Abstract
Leishmaniasis is a serious health problem in many countries, and continues expanding to new geographic areas including Europe and USA. This disease, caused by parasites of Leishmania spp. and transmitted by phlebotomine sand flies, causes up to 1.3 million new cases each year and despite efforts toward its functional dissection and treatment it causes 20–50 thousands deaths annually. Dependence of susceptibility to leishmaniasis on sex and host's genes was observed in humans and in mouse models. Several laboratories defined in mice a number of Lmr (Leishmania major response) genetic loci that control functional and pathological components of the response to and outcome of L. major infection. However, the development of its most aggressive form, visceral leishmaniasis, which is lethal if untreated, is not yet understood. Visceral leishmaniasis is caused by infection and inflammation of internal organs. Therefore, we analyzed the genetics of parasite load, spread to internal organs, and ensuing visceral pathology. Using a new PCR-based method of quantification of parasites in tissues we describe a network-like set of interacting genetic loci that control parasite load in different organs. Quantification of Leishmania parasites in lymph nodes, spleen and liver from infected F2 hybrids between BALB/c and recombinant congenic strains CcS-9 and CcS-16 allowed us to map two novel parasite load controlling Leishmania major response loci, Lmr24 and Lmr27. We also detected parasite-controlling role of the previously described loci Lmr4, Lmr11, Lmr13, Lmr14, Lmr15, and Lmr25, and describe 8 genetic interactions between them. Lmr14, Lmr15, Lmr25, and Lmr27 controlled parasite load in liver and lymph nodes. In addition, Leishmania burden in lymph nodes but not liver was influenced by Lmr4 and Lmr24. In spleen, parasite load was controlled by Lmr11 and Lmr13. We detected a strong effect of sex on some of these genes. We also mapped additional genes controlling splenomegaly and hepatomegaly. This resulted in a systematized insight into genetic control of spread and load of Leishmania parasites and visceral pathology in the mammalian organism.
Introduction
Leishmaniasis is a neglected tropical disease, which belongs to the top health problems because it is endemic in 98 countries in Asia, Africa, the Americas, and the Mediterranean region (1–3) and is gradually expanding to new areas, including Central Europe and USA (2, 4–9). The disease occurs in cutaneous, mucocutaneous, and visceral forms (9). It is caused by the protozoan intracellular parasite Leishmania transmitted by Phlebotomus spp. in the Old World and Lutzomyia spp. in the New World. The parasite can infect about 70 species of vertebrates, including humans (10–13). In addition, there are specific groups of asymptomatic infection (14), and post-kala-azar dermal leishmaniasis (15). Visceral leishmaniasis is fatal in more than 95% of cases if left untreated (9). Up to 1.3 million new cases occur annually: 300 000 are visceral and 1 million are cutaneous and mucocutaneous and about 20–50 thousands patients die (13). In the infected mammalian organism, Leishmania parasites invade “professional phagocytes,” including monocytes, macrophages, and neutrophils and can also reside in dendritic cells (DC) (16), immature myeloid precursor cells, hepatocytes, and fibroblasts; the parasite can also enter sialoadhesin-positive stromal macrophages (17).
Treatment of leishmaniasis is difficult because of the lack of reliable drugs. Existing leishmanicidal agents show severe side effects. In addition, the treatment is costly and not readily available to a majority of patients. In spite of numerous attempts to develop vaccination against leishmaniasis, there are still no safe and effective vaccines suitable for humans (18, 19). Clinical form and susceptibility to leishmaniasis are dependent on parasite species, environmental and social factors, and also on nutrition and genotype of the host (3, 10, 16, 20).
Parasite load is one of the most important parameters of leishmaniasis determining the course of infection and the degree of susceptibility. However, the information about genetic control of parasite load remains incomplete and fragmented; there is no systematic description of the control of parasite load in combination with other pathological parameters and influence of sex on these genes is not known for any of the studied Leishmania species. The use of mouse models in studies of selected candidate genes and also for hypothesis-generating genome-wide association and linkage analysis, revealed several genes and loci controlling parasite burden (16, 21–23) (Table 1). However, quantification of parasites had been a laborious task providing inaccurate results due to technical problems. These problems were reduced with development of sensitive PCR-based assays (18), which permitted to perform genome wide-search (21, 23).
Table 1
| Parasite | Organ | Gene/Locus | References |
|---|---|---|---|
| L. donovani | Spleen | Slc11a1 (weak effect), H2, Lyst | (24, 25) |
| Liver | Slc11a1, H2, Ir2 | (25–28) | |
| Bone marrow | H2 | (25) | |
| L. mexicana | Spleen | Slc11a1, H2 | (29) |
| Liver | Slc11a1, H2 | (29) | |
| L. infantum | Spleen | Slc11a1, H2 | (25) |
| Liver | Slc11a1, H2 | (25) | |
| Bone marrow | H2 | (25) | |
| L. major | Spleen | Lmr5 | (21) |
| Skin | Dice1.2 | (30) | |
| Lymph nodes | Lmr20 | (21) | |
| L. tropica | Spleen | Ltr3, Ltr6 | (23) |
| Liver | Ltr2, Ltr4, Ltr8 | (23) | |
| Lymph nodes | Ltr1, Ltr4 | (23) |
Genetic control of parasite load in mouse leishmaniasis.
Leishmania species and strains used in described experiments: (21), L. major (LV 561 (MHOM/IL/67/LRC-L137 JERICHO II); (23), L. tropica (MHOM/1999/TR/SU23); (24), L. donovani 2S (MHOM/SD/61/2S); (25), L. infantum, zymodem MON-1, L. donovani L82 (LV9) (MHOM/ET/67/L82); (26), L. donovani L82 (LV9) (MHOM/ET/67/L82); (27), L. donovani L82 (LV9) (MHOM/ET/67/L82); (28), L. donovani, 3S (MHOM/SD/62/3S); (29), L. mexicana (MNYC/B2/62/M379); (30) L. major (WHOM/IR/-/173).
We explored genetic control of parasite load in different organs after L. major infection. This parasite is the predominant causative agent of human cutaneous leishmaniasis in the Old World, in rare cases it can visceralize in an immunocompromised (HIV infected) host (31), but L. major strain (MRHOM/IR/75/ER) was described to visceralize also in an immunocompetent individual (32). Instances of L. major visceralization in non-immunocompromised people may suggest the presence of genetic factors determining extreme forms of high susceptibility to L. major infection. Infection by L. major in mouse is controlled by multiple genes. These multiple genes-loci have been mapped in three different resistant strains—C57BL/10Sn (B10.D2), C57BL/6 and STS—using the susceptible strain BALB/c for mapping in each case (16, 22); in cross with B10.D2 using for infection L. major (strain WHOM/IR/-/173), in crosses with C57BL/6 and STS L. major V121 (the cloned line V121 derived from MHOM/IL/67/Jericho II) and L. major strain (LV 561 (MHOM/IL/67/LRC-L137 JERICHO II), respectively. These experiments revealed 26 Lmr (Leishmania major response) and 5 Lmrq (Leishmania major resistance QTL) loci that determine skin lesion size, splenomegaly, hepatomegaly, cytokine levels in blood serum, eosinophil infiltration into lymphatic nodes, and parasite numbers in different organs (16, 21, 22, 33). Several loci (Lmr4/Lmrq1, Lmr5/Lmrq3, Lmr6/Lmrq4, and Lmr12/Dice1b) detected in crosses with resistant strains STS and B10.D2 overlap, which might indicate in these loci not only general response across different mouse strains, but also general response to different L. major strains.
In the present study, we tested parasite load and dissemination in F2 hybrids of BALB/c and recombinant congenic mouse strains CcS-9 and CcS-16, infected by L. major LV561. The disease development during previous experiments showed no significant difference between females and males of the CcS-16 strain after infection with L. major (34); however the influence of sex was present in CcS-9 strain (33). These differences determined selection of sex of mice used in the present F2 hybrid study. The current study aims to provide a first systematic a genome wide search description of the genetic control of parasite load in mammalian organs after L. major infection.
Materials and Methods
Mice
Recombinant congenic (RC) strains of mice of the BALB/c-c-STS/Dem (CcS/Dem) series, containing random distinct segments of 12.5% STS/A (STS) genes on the background of BALB/cHeA (BALB/c) genome (35), exhibit various susceptibility to Leishmania infection and proved to be a powerful tool in research of genetic control of the disease (16, 21, 23, 33, 36, 37). BALB/c is a standard inbred mouse strain, STS is an inbred mouse strain of Swiss origin. The parts of CcS-9 and CcS-16 genomes inherited from the BALB/c or STS parents were defined (35).
At the time of experiments, mice of CcS-9 strain were in the 40 generation of inbreeding, and therefore highly homozygous. F2 hybrids between CcS-9 and BALB/c (age 11–21 weeks at the time of infection, mean and median age 14.8 and 15 weeks, respectively) were produced at the Institute of Molecular Genetics; 254 F2 hybrids between BALB/c and CcS-9 comprised 139 females and 115 males. Mice were tested in three independent experimental groups; male and female mice were placed into separate rooms, and males were caged individually.
In CcS-16 experiments, only females were used due to absence of sex differences in previous experiments. Mice of CcS-16 strain were in the generation 37 of inbreeding, and therefore highly homozygous. We produced 577 female F2 hybrids between CcS-16 and BALB/c (age at the time of infection, 14–17 weeks) and tested them in four independent experiments.
Mice were kept in the animal facility of Institute of Molecular Genetics AS CR.
Parasites
Leishmania major LV 561 (MHOM/IL/67/LRC-L137 JERICHO II) was maintained in rump lesions of BALB/c females. Amastigotes were transformed to promastigotes using SNB-9 (38); 107 promastigotes from 6 days old subculture 2 were inoculated in 50 μl sterile saline s.c. into mouse rump (39).
Disease Phenotype
The size of the primary skin lesions was measured weekly using a Vernier caliper gauge. Mice were euthanized 8 weeks after infection and body, spleen, and liver weights were recorded. The blood, spleen, liver, and inguinal lymph nodes were collected for the further analysis. Splenomegaly (enlargement of the spleen) and hepatomegaly (enlargement of the liver) were calculated as organ-to-body weight ratio × 1000. Parasite load was presented in relative units as concentration of parasite DNA in ng per 1 μl.
Cytokine and IgE Levels
IgE, IL-4, IL-10, IL-12, IL-13, and IFNγ levels in serum were determined using the primary and secondary monoclonal antibodies (IgE: R35-72, R35-118, IL-4: 11B11, BVD6-24G2, IL-10: JES5-2A5, JES5-16E3, IL-12: C15.6, C17.8, IFNγ: R4-6A2, XMG1.2) and standards from Pharmingen (San Diego, CA, USA) (purified mIgE: C38-2, recombinant mouse IL-4, mIL-10, mIL-12 p70 heterodimer, and mIFNγ). The enzyme-linked immunosorbent assay (ELISA) was performed as recommended by Pharmingen. IL-13 level in serum were determined using the Murine IL-13 ELISA Development Kit 900-K207 (by PeproTech EC (London, United Kingdom), which contained both primary and secondary monoclonal antibodies and standard and the ELISA was performed as recommended by PeproTech EC. The IL-4, IL-10, IL-12, IL-13, IFNγ, and IgE levels were estimated using the curve fitter program KIM-E (Schoeller Pharma, Prague, Czech Republic).
Genotyping of F2 Hybrids
The frozen archive material of F2 crosses genotyped in previous mapping experiments (33, 40) was used for measurement of parasite DNA and analysis of genetic linkage.
In CcS-9 experiment, STS derived segments were typed in F2 hybrids using 19 microsatellite markers on eight chromosomes: D2Mit148, D2Mit283, D4Mit7, D4Mit17, D4Mit23, D4Mit53, D4Mit172, D5Mit24, D5Mit143, D6Mit122, D6Mit274, D9Mit15, D11Mit141, D11Mit242, D11Nds10, D11Nds18, D16Mit19, D17Mit120, and D17Mit122 as described elsewhere (33).
In CcS-16 experiment, the segments of STS origin on 9 chromosomes were typed in F2 hybrids using 23 markers: D2Mit51, D2Mit102, D2Mit156, D2Mit283, D2Mit389, D2Nds3, D3Mit11, D3Mit25, D4Mit153, D6Mit48, D6Mit320, D10Mit67, D10Mit103, D11Mit37, D11Mit139, D11Mit242, D16Mit126, D17Mit38, D17Mit130, D18Mit35, D18Mit40, D18Mit49, and D18Mit120 as described in Vladimirov et al. (40).
Measurement of Parasite Load in Organs
Total DNA was isolated from frozen lymph nodes, spleen, and liver samples, and parasite load was measured using PCR-ELISA according to the previously published protocol (41). Briefly, total DNA was isolated using a standard proteinase procedure (42). For detection of Leishmania parasite DNA in total DNA, PCR was performed using two primers (digoxigenin-labeled F 5′-ATT TTA CAC CAA CCC CCA GTT-3′ and biotin-labeled R 5′-GTG GGG GAG GGG CGT TCT-3′ (VBC Genomics Biosciences Research, Austria). The 120-bp fragment within the conserved region of the kinetoplast minicircle of Leishmania parasite was amplified. In each PCR reaction, 50 ng of extracted total DNA was used. As a positive control, 20 ng of L. major DNA per reaction was amplified as a highest concentration of the standard. In CcS-9 experiment, 26-cycle PCR reaction was used for quantification of parasites in lymph nodes and spleen, and 35 cycles for liver. In CcS-16 experiment, 24-cycle PCR reaction was used for quantification of parasites in lymph nodes; 28 cycles for spleen, and 33 cycles for liver. Parasite load was determined by measurement of the PCR product with the modified ELISA protocol (Pharmingen, San Diego, USA). Concentration of Leishmania DNA was measured at the ELISA Reader Tecan with the curve fitter program KIM-E (Schoeller Pharma, Prague, Czech Republic) using least squares-based linear regression analysis (21, 41).
Statistical Analysis
The role of genetic factors in control of parasite dissemination was estimated with ANOVA using Statistical for Windows 12.0 (StatSoft, Inc., Tulsa, OK, USA). Markers and interactions with P < 0.05 were combined in a single comparison. Genotype (marker), sex and age were fixed factors and the experiment was random factor. The time course of skin lesion development was evaluated on the basis of weekly measurements of lesion size in each mouse in weeks 4–8 after infection. Variance components and mixed model ANOVA of Statistica for Windows 12.0 (StatSoft, Inc., Tulsa, OK, USA) with marker as the fixed variable and the week of observation as the covariate have been used to evaluate the linkage.
When necessary, the original values of an analyzed parameter were transformed for normalization of the distribution as described in the legends to the Tables. For whole genome significance values (corrected P-values), the observed P-values (αT) were adjusted using the following formula (43):
In the present formula, G = 1.75 Morgan (the length of the segregating part of the genome: 12.5% of 14 M); C = 8 (number of chromosomes segregating in cross between CcS-9 and BALB/c) or C = 9 (between CcS-16 and BALB/c); ρ = 1.5 for F2 hybrids; h(T) = the observed statistic (F ratio).
The percent of the phenotypic variance explained by a certain locus or an interaction between loci was calculated subtracting the sums of squares of the model without this variable from the sum of squares of the full model and this difference, divided by the total regression sums of squares:
Results
The present studies revealed two novel Lmr loci, new functions of six previously mapped Lmr loci and described multiple and heterogeneous genetic effects influencing parasite dissemination into the host organism. Because the STS-derived regions in CcS-9 and CcS-16 are different, we detected in their respective F2 hybrids with BALB/c different loci controlling parasite load.
The CcS-9 Strain: Two Novel Lmr Loci, Multiple Interactions, and Sex Dependent Control of Parasite Load
In F2 hybrids prepared from the parental strain BALB/c and the RC strain CcS-9, the analysis of parasite load in inguinal lymph nodes, spleen and liver followed by linkage analysis, revealed both main effect loci and interactions of genes located on chromosomes 2, 4, 5, 6, 11, and 17. Two novel Lmr loci, Lmr24, and Lmr27 were detected in the CcS-9 experiments (Figure 1).
Figure 1
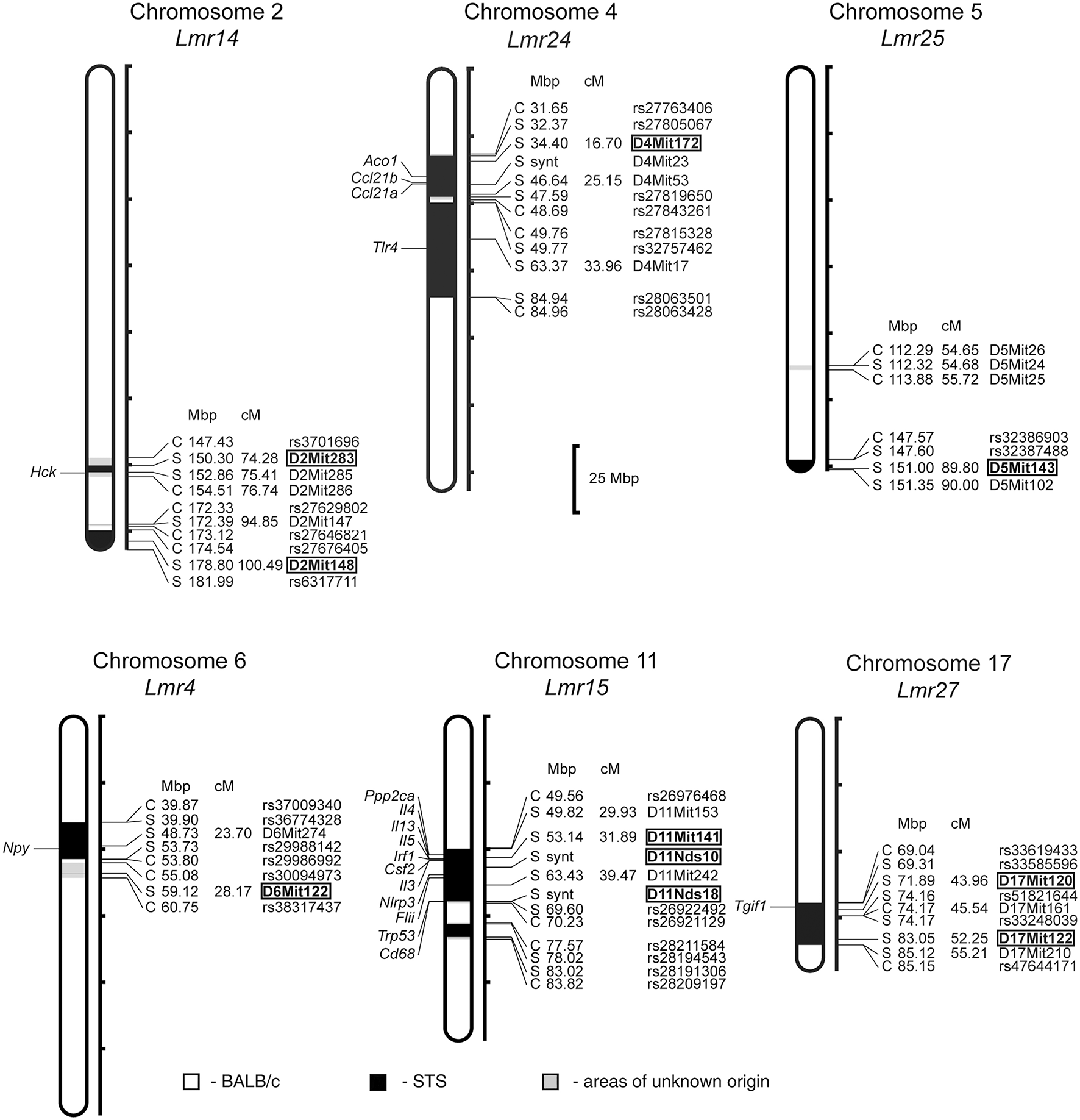
Positions of the loci controlling parasite load in organs of the RC strain CcS-9. The regions derived from the BALB/c strain are indicated as white; the regions of the STS origin are indicated as black; and the areas of undetermined origin are gray. Only the markers that determine the boundaries between BALB/c and STS-derived segments, and the markers used for statistical calculations are shown. Boxes indicate the markers that exhibited significant P-values, corrected for genome-wide search. The genes that participate in the infection-related immune processes (16, 17, 20, 22, 33) and are located within the present Lmr loci are listed next to their positions at the chromosomes. Abbreviations indicate genes related to the response to Leishmania ssp.: Hck, hemopoietic cell kinase; Aco1, aconitase 1; Ccl21a, chemokine (C-C motif) ligand 21A (serine); Ccl21b, chemokine (C-C motif) ligand 21B (leucine), Tlr4, toll-like receptor 4; Npy, neuropeptide Y; Ppp2ca, protein phosphatase 2, formerly 2A; catalytic subunit, alpha isoform; Il3, interleukin 3; Il4, interleukin 4; Il13, interleukin 13; Irf1, interferon regulatory factor 1; Csf2, colony stimulating factor 2 (granulocyte-macrophage); Nlrp3, NLR family, pyrin domain containing 3; Flii, flightless I actin binding protein; Trp53, transformation related protein 53; Cd68, CD68 antigen; Tgif1, TGFB-induced factor homeobox 1.
Both in males and females, a homozygous STS allele (SS) at the Lmr14 linked to the marker D2Mit283 determined the highest parasite load in inguinal lymph nodes interacting with homozygous STS alleles (SS) of the Lmr25 linked to D5Mit143 (corrected P = 0.0314, 2.1% of variance explained), and also with the Lmr24 linked to D4Mit172 (corrected P = 0.0247, 2% of variance explained) (Table 2).
Table 2
| Lymph nodes | D5Mit143 (Lmr25) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CS | SS | P-value | corr. P | % of explained variance | ||||||||
| D2Mit283 (Lmr14) | CC | n = 20 | 2.34 | 5.45 ± 0.22 | n = 21 | 1.33 | 4.89 ± 0.21 | n = 18 | 1.20 | 4.79 ± 0.23 | 0.00044 | 0.0314 | 2.1 |
| CS | n = 31 | 1.03 | 4.64 ± 0.19 | n = 57 | 2.19 | 5.39 ± 0.13 | n = 30 | 1.93 | 5.27 ± 0.18 | ||||
| SS | n = 24 | 1.09 | 4.69 ± 0.20 | n = 32 | 1.60 | 5.08 ± 0.17 | n = 20 | 2.26 | 5.42 ± 0.21 | ||||
| Lymph nodes | D2Mit283 (Lmr14) | ||||||||||||
| CC | CS | SS | P-value | corr. P | % of explained variance | ||||||||
| D4Mit172 (Lmr24) | CC | n = 15 | 1.25 | 4.83 ± 0.27 | n = 22 | 1.78 | 5.18 ± 0.21 | n = 16 | 1.70 | 5.14 ± 0.24 | 0.00034 | 0.0247 | 2.0 |
| CS | n = 28 | 1.47 | 4.99 ± 0.18 | n = 59 | 2.05 | 5.32 ± 0.12 | n = 36 | 1.01 | 4.61 ± 0.17 | ||||
| SS | n = 16 | 2.03 | 5.31 ± 0.23 | n = 37 | 1.20 | 4.79 ± 0.18 | n = 24 | 2.29 | 5.43 ± 0.20 | ||||
Interactions that control parasite load in lymph nodes of CcS-9 derived F2 hybrids of both sexes.
Means, standard error of mean (SE) and P-values were calculated by analysis of variance. The letters C and S indicate the BALB/c or STS allele, respectively; n indicates the number of mice in each group. Transformed means ± SE are shown next to non-transformed mean values in bold. The values of concentration of parasite DNA (ng/ml) in total DNA isolated from lymph nodes were transformed to obtain normal distribution: a natural logarithm of (value × 100). Only P-values significant after correction for genome-wide significance are shown.
Genetic control of parasite load in lymph nodes was strongly dependent on sex (Tables 3, 4): in interactions of sex with markers D11Nds10, D11Nds18, D11Mit141, D2Mit148, D17Mit122 - P < 10∧(−30); D11Nds10 and D17Mit122, as well as D6Mit122 and D17Mit122 - P < 10(∧−30). In females, the highest parasite load in lymph nodes were observed in mice homozygous for BALB/c (CC) allele in the Lmr15 linked to D11Nds10 (corrected P = 0.000696, 7.42% of variance explained) (Table 3A). In males, a homozygous STS allele (SS) of the Lmr14 linked to D2Mit148 (corrected P = 0.031, 10.5% of variance explained), and the novel locus Lmr27 linked to D17Mit122 (corrected P = 0.012, 15.3% of variance explained) determined higher parasite load in inguinal lymph nodes (Table 3B). Homozygotes in SS allele on the novel locus Lmr27 linked to D17Mit122 determined the highest parasite load in males in two interactions: with CC allele of the Lmr4 linked to D6Mit122 (corrected P = 0.00427, 7.4% of variance explained), and CS allele on the Lmr15 linked to D11Nds10 (corrected P = 0.0261, 6% of variance explained), respectively (Table 4).
Table 3A
| Lymph nodes | CC | CS | SS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | n | Mean | Transformed mean ± SE | n | Mean | Transformed mean ± SE | n | Mean | Transformed mean ± SE | P-value | corr. P | % of Explained variance |
| D11Mit141 (Lmr15) both sexes | 56 | 2.14 | 5.37 ± 0.15 | 127 | 1.56 | 5.05 ± 0.10 | 70 | 1.11 | 4.71 ± 0.13 | 0.00419 | 0.157 | 2.56 |
| D11Mit141 (Lmr15) females | 36 | 1.44 | 4.97 ± 0.17 | 69 | 1.02 | 4.62 ± 0.13 | 33 | 0.45 | 3.80 ± 0.18 | 0.000014 | 0.00102 | 6.38 |
| D11Mit141 (Lmr15) males | 20 | 3.18 | 5.76 ± 0.23 | 58 | 3.58 | 5.88 ± 0.14 | 37 | 2.99 | 5.70 ± 0.17 | 0.695 | 6.890 | 0.29 |
| D11Nds10 (Lmr15) both sexes | 58 | 2.22 | 5.40 ± 0.16 | 129 | 1.54 | 5.03 ± 0.09 | 66 | 1.13 | 4.73 ± 0.13 | 0.00395 | 0.149 | 3.01 |
| D11Nds10 (Lmr15) females | 36 | 1.58 | 5.06 ± 0.18 | 70 | 0.99 | 4.60 ± 0.13 | 32 | 0.45 | 3.80 ± 0.18 | 0.0000092 | 0.000696 | 7.42 |
| D11Nds10 males | 22 | 3.12 | 5.74 ± 0.26 | 59 | 3.11 | 5.74 ± 0.15 | 34 | 3.37 | 5.82 ± 0.19 | 0.935 | 7.811 | 2.5 |
| D11Nds18 (Lmr15) both sexes | 51 | 1.67 | 5.12 ± 0.16 | 136 | 1.79 | 5.19 ± 0.08 | 66 | 1.08 | 4.69 ± 0.12 | 0.00281 | 0.112 | 2.95 |
| D11Nds18 (Lmr15) females | 33 | 1.21 | 4.80 ± 0.16 | 73 | 0.97 | 4.58 ± 0.11 | 32 | 0.50 | 3.91 ± 0.16 | 0.00018 | 0.0101 | 4.2 |
| D11Nds18 (Lmr15) males | 18 | 1.86 | 5.23 ± 0.37 | 63 | 2.61 | 5.56 ± 0.19 | 34 | 4.13 | 6.02 ± 0.2 | 0.000175 | 0.0106 | 16.7 |
The Lmr15 controls parasite load in lymph nodes of CcS-9 derived females.
Means, standard error of mean (SE) and P-values were calculated by analysis of variance. The numbers in bold give the average non-transformed values. The letters C and S indicate the BALB/c or STS allele, respectively; n indicates the number of mice in each group. The values of concentration of parasite DNA (ng/ml) in total DNA isolated from lymph nodes were transformed to obtain normal distribution: a natural logarithm of (value × 100). Only P-values significant after correction for genome-wide significance are shown.
Table 3B
| Lymph nodes | CC | CS | SS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | n | Mean | Transformed mean ± SE | n | Mean | Transformed mean ± SE | n | Mean | Transformed mean ± SE | P value | corr. P | % of explained variance |
| D2Mit148 (Lmr14) both sexes | 78 | 1.72 | 5.15 ± 0.13 | 121 | 1.61 | 5.08 ± 0.11 | 54 | 1.68 | 5.13 ± 0.15 | 0.908 | 7.725 | 0.93 |
| D2Mit148 (Lmr14) females | 43 | 0.83 | 4.41 ± 0.14 | 69 | 0. 89 | 4.49 ± 0.12 | 26 | 0. 67 | 4.20 ± 0.18 | 0.411 | 5.220 | 0.39 |
| D2Mit148 (Lmr14) males | 35 | 3.04 | 5.72 ± 0.19 | 52 | 3.11 | 5.74 ± 0.14 | 28 | 16.20 | 7.39 ± 0.39 | 0.000616 | 0.03095 | 10.5 |
| D17Mit122 (Lmr27) both sexes | 74 | 1.61 | 5.08 ± 0.12 | 142 | 1.42 | 4.96 ± 0.10 | 37 | 2.04 | 5.32 ± 0.19 | 0.209 | 3.399 | 2.28 |
| D17Mit122 (Lmr27) females | 42 | 0.89 | 4.49 ± 0.15 | 77 | 0.77 | 4.35 ± 0.11 | 19 | 0.89 | 4.48 ± 0.21 | 0.678 | 6.813 | 0.17 |
| D17Mit122 (Lmr27) males | 32 | 3.44 | 5.84 ± 0.18 | 65 | 2.84 | 5.65 ± 0.13 | 18 | 15.56 | 7.35 ± 0.36 | 0.000206 | 0.0118 | 15.3 |
The Lmr14 and Lmr27 control parasite load in lymph nodes of CcS-9 derived males.
Means, standard error of mean (SE) and P-values were calculated by analysis of variance. The numbers in bold give the average non-transformed values. The letters C and S indicate the BALB/c or STS allele, respectively; n indicates the number of mice in each group. The values of concentration of parasite DNA (ng/ml) in total DNA isolated from lymph nodes were transformed to obtain normal distribution: a natural logarithm of (value × 100). Only P-values significant after correction for genome-wide significance are shown.
Table 4
| Lymph nodes | D11Nds10 (Lmr15) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CS | SS | P value | corr. P | % of explained variance | ||||||||
| D17Mit122 (Lmr27) | CC | n = 7 | 2.50 | 5.52 ± 0.31 | n = 12 | 3.22 | 5.77 ± 0.24 | n = 13 | 5.06 | 6.23 ± 0.23 | 0.000339 | 0.0261 | 6.0 |
| CS | n = 11 | 3.97 | 5.98 ± 0.25 | n = 37 | 2.92 | 5.68 ± 0.14 | n = 17 | 2.00 | 5.30 ± 0.20 | ||||
| SS | n = 4 | 4.68 | 6.15 ± 0.41 | n = 10 | 52.10 | 8.56 ± 0.26 | n = 4 | 15.53 | 7.35 ± 0.41 | ||||
| Lymph nodes | D6Mit122 (Lmr4) | ||||||||||||
| CC | CS | SS | P-value | corr. P | % of explained variance | ||||||||
| D17Mit122 (Lmr27) | CC | n = 10 | 5.21 | 6.26 ± 0.26 | n = 13 | 2.76 | 5.62 ± 0.23 | n = 9 | 2.84 | 5.65 ± 0.23 | 0.0000467 | 0.00427 | 7.4 |
| CS | n = 17 | 2.35 | 5.46 ± 0.20 | n = 35 | 4.26 | 6.05 ± 0.14 | n = 13 | 2.31 | 5.44 ± 0.20 | ||||
| SS | n = 5 | 73.38 | 8.90 ± 0.37 | n = 9 | 5.65 | 6.34 ± 0.28 | n = 4 | 9.13 | 6.82 ± 0.41 | ||||
Interactions that control parasite load in lymph nodes of CcS-9 derived males.
Means, standard error of mean (SE) and P-values were calculated by analysis of variance. The letters C and S indicate the BALB/c or STS allele, respectively; n indicates the number of mice in each group. Transformed means ± SE are shown next to non-transformed mean values in bold. The values of concentration of parasite DNA (ng/ml) in total DNA isolated from lymph nodes were transformed to obtain normal distribution: a natural logarithm of (value × 100). Only P-values significant after correction for genome-wide significance are shown.
No sex differences were detected in control of parasite load in liver. Parasite load in liver were therefore calculated for total group (both males and females), and revealed the effect of the CC allele on Lmr15 linked to D11Mit141 (corrected P = 0.0037, 8.6% of variance explained) and D11Nds10 (corrected P = 0.0011, 9.6% of variance explained) in control of higher parasite burden (Table 5). Parasites load in liver was also controlled by an interaction between the novel locus Lmr27 linked to D17Mit120 and Lmr14 linked to D2Mit283 (corrected P = 0.0184, 6.9% of variance explained) (Table 6) and by an interaction of Lmr27 linked to D17Mit122 with Lmr25 linked to D5Mit143 (suggestive linkage; corrected P = 0.0640, 6.2% of variance explained). Lowest parasite load are linked with the interaction of homozygotes in STS (SS) allele at Lmr25 with the homozygotes in BALB/c (CC) allele at Lmr27.
Table 5
| Liver | CC | CS | SS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | n | Mean | Transformed mean ± SE | n | Mean | Transformed mean ± SE | n | Mean | Transformed mean ± SE | P-value | corr. P | % of explained variance |
| D11Mit141 (Lmr15) | 56 | 3.43 | 5.84 ± 0.11 | 128 | 2.90 | 5.67 ± 0.07 | 69 | 1.87 | 5.23 ± 0.10 | 0.000061 | 0.00373 | 8.6 |
| D11Nds10 (Lmr15) | 58 | 3.36 | 5.82 ± 0.12 | 130 | 3.06 | 5.72 ± 0.08 | 65 | 1.87 | 5.23 ± 0.10 | 0.000015 | 0.00106 | 9.6 |
The Lmr15 controls parasite load in liver of CcS-9 derived F2 hybrids of both sexes.
Means, standard error of mean (SE) and P-values were calculated by analysis of variance. The numbers in bold give the average non-transformed values. The letters C and S indicate the BALB/c or STS allele, respectively; n indicates the number of mice in each group. The values of concentration of parasite DNA (ng/ml) in total DNA isolated from liver were transformed to obtain normal distribution: a natural logarithm of (value × 100). Only P-values significant after correction for genome-wide significance are shown.
Table 6
| Liver | D2Mit283 (Lmr14) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CS | SS | P value | corr. P | % of explained variance | ||||||||
| D17Mit120 (Lmr27) | CC | n = 17 | 1.99 | 5.29 ± 0.19 | n = 29 | 2.75 | 5.62 ± 0.15 | n = 27 | 3.29 | 5.80 ± 0.15 | |||
| CS | n = 31 | 3.56 | 5.87 ± 0.14 | n = 63 | 3.08 | 5.73 ± 0.10 | n = 44 | 1.78 | 5.18 ± 0.12 | 0.00024 | 0.0184 | 6.9 | |
| SS | n = 12 | 1.91 | 5.25 ± 0.22 | n = 25 | 2.67 | 5.59 ± 0.15 | n = 5 | 3.44 | 5.84 ± 0.35 | ||||
| Liver | D5Mit143 (Lmr25) | ||||||||||||
| CC | CS | SS | P-value | corr. P | % of explained variance | ||||||||
| CC | n = 30 | 3.17 | 5.76 ± 0.13 | n = 28 | 3.93 | 5.97 ± 0.14 | n = 16 | 1.57 | 5.05 ± 0.19 | ||||
| D17Mit122 (Lmr27) | CS | n = 36 | 2.80 | 5.63 ± 0.13 | n = 64 | 2.98 | 5.70 ± 0.10 | n = 41 | 2.54 | 5.54 ± 0.12 | 0.00096 | 0.0640 | 6.2 |
| SS | n = 9 | 2.21 | 5.40 ± 0.26 | n = 18 | 2.02 | 5.31 ± 0.20 | n = 11 | 3.85 | 5.95 ± 0.23 | ||||
Interactions that control parasite load in liver of CcS-9 derived F2 hybrids of both sexes.
Means, standard error of mean (SE) and P-values were calculated by analysis of variance. The letters C and S indicate the BALB/c or STS allele, respectively; n indicates the number of mice in each group. Transformed means ± SE are shown next to non-transformed mean values in bold. The values of concentration of parasite DNA (ng/ml) in total DNA isolated from liver were transformed to obtain normal distribution: a natural logarithm of (value × 100). Only P-values significant after correction for genome-wide significance are shown.
No loci that determine parasite load in spleen were found in F2 hybrids derived from the CcS-9 strain.
Loci Controlling Organ Pathology and Immune Response in CcS-9
Analysis of different parameters of the disease indicated a novel Lmr locus which controls organ pathology and systemic immune response (Table 7). Lmr24 on chromosome 4 influenced splenomegaly, levels of IFNγ, and IL-4 in serum and development of skin lesions. Control of splenomegaly was linked with the marker D4Mit23 (corrected P = 0.0426, 4.76% of variance explained), skin lesion size (kinetics of development of skin lesions)—with D4Mit172 and D4Mit23 (corrected P = 0.00201 and 0.0226, respectively), level of IFNγ in serum—with D4Mit53 (corrected P = 0.000692, 8.08% of variance explained), and level of IL-4 in serum—with D4Mit53 (corrected P = 0.0132, 5.72% of variance explained). The STS allele of these markers was responsible for lower level of IFNγ and IL-4 in serum, but with larger splenomegaly and larger skin lesions.
Table 7
| Phenotype | Locus | Marker | Genotype | P-value | corr. P | % of explained variance | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CS | SS | |||||||||
| Splenomegaly | Lmr15 | D11Mit141 | 17.86 ± 0.74 (n = 56) | 17.29 ± 0.42 (n = 128) | 14.86 ± 0.57 (n = 70) | 0.00066 | 0.0313 | 9.16 | |||
| D11Nds10 | 18.07 ± 0.73 (n = 58) | 17.18 ± 0.42 (n = 130) | 14.77 ± 0.59 (n = 66) | 0.00047 | 0.0235 | 11.80 | |||||
| Lmr24 | D4Mit23 | 15.71 ± 0.64 (n = 57) | 15.83 ± 0.45 (n = 122) | 18.17 ± 0.54 (n = 75) | 0.00093 | 0.0426 | 4.76 | ||||
| Hepatomegaly | Lmr15 | D11Nds18 | 64.02 ± 1.03 (n = 51) | 63.05 ± 0.60 (n = 136) | 58.50 ± 0.90 (n = 66) | 0.000029 | 0.00191 | 9.76 | |||
| Lesion size (in 7th week after infection) | Lmr15 | D11Mit141 | 83.68 ± 4.92 (n = 56) | 69.72 ± 3.00 (n = 128) | 55.68 ± 4.23 (n = 70) | 0.000095 | 0.00555 | 8.64 | |||
| D11Mit242 | 84.81 ± 5.00 (n = 51) | 69.18 ± 2.85 (n = 139) | 56.23 ± 4.00 (n = 64) | 0.000044 | 0.00274 | 7.55 | |||||
| D11Nds18 | 90.07 ± 5.55 (n = 51) | 69.47 ± 2.88 (n = 137) | 55.87 ± 3.93 (n = 66) | 0.0000035 | 0.000269 | 10.06 | |||||
| D11Nds10 | 86.07 ± 4.64 (n = 58) | 68.63 ± 2.91 (n = 130) | 55.25 ± 4.36 (n = 66) | 0.000010 | 0.000722 | 12.04 | |||||
| Lesion size (in 8th week after infection) | Lmr15 | D11Nds10 | 101.56 ± 5.62 (n = 58) | 88.47 ± 3.25 (n = 130) | 73.30 ± 4.95 (n = 66) | 0.00058 | 0.0282 | 9.37 | |||
| D11Nds18 | 100.93 ± 5.11 (n = 51) | 87.94 ± 3.18 (n = 137) | 76.31 ± 4.36 (n = 66) | 0.00093 | 0.0424 | 4.21 | |||||
| Lesion size (kinetics) | Lmr15 | D11Nds18 | 32.77 | 5.72 ± 0.13 (n = 51) | 28.37 | 5.33 ± 0.08 (n = 137) | 25.54 | 5.05 ± 0.11 (n = 66) | 0.00060 | 0.0282 | – |
| D11Nds10 | 31.33 | 5.60 ± 0.12 (n = 58) | 29.09 | 5.39 ± 0.08 (n = 130) | 24.91 | 4.99 ± 0.11 (n = 66) | 0.00090 | 0.0406 | – | ||
| Lmr24 | D4Mit172 | 27.19 | 5.21 ± 0.13 (n = 53) | 26.40 | 5.14 ± 0.08 (n = 124) | 32.92 | 5.74 ± 0.11 (n = 77) | 0.000032 | 0.00201 | – | |
| D4Mit23 | 26.97 | 5.19 ± 0.12 (n = 57) | 26.87 | 5.18 ± 0.08 (n = 122) | 32.38 | 5.69 ± 0.11 (n = 75) | 0.00047 | 0.0226 | – | ||
| Level of IgE in serum | Lmr15 | D11Mit141 | 35.09 | 3.12 ± 0.10 (n = 56) | 25.26 | 2.81 ± 0.07 (n = 127) | 15.29 | 2.39 ± 0.09 (n = 70) | 0.00000052 | 0.0000460 | 10.93 |
| D11Mit242 | 33.89 | 3.09 ± 0.11 (n = 51) | 25.78 | 2.83 ± 0.07 (n = 138) | 17.70 | 2.51 ± 0.10 (n = 64) | 0.00028 | 0.0148 | 6.23 | ||
| D11Nds10 | 36.94 | 3.17 ± 0.11 (n = 58) | 25.96 | 2.84 ± 0.07 (n = 129) | 13.72 | 2.31 ± 0.11 (n = 66) | 0.00000013 | 0.0000129 | 13.00 | ||
| Level of INFγ in serum | Lmr24 | D4Mit53 | 13.20 | 2.81 ± 0.14 (n = 59) | 5.93 | 2.04 ± 0.09 (n = 120) | 6.58 | 2.12 ± 0.12 (n = 74) | 0.0000097 | 0.000692 | 8.08 |
| Level of IL-4 in serum | Lmr24 | D4Mit53 | 1.79 | 0.47 ± 0.06 (n = 59) | 1.20 | 0.17 ± 0.05 (n = 120) | 1.26 | 0.21 ± 0.06 (n = 74) | 0.00025 | 0.0132 | 5.72 |
| D11Nds10 | 1.65 | 0.42 ± 0.07 (n = 58) | 1.48 | 0.34 ± 0.04 (n = 129) | 1.10 | 0.09 ± 0.06 (n = 66) | 0.00046 | 0.0230 | 6.73 | ||
| Level of IL-13 in serum | Lmr14 | D2Mit283 | 518.66 | 22.77 ± 1.18 (n = 44) | 551.08 | 23.48 ± 0.84 (n = 82) | 830.30 | 28.81 ± 1.24 (n = 57) | 0.00027 | 0.0146 | 10.76 |
Loci controlling organ pathology and immunological parameters in Leishmania major-infected F2 hybrids between CcS-9 and BALB/c.
Only linkages significant at whole genome level are given. In order to obtain normal distribution required for analysis of variance, the following transformations were used: lesion size (mm2)—power of 0.5; IgE in serum (μg/ml)—power of 0.32; IFN-γ in serum (ng/ml)—power of 0.4; IL-4 in serum (ng/ml)—Box-Cox transformation: [(x∧(−0.720642))−1]/(−0.720642), IL-13 in serum (pg/ml)—power of 0.5. The numbers in bold give the average non-transformed values. C and S indicate the presence of BALB/c and STS allele, respectively; n indicates number of mice.
In addition to the novel locus, we detected new functions of previously mapped loci (Table 7). Lmr14 was detected in F2 hybrids between CcS-16 and BALB/c. It was previously shown that it controlled hepatomegaly, splenomegaly, level of IgE and IFNγ in serum, and also infiltration of eosinophils to inguinal lymph nodes (33, 40, 44, 45). The present study revealed the role of Lmr14 (marker D2Mit283) in control of level of IL-13 in serum (corrected P = 0.0146, 10.76% of variance explained). The STS allele of this marker was linked with higher level of IL-13 in serum. Lmr15 was detected in F2 hybrids between CcS-16 and BALB/c and it influences hepatomegaly, IFNγ level in serum and infiltration of eosinophils in inguinal lymph nodes (33, 40, 45). The present study revealed several additional effects of Lmr15: control of splenomegaly (markers D11Mit141 and D11Nds10, corrected P = 0.0313 and 0.0235, respectively, 9.16 and 11.80% of variance explained), lesion size in week 8 after infection (markers D11Nds10 and D11Nds18, corrected P = 0.0282 and 0.0424, respectively, 9.37 and 4.21% of variance explained), kinetics of development of skin lesions (markers D11Nds18 and D11Nds10, corrected P = 0.0282 and 0.0406, respectively) and level of IgE in serum (markers D11Mit141, D11Mit242, and D11Nds10, corrected P = 0.0000460, 0.0148 and 0.0000129, respectively, 10.93, 6.23, and 13.00% of variance explained). The STS allele of these markers was linked with lower level of IgE in serum and smaller splenomegaly and skin lesions.
In addition, we detected two interactions between loci which control different parameters of the infection (Table 8). Interaction between markers D11Nds10 (Lmr15) and D16Mit19 (Lmr18) controlled lesion size in 7th week after infection. BALB/c homozygosity at Lmr15 synergizes with BALB/c homozygosity at Lmr18 in enlarging size of skin lesions (corrected P = 0.044, 6.71% of variance explained). Interaction between marker D4Mit53 (Lmr24) and D6Mit122 (Lmr4) controlled level of IL-10 in serum. STS homozygosity at Lmr24 synergized with STS homozygosity at Lmr4 in increasing of level of IL-10 in serum (corrected P = 0.0156, 9.61% of variance explained. The control of organ pathology and immune response in CcS-9 was not sex dependent.
Table 8
| Lesion size in 7th week after infection | D16Mit19 (Lmr18) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CS | SS | P-value | corr. P | % of explained variance | ||||||||
| D11Nds10 (Lmr15) | CC | n = 99 | 111.45 ± 10.05 | n = 30 | 73.89 ± 5.61 | n = 19 | 66.25 ± 6.88 | 0.00064 | 0.044 | 6.71 | |||
| CS | n = 32 | 59.48 ± 5.32 | n = 66 | 68.33 ± 3.75 | n = 32 | 76.07 ± 5.40 | |||||||
| SS | n = 20 | 63.07 ± 6.80 | n = 36 | 55.56 ± 5.13 | n = 10 | 51.56 ± 9.47 | |||||||
| Level of IL-10 in serum | D4Mit53 (Lmr24) | ||||||||||||
| CC | CS | SS | Pvalue | corr. P | % of explained variance | ||||||||
| D6Mit122 (Lmr4) | CC | n = 11 | 1.52 | 0.32 ± 0.03 | n = 24 | 2.10 | 0.47 ± 0.02 | n = 15 | 2.01 | 0.45 ± 0.02 | 0.00020 | 0.0156 | 9.61 |
| CS | n = 27 | 2.12 | 0.47 ± 0.02 | n = 44 | 1.94 | 0.43 ± 0.01 | n = 31 | 1.92 | 0.43 ± 0.02 | ||||
| SS | n = 10 | 1.87 | 0.42 ± 0.03 | n = 20 | 1.93 | 0.43 ± 0.02 | n = 15 | 2.15 | 0.47 ± 0.02 | ||||
Interactions between loci that control lesion size and level of IL-10 in serum in Leishmania major-infected F2 hybrids between CcS-9 and BALB/c.
In order to obtain normal distribution required for analysis of variance, the following transformation was used for level of IL-10 in serum (ng/ml) – [(x∧(−1.372544)]-1)/(−1.372544). C and S indicate the presence of BALB/c and STS allele, respectively; n indicates number of mice. Bold: non-transformed values.
The CcS-16 Strain: One Novel Lmr Locus Confirmed, Control of Parasite Load Detected for Two Additional Loci
Only female mice were used in the study with F2 hybrids between the BALB/c parental mouse strain and the RC strain CcS-16, because previous analysis of response to L. major in different RC strains (34) showed no significant dependence on sex in CcS-16 mice (in contrast to the strain CcS-9 described above). We confirmed a presence of a novel parasite controlling cluster Lmr4 (Figures 1, 2; Table 9) on chromosome 6, which influences parasite load in lymph nodes. A homozygous BALB/c (CC) allele in the Lmr4 linked to D6Mit48 was responsible for higher parasite load (corrected P = 0.017, 4.95% of variance explained) (Table 9). The effect of Lmr13 at the chromosome 18 linked to D18Mit40 on parasite load in liver was opposite to that of Lmr4: the STS allele (SS) was responsible for higher parasite load (corrected P = 0.00056, 10.4% of variance explained) (Table 9). Parasite load in liver controls also an interaction between Lmr14 linked to D2Nds3 with Lmr13 linked to D18Mit120 (suggestive linkage, corrected P = 0.0777, 4.8% of explained variance) (Table 10). Two genotype combinations determine highest parasite load: STS (SS) homozygotes in Lmr13 with BALB/c (CC) homozygotes in Lmr14, and STS (SS) homozygotes in Lmr14 with BALB/c (CC) homozygotes in Lmr13. A homozygous STS allele of Lmr13 linked to the markers D18Mit35 (corrected P = 0.0073, 4.85% of variance explained) and D18Mit120 (corrected P = 0.019, 5.18% of variance explained) determined the higher load of parasites in spleen (Table 9). Parasite load in spleen was also controlled by Lmr13 linked to D18Mit49 in interaction with Lmr11 linked to D3Mit11 (corrected P = 0.0295, 3.4% of variance explained). Highest parasite load was observed in combination of STS (SS) homozygotes in Lmr13 and BALB/c (CC) homozygotes in Lmr11 (Table 10).
Figure 2
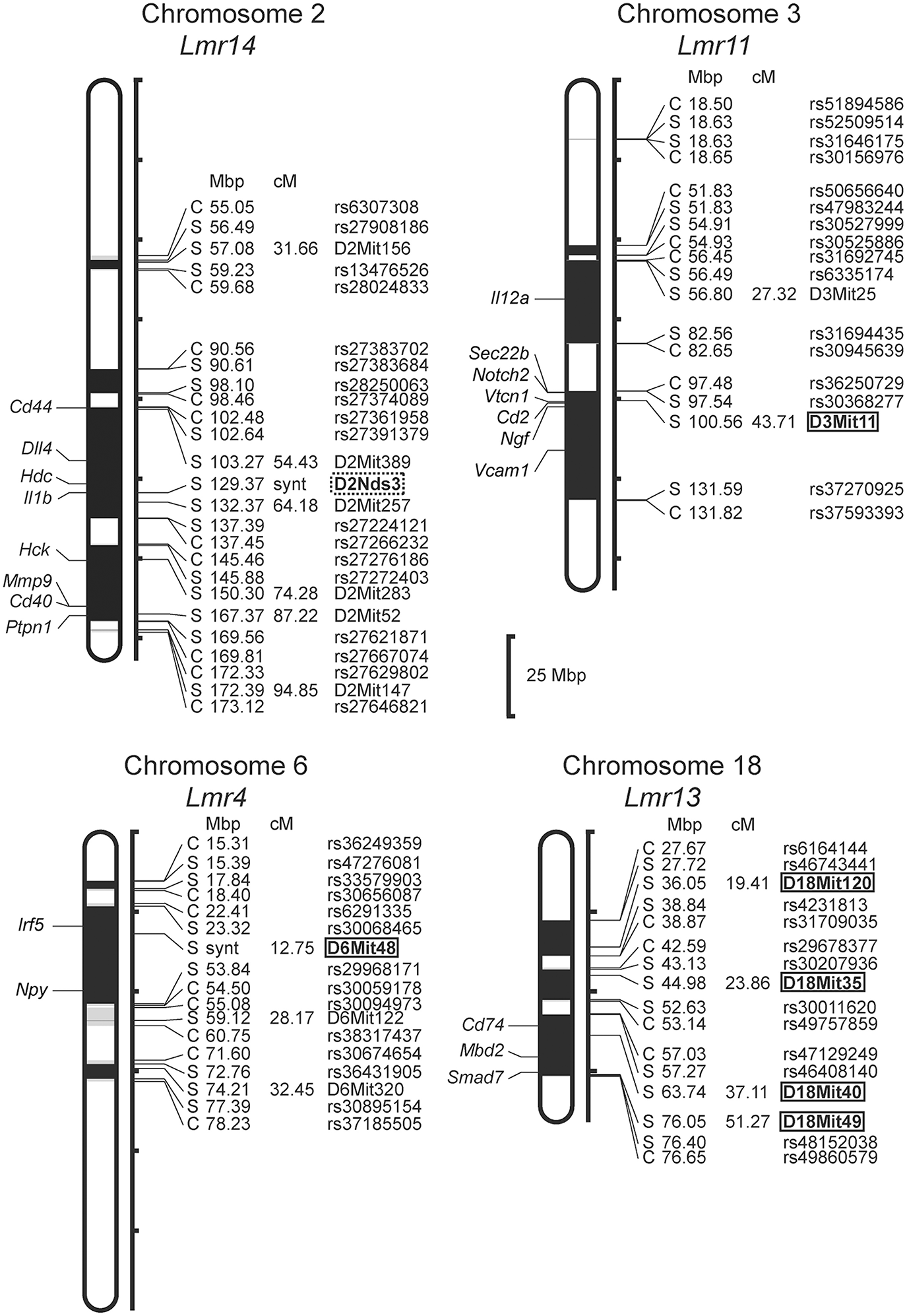
Positions of the loci controlling parasite load in organs of the RC strain CcS-16. The regions derived from the BALB/c strain are indicated as white; the regions of the STS origin are indicated as black; and the areas of undetermined origin are gray. Only the markers that determine the boundaries between BALB/c and STS-derived segments, and the markers used for statistical calculations are shown. Boxes indicate the markers that exhibited significant P-values, corrected for genome-wide search, dotted box—suggestive linkage. The genes that participate in the infection-related immune processes (16, 17, 20, 22, 23) and are located within the present Lmr loci are listed next to their positions at the chromosomes. Abbreviations indicate genes related to the response to Leishmania ssp.: Cd44, CD44 antigen; Dll4, delta like canonical Notch ligand 4; Hdc, histidine decarboxylase; Il1b, interleukin 1 beta; Hck, hemopoietic cell kinase; Mmp9, matrix metallopeptidase 9; Cd40, CD40 antigen; Ptpn1, protein tyrosine phosphatase, non-receptor type 1; Il12a, interleukin 12a; Sec22b, SEC22 homolog B, vesicle trafficking protein; Notch2, notch 2; Vtcn1, V-set domain containing T cell activation inhibitor 1; Cd2, CD2 antigen; Ngf, nerve growth factor; Vcam1, vascular cell adhesion molecule 1; Irf5, interferon regulatory factor 5; Npy, neuropeptide Y; Cd74, CD74 antigen [invariant polypeptide of major histocompatibility complex, class II antigen-associated)], Mbd2, methyl-CpG binding domain protein 2; Smad7, SMAD family member 7.
Table 9
| CC | CS | SS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | n | Mean | Transformed mean ± SE | n | Mean | Transformed mean ± SE | n | Mean | Transformed mean ± SE | P-value | corr. P | % of explained variance |
| Lymph nodes | ||||||||||||
| D6Mit48 (Lmr4) | 104 | 1.98 | 5.29 ± 0.12 | 230 | 1.16 | 4.75 ± 0.09 | 109 | 1.12 | 4.72 ± 0.12 | 0.00032 | 0.017 | 4.95 |
| Liver | ||||||||||||
| D18Mit40 (Lmr13) | 92 | 0.52 | 3.96 ± 0.12 | 178 | 0.43 | 3.75 ± 0.09 | 71 | 1.03 | 4.63 ± 0.16 | 0.0000078 | 0.00056 | 10.4 |
| Spleen | ||||||||||||
| D18Mit35 (Lmr13) | 136 | 0.52 | 3.95 ± 0.13 | 274 | 0.87 | 4.46 ± 0.09 | 133 | 1.07 | 4.67 ± 0.12 | 0.00013 | 0.0073 | 4.85 |
| D18Mit120 (Lmr13) | 133 | 0.52 | 3.95 ± 0.13 | 284 | 0.87 | 4.47 ± 0.09 | 130 | 0.99 | 4.59 ± 0.12 | 0.00038 | 0.019 | 5.18 |
The Lmr4 controls parasite load in lymph nodes, and Lmr13 controls parasite load in liver and spleen of CcS-16 derived F2 hybrids.
Means, standard error of mean (SE) and P-values were calculated by analysis of variance. The letters C and S indicate the BALB/c or STS allele, respectively; n indicates the number of mice in each group. The numbers in bold give the average non-transformed values. The values of concentration of parasite DNA (ng/ml) in total DNA isolated from lymph nodes, liver and spleen were transformed to obtain normal distribution: a natural logarithm of a (value *100). Only P-values significant after correction for genome-wide significance are shown.
Table 10
| Spleen | D3Mit11 (Lmr11) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CS | SS | P-value | corr. P | % of explained variance | ||||||||
| D18Mit49 (Lmr13) | CC | n = 27 | 0.40 | 3.68 ± 0.25 | n = 78 | 0.93 | 4.53 ± 0.14 | n = 37 | 0.63 | 4.14 ± 0.21 | |||
| CS | n = 59 | 0.78 | 4.35 ± 0.17 | n = 175 | 0.83 | 4.42 ± 0.10 | n = 48 | 0.84 | 4.43 ± 0.18 | 0.00042 | 0.0295 | 3.4 | |
| SS | n = 35 | 1.45 | 4.98 ± 0.21 | n = 53 | 0.59 | 4.07 ± 0.17 | n = 34 | 0.99 | 4.59 ± 0.22 | ||||
| Liver | D2Nds3 (Lmr14) | ||||||||||||
| CC | CS | SS | Pvalue | corr. P | % of explained variance | ||||||||
| CC | n = 28 | 0.48 | 3.87 ± 0.18 | n = 37 | 0.49 | 3.88 ± 0.15 | n = 15 | 0.80 | 4.38 ± 0.24 | ||||
| D18Mit120 (Lmr13) | CS | n = 38 | 0.37 | 3.61 ± 0.15 | n = 99 | 0.45 | 3.81 ± 0.10 | n = 41 | 0.53 | 3.96 ± 0.15 | 0.0012 | 0.0777 | 4.8 |
| SS | n = 15 | 0.97 | 4.58 ± 0.24 | n = 41 | 0.77 | 4.35 ± 0.15 | n = 22 | 0.37 | 3.62 ± 0.20 | ||||
Interactions that control parasite load in spleen and liver of CcS-16 derived F2 hybrids.
Means, standard error of mean (SE) and P-values were calculated by analysis of variance. The letters C and S indicate the BALB/c or STS allele, respectively; n indicates the number of mice in each group. Transformed means ± SE are shown next to mean non-transformed values in bold. The values of concentration of parasite DNA (ng/ml) in total DNA isolated from spleen and liver were transformed to obtain normal distribution: a natural logarithm of (a value*100). Only P-values significant after correction for genome-wide significance are shown.
Discussion
Parasite load and dissemination in visceral organs of infected host belong to the most important parameters in leishmaniasis. Elucidation of host determinants of parasite control could help to better understand these critical disease mechanisms. We have therefore addressed the following questions: genetic control of parasite load in organs after L. major infection, analysis of relationship among parasite-controlling genes and the genes controlling organ pathology, and role of sex in control of parasite load. We defined two new genetic loci controlling parasite load (Lmr24 and Lmr27) and described that six previously mapped loci (Lmr4, Lmr11, Lmr13, Lmr14, Lmr15, and Lmr25) also control parasite load. This enabled us to show a genetic network controlling interaction between parasite and mammalian host.
Control of Parasite Load in Organs: Multiple Genes and Distinct Organ Specificities That Operate in a Network With Many Gene-Gene Interactions
Visceral leishmaniasis is a life-threatening disease (9, 13) and hence understanding parasite spread to organs is essential. Although many of its host, vector, and parasite determinant have been described (46), the role of the host in control of parasite visceralization remains largely unknown. The importance of host determinants that can limit leishmaniasis to a cutaneous form or allow progressive visceral pathology is stressed by the observation that even though L. major causes mostly only cutaneous pathology in human, cases of visceralization are also known (31, 32). We have therefore studied parasite load after L. major infection in three organs: lymph nodes, spleen and liver, and found that parasite load in each tested organ is controlled by multiple genes. In this work, we report six loci (Lmr4, Lmr14, Lmr15, Lmr24, Lmr25, and Lmr27) that control parasite load in lymph nodes of the strains CcS-9 and CcS-16 (Table 11). Lmr4, Lmr14, and Lmr15 can operate independently from other genes (main effect loci), whereas Lmr24, Lmr25, and Lmr27 operate only in interaction with other loci (Figure 3). Previously, we described another locus, Lmr20, which influences parasite load in lymph nodes of the strain CcS-11 (21).
Table 11A
| Chr. | Lmr loci | Markers | Sex | Parasite control in organ | Response to parasite |
|---|---|---|---|---|---|
| 2 | Lmr14 | D2Mit148 | IL-13 in serum | ||
| D2Mit283 | M | Lymph nodes | Eosinophils in lymph nodes**# | ||
| 2 and 4 | Interaction, Lmr14 and Lmr24 | D2Mit283 | Lymph nodes | ||
| D4Mit172 | |||||
| 2 and 5 | Interaction, Lmr14 and Lmr25 | D2Mit283 | Lymph nodes | ||
| D5Mit143 | |||||
| 2 and 17 | Interaction, Lmr14 and Lmr27 | D2Mit283 | Liver | ||
| D17Mit120 | |||||
| 4 | Lmr24 | D4Mit23 | Skin lesion Splenomegaly IL-4, IFNγ in serum | ||
| D4Mit53 | |||||
| D4Mit172 | |||||
| 4 and 6 | Interaction, Lmr4 and Lmr24 | D4Mit53 | IL-10 in serum | ||
| D6Mit122 | |||||
| 5 | Lmr25 | D5Mit143 | Eosinophils in lymph nodes*** | ||
| 5 and 17 | Interaction, Lmr25 and Lmr27 | D5Mit143 | Liver* | ||
| D17Mit122 | |||||
| 6 and 17 | Interaction, Lmr4 and Lmr27 | D6Mit122 | M | Lymph nodes | |
| D17Mit122 | |||||
| 9 | Interaction, Lmr15 and Lmr26 | D9Mit15 D11Nds10 | M | Eosinophils in lymph nodes*** | |
| 11 | Lmr15 | D11Nds18 D11Nds10 D11Mit141 | Liver | Skin lesion Splenomegaly Hepatomegaly IL-4, IgE in serum | |
| F | Lymph nodes | ||||
| M | Lymph nodes | ||||
| 11 and 16 | Interaction, Lmr15 and Lmr18 | D11Nds10 D16Mit19 | Skin lesion | ||
| 11 and 17 | Interaction, Lmr15 and Lmr27 | D11Nds10 D17Mit122 | M | Lymph nodes | |
| 17 | Lmr27 | D17Mit122 | M | Lymph nodes |
Summary of loci that control parasite load detected in CcS-9 study.
Suggestive linkage.
Data from Slapničková et al. (33).
Interaction between sex and marker was not significant.
Significant only in males from the cross CcS-9 × BALB/c.
Table 11B
| Chr. | Lmr loci | Markers | Parasite control in organ | Immune response |
|---|---|---|---|---|
| 2 | Lmr14 | D2Mit52 D2Mit102 D2Mit283 D2Mit389 D2MitNds3 | Splenomegaly Hepatomegaly Spontaneous proliferation of splenocytes from infected mice IFNγ in serum | |
| 2 and 10 | Interaction, Lmr5, and Lmr14 | D2Mit102 D10Mit103 D2Mit389 | IL-12, IgE in serum | |
| 2 and 16 | Interaction, Lmr12 and Lmr14 | D2Mit283 D16Mit126 | TNFα in serum | |
| 2 and 18 | Interaction, Lmr13 and Lmr14 | D2Nds3 D18Mit120 | Liver* | TNFα in serum |
| 3 and 18 | Interaction, Lmr11 and Lmr13 | D3Mit11 D18Mit49 | Spleen | |
| 6 | Lmr4 | D6Mit48 | Lymph nodes | |
| 10 and 16 | Interaction, Lmr12 and Lmr19 | D10Mit65 D16Mit126 | Spontaneous proliferation of splenocytes from infected mice | |
| 11 | Lmr15 | D11Mit37 D11Mit 139 D11Mit 242 | Hepatomegaly IFNγ in serum | |
| 16 | Lmr12 | D16Mit126 | IL-4, IgE in serum | |
| 18 | Lmr13 | D18Mit35 D18Mit40 D18Mit120 | Spleen Liver | Lesion size IgE in serum |
Summary of loci that control parasite load and response to infection in CcS-16.
Figure 3
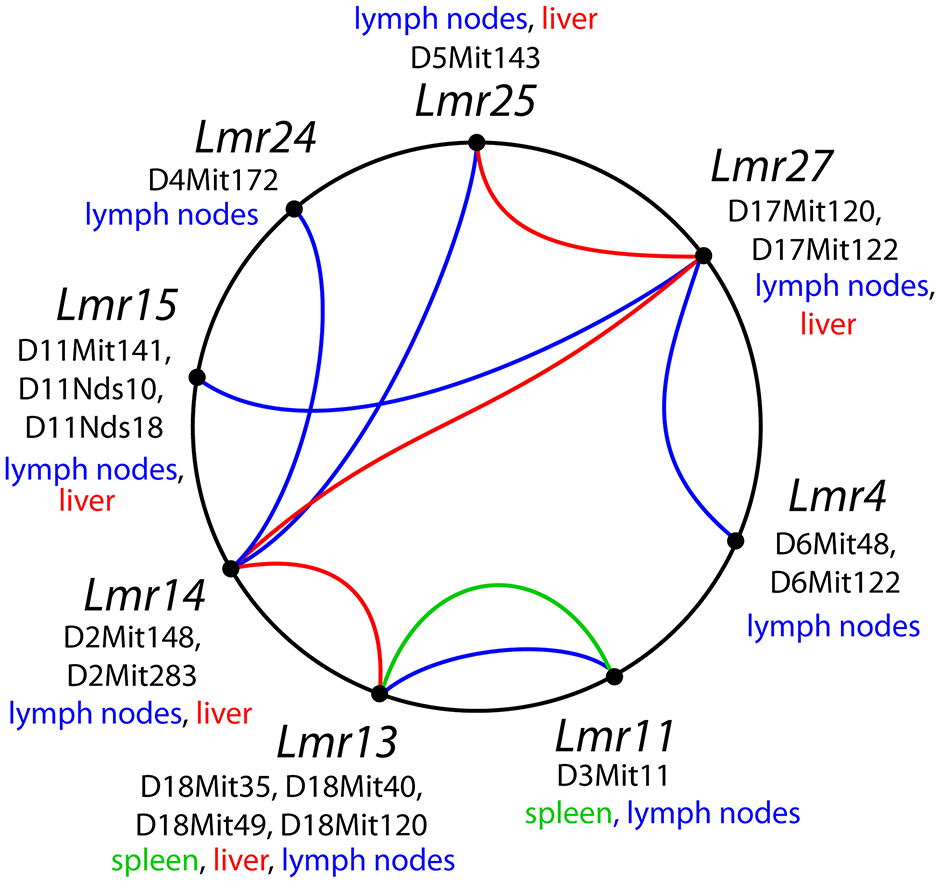
Interactions among loci controlling parasite load after L. major infection. The lines that connect related loci indicate interactions controlling a certain phenotype: parasite load in lymph nodes—blue color, in spleen—green, in liver—purple. Main gene effects of the Lmr13, Lmr14, Lmr15, Lmr28, and Lmr29 are also indicated.
We have found two loci that control parasite load in spleen of the strain CcS-16: Lmr13 and Lmr11 (Table 11B). Lmr13 operates as main effect locus, role of Lmr11 is visible only in interaction with Lmr13 (Tables 9, 10). No locus controlling parasite load in spleen was detected in the strain CcS-9 although spleens of this strain contain parasites (47). This might be caused by the fact that parasite load in spleen in the strain CcS-9 is controlled by multiple weak loci and our experiments lacked the power to detect them. Thus, with the previously detected Lmr5 controlling parasites in spleen in CcS-11 (21), there are three different loci controlling L. major load in spleen. Parasite load in liver is controlled by two main effect loci Lmr13 and Lmr15, operating in the strains CcS-16 and CcS-9, respectively, and three loci Lmr14, Lmr25 (suggestive linkage) and Lmr27 (in the strain CcS-9), whose effect is observed only in interaction.
Comparison of loci that control parasite burden in organs indicates organ specific control of parasite load, where dissemination of parasites to lymph nodes, spleen and liver were controlled by distinct sets of loci that only partly overlaps (Table 11). Some loci, such as Lmr13, Lmr14, Lmr15, Lmr25, and Lmr27 determined parasite load in two different organs, effects of others were limited to only one organ. Organ specific control was described also in candidate gene based studies of parasite burden after infection with L. donovani, where genes Slc11a1, H2 (25) and Lyst (24) control parasite load in spleen, whereas Slc11a1 (26), H2 (27, 28) and Ir2 (28) determine parasite load in liver. Genome-wide studies with L. tropica infection showed that parasite load in lymph nodes are determined by Ltr1 and Ltr4, in spleen by Ltr3 and Ltr6, and in liver by Ltr2, Ltr4, and Ltr8 (23) (Table 1). On the mechanistic level, organ-specific genetic control might be based on organ-specific immune response to Leishmania parasites (48). Some of the loci that determine parasite load in L. donovani (MHOM/SD/62/3S) and L. tropica (MHOM/1999/TR/SU23) overlap with the Lmr loci: Lmr13 that controls parasite load in liver and spleen co-localize with Ltr8 influencing parasite load in liver (23), Lmr14 (suggestive linkage) influencing in interaction with Lmr13 parasite load in liver overlaps with Ir2 (28) and Ltr2 (23) that control parasite load in liver after infection with L. donovani and L. tropica, respectively. Thus, similarly as H2 or Slc11a1, some parasite-controlling Lmr loci might affect several pathogens, probably reflecting immune responses effective against groups of infectious agents.
Relationship Between Control of Parasite Load, Organ Pathology, and Systemic Immune Response
We assessed relationship between parasite control, organ pathology, and systemic immune response (Table 11) and found a large heterogeneity in effects of controlling loci. Moreover, control of parasite load was linked with control of organ pathology in some loci, but not in others. Some loci, such as Lmr13 and Lmr14 carried by CcS-16, and Lmr15 and Lmr24 carried by CcS-9 controlled both parasite loads in organs, organ pathology and systemic immune response: Lmr13 determined parasite load in spleen and liver, skin lesions (40), IgE in serum (44), TNFα in serum (45); Lmr14 (CcS-16) influenced parasite load in liver (suggestive linkage), splenomegaly, hepatomegaly (40), IgE (44), IFNγ, IL-12, and TNFα in serum, and spontaneous proliferation of splenocytes from infected mice (45); Lmr15 (CcS-9) controlled parasite load in lymph nodes and in liver, infiltration of eosinophils into lymph nodes (33), skin lesions, hepatomegaly, IL-4 and IgE in serum; and Lmr24 controlled parasite load in lymph nodes, skin lesions, splenomegaly and IL-4, IL-10, and IFNγ in serum). Lmr14 carried by CcS-9 determined parasite load in lymph nodes and in liver and systemic immune response (IL-13 in serum), but no organ pathology. Lmr15 carried by CcS-16 did not influence parasite load, but controls hepatomegaly (40) and IFNγ in serum (45). Loci Lmr14 and Lmr15 might represent clusters of genes, their STS-derived segments in CcS-9 and CcS-16 overlap, but are not identical and they controlled different combinations of parameters in these strains (Tables 11A,B). Loci Lmr4, Lmr11, and Lmr27 controlled parasite load in organs, but neither organ pathology nor systemic immune response (Table 11). Locus Lmr25 controls parasite load in lymph nodes, infiltration of eosinophils to the lymph nodes, but no skin or visceral pathology. Locus Lmr18 influences only skin lesions. Lmr26 controls only infiltration of eosinophils to the lymph nodes (33), and locus Lmr5 carried by CcS-16, Lmr12 and Lmr19 control only systemic immune response to L. major (44, 45).
We have observed a discrepancy between genes controlling parasites in organs and genes controlling splenomegaly and hepatomegaly. In the strain CcS-9 parasite load in the liver was controlled by the loci Lmr14, Lmr15, and Lmr27, but only one of them, Lmr15, was involved in control of hepatomegaly. Similarly, in the strain CcS-16, parasite load in liver were controlled by the loci Lmr13 and Lmr14 (suggestive linkage), whereas hepatomegaly is determined by Lmr14 and Lmr15. In the strain CcS-9 we did not detect loci controlling parasite load in spleen (see above); splenomegaly was controlled by loci Lmr15 and Lmr24. In the strain CcS-16 parasite load in spleen was controlled by Lmr11 and Lmr13, but none of these loci was involved in control of splenomegaly, which was determined by Lmr14. The difference between the genes controlling parasite load and spread to an organ and those controlling pathology of this organ reflects the multiple facets of interaction between pathogen and host.
In visceral leishmaniasis, Leishmania amastigotes exist and proliferate in the mononuclear phagocytic system, especially spleen, liver and bone marrow. The response of immune system might lead either to parasite clearance or to unproductive inflammation resulting in organ hyperplasia (48, 49). These processes involve multiple steps that are regulated by different genes. The presented data are the first step in creating genetic model of visceralization, the most important pathology caused by Leishmania parasites, by identification the loci that control invasion of parasites and loci controlling the inflammatory response of the host.
Sex-Dependent Control of Parasite Load
Control of parasite load in inguinal lymph nodes is in many cases sex dependent. In the strain CcS-9, Lmr4, and Lmr27 controlled parasite load in males only; Lmr15 controlled parasite load both in females and males, but with the opposite direction: BALB/c (C) allele is linked with higher parasite load in females and lower parasite load in males. Sex has been found to influence susceptibility to many diseases, including leishmaniasis (50, 51) and genes controlling infections that are sex dependent have been observed with other infectious agents such as viruses (52, 53), bacteria (54), parasites (33, 55), fungi (56), and helminths (57). All the above mentioned loci are localized on autosomal chromosomes and thus are shared by both sexes, but the regulatory genome is sexually dimorphic (58). The regulatory genome includes steroid hormones responsive elements (59), sex-specific micro-RNA (60) and sexually dimorphic DNA methylation patterns, which vary significantly from tissue to tissue (61). Generally, the sex differences have complex functional structure and their explanation requires future molecular identification of the responsible genes.
Further studies are needed to show how these mechanisms influence frequent sex differences in human leishmaniasis. Human epidemiological studies demonstrated that women are less likely to develop leishmaniasis while men tend to be more susceptible (62), although there are exceptions (50, 63). Some epidemiological studies reported no significant sex differences in registered cases of cutaneous leishmaniasis caused by L. tropica (64) and L. major (65) between men and women. However, other studies revealed in male patients a higher incidence of cutaneous leishmaniasis caused by L. major and L. tropica (66, 67), L. major only (68), and also by L. guyanensis (69). Men were also more susceptible to visceral infection caused by L. donovani (70–72), L. infantum (73–76). As an exception to this general trend, the study in Afghanistan found that females developed more lesions and scars after L. tropica infection (77).
Our data do not allow conclusion of influence of sex on parasite load in organs of the strain CcS-16. We tested only females in this strain, because in previous experiments, they did not exhibit sex differences in lesion size (34), however CcS-16 might exhibit sex differences in parasite load.
Conclusions
The present study was focused on the genetic basis of one of the most important parameters of L. major caused leishmaniasis—parasite load in target organs. The study used a hypothesis-free experimental approach and recombinant congenic mouse strains to perform genome wide mapping of a complex system of genes that regulate dissemination of the parasite inside a mammalian organism and form a network-like structure (Figure 3). Host genes controlling L. major revealed a wide variety of heterogeneous effects that included distinct organ-specific control, single-gene effects, gene-gene interactions and sex dependent control. The presented results contribute to the understanding of genetic aspects of leishmaniasis. Mapping of these genes and subsequent identification of prospective candidate genes will allow their functional analysis. In addition, the obtained information allows making focused tests of human orthologous genes for their possible role in leishmaniasis and to elucidate pathogenesis and visceralization in individual patients.
Statements
Ethics statement
The experiments were performed in accordance with the European Union guidelines for work with animals under the Policy of Animal Protection Law (No.246/1992), and also with the regulations of the Ministry of Agriculture of the Czech Republic (No.207/2004). The experiments were approved by the Institutional Animal Care Committee of the Institute of Molecular Genetics AS CR and by Departmental Expert Committee for the Approval of Projects of Experiments on Animals of the Academy of Sciences of the Czech Republic (permissions 12/2002 and 89/2013).
Author contributions
TK and MČ conceived the study, performed experiments, interpreted the data and wrote the manuscript. VV performed experiments, analyzed the data and contributed to the writing of the manuscript. YS, HH, and MS performed the experiments. PD analyzed the data and contributed to the writing of the manuscript. ML conceived the study, interpreted data and wrote the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by the Czech Science Foundation (Grants GACR 14-30186S and GACR 16-22346S) the Academy of Sciences of the Czech Republic (RVO 68378050) and Ministry of Education, Youth, and Sports (grant LH12049).
Acknowledgments
MČ and YS are Ph. D. students of the Faculty of Science, Charles University, Prague, Czech Republic.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
- BALB
(Bagg and Albino) a standard inbred mouse strain
- CcS/Dem
BALB/c-c-STS/Dem (recombinant congenic strain)
- Dice1.2
determination of interleukin 4 commitment 1b
- H2
histocompatibility-2
- IL
interleukin
- IFNγ
interferon gamma
- IgE
immunoglobulin E
- Lmr
Leishmania major response
- Ltr
Leishmania tropica response
- PCR-ELISA
polymerase chain reaction enzyme linked immunosorbent assay
- QTL
quantitative trait locus
- RC
recombinant congenic
- Slc11a1, solute carrier family 11 (proton-coupled divalent metal ion transporters)
member 1
- SNB-9
saline-neopeptone-blood-9
- STS
an inbred mouse strain of Swiss origin.
Abbreviations
References
1.
DujardinJCCampinoLCañavateCDedetJPGradoniLSoteriadouKet al. Spread of vector-borne diseases and neglect of leishmaniasis, Europe. Emerg Infect Dis. (2008) 14:1013–8. 10.3201/eid1407.071589
2.
AlvarJVélezIDBernCHerreroMDesjeuxPCanoJet al. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE. (2012) 7:e35671. 10.1371/journal.pone.0035671
3.
Torres-GuerreroEQuintanilla-CedilloMRRuiz-EsmenjaudJArenasR. Leishmaniasis: a review. F1000Research. (2017) 6:750. 10.12688/f1000research.11120.1
4.
PetersenCA. Leishmaniasis, an emerging disease found in companion animals in the United States. Top Companion Anim Med. (2009) 24:182–8. 10.1053/j.tcam.2009.06.006
5.
GonzálezCWangOStrutzSEGonzález-SalazarCSánchez-CorderoVSarkarS. Climate change and risk of leishmaniasis in North America: predictions from ecological niche models of vector and reservoir species. PLoS Negl Trop Dis. (2010) 4:e585. 10.1371/journal.pntd.0000585
6.
ReadyPD. Leishmaniasis emergence in Europe. Eurosurveillance. (2010) 15:19505. 10.2807/ese.15.10.19505-en
7.
EschKJPetersenCA. Transmission and epidemiology of zoonotic protozoal diseases of companion animals. Clin Microbiol Rev. (2013) 26:58–85. 10.1128/CMR.00067–12
8.
DouvoyiannisMKhromachouTByersNHargreavesJMurrayHW. Cutaneous leishmaniasis in North Dakota. Clin Infect Dis. (2014) 59:e73–5. 10.1093/cid/ciu386
9.
WHO. Integrating Neglected Tropical Diseases in Global Health and Development. Fourth WHO Report on Neglected Tropical Diseases.Geneva: World Health Organization (2017).
10.
HerwaldtBL. Leishmaniasis. Lancet. (1999) 354:1191–9. 10.1016/S0140–6736(98)10178–2
11.
MachadoCMMartinsTCColturatoILeiteMSSimioneAJDe SouzaMPet al. Epidemiology of neglected tropical diseases in transplant recipients. Review of the literature and experience of a Brazilian HSCT center. Rev Inst Med Trop Sao Paulo. (2009) 51:309–24. 10.1590/S0036–46652009000600002
12.
PavliAMaltezouHC. Leishmaniasis, an emerging infection in travelers. Int J Infect Dis. (2010) 14:e1032–9. 10.1016/j.ijid.2010.06.019
13.
WHO. Investing to Overcome the Global Impact of Neglected Tropical Diseases: Third WHO Report on Neglected Diseases 2015.Geneva: World Health Organization (2015).
14.
PicadoAOstynBSinghSPUranwSHaskerERijalSet al. Risk factors for visceral leishmaniasis and asymptomatic Leishmania donovani infection in India and Nepal. PLoS ONE. (2014) 9:e87641. 10.1371/journal.pone.0087641
15.
WHO. Weekly epidemiological record: global leishmaniasis update, 2006–2015, a turning point in leishmaniasis surveillance. World Heal Organ. (2017) 92:557–72. 10.1186/1750-9378-2-15
16.
LipoldováMDemantP. Genetic susceptibility to infectious disease: Lessons from mouse models of leishmaniasis. Nat Rev Genet. (2006) 7:294–305. 10.1038/nrg1832
17.
TerrazasCATerrazasLIGómez-GarcíaL. Modulation of dendritic cell responses by parasites: a common strategy to survive. J Biomed Biotechnol. (2010) 2010:357106. 10.1155/2010/357106
18.
KobetsTGrekovILipoldovaM. Leishmaniasis: prevention, parasite detection and treatment. Curr Med Chem. (2012) 19:1443–74. 10.2174/092986712799828300
19.
GhorbaniMFarhoudiR. Leishmaniasis in humans: drug or vaccine therapy?Drug Des Devel Ther. (2018) 12:25–40. 10.2147/DDDT.S146521
20.
McMahon-PrattDAlexanderJ. Does the Leishmania major paradigm of pathogenesis and protection hold for New World cutaneous leishmaniases or the visceral disease?Immunol Rev. (2004) 201:206–24. 10.1111/j.0105–2896.2004.00190.x
21.
KureyIKobetsTHavelkováHSlapničkováMQuanLTrtkováKet al. Distinct genetic control of parasite elimination, dissemination, and disease after Leishmania major infection. Immunogenetics. (2009) 61:619–33. 10.1007/s00251–009-0392–9
22.
SakthianandeswarenAFooteSJHandmanE. The role of host genetics in leishmaniasis. Trends Parasitol. (2009) 25:383–91. 10.1016/j.pt.2009.05.004
23.
SohrabiYHavelkováHKobetsTŠímaMVolkovaVGrekovIet al. Mapping the genes for susceptibility and response to Leishmania tropica in mouse. PLoS Negl Trop Dis. (2013) 7:e2282. 10.1371/journal.pntd.0002282
24.
KirkpatrickCEFarrellJP. Leishmaniasis in beige mice. Infect Immun. (1982) 38:1208–16.
25.
LeclercqVLebastardMBelkaidYLouisJMilonG. The outcome of the parasitic process initiated by Leishmania infantum in laboratory mice: a tissue-dependent pattern controlled by the Lsh and MHC loci. J Immunol. (1996) 157:4537–45.
26.
BradleyDJTaylorBABlackwellJEvansEPFreemanJ. Regulation of Leishmania populations within the host. III. Mapping of the locus controlling susceptibility to visceral leishmaniasis in the mouse. Clin Exp Immunol. (1979) 37:7–14.
27.
BlackwellJFreemanJBradleyD. Influence of H-2 complex on acquired resistance to Leishmania donovani infection in mice. Nature. (1980) 283:72–4. 10.1038/283072a0
28.
DeTollaLJJr.SemprevivoLHPalczukNCPassmoreHC. Genetic control of acquired resistance to visceral leishmaniasis in mice. Immunogenetics. (1980) 10:353–61.
29.
RobertsMAlexanderJBlackwellJM. Influence of Lsh, H-2, and an H-11-linked gene on visceralization and metastasis associated with Leishmania mexicana infection in mice. Infect Immun. (1989) 57:875–81.
30.
BaguetAEplerJWenKWBixM. A Leishmania major response locus identified by interval-specific congenic mapping of a T helper type 2 cell bias-controlling quantitative trait locus. J Exp Med. (2004) 200:1605–12. 10.1084/jem.20040334
31.
KaramianMMotazedianMHMehrabaniDGholamiK. Leishmania major infection in a patient with visceral leishmaniasis: treatment with amphotericin B. Parasitol Res. (2007) 101:1431–4. 10.1007/s00436–007-0649-x
32.
MortazaviHMohebaliMTaslimiYSadeghipourPAnsariMKamyabKet al. Hoarseness as the presenting symptom of visceral leishmaniasis with muco-cutaneous lesions: a case report. Iran J Parasitol. (2015) 10:296–300.
33.
SlapničkováMVolkovaVCepičkováMKobetsTŠímaMSvobodováMet al. Gene-specific sex effects on eosinophil infiltration in leishmaniasis. Biol Sex Differ. (2016) 7:59. 10.1186/s13293–016-0117–3
34.
KobetsTHavelkováHGrekovIVolkovaVVojtíškováJSlapničkováMet al. Genetics of host response to Leishmania tropica in mice - different control of skin pathology, chemokine reaction, and invasion into spleen and liver. PLoS Negl Trop Dis. (2012) 6:e1667. 10.1371/journal.pntd.0001667
35.
StassenAPMGrootPCEppigJTDemantP. Genetic composition of the recombinant congenic strains. Mamm Genome. (1996) 7:55–810.1007/s003359900013
36.
DemantPLipoldováMSvobodováM. Resistance to Leishmania major in mice. Science. (1996) 274:1392a. 10.1126/science.274.5291.1392
37.
SohrabiYVolkovaVKobetsTHavelkováHKrayemISlapničkováMet al. Genetic regulation of guanylate-binding proteins 2b and 5 during leishmaniasis in mice. Front Immunol. (2018) 9:130. 10.3389/fimmu.2018.00130
38.
GrekovISvobodováMNohýnkováELipoldováM. Preparation of highly infective Leishmania promastigotes by cultivation on SNB-9 biphasic medium. J Microbiol Methods. (2011) 87:273–7. 10.1016/j.mimet.2011.08.012
39.
LipoldováMSvobodováMKrulováMHavelkováHBadalováJNohýnkováEet al. Susceptibility to Leishmania major infection in mice: Multiple loci and heterogeneity of immunopathological phenotypes. Genes Immun. (2000) 1:200–6. 10.1038/sj.gene.6363660
40.
VladimirovVBadalováJSvobodováMHavelkováHHartAAMBlaŽkováHet al. Different genetic control of cutaneous and visceral disease after Leishmania major infection in mice. Infect Immun. (2003) 71:2041–6. 10.1128/IAI.71.4.2041–2046.2003
41.
KobetsTBadalováJGrekovIHavelkováHSvobodováMLipoldováM. Leishmania parasite detection and quantification using PCR-ELISA. Nat Protoc. (2010) 5:1074–80. 10.1038/nprot.2010.68
42.
LairdPWZijderveldALindersKRudnickiMAJaenischRBernsA. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. (1991) 19:4293. 10.1093/nar/19.15.4293
43.
LanderESSchorkNJ. Genetic dissection of complex traits. Science. (1994) 265:2037–48. 10.1126/science.8091226
44.
BadalováJSvobodováMHavelkováHVladimirovVVojtíškováJEngováJet al. Separation and mapping of multiple genes that control IgE level in Leishmania major infected mice. Genes Immun. (2002) 3:187–95. 10.1038/sj.gene.6363838
45.
HavelkováHBadalováJSvobodováMVojtíškováJKureyIVladimirovVet al. Genetics of susceptibility to leishmaniasis in mice: four novel loci and functional heterogeneity of gene effects. Genes Immun. (2006) 7:220–33. 10.1038/sj.gene.6364290
46.
McCallLIZhangWWMatlashewskiG. Determinants for the development of visceral leishmaniasis disease. PLoS Pathog. (2013) 9:e1003053. 10.1371/journal.ppat.1003053
47.
LipoldováMSvobodováMHavelkováHKrulováMBadalováJNohýnkováEet al. Mouse genetic model for clinical and immunological heterogeneity of leishmaniasis. Immunogenetics. (2002) 54:174–83. 10.1007/s00251–002-0439–7
48.
de FreitasEOde LeorattiFMFreire-de-LimaCGMorrotAFeijóDF. The contribution of immune evasive mechanisms to parasite persistence in visceral leishmaniasis. Front Immunol. (2016) 7:153. 10.3389/fimmu.2016.00153
49.
VarmaNNaseemS. Hematologic changes in visceral leishmaniasis/kala azar. Indian J Hematol Blood Transfus. (2010) 26:78–82. 10.1007/s12288–010-0027–1
50.
SniderHLezama-DavilaCAlexanderJSatoskarAR. Sex hormones and modulation of immunity against leishmaniasis. Neuroimmunomodulation. (2009) 16:106–13. 10.1159/000180265
51.
BerninHLotterH. Sex bias in the outcome of human tropical infectious diseases: Influence of steroid hormones. J Infect Dis. (2014) 3:S107–13. 10.1093/infdis/jit610
52.
ButterfieldRJRoperRJRheinDMMelvoldRWHaynesLMaRZDoergeRWTeuscherC. Sex-specific quantitative trait loci govern susceptibility to Theiler's murine encephalomyelitis virus-induced demyelination. Genetics. (2003) 163:1041–6.
53.
BoivinGAPothlichetJSkameneEBrownEGLoredo-OstiJCSladekRet al. Mapping of clinical and expression quantitative trait loci in a sex-dependent effect of host susceptibility to mouse-adapted influenza H3N2/HK/1/68. J Immunol. (2012) 188:3949–60. 10.4049/jimmunol.1103320
54.
Min-OoGLindqvistLVaglenovAWangCFortinPLiYet al. Genetic control of susceptibility to pulmonary infection with Chlamydia pneumoniae in the mouse. Genes Immun. (2008) 9:383–8. 10.1038/sj.gene.6364450
55.
ŠímaMHavelkováHQuanLSvobodováMJarošíkováTVojtíškováJet al. Genetic control of resistance to Trypanosoma brucei brucei infection in mice. PLoS Negl Trop Dis. (2011) 5:e1173. 10.1371/journal.pntd.0001173
56.
CarrollSFLoredo OstiJCGuillotLMorganKQureshiST. Sex differences in the genetic architecture of susceptibility to Cryptococcus neoformans pulmonary infection. Genes Immun. (2008) 9:536–45. 10.1038/gene.2008.48
57.
HayesKSHagerRGrencisRK. Sex-dependent genetic effects on immune responses to a parasitic nematode. BMC Genomics. (2014) 15:193. 10.1186/1471–2164-15–193
58.
OberCLoiselDAGiladY. Sex-specific genetic architecture of human disease. Nat Rev Genet. (2008) 9:911–22. 10.1038/nrg2415
59.
CatoACBNestlAMinkS. Rapid actions of steroid receptors in cellular signaling pathways. Sci Signal. (2002) 43:1–36. 10.1126/stke.2002.138.re9
60.
FuentesNRoyAMishraVCabelloNSilveyraP. Sex-specific microRNA expression networks in an acute mouse model of ozone-induced lung inflammation. Biol Sex Differ. (2018) 9:18. 10.1186/s13293–018-0177–7
61.
McCormickHYoungPEHurSSJBooherKChungHet al. Isogenic mice exhibit sexually-dimorphic DNA methylation patterns across multiple tissues. BMC Genomics. (2017) 18:966. 10.1186/s12864–017-4350-x
62.
SchlagenhaufPChenLHWilsonMEFreedmanDOTchengDSchwartzEet al. Sex and gender differences in travel-associated disease. Clin Infect Dis. (2010) 50:826–32. 10.1086/650575
63.
Guerra-SilveiraFAbad-FranchF. Sex Bias in infectious disease epidemiology: patterns and processes. PLoS ONE. (2013) 8:e62390. 10.1371/journal.pone.0062390
64.
LayeghPMoghimanTAhmadian HoseiniSA. Children and cutaneous leishmaniasis: a clinical report and review. J Infect Dev Ctries. (2013) 7:614–7. 10.3855/jidc.2939
65.
BettaiebJToumiAChlifSChelghafBBoukthirAGharbiAet al. Prevalence and determinants of Leishmania major infection in emerging and old foci in Tunisia. Parasit Vect. (2014) 7:386. 10.1186/1756–3305-7–386
66.
GandacuDGlazerYAnisEKarakisIWarshavskyBSlaterPGrottoI. Resurgence of cutaneous leishmaniasis in Israel, 2001–2012. Emerg Infect Dis. (2014) 20:1605–11. 10.3201/eid2010.140182
67.
SpotinARouhaniSParviziP. The Associations of Leishmania major and Leishmania tropica aspects by focusing their morphological and molecular features on clinical appearances in Khuzestan province, Iran. Biomed Res Int. (2014) 2014:913510. 10.1155/2014/913510
68.
TalariSATalaeiRShajariGVakiliZTaghaviardakaniA. Childhood cutaneous leishmaniasis: report of 117 cases from Iran. Korean J Parasitol. (2006) 44:355–60. 10.3347/kjp.2006.44.4.355
69.
SoaresLAbad-FranchFFerrazG. Epidemiology of cutaneous leishmaniasis in central Amazonia: a comparison of sex-biased incidence among rural settlers and field biologists. Trop Med Int Health. (2014) 19:988–95. 10.1111/tmi.12337
70.
RijalSUranwSChappuisFPicadoAKhanalBPaudelISet al. Epidemiology of Leishmania donovani infection in high-transmission foci in Nepal. Trop Med Int Health. (2010) 15 (Suppl. 2):21–8. 10.1111/j.1365–3156.2010.02518.x
71.
SandanayakaRKahawitaIGamageASiribaddanaSAgampodiS. Emergence of cutaneous leishmaniasis in Polonnaruwa, Sri Lanka 2008–2011. Trop Med Int Health. (2014) 19:140–5. 10.1111/tmi.12232
72.
WondimenehYTakeleYAtnafuAFeredeGMuluyeD. Trend analysis of visceral leishmaniasis at Addis Zemen health center, northwest Ethiopia. Biomed Res Int. (2014) 2014:545393. 10.1155/2014/545393
73.
HarizanovRRainovaITzvetkovaNKaftandjievIBikovIMikovO. Geographical distribution and epidemiological characteristics of visceral leishmaniasis in Bulgaria, 1988 to 2012. Eurosurveillance. (2013) 18:20531. 10.2807/1560–7917.ES2013.18.29.20531
74.
LachaudLDedetJPMartyPFarautFBuffetPGangneuxJPet al. Surveillance of leishmaniases in France, 1999 to 2012. Eurosurveillance. (2013) 18:20534. 10.2807/1560–7917.ES2013.18.29.20534
75.
HerradorZGherasimAJimenezBCGranadosMSanMartín JVAparicioP. Epidemiological changes in leishmaniasis in Spain according to hospitalization-based records, 1997–2011: raising awareness towards leishmaniasis in Non-HIV patients. PLoS Negl Trop Dis. (2015) 9:e0003594. 10.1371/journal.pntd.0003594
76.
RodriguezNELimaIDDixitUGTurcotteEALockardRDBatra-SharmaHet al. Epidemiological and experimental evidence for sex-dependent differences in the outcome of Leishmania infantum infection. Am J Trop Med Hyg. (2018) 98:142–5. 10.4269/ajtmh.17–0563
77.
ReithingerRMohsenMAadilKSidiqiMErasmusPColemanPG. Anthroponotic cutaneous leishmaniasis, Kabul, Afghanistan. Emerg Infect Dis. (2003) 9:727–9. 10.3201/eid0906.030026
Summary
Keywords
Leishmania major, visceral leishmaniasis, parasite load, PCR-ELISA, susceptibility to Infection, QTL, mouse model, sex influence
Citation
Kobets T, Čepičková M, Volkova V, Sohrabi Y, Havelková H, Svobodová M, Demant P and Lipoldová M (2019) Novel Loci Controlling Parasite Load in Organs of Mice Infected With Leishmania major, Their Interactions and Sex Influence. Front. Immunol. 10:1083. doi: 10.3389/fimmu.2019.01083
Received
23 October 2018
Accepted
29 April 2019
Published
07 June 2019
Volume
10 - 2019
Edited by
Tamás Laskay, Universität zu Lübeck, Germany
Reviewed by
Saleh Ibrahim, Universität zu Lübeck, Germany; Ramona Hurdayal, University of Cape Town, South Africa
Updates
Copyright
© 2019 Kobets, Čepičková, Volkova, Sohrabi, Havelková, Svobodová, Demant and Lipoldová.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marie Lipoldová lipoldova@img.cas.cz
This article was submitted to Microbial Immunology, a section of the journal Frontiers in Immunology
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.