- 1Nephrology and Urology Research Affinity Group, The Abigail Wexner Research Institute at Nationwide Children's Hospital, Columbus, OH, United States
- 2Center of Clinical and Translational Research, The Abigail Wexner Research Institute at Nationwide Children's Hospital, Columbus, OH, United States
- 3Division of Nephrology, Nationwide Children's Hospital, Columbus, OH, United States
- 4Division of Urology, Nationwide Children's Hospital, Columbus, OH, United States
The lower urinary tract is routinely exposed to microbes residing in the gastrointestinal tract, yet the urothelium resists invasive infections by gut microorganisms. This infection resistance is attributed to innate defenses in the bladder urothelium, kidney epithelium, and resident or circulating immune cells. In recent years, surmounting evidence suggests that these cell types produce and secrete soluble host defense peptides, including members of the Ribonuclease (RNase) A Superfamily, to combat invasive bacterial challenge. While some of these peptides, including RNase 4 and RNase 7, are abundantly produced by epithelial cells, the expression of others, like RNase 3 and RNase 6, increase at infection sites with immune cell recruitment. The objective of this mini-review is to highlight recent evidence showing the biological importance and responses of RNase A Superfamily members to infection in the kidney and bladder.
Introduction
Urinary tract infections (UTIs) are one of the most common infections encountered in clinical medicine (1). Nearly half of all women develop one or more UTIs requiring antimicrobial therapy (2, 3). Specific subpopulations have a heightened UTI susceptibility, including pregnant women, people with diabetics, the elderly, people with acquired immunodeficiency diseases, people with structural urologic anomalies, and those who must perform bladder catheterization. Although UTI is not routinely associated with significant acute health morbidities, pregnant women who develop UTI have an increased risk for premature delivery and/or fetal mortality (4). In the elderly, urosepsis is a significant source of mortality (5). Long-term UTI complications include kidney scarring, hypertension, and chronic kidney disease. Thus, UTIs have a significant burden on healthcare resources, with annual costs for UTI management exceeding $2.5 billion United States' dollars (2, 6–8).
Uropathogenic Escherichia coli (UPEC), strains of E. coli that have adapted to live in extraintestinal niches and cause disease, cause the majority of UTIs (9, 10). UPEC originate from the fecal microbiota, spread across the perineum, ascend the urethra, and invade the bladder. The microbial virulence of UPEC has been linked to many factors that have been previously reviewed (11–13). The most prominent virulence factor are Type I fimbriae, which are adhesion organelles capped by the mannose-binding protein FimH. Type I fimbrae facilitate UPEC attachment to superficial bladder epithelial cells by binding to a matrix of uroplakin proteins (12). After binding, UPEC invade the urothelium and establish a state of commensalism or cause an invasive infection that triggers the activation of innate immune defenses, cellular injury, epithelial proliferation and shedding, cytokine release, and leukocyte recruitment (14). If UPEC ascend from the bladder to the kidney, they concentrate in the collecting duct and attach to the luminal surfaces of intercalated cells. Recent evidence suggests that intercalated cells have a role in UTI defense (15, 16).
To cause a symptomatic infection, UPEC must overcome several innate host defense mechanisms. These include the unidirectional flow of urine and regular bladder emptying that minimize UPEC attachment, alterations in urinary ionic composition that prevent bacterial replication, uroepithelial barrier formation and exfoliation during infection, mucous production, bacterial expulsion, and the secretion of antibacterial peptides and proteins (AMPs) that directly kill invading pathogens or modulate immune defenses (17–19). AMPs that have been identified to prevent UTI include defensins, cathelicidin, lectins, metal binding proteins, and bactericidal peptides of the Ribonuclease (RNase) A Superfamily (20, 21). The following sections of this mini-review highlight published literature investigating the roles of RNase A Superfamily in urinary tract host defense.
The Ribonuclease A Superfamily
The RNase A Superfamily is a vertebrate-specific gene family that was initially discovered to encode eight human peptides and proteins. These cationic peptides (RNases 1–8) are enzymatically active and can be grouped into four host defense peptide lineages: (1) eosinophil-produced RNases, (2) angiogenins, (3) RNase 6, and (4) RNase 7 and 8 (22–25). Nearly 15 years ago, five additional “non-canonical” ribonucleases were identified (RNase 9–13) that lack a catalytic domain and enzymatic activity (26, 27).
Each canonical RNase A peptide contains a signal peptide and a mature peptide containing 130–159 amino acid residues. Seven of the eight peptides possess eight cysteine residues, forming four disulfide bonds that confer a shared three-dimensional structure across family members. Each peptide also has a conserved catalytic motif (CKXXNTF) (28). Although the canonical peptides are enzymatically active, the catalytic activity may not be necessary for their immunomodulatory or antibacterial functions. While the catalytic motif is conserved, RNase A Superfamily peptides have significant sequence diversity, which may define each peptide's function(s) (21, 28).
Like other host defense peptides, the primary bactericidal mechanism of RNase A peptides is dependent on their ability to disrupt bacterial cell walls. This is driven by the peptide's net charge, amphipathicity, disulphide bonding, and secondary structure (29, 30). The peptide's bactericidal activity is primarily restricted to the amino terminus (31, 32). In addition to their membrane penetrating capability, RNase A peptides can interfere with bacterial attachment, translocate into bacterial cells to inhibit protein and/or DNA synthesis, or initiate signaling pathways important in innate immunity and inflammatory responses (19, 20). As recently reviewed, RNase A Superfamily members can act as chemoattractants, damage-associated molecular patterns (DAMPS or alarmins), immune cell activators, or opsonins. Also, they participate in extracellular RNA clearance (21, 22, 25, 28, 33–35). In the urinary tract, research has primarily focused on their bactericidal activity.
Epithelial-Produced Ribonucleases
RNase 4 and RNase 7 are produced by epithelial cells in the urinary tract. RNase 7 is produced by the urothelium of the ureter and bladder and secreted into the urinary stream. In the kidney, the collecting duct is the main source of RNase 4 and 7 production (Figure 1) (36, 37).
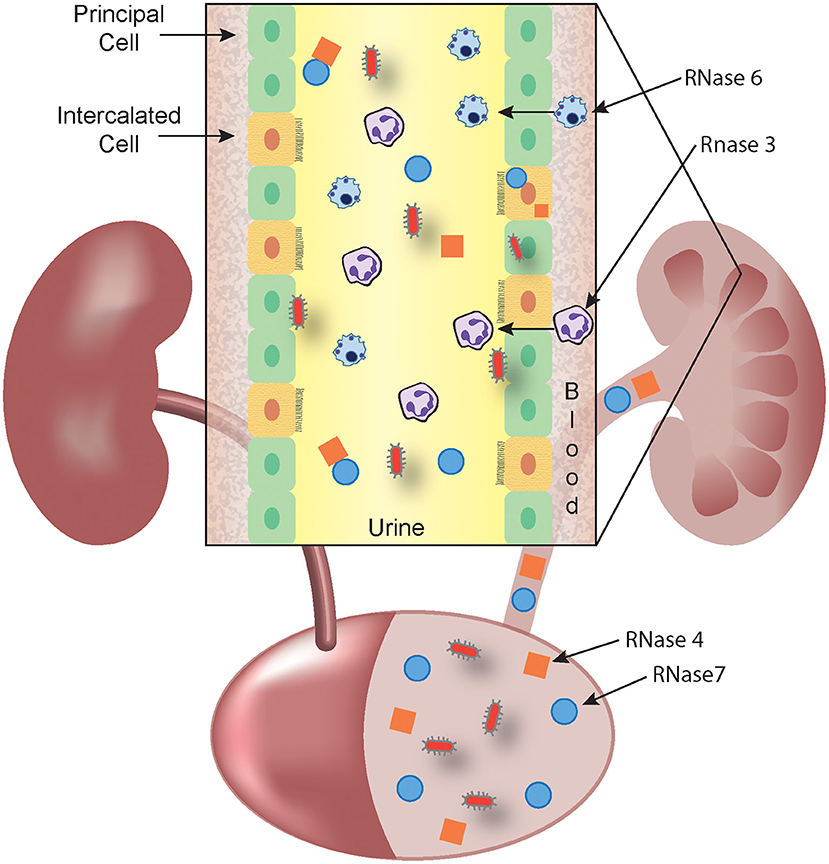
Figure 1. RNase A Superfamily members collaborate to prevent and eradicate UTI. Schematic representation showing that RNase 4 (orange squares) and RNase 7 (blue circles) are produced by the bladder urothelium and the kidney's collecting duct (inset) and released into the urine. In response to microbes (red), circulating leukocytes that harbor RNase 3 (eosinophils and neutrophils) and RNase 6 (monocytes and macrophages) exit the bloodstream and cross the urothelium to accumulate in the urine. The antimicrobial activity of RNase 3 and RNase 6 may be predominantly exerted at the intracellular level, following phagocytosis of microbes. Urinary, parenchymal, and leukocyte-produced RNases kill invading pathogens and facilitate bacterial clearance.
RNase 4 is one of the least studied members of the Ribonuclease A Superfamily. RNASE4 mRNA is expressed in multiple human tissues as well as circulating immune cells (38–41). In the urinary tract, the kidney's collecting duct is a source of RNase 4 production where it is regulated by insulin receptor activation and downstream phosphatidylinositol 3-kinase/AKT (PI3K/AKT) signaling (37, 42). RNase 4 is constitutively secreted into the urine and neutralization of urinary RNase 4's antimicrobial activity with RNase 4-specific antibodies facilitates UPEC replication, suggesting it plays a role in UTI defense. Recombinant RNase 4 peptide exhibits antimicrobial activity toward UPEC, including multi-drug resistant UPEC (37). It is possible that monocyte and macrophages are additional sources of RNase 4 in the urinary tract.
RNase 7, which was first identified in human skin, is the best example of an RNase that provides antibacterial defense (43, 44). RNase 7 possesses broad-spectrum antimicrobial activity against Gram-positive and Gram-negative urothopathogens (32, 45). In the urinary tract, RNase 7 is produced by the lower urinary tract and by the kidney's collecting tubules. Within the collecting tubules, RNase 7 is expressed by alpha and beta intercalated cells (36). RNase 7 is secreted into the urine and urinary concentrations increase with UTI to prevent infection (36, 45). Females and adolescent girls with recurrent UTI have suppressed urinary RNase 7 levels compared to controls without UTI (46). When human urine is incubated with RNase 7 neutralizing antibodies and inoculated with UPEC in vitro, bacterial growth significantly increases (36, 47). These findings indicate that decreased urinary RNase 7 concentrations are a UTI risk-factor and provide insight into why certain populations are more susceptible to UTI.
RNase 7 expression is restricted to humans and higher vertebrates (26). Genomes of the laboratory mouse or the rat do not contain sequences orthologous to RNase 7, which limited the capacity to assess RNase 7's bactericidal and immunomodulatory functions in vivo (26, 27, 30). However, our research group recently generated humanized RNase 7 transgenic mice. RNase 7 expression in these mice did not impact urothelial histology, endogenous innate immune profiles, or the diversity of the gastrointestinal microbiome. When subjected to UTI in vivo, these mice were significantly protected from UPEC, suggesting that RNase 7 could be a therapeutic target for protection against UTI (46).
Only limited evidence has evaluated the mechanisms that regulate RNase 7 expression. Published data shows that Toll-Like Receptor (TLR)-mediated pathways, the PI3K/AKT pathway, as well as the mitogen-activated protein kinase (MAPK) pathway regulate RNase 7 expression (48–51). Like RNase 4, our research team has shown that insulin enhances RNase 7 expression via PI3K/AKT activation to shield the urinary tract from UPEC (51). RNase 7's structure, immunomodulatory properties, and bactericidal activity have been recently reviewed (35, 44).
Leukocyte-Produced Ribonucleases
Eosinophil granule proteins were among the first leukocyte-produced RNases to demonstrate a role in innate immunity (52). Eosinophil Cationic Protein (RNase 3) is a highly basic, 21 kilodalton polypeptide originally isolated from eosinophilic granules (53). In the urinary tract, the expression, immunomodulatory properties, and anti-parasite activity of RNase 3 have been investigated in the setting of schistosomiasis. Urinary RNase 3 concentrations increase during schistosomiasis, and the magnitude of urinary RNase 3 is proportional to the intensity of infection (54, 55). Functionally, purified RNase 3 shows dose-dependent antihelminthic activity toward schistosomula, which represent the larval stage of the Schistosoma mansoni parasite (56). Subsequent work demonstrates that RNase 3 exhibits greater antimicrobial activity toward schistosomula than other eosinophilic granule proteins (57). To facilitate schistosomula killing, RNase 3 synergizes with the oxidative burst capacity of plasma membrane fractions derived from neutrophils and eosinophils (58).
While eosinophils are the most studied cellular source of RNase 3, eosinophils generally are not associated with the host response to bacterial UTI. However, studies in the literature argue that neutrophils are also a source of RNase 3, and this cell population is briskly recruited to the urinary space during UTI (59–61). When purified from eosinophils, RNase 3 exhibits antimicrobial activity toward laboratory strains of Staphyloccus aureus and E. coli (62). To our knowledge, RNase 3's bactericidal activity has not been tested against clinically relevant UPEC strains. Thus, further studies are needed that evaluate RNase 3's expression during bacterial UTI, its cellular source(s) of production, and its contribution to UPEC clearance.
Unlike RNase 3, RNase 6 has been investigated in the context of UTI (24). RNase 6 expression has been localized to human and murine monocytes, macrophages, and neutrophils that are recruited to the urinary tract during UPEC-UTI (Figure 1). Recombinant human and murine RNase 6 peptides exhibit potent, dose-dependent killing of Gram-negative and positive uropathogenic bacteria, comparable to human RNase 7 (24, 63). RNase 6 levels increase in urine from humans and mice with UTI, but its expression was largely retained as an intracellular pool, suggesting that RNase 6 may act intracellularly, following phagocytosis of microbes (24). Further studies are required to determine the contributions of RNase 6 to host immunity during UTI.
The Non-Canonical Ribonucleases
The less-studied non-canonical RNases, including RNases 9–13, lack the signature catalytic motif of the canonical RNases and do not require enzymatic activity to function (26). These RNases share 15–30% identity with the canonical RNases and contain the signal peptide as well as the three most conserved disulfide bonds. They lack the N-terminal region of mature RNases (21, 28, 64). These RNases are expressed in the epididymis of the male reproductive system and they may regulate sperm maturation and motility (65–67). Recombinant RNase 9 has bactericidal activity against E. coli (68). Additional investigation is warranted to define the role of these peptides in UTI defense and the prevention of sexually transmitted infections.
Discussion and Future Directions
As outlined by the published findings above, significant knowledge gaps remain defining the regulation of antimicrobial activity as well as the roles of antimicrobial RNases—both individually and collectively—in limiting the incidence and spread of UTI. Here, we outline these knowledge gaps and discuss the potential clinical applications of antimicrobial RNases during UTI.
Mechanisms of Antimicrobial Action
Most published studies of RNase function have focused on the structural elements of each peptide that are required for antimicrobial activity in vitro (31, 63, 69, 70). However, it is important that similar structure-function tests be undertaken in the context of eukaryotic cells, which serve as the natural source of RNase A Superfamily members. Recently, we demonstrated that human urothelial cells can be genetically modified to over-express RNase 7, which confers antimicrobial activity toward UPEC (46). This type of experimental approach affords the opportunity to test the relationship between RNase 7 structural elements and antimicrobial function in the context of its cellular source. It is conceivable that this experimental system may reveal key aspects of RNase regulation, including roles for post-translational modifications and protein-protein interactions.
Regulation of RNase Expression and Function
While emerging data point to the importance of cellular signal transduction pathways such as the insulin-PI3K/AKT pathway as regulators of RNase 4 and RNase 7 in the kidney or bladder, there are significant gaps in our understanding of this process or other processes that are activated during UTI and may impact RNase regulation (37, 51). Also, there are no published data regarding the transcriptional regulation and signal transduction pathways responsible for expression of leukocyte RNases during UTI.
In addition to their regulation at the level of mRNA and protein expression, evidence points to a key role for the RNase Inhibitor (RI) as a regulator of RNase function and cytotoxicity. RI is expressed by all mammalian cells and exhibits high-affinity binding with multiple members of the RNase A Superfamily (47, 71). RI complexes to RNase A proteins in the cytosolic compartment, attenuating or inhibiting their biological functions and protecting host cells from their cytotoxic RNase activity. Raines and colleagues have elegantly shown that RNase A peptides complexed to RI have limited cellular toxicity. In contrast, RI-elusive RNases degrade cellular RNA and trigger apoptosis (72, 73). Currently, there is only limited data evaluating the role of RI in the kidney or urinary tract. While no studies have evaluated the impact of RI on urothelial proliferation or cellular protection during UTI, we have shown that RI complexes to recombinant RNase 7 and abrogates it's antimicrobial activity against uropathogens (47). Similarly, RI complexes to RNase 7 in kidney tissue as well as in the urine sediment in vivo. In the context of acute or chronic UTI, the significance of this interaction remains unknown.
Specifically, are there events triggered by infection that modify the interaction between RNase 7 and RI? The interaction between RNase 7 and RI is dependent on the presence of intact disulfide bridges in RI, as disruption of these bridges triggers rapid dissociation of the RNase 7:RI complex (47). This has led us to speculate that the RNase 7's antimicrobial activity may be redox dependent (47). There is precedent for this among other AMPs, such as human β-defensin 1 and paneth cell α-defensin 6 (74, 75). Alternatively, we have found that neutrophil proteases can degrade RI, and this may be a mechanism for recruited inflammatory cells to augment local RNase activity during UTI (47). The precise roles of RNase:RI interactions and their regulation during UTI remain a significant knowledge gap. As research progresses, the benefits and risks of RI-interactions and evasion need to be carefully evaluated.
Roles of RNases During UTI in vivo
While the antimicrobial activity of RNases can be measured in vitro, the ultimate biological test of relevance is to over-express and delete antimicrobial RNases and demonstrate a consequence on host susceptibility to experimental UTI in laboratory animals. These experiments are challenging, since there are instances in which genes encoding human antimicrobial RNases have no orthologous mouse gene (RNASE7) or up to 15 paralogous mouse genes (RNASE3) (76). Moreover, there are instances in which the same cell type expresses multiple antimicrobial RNases, such as monocytes in the case of RNase 4 and RNase 6, or epithelial cells in the case of RNase 4 and RNase 7 (36, 37, 41, 77). These circumstances can lead to gene redundancy and lack of an overt phenotype when a single gene is deleted. Thus, we have taken the opposite approach, namely, to generate humanized RNASE7 transgenic mice. This gain of function experiment has allowed us to establish the consequences of RNASE7 expression on host immune functions and susceptibility to experimental UTI in vivo (46). Further studies are required to determine the function of RNases—both individually and collectively—in the setting of UTI, as well as to determine their mechanisms of action.
Impact of Uropathogens on RNase Expression and Activity
Up to this point, we have addressed the impact of RNases on microbes as unidirectional. However, it is likely that uropathogens influence the levels and activity of RNase peptides. For example, UPEC strains expressing the toxin Hemolysin A (HlyA) attenuate intracellular PI3K/AKT signaling, leading to decreased RNase 7 expression in vitro (51). Similarly, UPEC produced proteases like OmpT have the ability to degrade RNase 7 (78). Thus, additional studies are required to identify the ways in which microbes might regulate the expression and activity of antimicrobial RNases in the urinary tract.
Applications of RNases as Therapeutics
Interest has centered on use of AMPs as an antibiotic-independent mechanism of limiting, treating, and even preventing UTIs. This interest has been fueled by a growing concern regarding antibiotic overuse and its impact on rising bacterial antibiotic resistance rates (20, 79). AMPs may offer an alternate therapeutic strategy, with even some benefit over antibiotics given their efficacy at low concentrations, limited bacterial resistance patterns, and potential synergistic mechanisms of action with conventional antibiotics (19, 80–82).
However, multiple hurdles must be overcome to realize the direct therapeutic potential of RNase as AMPs in patients with UTI. This includes achieving the synthesis of large quantities of purified RNase peptides or peptide fragments, taking steps to assure stability and delivery, and ensuring that RNases do not exhibit significant toxicity toward host cells. Despite these formidable challenges, we envision future direct applications of antimicrobial RNases to people with highest UTI risk. In addition to such direct applications, we also envision circumstances in which endogenous pathways are pharmacologically triggered to induce RNase expression as an alternative strategy to prevent and even treat UTI. In sum, while many knowledge gaps remain, the urinary tract is an excellent venue to realize the biological significance and therapeutic applications of antimicrobial RNases and define their roles as novel therapies.
Author Contributions
BB, CC, and JS performed the literature review and wrote the manuscript together.
Funding
This work was supported by the National Institutes of Health (NIDDK) K08 DK102594 and R03 DK118306 (awarded to BB). CC was supported by the National Institutes of Health (NIDDK) K08 DK122119. JS was supported by the National Institutes of Health (NIDDK) R01 DK 114035 and R01 DK115737.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as A potential conflict of interest.
Acknowledgments
We acknowledge Ms. Lisa Feurer for assistance with the artwork in Figure 1.
References
1. Khoshnood S, Heidary M, Mirnejad R, Bahramian A, Sedighi M, Mirzaei H. Drug-resistant gram-negative uropathogens: a review. Biomed Pharmacother. (2017) 94:982–94. doi: 10.1016/j.biopha.2017.08.006
2. Foxman B. The epidemiology of urinary tract infection. Nat Rev Urol. (2010) 7:653–60. doi: 10.1038/nrurol.2010.190
3. Foxman B, Barlow R, D'Arcy H, Gillespie B, Sobel JD. Urinary tract infection: self-reported incidence and associated costs. Ann Epidemiol. (2000) 10:509–15. doi: 10.1016/S1047-2797(00)00072-7
4. Becknell B, Schober M, Korbel L, Spencer JD. The diagnosis, evaluation and treatment of acute and recurrent pediatric urinary tract infections. Expert Rev Anti Infect Ther. (2015) 13:81–90. doi: 10.1586/14787210.2015.986097
5. Klevens RM, Edwards JR, Richards CL Jr, Horan TC, Gaynes RP, Pollock DA, et al. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. (2007) 122:160–6. doi: 10.1177/003335490712200205
6. Schappert SM, Rechtsteiner EA. Ambulatory medical care utilization estimates for 2007. Vital Health Stat. (2011) 13:1–38.
7. Haque M, Sartelli M, McKimm J, Abu Bakar M. Health care-associated infections - an overview. Infect Drug Resist. (2018) 11:2321–33. doi: 10.2147/IDR.S177247
8. Simmering JE, Tang F, Cavanaugh JE, Polgreen LA, Polgreen PM. The increase in hospitalizations for urinary tract infections and the associated costs in the United States, 1998-2011. Open Forum Infect Dis. (2017) 4:ofw281. doi: 10.1093/ofid/ofw281
9. Russo TA, Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis. (2000) 181:1753–4. doi: 10.1086/315418
10. Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. (2015) 13:269–84. doi: 10.1038/nrmicro3432
11. Ragnarsdottir B, Svanborg C. Susceptibility to acute pyelonephritis or asymptomatic bacteriuria: host-pathogen interaction in urinary tract infections. Pediatr Nephrol. (2012) 27:2017–29. doi: 10.1007/s00467-011-2089-1
12. Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bad bugs and beleaguered bladders: interplay between uropathogenic Escherichia coli and innate host defenses. Proc Natl Acad Sci U.S.A. (2000) 97:8829–35. doi: 10.1073/pnas.97.16.8829
13. Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. (1998) 282:1494–7. doi: 10.1126/science.282.5393.1494
14. Weichhart T, Haidinger M, Horl WH, Saemann MD. Current concepts of molecular defence mechanisms operative during urinary tract infection. Eur J Clin Invest. (2008) 38(Suppl. 2):29–38. doi: 10.1111/j.1365-2362.2008.02006.x
15. Chassin C, Tourneur E, Bens M, Vandewalle A. A role for collecting duct epithelial cells in renal antibacterial defences. Cell Microbiol. (2011) 13:1107–13. doi: 10.1111/j.1462-5822.2011.01614.x
16. Paragas N, Kulkarni R, Werth M, Schmidt-Ott KM, Forster C, Deng R, et al. α-Intercalated cells defend the urinary system from bacterial infection. J Clin Invest. (2014) 124:2963–76. doi: 10.1172/JCI71630
17. Ching C, Schwartz L, Spencer JD, Becknell B. Innate immunity and urinary tract infection. Pediatr Nephrol. (2019) 1–10. doi: 10.1007/s00467-019-04269-9
18. Bien J, Sokolova O, Bozko P. Role of uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int J Nephrol. (2012) 2012:681473. doi: 10.1155/2012/681473
19. Spencer JD, Schwaderer AL, Becknell B, Watson J, Hains DS. The innate immune response during urinary tract infection and pyelonephritis. Pediatr Nephrol. (2014) 29:1139–49. doi: 10.1007/s00467-013-2513-9
20. Becknell B, Schwaderer A, Hains DS, Spencer JD. Amplifying renal immunity: the role of antimicrobial peptides in pyelonephritis. Nat Rev Nephrol. (2015) 11:642–55. doi: 10.1038/nrneph.2015.105
21. Schwartz L, Cohen A, Thomas J, Spencer JD. The immunomodulatory and antimicrobial properties of the vertebrate ribonuclease A superfamily. Vaccines. (2018) 6:76. doi: 10.3390/vaccines6040076
22. Rosenberg HF. RNase A ribonucleases and host defense: an evolving story. J Leukoc Biol. (2008) 83:1079–87. doi: 10.1189/jlb.1107725
23. Simanski M, Dressel S, Glaser R, Harder J. RNase 7 protects healthy skin from Staphylococcus aureus colonization. J Invest Dermatol. (2010) 130:2836–8. doi: 10.1038/jid.2010.217
24. Becknell B, Eichler TE, Beceiro S, Li B, Easterling RS, Carpenter AR, et al. Ribonucleases 6 and 7 have antimicrobial function in the human and murine urinary tract. Kidney Int. (2014) 87:151–61. doi: 10.1038/ki.2014.268
25. Sorrentino S. The eight human “canonical” ribonucleases: molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. (2010) 584:2194–200. doi: 10.1016/j.febslet.2010.04.018
26. Cho S, Beintema JJ, Zhang J. The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics. (2005) 85:208–20. doi: 10.1016/j.ygeno.2004.10.008
27. Goo SM, Cho S. The expansion and functional diversification of the mammalian ribonuclease a superfamily epitomizes the efficiency of multigene families at generating biological novelty. Genome Biol Evol. (2013) 5:2124–40. doi: 10.1093/gbe/evt161
28. Lu L, Li J, Moussaoui M, Boix E. Immune modulation by human secreted RNases at the extracellular space. Front Immunol. (2018) 9:1012. doi: 10.3389/fimmu.2018.01012
29. Koczera P, Martin L, Marx G, Schuerholz T. The ribonuclease A superfamily in humans: canonical RNases as the buttress of innate immunity. Int J Mol Sci. (2016) 17:E1278. doi: 10.3390/ijms17081278
30. Dyer KD, Rosenberg HF. The RNase a superfamily: generation of diversity and innate host defense. Mol Divers. (2006) 10:585–97. doi: 10.1007/s11030-006-9028-2
31. Torrent M, Pulido D, Valle J, Nogues MV, Andreu D, Boix E. Ribonucleases as a host-defence family: evidence of evolutionarily conserved antimicrobial activity at the N-terminus. Biochem J. (2013) 456:99–108. doi: 10.1042/BJ20130123
32. Wang H, Schwaderer AL, Kline J, Spencer JD, Kline D, Hains DS. Contribution of structural domains to the activity of ribonuclease 7 against uropathogenic bacteria. Antimicrob Agents Chemother. (2013) 57:766–74. doi: 10.1128/AAC.01378-12
33. Boix E, Nogues MV. Mammalian antimicrobial proteins and peptides: overview on the RNase A superfamily members involved in innate host defence. Mol Biosyst. (2007) 3:317–35. doi: 10.1039/b617527a
34. Gupta SK, Haigh BJ, Griffin FJ, Wheeler TT. The mammalian secreted RNases: mechanisms of action in host defence. Innate Immun. (2013) 19:86–97. doi: 10.1177/1753425912446955
35. Simanski M, Koten B, Schroder JM, Glaser R, Harder J. Antimicrobial RNases in cutaneous defense. J Innate Immun. (2012) 4:241–7. doi: 10.1159/000335029
36. Spencer JD, Schwaderer AL, Dirosario JD, McHugh KM, McGillivary G, Justice SS, et al. Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney Int. (2011) 80:174–80. doi: 10.1038/ki.2011.109
37. Murtha MJ, Eichler T, Bender K, Metheny J, Li B, Schwaderer AL, et al. Insulin receptor signaling regulates renal collecting duct and intercalated cell antibacterial defenses. J Clin Invest. (2018) 128:5634–46. doi: 10.1172/JCI98595
38. Hofsteenge J, Vicentini A, Zelenko O. Ribonuclease 4, an evolutionarily highly conserved member of the superfamily. Cell Mol Life Sci. (1998) 54:804–10. doi: 10.1007/s000180050209
39. Rosenberg HF, Dyer KD. Human ribonuclease 4 (RNase 4): coding sequence, chromosomal localization and identification of two distinct transcripts in human somatic tissues. Nucleic Acids Res. (1995) 23:4290–5. doi: 10.1093/nar/23.21.4290
40. Seno M, Futami J, Tsushima Y, Akutagawa K, Kosaka M, Tada H, et al. Molecular cloning and expression of human ribonuclease 4 cDNA. Biochim Biophys Acta. (1995) 1261:424–6. doi: 10.1016/0167-4781(95)00040-N
41. Egesten A, Dyer KD, Batten D, Domachowske JB, Rosenberg HF. Ribonucleases and host defense: identification, localization and gene expression in adherent monocytes in vitro. Biochim Biophys Acta. (1997) 1358:255–60. doi: 10.1016/S0167-4889(97)00081-5
42. Saxena V, Hains DS, Ketz J, Chanley M, Spencer JD, Becknell B, et al. Cell-specific qRT-PCR of renal epithelial cells reveals a novel innate immune signature in murine collecting duct. Am J Physiol Renal Physiol. (2018) 315:F812–23. doi: 10.1152/ajprenal.00512.2016
43. Rademacher F, Simanski M, Harder J. RNase 7 in cutaneous defense. Int J Mol Sci. (2016) 17:560. doi: 10.3390/ijms17040560
44. Becknell B, Spencer JD. A review of ribonuclease 7's structure, regulation, and contributions to host defense. Int J Mol Sci. (2016) 17:423. doi: 10.3390/ijms17030423
45. Spencer JD, Schwaderer AL, Wang H, Bartz J, Kline J, Eichler T, et al. Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney Int. (2013) 83:615–25. doi: 10.1038/ki.2012.410
46. Eichler T, Bender K, Murtha MJ, Schwartz L, Metheny J, Solden L, et al. Ribonuclease 7 shields the kidney and bladder from invasive uropathogenic Escherichia coli infection. J Am Soc Nephrol. (2019) 30:1385–97. doi: 10.1681/ASN.2018090929
47. Spencer JD, Schwaderer AL, Eichler T, Wang H, Kline J, Justice SS, et al. An endogenous ribonuclease inhibitor regulates the antimicrobial activity of ribonuclease 7 in the human urinary tract. Kidney Int. (2013) 85:1179–91. doi: 10.1038/ki.2013.395
48. Reithmayer K, Meyer KC, Kleditzsch P, Tiede S, Uppalapati SK, Gläser R, et al. Human hair follicle epithelium has an antimicrobial defence system that includes the inducible antimicrobial peptide psoriasin (S100A7) and RNase 7. Br J Dermatol. (2009) 161:78–89. doi: 10.1111/j.1365-2133.2009.09154.x
49. Mohammed I, Yeung A, Abedin A, Hopkinson A, Dua HS. Signalling pathways involved in ribonuclease-7 expression. Cell Mol Life Sci. (2011) 68:1941–52. doi: 10.1007/s00018-010-0540-2
50. Simanski M, Rademacher F, Schroder L, Schumacher HM, Glaser R, Harder J. IL-17A and IFN-γ synergistically induce RNase 7 expression via STAT3 in primary keratinocytes. PLoS ONE. (2013) 8:e59531. doi: 10.1371/journal.pone.0059531
51. Eichler TE, Becknell B, Easterling RS, Ingraham SE, Cohen DM, Schwaderer AL, et al. Insulin and the phosphatidylinositol 3-kinase signaling pathway regulate Ribonuclease 7 expression in the human urinary tract. Kidney Int. (2016) 90:568–79. doi: 10.1016/j.kint.2016.04.025
52. Acharya KR, Ackerman SJ. Eosinophil granule proteins: form and function. J Biol Chem. (2014) 289:17406–15. doi: 10.1074/jbc.R113.546218
53. Olsson I, Venge P, Spitznagel JK, Lehrer RI. Arginine-rich cationic proteins of human eosinophil granules: comparison of the constituents of eosinophilic and neutrophilic leukocytes. Lab Invest. (1977) 36:493–500.
54. Stecher CW, Fofana HKM, Madsen H, Wilson S, Keita AD, Landouré A, et al. Inflammation dynamics after praziquantel treatment of Schistosoma haematobium reflected by urinary eosinophil cationic protein. Trans R Soc Trop Med Hyg. (2017) 111:316–24. doi: 10.1093/trstmh/trx057
55. Leutscher PD, Reimert CM, Vennervald BJ, Ravaoalimalala VE, Ramarokoto CE, Serieye J, et al. Morbidity assessment in urinary schistosomiasis infection through ultrasonography and measurement of eosinophil cationic protein (ECP) in urine. Trop Med Int Health. (2000) 5:88–93. doi: 10.1046/j.1365-3156.2000.00522.x
56. McLaren DJ, McKean JR, Olsson I, Venges P, Kay AB. Morphological studies on the killing of schistosomula of Schistosoma mansoni by human eosinophil and neutrophil cationic proteins in vitro. Parasite Immunol. (1981) 3:359–73. doi: 10.1111/j.1365-3024.1981.tb00414.x
57. Ackerman SJ, Gleich GJ, Loegering DA, Richardson BA, Butterworth AE. Comparative toxicity of purified human eosinophil granule cationic proteins for schistosomula of Schistosoma mansoni. Am J Trop Med Hyg. (1985) 34:735–45. doi: 10.4269/ajtmh.1985.34.735
58. Yazdanbakhsh M, Tai PC, Spry CJ, Gleich GJ, Roos D. Synergism between eosinophil cationic protein and oxygen metabolites in killing of schistosomula of Schistosoma mansoni. J Immunol. (1987) 138:3443–7.
59. Sur S, Glitz DG, Kita H, Kujawa SM, Peterson EA, Weiler DA, et al. Localization of eosinophil-derived neurotoxin and eosinophil cationic protein in neutrophilic leukocytes. J Leukoc Biol. (1998) 63:715–22. doi: 10.1002/jlb.63.6.715
60. Ribas D, Fernandez-Carranco MC, Nabil H, Bobadilla P, Monteseirin J. Eosinophil cationic protein and histamine production by neutrophils from periodontitis patients. J Periodontol. (2017). doi: 10.1902/jop.2017.160679. [Epub ahead of print].
61. Monteseirin J, Vega A, Chacon P, Camacho MJ, El Bekay R, Asturias JA, et al. Neutrophils as a novel source of eosinophil cationic protein in IgE-mediated processes. J Immunol. (2007) 179:2634–41. doi: 10.4049/jimmunol.179.4.2634
62. Lehrer RI, Szklarek D, Barton A, Ganz T, Hamann KJ, Gleich GJ. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J Immunol. (1989) 142:4428–34.
63. Pulido D, Arranz-Trullen J, Prats-Ejarque G, Velázquez D, Torrent M, Moussaoui M, et al. Insights into the antimicrobial mechanism of action of human RNase6: structural determinants for bacterial cell agglutination and membrane permeation. Int J Mol Sci. (2016) 17:552. doi: 10.3390/ijms17040552
64. Devor EJ, Moffat-Wilson KA, Galbraith JJ. LOC 390443 (RNase 9) on chromosome 14q11.2 is related to the RNase A superfamily and contains a unique amino-terminal preproteinlike sequence. Hum Biol. (2004) 76:921–35. doi: 10.1353/hub.2005.0016
65. Liu J, Li J, Wang H, Zhang C, Li N, Lin Y, et al. Cloning, expression and location of RNase9 in human epididymis. BMC Res Notes. (2008) 1:111. doi: 10.1186/1756-0500-1-111
66. Zhu CF, Liu Q, Zhang L, Yuan HX, Zhen W, Zhang JS, et al. RNase9, an androgen-dependent member of the RNase A family, is specifically expressed in the rat epididymis. Biol Reprod. (2007) 76:63–73. doi: 10.1095/biolreprod.106.054635
67. Westmuckett AD, Nguyen EB, Herlea-Pana OM, Alvau A, Salicioni AM, Moore KL. Impaired sperm maturation in RNASE9 knockout mice. Biol Reprod. (2014) 90:120. doi: 10.1095/biolreprod.113.116863
68. Cheng GZ, Li JY, Li F, Wang HY, Shi GX. Human ribonuclease 9, a member of ribonuclease A superfamily, specifically expressed in epididymis, is a novel sperm-binding protein. Asian J Androl. (2009) 11:240–51. doi: 10.1038/aja.2008.30
69. Huang YC, Lin YM, Chang TW, Wu SJ, Lee YS, Chang MD, et al. The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity. J Biol Chem. (2007) 282:4626–33. doi: 10.1074/jbc.M607321200
70. Torrent M, Badia M, Moussaoui M, Sanchez D, Nogues MV, Boix E. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. (2010) 277:1713–25. doi: 10.1111/j.1742-4658.2010.07595.x
71. Abtin A, Eckhart L, Mildner M, Ghannadan M, Harder J, Schröder JM, et al. Degradation by stratum corneum proteases prevents endogenous RNase inhibitor from blocking antimicrobial activities of RNase 5 and RNase 7. J Invest Dermatol. (2009) 129:2193–201. doi: 10.1038/jid.2009.35
72. Haigis MC, Kurten EL, Raines RT. Ribonuclease inhibitor as an intracellular sentry. Nucleic Acids Res. (2003) 31:1024–32. doi: 10.1093/nar/gkg163
73. Rutkoski TJ, Raines RT. Evasion of ribonuclease inhibitor as a determinant of ribonuclease cytotoxicity. Curr Pharm Biotechnol. (2008) 9:185–9. doi: 10.2174/138920108784567344
74. Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1. Nature. (2011) 469:419–23. doi: 10.1038/nature09674
75. Schroeder BO, Ehmann D, Precht JC, Castillo PA, Küchler R, Berger J, et al. Paneth cell α-defensin 6 (HD-6) is an antimicrobial peptide. Mucosal Immunol. (2015) 8:661–71. doi: 10.1038/mi.2014.100
76. Zhang J, Dyer KD, Rosenberg HF. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc Natl Acad Sci USA. (2000) 97:4701–6. doi: 10.1073/pnas.080071397
77. Rosenberg HF, Dyer KD. Molecular cloning and characterization of a novel human ribonuclease (RNase k6): increasing diversity in the enlarging ribonuclease gene family. Nucleic Acids Res. (1996) 24:3507–13. doi: 10.1093/nar/24.18.3507
78. Desloges I, Taylor JA, Leclerc JM, Brannon JR, Portt A, Spencer JD, et al. Identification and characterization of OmpT-like proteases in uropathogenic Escherichia coli clinical isolates. Microbiol Open. (2019) 2019:e915. doi: 10.1101/594564
79. Goossens H, Ferech M, Vander Stichele R, Elseviers M, Group EP. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet. (2005) 365:579–87. doi: 10.1016/S0140-6736(05)70799-6
80. Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. (2003) 55:27–55. doi: 10.1124/pr.55.1.2
81. Zasloff M. Antimicrobial peptides in health and disease. N Engl J Med. (2002) 347:1199–200. doi: 10.1056/NEJMe020106
Keywords: Ribonuclease A Superfamily, urinary tract infection, innate immunity, pyelonephritis, antimicrobial peptides
Citation: Becknell B, Ching C and Spencer JD (2019) The Responses of the Ribonuclease A Superfamily to Urinary Tract Infection. Front. Immunol. 10:2786. doi: 10.3389/fimmu.2019.02786
Received: 18 August 2019; Accepted: 13 November 2019;
Published: 29 November 2019.
Edited by:
Ester Boix, Autonomous University of Barcelona, SpainReviewed by:
Peter Veranic, University of Ljubljana, SloveniaXiaocui He, La Jolla Institute for Immunology (LJI), United States
Lukas Martin, University Hospital RWTH Aachen, Germany
Copyright © 2019 Becknell, Ching and Spencer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John David Spencer, am9obi5zcGVuY2VyQG5hdGlvbndpZGVjaGlsZHJlbnMub3Jn
 Brian Becknell1,2,3
Brian Becknell1,2,3 Christina Ching
Christina Ching John David Spencer
John David Spencer