
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 28 May 2019
Sec. NK and Innate Lymphoid Cell Biology
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.01179
This article is part of the Research Topic In Memoriam of Professor Alessandro Moretta View all 9 articles
 Daniela Pende1†
Daniela Pende1† Michela Falco2†
Michela Falco2† Massimo Vitale1
Massimo Vitale1 Claudia Cantoni2,3
Claudia Cantoni2,3 Chiara Vitale1,4
Chiara Vitale1,4 Enrico Munari5
Enrico Munari5 Alice Bertaina6
Alice Bertaina6 Francesca Moretta7
Francesca Moretta7 Genny Del Zotto8
Genny Del Zotto8 Gabriella Pietra1,4
Gabriella Pietra1,4 Maria Cristina Mingari1,3
Maria Cristina Mingari1,3 Franco Locatelli9
Franco Locatelli9 Lorenzo Moretta10*
Lorenzo Moretta10*Natural killer (NK) cells contribute to the first line of defense against viruses and to the control of tumor growth and metastasis spread. The discovery of HLA class I specific inhibitory receptors, primarily of killer Ig-like receptors (KIRs), and of activating receptors has been fundamental to unravel NK cell function and the molecular mechanisms of tumor cell killing. Stemmed from the seminal discoveries in early ‘90s, in which Alessandro Moretta was the major actor, an extraordinary amount of research on KIR specificity, genetics, polymorphism, and repertoire has followed. These basic notions on NK cells and their receptors have been successfully translated to clinical applications, primarily to the haploidentical hematopoietic stem cell transplantation to cure otherwise fatal leukemia in patients with no HLA compatible donors. The finding that NK cells may express the PD-1 inhibitory checkpoint, particularly in cancer patients, may allow understanding how anti-PD-1 therapy could function also in case of HLA class Ineg tumors, usually susceptible to NK-mediated killing. This, together with the synergy of therapeutic anti-checkpoint monoclonal antibodies, including those directed against NKG2A or KIRs, emerging in recent or ongoing studies, opened new solid perspectives in cancer therapy.
The groundbreaking discoveries made by Alessandro Moretta left a very big footprint in Immunology, and, more generally, in Medicine (1, 2). Indeed, also now that “the music is over” (3) the Alessandro's legacy will remain, witnessed by the many human lives saved thanks to his seminal studies and his continuous efforts to translate discoveries to the benefit of patients with leukemia or solid tumors.
Since the beginning of his scientific career, Alessandro focused his research on two different issues that revealed to be fundamental for his future major accomplishments: on one side, the generation of monoclonal antibodies (mAbs) to functional surface molecules expressed by human T cells, and on the other side, the development of a highly efficient T cell cloning technique allowing clonal growth of virtually 100% T cells (4). While these early studies provided important information on co-receptor molecules inducing accessory signals for T cell activation and on the frequency and subset distribution of T cells endowed with given functional capabilities, they offered an invaluable tool for the subsequent development of techniques suitable for efficient cloning of natural killer (NK) cells. In turn, NK cell clones were fundamental for the discovery of both killer immunoglobulin-like receptors (KIRs) and natural cytotoxicity receptors (NCRs), the two most important steps toward unraveling NK cell function.
Notably, although NK cells were discovered in the mid ‘70s, the molecular mechanism(s) by which they discriminate between tumor and healthy cells remained obscure for long time. In late ‘80s, the “missing self-hypothesis,” proposed by Karre and Ljunggren, inspired the subsequent seminal discoveries in mice and humans in early ‘90s (5). The “missing self-hypothesis” stemmed from the observation that NK cells could kill a lymphoma cell line that had lost major histocompatibility (MHC) class I surface molecules, while the original MHC class Ipos were resistant to lysis. A conceivable interpretation was that NK cells are able to sense the absence of “self” MHC class I molecules on target cells. Indeed, MHC class I specific inhibitory receptors were discovered in parallel in humans and mice (6, 7). Surprisingly, human and mice receptors were found to belong to distinct molecular families, namely the Ig-superfamily and the lectin family, respectively. The inhibitory signals generated by the engagement with their MHC class I ligands resulted in NK cell inactivation. The prototypes of human leukocytes (HLA) class I specific receptors were two highly homologous molecules (p58.1 and p58.2) (8, 9), expressed by partially overlapping NK cell subsets. They were shown to recognize allotypic determinants shared by the two main groups of HLA-C alleles. While KIRs are expressed at late stages of NK cell differentiation, more immature NK cells express another important inhibitory receptor, the CD94:NKG2A heterodimer, specific for HLA-E (10). CD94:NKG2A is the first HLA class I specific receptor to be expressed during NK cell differentiation. At a further differentiation stage, it may be co-expressed with KIR(s), while it is lost by the most mature NK cells which express KIR(s) only (11). Notably, the pool of mature NK cells expresses at least one inhibitory receptor for self-HLA class I (whether KIR or NKG2A), thus avoiding autoreactivity. The very few NK cells lacking self-reactive receptors are anergic. This status is acquired during NK cell maturation by a process referred to as NK cell “licensing” or “education” (12, 13). Although HLA class I specific inhibitory receptors were fitting with the “missing self-hypothesis,” the understanding of the actual molecular mechanism(s) regulating NK cell function was incomplete, as the “on” signals inducing NK cell activation in the process of tumor cell lysis were unknown.
Again, the use of NK cell clones and the generation and selection of mAbs capable of modulating NK cell function allowed the identification of three novel activating receptors: NKp46 (14, 15), NKp44 (16, 17), and NKp30 (18), collectively named NCRs (19), playing a major role in tumor cell recognition and killing. In addition, Alessandro identified the function of other surface molecules acting as co-receptors, including p75/AIRM1, IRp60, 2B4, NTBA, and NKp80 (20–25), and could also determine the activating function of DNAM-1 (CD226) and its corresponding ligands (namely PVR and Nectin-2) (26). It became evident that NK cell activation is dependent on an array of receptors and co-receptors that interact with ligands overexpressed or expressed de novo on “stressed” cells and on tumor-transformed or virus-infected cells. While, in an autologous environment, healthy cells express HLA class I molecules that generate inhibitory signals via KIR or NKG2A, tumor- or virus-infected cells may display HLA down-regulation, allowing NK cell triggering via activating receptors and consequent target cell killing. In the case of viral infections that do not down-regulate HLA class I, the susceptibility to NK-mediated killing may be related to viral peptides that, upon binding to HLA molecules, could impair KIR engagement.
Altogether, these findings revealed that NK cell activation is under the control of inhibitory and activating receptors and their ligands on target cells, and thus receptor/ligand pairs could represent true checkpoints in the regulation of NK cell function (27). Notably, an important mechanism of tumor escape is the down-regulation of activating NK receptor expression, thus eluding the NK-mediated control of tumor growth and metastatic spread (28–30).
In humans, two main NK cell subsets were originally identified on the basis of the intensity of CD56 surface expression. The two subsets are differently distributed in blood and tissues: CD56dim are largely predominant in peripheral blood (PB), while CD56bright are much more abundant in tissues. CD56bright NK cells are relatively immature, express NKG2A and not KIR, are poorly cytolytic, secrete cytokines (primarily IFN-γ and TNF-α), and undergo intensive proliferation in response to IL-2 or IL-15. In contrast, CD56dim NK cells express NKG2A and/or KIR, are mature, display a strong cytolytic activity and cytokine secretion capability rapidly upon activation. Remarkably, on the basis of the surface expression of NKG2A and/or KIR, and other markers, CD56dim NK cells could be further subdivided in different subsets representative of distinct differentiation stages characterized by the progressive decrease of the proliferative capacity, paralleled by an increase of cytolytic activity (11, 31). The most mature, terminally differentiated, NK cells are KIRpos CD57pos CD16bright and may express the HLA-E specific activating receptor NKG2C. As recently revealed (also with the Alessandro's contribution), NKG2Cpos cells undergo expansion in CMV infections, displaying adaptive features and memory-like function (32–35).
During the last decade, cells belonging to the innate lymphoid cells (ILCs) were identified. They share with NK cells a common ID2pos lymphoid precursor. Absent or infrequent in PB of healthy individuals, they reside primarily in mucosal tissues, skin, and lymphoid organs (e.g., tonsils), where they participate to innate defense against pathogens and to tissue repair/regeneration (36–38). They are referred to as “helper” ILC, being non-cytolytic and producing typical sets of cytokines. While they will not be further discussed here, it is noteworthy that an important subset of ILC3 (the NCRpos ILC3) is characterized by the expression of NCR, the activating receptors originally described and characterized by Alessandro.
NK cells can migrate from blood to tissues or lymphoid organs. Their traffic is regulated by chemokines and their corresponding receptors, addressing different NK subsets to specific compartments or inflammatory sites. In addition, since CD34pos precursors, capable of differentiating toward NK cells, have been detected in tissues including liver (39), tonsils (40), thymus (41), and decidua (42), it is likely that some of the tissue resident NK cells may undergo differentiation from these precursors and, under the influence of specific tissue microenvironment, acquire unique functional properties.
While NK cells mediate a strong anti-tumor activity, their effectiveness may be greatly compromised by the suppressive microenvironment of different tumors. Suppression is mediated by a number of mechanisms, including release of soluble factors by tumor cells and by cells present in the microenvironment that have been attracted and/or “conditioned” by tumor cells. These cells include M2 macrophages, myeloid-derived suppressor cells (MDSC), T-reg and stromal cells (30). In addition, hypoxia, frequently occurring in tumor lesions, also contributes to the inhibition of immune effector cells. In the case of NK cells, the principal effect is the down-regulation of the activating NK receptors, thus rendering NK cells “disarmed” and unable to recognize specific ligands on tumor cells. Another related tumor-induced mechanism playing an important role in tumor escape is the expression of inhibitory checkpoints, primarily PD-1, in NK (and T) cells, and of the corresponding ligands in tumor cells (PD-L1 and PD-L2). The PD-1/PD-L1 interaction has been shown to induce NK cell inactivation (43). Notably, PD-1 expression in NK cells has been reported and functionally analyzed by Alessandro and coworkers (44). PD-1 will be briefly discussed, in the frame of novel immune-based strategies that appear very promising in tumor therapy. The important role of NK cells and their receptors in defense against tumors and leukemia is also witnessed by the great success achieved in the T-depleted, haploidentical hematopoietic stem cell transplantation (haplo-HSCT) setting to cure high-risk leukemia. The unthinkable benefit of this therapeutic approach is primarily related to the graft-vs.-leukemia effect of donor NK cells, arising from grafted stem cells and/or infused with the graft.
Thus, in less than 3 decades, a cell of the innate immunity that was substantially underscored (45) became an important tool to successfully treat otherwise incurable leukemia. Many of these major therapeutic achievements are based on seminal Alessandro's discoveries illustrated in this review.
While the main focus of our applied researches has been the involvement of KIRs in defense against tumors and leukemia, it is important to underline that KIRs and their polymorphisms have been shown to play a relevant role also in inflammation, infection, autoimmune diseases, and reproduction (46–48).
In late ‘80s, Alessandro Moretta came to Genoa from Lausanne with a “very special cell line”: the hybridoma producing GL183 mAb. Through this mAb he identified a novel 58-kD surface molecule expressed on a subset of NK cells and capable of modulating NK cell cytolytic function (8). The paper describing this molecule was published in 1990 and this was the beginning of a new exciting era that led to the discovery of various HLA class I specific receptors, using the same approach based on mice immunization with NK cell clones and on the hybridoma technology. Soon thereafter, by immunizing mice with a GL183neg NK cell clone, EB6 mAb was selected for its ability to react with a molecule distinct from GL183 but sharing most of its characteristics, as revealed by phenotypic, biochemical, and functional studies (9). Notably, the reactivity of the two mAbs allowed the identification of four NK cell subsets that could be extremely variable in terms of relative percentages among different individuals. Both mAbs could inhibit the cytolytic activity of selected NK clones (expressing either GL183 or EB6) in redirected killing assays, using the FcγRpos murine P815 target cell line. Interestingly, a striking correlation existed between the EB6pos GL183neg phenotype of NK cell clones and their ability to lyse normal allogeneic cells, such as PHA-blasts, in a previously defined type of alloreactivity (i.e., stimulator/responder combination in mixed lymphocyte reaction, MLR), termed “1 anti-A” or “group 1” (49–51). In the same years the seminal study by Ljunggren and Karre (an elaboration of the Karre's PhD thesis in early ‘80s) was published, formulating the “missing self” hypothesis (5). In humans, p58 molecules appeared to fit with this proposal, rendering NK cells capable of self/non-self discrimination. Then a close relationship could be determined between the expression of EB6- and GL183-reactive molecules, termed p58.1 and p58.2, respectively, and their specificity for different HLA-C alleles (52–54). Evidence was provided that HLA-C*03 and HLA-C*04 conferred selective protection from GL183pos and from EB6pos NK clones respectively (52, 55). These two specificities were extended to a series of HLA-C alleles related to HLA-C*03, characterized by Ser77-Asn80, and those related to HLA-C*04, characterized by Asn77-Lys80 (56). These two distinct groups of HLA-C alleles encompass virtually all the expressed HLA-C molecules. Thus, EB6pos GL183neg “group 1” NK clones could lyse allogeneic cells expressing only HLA-C alleles with Ser77-Asn80 motif, while EB6neg GL183pos “group 2” NK clones were alloreactive against target cells expressing only HLA-C alleles with Asn77-Lys80 motif. Importantly, treatment of NK clones with mAbs masking p58 molecules resulted in the killing of HLA-C protected cells, showing that this inhibitory receptor/ligand interaction was responsible for the blocking of target cell lysis (55). Consistent with these notions, NK clones co-expressing p58.1 and p58.2 molecules were inhibited by the interaction with any allogeneic target cells, thus being unable of any type of alloreactivity (57). In the same years, NKB1 was described as a 70 kDa glycoprotein, whose expression was detected by DX9 mAb, and DX9pos NK clones recognized a group of HLA-B alleles carrying the Bw4 public epitope (58, 59). Thus, analysis of NK cell clones, derived not only from different donors but also from single donors, allowed the detection of the great NK cell heterogeneity with extremely variable patterns of expression and co-expression of the various receptors with different HLA specificities. In 1992, studying in different donors the NK cell repertoire related to the ability to lyse allogeneic cells, five distinct allospecificities were described (60). While “group 1” and “group 2” had been previously defined, “group 3” could be later explained by the expression of p70 alone (detected by Z27 mAb), and “group 5” by the expression of both p58.1 and p70 (57). These data supported the notion that each HLA class I specific receptor could be physically and functionally independent (57, 61). Another important piece of history was added by the evidence that EB6 and GL183 mAb could also recognize activating receptors, named p50.1 and p50.2, respectively, according to their molecular mass (62). Subsequently, another activating receptor, termed p50.3, was identified by three PAX mAbs and was not stained by either GL183 or EB6 mAb (63). Differently from p58 molecules, found in all individuals, p50 receptors could be detected only in some donors. A crucial step was represented by the identification of the cDNAs coding for these receptors, demonstrating that they belong to the same molecular family (64). The p58 and p50 molecules were highly homologous in their extracellular regions formed by 2 Ig-like domains, while major differences existed in their transmembrane and cytoplasmic portions. Notably, while in p58 the transmembrane region included only non-polar residues, in p50 it contained the charged amino acid Lys. Moreover, whereas p58 displayed a 76–84 amino acid cytoplasmic tail containing two YxxL motifs (D/Ex8D/ExxYxxLx26YxxL), later referred to as immunoreceptor tyrosine-based inhibitory motifs (ITIMs), p50 was characterized by a shorter tail (39 amino acids) lacking YxxL (64–66). The cDNA encoding the Bw4 specific receptors, named NKB1/p70, was also identified, revealing that its extracellular region was composed by 3 Ig-like domains and that its cytoplasmic tail included ITIMs (65, 67). All the genes coding for these receptors were mapped on chromosome 19q13.4. Concomitantly, cloning of CD94-encoding gene revealed that CD94 is a type II protein of the C-type lectin superfamily and that this gene is located on chromosome 12 (68). NK cell clones expressing CD94bright phenotype did not lyse cell lines transfected with different HLA class I alleles, and their cytotoxicity could be restored by mAb-mediated masking of CD94 or HLA class I (69). Moreover, a functional ambivalence of CD94-associated surface antigens was described (see below) (70). Finally, an additional inhibitory NK receptor of the Ig-like superfamily was identified to be specific for HLA-A*03 and -A*011 allotypes (71, 72). In our laboratory the production of Q66 mAb allowed the characterization of this receptor, which appeared to be a disulphide-linked dimer and was termed p140. Importantly, KIRs (and NKG2A) were also detected in a subset of CD8pos αβ T cells (73) in which they could interfere with TCR-mediated cell activation. These T cells were found to represent oligoclonally or monoclonally expanded cell populations (74). Subsequent studies revealed that such KIRpos CD8pos cells were HLA-E restricted (75, 76).
All these data, published between 1990 and 1996, represent true milestones for our present knowledge of the HLA specific receptors. Alessandro pioneered these studies with the discovery of p58 receptors and gave a fundamental contribution to most of the other major findings. Tracking all the rational steps leading to these discoveries in a seminal Alessandro's review, published in Annual Review in Immunology in 1996, has been for us both moving and impressive (7). Indeed, his contributions still represent a complete and modern insight on NK cell biology.
Different research groups started to work on the NK cell recognition of HLA class I molecules, therefore receptors and encoding cDNAs were named differently, so that the need for a common nomenclature arose. Thus, in late ‘90s, the acronym KIR (killer-cell inhibitory receptor) was proposed to indicate the members of this novel family (77). Then, following the experimental evidences that some KIR receptors were able to transduce activating signals, it was suggested to preserve the KIR acronym simply changing the meaning of “I” into “immunoglobulin-like.” The KIR nomenclature was designed in order to reflect the structure and the function of the encoded molecules, as well as the nucleotide sequence similarity among the different KIR family members. Thus, the first 2 digits following KIR correspond to the number of the extracellular domains (2D and 3D), while the third digit provides information on the length of the cytoplasmic tail (L or S) and consequently reveals the protein function (inhibitory or activating, respectively). Moreover, the two pseudogenes are indicated with the letter “P” (namely, KIR2DP1 and KIR3DP1) (78). The establishment of a centralized, publicly accessible KIR database (IPD-KIR), collecting all the officially recognized nucleotide and protein sequences, has been of great help to all the immunologists involved in the study of KIR gene polymorphism (79). Temporally, the first KIR Nomenclature report coincided with the first release of the IPD-KIR database. Finally, like other immune cell surface proteins, a CD number has been assigned to KIR proteins (namely the CD158 series).
Reconnecting with what has been described in the previous paragraph, it is possible to attribute the KIR nomenclature to the molecules with the “old” denomination. Thus, p58.1 and p50.1 correspond to KIR2DL1 and KIR2DS1, p58.2 and p50.2 to KIR2DL2/L3 and KIR2DS2, p50.3 to KIR2DS4, NKB1/p70 to KIR3DL1, and p140/NKAT4 to KIR3DL2.
KIR gene family is characterized by an extraordinary high degree of diversity, which arises from variability in KIR gene content, due to differences in both presence/absence (P/A) of KIR genes and KIR gene copy number, and from allelic polymorphism (80–89). Several strategies have been developed to analyze KIR repertoires, including approaches able to analyze P/A of the different KIR genes, KIR gene copy number variations, and, more recently, a “whole” KIR allele typing by next generation sequencing (NGS) approach (87, 90–100) underlining the huge interest of the immunologists in the KIR field.
As previously mentioned, KIR gene family has been mapped on chromosome 19q13.4 where KIR genes, each spanning ~10–16 kb, are tightly arranged in a head-to-tail orientation. This family consists of 13 genes (KIR2DL1, KIR2DL2/L3, KIR2DL4, KIR2DL5A, KIR2DL5B, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DL1/S1, KIR3DL2, KIR3DL3) and 2 pseudogenes (KIR2DP1, and KIR3DP1) (82, 101, 102). With few exceptions, KIR haplotypes can be divided in two regions, termed centromeric (Cen) and telomeric (Tel), by the presence of a recombination hot spot (86, 87, 89). Four genes, named framework genes (i.e., KIR3DL3, KIR3DP1, KIR2DL4, and KIR3DL2), mark the beginning and the end of these two regions. Based on the different number and kind of KIR genes present, three different centromeric regions (referred to as Cen-A, Cen-B1, and Cen-B2) and two different telomeric regions (namely Tel-A and Tel-B) have been identified (Figure 1A). Despite the high level of KIR gene content variability described, two main groups of KIR haplotypes, termed as “A” and “B,” have been identified (46, 80, 87, 102–106). The A haplotypes have a fixed gene content and are composed by Cen-A and Tel-A regions (Figures 1A,B). They include inhibitory KIR genes coding for receptors recognizing HLA-C (KIR2DL1 and KIR2DL3), HLA-B (KIR3DL1), HLA-A (KIR3DL2), and HLA-G (KIR2DL4) molecules and only one activating KIR (KIR2DS4) that, in some A haplotypes, does not code for a surface receptor (102). The remaining KIR haplotypes are collectively referred to as B haplotypes, and comprise haplotypes carrying Cen-A/Tel-B, Cen-B/Tel-A, and Cen-B/Tel-B combinations (Figure 1B). They are characterized by the presence of at least one of the following KIR genes: KIR2DS2, KIR2DL2, KIR2DL5B, KIR2DS3, KIR3DS1, KIR2DL5A, KIR2DS5, and KIR2DS1. Consequently, KIR A haplotypes differ from each other predominantly on the basis of KIR allelic polymorphism, while KIR B haplotypes mainly by their gene content. Notably, although with different frequencies, A and B haplotypes are maintained within all human populations, strongly suggesting that they have distinct and complementary functions and that they are under balancing selection (46, 107, 108). Moreover, the peculiar structure of KIR locus, including the presence of several genes sharing exon/intron organization, having a high degree of homology, and displaying the same orientation, facilitates non-reciprocal recombination that promotes deletion or duplication of KIR genes (i.e., formation of truncated or extended haplotypes), as well as the generation of hybrid KIR genes (i.e., KIR alleles including exons derived by different KIR genes) (Figure 1C) (86, 87, 109–113).
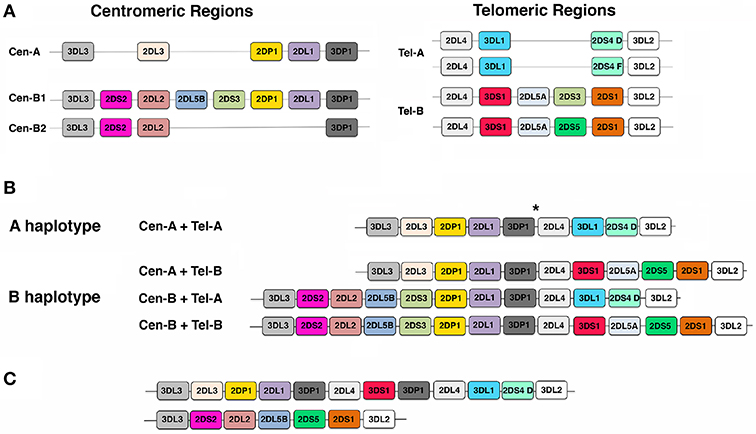
Figure 1. Gene organization of KIR locus. (A) A cartoon representation of the most frequent centromeric (Cen) and telomeric (Tel) regions detected in Caucasians. (B) Schematic pictures of KIR gene order in the A haplotypes and in 3 representative B haplotypes. * indicates the hot spot of recombination. (C) Examples of an extended and a truncated haplotype. Each colored box represents a KIR gene; for simplicity KIR gene names are reported in the boxes without the KIR acronym.
As previously reported, the first KIR cDNA sequences have been cloned in mid-90s. The great interest in the KIR field led to the identification, at the present, of ~1,000 alleles (https://www.ebi.ac.uk/ipd/kir/). This extremely high level of polymorphism, second only to that of HLA genes, arose from studies including different populations and relatively simple methodologies. Several allelic polymorphisms have been reported to impact KIR protein expression and function. Indeed, the presence of particular amino acid residues determining protein misfolding and resulting in the lack of surface receptor expression has been identified among KIR3DL1, KIR2DL1, and KIR2DL2 allotypes (95, 114–117). Moreover, KIR alleles characterized by a termination codon causing the premature end of the protein (namely KIR Null alleles) have been reported. Additionally, some KIR2DL4 and KIR2DS4 alleles are characterized by nucleotide deletions that, causing frameshift, lead to truncated polypeptides (102, 118, 119). Notably, different clades of alleles coding for surface receptors characterized by a low or high surface expression have been detected in KIR3DL1 and KIR2DL1 genes (95, 120, 121). Polymorphisms affecting KIR function may also cause differences in ligand affinity or in signal transduction. In this regard, it is well-established that KIR2DL1 allotypes coded by Cen-A regions are stronger receptors than those coded by Cen-B1 regions, and KIR2DL2 allotypes (coded by Cen-B regions) have a higher ligand affinity than KIR2DL3 allotypes (coded by Cen-A regions) (116, 122). In addition, the amino acid polymorphisms of KIR3DL1, the KIR receptor displaying the highest level of allelic variation, have been shown to impact not only its surface expression but also its affinity for HLA class I Bw4 bearing allotypes (123–126). Different amino acidic variations relevant for KIR signal transduction have been also reported in the exons coding for the transmembrane and cytoplasmic regions. In particular, the lack of charged residues, important for its association with signal-transducing polypeptides, has been described for a KIR2DS2 allele (127). In KIR2DL1, polymorphisms in the exons of the cytoplasmic tail, either causing the premature termination of translation at the end of the transmembrane domain or affecting the strength of inhibitory signal, have been reported (128, 129).
Thus, the extraordinary plasticity of the KIR gene family and the variability due to both KIR gene content and allelic polymorphism make unlikely that unrelated individuals share the same KIR genotype.
As mentioned above, the HLA class I ligands recognized by KIR2DL1, KIR2DL2/L3, KIR3DL1, and KIR3DL2 have been identified almost conjointly with the characterization of the receptors. Thereafter, the resolution of the crystal structures of KIRs in complex with their ligands, the development of several tools including soluble KIRs, KIR tetramers, HLA class I tetramers folded with selected peptides, and retrovirally transduced human cell lines (i.e., NKL, Jurkat) expressing only one KIR, allowed to refine KIR/KIR-ligand interactions (122, 130–134).
In Figure 2, the molecular structure of the inhibitory KIRs (iKIRs) and of their known cellular ligands is summarized. Using KIR2DL1-Fc (i.e., chimeric molecules comprising the extracellular portion of the receptor linked to the Fc region of human IgG1) in multiplex HLA class I binding assays, it has been possible to demonstrate that KIR2DL1 binds specifically, and with high avidity, to all HLA-C C2 but not HLA-C C1 or any HLA-A or HLA-B allotypes. The only exception is represented by KIR2DL1*022, a peculiar KIR2DL1 receptor carrying Lys44 (instead of Met44) that switches its ligand specificity to HLA-C C1 (129). By the use of KIR2DL1-Fc and plasmon resonance, it has been shown that several KIR2DL1 molecules recognize distinct HLA-C C2 allotypes with different avidities (the highest for HLA-C*15:02 and the lowest for HLA-C*04:01). As reported in early studies, and as expected on the basis of the homology in their extracellular domains, KIR2DL2 and KIR2DL3 bind a similar set of HLA-C ligands. Again, using KIR-Fc molecules it has been confirmed that KIR2DL2 and KIR2DL3 allotypes recognize, at high affinity, HLA-C C1 bearing epitope, and it has been demonstrated their ability to bind to two HLA-B allotypes (i.e., HLA-B*46:01 and HLA-B*73:01) that, being originated by crossing-over between HLA-C and HLA-B alleles, conserve the C1 epitope. Moreover, KIR2DL2 and KIR2DL3 display the capability to bind HLA-C C2 allotypes, even though with low affinity. Thus, in contrast to the simplicity of KIR2DL1 ligand recognition, a more complicated situation governs KIR2DL2 and KIR2DL3 ligand interactions (116, 122). The only known exception is represented by KIR2DL3*005 that, differing from other KIR2DL3 receptors, displays a HLA-C binding ability similar to that observed for KIR2DL2 allelic products (135). Increasing evidences indicate that both KIR2DL2/L3 and KIR2DL1, in addition to their ability to discriminate between C1 and C2 epitopes, bind numerous peptide/HLA-C combinations retaining a degree of peptide selectivity (136). Generally, KIR2DL2 and KIR2DL3 are characterized by a greater selectivity for peptide than KIR2DL1, and this ability seems to be particularly relevant for the recognition of the low affinity HLA-C C2 allotypes. Notably, the capability of KIR2DL1 to recognize HLA-C*08:02 (i.e., a HLA-C C1 allotype) presenting a restricted number of peptides has been recently reported (137). Taken together these studies underline how NK cells are able to sense, not only the down-regulation of HLA class I, but also alterations in HLA presented peptidome that may occur during viral infections or malignant transformations.
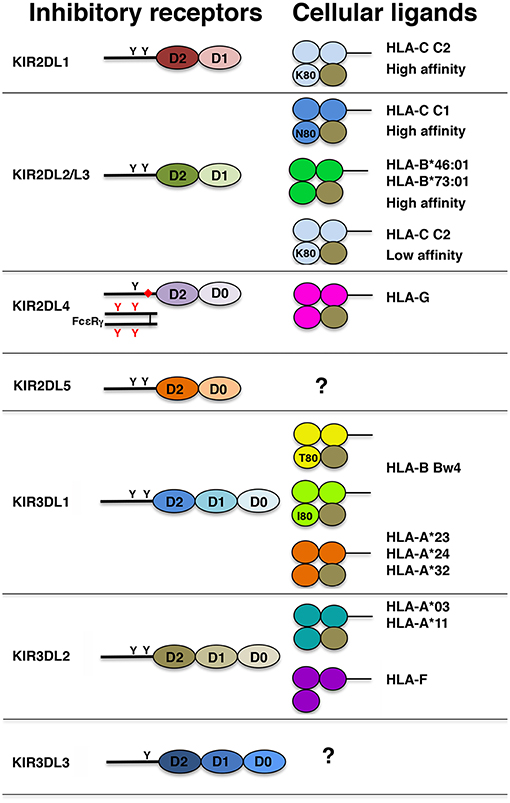
Figure 2. Schematic representation of the structure of iKIRs and their HLA class I ligands. Each Ig domain is depicted as an oval and, according to its structural features, labeled as D0, D1, and D2. Presence of ITIMs in KIR cytoplasmic tails and ITAMs in FcεR γ chain is indicated with black and red Y, respectively. The charged residue in KIR2DL4 transmembrane region is reported as a red diamond. Each domain of HLA class I molecules is represented as a colored circle and β2m as a brown circle. In HLA-C and HLA-B molecules, K80, N80, T80, and I80 indicate the presence, at residue 80, of lysine, asparagine, threonine, and isoleucine, respectively.
Between the two D0-D2 receptors, KIR2DL4 is the only one for which the ligand has been characterized. Differently from other iKIR2D receptors, KIR2DL4 binds to HLA-G, a non-classical HLA class I molecule that, in healthy cells, is restricted to the trophoblast cells invading the maternal decidua during early pregnancy. This receptor contains only one ITIM in the long cytoplasmic tail and a charged residue in the transmembrane region allowing its association with the γ chain of FcεR. These features render this receptor a peculiar member of the KIR family displaying an unusual hybrid structure and sharing characteristics with both inhibitory and activating KIRs. Functional studies revealed that KIR2DL4 is characterized by a weak inhibitory potential and that its engagement with soluble ligand results in strong cytokine release in the absence of killing (138, 139).
It is well-established that KIR3DL1 binds not only to HLA-B but also to some HLA-A Bw4 bearing allotypes. Indeed, it has been reported that among Bw4pos HLA-A allotypes, HLA-A*24:02, -A*32:01, -A*23:01, but not HLA-A*25:01, are able not only to inhibit but also to educate KIR3DL1pos NK cells (140). Among the five residues of α1 helix defining the Bw4 motif, the Ile/Thr dimorphism at residue 80 has been proposed as a marker of KIR3DL1 ligand affinity. Recent studies clearly indicate that, although many high affinity ligands possess Ile80 (Bw4 I80), KIR3DL1/HLA Bw4pos allotype interactions are very complex and strongly dependent on the avidity and surface expression of both receptor and ligand. Indeed, functional analyses provided clear evidences that both KIR3DL1 polymorphism and ligand variability strongly affect the avidity of KIR/KIR-L combinations sharply influencing NK cell function (123, 126, 141). As previously mentioned, KIR3DL2 allotypes bind two HLA-A allotypes (HLA-A*03 and -A*11) (71, 72). Functional analyses revealed that this receptor is characterized by low inhibitory capacity and its ligand interaction is highly dependent on the HLA-A bound peptide (132). Recently, it has been demonstrated that KIR3DL2 also recognizes the non-classical HLA class I molecule HLA-F at open conformation (142). Another relevant function of this iKIR is represented by its ability to function as sensor for pathogen-associated molecular patterns. Indeed, KIR3DL2 has been shown to bind CpG oligodeoxynucleotides (ODNs), and its D0 domain is primarily involved in ODN recognition (143). Remarkably, interaction between KIR3DL2 and ODN did not result in the delivery of inhibitory signals but in a sharp down-modulation of its surface expression, and in the induction of cytokine release underlining the antimicrobial role of NK cells in the course of infection.
From a structural point of view, in addition to a short cytoplasmic tail lacking ITIMs, a common feature of activating KIR (aKIRs) is the presence, in their trasmembrane region, of a charged residue (Lys) that allows their association with the signaling adaptor protein KARAP/DAP12 containing immunoreceptor tyrosine-based activating motifs (ITAMs) (144, 145). With the exception of KIR2DS1, the aKIR ligands remained elusive for a long time. Indeed, although the extracellular domains of some aKIRs display a high sequence homology with those of some iKIRs (namely, KIR2DS1-KIR2DL1, KIR2DS2-KIR2DL2, and KIR3DS1-KIR3DL1 pairs), it has been difficult to demonstrate aKIR/HLA class I interactions. Figure 3 shows the different aKIR/ligand pairs identified so far.
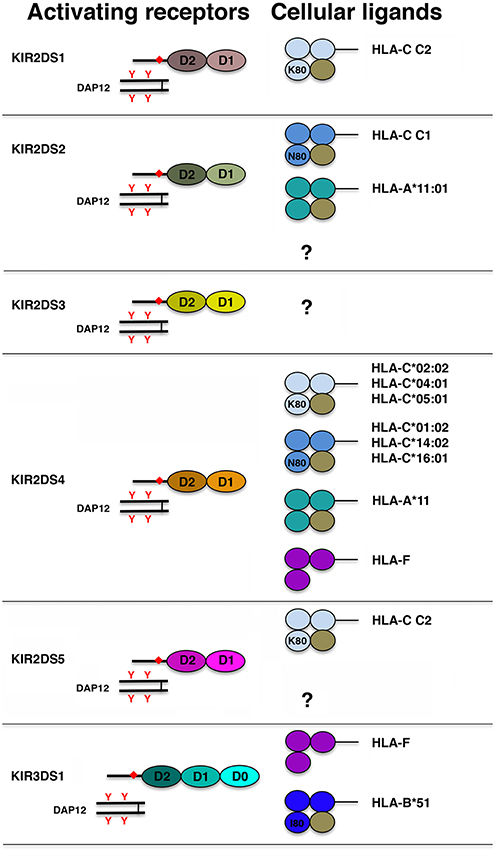
Figure 3. Schematic representation of the structure of aKIR and their HLA class I ligand. Each Ig domain is depicted as an oval and, according to its structural features, labeled as D0, D1, and D2. Presence of a charged residue in the KIR transmembrane region and of ITAMs in DAP12 cytoplasmic tail is indicated with orange diamond and orange Y, respectively. Each domain of HLA class I molecules is represented as a colored circle and β2m as a brown circle. In HLA-C and HLA-B molecules, K80, N80, and I80 indicate the presence, at residue 80, of lysine, asparagine, and isoleucine, respectively.
The best characterized aKIR is KIR2DS1. Functional analyses using in vitro expanded KIR2DS1pos NK cell clones, as well as KIR2DS1 soluble receptor and KIR2DS1 tetramer binding analyses, clearly demonstrated that this aKIR recognizes HLA-C C2 allotypes, although with an affinity lower than that of KIR2DL1 (i.e., its inhibitory counterpart) (62, 129, 146–149). Moreover, using both site-directed mutagenesis approaches and soluble receptors, it has been established that the residue at position 70 (Lys70 or Arg70) sharply affects the ligand affinity of KIR2DS1 molecules (146). Notably, a recent paper reported that modulation of HLA-C by certain strains of HCMV is required for a potent KIR2DS1-mediated NK cell activation. This strongly suggests a role of this aKIR in viral recognition (150). The HLA class I ligands recognized by KIR2DS2 have been identified only recently, and they include HLA-C C1 allotypes and HLA-A*11:01 (133, 147, 151). Remarkably, this aKIR/KIR-L interaction is highly peptide-dependent. In particular, the capability of KIR2DS2 to directly recognize peptides from HCV helicase presented by HLA-C*01:02 (an HLA-C C1 allotype) may explain, at least in part, the protective effect of this aKIR in chronic hepatitis C infection (152). In addition, it has been also reported that KIR2DS2 recognizes, on several human carcinoma cell lines, a still undefined, β2m-independent cell surface protein (153). Correlative studies reported that HIV-infected individuals carrying both KIR3DS1 and HLA-B Bw4 I80 display slower progression to AIDS and lower viral load. While these data could suggest KIR3DS1 interaction with HLA-B Bw4 I80 allotypes, the KIR3DS1 ligands have remained indefinite until recently, and they are represented by the open conformation of the non-classical HLA-F molecule and by the HLA-B*51, an HLA-B Bw4 I80 allotype (154–156).
KIR2DS3, KIR2DS4, and KIR2DS5 have no inhibitory counterparts. Most likely because KIR2DS3 does not seem to be expressed at the cell surface (157), it is still a receptor with unknown function and unknown ligand. On the contrary, the majority of KIR2DS4 and KIR2DS5 alleles code for surface activating receptors (63, 158). KIR2DS4 binds a minority of HLA-C allotypes carrying either C1 or C2 epitopes as well as HLA-A*11 (159). Recently, a highly peptide dependent ligand recognition has been reported also for KIR2DS4. This aKIR has been shown to recognize HLA-C*05:01 presenting a peptide derived from the highly conserved bacterial recombinase A, thus providing evidence that it is involved in defense against bacterial infections (160).
As previously reported, KIR2DS5 can be located either in the centromeric half of some KIR haplotypes (i.e., Cen-B1 region) or in the telomeric half (namely Tel-B region). With the only exception of KIR2DS5*005, KIR2DS5 centromeric alleles differ from the telomeric ones. Notably, four out of five KIR2DS5 centromeric alleles but only two out of six KIR2DS5 telomeric alleles code for receptors recognizing several HLA-C C2 molecules (161).
During their development, NK cells go through a process termed “education,” involving the interaction between inhibitory NK receptor (iNKR) and self HLA class I molecules. Although different models have been proposed, a general rule for education is that the strength of inhibitory interaction(s) dictates the efficiency of NK effector function (12, 13, 162, 163). A prominent role is played by the highly diversified system represented by the combinations between inhibitory KIR with their KIR-L (iKIR/KIR-L pairs). Another relevant iNKR is represented by the CD94:NKG2A heterodimer, a type II protein belonging to the C-type lectin superfamily, which recognizes the non-classical HLA class I molecule HLA-E (10). Since HLA-E binds peptides cleaved from the leader sequences (from −22 to −14 residues) of HLA-A, -B, or -C, it has been generally considered as a sensor of the overall amount of HLA class I molecules expressed on the cell surface. Recently, the Met/Thr (M/T) dimorphism at position −21 of the leader sequence of HLA-B (i.e., −21M and −21T) has been described to strongly impact the CD94:NKG2A/HLA-E interaction (164). Indeed, only a minority of HLA-B allotypes having −21M can supply HLA-E binding peptides, while the majority carrying −21T do not. Accordingly, individuals with −21M HLA-B could show higher HLA-E expression and more efficient NKG2Apos NK cells. The activating counterpart of CD94:NKG2A is represented by CD94:NKG2C heterodimer, which binds HLA-E as well, although with lower affinity.
Extremely variegated NK cell receptor repertoires can be observed among different individuals (7, 60, 80, 164–167). This diversity is primarily due to the high polymorphism of KIR and HLA class I genes, which segregate independently, leading to diverse compound genotypes (46). Another important feature of the repertoire is the clonal distribution of KIR and CD94:NKG2A. During education NK cells must engage at least one iNKR with cognate HLA class I to become fully functional, otherwise, they will be hypo-responsive. This process ensures that each NK cell produced in the body maintains tolerance to self. The hypothesis that NKR expression represents the result of a stochastic event, in which the various encoding genes are independently regulated, had been already discussed in Alessandro's review (7). From studies of NK cells at the clonal level, it was clear that each NK cell could express either one or more iNKR specific for self HLA (self-iNKR) as well as additional iNKR specific for non-self HLA. The presence of NK cells expressing a single self-iNKR confers an advantage to the host, since it allows to sense the loss or peptide-induced alteration of even a single HLA class I allotype, and thus to kill the damaged cell, through the mechanism of “missing self-recognition.”
While a strong interaction between iKIR and its KIR-L positively impacts NK cell education, the behavior of aKIR is opposite and induces down-regulation of NK cell responsiveness in the presence of a strong cognate ligand. This phenomenon has been described for KIR2DS1 and KIR3DS1. Thus, KIR2DS1pos NK cells are anergic in HLA-C C2/C2 individuals, while they are educated in C1/Cx ones (168). Similarly, KIR3DS1-mediated positive recognition of HLA-B*51 (Bw4 I80) could be detected in NK cell clones derived from Bw4 I80 negative donors, but not in those from Bw4 I80 positive donors (156).
Although genetics of KIR and HLA has the major impact on the receptor repertoire of the circulating NK cell pool, also environmental factors may play an important role. In this regard, the phenotypic analysis of NK cells derived from twins, who are genetically identical for KIR and HLA genes, has been particularly relevant (169). Indeed, twins' NK cell populations are similar but not identical to each other, especially in adult age. In this context, viral infections may impact the NK cell phenotype. In particular, HCMV shapes the receptor repertoire, driving to adaptive NK cell differentiation through epigenetic alterations (170–173). Thus, the expanded NK cell subset is characterized by the expression of CD94:NKG2C, mainly co-expressing KIR2DL specific for self HLA-C allotypes, and CD57, a marker of terminally differentiated stage (32, 33). Recent data revealed that the expansion and differentiation of adaptive NKG2Cpos NK cells could be determined by differentially recognition of distinct HCMV strains encoding variable UL40 peptides (174).
Nowadays, new technologies allow the identification of distinct subsets by the detection of multiple molecules in the whole NK cell population, even freshly derived from blood. Multi-parametric flow cytometry makes it possible the simultaneous use of numerous mAbs, detecting the pattern of different subpopulations carrying one or another iKIR, aKIR, CD94:NKG2A, and CD94:NKG2C. Table 1 describes the reactivity of some anti-KIR mAb, that can be used in combination to dissect various iKIR and aKIR. Moreover, mass cytometry by time-of-flight (CyTOF) technology allows the concomitant assessment of more than 30 parameters, revealing that at least 30.000 and even more distinct PB NK phenotypes can be detected in each individual (169).
NK cell education in pregnancy or in disease and the particular KIR/HLA combinations associated with either a protective role or a detrimental effect in placentation, infectious diseases, autoimmune and inflammatory diseases, and in cancer have recently been reviewed (46–48, 136, 175). Our group studied NK cell education in disease, particularly focusing on leukemia patients receiving HSCT (see below) or patients with X-linked lymphoproliferative disease 1 (XLP1). XLP1 is a primary immunodeficiency caused by mutations in SH2D1A, the gene encoding the signaling lymphocyte activation molecule (SLAM)-associated protein (SAP). Seminal studies coordinated by Alessandro provided evidence that, in the absence of SAP, 2B4 and NTB-A (belonging to SLAM family) co-receptors associate with protein tyrosine phosphatases thus delivering inhibitory instead of activating signals (22, 24, 176). This specific immune dysfunction in XLP1 patients causes the inability of NK cells to kill EBV-infected B cells (B-EBV), over-expressing their ligands (i.e., CD48 and NTB-A), with dramatic clinical consequences. Following this knowledge, we further analyzed the NK cell receptor repertoire of these patients, showing that substantial proportions of NK cells lacking any inhibitory receptor specific for self-HLA (self-NKRneg) are present and are fully functional, indicating that inhibitory 2B4 participates to NK cell education (177). Remarkably, self-NKRneg NK cells can efficiently kill CD48neg target cells, such as mature DC, with consequently defective antigen presentation, further exacerbating the immune defect of these patients.
As previously reported, the HLA class I specific inhibitory receptors are constitutively expressed by NK cells and finely regulate their function preventing NK-mediated damage to healthy tissues and allowing the elimination of tumors displaying defective HLA class I expression. Additional inhibitory checkpoints are inducible and play a wider physiological role in immune cell homeostasis. On the other hand, in tumors, they may severely compromise NK-mediated responses and promote tumor growth and metastasis. In particular, CTLA-4 and PD-1 are major regulators of immune responses and contribute to the maintenance of peripheral T cell tolerance. PD-1, a member of the Ig-superfamily, was first identified in T lymphocytes (178) and, more recently, in NK cells (44). In cancer, upon binding to its ligands (PD-L1 and/or PD-L2) expressed on tumor cells, PD-1 may impair the antitumor cytotoxicity, thus favoring tumor immune escape. Human PD-1bright NK cells have been detected in patients with CMV infections and tumors (44, 179). For example, in ovarian carcinoma patients, a sizable fraction of PD-1pos NK cells can be detected in PB, while larger percentages are present in the ascitic fluid of the same patients. Although PD-1 mRNA and protein are detectable at the cytoplasmic level, the mechanisms leading to its surface expression in human NK cells have not been defined yet. The detection of PD-1pos in NK cells associated to tumors suggests that signals delivered by the tumor or its microenvironment may be involved in its surface expression. Besides PD-1, several other inhibitory checkpoints expressed by NK cells have been detected, and have been recently reviewed (180). Briefly, CD96 and TIGIT belong to the same family of the activating receptor DNAM-1 (their common ligands, CD155 and CD112, are nectin-like molecules). TIGIT was found to be up-regulated in tumor-associated NK cells in colorectal cancer. KLRG1 is expressed by NK cells upon activation (as well as by other lymphoid and myeloid cell types). LAG-3 marks exhausted T cells infiltrating tumors, while its possible modulatory effect on NK cells has not been defined yet. TIM-3 binds primarily to Galectin-9 but also to other tumor-associated antigens. A recently identified inhibitory receptor which is likely to play an important role in controlling NK cell activation, proliferation and function is IL-1R8, a member of IL-1 receptor family and a component of the human IL-37 receptor. Mice lacking IL1R8 do not develop carcinogen-induced hepatocarcinoma (181). Thus, it is conceivable that IL-1R8 blocking may unleash NK cells and promote efficient antitumor responses.
As mentioned above, PD-1 may impair T cell responses against tumor cells. Importantly, immunotherapy with mAbs able to disrupt the PD-1/PD-L1 interaction proved to be highly effective, allowing unthinkable achievements in the cure of a fraction of tumor patients. In this context, the number of mAbs specific for PD-1 or PD-L1 approved for cancer therapy is growing fast and the spectrum of tumors for which the use of such agents is recommended includes non-small lung cancer, melanoma, urothelial cancers, kidney tumors, Hodgkin lymphoma, colorectal cancer, and hepatocellular carcinoma. While the use of such checkpoint inhibitors represents a real revolution in cancer treatment, in a still too large fraction of patients these agents are ineffective. Accordingly, a major goal is to predict the patient response to treatment, to avoid possible side effects and useless high costs of the therapy. The expression of PD-L1 on tumor cells undoubtedly represents a valuable biomarker. However, its predictive value is still unsatisfactory because of various limitations, including inter- and intra-tumor heterogeneous PD-L1 expression, use of PD-1/PD-L1 specific mAbs displaying a substantially different reactivity (182), different diagnostic material (biopsies vs. surgical specimens). Patient selection criteria are being standardized, for example, by defining the optimal number of biopsies that give results comparable to the whole tumor section, considered as the gold standard (183, 184). Given these limitations, research in progress is aimed at identifying additional checkpoints possibly to be used alone or in combination with PD-1/PD-L1 or CTLA-4.
After the launch of Innate-Pharma in Marseille, Alessandro's antibodies and his expertize were fundamental for the design of a program focalized on NK cells harnessing the blockade of inhibitory KIRs and NKG2A. Thus, three clinical grade mAbs were produced: Lirilumab (also called IPH2101, formerly 1-7F9) targeting KIRs, IPH4102 selectively recognizing KIR3DL2, and Monalizumab targeting NKG2A. These reagents are currently used in clinical trials to treat cancer patients. Thus, Lirilumab, a first-in-class humanized IgG4 mAb directed against a common epitope shared by KIR2D, disrupts the KIR/KIR-L interaction by rendering NK cells “alloreactive” and allows killing of tumor target cells. This antibody has been used in a phase I trial in association with Lenalidomide, for NK stimulation, to treat Multiple Myeloma. This combined treatment resulted to be safe, tolerable, and associated with signs of clinical efficacy (185). Because KIR3DL2 is highly expressed on cutaneous T cell lymphomas, including mycosis fungoides and Sézary syndrome (186), IPH4102 has been developed for the treatment of these diseases. Phase I trials provided evidence that the treatment is well-tolerated and efficacious. These encouraging results would support the large-scale clinical trials (187).
Recent studies identified NKG2A as an important checkpoint leading to remarkable antitumor effects in animal models (188–190). Encouraging results have also been obtained in a preliminary clinical trial in highly aggressive head and neck cancers (188). Notably, NKG2A, constitutively expressed by NK cells, can be induced also in T cells upon cytokine or antigen-induced activation (191, 192). Indeed, previous in vitro studies showed that mAb-mediated masking of NKG2A or KIRs, in both polyclonal and clonal NK or T cell populations, induced killing of HLA class Ipos tumor target cells (149, 188, 193–195). Vivier group also showed that the antitumor effects of anti-NKG2A mAb could be potentiated by the combined use of other therapeutic mAbs (188). Based on the expression of inhibitory checkpoint ligands on tumor cells, three therapeutic approaches have been proposed: (1) HLA-Epos tumors lacking the expression of other ligands for inhibitory checkpoints or tumor antigens: blocking of NKG2A could unleash both NK- and T-cell-mediated antitumor responses in a murine model; (2) tumors expressing both HLA-E and PD-L1: by the combined blocking of NKG2A and PD-1/PD-L1 axis, not only enhancement of NK and T cell cytotoxicity but also induction of T cell proliferation and establishment of T cell memory were detected; (3) tumors expressing HLA-E and specific tumor antigens (e.g., EGF-R): blockade of NKG2A increased the efficacy of anti-EGF-R by allowing a CD16-mediated efficient ADCC by mature, highly cytotoxic NK cells.
The discovery of both HLA class I specific inhibitory receptors and NK cell alloreactivity has been exploited, after a relatively short time interval, for the treatment of acute, high-risk leukemia in the haplo-HSCT setting. HSCT is the life-saving therapy for such leukemia with adverse molecular or cytogenetic characteristics, or with poor response to chemotherapy, or relapsing. However, an HLA 10/10 allelic matched donor, whether sibling or unrelated, is available only for two out of three patients, and this proportion can be even lower for patients belonging to certain ethnic groups. Thus, haplo-HSCT has been developed in the attempt to rescue these patients (196). In such transplantation setting, donor derived NK cells are defined as “alloreactive” when they express, as inhibitory receptor, exclusively KIR(s) specific for self-HLA class I allele(s) (KIR-L), allowing the elimination of patient cells missing that particular KIR-L. A seminal study by Ruggeri et al. (197) reported a 5-year survival probability of ~60% in adult AML patients in case of donor NK alloreactivity (KIR/KIR-L mismatch in graft vs. host direction), while survival was <5% in its absence. In 2002, Alessandro and some authors of the present contribution started a fruitful collaboration with Franco Locatelli, first at the University of Pavia and subsequently at the Ospedale Pediatrico Bambino Gesù in Roma (149, 198). In these studies, patients were represented by children and young adults (1–20 years) with acute high-risk ALL or AML. Notably, in the haplo-HSCT setting, most pediatric patients find an available donor, represented by one parent.
In view of the major relevance of NK alloreactivity, a great attention was paid by our group to the selection of the best possible donor, based primarily on the “perfect mismatch” (199) and on the size of alloreactive NK cell subset. To this end, a reliable method was developed to define the presence and the frequency of such subpopulation in potential donors (149). This is based on combined genotypic and phenotypic analyses to assess (i) the presence in the donor of KIR-L(s) absent in the recipient, (ii) donor's KIR genotype to evaluate the presence of KIR(s) specific for the mismatched KIR-L(s), and (iii) identification of the alloreactive subset using appropriate anti-KIR and anti-NKG2A mAb combinations (198). Leukemia patients, after the conditioning regimen, receive very high numbers (“megadoses”) of highly purified T-depleted CD34pos cells, isolated either from bone marrow (BM) or PB. Isolation from PB is preceded by donor treatment with G-CSF in order to “mobilize” CD34pos cells from BM. Notably, different from AML, adult high-risk ALL were poorly responding to haplo-HSCT even in the presence of donor NK alloreactivity. Thus, it was somewhat surprising to find opposite results in pediatric patients. Indeed, in case of NK alloreactivity, the survival rate was ~70% in ALL and ~40% in AML, while in the absence of NK alloreactivity, the percentages were ~35 and ~20%, respectively. Of note, in the transplantation setting with “pure CD34pos” cells, most deaths due to leukemia relapses or transplant-related mortality occurred early, within few weeks/months after transplantation. As mentioned above, NK cells, during their differentiation from HSC, first express CD94:NKG2A, while KIR are acquired only at later stages. Accordingly, the generation of alloreactive, mature NK cells requires a relatively long time interval (6–8 weeks). Thus, for several weeks, the absence of mature (alloreactive) NK cells together with the absence of adaptive immunity may be critical for the control of leukemia relapses and infections. The survival rate of leukemia patients receiving a CD34pos haplo-HSC could be considered satisfactory in view of the extremely poor prognosis of patients not receiving transplantation. However, the delayed appearance of alloreactive NK cells, conceivably explaining the occurrence of early relapses and infections, suggested, in 2010, the application of a new graft manipulation based on the depletion of αβ T lymphocytes and CD19pos B lymphocytes. Thanks to this novel approach, the graft infused contains, besides HSC, other cell types, including intermediate precursors (CD34neg), various myeloid cell types, mature NK cells, and γδ T lymphocytes. Notably, both NK and γδ T lymphocytes may exert a potent anti-leukemia activity and contribute to the control of infections. Accordingly, patients could immediately benefit of high numbers (30–40 × 106/kg) of donor mature NK cells and γδ T cells (3–5 × 106/kg). The clinical outcome in 80 children was excellent with an overall survival of ~70% not only in ALL, but also in AML (200). Surprisingly, NK alloreactivity did not appear to play a relevant role in the clinical outcome. The marked improvement of the survival rate in the absence of alloreactivity may be due, at least in part, to the additional selection criteria progressively applied to identify the most suitable among the available donors. These include the following positive characteristics: (i) KIR B/X genotype, (ii) HLA-C1pos/KIR2DS1pos for HLA-C2pos recipients, (iii) high absolute cell numbers of NK and γδT lymphocytes, and (iv) high expression of NKp46 and presence of NKG2C (198, 200). The presence of mature NK and γδ T from the beginning of the transplantation and their persistence in the recipient as efficient immune cells, due to the lack of GvHD prophylaxis, can explain the successful clinical results. In immunocompromised patients undergoing allogeneic HSCT, HCMV infection/reactivation frequently occurs (201). Several studies documented a role of HCMV in promoting rapid NK cell maturation and in the selective expansion of NKG2Cpos adaptive NK cells (34, 202, 203). This HCMV related effect on NK cell repertoire was also observed in pediatric patients receiving αβ T/B cell-depleted haplo-HSCT (204). Remarkably, a study revealed that, in NKG2Cneg/neg umbilical cord blood donors, HCMV infection promoted a maturation and expansion of NK cells expressing aKIRs (205). These findings suggest that, in the absence of NKG2C, aKIRs may sense HCMV infection and promote selective expansion of mature aKIRpos NK cells, underscoring the concept that aKIRs may sense microbial infections.
Taken together, the clinical results in haplo-HSCT indicate that cells belonging to innate immunity play a crucial role in the cure of patients with otherwise lethal leukemia and provide a solid background to further improve the survival rate and to implement novel NK-based therapies (e.g., CAR-NK). In this context, helper ILC, primarily ILC-3, may provide a tool to further improve early innate defense against pathogens after HSCT. Notably, it has been shown that ILC-3 development may be favored by given sources of HSC for transplantation (206).
There is little doubt that major advances in immunology and the exploitation of fundamental discoveries offered invaluable tools for the therapy of cancer. Indeed, the recent years witnessed unprecedented successes in the cure of different, high aggressive tumors and leukemia. In this context, NK cells and their receptors were shown to play an unexpected role. Thus, the discovery of KIRs provided the molecular basis for haplo-HSCT, which allowed the cure of patients with acute, high-risk leukemia for whom no HLA-compatible donors were available. These data also provided the first evidence that not only T and B lymphocytes, but also cells of the innate immunity may be exploited for tumor therapy. While checkpoint inhibitors targeting PD-1 or CTLA-4 represent the most effective tools to successfully treat a fraction of otherwise incurable cancer patients, their combined use with therapeutic antibodies blocking NKG2A and KIR may further extend the benefit to a large proportion of patients. Thus, the Alessandro's legacy, besides unraveling the molecular mechanisms of NK cell function, also extends to thousands of leukemia patients recovered from their otherwise fatal disease.
The continuous interest and evolution on KIR research is also witnessed by the organization of a KIR Workshop taking place every year and a half, allowing to share the most recent advances on KIRs in health and disease, with a fruitful exchange of ideas and knowledge in a friendly atmosphere. Last October, the KIR Workshop 2018 has been organized by our group, and Alessandro had begun to collaborate with us on its organization and realization. This meeting was dedicated to Alessandro.
LM, DP, and MF wrote, referenced the review, and prepared the figures. All authors contributed with their relevant published studies, cited in this review, and critically revised the manuscript.
This work was supported by grants awarded by: AIRC 5X1000, 2018 Project Code 21147 (to LM); AIRC IG 2017, Project Code 19920 (to LM); AIRC 5X1000, Project Code 21073; AIRC IG 2015, Project Code 16764 (to DP); AIRC IG 2014 project n. 15428 (to MV); 5X1000 Italian Ministry of Health 2015 (to MCM); Italian Ministry of Health, project Code RF-2016-02362288 (to GP).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Moretta L, Mantovani A. Alessandro Moretta 1953-2018. Nat Immunol. (2018) 19:315. doi: 10.1038/s41590-018-0075-5
2. Moretta L, Vivier E. Alessandro Moretta (1953-2018). Immunity. (2018). 48:601–2. doi: 10.1016/j.immuni.2018.03.021
3. Vivier E, Olive D. A Tribute to Alessandro Moretta (1953–2018). Living without Alessandro. Front. Immunol. (2018) 9:1292. doi: 10.3389/fimmu.2018.01292
4. Moretta A, Pantaleo G, Moretta L, Cerottini JC, Mingari MC. Direct demonstration of the clonogenic potential of every human peripheral blood T cell. Clonal analysis of HLA-DR expression and cytolytic activity. J Exp Med. (1983) 157:743–54. doi: 10.1084/jem.157.2.743
5. Ljunggren HG, Karre K. In search of the 'missing self': MHC molecules and NK cell recognition. Immunol Today. (1990) 11:237–44. doi: 10.1016/0167-5699(90)90097-S
6. Yokoyama WM, Seaman WE. The Ly-49 and NKR-P1 gene families encoding lectin-like receptors on natural killer cells: the NK gene complex. Annu Rev Immunol. (1993) 11:613–35. doi: 10.1146/annurev.immunol.11.1.613
7. Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, et al. Receptors for HLA class-I molecules in human natural killer cells. Annu Rev Immunol. (1996) 14:619–48. doi: 10.1146/annurev.immunol.14.1.619
8. Moretta A, Tambussi G, Bottino C, Tripodi G, Merli A, Ciccone E, et al. A novel surface antigen expressed by a subset of human CD3- CD16+ natural killer cells. Role in cell activation and regulation of cytolytic function. J Exp Med. (1990) 171:695–714. doi: 10.1084/jem.171.3.695
9. Moretta A, Bottino C, Pende D, Tripodi G, Tambussi G, Viale O, et al. Identification of four subsets of human CD3-CD16+ natural killer (NK) cells by the expression of clonally distributed functional surface molecules: correlation between subset assignment of NK clones and ability to mediate specific alloantigen recognition. J Exp Med. (1990) 172:1589–98. doi: 10.1084/jem.172.6.1589
10. Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. (1998) 391:795–9. doi: 10.1038/35869
11. Moretta L. Dissecting CD56dim human NK cells. Blood. (2010) 116:3689–91. doi: 10.1182/blood-2010-09-303057
12. Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. (2005) 436:709–13. doi: 10.1038/nature03847
13. Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. (2006) 25:331–42. doi: 10.1016/j.immuni.2006.06.013
14. Sivori S, Vitale M, Morelli L, Sanseverino L, Augugliaro R, Bottino C, et al. p46, a novel natural killer cell-specific surface molecule that mediates cell activation. J Exp Med. (1997) 186:1129–36. doi: 10.1084/jem.186.7.1129
15. Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L, et al. Molecular cloning of NKp46: a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J Exp Med. (1998) 188:953–60. doi: 10.1084/jem.188.5.953
16. Vitale M, Bottino C, Sivori S, Sanseverino L, Castriconi R, Marcenaro E, et al. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J Exp Med. (1998) 187:2065–72. doi: 10.1084/jem.187.12.2065
17. Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A, et al. NKp44, A triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J Exp Med. (1999) 189:787–96. doi: 10.1084/jem.189.5.787
18. Pende D, Parolini S, Pessino A, Sivori S, Augugliaro R, Morelli L, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. (1999) 190:1505–16. doi: 10.1084/jem.190.10.1505
19. Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. (2001) 19:197–223. doi: 10.1146/annurev.immunol.19.1.197
20. Falco M, Biassoni R, Bottino C, Vitale M, Sivori S, Augugliaro R, et al. Identification and molecular cloning of p75/AIRM1, a novel member of the sialoadhesin family that functions as an inhibitory receptor in human natural killer cells. J Exp Med. (1999) 190:793–802. doi: 10.1084/jem.190.6.793
21. Cantoni C, Bottino C, Augugliaro R, Morelli L, Marcenaro E, Castriconi R, et al. Molecular and functional characterization of IRp60, a member of the immunoglobulin superfamily that functions as an inhibitory receptor in human NK cells. Eur J Immunol. (1999) 29:3148–59. doi: 10.1002/(SICI)1521-4141(199910)29:10<3148::AID-IMMU3148>3.0.CO;2-L
22. Sivori S, Parolini S, Falco M, Marcenaro E, Biassoni R, Bottino C, et al. 2B4 functions as a co-receptor in human NK cell activation. Eur J Immunol. (2000) 30:787–93. doi: 10.1002/1521-4141(200003)30:3<787::AID-IMMU787>3.0.CO;2-I
23. Vitale M, Falco M, Castriconi R, Parolini S, Zambello R, Semenzato G, et al. Identification of NKp80, a novel triggering molecule expressed by human NK cells. Eur J Immunol. (2001) 31:233–42. doi: 10.1002/1521-4141(200101)31:1<233::AID-IMMU233>3.3.CO;2-W
24. Bottino C, Falco M, Parolini S, Marcenaro E, Augugliaro R, Sivori S, et al. NTB-A [correction of GNTB-A], a novel SH2D1A-associated surface molecule contributing to the inability of natural killer cells to kill Epstein-Barr virus-infected B cells in X-linked lymphoproliferative disease. J Exp Med. (2001) 194:235–46. doi: 10.1084/jem.194.3.235
25. Falco M, Marcenaro E, Romeo E, Bellora F, Marras D, Vely F, et al. Homophilic interaction of NTBA, a member of the CD2 molecular family: induction of cytotoxicity and cytokine release in human NK cells. Eur J Immunol. (2004) 34:1663–72. doi: 10.1002/eji.200424886
26. Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. (2003) 198:557–67. doi: 10.1084/jem.20030788
27. Moretta L, Bottino C, Pende D, Vitale M, Mingari MC, Moretta A. Different checkpoints in human NK-cell activation. Trends Immunol. (2004) 25:670–6. doi: 10.1016/j.it.2004.09.008
28. Costello RT, Sivori S, Marcenaro E, Lafage-Pochitaloff M, Mozziconacci MJ, Reviron D, et al. Defective expression and function of natural killer cell-triggering receptors in patients with acute myeloid leukemia. Blood. (2002) 99:3661–7. doi: 10.1182/blood.V99.10.3661
29. Stojanovic A, Correia MP, Cerwenka A. Shaping of NK cell responses by the tumor microenvironment. Cancer Microenviron. (2013) 6:135–46. doi: 10.1007/s12307-012-0125-8
30. Vitale M, Cantoni C, Pietra G, Mingari MC, Moretta L. Effect of tumor cells and tumor microenvironment on NK-cell function. Eur J Immunol. (2014) 44:1582–92. doi: 10.1002/eji.201344272
31. Romagnani C, Juelke K, Falco M, Morandi B, D'Agostino A, Costa R, et al. CD56brightCD16- killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol. (2007) 178:4947–55. doi: 10.4049/jimmunol.178.8.4947
32. Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. (2010) 116:3853–64. doi: 10.1182/blood-2010-04-281675
33. Hendricks DW, Balfour HH Jr, Dunmire SK, Schmeling DO, Hogquist KA, Lanier LL. Cutting edge: NKG2C(hi)CD57+ NK cells respond specifically to acute infection with cytomegalovirus and not Epstein-Barr virus. J Immunol. (2014) 192:4492–6. doi: 10.4049/jimmunol.1303211
34. Della Chiesa M, Falco M, Podesta M, Locatelli F, Moretta L, Frassoni F, et al. Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation: a role for human cytomegalovirus? Blood. (2012) 119:399–410. doi: 10.1182/blood-2011-08-372003
35. Muccio L, Falco M, Bertaina A, Locatelli F, Frassoni F, Sivori S, et al. Late development of FcεRγneg adaptive natural killer cells upon human cytomegalovirus reactivation in umbilical cord blood transplantation recipients. Front Immunol. (2018) 9:1050. doi: 10.3389/fimmu.2018.01050
36. Artis D, Spits H. The biology of innate lymphoid cells. Nature. (2015) 517:293–301. doi: 10.1038/nature14189
37. Diefenbach A, Colonna M, Romagnani C. The ILC world revisited. Immunity. (2017) 46:327–32. doi: 10.1016/j.immuni.2017.03.008
38. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell. (2018) 174:1054–66. doi: 10.1016/j.cell.2018.07.017
39. Poggi A, Sargiacomo M, Biassoni R, Pella N, Sivori S, Revello V, et al. Extrathymic differentiation of T lymphocytes and natural killer cells from human embryonic liver precursors. Proc Natl Acad Sci USA. (1993) 90:4465–9. doi: 10.1073/pnas.90.10.4465
40. Yu J, Freud AG, Caligiuri MA. Location and cellular stages of natural killer cell development. Trends Immunol. (2013) 34:573–82. doi: 10.1016/j.it.2013.07.005
41. Mingari MC, Vitale C, Cantoni C, Bellomo R, Ponte M, Schiavetti F, et al. Interleukin-15-induced maturation of human natural killer cells from early thymic precursors: selective expression of CD94/NKG2-A as the only HLA class I-specific inhibitory receptor. Eur J Immunol. (1997) 27:1374–80. doi: 10.1002/eji.1830270612
42. Vacca P, Vitale C, Montaldo E, Conte R, Cantoni C, Fulcheri E, et al. CD34+ hematopoietic precursors are present in human decidua and differentiate into natural killer cells upon interaction with stromal cells. Proc Natl Acad Sci USA. (2011) 108:2402–7. doi: 10.1073/pnas.1016257108
43. Vacca P, Munari E, Tumino N, Moretta F, Pietra G, Vitale M, et al. Human natural killer cells and other innate lymphoid cells in cancer: Friends or foes? Immunol Lett. (2018) 201:14–9. doi: 10.1016/j.imlet.2018.11.004
44. Pesce S, Greppi M, Tabellini G, Rampinelli F, Parolini S, Olive D, et al. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: a phenotypic and functional characterization. J Allergy Clin Immunol. (2017) 139:335–46 e3. doi: 10.1016/j.jaci.2016.04.025
46. Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. (2005) 5:201–14. doi: 10.1038/nri1570
47. Moffett A, Colucci F. Co-evolution of NK receptors and HLA ligands in humans is driven by reproduction. Immunol Rev. (2015) 267:283–97. doi: 10.1111/imr.12323
48. Boudreau JE, Hsu KC. Natural killer cell education in human health and disease. Curr Opin Immunol. (2018) 50:102–11. doi: 10.1016/j.coi.2017.11.003
49. Ciccone E, Viale O, Pende D, Malnati M, Biassoni R, Melioli G, et al. Specific lysis of allogeneic cells after activation of CD3- lymphocytes in mixed lymphocyte culture. J Exp Med. (1988) 168:2403–8. doi: 10.1084/jem.168.6.2403
50. Melioli G, Ciccone E, Mingari MC, Pende D, Viale O, Tambussi G, et al. Specific recognition by CD3- NK cells: a limiting dilution analysis of the frequency of alloreactive CD3- lymphocyte precursors. Int J Cancer Suppl. (1989) 4:56–7. doi: 10.1002/ijc.2910440715
51. Ciccone E, Pende D, Viale O, Tambussi G, Ferrini S, Biassoni R, et al. Specific recognition of human CD3-CD16+ natural killer cells requires the expression of an autosomic recessive gene on target cells. J Exp Med. (1990) 172:47–52. doi: 10.1084/jem.172.1.47
52. Ciccone E. Involvement of HLA class I alleles in natural killer (NK) cell-specific functions: expression of HLA-Cw3 confers selective protection from lysis by alloreactive NK clones displaying a defined specificity (specificity 2). J Exp Med. (1992) 176:963–71. doi: 10.1084/jem.176.4.963
53. Colonna M, Spies T, Strominger JL, Ciccone E, Moretta A, Moretta L, et al. Alloantigen recognition by two human natural killer cell clones is associated with HLA-C or a closely linked gene. Proc Natl Acad Sci USA. (1992) 89:7983–5. doi: 10.1073/pnas.89.17.7983
54. Christiansen FT, Witt CS, Ciccone E, Townend D, Pende D, Viale D, et al. Human natural killer (NK) alloreactivity and its association with the major histocompatibility complex: ancestral haplotypes encode particular NK-defined haplotypes. J Exp Med. (1993) 178:1033–9. doi: 10.1084/jem.178.3.1033
55. Moretta A, Vitale M, Bottino C, Orengo AM, Morelli L, Augugliaro R, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. (1993) 178:597–604. doi: 10.1084/jem.178.2.597
56. Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci USA. (1993) 90:12000–4. doi: 10.1073/pnas.90.24.12000
57. Vitale M, Sivori S, Pende D, Moretta L, Moretta A. Coexpression of two functionally independent p58 inhibitory receptors in human natural killer cell clones results in the inability to kill all normal allogeneic target cells. Proc Natl Acad Sci USA. (1995) 92:3536–40. doi: 10.1073/pnas.92.8.3536
58. Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med. (1994) 180:537–43. doi: 10.1084/jem.180.2.537
59. Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. (1995) 181:1133–44. doi: 10.1084/jem.181.3.1133
60. Ciccone E, Pende D, Viale O, Di Donato C, Tripodi G, Orengo AM, et al. Evidence of a natural killer (NK) cell repertoire for (allo) antigen recognition: definition of five distinct NK-determined allospecificities in humans. J Exp Med. (1992) 175:709–18. doi: 10.1084/jem.175.3.709
61. Lanier LL, Gumperz JE, Parham P, Melero I, Lopez-Botet M, Phillips JH. The NKB1 and HP-3E4 NK cells receptors are structurally distinct glycoproteins and independently recognize polymorphic HLA-B and HLA-C molecules. J Immunol. (1995) 154:3320–7.
62. Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, et al. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med. (1995) 182:875–84. doi: 10.1084/jem.182.3.875
63. Bottino C, Sivori S, Vitale M, Cantoni C, Falco M, Pende D, et al. A novel surface molecule homologous to the p58/p50 family of receptors is selectively expressed on a subset of human natural killer cells and induces both triggering of cell functions and proliferation. Eur J Immunol. (1996) 26:1816–24. doi: 10.1002/eji.1830260823
64. Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati MS, Vitale M, et al. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity. (1995) 2:439–49. doi: 10.1016/1074-7613(95)90025-X
65. Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. (1995) 268:405–8. doi: 10.1126/science.7716543
66. Biassoni R, Cantoni C, Falco M, Verdiani S, Bottino C, Vitale M, et al. The human leukocyte antigen (HLA)-C-specific “activatory” or “inhibitory” natural killer cell receptors display highly homologous extracellular domains but differ in their transmembrane and intracytoplasmic portions. J Exp Med. (1996) 183:645–50. doi: 10.1084/jem.183.2.645
67. D'Andrea A, Chang C, Franz-Bacon K, McClanahan T, Phillips JH, Lanier LL. Molecular cloning of NKB1. A natural killer cell receptor for HLA-B allotypes. J Immunol. (1995) 155:2306–10.
68. Chang C, Rodriguez A, Carretero M, Lopez-Botet M, Phillips JH, Lanier LL. Molecular characterization of human CD94: a type II membrane glycoprotein related to the C-type lectin superfamily. Eur J Immunol. (1995) 25:2433–7. doi: 10.1002/eji.1830250904
69. Moretta A, Vitale M, Sivori S, Bottino C, Morelli L, Augugliaro R, et al. Human natural killer cell receptors for HLA-class I molecules. Evidence that the Kp43 (CD94) molecule functions as receptor for HLA-B alleles. J Exp Med. (1994) 180:545–55. doi: 10.1084/jem.180.2.545
70. Perez-Villar JJ, Melero I, Rodriguez A, Carretero M, Aramburu J, Sivori S, et al. Functional ambivalence of the Kp43 (CD94) NK cell-associated surface antigen. J Immunol. (1995) 154:5779–88.
71. Pende D, Biassoni R, Cantoni C, Verdiani S, Falco M, di Donato C, et al. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J Exp Med. (1996) 184:505–18. doi: 10.1084/jem.184.2.505
72. Dohring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A1,2. J Immunol. (1996) 156:3098–101.
73. Mingari MC, Vitale C, Cambiaggi A, Schiavetti F, Melioli G, Ferrini S, et al. Cytolytic T lymphocytes displaying natural killer (NK)-like activity: expression of NK-related functional receptors for HLA class I molecules (p58 and CD94) and inhibitory effect on the TCR-mediated target cell lysis or lymphokine production. Int Immunol. (1995) 7:697–703. doi: 10.1093/intimm/7.4.697
74. Mingari MC, Schiavetti F, Ponte M, Vitale C, Maggi E, Romagnani S, et al. Human CD8+ T lymphocyte subsets that express HLA class I-specific inhibitory receptors represent oligoclonally or monoclonally expanded cell populations. Proc Natl Acad Sci USA. (1996) 93:12433–8. doi: 10.1073/pnas.93.22.12433
75. Pietra G, Romagnani C, Falco M, Vitale M, Castriconi R, Pende D, et al. The analysis of the natural killer-like activity of human cytolytic T lymphocytes revealed HLA-E as a novel target for TCR alpha/beta-mediated recognition. Eur J Immunol. (2001) 31:3687–93. doi: 10.1002/1521-4141(200112)31:12<3687::AID-IMMU3687>3.0.CO;2-C
76. Moretta L, Romagnani C, Pietra G, Moretta A, Mingari MC. NK-CTLs, a novel HLA-E-restricted T-cell subset. Trends Immunol. (2003) 24:136–43 doi: 10.1016/S1471-4906(03)00031-0
77. Long EO, Colonna M, Lanier LL. Inhibitory MHC class I receptors on NK and T cells: a standard nomenclature. Immunol Today. (1996) 17:100. doi: 10.1016/0167-5699(96)80590-1
78. Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Hum Immunol. (2003) 64:648–54. doi: 10.1016/S0198-8859(03)00067-3
79. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. (2015) 43:D423–31. doi: 10.1093/nar/gku1161
80. Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. (1997) 7:753–63. doi: 10.1016/S1074-7613(00)80394-5
81. Norman PJ, Stephens HA, Verity DH, Chandanayingyong D, Vaughan RW. Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics. (2001) 52:195–205. doi: 10.1007/s002510000281
82. Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. (2001) 15:363–74. doi: 10.1016/S1074-7613(01)00197-2
83. Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. (2002) 168:2307–15. doi: 10.4049/jimmunol.168.5.2307
84. Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. (2002) 20:217–51. doi: 10.1146/annurev.immunol.20.092501.134942
85. Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. (2002) 190:40–52. doi: 10.1034/j.1600-065X.2002.19004.x
86. Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE. (2010) 5:e15115. doi: 10.1371/journal.pone.0015115
87. Jiang W, Johnson C, Jayaraman J, Simecek N, Noble J, Moffatt MF, et al. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. (2012) 22:1845–54. doi: 10.1101/gr.137976.112
88. Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SG. IPD–the immuno polymorphism database. Nucleic Acids Res. (2013) 41:D1234–40. doi: 10.1093/nar/gks1140
89. Pyo CW, Wang R, Vu Q, Cereb N, Yang SY, Duh FM, et al. Recombinant structures expand and contract inter and intragenic diversification at the KIR locus. BMC Genomics. (2013) 14:89. doi: 10.1186/1471-2164-14-89
90. Vilches C, Castano J, Gomez-Lozano N, Estefania E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. (2007) 70:415–22. doi: 10.1111/j.1399-0039.2007.00923.x
91. Nong T, Saito K, Blair L, Tarsitani C, Lee JH. KIR genotyping by reverse sequence-specific oligonucleotide methodology. Tissue Antigens. (2007) 69(Suppl. 1):92–5. doi: 10.1111/j.1399-0039.2006.762_3.x
92. Houtchens KA, Nichols RJ, Ladner MB, Boal HE, Sollars C, Geraghty DE, et al. High-throughput killer cell immunoglobulin-like receptor genotyping by MALDI-TOF mass spectrometry with discovery of novel alleles. Immunogenetics. (2007) 59:525–37. doi: 10.1007/s00251-007-0222-x
93. Vendelbosch S, de Boer M, Gouw RA, Ho CK, Geissler J, Swelsen WT, et al. Extensive variation in gene copy number at the killer immunoglobulin-like receptor locus in humans. PLoS ONE. (2013) 8:e67619. doi: 10.1371/journal.pone.0067619
94. Roberts CH, Jiang W, Jayaraman J, Trowsdale J, Holland MJ, Traherne JA. Killer-cell Immunoglobulin-like Receptor gene linkage and copy number variation analysis by droplet digital PCR. Genome Med. (2014) 6:20. doi: 10.1186/gm537
95. Boudreau JE, Le Luduec JB, Hsu KC. Development of a novel multiplex PCR assay to detect functional subtypes of KIR3DL1 alleles. PLoS ONE. (2014) 9:e99543. doi: 10.1371/journal.pone.0099543
96. Jiang W, Johnson C, Simecek N, Lopez-Alvarez MR, Di D, Trowsdale J, et al. qKAT: a high-throughput qPCR method for KIR gene copy number and haplotype determination. Genome Med. (2016) 8:99. doi: 10.1186/s13073-016-0358-0
97. Norman PJ, Hollenbach JA, Nemat-Gorgani N, Marin WM, Norberg SJ, Ashouri E, et al. Defining KIR and HLA class I genotypes at highest resolution via high-throughput sequencing. Am J Hum Genet. (2016) 99:375–91. doi: 10.1016/j.ajhg.2016.06.023
98. Maniangou B, Legrand N, Alizadeh M, Guyet U, Willem C, David G, et al. Killer immunoglobulin-like receptor allele determination using next-generation sequencing technology. Front Immunol. (2017) 8:547. doi: 10.3389/fimmu.2017.00547
99. Luduec JL, Kudva A, Boudreau JE, Hsu KC. Novel multiplex PCR-SSP method for centromeric KIR allele discrimination. Sci Rep. (2018) 8:14853. doi: 10.1038/s41598-018-33135-1
100. Closa L, Vidal F, Herrero MJ, Caro JL. Design and validation of a multiplex KIR and HLA class I genotyping method using next generation sequencing. Front. Immunol. (2018) 9:2991. doi: 10.3389/fimmu.2018.02991
101. Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. (2000) 97:4778–83. doi: 10.1073/pnas.080588597
102. Hsu KC, Liu XR, Selvakumar A, Mickelson E, O'Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. (2002) 169:5118–29. doi: 10.4049/jimmunol.169.9.5118
103. Witt CS, Dewing C, Sayer DC, Uhrberg M, Parham P, Christiansen FT. Population frequencies and putative haplotypes of the killer cell immunoglobulin-like receptor sequences and evidence for recombination. Transplantation. (1999) 68:1784–9. doi: 10.1097/00007890-199912150-00024
104. Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The genomic context of natural killer receptor extended gene families. Immunol Rev. (2001) 181:20–38. doi: 10.1034/j.1600-065X.2001.1810102.x
105. Martin MP, Single RM, Wilson MJ, Trowsdale J, Carrington M. KIR haplotypes defined by segregation analysis in 59 Centre d'Etude Polymorphisme Humain (CEPH) families. Immunogenetics. (2008) 60:767–74. doi: 10.1007/s00251-008-0334-y
106. Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, et al. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. (2009) 19:757–69. doi: 10.1101/gr.085738.108
107. Parham P. Killer cell immunoglobulin-like receptor diversity: balancing signals in the natural killer cell response. Immunol Lett. (2004) 92:11–3. doi: 10.1016/j.imlet.2003.11.016
108. Parham P. Influence of KIR diversity on human immunity. Adv Exp Med Biol. (2005) 560:47–50. doi: 10.1007/0-387-24180-9_6
109. Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. (2003) 171:2192–5. doi: 10.4049/jimmunol.171.5.2192
110. Gomez-Lozano N, Estefania E, Williams F, Halfpenny I, Middleton D, Solis R, et al. The silent KIR3DP1 gene (CD158c) is transcribed and might encode a secreted receptor in a minority of humans, in whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 genes are duplicated. Eur J Immunol. (2005) 35:16–24. doi: 10.1002/eji.200425493
111. Traherne JA, Martin M, Ward R, Ohashi M, Pellett F, Gladman D, et al. Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet. (2010) 19:737–51. doi: 10.1093/hmg/ddp538
112. Ordonez D, Gomez-Lozano N, Rosales L, Vilches C. Molecular characterisation of KIR2DS2*005, a fusion gene associated with a shortened KIR haplotype. Genes Immun. (2011) 12:544–51. doi: 10.1038/gene.2011.35
113. Bono M, Pende D, Bertaina A, Moretta A, Della Chiesa M, Sivori S, et al. Analysis of KIR3DP1 polymorphism provides relevant information on centromeric KIR gene content. J Immunol. (2018) 201:1460–7. doi: 10.4049/jimmunol.1800564
114. Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. (2003) 171:6640–9. doi: 10.4049/jimmunol.171.12.6640
115. VandenBussche CJ, Dakshanamurthy S, Posch PE, Hurley CK. A single polymorphism disrupts the killer Ig-like receptor 2DL2/2DL3 D1 domain. J Immunol. (2006) 177:5347–57. doi: 10.4049/jimmunol.177.8.5347
116. Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, et al. Polymorphic HLA-C receptors balance the functional characteristics of KIR haplotypes. J Immunol. (2015) 195:3160–70. doi: 10.4049/jimmunol.1501358
117. Alicata C, Pende D, Meazza R, Canevali P, Loiacono F, Bertaina A, et al. Hematopoietic stem cell transplantation: improving alloreactive Bw4 donor selection by genotyping codon 86 of KIR3DL1/S1. Eur J Immunol. (2016) 46:1511–7. doi: 10.1002/eji.201546236
118. Martin AM, Freitas EM, Witt CS, Christiansen FT. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics. (2000) 51:268–80. doi: 10.1007/s002510050620
119. Goodridge JP, Lathbury LJ, Steiner NK, Shulse CN, Pullikotil P, Seidah NG, et al. Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. Eur J Immunol. (2007) 37:199–211. doi: 10.1002/eji.200636316
120. Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. (2001) 166:2992–3001. doi: 10.4049/jimmunol.166.5.2992
121. Dunphy SE, Guinan KJ, Chorcora CN, Jayaraman J, Traherne JA, Trowsdale J, et al. 2DL1, 2DL2 and 2DL3 all contribute to KIR phenotype variability on human NK cells. Genes Immun. (2015) 16:301–10. doi: 10.1038/gene.2015.15
122. Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. (2008) 180:3969–79. doi: 10.4049/jimmunol.180.6.3969
123. Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. (2005) 175:5222–9. doi: 10.4049/jimmunol.175.8.5222
124. Sharma D, Bastard K, Guethlein LA, Norman PJ, Yawata N, Yawata M, et al. Dimorphic motifs in D0 and D1+D2 domains of killer cell Ig-like receptor 3DL1 combine to form receptors with high, moderate, and no avidity for the complex of a peptide derived from HIV and HLA-A*2402. J Immunol. (2009) 183:4569–82. doi: 10.4049/jimmunol.0901734
125. Mulrooney TJ, Zhang AC, Goldgur Y, Boudreau JE, Hsu KC. KIR3DS1-Specific D0 domain polymorphisms disrupt KIR3DL1 surface expression and HLA binding. J Immunol. (2015) 195:1242–50. doi: 10.4049/jimmunol.1500243
126. Saunders PM, Pymm P, Pietra G, Hughes VA, Hitchen C, O'Connor GM, et al. Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J Exp Med. (2016) 213:791–807. doi: 10.1084/jem.20152023
127. Bottino C, Falco M, Sivori S, Moretta L, Moretta A, Biassoni R. Identification and molecular characterization of a natural mutant of the p50.2/KIR2DS2 activating NK receptor that fails to mediate NK cell triggering. Eur J Immunol. (2000) 30:3569–74. doi: 10.1002/1521-4141(200012)30:12<3569::AID-IMMU3569>3.0.CO;2-E
128. Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N, et al. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. (2009) 114:5182–90. doi: 10.1182/blood-2009-07-231977
129. Hilton HG, Norman PJ, Nemat-Gorgani N, Goyos A, Hollenbach JA, Henn BM, et al. Loss and gain of natural killer cell receptor function in an african hunter-gatherer population. PLoS Genet. (2015) 11:e1005439. doi: 10.1371/journal.pgen.1005439
130. Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. (2000) 405:537–43. doi: 10.1038/35014520
131. Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. (2001) 2:452–60. doi: 10.1038/87766
132. Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. (2004) 34:1673–9. doi: 10.1002/eji.200425089
133. Liu J, Xiao Z, Ko HL, Shen M, Ren EC. Activating killer cell immunoglobulin-like receptor 2DS2 binds to HLA-A*11. Proc Natl Acad Sci USA. (2014) 111:2662–7. doi: 10.1073/pnas.1322052111
134. Hilton HG, Moesta AK, Guethlein LA, Blokhuis J, Parham P, Norman PJ. The production of KIR-Fc fusion proteins and their use in a multiplex HLA class I binding assay. J Immunol Methods. (2015) 425:79–87. doi: 10.1016/j.jim.2015.06.012
135. Frazier WR, Steiner N, Hou L, Dakshanamurthy S, Hurley CK. Allelic variation in KIR2DL3 generates a KIR2DL2-like receptor with increased binding to its HLA-C ligand. J Immunol. (2013) 190:6198–208. doi: 10.4049/jimmunol.1300464
136. Hilton HG, Parham P. Missing or altered self: human NK cell receptors that recognize HLA-C. Immunogenetics. (2017) 69:567–79. doi: 10.1007/s00251-017-1001-y
137. Sim MJW, Malaker SA, Khan A, Stowell JM, Shabanowitz J, Peterson ME, et al. Canonical and cross-reactive binding of NK cell inhibitory receptors to HLA-C allotypes is dictated by peptides bound to HLA-C. Front. Immunol. (2017) 8:193. doi: 10.3389/fimmu.2017.00193
138. Selvakumar A, Steffens U, Dupont B. NK cell receptor gene of the KIR family with two IG domains but highest homology to KIR receptors with three IG domains. Tissue Antigens. (1996) 48(4 Pt 1):285–94. doi: 10.1111/j.1399-0039.1996.tb02647.x
139. Rajagopalan S, Fu J, Long EO. Cutting edge: induction of IFN-gamma production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J Immunol. (2001) 167:1877–81. doi: 10.4049/jimmunol.167.4.1877
140. Stern M, Ruggeri L, Capanni M, Mancusi A, Velardi A. Human leukocyte antigens A23, A24, and A32 but not A25 are ligands for KIR3DL1. Blood. (2008) 112:708–10. doi: 10.1182/blood-2008-02-137521
141. O'Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. (2007) 178:235–41 doi: 10.4049/jimmunol.178.1.235
142. Goodridge JP, Burian A, Lee N, Geraghty DE. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J Immunol. (2013) 191:3553–62. doi: 10.4049/jimmunol.1300081
143. Sivori S, Falco M, Carlomagno S, Romeo E, Soldani C, Bensussan A, et al. A novel KIR-associated function: evidence that CpG DNA uptake and shuttling to early endosomes is mediated by KIR3DL2. Blood. (2010) 116:1637–47. doi: 10.1182/blood-2009-12-256586
144. Olcese L, Cambiaggi A, Semenzato G, Bottino C, Moretta A, Vivier E. Human killer cell activatory receptors for MHC class I molecules are included in a multimeric complex expressed by natural killer cells. J Immunol. (1997) 158:5083–6.
145. Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. (1998) 391:703–7. doi: 10.1038/35642
146. Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, et al. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur J Immunol. (1997) 27:3095–9. doi: 10.1002/eji.1830271203
147. Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci USA. (2005) 102:13224–9. doi: 10.1073/pnas.0503594102
148. Chewning JH, Gudme CN, Hsu KC, Selvakumar A, Dupont B. KIR2DS1-Positive NK cells mediate alloresponse against the C2 HLA-KIR ligand group in vitro. J Immunol. (2007) 179:854–68. doi: 10.4049/jimmunol.179.2.854
149. Pende D, Marcenaro S, Falco M, Martini S, Bernardo ME, Montagna D, et al. Anti-leukemia activity of alloreactive NK cells in KIR ligand-mismatched haploidentical HSCT for pediatric patients: evaluation of the functional role of activating KIR and redefinition of inhibitory KIR specificity. Blood. (2009) 113:3119–29. doi: 10.1182/blood-2008-06-164103
150. van der Ploeg K, Chang C, Ivarsson MA, Moffett A, Wills MR, Trowsdale J. Modulation of human leukocyte antigen-C by human cytomegalovirus stimulates KIR2DS1 recognition by natural killer cells. Fronti. Immunol. (2017) 8:298. doi: 10.3389/fimmu.2017.00298
151. David G, Djaoud Z, Willem C, Legrand N, Rettman P, Gagne K, et al. Large spectrum of HLA-C recognition by killer Ig-like receptor (KIR)2DL2 and KIR2DL3 and restricted C1 specificity of KIR2DS2: dominant impact of KIR2DL2/KIR2DS2 on KIR2D NK cell repertoire formation. J Immunol. (2013) 191:4778–88. doi: 10.4049/jimmunol.1301580
152. Naiyer MM, Cassidy SA, Magri A, Cowton V, Chen K, Mansour S, et al. KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA-C. Sci Immunol. (2017) 2:eaal5296. doi: 10.1126/sciimmunol.aal5296
153. Thiruchelvam-Kyle L, Hoelsbrekken SE, Saether PC, Bjornsen EG, Pende D, Fossum S, et al. The activating human NK cell receptor KIR2DS2 recognizes a beta2-microglobulin-independent ligand on cancer cells. J Immunol. (2017) 198:2556–67. doi: 10.4049/jimmunol.1600930
154. Garcia-Beltran WF, Holzemer A, Martrus G, Chung AW, Pacheco Y, Simoneau CR, et al. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat Immunol. (2016) 17:1067–74. doi: 10.1038/ni.3513
155. Burian A, Wang KL, Finton KA, Lee N, Ishitani A, Strong RK, et al. HLA-F and MHC-I open conformers bind natural killer cell Ig-like receptor KIR3DS1. PLoS ONE. (2016) 11:e0163297. doi: 10.1371/journal.pone.0163297
156. Carlomagno S, Falco M, Bono M, Alicata C, Garbarino L, Mazzocco M, et al. KIR3DS1-mediated recognition of HLA-*B51: modulation of KIR3DS1 responsiveness by self HLA-B allotypes and effect on NK cell licensing. Front. Immunol. (2017) 8:581. doi: 10.3389/fimmu.2017.00581
157. VandenBussche CJ, Mulrooney TJ, Frazier WR, Dakshanamurthy S, Hurley CK. Dramatically reduced surface expression of NK cell receptor KIR2DS3 is attributed to multiple residues throughout the molecule. Genes Immun. (2009) 10:162–73. doi: 10.1038/gene.2008.91
158. Della Chiesa M, Romeo E, Falco M, Balsamo M, Augugliaro R, Moretta L, et al. Evidence that the KIR2DS5 gene codes for a surface receptor triggering natural killer cell function. Eur J Immunol. (2008) 38:2284–9. doi: 10.1002/eji.200838434
159. Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. (2009) 206:2557–72. doi: 10.1084/jem.20091010
160. Sim MJW, Rajagopalan S, Altmann DM, Boyton RJ, Sun PD, Long EO. (2019). HLA-C-restricted presentation of a conserved bacterial epitope to an innate NK cell receptor. biorxiv. doi: 10.1101/550889
161. Blokhuis JH, Hilton HG, Guethlein LA, Norman PJ, Nemat-Gorgani N, Nakimuli A, et al. KIR2DS5 allotypes that recognize the C2 epitope of HLA-C are common among Africans and absent from Europeans. Immun Inflamm Dis. (2017) 5:461–8. doi: 10.1002/iid3.178
162. Elliott JM, Yokoyama WM. Unifying concepts of MHC-dependent natural killer cell education. Trends Immunol. (2011) 32:364–72. doi: 10.1016/j.it.2011.06.001
163. Boudreau JE, Hsu KC. Natural killer cell education and the response to infection and cancer therapy: stay tuned. Trends Immunol. (2018) 39:222–39. doi: 10.1016/j.it.2017.12.001
164. Horowitz A, Djaoud Z, Nemat-Gorgani N, Blokhuis J, Hilton HG, Beziat V, et al. Class I HLA haplotypes form two schools that educate NK cells in different ways. Sci Immunol. (2016) 1:eaag1672. doi: 10.1126/sciimmunol.aag1672
165. Beziat V, Traherne JA, Liu LL, Jayaraman J, Enqvist M, Larsson S, et al. Influence of KIR gene copy number on natural killer cell education. Blood. (2013) 121:4703–7. doi: 10.1182/blood-2012-10-461442
166. Manser AR, Weinhold S, Uhrberg M. Human KIR repertoires: shaped by genetic diversity and evolution. Immunol Rev. (2015) 267:178–96. doi: 10.1111/imr.12316
167. Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The broad spectrum of human natural killer cell diversity. Immunity. (2017) 47:820–33. doi: 10.1016/j.immuni.2017.10.008
168. Fauriat C, Ivarsson MA, Ljunggren HG, Malmberg KJ, Michaelsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood. (2010) 115:1166–74. doi: 10.1182/blood-2009-09-245746
169. Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. (2013) 5:208ra145. doi: 10.1126/scitranslmed.3006702
170. Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. (2013) 121:2678–88. doi: 10.1182/blood-2012-10-459545
171. Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. (2015) 42:443–56. doi: 10.1016/j.immuni.2015.02.008
172. Pupuleku A, Costa-García M, Farré D, Hengel H, Angulo A, Muntasell A, et al. Elusive role of the CD94/NKG2C NK cell receptor in the response to cytomegalovirus: novel experimental observations in a reporter cell system. Front. Immunol. (2017) 8:1317. doi: 10.3389/fimmu.2017.01317
173. Hammer Q, Ruckert T, Romagnani C. Natural killer cell specificity for viral infections. Nat Immunol. (2018) 19:800–8. doi: 10.1038/s41590-018-0163-6
174. Hammer Q, Ruckert T, Borst EM, Dunst J, Haubner A, Durek P, et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol. (2018) 19:453–63. doi: 10.1038/s41590-018-0082-6
175. Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. (2013) 13:133–44. doi: 10.1038/nri3370
176. Parolini S, Bottino C, Falco M, Augugliaro R, Giliani S, Franceschini R, et al. X-linked lymphoproliferative disease. 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein-Barr virus-infected cells. J Exp Med. (2000) 192:337–46 doi: 10.1084/jem.192.3.337
177. Meazza R, Falco M, Marcenaro S, Loiacono F, Canevali P, Bellora F, et al. Inhibitory 2B4 contributes to NK cell education and immunological derangements in XLP1 patients. Eur J Immunol. (2017) 47:1051–61. doi: 10.1002/eji.201646885
178. Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. (1992) 11:3887–95 doi: 10.1002/j.1460-2075.1992.tb05481.x
179. Della Chiesa M, Pesce S, Muccio L, Carlomagno S, Sivori S, Moretta A, et al. Features of memory-like and PD-1(+) human NK cell subsets. Front Immunol. (2016) 7:351. doi: 10.3389/fimmu.2016.00351
180. Sivori S, Vacca P, Del Zotto G, Munari E, Mingari MC, Moretta L. Human NK cells: surface receptors, inhibitory checkpoints, and translational applications. Cell Mol Immunol. (2019) 16:430–41. doi: 10.1038/s41423-019-0206-4
181. Molgora M, Bonavita E, Ponzetta A, Riva F, Barbagallo M, Jaillon S, et al. IL-1R8 is a checkpoint in NK cells regulating anti-tumour and anti-viral activity. Nature. (2017) 551:110–4. doi: 10.1038/nature24293
182. Munari E, Rossi G, Zamboni G, Lunardi G, Marconi M, Sommaggio M, et al. PD-L1 Assays 22C3 and SP263 are not interchangeable in non-small cell lung cancer when considering clinically relevant cutoffs: an interclone evaluation by differently trained pathologists. Am J Surg Pathol. (2018) 42:1384–9. doi: 10.1097/PAS.0000000000001105
183. Munari E, Zamboni G, Marconi M, Sommaggio M, Brunelli M, Martignoni G, et al. PD-L1 expression heterogeneity in non-small cell lung cancer: evaluation of small biopsies reliability. Oncotarget. (2017) 8:90123–31. doi: 10.18632/oncotarget.21485
184. Munari E, Zamboni G, Lunardi G, Marchionni L, Marconi M, Sommaggio M, et al. PD-L1 Expression heterogeneity in non-small cell lung cancer: defining criteria for harmonization between biopsy specimens and whole sections. J Thorac Oncol. (2018) 13:1113–20. doi: 10.1016/j.jtho.2018.04.017
185. Benson DM, Jr., Cohen AD, Jagannath S, Munshi NC, Spitzer G, Hofmeister CC, et al. A phase I trial of the anti-KIR antibody IPH2101 and lenalidomide in patients with relapsed/refractory multiple myeloma. Clin Cancer Res. (2015) 21:4055–61. doi: 10.1158/1078-0432.CCR-15-0304
186. Bagot M, Moretta A, Sivori S, Biassoni R, Cantoni C, Bottino C, et al. CD4(+) cutaneous T-cell lymphoma cells express the p140-killer cell immunoglobulin-like receptor. Blood. (2001) 97:1388–91. doi: 10.1182/blood.V97.5.1388
187. Van Der Weyden C, Bagot M, Neeson P, Darcy PK, Prince HM. IPH4102, a monoclonal antibody directed against the immune receptor molecule KIR3DL2, for the treatment of cutaneous T-cell lymphoma. Expert Opin Investig Drugs. (2018) 27:691–7. doi: 10.1080/13543784.2018.1498081
188. Andre P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell. (2018) 175:1731–43 e13. doi: 10.1016/j.cell.2018.10.014
189. van Montfoort N, Borst L, Korrer MJ, Sluijter M, Marijt KA, Santegoets SJ, et al. NKG2A Blockade potentiates CD8 T cell immunity induced by cancer vaccines. Cell. (2018) 175:1744–55 e15. doi: 10.1016/j.cell.2018.10.028
190. Mingari MC, Pietra G, Moretta L. Immune checkpoint inhibitors: anti-NKG2A antibodies on board. Trends Immunol. (2019) 40:83–5. doi: 10.1016/j.it.2018.12.009
191. Mingari MC, Ponte M, Bertone S, Schiavetti F, Vitale C, Bellomo R, et al. HLA class I-specific inhibitory receptors in human T lymphocytes: interleukin 15-induced expression of CD94/NKG2A in superantigen- or alloantigen-activated CD8+ T cells. Proc Natl Acad Sci USA. (1998) 95:1172–7. doi: 10.1073/pnas.95.3.1172
192. Bertone S, Schiavetti F, Bellomo R, Vitale C, Ponte M, Moretta L, et al. Transforming growth factor-beta-induced expression of CD94/NKG2A inhibitory receptors in human T lymphocytes. Eur J Immunol. (1999) 29:23–9. doi: 10.1002/(SICI)1521-4141(199901)29:01<23::AID-IMMU23>3.0.CO;2-Y
193. Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain JF, de Smet C, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. (1997) 6:199–208. doi: 10.1016/S1074-7613(00)80426-4
194. Romagne F, Andre P, Spee P, Zahn S, Anfossi N, Gauthier L, et al. Preclinical characterization of 1-7F9, a novel human anti-KIR receptor therapeutic antibody that augments natural killer-mediated killing of tumor cells. Blood. (2009) 114:2667–77. doi: 10.1182/blood-2009-02-206532
195. Ruggeri L, Urbani E, Andre P, Mancusi A, Tosti A, Topini F, et al. Effects of anti-NKG2A antibody administration on leukemia and normal hematopoietic cells. Haematologica. (2016) 101:626–33. doi: 10.3324/haematol.2015.135301
196. Aversa F, Tabilio A, Velardi A, Cunningham I, Terenzi A, Falzetti F, et al. Treatment of high-risk acute leukemia with T-cell-depleted stem cells from related donors with one fully mismatched HLA haplotype. N Engl J Med. (1998) 339:1186–93. doi: 10.1056/NEJM199810223391702
197. Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. (2002) 295:2097–100. doi: 10.1126/science.1068440
198. Locatelli F, Pende D, Falco M, Della Chiesa M, Moretta A, Moretta L. NK cells mediate a crucial graft-versus-leukemia effect in haploidentical-HSCT to cure high-risk acute leukemia. Trends Immunol. (2018) 39:577–90. doi: 10.1016/j.it.2018.04.009
199. Karre K. Immunology. A perfect mismatch. Science. (2002) 295:2029–31. doi: 10.1126/science.1070538
200. Locatelli F, Merli P, Pagliara D, Li Pira G, Falco M, Pende D, et al. Outcome of children with acute leukemia given HLA-haploidentical HSCT after alphabeta T-cell and B-cell depletion. Blood. (2017) 130:677–85. doi: 10.1182/blood-2017-04-779769
201. Locatelli F, Bertaina A, Bertaina V, Merli P. Cytomegalovirus in hematopoietic stem cell transplant recipients - management of infection. Expert Rev Hematol. (2016) 9:1093–105. doi: 10.1080/17474086.2016.1242406
202. Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. (2011) 119:2665–74. doi: 10.1182/blood-2011-10-386995
203. Della Chiesa M, Falco M, Muccio L, Bertaina A, Locatelli F, Moretta A. Impact of HCMV infectionon NK cell development and function after HSCT. Front. Immunol. (2013) 4:458. doi: 10.3389/fimmu.2013.00458
204. Muccio L, Bertaina A, Falco M, Pende D, Meazza R, Lopez-Botet M, et al. Analysis of memory-like natural killer cells in human cytomegalovirus-infected children undergoing alphabeta+T and B cell-depleted hematopoietic stem cell transplantation for hematological malignancies. Haematologica. (2016) 101:371–81. doi: 10.3324/haematol.2015.134155
205. Della Chiesa M, Falco M, Bertaina A, Muccio L, Alicata C, Frassoni F, et al. Human Cytomegalovirus Infection Promotes Rapid Maturation of NK Cells Expressing Activating Killer Ig-Like Receptor in Patients Transplanted with NKG2C-/- Umbilical Cord Blood. J Immunol. (2014). 1921471–9. doi: 10.4049/jimmunol.1302053
Keywords: HLA class I, killer immunoglobulin-like receptors, KIR ligands, inhibitory checkpoints, NK alloreactivity, NK cell education, polymorphism
Citation: Pende D, Falco M, Vitale M, Cantoni C, Vitale C, Munari E, Bertaina A, Moretta F, Del Zotto G, Pietra G, Mingari MC, Locatelli F and Moretta L (2019) Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front. Immunol. 10:1179. doi: 10.3389/fimmu.2019.01179
Received: 24 March 2019; Accepted: 09 May 2019;
Published: 28 May 2019.
Edited by:
Eric Vivier, INSERM U1104 Centre d'Immunologie de Marseille-Luminy, FranceReviewed by:
Hans-Gustaf Ljunggren, Karolinska Institute (KI), SwedenCopyright © 2019 Pende, Falco, Vitale, Cantoni, Vitale, Munari, Bertaina, Moretta, Del Zotto, Pietra, Mingari, Locatelli and Moretta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorenzo Moretta, bG9yZW56by5tb3JldHRhQG9wYmcubmV0
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.