- 1Discipline of Infectious Diseases and Immunology, Faculty of Medicine and Health, Charles Perkins Centre, University of Sydney, Camperdown, NSW, Australia
- 2School of Microbiology, University College Cork, Cork, Ireland
- 3APC Microbiome Ireland, University College Cork, Cork, Ireland
The critical role of interferons (IFNs) in mediating the innate immune response to cytomegalovirus (CMV) infection is well established. However, in recent years the functional importance of the IFN-independent antiviral response has become clearer. IFN-independent, IFN regulatory factor 3 (IRF3)-dependent interferon-stimulated gene (ISG) regulation in the context of CMV infection was first documented 20 years ago. Since then several IFN-independent, IRF3-dependent ISGs have been characterized and found to be among the most influential in the innate response to CMV. These include virus inhibitory protein, endoplasmic reticulum-associated IFN-inducible (viperin), ISG15, members of the interferon inducible protein with tetratricopeptide repeats (IFIT) family, interferon-inducible transmembrane (IFITM) proteins and myxovirus resistance proteins A and B (MxA, MxB). IRF3-independent, IFN-independent activation of canonically IFN-dependent signaling pathways has also been documented, such as IFN-independent biphasic activation of signal transducer and activator of transcription 1 (STAT1) during infection of monocytes, differential roles of mitochondrial and peroxisomal mitochondrial antiviral-signaling protein (MAVS), and the ability of human CMV (HCMV) immediate early protein 1 (IE1) protein to reroute IL-6 signaling and activation of STAT1 and its associated ISGs. This review examines the role of identified IFN-independent ISGs in the antiviral response to CMV and describes pathways of IFN-independent innate immune response induction by CMV.
Introduction
HCMV has a 236 kbp double stranded DNA (dsDNA) genome, 165 genes (1) encoding up to 751 protein products (2), a 45–100% seroprevalence in the adult population (3–7), and remains a significant human pathogen particularly in those with an underdeveloped or suppressed immune system. Just as HCMV infection can profoundly alter the overall adaptive immune response (8–13), it also generates a powerful innate response. Key mediators of this innate response are IFNs. There are three types of IFN: type I (α, β, κ, ω, τ, and ε), type II (γ), and type III (λ1, λ2, λ3, λ4). Type I and II IFNs are the best characterized in the context of HCMV and their induction, antiviral roles as well as the viral antagonism of these processes have been extensively reviewed (14–19). A role for type III IFNs, in the innate response to HCMV and murine CMV (MCMV), whose pathogenesis closely parallels that of HCMV (20), has recently been elucidated (21–27).
The innate response to both HCMV and MCMV infection is initiated when virus is detected by pattern recognition receptors (PRRs) including toll-like receptors (TLRs) TLR2 (28–31) and TLR9 (32–34). Once virus has bound and entered cells, HCMV and MCMV can be detected by cytosolic DNA sensors such as IFI16 (35, 36), ZBP1/DAI (37–39) and cGAS (32, 40) that signal through the stimulator of IFN genes (STING). Each of these pathways culminates in activation and dimerization of IRF3 resulting in production of type I IFN (41–44). Type I IFN production is subsequently enhanced by upregulation of IRF7, an ISG that is also capable of dimerizing and activating the type I IFN promoter (45). HCMV and MCMV infection both trigger production of type II IFN from CD8+ T cells, CD4+ T cells and natural killer (NK) cells (46–48). HCMV even remodels the IFNγ locus (IFNG) for sustained IFNγ expression in NKG2Chi NK cells (49, 50). IFNλ production is induced by HCMV and MCMV infection (22) and these type III IFNs are themselves ISGs with production stimulated by IFNα and IFNβ treatment (51).
Key antiviral mediators of all IFN types are ISGs (52). Interferome, a database dedicated to chronicling all genes significantly regulated by IFN (changes ≥ 2-fold), identifies 12614 ISGs (53). Type I IFNs alone can trigger expression of more than 2,000 genes in humans, many of which are antiviral (54). Canonical induction of ISGs by type I, II, and III IFNs occurs by JAK/STAT signaling downstream of the type I IFN receptor (IFNAR1 + IFNAR2), the IFNγ receptor (IFNGR1 + IFNGR2) and the IFNλ receptor (IFNLR1 + IL10R2), respectively. The type I and II IFN receptors are widely expressed but type III IFN receptor expression is limited to epithelial cells (55, 56). ISGs stimulated by type I and III IFN contain an IFN stimulated response element (ISRE) in their promoter region that is bound by the activated transcription factor IFN stimulated gene factor 3 (ISGF3), comprised of phosphorylated STAT1 and STAT2 with IRF9 (55, 57–62), or by STAT2 homodimers associated with IRF9 (63–65). IFNγ induced ISG promoters contain γ-activated sequences (GAS) that are bound by STAT1 homodimers (66–70). However, upregulation of some ISG mRNAs in the early stages of HCMV infection (prior to DNA replication) are not inhibited by IFN neutralization (71, 72). Since this discovery, the body of literature demonstrating ISG induction independent of canonical IFN signaling pathways has been steadily expanding and those discussed in this review are summarized in Figure 1.
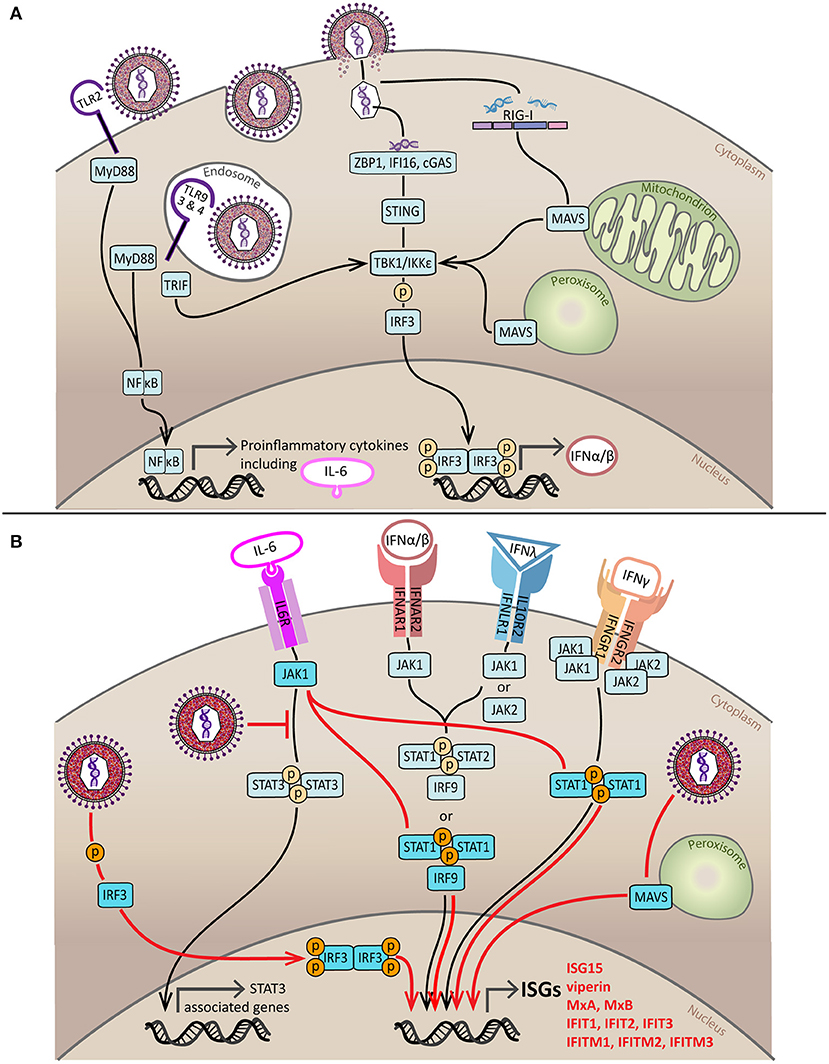
Figure 1. Induction and subversion of the innate IFN response by HCMV. (A) Sensing of HCMV by components of the innate immune response initiates production of IFNs and proinflammatory cytokines. HCMV is sensed by PRRs on the cell surface (TLR2) and in endosomes (TLR3, TLR4, and TLR9). Signaling from TLR2, TLR3, and TLR4 is through MyD88 and results in the activation and nuclear translocation NFκB, a transcription factor that stimulates expression of proinflammatory cytokines such as TNF, IL-8, IL-12, and IL-6. TLR9 and TLR4 signal through TRIF which causes activation by phosphorylation of IRF3 via TBK1/IKKε, activated IRF3 dimerizes and enters the nucleus to stimulate production of type I IFNs. HCMV infection can also be recognized by viral nucleic acid detectors in the cytoplasm; DNA sensors ZBP1, IFI16 and cGAS signal through ER-resident STING to activate TBK1/IKKε whilst the viral RNA sensor RIG-I activates TBK1/IKKε by signaling via MAVS located on the mitochondria or peroxisomes. The end result of both of these pathways is IRF3 phosphorylation, dimerization, nuclear translocation and production of type I IFNs. (B) IFN-dependent and IFN-independent pathways of ISG induction during HCMV infection. For IFN-dependent induction of ISGs to occur type I, type II and type III IFNs must bind to their cell surface receptors. Type I and III IFN receptors signal through various combinations of JAK proteins to phosphorylate STAT1 or STAT1 and STAT2 which form a complex referred to as ISGF3 with IRF9. ISGF3 then translocates to the nucleus where it binds to the ISRE to induce ISG production. The type II IFN receptor utilizes both JAK1 and JAK2 to phosphorylate STAT1, leading to its dimerization and nuclear translocation. Once in the nucleus, activated STAT1 dimers bind to GAS and stimulate ISG production. The three key pathways of HCMV-mediated IFN-independent ISG induction are indicated in red. Firstly, HCMV can directly activate IRF3; additionally, HCMV can sequester STAT3 and redirect the activated JAK1, created by IL-6 receptor binding, to phosphorylate STAT1; and finally peroxisomal MAVS may be able to trigger IFN-independent ISG expression at early times following infection. Black line = canonical IFN-dependent ISG induction pathway, red line = HCMV-induced, IFN-independent ISG induction pathway.
IFN-Independent ISG Production
Initial differential display analyses compared the susceptibility of genes upregulated early vs. late in infection to inhibition by IFN neutralizing antibodies and/or protein synthesis inhibitor cyclohexamide (CHX) (72). Three of these genes: IFIT2/ISG54/p54/cig42, IFIT3/ISG60/p60/cig49 and viperin/cig6, were upregulated by HCMV at 8 h post infection (hpi) and even accumulated following exposure to replication-incompetent ultraviolent-irradiated HCMV (UV-HCMV) (72). Blocking type I IFN with neutralizing antibodies failed to inhibit IFIT2, and IFIT3 induction, demonstrating that their upregulation was both IFN-independent and could be triggered by viral binding entry alone (72). A subsequent, broad mRNA analysis using oligonucleotide arrays found that levels of 258 mRNAs were altered more than 4-fold prior to initiation of HCMV DNA replication (71). IFIT2 and IFIT3 were among these quickly detected ISGs as were MxA, MxB, and ISG15 (71). The immediacy of this induction suggests a direct mechanism requiring few intermediary steps, indeed IFIT2, IFIT3, ISG15 (73) and viperin (72) upregulation can be detected 6 hpi with HCMV in the absence of de novo host and viral protein synthesis (cyclohexamide (CHX) treatment). This is also the case for IFIT1/ISG56/p56 (73) and indicates that this subset of ISGs may be induced/upregulated independently of IFN during HCMV infection.
IFN-Independent, IRF3-Dependent ISG Production
When searching for a mechanism underpinning IFN-independent ISG induction during CMV infection, initial studies turned to the powerful transcriptional regulator involved in IFN production, IRF3. Expression of constitutively active IRF3 in the absence of any viral stimulus could induce transcription of a subset of ISGs including IFIT1, IFIT2, IFIT3, ISG15, and viperin (74). IRF3-independent expression of these same ISGs was also observed during infection with other viruses: single stranded RNA (ssRNA) Newcastle disease virus (NDV) upregulated IFIT1, IFIT2 and ISG15 in cells that could respond to but were unable to produce type I IFN (75) and IFIT1 expression could be induced during ssRNA Sendai virus (SeV) infection by IRF3 nuclear translocation in cells unable to respond to type I IFN (76).
Studies using herpes simplex virus type 1 (HSV-1) demonstrated that IFIT1 expression could be driven by infection even in the presence of CHX in human fibroblasts (HFs) but could not be detected in the human epithelial osteosarcoma cell line U2OS (77). U2OS cells can respond to IFN but have defects in the STING signaling pathway (78) involved in IRF3 activation and dimerization in response to DNA sensing by IFI16, ZBP1/DAI, and cGAS (79–82). Furthermore, HSV-1 infection of IRF3−/−, IRF3−/−IRF9−/−, and IRF1−/− murine fibroblasts revealed that IRF3 was essential for generation of an antiviral state and IFIT2 expression in response to UV-HSV-1 (83). In the case of IFIT1, expression was directly induced by an IRF3-containing complex binding to its promoter region (77, 84).
In the context of HCMV infection, initiation of IFIT2 transcription was found to occur independently of STAT1 nuclear localization (85) and in the presence of CHX (86). Soon it emerged that expression of IFIT1, IFIT2, IFIT3 and ISG15 during HCMV could be IFN-independent but always required IRF3 activation (42, 73, 87). Subsequent studies revealed that viperin expression could be driven directly by HCMV glycoprotein B (gB), in an IFN-independent, IRF3/IRF1 dependent manner (88, 89). This aligns with data demonstrating that IRF3 translocation to the nucleus is a requirement for the IFN-independent induction of an antiviral state in response to UV-HCMV (87). In contrast, another transcription factor implicated in type I IFN production NFκB (90), remains cytosolic (91).
To interrogate the IFN-independent, IRF3-dependent response to HCMV HFs have been engineered (92, 93) to lack either IRF3 through expression of the nPro protein of bovine viral diarrhea virus (BVDV) (nPro/HFs) which binds and degrades IRF3 (94) or STAT1, by expression of the parainfluenza virus type 5 (PIV-5) V protein (V/HFs) which targets STAT1 for proteasomal degradation (95). These nPro/HFs and V/HFs were recently utilized, alongside IRF3 KO CRISPR/Cas9 HFs, to demonstrate that expression of viperin, ISG15, IFIT1, IFIT2, IFIT3, Mx1, and Mx2 mRNA during infection with HCMV can be induced in an IRF3-dependent, STAT1-independent manner (96). In fact, mRNA levels of IFIT1, IFIT2, and IFIT3 were as highly elevated in the absence of STAT1-mediated IFNAR signaling as in the parental HFs (96) underlining the capacity of such IFN-independent mechanisms to profoundly regulate ISG expression. Many of these IFN-independent, IRF3-dependent ISGs are among the most potently induced by CMV infection and examining the roles these genes play in the innate response to CMV is essential to understanding the ramifications of this non-canonical regulation.
Viperin
Viperin inhibits the egress and replication of many viruses (97–102). However, in the context of HCMV, viperin upregulation is proviral, initiated by infection to manipulate cellular metabolism and cause the accumulation of cytosolic lipids for use in production of the viral envelope (103). In trophoblasts, a cell type of particular clinical relevance due to their role in the transmission of congenital HCMV (104–106), viperin is required for efficient expression of immediate early viral genes (107). Viperin is also known to enhance type I IFN production in plasmacytoid dendritic cells (pDCs) by localizing to lipid rafts and acting as a scaffold for recruitment of interleukin-1 receptor-associated kinase 1 (IRAK1) and TNF receptor associated factor 6 (TRAF6) (108).
In addition, viperin has been identified to act in its capacity as a member of the radical S-adenosyl-L-methionine (SAM) superfamily of enzymes to facilitate conversion of cytidine triphosphate (CTP) to 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP) (109). Thus far, ddhCTP is known to act as a terminator of RNA synthesis by viral (Dengue and Zika) RNA-dependent RNA polymerases (109) and so investigations into its interaction with the HCMV encoded viral DNA polymerase are warranted. The viperin gene (RSAD2) lies in close proximity to the gene encoding cytidylate monophosphate kinase 2 (CMPK2) in the genome, suggesting a potential functional link to this pathway (109). Expression of CMPK2 is so closely linked to viperin that, following stimulation by IFN, viperin, CMPK2 and a long non-coding RNA (lncRNA) called lncRNA-CMPK2 are all co-transcribed (110). Interestingly, lncRNA-CMPK2 acts as a negative regulator of ISG expression (including ISG15, IFIT3 and IFITM1) (110). If IFN-independent, CMV-induced viperin upregulation also enhances expression of lncRNA-CMPK2, this could be a novel mechanism utilized by the virus to dampen the antiviral ISG response.
Furthermore, viperin has been demonstrated to be important for replication of Kaposi's sarcoma-associated herpesvirus (KSHV), a function attributed to the ability of viperin to catalyze oxidation of methionine in the viral DNA helicase, enhancing its expression and function (111). In this context, IFN-independent viperin upregulation by HCMV may be a way to ensure viral replication proceeds with maximum efficiency and thus the potential of viperin to modify the HCMV viral helicase-primase complex should be considered for further study. Overall, IFN-independent upregulation of viperin by HCMV seems to be a process initiated by the virus very early in infection to prepare the cell for its role as a virus-producing factory.
ISG15
ISG15 is a small ubiquitin-like protein that exists in three forms: (1) unconjugated within the cell, (2) conjugated within the cell (112, 113), and (3) secreted into serum (mainly by granulocytes) where it promotes NK maturation and IFNγ production (114). During HCMV infection accumulation of both free and conjugated ISG15 can be partially inhibited by interfering with the canonical IFNAR signaling pathway with a JAK inhibitor (115) but some IFN-independent, IRF3-dependent expression remains (96). Whilst the mechanisms by which ISG15 regulates CMV infection are currently unknown, it appears to possess antiviral activity as blocking ISG15 accumulation enhances viral replication (115) and HCMV antagonizes both the production of unconjugated ISG15 and ISGylation (115–118).
On the other hand, it is interesting to note that whilst in murine studies ISG15−/− mice are generally more sensitive to disseminated viral infections (119) human patients presenting with primary immunodeficiencies associated with defects in ISG15 expression are not (120). In fact, ISG15−/− fibroblasts isolated from such patients and primed with type I IFN were less susceptible to infection with HCMV than controls. This was attributed to the elevated levels of antiviral ISGs in these cells, a result of ISG15s ability to bind and stabilize the E3 ubiquitin ligase-like protein USP18, which acts as a negative regulator of the type I IFN response (120, 121).
It is also possible that HCMV manipulates levels of ISG15 to shift monocytes toward the mixed M1/M2 macrophage phenotype that is observed during infection (122) and hypothesized to enhance viral dissemination and persistence (123, 124). This is because in the absence of infection, ISG15 plays a role in the maintenance of mitochondrial homeostasis (125). Specifically, ISGylation of mitochondrial components can control mitochondrial function: reducing the rate of oxidative phosphorylation (OXPHOS) and causing a corresponding decrease in mitochondrial reactive oxygen species (ROS) (126). A reduction in levels of mitochondrial ROS alters macrophage polarization, shifting these cells toward a mixed M1/M2 phenotype (126).
IFITs
IFITs are ISGs with antiviral capabilities against flaviviruses, poxviruses, coronaviruses and papillomaviruses (127–130). A pan-viral mechanism of host defense mediated by IFITs is the sequestration of eukaryotic initiation factor (eIF3) by IFIT1 which slows the overall rate of cellular protein synthesis (76, 84, 131). A more specific strategy depends on the recognition and binding of viral RNA lacking 2′-O methylation of the 5′ RNA cap by IFIT1 (132). This binding ability is enhanced by association with IFIT2 and IFIT3 (133). Despite the fact that CMV replication takes place wholly within the nucleus, export of viral mRNAs does occur (134) and these may be sensed by IFITs. Another possibility is that IFITs may directly bind essential CMV proteins in the cytoplasm, as IFIT1 does to inhibit human papilloma virus (HPV) infection (135, 136). Although the mechanisms of the IFIT-mediated antiviral response to HCMV are still unclear, a significant reduction in titer has been reported when the virus is grown in IFIT1 overexpressing fetal astrocytes (137).
IFITMs
IFITM proteins are also implicated in the antiviral response against a wide range of viruses: orthomyxoviruses, flaviviruses, filoviruses, and coronaviruses often by blocking membrane fusion (127, 138–140). However, overexpression of IFITM1, IFITM2 and IFITM3 does not inhibit HCMV infection but rather results in a modest increase in the percentage of infected cells (141, 142). Short hairpin RNA (shRNA) knockdown of IFITM1 alone or in combination with IFITM2 and IFITM3 inhibits HCMV infection as they are required for successful formation of the HCMV virion assembly complex (vAC) and production of infectious progeny virions (142). It is interesting to note that despite this proviral role, IFITM proteins are noticeably downregulated at later stages of infection (48–72 hpi) (142).
Direct induction of IFITMs by HCMV may also contribute to the severe consequences of congenital infection as IFITM expression can inhibit the fusion of cytotrophoblast cells into the multinucleated syncytiotrophoblast, a structure at the interface between maternal and fetal tissue, essential for placental development (143).
MxA and MxB
The Mx proteins MxA and MxB are a family of dynamin-like GTPases first reported for their antiviral activity against influenza and are now well characterized in response to other viruses (144, 145). MxA is found in the cytosol and inhibits influenza virus infection through retention of the viral genome (146). On the other hand, MxB localizes to the cytoplasmic face of nuclear pores (147) and is able to inhibit HIV-1 replication by blocking nuclear viral genome accumulation (148, 149). Both MxA and MxB are highly upregulated by HCMV infection (73, 150) and it has recently been discovered that MxB overexpression inhibits replication of HSV-1, HSV-2, Kaposi's sarcoma-associated herpesvirus (KSHV), MCMV, and HCMV (151, 152). HSV-1 and MCMV inhibition manifested in a similar way to that of HIV-1, a block in the delivery of viral genome to the nucleus (151). However, in terms of the regions of protein at play, this mechanism was found to differ substantially with a requirement for GTP binding but not GTP hydrolysis (152, 153). Knockdown of MxB has also been implicated in stalling cell cycle progression (147) and it has been suggested that the HCMV virion protein pUL69 that contributes to the cell cycle arrest (154) does so via an interaction with MxB (155).
Alternate IFN-Independent Pathways of Innate Response Induction
Direct ISG induction by IRF3 is not the only pathway associated with the IFN-independent response to CMV. In human monocytes, IFN-independent, biphasic activation of STAT1 with differential phosphorylation at early (30 min) compared to late (24 h) time points post-HCMV infection appears to influence motility, migration, differentiation and polarization (156).
Regulation of mitochondrial activity is emerging as another IFN-independent innate response mediator. A number of years ago it was discovered that HCMV DNA could induce ISG expression in an IRF3-dependent, TLR-independent manner that involved TANK-binding kinase 1 (TBK1), IκB kinase epsilon [IKKε; originally called IKK-inducible (IKKi)], and mitochondrial antiviral-signaling protein (MAVS) (157). More recently, peroxisomal MAVS has been implicated in rapid type-I IFN-independent ISG (viperin, Mx2, IFIT3, IFIT2) expression (158). Conversely, mitochondrial MAVS appears to be involved in IFN-dependent ISG production (158). HCMV actively impairs mitochondrial MAVS signaling through the viral mitochondria-localized inhibitor of apoptosis (vMIA) and reduces type I IFN production (159). vMIA has also been found to localize to peroxisomes and induce their fragmentation by interaction with the cytoplasmic chaperone protein Pex19, hijacking the transport machinery of peroxisomal membrane proteins (160). This suggests that disabling IFN-independent ISG transcription induced by peroxisomal MAVS contributes to efficient CMV infection.
The HCMV immediate early gene 1 (IE1) is also capable of inducing expression of ISGs in the absence of IFN production. HCMV IE1 induces expression of IL-6 (161) which usually signals through JAK and STAT3 (162). However, IE1 binds and sequesters STAT3 (163), leaving JAK, already activated by IL-6, free to phosphorylate STAT1. Thus IE1 re-routes IL-6 signaling to activate STAT1 resulting in transcription of ISGs independently of IFN (164).
Functional Importance of IFN-Independent Innate Responses
Early studies examining IFN-independent induction of an antiviral state showed that treatment of human embryonic lung fibroblasts (HELFs) with UV-HCMV rendered these cells resistant to subsequent viral infection in the absence of detectable IFN production (91). Intriguingly, whilst high multiplicity of infection (MOI) UV-HCMV also induced an antiviral state in the HELFs, this required IFN production (91). Paladino et al. (91) proposed a model by which, when cells are exposed to limited numbers of virus particles (low MOI), induction of an internal antiviral state is sufficient to control infection, however, when many virus particles are present (high MOI), cells secrete IFN to protect neighboring cells too. The ability to induce an antiviral state in the absence of IFN production may be important in cells such as neurons, where inflammation is undesirable. In this respect, neurotropic arboviruses have been shown to induce protective type I IFN-independent, IRF3-dependent responses (165).
Recently, the power of IFN-independent innate responses to CMV has been illustrated by the finding that human macrophages co-cultured with HCMV-infected retinal pigment epithelial cells (RPEs) can limit viral replication and spread in a cell-cell contact dependent manner that could not be blocked by vaccinia-derived type I IFN binding protein B18R, nor by neutralizing antibodies against either IFNγ or TNFα (166). It has also been shown that HCMV virus particles pre-treated with HCMV-specific antibodies that do not replicate, nor express IE antigens, can enter human macrophages and induce an antiviral state that renders these cells less susceptible to subsequent HCMV infection independently of IFN production (167).
IFN-independent ISG induction can also be used to regulate the development of cells key to viral persistence and dissemination. In human monocytes infected with HCMV, ISGs are upregulated independently of IFN (4 hpi) that function to enhance monocyte motility and migration (156). This occurs in a STAT1-dependent manner that also suppresses transcription of anti-inflammatory M2-associated cytokines (IL-10 and CCL18), promoting polarization of macrophages toward a mixed M1/M2 phenotype (156). ISG15 was among the ISGs found to be upregulated in monocytes 4hpi with HCMV (156). ISG15 may contribute both directly and indirectly to the mixed M1/M2 macrophage phenotype, causing monocyte-specific upregulation of IL-10 (168) whilst simultaneously inducing production of M1 macrophage-stimulating cytokine IFNγ by NK and T cells (114).
Concluding Remarks
When considering the innate response to CMV infection, IFN and the ISG-mediated induction of an antiviral state are important first elements. The intention of this review has been to highlight the substantial body of literature accumulating around IFN-independent innate responses to CMV. IFN-independent induction of ISGs is an important phenomenon and ISGs produced via this pathway appear to play both pro- and anti-viral roles during infection. This complicates direct interrogation of the IFN response during viral infection and necessitates careful consideration of kinetics, as particular ISG may be upregulated directly by the virus, independently of IFN, to play a proviral role early in infection but later on, when IFN-dependent expression dominates, may antagonize infection.
When examining plaque number and size at 7 days post infection, we reported no difference in rates of CMV replication nor spread between cells unable to produce IFN (IRF3 degraded) and those that could not respond to IFN (lacking STAT1) (92), even though the latter cells still allowed viral induction of IFN-independent ISGs (96). Focusing on earlier time points, before loss of IFN production/signaling becomes the overwhelming factor affecting infection efficiency, may reveal more subtle differences conferred by abrogation of either IRF3 or STAT1 signaling.
A deeper understanding of the various functions of IFN-independent ISGs may enable their relative abundance to serve as a predictor of disease progression. For example, high levels of IL-6 have been correlated with CMV reactivation and poor prognosis for transplant patients (169–171); perhaps this is because ISGs produced by IE1 re-routing the IL-6 response to enhance infection. If this were the case, interference with STAT1 homodimer-mediated ISG expression may improve prognosis.
Since CMV infected, polarized macrophages are key mediators of T cell activation and proliferation (172), if IFN-independent ISGylation influences macrophage polarization then levels of ISG15 induced directly by CMV early in infection may provide an indication as to whether or not a robust T cell response will be generated.
It is also important to note that many of these ISGs, including viperin, IFIT2, IFIT3, Mx1 and ISG15 are defined as part of the 28 core mammalian ISGs i.e., produced in all nine mammalian species tested (54). It would therefore be prudent to determine whether their IFN independence is also conserved across species especially since rhesus CMV does not induce IRF3 activation nor the associated ISG expression (173).
Finally, with the IFN-independent nature of these ISGs becoming clear, caution should be exercised when using these ISGs as surrogate readouts for interferon signaling, as it is clear that they are also induced directly by viral infection.
Author Contributions
CA generated the initial draft of the manuscript. All other authors (BM, AA and BS) contributed to the subsequent writing and review of the manuscript.
Funding
This work was funded by National Health and Medical Research Project Grant funding (1063738) awarded to BS and AA. CA was the recipient of a Research Training Program Stipend (formerly Australian Postgraduate Award).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to acknowledge the many research groups who have contributed to a better understanding of the innate response to cytomegalovirus infection, particularly in the context of IFN-independence, and apologize to those whose work has not been cited due to space constraints.
References
1. Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, et al. Genetic content of wild-type human cytomegalovirus. J Gen Virol. (2004) 85(Pt 5):1301–12. doi: 10.1099/vir.0.79888-0
2. Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, et al. Decoding human cytomegalovirus. Science. (2012) 338:1088–93. doi: 10.1126/science.1227919
3. Mujtaba G, Shaukat S, Angez M, Alam MM, Hasan F, Zahoor Zaidi SS, et al. Seroprevalence of Human Cytomegalovirus (HCMV) infection in pregnant women and outcomes of pregnancies with active infection. J Pak Med Assoc. (2016) 66:1009–14.
4. Seale H, MacIntyre CR, Gidding HF, Backhouse JL, Dwyer DE, Gilbert L. National serosurvey of cytomegalovirus in Australia. Clin Vacc Immunol. (2006) 13:1181–4. doi: 10.1128/CVI.00203-06
5. Lachmann R, Loenenbach A, Waterboer T, Brenner N, Pawlita M, Michel A, et al. Cytomegalovirus (CMV) seroprevalence in the adult population of Germany. PLoS ONE. (2018) 13:e0200267. doi: 10.1371/journal.pone.0200267
6. Cannon MJ, Schmid DS, and Hyde TB Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. (2010) 20:202–13. doi: 10.1002/rmv.655
7. Cannon MJ, Griffiths PD, Aston V, Rawlinson WD. Universal newborn screening for congenital CMV infection: what is the evidence of potential benefit? Rev. Med. Virol. (2014) 24:291–307. doi: 10.1002/rmv.1790
8. Brodin P, Jojic V, Gao T, Bhattacharya S, Angel CJ, Furman D, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. (2015) 160:37–47. doi: 10.1016/j.cell.2014.12.020
9. Wallace DL, Masters JE, De Lara CM, Henson SM, Worth A, Zhang Y, et al. Human cytomegalovirus-specific CD8(+) T-cell expansions contain long-lived cells that retain functional capacity in both young and elderly subjects. Immunology. (2011) 132:27–38. doi: 10.1111/j.1365-2567.2010.03334.x
10. Weekes MP, Carmichael AJ, Wills MR, Mynard K, Sissons J. Human CD28(-)CD8(+) T cells contain greatly expanded functional virus-specific memory CTL clones. J Immunol. (1999) 162:7569–77.
11. van de Berg PJEJ, van Stijn A, ten Berge IJM, van Lier RAW A fingerprint left by cytomegalovirus infection in the human T cell compartment. J Clin Virol. (2008) 41:213–7. doi: 10.1016/j.jcv.2007.10.016
12. Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. (2005) 202:673–85. doi: 10.1084/jem.20050882
13. Looney RJ, Falsey A, Campbell D, Torres A, Kolassa J, Brower C, et al. Role of cytomegalovirus in the T cell changes seen in elderly individuals. Clin. Immunol. (1999) 90:213–9. doi: 10.1006/clim.1998.4638
14. Amsler L, Verweij M, and DeFilippis VR. The tiers and dimensions of evasion of the type I interferon response by human cytomegalovirus. J Mol Biol. (2013) 425:4857–71. doi: 10.1016/j.jmb.2013.08.023
15. Biolatti M, Gugliesi F, Dell'Oste V, Landolfo S. Modulation of the innate immune response by human cytomegalovirus. Infect Genet Evol. (2018) 64:105–14. doi: 10.1016/j.meegid.2018.06.025
16. Goodwin CM, Ciesla JH, Munger J. Who's driving? human cytomegalovirus, interferon, and NFκB signaling. Viruses. (2018) 10:E447. doi: 10.3390/v10090447
17. Galitska G, Biolatti M, Griffante G, Gugliesi F, Pasquero S, Dell'Oste V, et al. Catch me if you can: the arms race between human cytomegalovirus and the innate immune system. Fut Virol. (2019) 14:247–63. doi: 10.2217/fvl-2018-0189
18. Murray MJ, Peters NE, Reeves MB. Navigating the host cell response during entry into sites of latent cytomegalovirus infection. Pathogens. (2018) 7:30. doi: 10.3390/pathogens7010030
19. Rossini G, Cerboni C, Santoni A, Landini MP, Landolfo S, Gatti D, et al. Interplay between human cytomegalovirus and intrinsic/innate host responses: a complex bidirectional relationship 2012. Mediators Inflamm. (2012) 2012:607276. doi: 10.1155/2012/607276
20. Krmpotic A, Bubic I, Polic B, Lucin P, Jonjic S. Pathogenesis of murine cytomegalovirus infection. Microb Infect. (2003) 5:1263–77. doi: 10.1016/j.micinf.2003.09.007
21. Egli A, Levin A, Santer DM, Joyce M, O'Shea D, Thomas BS, et al. Immunomodulatory function of interleukin 28B during primary infection with cytomegalovirus. J Infect Dis. (2014) 210:717–27. doi: 10.1093/infdis/jiu144
22. Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, et al. IL-28A and IL-29 mediate antiproliferative and antiviral signals in intestinal epithelial cells and murine CMV infection increases colonic IL-28A expression. Am J Physiol Gastrointest Liver Physiol. (2005) 289:G960–8. doi: 10.1152/ajpgi.00126.2005
23. Annibali O, Piccioni L, Tomarchio V, Circhetta E, Sarlo C, Franceschini L, et al. Impact of IFN lambda 3/4 single nucleotide polymorphisms on the cytomegalovirus reactivation in autologous stem cell transplant patients. PLoS ONE. (2018) 13:e0200221. doi: 10.1371/journal.pone.0200221
24. Manuel O, Wójtowicz A, Bibert S, Mueller NJ, van Delden C, Hirsch HH, et al. Influence of IFNL3/4 polymorphisms on the incidence of cytomegalovirus infection after solid-organ transplantation. J Infect Dis. (2014) 211:906–14. doi: 10.1093/infdis/jiu557
25. Bibert S, Wojtowicz A, Taffe P, Manuel O, Bernasconi E, Furrer H, et al. The IFNL3/4 DeltaG variant increases susceptibility to cytomegalovirus retinitis among HIV-infected patients. Aids. (2014) 28:1885–9. doi: 10.1097/QAD.0000000000000379
26. Gimeno Brias S, Marsden M, Forbester J, Clement M, Brandt C, Harcourt K, et al. Interferon lambda is required for interferon gamma-expressing NK cell responses but does not afford antiviral protection during acute and persistent murine cytomegalovirus infection. PLoS ONE. (2018) 13:e0197596. doi: 10.1371/journal.pone.0197596
27. Ding S, Khoury-Hanold W, Iwasaki A, Robek MD. Epigenetic Reprogramming of the type III interferon response potentiates antiviral activity and suppresses tumor growth. PLoS Biol. (2014) 12:e1001758. doi: 10.1371/journal.pbio.1001758
28. Boehme KW, Singh J, Perry ST, Compton T. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein. J Virol B. (2004) 78:1202–11. doi: 10.1128/JVI.78.3.1202-1211.2004
29. Boehme KW, Guerrero M, Compton T. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J Immunol. (2006) 177:7094–102. doi: 10.4049/jimmunol.177.10.7094
30. Compton T, Kurt-Jones EA, Boehme KW, Belko J, Latz E, Golenbock DT, et al. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J Virol. (2003) 77:4588–96. doi: 10.1128/JVI.77.8.4588-4596.2003
31. Szomolanyi-Tsuda E, Liang X, Welsh RM, Kurt-Jones EA, and Finberg RW. Role for TLR2 in NK cell-mediated control of murine cytomegalovirus in vivo. J Virol. (2006) 80:4286–91. doi: 10.1128/JVI.80.9.4286-4291.2006
32. Paijo J, Döring M, Spanier J, Grabski E, Nooruzzaman M, Schmidt T, et al. cGAS senses human cytomegalovirus and induces type I interferon responses in human monocyte-derived cells. PLoS Pathog. (2016) 12:e1005546. doi: 10.1371/journal.ppat.1005546
33. Varani S, Cederarv M, Feld S, Tammik C, Frascaroli G, Landini MP, et al. Human cytomegalovirus differentially controls B cell and T cell responses through effects on plasmacytoid dendritic cells. J Immunol. (2007) 179:7767–76. doi: 10.4049/jimmunol.179.11.7767
34. Krug A, French AR, Barchet W. J., Fischer AA, Dzionek A, Pingel JT, et al. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. (2004) 21:107–19. doi: 10.1016/j.immuni.2004.06.007
35. Gariano GR. V., Dell'Oste, Bronzini M, Gatti D, Luganini A, De Andrea M, et al. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. (2012) 8:e1002498. doi: 10.1371/journal.ppat.1002498
36. Rolle S, De Andrea M, Gioia D, Lembo D, Hertel L, Landolfo S, et al. The interferon-inducible 204 gene is transcriptionally activated by mouse cytomegalovirus and is required for its replication. Virology. (2001) 286:249–55. doi: 10.1006/viro.2001.1021
37. DeFilippis VR, Alvarado D, Sali T, Rothenburg S, Früh K. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J Virol. (2010) 84:585–98. doi: 10.1128/JVI.01748-09
38. DeFilippis VR, Sali T, Alvarado D, White L, Bresnahan W, Früh KJ. Activation of the interferon response by human cytomegalovirus occurs via cytoplasmic double-stranded DNA but not glycoprotein B. J Virol. (2010) 84:8913–25. doi: 10.1128/JVI.00169-10
39. Upton JW, William KJ, Edward MS. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe. (2012) 11:290–7. doi: 10.1016/j.chom.2012.01.016
40. Lio CWJ, McDonald B, Takahashi M, Dhanwani R, Sharma N, Huang J, et al. cGAS-STING signaling regulates initial innate control of cytomegalovirus infection. J Virol. (2016) 90:7789–97. doi: 10.1128/JVI.01040-16
41. Au WC, Moore PA, Lowther W, Juang YT, Pitha PM. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. (1995) 92:11657–61. doi: 10.1073/pnas.92.25.11657
42. DeFilippis VR, Robinson B, Keck TM, Hansen SG, Nelson JA, Früh KJ. Interferon regulatory factor 3 is necessary for induction of antiviral genes during human cytomegalovirus infection. J Virol. (2006) 80:1032–7. doi: 10.1128/JVI.80.2.1032-1037.2006
43. Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. (2007) 448:501–5. doi: 10.1038/nature06013
44. Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STIN. Mol Cell. (2013) 51:226–35. doi: 10.1016/j.molcel.2013.05.022
45. Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. Embo J. (1998) 17:6660–9. doi: 10.1093/emboj/17.22.6660
46. Vieira Braga FA, Hertoghs KM, van Lier RA, van Gisbergen KP. Molecular characterization of HCMV-specific immune responses: Parallels between CD8(+) T cells, CD4(+) T cells, and NK cells. Eur J Immunol. (2015) 45:2433–45. doi: 10.1002/eji.201545495
47. Gamadia LE, Remmerswaal EBM, Weel JF, Bemelman F, van Lier RAW, Ten Berge IJM. Primary immune responses to human CMV: a critical role for IFN-γ-producing CD4+ T cells in protection against CMV disease. Blood. (2003) 101:2686–92. doi: 10.1182/blood-2002-08-2502
48. Quinn M, Turula H, Tandon M, Deslouches B, Moghbeli T, Snyder CM. Memory T cells specific for murine cytomegalovirus re-emerge after multiple challenges and recapitulate immunity in various adoptive transfer scenarios. J Immunol. (2015) 194:1726–36. doi: 10.4049/jimmunol.1402757
49. Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. (2012) 119:2665–74. doi: 10.1182/blood-2011-10-386995
50. Luetke-Eversloh M, Hammer Q, Durek P, Nordström K, Gasparoni G, Pink M, et al. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog. (2014) 10:e1004441. doi: 10.1371/journal.ppat.1004441
51. Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-λ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. (2006) 80:4501–9. doi: 10.1128/JVI.80.9.4501-4509.2006
52. Schoggins JW. Interferon-stimulated genes: what do they all do? Annu Rev Virol. (2019) 6:567–84. doi: 10.1146/annurev-virology-092818-015756
53. Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, et al. INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucl Acids Res. (2012). 41:D1040–6. doi: 10.1093/nar/gks1215
54. Shaw AE, Hughes J, Gu Q, Behdenna A, Singer JB, Dennis T, et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. (2017) 15:e2004086. doi: 10.1371/journal.pbio.2004086
55. Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J Virol. (2007) 81:7749. doi: 10.1128/JVI.02438-06
56. Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. (2008) 4:e1000017. doi: 10.1371/journal.ppat.1000017
57. Darnell JE Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. (1994) 264:1415–21. doi: 10.1126/science.8197455
58. Schindler C, Shuai K, Prezioso VR, Darnell JE Jr. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. (1992) 257:809–13. doi: 10.1126/science.1496401
59. Levy DE, Kessler DS, Pine R, Darnell JE Jr. Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. (1989) 3:1362–71. doi: 10.1101/gad.3.9.1362
60. Fu XY. A transcription factor with SH2 and SH3 domains is directly activated by an interferon alpha-induced cytoplasmic protein tyrosine kinase(s). Cell. (1992) 70:323–35. doi: 10.1016/0092-8674(92)90106-M
61. Levy DE, Kessler DS, Pine R, Reich N, Darnell JE Jr. Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. (1988) 2:383–93. doi: 10.1101/gad.2.4.383
62. Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. (2003) 4:69–77. doi: 10.1038/ni875
63. Poat B, Hazari S, Chandra PK, Gunduz F, Alvarez X, Balart LA, et al. Intracellular expression of IRF9 Stat fusion protein overcomes the defective Jak-Stat signaling and inhibits HCV RNA replication. Virol J. (2010) 7:265. doi: 10.1186/1743-422X-7-265
64. Kraus TA, Lau JF, Parisien JP, Horvath CM. A hybrid IRF9-STAT2 protein recapitulates interferon-stimulated gene expression and antiviral response. J Biol Chem. (2003) 278:13033–38. doi: 10.1074/jbc.M212972200
66. Shuai K, Schindler C, Prezioso V, Darnell J. Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science. (1992) 258:1808–12. doi: 10.1126/science.1281555
67. Decker T, Lew DJ, Mirkovitch J, Darnell JE Jr. Cytoplasmic activation of GAF, an IFN-gamma-regulated DNA-binding factor. Embo J. (1991) 10:927–32. doi: 10.1002/j.1460-2075.1991.tb08026.x
68. Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cytokine Res. (1997) 17:121–34. doi: 10.1089/jir.1997.17.121
69. Decker T, Lew DJ, Cheng YS, Levy DE, Darnell JE Jr. Interactions of alpha- and gamma-interferon in the transcriptional regulation of the gene encoding a guanylate-binding protein. Embo J. (1989) 8:2009–14. doi: 10.1002/j.1460-2075.1989.tb03608.x
70. Pearse RN, Feinman R, Ravetch JV. Characterization of the promoter of the human gene encoding the high-affinity IgG receptor: transcriptional induction by gamma-interferon is mediated through common DNA response elements. Proc Natl Acad Sci USA. (1991) 88:11305–9. doi: 10.1073/pnas.88.24.11305
71. Zhu H, Cong JP, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. (1998) 95:14470–5. doi: 10.1073/pnas.95.24.14470
72. Zhu H, Cong JP, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: Induction of interferon-responsive RNAs. Proc Natl Acad Sci USA. (1997) 94:13985–90. doi: 10.1073/pnas.94.25.13985
73. Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J Virol. (2001) 75:12319–30. doi: 10.1128/JVI.75.24.12319-12330.2001
74. Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. (2002) 76:5532–9. doi: 10.1128/JVI.76.11.5532-5539.2002
75. Wathelet MG, Berr PM, Huez GA. Regulation of gene expression by cytokines and virus in human cells lacking the type-I interferon locus. Eur J Biochem. (1992) 206:901–10. doi: 10.1111/j.1432-1033.1992.tb16999.x
76. Guo J, Peters KL, Sen GC. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology. (2000) 267:209–19. doi: 10.1006/viro.1999.0135
77. Mossman KL, Macgregor PF, Rozmus JJ, Goryachev AB, Edwards AM, Smiley JR. Herpes simplex virus triggers and then disarms a host antiviral response. J Virol. (2001) 75:750–8. doi: 10.1128/JVI.75.2.750-758.2001
78. Deschamps T, Kalamvoki M. Impaired STING pathway in human osteosarcoma U2OS cells contributes to the growth of ICP0-null mutant herpes simplex virus. J Virol. (2017) 91:e00006–17. doi: 10.1128/JVI.00006-17
79. Wu J, Sun L, Chen X, Du F, Shi H, Chen C, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNScience A. Science. (2013) 339:826–30. doi: 10.1126/science.1229963
80. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. (2013) 339:786–91. doi: 10.1126/science.1232458
81. Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMNature P. Nature. (2013) 503:530–4. doi: 10.1038/nature12640
82. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. (2015) 347:aaa2630. doi: 10.1126/science.aaa2630
83. Collins SE, Noyce RS, Mossman KL. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J Virol. (2004) 78:1706–17. doi: 10.1128/JVI.78.4.1706-1717.2004
84. Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. (2000) 19:6891–9. doi: 10.1093/emboj/19.24.6891
85. Navarro L, Mowen K, Rodems S, Weaver B, Reich N, Spector D, et al. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol Cell Biol. (1998) 18:3796–02. doi: 10.1128/MCB.18.7.3796
86. Preston CM, Harman AN, Nicholl MJ. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J Virol. (2001) 75:8909–16. doi: 10.1128/JVI.75.19.8909-8916.2001
87. Noyce RS, Collins SE, Mossman KL. Identification of a novel pathway essential for the immediate-early, interferon-independent antiviral response to enveloped virions. J Virol. (2006) 80:226–35. doi: 10.1128/JVI.80.1.226-235.2006
88. Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci USA. (2001) 98:15125–30. doi: 10.1073/pnas.011593298
89. Stirnweiss A, Ksienzyk A, Klages K, Rand U, Grashoff M, Hauser H, et al. IFN regulatory factor-1 bypasses IFN-mediated antiviral effects through viperin gene induction. J Immunol. (2010) 184:5179–85. doi: 10.4049/jimmunol.0902264
90. Jin J, Hu H, Li HS, Yu J, Xiao Y, Brittain GC, et al. Noncanonical NF-κB pathway controls the production of type I interferons in antiviral innate immunity. Immunity. (2014) 40:342–54. doi: 10.1016/j.immuni.2014.02.006
91. Paladino P, Cummings DT, Noyce RS, Mossman KL. The IFN-independent response to virus particle entry provides a first line of antiviral defense that is independent of TLRs and retinoic acid-inducible gene I. J Immunol. (2006) 177:8008–16. doi: 10.4049/jimmunol.177.11.8008
92. McSharry BP, Forbes SK, Avdic S, Randall RE, Wilkinson GW, Abendroth A, et al. Abrogation of the interferon response promotes more efficient human cytomegalovirus replication. J Virol. (2015) 89:1479–83. doi: 10.1128/JVI.02988-14
93. McSharry BP, Forbes SK, Cao JZ, Avdic S, Machala EA, Gottlieb DJ, et al. Human cytomegalovirus upregulates expression of the lectin galectin 9 via induction of beta interferon. J Virol. (2014) 88:10990–4. doi: 10.1128/JVI.01259-14
94. Hilton L, Moganeradj K, Zhang G, Chen YH, Randall RE, McCauley JW, et al. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J Virol. (2006) 80:11723–32. doi: 10.1128/JVI.01145-06
95. Andrejeva J, Young DF, Goodbourn S, Randall RE. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J Virol. (2002) 76:2159–67. doi: 10.1128/jvi.76.5.2159-2167.2002
96. Ashley CL, Abendroth A, McSharry BP, Slobedman B. Interferon-independent upregulation of interferon-stimulated genes during human cytomegalovirus infection is dependent on IRF3 expression. Viruses. (2019) 11:246. doi: 10.3390/v11030246
97. Van der Hoek KH, Eyre NS, Shue B, Khantisitthiporn O, Glab-Ampi K, Carr JM, et al. Viperin is an important host restriction factor in control of Zika virus infection. Sci Rep. (2017) 7:4475. doi: 10.1038/s41598-017-04138-1
98. Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. (2007) 2:96–105. doi: 10.1016/j.chom.2007.06.009
99. Rivieccio MA, Suh HS, Zhao Y, Zhao ML, Chin KC, Lee SC, et al. TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for viperin/cig5. J Immunol. (2006) 177:4735–41. doi: 10.4049/jimmunol.177.7.4735
100. Zhang Y, Burke CW, Ryman KD, Klimstra WB. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J Virol. (2007) 81:11246–55. doi: 10.1128/JVI.01282-07
101. Helbig KJ, Lau DT, Semendric L, Harley HA, Beard MR. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology. (2005) 42:702–10. doi: 10.1002/hep.20844
102. Helbig KJ, Beard MR. The role of viperin in the innate antiviral response. J Mol Biol. (2014) 426:1210–9. doi: 10.1016/j.jmb.2013.10.019
103. Seo JY, Cresswell P. Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog. (2013) 9:e1003497. doi: 10.1371/journal.ppat.1003497
104. Halwachs-Baumann G, Wilders-Truschnig M, Desoye G, Hahn T, Kiesel L, Klingel K, et al. Human trophoblast cells are permissive to the complete replicative cycle of human cytomegalovirus. J Virol. (1998) 72:7598–602.
105. Tabata T, Petitt M, Zydek M, Fang-Hoover J, Larocque N, Tsuge M, et al. Human cytomegalovirus infection interferes with the maintenance and differentiation of trophoblast progenitor cells of the human placenta. J Virol. (2015) 89:5134–47. doi: 10.1128/JVI.03674-14
106. Hemmings DG, Kilani R, Nykiforuk C, Preiksaitis J, Guilbert LJ. Permissive cytomegalovirus infection of primary villous term and first trimester trophoblasts. J Virol. (1998) 72:4970–9.
107. Wang B, Fang Y, Wu Y, Koga K, Osuga Y, Lv S, et al. Viperin is induced following toll-like receptor 3 (TLR3) ligation and has a virus-responsive function in human trophoblast cells. Placenta. (2015) 36:667–73. doi: 10.1016/j.placenta.2015.03.002
108. Saitoh T, Satoh T, Yamamoto N, Uematsu S, Takeuchi O, Kawai T, et al. Antiviral protein viperin promotes toll-like receptor 7- and toll-like receptor 9-mediated type i interferon production in plasmacytoid dendritic cells. Immunity. (2011) 34:352–63. doi: 10.1016/j.immuni.2011.03.010
109. Gizzi AS, Grove TL, Arnold JJ, Jose J, Jangra RK, Garforth SJ, et al. A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature. (2018) 558:610–4. doi: 10.1038/s41586-018-0238-4
110. Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucl Acids Res. (2014) 42:10668–80. doi: 10.1093/nar/gku713
111. Bai L, Dong J, Liu Z, Rao Y, Feng P, Lan K. Viperin catalyzes methionine oxidation to promote protein expression and function of helicases. Sci Adv. (2019) 5:eaax1031. doi: 10.1126/sciadv.aax1031
112. Blomstrom DC, Fahey D, Kutny R, Korant BD, Knight E Jr. Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J Biol Chem. (1986) 261:8811–6.
113. Haas AL, Ahrens P, Bright PM, Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem. (1987) 262:11315–23.
114. Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, et al. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. (2012) 337:1684–8. doi: 10.1126/science.1224026
115. Bianco C, Mohr I. Restriction of human cytomegalovirus replication by ISG15, a host effector regulated by cGAS-STING double-stranded-DNA sensing. J Virol. (2017) 91:e02483–16. doi: 10.1128/JVI.02483-16
116. Kim YJ, Kim ET, Kim YE, Lee MK, Kwon KM, Kim KI, et al. Consecutive inhibition of ISG15 expression and ISGylation by cytomegalovirus regulators. PLoS Pathog. (2016) 12:e1005850. doi: 10.1371/journal.ppat.1005850
117. Lee MK, Kim YJ, Kim YE, Han TH, Milbradt J, Marschall M, et al. Transmembrane protein pUL50 of human cytomegalovirus inhibits ISGylation by downregulating UBE1L. J Virol. (2018) 92:e00462–18. doi: 10.1128/JVI.00462-18
118. Zimmermann C, Büscher N, Krauter S, Krämer N, Wolfrum U, Sehn E, et al. The abundant tegument protein pUL25 of human cytomegalovirus prevents proteasomal degradation of pUL26 and supports its suppression of ISGylation. J Virol. (2018) 92:e01180–18. doi: 10.1128/JVI.01180-18
119. Schoggins JW. Interferon-stimulated genes: roles in viral pathogenesis. Curr Opin Virol. (2014) 6:40–6. doi: 10.1016/j.coviro.2014.03.006
120. Speer SD, Li Z, Buta S, Payelle-Brogard B, Qian L, Vigant F, et al. ISG15 deficiency and increased viral resistance in humans but not mice. Nat Commun. (2016) 7:11496. doi: 10.1038/ncomms11496
121. Malakhova OA, Yan M, Malakhov MP, Yuan Y, Ritchie KJ, Kim KI, et al. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. (2003) 17:455–60. doi: 10.1101/gad.1056303
122. Chan G, Bivins-Smith ER, Smith MS, Smith PM, Yurochko AD. Transcriptome analysis reveals human cytomegalovirus reprograms monocyte differentiation toward an M1 macrophage. J Immunol. (2008) 181:698–711. doi: 10.4049/jimmunol.181.1.698
123. Smith MS, Bentz GL, Alexander JS, Yurochko AD. Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J Virol. (2004) 78:4444–53. doi: 10.1128/JVI.78.9.4444-4453.2004
124. Chan G, Nogalski MT, Stevenson EV, Yurochko AD. Human cytomegalovirus induction of a unique signalsome during viral entry into monocytes mediates distinct functional changes: a strategy for viral dissemination. J Leukocyte Biol. (2012) 92:743–52. doi: 10.1189/jlb.0112040
125. Albert M, Bécares M, Falqui M, Fernández-Lozano C, Guerra S. ISG15, a small molecule with huge implications: regulation of mitochondrial homeostasis. Viruses. (2018) 10:629. doi: 10.3390/v10110629
126. Baldanta S, Fernández-Escobar M, Acín-Perez R, Albert M, Camafeita E, Jorge I, et al. ISG15 governs mitochondrial function in macrophages following vaccinia virus infection. PLoS Pathog. (2017) 13:e1006651. doi: 10.1371/journal.ppat.1006651
127. Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. (2012) 13:46. doi: 10.1038/nri3344
128. Vladimer GI, Górna MW, Superti-Furga G. IFITs: emerging roles as key anti-viral proteins. Front. Immunol. (2014) 5:94. doi: 10.3389/fimmu.2014.00094
129. Fensterl V, Sen GC. Interferon-induced Ifit proteins: their role in viral pathogenesis. J Virol. (2015) 89:2462–8. doi: 10.1128/JVI.02744-14
130. Mears HV, Sweeney TR. Better together: the role of IFIT protein-protein interactions in the antiviral response. J Gen Virol. (2018) 99:1463–77. doi: 10.1099/jgv.0.001149
131. Wang C, Pflugheber J, Sumpter R, Sodora DL, Hui D, Sen GC, et al. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol. (2003) 77:3898–912. doi: 10.1128/JVI.77.7.3898-3912.2003
132. Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, et al. 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. (2010) 468:452–6. doi: 10.1038/nature09489
133. Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, et al. IFIT1 is an antiviral protein that recognizes 5'-triphosphate RNA. Nat Immunol. (2011) 12:624–30. doi: 10.1038/ni.2048
134. Lischka P, Toth Z, Thomas M, Mueller R, Stamminger T. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol Cell Biol. (2006) 26:1631–43. doi: 10.1128/MCB.26.5.1631-1643.2006
135. Saikia P, Fensterl V, Sen GC. The inhibitory action of P56 on select functions of E1 mediates interferon's effect on human papillomavirus DNA replication. J Virol. (2010) 84:13036–9. doi: 10.1128/JVI.01194-10
136. Terenzi F, Saikia P, Sen GC. Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. Embo J. (2008) 27:3311–21. doi: 10.1038/emboj.2008.241
137. Zhang L, Wang B, Li L, Qian DM, Yu H, Xue ML, et al. Antiviral effects of IFIT1 in human cytomegalovirus-infected fetal astrocytes. J Med Virol. (2017) 89:672–84. doi: 10.1002/jmv.24674
138. Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol. (2013) 425:4937–55. doi: 10.1016/j.jmb.2013.09.024
139. Smith S, Weston S, Kellam P, Marsh M. IFITM proteins-cellular inhibitors of viral entry. Curr Opin Virol. (2014) 4:71–7. doi: 10.1016/j.coviro.2013.11.004
140. Liao Y, Goraya MU, Yuan X, Zhang B, Chiu SH, Chen JL. Functional involvement of interferon-inducible transmembrane proteins in antiviral immunity. Front Microbiol. (2019) 10:1097. doi: 10.3389/fmicb.2019.01097
141. Warren CJ, Griffin LM, Little AS, Huang IC, Farzan M, Pyeon D. The antiviral restriction factors IFITM1, 2 and 3 do not inhibit infection of human papillomavirus, cytomegalovirus and adenovirus. PLoS ONE. (2014) 9:e96579. doi: 10.1371/journal.pone.0096579
142. Xie M, Xuan B, Shan J, Pan D, Sun Y, Shan Z, et al. Human cytomegalovirus exploits interferon-induced transmembrane proteins to facilitate morphogenesis of the virion assembly compartment. J Virol. (2015) 89:3049–61. doi: 10.1128/JVI.03416-14
143. Buchrieser J, Degrelle SA, Couderc T, Nevers Q, Disson O, Manet C, et al. IFITM proteins inhibit placental syncytiotrophoblast formation and promote fetal demise. Science. (2019) 365:176–80.
144. Verhelst J, Hulpiau P, Saelens X. Mx proteins: antiviral gatekeepers that restrain the uninvited. Microbiol Mol Biol Rev. (2013) 77:551–66. doi: 10.1128/MMBR.00024-13
145. Haller O, Stertz S, Kochs G. The Mx GTPase family of interferon-induced antiviral proteins. Microb Infect. (2007) 9:1636–43. doi: 10.1016/j.micinf.2007.09.010
146. Xiao H, Killip MJ, Staeheli P, Randall RE, Jackson D. The human interferon-induced MxA protein inhibits early stages of influenza A virus infection by retaining the incoming viral genome in the cytoplasm. J Virol. (2013) 87:13053–8. doi: 10.1128/JVI.02220-13
147. King MC, Raposo G, Lemmon MA. Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc Natl Acad Sci USA. (2004) 101:8957–62. doi: 10.1073/pnas.0403167101
148. Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. (2013) 502:563–6. doi: 10.1038/nature12653
149. Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. (2013) 502:559–62. doi: 10.1038/nature12542
150. Pautasso S, Galitska G, Dell'Oste V, Biolatti M, Cagliani R, Forni D, et al. Strategy of human cytomegalovirus to escape interferon beta-induced APOBEC3G editing activity. J Virol. (2018) 92:e01224–18. doi: 10.1128/JVI.01224-18
151. Crameri M, Bauer M, Caduff N, Walker R, Steiner F, Franzoso FD, et al. MxB is an interferon-induced restriction factor of human herpesviruses. Nat Commun. (2018) 9:1980. doi: 10.1038/s41467-018-04379-2
152. Schilling M, Bulli L, Weigang S, Graf L, Naumann S, Patzina C, et al. Human MxB protein is a pan-herpesvirus restriction factor. J Virol. (2018) 92:e01056–18. doi: 10.1128/JVI.01056-18
153. Jaguva Vasudevan AA, Bähr A, Grothmann R, Singer A, Häussinger D, Zimmermann A, et al. MXB inhibits murine cytomegalovirus. Virology. (2018) 522:158–67. doi: 10.1016/j.virol.2018.07.017
154. Hayashi ML, Blankenship C, Shenk T. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc Natl Acad Sci USA. (2000) 97:2692–6. doi: 10.1073/pnas.050587597
155. Aoyagi M, Gaspar M, Shenk TE. Human cytomegalovirus UL69 protein facilitates translation by associating with the mRNA cap-binding complex and excluding 4EBP1. Proc Natl Acad Sci USA. (2010) 107:2640–5. doi: 10.1073/pnas.0914856107
156. Collins-McMillen D, Stevenson EV, Kim JH, Lee BJ, Cieply SJ, Nogalski MT, et al. Human cytomegalovirus utilizes a nontraditional signal transducer and activator of transcription 1 activation cascade via signaling through epidermal growth factor receptor and integrins to efficiently promote the motility, differentiation, and polarization of infected monocytes. J Virol. (2017) 91:e00622–17. doi: 10.1128/JVI.00622-17
157. Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, et al. A Toll-like receptor–independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. (2006) 7:40–8. doi: 10.1038/ni1282
158. Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. (2010) 141:668–81. doi: 10.1016/j.cell.2010.04.018
159. Castanier C, Garcin D, Vazquez A, Arnoult D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. (2010) 11:133–8. doi: 10.1038/embor.2009.258
160. Magalhães AC, Ferreira AR, Gomes S, Vieira M, Gouveia A, Valença I, et al. Peroxisomes are platforms for cytomegalovirus' evasion from the cellular immune response. Sci Rep. (2016) 6:26028. doi: 10.1038/srep26028
161. Geist LJ, Dai LY. Cytomegalovirus modulates interleukin-6 gene expression. Transplantation. (1996) 62:653–8. doi: 10.1097/00007890-199609150-00020
162. Zhong Z, Wen Z, Darnell JE Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. (1994) 264:95–8. doi: 10.1126/science.8140422
163. Reitsma JM, Sato H, Nevels M, Terhune SS, Paulus C. Human cytomegalovirus IE1 protein disrupts interleukin-6 signaling by sequestering STAT3 in the nucleus. (2013). J Virol. 87:10763–76. doi: 10.1128/JVI.01197-13
164. Harwardt T, Lukas S, Zenger M, Reitberger T, Danzer D, Übner T, et al. Human cytomegalovirus immediate-early 1 protein rewires upstream STAT3 to downstream STAT1 signaling switching an IL6-type to an IFNγ-like response. PLoS Pathog. (2016) 12:e1005748. doi: 10.1371/journal.ppat.1005748
165. Peltier DC, Lazear HM, Farmer JR, Diamond MS, Miller DJ. Neurotropic arboviruses induce interferon regulatory factor 3-mediated neuronal responses that are cytoprotective, interferon independent, and inhibited by Western equine encephalitis virus capsid. J Virol. (2013) 87:1821–33. doi: 10.1128/JVI.02858-12
166. Becker J, Kinast V, Döring M, Lipps C, Duran V, Spanier J, et al. Human monocyte-derived macrophages inhibit HCMV spread independent of classical antiviral cytokines. Virulence. (2018) 9:1669–84. doi: 10.1080/21505594.2018.1535785
167. Wu Z, Qin R, Wang L, Bosso M, Scherer M, Stamminger T, et al. Human cytomegalovirus particles treated with specific antibodies induce intrinsic and adaptive but not innate immune responses. J Virol. (2017) 91:e00678–17. doi: 10.1128/JVI.00678-17
168. Dos Santos PF, Van Weyenbergh J, Delgobo M, Oliveira Patricio D, Ferguson BJ, Guabiraba R, et al. ISG15-induced IL-10 is a novel anti-inflammatory myeloid axis disrupted during active tuberculosis. J Immunol. (2018) 200:1434–42. doi: 10.4049/jimmunol.1701120
169. Botto S, Streblow DN, DeFilippis V, White L, Kreklywich CN, Smith PP, et al. IL-6 in human cytomegalovirus secretome promotes angiogenesis and survival of endothelial cells through the stimulation of survivin. Blood. (2011) 117:352–61. doi: 10.1182/blood-2010-06-291245
170. Humar A. St., Louis P, Mazzulli T, McGeer A, Lipton J, Messner H, et al. Elevated serum cytokines are associated with cytomegalovirus infection and disease in bone marrow transplant recipients. J Infect Dis. (1999) 179:484–8. doi: 10.1086/314602
171. Limaye AP, Stapleton RD, Peng L, Gunn SR, Kimball LE, Hyzy R, et al. Effect of ganciclovir on IL-6 levels among cytomegalovirus-seropositive adults with critical illness: a randomized clinical trialganciclovir and IL-6 levels among CMV-seropositive critically Ill adultsganciclovir and IL-6 levels among CMV-seropositive critically Ill adults. JAMA. (2017) 318:731–40. doi: 10.1001/jama.2017.10569
172. Bayer C, Varani S, Wang L, Walther P, Zhou S, Straschewski S, et al. Human cytomegalovirus infection of M1 and M2 macrophages triggers inflammation and autologous T-cell proliferation. J Virol. (2013) 87:67–79. doi: 10.1128/JVI.01585-12
Keywords: interferon, cytomegalovirus, IFN-independent, ISG, herpes, innate immunity
Citation: Ashley CL, Abendroth A, McSharry BP and Slobedman B (2019) Interferon-Independent Innate Responses to Cytomegalovirus. Front. Immunol. 10:2751. doi: 10.3389/fimmu.2019.02751
Received: 20 August 2019; Accepted: 11 November 2019;
Published: 11 December 2019.
Edited by:
Soren R. Paludan, Aarhus University, DenmarkReviewed by:
Sarah Rowland-Jones, University of Oxford, United KingdomJohan Van Weyenbergh, KU Leuven, Belgium
Copyright © 2019 Ashley, Abendroth, McSharry and Slobedman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barry Slobedman, barry.slobedman@sydney.edu.au
†These authors have contributed equally to this work
 Caroline L. Ashley
Caroline L. Ashley Allison Abendroth
Allison Abendroth Brian P. McSharry
Brian P. McSharry Barry Slobedman1*†
Barry Slobedman1*†