- Department of Veterinary Pathobiology, Center for Veterinary Health Sciences, Oklahoma State University, Stillwater, OK, United States
Infection of cattle by bovine herpesvirus 1 (BoHV-1) can culminate in upper respiratory tract disorders, conjunctivitis, or genital disorders. Infection also consistently leads to transient immune-suppression. BoHV-1 is the number one infectious agent in cattle that is associated with abortions in cattle. BoHV-1, as other α-herpesvirinae subfamily members, establishes latency in sensory neurons. Stressful stimuli, mimicked by the synthetic corticosteroid dexamethasone, consistently induce reactivation from latency in latently infected calves and rabbits. Increased corticosteroid levels due to stress have a two-pronged effect on reactivation from latency by: (1) directly stimulating viral gene expression and replication, and (2) impairing antiviral immune responses, thus enhancing virus spread and transmission. BoHV-1 encodes several proteins, bICP0, bICP27, gG, UL49.5, and VP8, which interfere with key antiviral innate immune responses in the absence of other viral genes. Furthermore, the ability of BoHV-1 to infect lymphocytes and induce apoptosis, in particular CD4+ T cells, has negative impacts on immune responses during acute infection. BoHV-1 induced immune-suppression can initiate the poly-microbial disorder known as bovine respiratory disease complex, which costs the US cattle industry more than one billion dollars annually. Furthermore, interfering with antiviral responses may promote viral spread to ovaries and the developing fetus, thus enhancing reproductive issues associated with BoHV-1 infection of cows or pregnant cows. The focus of this review is to describe the known mechanisms, direct and indirect, by which BoHV-1 interferes with antiviral immune responses during the course of infection.
BoHV-1 is an Important Viral Pathogen
Bovine herpesvirus 1 (BoHV-1) is an α-herpesvirinae subfamily member that causes significant economical losses to the cattle industry (1). Three well-defined subtypes exist, BoHV-1.1, BoHV-1.2a, and BoHV-1.2b (2b) (2). Subtype 1 virus isolates are prevalent in Europe, North America, and South America: these subtypes are frequently detected in cattle suffering from infectious bovine rhinotracheitis (IBR) and the respiratory tract of aborted fetuses. Subtype 2a strains are prevalent in Brazil and are associated with respiratory and genital tract infections, including IBR, infectious pustular vulvovaginitis (IPV), balanopostitis (IPV), and abortions (3). Subtype 2b strains, which are frequently isolated in Australia or Europe (4), are associated with respiratory disease and IPV/IPB, but not abortion (3, 5). The seroprevelance of BoHV-1 ranges from 14 to 90% depending on the age of cattle and geographical location (6, 7). Serological testing and removal of infected animals has eliminated BoHV-1 from Denmark, Switzerland, and Austria (8).
BoHV-1 is the most frequently diagnosed cause of viral abortion in North American cattle (9). Exposure of a susceptible herd to BoHV-1 can result in abortion storms ranging from 25 to 60% of cows undergoing abortion. Commercially available modified live vaccines also induce abortions in pregnant cows. Furthermore, several studies concluded that naïve heifers vaccinated with an inactivated BoHV-1 vaccine are more likely to have a normal estrous cycle and significantly higher pregnancy rates relative to heifers vaccinated with a modified live (MLV) vaccine (9–13).
The incubation period for the genital forms of BoHV-1 is 2–6 day and initial clinical signs are frequent urination and a mild vaginal infection (14). It is also common to observe swollen vulva or small papules followed by erosions and ulcers on the mucosal surface. In bulls, similar lesions occur on the penis and prepuce. If secondary bacterial infections occur, inflammation of the uterus and transient infertility with purulent vaginal discharge occurs for several weeks. BoHV-1 infection, virulent field strains or modified live vaccines, of sero-negative heifers can target the ovary and corpus luteum during estrus and early in gestation (9).
Bovine respiratory disease complex (BRDC), a poly-microbial disease initiated by stress and/or virus infection, is the most economically important disease that affects beef and dairy cattle. Annual BRDC losses in the U.S. are ~$1 billion (15–18). A gram negative bacterium, Mannheimia haemolytica (MH), exists in the upper respiratory tract of healthy ruminants (19, 20). Following stressful stimuli or co-infections with other viruses (21), this commensal relationship is disrupted and MH becomes the predominant organism that causes life threatening bronchopneumonia in many BRDC cases (22–25). BoHV-1 infection frequently causes upper respiratory tract disease (26, 27), high fever, conjunctivitis, and erodes mucosal surfaces of the upper respiratory tract. Consequently, colonization of MH occurs in the lower respiratory tract (22, 23, 25), thus enhancing interactions between the MH leukotoxin, bovine peripheral blood mononuclear cells, and neutrophils (28, 29). Co-infection of calves with BoHV-1 and MH consistently leads to pneumonia (30). Finally, a BoHV-1 protein that is required for virus entry was identified as a significant BRDC susceptibility gene in Holsteins (31) confirming BoHV-1 is an important BRDC cofactor.
The BoHV-1 Latency-Reactivation Cycle is Important for Virus Transmission
Acute Infection Leads to High Levels of Virus Shedding
Acute BoHV-1 infection of cattle is initiated on mucosal surfaces and results in high levels of programmed cell death (32, 33). Acute infection leads to high levels of virus production and secretion in ocular, oral, nasal, or genital cavities for 7–10 days after infection. BoHV-1 gene expression during productive infection is operationally divided into three distinct phases: immediate early (IE), early (E), or late (L) (32, 33). IE gene expression is stimulated by VP16, a tegument protein (34, 35). Thus, IE mRNA expression does not require de novo protein synthesis. Two IE transcription units exist: IE transcription unit 1 (IEtu1) and IEtu2. IE transcription unit 1 (IEtu1) encodes two transcriptional regulatory proteins, bICP0 and bICP4, because a single IE transcript is differentially spliced and then translated into bICP0 or bICP4 (36–38). The bICP0 protein is also translated from an E mRNA (E2.6) because a separate E promoter drives expression of the bICP0 E transcript (36–39). The bICP0 protein has similar properties as HSV-1 encoded ICP0 (40), including a RING finger that is crucial for stimulating viral promoters and productive infection (41, 42). bICP4 is likely to possess similar functions as the HSV-1 encoded ICP4. bICP4 autoregulates the IEtu1 promoter, but activates the bICP0 E promoter.
E gene expression requires de novo protein expression, including bICP0 and bICP4, which transactivate E viral promoters. In general, the E proteins encode proteins that promote DNA synthesis. Example of early viral proteins include the DNA polymerase, thymidine kinase, small and large subunits of the ribonucleotide reductase, dUTPase, and origin binding protein. In general, the E proteins are non-structural.
The L genes are divided into two classes: Gamma-1 and Gamma-2 genes. Transcription of Gamma-1 genes requires de novo protein synthesis, including bICP0 and bICP4, but does not require viral DNA replication. Transcription of Gamma-2 genes requires de novo protein synthesis, including bICP0 and bICP4, and abundant expression requires viral DNA replication. In general, L proteins encode structural proteins and their synthesis culminates in virion assembly and release.
Summary of Latency-Reactivation Cycle
Viral particles enter the peripheral nervous system via cell-cell spread. If infection is initiated within the oral, nasal, or ocular cavity, the primary site for latency is sensory neurons in trigeminal ganglia (TG). Viral gene expression (43) and infectious virus (44) are detected in TG from 2 to 6 days after infection. Lytic gene expression is then extinguished, and surviving infected neurons harbor viral genomes (establishment of latency).
Abundant expression of the viral encoded latency related (LR) gene occurs in latently infected neurons, but infectious virus is not readily detected (maintenance of latency) (32, 33, 45–48). LR-RNA overlaps the bICP0 gene (49, 50), has two open reading frames (ORF1 and ORF2), two reading frames lacking an initiating ATG, and encodes two micro-RNAs. A LR mutant virus strain with three stop codons at the N-terminus of ORF2 has reduced virus shedding from the eye, TG, or tonsils of infected calves (44, 51, 52). LR-encoded proteins are expressed late during productive infection when infected with wild-type (wt) or LR-rescued virus, but have reduced or no expression after infection with the LR mutant virus (53, 54). Wt BoHV-1, but not the LR mutant virus, reactivates from latency (44). The anti-apoptosis activity of ORF2 (41, 55–57) and the micro-RNAs, which interfere with bICP0 expression (58) regulate the latency-reactivation cycle.
The synthetic corticosteroid dexamethasone (DEX) initiates reactivation from latency in latently infected calves or rabbits 100% of the time (27, 32, 33, 44, 47, 59). Within 6 h after latently infected calves are treated with DEX, viral regulatory proteins (ICP0 and VP16) (60, 61) and lytic cycle viral RNA expression are detected in TG neurons (62, 63). Within 3 h after DEX treatment, 11 cellular genes are induced more than 10-fold in TG (64). Pentraxin 3, a regulator of innate immunity and neuro-degeneration, is stimulated more than 30-fold at 3 or 6 h after DEX treatment. Two transcription factors, promyelocytic leukemia zinc finger (PLZF) and Slug are induced more than 15-fold 3 h after DEX treatment, which can enhance productive infection. Additional DEX induced transcription factors, SPDEF (Sam-pointed domain containing Ets transcription factor), Krüppel-like transcription factor 15 (KLF15), KLF4, KLF6, and GATA6, stimulate productive infection and certain key viral promoters. The finding that four KLF family members are stimulated during DEX induced reactivation from latency is intriguing because KLF family members resemble the Sp1 transcription factor family and both family of transcription factors interact with GC rich motifs, reviewed in Bieker (65) and Kaczynski et al. (66). The BoHV-1 genome is GC rich and many viral promoters contain Sp1 consensus binding sites and other GC rich motifs suggesting specific KLF transcription factors bind to viral sequences and stimulate viral transcription during early stages of reactivation from latency.
The IEtu1 promoter that drives bICP0 and bICP4 expression is stimulated by DEX and contains two consensus GR binding sites that are bound by the activated GR (67, 68). The GR and KLF15 are frequently expressed in the same TG neuron during reactivation and cooperatively stimulate productive infection and IEtu1 promoter activity. A host cellular factor 1 (HCF-1), which forms a complex with VP16 and Oct1 to bind to the IE enhancer core via the TAATGARAT motif, is important for GR mediated activation of the IEtu1 promoter suggesting glucocorticoid induction of viral reactivation may proceed via an HCF-1-GR mechanism in the absence of the viral IE activator VP16 (69). Stress-mediated activation of key viral promoters is predicted to be a very early event during reactivation from latency; then viral transactivators activate all other viral genes and virus production occurs. Hence, stress has a two-pronged effect on reactivation from latency by directly activating viral gene expression and indirectly enhancing viral spread via immunosuppression (70–72).
Immune Response to BoHV-1 Following Acute Infection
Cattle acutely infected with BoHV-1 develop an innate immune response (73–76); however, efficient virus replication and spread occurs. For example, virus neutralizing antibodies are detected after acute infection that recognize envelope glycoproteins, including gB, gC, gD, and gH (77, 78). Cytotoxic T cell responses to viral glycoproteins occur in cattle following infection (79–81). Infection of cultured cells also induces inflammasome formation (82), consistent with inflammation in the nasal cavity and upper respiratory tract during acute infection.
Although the host immune response clears virus after acute infection, viral infection impairs immune-recognition on several levels impairs: (1) cell-mediated immunity (83–86), (2) CD8+ T cell recognition of infected cells (68, 87–89), (3) CD4+ T cell functions because BoHV-1 infect these cells and rapidly inducing apoptosis after viral entry (90, 91), and (4) interferon responses (92–95). The known viral genes that antagonize immune responses are discussed below (see Figure 1 for a schematic that summarizes how viral genes impair immune responses).
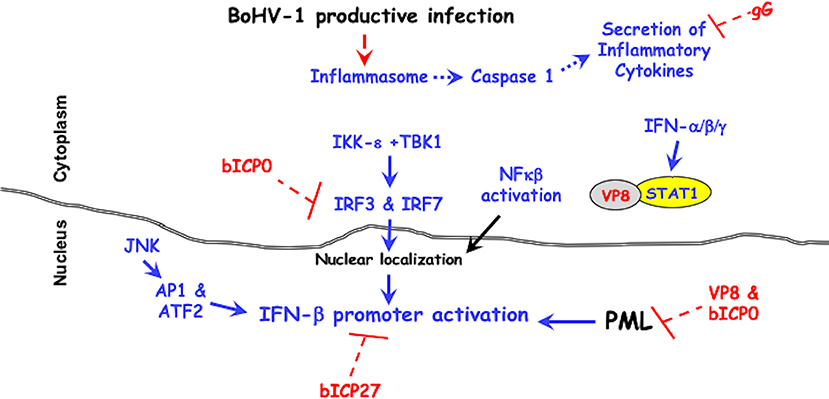
Figure 1. BoHV-1 encoded immune-evasion genes that promote productive infection. Cellular mechanisms leading to innate immune antiviral signaling pathways are denoted in blue. Red lettering denotes viral genes that counteract antiviral signaling pathways. It is well-established that two protein kinases (IKK-ε +TBK1) activate the transcription factors (IRF3 and IRF7), which are required for activating the IFN-β promoter (96–98). The JNK protein kinase (c-Jun N-terminal kinases) activates the AP1 (activating protein 1) and ATF2 (activating transcription factor 2), which are also required for activating the IFN-β promoter (96–98). For further details, see the text.
Viral Proteins Interfere With Innate Immune Responses and Immune-Surveillance
The amino-terminus of the bICP0 protein contains transcriptional activation domains, a nuclear localization signal (NLS) necessary for efficient transcriptional activation (99), and a C3HC4 zinc RING finger that is conserved in all ICP0 proteins (100, 101). Point mutations within the C3HC4 zinc RING finger domain of bICP0 interfere with transactivation of a simple viral promoter (99), stimulation of productive infection (41, 102), and reduces IFN-β promoter activity (92–95). bICP0 co-localizes with and disrupts the anti-viral promyelocytic leukemia (PML) protein-containing nuclear domains (41, 101). PML bodies are comprised of numerous proteins, which regulate the cell cycle, apoptosis, senescence, stress, DNA damage, and innate immune responses (103). Many DNA viruses reorganize or dissolve PML bodies, thus increasing viral replication. Interferon treatment increases components of PML bodies, Sp100, and PML for example (104, 105) and PML bodies increase beta-interferon (IFN-β) expression (106).
bICP0 inhibits IFN-β promoter activity in transient transfection studies (92, 94) by reducing IRF3 (interferon regulatory factor 3) protein levels. The RING finger of bICP0 (107) is an E3 ubiquitin ligase suggesting it mediates IRF3 degradation in a proteasome dependent manner. bICP0 also interacts with IRF7 and impairs activation of IFN-β promoter activity, but does not reduce IRF7 protein levels (94). IRF3 and IRF7 are transcription factors that stimulate IFN-β promoter activity (96–98). IRF3 directly binds several consensus DNA binding sites, including an ISRE (IFN response elements), and can activate IFN-stimulated promoters in the absence of IFN (108, 109). A recent study concluded PML regulates intrinsic and innate immune responses to HSV-1 infection, which is ablated by ICP0 (110). The ability of bICP0 to reduce IFN-β promoter activity correlates with IRF3 degradation, IRF7 interactions, and dissolving PML bodies.
The BoHV-1 bICP27 protein is expressed from an early promoter and based on similarity with the HSV-1 ICP27 is expected to shuttle RNA from the nucleus to the cytoplasm and regulate transcription (111). HSV-1 encoded ICP27 regulates IFN expression (112) by interfering with activation of the stimulator of interferon genes (STING) by tank binding protein kinase 1 (TBK1) (113). Interestingly, bICP27 reduces bovine IFN-β1 and IFN-β3 promoter activity in transfected cells (114). Bos Taurus encodes three functional IFN-β genes; all have anti-viral activity but each gene contains a unique promoter (115, 116).
Glycoprotein G (gG) promotes cell to cell spread (117) and maintains adherence of infected cells (118). gG is a unique viral glycoprotein because it can exist in three isoforms: a full-length membrane-bound form, a smaller membrane-bound form, and a secreted form. gG interferes with chemokine binding to their specific receptors and glycosaminoglycans (119). Although it is not known what role gG plays during acute infection of calves, the ability of chemokines to control the migratory patterns and positioning of immune cells (120) would likely be altered by gG.
The BoHV-1 UL49.5 ORF, also known as glycoprotein N (gN), is a 96 amino acid protein (121). The BoHV-1 and pseudorabies virus UL49.5 proteins interfere with processing of the transporter-associated antigen processing (TAP)-mediated transport of cytosolic peptides into the endoplasmic reticulum because UL49.5 renders the TAP complex susceptible to proteolytic degradation (122, 123). Peptide transport by TAP is crucial for MHC class I antigen presentation and recognition of infected cells by CD8+ T cells (122, 124–126). Infection of calves with a UL49.5 BoHV-1 mutant leads to increased levels of virus neutralizing antibody and cellular immune responses when compared to the parental wild-type virus (127).
VP8, the most abundant tegument protein in the virion, enhances growth in cultured cells and is required for pathogenesis in calves (128). VP8 interacts with DDB1 (DNA damaging-binding protein 1) that is associated with a E3 ubiquitin ligase complex (129), and remodels PML nuclear bodies (130). Recent studies demonstrated VP8 interacts with STAT1 (Signal transducer and activator of transcription 1) and prevents STAT1 from entering the nucleus (131). Stat1 is bound to the IFN-γ receptor and upon IFN-γ binding to its receptor (Jak1 and Jak2) phosphorylates specific tyrosine residues on STAT1. STAT1 subsequently enters the nucleus and stimulates GAS (IFN-γ activated sequences) setting off a second wave of IFN-γ (132). Following IFN-α or IFN-β stimulation, STAT1 forms a heterodimer with STAT2 and this heterodimer binds an ISRE element and activates transcription (133). VP8 also interferes with IFN-β signaling activity by reducing an interferon sensitive response element (ISRE) responsive promoter in transfected or infected cells. Thus, VP8 is a potent IFN antagonist that can interfere with host innate immune responses in the absence of de novo viral protein synthesis.
Conclusions/Discussion
BoHV-1 is a very successful pathogen because it encodes several genes that impair intrinsic and innate immune responses throughout productive infection (see Figure 1). VP8 is likely the initial anti-viral protein that impairs antiviral IFN responses because high levels of VP8 are present in the tegument of incoming viral particles. bICP0, which is encoded by the IEtu1 promoter, would be an early interferon antagonist. bICP27 via unknown mechanisms interferes with IFN-β promoter activation. Three late proteins (gG, UL49.5, and VP8) would further antagonize immune-recognition. In summary, the presence of viral proteins in the virion and expression of viral proteins throughout productive infection allows for high levels of virus production during acute infection and reactivation from latency in cattle.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This research was supported by grants from the USDA-NIFA Competitive Grants Program (2018-06668 and 2016-09370), National Institute of Neurological Disorders and Stroke of the NIH (R21NS102290), support from the Oklahoma Center for Respiratory and Infectious Diseases (National Institutes of Health Centers for Biomedical Research Excellence Grant # P20GM103648), and funds derived from the Sitlington Endowment.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Turin L, Russo S, Poli G. BHV-1: new molecular approaches to control a common and widespread infection. Molec Med. (1999) 5:261–84. doi: 10.1007/BF03402063
2. Metzler AE, Matile H, Gasman U, Engels M, Wyler R. European isolates of bovine herpesvirus 1: a comparison of restriction endonuclease sites, polypeptides and reactivity with monoclonal antibodies. Arch Virol. (1985) 85:57–69. doi: 10.1007/BF01317006
3. Oirschot JT. Bovine herpesvirus in semen of bulls and the risk of transmission: a brief overview. Vet Quart. (1995) 17:29–33. doi: 10.1080/01652176.1995.9694526
4. Edwards S, White H, Nixon P. A study of the predominant genotypes of bovine herpesvirus 1 isolated in the U.K. Vet Microbiol. (1990) 22:213–23. doi: 10.1016/0378-1135(90)90108-8
5. D'Arce RCF, Almedia RS, Silva TC, Spilki AC, Roehe PM, Ams CW. Restriction endonucleases and monoclonal antibody analysis of Brazilian isolates of bovine herpesvirus 1 and 5. Vet Microbiol. (2002) 88:315–34. doi: 10.1016/S0378-1135(02)00126-8
6. Straub OC. Infectious bovine rhinotracheitis virus. In: Dinter, Z and Morin, B editors. Virus Infections of Ruminants. Amsterdam: Elsevier. (1990). p. 71–109. doi: 10.1016/B978-0-444-87312-5.50020-5
7. Wuyckhuise L, Van Bosch J, Franken P, Hage J, Verhoeff J, Zimmer G. The prevalence of infectious bovine rhinotracheitis (IBR) in the Netherlands. Paper presented at: 18th World Buiatrics Congress, Bologna. (1994).
8. Ackermann M, Engals M. Pro and contra-IBR-eradication. Vet Microbiol. (2005) 113:293–302. doi: 10.1016/j.vetmic.2005.11.043
9. Chase C, Fulton RW, O'Toole D, Gillette B, Daly RF, Perry G, et al. Bovine herpesvirus 1 modified live vaccines for cattle reproduction: balancing protection with undesired effects. Vet Microbiol. (2017) 206:69–77. doi: 10.1016/j.vetmic.2017.03.016
10. Miller JM, Van der Matten MJ. Early embryonic death in heifers after inoculation with bovine herpesvirus-1 and reactivation of latency virus in reproductive tissues. Am J Vet Res. (1987) 48:1555–8.
11. O'Toole D, Miller MM, Cavender JL, Cornish TE. Pathology in practice. Vet Med Today. (2012) 241:189–91. doi: 10.2460/javma.241.2.189
12. O'Toole D, Corbett R. Letter to the editor, regarding Bovine herpesvirus 1 abortion and vaccination. J Vet Diagn Invest. (2013) 25:555. doi: 10.1177/1040638713496050
13. Perry G, Zimmerman AD, Daly RF, Butterbaugh RE, Rhoades J, Schultz D, et al. The effects of vaccination on serum hormone concentrations and conception rates in synchronized naive beef heifers. Theriongenology. (2013) 79:200–5. doi: 10.1016/j.theriogenology.2012.10.005
14. Yates WDG. A review of infectious bovine rhinotracheitis, shipping fever pneumonia, and viral-bacterial synergism in respiratory diesease of cattle. Can J Comp Med. (1982) 46:225–63.
15. National Agricultural Statistics Service (NASS). Agricultural Statistics Board. Washington, DC: U.S. Department of Agriculture (1996).
16. Edwards AJ. Respiratory diseases of feedlot cattle in teh central USA. Bovine Pract. (1996) 30:5–7.
17. Griffin D. Economic impact associated with respiratory disease in beef cattle. Vet Clin North Am Food Anim Pract. (1997) 13:367–77. doi: 10.1016/S0749-0720(15)30302-9
18. Kapil S, Basaraba RJ. Infectious bovine rhinotracheitis, parainfluenza-3, and respiratory coronavirus. Vet Clin North Am Food Anim Pract. (1997) 13:455–61. doi: 10.1016/S0749-0720(15)30308-X
19. Frank, GH. (ed.) Bacteria as Etiologic Agents in Bovine Respiratory Disease. College Station, TX: Texas A&M University Press (1984).
20. Songer, JG and Pos, KW. (ed.) The Genera Mannheimia and Pasteurella. St. Louis, MO: Elsevier Saunders (2005).
21. Rice JA, Carrasco-Medina L, Hodgins DC, Shewen PE. Mannheimia haemolytica and bovine respiratory disease. Anim Health Res Rev. (2008) 8:117–28. doi: 10.1017/S1466252307001375
22. Highlander SK. Molecular genetic analysis of virulence in Mannheimia (Pasteurella) haemolytica. Front Biosci. (2001) D1128–50. doi: 10.2741/A574
23. Highlander SK, Fedorova ND, Dusek DM, Panciera R, Alvarez LE, Renehart C. Inactivation of pasteurella (Mannheimia) haemolytica leukotoxin causes partial attenuation of virulence in a calf challenge model. Infect Immun. (2000) 68:3916–22. doi: 10.1128/IAI.68.7.3916-3922.2000
24. Shewen, PE and Hodkins, DC. (ed.) Pneumonic Pasteurellosis of Cattle. Cape Town: Oxford University Press (2004).
25. Zecchinon L, Frett T, Desmecht D. How mannheimia haemolytica defeats host defense through a kiss of death mechanism. Vet Res. (2005) 36:133–56. doi: 10.1051/vetres:2004065
26. Hodgson PD, Aich P, Manuja A, Hokamp K, Roche FM, Brinkman FS, et al. Effect of stress on viral-bacterial synergy in bovine respiratoryt disease: novel mechanisms to regulate inflammation. Comp Funct Genomics. (2005) 6:244–50. doi: 10.1002/cfg.474
27. Jones C, Chowdhury S. Bovine herpesvirus type 1 (BHV-1) is an important cofactor in the bovine respiratory disease complex. In: Cooper, VL and Broderson, B editors. Veterinary Clinics of North America, Food Animal Practice, Bovine Respiratory Disease. New York, NY: Elsevier, (2010). p. 303–21. doi: 10.1016/j.cvfa.2010.04.007
28. Leite F, Kuckleburg C, Atapattu D, Schulz R, Czuprynski CJ. BHV-1 infection and inflammatory cytokines amplify the interaction between mannheimia haemolytica lukotoxin with bovine peripheral blood mononuclear cells in vitro. Vet Immunol Immunopathol. (2004) 99:193–202. doi: 10.1016/j.vetimm.2004.02.004
29. Rivera-Rivas JJ, Kisiela D, Czuprynski CJ. Bovine herpesvirus type 1 infection of bovine bronchial epithelial cells increases neutrophil adhesion and activation. Vet Immunol Immunopathol. (2009) 131:167–76. doi: 10.1016/j.vetimm.2009.04.002
30. Yates WD, Babiuk LA, Jericho K W. Viral-bacterial pneumonia in calves: duration of the interaction between bovine herpesvirus 1 and pasteurella haemolytica. Can J Comp Med. (1983) 47:257–64.
31. Neibergs HL, Seabury CM, Wojtowicz AJ, Wang Z, Scraggs E, Kiser JN, et al. Susceptibility loci revealed for bovine respiratory disease complex in pre-weaned holstein calves. BMC Genomics. (2014) 15:1–19. doi: 10.1186/1471-2164-15-1164
32. Jones C. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv Virus Res. (1998) 51:81–133. doi: 10.1016/S0065-3527(08)60784-8
33. Jones C. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin Micro Rev. (2003) 16:79–95. doi: 10.1128/CMR.16.1.79-95.2003
34. Misra V, Bratanich AC, Carpenter D, O'Hare P. Protein and DNA elements involved in transactivation of the promoter of the bovine herpesvirus (BHV) 1 IE-1 transcription unit by the BHV alpha gene trans-inducing factor. J Virol. (1994) 68:4898–909.
35. Misra V, Walker S, Hayes S, O'Hare P. The bovine herpesvirus alpha gene trans-inducing factor activates transcription by mechanisms different from those of its herpes simplex virus type 1 counterpart VP16. J Virol. (1995) 69:5209–16.
36. Wirth UV, Fraefel C, Vogt B, Vlcek C, Paces V, Schwyzer M. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3' coterminal and encode a putative zinc finger transactivator protein. J Virol. (1992) 66:2763–72.
37. Wirth UV, Gunkel K, Engels M, Schwyzer M. Spatial and temporal distribution of bovine herpesvirus 1 transcripts. J Virol. (1989) 63:4882–9.
38. Wirth UV, Vogt B, Schwyzer M. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J Virol. (1991) 65:195–205.
39. Fraefel C, Zeng J, Choffat Y, Engels M, Schwyzer M, Ackermann M. Identification and zinc dependence of the bovine herpesvirus 1 transactivator protein BICP0. J Virol. (1994) 68:3154–62.
40. Boutell C, Everett RD. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol. (2013) 94:465–81. doi: 10.1099/vir.0.048900-0
41. Inman M, Zhang Y, Geiser V, Jones C. The zinc ring finger in the bICP0 protein encoded by bovine herpes virus-1 mediates toxicity and activates productive infection. J Gen Virol. (2001b) 82:483–92. doi: 10.1099/0022-1317-82-3-483
42. Saira S, Chowdhury S, Gaudreault N, Henderson G, Doster A, Jones C. The zinc RING finger of the bovine herpesvirus 1 encoded bICP0 protein is crucial for viral replication and virulence. J Virol. (2008) 82:12060–8. doi: 10.1128/JVI.01348-08
43. Schang L, Jones C. Analysis of bovine herpesvirus 1 transcripts during a primary infection of trigeminal ganglia of cattle. J Virol. (1997) 71:6786–95.
44. Inman M, Lovato L, Doster A, Jones C. A mutation in the latency related gene of bovine herpesvirus 1 interferes with the latency-reactivation cycle of latency in calves. J Virol. (2002) 76:6771–9. doi: 10.1128/JVI.76.13.6771-6779.2002
45. Jones C, Geiser V, Henderson G, Jiang Y, Meyer F, Perez S, et al. Functional analysis of bovine herpesvirus 1 (BHV-1) genes expressed during latency. Vet Micro. (2006) 113:199–210. doi: 10.1016/j.vetmic.2005.11.009
46. Kutish G, Mainprize T, Rock D. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J Virol. (1990) 64:5730–7.
47. Rock D, Lokensgard J, Lewis T, Kutish G. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J Virol. (1992) 66:2484–90.
48. Rock DL, Beam SL, Mayfield JE. Mapping bovine herpesvirus type 1 latency-related RNA in trigeminal ganglia of latently infected rabbits. J Virol. (1987) 61:3827–31.
49. Bratanich AC, Hanson ND, Jones C. The latency-related gene of bovine herpesvirus 1 inhibits the activity of immediate-early transcription unit 1. Virology. (1992) 191:988–91. doi: 10.1016/0042-6822(92)90278-W
50. Hossain A, Schang LM, Jones C. Identification of gene products encoded by the latency-related gene of bovine herpesvirus 1. J Virol. (1995) 69:5345–52.
51. Inman M, Lovato L, Doster A, Jones C. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J Virol. (2001a) 75:8507–15. doi: 10.1128/JVI.75.18.8507-8515.2001
52. Perez S, Inman M, Doster A, Jones C. Latency-related gene encoded by bovine herpesvirus 1 promotes virus growth and reactivation from latency in tonsils of infected calves. J Clin Micro. (2005) 43:393–401. doi: 10.1128/JCM.43.1.393-401.2005
53. Jiang Y, Inman M, Zhang Y, Posadas NA, Jones C. A mutation in the latency related gene of bovine herpesvirus 1 (BHV-1) inhibits protein expression of a protein from open reading frame 2 (ORF-2) and an adjacent reading frame during productive infection. J Virol. (2004) 78:3184–9. doi: 10.1128/JVI.78.6.3184-3189.2004
54. Meyer F, Perez S, Jiang Y, Zhou Y, Henderson G, Jones C. Identification of a novel protein encoded by the latency-related gene of bovine herpesvirus 1. J Neurovirology. (2007) 13:569–78. doi: 10.1080/13550280701620754
55. Ciacci-Zanella J, Stone M, Henderson G, Jones C. The latency-related gene of bovine herpesvirus 1 inhibits programmed cell death. J Virol. (1999) 73:9734–40.
56. Henderson G, Perng C-G, Nesburn A, Wechsler S, Jones C. The latency related gene of bovine herpesvirus 1 can suppress caspase 3 and caspase 9 during productive infection. J Neurovirol. (2004) 10:64–70. doi: 10.1080/13550280490261716
57. Shen W, Jones C. Open reading frame 2 encoded by the latency related gene of bovine herpesvirus 1 has anti-apoptosis activity in transiently transfected neuroblastoma cells. J Virol. (2008) 82:10940–5. doi: 10.1128/JVI.01289-08
58. Jaber T, Workman A, Jones C. Small noncoding RNAs encoded within the bovine herpesvirus 1 latency-related gene can reduce steady-state levels of infected cell protein 0 (bICP0). J Virol. (2010) 84:6297–307. doi: 10.1128/JVI.02639-09
59. Jones C, Newby TJ, Holt T, Doster A, Stone M, Ciacci-Zanella J, et al. Analysis of latency in cattle after inoculation with a temperature sensitive mutant of bovine herpesvirus 1 (RLB106). Vaccine. (2000) 18:3185–95. doi: 10.1016/S0264-410X(00)00106-7
60. Frizzo da Silva Kook LI, Doster A, Jones C. Bovine herpesvirus 1 regulatory proteins, bICP0 and VP16, are readily detected in trigeminal ganglionic neurons expressing the glucocorticoid receptor during the early stages of reactivation from latency. J Virol. (2013) 87:11214–22. doi: 10.1128/JVI.01737-13
61. Kook I, Doster A, Jones C. Bovine herpesvirus 1 regulatory proteins are detected in trigeminal ganglionic neurons during the early stages of stress-induced escape from latency. J Neurovirol. (2015a) 21:585–91. doi: 10.1007/s13365-015-0339-x
62. Winkler MT, Doster A, Jones C. Persistence and reactivation of bovine herpesvirus 1 in the tonsil of latently infected calves. J Virol. (2000) 74:5337–46. doi: 10.1128/JVI.74.11.5337-5346.2000
63. Winkler MT, Doster A, Sur JH, Jones C. Analysis of bovine trigeminal ganglia following infection with bovine herpesvirus 1. Vet Microbiol. (2002) 86:139–55. doi: 10.1016/S0378-1135(01)00498-9
64. Workman A, Eudy J, Smith L, Frizzo da Silva L, Sinani D, Bricker H, et al. Cellular transcription factors induced in trigeminal ganglia during dexamethasone-induced reactivation from latency stimulate bovine herpesvirus 1 productive infection and certain viral promoters. J Virol. (2012) 86:2459–73. doi: 10.1128/JVI.06143-11
65. Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. (2001) 276:34355–8. doi: 10.1074/jbc.R100043200
66. Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. (2003) 4:206.201. doi: 10.1186/gb-2003-4-2-206
67. El-Mayet FS, Sawant L, Thungunutla P, Jones C. Combinatorial effects of the glucocorticoid receptor and Krüppel-like transcription factor 15 on bovine herpesvirus 1 transcription and productive infection. J Virol. (2017) 91:e00904–17. doi: 10.1128/JVI.00904-17
68. Kook I, Henley C, Meyer F, Hoffmann F, Jones C. Bovine herpesvirus 1 productive infection and the immediate early transcription unit 1 are stimulated by the synthetic corticosteroid dexamethasone. Virology. (2015b) 484:377–85. doi: 10.1016/j.virol.2015.06.010
69. Sawant L, Kook I, Vogel JL, Kristie TM, Jones C. The cellular coactivator HCF-1 is required for glucocorticoid receptor-mediated transcription of bovine herpesvirus 1 immediate early genes. J Virol. (2018) 92:e00987–18. doi: 10.1128/JVI.00987-18
70. Barnes PJ. Anti-inflammatory actions of glucocorticoids: molecular mechanisms. Clin Sci. (1998) 94:557–72. doi: 10.1042/cs0940557
71. Funder JW. Glucocorticoids and mineralocorticoid receptors: biology and clinical relevance. Annu Rev Med. (1997) 48:231–40. doi: 10.1146/annurev.med.48.1.231
72. Schonevild OJLM, Gaemers IC, Lamers WH. Mechanisms of glucocorticoid signalling. Biochem Biophys Acta. (2004) 1680:114–28. doi: 10.1016/j.bbaexp.2004.09.004
73. Babiuk LA, Bielefeldt Ohamnn H, Gifford H, Czarniecki CW, Scialli VT, Hamilton EB. Effect of bovine alpha 1 interferon on bovine herpesvirus type 1 induced respiratory disease. J Gen Virol. (1985) 66:2383–94. doi: 10.1099/0022-1317-66-11-2383
74. Jensen J, Schulz RD. Bovine natuaral cell mediated cytotoxicity (NCMC): activation by cytokines. Vet Immunol Immunopathol. (1990) 24:113–29. doi: 10.1016/0165-2427(90)90014-J
75. Lawman MJP, Gifford G, Gyongyossy-Issa M, Dragan R, Heise J, Babiuk LA. Activity of polymorphonuclear (PMN) leukocytes diseases bovine herpesvirus-1 induced repiratory disease: Effect of recombinant bovine interferon alpha I. Antiviral Res. (1987) 8:225–38. doi: 10.1016/S0166-3542(87)80001-3
76. Straub OC, Ahl R. Lokale interferonbildung beim rind nach intranasaler infektion mit avirulentm IBR-IPV-virus und deren wirkung auf eine anschliebende inkection mit maul und klauenseuche-virus. Zbl Vet Med B. (1976) 23:470–82. doi: 10.1111/j.1439-0450.1976.tb01627.x
77. Marshall RL, Israel BA, Letchworth GJ. Monoclonal antibody analysis of bovine herpesvirus-1 glycoprotein antigenic areas relevant to natural infection. Virology. (1988) 165:338–47. doi: 10.1016/0042-6822(88)90578-8
78. Van Drunen Littel-van den Hurk S, Babiuk LA. Polypeptide specificity of the antibody reponse after primary and recurrent infection with bovine herpesvirus 1. J Clin Microbiol. (1986) 23:274–82.
79. Denis M, Slaoui M, Keil GM, Babiuk LA, Ernst E, Pastorer PP, et al. Identification of different target glycoproteins for bovine herpesvirus-1-specific cytotoxic T lymphocytes depending on the method of in vitro stimulation. Immunology. (1993) 78:7–13.
80. Huang Y, Babiuk LA, van Drunen Littel-van den Hurk S. Immunization with a bovine heprsvirus-1 glycoprotein B DNA vaccine induces cytotoxic T lymphocyte responses in mice and cattle. J Gen Virol. (2005) 88:887–98. doi: 10.1099/vir.0.80533-0
81. Van Drunen Littel-van den Hurk S. Cell-mediated immune responses induced by BHV-1: rational vaccine design. Expert Rev Vaccines. (2007) 6:369–80. doi: 10.1586/14760584.6.3.369
82. Wang J, Alexander J, Wiebe M, Jones C. Bovine herpesvirus 1 productive infection stimulates inflammasome formation and caspase 1 activity. Virus Res. (2014) 185:72–6. doi: 10.1016/j.virusres.2014.03.006
83. Carter JJ, Weinberg AD, Pollard A, Reeves R, Magnuson JA, Magnuson NS. Inhibition of T-lymphocyte mitogenic responses and effects on cell functions by bovine herpesvirus 1. J Virol. (1989) 63:1525–30.
84. Griebel P, Ohmann HB, Lawman MJ, Babiuk LA. The interaction between bovine herpesvirus type 1 and activated bovine T lymphocytes. J Gen Virol. (1990) 71:369–77. doi: 10.1099/0022-1317-71-2-369
85. Griebel P, Qualtiere L, Davis WC, Gee A, Bielefeldt Ohmann H, Lawman MJ, et al. T lymphocyte population dynamics and function following a primary bovine herpesvirus type-1 infection. Viral Immunol. (1987) 1:287–304. doi: 10.1089/vim.1987.1.287
86. Griebel PJ, Qualtiere L, Davis WC, Lawman MJ, Babiuk LA. Bovine peripheral blood leukocyte subpopulation dynamics following a primary bovine herpesvirus-1 infection. Viral Immunol. (1987) 1:267–86. doi: 10.1089/vim.1987.1.267
87. Hariharan MJ, Nataraj C, Srikumaran S. Down regulation of murine MHC class I expression by bovine herpesvirus 1. Viral Immunol. (1993) 6:273–84. doi: 10.1089/vim.1993.6.273
88. Hinkley S, Hill AB, Srikumaran S. Bovine herpesvirus-1 infection affects the peptide transport activity in bovine cells. Virus Res. (1998) 53:91–6. doi: 10.1016/S0168-1702(97)00128-7
89. Nataraj C, Eidmann S, Hariharan MJ, Sur JH, Perry GA, Srikumaran S. Bovine herpesvirus 1 downregulates the expression of bovine MHC class I molecules. Viral Immunol. (1997) 10:21–34. doi: 10.1089/vim.1997.10.21
90. Eskra L, Splitter GA. Bovine herpesvirus-1 infects activated CD4+ lymphocytes. J Gen Virol. (1997) 78:2159–66. doi: 10.1099/0022-1317-78-9-2159
91. Winkler MT, Doster A, Jones C. Bovine herpesvirus 1 can infect CD4(+) T lymphocytes and induce programmed cell death during acute infection of cattle. J Virol. (1999) 73:8657–68.
92. Henderson G, Zhang Y, Jones C. The bovine herpesvirus 1 gene encoding infected cell protein 0 (bICP0) can inhibit interferon-dependent transcription in the absence of other viral genes. J Gen Virol. (2005) 86:2697–702. doi: 10.1099/vir.0.81109-0
93. Jones C. Regulation of innate immune responses by bovine herpesvirus 1 and infected cell protein 0. Viruses. (2009) 1:255–75. doi: 10.3390/v1020255
94. Saira K, Zhou Y, Jones C. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) induces degradation of interferon response factor 3 (IRF3), and consequently inhibits beta interferon promoter activity. J Virol. (2007) 81:3077–86. doi: 10.1128/JVI.02064-06
95. Saira K, Jones C. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) associates with interferon regulatory factor 7 (IRF7), and consequently inhibits beta interferon promoter activity. J Virol. (2009) 83:3977–81. doi: 10.1128/JVI.02400-08
96. Goodbourn S, Zinn K, Maniatis T. Human beta-interferon gene expression is regulated by an inducible enhancer element. Cell. (1985) 41:509–20. doi: 10.1016/S0092-8674(85)80024-6
97. Honda K, Yanai H, Negishi H, Asagiri M, Saton M, Mizutani T, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. (2005) 434:772–7. doi: 10.1038/nature03464
98. Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-b enhancer in vivo. Mol Cell. (1998) 1:507–18. doi: 10.1016/S1097-2765(00)80051-9
99. Zhang Y, Jones C. Identification of functional domains within the bICP0 protein encoded by BHV-1. J Gen Virol. (2005) 86:879–86. doi: 10.1099/vir.0.80698-0
100. Everett RD. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays. (2000) 22:761–70. doi: 10.1002/1521-1878(200008)22:8<761::AID-BIES10>3.0.CO;2-A
101. Parkinson J, Everett RD. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol. (2000) 74:10006–17. doi: 10.1128/JVI.74.21.10006-10017.2000
102. Geiser V, Jones C. Stimulation of bovine herpesvirus 1 productive infection by the adneovirus E1A gene and a cell cycle regulatory gene, E2F-4. J Gen Virol. (2003) 84:929–38. doi: 10.1099/vir.0.18915-0
103. Scherer M, Stamminger T. Emerging role of PML nuclear bodies in innate immune signaling. J Virol. (2016) 90:5850–4. doi: 10.1128/JVI.01979-15
104. Chee AV, Pandolfi LP, Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J Virol. (2003) 77:7101–5. doi: 10.1128/JVI.77.12.7101-7105.2003
105. Regad T, Saib A, Lallemand-Breitenbach V, Pandolfi PP, de The H, Chelbi-Alix MK. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. Embo J. (2001) 20:3495–505. doi: 10.1093/emboj/20.13.3495
106. Chen Y, Wright J, Meng X, Leppard KN. Promyelocytic leukemia protein isoform II promotes transcription factor recruitment to activate interferon beta and interferon-responsive gene expression. Molec Cell Biol. (2015) 35:1660–72. doi: 10.1128/MCB.01478-14
107. Dia L, Zhang B, Fan J, Gao X, Sun S, Yang K, et al. Herpes virus proteins ICP0 and BICP0 can activate NF-kB by catalyzing IkBa ubiquitination. Cell Signal. (2005) 17:217–29. doi: 10.1016/j.cellsig.2004.07.003
108. Guo J, Peters KL, Sen GC. Induction of the human protein P56 by interferon, double stranded RNA, or virus infection. Virology. (2000) 267:209–19. doi: 10.1006/viro.1999.0135
109. Mossman KL, Macgregor PF, Rozmus JJ, Goryachev AB, Edwards AM, Smiley JR. Herpes simplex virus triggers and then disarms a host antiviral response. J Virol. (2001) 75:750–8. doi: 10.1128/JVI.75.2.750-758.2001
110. Alandijany T, Roberts APE, Con KL, Loney C, McFarlane S, Boutell C. Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PloS Pathog. (2018) 14:e1006769. doi: 10.1371/journal.ppat.1006769
111. Sandri-Goldin RM. The many roles of the highly interactive HSV protein ICP27, a key regulator of infection. Future Microbiol. (2011) 6:1261–77. doi: 10.2217/fmb.11.119
112. Johnson KE, Song B, Knipe DM. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology. (2008) 374:487–94. doi: 10.1016/j.virol.2008.01.001
113. Christensen M, Jensen SB, Miettinen JJ, Luecke S, Prabakaran T, Reinert LS, et al. HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. Embo J. (2016) 35:1385–99. doi: 10.15252/embj.201593458
114. da Silva LF, Jones C. The ICP27 protein encoded by bovine herpesvirus type 1 (bICP27) interferes with promoter activity of the bovine genes encoding beta interferon 1 (IFN-β1) and IFN-β3. Virus Res. (2012) 162–8. doi: 10.1016/j.virusres.2012.07.023
115. Valarcher J-F, Furze J, Wyld S, Cook R, Conzelman K-K, Taylor G. Role of alpha/beta interferons in the attenuation and immunogenecity of recombinant bovine respiratory syncitial viruses lacking NS proteins. J Virol. (2003) 77:8426–39. doi: 10.1128/JVI.77.15.8426-8439.2003
116. Wilson V, Jeffreys AJ, Barrie PA. A comparison of vertebrate interferon gene families deteced by hybridization with human interferon DNA. J Mol Biol. (1983) 166:457–75. doi: 10.1016/S0022-2836(83)80281-2
117. Nakamichi K, Ohara N, Kuroki D, Otsuka H. Bovine herpesvirus 1 glycoprotein G is required for viral growth by cell-to-cell infection. Virus Res. (2000) 68:175–81. doi: 10.1016/S0168-1702(00)00171-4
118. Nakanishi K, Maruyama M, Shibata T, Morishima N. Identification of a caspase-9 substrate and detection of its cleavage in programmed cell death during mouse development. J Biol Chem. (2001) 276:41237–44. doi: 10.1074/jbc.M105648200
119. Bryant NA, Davis-Poynter N, Vanderplasschen A, Alcami A. Glycoprotein G isoforms from some alphaherpesvirus function as broad-spectrum chemokine binding proteins. EMBO J. (2003) 22:833–46. doi: 10.1093/emboj/cdg092
120. Griffith J, Sokol CL, Luster AD. Chemokines and chemokine receptors: position cells for host defense and immunity. Ann Rev Immunol. (2014) 32:659–702. doi: 10.1146/annurev-immunol-032713-120145
121. Liang X, Tang M, Manns B, Babiuk LA, Zamb TJ. Identification and deletion mutagenesis of the bovine herpesvirus 1 dUTPase gene and a gene homologous to herpes simplex virus UL49.5. Virology. (1993) 195:42–50. doi: 10.1006/viro.1993.1344
122. Koppers-Lalic DE, Reits AJ, Ressing ME, Lipinska AD, Abele R, Koch J, et al. Varicelloviruses avoid T cell recognition by UL49.5-mediated inactivation of the transporter associated with antigen processing. Proc Nat Acad Sci. (2005) 102:5144–9. doi: 10.1073/pnas.0501463102
123. Lipinska AD, Koppers-Lalic D, Rychlowski M, Admiraal P, Rijsewijk FAM, Bienkowska-Szewczyk K, et al. Bovine herpesvirus 1 UL49.5 protein inhibits the transporter associated with antigen processing despite complex formation with glycoprotein M. J Virol. (2006) 81:5822–32. doi: 10.1128/JVI.02707-05
124. Ahn K, Meyer TH, Uebel S, Sempe P, Djaballah H, Yang Y, et al. Molecular mechanism and species specificity of TAP inhibition by herpes simplex virus ICP47. EMBO J. (1996) 15:3247–55. doi: 10.1002/j.1460-2075.1996.tb00689.x
125. Ambagala AP, Gopinath RS, Srikumaran S. Peptide transport activity of the transporter associated with antigen processing (TAP) is inhibited by an early protein of equine herpesvirus-1. J Gen Virol. (2004) 66:2383–94. doi: 10.1099/vir.0.19563-0
126. Hughes EA, Hammond C, Cresswell P. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteosome. Proc Nat Acad Sci. (1997) 94:1896–901. doi: 10.1073/pnas.94.5.1896
127. Wei H, He J, Paulsen DB, Chowdhury SI. Bovine herpesvirus type 1 (BHV-1) mutant lacking UL49.5 luminal domain residues 30-32 and cytoplasmic tail residues 80-96 induces more rapid onset of virus neutralizing antibody and cellular immune responses in calves than the wild-type strain Cooper. Vet Immunol Immunopathol. (2012) 147:223–9. doi: 10.1016/j.vetimm.2012.04.015
128. Lobanov VA, Maher-Sturgess SL, Snider MG, Lawman Z, Babiuk LA, van Drunen Littel-van den Hurk S. A UL47 gene deletion mutant of bovine herpesvirus type 1 exhibits impaired growth in cell culture and lack of virulence in cattle. J Virol. (2010) 84:445–58. doi: 10.1128/JVI.01544-09
129. Fruh M, Fruh K. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci STKE. (2006) 335:pe21. doi: 10.1126/stke.3352006pe21
130. Zhang K, Afroz S, Brownlie R, Snider M, van Drunen Littel-van den Hurk S. Regulation and function of phosphorylation on VP8, the major tegument protein of bovine herpesvirus 1. J Virol. (2015) 89:4598–611. doi: 10.1128/JVI.03180-14
131. Afroz S, Brownlie R, Fodje M, van Drunen Littel-van den Hurk S. VP8, the major tegument protein of bovine herpesvirus 1, interacts with cellular STAT1 and inhibits interferon beta signaling. J Virol. (2016) 90:4889–904. doi: 10.1128/JVI.00017-16
132. Sikorski K, Chmielewski S, Olejnik A, Wesoly JZ, Heemann U, Baumann M, et al. STAT1 as a central mediator of IFN-gamma and TLR4 signal integration in vascular dysfunction. Landes Biosci. (2012) 1:241–9. doi: 10.4161/jkst.22469
Keywords: bovine herpesvirus 1 (BoHV-1), immune evasion, VP8, infected cells protein 0 (bICP0), abortion, bovine respiratory disease complex
Citation: Jones C (2019) Bovine Herpesvirus 1 Counteracts Immune Responses and Immune-Surveillance to Enhance Pathogenesis and Virus Transmission. Front. Immunol. 10:1008. doi: 10.3389/fimmu.2019.01008
Received: 26 October 2018; Accepted: 18 April 2019;
Published: 07 May 2019.
Edited by:
Susmit Suvas, School of Medicine, Wayne State University, United StatesReviewed by:
Richard D. Dix, Georgia State University, United StatesJames Craig Forrest, University of Arkansas for Medical Sciences, United States
Homayon Ghiasi, Cedars-Sinai Medical Center, United States
Copyright © 2019 Jones. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clinton Jones, Y2xpbnQuam9uZXMxMEBva3N0YXRlLmVkdQ==
 Clinton Jones
Clinton Jones