- 1Institut Curie, PSL Research University, INSERM U932, Paris, France
- 2Université Paris Descartes, Paris, France
Circulating monocytes can infiltrate mucosal or inflamed tissues where they differentiate into either macrophages or dendritic cells. This paradigm is supported by numerous studies conducted in mice and in different in vitro settings for human cells. Determining whether it holds true in vivo in humans is essential for the successful design of monocyte-targeting therapies. Despite limitations inherent to working with human samples, there is accumulating evidence of the existence of in vivo-generated monocyte-derived cells in humans. Here, we review recent studies showing the recruitment of human monocytes into tissues and their differentiation into macrophages or dendritic cells, in normal or pathological settings. We examine the methods available in human studies to demonstrate the monocytic origin of infiltrating cells. Finally, we review the functions of human monocyte-derived cells and how they might contribute to pathogeny.
Introduction
Numerous studies in mice have shown that monocytes circulate in the blood and are recruited to mucosal tissues or inflammation sites, where they can differentiate into monocyte-derived macrophages (mo-Mac) or monocyte-derived dendritic cells (mo-DC) (1, 2). In models of inflammatory disorders, monocyte-derived cells have been shown to exert a deleterious role, in particular by fueling the inflammation and inducing tissue damage. Blocking their recruitment to inflamed tissues, using nanoparticules that induce apoptosis (3) or that contain si-RNA against CCR2 (4), reduces inflammation and improves the pathogeny in mouse models of colitis, peritonitis, and atherosclerosis. Monocytes have therefore emerged in the past few years as an attractive therapeutic target.
However, findings from mouse models do not always translate to humans due to genetic, physiological, and environmental differences. In particular, whether mice represent an appropriate model for analyzing inflammatory responses and chronic inflammatory diseases has been controversial (5–7). Despite inherent limitations, observations in humans are therefore essential to complement mouse studies to fully understand monocyte biology and the contribution of monocyte-derived cells to inflammatory disorders.
Monocyte Life Cycle
Circulating monocytes are classified into three subsets based on the expression of the surface markers CD14 and CD16: “classical” CD14highCD16-monocytes (around 85% of monocytes), “intermediate” CD14+CD16+ monocytes (5–10%) and “non-classical” CD14-CD16high (5–10%) monocytes. The life cycle and relationship of these subsets has been the subject of recent in vivo studies.
Several lines of investigation point to a linear differentiation relationship between monocyte subsets. Using in vivo labeling with a short pulse of 6,6-2H2-glucose in healthy volunteers, two studies have reported the sequential enrichment in the blood of labeled CD14high monocytes, then CD14+CD16+ monocytes and finally CD16high monocytes (8, 9). Similarly, following autologous stem cell transplantation, CD14high monocytes reappeared first in the blood after 7 days, followed by CD14+CD16+ monocytes and then CD16high monocytes after 10 days (10). Moreover, after in vivo endotoxin challenge in healthy volunteers, monocytes disappeared from the circulation within 2 h with CD14high monocytes recovering after 4 h, then CD14+CD16+ monocytes and CD16high monocytes after 24 h (9, 11). Mathematical modeling indicated that CD14high monocytes have a short lifespan in the blood of 1–2 days, before differentiating into CD14+CD16+ monocytes or disappearing from the circulation (8, 9).
Consistent with this model, single-cell RNA-seq (scRNA-seq) analysis of blood monocytes showed that CD14+CD16+ monocytes are a heterogeneous population with mixed transcriptional profiles (12). In an independent scRNA-seq analysis, purified CD14highCD16- monocytes were shown to contain two subsets: one with a typical transcriptional profile of CD14high monocytes and one with a profile closer to CD16high monocytes, suggesting that part of the CD14high monocytes are already en route to differentiate before up-regulation of CD16 (13).
Collectively, these observations support the notion that CD14high monocytes represent the precursor population of both CD16high monocytes in blood and monocyte-derived cells in tissues.
Monocyte Recruitment Into Tissues
In mice, circulating monocytes leave the bloodstream to infiltrate mucosal tissues or inflamed sites, or to reside in the spleen. What is the evidence that the same scheme applies to humans?
Several studies have shown monocyte recruitment in the context of acute inflammation. In dialysis-induced bacterial peritonitis, CD14+ monocytes number was increased in the peritoneum 1 day after infection (14). In the acute inflammation model of skin blister, a high proportion of CD14+ cells was observed 24 h after blister formation, suggesting monocyte recruitment (15). Similarly, CD14+ cell number increased in the nasal mucosa 12 h after allergen challenge in a model of allergic rhinitis (16) and in the bronchoalveolar lavage 8 h following LPS inhalation (17). Furthermore, S100A8/9+ cells accumulated in the bronchial mucosa of patients who died from asthma attack as compared to non-atopic controls (16). Monocytes also infiltrate the heart following acute myocardial infarction, as shown by the increase in CD14+CD16- and CD14+CD16+ cells as compared to heart tissue from donors who died of other causes (18). This strong influx of monocytes correlated with a decrease in the proportion of CD14+ cells in the bone marrow and spleen, suggesting that the spleen could be a monocyte reservoir in humans. This is consistent with the presence of bona fide monocytes in human spleen (19).
Monocytes also infiltrate tissues in chronic inflammation. In inflammatory bowel diseases (IBD) such as Crohn's disease (CD) and ulcerative colitis (UC), an increased infiltration of monocytes was described in the colon. Following injection of radiolabelled monocytes, radioactive CD14+ cells were detected in the intestine of patients with intestinal inflammation (20) and in joints of patients suffering from rheumatoid arthritis (RA) (21). Moreover, CD14highCD11chigh monocytes were increased in the inflamed mucosa of CD patients as compared to control samples (22–24). During multiple sclerosis (MS), infiltrating monocytes were detected in MS lesions (25). CD14+ CCR2+ CD16- MerTK- cells were identified in the synovial fluid from gouty arthritis patients, suggesting recruitment of monocytes (26). In cancer, monocytes were detected in lung tumors (27) and breast tumors (28) using scRNA-seq analysis.
Monocyte recruitment in steady-state tissues has also been evidenced in a few studies. Extravascular monocytes have been observed in lung from organ donors (29). Monocyte-derived cells have been described in non-diseased intestine, liver and skin (see below).
To conclude, there is ample evidence that, similarly to mice, human monocytes are recruited at steady state in tissues and mucosa to replenish the niche, and in acute and chronic inflammation. In these different contexts, monocytes will further differentiate.
Demonstrating the Monocytic Origin of Cells Isolated From Human Tissues
In mice, tracking monocyte fate from blood to tissues can be accomplished by adoptive transfer or genetic lineage tracing methods. These techniques are obviously not directly transposable to humans. When working with human samples, it is necessary to use alternative approaches to demonstrate the monocytic origin of tissue myeloid cells.
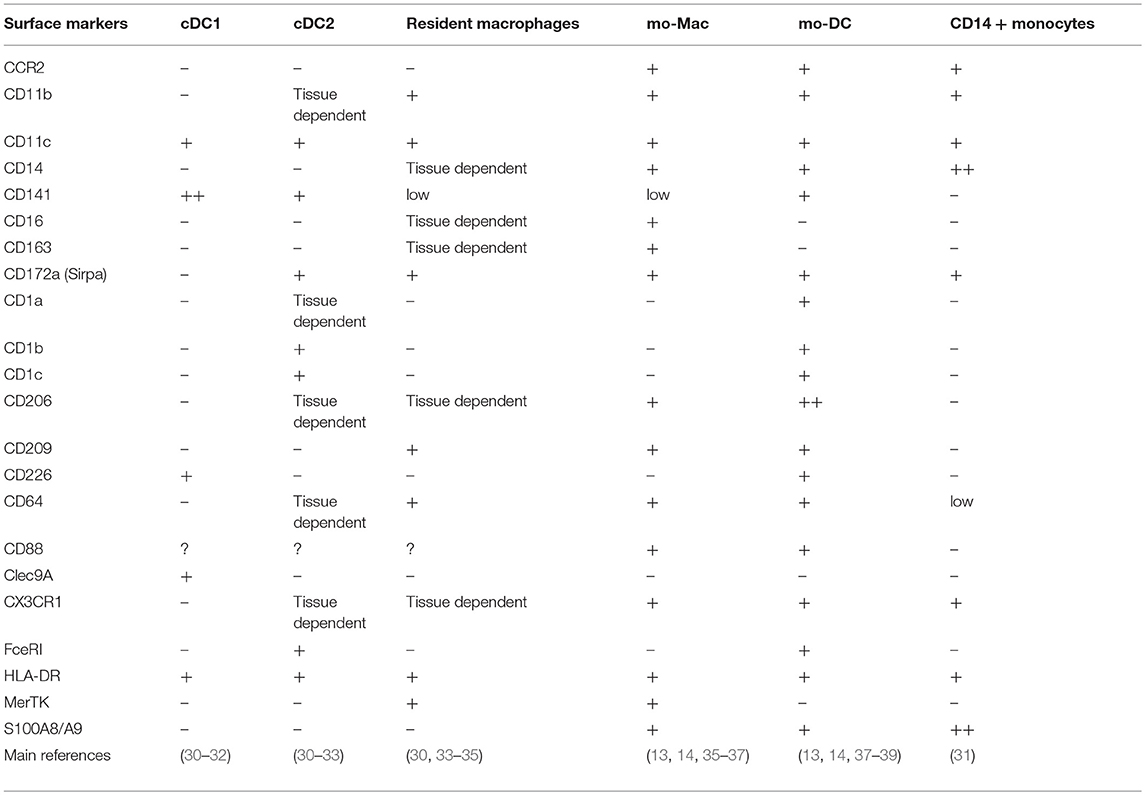
Phenotyping is the most widely used technique. It relies on the postulate that phenotypic markers expressed by blood monocytes will persist in tissues after their differentiation, allowing the identification of monocyte-derived cells. CD14, CCR2, and CX3CR1 are some of the most commonly used markers (Table 1). However, these molecules are not exclusive of monocyte-derived cells. CD14 is also expressed by tissue macrophages derived from embryonic precursors (40). CX3CR1 is highly expressed by microglia (1) and pre-DC (12). Finally, a population of pre-DC-derived CCR2+ DC has been described in mouse intestine (41).
Recently, S100A8/9 (calprotectin, an intracellular protein) has been proposed as a reliable marker for monocytes and monocyte-derived cells (34). S100A8/9 was detected both at the mRNA and protein levels in circulating monocytes (35, 42, 43). In the intestine, S100A8/9 expression in DC inversely correlated with Flt3 expression and S100A8/9 was highly expressed in short-lived myeloid cells, but not in long-lived macrophages (34). Collectively, this evidence suggests that S100A8/9 can be used as a marker for monocyte-derived cells.
Phenotyping is an easy way to characterize cellular identity. However, most markers are not specific to monocytes and monocyte-derived cells, and are tissue-dependent. Analyzing a combination of markers can increase the robustness of this approach.
Chimerism in the context of transplantation is another method to demonstrate a monocytic origin. This approach is based on the fact that following transplantation, cells from the recipient will repopulate the transplanted tissue while long-lived cells remain of donor origin, resulting in cellular chimerism. Resident macrophages derived from embryonic precursors are self-maintaining (1). Studying the replacement of tissue resident macrophages by monocyte-derived cells can be performed by analyzing markers of donor-recipient mismatch. For example, cells derived from the recipient's monocytes were evidenced in transplanted heart using in situ hybridization for Y chromosome (36). Cells derived from the donor or the recipient have also been distinguished by HLA-mismatch in the intestine (34, 35).
This method is an elegant way to analyse monocyte recruitment and differentiation in human tissues. However, it is not broadly available due to restricted access to transplanted tissue samples.
Another procedure to analyse monocyte fate is labeling, ex vivo or in vivo. For example, autologous monocytes were radiolabelled ex vivo and re-injected to patients with IBD (20) or RA (21). Monocytes can also be labeled in vivo through injection of ultra small particle iron oxide (USPIO). Labeled cells were observed in brain lesions of MS patients (25). This method allows a direct tracking of monocytes. However, it remains unconventional as it requires very specific procedures and ethics agreements.
Specific gene expression signatures can also be used to infer developmental origin. This approach is based on the idea that ontogeny will leave a transcriptomic imprint in monocyte-derived cells. One method is to perform comparative transcriptomic analysis, including blood monocytes as a reference. As an example, intestinal CD103-SIRPa+ cells (44) or CD14+ cells (34) clustered with blood monocytes, suggesting that these populations are related. Another method is to use transcriptional signatures specific of mo-Mac and mo-DC for enrichment analysis. scRNA-seq data of macrophages found in human fibrotic lung was annotated with signatures from bulk RNA-seq data from mouse macrophages and DC (45). This analysis revealed that pro-fibrotic macrophages may originate from monocytes. Similarly, scRNA-seq data from human ascite myeloid cells was analyzed using gene signatures generated from bulk RNA-seq of tissue-derived and in vitro-generated mo-DC and mo-Mac (46).
This approach has the advantage of being unbiased and requires no prior knowledge of putative “marker genes.” However, it is essential to use robust transcriptomic signatures as a reference.
The monocytic origin of tissue macrophages and DC can be demonstrated using various techniques. As none of these methods can lead to definitive conclusions, it is necessary to combine them to provide strong evidence that the cells of interest derive from monocytes.
In vivo Differentiation Into MO-Mac and MO-DC
Human monocytes can be differentiated in vitro into mo-Mac or mo-DC in various culture conditions. However, do monocytes have the capacity to differentiate into both macrophages and DC in vivo in humans?
How to Distinguish mo-Mac From mo-DC?
Distinguishing DC from macrophages by phenotyping is challenging as they share a lot of markers. MerTK, CD68, CD163, and the transcription factor MAFB are considered robust markers of macrophages, while DC express CD1a, CD1b, FcεRI, and CD226 (Table 1). Other techniques can help confirming cell identity. Analyzing cell morphology is a robust method for this (14, 36, 37, 47). Macrophages are large cells containing many phagocytic vesicles. By contrast, DC are smaller and display dendrites on their surface. Finally, transcriptomic approaches can also be used to distinguish mo-Mac from mo-DC, for instance by performing enrichment analysis for transcriptional signatures specific of DC and macrophages (34, 44, 46, 48–50).
Identification of mo-Mac and mo-DC in Human Tissues
Many studies describe the presence of mo-Mac in different tissues at steady state. In the small intestine, two subsets of macrophages were replaced 3 weeks following transplantation by recipient cells (35), demonstrating that they derive from monocytes. Monocytes also participate in the replenishment of skin macrophages (33). Following allogeneic hematopoietic stem cell transplantation, CD14+ cells reappeared after 8 days in blood and after 12 days in normal skin. This sequential detection of CD14+ cells suggested that blood monocytes give rise to skin CD14+ macrophages (33). Monocyte differentiation into mo-Mac also occurs in tissues with lower self-renewal capacities. For example, mo-Mac were identified in the heart (36), lung (43), and liver (51).
Furthermore, mo-Mac have been described in different inflammatory settings. The increased presence of macrophages has been observed in the colon of IBD patients (52, 53). CCR2 expression on their surface suggested their monocytic origin. In the cantharidin-induced skin blister model, following monocyte recruitment, HLADR+CD14+CD16+ cells increase their expression of CD163 suggesting that they differentiate into mo-Mac (15). In dialysis-induced peritonitis, CCR2+ mo-Mac are increased in the peritoneal cavity as compared to normal dialysis (14). Finally, mo-Mac are detected in tumors. In glioma patients, mo-Mac were identified in the brain by CX3CR1 expression and transcriptomic analysis (49). In melanoma, scRNAseq analysis evidenced one population expressing both macrophage and monocyte genes suggesting the presence of mo-Mac (48).
Similar findings apply to mo-DC. At steady state, mo-DC are mainly described in the intestine. Intestinal SIRPa+CD103- DCs (44) and SIRPa+CD103-CD14+ DCs (34) are transcriptionally related to blood monocytes. Moreover, S100A8/9 expression suggested that this population derives from monocytes (34). The presence mo-DC expressing CCR2 and S100A8/9 has also been suggested in non-diseased lung (54). In CD, CD14+CD163-MerTK- cells from inflamed gut exhibited a typical DC morphology and scRNA-seq showed signatures of monocyte lineage, suggesting that these cells are mo-DC (47). CCR2+ DC were also evidenced in the peritoneum at steady-state (12).
There is also evidence of monocyte differentiation into mo-DC in an inflammatory context. In atopic dermatitis and psoriasis, early studies have suggested monocyte differentiation into mo-DC (55–57). An increased proportion of “inflammatory” DC was found in atopic dermatitis and psoriasis patients in comparison to healthy skin (55). Their phenotype (CD1a+ FceRI+ CD206+) is reminiscent of that of mo-DC identified in subsequent studies. Similarly, DC displaying a phenotype consistent with mo-DC were observed in pleural effusions of tuberculosis patients (58) and evidenced in colorectal and breast tumors (38, 50). In lung cancer, phenotypic analysis as well as transcriptomic signatures in scRNAseq data suggested the presence of mo-DC (27, 38, 59). Using gene signatures, peritoneal ascite DC from ovarian cancer patients were also identified as mo-DC (46).
Collectively, these observations relying on phenotypic, morphological and transcriptomic analysis support the notion that human monocytes can differentiate in vivo into both macrophages and DC.
Functional Properties of MO-Mac and MO-DC
Classical DC and tissue resident macrophages play major roles in the initiation and resolution of immune responses. Do the DC and macrophages derived from monocytes display the same functions as their classical counterparts, or have specific functional properties?
Secretion of Soluble Mediators
A major property of myeloid cells is the secretion of soluble mediators. Mo-DC and mo-Mac have been reported as strong producers of pro-inflammatory cytokines such as TNFα and IL1β. Mo-Mac from healthy intestine or from the colon of CD and UC patients secreted higher levels of TNFα as compared to their tissue resident counterparts, with or without ex vivo restimulation (35, 52, 60). Mo-DC from the inflamed colon of CD patients produced high levels of IL1β (36, 50). Peritoneal ascites mo-DC and mo-Mac also secrete high levels of TNFα and IL1β (37). Furthermore, heart mo-Mac are potent producers of IL1β in contrast to CCR2- tissue resident macrophages (36).
IL23 secretion is more specific to mo-DC. Mo-DC from the inflamed intestine of CD patients or from peritoneal ascites of cancer patients secreted significant levels of IL23 with (37) or without ex vivo restimulation (47). Similar results were obtained with mo-DC from pleural effusions of tuberculosis patients restimulated ex vivo with Mycobacterium tuberculosis (58). Although IL23 seems to be mainly produced by mo-DC, it has also been reported for mo-Mac from CD patients (52). By contrast, IL12 is produced by mo-DC but not mo-Mac (46).
Finally, a few studies reported the production of the anti-inflammatory cytokine IL-10 by mo-Mac. In the context of CD, mo-Mac from inflamed colon secreted high levels of IL-10 with (52) or without restimulation (60). Il10 mRNA levels were upregulated in mo-Mac from CD patients after 24 h of culture without any stimulatory signal (36). Finally, mo-Mac from glioma were enriched for Il10 expression (49).
Taken together, these findings indicate that mo-DC and mo-Mac are strong producers of pro-inflammatory cytokines, which are essential for the recruitment of immune cells at an injured site and for the initiation of immune responses. However, in chronic inflammation, cytokine secretion by mo-DC and mo-Mac is exacerbated and contributes to the pathogenesis.
Fibrosis
Macrophages are key actors in wound healing and tissue repair by secreting growth factors for fibroblasts. The dysregulation of this mechanism can lead to fibrosis (61). There is evidence that mo-Mac participate in fibrosis. CX3CR1+ mo-Mac expressing pro-fibrotic Platelet Derived Growth Factor AA (PDGFAA) accumulated in fibrotic regions of lungs in comparison with non-fibrotic regions (45). Mo-Mac from cardiomyopathic heart expressed genes coding for growth factors and extracellular matrix components known to be involved in fibrosis (36). Moreover, these mo-Mac accumulated in scar or fibrotic tissues regions.
CD8 T Cell Responses
Few studies have investigated the role of monocyte-derived cells in CD8 T cell responses. Both mo-DC and mo-Mac isolated from peritoneal ascites were able to cross-present antigens using a non-conventional intracellular pathway (46), consistent with another study using peritoneal mo-Mac and mo-DC from peritoneal dialysis (14). Of note, the ability to cross-present was specific of mo-Mac as compared to lymphoid organ macrophages (62). However, only mo-DC could provide costimulatory signals for the differentiation of effector cytotoxic CD8 T cells (46).
CD4 T Cell Responses
One of the major roles of classical DC is to orient CD4 T cell polarization. Several studies have shown that mo-DC, but not mo-Mac, have the same property. Mo-DC isolated from synovial fluid of RA patients were better activators of CD4 T cell proliferation than mo-Mac from the same environment and induced Th17 polarization (37, 63). Th17 polarization was also induced by mo-DC from pleural effusions of tuberculosis patients (58) and from the inflamed colon of CD patients (47). Of note, mo-DC from synovial fluid of RA patients and from pleural effusions of tuberculosis patients were able to induce the proliferation of autologous CD4 T cells, showing that they can present antigens that were captured in vivo (58, 63). This Th17 polarization was associated with high secretion of IL-23 which is known to promote Th17 cells (37, 47, 58). In other studies, mo-DC from healthy small intestine or inflamed mucosa of CD patients preferentially induced Th1 polarization over Th17 (44, 52). This IFNγ production contributed to the pathogenesis of CD (52).
Finally, mo-DC from synovial fluid of RA patients and from peritoneal ascites induced CXCL13 secretion by CD4 T cells, suggesting Tfh polarization (64).
Collectively, these observations underline the capacity of mo-DC to polarize naïve CD4 T cells. In particular, Th17 polarization could contribute to the maintenance of chronic inflammation in Th17-driven pathologies such as RA and IBD.
In conclusion, mo-DC and mo-Mac share with their classical counterparts some of their hallmark functions (T cell stimulation for DC and tissue repair for macrophages). Ontogeny seems to influence mostly cytokine secretion, with mo-DC and mo-Mac being stronger producers of pro-inflammatory mediators than classical DC or resident macrophages from the same tissues. More studies using side-by-side comparisons will be needed to confirm this mixed functional profile.
Conclusion
Despite methodological limitations inherent to human samples, numerous studies support the notion that human monocytes can differentiate in vivo into DC or macrophages. This process occurs at steady state to replenish the niche, but can also play a major role in the initiation and maintenance of chronic inflammatory diseases.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was funded by INSERM and Agence Nationale de la Recherche (ANR-10-LABX-0043 and ANR-17-CE15-0011-01).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Ginhoux F, Guilliams M. tissue-resident macrophage ontogeny and homeostasis. Immunity. (2016) 44:439–49. doi: 10.1016/j.immuni.2016.02.024
2. Jakubzick CV, Randolph GJ, Henson PM. Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol. (2017) 17:349–62. doi: 10.1038/nri.2017.28
3. Getts DR, Terry RL, Getts MT, Deffrasnes C, Müller M, van Vreden C, et al. Therapeutic inflammatory monocyte modulation using immune-modifying microparticles. Sci Transl Med. (2014) 6:219ra7. doi: 10.1126/scitranslmed.3007563
4. Leuschner F, Dutta P, Gorbatov R, Novobrantseva TI, Donahoe JS, Courties G, et al. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat Biotechnol. (2011) 29:1005–10. doi: 10.1038/nbt.1989
5. Godec J, Tan Y, Liberzon A, Tamayo P, Bhattacharya S, Butte AJ, et al. Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity. (2016) 44:194–206. doi: 10.1016/j.immuni.2015.12.006
6. Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA. (2013) 110:3507–12. doi: 10.1073/pnas.1222878110
7. Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci USA. (2015) 112:1167–72. doi: 10.1073/pnas.1401965111
8. Tak T, Drylewicz J, Conemans L, de Boer RJ, Koenderman L, Borghans JAM, et al. Circulatory and maturation kinetics of human monocyte subsets in vivo. Blood. (2017) 130:1474–7. doi: 10.1182/blood-2017-03-771261
9. Patel AA, Zhang Y, Fullerton JN, Boelen L, Rongvaux A, Maini AA, et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med. (2017) 214:1913–23. doi: 10.1084/jem.20170355
10. Rogacev KS, Zawada AM, Hundsdorfer J, Achenbach M, Held G, Fliser D, et al. Immunosuppression and monocyte subsets. Nephrol Dial Transplant. (2015) 30:143–53. doi: 10.1093/ndt/gfu315
11. Thaler B, Hohensinner PJ, Krychtiuk KA, Matzneller P, Koller L, Brekalo M, et al. Differential in vivo activation of monocyte subsets during low-grade inflammation through experimental endotoxemia in humans. Sci Rep. (2016) 6:30162. doi: 10.1038/srep30162
12. Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. (2017) 356:eaah4573. doi: 10.1126/science.aah4573
13. Goudot C, Coillard A, Villani A-C, Gueguen P, Cros A, Sarkizova S, et al. Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages. Immunity. (2017) 47:582–596.e6. doi: 10.1016/j.immuni.2017.08.016
14. Liao C-T, Andrews R, Wallace LE, Khan MWA, Kift-Morgan A, Topley N, et al. Peritoneal macrophage heterogeneity is associated with different peritoneal dialysis outcomes. Kidney Int. (2017) 91:1088–103. doi: 10.1016/j.kint.2016.10.030
15. Jenner W, Motwani M, Veighey K, Newson J, Audzevich T, Nicolaou A, et al. Characterisation of leukocytes in a human skin blister model of acute inflammation and resolution. PLoS ONE. (2014) 9:e89375. doi: 10.1371/journal.pone.0089375
16. Eguíluz-Gracia I, Bosco A, Dollner R, Melum GR, Lexberg MH, Jones AC, et al. Rapid recruitment of CD14(+) monocytes in experimentally induced allergic rhinitis in human subjects. J Allergy Clin Immunol. (2016) 137:1872–81.e12. doi: 10.1016/j.jaci.2015.11.025
17. Jardine L, Wiscombe S, Reynolds G, McDonald D, Fuller A, Green K, et al. Lipopolysaccharide inhalation recruits monocytes and dendritic cell subsets to the alveolar airspace. Nat Commun. (2019) 10:1999. doi: 10.1038/s41467-019-09913-4
18. van der Laan AM, Ter Horst EN, Delewi R, Begieneman MPV, Krijnen PAJ, Hirsch A, et al. Monocyte subset accumulation in the human heart following acute myocardial infarction and the role of the spleen as monocyte reservoir. Eur Heart J. (2014) 35:376–85. doi: 10.1093/eurheartj/eht331
19. Nagelkerke SQ, Bruggeman CW, den Haan JMM, Mul EPJ, van den Berg TK, van Bruggen R, et al. Red pulp macrophages in the human spleen are a distinct cell population with a unique expression of Fc-\gamma receptors. Blood Adv. (2018) 2:941–53. doi: 10.1182/bloodadvances.2017015008
20. Grimm MC, Pullman WE, Bennett GM, Sullivan PJ, Pavli P, Doe WF. Direct evidence of monocyte recruitment to inflammatory bowel disease mucosa. J Gastroenterol Hepatol. (1995) 10:387–95. doi: 10.1111/j.1440-1746.1995.tb01589.x
21. Thurlings RM, Wijbrandts CA, Bennink RJ, Dohmen SE, Voermans C, Wouters D, et al. Monocyte scintigraphy in rheumatoid arthritis: the dynamics of monocyte migration in immune-mediated inflammatory disease. PLoS ONE. (2009) 4:e7865. doi: 10.1371/journal.pone.0007865
22. Baba N, Van VQ, Wakahara K, Rubio M, Fortin G, Panzini B, et al. CD47 fusion protein targets CD172a+ cells in Crohn's disease and dampens the production of IL-1β and TNF. J Exp Med. (2013) 210:1251–63. doi: 10.1084/jem.20122037
23. Jones G-R, Bain CC, Fenton TM, Kelly A, Brown SL, Ivens AC, et al. Dynamics of colon monocyte and macrophage activation during colitis. Front Immunol. (2018) 9:2764. doi: 10.3389/fimmu.2018.02764
24. Thiesen S, Janciauskiene S, Uronen-Hansson H, Agace W, Högerkorp C-M, Spee P, et al. CD14(hi)HLA-DR(dim) macrophages, with a resemblance to classical blood monocytes, dominate inflamed mucosa in Crohn's disease. J Leukoc Biol. (2014) 95:531–41. doi: 10.1189/jlb.0113021
25. Dousset V, Brochet B, Deloire MSA, Lagoarde L, Barroso B, Caille J-M, et al. MR imaging of relapsing multiple sclerosis patients using ultra-small-particle iron oxide and compared with gadolinium. Am J Neuroradiol. (2006) 27:1000–5.
26. Jeong JH, Hong S, Kwon OC, Ghang B, Hwang I, Kim Y-G, et al. CD14+ cells with the phenotype of infiltrated monocytes consist of distinct populations characterized by anti-inflammatory as well as pro-inflammatory activity in gouty arthritis. Front Immunol. (2017) 8:1260. doi: 10.3389/fimmu.2017.01260
27. Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. (2019) doi: 10.1016/j.immuni.2019.03.009
28. Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. (2018) 174:1293–308.e36. doi: 10.1016/j.cell.2018.05.060
29. Desch AN, Gibbings SL, Goyal R, Kolde R, Bednarek J, Bruno T, et al. Flow cytometric analysis of mononuclear phagocytes in nondiseased human lung and lung-draining lymph nodes. Am J Respir Crit Care Med. (2016) 193:614–26. doi: 10.1164/rccm.201507-1376OC
30. Segura E, Valladeau-Guilemond J, Donnadieu M-H, Sastre-Garau X, Soumelis V, Amigorena S. Characterization of resident and migratory dendritic cells in human lymph nodes. J Exp Med. (2012) 209:653–660. doi: 10.1084/jem.20111457
31. Alcántara-Hernández M, Leylek R, Wagar LE, Engleman EG, Keler T, Marinkovich MP, et al. High-dimensional phenotypic mapping of human dendritic cells reveals interindividual variation and tissue specialization. Immunity. (2017) 47:1037–50.e6. doi: 10.1016/j.immuni.2017.11.001
32. Heidkamp GF, Sander J, Lehmann CHK, Heger L, Eissing N, Baranska A, et al. Human lymphoid organ dendritic cell identity is predominantly dictated by ontogeny, not tissue microenvironment. Sci Immunol. (2016) 1:eaai7677. doi: 10.1126/sciimmunol.aai7677
33. McGovern N, Schlitzer A, Gunawan M, Jardine L, Shin A, Poyner E, et al. Human dermal CD14+ cells are a transient population of monocyte-derived macrophages. Immunity. (2014) 41:465–77. doi: 10.1016/j.immuni.2014.08.006
34. Richter L, Landsverk OJB, Atlasy N, Bujko A, Yaqub S, Horneland R, et al. Transcriptional profiling reveals monocyte-related macrophages phenotypically resembling DC in human intestine. Mucosal Immunol. (2018) 11:1512–23. doi: 10.1038/s41385-018-0060-1
35. Bujko A, Atlasy N, Landsverk OJB, Richter L, Yaqub S, Horneland R, et al. Transcriptional and functional profiling defines human small intestinal macrophage subsets. J Exp Med. (2018) 215:441–58. doi: 10.1084/jem.20170057
36. Bajpai G, Schneider C, Wong N, Bredemeyer A, Hulsmans M, Nahrendorf M, et al. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat Med. (2018) 24:1234–45. doi: 10.1038/s41591-018-0059-x
37. Segura E, Touzot M, Bohineust A, Cappuccio A, Chiocchia G, Hosmalin A, et al. Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity. (2013) 38:336–48. doi: 10.1016/j.immuni.2012.10.018
38. Laoui D, Keirsse J, Morias Y, Van Overmeire E, Geeraerts X, Elkrim Y, et al. The tumour microenvironment harbours ontogenically distinct dendritic cell populations with opposing effects on tumour immunity. Nat Commun. (2016) 7:13720. doi: 10.1038/ncomms13720
39. Baharom F, Thomas S, Rankin G, Lepzien R, Pourazar J, Behndig AF, et al. Dendritic cells and monocytes with distinct inflammatory responses reside in lung mucosa of healthy humans. J Immunol. (2016) 196:4498–509. doi: 10.4049/jimmunol.1600071
40. Bain CC, Scott CL, Uronen-Hansson H, Gudjonsson S, Jansson O, Grip O, et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. (2013) 6:498–510. doi: 10.1038/mi.2012.89
41. Scott CL, Bain CC, Wright PB, Sichien D, Kotarsky K, Persson EK, et al. CCR2(+)CD103(-) intestinal dendritic cells develop from DC-committed precursors and induce interleukin-17 production by T cells. Mucosal Immunol. (2015) 8:327–39. doi: 10.1038/mi.2014.70
42. Eguíluz-Gracia I, Malmstrom K, Dheyauldeen SA, Lohi J, Sajantila A, Aaløkken R, et al. Monocytes accumulate in the airways of children with fatal asthma. Clin Exp Allergy. (2018) 48:1631–9. doi: 10.1111/cea.13265
43. Eguíluz-Gracia I, Schultz HHL, Sikkeland LIB, Danilova E, Holm AM, Pronk CJH, et al. Long-term persistence of human donor alveolar macrophages in lung transplant recipients. Thorax. (2016) 71:1006–11. doi: 10.1136/thoraxjnl-2016-208292
44. Watchmaker PB, Lahl K, Lee M, Baumjohann D, Morton J, Kim SJ, et al. Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat Immunol. (2014) 15:98–108. doi: 10.1038/ni.2768
45. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. (2019) 20:163–72. doi: 10.1038/s41590-018-0276-y
46. Tang-Huau T-L, Gueguen P, Goudot C, Durand M, Bohec M, Baulande S, et al. Human in vivo-generated monocyte-derived dendritic cells and macrophages cross-present antigens through a vacuolar pathway. Nat Commun. (2018) 9:2570. doi: 10.1038/s41467-018-04985-0
47. Chapuy L, Bsat M, Sarkizova S, Rubio M, Therrien A, Wassef E, et al. Two distinct colonic CD14+ subsets characterized by single-cell RNA profiling in Crohn's disease. Mucosal Immunol. (2019) 12:703–19. doi: 10.1038/s41385-018-0126-0
48. Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. (2018) 175:998–1013.e20. doi: 10.1016/j.cell.2018.10.038
49. Müller S, Kohanbash G, Liu SJ, Alvarado B, Carrera D, Bhaduri A, et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. (2017) 18:234. doi: 10.1186/s13059-017-1362-4
50. Michea P, Noël F, Zakine E, Czerwinska U, Sirven P, Abouzid O, et al. Adjustment of dendritic cells to the breast-cancer microenvironment is subset specific. Nat Immunol. (2018) 19:885–97. doi: 10.1038/s41590-018-0145-8
51. MacParland SA, Liu JC, Ma X-Z, Innes BT, Bartczak AM, Gage BK, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. (2018) 9:4383. doi: 10.1038/s41467-018-06318-7
52. Kamada N, Hisamatsu T, Okamoto S, Chinen H, Kobayashi T, Sato T, et al. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J Clin Invest. (2008) 118:2269–80. doi: 10.1172/JCI34610
53. Bernardo D, Marin AC, Fernández-Tomé S, Montalban-Arques A, Carrasco A, Tristán E, et al. Human intestinal pro-inflammatory CD11chighCCR2+CX3CR1+ macrophages, but not their tolerogenic CD11c-CCR2-CX3CR1- counterparts, are expanded in inflammatory bowel disease. Mucosal Immunol. (2018) 11:1114–26. doi: 10.1038/s41385-018-0030-7
54. Patel VI, Booth JL, Duggan ES, Cate S, White VL, Hutchings D, et al. Transcriptional classification and functional characterization of human airway macrophage and dendritic cell subsets. J Immunol. (2017) 198:1183–201. doi: 10.4049/jimmunol.1600777
55. Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, Whynot J, Novitskaya I, Cardinale I, et al. Major differences in inflammatory dendritic cells and their products distinguish atopic dermatitis from psoriasis. J Allergy Clin Immunol. (2007) 119:1210–7. doi: 10.1016/j.jaci.2007.03.006
56. Zaba LC, Fuentes-Duculan J, Eungdamrong NJ, Abello MV, Novitskaya I, Pierson KC, et al. Psoriasis is characterized by accumulation of immunostimulatory and Th1/Th17 cell-polarizing myeloid dendritic cells. J Invest Dermatol. (2009) 129:79–88. doi: 10.1038/jid.2008.194
57. Wollenberg A, Kraft S, Hanau D, Bieber T. Immunomorphological and ultrastructural characterization of Langerhans cells and a novel, inflammatory dendritic epidermal cell. (IDEC) population in lesional skin of atopic eczema. J Invest Dermatol. (1996) 106:446–53. doi: 10.1111/1523-1747.ep12343596
58. Liu Y, Wang R, Jiang J, Cao Z, Zhai F, Sun W, et al. A subset of CD1c+ dendritic cells is increased in patients with tuberculosis and promotes Th17 cell polarization. Tuberculosis. (2018) 113:189–99. doi: 10.1016/j.tube.2018.10.007
59. Lavin Y, Kobayashi S, Leader A, Amir E-AD, Elefant N, Bigenwald C, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. (2017) 169:750–65.e17. doi: 10.1016/j.cell.2017.04.014
60. González-Domínguez É, Samaniego R, Flores-Sevilla JL, Campos-Campos SF, Gómez-Campos G, Salas A, et al. CD163L1 and CLEC5A discriminate subsets of human resident and inflammatory macrophages in vivo. J Leukoc Biol. (2015) 98:453–66. doi: 10.1189/jlb.3HI1114-531R
61. Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. (2016) 44:450–62. doi: 10.1016/j.immuni.2016.02.015
62. Segura E, Durand M, Amigorena S. Similar antigen cross-presentation capacity and phagocytic functions in all freshly isolated human lymphoid organ-resident dendritic cells. J Exp Med. (2013) 210:1035–47. doi: 10.1084/jem.20121103
63. Moret FM, Hack CE, van der Wurff-Jacobs KMG, de Jager W, Radstake TRDJ, Lafeber FPJG, et al. Intra-articular CD1c-expressing myeloid dendritic cells from rheumatoid arthritis patients express a unique set of T cell-attracting chemokines and spontaneously induce Th1, Th17 and Th2 cell activity. Arthritis Res Ther. (2013) 15:R155. doi: 10.1186/ar4338
Keywords: human, monocyte, macrophage, dendritic cell (DC), inflammation
Citation: Coillard A and Segura E (2019) In vivo Differentiation of Human Monocytes. Front. Immunol. 10:1907. doi: 10.3389/fimmu.2019.01907
Received: 03 May 2019; Accepted: 29 July 2019;
Published: 13 August 2019.
Edited by:
Florent Ginhoux, Singapore Immunology Network (A*STAR), SingaporeReviewed by:
Angel L. Corbi, Spanish National Research Council (CSIC), SpainFalk Nimmerjahn, University of Erlangen Nuremberg, Germany
Copyright © 2019 Coillard and Segura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elodie Segura, ZWxvZGllLnNlZ3VyYSYjeDAwMDQwO2N1cmllLmZy
 Alice Coillard
Alice Coillard Elodie Segura
Elodie Segura