- 1Department of Internal Medicine, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
- 2Key Laboratory of Rheumatology and Clinical Immunology, Department of Rheumatology and Clinical Immunology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Ministry of Education, Beijing, China
- 3Department of Neurology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
- 4Department of Clinical Laboratory, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
Background: Neuromyelitis optica spectrum disorder (NMOSD) is a neurological complication of primary Sjögren's syndrome (pSS).
Objective: We aimed to explore potential serological differences between pSS patients with and without NMOSD.
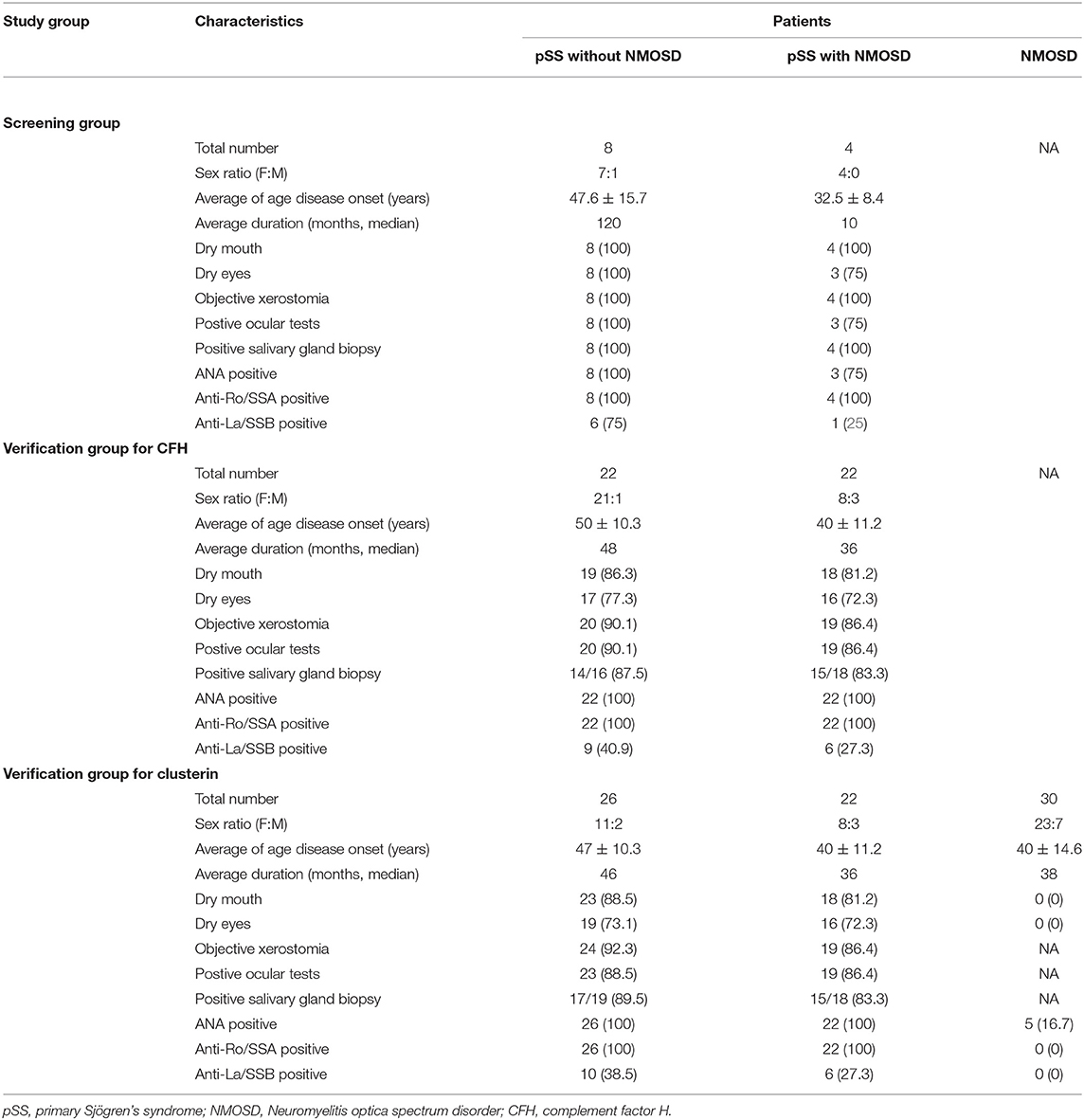
Methods: There were 4 pSS patients with NMOSD and 8 pSS patients without NMOSD enrolled as the screening group for two-dimensional difference gel electrophoresis (DIGE) analysis. Then differential expressed protein spots between groups were identified by MALDI-TOF/TOF MS. The levels of the identified potential biomarkers were verified by ELISA in a second independent cohort including 22 pSS patients with NMOSD, 26 pSS without NMOSD and 30 NMOSD patients.
Results: Nine proteins were identified significantly differently expressed (more than 1.5-fold, p < 0.05) between these two groups. Serum levels of clusterin and complement factor H (CFH) were further verified by ELISA. Results showed that the serum clusterin was significantly higher in NMOSD with pSS than without (298.33 ± 184.52 vs. 173.49 ± 63.03 ng/ml, p < 0.01), while the levels of CFH were lower in pSS patients with NMOSD than without (24.19 ± 1.79 vs. 25.87 ± 3.98 ng/ml, p < 0.01).
Conclusion: This is the first study of serological comparative proteomics between pSS patients with and without NMOSD. Serum clusterin and CFH might be potential biomarkers for pSS patients with NMOSD and play important role in the pathogenesis of the disease but needs further verification.
Introduction
Primary Sjögren's syndrome (pSS) is a systemic autoimmune disease characterized by dry eyes and dry mouth. About 40% of pSS patients were found to be complicated with neuromyelitis optica spectrum disorder (NMOSD) (1). NMOSD is an inflammatory demyelinating spectrum disorder of the central nervous system characterized by severe attacks of optic neuritis and myelitis. However, the pathogenesis of pSS complicated with NMOSD has not been elucidated yet (2).
Generally, the autoimmune response appears to be more intense in the NMO group with SS (3). The prognosis of NMOSD patients complicated with autoimmune diseases was considered to be more poor than NMOSD patients without (4–6). Therefore, early diagnosis and treatment of pSS patients with NMOSD will improve their prognosis. However, the diagnosis of pSS patients with NMOSD, especially NMOSD, was confronted with problems that the specific serum biomarker, anti-aquaporin4 (AQP4) antibody was not sensitive enough (7). The similarity of sensitivity and specificity of AQP4 antibody in idiopathic NMOSD patients and NMOSD associated with autoimmune diseases is indicative of two distinct diseases with sequential or simultaneous incidences (8). Moreover, the symptoms of the central nervous system behave before mucosal symptoms and detection of anti-SSA/SSB antibody in pSS patients with NMOSD (9–11), which may result in ignoring the existence and treatment of pSS. Exploring serum biomarker of pSS with NMOSD will not only be useful for making its clinical decision but also help further illustrate the pathogenesis.
As we know, proteomics has been widely used in exploring diagnosis and prognosis biomarkers of many diseases. Previously, we have applied proteomics in establishing classification tree models and differential diagnosis biomarkers for autoimmune disease (12–15). Recently, we illustrated the pathogenesis of primary biliary cholangitis by using label-free proteomics (16). Among these researches, we found that the two-dimensional difference gel electrophoresis (DIGE) is a more sensitive and high-throughput based two-dimensional gel electrophoresis (2-DE) method (17).
In this research, we aimed to investigate serological differences between pSS patients with and without NMOSD by using DIGE, to established important biomarkers or primarily demonstrated the pathogenesis of pSS patients with NMOSD.
Materials and Methods
Eligibility Criteria and Study Design
Serum samples were collected at Peking Union Medical College Hospital from June 2013 to December 2013. Participants met diagnosis criteria: pSS and NMOSD were diagnosed according to the revised version of classification criteria for pSS proposed by the American-European Consensus Group (18) and the Wingerchuk criteria (1, 19), respectively, and not receive immunosuppressive treatment could be included in this study. Four pSS patients with NMOSD and eight pSS patients without NMOSD were selected as the screening group for DIGE analysis. A second independent cohort including 22 pSS patients with NMOSD, 26 pSS without NMOSD, and 30 NMOSD patients was recruited for the verification of the DIGE results (Table 1). The Ethics Committee of the Peking Union Medical College Hospital approved this study, and all methods were performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from all participants.
Sample Collection and Preparation for 2-DE Analysis
Blood samples were collected from the cubital vein under fasting conditions and dispensed into 5-mL pro-coagulation tubes with gel (Becton, Dickinson and Company, UK). Blood samples were centrifuged at 1,000 × g for 5 min within 6 h at 4°C. Sera were stored at −80°C until use.
High abundant proteins from the serum samples included for DIGE analysis were removed by applying the Agilent Human 14 Multiple Affinity Removal System Column (Agilent Technologies, CA, USA) according to the manufacturer's protocol. The 14 most abundant proteins (albumin, immunoglobulin G, α1-antitrypsin, immunoglobulin A, transferrin, haptoglobin, fibrinogen, α2-macroglobulin, α1-acid glycoprotein, immunoglobulin M, apolipoprotein A-1, apolipoprotein A-II, complement C3, and transthyretin) were removed by this method. Then protein concentrations of the depleted serum samples were detected with the Bradford assay (Bio-Rad, USA).
DIGE
Depleted serum samples from the patients of screening group were analyzed by DIGE, which was performed according to our previous research (14). Briefly, 50 μg of each sample were pooled and used as an internal standard that will run on each gel to control gel-to-gel variation. Then protein extracts from pSS without NMOSD, pSS with NMOSD, and the internal standard were labeled with DIGE fluors, Cy3, Cy5, and Cy2 (GE Healthcare, NJ, USA), respectively, according to the manufacturer's protocols (20). The Cy2, Cy3, and Cy5 labeled samples were loaded onto a 13 cm, pH 3–10 Immobiline Drystrips (GE Healthcare), with the setting of electrophoresis conditions as follow: 30 V for 12 h, 500 V for 1 h, 1,000 V for 1 h, 8,000 V for 8 h, and 500 V for 4 h. After equilibration, the strips were loaded on a 12.5% polyacrylamide gel for second separation with the setting of electrophoresis, 15 mA for 10 min, and then at 30 mA until the samples were 5 mm from the bottom of the gel. The gel was then scanned by Typhoon 9400 (GE Healthcare). DeCyder 6.5 sofeware (GE Healthcare) was used for image analysis. The dye intensity of protein spots from pSS patients with and without NMOSD were compared by Biological Variation Analysis module using a t-test, and p-values < 0.05 were considered as significant. Spots that were significantly different between groups (more than 1.5-fold) were selected and identified.
One milligram of internal standard was run on a 2-DE gel to gain all the selected spots. After destained, dehydrated and proteins digested, gel pieces of these spots were prepared for protein identification. A 4800 Plus MALDI TOF/TOFTM Analyzer (AB SCIEX, USA) was used for protein identification. Bioinformation including MS/MS queries were run on the MASCOT search engine 2.2 (Matrix Science).
ELISA Validation
ELISA was used to verify the reliability of DIGE results. In particular, we investigated the expression of clusterin and complement factor H (CFH). The validation experiments were performed using the commercially available ELISA Kit for human clusterin (Abcam, Cambridge, UK) and CFH (Abcam, Cambridge, UK). Twenty-six pSS patients without NMOSD, 22 pSS patients with NMOSD, and 30 NMOSD patients were used to determine the level of clusterin. And 22 pSS patients without NMOSD and 22 pSS patients with NMOSD were used to determining the level of CFH. For the quantification of clusterin and CFH, 10 μL of each sample was diluted 1:500 and 1:1,000 with sample diluent, respectively. Serum samples and protein standards were added to pre-designated wells on the 96-well microtiter plate. The assay was performed according to the manufacturer's specifications. The absorbance was read by a microplate reader at 450 nm. And the assay results were analyzed using the GraphPad Prism software and SPSS 18.0 software package (IBM, Armonk, NY, USA).
Statistical Analysis
The data were expressed as the mean ± SD and were analyzed using SPSS statistical software (IBM, Armonk, NY, USA). Student's t-tests were used to compare pairs of means. The level of significance was set as p < 0.05. The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Results
Screening Protein Spots With Potential Value
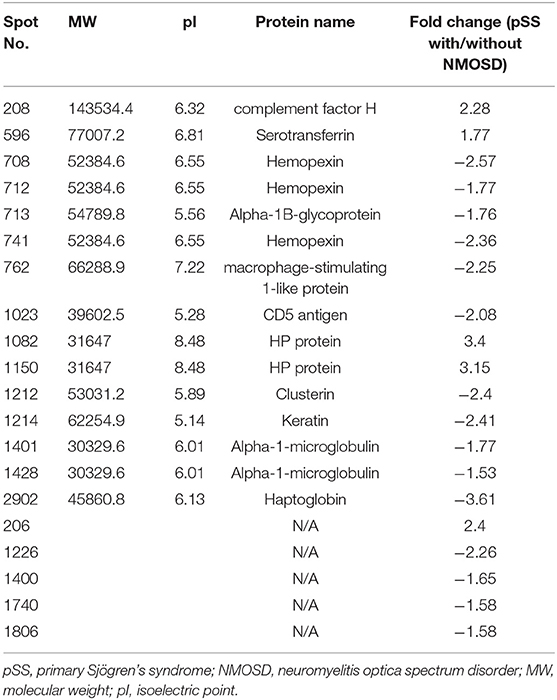
Firstly, the proteins that present in all gels from one group were selected. Secondly, the abundance of these protein spots was compared between pSS patients with and without NMOSD, and a total of 206 protein spots were found significantly differently expressed in the screening group. Thirdly, the relative abundance of proteins spots with potential value was estimated 1.5 times higher or lower than expressed in the other group. Thirty-two of the 206 proteins spots were found to be of potential value (Figure 1). Among them, the abundance of 16 protein spots was increased in pSS without NMOSD, while the other 16 protein spots decreased.
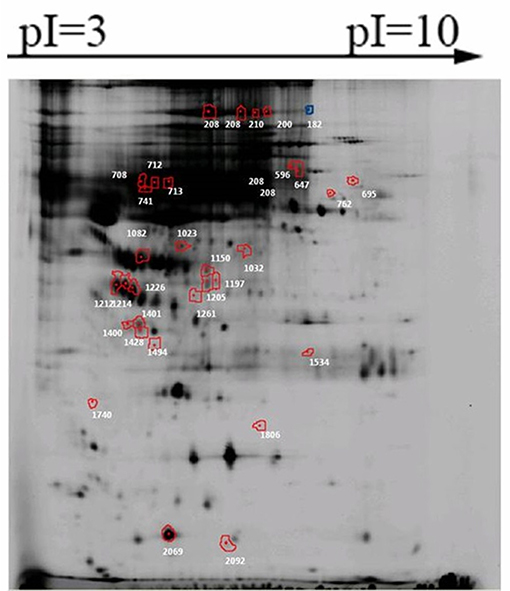
Figure 1. Significantly differently expressed protein spots between pSS patients with and without NMOSD. Proteins of each spot represented were listed in Table 2.
Identification of Protein Spots With Potential Value by MALDI-TOF/TOF MS
Only 20 out of the 32 protein spots were found in 2-DE gels, and 9 candidate proteins were identified: complement factor H (CFH), hemopexin, alpha-1B-glycoprotein, putative macrophage-stimulating 1-like protein, CD5 antigen-like OS, HP protein, clusterin, keratin (type I cytoskeletal 9 OS), alpha-1-microglobulin (Table 2). Based on a review of related literature, CFH and clusterin might be related to immune diseases, and thus they were chosen for further verification.
Verification of Serum Clusterin and CFH
The levels of clusterin and CFH in sera were quantified by ELISA to confirm the altered expression that was revealed by proteomic analysis and to validate their potentials as biomarkers of pSS with NMOSD. The results of DIGE were validated firstly. In the screening group, the level of clusterin was the same with DIGE while the level of CFH was not. Serum levels of clusterin and CFH were then determined in the verification group by ELISA. Results showed that serum clusterin was higher in pSS without NMOSD than with NMOSD but not significant (307.26 ± 140.26 vs. 298.33 ± 184.52 ng/ml, p = 1.00). However, serum clusterin was significantly higher in NMOSD with pSS than without (298.33 ± 184.52 vs. 173.49 ± 63.03 ng/ml, p < 0.01). On the other hand, the levels of CFH were lower in pSS patients with NMOSD than without NMOSD (24.19 ± 1.79 vs. 25.87 ± 3.98 ng/ml, p < 0.01). ELISA results for serum levels of clusterin and CFH in the verification group are shown in Figure 2.
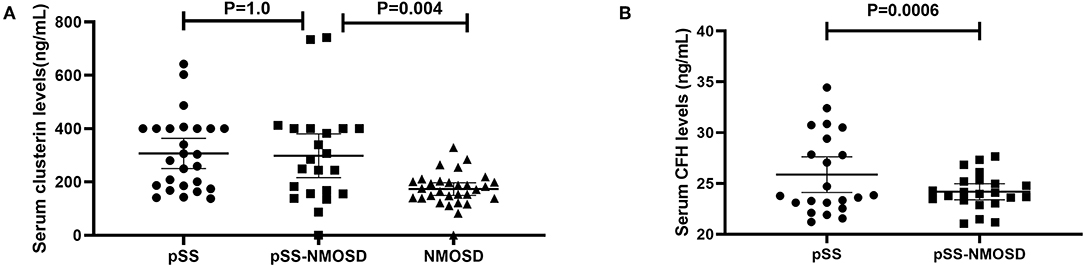
Figure 2. Verification of clusterin and CFH by ELISA asssys. (A) validation of serum clusterin levels; (B) validation of serum CFH levels.
In statistical analysis, the period that onset of disease to diagnosis was significantly different between SS with and without NMOSD in the screening group, while the age on disease onset was not. The SS with NMOSD, without NMOSD, and the NMOSD patients of the confirmation group were matched for most of the clinical condition, especially in the age of disease onset. We, therefore, considered that the differences we found come from disease heterogeneity.
Discussion
In this research, DIGE combined with MALDI-TOF/TOF MS was applied to compare the protein pattern of serum from pSS patients with and without NMOSD. There were 9 proteins found to be significantly differently expressed between groups and could be potential biomarkers for pSS with NMOSD. The serum levels of alpha-1B-glycoprotein, alpha-1-microglobulin, CD5 antigen-like OS, clusterin, hemopexin, type I cytoskeletal 9, and putative macrophage-stimulating 1-like protein were lower in pSS patients with NMOSD than without NMOSD, while CFH and HP protein OS were higher in pSS patients with NMOSD. Based on literature review, clusterin and CFH, which may be relevant to related diseases, were further verified in this study.
Clusterin is a multifaceted protein functioning at the crossroads of inflammation and autoimmune diseases. The main form of clusterin is a secreted heterologous protein with a molecular weight of 80 kDa (21). In our study, serum clusterin was higher in pSS without NMOSD than with NMOSD but not significant. However, serum clusterin was significantly higher in NMOSD with pSS than without (Figure 2). Therefore, clusterin might be a potential biomarker that can differentiate NMOSD with and without pSS. Recent researches showed that clusterin could be found in saliva, tears, and salivary glands of pSS patients and participate in the pathogenesis of exocrine involvement (22–24), which suggest that increased clusterin might participate in the pathogenesis of NMOSD to complicated with pSS.
For the pathogenesis of NMOSD, binding of NMO-IgG to its target AQP4 initiates several immune processes. Often, the binding could readily activate the complement system, subsequently, amplify inflammation, and disrupt the blood-brain barrier, leading to astrocyte injury and causing secondary demyelination and neuronal loss (25). CFH is an important regulator in complement activation alternative pathways. It also acts as a co-factor for factor I to regulate the C3 and C5 convertases. Shi et al. (26) reported that engineered neural stem cells with CFH can attenuate inflammatory infiltration and immune-mediated damage of astrocytes. Moreover, Pohl et al. (27) reported that only T cells were seen in the lesions of destructive astrocyte from NMO patients. Of note, the T cells in the lesions showed signs of activation. Since T cell activation was regulated by the decreased expression of CFH in microglia, decreased CFH might be a risk factor for NMOSD. Interestingly, the levels of CFH were lower in pSS patients with NMOSD than without NMOSD (Figure 2), which is consistent with the assumption that decreased CFH is a risk factor for NMOSD.
There are three limitations in the current study. Firstly, observing the levels change of clusterin and CFH during treatment will support our finding to an extent. However, the rate of lost to follow-up is high that we could not perform this study. Secondly, comparing the levels change of selected proteins in cerebrospinal fluid among groups, which might act as an important confirmation of their pathogenesis role, were not done owing to ethical issues. Thirdly, the SS with NMOSD is a rare disease that limits the inclusion of related patients. The current sample size could help access a statically significant result, but not enough for generating a stable statistical result.
In conclusion, this is the first serum comparative proteomics focusing on the difference between pSS patients with and without NMOSD. Our results confirm that DIGE is a useful tool for such comparison. Increased clusterin and decreased CFH in pSS patients with NMOSD were potential biomarkers and related to the pathogenesis of these diseases. Further researches with larger sample size and more subgroup of patients are required to verify the value of these candidate biomarkers and their clinical significance.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
This study was carried out in accordance with the recommendations of the Ethics Committee of the Peking Union Medical College Hospital with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the the Ethics Committee of the Peking Union Medical College Hospital.
Author Contributions
YL and YZ designed the research. LQ and CD performed the experiments. QW, WZ, YF, and YX recruited patients. CD wrote the manuscript and YL approved the version to be published.
Funding
This work was supported by the National Natural Science Foundation of China Grants [grant number 81671618], the Capital Health Research and Development of Special Fund [grant number 2014-1-4011], and the CAMS Initiative for Innovative Medicine [grant number 2017-I2M-3-001].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Wingerchuk DM, Lennon VA, Lucchinetti CF, Pittock SJ, Weinshenker BG. The spectrum of neuromyelitis optica. Lancet Neurol. (2007) 6:805–15. doi: 10.1016/S1474-4422(07)70216-8
2. Kahlenberg JM. Neuromyelitis optica spectrum disorder as an initial presentation of primary Sjogren's syndrome. Semin Arthritis Rheum. (2011) 40:343–8. doi: 10.1016/j.semarthrit.2010.05.005
3. Zhong YH, Zhong ZG, Zhou Z, Ma ZY, Qiu MY, Peng FH, et al. Comparisons of presentations and outcomes of neuromyelitis optica patients with and without Sjogren's syndrome. Neurol Sci. (2017) 38:271–7. doi: 10.1007/s10072-016-2751-2
4. Wingerchuk DM, Weinshenker BG. Neuromyelitis optica: clinical predictors of a relapsing course and survival. Neurology. (2003) 60:848–53. doi: 10.1212/01.wnl.0000049912.02954.2c
5. Delalande S, de Seze J, Fauchais AL, Hachulla E, Stojkovic T, Ferriby D, et al. Neurologic manifestations in primary Sjogren syndrome: a study of 82 patients. Medicine. (2004) 83:280–91. doi: 10.1097/01.md.0000141099.53742.16
6. Chourkani N, El Moutawakil B, Sibai M, Bourezgui M, Rafai MA, Slassi I. [Primary Sjogren's syndrome and neuromyelitis optica]. Rev Med Interne. (2010) 31:e13–5. doi: 10.1016/j.revmed.2009.08.018
7. Dellavance A, Alvarenga RR, Rodrigues SH, Kok F, de Souza AW, Andrade LE. Anti-aquaporin-4 antibodies in the context of assorted immune-mediated diseases. Eur J Neurol. (2012) 19:248–52. doi: 10.1111/j.1468-1331.2011.03479.x
8. Birnbaum J, Atri NM, Baer AN, Cimbro R, Montagne J, Casciola-Rosen L. Relationship between neuromyelitis optica spectrum disorder and Sjogren's syndrome: central nervous system extraglandular disease or unrelated, co-occurring autoimmunity? Arthritis Care Res. (2017) 69:1069–75. doi: 10.1002/acr.23107
9. Alhomoud IA, Bohlega SA, Alkawi MZ, Alsemari AM, Omer SM, Alsenani FM. Primary Sjogren's syndrome with central nervous system involvement. Saudi Med J. (2009) 30:1067–72.
10. Min JH, Kim HJ, Kim BJ, Lee KW, Sunwoo IN, Kim SM, et al. Brain abnormalities in Sjogren syndrome with recurrent CNS manifestations: association with neuromyelitis optica. Mult Scler. (2009) 15:1069–76. doi: 10.1177/1352458509106228
11. Qiao L, Wang Q, Fei Y, Zhang W, Xu Y, Zhang Y, et al. The clinical characteristics of primary Sjogren's syndrome with neuromyelitis optica spectrum disorder in China: a STROBE-compliant article. Medicine. (2015) 94:e1145. doi: 10.1097/MD.0000000000001145
12. Hu CJ, Song G, Huang W, Liu GZ, Deng CW, Zeng HP, et al. Identification of new autoantigens for primary biliary cirrhosis using human proteome microarrays. Mol Cell Proteomics. (2012) 11:669–80. doi: 10.1074/mcp.M111.015529
13. Yan Z, Chaojun H, Chuiwen D, Xiaomei L, Xin Z, Yongzhe L, et al. Establishing serological classification tree model in rheumatoid arthritis using combination of MALDI-TOF-MS and magnetic beads. Clin Exp Med. (2015) 15:19–23. doi: 10.1007/s10238-013-0265-2
14. Deng C, Lin M, Hu C, Li Y, Gao Y, Cheng X, et al. Exploring serological classification tree model of active pulmonary tuberculosis by magnetic beads pretreatment and MALDI-TOF MS analysis. Scand J Immunol. (2011) 74:397–405. doi: 10.1111/j.1365-3083.2011.02590.x
15. Baldini C, Cecchettini A, Gallo A, Bombardieri S. Updates on Sjogren's syndrome: from proteomics to protein biomarkers. Expert Rev Proteomics. (2017) 14:491–8. doi: 10.1080/14789450.2017.1333904
16. Deng CW, Wang L, Fei YY, Hu CJ, Yang YJ, Peng LY, et al. Exploring pathogenesis of primary biliary cholangitis by proteomics: a pilot study. World J Gastroenterol. (2017) 23:8489–99. doi: 10.3748/wjg.v23.i48.8489
17. Deng C, Hu C, Wang L, Zhang S, Li P, Wu Z, et al. Serological comparative proteomics analysis of mitochondrial autoantibody-negative and -positive primary biliary cirrhosis. Electrophoresis. (2015) 36:1588–95. doi: 10.1002/elps.201400342
18. Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE, et al. Classification criteria for Sjogren's syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis. (2002) 61:554–8. doi: 10.1136/ard.61.6.554
19. Wingerchuk DM, Lennon VA, Pittock SJ, Lucchinetti CF, Weinshenker BG. Revised diagnostic criteria for neuromyelitis optica. Neurology. (2006) 66:1485–9. doi: 10.1212/01.wnl.0000216139.44259.74
20. Byrne JC, Downes MR, O'Donoghue N, O'Keane C, O'Neill A, Fan Y, et al. 2D-DIGE as a strategy to identify serum markers for the progression of prostate cancer. J Proteome Res. (2009) 8:942–57. doi: 10.1021/pr800570s
21. Murphy BF, Kirszbaum L, Walker ID, d'Apice AJ. SP-40,40, a newly identified normal human serum protein found in the SC5b-9 complex of complement and in the immune deposits in glomerulonephritis. J Clin Invest. (1988) 81:1858–64. doi: 10.1172/JCI113531
22. Cuida M, Legler DW, Eidsheim M, Jonsson R. Complement regulatory proteins in the salivary glands and saliva of Sjogren's syndrome patients and healthy subjects. Clin Exp Rheumatol. (1997) 15:615–23.
23. Delaleu N, Mydel P, Kwee I, Brun JG, Jonsson MV, Jonsson R. High fidelity between saliva proteomics and the biologic state of salivary glands defines biomarker signatures for primary Sjogren's syndrome. Arthritis Rheumatol. (2015) 67:1084–95. doi: 10.1002/art.39015
24. Li B, Sheng M, Li J, Yan G, Lin A, Li M, et al. Tear proteomic analysis of Sjogren syndrome patients with dry eye syndrome by two-dimensional-nano-liquid chromatography coupled with tandem mass spectrometry. Sci Rep. (2014) 4:5772. doi: 10.1038/srep05772
25. Vincent T, Saikali P, Cayrol R, Roth AD, Bar-Or A, Prat A, et al. Functional consequences of neuromyelitis optica-IgG astrocyte interactions on blood-brain barrier permeability and granulocyte recruitment. J Immunol. (2008) 181:5730–7. doi: 10.4049/jimmunol.181.8.5730
26. Shi K, Wang Z, Liu Y, Gong Y, Fu Y, Li S, et al. CFHR1-modified neural stem cells ameliorated brain injury in a mouse model of neuromyelitis optica spectrum disorders. J Immunol. (2016) 197:3471–80. doi: 10.4049/jimmunol.1600135
Keywords: primary Sjögren's syndrome, proteome, neuromyelitis optica spectrum disorder, clusterin, complement factor H
Citation: Qiao L, Deng C, Wang Q, Zhang W, Fei Y, Xu Y, Zhao Y and Li Y (2019) Serum Clusterin and Complement Factor H May Be Biomarkers Differentiate Primary Sjögren's Syndrome With and Without Neuromyelitis Optica Spectrum Disorder. Front. Immunol. 10:2527. doi: 10.3389/fimmu.2019.02527
Received: 13 August 2018; Accepted: 10 October 2019;
Published: 25 October 2019.
Edited by:
Jorge Matias-Guiu, Complutense University of Madrid, SpainReviewed by:
Riwanti Estiasari, University of Indonesia, IndonesiaRicardo Constantino Ginestal, Hospital Clínico San Carlos, Spain
Copyright © 2019 Qiao, Deng, Wang, Zhang, Fei, Xu, Zhao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Zhao, emhhb3lhbl9wdW1jaDIwMDImI3gwMDA0MDthbGl5dW4uY29t; Yongzhe Li, eW9uZ3poZWxpcHVtY2gmI3gwMDA0MDsxMjYuY29t
†These authors are considered as co-first author
 Lin Qiao
Lin Qiao Chuiwen Deng
Chuiwen Deng Qian Wang2
Qian Wang2