- Infectious Disease Biology Group, Institute of Life Sciences (ILS), Bhubaneswar, India
Human cytomegalovirus (HCMV) is the most common cause of congenital infections and is an important pathogen in immunocompromised individuals. Despite a robust host immune system, HCMV able to replicate, evade host defenses, establish latency for life. A significant portion of HCMV genome dedicated to encode gene products for modulation of host immune response. Growing number of HCMV gene products are being recognized to play role in immune evasion. Information on viral immune evasion mechanisms by which HCMV persists in host will be useful in devising antiviral intervention strategies and development of new vaccines. This minireview provides a brief overview of immune evasion strategy adapted by HCMV by utilizing its gene products in modulation of host immune response.
Introduction
The human cytomegalovirus (HCMV) is a ubiquitous β-herpesvirus that establishes lifelong persistent infection following introduction to an immunocompetent host. Primary infection in a healthy individual leads to mild febrile illness, whereas HCMV causes serious complications in immunosuppressed subjects, especially in transplant recipients and in immunocompromised patients (1, 2). Human cytomegalovirus is the most common cause of congenital infections leading to neurodevelopmental sequelae. Each year, 20,000–40,000 children are born with congenital human CMV infection in the US, of which 10–15% develops permanent sequelae including sensorineural hearing loss (3–5). Furthermore, substantial fraction of the asymptomatic children develops late onset hearing loss. In an attempt to reduce these disabilities and loss of life, as well as the associated economic cost, the Institute of Medicine of National Academy of Sciences, USA have ranked the development of HCMV vaccine as a highest priority (6, 7).
Decades of research on cytomegalovirus has provided novel insight in understanding the host immune response and evasion strategies adapted by the virus. HCMV has dedicated more than half of its genome encoding for modulation of host response to infection (8, 9). This mini-review article discusses on current understanding of HCMV gene products in modulation of host immune response with an emphases on the immune evasion by interference in antigen presentation and activation of NK cells, viral strain diversity and superinfection in immune subject.
Modulation of Immune Responses by HCMV Gene Products
The virus has co-evolved with its host organism for 200 million years (9, 10). HCMV has a large genome size of 236 kb with unique long (UL) and unique short (US) regions flanked by terminal repeats and internal repeats. The genome has been annotated and encodes 167 gene products, as well as non-coding RNAs, microRNAs, and with an extensive alternate mRNA splicing. However, recent report suggested that HCMV encode to have more than 750 translated ORFs (11). More than 40 HCMV gene products are recognized to have a role in modulating the host immune response following infection (12, 13). Both the innate and adaptive arms of the immune system play a crucial role in controlling HCMV infection (12, 14). Despite a robust host immune system, HCMV is able to establish latency and once infected the HCMV remains in the host for life. The virus remains latent in the myeloid progenitor cells during its dormant phase; however on stimulation, or when the immune system is suppressed, the virus can once again become active (15). The battle between the host immune system and the virus continues throughout life, with HCMV having evolved multiple mechanisms to evade the host immune response. The divergence of the immune response and incomplete viral control may be attributed to the diversity of immune modulators encoded by HCMV gene products [Figure 1, Table 1]. Many of these gene products are homologs of host genes involved in the immune response.
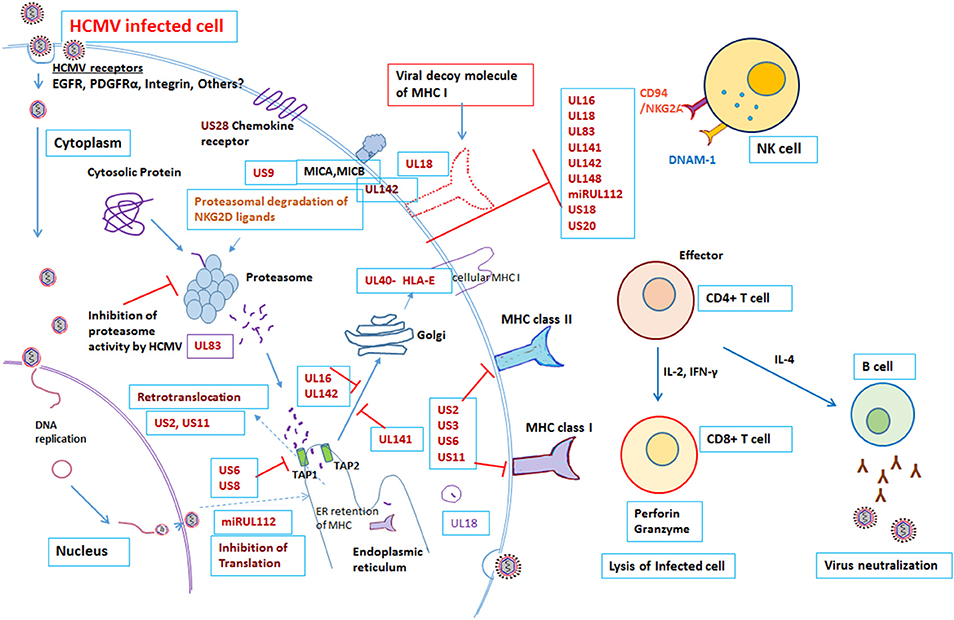
Figure 1. Modulation of Immune response by human cytomegalovirus. Overview of the interactions between HCMV and the immune system. Red “T” bars indicate inhibition. Blue arrows indicate activation. Detail mechanisms explained in the text.
To eliminate the virus, the host needs to have an effective immune system. After viral infection, host antigen presenting cells must present viral antigen to the immune cells in order to stimulate effector cells to eliminate the virus. However, HCMV has devised strategies to limit this presentation. NK cells are normally responsible for immediate control of viral infections; however, there are number of HCMV gene products that block NK cell mediated recognition. Approximately, there are 12 HCMV gene products, US20, UL16, UL17, UL18, UL40, UL43, UL140, UL83, UL141-UL144, and UL148, known to control NK cell modulation (Table 1). HCMV UL16, UL17, UL40, UL140, and UL142 genes all encode products that down-regulate NK cell activity by imitating the host HLA class I. For example, UL40 encodes a canonical ligand for HLA-E and negatively regulates NK cells, which results in down-regulation of activating ligand CD155 (20). Individuals with impaired NK cell function, succumbs to severe herpesvirus infections (31). In addition, HCMV gene products UL18 and UL83 (pp65) encode for an MHC-I homolog, modulate expression of other HCMV genes and inhibit NK cell lysis (12, 20). Furthermore, the HCMV microRNA miR-UL122 acts to suppress host MICB surface expression (13, 20, 29).
As is a common characteristic of herpes viruses, HCMV is able to interfere with the class I MHC molecule involved in antigen presentation to CD8+ T cells. HCMV establishes persistent infection by producing host homologous molecules that prevent recognition and interfere with antigen presentation, subverting the cytotoxic T lymphocytes (CTLs). Viral antigens are normally presented by the MHC class I proteins on the infected cell surface. HCMV gene products obstruct peptide translocation to the ER lumen and stimulate degradation of the MHC class I proteins before they can reach the cell surface. For example, the HCMV US3 gene product degrades the MHC class I heavy chain by interacting with Tapasin and retaining the class I molecule at the site of synthesis, in the ER. In addition, the US2 and US11 gene products relocate the heavy chain of MHC class I into the ER for proteosomal degradation. Similarly, another gene product of HCMV, US6, prevents peptide loading by inhibiting the binding of ATP to TAP, thereby preventing the transport of peptides through the TAP pore. The combined functions of the HCMV gene products US2, US3, US6, and US11, therefore, lead to peptide transport blockade, retention of MHC class I in the ER and ultimately proteasomal degradation. In addition, the gene product US2 interferes with MHC class II signal transduction by degradation of MHC class II proteins. US2 targets the class II DR and DMα chains for degradation in the cytosol, thereby preventing antigen presentation to CD4+ T lymphocytes (12, 14, 16, 32).
In addition to the above, the HCMV UL83 gene product, pp65 blocks the processing of immediate early-1 in the proteasome by phosphorylation. Besides, the tegument Protein UL82 evades antiviral immunity by inhibiting stimulator of interferon (STING) signaling (21) and may be responsible for induction of latency (15, 22). Recently, Nightingale et al. reported that the HCMV gene product UL145 facilitates degradation of the antiviral factor helicase like transcription factor (HLTF) by recruiting the host Cullin4 E3 ligase complex, and captures Cullin3 to invoke the strategy of immune evasion (27). Additionally, the HCMV late gene product UL111A encodes cmvIL-10, a homolog of human IL-10, which is expressed during viral latency, and causes a state of immune suppression (23). The cytokine Interleukin-10 has an immunosuppressive role on several effector cells of the immune system. The HCMV gene product cmvIL-10 exerts an immunosuppressive effect on the host by modulating the expression of the MHC class I and II molecules and interfering with dendritic cell (DC) function (24). In a murine model of CMV, following productive infection with CMV both in vitro and ex vivo, the virus reduced the expression of MHC as well as co-stimulation of DC. This eventually led to loss of expression of IL-2 and IL-12 and hindrance of DC differentiation (33–35). A recent report by Wang et al also demonstrated that the HCMV UL148 gene product suppresses co-stimulation and expression of the cell adhesion molecule CD58, endorsing cellular immune defense evasion by impairing NK and T cell activation (28). This work was further supported by HCMV UL148 mediated tropism and immune evasion by unfolded protein response (36). In Rhesus model, Rh159, a homolog of HCMV UL148 involved in retention of distinct set of costimulatory molecules and involved in NK cell evasion (37). HCMV UL148 gene products encode for avoidance of killing of HCMV infected cells from NK cells by down regulating MICA (38).
HCMV possesses a unique challenge, as it is able to super-infect in a subject already infected with the virus, even in the presence of a strong specific immune response. Several studies have demonstrated congenital HCMV infection in offspring of immune mothers because of reinfection with a different strain of virus (39–43). Further, congenital infected infant born to immune mother may develop sequelae similar to infants born to mother with primary infection during pregnancy. It has also been observed that infection with more than one strain of HCMV is common in nature (39, 40, 44). HCMV strain polymorphism could contribute to immune evasion. Since HCMV glycoproteins are highly polymorphic, antibody response to one strain may not efficiently neutralize infection with a different strain and this could enable to superinfection (45–47). In addition to interference in antigen presentation, the CMV gene products US2, US3, US6, and US11 encode for human homologs that interfere with the function of CD8+ T cells. This allowed viral replication and super-infection with a different strain of virus in a rhesus macaque model. This was confirmed, by the observation that US2-11 mutant virus, although able to produce infection, was unable to super-infect (17). However, further studies are needed to decipher the detailed mechanisms of the CTL response in contending with the combined action of these HCMV gene products. The large genome size of HCMV enables it to utilize an array of genes for host immune evasion, which allows long-term association and adaption of the virus in the host. In an immunocompetent host, viral latency is critical for its survival. After primary infection, the virus persists for a lifetime regardless of pre-existing immunity. During latency, the viral genome is maintained in the host without active replication and retains the capacity to reactivate in response to activation signals (48). Studies have linked various latency-associated determinants to HCMV latency (15), however, the detailed mechanisms of immune evasion during latency and how the virus persists in the host for life remains elusive. Deciphering these mechanisms could provide clues to allow us to prevent reactivation of this latent virus in congenital and transplant setup. Further, a note of caution is required; HCMV is strictly species specific. Since much of our understanding on cytomegalovirus biology is derived from in vitro cell culture studies and animal models, it is necessary to test these immune evasion functions in the appropriate setting. For instance, the UL18 gene product of HCMV encoding an MHC class I homolog was proposed to block NK cell activity by binding with KIR receptors; however, later studies have found it to enhance killing of infected fibroblasts by NK cells (12).
Further, extensive genetic variability has been observed in clinical isolate of HCMV (4, 40, 49–51), and even within a single host (4, 44, 52–54). High throughput sequencing of HCMV clinical isolates reveals that intrahost HCMV populations were as variable as seen in RNA virus quasispecies (52, 53). Viral strain diversity, differences in culture systems and population heterogeneity, make the generalization of genetic information difficult. In addition, a recent report showed that HCMV seroprevalence is related to a shift in immune phenotype along an age axis (55). This immunotypes varies in younger vs. elder individuals (55–57). In due course of evolution with the host, HCMV has been significant in shaping host immune system (57). HCMV also affects the host in response to infection with other pathogen. In HCMV seropositive children and in aging individuals have negative impact to Influenza; however, in younger individuals HCMV infection enhance immune response to influenza (58). Viral strain diversity could limit effective antiviral function, and the evasion strategy adapted by HCMV further complicates the development of an effective vaccine (45, 59). This underscores the need for large-scale genetic and immunological profiling studies, which could provide a decisive correlation on the nature of protective immune responses (56, 59–61).
HCMV has devised multiple strategies to interfere with antigen presentations and escape from CTL response, but this does not abrogate with the development of CTL response by host. This underscores the critical role of CD8+ T cells in HCMV infected cells as targets for immune clearance. Studies from adaptive transfer of HCMV specific CTL, in bone marrow transplant subjects, provide protection from HCMV disease (62). The complex interaction between the HCMV immune-evasins and host factors contributes to the levels of viral persistence in host (63). Information on viral immune evasion mechanisms by which HCMV persists in host will be useful in devising antiviral intervention strategies and development of new vaccines. Deletion of immune evasions could be a novel strategy for virus attenuation for vaccine candidate without compromising CD8 T cell response (64). Hansen et al reported that Simian immunodeficiency virus (SIV) protein expressing rhesus cytomegalovirus vector elicits SIV specific CD8+ T cells which recognizes unusual, diverse epitopes and results in immune clearance (65, 66). Thus, CMV vectors, genetically altered for diverse CD8+ T cell response could be useful for effective prophylactic and therapeutic vaccination (9, 65–68). Further, this could be useful in ultimately designing an effective vaccine that could protect primary as well as reinfections.
Conclusions
In conclusion, human cytomegalovirus is a master of disguise. HCMV has evolved mechanisms to replicate and evade the host immune system by targeting the host cell machinery. Information on the host cell receptor targeted by this virus and the mechanisms utilized to operate cellular processes and evade the host immune system will provide clues to viral pathogenesis. An increasing number of HCMV gene products have been reported to play roles in immune evasion. These gene products sophistically orchestrate to modulate the host immune system, thereby allowing persistent and latent infection and life-long existence in the host. Information on viral escape mechanisms will be useful in rational design of antiviral drugs and should bring us one step closer to development of an effective vaccine.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
AP was supported by grant from Department of Science and Technology, Science Engineering Research Board (Grant no. YSS/2015/00389), Govt. of India. The Institute of Life Sciences, Bhubaneswar is supported by core funds made available by Department of Biotechnology, Government of India.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I would like to apologize to those authors whose work could not be cited due to space limitations.
References
1. Manicklal S, Emery VC, Lazzarotto T, Boppana SB, Gupta RK. The “silent” global burden of congenital cytomegalovirus. Clin Microbiol Rev. (2013) 26:86–102. doi: 10.1128/CMR.00062-12
2. Griffiths P, Baraniak I, Reeves M. The pathogenesis of human cytomegalovirus. J Pathol. (2015) 235:288–97. doi: 10.1002/path.4437
3. Schleiss MR, Permar SR, Plotkin SA. Progress toward development of a vaccine against congenital cytomegalovirus infection. Clin Vaccine Immunol. (2017) 24:e00268–17. doi: 10.1128/CVI.00268-17.
4. Ross SA, Novak Z, Pati SK, Patro RK, Blumenthal J, Danthuluri VR, et al. Mixed infection and strain diversity in congenital cytomegalovirus infection. J Infect Dis. (2011) 204:1003–7. doi: 10.1093/infdis/jir457
5. Fowler KB, Boppana SB. Congenital cytomegalovirus infection. Semin Perinatol. (2018) 42:149–154. doi: 10.1053/j.semperi.2018.02.002
6. Britt WJ. Congenital human cytomegalovirus infection and the enigma of maternal immunity. J Virol. (2017) 91:e02392–16. doi: 10.1128/JVI.02392-16
7. Institute of Medicine Disivion of Health Promotion and Disease Prevention. Vaccines for the 21st Century: a Tool for Decision Making. IOM Committee to Study National Priorities for Vaccine Development. Washington, DC: National Academies Press (2000).
8. Benedict CA. A CMV vaccine: TREATing despite the TRICKs. Exp Rev Vacc. (2013) 12: 1235–1237. doi: 10.1586/14760584.2013.844653
9. Barry PA. Exploiting viral natural history for vaccine development. Med Microbiol Immunol. (2015) 204:255–62. doi: 10.1007/s00430-015-0406-1
10. McGeoch DJ, Cook S, Dolan A, Jamieson FE, Telford EA. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. (1995) 247:443–58. doi: 10.1006/jmbi.1995.0152
11. Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, et al. Decoding human cytomegalovirus. Science. (2012) 338:1088–93. doi: 10.1126/science.1227919
12. Mocarski ES Jr, Shenk T, Griffiths PD, Pass RF. Cytomegalovirus. In: Knipe DM and Howley PM, editors. Fields Virology, 6th ed. Philadelphia, PA: Wolters Kluwer, Lippincott Williams & Wilkins (2013) pp. 1960–2014.
13. Stern-Ginossar N, Saleh N, Goldberg MD, Prichard M, Wolf DG, Mandelboim O. Analysis of human cytomegalovirus-encoded microRNA activity during infection. J Virol. (2009) 83:10684–93. doi: 10.1128/JVI.01292-09
14. Jackson SE, Mason GM, Wills MR. Human cytomegalovirus immunity and immune evasion. Virus Res. (2011) 157:151–60. doi: 10.1016/j.virusres.2010.10.031
15. Goodrum F. Human cytomegalovirus latency: approaching the gordian knot. Annu Rev Virol. (2016) 3:333–357. doi: 10.1146/annurev-virology-110615-042422
16. Gilbert MJ, Riddell SR, Plachter B, Greenberg PD. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature. (1996) 383:720–2. doi: 10.1038/383720a0
17. Hansen SG, Powers CJ, Richards R, Ventura AB, Ford JC, Siess D, et al. HCMV Evasion of CD8+ T Cells Is Critical for Superinfection by Cytomegalovirus. Science. (2010) 328:102–6 doi: 10.1126/science.1185350
18. Tortorella D, Gewurz BE, Furman MH, Schust DJ, Ploegh HL. Viral subversion of the immune system. Annu Rev Immunol. (2000) 18:861–926. doi: 10.1146/annurev.immunol.18.1.861
19. Charpak-Amikam Y, Kubsch T, Seidel E, Oiknine-Djian E, Cavaletto N, Yamin R, et al. Human cytomegalovirus escapes immune recognition by NK cells through the downregulation of B7-H6 by the viral genes US18 and US20. Sci Rep. (2017) 7:8661. doi: 10.1038/s41598-017-08866-2
20. Patel M, Vlahava VM, Forbes SK, Fielding CA, Stanton RJ, Wang ECY. HCMV-encoded NK modulators: lessons from in vitro and in vivo genetic variation. Front Immunol. (2018) 9:2214. doi: 10.3389/fimmu.2018.02214
21. Fu YZ, Su S, Gao YQ, Wang PP, Huang ZF, Hu MM, et al. Human cytomegalovirus tegument protein UL82 inhibits STING-mediated signaling to evade antiviral immunity. Cell Host Microbe. (2017) 21:231–243. doi: 10.1016/j.chom.2017.01.001
22. Kalejta RF, Shenk T. The Human Cytomegalovirus UL82 Gene Product (pp71) Accelerates Progression through the G1 Phase of the Cell Cycle. J Virol. (2003) 77:3451–9. doi: 10.1128/JVI.77.6.3451-3459.2003
23. Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc Natl Acad Sci USA. (2000) 97:1695–700. doi: 10.1073/pnas.97.4.1695
24. Chang WL, Baumgarth N, Yu D, Barry PA. Human cytomegalovirus-encoded interleukin-10 homolog inhibits maturation of dendritic cells and alters their functionality. J Virol. (2004) 78:8720–31. doi: 10.1128/JVI.78.16.8720-8731.2004
25. Prichard MN, Britt WJ, Daily SL, Hartline CB, Kern ER. Human cytomegalovirus UL97 Kinase is required for the normal intranuclear distribution of pp65 and virion morphogenesis. J Virol. (2005) 79:15494–502. doi: 10.1128/JVI.79.24.15494-15502.2005
26. Yoo YD, Chiou CJ, Choi KS, Yi Y, Michelson S, Kim S, et al. The IE2 refulatory protein of human cytomegalovirus induces expression of the human transforming growth factor beta 1 gene through an Egr-1 binding site. J Virol. (1996) 70:7062–70.
27. Nightingale K, Lin KM, Ravenhill BJ, Davies C, Nobre L, Fielding CA, et al. High-definition analysis of host protein stability during human cytomegalovirus infection reveals antiviral factors and viral evasion mechanisms. Cell Host Microbe. (2018) 24:447–460.e11. doi: 10.1016/j.chom.2018.07.011
28. Wang ECY, Pjechova M, Nightingale K, Vlahava VM, Patel M, Ruckova E, et al. Suppression of costimulation by human cytomegalovirus promotes evasion of cellular immune defenses. Proc Natl Acad Sci USA. (2018) 115:4998–5003. doi: 10.1073/pnas.1720950115
29. Nachmani D, Lankry D, Wolf DG, Mandelboim O. The human cytomegalovirus microRNA miRUL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat Immunol. (2010) 11:806–813. doi: 10.1038/ni.1916
30. De Pelsmaeker S, Romero N, Vitale M, Favoreel HW. Herpesvirus evasion of natural killer cells. J Virol. (2018) 92:e02105-17. doi: 10.1128/JVI.02105-17
31. Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. (1989) 320:1731–5. doi: 10.1056/NEJM198906293202605
32. Ploegh HL. Viral strategies of immune evasion. Science. (1998) 10:248–53. doi: 10.1126/science.280.5361.248
33. Andrews DM, Andoniou CE, Granucci F, Ricciardi-Castagnoli P, Degli-Esposti MA. Infection of dendritic cells by murine cytomegalovirus induces functional paralysis. Nat Immunol. (2001) 2:1077–84. doi: 10.1038/ni724
34. Moutaftsi M1, Mehl AM, Borysiewicz LK, Tabi Z. Human cytomegalovirus inhibits maturation and impairs function of monocyte-derived dendritic cells. Blood. (2002) 99:2913–21. doi: 10.1182/blood.V99.8.2913
35. Gredmark-Russ S, Söderberg-Nauclér C. Dendritic cell biology in human cytomegalovirus infection and the clinical consequences for host immunity and pathology. Virulence. (2012) 3:621–34. doi: 10.4161/viru.22239
36. Siddiquey MNA, Zhang H, Nguyen CC, Domma AJ, Kamil JP. The human cytomegalovirus endoplasmic reticulum-resident glycoprotein UL148 activates the unfolded protein response. J Virol. (2018) 92:e00896–18. doi: 10.1128/JVI.00896-18
37. Sturgill ER, Malouli D, Hansen SG, Burwitz BJ, Seo S, Schneider CL, et al. Natural Killer Cell Evasion Is Essential for Infection by Rhesus Cytomegalovirus. PLoS Pathog. (2016) 12:e1005868. doi: 10.1371/journal.ppat.1005868
38. Dassa L, Seidel E, Oiknine-Djian E, Yamin R, Wolf DG, Le-Trilling VTK, et al. The human cytomegalovirus protein UL148A downregulates the NK cell-activating ligand MICA to avoid NK cell attack. J Virol. (2018) 92:e00162–18. doi: 10.1128/JVI.00162-18
39. Boppana SB, Rivera LB, Fowler KB, Mach M, Britt WJ. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N Engl J Med. (2001) 344:1366–71. doi: 10.1056/NEJM200105033441804
40. Novak Z, Ross SA, Patro RK, Pati SK, Kumbla RA, Brice S, et al. Cytomegalovirus strain diversity in seropositive women. J Clin Microbiol. (2008) 46:882–6. doi: 10.1128/JCM.01079-07
41. Yamamoto AY, Mussi-Pinhata MM, Boppana SB, Novak Z, Wagatsuma VM, Oliveira Pde F, et al. Human cytomegalovirus reinfection is associated with intrauterine transmission in a highly cytomegalovirus-immune maternal population. Am J Obstet Gynecol. (2010) 202:297.e1-8 doi: 10.1016/j.ajog.2009.11.018
42. Gaytant MA, Rours GI, Steegers EA, Galama JM, Semmekrot BA. Congenital cytomegalovirus infection after recurrent infection: case reports and review of the literature. Eur J Pediatr. (2003) 162:248–53.
43. Ikuta K, Minematsu T, Inoue N, Kubo T, Asano K, Ishibashi K, Imamura T, Nakai H, Yoshikawa T, Moriuchi H, Fujiwara S, Koyano S, Suzutani T. Cytomegalovirus (CMV) glycoprotein H-based serological analysis in Japanese healthy pregnant women, and in neonates with congenital CMV infection and their mothers. J Clin Virol. (2013) 58:474–8. doi: 10.1016/j.jcv.2013.07.004
44. Pati SK, Pinninti S, Novak Z, Chowdhury N, Patro RK, Fowler K, et al. NIDCD CHIMES Study Investigators. Genotypic diversity and mixed infection in newborn disease and hearing loss in congenital cytomegalovirus infection. Pediatr Infect Dis J. (2013) 32:1050–4. doi: 10.1097/INF.0b013e31829bb0b9
45. Gardner TJ, Tortorella D. Virion glycoprotein-mediated immune evasion by human cytomegalovirus: a sticky virus makes a slick getaway. Microbiol Mol Biol Rev. (2016) 80:663–77. doi: 10.1128/MMBR.00018-16
46. Burkhardt C, Himmelein S, Britt W, Winkler T, Mach M. Glycoprotein N subtypes of human cytomegalovirus induce a strain-specific antibody response during natural infection. J Gen Virol. (2009) 90(Pt 8):1951–61. doi: 10.1099/vir.0.010967-0
47. Pati SK, Novak Z, Purser M, Arora N, Mach M, Britt WJ, Boppana SB. Strain-specific neutralizing antibody responses against human cytomegalovirus envelope glycoprotein N. Clin Vaccine Immunol. (2012) 19:909–13. doi: 10.1128/CVI.00092-12
48. Söderberg-Nauclér C, Fish KN, Nelson JA. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. (1997)91:119–26. doi: 10.1016/S0092-8674(01)80014-3
49. Prichard MN, Penfold ME, Duke GM, Spaete RR, Kemble GW. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev Med Virol. (2001) 11:191–200. doi: 10.1002/rmv.315
50. Patro AR, Dar L, Pati SK, Agarwal SK, Guleria S, Broor S, et al. Human cytomegalovirus UL73 polymorphisms among renal transplant recipients in India. BMC Infect Dis. (2014)14:66. doi: 10.1186/1471-2334-14-S3-P66
51. Arav-Boger R. Strain variation and disease severity in congenital cytomegalovirus infection: In search of a viral marker. Infect Dis Clin North Am. (2015) 29:401–14. doi: 10.1016/j.idc.2015.05.009
52. Renzette N, Pokalyuk C, Gibson L, Bhattacharjee B, Schleiss MR, Hamprecht K, et al. Limits and patterns of cytomegalovirus genomic diversity in humans. Proc Natl Acad Sci USA. (2015) 112:E4120–8. doi: 10.1073/pnas.1501880112
53. Sackman AM, Pfeifer SP, Kowalik TF, Jensen JD. On the demographic and selective forces shaping patterns of human cytomegalovirus variation within hosts. Pathogens. (2018) 7:E16. doi: 10.3390/pathogens7010016
54. Cudini J, Roy S, Houldcroft CJ, Bryant JM, Depledge DP, Tutill H, et al. Human cytomegalovirus haplotype reconstruction reveals high diversity due to superinfection and evidence of within-host recombination. Proc Natl Acad Sci USA. (2019) 116:5693–8. doi: 10.1073/pnas.1818130116
55. Kaczorowski KJ, Shekhar K, Nkulikiyimfura D, Dekker CL, Maecker H, Davis MM, et al. Continuous immunotypes describe human immune variation and predict diverse responses. Proc Natl Acad Sci USA. (2017) 114:E6097–106. doi: 10.1073/pnas.1705065114
56. Davis MM, Brodin P. Rebooting human immunology. Annu Rev Immunol. (2018) 36:843–64. doi: 10.1146/annurev-immunol-042617-053206
57. Picarda G, Benedict CA. Cytomegalovirus: shape-shifting the immune system. J Immunol. (2018) 200:3881–9. doi: 10.4049/jimmunol.1800171
58. Furman D, Jojic V, Sharma S, Shen-Orr SS, Angel CJ, Onengut-Gumuscu S, et al. Cytomegalovirus infection enhances the immune response to influenza. Sci Transl Med. (2015) 7:28. doi: 10.1126/scitranslmed.aaa2293
59. Britt WJ. Maternal immunity and the natural history of congenital human cytomegalovirus infection. Viruses. (2018) 10:E405. doi: 10.3390/v10080405
60. Prober CG, Enright AM. Congenital cytomegalovirus (CMV) infections: hats off to Alabama. J Pediatr. (2003)143:4–6. doi: 10.1016/S0022-3476(03)00290-7
61. Liston A, Goris A. The origins of diversity in human immunity. Nat Immunol. (2018) 19:209–210. doi: 10.1038/s41590-018-0047-9
62. Cobbold M, Khan N, Pourgheysari B, Tauro S, McDonald D, Osman H, et al. Adoptive transfer of cytomegalovirus-specific CTL to stem cell transplant patients after selection by HLA-peptide tetramers. J Exp Med. (2005) 202:379–86. doi: 10.1084/jem.20040613
63. Ameres S, Besold K, Plachter B, Moosmann A. CD8 T cell-evasive functions of human cytomegalovirus display pervasive MHC allele specificity, complementarity, and cooperativity. J Immunol. (2014) 192:5894–905. doi: 10.4049/jimmunol.1302281
64. Goodier MR, Jonjić S, Riley EM, Juranić Lisnić V. CMV and natural killer cells: shaping the response to vaccination. Eur J Immunol. (2018) 48:50–65. doi: 10.1002/eji.201646762
65. Hansen SG, Sacha JB, Hughes CM, Ford JC, Burwitz BJ, Scholz I, et al. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science. (2013) 340:1237874. doi: 10.1126/science.1237874
66. Früh K, Picker L. CD8+ T cell programming by cytomegalovirus vectors: applications in prophylactic and therapeutic vaccination. Curr Opin Immunol. (2017) 47:52–6. doi: 10.1016/j.coi.2017.06.010
67. Hill AB. The immune response to CMV infection and vaccination in mice, monkeys and humans: recent developments. Curr Opin Virol. (2018) 28:161–66. doi: 10.1016/j.coviro.2018.01.006
Keywords: HCMV (human cytomegalovirus), immune evasion, pathogenesis, superinfection, vaccine
Citation: Patro ARK (2019) Subversion of Immune Response by Human Cytomegalovirus. Front. Immunol. 10:1155. doi: 10.3389/fimmu.2019.01155
Received: 28 November 2018; Accepted: 07 May 2019;
Published: 10 June 2019.
Edited by:
Richard D. Dix, Georgia State University, United StatesReviewed by:
Edward Mocarski, Emory University, United StatesSarah Rowland-Jones, University of Oxford, United Kingdom
Copyright © 2019 Patro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. Raj Kumar Patro, cmFqa3VtYXJwYXRyb0B5YWhvby5jb20=
 A. Raj Kumar Patro
A. Raj Kumar Patro