
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 10 October 2019
Sec. Comparative Immunology
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.02292
This article is part of the Research TopicPopulation and action mechanism of immune cells in fishView all 6 articles
The immune system is composed of two subsystems—the innate immune system and the adaptive immune system. The innate immune system is the first to respond to pathogens and does not retain memory of previous responses. Innate immune responses are evolutionarily older than adaptive responses and elements of innate immunity can be found in all multicellular organisms. If a pathogen persists, the adaptive immune system will engage the pathogen with specificity and memory. Several components of the adaptive system including immunoglobulins (Igs), T cell receptors (TCR), and major histocompatibility complex (MHC), are assumed to have arisen in the first jawed vertebrates—the Gnathostomata. This review will discuss and compare components of both the innate and adaptive immune systems in Gnathostomes, particularly in Chondrichthyes (cartilaginous fish) and in Osteichthyes [bony fish: the Actinopterygii (ray-finned fish) and the Sarcopterygii (lobe-finned fish)]. While many elements of both the innate and adaptive immune systems are conserved within these species and with higher level vertebrates, some elements have marked differences. Components of the innate immune system covered here include physical barriers, such as the skin and gastrointestinal tract, cellular components, such as pattern recognition receptors and immune cells including macrophages and neutrophils, and humoral components, such as the complement system. Components of the adaptive system covered include the fundamental cells and molecules of adaptive immunity: B lymphocytes (B cells), T lymphocytes (T cells), immunoglobulins (Igs), and major histocompatibility complex (MHC). Comparative studies in fish such as those discussed here are essential for developing a comprehensive understanding of the evolution of the immune system.
The vertebrate immune system is divided into 2 subsystems—the innate immune system and the adaptive immune system. The innate immune system is the first to respond to initial infection and disease and does not retain memory of previous responses. Components of the innate immune system include physical barriers such as the skin, cellular processes such as phagocytosis and humoral components such as soluble proteins (1). If a pathogen persists, despite the innate immune defenses, the adaptive immune system is recruited. The adaptive immune system is highly specific to a particular antigen and can provide long-lasting immunity (2). While the innate immune system is assumed to have arisen >600 million years ago (MYA), specific components of the adaptive immune system, including immunoglobulins (Igs), T cell receptors (TCR), and major histocompatibility complex (MHC), are comparatively newer and are assumed to have arisen approximately 450 MYA in the first jawed vertebrates (i.e., Gnathostomata) (3–5). In order to understand the evolution and functionality of the immune system in jawed vertebrates, a comparative analysis of the key branches of Gnathostomata (Chondrichthyes, Actinopterygii, and Sarcopterygii) is required.
Gnathostomes are subdivided into Chondrichthyes (cartilaginous fishes) and Osteichthyes (bony fishes). They diverged from a jawless common ancestor with the lineage leading to other bony vertebrates. While jawless fish have an adaptive immune system based on variable lymphocyte receptors (VLRs), B-like and T-like cells, Gnathostomes are the most distantly related group to mammals that have an adaptive immune system based on Igs, TCR, and MHC (3, 6).
There are over 1,000 species of cartilaginous fish, which are divided into two subclasses: Elasmobranchii (sharks, rays, skates, and sawfish) and Holocephali (chimeras) (7). The Osteichthyes are a diverse group of fish that have skeletons composed of calcified bone rather than cartilage and consist of over 40,000 species of fish (8). They are subdivided into two classes, the Actinopterygii (ray-finned fish) and the Sarcopterygii (lobe-finned fish) (Figure 1). The Actinopterygii have fins that are composed of webs of skin supported by bony spines, known as lepidotrichia. Ray-finned fish comprise 99% of the Osteichthyes, of which 96% are from the infraclass Teleostei (9, 12). Due to the large number of teleost species, as well their economic importance, there have been many genomic and functional immunological studies completed on teleost fish. The Sarcopterygii possess fleshy, lobed, paired fins, joined to the body by a single bone and are comprised of Actinistia (coelacanths) and Dipnoi (lungfish) (4). The majority of immunological studies on the cartilaginous fish and lobe-finned fish are genomic analyses, with very few functional studies. However, due to their unique position in the evolution of adaptive immunity, more functional studies are now being applied to cartilaginous fish. While there are several reviews that examine the innate or adaptive immune systems of Chondrichthyes and Actinopterygii, and some studies on Sarcopterygii (3, 13, 14), a comprehensive comparison of both the innate and adaptive immune systems in all 3 classes of fish is lacking. Thus here, we will endeavor to provide a comprehensive comparison of the innate and adaptive immune systems in cartilaginous fish, lobe-finned fish (focusing on coelacanths and lungfish), and ray-finned fish, with a focus on Teleost fish.
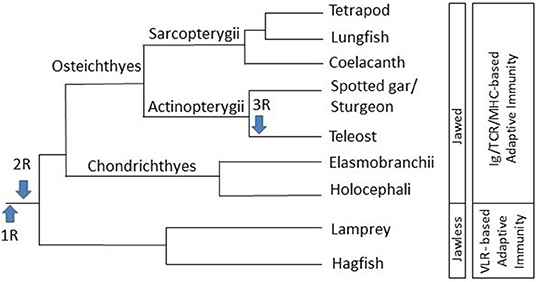
Figure 1. Schematic diagram of the evolution of jawed vertebrates and the immune system. Information sourced from multiple phylogenetic analyses (3, 4, 6, 9–11). R: genome duplication event.
The innate immune system is the first to respond to an initial infection and/or disease. Elements of the innate immune response can be found in all multicellular organisms (14). The innate immune system can be categorized into three defense mechanisms: (1) physical barriers, (2) cellular components, and (3) humoral responses (15). As will be discussed, the functions of these defense mechanisms are highly conserved between fish and mammals.
The first lines of defense in the fish innate immune system are physical barriers that prevent the entry of pathogens, which includes the skin (e.g., scales and mucus), gills, and epithelial layer of the gastrointestinal tract (15). One of the first physical barriers encountered by a pathogen is the skin. Fish are constantly immersed in an aquatic environment and as a result are continuously exposed to potential pathogens or other harmful agents. Therefore, the skin is extremely important in early prevention of pathogen invasion. Teleost skin has been shown to contain skin-associated lymphoid tissue (SALT) that consists of multiple cell types including secretory cells (e.g., goblet cells), lymphocytes (B and T cells), granulocytes, macrophages, and Langerhans-like cells (16, 17). In most teleost fish, the dermis layer of the skin consists of solid, bony scales known as leptoid scales. Interestingly, some teleost species, such as the catfish, have lost their scales during the course of evolution and instead some catfish species have regressed to having bony dermal plates covering their skin (18). The skin of cartilaginous fish also contains many cell types, including melanocytes, lymphocytes, macrophages, and granular leukocytes (19). The scales of cartilaginous fish are called placoid scales, also known as denticles (16). The skin of lobe-finned fish contains keratinocytes, granulocytes and B cells (20). Lobe-finned fish have cosmoid scales that includes a layer of dense, lamellar bone called isopedine. An equally important function of the skin is the ability to secrete mucus, which acts as both a physical barrier, by trapping pathogens, and a chemical barrier (16). Mucus from teleost fish contains a combination of lectins, lysozymes, complement proteins, and antimicrobial peptides (AMPs); all of which play a critical role in neutralizing pathogens (16, 21). While we hypothesize that skin mucus from both cartilaginous fish and lobe-finned fish contains these compounds as well, it has not been as extensively explored as in teleost fish. Supporting this hypothesis are studies showing that a transcript for a lectin, pentraxin, was found in the skin mucus of the common skate (Raja kenojei), while AMPs, including histones and S100 proteins, were found in the skin mucus of the African Lungfish (Protopterus dolloi) (20, 22).
In addition to being involved in osmotic balance and gas exchange, the gills are also an important physical barrier, having both innate and adaptive immune components. The physical barrier of the gills consists of the gill epithelium, a glycocalyx layer, and a mucus layer. In teleost fish, the interbranchial septum is reduced and contains a single caudal opening of the operculum, rather than multiple openings while in cartilaginous fish, the gills are supported for almost their entire length by an interbranchial septum with multiple branchial slits or gill openings (23). Immune cells, including macrophages, neutrophils and eosinophilic granulocytes have been observed in the gill associated lymphoid tissues (GIALT) of teleost fish (24). Lymphocytes have been identified in the gills of several teleost species (25, 26) and of the nurse shark (Ginglymostoma cirratum) (27). For example, B cells and T cells have been identified in the gills of rainbow trout (Oncorhynchus mykiss) and channel catfish (Ictalurus punctatus) while a specific B cell Ig transcript was observed in the gills of nurse shark (see adaptive immune section for a discussion on B cells, Ig, and T cells). Microbes present in the mucosal surface of the GIALT have been found to induce specific immunoglobulin producing B cells (28).
The gastrointestinal (GI) tract facilitates the absorption of nutrients, while preventing pathogen invasion through its epithelium. If a pathogen is ingested, it will encounter the GI tract, which, like the skin and gills, contains both innate and adaptive immune cellular components. Gut associated lymphoid tissue (GALT) can be found in both bony and cartilaginous fish; however, unlike in mammals, it is not highly organized but is composed of a diffuse network of myeloid and lymphoid cells. The intestine of teleost fish, especially the posterior segment, contains both innate and adaptive immune cells including macrophages, mast/eosinophilic granule cells, dendritic cells, B cells, and T cells (24, 29). Anal administration of Vibrio anguillarum to carp (Cyprinus carpio) and intraperitoneal injection of V. anguillarum to sea bass (Dicentrarchus labrax) resulted in the production of B-cells and Igs in the gut (30, 31). T cells have also been identified in the GALT of several teleost species (32–34). In teleost fish, as in mammals, the gut microbiota plays a major role in the development and maturation of the GALT, which in turn mediates its immune response (35, 36). For example, resident microbiota stimulates intestinal epithelial cell proliferation in the developing zebrafish intestine, while absence of microbiota prevents differentiation of the GI tract (37, 38). Dietary administration of probiotics to the gilthead seabream (Sparus aurata) enhanced the intestinal microbiota and increased expression of various immune genes in the intestine including MHCII and TNF-α, while administration of probiotics to the Nile Tilapia (Oreochromis niloticus) and rainbow trout promoted greater development of the intestine, as measured by villous height, and increased the population of intestinal granulocytes (39–41). Lymphoid aggregates, as well as macrophages and granular cells, have been found in the spiral valve of various shark and ray species (42, 43). Lymphocytes and macrophages appear in the gut of the Dogfish shark at hatching and their numbers increase with age, as determined by histological analysis (44). In addition, cytoplasmic Ig has been identified in some intraepithelial lymphoid cells of the shark gut and two Igs (one of high molecular weight and one of low molecular weight) were observed in the intestinal mucosa of the skate (Raja kenojei), although the exact Igs are unknown without the development of antibodies specific to detect cartilaginous fish Igs/proteins (42, 45). Large accumulations of lymphoid cells have been found in the gut of the Australian lungfish (Neoceratodus forsteri); however the cellular and molecular composition of these lymphoid masses is currently unknown (46). While there has been extensive research on the GALT of teleost fish, likely due to their economic importance, there are limited studies on the GALT of cartilaginous and lobe-finned fish and most are histological studies. It is unknown how the GALT in these species respond to infection and if it is in a similar manner as teleost fish and mammals. In addition, while the gut microbiome of some shark species has been identified (47), it is unknown how the microbiota effects the development of the GALT and its immune response in both cartilaginous fish and lobe-finned fish.
If a pathogen passes through the physical barriers, it will encounter the cellular and humoral aspects of the innate immune system. The cellular components of the fish innate immune system consist of many different cell types such as monocytes/macrophages, granulocytes such as mast/eosinophilic granule cells and neutrophils, dendritic cells, and natural killer cells. In bony fish, the primary sites for leukocyte production are the anterior (or head) kidney and thymus, while in cartilaginous fish, the primary sites include the epigonal organ, Leydig organ, thymus, and spleen (48). Analyses of possible sites of leukocyte production (such as the kidney and/or gonads) have yet to be studied in lobe-finned fish (49). Knowing the site of hematopoiesis in lobe-finned fish would allow for isolation of these cells and experiments that would lead to a better understanding of immune cells in these species.
When an innate immune cell encounters a pathogen, it will recognize a pathogen-associated molecular pattern (PAMP) found on the pathogen. Once recognized, the innate immune cell will become activated and can participate in several responses depending on their cell subtype including, but not limited to, phagocytosis and subsequent destruction of the pathogen, production of various cytokines and activation of the adaptive immune system via antigen presentation along with cytokine stimulation.
Monocytes/macrophages and neutrophils are the first to arrive and respond to initial infection. Macrophages are derived from hematopoietic progenitors which differentiate via circulating monocytes or via tissue resident macrophages. Differentiation of vertebrate macrophages is controlled by engagement of the colony-stimulating factor 1 receptor (CSF1R) (50). CSF1R has been characterized in several teleost species, and has been identified in the elephant shark (Callorhinchus milii) genome (51–54). Macrophages play a role in both the innate and adaptive immune systems and are key players during inflammation and pathogen infection, as well as in tissue homeostasis. In the innate immune system, macrophages of several teleost fish species have been demonstrated to destroy pathogens through phagocytosis, the production of reactive oxygen species (ROS) and nitric oxide (NO), and the release of several inflammatory cytokines and chemokines, similar to mammalian macrophages [reviewed in (55–57)]. In the adaptive immune system, macrophages are one type of professional antigen presenting cell (pAPC) that can present phagocytosed materials to the T lymphocytes of the adaptive immune system through a process termed antigen presentation. Macrophages in cartilaginous fish have not been studied as in depth as in teleost fish, however, it is known that nurse shark macrophages exhibit spontaneous cytotoxicity, a nonphagocytic killing mechanism (58). Lungfish macrophages are described to have typical vertebrate macrophage morphology (59, 60). Very few functional studies have been completed in lungfish, however, one study found that injection of lipopolysaccharide (LPS) did not change the number of macrophages in the coelomic cavity, as was expected (59). Similar to mammals, functionally distinct subpopulations of macrophages exist in bony fish. M1 (classically activated macrophages) are characterized by production of pro-inflammatory cytokines such as TNFα and IL-1β and production of ROS and NO, whereas M2 (alternatively activated macrophages) are linked to immunosuppression, wound repair and increased levels of arginase and anti-inflammatory cytokines such as interleukin (IL)-10 (55, 57, 61). The best characterized macrophage phenotype in teleost fish is comparable to M1 macrophages where they can destroy pathogens via acidification, nutrient restriction, production of reactive intermediates and various cytokines and chemokines (55–57). Macrophages, as well as virtually all immune cells, are able to communicate with each other via cell-derived extracellular vesicles (EVs) which contain and deliver messenger RNA (mRNAs), microRNA (miRNAs) and proteins (62, 63). While in recent years, EVs have been extensively studied in mammals, very few studies exist in fish. In one fish study, Atlantic salmon (Salmo salar) head kidney leukocytes were stimulated with CpG oligonucleotides which caused the release of EVs that contained mRNA and miRNA, as well as a protein composition similar to mammals including MHC I and MHC II molecules (64). The secretion of EVs was not induced by CpG in a splenocyte culture (containing mostly B cells) suggesting that the EVs were likely produced by macrophages or dendritic cells in the head kidney leukocyte culture (64). The existence of M1 and M2 cell populations, as well as EVs, have yet to be examined in cartilaginous and lobe-finned fish.
The most abundant granulocytes in bony fish are neutrophils, and like macrophages, neutrophils are critical to the innate defense against pathogens (65). Neutrophils exhibit potent antimicrobial responses through various intracellular and extracellular mechanisms including the release of granules containing cytotoxic and antimicrobial enzymes, the release of neutrophil extracellular traps (NETs), phagocytosis and the production of ROS and NO [reviewed in (57, 65)]. Some bony fish granulocytes have a similar appearance to that of mammalian cells (neutrophils) or avian cells (heterophils). Fish granulocytes exhibit a wide variation in morphology, numbers and types of cells between species causing much confusion regarding their nomenclature. For example, a study by Tavares-Dias et al. (66) identified only one type of neutrophil in channel catfish, while a study by Cannon et al. (67) reported heterophils instead of neutrophils. Granulocytes in cartilaginous fish are classified in three types based on size, shape, and staining properties. G1 granulocytes, referred to as heterophils or fine eosinophilic granulocytes, are often the most common granulocyte in cartilaginous fish. Their numbers can range from 20 to 50% of the total leukocytes in the blood, depending on species (68). G2 granulocytes resemble mammalian neutrophils, while G3 are referred to as coarse eosinophilic granulocytes (68, 69). G3 is more commonly seen in cartilaginous fish, compared to bony fish (68). Not all species of cartilaginous fish exhibit all three types of granulocytes; for example, only G1 and G3 granulocytes have been found in Thornback rays (Raya clavate) and small eyed rays (Raja microcellata) (68). In the African lungfish (P. dolloi), two types of granulocytes were identified in the South American lungfish (Lepidosiren paradoxa), three granulocyte types were identified based on Giemsa-staining and granule size (eosinophilic I, eosinophilic II and basophilic type) (70) and in the Australian lungfish (N. forsteri) four types of granulocytes have been described (basophil, neutrophils, large eosinophils and small eosinophils) (71).
Initiation of the innate immune response begins when germline-encoded intracellular or extracellular pattern recognition receptors (PRRs) of an immune cell bind to a PAMP found on a pathogen, such as bacteria-derived LPS, viral RNA, bacterial DNA, or a danger-associated molecular pattern (DAMP) found on proteins or other biomolecules that are released from stressed cells or injured cells. All PRRs have a domain for recognizing the PAMP that is coupled to a domain that interacts with downstream signaling molecules (72). In mammals, PRRs can be classified into at least five major groups: Toll-like receptors (TLRs), retinoic acid inducible gene I (RIG-I)-like receptors (RLRs), C-type lectins (CLRs), the nucleotide-binding domain, leucine-rich repeat containing proteins (NLRs), and absent in melanoma (AIM)-like receptors (73). Many homologs of mammalian PRRs have been identified in fish.
TLRs were the first PRRs to be discovered in fish and therefore have been the most extensively studied. To date there have been 13 TLRs identified in mammals, whereas over 20 have been identified in different fish species (73–76). A comparison of the TLRs found in mammals, cartilaginous fish, ray-finned fish and lobe-finned fish, as well as their ligands (in mammals and when known in bony fish) can be found in Table 1. Some mammalian orthologs of TLRs have not been identified in fish, whereas some TLRs, including soluble TLR5 (sTLR5), TLR13, TLR14, and TLR18-28 are “fish-specific” (77). For example, a sTLR5 has been identified in bony fish, including rainbow trout, and Atlantic salmon, whereas no sTLR5 has been found in mammalian genomes (82, 90, 91). Interestingly, TLR5, as well as TLR1, TLR2, and TLR6, are missing from the Atlantic cod (Gadus morhua) genome (87, 88). Some bony fish, including the zebrafish (Danio rerio), the Dabry's sturgeon (Acipenser dabryanus) and the yellow catfish (Pelteobagrus hydrophila), possess TLR4-like genes, while TLR4 is absent in other bony fish species, as well as absent in coelacanths and cartilaginous fish (56, 85, 86). TLR4 in fish, however, does not possess the ability to recognize LPS as it does in mammals (56). TLR27 was first identified and thought to only be found in the coelacanth genome but has since been identified in the spotted gar (Lepisosteus oculatus) and elephant shark (78, 89). TLR2, TLR3, TLR6, and TLR9 have been identified in the gray bamboo shark (Chiloscyllium griseum) genome whereas no TLR6 or TLR10 homolog has been identified in teleost fish. In addition, a novel TLR with sequence similarity to TLR4 and TLR13 in mammals, and TLR21 in teleost fish, has been identified in the whale shark (Rhincodon typus) (80, 83).
Due to genome duplication events, several paralogs of various TLRs exist in fish. Two rounds of genome duplication (1R and 2R) are thought to have occurred early in vertebrate evolution, before the Cyclostome/Gnathostome divergence, ~500–800 MYA (Figure 1) (10, 11). Evidence, such as an increase in the number of Hox gene clusters, indicates that an additional genome duplication event (3R) occurred early in the teleost lineage, after it split from the lobe-finned lineage 325–350 MYA, while an additional round of genome duplication (4R) occurred in some fish species, including salmonids, thus leading to several paralogs of genes, including TLRs (79, 92). Paralogous TLR4 and TLR8 genes have been identified in zebrafish (D. rerio) (81, 93), TLR8 in rainbow trout (76) and TLR3 and TLR7 in common carp (C. carpio) (84), while multiple copies of TLR7, TLR8, TLR9, TLR22, and TLR25 have been identified in the Atlantic cod (87). The high number and large diversity of fish TLRs are likely derived from their distinct and diverse evolutionary history and environments that they occupy [reviewed in (77)].
In addition to TLRs, differences in several other PRRs between ray-finned, lobe-finned and cartilaginous fish have been noted. While AIM has not been identified in teleost or cartilaginous fish, two HIN200 domains, a PAMP-recognizing protein domain characteristic of AIM in mammals, were discovered in the coelacanth genome (78, 94). A group of unique NLRs possessing a C-terminal B30.2 domain has been identified in teleost fish, but is missing from the coelacanth genome (78). Additionally, novel immune-type receptors (NITRs) which have been studied extensively in ray-finned fishes are missing from the coelacanth genome (78). While all three RIG-I-like receptors have been characterized in teleost fish, only RIG-I and MDA5 have been identified in the elephant shark and coelacanth genomes (54, 78). However, as more high quality, well-assembled, and annotated genomes become available for additional cartilaginous and lobe-finned fish, additional NITRs may be identified. These differences indicate that not only is pathogen recognition quite diverse in fish, it can also be lineage-specific.
Phagocytosis is one of the most ancient and universal tools of defense against foreign material. This mechanism of defense is observed even in unicellular eukaryotes, predating complex multicellular life (57, 95–98). Binding of a pathogen to a PRR triggers phagocytosis in cells termed phagocytes. These include macrophages, monocytes, neutrophils and dendritic cells and are found in both bony and cartilaginous fish (57, 95–98). Recently, the existence of B cells with phagocytic ability was discovered in various teleost fish species including rainbow trout, Atlantic salmon, and Atlantic cod (99, 100). It is unknown if cartilaginous fish and lobe-finned fish have phagocytic B cells. After engulfment, the phagosome, containing the pathogen, binds to a lysosome, forming a phagolysosome, where the pathogen is killed by various means including the production of ROS and NO (57). Studies in shark, skate, lungfish and teleost fish have demonstrated both ROS and NO production in various leukocytes (65, 101).
Humoral responses are mediated by macromolecules produced by cells and released into the extracellular fluids following infection by a pathogen. Some of the most studied humoral components in fish include the complement system, lysozyme, antimicrobial peptides, and acute phase proteins. These components have many different functions including the promotion of inflammation and phagocytosis and direct bactericidal effects.
The complement system is a cascade of serum proteins that act cooperatively to mediate defense mechanisms including the elimination of pathogens through opsonization and phagocytosis and the promotion of the inflammatory response. The mammalian complement system is composed of ~30 proteins that make up three activation pathways: the classical pathway, activated by antibody-antigen complexes and thus a bridge between innate and adaptive immunity; the alternative pathway, which is independent of antibodies and activated directly by pathogens; and the lectin pathway which is activated by the binding of the mannose-binding lectin (MBL), or ficolin, to mannose (or other sugar) residues present on the pathogen surface (102). Figure 2 illustrates these three pathways, along with some of the associated proteins. Ultimately, these pathways induce activation of the C3 convertase, which cleaves inactive C3 into C3a, an anaphylatoxin that acts as a chemotactic factor and aids in inflammation, and C3b, which acts as an opsonin, as well as an activator of downstream complement proteins leading to the formation of the membrane attack complex (103, 104).
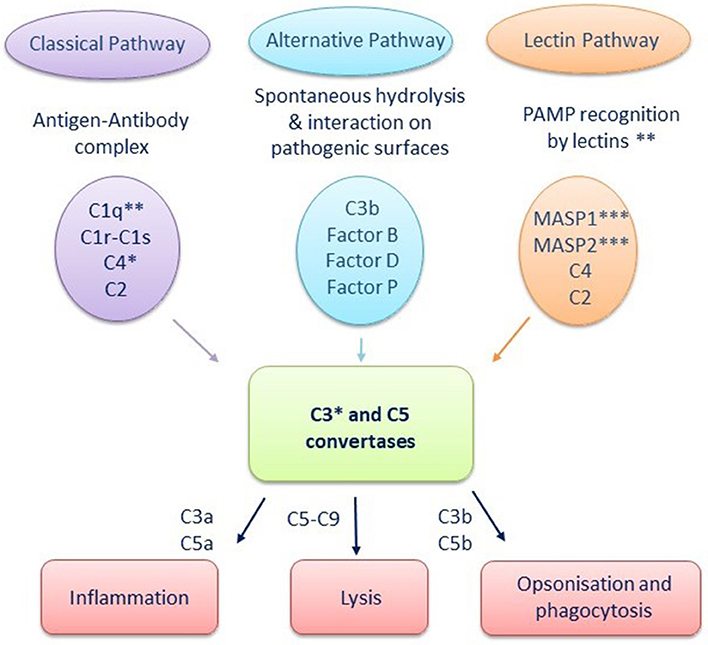
Figure 2. The three complement pathways with associated proteins. *Multiple C3 and/or C4 isoforms in some teleost and cartilaginous fish species. **Absence of mannose-binding lectin (MBL), ficolin, C1qA, and C1qC from genomes of any cartilaginous fish studied to date ***MASP2 transcript with no serine protease domain in hammerhead shark genome; MASP2 missing from elephant shark, little skate, and catshark genomes (102–109).
Most of the mammalian complement components have homologs in various teleost species, including rainbow trout (105), zebrafish (110), and channel catfish (111), among many others, and their functions have been well-characterized [reviewed in (56, 112)]. Similarly, components of all three pathways have been characterized in several cartilaginous fish species, where they have been found to have hemolytic properties (113–116). Furthermore, genes encoding complement components have been identified in lungfish (117, 118) and in the coelacanth genome (78). These studies in different fish classes/subclasses suggest that some components of the complement system are evolutionarily conserved and similar to those of higher vertebrates (102).
However, not all fish species contain all three pathways. MBL and ficolin genes have not been identified in any cartilaginous fish studied to date, while MASP2 transcripts are lacking in the elephant shark, little skate (Leucoraja erinacea) and catshark (Scyliorhinus canicular) (54, 106, 107). In addition, the hammerhead shark contains a MASP2 transcript that contains no serine protease domain, which is necessary to initiate the lectin pathway. This data suggests that the lectin pathway may not be present in cartilaginous fish (106).
Furthermore, some fish species contain multiple forms of various complement factors. Multiple C3 forms have been identified in teleost fish and cartilaginous fish. For example, rainbow trout have three C3 forms, common carp have eight, and gilthead seabream (S. aurata) have five (108, 109, 119), with each form demonstrating different binding efficiencies and functions. Two C3 variants have been described in the nurse shark and the small-spotted catshark, while two C4 gene haven been identified in the elephant shark and hammerhead shark (54, 106, 107). This structural and functional diversity suggests that these fish may have an increased capacity to recognize and destroy a broader range of pathogens compared to those with fewer forms, although this remains to be demonstrated.
Lysozyme is a lytic enzyme that acts on the peptidoglycan layer of bacterial cell walls by hydrolyzing 1–4 β-linked glycoside bonds resulting in lysis of the bacterium. It is also involved in other defenses such as opsonization and phagocytosis and activation of the complement system (120–122). Two types of lysozyme have been described in vertebrates: chicken (c)-type and goose (g)-type.
Lysozyme is one of the most studied innate immune components in fish. C-type and g-type lysozymes have been reported in several teleost species where they are found in neutrophils, monocytes and to a lesser extent in macrophages of several tissues (e.g., liver, kidney, spleen, gills) and in mucus (120, 123, 124). Recombinant (r-) c-type and g-type lysozymes have been found to have high bacteriolytic activity against a variety of pathogens of teleost fish such as V. anguillarum, Aeromonas hydrophila, and Micrococcus lysodeikticus (125, 126). A sequence homology search of the Atlantic cod genome revealed an absence of c-type lysozyme genes; however, four g-type lysozyme genes were identified in several different tissues (102). Intraperitoneal injection of Francisella noatunensis, an intracellular bacterium that commonly infects cod, stimulated the expression of two of the g-type lysozyme genes in the head kidney (127). The presence of multiple g-type lysozymes may compensate for the lack of c-type lysozymes in the Atlantic cod (127). The presence of lysozyme in the lymphomyeloid tissues of several cartilaginous fish was first discovered in 1979 (128). A genomic investigation by Venkatesh et al. failed to identify g-type lysozyme in the elephant shark genome, however c-type lysozyme was identified (54). This c-type lysozyme was characterized in the nurse shark and found to hydrolyze the cell wall of M. lysodeikticus and inhibit the growth of Gram-positive bacteria, suggesting a similar function for lysozyme as in teleost fish and higher vertebrates (129). In addition, two g-type lysozyme genes were discovered in the coelacanth genome, although no functional studies on lysozymes have been completed in coelacanth or lungfish to date (130). Collectively, these studies suggest that the function of lysozyme is similar in both bony and cartilaginous fish.
AMPs, also known as host defense peptides, that are generally oligopeptides with a varying number of amino acids that are generally positively charged and play a major role in the innate immune system. AMPs protect against a variety of pathogens via disruptive or pore-forming actions against bacterial membranes. Over 90 fish AMPs have been identified and are characterized as β-defensins, cathelicidins, hepcidins, histone-derived peptides and fish-specific piscidins. Several of these AMPs have been cloned and subsequent functional studies have demonstrated antiviral and antibacterial activities against a variety of pathogens, demonstrating that AMPs from teleost fish exhibit many if not all of the characteristics of other vertebrate AMPs (131–134). For example, β-defensin has been characterized in gilthead seabream, where it demonstrated antimicrobial activity against V. anguillarum, while in Nile tilapia (O. niloticus) β-defensin has shown an inhibitory effect on the growth of Escherichia coli DH5α and Streptococcus agalactiae (135). Two cathelicidin genes have been identified in rainbow trout where they displayed activities against bacteria including V. anguillarum and P. damselae (136) while in Atlantic salmon, cathelicidin has demonstrated microbicidal properties against V. anguillarum (137). Unlike the comprehensive studies conducted on AMPs in teleost fish, research into shark and lobe-finned fish AMPs has not been as extensive. Two AMPs have been isolated from the dogfish shark (Squalus acanthias), transferrin (138) and squalamine (139), which were found to have potent bactericidal activity against both Gram-negative and Gram-positive bacteria. In addition, the AMP Kenojeninin I, has been isolated from the skin of fermented skate (R. kenojei) and was found to have inhibitory effects on Bacillus subtilis, E. coli and Saccharomyces cerevisiae (140). A recent study by Heimroth et al. (20) identified an increase in proteins with known antimicrobial function including histones and S100 proteins in skin mucus of the lungfish P. dollo during terrestrialization.
In both fish and mammals, tissue injury, infection and inflammation induce immune cells, such as macrophages, to secrete various cytokines into the bloodstream, which stimulate hepatocytes to produce and release acute phase proteins (APPs) (141, 142). APPs are classified based on the extent to which their concentrations change (minor, intermediate, or major) and the direction of change (positive or negative). They are involved in a variety of defense activities and include coagulation factors, such as fibrinogen and prothrombin, transport proteins such as ferritin, complement components, C-reactive protein (CRP) and serum amyloid proteins (SAP) [reviewed in (143)]. APPs are well-conserved in arthropods, fish, amphibians, and mammals (144). CRP and SAP are considered major APPs (e.g., their concentrations may increase up to 1,000-fold) and are the most extensively studied APPs in fish. They are members of the pentraxin family of APPs, are present in the body fluids of vertebrates and invertebrates, and are commonly associated with the acute phase response of inflammation (143). In addition to inflammation, CRP and SAP have been shown to activate the complement pathways and play a role in the clearance of apoptotic cells (143, 145).
Both CRP and SAP have been identified in several teleost species (146–148) where their levels in the serum have been shown to increase in response to various inflammation-inducing stimuli (149–152). For example, CRP and SAP expression in Atlantic salmon head kidney leukocytes are upregulated in response to r-IL-Iβ and r-IFNγ, two cytokines that stimulate APP production in mammals, suggesting that the acute phase response is evolutionarily conserved (151). Both CRP and SAP have also been identified in several different cartilaginous fish (153–155). CRP and SAP isolated from the serum of iridescent shark (Pangasianodon hypophthalmus) was found to agglutinate Edwardsiella ictaluri and A. hydrophila (156). Moreover, increased levels of CRP were found in the serum of sharks inhabiting a highly industrialized harbor estuary where exposure to polycyclic aromatic hydrocarbons (PAHs) and other contaminates was likely to lead to an inflammatory response (155). As well, transcriptome analysis of the Indonesian coelacanth, Latimeria menadoensis, genome identified SAP encoding transcripts (157), however, to our knowledge, no other studies examining CRP or SAP in coelacanths or lungfish have been reported.
If a pathogen persists, despite the innate immune defenses, the adaptive immune system will be activated. As previously stated, while jawless fish have an adaptive immune system based on VLRs, B-like and T-like cells, several components of the adaptive immune system, including Igs [also known as antibodies (Ab)], TCR and MHC, are believed to have arisen in the first jawed vertebrates (3, 6).
Like the innate immune system, the adaptive immune system includes both humoral and cellular components. B cells are key elements of the humoral adaptive immune response. The main role of B cells is to produce high affinity Ig against foreign antigen, and to act as a pAPC to present processed antigen to activate T cells. Abs occur in two forms: a soluble form that is secreted from the cell and a membrane-bound form that, in combination with the signaling molecules Igα/Igβ, forms the B cell receptor (BCR). T cells are key elements of cellular adaptive immunity. The T cell receptor (TCR) is always membrane bound and once stimulated via interaction with antigen presented by the pAPC, in the presence of co-stimulation, the T cell can be activated to function as a helper (CD4+) T cell, a regulatory (CD4+) T cell or a cytotoxic (CD8+) T cell.
Antigen-specificity of B cells and T cells is determined by their BCR or TCR, respectively, which are formed from somatic recombination of variable (V), diversity (D) and joining (J) gene segments (Figure 3A), produced by the DNA-recombination ability of the RAG 1 and 2 enzymes and TdT (163, 164). RAG 1/2 and TdT enzymes, as well as the gene segments V, D, and J are present in all classes of jawed vertebrates [reviewed in (158, 165)]. This results in a highly diverse repertoire of BCRs and TCRs able to recognize innumerable different specific antigens and is unique to the adaptive immune system. Due to the random nature of the VDJ recombination, some BCRs and TCRs produced may recognize self-antigens as foreign. Therefore, developing B and T cells will undergo negative selection to ensure only cells that recognize foreign antigen survive. Negative selection occurs when a B cell recognizes self-antigen, inducing apoptosis or receptor editing, while positive selection occurs through antigen-independent signaling involving the BCR. In the case of T cells, a double positive T cell (CD4+ and CD8+) must bind MHC I or MHC II complex to be positively selected, which will induce the surviving T cell to become a CD8+ or CD4+ T cell, respectively. Negative selection occurs when a double positive T cells binds to MHC I or II with a high enough affinity to receive an apoptotic signal. While VDJ recombination has been characterized in fish [reviewed in (158, 165)], the process of negative and positive selection of developing B and T cells has not been fully elucidated, although these processes possibly occur in a similar manner as mammals. For example, double positive T cells were observed in the thymic cortex of sea bass, while single CD4+ or CD8 α+ cells were found in the thymic medulla, similar to that of mammals (166). There is little to no research on negative and positive selection of developing B and T cells in cartilaginous and lobe-finned fish, while there is very limited research in teleost fish. Studies into the regulation of autoimmunity would be valuable to better understand the mechanisms of negative selection in fish. The development of antibodies that specifically detect fish proteins, such as CD4 and CD8, is necessary to fully understand the homing and recirculation of B and T cells in fish.
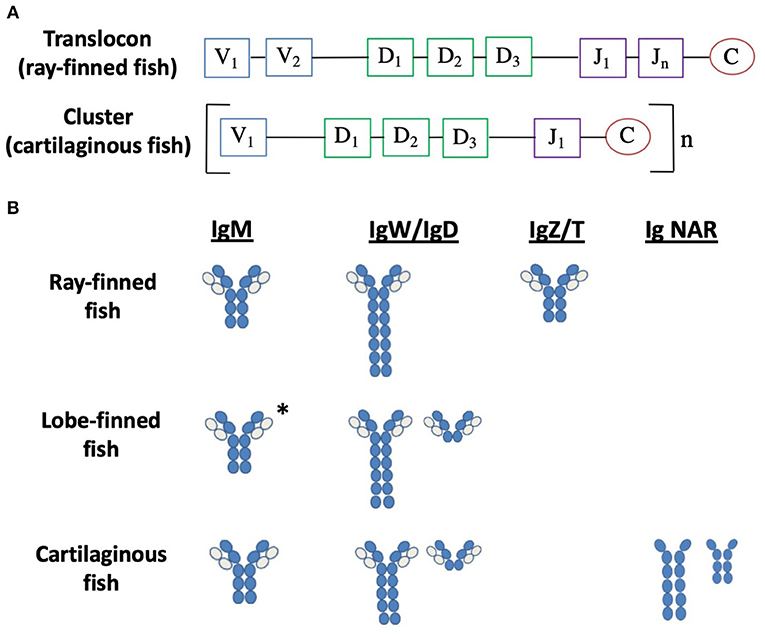
Figure 3. Antibody diversity and isotypes are divergent in fish. (A) Arrangement of the heavy chain loci in bony fish and cartilaginous fish. V represents variable segments, D represents diversity segments, J represents joining segments and C represents constant domains. (B) Examples of the immunoglobulin isotypes in fish. Dark blue circles represent heavy chain domains, light blue circles represent light chain domains (158–162). *IgM for lungfish only; no IgM in the coelacanth.
The BCR includes the membrane-bound antibody (Ab) and the Ig-α/Ig-β (CD79a/b) heterodimer, which is involved in signal transduction. Ab proteins are comprised of two heavy chains (IgH) and two light chains (IgL) held together by disulfide bonds forming a “Y” shaped quaternary structure (158). Both IgH and IgL chains contain one N-terminal variable domain (VH and VL) and one or more C-terminal constant domains (CH and CL). The arms of the “Y” are composed of one constant and one variable domain from each heavy and light chain and are the site of antigen binding, called the Fab region (fragment, antigen-binding). The base of the “Y” is composed of two heavy chain constant domains and is referred to as the Fc (fragment, crystallizable) region. The Fc region mediates the effector functions of the antibody by binding to a specific class of Fc receptors (and other molecules such as complement proteins) with the IgH categorizing them into specific isotypes. The variable regions of the heavy and light chain loci are assembled via somatic gene rearrangement from an array of multiple V, D, and J segments during B cell development, allowing each B cell to produce a unique Ab. In response to antigen, in combination with helper T cell interactions, B cells will secrete antigen–specific Abs. Three classes of Ab have been identified in both teleost fish and cartilaginous fish: IgM, IgD, and IgZ/T in teleost fish, and IgM, IgW, and IgNAR in cartilaginous fish, presumably each with different effector functions. In lungfish, IgM, IgW, and IgN have been identified, while in coelacanths two forms of IgW has been discovered (Figure 3B) [reviewed in (158)].
Similar to all vertebrates (except cartilaginous fish), the IgH genes of teleost fish are arranged in a translocon configuration of which multiple V segments are found upstream of several D and J segments, followed by C segments (Vn-Dn-Jn-C) (Figure 3A) (159). Depending on the species, differences may occur such as duplication of individual V, D, or J segments, or tandem duplication of C domain exons such as that found in Atlantic salmon and zebrafish (167, 168). Instead of the single translocon locus, the IgH loci of cartilaginous fish adopt a multiple mini-cluster organization, with each cluster consisting of one V, two or three Ds, and one J, followed by one set of C region exons for a specific isotype (Figure 3A) (169). The clusters can be repeated as many as 100 times in the genome, depending on the species. While most clusters are capable of rearrangement, some clusters are partially (VD-J) or fully recombined (VDJ or VJ) in the germline, a rearrangement that is unique to cartilaginous fish (170). IgH genes in lungfish are organized in a transiting form, having both cluster (like cartilaginous fish) and translocon (like teleost fish) configurations (160).
IgM is the most ancient antibody class found in all jawed vertebrates; with the exception of coelacanths, which is the only known jawed vertebrate that does not contain IgM in the genome (171–173). IgM is the most prevalent Ab in both bony and cartilaginous fish plasma and can be found in both secreted and transmembrane forms. It shares a similar function in all jawed vertebrates, which includes mediating opsonization, antibody-dependent cell-mediated cytotoxicity, and complement activation, and thus contributes to both innate and adaptive immune responses (58, 102, 174, 175).
In teleost fish, IgM is multimerized into a tetrameric form, although there have been reports of a monomeric IgM form in some teleost species (176, 177). Due to an alternative splicing pathway, the transmembrane form of IgM is one domain shorter than the secreted form in teleost fish, resulting in a shortened IgM receptor on the B cell surface (178). The lack of this domain does not interfere with the ability to interact with Igα/Igβ signaling molecules (179). The J chain, which is required for IgM polymerization and secretion into the mucosa, has not been found in teleost fish, and therefore, tetrameric IgM is polymerized by interchain disulfide bonds (180). IgM is the only teleost isotype for which sub-isotypes have been identified. Two sub-isotypes of IgM have been identified in Atlantic salmon and brown trout (Salmo trutta), reflecting the pseudotetraploid state of salmonid genomes (181, 182).
In cartilaginous fish, IgM accounts for more than 50% of serum protein (158). Both the secreted and transmembrane forms of IgM contain four C domains, except in the neonatal nurse shark, where a subclass of IgM (IgM1gj) found in high amounts in the serum has only 3 C domains (169). IgM in the serum of cartilaginous fish is found in two different states, a monomeric 7S and pentameric 19S, which are present in approximately equal amounts (183). Pentameric IgM serves as the first line of defense, while 7S is produced later (183). Both 7S and 19S IgM play a role in cytotoxicity reactions via phagocytosis (58). In some cartilaginous fish species, such as the nurse shark, the J chain is present in pentameric IgM, although it may not be involved with IgM secretion, unlike the J chain in mammalian IgM (161, 184).
In contrast to the coelacanth, which does not contain IgM in the genome, lungfish species express multiple diverse IgM genes which vary among species (160, 185). For example, the West African lungfish has three IgM isotypes, while the spotted lungfish (P. dolloi) has two. Recently the J chain was identified in the spotted lungfish (186).
IgD is found in many vertebrate classes, including teleost fish and acipenseriformes (a group of fish that phylogenetically links elasmobranches, teleosts, and sturgeons). It is orthologous to IgW (also known as IgX, IgNARC, or IgR depending on the species), which is found only in cartilaginous fish (187, 188), lungfish, and coelacanths (172, 173, 185), suggesting that IgD/IgW is as phylogenetically old as IgM (189, 190). The function of IgW and IgD, however, is poorly understood in both fish and mammals.
Teleost fish contain many forms of IgD, with constant domains ranging from 2 to 16 (191–193). IgD has only been found in a transmembrane form, with the exception of the channel catfish and the Japanese puffer (Takifugu rubripes), which contain both membrane and secretory forms (159). Teleost IgD is unique in that it is a hybrid of the CHμ1 domain followed by a varying number of CH-δ domains, depending on the species (194–197). The IgD heavy chain has not been identified in any teleost fish without the CHμ1 domain (195, 198, 199). IgD is co-expressed with IgM in most teleost fish, although they are absent in channel catfish and rainbow trout. Three different types of IgD+ cells have been identified in catfish: small IgM+/IgD+ B cells, larger IgM–/IgD+ B cells and granular cells containing exogenous IgD via a putative IgD-receptor. In rainbow trout, the ratio of IgD to IgM in the gills is much higher than other tissues. As well, an IgM–/IgD+ B cell subset has been found mainly expressed in the gills, indicating a role for IgD in the gills (191–193).
IgW in cartilaginous fish contains six to eight C domain exons, in addition to the V, D, and J segments. Multiple splice forms of IgW exist in cartilaginous fish other than the original six C domains (IgW-long), including a two C domain (IgW-short) form and a four C domain form (188, 200, 201). A V-less form of IgW has been identified in both the spiny dogfish (S. acanthias) and the nurse shark but represents only 8% of the IgW transcripts analyzed (200).
Two IgW transcripts have been identified in the African lungfish (160). Similar to cartilaginous fish, lungfish IgW can be found in a long form, consisting of seven C domains (homologous to IgW-long) or a short form, consisting of two C domains (160, 185). Two distinct loci for IgW have also been discovered in the Indonesian and African coelacanth (Latimeria chalumnae) (173). It remains unknown if the short and long forms of IgW found in cartilaginous fish and in lungfish have different effector functions and if the functions of IgD/IgW are species specific.
IgNAR (new/nurse shark antigen receptor) is a heavy-chain only Ig found only in sharks. Each chain of IgNAR contains a single-domain V region that can bind to antigen independently (202). IgNAR exists in both long and short forms, which can vary between species (183). The long transmembrane and secreted forms consist of five C domains while the short transmembrane form consists of three C domains (184, 203). Serum levels of IgNAR are much lower than IgM and it is unknown if the J chain is required for IgNAR multimer formation (8).
The immunoglobulin IgT/Z is produced only in bony fish and was first identified in rainbow trout (IgT) and zebrafish (IgZ) (167, 199). In most bony fish characterized to date, IgT/Z contain four C domains, although this is known to vary in a number of species (204–206). While only a few studies have been performed, it is thought that IgT is specialized for mucosal immunity and functions analogously to mammalian IgA. For example, the concentration of IgT/Z in the serum of rainbow trout is much lower than that of IgM, the ratio of IgT/Z:IgM is 63 times higher in the gut than in the serum (207). This study also demonstrated that, following intestinal parasitic infection, the number of IgT+ B cells increased in the gut, but the number of IgM+ B cells in the gut did not change (207). In addition, IgT+ B cells are also found in teleost skin associated lymphoid tissue (SALT) where they secrete IgT into skin mucus (17).
High-throughput sequencing of two species of African lungfish (P. dolloi and P. annectens), followed by Southern blot, identified two unique Ig isotypes in lungfish; these include 3 IgN isotypes (IgN1 found only in P. dolloi while IgN2 and IgN3 found only in P. annectens) and IgQ (found only in P. annectens) (160). Both IgN and IgQ are thought to originate from the IgW lineage (160).
Both bony and cartilaginous fish lack bone marrow, the main site of hematopoiesis in mammals, and germinal centers (GC), specialized sites where mature B cells proliferate, differentiate, and selection of high affinity BCR occurs in mammals. Instead, in teleost fish, the main site of hematopoiesis is the anterior (or head) kidney. Progenitor B cells and plasma cells are found in the anterior kidney, while mature B cells and plasma blasts are found in the posterior kidney and in the spleen (208, 209). Evidence for B cell development in the anterior kidney is supported by expression of RAG-1/2 and terminal deoxynucleotidyl transferase (TdT), and the resulting development of immature B cells with membrane Ig on their surface. It is proposed that mature B cells are released into the blood where they encounter antigen and mature into plasma blasts or plasma cells. Plasma cells then migrate back to the anterior kidney where they may become long-lived plasma cells, supporting the storage of Ig-secreting cells (208, 210). However, more work is required to fully elucidate the mechanisms regulating homing of B cells in fish. The spleen is considered the only secondary lymphoid organ (SLO) in teleost fish, where expression of AID (see below) has been observed, suggesting that the spleen is the site for antigen stimulation (211).
In cartilaginous fish, the Leydig organ, a gland-like structure associated with the esophagus, and the epigonal organ, a structure physically attached to the gonads with a similar structure and organization as the Leydig organ, are the main sites of hematopoiesis and B cell production (48). Lymphocytes of various sizes are abundant in these organs and form a loose follicle-like aggregate with scattered plasma cells (212). While most cartilaginous species have both organs, some species only have one, such as the nurse shark, which only has an epigonal organ (48). Like bony fish, RAG1 and TdT expression in the epigonal organ provides evidence that it is a site of B cell development (213). Additionally, hematopoietic transcription factors important in B and T cell development are expressed in the Leydig and epigonal organ of the embryonic clearnose skate (Raja eglanteria) (214). The spleen of cartilaginous fish contains well-defined white pulp (WP) and red pulp (RP) regions and is considered a SLO. The WP consists of lymphocytes and mature and developing plasma cells, while the RP consists of macrophages, erythrocytes and plasma cells (213, 215). Antigen stimulation, leading to Ab synthesis, has been described in the cartilaginous fish spleen (213, 215). As previously stated, analysis of possible hematopoietic organs (kidney and/or gonads) in lobe-finned fish has yet to be completed (49). Structural analysis of the African lungfish spleen identified characteristics of a secondary lymphoid organ; the red pulp is likely the site of erythropoiesis, as well as site of plasma cell differentiation, similar to cartilaginous fish, as evidenced by mature and immature plasma cells (49). The WP appears to be involved in immune responses (49).
Both bony and cartilaginous fish have been shown to develop immunological memory (i.e., the ability to respond more rapidly and effectively to a pathogen that has been previously encountered). One of the first studies to identify immunological memory in fish was in rainbow trout where it was demonstrated that the secondary response to trinitrophenylated-keyhole limpet hemocyannin (TNP-KLH) was faster and of a larger magnitude than the primary response, as determined by ELISA (216). Several other studies in fish, including rainbow trout and turbot (Psetta maxima), have since shown that neutralizing Ab can be induced against a variety of viral, bacterial and parasitic pathogens and vaccines (217, 218). However, the response time of teleost IgM is much slower than in mammals, taking 3–4 weeks after immunization before specific titers are detected. Interestingly, some fish species, such as the Atlantic cod, do not appear to produce a specific antibody response upon immunization, despite high levels of serum Abs (219). This is likely due to the lack of MHC II genes and gene products in the Atlantic cod (87, 220).
Similar to teleost fish, the immune response time of IgM in cartilaginous fish is much longer than in mammals. Dooley and Flajnik completed a 3 year-long immunization study in the nurse shark (183). The results demonstrated that, following immunization, pentameric IgM, which localizes mainly in the plasma, was induced before other isotypes, but with a low-affinity interaction with antigens. The results also demonstrated that monomeric IgM, which is capable of entering tissues, appeared after pentameric IgM and was the main Ig involved in antigen-specific responses. A significant increase in antigen-specific IgNAR titers was also observed with a high specificity to antigen following immunization. It can take up to 28 months before the antigen-specific titer levels return to pre-immunization levels once the Ig response has reached a plateau (183). Memory was demonstrated for both monomeric IgM and IgNAR as re-immunization after a decrease in titer induced a quicker response than the primary immunization (183).
Activation-induced cytidine deaminase (AID) is an enzyme that mediates somatic hypermutation (SHM) [i.e., a process that fine tunes the Ig, increasing its affinity (affinity maturation)], and mediates class switch recombination (CSR) (i.e., a process whereby the constant region of an antibody heavy chain is changed to a different isotype, thus changing its effector function) (221). AID in fish was first reported in channel catfish, and has since been reported in many other fish species (222, 223). Teleost fish AID differs from mammals in that it has a longer cytidine deaminase motif and substitutions in its carboxy-terminal region (224). Catfish and zebrafish AID have been demonstrated to mediate SHM in mouse fibroblasts (NIH3T3PI19) (225), while zebrafish AID can efficiently deaminate methylated deoxycytidines (226). In addition, the biochemical properties of AID from the sea lamprey, nurse shark, tetraodon, and coelacanth were recently characterized where it was found that these AIDs exhibit unique substrate specificities and optimal temperature tolerances while the lethargic enzymatic rate and affinity for ssDNA of AID are conserved (227). However, a search of the African lungfish mucosal lymphoid tissue transcriptome for AID found no evidence of expression using cartilaginous fish, teleost fish, or tetrapod AID sequences for comparison suggesting that the African lungfish may have lost AID expression in its genome (228). In addition, no AID was found in the African lungfish using RT-qPCR (228). However, other members of the apolipoprotein B mRNA-editing catalytic polypeptide (APOBEC) family (to which AID belongs) were found to be expressed in the African lungfish (228).
Affinity maturation is generated during immune responses in bony fish, as evidenced by the replacement of low-affinity Ab by intermediate-affinity Ab and eventually by high-affinity Ab in rainbow trout (229). The affinity maturation response in fish is much less efficient than mammals, likely due to the absence of GCs. Affinity maturation was also reported in the nurse shark, where purified monomeric IgM showed an increase in the intrinsic association constant to a 3H-ε-DNP-l-lysine ligand over a 20 month period (230). IgNAR also exhibits affinity maturation, as demonstrated by a correlation between somatic mutations and increased binding affinity in IgNAR clones from immune tissues of a hyperimmunized nurse shark (231). The affinity of pentameric IgM, however, does not increase during an immune response (183).
Although teleost fish express AID, they lack class switch recombination (CSR), likely due in part to the structure of the IgH gene (225). However, AID from teleost fish, specifically zebrafish, Japanese puffer and catfish can catalyze CSR in vitro in mammalian AID−/− lymphocytes suggesting that teleost AID has the full catalytic functions capable for CSR (225, 232). Although it was once thought that cartilaginous fish were also incapable of CSR due to the cluster organization of their genes, it is now known that they can undergo an “unconventional” type of CSR among different IgM clusters and between IgW and IgM clusters (233).
A major function of B cells, as well as other pAPCs such as macrophages and dendritic cells, is to process and present antigen to activate T cells. T cells, however, will only recognize antigen fragments that are bound to MHC I or MHC II, cell surface proteins found on pAPCs. While the structure of MHC is conserved over various species, the genes encoding MHC demonstrate a high degree of polymorphism in mammals, lobe-finned fish, and ray-finned fish and cartilaginous fish, allowing different repertoires of peptides to be presented (234–236). In most teleost fish, MHC class I and II reside on different chromosomes, while in cartilaginous fish, and all other vertebrates, MHC I and II are found on the same chromosome (237–239). Interestingly, while MHC I and II are conserved in most jawed vertebrates, Gadiformes, such as the Atlantic cod, have lost the genes for MHC II and CD4, a co-receptor on T cells that interacts with MHC II (87, 88, 220, 240). The Atlantic cod does, however, contain more genes related to the MHC I component of the immune system, as well as the expansion of some TLR clades, compared to other vertebrates, which may help compensate for the missing MHC II and CD4 (87, 220).
Antigens that are to be presented by MHC I are processed via the immunoproteasome and transferred to the endoplasmic reticulum (ER) by transporter associated with antigen processing (TAP) where they associate with MHC I and are eventually transported to the cell membrane. MHC I is ubiquitously expressed in various tissues in teleost and cartilaginous fish including spleen and head kidney (239, 241, 242). In addition, β2 microglobulin, which is associated with MHC I, has been isolated in several teleost fish, as well as the nurse shark and sandbar shark (Carcharhinus plumbeus) (243–245). MHC-I related immunoproteasomes, as well as TAP genes, have also been identified in both bony and cartilaginous fish (234). While there have only been a few studies examining MHC I in lobe-finned fishes, MHC class I genes, including α1, α2, and α3, have been sequenced from blood of the African lungfish and muscle and skin of the West Indian Ocean coelacanth (L. chalumnae) (246, 247). Additionally, lmp1 and lmp2, catalytic subunits of the immunoproteasome, have been characterized in the African lungfish and were found to be induced in primary lung and kidney cell cultures by the synthetic dsRNA polyinosinic-polycytidylic acid [poly (I:C)] (248).
Antigens that are to be presented by MHC II are endocytosed, digested in lysosomes and loaded onto MHC II molecules prior to their migration to the cell surface. MHC II genes have been identified in teleost fish, cartilaginous fish, and the African coelacanth (173, 249, 250). Teleost MHC class II genes can be organized into three groups based on sequence features such as insertions and deletions (250). It has been shown that MHC class II affects resistance to bacterial pathogens, including Aeromonas salmonicida in Atlantic salmon (251). Likewise, challenge with Vibrio harveyi increased expression of MHC II B mRNA in the gill, liver, and spleen of the white bamboo shark (Chiloscyllium plagiosum), similar to teleost fish (252). The identification and characterization of MHC I and II genes in both bony and cartilaginous fish, with the exception of the Gadiformes lineage, suggests that MHC is generally well conserved in these species.
T cells possess a TCR which recognizes a specific antigen and is formed using RAG-mediated V(D)J rearrangement for the development of diverse repertoires. However, unlike the BCR, the TCR is always membrane bound and only recognizes antigen when presented in the context of MHC I or II (3). T cells are classified into 2 main populations: CD8+ cytotoxic T-cells (Tc) which interact with MHC class I and CD4+ helper T cells (Th) which interact with MHC class II. In addition to MHC, all TCR possess a CD3 complex and recognize co-stimulatory (e.g., CD28) and co-inhibitory (e.g., CTLA-4) molecules. In both bony and cartilaginous fish, and similar to mammals, T cells are produced in the thymus. Research in sea bass detected T cells in the developing GALT at the same time as in the thymus, suggesting that the gut may also be a primary lymphoid organ for T cells in bony fish (33).
TCRs are type I transmembrane glycoproteins with extracellular V and C Ig domains and a short cytoplasmic tail (Figure 4). This structure is conserved in almost all vertebrates (255). The TCR is found in two forms: a heterodimer of α and β chains (αβ-TCR) or a heterodimer of γ and δ chains (γδ-TCR), linked by disulphide bonds. Most T cells contain the αβ-TCR, while γδ-T cells account for 1–10% of T cells in the blood of mammals, and 8–20% of total lymphocytes in various tissues of the zebrafish (256). In situ hybridization experiments in the nurse shark identified higher levels of TCR α and β in the central cortex of the thymus but weaker expression in the medulla and subcapsular region. Expression of TCR γ and δ were also high in central cortical cells but were most highly expressed in the subcapsular region. TCR δ was the most highly expressed TCR chain in the medulla (27).
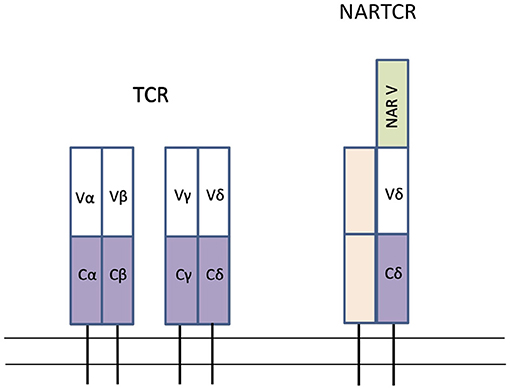
Figure 4. Schematic of the conventional TCR forms found in all Gnathostomes and NARTCR found in cartilaginous fish. Rectangles represent Ig super-family domains; V represent variable domains (white), C represents constant domains (purple) and NAR V represents extra variable domain in NARTCR (green). Modified from Criscitiello et al. (162), Roux et al. (253), and Criscitiello (254).
The genes for TCR-α, β, γ, and δ are diversified using V(D)J rearrangement and have been identified in teleost fish, cartilaginous fish, and coelacanths (173, 255, 257, 258). In both bony fish and cartilaginous fish, the TCR gene segments are in the translocon arrangement, similar to mammals (27, 259). While the structure of TCR is generally well-conserved among all vertebrates, there are several unusual aspects of the cartilaginous fish TCR. Two forms of TCR-δ have been identified in cartilaginous fish, one form having an extra V domain that is closely related to IgNAR (and thus given the name NARTCR) (162, 253). It is hypothesized that the NARTCR-δ chain dimerizes with a TCR-γ chain that lacks the additional domain and therefore NARTCR-δ V domain does not have a binding partner (254). Another unique aspect of shark TCR is that some TCR-δ chains may be formed from a trans-rearrangement of Ig heavy chain V segments with D, J, and C segments of TCR-δ (162). Finally, sharks use AID and SHM to diversify the shark TCR-γ and -α chains (27, 259). SHM is not known to diversify the TCR in any other vertebrates (27, 259). Figure 4 depicts a schematic of a conventional TCR found in gnathostomes and a NARTCR found in cartilaginous fish.
The αβ subtypes can be further divided into helper CD4+ cells (Th) or cytotoxic CD8+ cells (Tc). CD4+ T cells are stimulated by peptides presented via MHC-II molecules and, when activated, CD4+ T cells release cytokines that can activate and regulate responses elicited by the antigen (260). The CD4 molecule is a single protein with four extracellular Ig-like domains and a cytoplasmic tail containing a CxC motif which interacts with the tyrosine kinase Lck, initiating intracellular signaling (261). While tetrapods contain a single CD4 molecule with four Ig domains, two types of CD4 molecules have been described in bony fish: CD4-1 containing four Ig domains and CD4-2 which contains either two or three Ig domains, depending on the species (255). In addition, salmonids contain two CD4-2 molecules (CD4-2a and CD4-2b) (262). An early study of the elephant shark genome suggested that CD4, as well as CD4 associated genes involved in the differentiation (RORC, FOXP3) and function (IL-4, IL-5, IL-13, IL-9, IL-21) of CD4+ cell lineages were missing from the genome (54). Although this study identified several CD4/Lag3-like molecules, they lacked the C-terminal intracellular CxC motif required for interacting with Lcks suggesting that CD4 is absent or not functional in the cartilaginous fish genome. However, a more recent genomic study by Redmond et al. (263) in the Small-spotted catshark used newly available sequence datasets and found putative sequences for CD4 T-cell associated genes including IL-4/IL-13, IL-21, IL-23, IL-27, IL-6Ra, IL-12R, and FOXP3, suggesting that cartilaginous fish do in fact have CD4 T-cell subsets, although more work is still required to fully understand the T cell subsets present in cartilaginous fish, as well as their biological roles (263).
CD8+ T cells are activated by peptides presented via MHC-I molecules and secrete cytotoxins such as perforin and granzymes that initiate apoptosis in the target cells. The CD8 molecule can be in one of two forms: a homodimer formed from two α-chains (CD8αα) or a heterodimer formed from one α- and one β- chain (CD8αβ) (264). Both CD8 chains have been characterized in multiple teleost fish and cartilaginous fish (27, 54, 265, 266). Teleost and cartilaginous fish CD8 exhibit an extracellular Ig-like domain, but the domain has a CxH motif in the cytoplasmic tail, instead of the CxC motif found in mammals, suggesting that CxH represents a primordial Lck binding site (162, 267).
T-cell activation is triggered via antigen:MHC recognition by the TCR and mediated via CD3. All TCR have a short cytoplasmic tail and therefore need to partner with CD3, a complex of transmembrane proteins with intracellular domains containing the conserved motif known as immunoreceptor tyrosine-based activation motif (ITAM). Characterization studies of CD3 in teleost fish have identified a conserved structure of CD3 between teleost fish and mammals (268, 269). Genes encoding the CD3 chains have been annotated in the elephant shark genome and were recently cloned in the small-spotted catshark (S. canicula) where two copies of CD3 were observed (54, 270). Three CD3 chains have also been identified in the coelacanth genome (173). The sequence homology of all 3 chains encoded in the coelacanth genome were distinct from other fishes but grouped together with the corresponding molecules found in avians and mammals (173).
The initial interaction of TCR/MHC/peptide is not sufficient to fully induce activation of naïve T cells and therefore T cells require additional co-stimulatory signals. This is provided by the interactions between CD28, a co-stimulatory factor expressed on T cells, and B7.1 (CD80) and B7.2 (CD86) ligands on the APC. In contrast, binding of B7.1 and B7.2 to CTLA4, a powerful negative regulator of T cell activation, exerts an inhibitory effect on T cell activation. Both CD28 and CLTA4, as well as orthologs of B7.1 and B7.2, have been identified in several teleost species (271–273). The binding sites for B7.1 and B7.2 are conserved in teleost fish CD28 and CTLA4, indicating that CD28 and CTLA4 recognize a B7-like receptor (271). In addition, viral infection in rainbow trout increases CTLA-4 expression, while CD28 remains constitutively expressed, similar to mammals, suggesting that these molecules may have similar roles as their mammalian orthologs (271). Putative CD28, CLA-4, and B7 genes have been annotated in the elephant shark genome, while CD28 has been identified in the coelacanth genome, however the function of these co-receptors in many fish species remains to be fully investigated (54, 173).
Upon activation of CD4+ cells, naïve cells can differentiate into specific subsets including Th1, Th2, Th17, and inducible T-regulatory (Treg) cells; each subset defined by their cytokine production (274). Activation of CD8+ cells induces differentiation into cytotoxic effector cells which release cytotoxins that induce apoptosis of the target cell.
The structures of several orthologs and paralogs of Th cytokines, as well as their functions, have been characterized in both teleost fish and cartilaginous fish and are reviewed in Secombes et al. (275, 276) and Secombes and Wang (277). In brief, two forms of IFNγ, produced by Th1 cells, IFNγ, and IFNγ rel, have been identified in teleost fish including Atlantic salmon, rainbow trout, and ginbuna crucian carp (Carassius carassius), while one form has been identified in fugu (278–280). Recombinant IFNγ (r-IFNγ) was found to increase the expression of anti-viral and inflammation-relevant genes, as well as increase ROS and NO production in zebrafish, rainbow trout and goldfish macrophages, indicating a similar function as mammalian IFNγ (281, 282). A single of copy IFNγ has been identified in the Elephant shark genome (54). Three Il-4/13 genes (IL-4/13A, IL-4/13B1, and IL-4/13B2), produced by Th cells, have been characterized in salmonids (276, 283, 284). Intraperitoneal injection of r-IL-4/13A in zebrafish increased the number of IgZ+ B cells circulating in the blood, compared to a PBS control injection (285), while r-IL-4/13A in rainbow trout modulates the expression of a number of Th2 genes (286). While Venkatesh et al. (54) found no IL-4/13 genes in the elephant shark genome, subsequent interrogation of the genome by Dijkstra (287) found three putative IL-4/13 genes. In addition, Redmond et al. identified a IL-4/IL-13 gene in the small spotted catshark genome (263). Analysis of the coelacanth genome failed to identify Il-4 (173). The IL-17 family in teleost fish, produced by Th17 cells, has several members (A-F) which are structurally related to orthologous proteins in mammals (288, 289). Two homologs of the IL-17 family, IL-17B and IL-17D have been identified in teleost fish, as well as several isoforms of molecules termed IL-17A/F1-3, IL-17C, and IL-17E (290). r-IL-17A/F2 induced the expression of antibacterial peptide β-defensin-3 and the pro-inflammatory cytokines IL-6 and IL-8 in rainbow trout splenocytes, suggesting its role in antibacterial defenses (289). Several IL-17 family members have been found in a cartilaginous fish genome (C. milii) including IL-17A/F, IL-17B, IL-17C, and IL-17D (275). One copy of the IL-10 gene, produced by Treg cells, is found in most species of teleost fish, although two copies have been identified in rainbow trout and European common carp (291, 292). Sequences with homology to IL-10 were found in the spiny dogfish (S. acanthias), elephant shark, and coelacanth genomes (173, 275). These studies, among many other fish cytokine studies, indicate that the structure of cytokines released from Th cells is relatively conserved between ray-finned fish, lobe-finned fish, and cartilaginous fish.
Cytotoxic T cells kill their targets via two mechanisms: the secretory and non-secretory pathways, both of which induce apoptosis. The secretory pathway releases granular toxins such as perforin and serine proteases called granzymes which work together to induce apoptosis (293). The non-secretory pathway involves the engagement of target-cell death receptors, such as Fas, located on the cell surface of the cytotoxic T cells, which results in caspase-dependent apoptosis (294).
The secretory pathway has been identified in many different fish species. A perforin-like molecule has been characterized in several teleost species (295, 296). The killing function of α/β TCR alloantigen specific cytotoxic clones was inhibited in channel catfish by treatment with concanamycin A, a perforin inhibitor (297). Similarly, treatment of ginbuna crucian carp CD8α+ lymphocytes with concanamycin A partially inhibited their function in a dose dependent manner, suggesting that the perforin-mediated pathway in teleost fish is similar to that of higher vertebrates (298). Granzyme has also been recently identified in ginbuna crucian carp (gcGzm) and has a similar primary structure to that of mammals (299). Expression of gcGzm mRNA was greatly enhanced by allo-sensitization and infection with Edwardsiella tarda, indicating that gcGzm is involved in cell mediated immunity (299). In spite of the absence of CD4 and associated CD4 genes, many cytotoxic T cell related genes, including perforin and granzyme, have been identified in the elephant shark genome, suggesting that such cell types are present in cartilaginous fish (54).
While the non-secretory pathway has not been as thoroughly studied in fish as the secretory pathway, the FasL protein has been identified in channel catfish, tilapia (O. niloticus), and gilthead sea bream (300–302). Recombinant FasL protein from Japanese flounder (Paralichthys olivaceus) induced apoptosis in a flounder cell line, indicating that fish possess a similar Fas ligand system (303). FasL has yet to be identified in cartilaginous and lobe-finned fish.
Comparative studies in fish help to reveal the evolutionary history of the immune system. Whereas, innate immunity is present in all multicellular organisms, an adaptive immune system, based on VLRs, B-like, and T-like cells is found in jawless vertebrate, while an adaptive immune system, based on an Ig/TCR/MHC system, evolved with the appearance of jawed vertebrates. Research on the fish immune systems is continuously on the rise, however there is still much to be discovered. For example, there is limited information on TLR ligands, especially in cartilaginous and lobe-finned fish, as well as limited information on complement proteins in lobe-finned fish. In order to gain a better understanding of the lobe-finned fish immune system, the site(s) of hematopoiesis must be determined. There is also limited knowledge, compared to mammals, on the homing and recirculation of B and T cells in ray-finned fish, cartilaginous fish, and lobe-finned fish. Much of this knowledge will only be gained with the development of the appropriate reagents and techniques. The development of cell lines for cartilaginous and lobe-finned fish will aid in determining basic cell biology, one of the first steps in understanding the immune system. Many comparative fish immunology studies are genome-based, and fish genomes are often not well-assembled and/or annotated. The development of high-quality, well-assembled, and annotated genomes in fish species will allow the identification of more immune-relevant transcripts, such as NITRs. In addition, the lack of protein-specific antibodies for fish is hindering many research avenues, such as flow cytometry and cell-specific analyses. A comprehensive understanding of the evolution of the immune system will continue to develop as more comparative research on cartilaginous fish, lobed-finned fish, and ray-fined fish is completed.
NS reviewed the literature, generated the figures, and wrote the manuscript. SC and MR revised and edited the manuscript.
NS was supported by an NSERC PGS D fellowship, MR was supported by NSERC Discovery Grant 341304-2012, and SC was supported by NSERC Discovery Grant 2017-04630 and a Memorial University of Newfoundland Seed grant.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Riera Romo M, Perez-Martinez D, Castillo Ferrer C. Innate immunity in vertebrates: an overview. Immunology. (2016) 148:125–39. doi: 10.1111/imm.12597
2. Alberts B, Johnson A, Lewis J. Chapter 24: The adaptive immune system. In: Molecular Biology of the Cell. 4th ed. New York, NY: Garland Science (2002).
3. Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. (2010) 11:47–59. doi: 10.1038/nrg2703
4. Brazeau MD, Friedman M. The origin and early phylogenetic history of jawed vertebrates. Nature. (2015) 520:490–7. doi: 10.1038/nature14438
5. Buchmann K. Evolution of innate immunity: clues from invertebrates via fish to mammals. Front Immunol. (2014) 5:459. doi: 10.3389/fimmu.2014.00459
6. Flajnik MF. A cold-blooded view of adaptive immunity. Nat Rev Immunol. (2018) 18:438–53. doi: 10.1038/s41577-018-0003-9
7. Inoue JG, Miya M, Lam K, Tay BH, Danks JA, Bell J, et al. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Mol Biol Evol. (2010) 27:2576–86. doi: 10.1093/molbev/msq147
8. Pettinello R, Dooley H. The immunoglobulins of cold-blooded vertebrates. Biomolecules. (2014) 4:1045–69. doi: 10.3390/biom4041045
9. Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, et al. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci USA. (2012) 109:13698–703. doi: 10.1073/pnas.1206625109
10. Panopoulou G, Poustka AJ. Timing and mechanism of ancient vertebrate genome duplications – the adventure of a hypothesis. Trends Genet. (2005) 21:559–67. doi: 10.1016/j.tig.2005.08.004
11. Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. (2009) 26:47–59. doi: 10.1093/molbev/msn222
12. Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, et al. The Atlantic salmon genome provides insights into rediploidization. Nature. (2016) 533:200–5. doi: 10.1038/nature17164
13. Buonocore F, Gerdol M. Alternative adaptive immunity strategies: coelacanth, cod and shark immunity. Mol Immunol. (2016) 69:157–69. doi: 10.1016/j.molimm.2015.09.003
14. Whyte SK. The innate immune response of finfish–a review of current knowledge. Fish Shellfish Immunol. (2007) 23:1127–51. doi: 10.1016/j.fsi.2007.06.005
15. Magnadottir B. Innate immunity of fish (overview). Fish Shellfish Immunol. (2006) 20:137–51. doi: 10.1016/j.fsi.2004.09.006
16. Esteban MÁ. An overview of the immunological defenses in fish skin. ISRN Immunol. (2012) 2012:853470. doi: 10.5402/2012/853470
17. Xu Z, Parra D, Gomez D, Salinas I, Zhang YA, von Gersdorff Jorgensen L, et al. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc Natl Acad Sci USA. (2013) 110:13097–102. doi: 10.1073/pnas.1304319110
18. Liu Z, Liu S, Yao J, Bao L, Zhang J, Li Y, et al. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat Commun. (2016) 7:11757. doi: 10.1038/ncomms11757
19. Meyer W, Seegers U. Basics of skin structure and function in elasmobranchs: a review. J Fish Biol. (2012) 80:1940–67. doi: 10.1111/j.1095-8649.2011.03207.x
20. Heimroth RD, Casadei E, Salinas I. Effects of experimental terrestrialization on the skin mucus proteome of African lungfish (Protopterus dolloi). Front Immunol. (2018) 9:1259. doi: 10.3389/fimmu.2018.01259
21. Fast MD, Sims DE, Burka JF, Mustafa A, Ross NW. Skin morphology and humoral non-specific defence parameters of mucus and plasma in rainbow trout, coho and Atlantic salmon. Comp Biochem Physiol A Mol Integr Physiol. (2002) 132:645–57. doi: 10.1016/S1095-6433(02)00109-5
22. Tsutsui S, Yamaguchi M, Hirasawa A, Nakamura O, Watanabe T. Common skate (Raja kenojei) secretes pentraxin into the cutaneous secretion: the first skin mucus lectin in cartilaginous fish. J Biochem. (2009) 146:295–306. doi: 10.1093/jb/mvp069
23. Wilson JM, Laurent P. Fish gill morphology: inside out. J Exp Zool. (2002) 293:192–213. doi: 10.1002/jez.10124
24. Salinas I. The mucosal immune system of teleost fish. Biology. (2015) 4:525–39. doi: 10.3390/biology4030525
25. Haugarvoll E, Bjerkas I, Nowak BF, Hordvik I, Koppang EO. Identification and characterization of a novel intraepithelial lymphoid tissue in the gills of Atlantic salmon. J Anat. (2008) 213:202–9. doi: 10.1111/j.1469-7580.2008.00943.x
26. Nunez Ortiz N, Gerdol M, Stocchi V, Marozzi C, Randelli E, Bernini C, et al. T cell transcripts and T cell activities in the gills of the teleost fish sea bass (Dicentrarchus labrax). Dev Comp Immunol. (2014) 47:309–18. doi: 10.1016/j.dci.2014.07.015
27. Criscitiello MF, Ohta Y, Saltis M, McKinney EC, Flajnik MF. Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J Immunol. (2010) 184:6950–60. doi: 10.4049/jimmunol.0902774
28. Sepahi A, Cordero H, Goldfine H, Esteban MA, Salinas I. Symbiont-derived sphingolipids modulate mucosal homeostasis and B cells in teleost fish. Sci Rep. (2016) 6:39054. doi: 10.1038/srep39054
29. Tafalla C, Leal E, Yamaguchi T, Fischer U. T cell immunity in the teleost digestive tract. Dev Comp Immunol. (2016) 64:167–77. doi: 10.1016/j.dci.2016.02.019
30. Rombout JW, Blok LJ, Lamers CH, Egberts E. immunization of carp (Cyprinus carpio) with a Vibrio anguillarum bacterin: indications for a common mucosal immune system. Dev Comp Immunol. (1986) 10:341–51. doi: 10.1016/0145-305X(86)90024-8
31. Dos Santos NMS, Taverne-Thiele J, Barnes AC, Ellis AE, Rombout Jan H. Kinetics of juvenile sea bass (Dicentrarchus labrax, L.) systemic and mucosal antibody secreting cell response to different antigens (Photobacterium damselae spp. piscicida, Vibrio anguillarum and DNP). Fish Shellfish Immunol. (2001) 11:317–31. doi: 10.1006/fsim.2000.0320
32. Rombout JH, Joosten PH, Engelsma MY, Vos AP, Taverne N, Taverne-Thiele JJ. Indications for a distinct putative T cell population in mucosal tissue of carp (Cyprinus carpio L.). Dev Comp Immunol. (1998) 22:63–77. doi: 10.1016/S0145-305X(97)00048-7
33. Picchietti S, Terribili FR, Mastrolia L, Scapigliati G, Abelli L. Expression of lymphocyte antigenic determinants in developing gut-associated lymphoid tissue of the sea bass Dicentrarchus labrax (L.). Anat Embryol. (1997) 196:457–63. doi: 10.1007/s004290050113
34. Picchietti S, Guerra L, Bertoni F, Randelli E, Belardinelli MC, Buonocore F, et al. Intestinal T cells of Dicentrarchus labrax (L.): gene expression and functional studies. Fish Shellfish Immunol. (2011) 30:609–17. doi: 10.1016/j.fsi.2010.12.006
35. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. (2012) 336:1268–73. doi: 10.1126/science.1223490
36. Wang AR, Ran C, Ring à E, Zhou ZG. Progress in fish gastrointestinal microbiota research. Rev Aquacult. (2018) 10:626–40. doi: 10.1111/raq.12191
37. Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci USA. (2011) 108(Suppl. 1):4570–7. doi: 10.1073/pnas.1000072107
38. Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol. (2006) 297:374–86. doi: 10.1016/j.ydbio.2006.05.006
39. Merrifield DL, Harper GM, Dimitroglou A, Ringà E, Davies SJ. Possible influence of probiotic adhesion to intestinal mucosa on the activity and morphology of rainbow trout (Oncorhynchus mykiss) enterocytes. Aquacult Res. (2010) 41:1268–72. doi: 10.1111/j.1365-2109.2009.02397.x
40. Pirarat N, Pinpimai K, Endo M, Katagiri T, Ponpornpisit A, Chansue N, et al. Modulation of intestinal morphology and immunity in nile tilapia (Oreochromis niloticus) by Lactobacillus rhamnosus GG. Res Vet Sci. (2011) 91:e92–7. doi: 10.1016/j.rvsc.2011.02.014
41. Tapia-Paniagua S, Reyes-Becerril M, Ascencio-Valle F, Esteban M, Clavijo E, Balebona MC, et al. Modulation of the intestinal microbiota and immune system of farmed Sparus aurata by the administration of the yeast Debaryomyces hansenii L2 in conjunction with inulin. J Aquac Res Dev. (2011) 2011:S1012. doi: 10.4172/2155-9546.S1-012
42. Tomonaga S, Kobayashi K, Hagiwara K, Yamaguchi K, Awaya K. Gut-associated lymphoid tissue in elasmobranchs. Zool Sci. (1986) 3:453.
43. Hart S, Wrathmell AB, Harris JE, Doggett TA. Gut-associated lymphoid tissue (GALT) in the common dogfish Scyliorhinus canicula L.: an ultrastructural study. J Mar Biol Assoc. (1987) 67:639–45. doi: 10.1017/S002531540002734X
44. Hart S, Wrathmell AB, Harris JE. Ontogeny of gut-associated lymphoid tissue (GALT) in the dogfish Scyliorhinus canicula L. Vet Immunol Immunopathol. (1986) 12:107–16. doi: 10.1016/0165-2427(86)90115-7
45. Tomonaga S, Kobayashi K, Kajii T, Awaya K. Two populations of immunoglobulin-forming cells in the skate, Raja kenojei: their distribution and characterization. Dev Comp Immunol. (1984) 8:803–12. doi: 10.1016/0145-305X(84)90063-6
46. Hassanpour M, Joss J, Mohammad MG. Functional analyses of lymphocytes and granulocytes isolated from the thymus, spiral valve intestine, spleen, and kidney of juvenile Australian lungfish, Neoceratodus forsteri. Fish Shellfish Immunol. (2013) 35:107–14. doi: 10.1016/j.fsi.2013.04.006
47. Givens CE, Ransom B, Bano N, Hollibaugh JT. Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar Ecol Prog Ser. (2015) 518:209–23. doi: 10.3354/meps11034
48. Luer CA, Walsh CJ, Bodine AB. Sites of immune cell production in elasmobranch fishes. Lymphomyeloid tissues and organs. In: Smith SL, Sim R, Flajnik M, editors. Immunobiology of the Shark. Boca Raton, FL: CRC Press (2014). p. 79.
49. Icardo JM, Wong WP, Colvee E, Loong AM, Ip YK. The spleen of the African lungfish Protopterus annectens: freshwater and aestivation. Cell Tissue Res. (2012) 350:143–56. doi: 10.1007/s00441-012-1462-0
50. Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol. (2008) 8:533–44. doi: 10.1038/nri2356
51. Wang T, Hanington PC, Belosevic M, Secombes CJ. Two macrophage colony-stimulating factor genes exist in fish that differ in gene organization and are differentially expressed. J Immunol. (2008) 181:3310–22. doi: 10.4049/jimmunol.181.5.3310
52. Hanington PC, Wang T, Secombes CJ, Belosevic M. Growth factors of lower vertebrates: characterization of goldfish (Carassius auratus L.) macrophage colony-stimulating factor-1. J Biol Chem. (2007) 282:31865–72. doi: 10.1074/jbc.M706278200
53. Roca FJ, Sepulcre MA, Lopez-Castejon G, Meseguer J, Mulero V. The colony-stimulating factor-1 receptor is a specific marker of macrophages from the bony fish gilthead seabream. Mol Immunol. (2006) 43:1418–23. doi: 10.1016/j.molimm.2005.07.028
54. Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. (2014) 505:174–9. doi: 10.1038/nature12826
55. Hodgkinson JW, Grayfer L, Belosevic M. Biology of bony fish macrophages. Biology. (2015) 4:881–906. doi: 10.3390/biology4040881
56. Grayfer L, Kerimoglu B, Yaparla A, Hodgkinson JW, Xie J, Belosevic M. Mechanisms of fish macrophage antimicrobial immunity. Front Immunol. (2018) 9:1105. doi: 10.3389/fimmu.2018.01105
57. Neumann NF, Stafford JL, Barreda D, Ainsworth AJ, Belosevic M. Antimicrobial mechanisms of fish phagocytes and their role in host defense. Dev Comp Immunol. (2001) 25:807–25. doi: 10.1016/S0145-305X(01)00037-4
58. McKinney EC, Flajnik MF. IgM-mediated opsonization and cytotoxicity in the shark. J Leukoc Biol. (1997) 61:141–6. doi: 10.1002/jlb.61.2.141
59. Hine PM, Lester R, Wain JM. Observations on the blood of the Australian lungfish, Neoceratodus-Forsteri Klefft. 1. Ultrastructure of granulocytes, monocytes and thrombocytes. Aust J Zool. (1990) 38:131–44. doi: 10.1071/ZO9900131
60. Ribeiro ML, DaMatta RA, Diniz JA, de Souza W, do Nascimento JL, de Carvalho TM. Blood and inflammatory cells of the lungfish Lepidosiren paradoxa. Fish Shellfish Immunol. (2007) 23:178–87. doi: 10.1016/j.fsi.2006.10.005
61. Joerink M, Ribeiro CM, Stet RJ, Hermsen T, Savelkoul HF, Wiegertjes GF. Head kidney-derived macrophages of common carp (Cyprinus carpio L.) show plasticity and functional polarization upon differential stimulation. J Immunol. (2006) 177:61–9. doi: 10.4049/jimmunol.177.1.61
62. Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. (2014) 14:195–208. doi: 10.1038/nri3622
63. Groot Kormelink T, Mol S, de Jong EC, Wauben MHM. The role of extracellular vesicles when innate meets adaptive. Semin Immunopathol. (2018) 40:439–52. doi: 10.1007/s00281-018-0681-1
64. Iliev DB, Jorgensen SM, Rode M, Krasnov A, Harneshaug I, Jorgensen JB. CpG-induced secretion of MHCIIbeta and exosomes from salmon (Salmo salar) APCs. Dev Comp Immunol. (2010) 34:29–41. doi: 10.1016/j.dci.2009.07.009
65. Havixbeck JJ, Barreda DR. Neutrophil development, migration, and function in teleost fish. Biology. (2015) 4:715–34. doi: 10.3390/biology4040715
66. Tavares-Dias M, de Moraes FR. Leukocyte and thrombocyte reference values for channel catfish (Ictalurus punctatus Raf), with an assessment of morphologic, cytochemical, and ultrastructural features. Vet Clin Pathol. (2007) 36:49–54. doi: 10.1111/j.1939-165X.2007.tb00181.x
67. Cannon MS, Mollenhauer HH, Eurell TE, Lewis DH, Cannon AM, Tompkins C. An ultrastructural study of the leukocytes of the channel catfish, Ictalurus punctatus. J Morphol. (1980) 164:1–23. doi: 10.1002/jmor.1051640102
68. Campbell T. Peripheral blood of fish. In: Campbell TW, editor. Exotic Animal Hematology and Cytology. 4th ed. Wiley Blackwell (2015). p. 97–114. doi: 10.1002/9781118993705
69. Grant KR. Fish hematology and associated disorders. Clin Lab Med. (2015) 35:681–701. doi: 10.1016/j.cll.2015.05.015
70. Bielek E, Strauss B. Ultrastructure of the granulocytes of the South American lungfish, Lepidosiren paradoxa: morphogenesis and comparison to other leucocytes. J Morphol. (1993) 218:29–41. doi: 10.1002/jmor.1052180103
71. Ward JW. Hematological studies on the Australian lungfish, Neoceratodus forsteri. Copeia. (1969) 1969:633–5. doi: 10.2307/1441954
72. Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. (2009) 22:240–73. doi: 10.1128/CMR.00046-08
73. Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. (2009) 21:317–37. doi: 10.1093/intimm/dxp017
74. Krishnan J, Selvarajoo K, Tsuchiya M, Lee G, Choi S. Toll-like receptor signal transduction. Exp Mol Med. (2007) 39:421–38. doi: 10.1038/emm.2007.47
75. Zhang J, Kong X, Zhou C, Li L, Nie G, Li X. Toll-like receptor recognition of bacteria in fish: ligand specificity and signal pathways. Fish Shellfish Immunol. (2014) 41:380–8. doi: 10.1016/j.fsi.2014.09.022
76. Palti Y, Gahr SA, Purcell MK, Hadidi S, Rexroad CE III, Wiens GD. Identification, characterization and genetic mapping of TLR7, TLR8a1 and TLR8a2 genes in rainbow trout (Oncorhynchus mykiss). Dev Comp Immunol. (2010) 34:219–33. doi: 10.1016/j.dci.2009.10.002
77. Nie L, Cai S, Shao J, Chen J. Toll-like receptors, associated biological roles, and signaling networks in non-mammals. Front Immunol. (2018) 9:1523. doi: 10.3389/fimmu.2018.01523
78. Boudinot P, Zou J, Ota T, Buonocore F, Scapigliati G, Canapa A, et al. A tetrapod-like repertoire of innate immune receptors and effectors for coelacanths. J Exp Zool B Mol Dev Evol. (2014) 322:415–37. doi: 10.1002/jez.b.22559
79. Palti Y. Toll-like receptors in bony fish: from genomics to function. Dev Comp Immunol. (2011) 35:1263–72. doi: 10.1016/j.dci.2011.03.006
80. Read TD, Petit RA 3rd, Joseph SJ, Alam MT, Weil MR, Ahmad M, et al. Draft sequencing and assembly of the genome of the world's largest fish, the whale shark: Rhincodon typus Smith 1828. BMC Genomics. (2017) 18:532. doi: 10.1186/s12864-017-4138-z
81. Jault C, Pichon L, Chluba J. Toll-like receptor gene family and TIR-domain adapters in Danio rerio. Mol Immunol. (2004) 40:759–71. doi: 10.1016/j.molimm.2003.10.001
82. Oshiumi H, Tsujita T, Shida K, Matsumoto M, Ikeo K, Seya T. Prediction of the prototype of the human Toll-like receptor gene family from the pufferfish, Fugu rubripes, genome. Immunogenetics. (2003) 54:791–800. doi: 10.1007/s00251-002-0519-8
83. Krishnaswamy Gopalan T, Gururaj P, Gupta R, Gopal DR, Rajesh P, Chidambaram B, et al. Transcriptome profiling reveals higher vertebrate orthologous of intra-cytoplasmic pattern recognition receptors in grey bamboo shark. PLoS ONE. (2014) 9:e100018. doi: 10.1371/journal.pone.0100018
84. Kongchum P, Palti Y, Hallerman EM, Hulata G, David L. SNP discovery and development of genetic markers for mapping innate immune response genes in common carp (Cyprinus carpio). Fish Shellfish Immunol. (2010) 29:356–61. doi: 10.1016/j.fsi.2010.04.013
85. Han P, Wang S, Zhang Q, Zhang S, Shao R, Xu W, et al. Molecular characterization and expression analysis of TLR1 and TLR4 from the endangered fish Dabry's sturgeon (Acipenser dabryanus). Dev Comp Immunol. (2018) 86:180–8. doi: 10.1016/j.dci.2018.05.009
86. Zhang XT, Zhang GR, Shi ZC, Yuan YJ, Zheng H, Lin L, et al. Expression analysis of nine Toll-like receptors in yellow catfish (Pelteobagrus fulvidraco) responding to Aeromonas hydrophila challenge. Fish Shellfish Immunol. (2017) 63:384–93. doi: 10.1016/j.fsi.2017.02.021
87. Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrom M, Gregers TF, et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature. (2011) 477:207–10. doi: 10.1038/nature10342
88. Solbakken MH, Torresen OK, Nederbragt AJ, Seppola M, Gregers TF, Jakobsen KS, et al. Evolutionary redesign of the Atlantic cod (Gadus morhua L.) Toll-like receptor repertoire by gene losses and expansions. Sci Rep. (2016) 6:25211. doi: 10.1038/srep25211
89. Wang J, Zhang Z, Liu J, Li F, Chang F, Fu H, et al. Structural characterization and evolutionary analysis of fish-specific TLR27. Fish Shellfish Immunol. (2015) 45:940–5. doi: 10.1016/j.fsi.2015.06.017
90. Tsujita T, Tsukada H, Nakao M, Oshiumi H, Matsumoto M, Seya T. Sensing bacterial flagellin by membrane and soluble orthologs of Toll-like receptor 5 in rainbow trout (Onchorhynchus mikiss). J Biol Chem. (2004) 279:48588–97. doi: 10.1074/jbc.M407634200
91. Tsoi S, Park KC, Kay HH, O'Brien TJ, Podor E, Sun G, et al. Identification of a transcript encoding a soluble form of toll-like receptor 5 (TLR5) in Atlantic salmon during Aeromonas salmonicida infection. Vet Immunol Immunopathol. (2006) 109:183–7. doi: 10.1016/j.vetimm.2005.05.013
92. Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays. (2005) 27:937–45. doi: 10.1002/bies.20293
93. Meijer AH, Gabby Krens SF, Medina Rodriguez IA, He S, Bitter W, Ewa Snaar-Jagalska B, et al. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol Immunol. (2004) 40:773–83. doi: 10.1016/j.molimm.2003.10.003
94. Hansen JD, Vojtech LN, Laing KJ. Sensing disease and danger: a survey of vertebrate PRRs and their origins. Dev Comp Immunol. (2011) 35:886–97. doi: 10.1016/j.dci.2011.01.008
95. Braun-Nesje R, Kaplan G, Seljelid R. Rainbow trout macrophages in vitro: morphology and phagocytic activity. Dev Comp Immunol. (1982) 6:281–91. doi: 10.1016/S0145-305X(82)80011-6
96. Esteban MA, Cuesta A, Chaves-Pozo E, Meseguer J. Phagocytosis in teleosts. Implications of the new cells involved. Biology. (2015) 4:907–22. doi: 10.3390/biology4040907
97. Walsh CJ, Luer CA. Comparative phagocytic and pinocytic activities of leucocytes from peripheral blood and lymphomyeloid tissues of the nurse shark (Ginglymostoma cirratum Bonaterre) and the clearnose skate (Raja eglanteria Bosc). Fish Shellfish Immunol. (1998) 8:197–215. doi: 10.1006/fsim.1997.0137
98. Hyder SL, Cayer ML, Pettey CL. Cell types in peripheral blood of the nurse shark: an approach to structure and function. Tissue Cell. (1983) 15:437–55. doi: 10.1016/0040-8166(83)90075-7
99. Overland HS, Pettersen EF, Ronneseth A, Wergeland HI. Phagocytosis by B-cells and neutrophils in Atlantic salmon (Salmo salar L.) and Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol. (2010) 28:193–204. doi: 10.1016/j.fsi.2009.10.021
100. Li J, Barreda DR, Zhang YA, Boshra H, Gelman AE, Lapatra S, et al. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat Immunol. (2006) 7:1116–24. doi: 10.1038/ni1389
101. Walsh CJ, Toranto JD, Gilliland CT, Noyes DR, Bodine AB, Luer CA. Nitric oxide production by nurse shark (Ginglymostoma cirratum) and clearnose skate (Raja eglanteria) peripheral blood leucocytes. Fish Shellfish Immunol. (2006) 20:40–6. doi: 10.1016/j.fsi.2005.03.011
102. Boshra H, Li J, Sunyer JO. Recent advances on the complement system of teleost fish. Fish Shellfish Immunol. (2006) 20:239–62. doi: 10.1016/j.fsi.2005.04.004
103. Noris M, Remuzzi G. Overview of complement activation and regulation. Semin Nephrol. (2013) 33:479–92. doi: 10.1016/j.semnephrol.2013.08.001
104. Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res. (2010) 20:34–50. doi: 10.1038/cr.2009.139
105. Chondrou MP, Mastellos D, Zarkadis IK. cDNA cloning and phylogenetic analysis of the sixth complement component in rainbow trout. Mol Immunol. (2006) 43:1080–7. doi: 10.1016/j.molimm.2005.07.036
106. Goshima M, Sekiguchi R, Matsushita M, Nonaka M. The complement system of elasmobranches revealed by liver transcriptome analysis of a hammerhead shark, Sphyrna zygaena. Dev Comp Immunol. (2016) 61:13–24. doi: 10.1016/j.dci.2016.03.009
107. Smith SL, Nonaka M. Shark complement: genes, proteins and function. In: Smith SL, Sim RB, Flajnik MF, editors. Immunobiology of the Shark. 1st ed. New York, NY: CRC Press (2015). p. 143.
108. Nonaka M, Nonaka M, Irie M, Tanabe K, Kaidoh T, Natsuume-Sakai S, et al. Identification and characterization of a variant of the third component of complement (C3) in rainbow trout (Salmo gairdneri) serum. J Biol Chem. (1985) 260:809–15.
109. Sunyer JO, Tort L, Lambris JD. Diversity of the third form of complement, C3, in fish: functional characterization of five forms of C3 in the diploid fish Sparus aurata. Biochem J. (1997) 326(Pt 3):877–81. doi: 10.1042/bj3260877
110. Wang Z, Zhang S, Wang G, An Y. Complement activity in the egg cytosol of zebrafish Danio rerio: evidence for the defense role of maternal complement components. PLoS ONE. (2008) 3:e1463. doi: 10.1371/journal.pone.0001463
111. Jenkins JA, Ourth DD. Opsonic effect of the alternative complement pathway on channel catfish peripheral blood phagocytes. Vet Immunol Immunopathol. (1993) 39:447–59. doi: 10.1016/0165-2427(93)90074-E
112. Rebl A, Goldammer T. Under control: the innate immunity of fish from the inhibitors' perspective. Fish Shellfish Immunol. (2018) 77:328–49. doi: 10.1016/j.fsi.2018.04.016
113. Dodds AW, Smith SL, Levine RP, Willis AC. Isolation and initial characterisation of complement components C3 and C4 of the nurse shark and the channel catfish. Dev Comp Immunol. (1998) 22:207–16. doi: 10.1016/S0145-305X(98)00002-0
114. Graham M, Shin DH, Smith SL. Molecular and expression analysis of complement component C5 in the nurse shark (Ginglymostoma cirratum) and its predicted functional role. Fish Shellfish Immunol. (2009) 27:40–9. doi: 10.1016/j.fsi.2009.04.001
115. Aybar L, Shin DH, Smith SL. Molecular characterization of the alpha subunit of complement component C8 (GcC8alpha) in the nurse shark (Ginglymostoma cirratum). Fish Shellfish Immunol. (2009) 27:397–406. doi: 10.1016/j.fsi.2009.05.020
116. Kimura A, Nonaka M. Molecular cloning of the terminal complement components C6 and C8β of cartilaginous fish. Fish Shellfish Immunol. (2009) 27:768–72. doi: 10.1016/j.fsi.2009.08.008
117. Sato A, Sultmann H, Mayer WE, Figueroa F, Tichy H, Klein J. cDNA sequence coding for the α'-chain of the third complement component in the African lungfish. Scand J Immunol. (1999) 49:367–75. doi: 10.1046/j.1365-3083.1999.00512.x
118. Loong AM, Hiong KC, Wong WP, Chew SF, Ip YK. Differential gene expression in the liver of the African lungfish, Protopterus annectens, after 6 days of estivation in air. J Comp Physiol B. (2012) 182:231–45. doi: 10.1007/s00360-011-0613-z
119. Nakao M, Mutsuro J, Obo R, Fujiki K, Nonaka M, Yano T. Molecular cloning and protein analysis of divergent forms of the complement component C3 from a bony fish, the common carp (Cyprinus carpio): presence of variants lacking the catalytic histidine. Eur J Immunol. (2000) 30:858–66. doi: 10.1002/1521-4141(200003)30:3<858::AID-IMMU858>3.0.CO;2-M
120. Saurabh S, Sahoo PK. Lysozyme: an important defence molecule of fish innate immune system. Aquacult Res. (2008) 39:223–39. doi: 10.1111/j.1365-2109.2007.01883.x
121. Grinde B. A lysozyme isolated from rainbow trout acts on mastitis pathogens. FEMS Microbiol Lett. (1989) 51:179–82. doi: 10.1111/j.1574-6968.1989.tb03441.x
122. Jolles P, Jolles J. What's new in lysozyme research? Always a model system, today as yesterday. Mol Cell Biochem. (1984) 63:165–89. doi: 10.1007/BF00285225
123. Hikima J, Minagawa S, Hirono I, Aoki T. Molecular cloning, expression and evolution of the Japanese flounder goose-type lysozyme gene, and the lytic activity of its recombinant protein. Biochim Biophys Acta. (2001) 1520:35–44. doi: 10.1016/S0167-4781(01)00248-2
124. Myrnes B, Seppola M, Johansen A, Overbo K, Callewaert L, Vanderkelen L, et al. Enzyme characterisation and gene expression profiling of Atlantic salmon chicken- and goose-type lysozymes. Dev Comp Immunol. (2013) 40:11–9. doi: 10.1016/j.dci.2013.01.010
125. Yin ZX, He JG, Deng WX, Chan SM. Molecular cloning, expression of orange-spotted grouper goose-type lysozyme cDNA, and lytic activity of its recombinant protein. Dis Aquat Organ. (2003) 55:117–23. doi: 10.3354/dao055117
126. Buonocore F, Randelli E, Trisolino P, Facchiano A, de Pascale D, Scapigliati G. Molecular characterization, gene structure and antibacterial activity of a g-type lysozyme from the European sea bass (Dicentrarchus labrax L.). Mol Immunol. (2014) 62:10–8. doi: 10.1016/j.molimm.2014.05.009
127. Seppola M, Bakkemo KR, Mikkelsen H, Myrnes B, Helland R, Irwin DM, et al. Multiple specialised goose-type lysozymes potentially compensate for an exceptional lack of chicken-type lysozymes in Atlantic cod. Sci Rep. (2016) 6:28318. doi: 10.1038/srep28318
128. Lundblad G, Fänge R, Slettengren K, Lind J. Lysozyme, chitinase and exo-N-acetyl-β-D-glucosaminidase (NAGase) in lymphomyeloid tissue of marine fishes. Mar Biol. (1979) 53:311. doi: 10.1007/BF00391613
129. Hinds Vaughan N, Smith SL. Isolation and characterization of a c-type lysozyme from the nurse shark. Fish Shellfish Immunol. (2013) 35:1824–8. doi: 10.1016/j.fsi.2013.09.001
130. Irwin DM. Evolution of the vertebrate goose-type lysozyme gene family. BMC Evol Biol. (2014) 14:188. doi: 10.1186/s12862-014-0188-x
131. Lazarovici P, Primor N, Loew LM. Purification and pore-forming activity of two hydrophobic polypeptides from the secretion of the Red Sea Moses sole (Pardachirus marmoratus). J Biol Chem. (1986) 261:16704–13.
132. Lemaitre C, Orange N, Saglio P, Saint N, Gagnon J, Molle G. Characterization and ion channel activities of novel antibacterial proteins from the skin mucosa of carp (Cyprinus carpio). Eur J Biochem. (1996) 240:143–9. doi: 10.1111/j.1432-1033.1996.0143h.x
133. Cuesta A, Meseguer J, Esteban MA. Molecular and functional characterization of the gilthead seabream β-defensin demonstrate its chemotactic and antimicrobial activity. Mol Immunol. (2011) 48:1432–8. doi: 10.1016/j.molimm.2011.03.022
134. Browne MJ, Feng CY, Booth V, Rise ML. Characterization and expression studies of Gaduscidin-1 and Gaduscidin-2; paralogous antimicrobial peptide-like transcripts from Atlantic cod (Gadus morhua). Dev Comp Immunol. (2011) 35:399–408. doi: 10.1016/j.dci.2010.11.010
135. Dong HT, Nguyen VV, Le HD, Sangsuriya P, Jitrakorn S, Saksmerprome V, et al. Naturally concurrent infections of bacterial and viral pathogens in disease outbreaks in cultured Nile tilapia (Oreochromis niloticus) farms. Aquaculture. (2015) 448:427–35. doi: 10.1016/j.aquaculture.2015.06.027
136. Chang CI, Zhang YA, Zou J, Nie P, Secombes CJ. Two cathelicidin genes are present in both rainbow trout (Oncorhynchus mykiss) and Atlantic salmon (Salmo salar). Antimicrob Agents Chemother. (2006) 50:185–95. doi: 10.1128/AAC.50.1.185-195.2006
137. Bridle A, Nosworthy E, Polinski M, Nowak B. Evidence of an antimicrobial-immunomodulatory role of Atlantic salmon cathelicidins during infection with Yersinia ruckeri. PLoS ONE. (2011) 6:e23417. doi: 10.1371/journal.pone.0023417
138. Davies DH, Lawson R, Burch SJ, Hanson JE. Evolutionary relationships of a “primitive” shark (Heterodontus) assessed by micro-complement fixation of serum transferrin. J Mol Evol. (1987) 25:74–80. doi: 10.1007/BF02100043
139. Moore KS, Wehrli S, Roder H, Rogers M, Forrest JN Jr, McCrimmon D, et al. Squalamine: an aminosterol antibiotic from the shark. Proc Natl Acad Sci USA. (1993) 90:1354–8. doi: 10.1073/pnas.90.4.1354
140. Cho SH, Lee BD, An H, Eun JB, Kenojeinin I. Antimicrobial peptide isolated from the skin of the fermented skate, Raja kenojei. Peptides. (2005) 26:581–7. doi: 10.1016/j.peptides.2004.11.011
141. Roy S, Kumar V, Kumar V, Behera BK. Acute phase proteins and their potential role as an indicator for fish health and in diagnosis of fish diseases. Protein Pept Lett. (2017) 24:78–89. doi: 10.2174/0929866524666161121142221
142. Charlie-Silva I, Klein A, Gomes JMM, Prado EJR, Moraes AC, Eto SF, et al. Acute-phase proteins during inflammatory reaction by bacterial infection: fish-model. Sci Rep. (2019) 9:4776. doi: 10.1038/s41598-019-41312-z
143. Bayne CJ, Gerwick L. The acute phase response and innate immunity of fish. Dev Comp Immunol. (2001) 25:725–43. doi: 10.1016/S0145-305X(01)00033-7
144. Garlanda C, Bottazzi B, Bastone A, Mantovani A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu Rev Immunol. (2005) 23:337–66. doi: 10.1146/annurev.immunol.23.021704.115756
145. Nauta AJ, Bottazzi B, Mantovani A, Salvatori G, Kishore U, Schwaeble WJ, et al. Biochemical and functional characterization of the interaction between pentraxin 3 and C1q. Eur J Immunol. (2003) 33:465–73. doi: 10.1002/immu.200310022
146. Hoover GJ, el-Mowafi A, Simko E, Kocal TE, Ferguson HW, Hayes MA. Plasma proteins of rainbow trout (Oncorhynchus mykiss) isolated by binding to lipopolysaccharide from Aeromonas salmonicida. Comp Biochem Physiol B Biochem Mol Biol. (1998) 120:559–69. doi: 10.1016/S0305-0491(98)10042-1
147. Lund V, Olafsen JA. A comparative study of pentraxin-like proteins in different fish species. Dev Comp Immunol. (1998) 22:185–94. doi: 10.1016/S0145-305X(97)00051-7
148. Kovacevic N, Hagen MO, Xie J, Belosevic M. The analysis of the acute phase response during the course of Trypanosoma carassii infection in the goldfish (Carassius auratus L.). Dev Comp Immunol. (2015) 53:112–22. doi: 10.1016/j.dci.2015.06.009
149. Liu Y, Iwasaki T, Watarai S, Kodama H. Effect of turpentine oil on C-reactive protein (CRP) production in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. (2004) 17:203–10. doi: 10.1016/j.fsi.2004.03.003
150. Kodama H, Matsuoka Y, Tanaka Y, Liu Y, Iwasaki T, Watarai S. Changes of C-reactive protein levels in rainbow trout (Oncorhynchus mykiss) sera after exposure to anti-ectoparasitic chemicals used in aquaculture. Fish Shellfish Immunol. (2004) 16:589–97. doi: 10.1016/j.fsi.2003.09.007
151. Lee PT, Bird S, Zou J, Martin SAM. Phylogeny and expression analysis of C-reactive protein (CRP) and serum amyloid-P (SAP) like genes reveal two distinct groups in fish. Fish Shellfish Immunol. (2017) 65:42–51. doi: 10.1016/j.fsi.2017.03.037
152. MacCarthy EM, Burns I, Irnazarow I, Polwart A, Greenhough TJ, Shrive AK, et al. Serum CRP-like protein profile in common carp Cyprinus carpio challenged with Aeromonas hydrophila and Escherichia coli lipopolysaccharide. Dev Comp Immunol. (2008) 32:1281–9. doi: 10.1016/j.dci.2008.04.004
153. Robey FA, Tanaka T, Liu TY. Isolation and characterization of two major serum proteins from the dogfish, Mustelus canis, C-reactive protein and amyloid P component. J Biol Chem. (1983) 258:3889–94.
154. Krol L, Allender M, Cray C, George R. Plasma proteins and selected acute-phase proteins in the whitespotted bamboo shark (Chiloscyllium plagiosum). J Zoo Wildl Med. (2014) 45:782–6. doi: 10.1638/2013-0303.1
155. Karsten AH, Rice CD. c-Reactive protein levels as a biomarker of inflammation and stress in the Atlantic sharpnose shark (Rhizoprionodon terraenovae) from three southeastern USA estuaries. Mar Environ Res. (2004) 58:747–51. doi: 10.1016/j.marenvres.2004.03.089
156. Huong Giang DT, Van Driessche E, Vandenberghe I, Devreese B, Beeckmans S. Isolation and characterization of SAP and CRP, two pentraxins from Pangasianodon (Pangasius) hypophthalmus. Fish Shellfish Immunol. (2010) 28:743–53. doi: 10.1016/j.fsi.2010.01.007
157. Pallavicini A, Canapa A, Barucca M, Alfoldi J, Biscotti MA, Buonocore F, et al. Analysis of the transcriptome of the Indonesian coelacanth Latimeria menadoensis. BMC Genomics. (2013) 14:538. doi: 10.1186/1471-2164-14-538
158. Mashoof S, Criscitiello MF. Fish immunoglobulins. Biology. (2016) 5:45. doi: 10.3390/biology5040045
159. Hikima J, Jung TS, Aoki T. Immunoglobulin genes and their transcriptional control in teleosts. Dev Comp Immunol. (2011) 35:924–36. doi: 10.1016/j.dci.2010.10.011
160. Zhang T, Tacchi L, Wei Z, Zhao Y, Salinas I. Intraclass diversification of immunoglobulin heavy chain genes in the African lungfish. Immunogenetics. (2014) 66:335–51. doi: 10.1007/s00251-014-0769-2
161. Hohman VS, Stewart SE, Rumfelt LL, Greenberg AS, Avila DW, Flajnik MF, et al. J chain in the nurse shark: implications for function in a lower vertebrate. J Immunol. (2003) 170:6016–23. doi: 10.4049/jimmunol.170.12.6016
162. Criscitiello MF, Saltis M, Flajnik MF. An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc Natl Acad Sci USA. (2006) 103:5036–41. doi: 10.1073/pnas.0507074103
163. Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. (2004) 4:123–32. doi: 10.1038/nri1292
164. Alt FW, Rathbun G, Oltz E, Taccioli G, Shinkai Y. Function and control of recombination-activating gene activity. Ann N Y Acad Sci. (1992) 651:277–94. doi: 10.1111/j.1749-6632.1992.tb24626.x
165. Hsu E. V(D)J recombination: of mice and sharks. Adv Exp Med Biol. (2009) 650:166–79. doi: 10.1007/978-1-4419-0296-2_14
166. Picchietti S, Guerra L, Buonocore F, Randelli E, Fausto AM, Abelli L. Lymphocyte differentiation in sea bass thymus: CD4 and CD8-alpha gene expression studies. Fish Shellfish Immunol. (2009) 27:50–6. doi: 10.1016/j.fsi.2009.04.003
167. Danilova N, Bussmann J, Jekosch K, Steiner LA. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol. (2005) 6:295–302. doi: 10.1038/ni1166
168. Yasuike M, de Boer J, von Schalburg KR, Cooper GA, McKinnel L, Messmer A, et al. Evolution of duplicated IgH loci in Atlantic salmon, Salmo salar. BMC Genomics. (2010) 11:486. doi: 10.1186/1471-2164-11-486
169. Rumfelt LL, Avila D, Diaz M, Bartl S, McKinney EC, Flajnik MF. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc Natl Acad Sci USA. (2001) 98:1775–80. doi: 10.1073/pnas.98.4.1775
170. Lee V, Huang JL, Lui MF, Malecek K, Ohta Y, Mooers A, et al. The evolution of multiple isotypic IgM heavy chain genes in the shark. J Immunol. (2008) 180:7461–70. doi: 10.4049/jimmunol.180.11.7461
171. Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol. (2002) 2:688–98. doi: 10.1038/nri889
172. Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, Maccallum I, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. (2013) 496:311–6. doi: 10.1038/nature12027
173. Saha NR, Ota T, Litman GW, Hansen J, Parra Z, Hsu E, et al. Genome complexity in the coelacanth is reflected in its adaptive immune system. J Exp Zool B Mol Dev Evol. (2014) 322:438–63. doi: 10.1002/jez.b.22558
174. Boshra H, Gelman AE, Sunyer JO. Structural and functional characterization of complement C4 and C1s-like molecules in teleost fish: insights into the evolution of classical and alternative pathways. J Immunol. (2004) 173:349–59. doi: 10.4049/jimmunol.173.1.349
175. Haynes L, Fuller L, McKinney EC. Fc receptor for shark IgM. Dev Comp Immunol. (1988) 12:561–71. doi: 10.1016/0145-305X(88)90072-9
176. Clem LW. Phylogeny of immunoglobulin structure and function. IV. Immunoglobulins of the giant grouper, Epinephelus itaira. J Biol Chem. (1971) 246:9–15.
177. Elcombe BM, Chang RJ, Taves CJ, Winkelhake JL. Evolution of antibody structure and effector functions: comparative hemolytic activities of monomeric and tetrameric IgM from rainbow trout, Salmo gairdnerii. Comp Biochem Physiol B. (1985) 80:697–706. doi: 10.1016/0305-0491(85)90448-1
178. Ross DA, Wilson MR, Miller NW, Clem LW, Warr GW. Evolutionary variation of immunoglobulin mu heavy chain RNA processing pathways: origins, effects, and implications. Immunol Rev. (1998) 166:143–51. doi: 10.1111/j.1600-065X.1998.tb01259.x
179. Sahoo M, Edholm ES, Stafford JL, Bengten E, Miller NW, Wilson M. B cell receptor accessory molecules in the channel catfish, Ictalurus punctatus. Dev Comp Immunol. (2008) 32:1385–97. doi: 10.1016/j.dci.2008.05.008
180. Castro CD, Flajnik MF. Putting J chain back on the map: how might its expression define plasma cell development? J Immunol. (2014) 193:3248–55. doi: 10.4049/jimmunol.1400531
181. Hordvik I, Berven FS, Solem ST, Hatten F, Endresen C. Analysis of two IgM isotypes in Atlantic salmon and brown trout. Mol Immunol. (2002) 39:313–21. doi: 10.1016/S0161-5890(02)00114-1
182. Hordvik I, Voie AM, Glette J, Male R, Endresen C. Cloning and sequence analysis of two isotypic IgM heavy chain genes from Atlantic salmon, Salmo salar L. Eur J Immunol. (1992) 22:2957–62. doi: 10.1002/eji.1830221130
183. Dooley H, Flajnik MF. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur J Immunol. (2005) 35:936–45. doi: 10.1002/eji.200425760
184. Dooley H, Flajnik MF. Antibody repertoire development in cartilaginous fish. Dev Comp Immunol. (2006) 30:43–56. doi: 10.1016/j.dci.2005.06.022
185. Ota T, Rast JP, Litman GW, Amemiya CT. Lineage-restricted retention of a primitive immunoglobulin heavy chain isotype within the Dipnoi reveals an evolutionary paradox. Proc Natl Acad Sci USA. (2003) 100:2501–6. doi: 10.1073/pnas.0538029100
186. Tacchi L, Larragoite E, Salinas I. Discovery of J chain in African lungfish (Protopterus dolloi, Sarcopterygii) using high throughput transcriptome sequencing: implications in mucosal immunity. PLoS ONE. (2013) 8:e70650. doi: 10.1371/journal.pone.0070650
187. Anderson MK, Strong SJ, Litman RT, Luer CA, Amemiya CT, Rast JP, et al. A long form of the skate IgX gene exhibits a striking resemblance to the new shark IgW and IgNARC genes. Immunogenetics. (1999) 49:56–67. doi: 10.1007/s002510050463
188. Kobayashi K, Tomonaga S, Kajii T. A second class of immunoglobulin other than IgM present in the serum of a cartilaginous fish, the skate, Raja kenojei: isolation and characterization. Mol Immunol. (1984) 21:397–404. doi: 10.1016/0161-5890(84)90037-3
189. Zhu L, Yan Z, Feng M, Peng D, Guo Y, Hu X, et al. Identification of sturgeon IgD bridges the evolutionary gap between elasmobranchs and teleosts. Dev Comp Immunol. (2014) 42:138–47. doi: 10.1016/j.dci.2013.08.020
190. Ohta Y, Flajnik M. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc Natl Acad Sci USA. (2006) 103:10723–8. doi: 10.1073/pnas.0601407103
191. Edholm ES, Bengten E, Wilson M. Insights into the function of IgD. Dev Comp Immunol. (2011) 35:1309–16. doi: 10.1016/j.dci.2011.03.002
192. Ramirez-Gomez F, Greene W, Rego K, Hansen JD, Costa G, Kataria P, et al. Discovery and characterization of secretory IgD in rainbow trout: secretory IgD is produced through a novel splicing mechanism. J Immunol. (2012) 188:1341–9. doi: 10.4049/jimmunol.1101938
193. Parra D, Korytar T, Takizawa F, Sunyer JO. B cells and their role in the teleost gut. Dev Comp Immunol. (2016) 64:150–66. doi: 10.1016/j.dci.2016.03.013
194. Bengten E, Quiniou SM, Stuge TB, Katagiri T, Miller NW, Clem LW, et al. The IgH locus of the channel catfish, Ictalurus punctatus, contains multiple constant region gene sequences: different genes encode heavy chains of membrane and secreted IgD. J Immunol. (2002) 169:2488–97. doi: 10.4049/jimmunol.169.5.2488
195. Stenvik J, Jorgensen TO. Immunoglobulin D (IgD) of Atlantic cod has a unique structure. Immunogenetics. (2000) 51:452–61. doi: 10.1007/s002510050644
196. Srisapoome P, Ohira T, Hirono I, Aoki T. Genes of the constant regions of functional immunoglobulin heavy chain of Japanese flounder, Paralichthys olivaceus. Immunogenetics. (2004) 56:292–300. doi: 10.1007/s00251-004-0689-7
197. Xiao FS, Wang YP, Yan W, Chang MX, Yao WJ, Xu QQ, et al. Ig heavy chain genes and their locus in grass carp Ctenopharyngodon idella. Fish Shellfish Immunol. (2010) 29:594–9. doi: 10.1016/j.fsi.2010.06.004
198. Fillatreau S, Six A, Magadan S, Castro R, Sunyer JO, Boudinot P. The astonishing diversity of Ig classes and B cell repertoires in teleost fish. Front Immunol. (2013) 4:28. doi: 10.3389/fimmu.2013.00028
199. Hansen JD, Landis ED, Phillips RB. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: implications for a distinctive B cell developmental pathway in teleost fish. Proc Natl Acad Sci USA. (2005) 102:6919–24. doi: 10.1073/pnas.0500027102
200. Smith LE, Crouch K, Cao W, Muller MR, Wu L, Steven J, et al. Characterization of the immunoglobulin repertoire of the spiny dogfish (Squalus acanthias). Dev Comp Immunol. (2012) 36:665–79. doi: 10.1016/j.dci.2011.10.007
201. Rumfelt LL, Diaz M, Lohr RL, Mochon E, Flajnik MF. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J Immunol. (2004) 173:1129–39. doi: 10.4049/jimmunol.173.2.1129
202. Hsu E. Assembly and expression of shark Ig genes. J Immunol. (2016) 196:3517–23. doi: 10.4049/jimmunol.1600164
203. Greenberg AS, Hughes AL, Guo J, Avila D, McKinney EC, Flajnik MF. A novel “chimeric” antibody class in cartilaginous fish: IgM may not be the primordial immunoglobulin. Eur J Immunol. (1996) 26:1123–9. doi: 10.1002/eji.1830260525
204. Salinas I, Zhang YA, Sunyer JO. Mucosal immunoglobulins and B cells of teleost fish. Dev Comp Immunol. (2011) 35:1346–65. doi: 10.1016/j.dci.2011.11.009
205. Savan R, Aman A, Nakao M, Watanuki H, Sakai M. Discovery of a novel immunoglobulin heavy chain gene chimera from common carp (Cyprinus carpio L.). Immunogenetics. (2005) 57:458–63. doi: 10.1007/s00251-005-0015-z
206. Gambon-Deza F, Sanchez-Espinel C, Magadan-Mompo S. Presence of an unique IgT on the IGH locus in three-spined stickleback fish (Gasterosteus aculeatus) and the very recent generation of a repertoire of VH genes. Dev Comp Immunol. (2010) 34:114–22. doi: 10.1016/j.dci.2009.08.011
207. Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. (2010) 11:827–35. doi: 10.1038/ni.1913
208. Zwollo P, Cole S, Bromage E, Kaattari S. B cell heterogeneity in the teleost kidney: evidence for a maturation gradient from anterior to posterior kidney. J Immunol. (2005) 174:6608–16. doi: 10.4049/jimmunol.174.11.6608
209. Zwollo P. Dissecting teleost B cell differentiation using transcription factors. Dev Comp Immunol. (2011) 35:898–905. doi: 10.1016/j.dci.2011.01.009
210. Bromage ES, Kaattari IM, Zwollo P, Kaattari SL. Plasmablast and plasma cell production and distribution in trout immune tissues. J Immunol. (2004) 173:7317–23. doi: 10.4049/jimmunol.173.12.7317
211. Saunders HL, Oko AL, Scott AN, Fan CW, Magor BG. The cellular context of AID expressing cells in fish lymphoid tissues. Dev Comp Immunol. (2010) 34:669–76. doi: 10.1016/j.dci.2010.01.013
212. Fänge R. Lymphomyeloid tissues and blood cell morphology in elasmobranchs. Arch Biol. (1987) 98:187.
213. Rumfelt LL, McKinney EC, Taylor E, Flajnik MF. The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand J Immunol. (2002) 56:130–48. doi: 10.1046/j.1365-3083.2002.01116.x
214. Anderson MK, Pant R, Miracle AL, Sun X, Luer CA, Walsh CJ, et al. Evolutionary origins of lymphocytes: ensembles of T cell and B cell transcriptional regulators in a cartilaginous fish. J Immunol. (2004) 172:5851–60. doi: 10.4049/jimmunol.172.10.5851
215. Zapata AG, Torroba M, SacedÃ3n R, Varas A, Vicente A. Structure of the lymphoid organs of elasmobranchs. J Exp Zool. (1996) 275:125–43. doi: 10.1002/(SICI)1097-010X(19960601/15)275:2/3<125::AID-JEZ6>3.0.CO;2-F
216. Arkoosh MR, Kaattari SL. Development of immunological memory in rainbow trout (Oncorhynchus mykiss). I. An immunochemical and cellular analysis of the B cell response. Dev Comp Immunol. (1991) 15:279–93. doi: 10.1016/0145-305X(91)90021-P
217. Sanmartin ML, Parama A, Castro R, Cabaleiro S, Leiro J, Lamas J, et al. Vaccination of turbot, Psetta maxima (L.), against the protozoan parasite Philasterides dicentrarchi: effects on antibody production and protection. J Fish Dis. (2008) 31:135–40. doi: 10.1111/j.1365-2761.2007.00876.x
218. Raida MK, Nylen J, Holten-Andersen L, Buchmann K. Association between plasma antibody response and protection in rainbow trout Oncorhynchus mykiss immersion vaccinated against Yersinia ruckeri. PLoS ONE. (2011) 6:e18832. doi: 10.1371/journal.pone.0018832
219. Pilstrom L, Warr GW, Stromberg S. Why is the antibody response of Atlantic cod so poor? The search for a genetic explanation. Fish Sci. (2005) 71:961–71. doi: 10.1111/j.1444-2906.2005.01052.x
220. Star B, Jentoft S. Why does the immune system of Atlantic cod lack MHC II? Bioessays. (2012) 34:648–51. doi: 10.1002/bies.201200005
221. Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol. (2004) 16:257–75. doi: 10.1016/j.smim.2004.08.005
222. Magor BG. Antibody affinity maturation in fishes-our current understanding. Biology. (2015) 4:512–24. doi: 10.3390/biology4030512
223. Saunders HL, Magor BG. Cloning and expression of the AID gene in the channel catfish. Dev Comp Immunol. (2004) 28:657–63. doi: 10.1016/j.dci.2004.01.002
224. Zhao Y, Pan-Hammarstrom Q, Zhao Z, Hammarstrom L. Identification of the activation-induced cytidine deaminase gene from zebrafish: an evolutionary analysis. Dev Comp Immunol. (2005) 29:61–71. doi: 10.1016/j.dci.2004.05.005
225. Wakae K, Magor BG, Saunders H, Nagaoka H, Kawamura A, Kinoshita K, et al. Evolution of class switch recombination function in fish activation-induced cytidine deaminase, AID. Int Immunol. (2006) 18:41–7. doi: 10.1093/intimm/dxh347
226. Abdouni H, King JJ, Suliman M, Quinlan M, Fifield H, Larijani M. Zebrafish AID is capable of deaminating methylated deoxycytidines. Nucleic Acids Res. (2013) 41:5457–68. doi: 10.1093/nar/gkt212
227. Quinlan EM, King JJ, Amemiya CT, Hsu E, Larijani M. Biochemical regulatory features of activation-induced cytidine deaminase remain conserved from lampreys to humans. Mol Cell Biol. (2017) 37:e00077-17. doi: 10.1128/MCB.00077-17
228. Tacchi L, Larragoite ET, Munoz P, Amemiya CT, Salinas I. African lungfish reveal the evolutionary origins of organized mucosal lymphoid tissue in vertebrates. Curr Biol. (2015) 25:2417–24. doi: 10.1016/j.cub.2015.07.066
229. Ye J, Bromage ES, Kaattari SL. The strength of B cell interaction with antigen determines the degree of IgM polymerization. J Immunol. (2010) 184:844–50. doi: 10.4049/jimmunol.0902364
230. Sigel MM, Voss EW Jr, Rudikoff S. Binding properties of shark immunoglobulins. Comp Biochem Physiol A Comp Physiol. (1972) 42:249–59. doi: 10.1016/0300-9629(72)90384-2
231. Dooley H, Stanfield RL, Brady RA, Flajnik MF. First molecular and biochemical analysis of an in vivo affinity maturation in an ectothermic vertebrate. Proc Natl Acad Sci USA. (2006) 103:1846–51. doi: 10.1073/pnas.0508341103
232. Barreto VM, Pan-Hammarstrom Q, Zhao Y, Hammarstrom L, Misulovin Z, Nussenzweig MC. AID from bony fish catalyzes class switch recombination. J Exp Med. (2005) 202:733–8. doi: 10.1084/jem.20051378
233. Zhu C, Lee V, Finn A, Senger K, Zarrin AA, Du Pasquier L, et al. Origin of immunoglobulin isotype switching. Curr Biol. (2012) 22:872–80. doi: 10.1016/j.cub.2012.03.060
234. Shen T, Lei M, Wang J, He X, Li X, Li J. Molecular cloning, organization, expression and 3D structural analysis of the MHC class Ia gene in the whitespotted bamboo shark (Chiloscyllium plagiosum). Vet Immunol Immunopathol. (2014) 157:111–8. doi: 10.1016/j.vetimm.2013.10.012
235. Rock KL, Reits E, Neefjes J. Present yourself! By MHC class I and MHC class II molecules. Trends Immunol. (2016) 37:724–37. doi: 10.1016/j.it.2016.08.010
236. Dirscherl H, McConnell SC, Yoder JA, de Jong JL. The MHC class I genes of zebrafish. Dev Comp Immunol. (2014) 46:11–23. doi: 10.1016/j.dci.2014.02.018
237. Kuroda N, Figueroa F, O'hUigin C, Klein J. Evidence that the separation of MHC class II from class I loci in the zebrafish, Danio rerio, occurred by translocation. Immunogenetics. (2002) 54:418–30. doi: 10.1007/s00251-002-0473-5
238. Flajnik MF, Kasahara M. Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity. (2001) 15:351–62. doi: 10.1016/S1074-7613(01)00198-4
239. Stet RJ, Kruiswijk CP, Dixon B. Major histocompatibility lineages and immune gene function in teleost fishes: the road not taken. Crit Rev Immunol. (2003) 23:441–71. doi: 10.1615/CritRevImmunol.v23.i56.50
240. Solbakken MH, Rise ML, Jakobsen KS, Jentoft S. Successive losses of central immune genes characterize the gadiformes' alternate immunity. Genome Biol Evol. (2016) 8:3508–15. doi: 10.1093/gbe/evw250
241. Wang C, Perera TV, Ford HL, Dascher CC. Characterization of a divergent non-classical MHC class I gene in sharks. Immunogenetics. (2003) 55:57–61. doi: 10.1007/s00251-003-0542-4
242. Grimholt U, Drablos F, Jorgensen SM, Hoyheim B, Stet RJ. The major histocompatibility class I locus in Atlantic salmon (Salmo salar L.): polymorphism, linkage analysis and protein modelling. Immunogenetics. (2002) 54:570–81. doi: 10.1007/s00251-002-0499-8
243. Magor KE, Shum BP, Parham P. The β2-microglobulin locus of rainbow trout (Oncorhynchus mykiss) contains three polymorphic genes. J Immunol. (2004) 172:3635–43. doi: 10.4049/jimmunol.172.6.3635
244. Chen H, Kshirsagar S, Jensen I, Lau K, Simonson C, Schluter SF. Characterization of arrangement and expression of the β-2 microglobulin locus in the sandbar and nurse shark. Dev Comp Immunol. (2010) 34:189–95. doi: 10.1016/j.dci.2009.09.008
245. Lu S, Yao S, Chen R, Zhang N, Chen J, Gao F, et al. Expression, purification, crystallization and preliminary X-ray diffraction analysis of nurse shark β2-microglobulin. Acta Crystallogr Sect F Struct Biol Cryst Commun. (2012) 68(Pt 4):460–3. doi: 10.1107/S1744309112006811
246. Sato A, Sultmann H, Mayer WE, Klein J. Mhc class I gene of African lungfish. Immunogenetics. (2000) 51:491–5. doi: 10.1007/s002510050649
247. Betz UA, Mayer WE, Klein J. Major histocompatibility complex class I genes of the coelacanth Latimeria chalumnae. Proc Natl Acad Sci USA. (1994) 91:11065–9. doi: 10.1073/pnas.91.23.11065
248. Tacchi L, Misra M, Salinas I. Anti-viral immune responses in a primitive lung: characterization and expression analysis of interferon-inducible immunoproteasome subunits LMP2, LMP7 and MECL-1 in a sarcopterygian fish, the Nigerian spotted lungfish (Protopterus dolloi). Dev Comp Immunol. (2013) 41:657–65. doi: 10.1016/j.dci.2013.07.023
249. Bartl S, Weissman IL. Isolation and characterization of major histocompatibility complex class IIB genes from the nurse shark. Proc Natl Acad Sci USA. (1994) 91:262–6. doi: 10.1073/pnas.91.1.262
250. Dijkstra JM, Grimholt U, Leong J, Koop BF, Hashimoto K. Comprehensive analysis of MHC class II genes in teleost fish genomes reveals dispensability of the peptide-loading DM system in a large part of vertebrates. BMC Evol Biol. (2013) 13:260. doi: 10.1186/1471-2148-13-260
251. Kjøglum S, Larsen S, Bakke HG, Grimholt U. The effect of specific MHC class I and class II combinations on resistance to furunculosis in Atlantic Salmon (Salmo salar). Scand J Immunol. (2008) 67:160–8. doi: 10.1111/j.1365-3083.2007.02052.x
252. Ma Q, Su YQ, Wang J, Zhuang ZM, Tang QS. Molecular cloning and expression analysis of major histocompatibility complex class IIB gene of the Whitespotted bambooshark (Chiloscyllium plagiosum). Fish Physiol Biochem. (2013) 39:131–42. doi: 10.1007/s10695-012-9685-2
253. Roux KH, Greenberg AS, Greene L, Strelets L, Avila D, McKinney EC, et al. Structural analysis of the nurse shark (new) antigen receptor (NAR): molecular convergence of NAR and unusual mammalian immunoglobulins. Proc Natl Acad Sci USA. (1998) 95:11804–9. doi: 10.1073/pnas.95.20.11804
254. Criscitiello MF. Shark T-cell receptors. In: Smith S, Sim R, Flajnik M, editors, Immunobiology of the Shark. 1st ed. Boca Raton, FL: CRC Press (2014). p. 237.
255. Castro R, Bernard D, Lefranc MP, Six A, Benmansour A, Boudinot P. T cell diversity and TcR repertoires in teleost fish. Fish Shellfish Immunol. (2011) 31:644–54. doi: 10.1016/j.fsi.2010.08.016
256. Wan F, Hu C, Ma J, Gao K, Xiang L, Shao J. Characterization of γδ T cells from zebrafish provides insights into their important role in adaptive humoral immunity. Front Immunol. (2016) 7:675. doi: 10.3389/fimmu.2016.00675
257. Magadan S, Sunyer OJ, Boudinot P. Unique features of fish immune repertoires: particularities of adaptive immunity within the largest group of vertebrates. Results Probl Cell Differ. (2015) 57:235–64. doi: 10.1007/978-3-319-20819-0_10
258. Yazawa R, Cooper GA, Hunt P, Beetz-Sargent M, Robb A, Conrad M, et al. Striking antigen recognition diversity in the Atlantic salmon T-cell receptor alpha/delta locus. Dev Comp Immunol. (2008) 32:204–12. doi: 10.1016/j.dci.2007.05.002
259. Chen H, Kshirsagar S, Jensen I, Lau K, Covarrubias R, Schluter SF, et al. Characterization of arrangement and expression of the T cell receptor γ locus in the sandbar shark. Proc Natl Acad Sci USA. (2009) 106:8591–6. doi: 10.1073/pnas.0811283106
260. Alberts B, Johnson A, Lewis J, et al. Helper T cells and lymphocyte activation. In: Molecular Biology of the Cell. 4th Edn. New York, NY: Garland Science (2002). Available Online at: https://www.ncbi.nlm.nih.gov/books/NBK26827/
261. Gratton S, Haughn L, Sekaly RP, Julius M. The extracellular domain of CD4 regulates the initiation of T cell activation. Mol Immunol. (2000) 37:213–9. doi: 10.1016/S0161-5890(00)00046-8
262. Moore LJ, Dijkstra JM, Koppang EO, Hordvik I. CD4 homologues in Atlantic salmon. Fish Shellfish Immunol. (2009) 26:10–8. doi: 10.1016/j.fsi.2008.09.019
263. Redmond AK, Macqueen DJ, Dooley H. Phylotranscriptomics suggests the jawed vertebrate ancestor could generate diverse helper and regulatory T cell subsets. BMC Evol Biol. (2018) 18:169. doi: 10.1186/s12862-018-1290-2
264. Cole DK, Gao GF. CD8: adhesion molecule, co-receptor and immuno-modulator. Cell Mol Immunol. (2004) 1:81–8. Available online at: http://www.cmi.ustc.edu.cn/1/2/81.pdf
265. Forlenza M, de Carvalho Dias JD, Vesely T, Pokorova D, Savelkoul HF, Wiegertjes GF. Transcription of signal-3 cytokines, IL-12 and IFNαβ, coincides with the timing of CD8αβ up-regulation during viral infection of common carp (Cyprinus carpio L). Mol Immunol. (2008) 45:1531–47. doi: 10.1016/j.molimm.2007.10.010
266. Somamoto T, Yoshiura Y, Sato A, Nakao M, Nakanishi T, Okamoto N, et al. Expression profiles of TCRbeta and CD8alpha mRNA correlate with virus-specific cell-mediated cytotoxic activity in ginbuna crucian carp. Virology. (2006) 348:370–7. doi: 10.1016/j.virol.2006.01.019
267. Quiniou SM, Sahoo M, Edholm ES, Bengten E, Wilson M. Channel catfish CD8α and CD8β co-receptors: characterization, expression and polymorphism. Fish Shellfish Immunol. (2011) 30:894–901. doi: 10.1016/j.fsi.2011.01.011
268. Overgard AC, Hordvik I, Nerland AH, Eikeland G, Patel S. Cloning and expression analysis of Atlantic halibut (Hippoglossus hippoglossus) CD3 genes. Fish Shellfish Immunol. (2009) 27:707–13. doi: 10.1016/j.fsi.2009.08.011
269. Shang N, Sun XF, Hu W, Wang YP, Guo QL. Molecular cloning and characterization of common carp (Cyprinus carpio L.) TCRγ and CD3γ/δ chains. Fish Shellfish Immunol. (2008) 24:412–25. doi: 10.1016/j.fsi.2007.12.007
270. Pettinello R, Redmond AK, Secombes CJ, Macqueen DJ, Dooley H. Evolutionary history of the T cell receptor complex as revealed by small-spotted catshark (Scyliorhinus canicula). Dev Comp Immunol. (2017) 74:125–35. doi: 10.1016/j.dci.2017.04.015
271. Bernard D, Riteau B, Hansen JD, Phillips RB, Michel F, Boudinot P, et al. Costimulatory receptors in a teleost fish: typical CD28, elusive CTLA4. J Immunol. (2006) 176:4191–200. doi: 10.4049/jimmunol.176.7.4191
272. Hansen JD, Du Pasquier L, Lefranc MP, Lopez V, Benmansour A, Boudinot P. The B7 family of immunoregulatory receptors: a comparative and evolutionary perspective. Mol Immunol. (2009) 46:457–72. doi: 10.1016/j.molimm.2008.10.007
273. Sugamata R, Suetake H, Kikuchi K, Suzuki Y. Teleost B7 expressed on monocytes regulates T cell responses. J Immunol. (2009) 182:6799–806. doi: 10.4049/jimmunol.0803371
274. Luckheeram RV, Zhou R, Verma AD, Xia B. CD4(+)T cells: differentiation and functions. Clin Dev Immunol. (2012) 2012:925135. doi: 10.1155/2012/925135
275. Secombes CJ, Zou J, Bird S. Cytokines of cartilaginous fish. In: Smith SL, Sim RB, Flajnik MF, editors. Immunobiology of the Shark. Boca Raton, FL: CRC Press (2014). p. 123–42.
276. Secombes CJ, Wang T, Bird S. Vertebrate cytokines and their evolution. In: Malagoli, editor. The Evolution of the Immune System. Cambridge, MA: Academic Press (2016). p. 87–150. doi: 10.1016/B978-0-12-801975-7.00005-0
277. Secombes CJ, Wang T. The innate and adaptive immune system of fish. In: Austin B, editor. Infectious Disease in Aquaculture: Prevention and Control. (2012). p. 3–68. doi: 10.1533/9780857095732.1.3
278. Zou J, Gorgoglione B, Taylor NG, Summathed T, Lee PT, Panigrahi A, et al. Salmonids have an extraordinary complex type I IFN system: characterization of the IFN locus in rainbow trout Oncorhynchus mykiss reveals two novel IFN subgroups. J Immunol. (2014) 193:2273–86. doi: 10.4049/jimmunol.1301796
279. Robertsen B, Bergan V, Rokenes T, Larsen R, Albuquerque A. Atlantic salmon interferon genes: cloning, sequence analysis, expression, and biological activity. J Interferon Cytokine Res. (2003) 23:601–12. doi: 10.1089/107999003322485107
280. Yabu T, Toda H, Shibasaki Y, Araki K, Yamashita M, Anzai H, et al. Antiviral protection mechanisms mediated by ginbuna crucian carp interferon gamma isoforms 1 and 2 through two distinct interferon gamma-receptors. J Biochem. (2011) 150:635–48. doi: 10.1093/jb/mvr108
281. Zou J, Carrington A, Collet B, Dijkstra JM, Yoshiura Y, Bols N, et al. Identification and bioactivities of IFN-γ in rainbow trout Oncorhynchus mykiss: the first Th1-type cytokine characterized functionally in fish. J Immunol. (2005) 175:2484–94. doi: 10.4049/jimmunol.175.4.2484
282. Lopez-Munoz A, Roca FJ, Meseguer J, Mulero V. New insights into the evolution of IFNs: zebrafish group II IFNs induce a rapid and transient expression of IFN-dependent genes and display powerful antiviral activities. J Immunol. (2009) 182:3440–9. doi: 10.4049/jimmunol.0802528
283. Yang ZJ, Li CH, Chen J, Zhang H, Li MY, Chen J. Molecular characterization of an interleukin-4/13B homolog in grass carp (Ctenopharyngodon idella) and its role in fish against Aeromonas hydrophila infection. Fish Shellfish Immunol. (2016) 57:136–47. doi: 10.1016/j.fsi.2016.08.022
284. Li JH, Shao JZ, Xiang LX, Wen Y. Cloning, characterization and expression analysis of pufferfish interleukin-4 cDNA: the first evidence of Th2-type cytokine in fish. Mol Immunol. (2007) 44:2078–86. doi: 10.1016/j.molimm.2006.09.010
285. Hu YL, Xiang LX, Shao JZ. Identification and characterization of a novel immunoglobulin Z isotype in zebrafish: implications for a distinct B cell receptor in lower vertebrates. Mol Immunol. (2010) 47:738–46. doi: 10.1016/j.molimm.2009.10.010
286. Wang T, Johansson P, Abos B, Holt A, Tafalla C, Jiang Y, et al. First in-depth analysis of the novel Th2-type cytokines in salmonid fish reveals distinct patterns of expression and modulation but overlapping bioactivities. Oncotarget. (2016) 7:10917–46. doi: 10.18632/oncotarget.7295
287. Dijkstra JM. TH2 and Treg candidate genes in elephant shark. Nature. (2014) 511:E7–9. doi: 10.1038/nature13446
288. Wang T, Martin SA, Secombes CJ. Two interleukin-17C-like genes exist in rainbow trout Oncorhynchus mykiss that are differentially expressed and modulated. Dev Comp Immunol. (2010) 34:491–500. doi: 10.1016/j.dci.2009.11.011
289. Secombes CJ, Wang T, Bird S. The interleukins of fish. Dev Comp Immunol. (2011) 35:1336–45. doi: 10.1016/j.dci.2011.05.001
290. Zou J, Secombes CJ. The function of fish cytokines. Biology. (2016) 5:e23. doi: 10.3390/biology5020023
291. Harun NO, Wang T, Secombes CJ. Gene expression profiling in naive and vaccinated rainbow trout after Yersinia ruckeri infection: insights into the mechanisms of protection seen in vaccinated fish. Vaccine. (2011) 29:4388–99. doi: 10.1016/j.vaccine.2011.04.003
292. Piazzon MC, Wentzel AS, Wiegerties GF, Forlenza M. Carp Il10a and Il10b exert identical biological activities in vitro, but are differentially regulated in vivo. Dev Comp Immunol. (2017) 67:350–60. doi: 10.1016/j.dci.2016.08.016
293. Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. (2007) 35:495–516. doi: 10.1080/01926230701320337
294. Janeway CA Jr, Travers P, Walport M, Shlomchik MJ. T cell-mediated cytotoxicity. In: Immunobiology: The Immune System in Health and Disease. 5th Edn. New York, NY: Garland Science (2001).
295. Hwang JY, Ohira T, Hirono I, Aoki T. A pore-forming protein, perforin, from a non-mammalian organism, Japanese flounder, Paralichthys olivaceus. Immunogenetics. (2004) 56:360–7. doi: 10.1007/s00251-004-0688-8
296. Praveen K, Evans DL, Jaso-Friedmann L. Evidence for the existence of granzyme-like serine proteases in teleost cytotoxic cells. J Mol Evol. (2004) 58:449–59. doi: 10.1007/s00239-003-2566-7
297. Zhou H, Stuge TB, Miller NW, Bengten E, Naftel JP, Bernanke JM, et al. Heterogeneity of channel catfish CTL with respect to target recognition and cytotoxic mechanisms employed. J Immunol. (2001) 167:1325–32. doi: 10.4049/jimmunol.167.3.1325
298. Toda H, Araki K, Moritomo T, Nakanishi T. Perforin-dependent cytotoxic mechanism in killing by CD8 positive T cells in ginbuna crucian carp, Carassius auratus langsdorfii. Dev Comp Immunol. (2011) 35:88–93. doi: 10.1016/j.dci.2010.08.010
299. Matsuura Y, Yabu T, Shiba H, Moritomo T, Nakanishi T. Identification of a novel fish granzyme involved in cell-mediated immunity. Dev Comp Immunol. (2014) 46:499–507. doi: 10.1016/j.dci.2014.06.006
300. Cuesta A, Esteban MA, Meseguer J. Identification of a FasL-like molecule in leucocytes of the teleost fish gilthead seabream (Sparus aurata L.). Dev Comp Immunol. (2003) 27:21–7. doi: 10.1016/S0145-305X(02)00041-1
301. Long S, Wilson M, Bengten E, Clem LW, Miller NW, Chinchar VG. Identification and characterization of a FasL-like protein and cDNAs encoding the channel catfish death-inducing signaling complex. Immunogenetics. (2004) 56:518–30. doi: 10.1007/s00251-004-0701-2
302. Ma TY, Wu JY, Gao XK, Wang JY, Zhan XL, Li WS. Molecular cloning, functional identification and expressional analyses of FasL in Tilapia, Oreochromis niloticus. Dev Comp Immunol. (2014) 46:448–60. doi: 10.1016/j.dci.2014.06.003
Keywords: innate immunity, adaptive immunity, chondrichthyes, actinopterygii, sarcopterygii
Citation: Smith NC, Rise ML and Christian SL (2019) A Comparison of the Innate and Adaptive Immune Systems in Cartilaginous Fish, Ray-Finned Fish, and Lobe-Finned Fish. Front. Immunol. 10:2292. doi: 10.3389/fimmu.2019.02292
Received: 31 May 2019; Accepted: 10 September 2019;
Published: 10 October 2019.
Edited by:
Tiehui Wang, University of Aberdeen, United KingdomReviewed by:
Helen Dooley, University of Maryland, Baltimore, United StatesCopyright © 2019 Smith, Rise and Christian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sherri L. Christian, c2hlcnJpLmNocmlzdGlhbkBtdW4uY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.