- 1Immunogenetics Laboratory, Shenzhen Blood Center, Shenzhen, China
- 2School of Ophthalmology and Optometry, Shenzhen Eye Hospital, Shenzhen University, Shenzhen, China
- 3Basic Science Program, Frederick National Laboratory for Cancer Research, Frederick, MD, United States
- 4Department of Hematology, Shenzhen Second People's Hospital, Shenzhen, China
- 5Central Laboratory, Baoan Maternal and Child Health Hospital, Shenzhen, China
- 6Research and Development Department, Shenzhen Hank Bioengineering Institute, Shenzhen, China
- 7Ragon Institute of MGH MIT and Harvard, Cambridge, MA, United States
- 8Division of Biomedical Informatics and Personalized Medicine, Department of Microbiology and Immunology, University of Colorado Anschutz Medical Campus, Aurora, CO, United States
Interactions of human natural killer (NK) cell inhibitory receptors with polymorphic HLA-A, -B and -C molecules educate NK cells for immune surveillance against tumor cells. The KIR A haplotype encodes a distinctive set of HLA-specific NK cell inhibiting receptors having strong influence on immunity. We observed higher frequency of KIR A homozygosity among 745 healthy Chinese Southern Han than 836 adult patients representing three types of leukemia: ALL (OR = 0.68, 95% CI = 0.52–0.89, p = 0.004), AML (OR = 0.76, 95% CI = 0.59–0.98, p = 0.034), and CML (OR = 0.72 95% CI = 0.51–1.0, ns). We observed the same trend for NHL (OR = 0.47 95% CI = 0.26–0.88 p = 0.017). For ALL, the protective effect of the KIR AA genotype was greater in the presence of KIR ligands C1 (Pc = 0.01) and Bw4 (Pc = 0.001), which are tightly linked in East Asians. By contrast, the C2 ligand strengthened protection from CML (Pc = 0.004). NK cells isolated from KIR AA individuals were significantly more cytotoxic toward leukemic cells than those from other KIR genotypes (p < 0.0001). These data suggest KIR allotypes encoded by East Asian KIR A haplotypes are strongly inhibitory, arming NK cells to respond to leukemogenic cells having altered HLA expression. Thus, the study of populations with distinct KIR and HLA distributions enlightens understanding of immune mechanisms that significantly impact leukemia pathogenesis.
Introduction
Natural killer (NK) cells can detect and eliminate leukemia cells (1). To perform this function, NK cells express multiple inhibitory and activating receptors specific for the HLA class I proteins that are expressed on tissue cells (2–4). Polymorphic inhibitory receptors educate NK cells, enabling them to kill tumors or infected cells having altered or reduced HLA class I expression. In turn, the activating receptors can complement this function by identifying foreign or neo-antigens (5–7). Study of NK cell receptor polymorphism is thus critical for understanding the anti-tumor effects of NK cells, and to facilitate their subsequent manipulation for use in immunotherapy (8).
Killer cell immunoglobulin-like receptors (KIR) are the most polymorphic of the human NK cell receptors, and are characterized by extreme variation in gene number (9). The KIR locus comprises up to 13 functional genes, KIR2DL1-5, KIR2DS1-5, KIR3DL1/S1, and KIR3DL2-3, where those designated with an “L” suffix encode inhibitory receptors and those with an “S” suffix encode activating receptors. Previous population, transplantation, and disease association studies have identified two functionally distinct groups of KIR haplotypes that are present in all populations (10, 11). KIR A haplotypes express four inhibitory receptors specific for polymorphic HLA class I (KIR2DL1, KIR2DL3, KIR3DL1, KIR3DL2), plus zero or one activating receptor (KIR2DS4) also specific for HLA class I. Multiple distinct KIR B haplotypes express fewer inhibitory receptors but more activating receptors than the KIR A haplotypes (12).
Although previous examinations of the role of KIR diversity in leukemia control have yielded potentially conflicting results (13), a common feature spanning multiple different populations and leukemia subtypes (14–19) is they are consistent with the KIR A haplotype being more frequent in healthy controls than in the patient groups. Because AA homozygous individuals possess all four inhibitory receptors and few activating receptors specific for HLA class I, their NK cells may therefore be strongly educated to detect any changes in HLA expression that can occur on leukemia cells (20). However, both the frequency of the KIR A haplotype, and the abundance of KIR ligands are highly variable across human populations (21, 22). In this context, examination of specific well-defined human populations is critical for understanding the role of KIR in leukemia control. Here we interrogate such effects in three leukemia subtypes sampled in a large cohort from Southern China, where the KIR A haplotype is very frequent.
Materials and Methods
We studied 836 adult ALL, AML or CML patients, 225 pediatric leukemia patients and 745 healthy adult blood donors. We also studied 46 adult NHL patients, all having diffuse large B-cell lymphoma. The mean age of the adult patients analyzed was 31.4 years, controls 32.7 years, and pediatric cases 11.2 years. The patients were recruited from the hematopoietic stem cell transplantation (HSCT) program in the Shenzhen Blood Center, from August 1999 through June 2015. Leukemia diagnosis followed the French-American-British (FAB) classification, based on morphological observations. CML patients were not included if they had elected to take TKI (Imatinib) therapy instead of transplant. The CML patients included were from a study published in China (23). The controls were all unrelated healthy blood donors from Shenzhen Blood Center. All subjects are self-identified as Chinese Southern Han. Written informed consent was obtained from all study participants, and from the parents of all pediatric leukemia patients. The cohorts are described in Supplemental Figure 1.
KIR and HLA Genotyping
The presence or absence of all 13 functional KIR genes and two pseudogenes was determined from genomic DNA using the PCR-SSP based KIR Ready Gene kit (Inno-Train Diagnostik GmbH, Kronberg Im Taunus, Germany). It is difficult to distinguish KIR AB heterozygotes from KIR BB homozygotes, but the KIR AA genotype is readily identifiable (9, 24). Thus, individuals were designated as homozygous for the KIR A haplotype if they tested positive for KIR2DL1, KIR2DL3, KIR2DL4, KIR2DS4, KIR3DL1, KIR3DL2, KIR3DL3, KIR2DP1, KIR3DP1, and no other KIR. KIR2DL3 and KIR3DL1/S1 alleles were determined by Sanger sequencing from a subset of 306 controls, as described (25–27). HLA class I genes were sequenced as previously described (28), and assigned genotypes at two field (distinct polypeptide sequence) resolution using the ASSIGN4.7 software (Conexio Genomics, Applecross, Australia).
Statistical Analysis
For comparison of KIR AA frequencies, data from other populations were obtained from Allelefrequencies.net (21). The database was accessed in August 2018 and all populations having KIR AA genotype frequency data were used (N = 154). The populations were divided into major geographic origins as indicated (Amerindian, East Asian, European, Oceanian, sub-Saharan African) (21). The frequencies of KIR AA genotypes were compared across these major population groups using ANOVA, and, when comparing two populations, using unpaired t-tests for comparison of means. The tests were performed using GraphPad software (https://www.graphpad.com/). Odds ratio and confidence intervals were calculated using MedCalc (https://www.medcalc.org/calc/). Where appropriate, the analyses were corrected for multiple comparisons using the Bonferroni method.
NK Cell Cytotoxicity Assays
A total of 15 healthy Chinese Southern Han subjects (six of them with KIR AA genotype) were recruited from Shenzhen Blood Center. PBMC were separated by density gradient from 10 ml of peripheral blood and cultured for 13–14 days using the HANK cell in vitro preparation kit (Hank Bioengineering, Shenzhen, China). This process uses irradiated feeder cells expressing IL15 to promote NK cell expansion as described (29) for use in immunotherapy (30, 31), and we use it here to produce NK cells for in vitro assay. The expansion process increases the number and activity of NK cells but does not affect KIR expression (29, 32, 33). After this process, the mean purity of CD56+CD3− cells was 86 ± 4.5%. Antibodies used were: UCHT1 mouse anti-human CD3-FITC and B159 mouse anti-human CD56-APC (BD Biosciences, CA, USA). Target cells (1 × 106) were incubated for 10 min at 37°C in 1 mL PBS containing 2.5 μmol/L CFSE and then washed in PBS. Target cells were K562, which is a leukemia derived cell line that lacks HLA class I expression, and seven primary leukemic blasts from ALL and AML patients. NK cells and target cells were combined at an effector-target-ratio of 20:1, and incubated in RPMI-1640 complete medium for 4 h at 37°C with 5% CO2. Dead cells were stained using 7-AAD (BD Biosciences, CA, USA) for 15 min as described (34) and measured using a BD ACCURI C6 flow cytometer (BD Biosciences, CA, USA). Spontaneous lysis of target cells was measured by including a control without NK cells, and this value was subtracted from the total percentage of specific target cell lysis. All experiments were performed in duplicate.
Detection of CD107a Expression
A total of 18 healthy Chinese Southern Han individuals (11 of them with KIR AA genotype) were recruited from Shenzhen Blood Center. NK cells were isolated as above and incubated with K562 cells at a ratio of 5:1 for 5 h at 37°C. Optimal expression of CD107a on the cell surface of degranulating NK cells occurs between 4~6 h post stimulation (35). After the first 1 h, 4 μL monensin (BD Biosciences, CA, USA) was added to each 6 mL culture. The cells were then incubated with 2.5 uL human Fc block (BD Biosciences, CA, USA) at 25°C for 15 min, then stained with CD56, CD3, and CD107a mAbs. Antibodies used were: UCHT1 mouse anti-human CD3-FITC, H4A3 mouse anti-human CD107a-PECy5, and B159 mouse anti-human CD56-APC (all BD Biosciences, CA, USA). Flow cytometric analysis was performed using an ACCURI C6 machine (BD Biosciences, CA, USA) Surface expression of CD107a was assessed in CD56+CD3− cells (36). To detect spontaneous degranulation, a control sample without target cells was included in every experiment and this information was used for background subtraction of the results.
Results
KIR A Homozygosity Protects Chinese Han From Leukemia and Non-Hodgkin's Lymphoma
The KIR A haplotype is found in all human populations to varying frequency and is associated with NK cell function in protection from infectious diseases and in susceptibility to preeclampsia (10, 37). Because NK cells also function in tumor control, we tested whether KIR A affects the probability of developing leukemia. We first demonstrated that the frequency of individuals who are homozygous for the KIR A haplotype is significantly greater in East Asians than in other populations (p < 0.0001: Figure 1). We therefore examined a Chinese Southern Han cohort. In this cohort, the frequency of the KIR AA genotype is significantly higher among healthy individuals (55.3%) than adult leukemia patients (47.1%: OR = 0.72, 95% CI = 0.59–0.88, p = 0.0012) or pediatric leukemia patients (44.9%: OR = 0.6, 95% CI = 0.48–0.89, p = 0.006) (Figure 2A). When analyzed by disease subgroup, the KIR AA genotype was more frequent in healthy controls than in ALL, AML or CML patients (Figure 2B). For adult ALL (OR = 0.68, 95% CI = 0.52–0.89, p = 0.004) and AML (OR = 0.76, 95% CI = 0.59–0.98, p = 0.034), and pediatric ALL (OR = 0.64, 95% CI = 0.45–0.92, p = 0.017) the differences are statistically significant (Figure 2B). Similar trends were observed for CML and pediatric AML but did not reach statistical significance (p = 0.052 and p = 0.1, respectively), likely due to smaller sample sizes. We also analyzed non-Hodgkin's lymphoma (NHL) and again observed a protective effect of the KIR AA genotype (OR = 0.47, 95% CI = 0.26–0.88, p = 0.017), which was statistically significant in spite of the small sample size (N = 46: Figure 2A). The consistent reduction of frequency in all the disease groups analyzed strongly suggests Chinese Southern Han individuals having the KIR AA genotype are protected from developing leukemia or lymphoma.
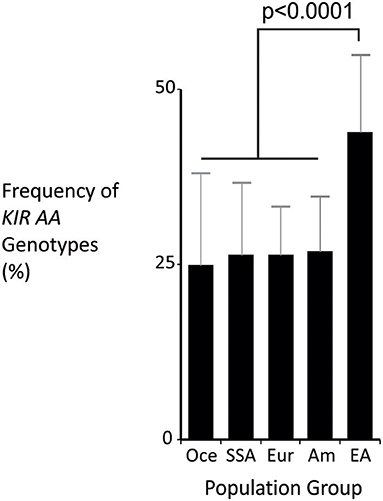
Figure 1. The KIR AA genotype is most frequent in East Asia. Mean frequencies of the KIR AA genotype from multiple populations representing major world groups: Oce—Oceanian (13 populations), SSA –sub-Saharan African (13 populations), Eur—European (60 populations), Am—Amerindian (24 populations), EA—East Asian (31 populations). Data were obtained from allelefrequencies.net (21). ANOVA test for between group differences p = 2.1 × 10−16. The p-value shown is from t-test of comparison of means: East Asian is significantly different to every other group, with p < 0.001 for each comparison. Vertical bars are S.D.
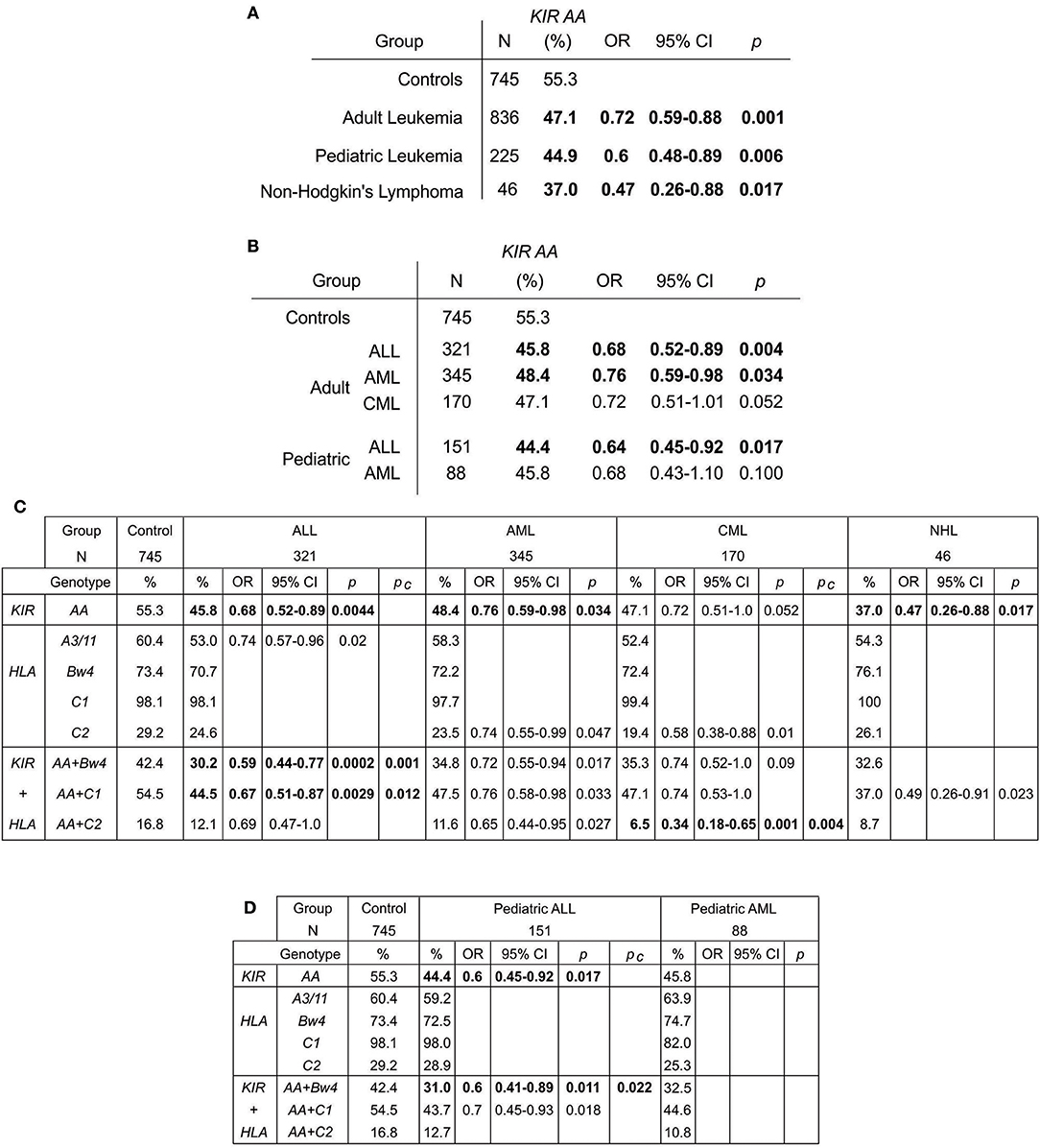
Figure 2. The KIR AA genotype associates with protection against leukemia. (A,B) Odds ratios and statistical significance obtained by comparing the KIR AA genotype frequencies of healthy controls with (A) adult and pediatric leukemia cases and non-Hodgkin's lymphoma, and (B) with the subtypes of leukemia. Statistically significant values are indicated in bold. (C). Genotype frequencies, odds ratios and statistical significance obtained by comparing healthy controls (left), with three leukemia types, ALL, AML and CML (center), and NHL (right). The genotypes are: KIR AA genotype (top), the four KIR ligands (center) and KIR AA genotype in combination with each ligand (bottom). Only those results having 95% CI that do not encompass 1 are shown. Pc–Bonferroni correction factors used were x4 for ligands and x3 for ligand+KIR AA. Bold indicates p-value remains significant after this correction. (D) Shows the equivalent values obtained for pediatric ALL and AML.
In KIR A Homozygotes, KIR Ligands Differentially Protect From Leukemia Subtypes
To further explore our findings, we analyzed the HLA class I ligands for KIR. HLA class I allele frequencies can vary significantly between the ethnic groups and geographic regions of China, for example between Northern and Southern Han (38, 39). Validating the equivalent compositions of our disease and control subjects, there were no significant differences in HLA-A, -B or -C allele or haplotype frequency spectra between controls and the full patient group (Supplemental Figures 2A,B) or from previously published frequencies of HLA class I alleles of Chinese Southern Han (40). Neither was there a difference between groups in the frequencies of the −21M variant of HLA-B (Supplemental Figure 2C), which can influence NK cell education (41).
KIR bind differentially to specific amino acid motifs present on non-overlapping subsets of HLA class I variants (allotypes). The HLA allotypes are HLA-A*03 and -A*11 (the A3/11 motif: KIR3DL2 ligands), HLA-A and -B allotypes that contain the Bw4 motif (KIR3DL1 ligands), HLA-B and -C allotypes containing the C1 motif (primarily KIR2DL2/3 ligands), and HLA-C allotypes containing the C2 motif (primarily KIR2DL1 ligands). We observed trends for reduction of A3/11 frequency in the adult ALL patients (60.4 vs. 53%, p = 0.02: Figure 2C) and reduction of C2 frequency in the adult AML (29.2 vs. 23.5%, p = 0.047) and CML (29.2 vs. 19.4%, p = 0.01) patients relative to controls. However, these findings did not remain significant after accounting for multiple testing by correction for the four ligands analyzed (Figure 2C). By contrast, combinatorial analysis of KIR and their HLA class I ligands revealed clear distinctions between the three leukemia subsets (Figures 2C,D). The KIR AA plus Bw4 compound genotype was more frequent in healthy individuals (42.4%) than in subjects with ALL (30.2% adult, p = 0.0002, and 31.0% pediatric p = 0.011) or AML (34.8% adult, p = 0.017 and 32.5% pediatric, ns). Similarly, the KIR AA plus C1 compound genotype was significantly more frequent amongst the healthy controls (54.5%) than either the ALL (44.5% adult, p = 0.003 and 43.7% pediatric, p = 0.018) or AML (47.5% adult, p = 0.033 and 44.6% pediatric, ns) patients. For adult ALL, the strength of the effect was consistently greater when considering KIR in combination with HLA class I than when considering KIR genotype alone. These associations remained significant after correction for multiple testing (Pc = 0.001 Bw4+HLA, and Pc = 0.012 C1+HLA: Figure 2C). A reduced frequency of each of the three KIR/ligand combinations was observed in AML and NHL patients, but neither association remained significant after correction, suggesting there is likely limited improvement in protection when considering ligand over that offered by KIR alone.
Analysis of representative world populations revealed distinctively strong linkage disequilibrium between Bw4+HLA-B and C1+HLA-C in East Asians (Figure 3A). HLA-B*46, which is the only frequent HLA-B allele that expresses a C1 motif, is characteristic to East Asia, common in the Chinese Southern Han (~15%), and is linked to C1+HLA-C (21, 40). Also in the Chinese Southern Han, all KIR A haplotypes express KIR2DL3*001, which is an inhibitory receptor that interacts with C1+HLA-C (26). KIR2DL3*001 also predominates in Japanese, another East Asian population (44). Similarly, the majority of East Asian KIR A haplotypes (>70%) express either KIR3DL1*001 or KIR3DL1*015, which are strongly-inhibitory KIR3DL1 allotypes specific for Bw4+HLA (44). Of note, the non-expressed KIR3DL1*004 allele that is common in Europeans (25) is absent from East Asians (44–46). Thus, in addition to the high frequency of KIR A haplotypes, a given Chinese Southern Han KIR A haplotype is more than twice as likely to express KIR3DL1*001 or *015 than a European KIR A haplotype (Figure 3B). Taken together, these observations indicate that Chinese Southern Han having the KIR AA genotype are more likely to have multiple interactions between strongly inhibitory KIR and their corresponding HLA ligands than are European individuals, who are more commonly studied. Thus, these strongly inhibitory KIR/HLA ligand interactions may be important factors in the NK cell control of ALL and to a lesser degree AML, in Chinese Southern Han.
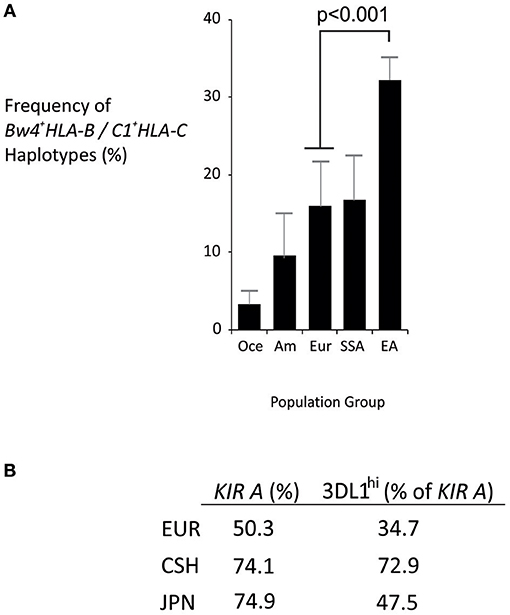
Figure 3. Distinctive HLA and KIR haplotypes in Chinese Southern Han. (A) Mean frequencies of Bw4+HLA-B/C1+HLA-C haplotypes from multiple populations representing major world groups, Oce—Oceanian (3 populations), Am—Amerindian (4 populations), Eur—European (4 populations), SSA—sub-Saharan African (6 populations), EA—East Asian (8 populations). Data were obtained from Solberg et al. (42) and haplotype frequencies calculated using the Expectation Maximization algorithm (43). Vertical bars are S.D. (B) Frequency of KIR A haplotypes in European (EUR), Chinese Southern Han (CSH) and Japanese (JPN) populations, and the percentage of KIR A haplotypes that express either KIR3DL1*001 or KIR3DL1*015 (KIR3DL1hi).
In contrast to ALL and AML, the strength of protection from developing CML offered by the KIR AA genotype was enhanced by the C2 ligand (OR = 0.34, 95% CI = 0.18–0.65, Pc = 0.004) to a much greater degree than by the C1/Bw4 ligands (Figure 2C). Although the sample size of the CML group is relatively small, the strength of the effect provides compelling evidence that Chinese Southern Han individuals having the KIR AA plus C2+HLA-C genotype are less likely to develop CML than those who do not have this compound genotype. KIR2DL1, which is encoded by the KIR A haplotype, has high specificity for C2+HLA-C, but the strength of inhibition resulting from the interaction varies across KIR2DL1 subtypes (47, 48). In East Asians, only two KIR2DL1 subtypes predominate: strongly inhibitory KIR2DL1*003 encoded by KIR A haplotypes, and weakly inhibitory KIR2DL1*004 encoded by KIR B haplotypes (44) (and ZD, PJN unpublished). NK cells expressing KIR2DL1*003 are highly effective at killing cells that have reduced C2+HLA-C expression, whereas those expressing KIR2DL1*004 are significantly weaker in this regard (47, 48). Thus, individuals who have the KIR AA genotype in this population likely have two genomic copies of the strongly inhibiting KIR2DL1 allotype, KIR2DL1*003, and should be best equipped at recognizing and eliminating tumor cells having aberrant C2+HLA-C expression. This could take the form of reduction or loss of HLA expression (49, 50) or presentation of tumor neoantigen peptides by the C2+HLA-C molecules, abrogating interaction with KIR2DL1 (5, 51). This result thus identifies a role for strongly functional KIR2DL1 allotypes in protection from CML in Chinese Han. This finding strikes strong parallel with several reproductive diseases including preeclampsia, where the KIR AA genotype in combination with C2 ligand impacts the risk of developing the disease (37), and highlights the need for study of populations in addition to Europeans for understanding mechanisms of protection from disease (52).
In summary, the KIR AA genotype confers differential protection against ALL, AML, CML and NHL; whereas the C2 ligand enhances KIR AA protection against CML, the C1 and Bw4 ligands enhance KIR AA protection against ALL, and the KIR AA genotype alone protects from AML and NHL.
NK Cells From KIR A Homozygous Individuals Are Strongly Cytotoxic to Leukemia Cells
To investigate whether individuals who are KIR AA+ have stronger responses to leukemia than other individuals do, we measured the cytotoxicity against leukemia cells, of NK cells from healthy individuals with and without KIR AA. We used standard assays for NK cell activity (30, 31, 35, 36) to study six healthy Chinese Southern Han subjects having the KIR AA genotype, and nine without it. NK cells from individuals with the KIR AA genotype demonstrated significantly greater cytotoxicity against the HLA class I negative K562 leukemia cell line, a standard target used to measure NK cell cytotoxicity, than NK cells isolated from individuals without the AA genotype (mean killing 65 vs. 49%, p = 0.002; Figure 4A). These data suggest that NK cells from KIR AA individuals are better educated to respond to loss of HLA class I expression than are those from individuals carrying other KIR genotypes.
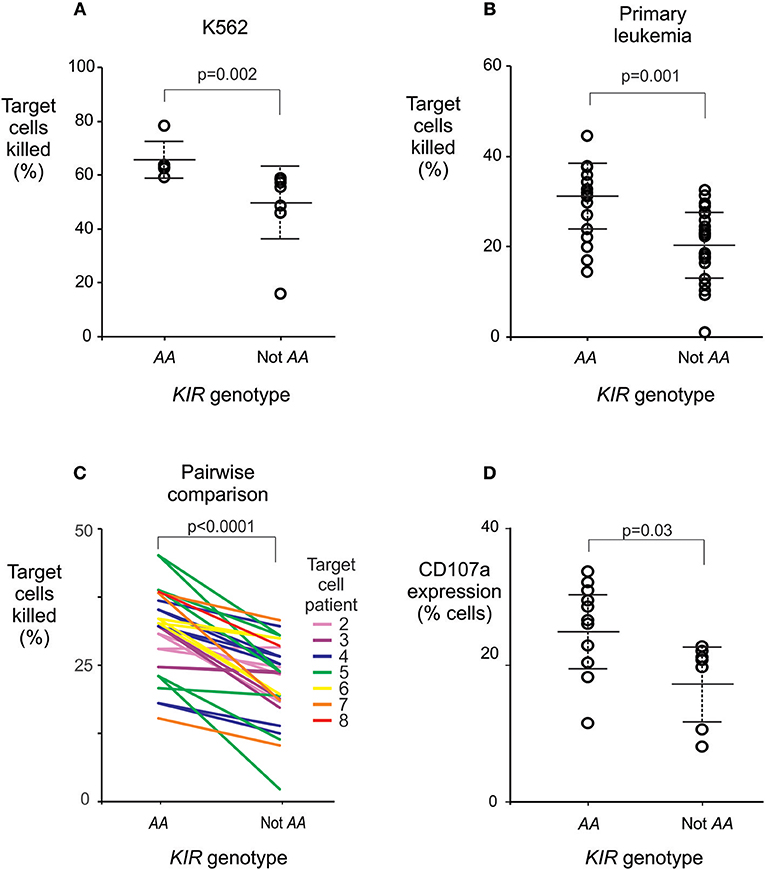
Figure 4. The KIR AA genotype correlates with enhanced potential of NK cells to lyse leukemic cells. (A) Mean % killing of K562 target cells by NK cells isolated from six healthy individuals with (left) and nine without (right) the KIR AA genotype. Each dot represents the mean result from a duplicated assay (Supplemental Figure 3E). P-value is from a Mann-Whitney U-test. (B) Mean % killing of primary leukemia cells using the same NK cells as (A). The primary leukemia targets were obtained from two ALL patients and five AML patients (Supplemental Figure 3). Each dot represents the mean result from a duplicated assay (Supplemental Figure 3). P-value is from a Mann-Whitney U-test. Six NK cell donors were tested against two targets, seven were tested against three targets, and two against four targets, as detailed in Supplemental Figure 3. (C) Pairwise comparisons of % killing by NK cells from KIR AA vs. non AA individuals against identical target leukemia cells. Each line represents one of the seven targets, incubated with NK cells from an AA+ donor (left) or an AA− donor (right), at the same time and conditions. Line colors indicate the seven individual targets as given in Supplemental Figure 3 (Target 1 is K562; not shown for clarity). P-value is from a paired t-test. (D) Shows CD107a expression by NK cells following incubation with K562 targets. The NK cells were isolated from a further 11 healthy individuals with (left) and seven without (right) the KIR AA genotype. P-value is from a Mann-Whitney U-test. Error bars are mean ± sd. The NK killing assays are described in Supplemental Figure 3.
Next, the same NK cells were tested for their cytotoxicity against primary leukemic cells, which were isolated from five patients with AML and two with ALL (Supplemental Figure 3). Again, NK cells isolated from individuals who have the KIR AA genotype had a stronger cytotoxic response than those isolated from non-AA individuals (29.9 vs. 20.5%, p = 0.001; Figure 4B). In line with previous observations on European subjects (53), we observed variability in the baseline killing of leukemia cells across NK cell donors. This may be due to differential education by donor HLA class I allotypes. Although all the donors and patients studied possess the C1 ligand (Supplemental Figure 3), the very high number of subjects required precludes from completely controlling for HLA variation. Nevertheless, a higher degree of cytotoxicity was consistently observed for NK cells from KIR AA+ donors as compared to KIR AA− donors regardless of the source of leukemia cell targets (p < 0.0001 by paired t-test; Figure 4C). This result was further validated in an independent set of 18 healthy individuals and a complementary method, which measured the cell surface expression of CD107a that accompanies degranulation of NK cells (35). Here, a mean of 25.4 % NK cells from KIRAA+ subjects expressed CD107a following contact with K562 cells, vs. 17.8% from non-AA subjects (p = 0.03: Figure 4D). In summary, these findings show that in Chinese Southern Han, NK cells from individuals with the KIR AA genotype exert greater cytotoxicity against leukemic cells than those from individuals having any other KIR genotype.
Discussion
KIR are highly polymorphic and the distribution of KIR haplotypes differs markedly across human populations (11). Because multiple previous studies were consistent with, but had not identified, a role for the KIR AA genotype in protection from leukemia, we studied a cohort of Chinese Southern Han from East Asia, where the KIR A haplotype is at highest frequency. We showed that possession of the KIR AA genotype protects individuals from developing either of three types of leukemia and one lymphoma analyzed. These results were further refined by considering the HLA ligands for KIR in the analysis. Importantly, in vitro functional assays demonstrated that NK cells from healthy individuals with KIR AA genotype had strong reactivity against leukemic cells.
Because there have been conflicting reports on the role of KIR in protection from leukemia, we sought to study a large number of patients and controls that were matched by clearly defined age and ethnicity. We studied over 1,000 patients and 745 controls and observed no significant difference in the overall frequency distribution of HLA class I alleles between the two groups or compared with previous studies of Chinese Southern Han (40). Because allele frequency spectra of these highly polymorphic genes characterize and distinguish human populations (42), this finding validates the self-defined ancestry of the cohorts as well as their suitability for use in these comparisons. We studied subjects from China, where the mean age for leukemia diagnosis is younger than that seen for Europeans (54–56), and compared them with healthy blood donors having a similar age distribution. The high frequency of KIR AA homozygous individuals observed in our control group is consistent with multiple previous studies of healthy Chinese Southern Han (19, 57–59). We also observed a difference in KIR AA frequency between AML patients and controls of similar magnitude as that seen in a previous study of Chinese subjects from Singapore (19). One caveat is that we studied patients selected as potential transplantation recipients, but we consider it unlikely that KIR/HLA genotype affects this initial decision. Moreover, our finding that possession of the KIR AA genotype consistently protects from at least three types of leukemia and one lymphoma was further validated by in vitro functional assays of NK cell cytotoxicity and lytic granule release.
Our results suggest that any contradictory findings in the association of KIR/HLA and leukemia that have been reported are likely to be driven by the relative population frequencies of the KIR A haplotypes and the KIR allotypes they encode. Many previous studies are concordant with the results presented here (14, 15, 17–19, 60), while others that differ could be explained by cryptic differences in ancestries of patients and controls, resulting in differences in KIR A haplotype frequencies, and the alleles present on those haplotypes (11, 26, 61–65). We hypothesize that the strongly inhibitory KIR allotypes that characterize East Asian KIR haplotypes efficiently educate NK cells to detect changes in HLA expression that can occur in leukemic cells (20, 44). These changes may include loss or reduction of HLA expression, or alterations to the peptide repertoire that may be detected by NK cells through KIR binding (5, 51). That the same strongly-inhibiting KIR allotypes are present in other populations, albeit to lower frequencies (11), suggests their effect on leukemia control will be similar, but much larger sample sizes will be required to detect them. We studied a population with relatively low genetic diversity, such that possession of the KIR A haplotype is accompanied by a uniform set of alleles of the component genes. High resolution analyses will likely be required to account for the substantial allelic diversity of any further population groups analyzed (66).
Although the KIR AA genotype appears to protect against leukemia and NHL in Chinese Southern Hans, there were differences in the HLA ligand required to strengthen this effect. These differences may be due to the distinct maturation stages and the non-overlapping subsets of lineage markers and fusion proteins expressed by the leukemia subtypes (67, 68). The clearest ligand effect was observed for CML, which is likely due to the relative uniformity of this leukemia class (69). The least defined ligand effect was observed for AML, likely due to the highly heterogeneous nature of this disease (70). Another ligand for inhibitory NK cell receptors is HLA-E, which can present peptide fragments derived from the expression of HLA-A, -B or -C. CD94/NKG2A educates and modulates NK cells through binding HLA-E, complementing the role of KIR and extending the capacity of NK cells to monitor HLA class I expression (41, 71). Although CD94/NKG2A is conserved, diversity across individuals occurs because only some HLA-B alleles encode the −21M variant peptide that binds to HLA-E. Accordingly, a recent study showed −21M HLA-B associated with increased survival for AML patients receiving immune therapy (72). However, as we observed no significant differences in HLA allele frequencies (Supplemental Figure 2), there were no significant differences in frequency of the HLA-B −21M variant between the patients and controls of our study. This indicates that HLA-B −21M polymorphism is unlikely to affect leukemia susceptibility in Chinese Southern Han.
Although the endpoint of our analysis was disease diagnosis, a limitation is that we were not able to perform functional tests of NK cells extracted from leukemia patients, or phenotypic analyses of HLA expression by leukemic cells. The exception is the K562 target, which is a leukemia cell known to lack HLA class I expression. Another limitation is that the relatively low sample numbers we obtained for the functional tests did not allow for thorough analysis of KIR ligand mismatches between NK cell donor and leukemia target cells. It will be important in future work to examine how changes in HLA expression or neo-antigen peptide presentation may affect the NK cell response to cells that could be leukemogenic, and ultimately how they may influence disease course. These kinds of experiments require pre-genotyping of large numbers of individuals, and will be greatly facilitated by involving populations where the respective genotypes are at high frequency, including the Chinese Southern Han.
In conclusion, this study enhances our understanding of the immune response to leukemia in the world's largest population group, shows the benefit of including specific well-defined populations in study of diseases involving highly polymorphic genes, and identifies a role for NK cell education by inhibitory KIR in control of leukemia. In light of recent studies that promote expansion beyond the usual European populations analyzed (73), the benefits of studying well-defined population groups is becoming increasingly apparent.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Ethics Statement
The study was approved by the ethics review board of Shenzhen Blood Center, Shenzhen, Guangdong, China. Written informed consent was obtained from all study participants.
Author Contributions
ZD, SC, JZha, WH, MC, and PN: experiment and analysis design. ZD, YQ, RC, JZhu, JZhe, MZ, GZ, LH, HZ, LL, and WZ: data generation and interpretation. ZD, YQ, QY, MM, XG, and PN: Data analysis. ZD, JZha, XG, MM, MC, and PN: Manuscript preparation. All authors: manuscript review and approval.
Conflict of Interest Statement
MZ is an employee of Shenzhen Hank Bioengineering Institute.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Number: 81373158). PN was supported by start-up funds from the University of Colorado and by NIH U01 AI090905 and U19 NS095774. YQ, MM, XG, and MC were supported by federal funds from the National Cancer Institute, National Institutes of Health (NIH) under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. YQ, MM, XG, and MC were also supported by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research. We thank the patients and marrow registry donors for generously providing blood samples for this study. Thanks to Lisbeth Guethlein and Jill Hollenbach for critical comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.01646/full#supplementary-material
References
1. Locatelli F, Pende D, Falco M, Della Chiesa M, Moretta A, Moretta L. NK cells mediate a crucial graft-versus-leukemia effect in haploidentical-HSCT to cure high-risk acute leukemia. Trends Immunol. (2018) 39:577–90. doi: 10.1016/j.it.2018.04.009
2. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. (2011) 331:44–9. doi: 10.1126/science.1198687
3. Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol. (2013) 31:227–58. doi: 10.1146/annurev-immunol-020711-075005
4. Saunders PM, Vivian JP, O'connor GM, Sullivan LC, Pymm P, Rossjohn J, et al. A bird's eye view of NK cell receptor interactions with their MHC class I ligands. Immunol Rev. (2015) 267:148–66. doi: 10.1111/imr.12319
5. Das J, Khakoo SI. NK cells: tuned by peptide? Immunol Rev. (2015) 267:214–27. doi: 10.1111/imr.12315
6. Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The broad spectrum of human natural killer cell diversity. Immunity. (2017) 47:820–33. doi: 10.1016/j.immuni.2017.10.008
7. Holzemer A, Garcia-Beltran WF, Altfeld M. Natural killer cell interactions with classical and non-classical human leukocyte antigen class I in HIV-1 infection. Front Immunol. (2017) 8:1496. doi: 10.3389/fimmu.2017.01496
8. Daher M, Rezvani K. Next generation natural killer cells for cancer immunotherapy: the promise of genetic engineering. Curr Opin Immunol. (2018) 51:146–53. doi: 10.1016/j.coi.2018.03.013
9. Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. (1997) 7:753–63. doi: 10.1016/S1074-7613(00)80394-5
10. Bashirova AA, Martin MP, Mcvicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet. (2006) 7:277–300. doi: 10.1146/annurev.genom.7.080505.115726
11. Beziat V, Hilton HG, Norman PJ, Traherne JA. Deciphering the killer-cell immunoglobulin-like receptor system at super-resolution for natural killer and T-cell biology. Immunology. (2017) 150:248–64. doi: 10.1111/imm.12684
12. Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. (2002) 190:40–52. doi: 10.1034/j.1600-065X.2002.19004.x
13. Augusto DG. The impact of KIR polymorphism on the risk of developing cancer: not as strong as imagined? Front Genet. (2016) 7:121. doi: 10.3389/fgene.2016.00121
14. Verheyden S, Bernier M, Demanet C. Identification of natural killer cell receptor phenotypes associated with leukemia. Leukemia. (2004) 18:2002–7. doi: 10.1038/sj.leu.2403525
15. Verheyden S, Demanet C. Susceptibility to myeloid and lymphoid leukemia is mediated by distinct inhibitory KIR-HLA ligand interactions. Leukemia. (2006) 20:1437–8. doi: 10.1038/sj.leu.2404279
16. Zhang Y, Wang B, Ye S, Liu S, Liu M, Shen C, et al. Killer cell immunoglobulin-like receptor gene polymorphisms in patients with leukemia: possible association with susceptibility to the disease. Leuk Res. (2010) 34:55–8. doi: 10.1016/j.leukres.2009.04.022
17. Vejbaesya S, Sae-Tam P, Khuhapinant A, Srinak D. Killer cell immunoglobulin-like receptors in Thai patients with leukemia and diffuse large B-cell lymphoma. Hum Immunol. (2014) 75:673–6. doi: 10.1016/j.humimm.2014.04.004
18. Misra MK, Prakash S, Moulik NR, Kumar A, Agrawal S. Genetic associations of killer immunoglobulin like receptors and class I human leukocyte antigens on childhood acute lymphoblastic leukemia among north Indians. Hum Immunol. (2016) 77:41–6. doi: 10.1016/j.humimm.2015.10.009
19. Shen M, Linn YC, Ren EC. KIR-HLA profiling shows presence of higher frequencies of strong inhibitory KIR-ligands among prognostically poor risk AML patients. Immunogenetics. (2016) 68:133–44. doi: 10.1007/s00251-015-0888-4
20. Reusing SB, Manser AR, Enczmann J, Mulder A, Claas FH, Carrington M, et al. Selective downregulation of HLA-C and HLA-E in childhood acute lymphoblastic leukaemia. Br J Haematol. (2016) 174:477–80. doi: 10.1111/bjh.13777
21. Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, Da Silva AL, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. (2015) 43:D784–8. doi: 10.1093/nar/gku1166
22. Guethlein LA, Norman PJ, Hilton HH, Parham P. Co-evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol Rev. (2015) 267:259–82. doi: 10.1111/imr.12326
23. Deng Z, Zhen J, Wang D, He L, Zou H. [Association of genetic polymorphisms of KIR-HLA system with chronic myeloid leukemia among ethnic Hans from southern China]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. (2017) 34:53–7. doi: 10.3760/cma.j.issn.1003-9406.2017.01.012
24. Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE. (2010) 5:e15115. doi: 10.1371/journal.pone.0015115
25. Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. (2007) 39:1092–9. doi: 10.1038/ng2111
26. Zhu FM, Jiang K, Lv QF, Zhang W, He J, Xu XG, et al. [Polymorphism of killer cell immunoglobulin-like receptor 2DL3 gene in Zhejiang Han population]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. (2007) 24:88–90. doi: 10.3760/j.issn:1003-9406.2007.01.021
27. Zhen J, He L, Xu Y, Zhao J, Yu Q, Zou H, et al. Allelic polymorphism of KIR2DL2/2DL3 in a southern Chinese population. Tissue Antigens. (2015) 86:362–7. doi: 10.1111/tan.12681
28. Zhen J, Wang D, He L, Zou H, Xu Y, Gao S, et al. Genetic profile of KIR and HLA in southern Chinese Han population. Hum Immunol. (2014) 75:59–64. doi: 10.1016/j.humimm.2013.09.006
29. Imai C, Iwamoto S, Campana D. Genetic modification of primary natural killer cells overcomes inhibitory signals and induces specific killing of leukemic cells. Blood. (2005) 106:376–83. doi: 10.1182/blood-2004-12-4797
30. Zhang M, Daniel S, Huang Y, Chancey C, Huang Q, Lei YF, et al. Anti-West Nile virus activity of in vitro expanded human primary natural killer cells. BMC Immunol. (2010) 11:3. doi: 10.1186/1471-2172-11-3
31. Qin Z, Chen J, Zeng J, Niu L, Xie S, Wang X, et al. Effect of NK cell immunotherapy on immune function in patients with hepatic carcinoma: a preliminary clinical study. Cancer Biol Ther. (2017) 18:323–30. doi: 10.1080/15384047.2017.1310346
32. Fujisaki H, Kakuda H, Shimasaki N, Imai C, Ma J, Lockey T, et al. Expansion of highly cytotoxic human natural killer cells for cancer cell therapy. Cancer Res. (2009) 69:4010–7. doi: 10.1158/0008-5472.CAN-08-3712
33. Romee R, Rosario M, Berrien-Elliott MM, Wagner JA, Jewell BA, Schappe T, et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci Transl Med. (2016) 8:357ra123. doi: 10.1126/scitranslmed.aaf2341
34. Schmid I, Krall WJ, Uittenbogaart CH, Braun J, Giorgi JV. Dead cell discrimination with 7-amino-actinomycin D in combination with dual color immunofluorescence in single laser flow cytometry. Cytometry. (1992) 13:204–8. doi: 10.1002/cyto.990130216
35. Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. (2004) 294:15–22. doi: 10.1016/j.jim.2004.08.008
36. Penack O, Gentilini C, Fischer L, Asemissen AM, Scheibenbogen C, Thiel E, et al. CD56dimCD16neg cells are responsible for natural cytotoxicity against tumor targets. Leukemia. (2005) 19:835–40. doi: 10.1038/sj.leu.2403704
37. Moffett A, Colucci F. Co-evolution of NK receptors and HLA ligands in humans is driven by reproduction. Immunol Rev. (2015) 267:283–97. doi: 10.1111/imr.12323
38. Xu S, Yin X, Li S, Jin W, Lou H, Yang L, et al. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet. (2009) 85:762–74. doi: 10.1016/j.ajhg.2009.10.015
39. Chen CH, Yang JH, Chiang CWK, Hsiung CN, Wu PE, Chang LC, et al. Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum Mol Genet. (2016) 25:5321–31. doi: 10.1093/hmg/ddw346
40. Trachtenberg E, Vinson M, Hayes E, Hsu YM, Houtchens K, Erlich H, et al. HLA class I (A, B, C) and class II (DRB1, DQA1, DQB1, DPB1) alleles and haplotypes in the Han from southern China. Tissue Antigens. (2007) 70:455–63. doi: 10.1111/j.1399-0039.2007.00932.x
41. Horowitz A, Djaoud Z, Nemat-Gorgani N, Blokhuis J, Hilton HG, Beziat V, et al. Class I HLA haplotypes form two schools that educate NK cells in different ways. Sci Immunol. (2016) 1:eaag1672. doi: 10.1126/sciimmunol.aag1672
42. Solberg OD, Mack SJ, Lancaster AK, Single RM, Tsai Y, Sanchez-Mazas A, et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum Immunol. (2008) 69:443–64. doi: 10.1016/j.humimm.2008.05.001
43. Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. (2010) 10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x
44. Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. (2006) 203:633–45. doi: 10.1084/jem.20051884
45. Tao SD, He YM, Ying YL, He J, Zhu FM, Lv HJ. KIR3DL1 genetic diversity and phenotypic variation in the Chinese Han population. Genes Immun. (2014) 15:8–15. doi: 10.1038/gene.2013.55
46. Deng Z, Zhen J, Zhu B, Zhang G, Yu Q, Wang D, et al. Allelic diversity of KIR3DL1/3DS1 in a southern Chinese population. Hum Immunol. (2015) 76:663–6. doi: 10.1016/j.humimm.2015.09.017
47. Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N, et al. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. (2009) 114:5182–90. doi: 10.1182/blood-2009-07-231977
48. Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, et al. Polymorphic HLA-C receptors balance the functional characteristics of KIR haplotypes. J Immunol. (2015) 195:3160–70. doi: 10.4049/jimmunol.1501358
49. Yu J, Heller G, Chewning J, Kim S, Yokoyama WM, Hsu KC. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J Immunol. (2007) 179:5977–89. doi: 10.4049/jimmunol.179.9.5977
50. Joncker NT, Fernandez NC, Treiner E, Vivier E, Raulet DH. NK cell responsiveness is tuned commensurate with the number of inhibitory receptors for self-MHC class I: the rheostat model. J Immunol. (2009) 182:4572–80. doi: 10.4049/jimmunol.0803900
51. Sim MJ, Malaker SA, Khan A, Stowell JM, Shabanowitz J, Peterson ME, et al. Canonical and cross-reactive binding of NK cell inhibitory receptors to HLA-C allotypes is dictated by peptides bound to HLA-C. Front Immunol. (2017) 8:193. doi: 10.3389/fimmu.2017.00193
52. Nakimuli A, Chazara O, Hiby SE, Farrell L, Tukwasibwe S, Jayaraman J, et al. A KIR B centromeric region present in Africans but not Europeans protects pregnant women from pre-eclampsia. Proc Natl Acad Sci USA. (2015) 112:845–50. doi: 10.1073/pnas.1413453112
53. Fauriat C, Andersson S, Bjorklund AT, Carlsten M, Schaffer M, Bjorkstrom NK, et al. Estimation of the size of the alloreactive NK cell repertoire: studies in individuals homozygous for the group A KIR haplotype. J Immunol. (2008) 181:6010–9. doi: 10.4049/jimmunol.181.9.6010
54. Cma CMA. [Guideline for management of chronic myeloid leukemia (2013)]. Zhonghua Xue Ye Xue Za Zhi. (2013) 34:464–70. doi: 10.3760/cma.j.issn.0253-2727.2013.05.021
55. He XF, Wang QR, Cen JN, Qiu HY, Sun AN, Chen SN, et al. [EVI1 expression, clinical and cytogenetical characteristics in 447 patients with acute myeloid leukemia]. Zhonghua Xue Ye Xue Za Zhi. (2016) 37:936–41. doi: 10.3760/cma.j.issn.0253-2727.2016.11.002
56. Yu WJ, Wang Y, Xu LP, Zhang XH, Liu KY, Huang XJ. [Influence of donor-recipient sex matching on outcomes of haploidentical hematopoietic stem cell transplantation for acute leukemia]. Zhonghua Xue Ye Xue Za Zhi. (2018) 39:398–403. doi: 10.3760/cma.j.issn.0253-2727.2018.05.010
57. Jiang K, Zhu FM, Lv QF, Yan LX. Distribution of killer cell immunoglobulin-like receptor genes in the Chinese Han population. Tissue Antigens. (2005) 65:556–63. doi: 10.1111/j.1399-0039.2005.00412.x
58. Wu GQ, Zhao YM, Lai XY, Yang KL, Zhu FM, Zhang W, et al. Distribution of killer-cell immunoglobulin-like receptor genes in Eastern mainland Chinese Han and Taiwanese Han populations. Tissue Antigens. (2009) 74:499–507. doi: 10.1111/j.1399-0039.2009.01366.x
59. Shi L, Shi L, Tao Y, Lin K, Liu S, Yu L, et al. Distribution of killer cell immunoglobulin-like receptor genes and combinations with HLA-C ligands in an isolated Han population in southwest China. Tissue Antigens. (2011) 78:60–4. doi: 10.1111/j.1399-0039.2011.01683.x
60. Pamuk GE, Tozkir H, Uyanik MS, Gurkan H, Duymaz J, Pamuk ON. Natural killer cell killer immunoglobulin-like gene receptor polymorphisms in non-Hodgkin lymphoma: possible association with clinical course. Leuk Lymphoma. (2015) 56:2902–7. doi: 10.3109/10428194.2015.1014361
61. Middleton D, Diler AS, Meenagh A, Sleator C, Gourraud PA. Killer immunoglobulin-like receptors (KIR2DL2 and/or KIR2DS2) in presence of their ligand (HLA-C1 group) protect against chronic myeloid leukaemia. Tissue Antigens. (2009) 73:553–60. doi: 10.1111/j.1399-0039.2009.01235.x
62. Almalte Z, Samarani S, Iannello A, Debbeche O, Duval M, Infante-Rivard C, et al. Novel associations between activating killer-cell immunoglobulin-like receptor genes and childhood leukemia. Blood. (2011) 118:1323–8. doi: 10.1182/blood-2010-10-313791
63. Babor F, Manser AR, Fischer JC, Scherenschlich N, Enczmann J, Chazara O, et al. KIR ligand C2 is associated with increased susceptibility to childhood ALL and confers an elevated risk for late relapse. Blood. (2014) 124:2248–51. doi: 10.1182/blood-2014-05-572065
64. De Smith AJ, Walsh KM, Ladner MB, Zhang S, Xiao C, Cohen F, et al. The role of KIR genes and their cognate HLA class I ligands in childhood acute lymphoblastic leukemia. Blood. (2014) 123:2497–503. doi: 10.1182/blood-2013-11-540625
65. Stringaris K, Marin D, Barrett AJ, Hills R, Sobieski C, Cao K, et al. KIR gene haplotype: an independent predictor of clinical outcome in MDS patients. Blood. (2016) 128:2819–23. doi: 10.1182/blood-2016-05-713099
66. Norman PJ, Hollenbach JA, Nemat-Gorgani N, Marin WM, Norberg SJ, Ashouri E, et al. Defining KIR and HLA class I genotypes at highest resolution via high-throughput sequencing. Am J Hum Genet. (2016) 99:375–91. doi: 10.1016/j.ajhg.2016.06.023
67. Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the international microarray innovations in leukemia study group. J Clin Oncol. (2010) 28:2529–37. doi: 10.1200/JCO.2009.23.4732
68. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. (2016) 127:2391–405. doi: 10.1182/blood-2016-03-643544
69. Chereda B, Melo JV. Natural course and biology of CML. Ann Hematol. (2015) 94(Suppl. 2):S107–21. doi: 10.1007/s00277-015-2325-z
70. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. (2013) 368:2059–74. doi: 10.1056/NEJMoa1301689
71. Braud VM, Allan DS, O'callaghan CA, Soderstrom K, D'andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. (1998) 391:795–9. doi: 10.1038/35869
72. Hallner A, Bernson E, Hussein BA, Ewald Sander F, Brune M, Aurelius J, et al. The HLA-B−21 dimorphism impacts on NK cell education and clinical outcome of immunotherapy in acute myeloid leukemia. Blood. (2019) 133:1479–88. doi: 10.1182/blood-2018-09-874990
Keywords: leukemia, AML, ALL, CML, NHL, HLA, KIR, Chinese Southern Han
Citation: Deng Z, Zhao J, Cai S, Qi Y, Yu Q, Martin MP, Gao X, Chen R, Zhuo J, Zhen J, Zhang M, Zhang G, He L, Zou H, Lu L, Zhu W, Hong W, Carrington M and Norman PJ (2019) Natural Killer Cells Offer Differential Protection From Leukemia in Chinese Southern Han. Front. Immunol. 10:1646. doi: 10.3389/fimmu.2019.01646
Received: 13 April 2019; Accepted: 02 July 2019;
Published: 16 July 2019.
Edited by:
Alexander David Barrow, The University of Melbourne, AustraliaReviewed by:
Amir Horowitz, Icahn School of Medicine at Mount Sinai, United StatesNicole Bernard, McGill University, Canada
Copyright © 2019 Deng, Zhao, Cai, Qi, Yu, Martin, Gao, Chen, Zhuo, Zhen, Zhang, Zhang, He, Zou, Lu, Zhu, Hong, Carrington and Norman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul J. Norman, paul.norman@ucdenver.edu
†These authors have contributed equally to this work
 Zhihui Deng1†
Zhihui Deng1† Mary Carrington
Mary Carrington Paul J. Norman
Paul J. Norman