- 1Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services, Bethesda, MD and Detroit, MI, United States
- 2Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, United States
- 3Department of Biochemistry, Microbiology and Immunology, Wayne State University School of Medicine, Detroit, MI, United States
- 4Department of Obstetrics and Gynecology, University of Michigan, Ann Arbor, MI, United States
- 5Department of Epidemiology and Biostatistics, Michigan State University, East Lansing, MI, United States
- 6Center for Molecular Medicine and Genetics, Wayne State University, Detroit, MI, United States
- 7Detroit Medical Center, Detroit, MI, United States
- 8Department of Obstetrics & Gynecology, Florida International University, Miami, FL, United States
- 9Department of Physiology, Wayne State University School of Medicine, Detroit, MI, United States
- 10Division of Obstetrics and Gynecology, Faculty of Medicine, Pontificia Universidad Católica de Chile, Santiago, Chile
- 11Center for Research and Innovation in Maternal-Fetal Medicine (CIMAF), Department of Obstetrics and Gynecology, Sótero del Río Hospital, Santiago, Chile
- 12Department of Computer Science, Wayne State University College of Engineering, Detroit, MI, United States
Pregnancy represents a unique immunological state in which the mother adapts to tolerate the semi-allogenic conceptus; yet, the cellular dynamics in the maternal circulation are poorly understood. Using exon-level expression profiling of up to six longitudinal whole blood samples from 49 pregnant women, we undertook a systems biology analysis of the cellular transcriptome dynamics and its correlation with the plasma proteome. We found that: (1) chromosome 14 was the most enriched in transcripts differentially expressed throughout normal pregnancy; (2) the strongest expression changes followed three distinct longitudinal patterns, with genes related to host immune response (e.g., MMP8, DEFA1B, DEFA4, and LTF) showing a steady increase in expression from 10 to 40 weeks of gestation; (3) multiple biological processes and pathways related to immunity and inflammation were modulated during gestation; (4) genes changing with gestation were among those specific to T cells, B cells, CD71+ erythroid cells, natural killer cells, and endothelial cells, as defined based on the GNF Gene Expression Atlas; (5) the average expression of mRNA signatures of T cells, B cells, and erythroid cells followed unique patterns during gestation; (6) the correlation between mRNA and protein abundance was higher for mRNAs that were differentially expressed throughout gestation than for those that were not, and significant mRNA-protein correlations were observed for genes part of the T-cell signature. In summary, unique changes in immune-related genes were discovered by longitudinally assessing the cellular transcriptome in the maternal circulation throughout normal pregnancy, and positive correlations were noted between the cellular transcriptome and plasma proteome for specific genes/proteins. These findings provide insights into the immunobiology of normal pregnancy.
Introduction
Pregnancy represents a unique immunological state in which the immune system of the mother undergoes adaptations that allow her to tolerate the semi-allogenic conceptus (1–3). Indeed, pregnancy is divided into three different immunological stages based on cytokine profiles (4). Pioneer studies indicated that, while the innate immune system is upregulated to protect the mother against infection and the fetus from rejection (5, 6), the adaptive immune response toward paternal/fetal antigens seems to be selectively suppressed [i.e., driven toward a T-helper (Th)2-like phenotype] (7–11). Specifically, the cellular components of the innate immune system in the maternal systemic circulation are activated as evidenced by increased numbers of monocytes and granulocytes (12, 13). Such innate immune cells display an activated phenotype, comparable to that observed in women with sepsis (13), and exhibit enhanced functionality (phagocytosis, respiratory burst activity, and cytokine production) (14–18). The humoral components of the innate immune system are also boosted during pregnancy (5). For example, complement components and acute phase proteins are increased in the circulation of pregnant women (19–24). In contrast to the innate immune system, the cellular (e.g., T cells and B cells) and humoral (e.g., antibodies) components of the adaptive immune system in the maternal circulation during normal pregnancy have received less attention.
The systemic intravascular inflammatory response during normal pregnancy is especially activated in women who experience the physiological process of labor at term (18, 25) and in those who undergo pregnancy complications such as preterm labor (18, 26, 27), preterm premature rupture of membranes (28), and preeclampsia (13, 17, 29–32). Therefore, the systemic immune response reflects both physiological and pathological processes, and the early detection of these changes may lead to the discovery of non-invasive biomarkers for obstetrical disease.
Herein, for the first time, we aimed to provide a roadmap of the modulations in the cellular transcriptome in maternal circulation during normal pregnancy. In addition, we assessed whether gestational mRNA changes of the cellular transcriptome correlate to those of the plasma proteome during normal pregnancy.
Materials and Methods
Study Design
We conducted a prospective longitudinal study that enrolled women attending the Center for Advanced Obstetrical Care and Research of the Perinatology Research Branch, NICHD/NIH/DHHS; the Detroit Medical Center, and Wayne State University School of Medicine. Based on this cohort, we designed a retrospective study that included 49 women with normal pregnancy who delivered at term and had 4–6 blood samples collected throughout gestation [median number of samples = 5, interquartile range (IQR) = 5–6] (n = 282). Blood samples were collected at the time of a prenatal visit, scheduled at 4-week intervals from the first or early second trimester until delivery in the following gestational age intervals: 8- <16, 16- <24, 24- <28, 28- <32, 32- <37, and >37 weeks. All patients provided written informed consent and the use of biological specimens, as well as clinical and ultrasound data, for research purposes were approved by the Institutional Review Boards of Wayne State University and NICHD. All experiments were performed in accordance with relevant guidelines and regulations.
RNA Extraction
RNA was isolated from PAXgene® Blood RNA collection tubes (BD Biosciences, San Jose, CA; Catalog #762165), as described in the PAXgene® Blood miRNA Kit Handbook. Purified RNA was quantified by UV spectrophotometry using the DropSense96® Microplate Spectrophotometer (Trinean, Gentbrugge, Belgium), and quality was assessed by microfluidics using the RNA ScreenTape on the Agilent 2200 TapeStation (Agilent Technologies, Wilmington, DE, USA).
Microarray Analysis
RNA was processed and hybridized to GeneChip™ Human Transcriptome Arrays 2.0 (P/N 902162) according to the Affymetrix GeneChip™ WT Pico Reagent Kit Users Guide (P/N 703262 Rev. 1) as follows: Biotinylated cDNA were prepared from 20–50 ng total RNA. Labeled cDNA were hybridized to the arrays in a GeneChip™ Hybridization Oven 640 by rotating at 60 rpm, 45°C for 16 h. Arrays were then washed and stained in the Affymetrix Fluidics Station 450 and scanned using the Affymetrix 3000 7G GeneChip™ Scanner with Autoloader. Raw intensity data were generated from array images using the Affymetrix AGCC software.
Data Analysis
Preprocessing
Affymetrix Human Transcriptome Arrays CEL files were preprocessed using Robust Multi-array Average (RMA) (33) implemented in the oligo package (34) and annotation from the hta20sttranscriptcluster.db package of Bioconductor (35). Since samples were profiled in several batches as a part of a larger study, correction for potential batch effects was performed using the removeBatchEffect function of the limma package in Bioconductor. After batch effect correction, data from the sample collected at the time of labor from the 21 women who had spontaneous term labor were removed to avoid confounding gestational age-related changes with those due to the onset of labor at term. The final analysis set of 261 transcriptomes was used in downstream analyses described below.
Expression Calling
Transcript clusters (typically one or two per unique gene) were deemed present in a given sample if one of its probesets (targeting a specific exon) was expressed above background (p-value for expression above background pDBAG <0.05) determined using the Transcriptome Analysis Console (version 4.0) (ThermoFisher Scientific). Genes were retained if deemed present in >25% of the 261 samples.
Differential Expression
Expression profiles were visually inspected to determine consistency of the data in sequential samples collected from the same woman. One of 261 samples consistently had the lowest value for a large fraction of the genes and was deemed as outlier and removed from further analysis. Linear mixed-effects models (36) were then used to fit log2 gene expression data as a function of gestational age (continuous) and included cubic polynomial terms of gestational age as fixed effects and a random intercept term for each woman. Significance p-values for the association of gene expression and gestational age were determined using likelihood ratio tests between a model with and without gestational age terms. A False Discovery Rate adjusted p-value (q-value) <0.1 and a fold change (FC) of >1.25 were required for significance. Fold change was determined as the ratio of the highest vs. lowest average expression from 10 to 40 weeks of gestation. Linear mixed-effects models were fit using the lmer function, while the likelihood ratio tests were performed using the anova function available in the lme4 R package (36).
Gene Ontology and Pathway Analysis
Gene ontology and pathway analysis was conducted using a hypergeometric test on Gene Ontology (GO) (37) and Developmental FunctionaL Annotation at Tufts (DFLAT) databases (38), as well as on Curated Gene Sets (C2) collection from the Molecular Signatures Database (MSigDB) database (39). In addition, enrichment tests were performed for tissue specificity and chromosomal locations of genes. Tissue-specific genes were defined as those with median expression 30 times higher in a given tissue than the median expression of all other tissues documented in the Gene Atlas (40) as previously described (41).
Unless otherwise stated, all enrichment analyses were based on a hypergeometric test and accounted for multiple testing with q < 0.05 being considered a significant result. In all enrichment analyses, the background gene list was defined as the compendium of genes deemed present in >25% of the samples.
Changes in Cell-Type Specific mRNA Signatures With Gestational Age
In this analysis, we tested whether previously reported cell-type specific mRNA signatures derived by single-cell RNA-Seq studies of placenta tissues (42) were modulated with advancing gestation in normal pregnancy. The 13 cell types identified by Tsang et al. (42) were: B cells, T cells, monocytes, cytotrophoblasts, syncytiotrophoblast, decidual cells, dendritic cells, endothelial cells, erythrocytes, Hofbauer cells, stromal cells, vascular smooth muscle cell, and extravillous trophoblasts. The mRNA signatures for these cell types were first quantified in each patient sample by averaging expression data over genes part of each signature. Before averaging, the data for each gene was first standardized by subtracting the mean and dividing by standard deviation of expression across term samples (>37 weeks). Cell-type specific expression averages were then fit as a function of gestational age using linear mixed-effects models, as described above for the analysis of data of individual genes.
Assessment of mRNA Protein Correlations
Maternal plasma abundance of 1,125 proteins in 71 samples collected from 16 of the women included in the current study were obtained from the S1 File of Erez et al. (43). The correlation between each mRNA and corresponding protein pair was assessed by fitting linear mixed-effects models with the response being the protein abundance and the predictor being the mRNA expression. These models included a random intercept term to account for the repeated observations from the same subject. The meaning of the mRNA coefficient in this model is change in log2 protein abundance for one unit change in log2 gene expression. The significance of the protein—mRNA correlation was assessed by the t-score for the regression line slope, and false discovery rate adjustment of resulting p-values was performed across all mRNA-protein pairs that were tested. A q-value <0.1 was considered a significant result.
Results
Longitudinal Patterns of the Cellular Transcriptome Throughout Normal Pregnancy
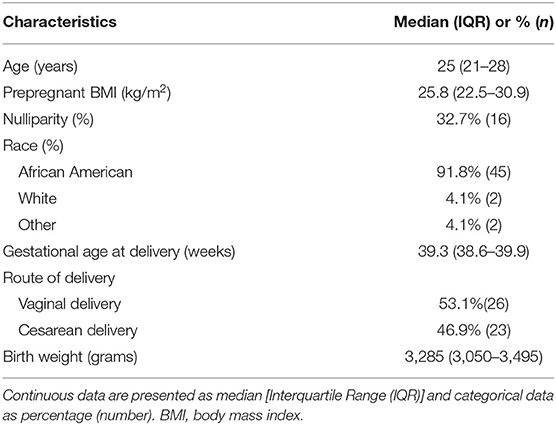
The mRNA profiles of longitudinal maternal blood samples were determined at exon level resolution by microarrays. The characteristics of the study population are shown in Table 1. A total of 26,458 protein-coding mRNA transcript clusters were expressed above background levels in at least 25% of the samples, as were 5,706 non-coding RNA transcript clusters. Analysis of longitudinal expression patterns identified 614 transcript clusters (510 coding and 104 non-coding) with significant expression modulation during gestation (q < 0.1 and minimum fold change of 1.25) (Supplementary File 1, Supplementary Figure 1). Significant transcripts were found on all chromosomes; yet, more differentially expressed transcripts than expected were observed on chromosome 14 (51/614 transcript clusters, odds ratio = 3.5, p < 0.0001; Figure 1), with 28/51 differentially expressed genes on this chromosome being annotated to immune processes. Chromosome 14 includes genes of critical importance for immunity (44); therefore, these data show that pregnancy has a strong effect on the maternal immune system.
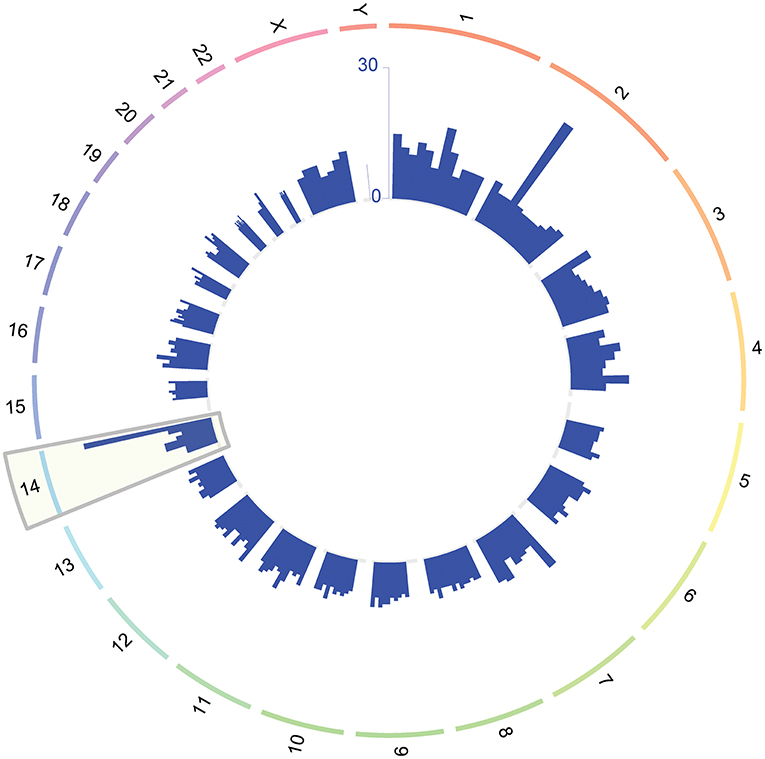
Figure 1. Chromosomal location of genes modulated throughout normal pregnancy. The outer circle represents the different chromosomes while the inner histograms show the number of differentially expressed genes binned by the genomic location within each chromosome. Chromosome 14 was the most enriched in differentially expressed genes throughout normal pregnancy (gray rectangle).
To define clusters of expression trajectories during gestation, we focused on 112 of the 614 significant transcript clusters that changed more than 1.5-fold from 10 to 40 weeks of gestation. Three distinct clusters of expression modulation emerged: genes that (1) steadily increased throughout gestation (89 genes; Figure 2, red cluster), (2) steadily decreased throughout gestation (12 genes; Figure 2, green cluster), and (3) decreased prior to mid-gestation followed by an increase (11 genes; Figure 2, blue cluster). These results indicate that the expression of the most highly modulated genes increases with advancing gestational age.
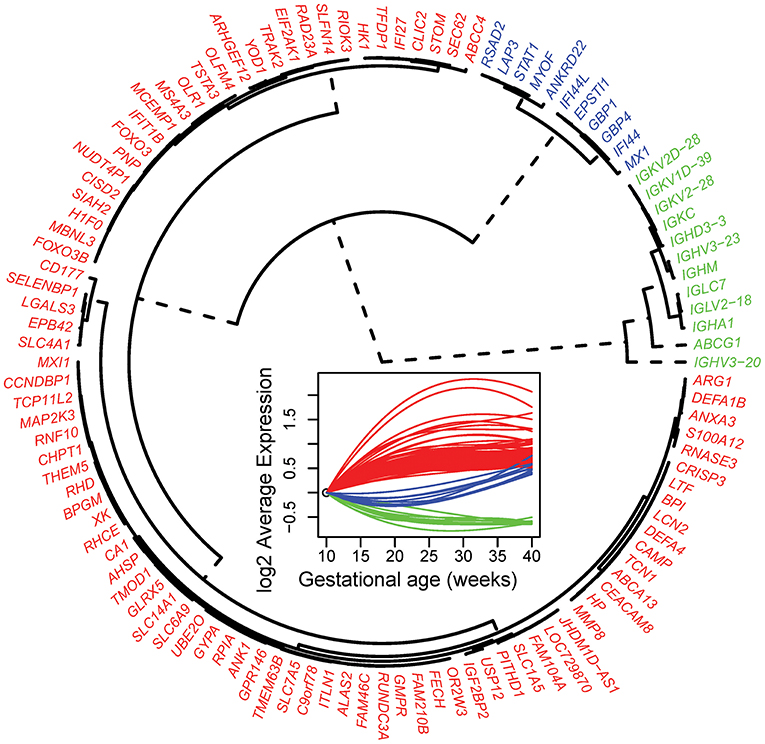
Figure 2. Clustering of average gene expression profiles throughout normal pregnancy. Average profiles of genes that change throughout normal pregnancy and have a fold change >1.5 were clustered using hierarchical clustering. The distance metric used in the clustering was 1-Pearson correlation. Three clusters were identified: Cluster 1 (red, 89 genes), Cluster 2 (green, 12 genes), and Cluster 3 (blue, 11 genes). Note that, in this figure, the average gene expression profiles vs. gestational age were reset so that their value is 0 at 10 weeks of gestation.
Of note, the 19 mRNA transcript clusters (corresponding to 16 unique genes) that changed more than 2-fold in expression during pregnancy all increased from 10 to 40 weeks of gestation (Figure 3, gray lines correspond to individual pregnancies and blue lines show the average expression). The expression of these 16 genes increased from early to late pregnancy and tended to plateau near term, with the exception of 2 genes (interferon-induced protein 27 and 44-like) (Figure 3). Several of these most highly modulated genes are related to host immune response (e.g., MMP8, DEFA1B, and DEFA4) (45, 46), again emphasizing the immune response adaptations during normal pregnancy.
Figure 3. Genes changing in expression >2-fold from 10 to 40 weeks of gestation. Gray lines represent log2 normalized gene expression in 4–6 samples for each of the 49 women. Blue lines correspond to the average expression determined by a polynomial fit by linear mixed-effects models.
Biological Processes, Pathways, and Immune Cell Signatures Associated With Advancing Gestation in Normal Pregnancy
We performed gene ontology enrichment analysis to interpret the changes in gene expression occurring throughout gestation. We identified 157 biological processes modulated during gestation, which included cellular and humoral immunity, defense response, response to external biotic stimulus (e.g., bacteria and viruses), regulation of lipid storage, interleukin-1beta production and secretion, and erythrocyte development, among others (Table 2). An additional 134 biological processes altered during gestation were found when querying the Developmental FunctionaL Annotation at Tufts (DFLAT) database, such as stress response, immune system development, cytokine response, and regulation of angiogenesis (Table 3).
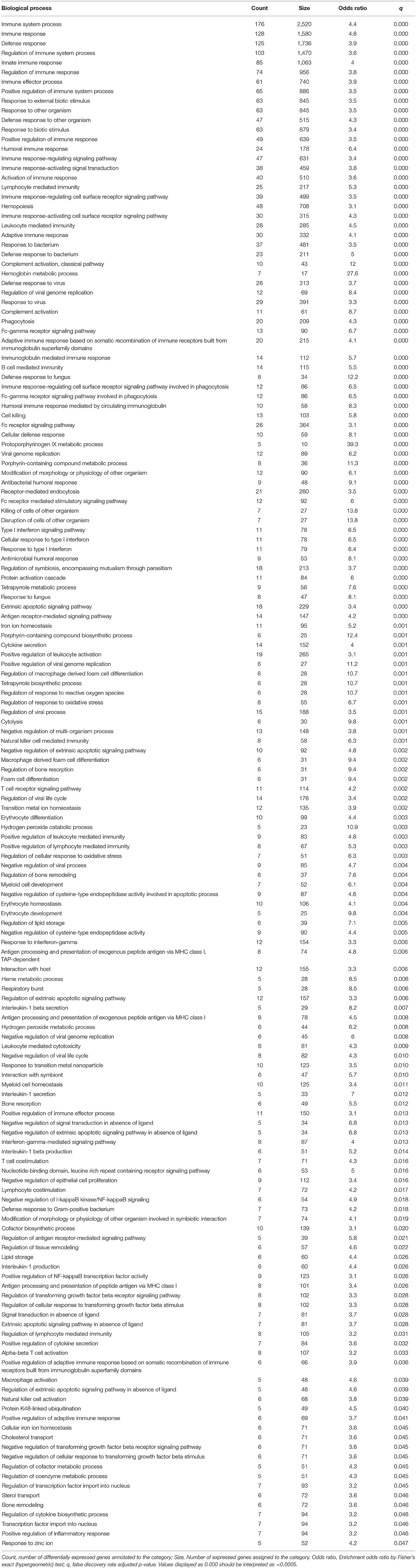
Table 2. Gene ontology biological processes enriched in genes differentially expressed with gestational age.

Table 3. DFLAT biological processes enriched in genes differentially expressed with gestational age.
Enrichment analyses were then expanded to canonical pathways and gene sets from the Molecular Signatures Database (MSigDB), and 53 such pathways were found to be associated with advancing gestation. These included the Reactome database (47) pathways: immune system, adaptive immune system, cytokine signaling in immune system, and immunoregulatory interactions between a lymphoid cell and a non-lymphoid cell, as well as the KEGG database (48) pathways: natural killer cell-mediated cytotoxicity, antigen processing and presentation, and graft vs. host disease (Table 4).
We then aimed at determining the origin of observed transcriptional activity by using the GNF Gene Expression Atlas to define genes predominantly expressed in specific human tissue or cell types, as previously described (41). This analysis revealed that gene sets specific to CD4+ and CD8+ T cells, CD71+ erythroid cells, CD105+ endothelial cells, and CD56+ NK cells, among others, were over-represented among the mRNAs that were modulated during gestation (q < 0.05) (Table 5). In addition, genes reported to be specific to fetal organs (liver, lung, and brain) and the placenta were also enriched among significant genes (q < 0.05) (Table 5). These data show that maternal and fetal cell-specific transcripts found in the maternal circulation are being modulated with advancing gestation.
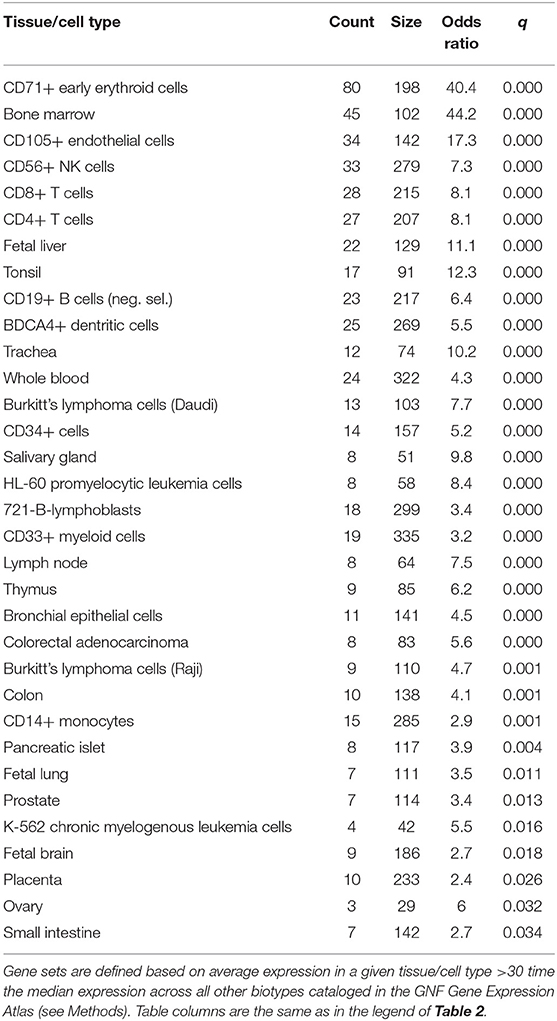
Table 5. Tissue or cell type-specific gene sets enriched in genes differentially expressed with gestational age.
The average abundance of cell type-specific gene sets recently defined using single-cell transcriptomics (42) were also analyzed for systematic changes with gestational age at blood draw in our cohort. This analysis revealed that expression of mRNAs specific to three cell subtypes were dynamically altered throughout normal pregnancy: (1) the T-cell-specific mRNA signature decreased from the first to second trimester, followed by a subsequent increase during the third trimester (Figure 4A); (2) the B-cell-specific mRNA signature decreased steadily throughout gestation (Figure 4B); and (3) the expression of genes specific to nucleated erythroid cells (HBZ, ALAS2, and AHSP) significantly increased as gestation progressed (Figure 4C). These findings demonstrate, for the first time, that single-cell RNA-Seq-derived signatures of erythroid cells change throughout normal gestation in maternal whole blood, while trends found for T cells and B cells were similar to those reported in whole blood (49) and by using cell-free RNA analysis (42).
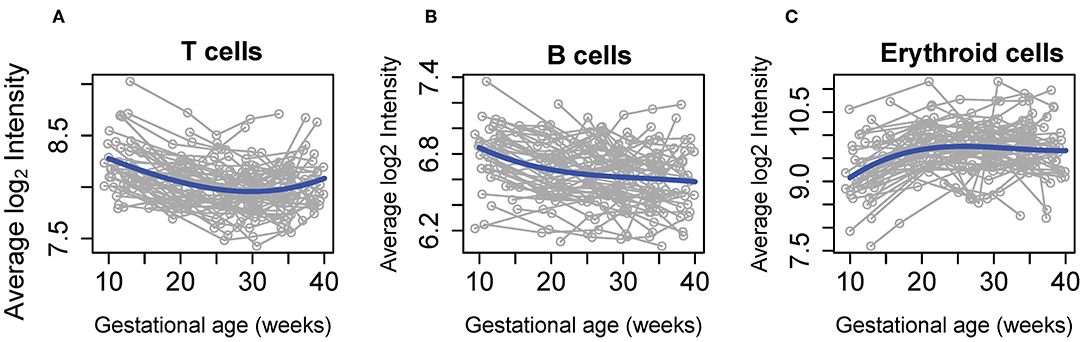
Figure 4. Meta-gene expression of specific cell types differentially regulated throughout normal pregnancy. The average expression of genes defined as specific for (A) T cell, (B) B cell, and (C) erythroid cell populations by Tsang et al. (42) are shown as a function of gestation. Blue lines correspond to the average expression estimated by linear mixed-effects models. The fold change in expression from 10 to 40 weeks was 1.2 for T cells and B cells and 1.6 for erythroid cells (all, p < 0.001).
Correlation Between the Cellular Transcriptome and Plasma Proteome Throughout Normal Pregnancy
Transcription does not always correlate with protein translation (50, 51). Therefore, we investigated the correlation between the mRNAs that were modulated throughout gestation and their corresponding protein abundance. Maternal plasma abundance of 1,125 proteins in 71 samples collected from 16 of the women included in the current study was previously reported (43, 52). First, we assessed the mRNA-protein correlation for 53 of the 614 transcript clusters that changed throughout gestation and for which abundance data for the corresponding protein were available. These correlations were compared to those of 1,011 mRNA-protein pairs that did not change with gestation. The mRNA-protein correlations were significantly higher for transcripts that changed throughout gestation compared to those that did not (Wilcoxon test for comparing t-scores of the linear regression slope obtained by linear mixed-effects models for each mRNA-protein pair, p = 0.01) (Figure 5). Among the 53 transcripts that changed throughout gestation, BPI, IGHG1, CXCL10, GNLY, and GZMA had a significant mRNA-protein correlation as assessed by both linear mixed-effects models and a naïve Spearman correlation test (q < 0.05 for both analyses) (Figure 6). Notably, two of these genes (GNLY and GZMA) were included in the T-cell-specific mRNA signature that was modulated overall throughout gestation.
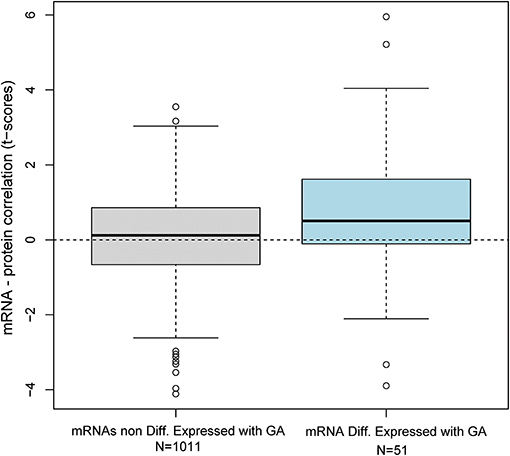
Figure 5. Distribution of mRNA-protein correlation t-scores. The correlation between mRNA and protein abundance was assessed by linear mixed-effects models using data collected from 71 samples provided by 16 women. The distribution of t-scores for the linear correlation slope is shown for 51 mRNAs differentially expressed with gestation and 1011 mRNAs that were not differentially expressed.
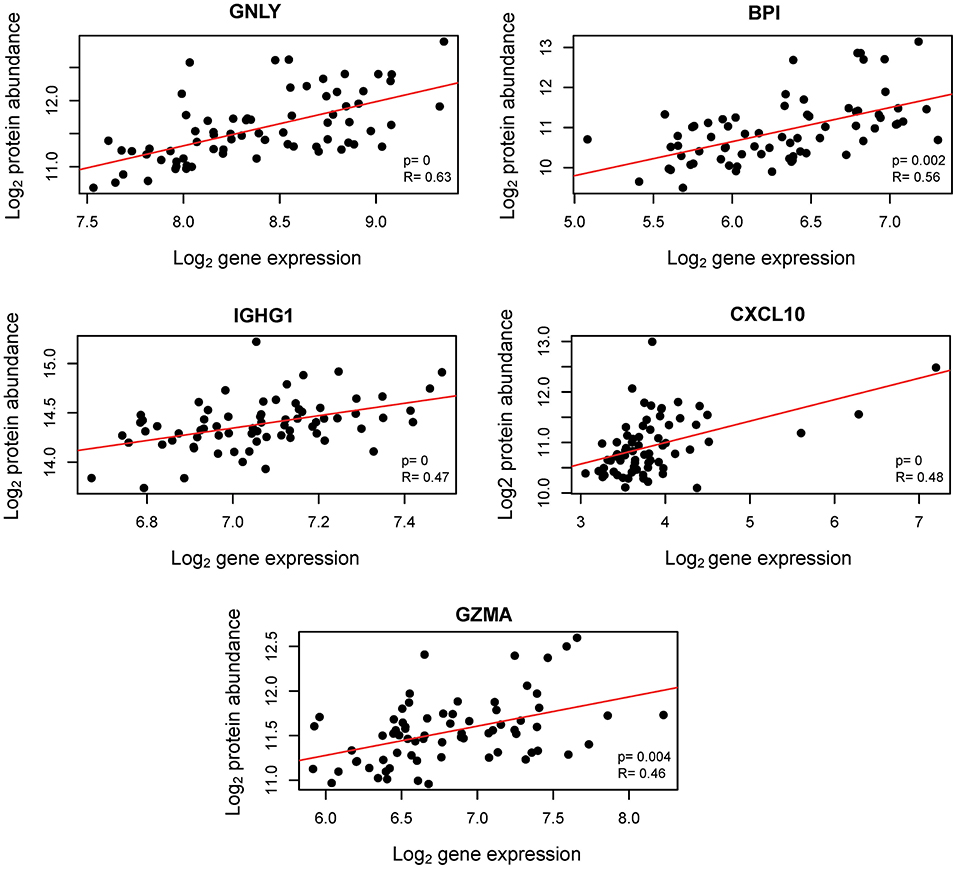
Figure 6. Correlation between cellular transcriptome and maternal plasma proteome throughout normal pregnancy. Aptamer-based protein abundance measurements are plotted against mRNA expression. R: naïve Spearman correlation coefficient; p: likelihood ratio test p-value from linear mixed-effects models assessing the linear correlation accounting for repeated measurements from the same subjects.
Discussion
Principal Findings of the Study
(1) Chromosome 14 was the most enriched in transcripts differentially expressed throughout normal pregnancy (51/613 mRNA clusters). (2) Among the most differentially expressed genes (q < 0.1, and fold change > 1.5), three distinct longitudinal patterns were observed: (i) steady increase throughout gestation (89 genes), (ii) steady decrease throughout gestation (12 genes), or (iii) decrease prior to mid-gestation followed by an increase (11 genes). (3) Sixteen genes, most of them related to host immune response mediators (e.g., MMP8, DEFA1B, DEFA4, LTF), displayed >2-fold change in expression and steadily increased from 10 to 40 weeks of gestation. (4) Approximately 300 biological processes and 53 pathways, many of which were related to immunity and inflammation, were enriched among the differentially expressed genes (q < 0.05). (5) Genes changing with gestation were among those specific to T cells, B cells, CD71+ erythroid cells, natural killer cells, and endothelial cells, as defined based on the GNF Gene Expression Atlas. (6) The meta-gene expression of mRNA signatures for T cells, B cells, and erythrocyte cells were significantly modulated throughout gestation, each following a unique pattern (p < 0.05). (7) The correlation between mRNA and protein abundance was higher for mRNAs that were differentially expressed throughout gestation than for those that were not (p = 0.01). (8) Significant and positive mRNA-protein correlations (q < 0.05) were observed for BPI, IGHG1, CXCL10, and two members of the T-cell mRNA signature (GNLY, GZMA). The expression trends and variability in expression of individual genes and meta-genes in normal pregnancy (nomograms) derived herein will be the basis for future studies aiming at developing biomarkers for obstetrical disease.
Transcriptomic Changes During Pregnancy
Previous studies have investigated the cellular (53, 54) and cell-free (55) transcriptome in the maternal circulation at different time points during normal pregnancy using either 3-prime-end biased microarrays or targeted approaches. The current study, however, is the first to quantify at exon-level resolution the cellular transcriptome during normal pregnancy in up to six samples per pregnancy. More than one-half (54%, 277/514) of the unique differentially expressed genes identified herein were also among the 2,321 genes (q < 0.1) reported by Heng et al. (54) to change from 17–23 to 27–33 weeks of gestation. Similarly, 47% (242/514) of the genes found in this study were among the 3,830 genes reported by Al-Garawi et al. (53) as changing from 10–18 to 30–38 weeks. The overlap between the genes reported as differentially expressed in these two studies and those identified herein is significant (Fisher's exact test p < 0.0001 for both). However, unlike in the two previous studies involving a pair of samples from each woman, the availability of four to six longitudinal samples per patient in this study enabled us to capture more complex expression trajectories in the window of 10–40 weeks of gestation, and to identify distinct clusters of such gene expression trajectories.
Compared to another recent study by Ngo et al. (55) that involved more frequent sampling than used herein, our study has the advantage of an unbiased assessment of the whole cellular transcriptome as opposed to a targeted assessment of genes that are placenta-, immune-, and fetal liver-specific. Of note, among the 14 immune-specific cell-free mRNAs selected by Ngo et al. (55) as best predictors of gestational age at blood draw, 11 were also identified in our study, with CEACAM8, DEFA4, LTF, and MMP8 being among those with highest fold change. Although our results are somewhat consistent with those reported by Ngo et al. (55), it should be noted that cellular and cell-free transcripts can follow different patterns in similar physiological and pathological processes (56).
Correlations Between the Cellular Transcriptome and the Plasma Proteome Throughout Normal Pregnancy
The finding that the maternal transcriptome features inflammation-related processes and pathways that are being activated in preparation for labor at term is in agreement with our previous studies in gestational tissues [cervix (57), myometrium (58), membranes (59)] and a similar longitudinal study of the maternal plasma proteome (52). In addition to finding several common biological processes that are modulated in both the maternal plasma proteome and cellular transcriptome (such as defense response, defense response to bacterium, defense response to fungus, regulation of bone resorption, leukocyte migration) we assessed, for the first time, the extent of the agreement in whole blood mRNA and protein changes with gestation in the same set of samples. Although the correlations between mRNA and protein expression reported in the literature are notoriously poor, recent studies showed that mRNA-protein correlation is higher for mRNAs that are differentially expressed in a given condition than for those that are not (51). Our finding that the correlation of mRNA-protein pairs is higher for transcripts changing with gestation than those who do not is therefore consistent with previous observations (51).
T Cells in the Maternal Circulation During Normal Pregnancy
Maternal T cells are implicated in the physiological processes occurring throughout gestation (60–63). Effector and activated T cells are found at the maternal-fetal interface before (64–70) and during (71–73) spontaneous labor at term, and these cells are associated with the timing of term parturition (74). Effector T cells are also found in the maternal blood prior to Shah et al. (75) and during (76) labor at term. In the current study, we demonstrated that the T-cell-specific mRNA expression in the maternal circulation was decreased prior to mid-gestation but upregulated from mid-gestation until term. Moreover, for two of 19 genes of this signature, there was a significant correlation between cellular mRNA and plasma proteomic profiles; this is consistent with recent cytomic and proteomic studies in the maternal circulation (77, 78). In addition, we recently showed the same u-shaped pattern of expression for the T-cell mRNA signature during gestation in a smaller set of patients profiled with RNA-Seq and qRT-PCR platforms (49). Together, these findings illustrate the importance of maternal T cells during normal pregnancy.
Alteration of systemic T-cell populations has also been implicated in preterm parturition (79–81), especially since aberrant activation of these cells can induce the onset of preterm labor (82, 83). On the other hand, the absence of T cells in a mouse model caused an increased susceptibility to endotoxin-induced preterm birth, which was reversed by adoptive transfer of CD4+ T cells (84). From a histopathological standpoint, T cells are detected in placental lesions related to maternal anti-fetal rejection such as villitis of unknown etiology (85–87), chronic chorioamnionitis (88), and chronic deciduitis (89), which have also been linked to the onset of term and preterm labor (88, 90–94). The chronic nature (95, 96) of these lesions has led our group to propose them as indicators of maternal anti-fetal rejection, which can lead to preterm labor or even fetal death (86, 90, 93, 94, 97–100). Future studies are needed to establish whether the early detection of T-cell alterations in the maternal circulation may identify pregnancies at risk for obstetrical disease such as preterm labor/birth and fetal death.
B Cells in the Maternal Circulation During Normal Pregnancy
Several studies have suggested a role for B cells in the maintenance and success of pregnancy (101–106). Circulating CD5+ (B1) B cells were shown to decrease during pregnancy, only returning to normal levels after parturition (107). This finding was later shown to occur in mice, where a decreased influx of newly generated B cells to the blood and spleen was observed while mature B cells were increased in uterine-draining lymph nodes (108). An expansion of marginal zone B cells also ensued (108, 109), which was proposed to participate in the production of protective antibodies during pregnancy (109, 110). Accordingly, maternal serum antibody concentrations increased concomitantly with B-cell population changes (109), possibly as a result of the anti-inflammatory microenvironment maintained at the maternal-fetal interface throughout most of pregnancy (111). Such antibody production has been considered the primary contribution of B cells to maternal-fetal tolerance during pregnancy (101).
Interleukin-10-producing regulatory B cells (Bregs) have also been described as important players during pregnancy (112, 113). Such adaptive immune cells increased in normal pregnancy in an hCG-dependent manner (113, 114) and suppressed effector T-cell cytokine production (113). Trophoblast cells facilitated the conversion of IL10-deficient B cells into IL10-expressing B cells (114), which is in line with a previous report showing that the adoptive transfer of Bregs restored maternal-fetal tolerance (112). Indeed, pregnant women treated with the B-cell-depleting treatment rituximab had a higher occurrence of pregnancy loss (115), although further investigation of this phenomenon is warranted (116).
In the current study, we showed that the B-cell-specific mRNA signature moderately decreased throughout pregnancy. Our observations correspond to a previous report indicating that the majority of maternal peripheral B-cell subsets are reduced in late gestation compared to the non-pregnant state (117), whereas Bregs are upregulated (117). The combined effects of such dynamic changes on the overall circulating B-cell mRNA signature are therefore minimal, as we have demonstrated here. Taken together, these findings suggest that, while total maternal peripheral B cells are mostly maintained, subset-specific changes occur throughout pregnancy.
Erythroid Cells in the Maternal Circulation During Normal Pregnancy
A constant bi-directional trafficking of maternal and fetal cells occurs during normal pregnancy (118–124). Indeed, cell-free fetal DNA is present in the maternal circulation throughout normal pregnancy (125–132) and its levels increased from mid to late gestation (128, 133–140). Increased concentrations of cell-free fetal DNA or numbers of fetal cells in the maternal circulation (fetal microchimerism) have been linked to pregnancy complications such as preterm labor (141–145), preeclampsia (146–150), and intrauterine growth restriction (149, 151, 152). In addition, sequencing cell-free fetal DNA in the maternal circulation may serve for non-invasive prenatal testing (153). Therefore, determining the impact that fetal cells and their released products (e.g., cell-free fetal DNA) may have on maternal health is critical, given that such cells can remain in the maternal circulation for decades (153, 154).
Fetal nucleated erythroid cells have been detected in the maternal blood (121, 155, 156) where they may be a source of cell-free fetal DNA (155). Previous reports showed that neonatal CD71+ erythroid cells have immunomodulatory functions on cord blood leukocytes (157–159), and that their direct contact with maternal peripheral immune cells increases the secretion of pro-inflammatory mediators by such cells (159). Therefore, it is likely that the trafficking of CD71+ erythroid cells from the fetus into the mother directly affects maternal immune responses (159). Nucleated erythroid cells have also been described in the placenta, where their presence is correlated with the number of such cells in the cord blood (160), and these cells also display immunomodulatory properties in vitro (161).
Herein, we showed that the erythroid cell-specific mRNA signature was upregulated throughout gestation in the maternal circulation. This finding is in line with previous reports showing that fetal microchimerism increases during pregnancy (162, 163). In addition, a recent study showed that CD71+ erythroid cells are increased in the maternal circulation throughout gestation, peaking during the third trimester and falling to baseline levels after delivery (164). Together, these findings illustrate that erythroid cells, most likely of fetal origin, are present in the maternal circulation and their transcriptome is modulated as gestation progresses. These data provide a possible mechanism whereby the developing fetus can modulate maternal immunity.
Conclusion
We have reported herein a detailed characterization of the longitudinal maternal whole blood transcriptomic changes in normal pregnancy. We have shown that these changes are genome-wide, yet we found that chromosome 14 was particularly enriched in genes modulated with advancing gestation. There was a significant overlap in expression changes described herein with those previously reported in whole blood analyses based on only two time points, while some of the most strongly modulated mRNAs identified herein were also previously reported as the best predictors of gestational age in cell-free RNA analyses of maternal blood. Our systems biology approach to the interpretation of these expression changes in the maternal cellular transcriptome during pregnancy revealed significant longitudinal patterns of expression for immune-related gene sets, such as those specific to T cells, B cells, and erythroid cells. Moreover, for the first time, we demonstrated positive correlations between the cellular transcriptome and plasma proteome for specific genes, including those expressed by T cells. The expression trajectories of protein coding and non-coding transcripts in normal pregnancy described herein may serve as references and hence enable the discovery of biomarkers for obstetrical disease.
Data Availability Statement
The raw and summarized microarrays gene expression data are available as a Gene Expression Omnibus series (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121974).
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Boards of Wayne State University and NICHD. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AT, RR, SH, and NG-L conceived the research. SH and RR supervised the enrollment of the patients and collection of samples. AT and NG-L carried out research and drafted the manuscript. GB contributed to data visualization and to the preparation of data submission to the Gene Expression Omnibus. AT analyzed data. AT, RR, and NG-L interpreted the data. SH, RR, PP, JK, and SB provided feedback on the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported, in part, by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS); and in part, with Federal funds from NICHD/NIH/DHHS under Contract No. HHSN275201300006C. AT and NG-L were also supported by the Perinatal Initiative of the Wayne State University School of Medicine. RR has contributed to this work as part of his official duties as an employee of the United States Federal Government.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor is currently co-organizing a Research Topic with one of the authors, NG-L, and confirms the absence of any other collaboration.
Acknowledgments
We thank Dan Lot and Dr. Susan Land for conducting the RNA extraction at the Applied Genomics Technology Center of Wayne State University in Detroit, Michigan. We acknowledge Dr. Christopher Krebs for conducting the microarray experiments at the DNA Sequencing Core at the University of Michigan in Ann Arbor, Michigan.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02863/full#supplementary-material
Supplementary File 1. Differential gene expression with gestational age. Each row corresponds to one of the 614 transcript clusters associated with gestational age. ID, Affymetrix transcript cluster identifier; SYMBOL, gene symbol; ENTREZ, Entrez gene database identifier; Name, Gene name; mRNA, mRNA identifier; Source, mRNA identifier source; locus.type, coding vs. non-protein coding assignment; Chromosome, chromosome number; strand, chromosome strand; start/stop, genomic coordinate start and stop for the transcript cluster; p-value, significance value for the polynomial relation between log gene expression and gestational age; FC, log2 fold change representing the log2 ratio of the highest and lowest fitted value from 10 to 40 weeks of gestation (see Figure 3). The sign is positive if value at 40 weeks is higher than the value at 10 weeks, and negative otherwise. adj.P.Value, False discovery adjusted p-value.
Supplementary Figure 1. Longitudinal gene expression profiles of genes associated with gestational age. Each figure shows data for one of the 614 transcript clusters associated with gestational age. The y-axis represents the log2 normalized gene expression, while the x-axis represents gestational age (weeks). Each line corresponds to one patient and each dot to one sample. The thick blue line represents the linear mixed effects model fit. The title in each plot, represents the transcript cluster identifier, gene symbol, gene name, p-value, and fold change. The meanings of these annotations are as in the legend of Supplementary File 1.
References
1. Medawar PB. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. Symp Soc Exp Biol. (1953) 7:320–8.
2. Wegmann TG, Lin H, Guilbert L, Mosmann TR. Bidirectional cytokine interactions in the maternal-fetal relationship: is successful pregnancy a TH2 phenomenon? Immunol Today. (1993) 14:353–6. doi: 10.1016/0167-5699(93)90235-D
3. Bonney EA, Matzinger P. The maternal immune system's interaction with circulating fetal cells. J Immunol. (1997) 158:40–7.
4. Mor G, Cardenas I. The immune system in pregnancy: a unique complexity. Am J Reprod Immunol. (2010) 63:425–33. doi: 10.1111/j.1600-0897.2010.00836.x
5. Sacks G, Sargent I, Redman C. An innate view of human pregnancy. Immunol Today. (1999) 20:114–8. doi: 10.1016/S0167-5699(98)01393-0
6. Sacks G, Sargent I, Redman C. Innate immunity in pregnancy. Immunol Today. (2000) 21:200–1. doi: 10.1016/S0167-5699(00)01615-7
7. Lin H, Mosmann TR, Guilbert L, Tuntipopipat S, Wegmann TG. Synthesis of T helper 2-type cytokines at the maternal-fetal interface. J Immunol. (1993) 151:4562–73.
8. Tafuri A, Alferink J, Moller P, Hammerling GJ, Arnold B. T cell awareness of paternal alloantigens during pregnancy. Science. (1995) 270:630–3. doi: 10.1126/science.270.5236.630
9. Marzi M, Vigano A, Trabattoni D, Villa ML, Salvaggio A, Clerici E, et al. Characterization of type 1 and type 2 cytokine production profile in physiologic and pathologic human pregnancy. Clin Exp Immunol. (1996) 106:127–33. doi: 10.1046/j.1365-2249.1996.d01-809.x
10. Ekerfelt C, Matthiesen L, Berg G, Ernerudh J. Paternal leukocytes selectively increase secretion of IL-4 in peripheral blood during normal pregnancies: demonstrated by a novel one-way MLC measuring cytokine secretion. Am J Reprod Immunol. (1997) 38:320–6. doi: 10.1111/j.1600-0897.1997.tb00307.x
11. Ekerfelt C, Matthiesen L, Berg G, Ernerudh J. Th2-deviation of fetus-specific T cells. Immunol Today. (1999) 20:534. doi: 10.1016/S0167-5699(99)01511-X
12. Efrati P, Presentey B, Margalith M, Rozenszajn L. Leukocytes of normal pregnant women. Obstet Gynecol. (1964) 23:429–32.
13. Sacks GP, Studena K, Sargent K, Redman CW. Normal pregnancy and preeclampsia both produce inflammatory changes in peripheral blood leukocytes akin to those of sepsis. Am J Obstet Gynecol. (1998) 179:80–6. doi: 10.1016/S0002-9378(98)70254-6
14. Koumandakis E, Koumandaki I, Kaklamani E, Sparos L, Aravantinos D, Trichopoulos D. Enhanced phagocytosis of mononuclear phagocytes in pregnancy. Br J Obstet Gynaecol. (1986) 93:1150–4. doi: 10.1111/j.1471-0528.1986.tb08636.x
15. Shibuya T, Izuchi K, Kuroiwa A, Okabe N, Shirakawa K. Study on nonspecific immunity in pregnant women: increased chemiluminescence response of peripheral blood phagocytes. Am J Reprod Immunol Microbiol. (1987) 15:19–23. doi: 10.1111/j.1600-0897.1987.tb00144.x
16. Naccasha N, Gervasi MT, Chaiworapongsa T, Berman S, Yoon BH, Maymon E, et al. Phenotypic and metabolic characteristics of monocytes and granulocytes in normal pregnancy and maternal infection. Am J Obstet Gynecol. (2001) 185:1118–23. doi: 10.1067/mob.2001.117682
17. Germain SJ, Sacks GP, Sooranna SR, Sargent IL, Redman CW. Systemic inflammatory priming in normal pregnancy and preeclampsia: the role of circulating syncytiotrophoblast microparticles. J Immunol. (2007) 178:5949–56. doi: 10.4049/jimmunol.178.9.5949
18. Zhang J, Shynlova O, Sabra S, Bang A, Briollais L, Lye SJ. Immunophenotyping and activation status of maternal peripheral blood leukocytes during pregnancy and labour, both term and preterm. J Cell Mol Med. (2017) 21:2386–402. doi: 10.1111/jcmm.13160
19. Kitzmiller JL, Stoneburner L, Yelenosky PF, Lucas WE. Serum complement in normal pregnancy and pre-eclampsia. Am J Obstet Gynecol. (1973) 117:312–5. doi: 10.1016/0002-9378(73)90031-8
20. Baines MG, Millar KG, Mills P. Studies of complement levels in normal human pregnancy. Obstet Gynecol. (1974) 43:806–10.
21. Stirling Y, Woolf L, North WR, Seghatchian MJ, Meade TW. Haemostasis in normal pregnancy. Thromb Haemost. (1984) 52:176–82. doi: 10.1055/s-0038-1661167
22. Hopkinson ND, Powell RJ. Classical complement activation induced by pregnancy: implications for management of connective tissue diseases. J Clin Pathol. (1992) 45:66–7. doi: 10.1136/jcp.45.1.66
23. Comeglio P, Fedi S, Liotta AA, Cellai AP, Chiarantini E, Prisco D, et al. Blood clotting activation during normal pregnancy. Thromb Res. (1996) 84:199–202. doi: 10.1016/0049-3848(96)00176-4
24. Richani K, Soto E, Romero R, Espinoza J, Chaiworapongsa T, Nien JK, et al. Normal pregnancy is characterized by systemic activation of the complement system. J Matern Fetal Neonatal Med. (2005) 17:239–45. doi: 10.1080/14767050500072722
25. Bardou M, Hadi T, Mace G, Pesant M, Debermont J, Barrichon M, et al. Systemic increase in human maternal circulating CD14+CD16- MCP-1+ monocytes as a marker of labor. Am J Obstet Gynecol. (2014) 210:e71–9. doi: 10.1016/j.ajog.2013.08.031
26. Gervasi MT, Chaiworapongsa T, Naccasha N, Blackwell S, Yoon BH, Maymon E, et al. Phenotypic and metabolic characteristics of maternal monocytes and granulocytes in preterm labor with intact membranes. Am J Obstet Gynecol. (2001) 185:1124–9. doi: 10.1067/mob.2001.117681
27. Paquette AG, Shynlova O, Kibschull M, Price ND, Lye SJ Global Alliance to Prevent P. Comparative analysis of gene expression in maternal peripheral blood and monocytes during spontaneous preterm labor. Am J Obstet Gynecol. (2018) 218:e341–5.e30. doi: 10.1016/j.ajog.2017.12.234
28. Gervasi MT, Chaiworapongsa T, Naccasha N, Pacora P, Berman S, Maymon E, et al. Maternal intravascular inflammation in preterm premature rupture of membranes. J Matern Fetal Neonatal Med. (2002) 11:171–5. doi: 10.1080/jmf.11.3.171.175
29. Barden A, Graham D, Beilin LJ, Ritchie J, Baker R, Walters BN, et al. Neutrophil CD11B expression and neutrophil activation in pre-eclampsia. Clin Sci. (1997) 92:37–44. doi: 10.1042/cs0920037
30. Redman CW, Sacks GP, Sargent IL. Preeclampsia: an excessive maternal inflammatory response to pregnancy. Am J Obstet Gynecol. (1999) 180(2 Pt 1):499–506. doi: 10.1016/S0002-9378(99)70239-5
31. Gervasi MT, Chaiworapongsa T, Pacora P, Naccasha N, Yoon BH, Maymon E, et al. Phenotypic and metabolic characteristics of monocytes and granulocytes in preeclampsia. Am J Obstet Gynecol. (2001) 185:792–7. doi: 10.1067/mob.2001.117311
32. Ogge G, Romero R, Chaiworapongsa T, Gervasi MT, Pacora P, Erez O, et al. Leukocytes of pregnant women with small-for-gestational age neonates have a different phenotypic and metabolic activity from those of women with preeclampsia. J Matern Fetal Neonatal Med. (2010) 23:476–87. doi: 10.3109/14767050903216033
33. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. (2003) 4:249–64. doi: 10.1093/biostatistics/4.2.249
34. Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. (2010) 26:2363–7. doi: 10.1093/bioinformatics/btq431
35. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. (2004) 5:R80. doi: 10.1186/gb-2004-5-10-r80
36. Bates D, Maechler M, Bolker B, Walker S. lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1-10. (2015). Available online at: http://CRAN.R-project.org/package=lme4
37. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. (2000) 25:25–9. doi: 10.1038/75556
38. Wick HC, Drabkin H, Ngu H, Sackman M, Fournier C, Haggett J, et al. DFLAT: functional annotation for human development. BMC Bioinformatics. (2014) 15:45. doi: 10.1186/1471-2105-15-45
39. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
40. Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. (2004) 101:6062–7. doi: 10.1073/pnas.0400782101
41. Hui L, Slonim DK, Wick HC, Johnson KL, Bianchi DW. The amniotic fluid transcriptome: a source of novel information about human fetal development. Obstet Gynecol. (2012) 119:111–8. doi: 10.1097/AOG.0b013e31823d4150
42. Tsang JCH, Vong JSL, Ji L, Poon LCY, Jiang P, Lui KO, et al. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc Natl Acad Sci USA. (2017) 114:E7786–95. doi: 10.1073/pnas.1710470114
43. Erez O, Romero R, Maymon E, Chaemsaithong P, Done B, Pacora P, et al. The prediction of late-onset preeclampsia: Results from a longitudinal proteomics study. PLoS ONE. (2017) 12:e0181468. doi: 10.1371/journal.pone.0181468
44. Heilig R, Eckenberg R, Petit JL, Fonknechten N, Da Silva C, Cattolico L, et al. The DNA sequence and analysis of human chromosome 14. Nature. (2003) 421:601–7. doi: 10.1038/nature01348
45. Fellermann K, Stange EF. Defensins – innate immunity at the epithelial frontier. Eur J Gastroenterol Hepatol. (2001) 13:771–6. doi: 10.1097/00042737-200107000-00003
46. Manicone AM, McGuire JK. Matrix metalloproteinases as modulators of inflammation. Semin Cell Dev Biol. (2008) 19:34–41. doi: 10.1016/j.semcdb.2007.07.003
47. Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. (2005) 33:D428–32. doi: 10.1093/nar/gki072
48. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. (1999) 27:29–34. doi: 10.1093/nar/27.1.29
49. Tarca AL, Romero R, Xu Z, Gomez-Lopez N, Erez O, Hsu CD, et al. Targeted expression profiling by RNA-Seq improves detection of cellular dynamics during pregnancy and identifies a role for T cells in term parturition. Sci Rep. (2019) 29:848. doi: 10.1038/s41598-018-36649-w
50. Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. (2009) 583:3966–73. doi: 10.1016/j.febslet.2009.10.036
51. Koussounadis A, Langdon SP, Um IH, Harrison DJ, Smith VA. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci Rep. (2015) 5:10775. doi: 10.1038/srep10775
52. Romero R, Erez O, Maymon E, Chaemsaithong P, Xu Z, Pacora P, et al. The maternal plasma proteome changes as a function of gestational age in normal pregnancy: a longitudinal study. Am J Obstet Gynecol. (2017) 217:e61–7.e21. doi: 10.1016/j.ajog.2017.02.037
53. Al-Garawi A, Carey VJ, Chhabra D, Mirzakhani H, Morrow J, Lasky-Su J, et al. The role of vitamin D in the transcriptional program of human pregnancy. PLoS ONE. (2016) 11:e0163832. doi: 10.1371/journal.pone.0163832
54. Heng YJ, Pennell CE, McDonald SW, Vinturache AE, Xu J, Lee MW, et al. Maternal whole blood gene expression at 18 and 28 weeks of gestation associated with spontaneous preterm birth in asymptomatic women. PLoS ONE. (2016) 11:e0155191. doi: 10.1371/journal.pone.0155191
55. Ngo TTM, Moufarrej MN, Rasmussen MH, Camunas-Soler J, Pan W, Okamoto J, et al. Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science. (2018) 360:1133–6. doi: 10.1126/science.aar3819
56. Savelyeva AV, Kuligina EV, Bariakin DN, Kozlov VV, Ryabchikova EI, Richter VA, et al. Variety of RNAs in peripheral blood cells, plasma, and plasma fractions. Biomed Res Int. (2017) 2017:7404912. doi: 10.1155/2017/7404912
57. Hassan SS, Romero R, Tarca AL, Draghici S, Pineles B, Bugrim A, et al. Signature pathways identified from gene expression profiles in the human uterine cervix before and after spontaneous term parturition. Am J Obstet Gynecol. (2007) 197:e251–7. doi: 10.1016/j.ajog.2007.07.008
58. Mittal P, Romero R, Tarca AL, Gonzalez J, Draghici S, Xu Y, et al. Characterization of the myometrial transcriptome and biological pathways of spontaneous human labor at term. J Perinat Med. (2010) 38:617–43. doi: 10.1515/jpm.2010.097
59. Haddad R, Tromp G, Kuivaniemi H, Chaiworapongsa T, Kim YM, Mazor M, et al. Human spontaneous labor without histologic chorioamnionitis is characterized by an acute inflammation gene expression signature. Am J Obstet Gynecol. (2006) 195:394.e1–24. doi: 10.1016/j.ajog.2005.08.057
60. Gomez-Lopez N, Guilbert LJ, Olson DM. Invasion of the leukocytes into the fetal-maternal interface during pregnancy. J Leukoc Biol. (2010) 88:625–33. doi: 10.1189/jlb.1209796
61. Bonney EA, Shepard MT, Bizargity P. Transient modification within a pool of CD4 T cells in the maternal spleen. Immunology. (2011) 134:270–80. doi: 10.1111/j.1365-2567.2011.03486.x
62. Gomez-Lopez N, St Louis D, Lehr MA, Sanchez-Rodriguez EN, Arenas-Hernandez M. Immune cells in term and preterm labor. Cell Mol Immunol. (2014) 11:571–81. doi: 10.1038/cmi.2014.46
63. Bonney EA. Alternative theories: pregnancy and immune tolerance. J Reprod Immunol. (2017) 123:65–71. doi: 10.1016/j.jri.2017.09.005
64. Sindram-Trujillo A, Scherjon S, Kanhai H, Roelen D, Claas F. Increased T-cell activation in decidua parietalis compared to decidua basalis in uncomplicated human term pregnancy. Am J Reprod Immunol. (2003) 49:261–8. doi: 10.1034/j.1600-0897.2003.00041.x
65. Sindram-Trujillo AP, Scherjon SA, van Hulst-van Miert PP, Kanhai HH, Roelen DL, Claas FH. Comparison of decidual leukocytes following spontaneous vaginal delivery and elective cesarean section in uncomplicated human term pregnancy. J Reprod Immunol. (2004) 62:125–37. doi: 10.1016/j.jri.2003.11.007
66. Tilburgs T, Roelen DL, van der Mast BJ, van Schip JJ, Kleijburg C, de Groot-Swings GM, et al. Differential distribution of CD4(+)CD25(bright) and CD8(+)CD28(-) T-cells in decidua and maternal blood during human pregnancy. Placenta. (2006) 27(Suppl A):S47–53. doi: 10.1016/j.placenta.2005.11.008
67. Tilburgs T, Scherjon SA, Roelen DL, Claas FH. Decidual CD8+CD28- T cells express CD103 but not perforin. Hum Immunol. (2009) 70:96–100. doi: 10.1016/j.humimm.2008.12.006
68. Tilburgs T, van der Mast BJ, Nagtzaam NM, Roelen DL, Scherjon SA, Claas FH. Expression of NK cell receptors on decidual T cells in human pregnancy. J Reprod Immunol. (2009) 80:22–32. doi: 10.1016/j.jri.2009.02.004
69. Tilburgs T, Schonkeren D, Eikmans M, Nagtzaam NM, Datema G, Swings GM, et al. Human decidual tissue contains differentiated CD8+ effector-memory T cells with unique properties. J Immunol. (2010) 185:4470–7. doi: 10.4049/jimmunol.0903597
70. Powell RM, Lissauer D, Tamblyn J, Beggs A, Cox P, Moss P, et al. Decidual T cells exhibit a highly differentiated phenotype and demonstrate potential fetal specificity and a strong transcriptional response to IFN. J Immunol. (2017) 199:3406–17. doi: 10.4049/jimmunol.1700114
71. Gomez-Lopez N, Vadillo-Perez L, Hernandez-Carbajal A, Godines-Enriquez M, Olson DM, Vadillo-Ortega F. Specific inflammatory microenvironments in the zones of the fetal membranes at term delivery. Am J Obstet Gynecol. (2011) 205:235.e15–24. doi: 10.1016/j.ajog.2011.04.019
72. Gomez-Lopez N, Hernandez-Santiago S, Lobb AP, Olson DM, Vadillo-Ortega F. Normal and premature rupture of fetal membranes at term delivery differ in regional chemotactic activity and related chemokine/cytokine production. Reprod Sci. (2013) 20:276–84. doi: 10.1177/1933719112452473
73. Gomez-Lopez N, Vega-Sanchez R, Castillo-Castrejon M, Romero R, Cubeiro-Arreola K, Vadillo-Ortega F. Evidence for a role for the adaptive immune response in human term parturition. Am J Reprod Immunol. (2013) 69:212–30. doi: 10.1111/aji.12074
74. Gomez-Lopez N, Olson DM, Robertson SA. Interleukin-6 controls uterine Th9 cells and CD8(+) T regulatory cells to accelerate parturition in mice. Immunol Cell Biol. (2016) 94:79–89. doi: 10.1038/icb.2015.63
75. Shah NM, Herasimtschuk AA, Boasso A, Benlahrech A, Fuchs D, Imami N, et al. Changes in T cell and dendritic cell phenotype from mid to late pregnancy are indicative of a shift from immune tolerance to immune activation. Front Immunol. (2017) 8:1138. doi: 10.3389/fimmu.2017.01138
76. Yuan M, Jordan F, McInnes IB, Harnett MM, Norman JE. Leukocytes are primed in peripheral blood for activation during term and preterm labour. Mol Hum Reprod. (2009) 15:713–24. doi: 10.1093/molehr/gap054
77. Aghaeepour N, Ganio EA, McIlwain D, Tsai AS, Tingle M, Van Gassen S, et al. An immune clock of human pregnancy. Sci Immunol. (2017) 2:eaan2946. doi: 10.1126/sciimmunol.aan2946
78. Aghaeepour N, Lehallier B, Baca Q, Ganio EA, Wong RJ, Ghaemi MS, et al. A proteomic clock of human pregnancy. Am J Obstet Gynecol. (2018) 218:347.e1–347.e14. doi: 10.1016/j.ajog.2017.12.208
79. Arenas-Hernandez M, Romero R, St Louis D, Hassan SS, Kaye EB, Gomez-Lopez N. An imbalance between innate and adaptive immune cells at the maternal-fetal interface occurs prior to endotoxin-induced preterm birth. Cell Mol Immunol. (2016) 13:462–73. doi: 10.1038/cmi.2015.22
80. St. Louis D, Romero R, Plazyo O, Arenas-Hernandez M, Panaitescu B, Xu Y, et al. Invariant NKT cell activation induces late preterm birth that is attenuated by rosiglitazone. J Immunol. (2016) 196:1044–59. doi: 10.4049/jimmunol.1501962
81. Gomez-Lopez N, Romero R, Arenas-Hernandez M, Schwenkel G, St Louis D, Hassan SS, et al. In vivo activation of invariant natural killer T cells induces systemic and local alterations in T-cell subsets prior to preterm birth. Clin Exp Immunol. (2017) 189:211–25. doi: 10.1111/cei.12968
82. Gomez-Lopez N, Romero R, Arenas-Hernandez M, Ahn H, Panaitescu B, Vadillo-Ortega F, et al. In vivo T-cell activation by a monoclonal alphaCD3epsilon antibody induces preterm labor and birth. Am J Reprod Immunol. (2016) 76:386–90. doi: 10.1111/aji.12562
83. Arenas-Hernandez M, Romero R, Xu Y, Panaitescu B, Garcia-Flores V, Miller D, et al. Effector and activated T cells induce preterm labor and birth that is prevented by treatment with progesterone. J Immunol. (2019) 202:2585–608. doi: 10.4049/jimmunol.1801350
84. Bizargity P, Del Rio R, Phillippe M, Teuscher C, Bonney EA. Resistance to lipopolysaccharide-induced preterm delivery mediated by regulatory T cell function in mice. Biol Reprod. (2009) 80:874–81. doi: 10.1095/biolreprod.108.074294
85. Kim JS, Romero R, Kim MR, Kim YM, Friel L, Espinoza J, et al. Involvement of Hofbauer cells and maternal T cells in villitis of unknown aetiology. Histopathology. (2008) 52:457–64. doi: 10.1111/j.1365-2559.2008.02964.x
86. Kim MJ, Romero R, Kim CJ, Tarca AL, Chhauy S, LaJeunesse C, et al. Villitis of unknown etiology is associated with a distinct pattern of chemokine up-regulation in the feto-maternal and placental compartments: implications for conjoint maternal allograft rejection and maternal anti-fetal graft-versus-host disease. J Immunol. (2009) 182:3919–27. doi: 10.4049/jimmunol.0803834
87. Ito Y, Matsuoka K, Uesato T, Sago H, Okamoto A, Nakazawa A, et al. Increased expression of perforin, granzyme B, and C5b-9 in villitis of unknown etiology. Placenta. (2015) 36:531–7. doi: 10.1016/j.placenta.2015.02.004
88. Kim CJ, Romero R, Kusanovic JP, Yoo W, Dong Z, Topping V, et al. The frequency, clinical significance, and pathological features of chronic chorioamnionitis: a lesion associated with spontaneous preterm birth. Mod Pathol. (2010) 23:1000–11. doi: 10.1038/modpathol.2010.73
89. Khong TY, Bendon RW, Qureshi F, Redline RW, Gould S, Stallmach T, et al. Chronic deciduitis in the placental basal plate: definition and interobserver reliability. Hum Pathol. (2000) 31:292–5. doi: 10.1016/S0046-8177(00)80241-5
90. Lee J, Romero R, Xu Y, Kim JS, Topping V, Yoo W, et al. A signature of maternal anti-fetal rejection in spontaneous preterm birth: chronic chorioamnionitis, anti-human leukocyte antigen antibodies, and C4d. PLoS ONE. (2011) 6:e16806. doi: 10.1371/journal.pone.0016806
91. Lee J, Kim JS, Park JW, Park CW, Park JS, Jun JK, et al. Chronic chorioamnionitis is the most common placental lesion in late preterm birth. Placenta. (2013) 34:681–9. doi: 10.1016/j.placenta.2013.04.014
92. Tamblyn JA, Lissauer DM, Powell R, Cox P, Kilby MD. The immunological basis of villitis of unknown etiology - review. Placenta. (2013) 34:846–55. doi: 10.1016/j.placenta.2013.07.002
93. Kim CJ, Romero R, Chaemsaithong P, Kim JS. Chronic inflammation of the placenta: definition, classification, pathogenesis, and clinical significance. Am J Obstet Gynecol. (2015) 213:S53–69. doi: 10.1016/j.ajog.2015.08.041
94. Maymon E, Romero R, Bhatti G, Chaemsaithong P, Gomez-Lopez N, Panaitescu B, et al. Chronic inflammatory lesions of the placenta are associated with an up-regulation of amniotic fluid CXCR3: a marker of allograft rejection. J Perinat Med. (2018) 46:123–37. doi: 10.1515/jpm-2017-0042
95. Lee J, Romero R, Xu Y, Kim JS, Park JY, Kusanovic JP, et al. Maternal HLA panel-reactive antibodies in early gestation positively correlate with chronic chorioamnionitis: evidence in support of the chronic nature of maternal anti-fetal rejection. Am J Reprod Immunol. (2011) 66:510–26. doi: 10.1111/j.1600-0897.2011.01066.x
96. Lee J, Romero R, Xu Y, Miranda J, Yoo W, Chaemsaithong P, et al. Detection of anti-HLA antibodies in maternal blood in the second trimester to identify patients at risk of antibody-mediated maternal anti-fetal rejection and spontaneous preterm delivery. Am J Reprod Immunol. (2013) 70:162–75. doi: 10.1111/aji.12141
97. Lee J, Romero R, Dong Z, Xu Y, Qureshi F, Jacques S, et al. Unexplained fetal death has a biological signature of maternal anti-fetal rejection: chronic chorioamnionitis and alloimmune anti-human leucocyte antigen antibodies. Histopathology. (2011) 59:928–38. doi: 10.1111/j.1365-2559.2011.04038.x
98. Ogge G, Romero R, Lee DC, Gotsch F, Than NG, Lee J, et al. Chronic chorioamnionitis displays distinct alterations of the amniotic fluid proteome. J Pathol. (2011) 223:553–65. doi: 10.1002/path.2825
99. Lannaman K, Romero R, Chaiworapongsa T, Kim YM, Korzeniewski SJ, Maymon E, et al. Fetal death: an extreme manifestation of maternal anti-fetal rejection. J Perinat Med. (2017) 45:851–68. doi: 10.1515/jpm-2017-0073
100. Romero R, Chaemsaithong P, Chaiyasit N, Docheva N, Dong Z, Kim CJ, et al. CXCL10 and IL-6: markers of two different forms of intra-amniotic inflammation in preterm labor. Am J Reprod Immunol. (2017) 78:e12685. doi: 10.1111/aji.12685
101. Muzzio D, Zenclussen AC, Jensen F. The role of B cells in pregnancy: the good and the bad. Am J Reprod Immunol. (2013) 69:408–12. doi: 10.1111/aji.12079
102. Zenclussen AC. Adaptive immune responses during pregnancy. Am J Reprod Immunol. (2013) 69:291–303. doi: 10.1111/aji.12097
103. Fettke F, Schumacher A, Costa SD, Zenclussen AC. B cells: the old new players in reproductive immunology. Front Immunol. (2014) 5:285. doi: 10.3389/fimmu.2014.00285
104. Muzzio DO, Soldati R, Rolle L, Zygmunt M, Zenclussen AC, Jensen F. B-1a B cells regulate T cell differentiation associated with pregnancy disturbances. Front Immunol. (2014) 5:6. doi: 10.3389/fimmu.2014.00006
105. Schumacher A, Ehrentraut S, Scharm M, Wang H, Hartig R, Morse HC 3rd, et al. Plasma cell alloantigen 1 and IL-10 secretion define two distinct peritoneal B1a B cell subsets with opposite functions, PC1(high) cells being protective and PC1(low) cells harmful for the growing fetus. Front Immunol. (2018) 9:1045. doi: 10.3389/fimmu.2018.01045
106. Leng Y, Romero R, Xu Y, Galaz J, Slutsky R, Arenas-Hernandez M, et al. Are B cells altered in the decidua of women with preterm or term labor? Am J Reprod Immunol. (2019) 81:e13102. doi: 10.1111/aji.13102
107. Bhat NM, Mithal A, Bieber MM, Herzenberg LA, Teng NN. Human CD5+ B lymphocytes (B-1 cells) decrease in peripheral blood during pregnancy. J Reprod Immunol. (1995) 28:53–60. doi: 10.1016/0165-0378(94)00907-O
108. Muzzio DO, Soldati R, Ehrhardt J, Utpatel K, Evert M, Zenclussen AC, et al. B cell development undergoes profound modifications and adaptations during pregnancy in mice. Biol Reprod. (2014) 91:115. doi: 10.1095/biolreprod.114.122366
109. Muzzio DO, Ziegler KB, Ehrhardt J, Zygmunt M, Jensen F. Marginal zone B cells emerge as a critical component of pregnancy well-being. Reproduction. (2016) 151:29–37. doi: 10.1530/REP-15-0274
110. Arck PC, Hecher K, Solano ME. B cells in pregnancy: functional promiscuity or tailored function? Biol Reprod. (2015) 92:12. doi: 10.1095/biolreprod.114.126110
111. Canellada A, Farber A, Zenclussen AC, Gentile T, Dokmetjian J, Keil A, et al. Interleukin regulation of asymmetric antibody synthesized by isolated placental B cells. Am J Reprod Immunol. (2002) 48:275–82. doi: 10.1034/j.1600-0897.2002.01125.x
112. Jensen F, Muzzio D, Soldati R, Fest S, Zenclussen AC. Regulatory B10 cells restore pregnancy tolerance in a mouse model. Biol Reprod. (2013) 89:90. doi: 10.1095/biolreprod.113.110791
113. Rolle L, Memarzadeh Tehran M, Morell-Garcia A, Raeva Y, Schumacher A, Hartig R, et al. Cutting edge: IL-10-producing regulatory B cells in early human pregnancy. Am J Reprod Immunol. (2013) 70:448–53. doi: 10.1111/aji.12157
114. Fettke F, Schumacher A, Canellada A, Toledo N, Bekeredjian-Ding I, Bondt A, et al. Maternal and fetal mechanisms of B cell regulation during pregnancy: human chorionic gonadotropin stimulates B cells to produce IL-10 while alpha-fetoprotein drives them into apoptosis. Front Immunol. (2016) 7:495. doi: 10.3389/fimmu.2016.00495
115. Chakravarty EF, Murray ER, Kelman A, Farmer P. Pregnancy outcomes after maternal exposure to rituximab. Blood. (2011) 117:1499–506. doi: 10.1182/blood-2010-07-295444
116. Das G, Damotte V, Gelfand JM, Bevan C, Cree BAC, Do L, et al. Rituximab before and during pregnancy: a systematic review, and a case series in MS and NMOSD. Neurol Neuroimmunol Neuroinflamm. (2018) 5:e453. doi: 10.1212/NXI.0000000000000453
117. Lima J, Martins C, Leandro MJ, Nunes G, Sousa MJ, Branco JC, et al. Characterization of B cells in healthy pregnant women from late pregnancy to post-partum: a prospective observational study. BMC Pregnancy Childbirth. (2016) 16:139. doi: 10.1186/s12884-016-0927-7
118. Desai RG, Creger WP. Maternofetal passage of leukocytes and platelets in man. Blood. (1963) 21:665–73. doi: 10.1182/blood.V21.6.665.665
119. Herzenberg LA, Bianchi DW, Schroder J, Cann HM, Iverson GM. Fetal cells in the blood of pregnant women: detection and enrichment by fluorescence-activated cell sorting. Proc Natl Acad Sci USA. (1979) 76:1453–5. doi: 10.1073/pnas.76.3.1453
120. Iverson GM, Bianchi DW, Cann HM, Herzenberg LA. Detection and isolation of fetal cells from maternal blood using the flourescence-activated cell sorter (FACS). Prenat Diagn. (1981) 1:61–73. doi: 10.1002/pd.1970010111
121. Wang JY, Zhen DK, Falco VM, Farina A, Zheng YL, Delli-Bovi LC, et al. Fetal nucleated erythrocyte recovery: fluorescence activated cell sorting-based positive selection using anti-gamma globin versus magnetic activated cell sorting using anti-CD45 depletion and anti-gamma globin positive selection. Cytometry. (2000) 39:224–30. doi: 10.1002/(SICI)1097-0320(20000301)39:3<224::AID-CYTO8>3.0.CO;2-J
122. Berry SM, Hassan SS, Russell E, Kukuruga D, Land S, Kaplan J. Association of maternal histocompatibility at class II HLA loci with maternal microchimerism in the fetus. Pediatr Res. (2004) 56:73–8. doi: 10.1203/01.PDR.0000129656.10005.A6
123. Jeanty C, Derderian SC, Mackenzie TC. Maternal-fetal cellular trafficking: clinical implications and consequences. Curr Opin Pediatr. (2014) 26:377–82. doi: 10.1097/MOP.0000000000000087
124. Boddy AM, Fortunato A, Wilson Sayres M, Aktipis A. Fetal microchimerism and maternal health: a review and evolutionary analysis of cooperation and conflict beyond the womb. BioEssays. (2015) 37:1106–18. doi: 10.1002/bies.201500059
125. Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. (1997) 350:485–7. doi: 10.1016/S0140-6736(97)02174-0
126. Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. (1998) 62:768–75. doi: 10.1086/301800
127. Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. (1999) 64:218–24. doi: 10.1086/302205
128. Khosrotehrani K, Wataganara T, Bianchi DW, Johnson KL. Fetal cell-free DNA circulates in the plasma of pregnant mice: relevance for animal models of fetomaternal trafficking. Hum Reprod. (2004) 19:2460–4. doi: 10.1093/humrep/deh445
129. Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat Diagn. (2013) 33:662–6. doi: 10.1002/pd.4119
130. Livergood MC, LeChien KA, Trudell AS. Obesity and cell-free DNA “no calls”: is there an optimal gestational age at time of sampling? Am J Obstet Gynecol. (2017) 216:e411–3.e419. doi: 10.1016/j.ajog.2017.01.011
131. Petersen AK, Cheung SW, Smith JL, Bi W, Ward PA, Peacock S, et al. Positive predictive value estimates for cell-free noninvasive prenatal screening from data of a large referral genetic diagnostic laboratory. Am J Obstet Gynecol. (2017) 217:e691–6. doi: 10.1016/j.ajog.2017.10.005
132. Yang Q, Du Z, Song Y, Gao S, Yu S, Zhu H, et al. Size-selective separation and overall-amplification of cell-free fetal DNA fragments using PCR-based enrichment. Sci Rep. (2017) 7:40936. doi: 10.1038/srep40936
133. Ariga H, Ohto H, Busch MP, Imamura S, Watson R, Reed W, et al. Kinetics of fetal cellular and cell-free DNA in the maternal circulation during and after pregnancy: implications for noninvasive prenatal diagnosis. Transfusion. (2001) 41:1524–30. doi: 10.1046/j.1537-2995.2001.41121524.x
134. Jimenez DF, Tarantal AF. Quantitative analysis of male fetal DNA in maternal serum of gravid rhesus monkeys (Macaca mulatta). Pediatr Res. (2003) 53:18–23. doi: 10.1203/00006450-200301000-00007
135. Birch L, English CA, O'Donoghue K, Barigye O, Fisk NM, Keer JT. Accurate and robust quantification of circulating fetal and total DNA in maternal plasma from 5 to 41 weeks of gestation. Clin Chem. (2005) 51:312–20. doi: 10.1373/clinchem.2004.042713
136. Majer S, Bauer M, Magnet E, Strele A, Giegerl E, Eder M, et al. Maternal urine for prenatal diagnosis–an analysis of cell-free fetal DNA in maternal urine and plasma in the third trimester. Prenat Diagn. (2007) 27:1219–23. doi: 10.1002/pd.1875
137. Mitsunaga F, Ueiwa M, Kamanaka Y, Morimoto M, Nakamura S. Fetal sex determination of macaque monkeys by a nested PCR using maternal plasma. Exp Anim. (2010) 59:255–60. doi: 10.1538/expanim.59.255
138. Phillippe M. Cell-free fetal DNA–a trigger for parturition. N Engl J Med. (2014) 370:2534–6. doi: 10.1056/NEJMcibr1404324
139. Herrera CA, Stoerker J, Carlquist J, Stoddard GJ, Jackson M, Esplin S, et al. Cell-free DNA, inflammation, and the initiation of spontaneous term labor. Am J Obstet Gynecol. (2017) 217:e581–3.e588. doi: 10.1016/j.ajog.2017.05.027
140. Phillippe M. The link between cell-free DNA, inflammation and the initiation of spontaneous labor at term. Am J Obstet Gynecol. (2017) 217:501–2. doi: 10.1016/j.ajog.2017.09.003
141. Leung TN, Zhang J, Lau TK, Hjelm NM, Lo YM. Maternal plasma fetal DNA as a marker for preterm labour. Lancet. (1998) 352:1904–5. doi: 10.1016/S0140-6736(05)60395-9
142. Farina A, LeShane ES, Romero R, Gomez R, Chaiworapongsa T, Rizzo N, et al. High levels of fetal cell-free DNA in maternal serum: a risk factor for spontaneous preterm delivery. Am J Obstetr Gynecol. (2005) 193:421–5. doi: 10.1016/j.ajog.2004.12.023
143. Jakobsen TR, Clausen FB, Rode L, Dziegiel MH, Tabor A. High levels of fetal DNA are associated with increased risk of spontaneous preterm delivery. Prenat Diagn. (2012) 32:840–5. doi: 10.1002/pd.3917
144. Dugoff L, Barberio A, Whittaker PG, Schwartz N, Sehdev H, Bastek JA. Cell-free DNA fetal fraction and preterm birth. Am J Obstet Gynecol. (2016) 215:e231–7. doi: 10.1016/j.ajog.2016.02.009
145. Gomez-Lopez N, Romero R, Schwenkel G, Garcia-Flores V, Panaitescu B, Varrey A, et al. Cell-free fetal DNA increases prior to labor at term and in a subset of preterm births. Reprod Sci. (2019). doi: 10.1007/s43032-019-00023-6
146. Holzgreve W, Ghezzi F, Di Naro E, Ganshirt D, Maymon E, Hahn S. Disturbed feto-maternal cell traffic in preeclampsia. Obstetr Gynecol. (1998) 91(5 Pt 1):669–72. doi: 10.1097/00006250-199805000-00005
147. Scharfe-Nugent A, Corr SC, Carpenter SB, Keogh L, Doyle B, Martin C, et al. TLR9 provokes inflammation in response to fetal DNA: mechanism for fetal loss in preterm birth and preeclampsia. J Immunol. (2012) 188:5706–12. doi: 10.4049/jimmunol.1103454
148. Munoz-Hernandez R, Medrano-Campillo P, Miranda ML, Macher HC, Praena-Fernandez JM, Vallejo-Vaz AJ, et al. Total and fetal circulating cell-free DNA, angiogenic, and antiangiogenic factors in preeclampsia and HELLP syndrome. Am J Hypertens. (2017) 30:673–82. doi: 10.1093/ajh/hpx024
149. Rafaeli-Yehudai T, Imterat M, Douvdevani A, Tirosh D, Benshalom-Tirosh N, Mastrolia SA, et al. Maternal total cell-free DNA in preeclampsia and fetal growth restriction: Evidence of differences in maternal response to abnormal implantation. PLoS ONE. (2018) 13:e0200360. doi: 10.1371/journal.pone.0200360
150. Rolnik DL, da Silva Costa F, Lee TJ, Schmid M, McLennan AC. Association between fetal fraction on cell-free DNA testing and first trimester markers for pre-eclampsia. Ultrasound Obstet Gynecol. (2018) 52:722–7. doi: 10.1002/uog.18993
151. Al-Mufti R, Lees C, Albaiges G, Hambley H, Nicolaides KH. Fetal cells in maternal blood of pregnancies with severe fetal growth restriction. Hum Reprod. (2000) 15:218–21. doi: 10.1093/humrep/15.1.218
152. Morano D, Rossi S, Lapucci C, Pittalis MC, Farina A. Cell-free DNA (cfDNA) fetal fraction in early- and late-onset fetal growth restriction. Mol Diagn Ther. (2018) 22:613–9. doi: 10.1007/s40291-018-0353-9
153. Bianchi DW, Chiu RWK. Sequencing of circulating cell-free DNA during pregnancy. N Engl J Med. (2018) 379:464–73. doi: 10.1056/NEJMra1705345
154. Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci USA. (1996) 93:705–8. doi: 10.1073/pnas.93.2.705
155. Bianchi DW, Flint AF, Pizzimenti MF, Knoll JH, Latt SA. Isolation of fetal DNA from nucleated erythrocytes in maternal blood. Proc Natl Acad Sci USA. (1990) 87:3279–83. doi: 10.1073/pnas.87.9.3279
156. Bianchi DW, Zickwolf GK, Yih MC, Flint AF, Geifman OH, Erikson MS, et al. Erythroid-specific antibodies enhance detection of fetal nucleated erythrocytes in maternal blood. Prenat Diagn. (1993) 13:293–300. doi: 10.1002/pd.1970130408
157. Elahi S, Ertelt JM, Kinder JM, Jiang TT, Zhang X, Xin L, et al. Immunosuppressive CD71+ erythroid cells compromise neonatal host defence against infection. Nature. (2013) 504:158–62. doi: 10.1038/nature12675
158. Gomez-Lopez N, Romero R, Xu Y, Miller D, Unkel RC, MacKenzie T, et al. Umbilical cord CD71+ erythroid cells are reduced in neonates born to women in spontaneous preterm labor. Am J Reprod Immunol. (2016) 76:280–4. doi: 10.1111/aji.12556
159. Miller D, Romero R, Unkel R, Xu Y, Vadillo-Ortega F, Hassan SS, et al. CD71+ erythroid cells from neonates born to women with preterm labor regulate cytokine and cellular responses. J Leukocyte Biol. (2018) 103:761–75. doi: 10.1002/JLB.5A0717-291RRR
160. Curtin WM, Shehata BM, Khuder SA, Robinson HB, Brost BC. The feasibility of using histologic placental sections to predict newborn nucleated red blood cell counts. Obstetr Gynecol. (2002) 100:305–10. doi: 10.1016/S0029-7844(02)02041-0
161. Delyea C, Bozorgmehr N, Koleva P, Dunsmore G, Shahbaz S, Huang V, et al. CD71(+) erythroid suppressor cells promote fetomaternal tolerance through arginase-2 and PDL-1. J Immunol. (2018) 200:4044–58. doi: 10.4049/jimmunol.1800113
162. Fujiki Y, Johnson KL, Tighiouart H, Peter I, Bianchi DW. Fetomaternal trafficking in the mouse increases as delivery approaches and is highest in the maternal lung. Biol Reprod. (2008) 79:841–8. doi: 10.1095/biolreprod.108.068973
163. Adams Waldorf KM, Gammill HS, Lucas J, Aydelotte TM, Leisenring WM, Lambert NC, et al. Dynamic changes in fetal microchimerism in maternal peripheral blood mononuclear cells, CD4+ and CD8+ cells in normal pregnancy. Placenta. (2010) 31:589–94. doi: 10.1016/j.placenta.2010.04.013
Keywords: B cells, biomarker, cytokines, erythroid cells, immunity, pregnancy, T cells
Citation: Gomez-Lopez N, Romero R, Hassan SS, Bhatti G, Berry SM, Kusanovic JP, Pacora P and Tarca AL (2019) The Cellular Transcriptome in the Maternal Circulation During Normal Pregnancy: A Longitudinal Study. Front. Immunol. 10:2863. doi: 10.3389/fimmu.2019.02863
Received: 01 August 2019; Accepted: 21 November 2019;
Published: 17 December 2019.
Edited by:
Ana Claudia Zenclussen, University Hospital Magdeburg, GermanyCopyright © 2019 Gomez-Lopez, Romero, Hassan, Bhatti, Berry, Kusanovic, Pacora and Tarca. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Romero, cHJiY2hpZWZzdGFmZiYjeDAwMDQwO21lZC53YXluZS5lZHU=; Adi L. Tarca, YXRhcmNhJiN4MDAwNDA7bWVkLndheW5lLmVkdQ==
 Nardhy Gomez-Lopez
Nardhy Gomez-Lopez Roberto Romero1,4,5,6,7,8*
Roberto Romero1,4,5,6,7,8* Stanley M. Berry
Stanley M. Berry Adi L. Tarca
Adi L. Tarca