- 1Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, WA, United States
- 2School of Life Sciences, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
- 3Precision Medicine Unit, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland
- 4U. S. Military HIV Research Program, Walter Reed Army Institute of Research, Silver Spring, MD, United States
- 5Henry M. Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD, United States
Fc receptor (FcR) genes collectively have copy number and allelic polymorphisms that have been implicated in multiple inflammatory and autoimmune diseases. This variation might also be involved in etiology of infectious diseases. The protective role of Fc-mediated antibody-function in HIV-1 immunity has led to the investigation of specific polymorphisms in FcR genes on acquisition, disease progression, and vaccine efficacy in natural history cohorts. The purpose of this review is not only to explore these known HIV-1 host genetic associations, but also to re-evaluate them in the context of genome-wide data. In the current era of effective anti-retroviral therapy, the potential impact of such variation on post-treatment cohorts cannot go unheeded and is discussed here in the light of current findings. Specific polymorphisms associating with HIV-1 pathogenesis have previously been genotyped by assays that captured only the single-nucleotide polymorphism (SNP) of interest without relative information of neighboring variants. With recent technological advances, variation within these genes can now be characterized using next-generation sequencing, allowing precise annotation of the whole chromosomal region. We herein also discuss updates in the annotation of common FcR variants that have been previously associated with HIV-1 pathogenesis.
Introduction
Fc receptors comprise a class of cell surface receptors expressed on various hematopoietic cells that bind to the Fc portion of antibodies to form immune complexes and recruit the complement and/or effector system to defend the body against pathogens. The Fc receptors are classified based on their binding to the Fc domain of immunoglobulin (Ig). The most abundant Ig in serum is IgG which can bind to different classes of FcγR. Other types of FcR including FcεR, Fcα/μR, and FcαR1 are receptors for other Ig classes such as IgE, IgM, and IgA.
More recently vaccine studies in infectious diseases point to a critical role of non-neutralizing antibody functions, which is the ability of an antibody to interact with other immune components and effector cells via their Fc portions to mediate killing or control of the pathogen. These mechanisms include, but are not limited to, antibody dependent cellular cytotoxicity (ADCC), antibody-dependent cell-mediated virus inhibition (ADCVI), antibody dependent cellular phagocytosis (ADCP), and antibody dependent complement deposition (ADCD) (1, 2). These functions are mediated by three distinct classes of FcγRs that are expressed on most human immune cells, with varying levels of expression dependent on cell type such as monocytes, macrophages, natural killer cells, eosinophils, neutrophils, B cells but not on T cells (3). These receptors include FcγRI (CD64), FcγRIIa/b/c (CD32), and FcγRIIIa/b (CD16), which bind the different IgG subclasses with varying proficiency, and can cause either activation or inhibition of the effector cell.
Polymorphisms in the FcγR have been shown to affect binding affinity to the Fc region of IgG and can trigger a range of effector and immunoregulatory functions. Such variation has been shown to play a crucial role in the pathogenesis of a range of chronic inflammatory and autoimmune diseases, as well as susceptibility to infectious pathogens (4, 5). An overview of FcγR biology has been recently summarized and so our focus for this review will be to evaluate the effect of genetic variation in the human Fcγ receptors and their role specifically in HIV-1 disease pathogenesis (6). Although there is evidence that the neonatal Fc receptor (FcRn), an MHC class I-related molecule expressed on many cells, functions in HIV-1 vaccination and infection (7, 8), no significant genetic variation has been identified for this locus, and we have not included it in this review.
Fcγ Receptor Genetic Diversity
The Fcγ receptors are encoded by the FCGR genes located on chromosome 1 in humans, including five FCGRs in a tandem arrangement within ~200 kb of genomic sequence (Figure 1A). A sixth gene, FCGR1A, is located ~12 Mb distant from the five gene cluster. Genetic variation at the FCGR gene cluster bears similarity to the Killer Ig-like receptor (KIR) region which is shown in comparison to emphasize both the types and extent of copy number and allelic variation (Figures 1A,B) (9). Like KIR, the genes in the FCGR cluster are arranged in haplotypes containing both invariant framework and copy number variant genes. An examination of total nucleotide variation in FCGR from a recent genome build indicates extensive depths of SNP variation, similar in overall extent to KIR and the vast majority of which has not been functionally characterized (Figures 1B,C). Both gene families encode receptors for other central components of the immune response (KIR and MHC class I; FCGR and IgG constant domains) placing them in distinct roles but perhaps of equivalent importance in investigations of host genetics and its relationship to immune function. Given the parallels in significance and the similar physical characteristics of both copy number variation and allelic polymorphism, a major difference is that the allelic variations for FCGR genes have been less examined, curated and annotated. This review in part is attempting to address this deficit as an organizing framework of characterized variation possibly guided by established methods for structural and allelic annotation as currently employed for the KIR system (10).
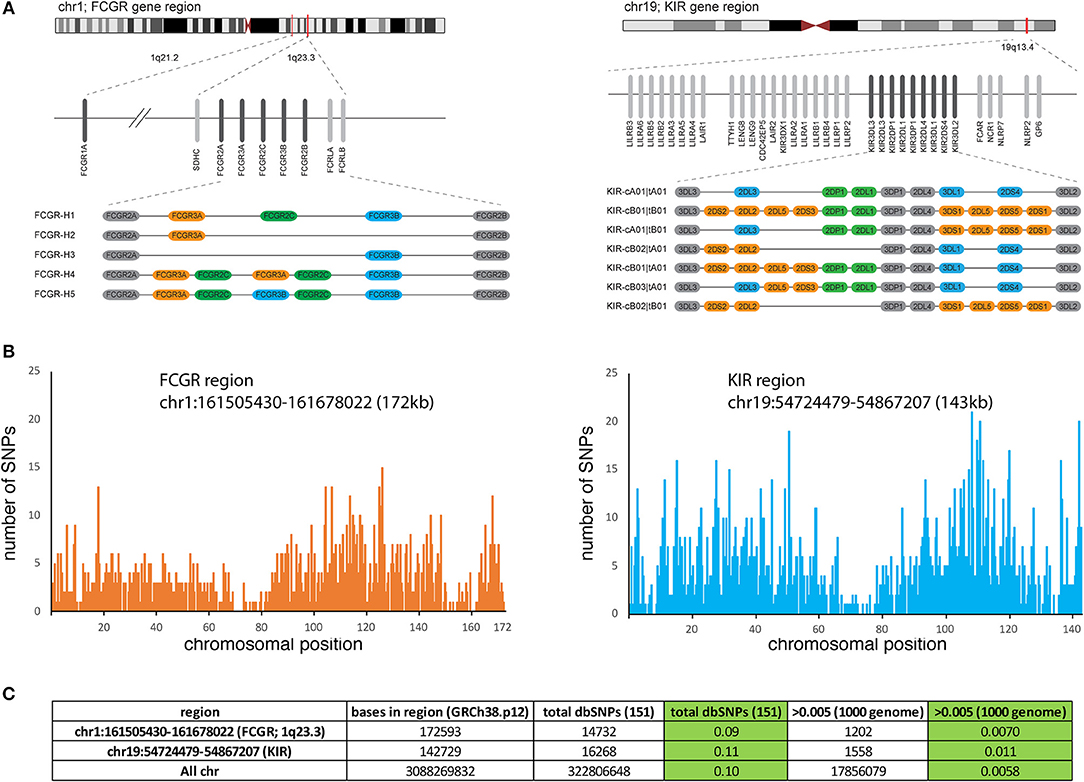
Figure 1. Structure and polymorphism of the FCGR genes. (A) Approximate chromosomal locations of the FCGR gene complex containing 5 FCGR genes and the FCGR1A gene located at ~12 Mb centromeric to the FCGR cluster. In comparison, the KIR gene family is illustrated immediately to the right including adjacent genes (FCAR) and gene clusters. For both FCGR and KIR, alternative haplotypes identified in populations are illustrated with the colored bars depicting genes present in a subset of haplotypes and shaded bars depicting genes present in all haplotypes (framework genes). Figure depicting KIR has been reproduced with permission from the Oxford University Press (9). (B) The plots show the abundance of SNPs at each position of the indicated regions from chr1 and chr19, using data derived from the UCSC genome browser. (C) Approximate number of SNPs in both regions are listed including a summary from all chromosomes for comparison.
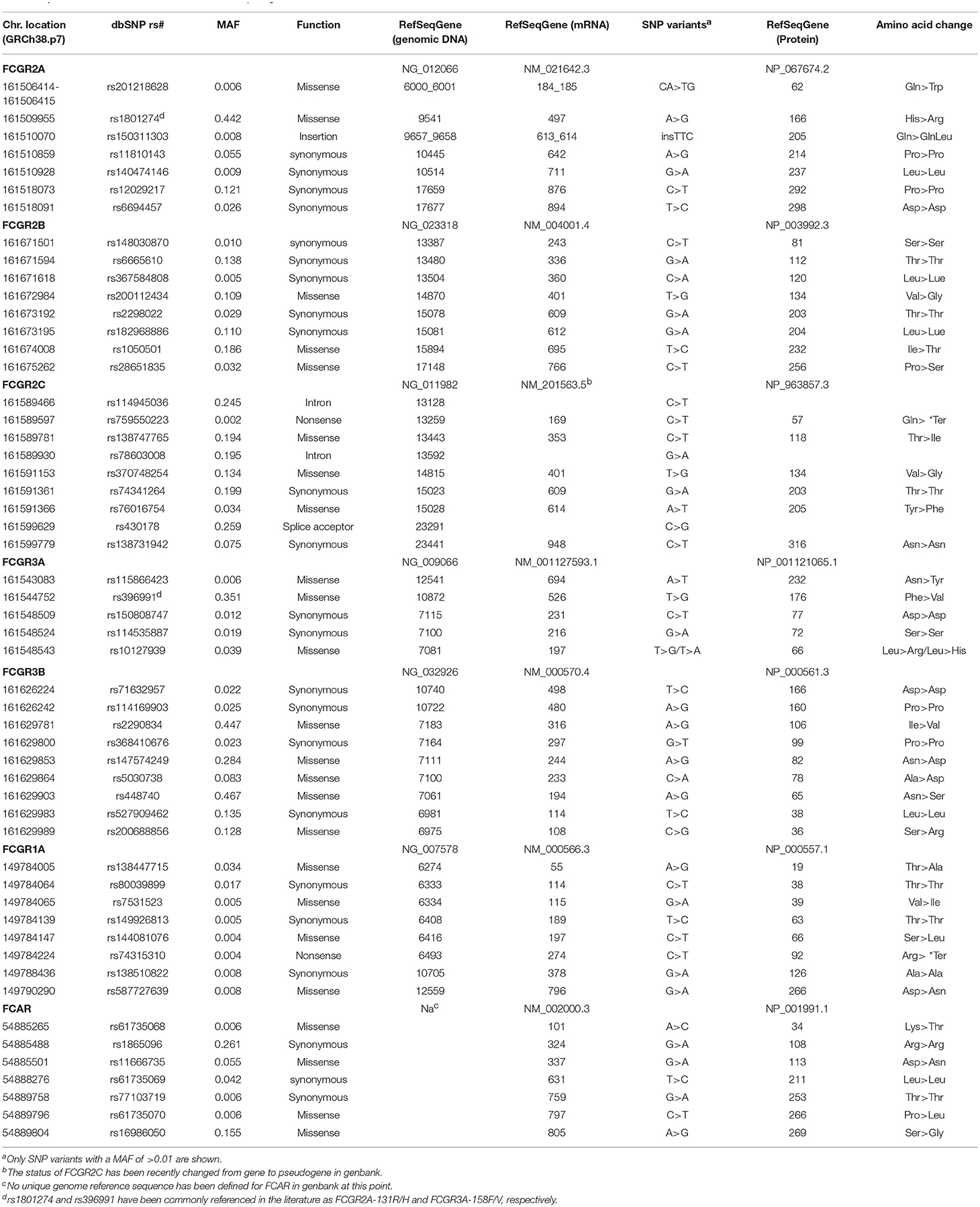
In the FCGR family of six genes, several nonsynonymous single nucleotide polymorphisms (SNP), SNPs encoding altered splice sites, and copy number variants (CNVs) encoding addition, or deletion of one or more gene have been functionally characterized (Figure 1, Table 1). Variation in the FCGR1 gene is limited, with the most frequent minor allele characterized at < 4%, but there is considerable diversity in the other FCGR genes. The most studied SNPs in the FCGR genes over the past two decades have centered on nonsynonymous substitutions that contribute to differential binding affinity for subclasses of IgG. FCGR2A has two allele variants encoding arginine or histidine at amino acid position 166 (rs1801274), with the latter resulting in a higher affinity for IgG1, and IgG2 (11–13). FCGR3A also has two common allele variants differing by a single SNP, altering codon 176 from phenylalanine to valine (rs396991) resulting in a higher affinity for IgG1, IgG2, IgG3, and IgG4 for the 176V variant (11, 12, 14). This stronger binding affinity is associated with functional capacity of the receptor in different experimental and clinical contexts (15–17). Two adjacent nonsynonymous SNPs in FCGR2A (together altering codon 63 from Q to W; rs201218628) have been studied, although their functional consequence is less clear, and their frequency is rare (18). FCGR2B has an isoleucine to threonine change at position 232 (rs1050501), which alters the transmembrane region, with the 232T allele inhibiting the association of FCGR2B with lipid rafts in a human B cell line measuring downstream function (19). The frequency of this alteration is low at ~1% in Caucasians, and while more prevalent among African Americans and Asians (5–11%) has been less studied.
A major studied polymorphism in the FCGR2C gene is a SNP in exon 3 (rs759550223) that encodes a glutamine or a stop codon resulting in the presence or absence of protein expression (20–22). The frequency of the minor allele varies between populations, and studies have suggested it is expressed on NK cells and is capable of inducing ADCC after receptor cross-linking on purified NK cells as measured by their ability to lyse the target P815 cell line (20, 23). In addition, both alleles have been associated with both null and surface expression on NK cells as measured by anti-FCGR2B/C specific mAb 2B6 (22). However, there may be some confusion regarding this SNP as it is identical to rs10917661, which is assigned to FCGR2B, in a reference SNP identification (rs id) segment where the two genes have identical sequences except at the variant position. Sequence identity between FCGR2B and FCGR2C may lead to incorrect assignment of SNPs to these two loci. Also, rs759550223 has a very low minor allele frequency defined in the SNP database (dbSNP), and the minor allele assigned is identical to the FCGR2B-derived sequence, suggesting the possibility that the SNP has been falsely generated by a combination of variants between two distinct loci. FCGR2C has been previously reported to have arisen from an unequal recombination between the FCGR2A and FCGR2B genes, and encoded a functional molecule that exhibited differential expression in natural killer cells (21, 24, 25). However, FCGR2C is also classified as a gene/pseudogene in the NCBI gene database. These inconsistencies further emphasize the need of validation of the FCGR genes by a combination of methods such as next generation sequencing (NGS) technologies as discussed below, in addition to precise curating of allelic variation and flow cytometry phenotyping.
A triallelic nonsynonymous SNP at codon 66 (66R, 66L, 66H; rs10127939) of the FCGR3A gene has also been found to affect affinity for immune complexes (ICs), with the FCGR3A-66R and 66H alleles exhibiting higher affinity (26). Other nonsynonymous SNPs have been identified but none have been characterized functionally or in association analyses. FCRG3B polymorphisms were first described as the human neutrophil antigen (HNA)-1 system (27, 28). The three major HNA-1 variants have differential affinity for IgG1 and IgG3, with the higher affinity HNA-1a and lower affinity HNA-1b differing at 4 nonsynonymous codon positions (rs2290834, rs200688856, rs448740, rs147574249). Consistent with this differential affinity, phagocytosis was lower with HNA-1b through analysis of antibacterial IgG subclass antibodies and with IgG1 and IgG3 anti-Rhesus D (29, 30). A third isoform, termed HNA-1c, of unknown function is identical to the HNA-1b isoform except at the rs5030738 polymorphic site, where it encodes an asparagine rather than alanine residue (31). Other variants of the HNA-1 antigen system have also been described but to date no functional or association studies interrogating them have been reported (32, 33).
CNV is a hallmark of multicopy gene family genomic regions, including notably among them, those encoding immune response genes (34). In the FCGR region, CNVs include at least five haplotypes with varying combinations of deletions and duplications of the FCGR2C, FCGR3A, and FCGR3B genes, flanked by the invariant framework FCGR2A and FCGR2B genes (Figure 1A, left) (34–36). FCGR-H1 forms the most common among these haplotypes, containing the five loci, with FCGR-H2 being the most commonly observed CNV [equivalent to CNR1 in Nederer et al. (36)] and FCGR-H3 less prevalent (equivalent to CNR2 or CNR3). Although variants FCGR-H4 and -H5 have not been explicitly described, they form predicted reciprocal structures of the H2 and H3 deletion variants (34). Individuals with FCGR gene copy numbers that may be consistent with those structures have been described (22, 34–38). As discussed above, older genotyping methods focusing on specific regions of the genes may have misassigned SNPs, and the lack of a standard nomenclature of common FGCR coding variants may lead to misinterpretation when comparing different studies. Newer NGS technologies have allowed for updated annotation of the FCGR genes as per the current Human genome database reference hg38. SNPs with a minor allele frequency >0.01 are shown in Table 1.
FCGR Variation and HIV-1 Disease Pathogenesis
The first report of the effect of polymorphisms in the FCGR genes on HIV-1 disease progression was in two natural history HIV-1 cohorts consisting of anti-retroviral therapy (ART) naïve individuals (39). Since then, functional SNPs in the FCGR2A (rs1801274) and FCGR3A (rs396991) genes that affect binding affinity to the Fc domain of IgG have been evaluated in the context of HIV-1 acquisition, disease progression, and vaccine efficacy. Now that most HIV-1 infected individuals are on ART, there is an opportunity to evaluate disease outcomes after ART initiation. With increased high-throughput sequencing, targeted SNP genotyping is being replaced by whole gene and genome sequencing. This gives the opportunity to evaluate previous host genetic findings in the light of genome wide findings and also examine other SNPs in nearby genes. We will discuss the effects of genetic polymorphisms in the FCGR genes and their impact on HIV-1 disease progression, acquisition, post-ART and vaccine outcomes in the next sections.
FCGR Polymorphisms and HIV-1 Disease Progression
Candidate Gene Studies
Forthal et al. identified an association between the FCGR2A low binding RR (rs1801274) genotype and a faster rate of CD4+ T cell decline and progression to AIDS using samples and data from the Multicenter AIDS Cohort Study (MACS) consisting of more than 500 HIV-1 infected males of mostly Western European ancestry (40). Paradoxically, the same RR genotype was also found to associate with a decreased risk of Pneumocystis jiroveci (carinii) pneumonia, an AIDS defining illness, when compared to the HH genotype in the same cohort. At the functional level, cells from RR homozygous carriers demonstrated less efficient phagocytosis of HIV-1/IgG complexes. There was no association of the FCGR2A genotype with viral load setpoint (spVL), defined as the number of HIV-1 RNA copies/ml in a plasma sample collected 18 months after the first seropositive test. The absence of association of FCGR2A variation with spontaneous viral load control was also confirmed in an HIV-1 seroconverting cohort including 253 Kenyan women, in which the associations with disease progression and CD4+ T cell decline were not replicated (41).
No association was observed by Forthal et al. in the MACS cohort between a specific FCGR3A genotype (rs396991) and spontaneous viral control or disease progression (40). Similarly, Weis et al. did not identify any significant genetic associations of FCGR3A variation with disease progression or spVL in the Kenyan's women cohort (41). There is one report of the VV genotype of FCGR3A being overrepresented in 43 untreated controllers compared to 59 HIV positive progressors on ART (42). However, since the HIV positive progressors were on ART, analyses with measures of spVL or CD4+ T cell counts could not be performed and this finding remains inconclusive.
A more consistent finding has been reported by two independent groups showing the association between the FCGR3A FF genotype and decreased risk of Kaposi's sarcoma (KS) (39, 40). In the first study, FCGR3A genotyping was performed in two small cohorts consisting of 119 and 131 HIV-1 infected males of Western European ancestry. A significant association with protection was identified in each cohort independently and in the combined analysis. Forthal et al. replicated this finding in the MACS cohort. KS is the most frequent malignant condition associated with HIV-1 related immunosuppression, and alterations in the cytokine balance have been suggested to play a critical role in its pathogenesis. Differences in genotype have been shown to alter IgG binding that could influence cytokine levels, with the V allele having higher affinity than the F. The authors concluded that FF homozygous individuals might be at lower risk of KS because of a less vigorous proinflammatory response. The VV genotype has been associated with an increased risk of cryptococcal disease in 164 HIV-1 infected men, again in the MACS cohort (43). Of note, this observation extends beyond HIV-1 infection, because the VV genotype was previously associated with cryptococcal disease in non-HIV-infected individuals in a separate study (44).
Genome-Wide Testing
The associations with HIV-1 natural history described above were tested using a candidate gene study design in cohorts with relatively small sample size. The current availability of genome-wide genotyping and sequencing data provides an opportunity to reassess the potential involvement of FCGR variation in HIV-1 disease in larger cohorts, by applying more stringent standards for significance level and including robust population stratification (45). We therefore accessed previously published data generated from cohorts and studies that contributed to the International Collaboration for the Genomics of HIV (ICGH) (46–48) and assessed genetic associations with HIV-1 disease outcomes in the FCGR2A and FCGR3A regions.
The potential associations between FCGR2A or FCGR3A variants and spVL were evaluated using a fixed-effect inverse-variance weighted meta-analysis across cohorts, including a total of 7,266 HIV positive patients of Western European ancestry. We tested all common polymorphisms (minor allele frequency >5%) in a 50 kb window around the gene. In line with previous studies, no significant association with spVL was observed. An additional analysis was performed in a subset of ICGH, consisting of 467 long-term non-progressors (individuals with CD4+ T cell counts consistently above 500 cells/mm (3) for >10 years without treatment) and 517 rapid progressors (individuals with two or more CD4+ T cell counts below 300 cells/mm (3) within 3 years after the last seronegative test result). We were unable to replicate findings by Forthal et al. and did not observe associations with HIV-1 progression (40). These analyses included the FCGR2A rs1801274 polymorphism; however, the FCGR3A polymorphism rs396991 was not available from the ICGH meta-analysis as it is not directly genotyped on most genotyping arrays and could not be reliably imputed. Thus, in order to evaluate its association with HIV-1 spVL, we reassessed a standard genome-wide association study (GWAS) using exome sequencing data from 395 individuals of European descent in the Swiss HIV Cohort Study (SHCS), following the procedures described in McLaren et al. (48). This analysis did not show any association between rs396991 and HIV-1 spVL (p = 0.21). Additionally, the rs1801274 did not show any association with spVL (p = 0.54) in the same cohort (Figure 2). The inability to replicate previous findings could be attributed to differences in sample size, clinical definition and statistical rigor employed. Globally, these new analyses confirm the previous findings that common human genetic variants in FCGR2A or FCGR3A are not associated with spontaneous control of HIV-1 infection.
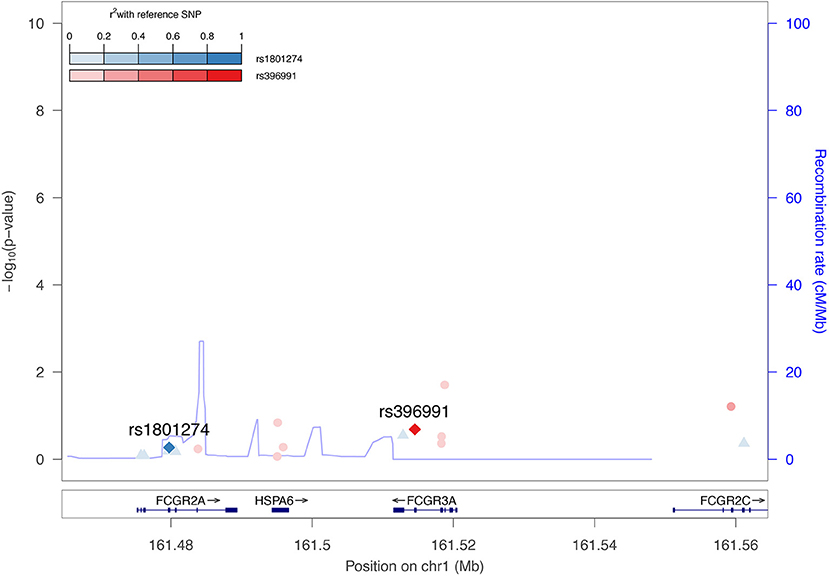
Figure 2. No association between FCGR2A or FCGR3A polymorphisms on HIV-1 set point viral load. Regional association plot highlighting the association between the FCGR2A (rs1801274) and FCGR3A (rs396991) polymorphisms and HIV-1 spVL across 395 exome sequenced patients (48). Color intensities represent the linkage disequilibrium (r2) of other SNPs in the region with rs1801287 and rs396991, respectively. The blue line indicates the estimated recombination rate in cM/Mb from The International HapMap Consortium (2007).
Influence of FCGR Diversity on HIV-1 Acquisition
There are no conclusive studies reporting associations of FCGR polymorphisms with HIV-1 acquisition. Two independent mother-to-child transmission cohorts reported contrasting findings of FCGR2A genotypes associating with increased infection risk in children with the high-affinity HH (rs1801274) genotype (49, 50).
Here again, we evaluated potential associations between FCGR2A and FCGR3A variants and susceptibility to HIV-1 infection by accessing the results of a previous GWAS of HIV-1 acquisition that compared 6,300 HIV-1 infected individuals and 7,200 controls of European ancestry (46). The rs1801274 polymorphism did not show any sign of association with HIV-1 acquisition (p = 0.81), and all other tested polymorphisms in a 50 kb window around both FCGR2A and FCGR3A were also non-significant after correction for multiple testing. The FCGR3A SNP (rs396991) was not included on the genotyping chip and could not be reliably imputed and so was not tested directly. This analysis in the largest acquisition cohort published to date adds substantial evidence to the lack of involvement of common FCGR2A and FCGR3A polymorphisms in HIV-1 acquisition.
Role of FCGR Polymorphisms on Outcomes After ART Initiation
Variation in genotype and expression of host genes is well established to impact HIV-1 susceptibility and disease progression in ART- naïve individuals (47). Initiation of ART in acute HIV-1 infection can limit establishment of viral reservoirs and induces post-treatment control in some individuals (51, 52). Host variation that influences viral reservoir size or reactivation during ART has not been definitively studied and has potential to significantly advance HIV cure research. There is at least one report indicating that broadly neutralizing antibodies (bNAbs) can interfere with establishment of a silent reservoir by Fc-FcR mediated mechanisms in humanized mice when administered early in the infection (53). Recently, Descours et al. identified CD32a (FCGR2A) as a marker of latently infected CD4 T cells (54). Given the previous associations of FCGR2A with HIV-1 disease pathogenesis, we hypothesized that polymorphisms in this gene might affect the size of the viral reservoir in patients that went on ART early in acute infection (55, 56). We examined genetic variation in the FCGR2A gene, characterizing polymorphisms in 436 ART-suppressed patients from the RV254 cohort. We screened for 18 variants in the extracellular domains of FCGR2A including rs1801274 and did not find associations with total or integrated HIV DNA (p > 0.05) (Figure 3). Surprisingly, recent reports from several independent groups confirm that they were unable to replicate the original findings from the Descours et al. study, showing that associations with post-ART control continue to be elusive (57–60).
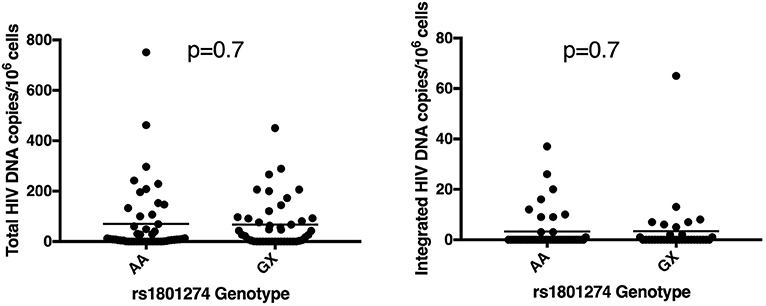
Figure 3. Polymorphism in FCGR2A (CD32a) does not associate with reservoir size. The rs1801274 SNP variant did not associate with levels of total or integrated HIV DNA, determined in N = 93 and N = 78 of the patients, respectively (55, 56).
Effect of FCGR Variation on HIV-1 Vaccine Efficacy
Variation in host genes can impact vaccine outcomes, and other than HLA, the only other gene to impact HIV-1 vaccine efficacy was in the FCGR locus (61–63). Two studies of HIV-1 vaccine efficacy revealed a remarkable coincidence of FCGR polymorphism associated with opposing directions for efficacy, suggesting that the effect of FCGR genetic variation may be specific to vaccine regimens. The FCGR2C association study of Li et al. (62), used a direct sequencing approach to identify FCGR2C SNPs that associated with vaccine efficacy (VE) against HIV-1 in the RV144 vaccine trial, that showed modest efficacy (64). Individuals with at least one minor allele of three FCGR2C SNPs (rs114945036, rs138747765, and rs78603008) had a vaccine efficacy of 64% against any HIV-1 subtype and 91% against the CRF01-AE subtype with the protective 169K HIV-1 variant identified previously by sieve analysis (65). Although the functional mechanisms underlying the association were not revealed in this study, a subsequent examination of the FCGR2C SNPs showed rs114945036 correlated with expression levels of FCGR2A/C (66). This effect was found across different populations and was specific to the rs114945036 SNP located in the intron. Further, rs114945036 also associated with the expression of the Fc receptor-like A (FCRLA) gene, an FCGR related gene located within a gene cluster adjacent to FCGR (see Figure 1A). These results suggest that the FCGR expression is either influenced by this SNP through an undefined mechanism, or is in linkage with other causal variants that directly affect expression levels.
In the second study, four FCGR2C SNPs significantly modified the hazard ratio in the HVTN505 trial that did not show protection against HIV-1 acquisition (62). Three of the SNPs were common with those previously identified in RV144. In contrast to the RV144 study, in HVTN505 among the recipients carrying the FCGR2C minor alleles, HIV-1 acquisition risk was higher in the vaccine group than in the placebo group, in precisely the opposite direction of that observed in RV144 (efficacy against HIV-1 acquisition hazard ratios (HR) of 9.79 (p = 0.035) and 0.36 (p = 0.04), respectively). It is not clear how polymorphisms in a pseudogene functions during HIV-1 vaccination and their associations with FCGR expression may provide a novel avenue for further investigation.
Two additional studies of outcomes in Vax004, a trial testing recombinant gp120 vaccination in preventing sexually acquired HIV infection, also implicated FCGR variation in HIV infection and vaccine efficacy. Both studies tested the classical FCGR2A or FCGR3A variants comprised of the FCGR2A-R/H (rs1801274) and FCGR3A-F/V (rs396991) alleles. The first study found that lower affinity receptors (FCG2A-RR or HR and FCGR3A-FF) were associated with higher serum ADCVI activity, which itself predicted the rate of infection (67). A second study by the same group, showed the FCGR3A-VV genotype distinguished the lowest behavioral risk group from the high-risk behavioral group (68). The low risk group had a higher infection rate than low risk vaccinees with one or two F alleles (HR = 3.52; p = 0.002) while the high-risk group showed no association. Functional studies may be directed by these findings to interrogate quantitative and qualitative effects on FCGRs and associated antibody production. At a minimum, the intersection of these studies suggests that the impact of FCGR genetic variations on vaccine efficacy should be further investigated.
FCGR and IGHG—Future Directions For Genetic Analysis
It is apparent that the genomic complexity of the FCGR region presents a major challenge for uncovering the underlying causal FCGR variants. Different FCGR have distinct functions and mechanisms of regulation but share highly similar sequences. While FCGR genetic variations are clearly linked to host defense against infectious diseases and other important immune functions as discussed, current approaches measure only a small portion of the existing FCGR variation. The HVTN505 and RV144 studies referenced above were by far the most comprehensive in that regard, measuring ~10 kb of the FCGR region, including functional exons encoding external protein domains and flanking intron sequences from the five FCGR genes. However, the complete FCGR region extends over 200 kb leaving open the likelihood of additional causal variation (Figure 1A). Indeed, the lack of phasing of the over 20,000 SNPs documented in the FCGR region significantly limits its direct utility for association analysis and the ultimate goal of identifying causal variants (69). Complete haplotype-resolved FCGR genomic sequences across human populations by approaches such as those used for defining variability in the KIR region may be necessary in order to provide a complete analysis of these loci (70–72).
When considering FCGR host genetics and its relationship to HIV-1 susceptibility and vaccine efficacy—or any association with disease—a natural but not often considered extension to host genetic association studies interrogating FCGR variability lies within the human immunoglobulin constant heavy G chain (IGHG) gene region on chromosomal segment 14q32.3 (73). This region encoding the human IG heavy constant genes (IGHG3, IGHG1, IGHG2, IGHA2, and the IGH locus on chromosome 14) provides access to a system for understanding immunogenicity of the polymorphic IG chains (74, 75). The evident functional relationship between FCGR and IgG constant region variability, which itself is substantial (74), argues strongly for host genetic studies of FCGR to be paired with analysis of IGHG. Although little genomic characterization for the IGHG system is available at present, we anticipate that NGS technologies will rapidly fill that void. Of course, once provided with high quality and high resolution data, significant effort will need to be invested in new sophisticated analytical approaches examining multiple factors simultaneously to find the causal variation revealing operative biological mechanisms.
Concluding Remarks
Disease pathogenesis of HIV-1 has been shown to be modulated by allelic variants in the FCGR genes. However, such findings have not always been robust, as they were not replicated or in some cases were contradictory. There is however considerable interest in the role of Fc-mediated antiviral functions such as ADCC, ADCP, ADCD, and ADCVI in protective immunity against HIV-1 (76). Host genetics of the Fc receptors that bind to the Fc domain of the IgG antibody might modulate the functional antiviral antibody responses to HIV-1 vaccination. ADCC was previously identified as a correlate of protection in the RV144 human efficacy trial (64). More recently, ADCP has been shown to correlate with protection against acquisition of SIV/SHIV/HIV-1 in multiple preclinical and human efficacy trials (77–80). Given such associations, it would be critical to investigate association of host variation in FCGR genes and such Fc-mediated antiviral functions that are now being generated using technologies such as systems serology (1). Such findings might shed light on the role of Fc gene and receptor genotypes on HIV-1 disease pathogenesis.
Author Contributions
RT conceptualized, organized the content of the review and wrote sections Introduction, FCGR polymorphisms and HIV-1 disease progression, Influence of FCGR diversity on HIV-1 acquisition, Role of FCGR polymorphisms on outcomes after ART initiation, and Concluding Remarks. DG contributed to sections Fcγ receptor genetic diversity, Effect of FCGR variation on HIV-1 vaccine efficacy, and FCGR and IGHG–future directions for genetic analysis. JF and CT drafted sections FCGR polymorphisms and HIV-1 disease progression, Influence of FCGR diversity on HIV-1 acquisition. All authors participated in editing and revising the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge the RV254 study group led by Dr. Jintanat Ananworanich and Dr. Sodsai Tovanabutra, MHRP for providing samples, clinical data, and viral reservoir measurements. We thank Dr. Vicky Polonis, MHRP and Mr. Philip Ehrenberg for thoughtful comments and suggestions. We thank Ms. Aviva Geretz, MHRP for reviewing the compiled FCGR polymorphisms. This work was supported by a cooperative agreement (W81XWH-18-2-0040) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U. S. Department of Defense (DOD). This research was funded, in part, by the U. S. National Institute of Allergy and Infectious Disease. The views expressed are those of the authors and should not be construed to represent the positions of the U. S. Army or the DOD.
References
1. Chung AW, Alter G. Systems serology: profiling vaccine induced humoral immunity against HIV. Retrovirology. (2017) 14:57. doi: 10.1186/s12977-017-0380-3
2. Forthal DN, Moog C. Fc receptor-mediated antiviral antibodies. Curr Opin HIV AIDS. (2009) 4:388–93. doi: 10.1097/COH.0b013e32832f0a89
3. Cocklin SL, Schmitz JE. The role of Fc receptors in HIV infection and vaccine efficacy. Curr Opin HIV AIDS. (2014) 9:257–62. doi: 10.1097/coh.0000000000000051
4. Bournazos S, Woof JM, Hart SP, Dransfield I. Functional and clinical consequences of Fc receptor polymorphic and copy number variants. Clin Exp Immunol. (2009) 157:244–54. doi: 10.1111/j.1365-2249.2009.03980.x
5. Li X, Gibson AW, Kimberly RP. Human FcR polymorphism and disease. Curr Top Microbiol Immunol. (2014) 382:275–302. doi: 10.1007/978-3-319-07911-0_13.
6. Bournazos S, Ravetch JV. Fcgamma receptor function and the design of vaccination strategies. Immunity. (2017) 47:224–33. doi: 10.1016/j.immuni.2017.07.009
7. Kratochvil S, McKay PF, Chung AW, Kent SJ, Gilmour J, Shattock RJ. Immunoglobulin G1 allotype influences antibody subclass distribution in response to HIV gp140 vaccination. Front Immunol. 8:1883. (2017) doi: 10.3389/fimmu.2017.01883
8. Gupta S, Gach JS, Becerra JC, Phan TB, Pudney J, Moldoveanu Z, et al. The Neonatal Fc receptor (FcRn) enhances human immunodeficiency virus type 1 (HIV-1) transcytosis across epithelial cells. PLoS Pathog. (2013) 9:e1003776. doi: 10.1371/journal.ppat.1003776
9. Shen S, Pyo CW, Vu Q, Wang R, Geraghty DE. The essential detail: the genetics and genomics of the primate immune response. ILAR J. (2013) 54:181–95. doi: 10.1093/ilar/ilt043
10. Misra MK, Augusto DG, Martin GM, Nemat-Gorgani N, Sauter J, Hofmann JA, et al. Report from the Killer-cell Immunoglobulin-like Receptors (KIR) component of the 17th International HLA and Immunogenetics Workshop. Hum Immunol. (2018) 79:825–33 doi: 10.1016/j.humimm.2018.10.003
11. Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood. (2012) 119:5640–9. doi: 10.1182/blood-2012-01-380121
12. Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. (2009) 113:3716–25. doi: 10.1182/blood-2008-09-179754
13. Warmerdam PA, van de Winkel JG, Gosselin EJ, Capel PJ. Molecular basis for a polymorphism of human Fc gamma receptor II (CD32). J Exp Med. (1990) 172:19–25.
14. Ravetch JV, Perussia B. Alternative membrane forms of Fc gamma RIII(CD16) on human natural killer cells and neutrophils. Cell type-specific expression of two genes that differ in single nucleotide substitutions. J Exp Med. (1989) 170:481–97.
15. Seret G, Hanrotel C, Bendaoud B, Le Meur Y, Renaudineau Y. Homozygous FCGR3A-158F mutation is associated with delayed B-cell depletion following rituximab but with preserved efficacy in a patient with refractory lupus nephritis. Clini Kidney J. (2013) 6:74–6. doi: 10.1093/ckj/sfs162
16. Shimizu, Tanaka Y, Tazawa H, Verma S, Onoe T, Ishiyama K, et al. Fc-gamma receptor polymorphisms predispose patients to infectious complications after liver transplantation. Am J Transplant. (2016) 16:625–33. doi: 10.1111/ajt.13492
17. Taylor RJ, Saloura V, Jain A, Goloubeva O, Wong S, Kronsberg S, et al. Ex vivo antibody-dependent cellular cytotoxicity inducibility predicts efficacy of cetuximab. Cancer Immunol Res. (2015) 3:567–74. doi: 10.1158/2326-6066.cir-14-0188
18. Flinsenberg TW, Janssen WJ, Herczenik E, Boross P, Nederend M, Jongeneel LH, et al. A novel FcgammaRIIa Q27W gene variant is associated with common variable immune deficiency through defective FcgammaRIIa downstream signaling. Clin Immunol. (2014) 155:108–17. doi: 10.1016/j.clim.2014.09.006
19. Kono H, Kyogoku C, Suzuki T, Tsuchiya N, Honda H, Yamamoto K, et al. FcgammaRIIB Ile232Thr transmembrane polymorphism associated with human systemic lupus erythematosus decreases affinity to lipid rafts and attenuates inhibitory effects on B cell receptor signaling. Hum Mol Genet. (2005) 14:2881–92. doi: 10.1093/hmg/ddi320
20. Ernst LK, Metes D, Herberman RB, Morel PA. Allelic polymorphisms in the FcgammaRIIC gene can influence its function on normal human natural killer cells. J Mol Med. (2002) 80:248–57. doi: 10.1007/s00109-001-0294-2
21. Metes D, Ernst LK, Chambers WH, Sulica A, Herberman RB, Morel PA. Expression of functional CD32 molecules on human NK cells is determined by an allelic polymorphism of the FcgammaRIIC gene. Blood. (1998) 91:2369–80.
22. Nagelkerke SQ, Tacke CE, Breunis WB, Geissler J, Sins JW, Appelhof B, et al. Nonallelic homologous recombination of the FCGR2/3 locus results in copy number variation and novel chimeric FCGR2 genes with aberrant functional expression. Genes Immun. (2015) 16:422–9. doi: 10.1038/gene.2015.25
23. van der Heijden J, Breunis WB, Geissler J, de Boer M, van den Berg TK, Kuijpers TW. Phenotypic variation in IgG receptors by nonclassical FCGR2C alleles. J Immunol. (2012) 188:1318–24. doi: 10.4049/jimmunol.1003945
24. Metes D, Galatiuc C, Moldovan I, Morel PA, Chambers WH, DeLeo AB, et al. Expression and function of Fc gamma RII on human natural killer cells. Nat Immun. (1994) 13:289–300.
25. Qiu WQ, de Bruin D, Brownstein BH, Pearse R, Ravetch JV. Organization of the human and mouse low-affinity Fc gamma R genes: duplication and recombination. Science. (1990) 248:732–5.
26. de Haas M, Koene HR, Kleijer M, de Vries E, Simsek S, van Tol MJ, et al. A triallelic Fc gamma receptor type IIIA polymorphism influences the binding of human IgG by NK cell Fc gamma RIIIa. J Immunol. (1996) 156:2948–55.
27. Ory PA, Clark MR, Kwoh EE, Clarkson SB, Goldstein IM. Sequences of complementary DNAs that encode the NA1 and NA2 forms of Fc receptor III on human neutrophils. J Clin Invest. (1989) 84:1688–91. doi: 10.1172/jci114350
28. Ory PA, Goldstein IM, Kwoh EE, Clarkson SB. Characterization of polymorphic forms of Fc receptor III on human neutrophils. J Clin Invest. (1989) 83:1676–81. doi: 10.1172/jci114067
29. Bredius RG, Fijen CA, De Haas M, Kuijper EJ, Weening RS, Van de Winkel JG, et al. Role of neutrophil Fc gamma RIIa (CD32) and Fc gamma RIIIb (CD16) polymorphic forms in phagocytosis of human IgG1- and IgG3-opsonized bacteria and erythrocytes. Immunology. (1994) 83:624–30.
30. Salmon JE, Edberg JC, Kimberly RP. Fc gamma receptor III on human neutrophils. Allelic variants have functionally distinct capacities. J Clin Invest. (1990) 85:1287–95. doi: 10.1172/jci114566
31. Bux J, Stein EL, Bierling P, Fromont P, Clay M, Stroncek D, et al. Characterization of a new alloantigen (SH) on the human neutrophil Fc gamma receptor IIIb. Blood. (1997) 89:1027–34.
32. Flesch BK, Doose S, Siebert R, Ntambi E, Neppert J. FCGR3 variants and expression of human neutrophil antigen-1a,−1b, and−1c in the populations of northern Germany and Uganda. Transfusion. (2002) 42:469–75. doi: 10.1046/j.1525-1438.2002.00087.x
33. Reil A, Sachs UJ, Siahanidou T, Flesch BK, Bux J. HNA-1d: a new human neutrophil antigen located on Fcgamma receptor IIIb associated with neonatal immune neutropenia. Transfusion. (2013) 53:2145–51. doi: 10.1111/trf.12086
34. Hollox EJ, Hoh BP. Human gene copy number variation and infectious disease. Human Genet. (2014) 133:1217–33. doi: 10.1007/s00439-014-1457-x
35. Breunis WB, van Mirre E, Geissler J, Laddach N, Wolbink G, van der Schoot E, et al. Copy number variation at the FCGR locus includes FCGR3A, FCGR2C, and FCGR3B but not FCGR2A and FCGR2B. Human Mutat. (2009) 30:E640–50. doi: 10.1002/humu.20997
36. Niederer HA, Willcocks LC, Rayner TF, Yang W, Lau YL, Williams TN, et al. Copy number, linkage disequilibrium and disease association in the FCGR locus. Hum Mol Genet. (2010) 19:3282–94. doi: 10.1093/hmg/ddq216
37. Hargreaves CE, Rose-Zerilli MJ, Machado LR, Iriyama C, Hollox EJ, Cragg MS, et al. Fcgamma receptors: genetic variation, function, and disease. Immunol Rev. (2015) 268:6–24. doi: 10.1111/imr.12341
38. Machado LR, Hardwick RJ, Bowdrey J, Bogle H, Knowles TJ, Sironi M, et al. Evolutionary history of copy-number-variable locus for the low-affinity Fcgamma receptor: mutation rate, autoimmune disease, and the legacy of helminth infection. Am J Hum Genet. (2012) 90:973–85. doi: 10.1016/j.ajhg.2012.04.018
39. Lehrnbecher TL, Foster CB, Zhu S, Venzon D, Steinberg SM, Wyvill K, et al. Variant genotypes of FcgammaRIIIA influence the development of Kaposi's sarcoma in HIV-infected men. Blood. (2000) 95:2386–90.
40. Forthal DN, Landucci G, Bream J, Jacobson LP, Phan TB, Montoya B. FcgammaRIIa genotype predicts progression of HIV infection. J Immunol. (2007) 179:7916–23. doi: 10.4049/jimmunol.179.11.7916
41. Weis JF, McClelland RS, Jaoko W, Mandaliya KN, Overbaugh J, Graham SM, et al. Short communication: Fc gamma receptors IIa and IIIa genetic polymorphisms do not predict HIV-1 disease progression in Kenyan women. AIDS Res Hum Retroviruses. (2015) 31:288–92. doi: 10.1089/AID.2014.0209
42. Poonia B, Kijak GH, Pauza CD. High affinity allele for the gene of FCGR3A is risk factor for HIV infection and progression. PLoS ONE. (2010) 5:e15562. doi: 10.1371/journal.pone.0015562
43. Rohatgi S, Gohil S, Kuniholm MH, Schultz H, Dufaud C, Armour KL, et al. Fc gamma receptor 3A polymorphism and risk for HIV-associated cryptococcal disease. MBio. (2013) 4:e00573–13. doi: 10.1128/mBio.00573-13
44. Meletiadis J, Walsh TJ, Choi EH, Pappas PG, Ennis D, Douglas J, et al. Study of common functional genetic polymorphisms of FCGR2A, 3A and 3B genes and the risk for cryptococcosis in HIV-uninfected patients. Med Mycol. (2007) 45:513–8. doi: 10.1080/13693780701390140
45. Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, et al. STrengthening the REporting of Genetic Association Studies (STREGA): an extension of the STROBE statement. PLoS Med. (2009) 6:e22. doi: 10.1371/journal.pmed.1000022
46. McLaren PJ, Coulonges C, Ripke S, van den Berg L, Buchbinder S, Carrington M, et al. Association study of common genetic variants and HIV-1 acquisition in 6,300 infected cases and 7,200 controls. PLoS Pathog. (2013) 9:e1003515. doi: 10.1371/journal.ppat.1003515
47. McLaren PJ, Carrington M. The impact of host genetic variation on infection with HIV-1. Nat Immunol. (2015) 16:577–83. doi: 10.1038/ni.3147
48. McLaren PJ, Pulit SL, Gurdasani D, Bartha I, Shea PR, Pomilla C, et al. Evaluating the impact of functional genetic variation on HIV-1 control. J Infect Dis. (2017) 216:1063–9. doi: 10.1093/infdis/jix470
49. Brouwer KC, Lal RB, Mirel LB, Yang C, van Eijk AM, Ayisi J, et al. Polymorphism of Fc receptor IIa for IgG in infants is associated with susceptibility to perinatal HIV-1 infection. AIDS. (2004) 18:1187–94. doi: 10.1097/00002030-200405210-00012
50. Milligan C, Richardson BA, John-Stewart G, Nduati R, Overbaugh J. FCGR2A and FCGR3A genotypes in human immunodeficiency virus mother-to-child transmission. Open Forum Infect Dis. (2015) 2:ofv149. doi: 10.1093/ofid/ofv149
51. Ananworanich J, Chomont N, Eller LA, Kroon E, Tovanabutra S, Bose M, et al. HIV DNA set point is rapidly established in acute HIV infection and dramatically reduced by early ART. EBioMedicine. (2016) 11:68–72. doi: 10.1016/j.ebiom.2016.07.024
52. Saez-Cirion A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, et al. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog. (2013) 9:e1003211. doi: 10.1371/journal.ppat.1003211
53. Halper-Stromberg A, Lu CL, Klein F, Horwitz JA, Bournazos S, Nogueira L, et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell. (2014) 158:989–99. doi: 10.1016/j.cell.2014.07.043
54. Descours B, Petitjean G, López-Zaragoza JL, Bruel T, Raffel R, Psomas C, et al. CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature. (2017) 543:564–7. doi: 10.1038/nature21710
55. De Souza MS, Phanuphak N, Pinyakorn S, Trichavaroj R, Pattanachaiwit S, Chomchey N, et al. Impact of nucleic acid testing relative to antigen/antibody combination immunoassay on the detection of acute HIV infection. AIDS. (2015) 29:793–800. doi: 10.1097/QAD.0000000000000616
56. Ananworanich J, Fletcher JL, Pinyakorn S, van Griensven F, Vandergeeten C, Schuetz A, et al. A novel acute HIV infection staging system based on 4th generation immunoassay. Retrovirology. (2013) 10:56. doi: 10.1186/1742-4690-10-56
57. Abdel-Mohsen M, Kuri-Cervantes L, Grau-Exposito J, Spivak AM, Nell RA, Tomescu C, et al. CD32 is expressed on cells with transcriptionally active HIV but does not enrich for HIV DNA in resting T cells. Sci Transl Med. (2018) 10:eaar6759. doi: 10.1126/scitranslmed.aar6759
58. Osuna CE, Lim SY, Kublin JL, Apps R, Chen E, Mota TM, et al. Evidence that CD32a does not mark the HIV-1 latent reservoir. Nature. (2018) 561:E20–8. doi: 10.1038/s41586-018-0495-2
59. Bertagnolli LN, White JA, Simonetti FR, Beg SA, Lai J, Tomescu C, et al. The role of CD32 during HIV-1 infection. Nature. (2018) 561:E17–9. doi: 10.1038/s41586-018-0494-3
60. Perez L, Anderson J, Chipman J, Thorkelson A, Chun TW, Moir S, et al. Conflicting evidence for HIV enrichment in CD32(+) CD4 T cells. Nature. (2018) 561, E9–16. doi: 10.1038/s41586-018-0493-4
61. Prentice HA, Tomaras GD, Geraghty DE, Apps R, Fong Y, Ehrenberg PK, et al. HLA class II genes modulate vaccine-induced antibody responses to affect HIV-1 acquisition. Sci Transl Med. (2015) 7:296ra112. doi: 10.1126/scitranslmed.aab4005
62. Li SS, Gilbert PB, Tomaras GD, Kijak G, Ferrari G, Thomas R, et al. FCGR2C polymorphisms associate with HIV-1 vaccine protection in RV144 trial. J Clin Invest. (2014) 124:3879–90. doi: 10.1172/JCI75539
63. Gartland AJ, Li S, McNevin J, Tomaras GD, Gottardo R, Janes H, et al. Analysis of HLA A*02 association with vaccine efficacy in the RV144 HIV-1 vaccine trial. J Virol. (2014) 88:8242–55. doi: 10.1128/JVI.01164-14
64. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. (2009) 361:2209–20. doi: 10.1056/NEJMoa0908492
65. Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. (2012) 490:417–20. doi: 10.1038/nature11519
66. Peng X, Li SS, Gilbert PB, Geraghty DE, Katze MG. FCGR2C polymorphisms associated with HIV-1 vaccine protection are linked to altered gene expression of Fc-gamma receptors in human B cells. PLoS ONE. (2016) 11:e0152425. doi: 10.1371/journal.pone.0152425
67. Forthal DN, Gilbert PB, Landucci G, Phan T. Recombinant gp120 vaccine-induced antibodies inhibit clinical strains of HIV-1 in the presence of Fc receptor-bearing effector cells and correlate inversely with HIV infection rate. J Immunol. (2007) 178:6596–603.
68. Forthal DN, Gabriel EE, Wang A, Landucci G, Phan TB. Association of Fcgamma receptor IIIa genotype with the rate of HIV infection after gp120 vaccination. Blood. (2012) 120:2836–42. doi: 10.1182/blood-2012-05-431361
69. EMBL-EBI. Ensembl Variation Database. (2018). Avaliable online at: https://www.ensembl.org/info/genome/variation/index.html
70. Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE. (2010) 5:e15115. doi: 10.1371/journal.pone.0015115
71. Pyo CW, Wang R, Vu Q, Cereb N, Yang SY, Duh FM, et al. Recombinant structures expand and contract inter and intragenic diversification at the KIR locus. BMC Genomics. (2013) 14:89. doi: 10.1186/1471-2164-14-89
72. Roe D, Vierra-Green C, Pyo CW, Eng K, Hall R, Kuang R, et al. Revealing complete complex KIR haplotypes phased by long-read sequencing technology. Genes Immun. (2017) 18:127–34. doi: 10.1038/gene.2017.10
73. Oxelius VA, Pandey JP. Human immunoglobulin constant heavy G chain (IGHG) (Fcgamma) (GM) genes, defining innate variants of IgG molecules and B cells, have impact on disease and therapy. Clin Immunol. (2013) 149:475–86. doi: 10.1016/j.clim.2013.10.003
74. Lefranc MP, Lefranc G. Human Gm, Km, and Am allotypes and their molecular characterization: a remarkable demonstration of polymorphism. Methods Mol Biol. (2012) 882:635–80. doi: 10.1007/978-1-61779-842-9_34
75. Pandey JP, Li Z. The forgotten tale of immunoglobulin allotypes in cancer risk and treatment. Exper Hematol Oncol. (2013) 2:6. doi: 10.1186/2162-3619-2-6
76. Lewis GK. Role of Fc-mediated antibody function in protective immunity against HIV-1. Immunology. (2014) 142:46–57. doi: 10.1111/imm.12232
77. Barouch DH, Alter G, Broge T, Linde C, Ackerman ME, Brown EP, et al. Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science. (2015) 349:320–4. doi: 10.1126/science.aab3886
78. Barouch DH, Tomaka FL, Wegmann F, Stieh DJ, Alter G, Robb ML, et al. Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13-19). Lancet. (2018) 392:232–43. doi: 10.1016/S0140-6736(18)31364-3
79. Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, et al. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell. (2013) 155:531–9. doi: 10.1016/j.cell.2013.09.061
Keywords: next-generation sequencing, polymorphism, disease association, Fc receptors, HIV-1
Citation: Geraghty DE, Thorball CW, Fellay J and Thomas R (2019) Effect of Fc Receptor Genetic Diversity on HIV-1 Disease Pathogenesis. Front. Immunol. 10:970. doi: 10.3389/fimmu.2019.00970
Received: 19 January 2019; Accepted: 15 April 2019;
Published: 09 May 2019.
Edited by:
R. Keith Reeves, Harvard Medical School, United StatesReviewed by:
Rebecca M. Lynch, George Washington University, United StatesJennifer Manuzak, University of Miami, United States
Copyright © 2019 Geraghty, Thorball, Fellay and Thomas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rasmi Thomas, cnRob21hc0BoaXZyZXNlYXJjaC5vcmc=
 Daniel E. Geraghty
Daniel E. Geraghty Christian W. Thorball
Christian W. Thorball Jacques Fellay
Jacques Fellay Rasmi Thomas
Rasmi Thomas