- 1Division of Pathology, Singapore General Hospital, Singapore, Singapore
- 2A. Menarini Biomarkers Singapore Pte Ltd., Singapore, Singapore
- 3Duke-NUS Medical School, Singapore, Singapore
- 4Translational Immunology Institute (TII), SingHealth DukeNUS Academic Medical Centre, Singapore, Singapore
- 5Temasek Polytechnic, Singapore, Singapore
- 6Department of Medical Science, University of Turin and Fondazione Ricerca Molinette, Turin, Italy
- 7Division of Medicine, Singapore General Hospital, Singapore, Singapore
- 8Centre for Quantitative Medicine, Duke-NUS Medical School, Singapore, Singapore
- 9Division of Medical Oncology, National Cancer Centre Singapore, Singapore, Singapore
- 10Department of Hepatopancreatobiliary and Transplant Surgery, Singapore General Hospital, Singapore, Singapore
- 11Institute of Molecular Cell Biology, Agency of Science, Technology and Research (A*STAR), Singapore, Singapore
Background: CD38 is involved in the adenosine pathway, which represents one of the immunosuppressive mechanisms in cancer. CD38 is broadly expressed across immune cell subsets, including human macrophages differentiated in vitro from monocytes, but expression by tissue-resident macrophages remains to be demonstrated.
Methods: Tissue samples were obtained from 66 patients with hepatocellular carcinoma (HCC) from Singapore and analyzed using immunohistochemistry. Tumor-infiltrating leukocytes (TILs) were further examined using DEPArray™, and the phenotype of freshly isolated TILs was determined using flow cytometry.
Results: CD38 was frequently co-expressed with the macrophage-specific marker CD68. CD38+CD68+ macrophage density was associated with improved prognosis after surgery, while total CD68+ macrophage density was associated with poor prognosis. DEPArray™ analysis revealed the presence of large (>10 μm), irregularly shaped CD45+CD14+ cells that resembled macrophages, with concurrent CD38+ expression. Flow cytometry also revealed that majority of CD14+HLA-DR+ cells expressed CD38.
Conclusion: CD38 expression was clearly demonstrated on human macrophages in an in vivo setting. The positive association identified between CD38+ macrophage density and prognosis may have implications for routine diagnostic work.
Introduction
Human hepatocellular carcinoma (HCC) is the most common primary malignancy of the liver (1, 2), and represents a severe, worldwide threat to human health and quality of life. Patient survival after surgery remains relatively low, with 5-year survival rates after resection for early-stage disease ranging between 17 and 53%, and recurrence rates being as high as 70% (2–4). Therefore, it is important to identify biomarkers that reliably distinguish patients at high risk of recurrence. HCC progression is known to be driven by chronic inflammation, which may arise from viral infection, hemochromatosis and alcoholic or non-alcoholic steatosis (5). The innate immune system also appears to serve a key role in the progression of HCC. For example, tumor-associated macrophages (TAMs) are widely considered to be correlated with poor prognosis in patients with HCC (6–9).
CD38 is a multifunctional, membrane-associated ectozyme belonging to the ribosyl cyclase family, and is widely expressed across the immune system. This molecule was initially discovered on the surfaces of thymocytes and T cells. Subsequently, it was found to be expressed by other types of immune cell and certain non-lymphoid tissues, including brain, eye, gut and prostate tissues (10). CD38 cleaves NAD+ and NADP+ to generate the Ca2+-mobilizing compounds adenosine diphosphate ribose (ADPR), cyclic ADPR (cADPR), and nicotinic acid adenine dinucleotide phosphate (NAADP) (10). It also functions as a receptor, mediating the transduction of signals associated with activation and proliferation. These dual enzymatic and receptorial functions mean that CD38 is involved in a diverse range of cellular activities. In the context of the immune system, CD38 is known to induce pro-inflammatory and regulatory cytokine production in monocytes and dendritic cells, and to induce proliferation and inhibit apoptosis in lymphocytes (11). Downregulation of CD38 alters the migration of neutrophils and monocytes (12), and impairs the innate immune response against Listeria monocytogenes infection (13). CD38 is also known to be present in monocytes, where it acts as a co-accessory molecule for MHC Class II-induced T lymphocyte activation by superantigen (14).
In addition, CD38 is involved in the adenosinergic pathway via its NADase function. This pathway is of significant interest to the field of cancer immunotherapy, as it represents a major alternative immunosuppressive mechanism to the PD-1/PD-L1 pathway. Here, the CD38-CD203a (also known as ENPP1 or PC1)-CD73 salvage pathway generates adenosine through the degradation of pyridine metabolites, such as NAD+. Specifically, CD38 hydrolyses NAD+ to ADPR, and CD203a further degrades ADPR to adenosine monophosphate (AMP). Following this, CD73 dephosphorylates AMP to adenosine (15–17). Tumor proliferation, survival, adhesion and migration may be regulated through the adenosine pathway. For example, in immune cells, adenosine molecules hamper vital effector cell functions and may be involved in mediating T cell exhaustion (18). A recent study using a murine lung cancer model reported that CD38 expression on cancer cells was upregulated in response to PD-1/PD-L1 blockade, resulting in the inhibition of CD8+ T-cell function via adenosine receptor signaling (19).
Our group recently established the relevance of CD38 to HCC by identifying a correlation between CD38+ tumor-infiltrating leukocyte (TIL) density and improved patient survival (20). We studied the expression of CD38 on lymphocytes, Natural Killer (NK) cells, NKT cells and monocytes, but not on macrophages. Indeed, CD38 expression has previously been reported on all major leukocyte populations, including B cells, T cells, NK cells, monocytes and dendritic cells (10). However, with respect to macrophages, data concerning CD38 expression is limited. Upregulation of CD38 has been observed on murine and human macrophages following in vitro stimulation with IFN-γ ± lipopolysaccharide (LPS), suggesting an association between CD38 and the pro-inflammatory M1 state (21, 22). However, in vivo expression of CD38 by tissue-resident macrophages remains to be demonstrated, and in vitro cultures may not represent in vivo conditions accurately. Considering the known correlation between TAMs and poor prognosis in HCC (6–9), this requires further investigation.
In the present study, we sought to ascertain the in vivo expression of CD38 on macrophages from tumor tissues obtained from patients with HCC. Using immunohistochemistry (IHC) and Multiplex immunofluorescence (mIF), we confirmed the co-expression of CD38 with the macrophage-specific marker CD68. Notably, through Kaplan-Meier survival analysis and multivariate Cox regression, we established that the presence of CD38+CD68+ macrophages predicted improved prognosis after surgery, while total CD68+ macrophage density was associated with poor prognosis. In vitro functional studies using THP-1 derived macrophages revealed CD38 expression to be associated with the M1 state, characterized by CD80 expression and secretion of TNFα and IL-6, all of which contribute to the anti-tumor immune response. Using the DEPArray™, an automated platform that is able to identify and recover single cells with high resolution and purity, we visualized the expression of CD38 on macrophage-like cells isolated from HCC tumors. Finally, using flow cytometry, the co-expression of CD38 with CD14+HLA-DR+ cells was further confirmed.
Materials and Methods
Patients and Tumor Samples
A total of 66 archival formalin-fixed, paraffin-embedded (FFPE) specimens from patients diagnosed with HCC between January 2001 and December 2011 at the Department of Anatomical Pathology, Division of Pathology, Singapore General Hospital, were analyzed. All samples were obtained prior to chemo- or radiotherapy. Clinicopathological parameters, including patient age, tumor size, histologic growth pattern, grade, subtype, lymphovascular invasion and axillary lymph node status, were reviewed (Supplementary Table 1). Tumors were staged and graded according to the AJCC staging system (23) and the Edmonson-Steiner grading system (24). Ishak fibrosis scoring (25, 26) was adopted to evaluate the fibrosis status of the non-neoplastic liver, as documented in the pathological diagnostic reports. The Centralized Institutional Review Board of SingHealth provided ethical approval for the use of patient materials in this study (CIRB ref: 2009/907/B).
Tissue Microarray Construction
Tumor regions for tissue microarray (TMA) construction were selected based on pathological assessment, with >50% of the sample representing tumor area. For each sample, two or three representative tumor cores of 1 mm diameter were transferred from donor FFPE tissue blocks to recipient TMA blocks using a MTA-1 Manual Tissue Arrayer (Beecher Instruments, Inc., Sun Prairie, WI, USA).
Multiplex Immunofluorescence Analysis of TMAs
Multiplex immunofluorescence (mIF) was performed using an Opal Multiplex fIHC kit (PerkinElmer, Inc., Waltham, MA, USA) as previously described by our group and other studies (20, 20, 27–36), on FFPE tissue sections. TMA sections of 4 μm thickness were incubated with primary antibodies against CD38 and CD68, followed by appropriate secondary antibodies (Supplementary Table 2). Following this, a fluorophore-conjugated tyramide signal amplification buffer (PerkinElmer, Inc.) was applied, and DAPI was used as a nuclear counterstain. Images were acquired using a Vectra 3 pathology imaging system microscope (PerkinElmer, Inc.) and analyzed using inForm software (version 2.4.1; PerkinElmer, Inc.) (28, 37, 38). Maximally selected rank statistics (39) were applied using the maxstat R package to find optimal cut-off points for variables with good survival outcome prediction (Cutoff for CD38+ macrophages = 56.9%, whereas cutoff for CD68 immune infiltrates = 19.2%).
In vitro THP-1 Functional Study
THP-1 cells were cultured and polarized based on a previously published protocol (21). In brief, cells were cultured in complete RPMI 1640 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS) (Gibco), 1% penicillin/streptomycin (Gibco), and 50 μM 2-mercaptoethanol (Gibco). Cells were then plated in 24-well plates at a density of 5 × 105 cells/ml, differentiated with 50 ng phorbol myristate acetate (Sigma-Aldrich) for 24 h, washed, and cultured in fresh medium for 48 h. Macrophages were polarized to the M1 state with 20 ng/ml IFN-γ (R&D) and 100 ng/ml LPS (Sigma-Aldrich), or M2 state with 20 ng/ml IL-4 (R&D). After 24 h, cells were harvested for analysis by flow cytometry. Culture supernatant was collected for analysis of cytokines by ELISA.
ELISA
ELISA was performed to detect IL-6, TNFα, IL-10, EBI3 (IL-35 subunit), and IL-12(p70). IL-6, TNFα and IL-10 kits were purchased from eBioscience (San Diego, CA, USA); EBI3 kit was purchased from R&D Systems, Inc. (Minneapolis, MN, USA); IL-12(p70) kit was purchased from RayBiotech (Norcross, GA, USA). Wells of microtiter plates were coated (18 h, room temperature) with respective capture antibody in 100 μl of coating buffer and were then blocked with 1% BSA in PBS (reagent diluent) for 1 h at room temperature. Samples or standards (100 μl) were loaded in duplicates and incubated for 2 h at room temperature, followed by the addition of 100 μl detection antibody for additional 2 h at room temperature. HRP-conjugated with Strepavidin (1:20,000) in reagent diluent was added (20 min, room temperature) and the reaction was visualized by the addition of 50 μl chromogenic substrate (TMB) for 10 min. The reaction was stopped with 100 μl H2SO4 and absorbance at 450 nm was measured using ELISA microplate reader (Perkin Elmer's Enspire 2300). Plates were washed three times with washing buffer (PBS, pH 7.4, containing 0.05% (v/v) Tween 20) after each step. As a reference for quantification, individual standard curves were established by serial dilution of respective recombinant cytokines.
Tissue Dissociation and Isolation of Leukocytes
Liver tissue was cut into fine pieces and digested with 0.5 mg/ml Collagenase IV (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 0.05 mg/ml DNAse I (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in complete RPMI (Gibco; Thermo Fisher Scientific, Inc.) for 30 min at 37°C. Digested tissue was filtered using a 70 μm cell strainer. Cells were pelleted and treated with red blood cell lysis buffer (G-Biosciences, St Louis, MO, USA) for 5 min at room temperature. Cell debris was removed using Debris Removal Solution (Miltenyi Biotec, Ltd., Bergisch Gladbach, Germany) and tissue-infiltrating leukocytes were subsequently isolated using CD45 (TIL) MicroBeads, Human (Miltenyi Biotec, Ltd.), according to the manufacturer's protocol.
Flow Cytometry
Cells were incubated with Fixable Viability Dye eFluor™ 455UV (eBioscience; Thermo Fisher Scientific, Inc.) for 30 min at 4°C for live/dead discrimination. Fc receptors were blocked using Human TruStain FcX™ (BioLegend; San Diego, CA, USA) for 5 min at room temperature. Cell surfaces were labeled with antibodies against markers of interest (refer to Supplementary Table 3 for antibody panel) for 30 min at 4°C. Matched isotype controls were included for antibodies against CD38. Single color compensation controls were prepared using Ultracomp eBeads (eBioscience; Thermo Fisher Scientific, Inc.). Samples were read in an LSRFortessa™ flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Data analysis was performed using FlowJo® V10 software (FlowJo LLC, Ashland, OR, USA).
DEPArray™
Dissociated leukocytes were Fc-blocked using Human TruStain FcX™ (BioLegend; San Diego, CA, USA) and their surfaces labeled with antibodies against CD45, CD14, and CD38 (refer to Supplementary Table 3 for antibody panel). Cells were fixed with PFA and washed with SB115 buffer (Menarini Silicon Biosystems, Castel Maggiore, Italy). Approximately 10,000 cells were loaded into the DEPArray™ cartridge (Menarini Silicon Biosystems) for imaging, and were visually inspected to confirm the expression of markers of interest.
Validation, follow-Up, and Statistical Analysis
Follow-up data were obtained from medical records. Disease-free survival (DFS) and overall survival (OS) were defined as the time from diagnosis to recurrence or death/date of last follow-up, respectively. Statistical analysis was performed using RStudio 1.1.456 running R 3.5.0 (40, 41) (R-core Team, R Foundation for Statistical Computing, Vienna, Austria) and SPSS 23.0 for Windows (IBM Corp., Armonk, NY, USA). Multivariate Cox regression was performed to evaluate the effect of selected variables on survival after adjusting for clinicopathological parameters, including for tumor size and stage. P < 0.05 indicates a statistically significance difference.
Results
Expression of CD38 by Tumor-Associated Macrophages (TAMs) in HCC Is Visualized by Immunohistochemistry (IHC)
IHC staining of FFPE tumor sections revealed the expression of CD38 by cells residing in the liver sinusoids. As shown in Figure 1A, cells that were stained had an irregular shape with many cytoplasmic extensions. We therefore hypothesized that these CD38-expressing cells may be macrophages, based on both their morphology and anatomical location (42). Moreover, previous studies have demonstrated that CD38 is expressed on macrophages in animal models (13, 22, 43, 44) and that CD38 expression on macrophages is correlated with inflammation in humans (21). Thus, we performed Opal-Vectra multiplex IHC on the specimens, using protocols previously optimized and reported by our group and other laboratories (20, 27, 30–35), to identify the co-expression of CD68 and CD38 in liver immune cells. CD38 and CD68 were frequently co-expressed by the same cells (Figures 1B–D), with 62.9 ± 19.2% of CD38+ cells bearing the macrophage marker CD68 and 32.3 ± 24.1% of CD68+ cells being CD38+ (Figures 1E,F). This suggested that a large proportion of CD38+ cells were macrophages.
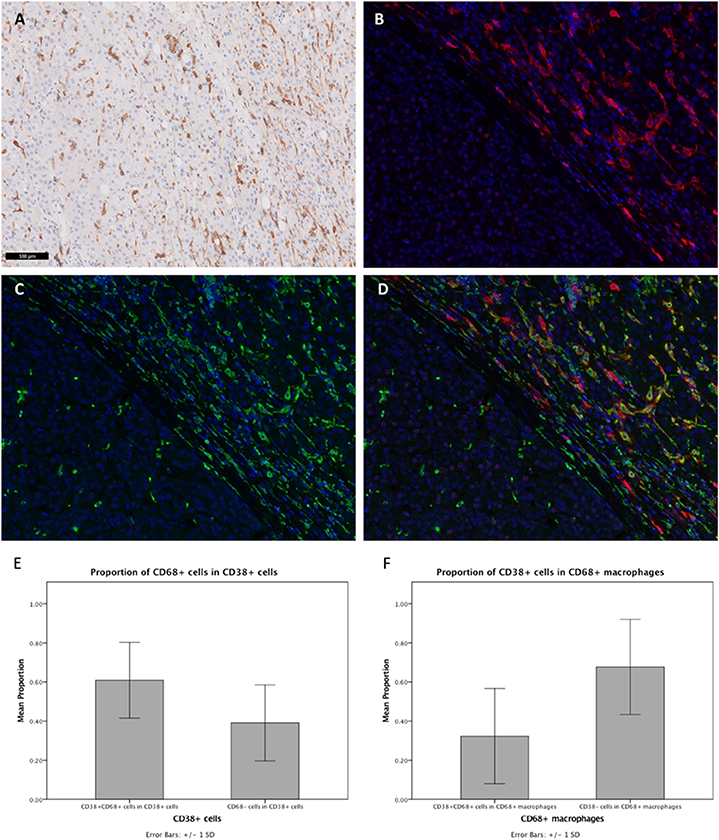
Figure 1. Expression of CD38 by TAMs from HCC patients was visualized by IHC. (A) Conventional IHC staining of a representative FFPE tumor section revealing the presence of macrophage-like cells (brown) in the liver sinusoids. (B,C) Immunofluorescent staining for CD38 (red) and macrophage marker CD68 (green), respectively, on tumor section. (D) Merged image showing co-localization of CD38 (red) and CD68 (green) in the tumor microenvironment. Images are presented at 40× magnification. (E) Proportions of CD38+CD68+ and CD38+CD68− leukocytes. (F) Proportions of CD38+CD68+ and CD38−CD68+ macrophages. Data is presented as the mean ± standard deviation.
High CD38+ Macrophage Density Is Associated With Improved Survival in HCC
Next, we investigated whether CD38+ macrophage density in tumors affected survival outcome in patients with HCC. Patients with high density of CD38+CD68+ macrophages experienced significantly improved overall survival (OS) compared with those with low density of CD38+CD68+ macrophages (P = 0.0367; Figure 2A); in contrast, disease free survival (DFS) did not differ significantly between groups (P = 0.2400; Figure 2B). Multivariate analysis further supported the association between a high density of intratumoral CD38+ macrophages and significantly improved OS (HR = 0.41; 95% CI 0.18–0.93; P = 0.0322; Table 1).
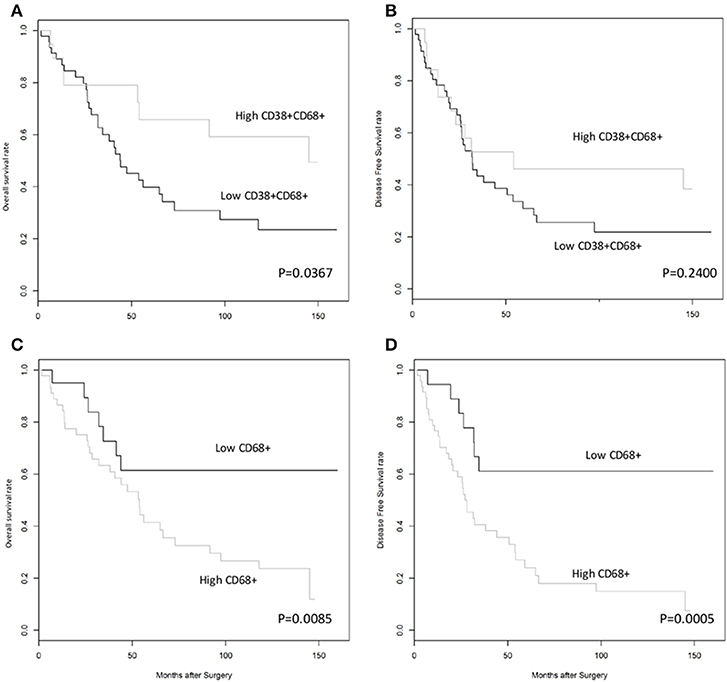
Figure 2. High density of CD38+, but not total, CD68+ TAMs was associated with improved overall survival rates in patients with HCC. (A,B) Kaplan-Meier curves showing overall and disease free survival rates, respectively, for patients grouped according to high or low density of CD38+CD68+ macrophages in the tumor microenvironment. (C,D) Kaplan-Meier curves showing overall and disease free survival rates, respectively, for patients grouped according to high or low density of CD68+ total macrophages in the tumor microenvironment. Cut-offs for high/low density of CD38+CD68+ macrophages (0.5690) and CD68+ total macrophages (0.1918) were based on optimal values calculated using statistical software.
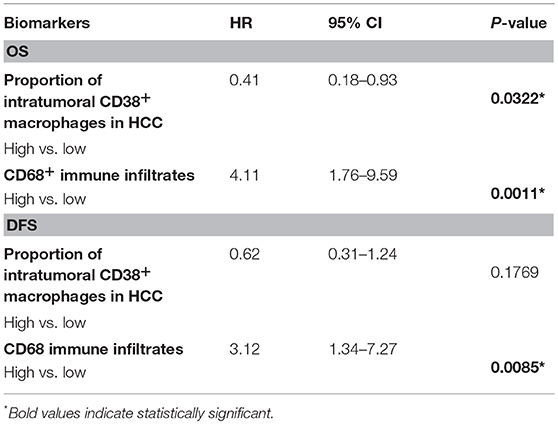
Table 1. Multivariate analysis of the effect of intratumoral macrophage density on OS and DFS, adjusted for tumor size, grade, age, and lymph node status.
The effect of total CD68+ macrophages on prognosis was also investigated. Notably, high CD68+ macrophage density was associated with reduced OS (HR = 3.12; 95% CI 1.34–7.27; P = 0.0085) and DFS (HR = 4.44; 95% CI 1.92–10.26; P = 0.0005) in HCC patients (Figures 2C,D, respectively). Multivariate analysis also revealed that CD68+ macrophage density was an independent prognostic marker for reduced OS (HR = 3.01; 95% CI 1.27–7.16; P = 0.0124) and DFS (HR = 4.11; 95% CI 1.76–9.59; P = 0.0011) after adjusting for clinicopathological parameters (Table 1).
CD38 Up-regulation Was Associated With M1 Macrophages, Characterized by Expression of Co-stimulatory CD80 and a Pro-inflammatory Cytokine Profile
To ascertain the function of CD38+ macrophages, we took advantage of the THP-1 cell line to investigate surface expression and cytokine secretion of CD38+ macrophages under in vitro culture conditions. THP-1 cells were polarized into M1/M2 macrophages that showed differential expression of CD80 and DC-SIGN (Figures 3A,B), consistent with the results of a previous study (45). Notably, robust expression of CD80 was observed on M1 macrophages but not M0 or M2. Additionally, we found up-regulation of CD38 on M1 macrophages (Figures 3C,D), concomitant with pro-inflammatory IL-6 and TNFα secretion (Figure 3E). By contrast, M0 or M2 macrophages did not secrete IL-6 or TNF-α (Figure 3E). As compared to IL-6 (786.7 ± 117.1 pg/ml) and TNFα (18.2 ± 1.9 ng/ml), the level of IL-35 secretion was minimal (82.0 ± 4.2 pg/ml) and there was no detectable IL-10 and IL-12(p70) secretion from M1 macrophages (Figure 3F). Altogether, our results suggest that CD38+ macrophages are associated with an M1 phenotype that is characterized by higher CD80 expression and a pro-inflammatory cytokine profile.
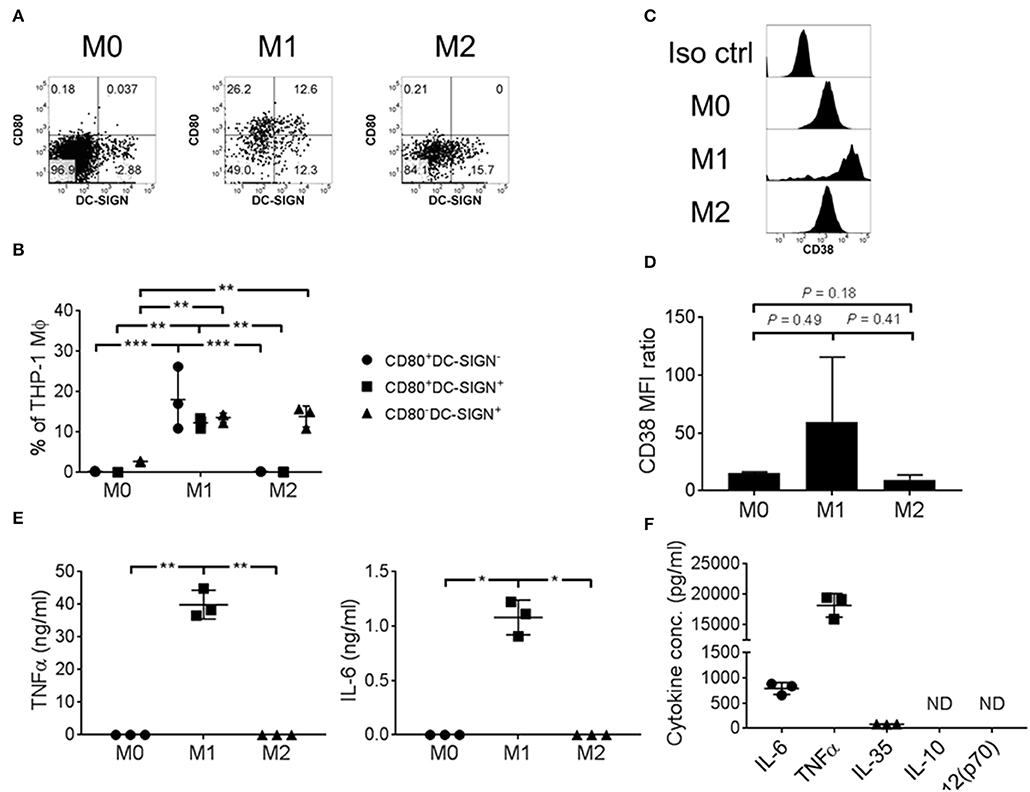
Figure 3. THP-1 derived M1 macrophages expressed CD38 and secreted pro-inflammatory cytokines. THP-1 cells were polarized into M0/M1/M2 macrophages with differential surface expression of (A,B) CD80 and/or DC-SIGN; (C,D) CD38. Representative flow cytometry data are shown. MFI: geometric mean fluorescent intensity. (E) IL-6 and TNFα cytokine secretion, as determined by ELISA using culture supernatant. The data represents the means ± s.d. (F) Cytokine secretion profile of M1 macrophages, as determined by ELISA using culture supernatant. ND: not detected. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Confirmation of CD38 Expression on Macrophages by DEPArray™ and Flow Cytometry
To further confirm the expression of CD38 on TAMs from patients with HCC, tumor-infiltrating leukocytes (TILs) were examined using the DEPArray™, which is an automated platform capable of visualizing and isolating single cells with high resolution and purity. Images captured by the DEPArray™ revealed the presence of large (>10 μm), irregularly shaped CD45+CD14+ cells that resembled macrophages (Figure 4). CD38 expression was clearly detected on the surfaces of these cells. Next, flow cytometry was performed to analyse TILs in greater detail. Monocytes/macrophages were identified in tumor tissues from four HCC patients using the gating strategy shown in Figure 5A. The vast majority of CD14+HLA-DR+ cells expressed CD38 (84.9 ± 7.4%; Figure 5B), indicating enrichment of CD38+ monocytes/macrophages in the tumors of these four patients. In terms of total CD38-expressing leuckocytes, monocytes/macrophages constituted only 34.6 ± 26.6% (Figure 5B), indicating the presence of other CD38-expressing immune lineages.
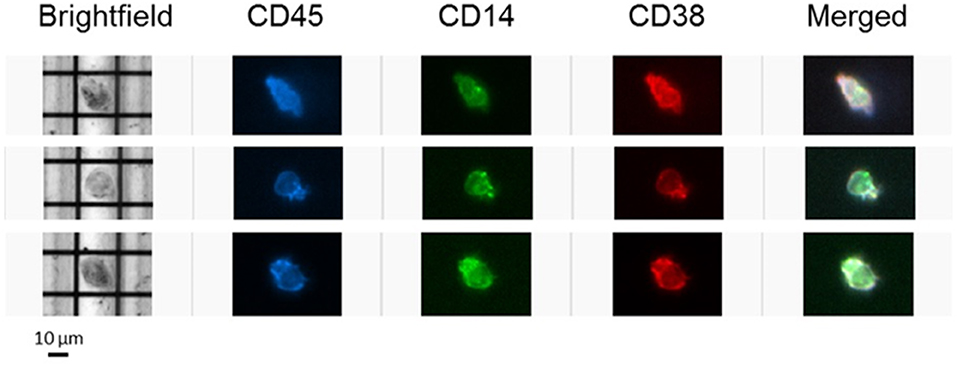
Figure 4. CD38-expressing tumor-infiltrating leukocytes were visualized using the DEPArray™ platform. Three single cells resembling macrophages are presented. Cells are large (>10 μm), irregularly shaped and express myeloid marker CD14.
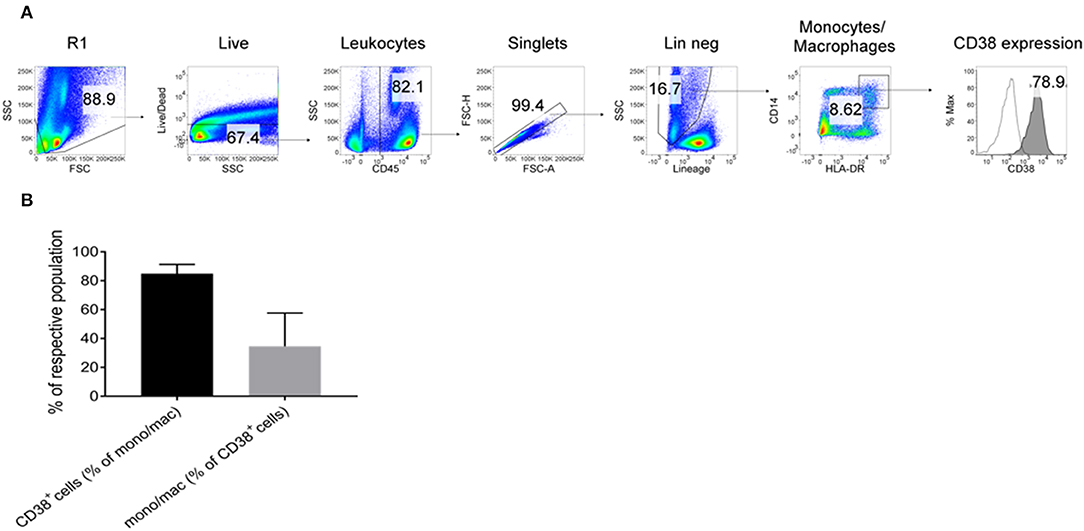
Figure 5. Expression of CD38+ by tumor-infiltrating monocytes/macrophages further confirmed by flow cytometry. (A) Tumor-infiltrating leukocytes from four HCC patients were analyzed. Lineage+ cells (CD3/CD7/CD19/CD20/CD56) were excluded in a dump channel. Monocytes/macrophages were identified by co-expression of CD14 and HLA-DR. Expression of CD38 (filled histogram) was determined with respect to the isotype control antibody (clear histogram). (B) % of monocytes/macrophages that expressed CD38 (black column) and % of CD38+ leukocytes that were monocytes/macrophages (gray column). The data represents the means ± s.d.
Discussion
Through multiplex IF and conventional IHC, our group was able to identify and enumerate CD68+ TAMs present in the tumor samples from HCC patients. We established that CD68+ TAM density was correlated with poor prognosis in HCC, in line with the finding of a previous report (6). The tumor microenvironment is known to be dominated by cytokines and growth factors that establish a Th2-type, anti-inflammatory immune environment that favors the survival of tumor cells. Under such environmental influences, TAMs are driven toward a M2 phenotype. This transition is accompanied by increased pro-tumoral activities, including the suppression of the adaptive immune response and the promotion of cancer proliferation, angiogenesis and extracellular matrix remodeling (46).
To the best of our knowledge, the present study represents the first instance that CD38 expression has been demonstrated on human macrophages in an in vivo setting. Previously, CD38 expression was reported on macrophages isolated from mice (13, 47), on a murine macrophage cell line (48) and on human macrophages under ex vivo conditions (21). We initially observed the co-localization of CD38 and CD68 signals in the sinusoids of liver sections, and subsequently from freshly isolated TILs. CD38 was visually confirmed on macrophage-like CD14+ cells using DEPArray™, and also on CD14+HLA-DR+ monocytes/macrophages using flow cytometry. Notably, statistical analysis of the IHC data revealed that CD38+CD68+ macrophage density was positively associated with improved prognosis, in direct contrast to total CD68+ macrophage density. This suggested the existence of functionally-distinct macrophage subsets, with CD38 marking a subpopulation of pro-inflammatory macrophages (21) that may play a role in tumor suppression. Supporting this hypothesis was data from our in vitro functional study of THP-1 cells polarized to M1 or M2 macrophages. Specifically, M1 macrophages were distinguished by higher expression of CD80—a costimulatory molecule that signals through CD28 to amplify T cell activation and contributes to anti-tumor immunity (49, 50). We also found that CD38 was up-regulated on M1 macrophages, concomitant with a pro-inflammatory cytokine profile characterized by robust IL-6 and TNFα secretion, with minimal IL-35 secretion and no secretion of IL-10 and IL-12 cytokines. By contrast, M0 or M2 macrophages did not produce detectable levels of all cytokines examined. A previous study reported that deletion of IL6 or TNFα in a mouse model of HCC accelerated tumor development (51). Altogether, our results suggest that CD38+ macrophages are associated with an M1 phenotype that is characterized by higher CD80 expression and a pro-inflammatory cytokine profile, all of which contribute to the anti-tumor immune response. This effect may account for the improved prognosis of HCC patients with higher proportion of tumor-infiltrating CD38+ macrophages. To further appreciate the complexities of TAMs in liver cancer, a future single cell study could be conducted. In view of this, the DEPArray™ platform may be useful for isolating single cells with high purity, as required for sensitive downstream genomic and expression analyses (52–54).
It is important to note that CD38 is involved in the production of adenosine from NAD+. Adenosine is known to be an inhibitor of effector T cells and is produced by various regulatory cells expressing CD38, including myeloid-derived suppressor cells, mesenchymal stem cells and NK cells (55). In view of this, the expression of CD38 on tumor-infiltrating macrophages in the present study raises the possibility of participation in this mode of immunosuppression. This suggests the presence of a complex interplay between the inflammatory response and immune suppression via adenosine production.
When flow cytometry was used to investigate TILs from four patients with HCC, high levels of CD38 expression were found among the monocytes/macrophages. However, these monocytes/macrophages represented only 34.6% of total CD38+ leukocytes, which indicated the presence of other CD38+ immune lineages. In our previous study concerning TILs in HCC, we reported expression of CD38 on lymphocytes, NK cells, NKT cells and monocytes (20). Such broad expression was in line with the reported ubiquity of CD38 across the immune system (10).
Of interest to us in future studies is CD157, a paralogue of CD38 derived from gene duplication. CD157 also possesses dual receptorial and NADase functions and is widespread across lymphoid tissues, including macrophages (56). Given the high level of sequence similarity between CD38 and CD157, it is important to establish whether CD157+ macrophages also predict improved prognosis in patients with HCC.
In conclusion, this study confirmed the expression of CD38 by human macrophages in vivo, and established that CD38+ macrophages are correlated with improved prognosis after surgery. The function of CD38 in myeloid cells warrants further study, and this marker may be utilized in routine diagnostic work in the era of cancer immunotherapy.
Ethics Statement
The Centralized Institutional Review Board of SingHealth provided ethical approval for the use of patient materials in this study (CIRB ref: 2009/907/B).
Author Contributions
JY, TSL and TKL conceived and directed the study. JY and TSL supervised the research. JHL, HN, and CL collated and interpreted the data and performed biostatistical analysis. XS and JL constructed TMAs and performed IHC. HL performed additional biostatistical analysis. HN, XS, and J-KK performed immunohistochemical scoring. FM, KS, CO, TL, WL, and VC contributed to the scientific content of the study. SC and HT provided scientific inputs from Oncology perspectives. SL and CC provided scientific inputs from Surgery perspectives. HN, JHL, and JY drafted the manuscript with the assistance and final approval of all authors.
Funding
This research was partially funded by the Center Grant (CG) grant of Singapore General Hospital (NMRC/CG/M011/2017_SGH) and the AM-ETHOS Duke-NUS Medical Student Fellowship Award (AM-ETHOS01/FY2018/10-A10). This work was also supported by the National Medical Research Council (NMRC), Singapore (ref numbers: TCR15Jun006, CIRG16may048, CSAS16Nov006, CSASI17may003, and LCG17MAY003).
Conflict of Interest Statement
JHL and TSL are employees of A. Menarini Biomarkers Singapore Pte Ltd. FM received research support from Janssen Pharmaceuticals, Celgene, Tusk Therapeutics and Centrose, and served on advisory boards for Centrose, Tusk Therapeutics, Jenssen, Takeda and Sanofi.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Prof. Paola Ricciardi-Castagnoli of Toscana Life Sciences Foundation (TLS) and Dr. Alice Bridges for critical review of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02093/full#supplementary-material
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. (2015) 136:E359–86. doi: 10.1002/ijc.29210
2. Zheng J, Chou JF, Gonen M, Vachharajani N, Chapman WC, Majella Doyle MB, et al. Prediction of hepatocellular carcinoma recurrence beyond milan criteria after resection: validation of a clinical risk score in an international cohort. Ann Surg. (2017) 266:693–701. doi: 10.1097/SLA.0000000000002360
3. Chen XP, Qiu FZ, Wu ZD, Zhang ZW, Huang ZY, Chen YF. Long-term outcome of resection of large hepatocellular carcinoma. Br J Surg. (2006) 93:600–6. doi: 10.1002/bjs.5335
4. Ruan DY, Lin ZX, Wang TT, Zhao H, Wu DH, Chen J, et al. Nomogram for preoperative estimation of long-term survival of patients who underwent curative resection with hepatocellular carcinoma beyond Barcelona clinic liver cancer stage A1. Oncotarget. (2016) 7:61378–89. doi: 10.18632/oncotarget.11358
5. Nikolaou K, Sarris M, Talianidis I. Molecular pathways: the complex roles of inflammation pathways in the development and treatment of liver cancer. Clin Cancer Res. (2013) 19:2810–6. doi: 10.1158/1078-0432.CCR-12-1961
6. Ding T, Xu J, Wang F, Shi M, Zhang Y, Li SP, et al. High tumor-infiltrating macrophage density predicts poor prognosis in patients with primary hepatocellular carcinoma after resection. Hum Pathol. (2009) 40:381–9. doi: 10.1016/j.humpath.2008.08.011
7. Lanaya H, Natarajan A, Komposch K, Li L, Amberg N, Chen L, et al. EGFR has a tumour-promoting role in liver macrophages during hepatocellular carcinoma formation. Nat Cell Biol. (2014) 16:972–7. doi: 10.1038/ncb3031
8. Yeung OW, Lo CM, Ling CC, Qi X, Geng W, Li CX, et al. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol. (2015) 62:607–16. doi: 10.1016/j.jhep.2014.10.029
9. Zhu XD, Zhang JB, Zhuang PY, Zhu HG, Zhang W, Xiong YQ, et al. High expression of macrophage colony-stimulating factor in peritumoral liver tissue is associated with poor survival after curative resection of hepatocellular carcinoma. J Clin Oncol. (2008) 26:2707–16. doi: 10.1200/JCO.2007.15.6521
10. Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, et al. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev. (2008) 88:841–86. doi: 10.1152/physrev.00035.2007
11. March S, Graupera M, Rosa Sarrias M, Lozano F, Pizcueta P, Bosch J, et al. Identification and functional characterization of the hepatic stellate cell CD38 cell surface molecule. Am J Pathol. (2007) 170:176–87. doi: 10.2353/ajpath.2007.051212
12. Lund FE, Cockayne DA, Randall TD, Solvason N, Schuber F, Howard MC. CD38: a new paradigm in lymphocyte activation and signal transduction. Immunol Rev. (1998) 161:79–93. doi: 10.1111/j.1600-065X.1998.tb01573.x
13. Lischke T, Heesch K, Schumacher V, Schneider M, Haag F, Koch-Nolte F, et al. CD38 controls the innate immune response against Listeria monocytogenes. Infect Immun. (2013) 81:4091–9. doi: 10.1128/IAI.00340-13
14. Zilber M-T, Gregory S, Mallone R, Deaglio S, Malavasi F, Charron D, et al. CD38 expressed on human monocytes: A coaccessory molecule in the superantigen-induced proliferation. Proc Natl Acad Sci USA. (2000) 97:2840–5. doi: 10.1073/pnas.050583197
15. Horenstein AL, Chillemi A, Quarona V, Zito A, Roato I, Morandi F, et al. NAD+-metabolizing ectoenzymes in remodeling tumor-host interactions: the human myeloma model. Cells. (2015) 4:520–37. doi: 10.3390/cells4030520
16. Vaisitti T, Audrito V, Serra S, Bologna C, Brusa D, Malavasi F, et al. NAD+-metabolizing ecto-enzymes shape tumor-host interactions: the chronic lymphocytic leukemia model. FEBS Lett. (2011) 585:1514–20. doi: 10.1016/j.febslet.2011.04.036
17. Malavasi F, Deaglio S, Damle R, Cutrona G, Ferrarini M, Chiorazzi N. CD38 and chronic lymphocytic leukemia: a decade later. Blood. (2011) 118:3470–8. doi: 10.1182/blood-2011-06-275610
18. Hilchey SP, Kobie JJ, Cochran MR, Secor-Socha S, Wang JC, Hyrien O, et al. Human follicular lymphoma CD39+-infiltrating T cells contribute to adenosine-mediated T cell hyporesponsiveness. J Immunol. (2009) 183:6157–66. doi: 10.4049/jimmunol.0900475
19. Chen L, Diao L, Yang Y, Yi X, Rodriguez BL, Li Y, et al. CD38-mediated immunosuppression as a mechanism of tumor cell escape from PD-1/PD-L1 blockade. Cancer Discov. (2018) 8:1156–75. doi: 10.1158/2159-8290.CD-17-1033
20. Garnelo M, Tan A, Her Z, Yeong J, Lim CJ, Chen J, et al. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut. (2017) 66:342–51. doi: 10.1136/gutjnl-2015-310814
21. Amici SA, Young NA, Narvaez-Miranda J, Jablonski KA, Arcos J, Rosas L, et al. CD38 is robustly induced in human macrophages and monocytes in inflammatory conditions. Front Immunol. (2018) 9:1593. doi: 10.3389/fimmu.2018.01593
22. Jablonski KA, Amici SA, Webb LM, Ruiz-Rosado Jde D, Popovich PG, Partida-Sanchez S, et al. Novel markers to delineate murine M1 and M2 macrophages. PLoS ONE. (2015) 10:e0145342. doi: 10.1371/journal.pone.0145342
23. Henderson JM, Sherman M, Tavill A, Abecassis M, Chejfec G, Gramlich T. AHPBA/AJCC consensus conference on staging of hepatocellular carcinoma: consensus statement. HPB. (2003) 5:243–50. doi: 10.1080/13651820310015833
24. Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. (1954) 7:462–503.
25. Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, et al. Histological grading and staging of chronic hepatitis. J Hepatol. (1995) 22:696–9. doi: 10.1016/0168-8278(95)80226-6
26. Goodman ZD. Grading and staging systems for inflammation and fibrosis in chronic liver diseases. J Hepatol. (2007) 47:598–607. doi: 10.1016/j.jhep.2007.07.006
27. Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. (2014) 70:46–58. doi: 10.1016/j.ymeth.2014.08.016
28. Abel EJ, Bauman TM, Weiker M, Shi F, Downs TM, Jarrard DF, et al. Analysis and validation of tissue biomarkers for renal cell carcinoma using automated high-throughput evaluation of protein expression. Human Pathol. (2014) 45:1092–9. doi: 10.1016/j.humpath.2014.01.008
29. Lovisa S, LeBleu VS, Tampe B, Sugimoto H, Vadnagara K, Carstens JL, et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med. (2015) 21:998–1009. doi: 10.1038/nm.3902
30. Yeong J, Thike AA, Lim JC, Lee B, Li H, Wong SC, et al. Higher densities of Foxp3+ regulatory T cells are associated with better prognosis in triple-negative breast cancer. Breast Cancer Res Treat. (2017) 163:21–35. doi: 10.1007/s10549-017-4161-4
31. Lim JCT, Yeong JPS, Lim CJ, Ong CCH, Chew VSP, Ahmed SS, et al. An automated staining protocol for 7-colour immunofluorescence of human tissue sections for diagnostic and prognostic use. J Royal Coll Pathol Aust. 50:333–41. doi: 10.1016/j.pathol.2017.11.087
32. Esbona K, Inman D, Saha S, Jeffery J, Schedin P, Wilke L, et al. COX-2 modulates mammary tumor progression in response to collagen density. Breast Cancer Res. (2016) 18:35. doi: 10.1186/s13058-016-0695-3
33. Mlecnik B, Bindea G, Kirilovsky A, Angell HK, Obenauf AC, Tosolini M, et al. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Sci Transl Med. (2016) 8:327ra26. doi: 10.1126/scitranslmed.aad6352
34. Nghiem PT, Bhatia S, Lipson EJ, Kudchadkar RR, Miller NJ, Annamalai L, et al. PD-1 blockade with pembrolizumab in advanced merkel-cell carcinoma. New Engl J Med. (2016) 374:2542–52. doi: 10.1056/NEJMoa1603702
35. Feng Z, Jensen SM, Messenheimer DJ, Farhad M, Neuberger M, Bifulco CB, et al. Multispectral Imaging of T and B cells in murine spleen and tumor. J Immunol. (2016) 196:3943–50. doi: 10.4049/jimmunol.1502635
36. Yeong J, Lim JCT, Lee B, Li H, Chia N, Ong CCH, et al. High Densities of Tumor-Associated Plasma Cells Predict Improved Prognosis in Triple Negative Breast Cancer. Front Immunol. (2018) 9:1209. doi: 10.3389/fimmu.2018.01209
37. Fiore C, Bailey D, Conlon N, Wu X, Martin N, Fiorentino M, et al. Utility of multispectral imaging in automated quantitative scoring of immunohistochemistry. J Clin Pathol. (2012) 65:496–502. doi: 10.1136/jclinpath-2012-200734
38. Feng Z, Bethmann D, Kappler M, Ballesteros-Merino C, Eckert A, Bell RB, et al. Multiparametric immune profiling in HPV– oral squamous cell cancer. JCI Insight. (2017) 2:93652. doi: 10.1172/jci.insight.93652
39. Lausen B, Schumacher M. Maximally selected rank statistics. Biometrics. (1992) 48:73–85. doi: 10.2307/2532740
41. R Foundation for Statistical Computing. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing (2016).
42. Smedsrod B, De Bleser PJ, Braet F, Lovisetti P, Vanderkerken K, Wisse E, et al. Cell biology of liver endothelial and Kupffer cells. Gut. (1994) 35:1509–16. doi: 10.1136/gut.35.11.1509
43. Botta D, Rivero-Nava L, Lund F. The NAD glycohydrolase CD38 regulates macrophage effector function and defense against Listeria monocytogenes. (INC7P.409). J Immunol. (2014) 192(Suppl. 1):186.10.
44. Schneider M, Schumacher V, Lischke T, Lucke K, Meyer-Schwesinger C, Velden J, et al. CD38 is expressed on inflammatory cells of the intestine and promotes intestinal inflammation. PLoS ONE. (2015) 10:e0126007. doi: 10.1371/journal.pone.0126007
45. Tarique AA, Logan J, Thomas E, Holt PG, Sly PD, Fantino E. Phenotypic, functional, and plasticity features of classical and alternatively activated human macrophages. Am J Respir Cell Mol Biol. (2015) 53:676–88. doi: 10.1165/rcmb.2015-0012OC
46. Tahmasebi Birgani M, Carloni V. Tumor Microenvironment, a Paradigm in Hepatocellular Carcinoma Progression and Therapy. Int J Mol Sci. (2017) 18:E405. doi: 10.3390/ijms18020405
47. Kang J, Park KH, Kim JJ, Jo EK, Han MK, Kim UH. The role of CD38 in Fcgamma receptor (FcgammaR)-mediated phagocytosis in murine macrophages. J Biol Chem. (2012) 287:14502–14. doi: 10.1074/jbc.M111.329003
48. Lee HC. Cyclic ADP-ribose and nicotinic acid adenine dinucleotide phosphate (NAADP) as messengers for calcium mobilization. J Biol Chem. (2012) 287:31633–40. doi: 10.1074/jbc.R112.349464
49. Driessens G, Kline J, Gajewski TF. Costimulatory and coinhibitory receptors in anti-tumor immunity. Immunol Rev. (2009) 229:126–44. doi: 10.1111/j.1600-065X.2009.00771.x
50. Haile ST, Horn LA, Ostrand-Rosenberg S. A soluble form of CD80 enhances antitumor immunity by neutralizing programmed death ligand-1 and simultaneously providing costimulation. Cancer Immunol Res. (2014) 2:610–5. doi: 10.1158/2326-6066.CIR-13-0204
51. Ji T, Li G, Chen J, Zhao J, Li X, Lin H, et al. Distinct role of interleukin-6 and tumor necrosis factor receptor-1 in oval cell- mediated liver regeneration and inflammation-associated hepatocarcinogenesis. Oncotarget. (2016) 7:66635–46. doi: 10.18632/oncotarget.11365
52. Fontana F, Rapone C, Bregola G, Aversa R, de Meo A, Signorini G, et al. Isolation and genetic analysis of pure cells from forensic biological mixtures: The precision of a digital approach. Forensic Sci Int Genet. (2017) 29:225–41. doi: 10.1016/j.fsigen.2017.04.023
53. Paolillo C, Mu Z, Rossi G, Schiewer MJ, Nguyen T, Austin L, et al. Detection of activating estrogen receptor gene (ESR1) mutations in single circulating tumor cells. Clin Cancer Res. (2017) 23:6086–93. doi: 10.1158/1078-0432.CCR-17-1173
54. Shaw JA, Guttery DS, Hills A, Fernandez-Garcia D, Page K, Rosales BM, et al. Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high circulating tumor cell counts. Clin Cancer Res. (2017) 23:88–96. doi: 10.1158/1078-0432.CCR-16-0825
55. Morandi F, Horenstein AL, Rizzo R, Malavasi F. The role of extracellular adenosine generation in the development of autoimmune diseases. Mediators Inflamm. (2018) 2018:7019398. doi: 10.1155/2018/7019398
Keywords: macrophage, CD38, marker, prognosis, cancer, hepatocellular carcinoma
Citation: Lam JH, Ng HHM, Lim CJ, Sim XN, Malavasi F, Li H, Loh JJH, Sabai K, Kim J-K, Ong CCH, Loh T, Leow WQ, Choo SP, Toh HC, Lee SY, Chan CY, Chew V, Lim TS, Yeong J and Lim TKH (2019) Expression of CD38 on Macrophages Predicts Improved Prognosis in Hepatocellular Carcinoma. Front. Immunol. 10:2093. doi: 10.3389/fimmu.2019.02093
Received: 09 December 2018; Accepted: 20 August 2019;
Published: 04 September 2019.
Edited by:
Katy Rezvani, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Pin Wu, Zhejiang University, ChinaXin-Yuan Guan, The University of Hong Kong, Hong Kong
Copyright © 2019 Lam, Ng, Lim, Sim, Malavasi, Li, Loh, Sabai, Kim, Ong, Loh, Leow, Choo, Toh, Lee, Chan, Chew, Lim, Yeong and Lim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tong Seng Lim, dG9uZ3NlbmcubGltQG1iaW9tYXJrZXJzLmNvbQ==; Joe Yeong, Sm9lLnllb25nLnAuc0BzZ2guY29tLnNn; Tony Kiat Hon Lim, bGltLmtpYXQuaG9uQHNpbmdoZWFsdGguY29tLnNn
†These authors have contributed equally to this work
 Jian Hang Lam
Jian Hang Lam Harry Ho Man Ng
Harry Ho Man Ng Chun Jye Lim4
Chun Jye Lim4 Xin Ni Sim
Xin Ni Sim Fabio Malavasi
Fabio Malavasi Joo-Kyung Kim
Joo-Kyung Kim Clara Chong Hui Ong
Clara Chong Hui Ong Tracy Loh
Tracy Loh Tong Seng Lim
Tong Seng Lim Joe Yeong
Joe Yeong