01546-15
33. Strauss-Albee DM, Horowitz A, Parham P, Blish CA. Coordinated regulation of NK receptor expression in the maturing human immune system. J Immunol. (2014) 193:4871–9. doi: 10.4049/jimmunol.1401821
34. Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. (2004) 104:3664–71. doi: 10.1182/blood-2004-05-2058
35. Bjorkstrom NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, et al. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med. (2011) 208:13–21. doi: 10.1084/jem.20100762
36. Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. (2013) 121:2678–88. doi: 10.1182/blood-2012-10-459545
Keywords: malaria, Plasmodium falciparum, controlled human malaria infection, NK cells, NKp30, innate immunity
Citation: Walk J and Sauerwein RW (2019) Activatory Receptor NKp30 Predicts NK Cell Activation During Controlled Human Malaria Infection. Front. Immunol. 10:2864. doi: 10.3389/fimmu.2019.02864
Received: 29 May 2019; Accepted: 21 November 2019;
Published: 10 December 2019.
Edited by:
Laurel L. Lenz, University of Colorado School of Medicine, United StatesReviewed by:
Jianzhu Chen, Massachusetts Institute of Technology, United StatesEric O. Long, National Institute of Allergy and Infectious Diseases (NIAID), United States
Copyright © 2019 Walk and Sauerwein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jona Walk, am9uYS53YWxrQHJhZGJvdWR1bWMubmw=; Robert W. Sauerwein, Ui5TYXVlcndlaW5AdHJvcGlxLm5s
 Jona Walk
Jona Walk Robert W. Sauerwein*
Robert W. Sauerwein*