
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 21 November 2019
Sec. T Cell Biology
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.02710
The invariant NKT (iNKT) cells recognize glycolipid antigens presented by the non-classical MHC like molecule CD1d. They represent an innate T-cell lineage with the ability to rapidly produce a variety of cytokines in response to agonist stimulation to bridge innate and adaptive immunity. In thymus, most iNKT cells complete their maturation and differentiate to multiple effector lineages such as iNKT-1, iNKT-2, and iNKT-17 cells that possess the capability to produce IFNγ, IL-4, and IL-17A, respectively, and play distinct roles in immune responses and diseases. Mechanisms that control iNKT lineage fate decisions are still not well understood. Evidence has revealed critical roles of Foxo1 of the forkhead box O1 subfamily of transcription factors in the immune system. However, its role in iNKT cells has been unknown. In this report, we demonstrate that deletion of Foxo1 causes severe decreases of iNKT cell total numbers due to impairment of late but not early iNKT cell development. Deficiency of Foxo1 results in decreases of iNKT-1 but increases of iNKT-17 cells. Our data reveal that Foxo1 controls iNKT effector lineage fate decision by promoting iNKT-1 but suppressing iNKT-17 lineages.
iNKT cells express TCRs that contain an invariant TCRVα14-Jα13 (iVα14) chain paired with limited numbers of TCRβ chain that recognize glycolipid antigens presented by the non-classic MHC-I like molecule CD1d. They play important roles in immune responses and tissue repair but also contribute to the pathogenesis of diseases (1–4). The iVα14 TCR signaling via multiple pathways such as RasGRP1-Ras-Erk1/2 pathway, the PKCθ-NFκB pathway, and mTOR is crucial for iNKT cell development in the thymus (2, 5–12). iNKT cells are traditionally defined into four developmental stages in the thymus: stage 0 (CD24+CD44−NK1.1−), stage 1 (CD24−CD44−NK1.1−), stage 2 (CD24−CD44+NK1.1−), and stage 3 (CD24−CD44+NK1.1+) (13, 14). Recently, iNKT cells have also been defined into multiple effector lineages based on expression of specific transcription factors and cytokines. These effector lineages identified thus far include IFN-γ-producing iNKT1, IL-4-producing iNKT2, IL-17A-producing iNKT17, iNKT10, Tfh-like iNKTfh, and Treg-like iNKT cells. iNKT1, iNKT2, iNKT17, and iNKTfh cells express T-bet, Gata3, RORγt, and Bcl6 that correspond to master transcription factors in individual T helper lineages, respectively (15–23). Stage 3 iNKT cells are virtually all iNKT1 cells (15, 24, 25), while iNKT17 cells reside in stage 2 CD44+NK1.1− neuropilin-1+ ICOS+ cells (8, 18, 26–28). iNKT2 cells can be found in both stage 1 and stage 2 iNKT cells (28). Individual iNKT cell lineages exert distinct functions in immune responses and diseases. iNKT cell derived IL-4 participates in regulation of early germinal center B cell responses (29), liver regeneration and repair after injury (30), and adipocyte homeostasis (31). iNKT cell derived IL17 exacerbates airway hypersensitivity by recruiting neutrophils and inducing smooth muscle contraction (8, 20). iNKTfh cells secrets IL-21 and provides direct cognate help to antigen-specific B cells (16). Different from conventional αβT cells, most iNKT cells complete their effector lineage differentiation in the thymus and their relative abundance is influenced by genetic backgrounds, environmental factors, and intrinsic signal, metabolic, and transcriptional pathways. The mechanisms that control iNKT cell effector lineage fate decision are still not fully understood.
Foxo1 belongs to the Forkhead box “O” subfamily of transcription factors binding to a consensus DNA sequence 5′-(A/C)AA(C/T)A to regulate transcription of many genes involved in cell survival, cell cycle, differentiation, and cancer (32). Many mechanisms have evolved to regulate Foxo1 at both transcriptional and post transcriptional levels. A key mechanism is its phosphorylation by Akt and serum/glucocorticoid regulated kinase 1 (SGK1) on different sites that leads to its export from the nucleus and sequestration in the cytosol due to increased association to 14-3-3 proteins (33). Many studies have revealed important roles of the Foxo subfamily in the immune system (32, 34). Foxo1 promotes production of inflammatory cytokines in and regulates migration of dendritic cells (35, 36), is essential for B cell development and Ig class-switch (37–39), and regulates T cell differentiation and function (34). It controls T cell survival and homing via increasing IL7 receptor expression, CCR7, and L-selectin expression (40), plays important roles for regulatory T cell development and function (41–43), negatively controls Th17 cell differentiation (44–47) and Tfh differentiation (48–50), promotes Th9 responses (51–53), and regulates CD8 T cell effetor/memory differentiation (54–56). We report here that deficiency of Foxo1 does not grossly affect early iNKT cell development but severely impacts late iNKT cell development, manifested by severe reduction of iNKT1 cells but increases of iNKT17 cells. Our data suggest that Foxo1 is a critical regulator that controls iNKT1 and iNKT17 effector fate decision.
Foxo1f/f and Cd2iCre mice were purchased from the Jackson Laboratory (57, 58). Foxo1f/f had further backcrossed to C57BL6/J background. Two to ten-week-old Foxo1f/f-Cd2iCre and Foxo1f/f or Cd2iCre littermate control mice were used for the experiments. For each experiment, one pair of test and control mice of the same sex littermates examined. All mouse experiments were performed according to protocols approved by the Duke University Institute Animal Care and Use Committee.
Ten millions of viable CD4+CD8+ thymocytes from age- and sex-matched Foxo1f/f and Foxo1f/f -Cd2iCre mice were sorted on MoFlo Cell Sorter (Beckman Coulter) with post-sort purity of 98%. Genomic DNAs were extracted with phenol/chloroform, precipitated with 70% ethanol, dissolved in TE buffer (10 mM Tris-0.5 mM EDTA [pH 8]), and utilized as templates for PCR reaction. The forward and reverse primers for Foxo1 were 5′-ACCACTCTGGACGGCATACT-3′ and 5′-TAACATAAAGGGAGATGAAGCA-3′, respectively. These two primers flank one of the two Loxp sites in the Foxo1 locus so that they did not amply a PCR product after Cre mediated recombination. Primers for Dgkz as control were 5′-AGAAAGCTGATCCCCCACAT-3′ (forward) and 5′-AGAGAGCGTCCTTCAAGAGG-3′ (reverse). PCR products were visualized after electrophoresis in 1% agarose gel.
Thymocytes, splenocytes, and liver mononuclear cells were prepared according to published protocols (9, 10). Cells were stained for surface markers with appropriate fluorochrome-conjugated antibodies in PBS containing 2% FBS on ice for 30 min followed by intracellular staining of transcription factors using the BD Bioscience Transcription Factor Buffer Set or Ki67 using the BD Bioscience Cytofix/Cytoperm™ solution according to the manufacturer's protocols. Data were collected using a BD LSRFortessa™ cytometer (BD Biosciences). PE- or allophycocyanin-labeled PBS57-loaded CD1d tetramers (CD1dTet) were provided by the NIH Tetramer Core Facility. Fluorochrome-conjugated anti-CD45.2 (clone 104), CD45.1 (A20), TCR-β (clone H57-597), NK1.1 (clone PK136), CD44 (clone IM7), CD24 (clone M1/69), CD11b (clone M170), CD11c (clone N418), F4/80 (clone BM8), B220 (clone RA3-6B2), TER119/Erythroid Cells (clone TER-119), CD4 (clone GK1.5), CD8a (clone 53-6.7), ICOS (clone C398.4A), T-bet (clone 4B10), IL7Rα (clone SB/199) were purchased from Biolegend; anti-GATA3 (clone L50-823), CD122 (clone TM-b1), RORγt (clone Q31-378), Streptavidin (BV711), and Ki67 were purchased from BD Biosciences; anti-PLZF (clone Mags.21F7) was purchased from eBioscience. Cell death was identified using the Live/Dead™ Fixable Violet Dead Cell Stain (Thermo Fisher Scientific). Reactive oxygen species (ROS) were detected with 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) (ThermoFisher). Goat anti-mouse IgG (H+L) antibody (Alexa Fluor 568) for detection of the anti-Ki67 antibody was purchased from Thermo Fisher Scientific. Data were analyzed using the FlowJo Version 9.2 software (Tree Star).
CD45.1+CD45.2+ WT mice in C57BL/6 background were irradiated with a single dose of 800 rad X-Ray and intravenously injected with 10–15 million of a mixture of BM cells from CD45.1+ WT mice and CD45.2+ Foxo1f/f -Cd2iCre mice at 1:1 ratio. Recipient mice were euthanized and analyzed 8 weeks later.
Data were presented as mean ± SEM and analyzed for statistical differences using the Prism 5/GraphPad software. Comparisons were made using the two-tailed paired or unpaired Student t-test. Each pair represents age- and sex-matched littermates that were examined in a single experiment and is indicated by a connecting line between test and control mice. P-values less than 0.05 were considered significant (*p < 0.05, **p < 0.01, ***p < 0.001).
To investigate the role of Foxo1 in iNKT cell development, we bred Foxo1f/f mice (57) with hCD2-iCre (Cd2iCre) mice (58) to generate Foxo1f/f -Cd2iCre (Foxo1KO) mice. CD2iCre induces gene ablation of floxed genes in both T cells and B cells (58) and in CD4+CD8+ double positive (DP) thymocytes (Figure 1A). We used TCRβ and PBS-57 loaded CD1d tetramer (CD1dTet) to detect iNKT cells. In Foxo1f/f -Cd2iCre mice, TCRβ+CD1dTet+ iNKT cell percentages and numbers were decreased 64.41% and 68.56%, respectively in the thymus compared with Foxo1f/f controls (Figures 1B–D). Moreover, iNKT cell percentages and numbers were also reduced in the spleen and liver in Foxo1f/f -Cd2iCre mice with the exception of splenic iNKT cells in one Foxo1f/f -Cd2iCre mice due severe splenomegaly likely caused by defective function of regulatory T cells. In contrast, CD4+CD8− single positive (SP) TCRβ+ and CD4−CD8+ SP TCRβ+ thymocyte numbers were similar between control and Foxo1KO mice (Figure 1E). Thus, Foxo1 deficiency resulted in severe impairment of iNKT cell but not conventional αβT cell development.
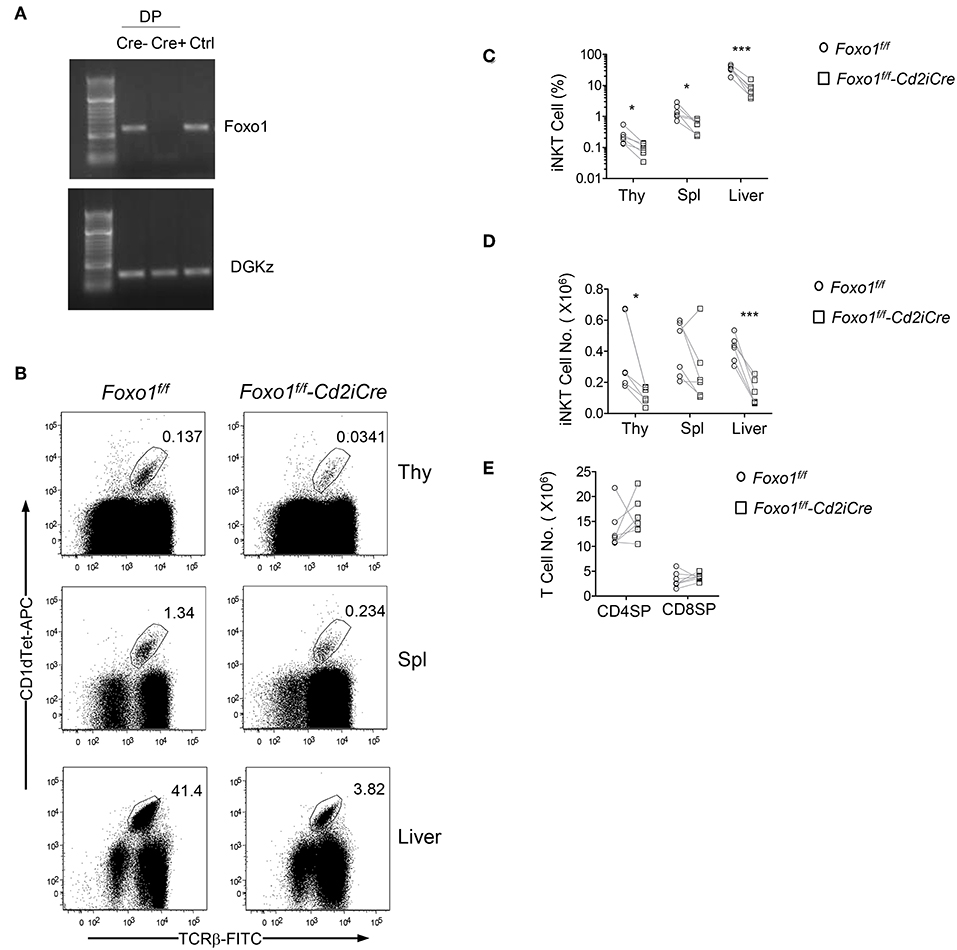
Figure 1. Severe decreases of iNKT cells in Foxo1f/f -Cd2iCre mice. Six to ten weeks old Foxo1f/f -Cd2iCre (Foxo1KO) and Foxo1f/f or Cd2iCre controls (Ctrl) mice were analyzed for iNKT cells by flow cytometry. (A) Assessment of Cre mediated deletion in Foxo1 gene in DP thymocytes. Cre mediated recombination causes deletion of the PCR template. Cre–: Foxo1f/f; Cre+: Foxo1f/f -Cd2iCre; Ctrl: tail DNA from Foxo1f/f -Cd2iCre mice. (B) TCRβ and CD1dTet staining of thymocytes, splenocytes, and liver mononuclear cells (MNCs). Live gated Lin-singlets are shown. (C) Percentages of iNKT cells in the indicated organ. (D) Numbers of iNKT cells in the indicated organs. (E) TCRβ+ SP thymocyte numbers. FACS plots are representative of five experiments. Scatter plots are pooled from five experiments (C,D) or seven experiments (E). Each circle and square represents one WT and one Foxo1KO mice, respectively. Connecting lines indicate individual pairs of sex- and age-matched test and control mice in each experiment. One pair of mice was examined in each experiment. *p < 0.05; ***p < 0.001 determined by two-tail pair-wised Student t-test.
Because Foxo1 was ablated in both T and B cell lineages and Foxo1 deficiency causes severe B cell developmental defects, impairment of regulatory T cell function, and Th lineage and CD8 effector/memory differentiation (32, 59), the severe decreases of iNKT cells could result from change of environment in these mice. To determine if abnormal iNKT cell development in Foxo1f/f -Cd2iCre mice was autonomous, we generated mixed bone marrow (BM) irradiation chimeric mice by injecting a mixture of CD45.1+ WT and CD45.2+ Foxo1f/f -Cd2iCre BM cells at a 1:1 ratio into sublethally irradiated CD45.1+CD45.2+ recipient mice. Two months after reconstitution, recipient mice contained about 1:1 ratio of CD45.2+ and CD45.1+ CD11b+Ly6G+Ly6C− neutrophils in the spleen (Figures 2A,B) suggesting equal reconstitution of these two types of hematopoietic stem cells (HSCs). Additionally, the ratios of CD45.2+ to CD45.1+ CD4+CD8+ double positive (DP) thymocytes, the immediate precursors of iNKT cells, and non-iNKT TCRβ+ conventional T cells were also 1:1 (Figures 2C,D,F). However, the CD45.2+ Foxo1KO to CD45.1 WT ratios for iNKT cells were drastically decreased to 0.2:1 (Figures 2D,F). In contrast, the ratio for CD1dTet−TCRβ+ conventional αβT cells in the thymus were not decreased (Figures 2D,F). Consistent with severe defects in iNKT cell generation, very few iNKT cells in the spleen and liver in the recipients were derived from Foxo1KO HSCs (Figures 2E,F). Thus, Foxo1 deficiency intrinsically inhibited iNKT but not conventional αβT cell development.
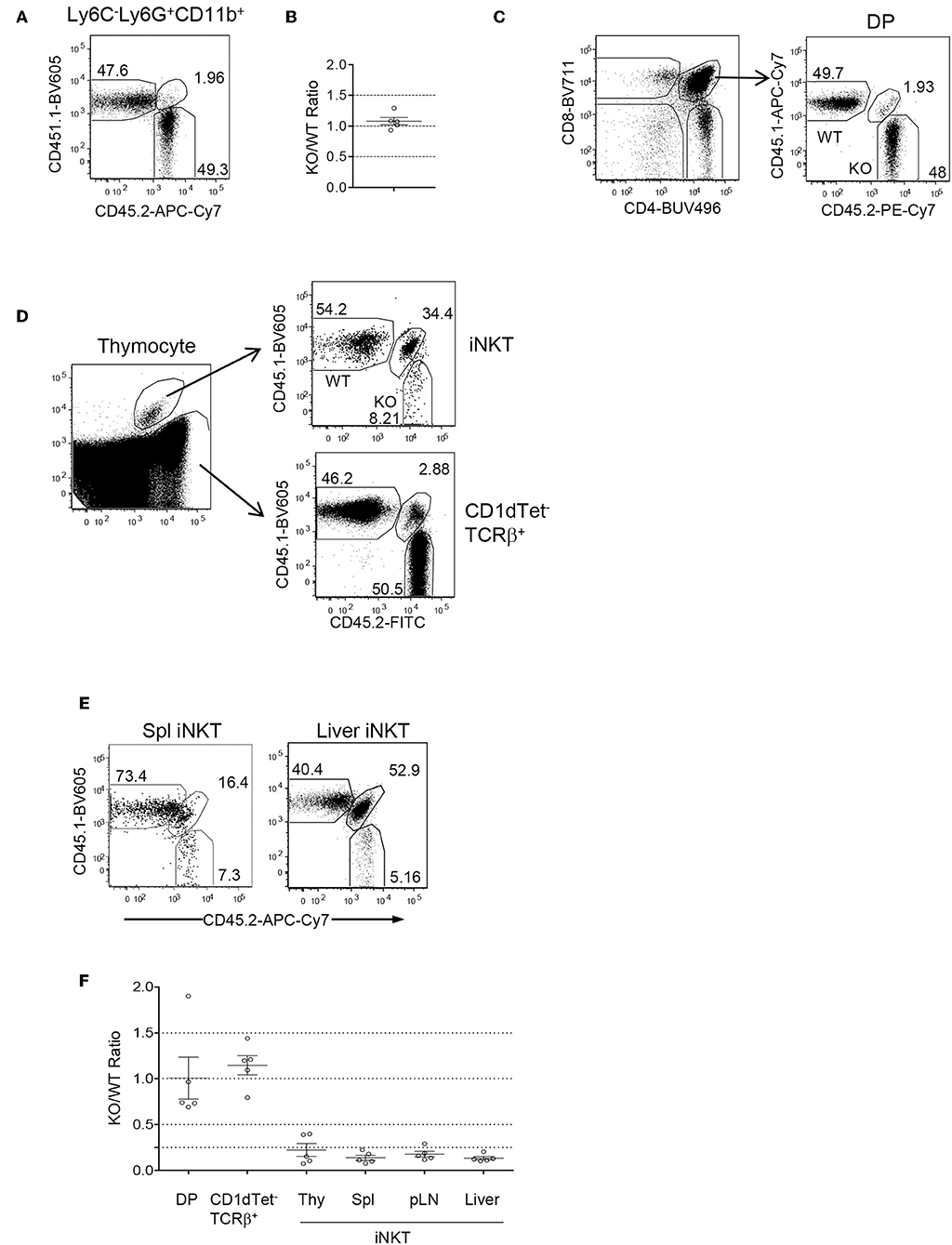
Figure 2. Intrinsic control of iNKT cell development by Foxo1. CD45.1+CD45.2+ WT mice were irradiated with 800 rad X-ray and intravenously injected with a mixture of CD45.1+ WT and CD45.2+ Foxo1f/f -Cd2iCre BM cells at a 1:1 ratio. Recipient mice were euthanized and analyzed 2 months after reconstitution. (A) Representative FACS plots showing CD45.1 and CD45.2 staining in splenic Ly6C−Ly6G+CD11b+ neutrophils. (B) Scatter plots showing ratios of CD45.2+ to CD45.1+ neutrophils. (C) Representative FACS plots showing CD4 and CD8 staining of live gated thymocytes (left panel) and CD45.1 and 45.2 staining of CD4+CD8+ DP thymocytes (right panel). (D) Representative FACS plot showing CD1dTet and TCRβ staining (left panel) of live gated Lin-thymocytes and CD45.1 and 45.2 staining of gated CD1dTet+TCRβ+ iNKT cells (top right panel) and CD1dTet−TCRβ+ non-iNKT cells (bottom right panel). (E) Representative FACS plot showing CD45.1 and 45.2 staining of live gated CD1dTet+TCRβ+ iNKT cells from the spleen and liver. (F) CD45.2/CD45.1 ratios of indicated populations of cells. Data shown are representative or pooled from five experiments. One chimeric mouse was examined in each experiment.
iNKT cell development in the thymus is traditionally defined sequentially into CD24+CD44−NK1.1− stage 0, CD24−CD44−NK1.1− stage 1, CD24−CD44+NK1.1− stage 2, and CD24−CD44+NK1.1+ stage 3 (13, 14). In Foxo1f/f -Cd2iCre mice, stage 0 iNKT cell percentages were increased but numbers were not altered compared with control mice (Figures 3A,B). Stages 1 and 2 iNKT cell percentages displayed increased trend, although not statistically significant. However, their numbers were not obviously altered (Figures 3A,C). Importantly, stage 3 iNKT cell percentages and numbers were both decreased in Foxo1f/f -Cd2iCre thymus (Figures 3A,C). In Foxo1KO spleen, stage 2 iNKT cell numbers were similar to WT control but stage 3 showed decreased except one mouse (Figure 3D), which contained increased stage 3 iNKT cells due to considerable enlargement of the spleen that was likely caused defects in regulatory T cells and lymphoproliferative disorders. In Foxo1KO liver, stage 2 iNKT cell numbers were also similar to WT control but stage 3 iNKT cells were obviously decreased (Figure 3E). Thus, Foxo1 deficiency appears to selectively affect late stage iNKT cell development.
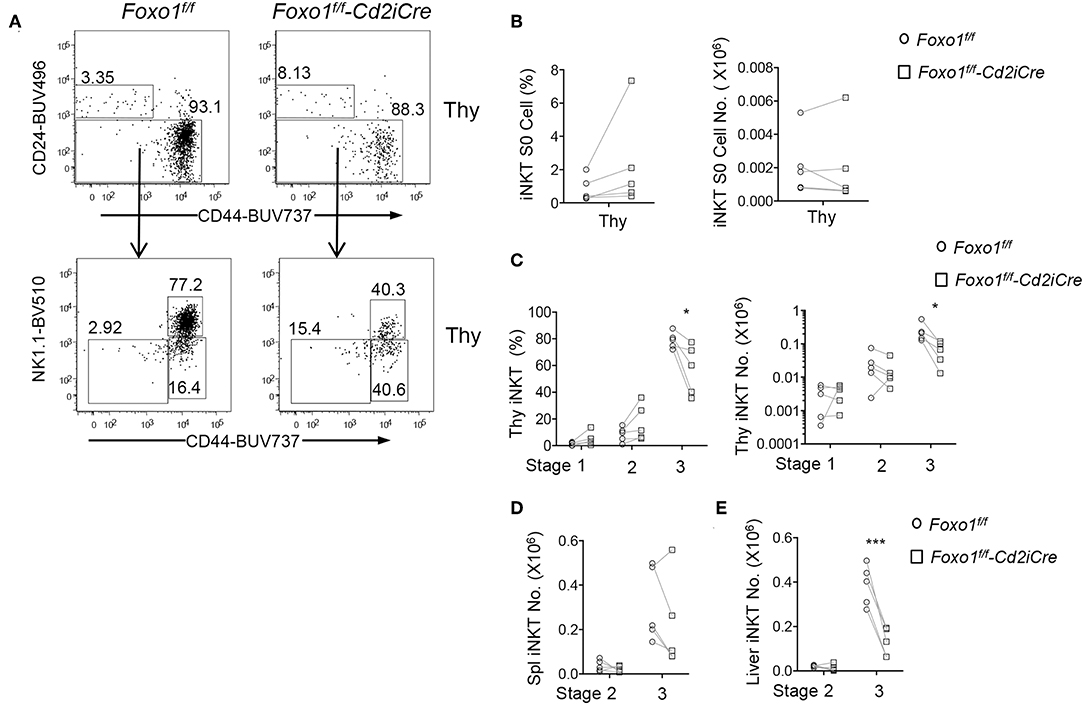
Figure 3. Impaired late stage iNKT cell development in Foxo1f/f -Cd2iCre mice. Six to ten weeks old Foxo1f/f -Cd2iCre and control mice were analyzed similar to Figure 1 with additional antibodies for CD24, CD44, NK1.1, and CD11b, CD11, F4/80, Ter119 for lineage gating. (A) Top panels: CD24 vs. CD44 staining of live gated Lin− iNKT cells in the thymus. Bottom panels, CD44 vs. NK1.1 staining on CD24− iNKT cells. (B) Stage 0 iNKT percentages and numbers in the thymus. (C) Stages 1, 2, and 3 iNKT cell percentages and numbers in the thymus. (D) Stages 2 and 3 iNKT cell numbers in the spleen. (E) Stages 2 and 3 iNKT cell numbers in the liver. Data shown are representative or pooled from five experiments. Connecting lines indicate individual pairs of sex- and age-matched test and control mice in each experiment. *p < 0.05; ***p < 0.001 determined by two-tail pair-wised Student t-test.
We further examined whether Foxo1 intrinsically promotes late stage iNKT cell maturation using the mixed BM chimeric mice described in Figure 2. Within CD45.2+ Foxo1KO iNKT cells, the percentages of stages 0, 1, and 2 were increased 11.08, 2.54, and 1.87 folds compared with CD45.1+ WT iNKT cells, respectively; but stage 3 were drastically decreased (Figures 4A,B), indicating an intrinsic role of Foxo1 for iNKT cell terminal maturation to stage 3. Because iNKT cells were predominantly stage 3 and its dramatic decreases could affect the percentages of stage 0–2 iNKT cells, we further calculated CD45.2 Foxo1KO/CD45.1 WT ratios of each stage of iNKT cells in individual recipient mice. As shown in Figures 4C,D, the ratios of CD45.2/CD45.1 of stage 0 iNKT cell were increased to 3.4:1 compared with the ratios of DP thymocytes. But the ratios of stages 1, 2, and 3 were progressively decreased from 0.60:1, to 0.35:1, and finally to 0.073:1. These results suggested that Foxo1 deficiency exerted weak inhibition on stage 1 and stage 2 iNKT cell maturation but much strong impact on stage 3 iNKT cell maturation. The increases of stage 0 ratios could be caused by enhanced early iNKT cell generation and/or blockade of stage 0 to stage 1 iNKT cell transition in the absence of Foxo1. Thus, Foxo1 intrinsically promoted iNKT cell maturation, particularly late stage maturation.
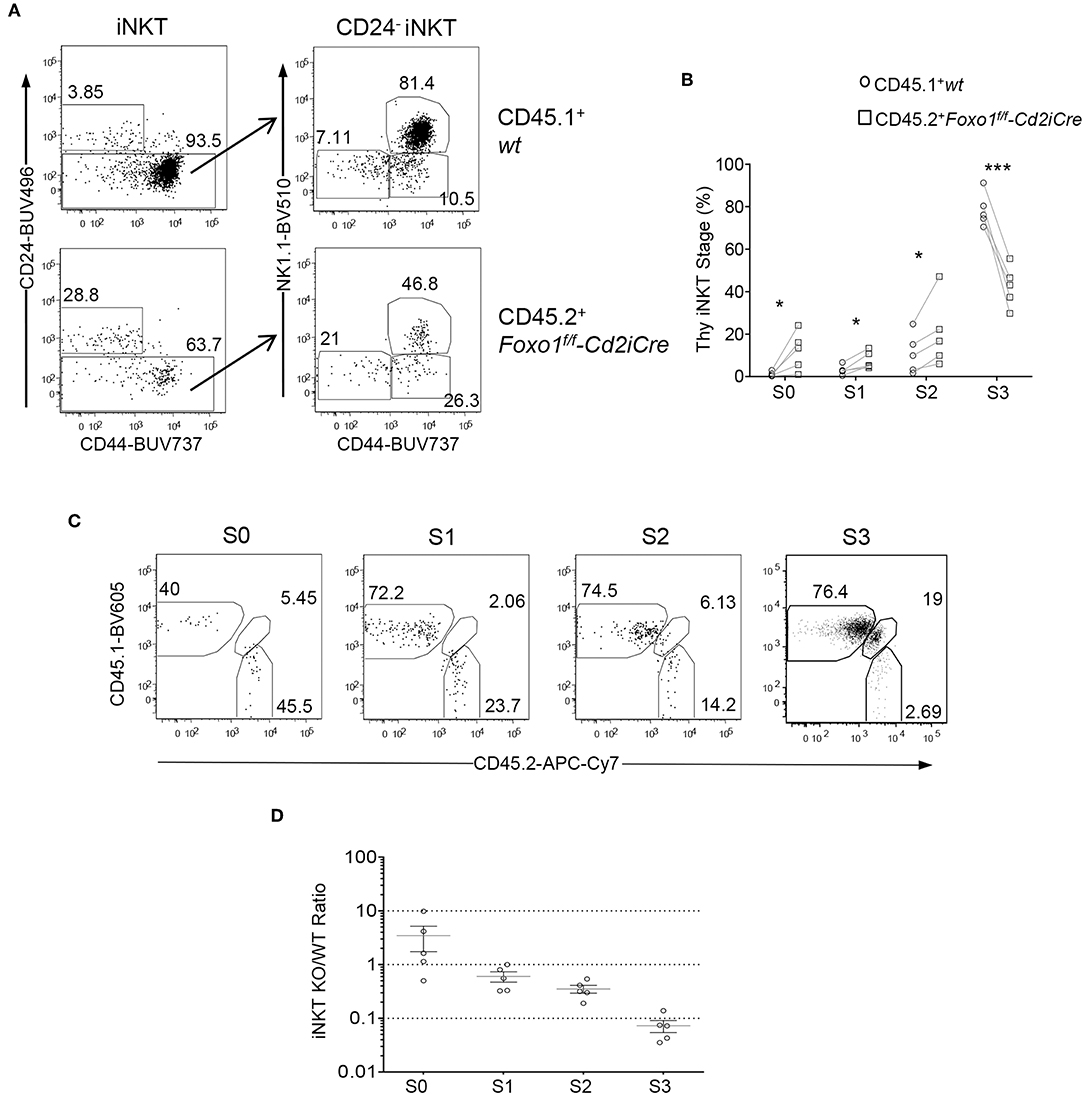
Figure 4. Intrinsic control of late stage iNKT cell development by Foxo1. Thymocytes and splenocytes and liver MNCs from mixed BM chimeric mice as described in Figure 2 were similarly analyzed. (A) Representative FACS plots showing CD24 and CD44 staining in live gated Lin-CD45.1+ WT and CD45.2+ Foxo1KO thymic iNKT cells (left panels) and CD44 and NK1.1 staining in the CD24− population of iNKT cells. (B) Scatter plots showing percentages of iNKT cell developmental stages. (C) Representative FACS plots showing CD45.1 and CD45.2 staining on stage 0–3 iNKT cells. (D) Scatter plots showing CD45.2/CD45.1 ratios of individual iNKT cell developmental stages. Connecting lines indicate CD45.1 WT and CD45.2 Foxo1KO iNKT cells in individual mice. One chimeric mouse was examined in each experiment. *p < 0.05; ***p < 0.001 determined by two-tail pair-wised Student t-test.
iNKT cells differentiate to multiple effector lineages in the thymus that can be defined based expression transcriptional factors/repressors PLZF, GATA3, T-bet, and RORγt (15). Within iNKT cells from Foxo1f/f -Cd2iCre thymus, the percentages of PLZF+GATA3+ iNKT2 and RORγt+T-bet− iNKT17 cells were increased but RORγt−T-bet+ iNKT1 cells were decreased (Figures 5A,B). Thus, Foxo1 deficiency greatly inhibited iNKT1 development, which was consistent with severe decreases of stage 3 iNKT cells that are mostly iNKT1 cells.
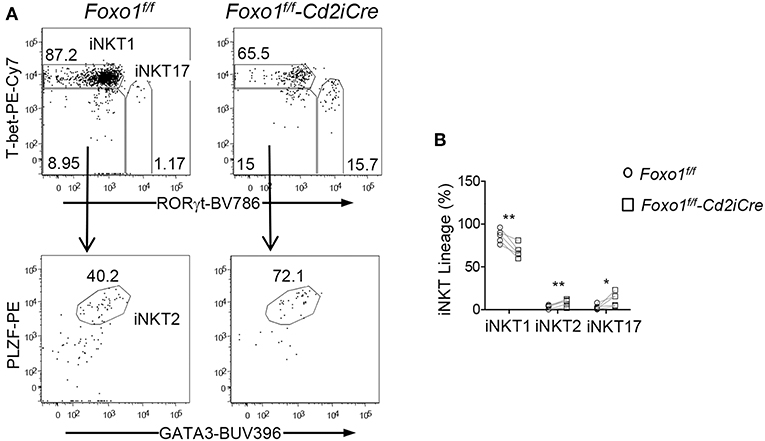
Figure 5. Selective impairment of iNKT1 differentiation in Foxo1 deficient mice. (A) Top panels: T-bet and RORγt staining of thymic iNKT cells. Bottom panels: PLZF and GATA3 staining of RORγt−T-bet− iNKT cells. (B) Percentages of iNKT effector lineages. Data shown are representative or pooled from five experiments. Connecting lines indicate individual pairs of sex- and age-matched test and control mice in each experiment. One pair of mice was examined in each experiment. *p < 0.05; **p < 0.01 determined by two-tail pair-wised Student t-test.
Because iNKT effector lineages may compete with each other during development and are influenced by thymic environment, we further examined whether Foxo1 intrinsically controls iNKT cell effector lineage differentiation using the mixed BM chimeric mice described in Figure 2. Similar to data shown in Figure 5, the percentages of iNKT1 were decreased but iNKT17 and, to certain extent, iNKT2 were increased within CD45.2+ Foxo1KO iNKT cells (Figures 6A,B). Additionally, the Foxo1KO to WT ratios of iNKT1, iNKT2, and iNKT17 cells were 0.077:1, 0.36:1, and 1.1:1, respectively (Figure 6C). Because the Foxo1KO/WT iNKT17 ratios (1.1:1) were greater than stage 2 iNKT ratios (0.35:1, Figure 4D), it suggested that Foxo1KO iNKT17 differentiation was enhanced and that Foxo1 deficiency intrinsically inhibited iNKT1 but promoted iNKT17 differentiation.
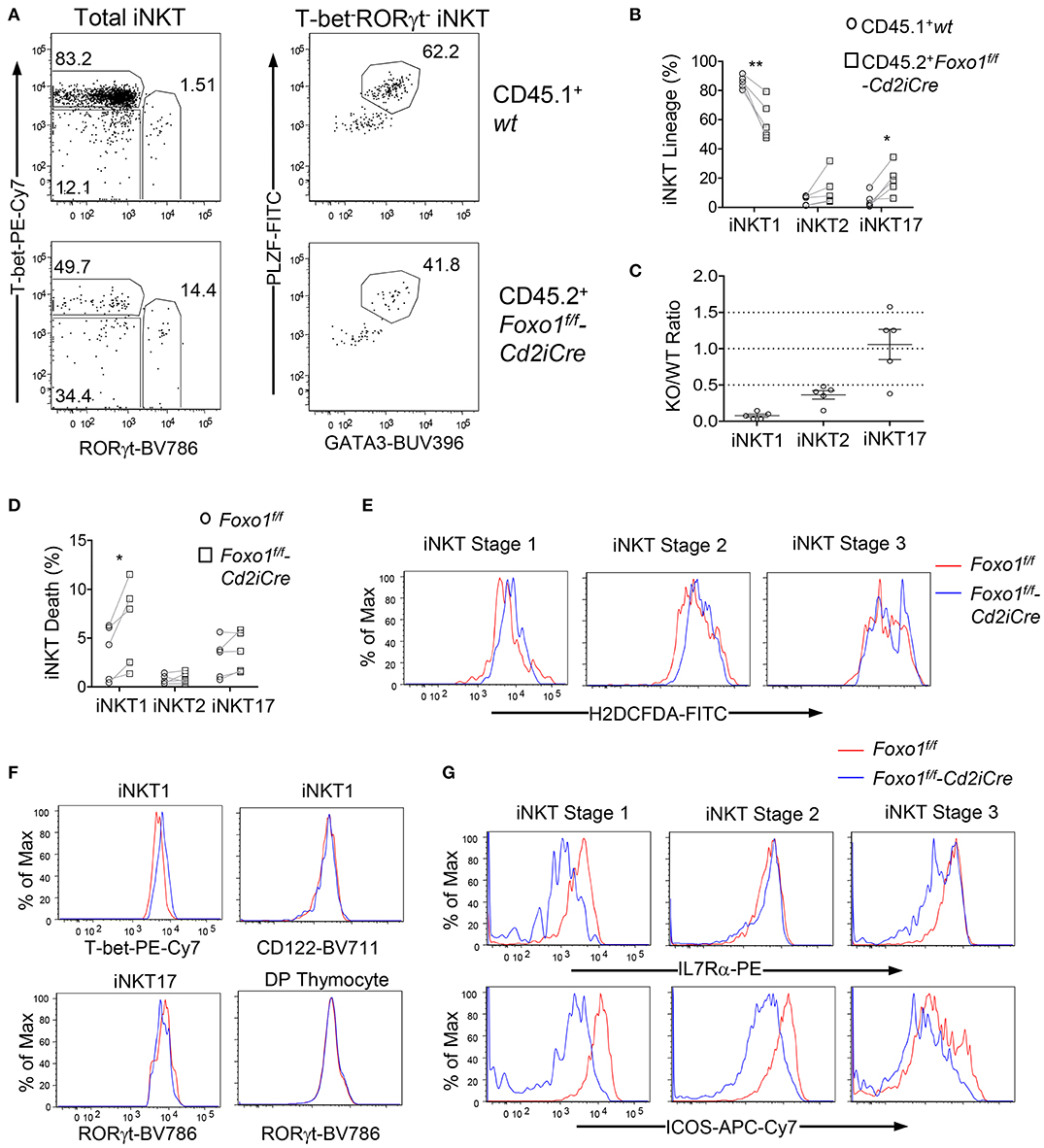
Figure 6. Intrinsic control of iNKT cell effector lineage differentiation by Foxo1. (A–C) Thymocytes from mixed BM chimeric mice as described in Figure 2 were similarly analyzed. (A) Representative FACS plots showing RORγt and T-bet staining in gated CD45.1+ WT and CD45.2+ Foxo1KO iNKT cells (left panels) and PLZF and GATA3 staining in gated RORγt−T-bet− iNKT cells. (B) Scatter plots showing percentages of iNKT effector lineages. (C) Scatter plots showing CD45.2 Foxo1KO/CD45.1WT ratios of individual iNKT effector lineages. Data shown are representative or pooled from five experiments. Connecting lines indicate CD45.1 WT and CD45.2 Foxo1KO iNKT cells in individual mice. One chimeric mouse was examined in each experiment.*p < 0.05; **p < 0.01 determined by two-tail pair-wised Student t-test. (D–G) Two to three weeks old Foxo1f/f -Cd2iCre and Foxo1f/f mice were analyzed for thymic iNKT cells by flow cytometry. (D) Death rates of iNKT effector lineages detected by Live/Dead™ Fixable Violet Dead Cell Stain. N = 5. *p < 0.05 determined by two-tail pair-wised Student t-test. (E) Overlaid histograms show ROS levels in stages 1–3 iNKT cells. (F) Overlaid histograms show T-bet and CD122 levels in iNKT1 cells and RORγt levels in iNKT17 cells and in DP thymocytes. (G) Overlaid histograms show IL7Rα and ICOS levels in stages 1–3 iNKT cells. Data in (E–G) are representative of three experiments.
In Foxo1KO mice, iNKT1 cells but not iNKT2 or iNKT17 cells were prone to death compared with WT iNKT cells (Figure 6D). ROS levels were weakly increased in stage 3 iNKT cells (Figure 6E), which might contribute increased death and decreases of iNKT1 cells as most stage 3 iNKT cells are iNKT1 cells. Foxo1 inhibits T-bet expression in NK cells (60). Within Foxo1KO iNKT1 cells, T-bet expression was increased compared with WT controls, suggesting that Foxo1 also inhibited T-bet expression in iNKT cells (Figure 6F). Additionally, CD122, the IL2/15Rβ chain that mediates IL15 signal to positively regulate T-bet expression and to promote iNKT1 differentiation and survival (61, 62), was expressed at similar levels between WT and Foxo1KO iNKT1 cells (Figure 6F), suggesting that the reduction of Foxo1KO iNKT1 cells was unlikely due to altered T-bet or CD122 expression. RORγt is crucial iNKT17 differentiation (18). RORγt levels were not obviously changed in Foxo1KO iNKT17 cells or in DP thymocytes (Figure 6F). IL7 receptor (IL7R) signal and ICOS costimulatory signal promote iNKT17 lineage differentiation/homeostasis (8, 61, 63). However, both IL7Rα and ICOS levels were reduced in iNKT cells at different stages (Figure 6G), suggesting that Foxo1 deficiency alleviated the requirement of these signals for iNKT17 cells and might promote iNKT17 lineage via other mechanisms.
iNKT cells differentiate to multiple effector lineages that play distinct roles in immune responses and diseases. Evidence suggests that iNKT1 and iNKT17 are competing lineages during iNKT effector differentiation. Both intrinsic program and environmental factors are both involved in control the balance between iNKT1 and iNKT17 cells (8, 15, 63–67). In this report, we demonstrated that deficiency of Foxo1 intrinsically inhibited iNKT1 but enhanced iNKT17 differentiation without obviously affecting early iNKT cell development, suggesting that Foxo1 controls iNKT effector lineage fate decision.
In Foxo1f/f -Cd2iCre mice, stage 0, 1, and 2 iNKT cell numbers are not obviously different from WT control mice and only stage 3 iNKT cell numbers and percentages were decreased. In mixed BM chimeric mice reconstituted with Foxo1f/f -Cd2iCre and congenic WT BM cells, Foxo1KO to WT ratios of stage 0 iNKT cells are slightly increased compared with the ratios of CD4+CD8+ thymocytes. Thus, Foxo1 is dispensable for early iNKT cell development. However, other members of the Foxo subfamily are also expressed in developing thymocytes, our data do not rule out the possibility that those factors may function redundantly with Foxo1 and compensate for its deficiency during early iNKT cell development. Further studies with compound deficiency of Foxo1 and other family members are needed to fully rule out a role of Foxo1 in early iNKT cell development.
Foxo1 deficiency leads to considerable decreases of CD44+NK1.1+T-bet+ iNKT1 cells but increases of CD44+NK1.1−RORγt+ iNKT17 cells. iNKT1 cell percentages and numbers are both decreased, revealed a positive role of Foxo1 in promoting iNKT1 differentiation/ maintenance. The increases of iNKT17 cells in the absence of Foxo1 are not solely resulted from decreases of iNKT1 cells and consequent increases of iNKT17 ratios. The Foxo1KO to WT ratios of iNKT17 cells are significantly overrepresented compared with those of stage 1 and 2 iNKT cells in mixed BM chimeric mice, indicating that Foxo1 negatively controls iNKT17 differentiation. Thus, Foxo1 plays an intrinsic role in controlling iNKT1/17 effector fate decision and may rheostat the balance between these two iNKT effector lineages.
Numerous mechanisms have been found to control iNKT1/17 lineage differentiation. Many transcription factors and regulators such as cMAF, Runx1, MED2/3, and NKAP are important for iNKT17 differentiation (68–74), while the epigenetic modifiers of the TET-family dioxygenases inhibit iNKT17 but promote iNKT1 differentiation (67). Foxo1 deficient iNKT1 but not iNKT2/17 cells were prone to death and contained increased ROS, suggesting that Foxo1 might regulate ROS production to promote iNKT1 cell survival. IL15R signal is critical for iNKT1 cell differentiation and homeostasis in part by increasing T-bet expression (61, 62). We have found IL15Rβ chain expression is not reduced in Foxo1KO iNKT1 cells. Thus, Foxo1 deficiency may cause iNKT1 defect via mechanisms other than altered expression of T-bet or CD122. IL7R signal and ICOS costimulatory signal promote iNKT17 cell homeostasis (8, 61, 63). Both IL7Rα and ICOS levels were decreased in Foxo1KO iNKT cells. In conventional T cells, Foxo1 binds to Il7ra and Icos loci to promote their transcription (48, 75). Foxo1 may function similarly to promote expression of these molecules in iNKT cells. However, it is unlikely that the decreased expression of IL7R and ICOS causes enhanced iNKT17 differentiation in Foxo1KO mice. Although ICOS and IL7R are not crucial for iNKT1 differentiation, our data do not rule out the possibility that decreased expression of these molecules may impair iNKT1 lineage differentiation/homeostasis in the context of Foxo1 deficiency.
Foxo1 inhibits Th17 differentiation and IL-17A expression by directly suppressing transcription of RORγt and IL-23R expression and by interacting with RORγt (44, 47, 76). Although such mechanisms could operate similarly iNKT cells, we do not observe increased RORγt protein levels in Foxo1KO iNKT17 cells. It is interesting to note that Foxo1 inhibits T-bet mediated effector differentiation of CD8 T cells and Th1 differentiation (55, 77) and binds to the Tbx21 promoter to inhibit T-bet expression in NK cells (60). In Foxo1 deficient iNKT1 cells, T-bet levels were increased, suggesting that Foxo1 may also directly suppress T-bet expression in iNKT cells. However, increased T-bet expression should not be the reason for deficiency of iNKT1 cells in Foxo1KO mice.
Foxo1 is regulated by multiple mechanisms. Akt, SGK1, and the serine/threonine kinase CK2 phosphorylate Foxo1 to inhibit its nuclear localization and activity. mTOR regulates iNKT1 and iNKT17 differentiation via mTOR complex 1 (mTORC1) and mTORC2 and their tight regulation by the tumor suppressor TSC1 ensures proper iNKT1/17 balance (8, 66, 78, 79). mTORC2 phosphorylates Akt and SGK1 to enhance their enzyme activities (80–82). Deficiency of either Akt2 or mTORC2 causes decreases of iNKT17 cells, which correlates with decreased Foxo1 phosphorylation or increased Foxo1 nuclear localization (66, 83). The data we report here provide direct evidence that Foxo1 plays critical roles in iNKT cell development and effector lineage differentiation. Although the importance of SGK1 and CK2 in iNKT cells has been unclear, both SGK1 and CK2 promote Th17 differentiation at least partially through inhibition of Foxo1 (45, 84, 85). Our data together with these observations indicate that Foxo1 may integrate signals from mTORC2/Akt/SGK1 and other enzymes to control iNKT cell effector lineage fate decision.
The datasets generated for this study are available on request to the corresponding author.
The protocol was approved by the Duke University Institute Animal Care and Use Committee.
HT and LL designed and performed experiments, analyzed data, and were involved in manuscript preparation. YG and ZW were involved in data analyses and manuscript preparation. X-PZ conceived the project, designed experiments, analyzed data, and wrote the manuscript.
This study was supported by the National Institutes of Health (AI079088 and AI101206) to X-PZ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the NIH Tetramer Core Facility for CD1d tetramers and the flow cytometry core facility at Duke University for services.
1. Chandra S, Kronenberg M. Activation and function of iNKT and MAIT cells. Adv Immunol. (2015) 127:145–201. doi: 10.1016/bs.ai.2015.03.003
2. Yang W, Gorentla B, Zhong XP, Shin J. mTOR and its tight regulation for iNKT cell development and effector function. Mol Immunol. (2015) 68:536–45. doi: 10.1016/j.molimm.2015.07.022
3. Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. (2013) 13:101–17. doi: 10.1038/nri3369
4. Wu L, Van Kaer L. Natural killer T cells in health and disease. Front Biosci. (2011) 3:236–51. doi: 10.2741/s148
5. Zhang J, Bedel R, Krovi SH, Tuttle KD, Zhang B, Gross J, et al. Mutation of the Traj18 gene segment using TALENs to generate Natural Killer T cell deficient mice. Sci Rep. (2016) 6:27375. doi: 10.1038/srep27375
6. Dashtsoodol N, Shigeura T, Ozawa R, Harada M, Kojo S, Watanabe T, et al. Generation of novel Traj18-deficient mice lacking Valpha14 natural killer T cells with an undisturbed T cell receptor alpha-chain repertoire. PLoS ONE. (2016) 11:e0153347. doi: 10.1371/journal.pone.0153347
7. Chandra S, Zhao M, Budelsky A, de Mingo Pulido A, Day J, Fu Z, et al. A new mouse strain for the analysis of invariant NKT cell function. Nat Immunol. (2015) 16:799–800. doi: 10.1038/ni.3203
8. Wu J, Yang J, Yang K, Wang H, Gorentla B, Shin J, et al. iNKT cells require TSC1 for terminal maturation and effector lineage fate decisions. J Clin Invest. (2014) 124:1685–98. doi: 10.1172/JCI69780
9. Shin J, Wang S, Deng W, Wu J, Gao J, Zhong XP. Mechanistic target of rapamycin complex 1 is critical for invariant natural killer T-cell development and effector function. Proc Natl Acad Sci USA. (2014) 111:E776–83. doi: 10.1073/pnas.1315435111
10. Shen S, Wu J, Srivatsan S, Gorentla BK, Shin J, Xu L, et al. Tight regulation of diacylglycerol-mediated signaling is critical for proper invariant NKT cell development. J Immunol. (2011) 187:2122–9. doi: 10.4049/jimmunol.1100495
11. Shen S, Chen Y, Gorentla BK, Lu J, Stone JC, Zhong XP. Critical roles of RasGRP1 for invariant NKT cell development. J Immunol. (2011) 187:4467–73. doi: 10.4049/jimmunol.1003798
12. Hu T, Gimferrer I, Simmons A, Wiest D, Alberola-Ila J. The Ras/MAPK pathway is required for generation of iNKT cells. PLoS ONE. (2011) 6:e19890. doi: 10.1371/journal.pone.0019890
13. Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. (2007) 25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711
14. Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. (2010) 11:197–206. doi: 10.1038/ni.1841
15. Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. (2013) 14:1146–54. doi: 10.1038/ni.2731
16. Chang PP, Barral P, Fitch J, Pratama A, Ma CS, Kallies A, et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol. (2011) 13:35–43. doi: 10.1038/ni.2166
17. Rampuria P, Lang ML. CD1d-dependent expansion of NKT follicular helper cells in vivo and in vitro is a product of cellular proliferation and differentiation. Int Immunol. (2015) 27:253–63. doi: 10.1093/intimm/dxv007
18. Michel ML, Mendes-da-Cruz D, Keller AC, Lochner M, Schneider E, Dy M, et al. Critical role of ROR-gammat in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci USA. (2008) 105:19845–50. doi: 10.1073/pnas.0806472105
19. Matsuda Y, Toda M, Kato T, Kuribayashi K, Kakimi K. Fulminant liver failure triggered by therapeutic antibody treatment in a mouse model. Int J Oncol. (2006) 29:1119–25. doi: 10.3892/ijo.29.5.1119
20. Michel ML, Keller AC, Paget C, Fujio M, Trottein F, Savage PB, et al. Identification of an IL-17-producing NK1.1 (neg) iNKT cell population involved in airway neutrophilia. J Exp Med. (2007) 204:995–1001. doi: 10.1084/jem.20061551
21. Tonti E, Fedeli M, Napolitano A, Iannacone M, von Andrian UH, Guidotti LG, et al. Follicular helper NKT cells induce limited B cell responses and germinal center formation in the absence of CD4+ T cell help. J Immunol. (2012) 188:3217–22. doi: 10.4049/jimmunol.1103501
22. Lynch L, Michelet X, Zhang S, Brennan PJ, Moseman A, Lester C, et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T (reg) cells and macrophages in adipose tissue. Nat Immunol. (2015) 16:85–95. doi: 10.1038/ni.3047
23. Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest. (2014) 124:3725–40. doi: 10.1172/JCI72308
24. Matsuda JL, Zhang Q, Ndonye R, Richardson SK, Howell AR, Gapin L. T-bet concomitantly controls migration, survival, and effector functions during the development of Valpha14i NKT cells. Blood. (2006) 107:2797–805. doi: 10.1182/blood-2005-08-3103
25. Kim HY, Pichavant M, Matangkasombut P, Koh YI, Savage PB, DeKruyff RH, et al. The development of airway hyperreactivity in T-bet-deficient mice requires CD1d-restricted NKT cells. J Immunol. (2009) 182:3252–3261. doi: 10.4049/jimmunol.0803339
26. Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4−NK1.1− NKT cell population. Proc Natl Acad Sci USA. (2008) 105:11287–92. doi: 10.1073/pnas.0801631105
27. Milpied P, Massot B, Renand A, Diem S, Herbelin A, Leite-de-Moraes M, et al. IL-17-producing invariant NKT cells in lymphoid organs are recent thymic emigrants identified by neuropilin-1 expression. Blood. (2011) 118:2993–3002. doi: 10.1182/blood-2011-01-329268
28. Watarai H, Sekine-Kondo E, Shigeura T, Motomura Y, Yasuda T, Satoh R, et al. Development and function of invariant natural killer T cells producing T (h) 2- and T (h) 17-cytokines. PLoS Biol. (2012) 10:e1001255. doi: 10.1371/journal.pbio.1001255
29. Gaya M, Barral P, Burbage M, Aggarwal S, Montaner B, Warren Navia A, et al. Initiation of antiviral B cell immunity relies on innate signals from spatially positioned NKT cells. Cell. (2018) 172:517–33 e520. doi: 10.1016/j.cell.2017.11.036
30. Liew PX, Lee WY, Kubes P. iNKT cells orchestrate a switch from inflammation to resolution of sterile liver injury. Immunity. (2017) 47:752–65 e755. doi: 10.1016/j.immuni.2017.09.016
31. Lynch L, Nowak M, Varghese B, Clark J, Hogan AE, Toxavidis V, et al. Adipose tissue invariant NKT cells protect against diet-induced obesity and metabolic disorder through regulatory cytokine production. Immunity. (2012) 37:574–87. doi: 10.1016/j.immuni.2012.06.016
32. Cabrera-Ortega AA, Feinberg D, Liang Y, Rossa C Jr, Graves DT. The role of Forkhead Box 1 (FOXO1) in the immune system: dendritic cells, T cells, B cells, and hematopoietic stem cells. Crit Rev Immunol. (2017) 37:1–13. doi: 10.1615/CritRevImmunol.2017019636
33. Silhan J, Vacha P, Strnadova P, Vecer J, Herman P, Sulc M. 14-3-3 protein masks the DNA binding interface of forkhead transcription factor FOXO4. J Biol Chem. (2009) 284:19349–60. doi: 10.1074/jbc.M109.002725
34. Ouyang W, Li MO. Foxo: in command of T lymphocyte homeostasis and tolerance. Trends Immunol. (2011) 32:26–33. doi: 10.1016/j.it.2010.10.005
35. Xiao W, Dong G, Pacios S, Alnammary M, Barger LA, Wang Y, et al. FOXO1 deletion reduces dendritic cell function and enhances susceptibility to periodontitis. Am J Pathol. (2015) 185:1085–1093. doi: 10.1016/j.ajpath.2014.12.006
36. Dong G, Wang Y, Xiao W, Pacios Pujado S, Xu F, Tian C, et al. FOXO1 regulates dendritic cell activity through ICAM-1 and CCR7. J Immunol. (2015) 194:3745–3755. doi: 10.4049/jimmunol.1401754
37. Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. (2008) 9:613–22. doi: 10.1038/ni.1612
38. Herzog S, Hug E, Meixlsperger S, Paik JH, DePinho RA, Reth M, et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat Immunol. (2008) 9:623–31. doi: 10.1038/ni.1616
39. Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. (2010) 11:635–43. doi: 10.1038/ni.1891
40. Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. (2009) 10:176–84. doi: 10.1038/ni.1689
41. Luo CT, Liao W, Dadi S, Toure A, Li MO. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature. (2016) 529:532–6. doi: 10.1038/nature16486
42. Kerdiles YM, Stone EL, Beisner DL, McGargill MA, Ch'en IL, Stockmann C, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. (2010) 33:890–904. doi: 10.1016/j.immuni.2010.12.002
43. Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. (2010) 11:618–27. doi: 10.1038/ni.1884
44. Ichiyama K, Gonzalez-Martin A, Kim BS, Jin HY, Jin W, Xu W, et al. The microRNA-183-96-182 cluster promotes T helper 17 cell pathogenicity by negatively regulating transcription factor Foxo1 expression. Immunity. (2016) 44:1284–98. doi: 10.1016/j.immuni.2016.05.015
45. Gibson SA, Yang W, Yan Z, Qin H, Benveniste EN. CK2 controls Th17 and regulatory T cell differentiation through inhibition of FoxO1. J Immunol. (2018) 201:383–92. doi: 10.4049/jimmunol.1701592
46. Liu L, Liu Y, Yuan M, Xu L, Sun H. Elevated expression of microRNA-873 facilitates Th17 differentiation by targeting forkhead box O1 (Foxo1) in the pathogenesis of systemic lupus erythematosus. Biochem Biophys Res Commun. (2017) 492:453–60. doi: 10.1016/j.bbrc.2017.08.075
47. Laine A, Martin B, Luka M, Mir L, Auffray C, Lucas B, et al. Foxo1 is a T cell-intrinsic inhibitor of the RORgammat-Th17 program. J Immunol. (2015) 195:1791–803. doi: 10.4049/jimmunol.1500849
48. Stone EL, Pepper M, Katayama CD, Kerdiles YM, Lai CY, Emslie E, et al. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity. (2015) 42:239–51. doi: 10.1016/j.immuni.2015.01.017
49. Weber JP, Fuhrmann F, Feist RK, Lahmann A, Al Baz MS, Gentz LJ, et al. ICOS maintains the T follicular helper cell phenotype by down-regulating Kruppel-like factor 2. J Exp Med. (2015) 212:217–33. doi: 10.1084/jem.20141432
50. Zeng H, Cohen S, Guy C, Shrestha S, Neale G, Brown SA, et al. mTORC1 and mTORC2 kinase signaling and glucose metabolism drive follicular helper T cell differentiation. Immunity. (2016) 45:540–54. doi: 10.1016/j.immuni.2016.08.017
51. Buttrick TS, Wang W, Yung C, Trieu KG, Patel K, Khoury SJ, et al. Foxo1 promotes Th9 cell differentiation and airway allergy. Sci Rep. (2018) 8:818. doi: 10.1038/s41598-018-19315-z
52. Malik S, Sadhu S, Elesela S, Pandey RP, Chawla AS, Sharma D, et al. Transcription factor Foxo1 is essential for IL-9 induction in T helper cells. Nat Commun. (2017) 8:815. doi: 10.1038/s41467-017-00674-6
53. Bi E, Ma X, Lu Y, Yang M, Wang Q, Xue G, et al. Foxo1 and Foxp1 play opposing roles in regulating the differentiation and antitumor activity of Th9 cells programmed by IL-7. Sci Signal. (2017) 10:eaak9741. doi: 10.1126/scisignal.aak9741
54. Zhang L, Tschumi BO, Lopez-Mejia IC, Oberle SG, Meyer M, Samson G. Mammalian target of rapamycin complex 2 controls CD8 T cell memory differentiation in a Foxo1-dependent manner. Cell Rep. (2016) 14:1206–17. doi: 10.1016/j.celrep.2015.12.095
55. Rao RR, Li Q, Gubbels Bupp MR, Shrikant PA. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8(+) T cell differentiation. Immunity. (2012) 36:374–87. doi: 10.1016/j.immuni.2012.01.015
56. Kim MV, Ouyang W, Liao W, Zhang MQ, Li MO. The transcription factor Foxo1 controls central-memory CD8+ Tcell responses to infection. Immunity. (2013) 39:286–97. doi: 10.1016/j.immuni.2013.07.013
57. Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. (2007) 128:309–23. doi: 10.1016/j.cell.2006.12.029
58. de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. (2003) 33:314–25. doi: 10.1002/immu.200310005
59. Luo CT, Li MO. Transcriptional control of regulatory T cell development and function. (2013) Trends Immunol. 34:531–9. doi: 10.1016/j.it.2013.08.003
60. Du X, de Almeida P, Manieri N, de Almeida Nagata D, Wu TD, Harden Bowles K, et al. CD226 regulates natural killer cell antitumor responses via phosphorylation-mediated inactivation of transcription factor FOXO1. Proc Natl Acad Sci USA. (2018) 115:E11731–40. doi: 10.1073/pnas.1814052115
61. Matsuda JL, Gapin L, Sidobre S, Kieper WC, Tan JT, Ceredig R, et al. Homeostasis of V alpha 14i NKT cells. Nat Immunol. (2002) 3:966–74. doi: 10.1038/ni837
62. Gordy LE, Bezbradica JS, Flyak AI, Spencer CT, Dunkle A, Sun J, et al. IL-15 regulates homeostasis and terminal maturation of NKT cells. J Immunol. (2011) 187:6335–45. doi: 10.4049/jimmunol.1003965
63. Webster KE, Kim HO, Kyparissoudis K, Corpuz TM, Pinget GV, Uldrich AP, et al. IL-17-producing NKT cells depend exclusively on IL-7 for homeostasis and survival. Mucosal Immunol. (2014) 7:1058–67. doi: 10.1038/mi.2013.122
64. Castillo EF, Acero LF, Stonier SW, Zhou D, Schluns KS. Thymic and peripheral microenvironments differentially mediate development and maturation of iNKT cells by IL-15 transpresentation. Blood. (2010) 116:2494–503. doi: 10.1182/blood-2010-03-277103
65. Monteiro M, Almeida CF, Agua-Doce A, Graca L. Induced IL-17-producing invariant NKT cells require activation in presence of TGF-beta and IL-1beta. J Immunol. (2013) 190:805–11. doi: 10.4049/jimmunol.1201010
66. Wei J, Yang K, Chi H. Cutting edge: Discrete functions of mTOR signaling in invariant NKT cell development and NKT17 fate decision. J Immunol. (2014) 193:4297–301. doi: 10.4049/jimmunol.1402042
67. Tsagaratou A, Gonzalez-Avalos E, Rautio S, Scott-Browne JP, Togher S, Pastor WA, et al. TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells. Nat Immunol. (2017) 18:45–53. doi: 10.1038/ni.3630
68. Thapa P, Manso B, Chung JY, Romera Arocha S, Xue HH, Angelo DBS, et al. The differentiation of ROR-gammat expressing iNKT17 cells is orchestrated by Runx1. Sci Rep. (2017) 7:7018. doi: 10.1038/s41598-017-07365-8
69. Yu JS, Hamada M, Ohtsuka S, Yoh K, Takahashi S, Miaw SC. Differentiation of IL-17-producing invariant natural killer T cells requires expression of the transcription factor c-Maf. Front Immunol. (2017) 8:1399. doi: 10.3389/fimmu.2017.01399
70. Thapa P, Chen MW, McWilliams DC, Belmonte P, Constans M, Sant'Angelo DB, et al. NKAP regulates invariant NKT cell proliferation and differentiation into ROR-gammat-expressing NKT17 cells. J Immunol. (2016) 196:4987–98. doi: 10.4049/jimmunol.1501653
71. Yue X, Izcue A, Borggrefe T. Essential role of Mediator subunit Med1 in invariant natural killer T-cell development. Proc Natl Acad Sci USA. (2011) 108:17105–10. doi: 10.1073/pnas.1109095108
72. Yu S, Cantorna MT. The vitamin D receptor is required for iNKT cell development. Proc Natl Acad Sci USA. (2008) 105:5207–12. doi: 10.1073/pnas.0711558105
73. Thapa P, Romero Arocha S, Chung JY, Sant'Angelo DB, Shapiro VS. Histone deacetylase 3 is required for iNKT cell development. Sci Rep. (2017) 7:5784. doi: 10.1038/s41598-017-06102-5
74. Xu Y, Sun Y, Shen H, Dai Y, Liu H, Li R, et al. Regulation of the terminal maturation of iNKT cells by mediator complex subunit 23. Nat Commun. (2018) 9:3875. doi: 10.1038/s41467-018-06372-1
75. Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. (2009) 30:358–71. doi: 10.1016/j.immuni.2009.02.003
76. Yang X, He Q, Guo Z, Xiong F, Li Y, Pan Y, et al. MicroRNA-425 facilitates pathogenic Th17 cell differentiation by targeting forkhead box O1 (Foxo1) and is associated with inflammatory bowel disease. Biochem Biophys Res Commun. (2018) 496:352–8. doi: 10.1016/j.bbrc.2018.01.055
77. Harada Y, Harada Y, Elly C, Ying G, Paik JH, DePinho RA, et al. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J Exp Med. (2010) 207:1381–91. doi: 10.1084/jem.20100004
78. Lee YJ, Starrett GJ, Lee ST, Yang R, Henzler CM, Jameson SC, et al. Lineage-specific effector signatures of invariant NKT cells are shared amongst gammadelta T, innate lymphoid, and Th cells. J Immunol. (2016) 197:1460–70. doi: 10.4049/jimmunol.1600643
79. Wu J, Shin J, Xie D, Wang H, Gao J, Zhong XP. Tuberous sclerosis 1 promotes invariant NKT cell anergy and inhibits invariant NKT cell-mediated antitumor immunity. J Immunol. (2014) 192:2643–50. doi: 10.4049/jimmunol.1302076
80. Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem J. (2008) 416:375–85. doi: 10.1042/BJ20081668
81. Dibble CC, Asara JM, Manning BD. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol. (2009). 29:5657–70. doi: 10.1128/MCB.00735-09
82. Heikamp EB, Patel CH, Collins S, Waickman A, Oh MH, Sun IH, et al. The AGC kinase SGK1 regulates TH1 and TH2 differentiation downstream of the mTORC2 complex. Nat Immunol. (2014). 15:457–64. doi: 10.1038/ni.2867
83. Niu L, Xuan X, Wang J, Li L, Yang D, Jing Y, et al. Akt2 regulates the differentiation and function of NKT17 cells via FoxO-1-ICOS axis. Front Immunol. (2018). 9:1940. doi: 10.3389/fimmu.2018.01940
84. Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, et al. Sodium chloride drives autoimmune disease by the induction of pathogenic Th17 cells. Nature. (2013). 496:518–22. doi: 10.1038/nature11868
Keywords: invariant NK T cells, iNKT cells, Foxo1, T-bet, RORγt, T cell development, IFNγ, IL-17A
Citation: Tao H, Li L, Gao Y, Wang Z and Zhong X-P (2019) Differential Control of iNKT Cell Effector Lineage Differentiation by the Forkhead Box Protein O1 (Foxo1) Transcription Factor. Front. Immunol. 10:2710. doi: 10.3389/fimmu.2019.02710
Received: 23 April 2019; Accepted: 04 November 2019;
Published: 21 November 2019.
Edited by:
Yun-Cai Liu, Tsinghua University, ChinaReviewed by:
Jinfang Zhu, National Institute of Allergy and Infectious Diseases (NIAID), United StatesCopyright © 2019 Tao, Li, Gao, Wang and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Ping Zhong, eGlhb3BpbmcuemhvbmdAZHVrZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.