- 1Infectious Diseases Unit, I. Department of Medicine, University Medical Center Hamburg-Eppendorf, Hamburg, Germany
- 2German Center for Infection Research (DZIF), Partner Site Hamburg-Lübeck-Borstel-Riems, Hamburg, Germany
- 3Division of Vaccine Discovery, La Jolla Institute for Immunology, La Jolla, CA, United States
- 4Department of Medicine, Division of Infectious Diseases, University of California, San Diego (UCSD), La Jolla, CA, United States
- 5Protozoa Immunology, Bernhard-Nocht-Institute of Tropical Medicine, Hamburg, Germany
Control of malaria is an important global health issue and there is still an urgent need for the development of an effective prophylactic vaccine. Multiple studies have provided strong evidence that Plasmodium falciparum-specific MHC class I-restricted CD8+ T cells are important for sterile protection against Plasmodium falciparum infection. Here, we present an interactive epitope map of all P. falciparum-specific CD8+ T cell epitopes published to date, based on a comprehensive data base (IEDB), and literature search. The majority of the described P. falciparum-specific CD8+ T cells were directed against the antigens CSP, TRAP, AMA1, and LSA1. Notably, most of the epitopes were discovered in vaccine trials conducted with malaria-naïve volunteers. Only few immunological studies of P. falciparum-specific CD8+ T cell epitopes detected in patients suffering from acute malaria or in people living in malaria endemic areas have been published. Further detailed immunological mappings of P. falciparum-specific epitopes of a broader range of P. falciparum proteins in different settings and with different disease status are needed to gain a more comprehensive understanding of the role of CD8+ T cell responses for protection, and to better guide vaccine design and to study their efficacy.
Introduction
Malaria remains one of the pressing global health issues with ~216 million cases per year (1). The most virulent Plasmodium species, Plasmodium falciparum (P. falciparum), accounts for the vast majority of the 445,000 deaths which occurred in 2016 due to malaria. Rising rates of drug (parasite-related) and insecticide (vector-related) resistance underscore the urgent need for an effective vaccine (2). The feasibility of achieving sterile protection by vaccination has already been demonstrated more than 50 years ago by experiments that could induce sterile immunity using irradiated sporozoites in mice (3) and in humans (4, 5). But the immunization of large numbers of individuals using sporozoites remains challenging and there is limited evidence for durable, high-level efficacy against natural exposure (6, 7). So far, the development of a subunit vaccine (that contains only the antigenic parts of a pathogen) has not succeeded despite decades of research (8, 9). In order to achieve sterile protection against P. falciparum infection, a better understanding of host-parasite interaction, and correlation of a protective P. falciparum-specific immune response is necessary. In particular, detailed knowledge of the P. falciparum-specific CD8+ T cell epitope repertoire is needed to further optimize vaccine design and to improve immune monitoring of future clinical malaria vaccine trials. In this review, we summarize and discuss all of the P. falciparum-specific CD8+ T cell epitopes in humans that have been identified so far, their potential significance as well as potential knowledge gaps.
The RTS,S Vaccine and CSP-Specific CD8+ T Cells
There are currently several vaccine trials under way. However, recently published trials could only demonstrate suboptimal effectivity (10, 11). A malaria subunit vaccine, called the RTS,S vaccine (Mosquirix) targeting the CSP protein is the most advanced vaccine (12). Unfortunately vaccine efficacy was only modest (13), waned over time, and was not detectable any more three years after vaccination (11). Elevated IFNγ-levels after antigen-specific short-term cultivation and re-stimulation were detected by ELISPOT in subjects vaccinated with the RTS,S and CSP constructs and their levels decreased with waning protection (14). These studies demonstrate the importance of P. falciparum-specific T cells and the need to improve future strategies to induce a strong, broad and long-lasting vaccine-induced P. falciparum-specific CD8+ T cell response (15).
Challenges of Malaria-Vaccine Development
Some of the particular obstacles of the development of a protective malaria vaccine are:
a) The large size of the 23-megabase nuclear genome which consists of 14 chromosomes encoding for about 5,300 genes (16),
b) the great genetic heterogeneity of some regions of those genes, and
c) the complex life cycle and expression pattern of these genes (17),
rendering malaria vaccine design more complicated than for most viral and bacterial infections.
A detailed understanding of the life cycle of the parasites and determination of the immune responses that confer protection after natural infection will provide important insights for future vaccine development. Within the human host, two main malaria life cycle stages are completed (18): the pre-erythrocytic stage and the blood stage. The pre-erythrocytic stage can be divided into an early sporozoite stage and the liver stage. During the early sporozoite stage, sporozoites are injected into the skin, enter the blood stream, and consecutively infect hepatocytes. This stage lasts minutes to hours (19). The ensuing liver stage consists of asexual replication and the maturation of sporozoites into schizonts within the hepatocytes, which lasts for ~1 week in humans in P. falciparum malaria (20). The subsequent blood stage is initiated with the release of merozoites that infect red blood cells (RBCs) and is the period in which clinical symptoms occur. These symptoms are partly induced by excessive host immune responses [reviewed in Artavanis-Tsakonas et al. (21)]. A small number of parasites in the blood develop into sexual-stage gametocytes, which can be taken up by the mosquito and continue the cycle of infection. The proteins that are expressed by plasmodia are life cycle stage specific: During the liver stage different (surface-) proteins like the circumsporozoite protein (CSP), liver stage antigens (LSA) and thrombospondin-related anonymous protein (TRAP) are expressed, while the merozoite surface protein (MSP) are expressed during the blood stage. The apical membrane antigen 1 (AMA1) is present in both stages (22). P. falciparum-specific T cells are primed within different immunological environments which might influence breadth and quality of the P. falciparum-specific response against the different antigens. It has been proposed that parasite-specific CD8+ T cells are mostly primed during the liver stage, while P. falciparum-specific CD4+ T cells enable clearance of the blood stage infection (23). CD8+ T cells are probably not important effector cells during the blood stage because the parasite-infected human erythrocytes do not display MHC class I molecules (24).
Nevertheless, naturally acquired immunity against malaria is primarily directed against blood stage antigens (mainly mediated via antibodies) and therefore prevents clinical disease but not the infection of hepatocytes (6, 7, 25), suggesting that there is no, or only little naturally acquired immunity against the pre-erythrocytic stage (26, 27). Of note, the pre-erythrocytic stage represents a bottleneck for the parasite (and is an ideal target for the host specific immune response) because of the relatively low number of parasites passing through this phase (~100 sporozoites, only) (28). A strong, specific-CD8+ T cell response directed against the liver stage would optimally prevent the parasite's transition to the blood stage and clinical symptoms and disease could be avoided (15).
The P. falciparum-Specific CD8+ T Cell Response
Plasmodia, like other pathogens, induce a variety of immune effector responses: P. falciparum-specific antibodies, parasite-specific CD8+, and CD4+ T cells, as well as certain cytokines have been implicated as important effectors [reviewed in Stevenson and Riley (29) and Dobaño and Moncunill (30)]. The current review will focus on human P. falciparum-specific CD8+ T cells since there is strong experimental evidence in rodent and primate models as well as in humans that CD8+ T cells play a major role in providing protection:
• Sterile immunity using irradiated sporozoites in mice could be induced. This immunity was specific against the liver phase and later inoculation with parasitized red blood cells led to infection (3).
• Mice and rhesus monkeys were protected by immunization with attenuated sporozoites. This immunity was abrogated by experimental depletion of CD8+ T cells (31–34).
• The transfer of CSP-specific CD8+ T cell clones, as well as the transfer of a defined CD8+ T cell epitope conferred protection to malaria-naïve mice (35–39). Likewise, TRAP-peptides from P. berghei and a novel liver stage antigen MIF-4-like protein peptide Kb-17 have been able to elicit a CD8+ T cell-dependent response against murine malaria (40, 41).
• Immunity provided by antibodies was shown to be suboptimal most likely because blood-stage surface antigens show great variability (42, 43). Mouse models demonstrated that a robust CD4+ and CD8+ T cell response improves immunity since these T cell responses target internal antigens that are more likely to be conserved (44–46).
• Mouse models could show that induction of extremely high numbers of memory CD8+ T cells were a prerequisite for solid, sterile protection (47, 48).
• CD8+ T cells have shown to be important effectors that form clusters around infected hepatocytes and destroy them (49, 50). The importance of liver resident CD8+ T cells in protective immunity induced by attenuated P. berghei sporozoites has also been reported (51, 52). Mounting evidence suggests that effector CD8+ T cells eliminate the parasites without direct contact with infected hepatocytes via cytokine release (53). This model is also supported by the lymphogenic features of the liver (53).
• In humans, the inoculation with intact sporozoites (that were not attenuated) led to an increase of parasite-specific pluripotent effector memory T cells (54). The levels of P. falciparum-specific CD8+ T cells were also higher in those subjects who were protected after immunization with irradiated sporozoites compared to unprotected subjects (55).
• The polymorphism of CSP is primarily located in the region of identified CTL and T helper epitopes (56).
• An association with HLA class I and the course of the disease has been described showing that HLA-B*53, was associated with resistance to severe malaria (57). MHC class I-dependent presentation of antigens in P. berghei malaria was also demonstrated in the mouse model (58).
• Humans immunized with irradiated sporozoites or naturally exposed to malaria can generate a CTL response to pre-erythrocytic-stage antigens (8, 59–63).
• In mouse models, an excessively strong CD8+ T cell response has been associated with the development of cerebral malaria (64), and a deeper understanding of the CD8+ T cell repertoire may have implications beyond vaccine development and could be relevant for the clinical course of this disease in humans.
The mechanism of protection by CD8+ T cells is thought to be partly cytokine-mediated by interferon-γ (IFNγ) (31) and tumor necrosis factor (TNF) [reviewed in Dobaño and Moncunill (30)] that both inhibit parasite development. Perforin and granzymes kill infected hepatocytes through direct lysis (39), (65–68). IFNγ responses directed against pre-erythrocytic stage antigens of P. falciparum are associated with higher hemoglobin levels and significantly reduced prevalence of severe anemia (69) and are associated with significant resistance to re-infection (70, 71). It could also be shown that age-related cumulative exposure to P. falciparum increases the frequency of IFNγ responses (72, 73). This demonstrates the importance of IFNγ production for protection against malaria.
Challenges of Investigating the P. falciparum-Specific CD8+ T Cell Response
So far, the breadth and specificity of the parasite-specific CD8+ T cell response in malaria has only been poorly described. One problem of the mapping of P. falciparum-specific T cell responses is the comparatively low ex vivo frequency of circulating peripheral P. falciparum-specific T cells (74, 75). There are several proposed hypotheses to explain low frequencies and responses in peripheral blood samples:
• Lymphopenia occurs during acute malaria infection [reviewed in Scholzen and Sauerwein (76)]. T lymphocytes are thought to migrate into different tissues, e.g., the liver (8, 77).
• There is a high genetic diversity between different P. falciparum strains with high mutation rates of several antigenic regions of the genome that complicates detection of T cells (Supplementary Figures 1–4). Additionally, no optimized consensus sequence for screening purposes of T cell responses against malaria proteins has been designed as of now (78–80). This extreme genetic diversity, particularly of the surface antigens, makes the detection of P. falciparum-specific T cells difficult (78–80).
• The immunoregulatory effects of a blood stage infection are thought to influence the priming of an adaptive immunity against the pre-erythrocytic stage (81–83).
• P. falciparum contains thousands of antigens that could serve as potential T cell epitopes. This sheer breadth of epitopes primed during an acute infection could also explain the low frequencies of specific T cells against an individual T cell epitope (84).
• The inability to generate new responses against emerging variants (original antigenic sin), is a phenomenon that has been described for several pathogens like influenza, HIV, and the hepatitis B virus (85–91). Malaria can be caused by infection with multiple Plasmodium strains (92, 93) and limited responsiveness of CD8+ T cells in coinfection with different Plasmodium strains could be shown (94) similar to the limited cross-genotype responsiveness after infection with different HCV genotypes (95).
• The human immune T cell response is modulated and dampened by the parasite through several mechanisms (83, 96). Walther et al. showed that regulatory T cells were rapidly induced following blood-stage infection and were associated with a burst of TGF-β production, decreased proinflammatory cytokine production, and decreased antigen-specific immune responses (96). Mackroth et al. demonstrated that acute P. falciparum malaria induced P. falciparum-specific PD1+CTLA4+CD4+ T effector cells that co-produced IFNγ and IL-10 and inhibited other CD4+ T cells (83). In rodent malaria it could be shown that PD-1 mediates up to 95% reduction in numbers and functional capacity of parasite-specific CD8+ T cells (97).
• Evasion mechanisms like Kupffer cell apoptosis and reduced expression of major histocompatibility complex (MHC)-I also resulted in T cell tolerance (98). Additionally, a reduced APC (antigen presenting cell) function of the Kupffer cells from sporozoite-infected mice was shown which might also explain the comparatively low magnitude of P. falciparum-specific T cell responses detected in malaria patients (99).
The Role of HLA Alleles in Malaria
Certain human leukocyte antigens (HLAs) like HLA-B53, DRB*13:02, and DQB*05:01 are associated with protection from severe malaria (84, 100). This suggests that selection of particular epitopes might be associated with better control of parasitaemia. Gabonese children carrying the HLA class II allele DQB1*0501 had a higher frequency of IFNγ-responses to LSA1 T cell epitopes, compared with non-carriers, and were better protected against malaria anemia and re-infections (100).
APFISAVAA (LSA3), EPKDEIVEV (LSA3), and KPIVQYDNF (LSA1) are the only known CD8+ T cell epitopes with HLA-B53 restriction and are located on the liver stage antigen 1 (LSA1) and the liver stage antigen 3 (LSA3) (23, 63, 100–103).
The genes coding the HLA molecules are the most polymorphic genes in humans and several observations indicate that this is due to selective immunological pressure (84). This polymorphism leads to varying degrees of protection against infectious pathogens (but also to association between MHC types and autoimmune diseases) (104). P. falciparum infection has been a major selective force in human evolution, especially in West Africa, where the HLA alleles HLA-A*53, DRB1*13:02, and DQB1*05:01 are more frequent than in the rest of the world (allelefrequencies.net) (105). But P. falciparum contains thousands of different potential T cell epitopes and multiple immune responses are evoked by a single infection (74). This implies that most of these responses are possibly of limited protective value and only a few epitopes will elicit strong, long lasting and protective immune answers (84). However, longitudinal studies about the relationship between HLA alleles, cytokine patterns, and the outcome of P. falciparum infection are rare (100) and other protective HLA alleles have not yet been identified in humans.
Methods for Detection of Antigen-Specific T Cells
Over the last 20 years, the methodologies of T cell detection have undergone considerable changes. P. falciparum-specific T cells have been detected by a number of different assays that differ by practicability, price, sensitivity, and functional read-out. Different assays have different advantages and disadvantages, which has to be taken into account when comparing results of different studies that have characterized P. falciparum-specific T cell responses over the last decades. The most commonly used techniques for T cell analysis in malaria are shown in Table 1.
Potential difficulties of the generally low frequencies of the P. falciparum-specific T cells can be overcome by in vitro expansion and short-term culture methods. Numerous studies have cultivated PBMCs with the particular antigen for a few days up to 2 weeks to allow expansion of the specific T cells (74, 121). However, in vitro expansion alters the T cell function and phenotype to some extent. It is also difficult to extrapolate the original specific ex vivo T cell frequency after culture. An alternative approach to increase low frequencies of antigen-specific T cells is the quantitative pre-enrichment of target cells via magnetic cell separation (118). This approach allows phenotypic assessment of the specific T cells but has not yet been used in the context of malaria CD8+ T cell research.
Resources for P. falciparum-Specific Epitope Research and in silico Analysis
To date, there are several online P. falciparum-specific resources available on the internet that provide immunological information and/or data related to plasmodial antigens, or that offered links to clinical trial data. This section provides a brief overview of these important resources.
The IEDB: An Online Epitope Resource
The Immune Epitope Database and Analysis Resource (IEDB.org) was created by the National Institute of Allergy and Infectious Disease (NIAID) to provide the scientific community with a repository of freely accessible immune epitope data (122, 123). The IEDB contains data captured from the published literature, as well as data submitted through NIAID's high-throughput epitope discovery efforts. The IEDB includes antibody and T cell data from human, non-human primate, and rodent hosts, as well as other animal species, and encompasses epitopes associated with all infectious diseases, allergy, autoimmunity and transplantation and/or alloreactivity. In addition to antibody and T cell response data, the IEDB also contains MHC binding and MHC ligand elution data (mass spectrometry).
It thus provides a unique resource to inventory and analyze immunological data for a given pathogen or disease and has been utilized to produce several meta analyses including a 2009 analysis of epitopes derived from Plasmodium (110).
Currently, the IEDB contains 3,000 unique epitopes derived from the genus Plasmodium. Of these data, species causing malaria, P. falciparum, P. vivax, and P. malariae, represent 88% of the total, with P. falciparum-specific determinants most prominent among them (2,332). P. falciparum-specific data include epitopes defined for more than 160 antigens, including both protein and non-protein (cell surface glycolipids) targets. Data related to human disease are most numerous, with 1,485 epitopes defined from 126 different plasmodial antigens and reported from 295 references. These were data derived from more than 3,900 immunoassays (2,205 T cell and 1,693 antibody). Data generated using murine models of disease (710 epitopes) and non-human primates (104 epitopes) are also available. At the level of immune response phenotype, there are currently 774 human antibody epitopes from 120 antigens and 843 T cell epitopes from 32 antigens, including linear and non-linear antibody epitopes, and class I (147) and class II (596). Of the 147 MHC class I/ CD8+ T cell epitopes, we included 132 epitopes in this review. Fifteen epitopes were excluded because they were longer than 20 amino acids (AA).
In addition to its epitope data repository, the IEDB hosts an array of epitope prediction and analysis tools, including those for T cell class I and II prediction (tools.iedb.org/main), such as TepiTool (124), the T cell class I pMHC immunogenicity predictor (125), and the CD4+ T cell immunogenicity prediction tool (126). Useful among the other analysis resources are the Immunome Browser, which maps epitope prominence along the antigen, the Epitope Cluster Analysis Tool 2.0, and the Epitope Conservation Analysis tool (127).
Other P. falciparum-Specific Online Resources
Informatic sites containing plasmodial antigen sequence and/or structural data include the Plasmodium Genomics Resource (plasmodb.org/plasmo) (128) and the Plasmodium vivax Structural Databank (PvaxDB) (scfbio-iitd.res.in/PvaxDB) (129). The Rodent Malaria genetically modified Parasites (RMgmDB) (pberghei.eu/index.php) site (130) contains genetically modified rodent malaria parasites that have been generated by many labs worldwide. The Malaria Data site (ebi.ac.uk/chembl/malaria) provides a searchable and downloadable public resource for targets, compounds, assays, and other data related to malaria research (131). Similarly, the Malaria Immunology database (malarimdb.org/) provides users an overview of the roles of the various immunological, metabolic vascular, and erythrocytic factors involved in blood-stage malaria and includes human patient data, as well as data from murine models of disease (132). The Malaria Atlas Project (map.ox.ac.uk) enables users to download, visualize and manipulate global parasite rate survey data (133). MalariaGen (malariagen.net) is a genomic epidemiology network for next generation DNA sequencing tools and technologies (134).
Comprehensive Review of P. falciparum-Specific CD8+ T Cell Responses
While a great number of P. falciparum-specific T cell responses can be searched in the IEDB (122), a comprehensive review providing an overview of discovered CD8+ T cell epitopes in P. falciparum and discussing them in context is lacking, to date. Therefore, we conducted an exhaustive database (www.iedb.org) (Terms: P. falciparum; MHC Class I; Humans) and literature review (PubMed)1 (Terms: P.falciparum; CD8+ T cells; epitopes; cytotoxic T cells; MHC class I) and summarized our findings in form of an epitope map (Figures 1–7). We defined an epitope as a P. falciparum-specific peptide of 20 AA (amino acids) or less that elicited a CD8+ T cell response. Fifteen epitopes that had shown to be restricted to MHC class I, but were longer than 20 AA, were excluded from our review (106, 116). Epitopes described for other Plasmodium species or T cell responses primed and detected in murine malaria infection were similarly not included in the current overview. Likewise, P. falciparum-specific CD4+ T cell responses were also not listed. The relative pattern of immune dominance or relative breadth and quality (e.g., cytokine pattern) of the response measured against each of the epitopes was also not assessed.

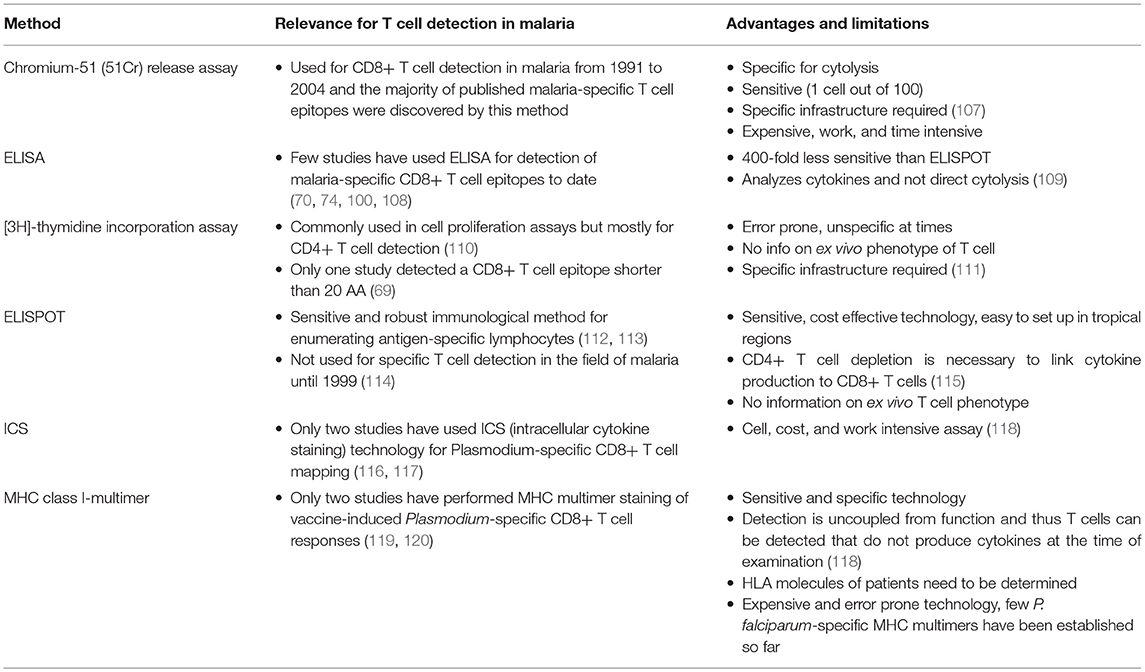
Figure 1. (A) Schematic draft of P. falciparum CSP. The signal sequence (SS) and region one (RI) are localized within the N-terminus. The central repeat region is a region of NANP (dark blue) and NVDP (light blue) repeats. The C-terminus (green shades) is linked to the repeat region and contains an α1-helix thrombospondin type-1 repeat (αTSR). The αTSR contains the overlapping Th2-Region (Th2R), region II+ (RII+), Th3- Region (Th3R), and CS.T3. The glycosylphosphatidylinositol (GPI) attachment site is an anchor for the protein to the sporozoite plasma membrane (135, 136). The RTS,S vaccine contains a part of the central repeat region and the C-terminus. (B) Epitope map of CSP. All MHC class I epitopes that can be found for the CSP (circumsporozoite protein) are marked in this epitope map (epitopes longer than 20 AA were excluded). Dark blue boxes: naturally exposed volunteers; blue boxes: volunteers who were immunized with attenuated sporozoites; light blue boxes: malaria-naïve volunteers immunized with P. falciparum antigens. Red font color: response frequency of 40% or greater (when more than 4 subjects were tested). The small number next to the epitope is a link to the reference that published the epitope (you will be forwarded to the journal website by clicking onto the link). Within each box the according MHC type is marked and variants within the epitope sequence are indicated by a dash. Most epitopes that were detected within the N-terminus of CSP were found by Sedegah et al. (137). Within the central tandem repeat region no CD8+ T cell epitopes have been found so far. The C-terminus is a very immunogenic region of this protein and contains most of the epitopes.
From the 147 CD8+ T cell epitopes found in the IEDB, we included 132. These epitopes were described in 36 different publications. Additional information about the studies, like the methods that were used, the experimentally assessed HLA restriction (if available) and the study population that was tested, was included for each epitope in Supplementary Table 1. We distinguished between four different types of study populations that were tested for specific CD8+ T cells:
1. Malaria-naïve volunteers immunized with P. falciparum antigens in a vaccine trial
2. Malaria-naïve volunteers immunized with attenuated sporozoites
3. Volunteers from a malaria endemic area naturally exposed to P. falciparum
4. Volunteers with known P. falciparum infection (acute or post clinical malaria).
The epitopes are then discussed in the context of the protein structure and their role in pathogenesis, as well as in terms of overall reported immunodominance. For this, response frequency (RF) data was used. RF represents the number of respondents over the number of subjects tested, when provided by the investigator for the respective assay. It has to be taken into account that some studies only included volunteers that carry the MHC allele HLA-A*02 (141). In this case the RF is not representative for a MHC diverse population.
The assessment of the breadth and specificity of a P. falciparum-specific immune response will be a useful tool for the field and provide better knowledge of the P. falciparum-specific T cell response on an epitope level. This will likely be crucial for the understanding of the exact role of CD8+ T cells for control of disease, for vaccine design and for monitoring of the CD8+ T cell response in future vaccine trials (142). Of the more than 5,000 P. falciparum proteins, P. falciparum-specific CD8+ T cell epitopes have only been described thus far for the following nine antigens:
1. Circumsporozoite protein (CSP)
2. Thrombospondin-related anonymous protein (TRAP)
3. Apical membrane antigen 1 (AMA1)
4. Liver stage antigen 1 (LSA1)
5. Liver stage antigen 3 (LSA3)
6. Circumsporozoite-related antigen (EXP1)
7. Merozoite surface protein 1 (MSP1)
8. Sexual stage specific protein 16 (Pfs16)
9. Sporozoite threonine and asparagine-rich protein (STARP).
The majority of published MHC class I restricted CD8+ T cell epitopes was detected on CSP, with 55 known epitopes. This corresponds to 42% of all CD8+ T cell epitopes. This is followed by TRAP with 28 CD8+ T cell epitopes (21%). Nineteen epitopes on AMA1, 13 epitopes on LSA1, 7 epitopes on LSA3, and 6 epitopes on EXP1 that are specific for MHC class I/ CD8+ T cells had been detected. For the antigens MSP1, Pfs16, and STARP only one CD8+ T cell epitope, respectively, has been found (63, 69, 74, 143).
The average response frequency of all assays combined (when provided) was 23% [2.2–100% (74)] (total number of assays: 228). An average of 2.4 [1–11 (144)] subjects responded and the average size of a study population was 10.3 subjects [1–45 (74)]. Looking only at the studies that included malaria-naïve volunteers (total number of assays: 97), an average of 26.8% [5.3–100% (145)] responded in an average group of 8.2 [1–19 (145)] subjects. The study cohorts with volunteers from a malaria endemic area had an average response frequency of 16.0% [2.2–100% (74)] (total number of assays: 91). The average size of these cohorts was 15.4 study subjects [1–45 (74)]. The study group of volunteers immunized with attenuated sporozoites was a lot smaller with an average of 3 [1–4 (146)] subjects tested. The average RF was 72.6% [50–100% (147)] (total number of assays: 40).
In the following section MHC class I-restricted, P. falciparum-specific CD8+ T cell epitopes that were published at least in three different publications and/or showed a response frequency of 40% or greater or other important features will be listed according to each of these proteins. Supplementary Table 1, which gives a more comprehensive overview, lists every CD8+ T cell epitope that has been published so far.
Circumsporozoite Protein (CSP)
The circumsporozoite protein (CSP) is an important protein of the pre-erythrocytic stage of the parasite. It is the most abundant surface protein on sporozoites and has important roles in the Plasmodium life cycle (148). The function of CSP has been investigated in detail in P. berghei infection where it is essential for sporulation, gliding mobility and binding to hepatocytes (149–151). Via CSP, the sporozoites recognize heparan sulfate proteoglycans (HSPGs) which are glycoproteins on the surface of hepatocytes that allow cell interactions (152). The function of CSP in P. falciparum malaria is less well studied, but binding to hepatocytes has also been demonstrated (152).
The CSP protein is 397 amino acids in length and shows a similar structure among different Plasmodium species, suggesting conserved essential roles for each domain. The protein consists of the N-terminus, the central tandem repeat region and the C-terminus (Figure 1A) (153).
The N-terminus, and in particular the first 78 amino acids (AA), is a very well conserved part of the CSP (Supplementary Figure 1). It contains region I (RI), a five AA sequence (KLKQP) conserved in almost all Plasmodium species (154). The N-terminus is important for binding to hepatocytes (152, 155). Cleavage occurs at RI by a parasite protease upon contact with host hepatocytes and leads to a change of an “adhesive” to an “invasive” behavior (156, 157). Antibodies to RI can inhibit cleavage of CSP and therefore impair invasion in vitro, as well as in vivo (158).
The following epitopes localized within the N-terminus have been described to date (Figure 1B): The epitope MMRKLAILSV shows a high response frequency (RF) of 40% (6/15) in a study with volunteers from a malaria endemic area (159). The recognition of the peptide was demonstrated via an IFNγ-specific ELISA. It was also recognized by malaria-naïve volunteers immunized with a plasmid DNA vaccine (121, 144). 35.7% (5/14) to 36.4% (4/11) of the volunteers recognized the epitope. The overlapping epitope MMRKLAILSVSSFLFVEALF also had a high RF of 78.6% (11/14) in immunized but malaria-naïve volunteers (144, 159). The overlapping epitope ILSVSSFLFV showed response frequencies of 50% (1/2) to 71.4% (10/14) in studies with malaria-naïve volunteers who were immunized with CSP, but the epitope could also be detected in inhabitants from a malaria endemic-area [RF: 5.9% (1/17) to 8.3% (3/36)] (74, 121, 141, 144).
Within the N-terminus of CSP Sedegah et al. detected responses directed against 15 CD8+ T cell epitopes in 2013 (137). They performed an IFNγ-specific ELISPOT after adenovirus vector-based immunization of malaria-naïve volunteers with CSP and AMA1. Six to ten subjects were tested, and RF varied from 10% (1/10) to 40% (4/10).
The epitope LRKPKHKKL was detected in both, vaccinated malaria-naïve volunteers and volunteers from an endemic area and was restricted to HLA-B8 (63, 121, 145). LRKPKHKKL showed a RF of 18.2% (2/11) in a malaria-endemic study population (63).
The tandem repeat region located in the center of the protein is an immunodominant B cell region and most of the described P. falciparum-specific antibody-epitopes are located here (160, 161) (Figure 1A). This region comprises of the tetra peptide repeats NANP and NVDP. Notably, no P. falciparum-specific CD8+ T cell epitopes have been detected in this region, but several CD4+ T cell epitopes of different length have been found in this area (162–166).
Finally, the C-terminus contains a linker followed by an α-thrombospondin type-1 repeat (αTSR) domain (153, 167) (Figure 1A). Within the αTSR domain the region II-plus is located. This is an 18-amino acid sequence that also mediates adhesion of sporozoites to the heparan sulfate proteoglycans (HSPGs) (168, 169). CSP is anchored to the sporozoite plasma membrane by a glycosylphosphatidylinositol (GPI) attachment site at its C-terminus (170). Lymphoproliferative assays in naturally exposed patients have shown three immunodominant T cell epitope domains located within the C-terminus, called Th2R, Th3R, and CS.T3 (171).
Within the C-terminus of the protein, the following epitopes have been described (Figure 1B):
The epitope MPNDPNRNV was tested in malaria-naïve volunteers as well as in subjects from a malaria-endemic area. RF varied from 1/10 (10%) to 1/3 (33%) in malaria-naïve volunteers immunized with CSP (106, 121, 144, 172). The peptide is restricted by different HLA molecules: HLA-A*02:01, HLA-B7, and HLA-B35. Of note, HLA-B7 and HLA-B35 both belong to the HLA-B7 supertype (173). In volunteers from a malaria-endemic area 16.7% (1/6) to 100% (1/1) recognized the peptide (60, 63, 101).
Two epitopes are located within the Th2R region: EPSDKHIKEY, restricted to HLA-A*01:01 was only tested in malaria-naïve volunteers who were immunized with plasmid DNA or irradiated sporozoites (106, 121, 144, 174). RF varied from 10% (1/10) to 100% (2/2).
Wang et al. detected the epitope IKEYLNKIQNSLSTEWSPCS and RF was 35.7% (5/14) to 100% (8/8) (106, 144). The peptide Y/KLN/KIQ/KNSL/I is the best characterized T cell epitope and was described in 8 publications (106, 108, 119, 121, 141, 144, 145, 159, 175). It is also the most polymorph epitope within CSP (Supplementary Table 1). YLNKIQNSL was recognized in 23.5% (4/17) volunteers from a malaria-endemic area. In malaria-naïve volunteers who were immunized with a subunit vaccine RF was 13.3 (2/15) to 100% (5/5). YLKKIKNSL, YLKKIQNSL, and KLKKIKNSI were all detected in a malaria-endemic study population. RF for YLKKIKNSL was 60% (9/15) to 100% (1/1), for YLKKIQNSL 33.3% (5/15) and to KLKKIKNSI responded 41.2% (7/17) of the study population. The epitope VTCGNGIQVR is located within the region II-plus. This epitope was tested in subunit vaccine trials [RF = 10% (1/10) to 57.1% (4/7)], with irradiated sporozoites [RF = 100% (4/4)] and naturally exposed volunteers [RF = 17.8% (8/45) to 41.7% (5/12)] (74, 102, 121, 144). The epitope is restricted to HLA-A*03:01 and HLA-A*11 (121).
KPKDELDY, restricted to HLA-B*35:01 was detected in malaria-naïve [RF = 100% (1/1)] and naturally exposed subjects (RF = 12.5% (1/8) to 20% (3/15)] (23, 63, 94, 101, 121, 176). KSKDELDY had a RF of 10% (1/10) to 12.5% (1/8) in a naturally exposed population.
GLIMVLSFL was tested in subunit vaccine trials [RF = 6.7% (1/15) to 78.6% (11/14)], with irradiated sporozoites [RF = 100% (2/2)] and naturally exposed volunteers [RF = 9.1% (1/11) to 11.1% (4/36)] (74, 121, 145).
The CSP is the most frequently investigated antigen in terms of the number of different studies and number of described T cell epitopes. This is probably due to the fact that CSP was the first of the malarial proteins to be cloned, and as a result, significant efforts have been undertaken to investigate this protein (177). Ever since, CSP has been used as antigen for several vaccine trials so far (12, 178). The RTS,S vaccine contains half of the central repeat region and the entire C-terminus of CSP (Figure 1A) (12).
Thrombospondin-Related Anonymous Protein (TRAP)
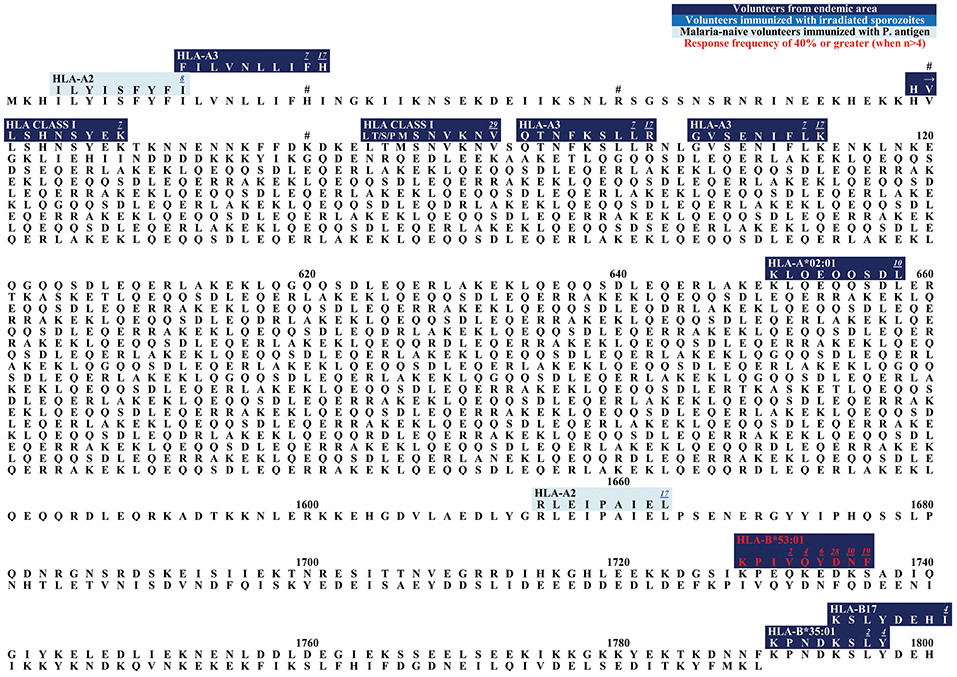
TRAP is a 559 AA long protein that induces antibody and T cell responses (179). This transmembrane protein is localized in the micronemes (specialized secretory organelles) and on the surface of sporozoites (180). It is important in the infection of hepatocytes (181, 182), has a function during the invasion of mosquito salivary glands, and TRAP also plays a role in sporozoite motility (140).
The N-terminus of TRAP contains the A domain and thrombospondin type-I repeat (TSR) which are two adhesive areas that can bind to heparan sulfate proteoglycans (HSPG) (181, 182) (Figure 2A). Upon contact with the host cell, TRAP is released from the micronemes and allows these adhesive domains to interact with the hepatocytes (181–184). Presumably, other non-HSPG-receptors also play a role for hepatocyte invasion (181).
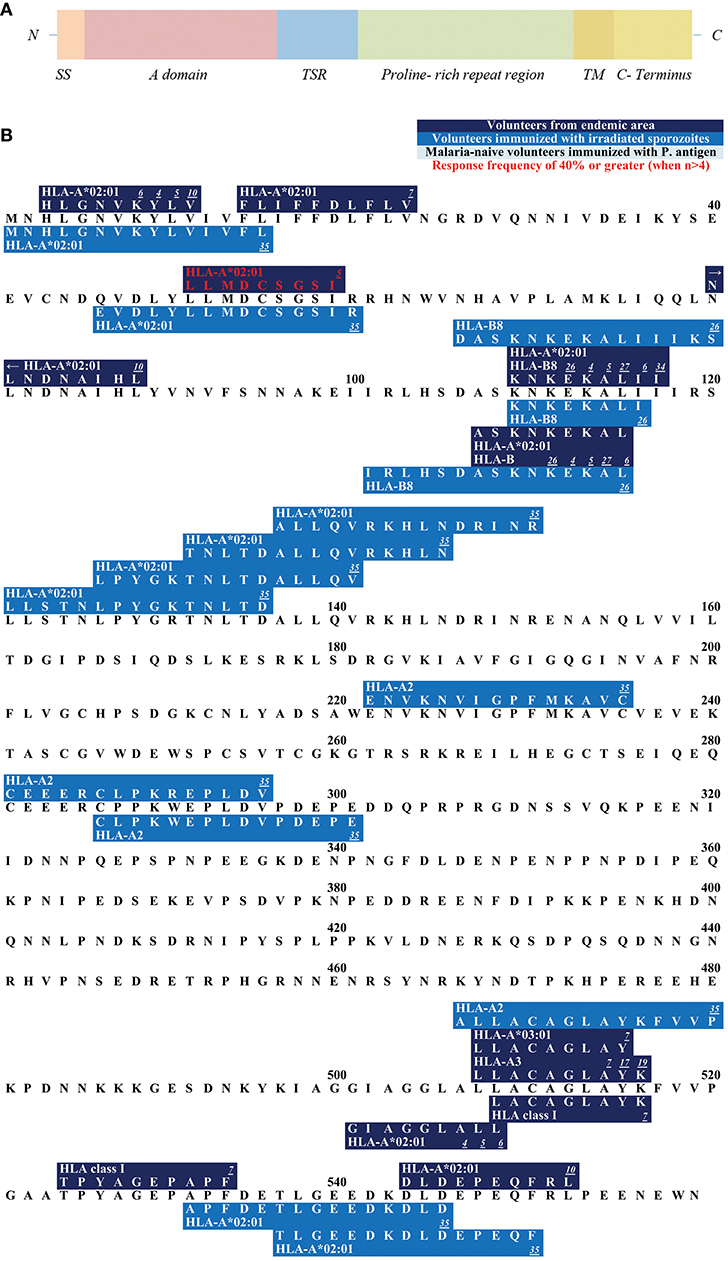
Figure 2. (A) Schematic draft of P. falciparum TRAP. This schematic draft shows the signal sequence (SS) at the N- terminus containing two adhesive domains i.e., the A domain (von Willebrand factor A- like domain) and the TSR (thrombospondin type-I repeat) domain (138, 139). This is followed by a proline-rich repeat region, a transmembrane domain (TM) and finally the acidic C-terminus located in the cytosol (140). (B) Epitope map of TRAP. Most epitopes within TRAP have been detected in studies that worked with malaria-naïve volunteers who were immunized with attenuated sporozoites.
The C-terminus of TRAP remains intracellularly located and is attached to the sporozoite motor system (169) (Figure 2A). The N-terminus and the intracellular C-terminus form a moving junction and allow gliding without cilia or flagella or major changes in the cell morphology (185, 186).
The N-terminus of TRAP contains 11 different epitopes (Figure 2B). HLGNVKYLV was tested in HLA-A*02:01 positive study participants who were naturally exposed to malaria. RF varied from 5.9% (1/17) to 100% (1/1). 11.1% (4/36) of volunteers from a malaria-endemic area and 50% (1/2) of volunteers immunized by irradiated sporozoites responded to the peptide (60, 63, 141).
LLMDCSGSI has shown to be restricted to HLA-A*02:01 and 44.4% (4/9) of volunteers naturally exposed to malaria responded to this epitope (60).
ASKNKEKAL was restricted by HLA-A*02:01 and HLA-B (60, 63, 101, 147, 187) and was recognized by 9.1% (11/1) to 33.3% (2/6) in naturally-exposed volunteers and by 100% (1/1) of volunteers immunized with irradiated sporozoites.
RF scores show that 28.6% (2/7) to 100% (1/1) of subjects from a malaria-endemic area responded to GIAGGLALL (60, 63, 101) which is located near the C-terminus. LLACAGLAYK was detected in two different study populations: in naturally-exposed volunteers [RF = 22.2% (2/9)] and in study participants who had been experimentally immunized with irradiated sporozoites [75% (3/4)] (74, 145). LLACAGLAYK was also detected by malaria-naïve subjects immunized with TRAP [5.3% (1/19)].
Apical Membrane Antigen 1 (AMA1)
AMA1 is both blood stage as well as a sporozoite stage antigen (22). It is important for invasion of erythrocytes, which makes AMA1 a target used as blood stage vaccine candidate (22, 188–191). Invasion of erythrocytes is a fast and complex process that takes a series of distinct steps. Firstly, merozoites, the invasive form of Plasmodia, attach to the host cell. Secondly, reorientation of the merozoites brings the apical end of the parasite toward the host cell. Finally, the erythrocytes are invaded (192–194).
In order to enter red blood cells (RBC), merozoites release specialized proteins out of secretory organelles called micronemes and rhoptries. AMA1 is one of the proteins, that are released to the surface of merozoites prior to invasion (188, 189, 195). A tight junction between merozoites and RBC is formed with an AMA1 and Rhoptry neck protein complex (RON) (190). The RON protein complex is initially stored within the rhoptries and then inserted into the RBC membrane (22). This junction then allows invagination of the host cell (191).
AMA1 is a 622 AA long protein. At its N-terminus, AMA1 consists of a propeptide that is removed before translocation of the protein onto the merozoite's surface (Figure 3A). This cleavage turns the precursor protein (83 kDa) into the mature 62-kDa form. The mature form has structural features of a type 1 integral membrane protein (188, 189, 195).

Figure 3. (A) Schematic draft of P. falciparum AMA1. This schematic draft shows the signal sequence (SS) toward the N- terminus followed by the propeptide which is cleaved off during the maturing process. Domain I, II, and III build the ectodomain of AMA1 with a hydrophobic cleft and a PAN fold in the tertiary structure. The transmembrane domain (TM) as well as the cytoplasmic domain (CPD) are located within the C-terminus. (B) Epitope map of AMA1. Only epitopes that were tested in malaria-naïve study cohorts immunized with P. falciparum antigens have been published for AMA1. NEVVKEEY is the only epitope within this antigen that reached a response RF (response frequency) of 40% or greater.
The ectodomain consists of three domains that is the extracellular part of AMA1 (196). Domain I and II show a high level of sequence conservation across Apicomplexa, suggesting a conserved function for these domains, whereas domain III is divergent (197). Domain I and II contain a PAN fold, a flexible loop structure with disulfide bonds. These two domains fold together and form a large hydrophobic cleft on the surface of AMA1 (198). This cleft is speculated to be a ligand-binding pocket (198). It is likely that the flexible and polymorphic loops protect the conserved hydrophobic cleft from detection by the host immune system (197).
All published epitopes on AMA1 were discovered in studies with malaria-naïve vaccines (Figure 3B).
The majority of epitopes has been described and published by Sedegah et al. (172). The malaria-naïve study population was immunized with the NMRC-M3V-Ad-PfCA vaccine, which is a combination of two separate recombinant adenovirus 5 constructs, one expressing P. falciparum CSP and the other expressing full-length P. falciparum AMA1 (both strain 3D7). In this study, the P. falciparum-specific CD8+ T cells were detected by ELISPOT. Five subjects were tested, and the response frequencies varied from 20% (1/5) to 40% (2/5). TLDEMRHFY and NEVVVKEEY are the only CD8+ T cell epitopes that were detected with tetramer-technology after immunization with the NMRC-M3V-Ad-PfCA vaccine (120). TLDEMRHFY had a response frequency of 66.7% (2/3) and NEVVVKEEY of 100% (1/1). Other epitopes were detected after immunization with DNA plasmids and adeno-viral vectors of CSP and AMA1 (117, 178).
Liver Stage Antigen 1 and 3 (LSA1, LSA3)
The function of liver stage antigen 1 (LSA1) in P. falciparum malaria is not clear. The protein, which consists of 1,162 AA (Strain: 3D7) (199), is only expressed during the liver stage and detectable in the parasitophorous vacuole (200). The potential importance of LSA1 as a vaccine candidate was suggested by the observation that the epitope KPIVQYDNF restricted by HLA-B53 is associated with resistance to severe malaria (84) (Figure 4). RF varied from 23.1% (3/13) to 50% (3/6) in study participants who were naturally exposed to malaria (23, 63, 70, 100–102).
Two epitopes restricted by the protective HLA-B53 alleles have also been found in LSA3 (Figure 5): EPKDEIVEV (103) and APFISAVAA (102). Response frequency of EPKDEIVEV was 16.7% (1/6) and APFISAVAA showed a RF of 28.6% (2/7). Both epitopes were detected in a naturally exposed study population.
Circumsporozoite-Related Protein (EXP1)
Inside the host cell, malaria parasites mature within the parasitophorous vacuole (PV) separated from the cytosol by the PV membrane (PVM) (201). The PVM serves as an interface between parasite and host with important functions like nutrient acquisition, host-cell remodeling, waste disposal, environmental sensing, and protection of the parasite (202). Within the PVM, the circumsporozoite-related protein (EXP1) is a component that is highly expressed during the blood and liver stage (203). Although this protein is likely to be essential, its exact function in the P. falciparum malaria life cycle is far from clear. EXP1 is transported out of the parasite into the parasitophorous vacuole membrane (204, 205) after invasion of host cells (206). It is then integrated in the PVM with the C-terminus exposed to the host cell cytosol and the N-terminus in the lumen of the PVM (207). EXP1 has been described to act as a glutathione S-transferase (GST) that protects the parasites from oxidative stress (208) and its enzymatic activity is inhibited by artesunate, a frontline antimalarial drug (208). EXP1 seems also to be essential for asexual development and the progression across the RBC cycle because the gene is one of the most abundantly transcribed loci during the ring and early trophozoite stages. This is the asexual growth phase inside erythrocytes (209, 210).
EXP-1 is a 162 AA long protein consisting out of a signal sequence (AA 1–23), an N-terminus (AA 23–79), a transmembrane domain (AA 79–101), and a C-terminus (AA 101–162) (211). Six epitopes within this protein have been discovered (Figure 6).
All epitopes were detected in subjects from an endemic area. Interestingly, no epitope within the N- or C-terminus has been described so far.
Merozoite Surface Protein 1 (MSP1)
In the blood phase, the invasion of human erythrocytes is mediated through different proteins. Merozoite surface proteins (MSPs) are thought to be the primary ligands responsible for the low-affinity interactions between the merozoites and the erythrocytes [reviewed in Cowman and Crabb (193)]. The most abundant MSP is the glycosylphosphatidylinositol (GPI)-anchored MSP1 (212). Merozoite surface proteins present themselves as promising vaccine candidates because of their location on the parasite surface and thus exposure to the host immune system. It has been demonstrated that antibodies against MSPs are able to decrease parasitemia in vivo (213, 214). Within MSP1, which consists of 1,720 AA (Sequence 3D7) (199), only one CD8+ T cell epitope has been described so far (Figure 7): KLKEFIPKV is restricted by HLA-A*02:01 and was detected via secreted MHC/mass spectrometry ligand presentation in monochain transgenic mice (143).
Sexual Stage-Specific Protein (Pfs16)
A small proportion of the asexual parasite population enters the sexual pathway and develops into male and female sexual forms called gametocytes, which are essential for transmission to the mosquito vector (215). These gametocytes are inside the parasitophorous vacuole membrane (PVM) and the sexual stage-specific protein (Pfs16), which is 157 AA in length (strain NF54) (199), is part of that membrane and also required for gametocyte maturation (216–219). So far, only one CD8+ T cell epitope has been detected within Pfs16 (Figure 7). MPLETQLAI had a response frequency of 100% (4/4) in subjects immunized with irradiated sporozoites. It was also tested in volunteers from a malaria endemic area [RF: 6.7% (3/45)−30% (3/10)] (69, 74).
Sporozoite Threonine and Asparagine-Rich Protein (STARP)
Sporozoite threonine and asparagine-rich protein (STARP) is expressed on the surface of sporozoites that invade hepatic cells, which suggests that it plays a role during parasite entry into the hepatic cell and infection (220). The STARP protein, which consists of 594 AA (strain: 3D7) (199), has been considered to be a potential pre-erythrocytic vaccine candidate because antibodies to this antigen could block P. falciparum sporozoite invasion of hepatocytes (221). So far, only one epitope has been located within STARP (Figure 7). MINAYLDKL is restricted by HLA-A*02:01 and *02:02. All studies that were conducted included subjects from endemic areas. Response frequency varied from 11.8% (2/17) to 75% (6/8) (60, 101, 141).
Discussion
Here we present a comprehensive overview of all human P. falciparum-specific CD8+ T cell epitopes published so far, and show their role in the Plasmodium life cycle, potential importance for vaccine development, point out areas where there are still knowledge gaps and outline future directions.
We summarize the data of 132 different P. falciparum-specific CD8+ T cell epitopes that have been described in the literature, the respective study population and response frequency, methodology used for detection, and corresponding restricting HLA-molecule.
We hope, that this review will be used as a tool for the selection of suitable epitopes for MHC tetramer synthesis in order to investigate the phenotype of P. falciparum-specific CD8+ T cells, for immunomonitoring in vaccine trials, and finally to assist researcher interested in the development of a subunit vaccine. The different and detailed epitope maps give an overview of all epitopes within the antigens (Figures 1–7). This shows the immunodominant areas of a protein as well as regions that do not elicit a CD8+ T cell immune response.
Notably, the majority of the described P. falciparum-specific CD8+ T cell epitopes were discovered in human vaccine trials conducted in malaria-naïve volunteers (121, 145, 172). It has to be taken into account that vaccination with only one Plasmodium strain [namely strain 3D7 and NF54 (137, 175, 222)] does not represent a natural malaria infection which can be caused by several strains which might affect the priming and shaping of the T cell response (93, 223). Single-strain vaccines against intense heterogeneous parasite exposure are most likely not going to be successful (15). Further vaccine trials should target more than one antigen from different life-cycle stages to elicit broader immune responses (15, 178). Since to date only a few malaria antigens have been used in vaccine studies, epitopes located within these specific regions are overrepresented in the literature. This demonstrates that the choice of antigens in vaccine trials influences our knowledge about epitopes, but our knowledge about epitopes should also be considered for future vaccine designs. Next to eliciting stronger immune responses, it is widely believed that future malaria vaccines should include a larger number of epitopes in order to show broader protection (15).
In most studies summarized in this review, only small and partly pre-selected (e.g. for HLA-A*2) study populations were tested (an average of 10.3 subjects). Notably, the response frequencies varied between the different study populations. The group that was immunized with irradiated sporozoites had the highest response frequency (72.6%). The group that was immunized with parts of the plasmodial antigens showed a response frequency of 26.8% and the volunteers from an endemic area had a RF of 16.0% only. The very high RF after immunization with irradiated sporozoites might explain why this is the most effective form of vaccination so far (15).
Only few studies used comprehensive, overlapping peptide libraries for the few antigens tested and mostly already known or in silico predicted peptides were utilized (106, 222). Many studies worked with peptide pools and single peptides could not be identified (178, 222). A number of studies focused on HLA-A*2 restricted epitopes, which is the most common expressed Caucasian HLA molecule but not that common in Africa (141, 146). Epitopes recognized in the context of a more diverse set of HLA molecules need to be defined (110).
Furthermore, there is a need for new vaccine targets because most of the known targets have been the same over the last decades (224). For example, only one CD8+ T cell epitope has been defined for an antigen from the sexual stage (Pfs16) (which could potentially prevent transmission by the mosquito) (74). Hill et al. suggest that the search for an association between the ability to respond to a particular malaria antigen or epitope with protective HLA types could be useful to identify an antigen that confers naturally acquired immunity and use it as an antigen for vaccine trials (84).
Indeed, the missing knowledge (and attention paid to) the discovery of new specific malaria epitopes combined with the overall low frequency of P. falciparum-specific T cells in the peripheral blood has led to the fact that most immunological studies rather analyzed the function and phenotype of bulk CD8+ T cells (225, 226). Studies on the phenotype and function of P. falciparum-specific T cells are largely missing. Fine mapping of the optimal length of the CD8+ T cell epitopes and investigation of their HLA restriction will allow the synthesis of suitable MHC tetramers for phenotypical analysis, most likely by employing further in vitro column tetramer-enrichment techniques (227).
The majority of studies on the breadth and specificity of the P. falciparum-specific T cell answer was published before 2000 (60, 63, 159). For this and other reasons only few studies used novel and sensitive technologies like ICS, MHC tetramer or even ELISPOT assays (117, 119, 137). Indeed, the last experimental mapping paper was published in 2016 (117, 119). This mirrors the fact that the most intense immunological investigations in the field of malaria epitope discovery took place in the late-1980s and 1990s (110).
Apart from the multicolor flow ICS analysis of vaccine induced P. falciparum-specific responses in a couple of vaccine studies, few studies have looked at other effector cytokines or the degree of multifunctionality of antiparasitical cytokines in malaria patients e.g., using multicolor flow cytometry based ICS (228, 229).
As future direction (Table 2), mass spectrometry studies e.g., of dendritic cells exposed to malaria parasites, or hepatocytes infected with liver stage antigen in order to identify further protective epitopes will potentially be another highly useful tool for epitope discovery (231).
Interestingly, many of the described P. falciparum-specific T cell responses are situated in structurally and functionally important regions. For example: the epitopes that have been described for EXP1, are mostly located within the transmembrane domain but not in the C- nor N-terminus (74, 145). Another example is the epitope LRKPKHKKL within the CSP (63, 121, 145) that overlaps with region I, a region which is conserved in almost all Plasmodium species and has an important function for hepatocyte invasion (152). Analysis of these and further interactions might be important to assess the role of T cell escape and fitness cost of certain epitopes for future vaccine design.
Supplementary Figures 1–4 depict the entropy blots of the proteins CSP, TRAP, AMA1, and EXP1. This Shannon Entropy-one calculation by the HIV Sequence database compares different sequences of the proteins found on Uniprot, altogether demonstrating the high diversity of the different malaria antigens and strains (199) but also showing the lack of antigen sequencing. For example, only 12 different sequences could be found in open resources for the P. falciparum antigen EXP1 (199). Many regions within these antigens used for the entropy map are thought to have important physiological functions but, nevertheless, are polymorph. P. falciparum-specific peptides made with a consensus sequence selected out of the sequence of different P. falciparum strains could be used for future studies, ideally such a consensus sequence should be designed specifically for individual geographic areas. Studies that investigate the specific CD8+ T cell response should also test different variants from different Plasmodium strains if possible. González et al. e.g., tested the variants TLRKPKHKKL and KLRKPKHKKL, both restricted to HLA-A*02:01, in 17 subjects (141). RF for TLRKPKHKKL was 5.9% whereas for KLRKPKHKKL it was 17.6%. This demonstrates that small changes within the AA structure of an epitope can change the binding behavior and improve the priming of CD8+ T cells in context of vaccine design.
Most studies were performed either in the setting of a protective vaccine or in healthy exposed volunteers. Future studies should look at the kinetics of the priming of the CD8+ T cell response during a natural and acute malaria infection with longitudinal long-term follow up to understand the longevity of the P. falciparum-specific T cell response. These studies will be crucial to evaluate the difference between vaccine and naturally induced P. falciparum-specific T cell responses. Epitope data in the context of clinical stages (age, parasitaemia, complicated, or uncomplicated course of disease, etc.) during an acute disease are lacking because specific CD8+ T cells have rarely been investigated in malaria patients.
Even though a T cell response is mounted against a certain epitope does not mean that this response will correlate with protection. E.g., RAS-vaccines are likely to induce CD8+ T cell responses to many sporozoite proteins, but only a subset of these will be presented by liver stage infected hepatocytes which are the target of protective immunity (232). In different murine and human immunological studies, it could also be demonstrated that only certain epitopes correlate with protection (233–235). The identification of further protective epitopes is therefore necessary. At the same time any future malaria vaccine will have to render a broad population-wide coverage since a protective epitope might be presented by only certain HLA molecules.
Additionally, studies looking at the breadth, specificity, and functionality of tissue-resident, P. falciparum-specific T cells in humans are missing. Fine needle aspiration could be introduced as a useful tool to investigate liver-resident T cells because understanding of the CD8+ T cell response from peripheral blood samples has proven to be difficult (236).
The biggest effort in vaccine development was put into the assessment of the humoral response (237). Nevertheless, has this approach not lead to an effective vaccine (e.g., the RTS,S vaccine). The cellular response should therefore not be disregarded (8). On that note, it has become clear that a strong and long lasting CD8+ T cell response is dependent on CD4+ T cells (238, 239). The P. falciparum-specific CD4+ T cell response has so far been investigated in more detail with 596 epitopes published compared to the 147 published CD8+ T cell epitopes. This may simply reflect the technical ease of certain assays (lymphoproliferation for CD4+ T cell assessment compared to cytotoxicity assays for CD8+ T cells) but it could also reflect the focus of scientific investigations (110). It is also possible that this disparity is due to biological difficulties in the detection of P. falciparum-specific CD8+ T cells (as discussed above).
Further and more comprehensive studies using different cohorts, novel and more sensitive immunological techniques and assays and comprehensive, overlapping peptide sets for a number of different malaria proteins are warranted. In other words: the lacking knowledge of potential P. falciparum-specific CD8+ T cell epitopes is hampering optimization for a malaria vaccine.
In summary, the discovery of the full breadth and exact specificities of the P. falciparum-specific immune response on an epitope level is of utmost importance for the detailed understanding of the role of CD8+ T cells for control of disease and will give us the tools to optimize vaccine design, immune monitoring of future malaria vaccine trials and to better understand naturally acquired immunity.
Data Availability
All datasets generated for this study are included in the manuscript and/or the supplementary files.
Author Contributions
JH, TJ, and JSzW: conception. JH and JSzW: first draft. All authors: important contributions and proofread.
Funding
DZIF stipend to JH, DFG grants to TJ, and JSzW, NIH grant to AS and KV.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project has been funded by the Deutsche Forschungsgemeinschaft SFB841 (TJ and JSzW) and Deutsches Zentrum für Infektionsforschung DZIF (JH, TJ, JSzW). Further funding comes from federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Department of Health and Human Services (KV, AS) under Contract Number HHSN272201200010C (IEDB).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00397/full#supplementary-material
Supplementary Table 1. P. falciparum CD8+ T cell epitopes described in humans. All P. falciparum-specific CD8+ T cell epitopes are listed in this table. The reference number (1–35) refers to the number written in each epitope box on the epitope maps. The antigen, the study population, the method used, the response frequency as well as the allele name responding to a certain epitope can be found in this table. Response frequency (RF) represents the number of respondents over the number of subjects tested, when provided by the investigator for the respective assay. ND stands for “not determined” when the investigator did not share this information in the article.
Supplementary Figure 1. Entropy map of P. falciparum CSP. The entropy value was calculated for each AA position. For this calculation 100 different P. falciparum sequences were compared. The first 78 aminoacids are well conserved within the N-terminus. The repeat region differs in the number of NANP repeats. The C-terminus is very entropy-rich. We chose the first 100 sequences available on UniProt for this Shannon Entropy-one calculation by the HIV Sequence database.
Supplementary Figure 2. Entropy map of TRAP. The entropy value was calculated for each AA position. For this calculation six different P. falciparum sequences were compared. We chose six sequences available on UniProt for this Shannon Entropy-one calculation by the HIV Sequence database.
Supplementary Figure 3. Entropy map of P. falciparum AMA1. The entropy value was calculated for each AA position. For this calculation 680 different P. falciparum sequences of AMA1 were compared. We chose all sequences available on UniProt for this Shannon Entropy-one calculation by the HIV Sequence database.
Supplementary Figure 4. Entropy map of P. falciparum EXP1. The entropy value was calculated for each AA position. For this calculation 12 different P. falciparum sequences of EXP1 were compared. We chose all sequences available on UniProt for this Shannon Entropy-one calculation by the HIVSequence database.
Abbreviations
AA, amino acids; AMA1, apical membrane antigen; APC, antigen presenting cells; CSP, circumsporozoite protein; CTL, cytotoxic T lymphocytes; ELISA, enzyme linked immunosorbent assay; EXP1, exported protein one; GLURP, glutamate-rich protein; HSPGs, heparan sulfate proteoglycans; HLA, human leukocyte antigens; ICS, intracellular staining; IEDB, The Immune Epitope Database and Analysis Resource; IFNγ, interferon gamma; Il, interleukin; LSA, liver stage antigen; MHC, major histocompatibility complex; MSP, merozoite surface protein; P, Plasmodium; Pf, Plasmodium falciparum; PfEMP, erythrocyte membrane protein; Pfs16, sexual stage specific protein; RF, response frequency; STARP, sporozoite threonine and asparagine-rich protein; TNFα, tumornecrosis factor alpha; TRAP, thrombospondin-related antigen.
Footnotes
1. ^PubMed Help [Internet]. Bethesda, MD: National Center for Biotechnology Information (US); 2005-. Available online at: https://www.ncbi.nlm.nih.gov/books/NBK3827/. Home - PubMed – NCBI; https://www.ncbi.nlm.nih.gov/pubmed/. (Accessed: August 29, 2018).
References
2. Mvumbi DM, Kayembe JM, Situakibanza H, Bobanga TL, Nsibu CN, Mvumbi GL, et al. Falciparum malaria molecular drug resistance in the Democratic Republic of Congo: a systematic review. Malar J. (2015) 14:354. doi: 10.1186/s12936-015-0892-z
3. Nussenzweig RS, Vanderberg J, Most H, Orton C. Protective immunity produced by the injection of X-irradiated Sporozoites of Plasmodium berghei. Nature. (1967) 216:160–2. doi: 10.1038/216160a0
4. Clyde D F, Most H, McCarthy V C, Vanderberg JP. Immunization of man against sporozite-induced falciparum malaria. Am J Med Sci. (1973) 266:169–77. doi: 10.1097/00000441-197309000-00002
5. Clyde D F. Immunization of man against falciparum and vivax malaria by use of attenuated sporozoites. Am J Trop Med Hyg. (1975) 24:397–401. doi: 10.4269/ajtmh.1975.24.397
6. Langhorne J, Ndungu FM, Sponaas A-M, Marsh K. Immunity to malaria: more questions than answers. Nat Immunol. (2008) 9:725–32. doi: 10.1038/ni.f.205
7. Tran TM, Li S, Doumbo S, Doumtabe D, Huang CY, Dia S, et al. An intensive longitudinal cohort study of Malian children and adults reveals no evidence of acquired immunity to Plasmodium falciparum infection. Clin Infect Dis. (2013) 57:40–7. doi: 10.1093/cid/cit174
8. Epstein JE, Tewari K, Lyke KE, Sim BK, Billingsley PF, Laurens MB, et al. Live attenuated malaria vaccine designed to protect through hepatic CD8+ T cell immunity. Science. (2011) 334:475–80. doi: 10.1126/science.1211548
9. Hoffman SL, Goh LM, Luke TC, Schneider I, Le TP, Doolan DL, et al. Protection of humans against malaria by immunization with radiation-attenuated Plasmodium falciparum sporozoites. J Infect Dis. (2002) 185:1155–64. doi: 10.1086/339409
10. Ouattara A, Mu J, Takala-Harrison S, Saye R, Sagara I, Dicko A, et al. Lack of allele-specific efficacy of a bivalent AMA1 malaria vaccine. Malar J. (2010) 9:175. doi: 10.1186/1475-2875-9-175
11. Bejon P, White MT, Olotu A, Bojang K, Lusingu JP, Salim N, et al. Efficacy of RTS,S malaria vaccines: individual-participant pooled analysis of phase 2 data. Lancet Infect Dis. (2013) 13:319–27. doi: 10.1016/S1473-3099(13)70005-7
12. Casares S, Brumeanu T-D, Richie TL. The RTS,S malaria vaccine. Vaccine. (2010) 28:4880–94. doi: 10.1016/j.vaccine.2010.05.033
13. Agnandji ST, Lell B, Fernandes JF, Abossolo BP, Methogo BG, et al. A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. N Engl J Med. (2012) 367:2284–95. doi: 10.1056/NEJMoa1208394
14. Sun P, Schwenk R, White K, Stoute JA, Cohen J, Ballou WR, et al. Protective immunity induced with malaria vaccine, RTS,S, is linked to Plasmodium falciparum circumsporozoite protein-specific CD4+ and CD8+ T cells producing IFN-gamma. J Immunol. (2003) 171:6961–7. doi: 10.4049/jimmunol.171.12.6961
15. Cockburn IA, Seder RA. Malaria prevention: from immunological concepts to effective vaccines and protective antibodies. Nat Immunol. (2018) 19:1199–211. doi: 10.1038/s41590-018-0228-6
16. Gardner MJ, Shallom SJ, Carlton JM, Salzberg SL, Nene V, Shoaibi A, et al. Sequence of Plasmodium falciparum chromosomes 2:10, 11 and 14. Nature. (2002) 419:531–4. doi: 10.1038/nature01094
17. Crompton PD, Moebius J, Portugal S, Waisberg M, Hart G, Garver LS, et al. Malaria immunity in man and mosquito: insights into unsolved mysteries of a deadly infectious disease. Annu Rev Immunol. (2014) 32:157–87. doi: 10.1146/annurev-immunol-032713-120220
18. Anum D, Kusi KA, Ganeshan H, Hollingdale MR, Ofori MF, Koram KA, et al. Measuring naturally acquired ex vivo IFN-γ responses to Plasmodium falciparum cell-traversal protein for ookinetes and sporozoites (CelTOS) in Ghanaian adults. Malar J. (2015) 14:20. doi: 10.1186/s12936-014-0539-5
19. Yamauchi LM, Coppi A, Snounou G, Sinnis P. Plasmodium sporozoites trickle out of the injection site. Cell Microbiol. (2007) 9:1215–22. doi: 10.1111/j.1462-5822.2006.00861.x
20. Mota MM, Hafalla JC, Rodriguez A. Migration through host cells activates Plasmodium sporozoites for infection. Nat Med. (2002) 8:1318–22. doi: 10.1038/nm785
21. Artavanis-Tsakonas K, Tongren JE, Riley EM. The war between the malaria parasite and the immune system: immunity, immunoregulation and immunopathology. Clin Exp Immunol. (2003) 133:145–52. doi: 10.1046/j.1365-2249.2003.02174.x
22. Riglar DT, Richard D, Wilson DW, Boyle MJ, Dekiwadia C, Turnbull L, et al. Super-resolution dissection of coordinated events during malaria parasite invasion of the human erythrocyte. Cell Host Microbe. (2011) 9:9–20. doi: 10.1016/j.chom.2010.12.003
23. Hill VS. Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature. (1992) 360:434–9. doi: 10.1038/360434a0
24. Silvestre D, Kourilsky FM, Nicolai MG, Levy JP. Presence of HLA antigens on human reticulocytes as demonstrated by electron microscopy. Nature. (1970) 228:67–8. doi: 10.1038/228067a0
25. Sagara I, Sangaré D, Dolo G, Guindo A, Sissoko M, Sogoba M, et al. A high malaria reinfection rate in children and young adults living under a low entomological inoculation rate in a periurban area of Bamako, Mali. Am J Trop Med Hyg. (2002) 66:310–3. doi: 10.4269/ajtmh.2002.66.310
26. Sokhna CS, Faye FBK, Spiegel A, Dieng H, Trape JF. Rapid reappearance of Plasmodium falciparum after drug treatment among Senegalese adults exposed to moderate seasonal transmission. Am J Trop Med Hyg. (2001) 65:167–70. doi: 10.4269/ajtmh.2001.65.167
27. Owusu-Agyei S, Koram KA, Baird JK, Utz GC, Binka FN, Nkrumah FK, et al. Incidence of symptomatic and asymptomatic Plasmodium falciparum infection following curative therapy in adult residents of northern Ghana. Am J Trop Med Hyg. (2001) 65:197–203. doi: 10.4269/ajtmh.2001.65.197
28. Hafalla JCR, Sano G, Carvalho LH, Morrot A, Zavala F. Short-term antigen presentation and single clonal burst limit the magnitude of the CD8(+) T cell responses to malaria liver stages. Proc Natl Acad Sci USA. (2002) 99:11819–24. doi: 10.1073/pnas.182189999
29. Stevenson MM, Riley EM. Innate immunity to malaria. Nat Rev Immunol. (2004) 4:169–80. doi: 10.1038/nri1311
30. Doba-o C, Moncunill G. Naturally Acquired Immunity (NAI). In Encyclopedia of Malaria. New York, NY: Springer New York (2018). p. 1–15. doi: 10.1007/978-1-4614-8757-9_131-1
31. Schofield L, Villaquiran J, Ferreira A, Schellekens H, Nussenzweig R, Nussenzweig V. γ Interferon, CD8+ T cells and antibodies required for immunity to malaria sporozoites. Nature. (1987) 330:664–6. doi: 10.1038/330664a0
32. Weiss WR, Sedegah M, Beaudoin RL, Miller LH, Good MF. CD8+ T cells (cytotoxic/suppressors) are required for protection in mice immunized with malaria sporozoites. Proc Natl Acad Sci USA. (1988) 85:573–6. doi: 10.1073/pnas.85.2.573
33. Weiss WR, Jiang CG. Protective CD8+ T lymphocytes in primates immunized with malaria sporozoites. PLoS ONE. (2012) 7:e31247. doi: 10.1371/journal.pone.0031247
34. Doolan DL, Hoffman SL. The complexity of protective immunity against liver-stage malaria. J Immunol. (2000) 165:1453–62. doi: 10.4049/jimmunol.165.3.1453
35. Romero P, Maryanski JL, Corradin G, Nussenzweig RS, Nussenzweig V, Zavala F. Cloned cytotoxic T cells recognize an epitope in the circumsporozoite protein and protect against malaria. Nature. (1989) 341:323–6. doi: 10.1038/341323a0
36. Rodrigues MM, Cordey AS, Arreaza G, Corradin G, Romero P, Maryanski JL, et al. CD8+ cytolytic T cell clones derived against the Plasmodium yoelii circumsporozoite protein protect against malaria. Int Immunol. (1991) 3:579–85. doi: 10.1093/intimm/3.6.579
37. Weiss WR, Berzofsky JA, Houghten RA, Sedegah M, Hollindale M, Hoffman SL. A T cell clone directed at the circumsporozoite protein which protects mice against both Plasmodium yoelii and Plasmodium berghei. J Immunol. (1992) 149:2103–9.
38. Franke ED, Corradin G, Hoffman SL. Induction of protective CTL responses against the Plasmodium yoelii circumsporozoite protein by immunization with peptides. J Immunol. (1997) 159:3424–33.
39. Schneider J, Gilbert SC, Blanchard TJ, Hanke T, Robson KJ, Hannan CM, et al. Enhanced immunogenicity for CD8+ T cell induction and complete protective efficacy of malaria DNA vaccination by boosting with modified vaccinia virus Ankara. Nat Med. (1998) 4:397–402. doi: 10.1038/nm0498-397
40. Hafalla JCR, Bauza K, Friesen J, Gonzalez-Aseguinolaza G, Hill AVS, Matuschewski K. Identification of targets of CD8+ T cell responses to malaria liver stages by genome-wide epitope profiling. PLoS Pathog. (2013) 9:e1003303. doi: 10.1371/journal.ppat.1003303
41. Pichugin A, Zarling S, Perazzo L, Duffy PE, Ploegh HL, Krzych U. Identification of a novel CD8 T cell epitope derived from plasmodium berghei protective liver-stage antigen. Front Immunol. (2018) 9:91. doi: 10.3389/fimmu.2018.00091
42. Chan JA, Howell KB, Reiling L, Ataide R, Mackintosh CL, Fowkes FJ, et al. Targets of antibodies against Plasmodium falciparum-infected erythrocytes in malaria immunity. J Clin Invest. (2012) 122:3227–38. doi: 10.1172/JCI62182
43. Bull PC, Lowe BS, Kortok M, Molyneux CS, Newbold CI, Marsh K. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat Med. (1998) 4:358–60. doi: 10.1038/nm0398-358
44. Pinzon-Charry A, McPhun V, Kienzle V, Hirunpetcharat C, Engwerda C, McCarthy J, et al. Low doses of killed parasite in CpG elicit vigorous CD4+ T cell responses against blood-stage malaria in mice. J Clin Invest. (2010) 120:2967–78. doi: 10.1172/JCI39222
45. Süss G, Eichmann K, Kury E, Linke A, Langhorne J. Roles of CD4- and CD8-bearing T lymphocytes in the immune response to the erythrocytic stages of Plasmodium chabaudi. Infect Immun. (1988) 56:3081–8.
46. Imai T, Shen J, Chou B, Duan X, Tu L, Tetsutani K, et al. Involvement of CD8 + T cells in protective immunity against murine blood-stage infection with Plasmodium yoelii 17XL strain. Eur J Immunol. (2010) 40:1053–61. doi: 10.1002/eji.200939525
47. Schmidt NW, Butler NS, Badovinac VP, Harty JT. Extreme CD8 T cell requirements for anti-malarial liver-stage immunity following immunization with radiation attenuated sporozoites. PLoS Pathog. (2010) 6:e1000998. doi: 10.1371/journal.ppat.1000998
48. Van Braeckel-Budimir N, Harty JT. CD8 T-cell-mediated protection against liver-stage malaria: lessons from a mouse model. Front Microbiol. (2014) 5:272. doi: 10.3389/fmicb.2014.00272
49. Kimura K, Kimura D, Matsushima Y, Miyakoda M, Honma K, Yuda M, et al. CD8+ T cells specific for a malaria cytoplasmic antigen form clusters around infected hepatocytes and are protective at the liver stage of infection. Infect Immun. (2013) 81:3825–34. doi: 10.1128/IAI.00570-13
50. Cockburn IA, Amino R, Kelemen RK, Kuo SC, Tse SW, Radtke A, et al. In vivo imaging of CD8+ T cell-mediated elimination of malaria liver stages. Proc Natl Acad Sci USA. (2013) 110:9090–5. doi: 10.1073/pnas.1303858110
51. Guebre-Xabier M, Schwenk R, Krzych U. Memory phenotype CD8+ T cells persist in livers of mice protected against malaria by immunization with attenuated Plasmodium berghei sporozoites. Eur J Immunol. (1999) 29:3978–86. doi: 10.1002/(SICI)1521-4141(199912)29:12<3978::AID-IMMU3978>3.0.CO;2-0
52. Krzych U, Schwenk R, Guebre-Xabier M, Sun P, Palmer D, White K, et al. The role of intrahepatic lymphocytes in mediating protective immunity induced by attenuated Plasmodium berghei sporozoites. Immunol Rev. (2000) 174:123–34. doi: 10.1034/j.1600-0528.2002.00013h.x
53. Frevert U, Krzych U. Plasmodium cellular effector mechanisms and the hepatic microenvironment. Front Microbiol. (2015) 6:482. doi: 10.3389/fmicb.2015.00482
54. Roestenberg M, McCall M, Hopman J, Wiersma J, Luty AJ, van Gemert GJ, et al. Protection against a malaria challenge by sporozoite inoculation. N Engl J Med. (2009) 361:468–77. doi: 10.1056/NEJMoa0805832
55. Seder RA. Protection against malaria by intravenous immunization with a non-replicating sporozoite vaccine. Science. (2013) 341:1359–65. doi: 10.1126/science.1241800
56. Doolan DL, Saul AJ, Good MF. Geographically restricted heterogeneity of the Plasmodium falciparum circumsporozoite protein: relevance for vaccine development. Infect Immun. (1992) 60:675–82.
57. Hill AVS. HLA Associations with Malaria in Africa: Some Implications for MHC Evolution. In Molecular Evolution of the Major Histocompatibility Complex. Berlin, Heidelberg: Springer Berlin Heidelberg (1991). p. 403–20. doi: 10.1007/978-3-642-84622-9_33
58. White KL, Snyder HL, Krzych U. MHC class I-dependent presentation of exoerythrocytic antigens to CD8+ T lymphocytes is required for protective immunity against Plasmodium berghei. J Immunol. (1996) 156:3374–81.
59. Doolan DL, Houghten RA, Good MF. Location of human cytotoxic T cell epitopes within a polymorphic domain of the Plasmodium falciparum circumsporozoite protein. Int Immunol. (1991) 3:511–6. doi: 10.1093/intimm/3.6.511
60. Lalvani A, Hurt N, Aidoo M, Kibatala P, Tanner M, Hill AVS. Cytotoxic T lymphocytes to Plasmodium falciparum epitopes in an area of intense and perennial transmission in Tanzania. Eur J Immunol. (1996) 26:773–9. doi: 10.1002/eji.1830260408
61. Malik A, Egan JE, Houghten AR, Sadoff JC, Hoffman SL. Human cytotoxic T lymphocytes against the Plasmodium falciparum circumsporozoite protein. Proc Natl Acad Sci USA. (1991) 88:3300–04. doi: 10.1073/pnas.88.8.3300
62. Sedegah M. Naturally acquired CD8+ cytotoxic T lymphocytes against the Plasmodium falciparum circumsporozoite protein. J Immunol. (1992) 149:966–71.
63. Aidoo M. Identification of conserved antigenic components for a cytotoxic T lymphocyte-inducing vaccine against malaria. Lancet. (1995) 345:1003–7. doi: 10.1016/S0140-6736(95)90754-8
64. Belnoue E, Kayibanda M, Vigario AM, Deschemin JC, van Rooijen N, Viguier M, et al. On the pathogenic role of brain-sequestered CD8+ T cells in experimental cerebral malaria. J Immunol. (2002) 169:6369–75. doi: 10.4049/jimmunol.169.11.6369
65. Chen DH, Tigelaar RE, Weinbaum FI. Immunity to sporozoite-induced malaria infeciton in mice. I. The effect of immunization of T and B cell-deficient mice. J Immunol. (1977) 118:1322–7.
66. Hoffman SL, Oster CN, Mason C, Beier JC, Sherwood JA, Ballou WR, et al. Human lymphocyte proliferative response to a sporozoite T cell epitope correlates with resistance to falciparum malaria. J Immunol. (1989) 142:1299–303.
67. Verhave JP, Strickland GT, Jaffe HA, Ahmed A. Studies on the transfer of protective immunity with lymphoid cells from mice immune to malaria sporozoites. J Immunol. (1978) 121:1031–3.
68. Lalvani A, Brookes R, Hambleton S, Britton WJV, Hill A, McMichael AJ. Rapid effector function in CD8+ memory T cells. J Exp Med. (1997) 186:859–65. doi: 10.1084/jem.186.6.859
69. Ong'echa JM, Lal AA, Terlouw DJ, Ter Kuile FO, Kariuki SK, Udhayakumar V, et al. Association of interferon-γ responses to pre-erythrocytic stage vaccine candidate antigens of Plasmodium falciparum in young Kenyan children with improved hemoglobin levels: XV. Asembo bay cohort project. Am J Trop Med Hyg. (2003) 68:590–7. doi: 10.4269/ajtmh.2003.68.590
70. Luty AJ, Lell B, Schmidt-Ott R, Lehman LG, Luckner D, Greve B, et al. Interferon-γ responses are associated with resistance to reinfection with Plasmodium falciparum in young African children. J Infect Dis. (1999) 179:980–8. doi: 10.1086/314689
71. John CC, Moormann AM, Sumba PO, Ofulla AV, Pregibon DC, Kazura JW. Gamma interferon responses to Plasmodium falciparum liver-stage antigen 1 and thrombospondin-related adhesive protein and their relationship to age, transmission intensity, and protection against malaria. Infect Immun. (2004) 72:5135–42. doi: 10.1128/IAI.72.9.5135-5142.2004
72. Bucci K, Kastens W, Hollingdale MR, Shankar A, Alpers MP, King CL, et al. Influence of age and HLA type on interferon-gamma (IFN-gamma) responses to a naturally occurring polymorphic epitope of Plasmodium falciparum liver stage antigen-1 (LSA-1). Clin Exp Immunol. (2000) 122:94–100. doi: 10.1046/j.1365-2249.2000.01346.x
73. Chelimo K, Sumba PO, Kazura JW, Ofula AV, John CC. Interferon-gamma responses to Plasmodium falciparum liver-stage antigen-1 and merozoite-surface protein-1 increase with age in children in a malaria holoendemic area of western Kenya. Malar J. (2003) 2:37. doi: 10.1186/1475-2875-2-37
74. Doolan DL, Hoffman SL, Southwood S, Wentworth PA, Sidney J, Chesnut RW, et al. Degenerate cytotoxic T cell epitopes from P. falciparum restricted by multiple HLA-A and HLA-B supertype alleles. Immunity. (1997) 7:97–112. doi: 10.1016/S1074-7613(00)80513-0
75. Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci USA. (2003) 100:9952–7. doi: 10.1073/pnas.1633254100
76. Scholzen A, Sauerwein RW. Immune activation and induction of memory: Lessons learned from controlled human malaria infection with Plasmodium falciparum. Parasitology. (2016) 143:224–35. doi: 10.1017/S0031182015000761
77. Chakravarty S, Cockburn IA, Kuk S, Overstreet MG, Sacci JB, Zavala F. CD8+ T lymphocytes protective against malaria liver stages are primed in skin-draining lymph nodes. Nat Med. (2007) 13:1035–41. doi: 10.1038/nm1628
78. Mu J, Awadalla P, Duan J, McGee KM, Joy DA, McVean GA, et al. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. (2005) 3:e335. doi: 10.1371/journal.pbio.0030335
79. Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. (2007) 39:113–9. doi: 10.1038/ng1930
80. Jiang H, Yi M, Mu J, Zhang L, Ivens A, Klimczak LJ, et al. Detection of genome-wide polymorphisms in the AT-rich Plasmodium falciparum genome using a high-density microarray. BMC Genom. (2008) 9:398. doi: 10.1186/1471-2164-9-398
81. Oca-a-Morgner C, Mota MM, Rodriguez A. Malaria blood stage suppression of liver stage immunity by dendritic cells. J Exp Med. (2003) 197:143–51. doi: 10.1084/jem.20021072
82. Keitany GJ, Kim KS, Krishnamurty AT, Hondowicz BD, Hahn WO, Dambrauskas N, et al. Blood stage malaria disrupts humoral immunity to the pre-erythrocytic stage circumsporozoite protein. Cell Rep. (2016) 17:3193–205. doi: 10.1016/j.celrep.2016.11.060
83. Mackroth MS, Abel A, Steeg C, Schulze Zur Wiesch J, Jacobs T. Acute malaria induces PD1 + CTLA4 + effector T cells with cell-extrinsic suppressor function. PLoS Pathog. (2016) 12:e1005909. doi: 10.1371/journal.ppat.1005909
84. Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, et al. Common West African HLA antigens are associated with protection from severe malaria. Nature. (1991) 352:595–600. doi: 10.1038/352595a0
85. Francis T. Influenza: the new acquayantance. Ann Intern Med. (1953) 39:203–21. doi: 10.7326/0003-4819-39-2-203
86. Fazekas de St Groth, Webster RG. Disquisitions of original antigenic sin. I. Evidence in man. J Exp Med. (1966) 124:331–45. doi: 10.1084/jem.124.3.331
87. Fazekas de St Groth, Webster RG. Disquisitions on original antigenic sin. II. Proof in lower creatures. J Exp Med. (1966) 124:347–61. doi: 10.1084/jem.124.3.347
88. Klenerman P, Zinkernagel RM. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature. (1998) 394:482–5. doi: 10.1038/28860
89. Ciurea A, Klenerman P, Hunziker L, Horvath E, Senn BM, Ochsenbein AF, et al. Viral persistence in vivo through selection of neutralizing antibody-escape variants. Proc Natl Acad Sci. (2000) 97:2749–54. doi: 10.1073/pnas.040558797
90. Bertoletti A, Sette A, Chisari FV, Penna A, Levrero M, De Carli M, et al. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature. (1994) 369:407–10. doi: 10.1038/369407a0
91. Klenerman P. Cytotoxic T-cell activity antagonized by naturally occurring HIV-1 Gag variants. Nature. (1994) 369:403–7. doi: 10.1038/369403a0
92. Conway DJ, Greenwood BM, McBride JS. The epidemiology of multiple-clone Plasmodium falciparum infections in Gambian patients. Parasitology. (1991) 103(Pt 1):1–6. doi: 10.1017/S0031182000059217
93. Zhong D, Afrane Y, Githeko A, Yang Z, Cui L, Menge DM, et al. Plasmodium falciparum genetic diversity in Western Kenya highlands. Am J Trop Med Hyg. (2007) 77:1043–50. doi: 10.4269/ajtmh.2007.77.1043
94. Gilbert SC, Plebanski M, Gupta S, Morris J, Cox M, Aidoo M, et al. Association of malaria parasite population structure, HLA, and immunological antagonism. Science. (1998) 279:1173–7. doi: 10.1126/science.279.5354.1173
95. Schulze Zur Wiesch J, Lauer GM, Timm J, Kuntzen T, Neukamm M, Berical A, et al. Immunologic evidence for lack of heterologous protection following resolution of HCV in patients with non genotype 1 infection. Blood. (2007) 110:1559–69. doi: 10.1182/blood-2007-01-069583
96. Walther M, Tongren JE, Andrews L, Korbel D, King E, Fletcher H, et al. Upregulation of TGF-β, FOXP3, and CD4+CD25+ Regulatory T cells correlates with more rapid parasite growth in human malaria infection. Immunity. (2005) 23:287–96. doi: 10.1016/j.immuni.2005.08.006
97. Horne-Debets JM, Faleiro R, Karunarathne DS, Liu XQ, Lineburg KE, Poh CM, et al. PD-1 dependent exhaustion of CD8+ T cells drives chronic malaria. Cell Rep. (2013) 5:5. doi: 10.1016/j.celrep.2013.11.002
98. Belachew EB. Immune response and evasion mechanisms of Plasmodium falciparum parasites. J Immunol Res. (2018) 2018:1–6. doi: 10.1155/2018/6529681
99. Steers N, Schwenk R, Bacon DJ, Berenzon D, Williams J, Krzych U. The immune status of Kupffer cells profoundly influences their responses to infectious Plasmodium berghei sporozoites. Eur J Immunol. (2005) 35:2335–46. doi: 10.1002/eji.200425680
100. May J, Lell B, Luty AJF, Meyer CG, Kremsner PG. HLA-DQB1*0501–restricted Th1 type immune responses to Plasmodium falciparum liver stage antigen 1 protect against malaria anemia and reinfections. J Infect Dis. (2001) 183:168–72. doi: 10.1086/317642
101. Plebanski M, Aidoo M, Whittle HCV, Hill A. Precursor frequency analysis of cytotoxic T lymphocytes to pre-erythrocytic antigens of Plasmodium falciparum in West Africa. J Immunol. (1997) 158:2849–55.
102. Dodoo D, Hollingdale MR, Anum D, Koram KA, Gyan B, Akanmori BD, et al. Measuring naturally acquired immune responses to candidate malaria vaccine antigens in Ghanaian adults. Malar J. (2011) 10:168. doi: 10.1186/1475-2875-10-168
103. Aidoo M, Lalvani A, Gilbert SC, Hu JT, Daubersies P, Hurt N, et al. Cytotoxic T-lymphocyte epitopes for HLA-B53 and other HLA types in the malaria vaccine candidate liver-stage antigen 3. Infect Immun. (2000) 68:227–32. doi: 10.1128/IAI.68.1.227-232.2000
104. Sinha AA, Lopez MT, McDevitt HO. Autoimmune diseases: the failure of self tolerance. Science. (1990) 248:1380–8. doi: 10.1126/science.1972595
105. The Allele Frequency Net Database. Available online at: http://allelefrequencies.net/. (Accessed: November 14, 2018).
106. Wang R, Epstein J, Charoenvit Y, Baraceros FM, Rahardjo N, Gay T, et al. Induction in humans of CD8+ and CD4+ T cell and antibody responses by sequential immunization with malaria DNA and recombinant protein. J Immunol. (2004) 172:5561–9. doi: 10.4049/jimmunol.172.9.5561
107. Meierhoff G, Ott PA, Lehmann PV, Schloot NC. Cytokine detection by ELISPOT: relevance for immunological studies in type 1 diabetes. Diabetes Metab Res Rev. (2002) 18:367–80. doi: 10.1002/dmrr.320
108. Prato S, Fleming J, Schmidt CW, Corradin G, Lopez JA. Cross-presentation of a human malaria CTL epitope is conformation dependent. Mol Immunol. (2006) 43:2031–6. doi: 10.1016/j.molimm.2005.12.014
110. Vaughan K, Blythe M, Greenbaum J, Zhang Q, Peters B, Doolan DL, et al. Meta-analysis of immune epitope data for all Plasmodia: overview and applications for malarial immunobiology and vaccine-related issues. Parasite Immunol. (2009) 31:78–97. doi: 10.1111/j.1365-3024.2008.01077.x
111. Maurer HR. Potential pitfalls of [3 H]Thymidine techniques to measure cell proliferation. Cell Prolif . (1981) 14:111–20. doi: 10.1111/j.1365-2184.1981.tb00516.x
112. Czerkinsky CC, Nilsson LA, Nygren H, Ouchterlony O, Tarkowski A. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. J Immunol Methods. (1983) 65:109–21. doi: 10.1016/0022-1759(83)90308-3
113. Streeck H, Frahm N, Walker BD. The role of IFN-γ Elispot assay in HIV vaccine research. Nat Protoc. (2009) 4:461–9. doi: 10.1038/nprot.2009.7
114. Flanagan KL. Broadly distributed T cell reactivity, with no immunodominant loci, to the pre-erythrocytic antigen thrombospondin-related adhesive protein of Plasmodium falciparum in West Africans. Eur J Immunol. (1999) 29:1943–54. doi: 10.1002/(SICI)1521-4141(199906)29:06<1943::AID-IMMU1943>3.0.CO;2-1
115. Ranieri E, Popescu I, Gigante M. CTL ELISPOT assay. Methods Mol Biol. (2014) 1386:75–86. doi: 10.1007/978-1-4939-1158-5_6
116. Brice GT, Graber NL, Hoffman SL, Doolan DL. Expression of the chemokine MIG is a sensitive and predictive marker for antigen-specific, genetically restricted IFN-γ production and IFN-γ-secreting cells. J Immunol Methods. (2001) 257:55–69. doi: 10.1016/S0022-1759(01)00446-X
117. Sedegah M. Vaccine strain-specificity of protective HLA-restricted class 1 P. falciparum epitopes. PLoS ONE. (2016) 11:1–14. doi: 10.1371/journal.pone.0163026
118. Bacher P, Scheffold A. Flow-cytometric analysis of rare. Cytom. (2013) 83A:692–701. doi: 10.1002/cyto.a.22317
119. Li X, Huang J, Zhang M, Funakoshi R, Sheetij D, Spaccapelo R, et al. Human CD8+ T cells mediate protective immunity induced by a human malaria vaccine in human immune system mice. Vaccine. (2016) 34:4501–6. doi: 10.1016/j.vaccine.2016.08.006
120. Schwenk R, Banania G, Epstein J, Kim Y, Peters B, Belmonte M, et al. Ex vivo tetramer staining and cell surface phenotyping for early activation markers CD38 and HLA-DR to enumerate and characterize malaria antigen-specific CD8+ T-cells induced in human volunteers immunized with a Plasmodium falciparum adenovirus-vectored. Malar J. (2013) 12:376. doi: 10.1186/1475-2875-12-376
121. Wang R, Doolan DL, Le TP, Hedstrom RC, Coonan KM, Charoenvit Y, et al. Induction of antigen-specific cytotoxic T lymphocytes in humans by a malaria DNA vaccine. Science. (1998) 282:476–80. doi: 10.1126/science.282.5388.476
122. Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. (2015) 43:D405–12. doi: 10.1093/nar/gku938
123. Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. (2005) 3:e91. doi: 10.1371/journal.pbio.0030091
124. Paul S, Sidney J, Sette A, Peters B. TepiTool: a pipeline for computational prediction of T cell epitope candidates. Curr Protoc Immunol. (2016) 114:18.19.1–18.19.24. doi: 10.1002/cpim.12
125. Calis JJ, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, Sette A, et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol. (2013) 9:e1003266. doi: 10.1371/journal.pcbi.1003266
126. Paul S, Paul S, Arlehamn CSL, Scriba TJ, Dillon MBC, Oseroff C, Hinz D, et al. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J Immunol Methods. (2015) 422:28–34. doi: 10.1016/j.jim.2015.03.022
127. Bui HH, Sidney J, Li W, Fusseder N, Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinformatics. (2007) 8:361. doi: 10.1186/1471-2105-8-361
128. PlasmoDB : The Plasmodium Genomics Resource. Available online at: http://plasmodb.org/plasmo/. (Accessed: November 13, 2018).
129. PvaxDB | SCFBio. Available online at: https://www.scfbio-iitd.res.in/PvaxDB. (Accessed: November 13, 2018).
130. RMgmDB - Rodent Malaria Genetically Modified Parasites - P. berghei. Available online at: https://www.pberghei.eu/index.php. (Accessed: November 13, 2018).
131. Malaria Data. Available online at: https://www.ebi.ac.uk/chembl/malaria/. (Accessed: November 13, 2018).
132. Welcome, at MalarImDB | malarimdb,.org. Available online at: http://www.malarimdb.org/. (Accessed: November 13, 2018).
133. Welcome to the Malaria Atlas Project - MAP. Available online at: https://map.ox.ac.uk/. (Accessed: 13-Nov-2018).
134. Contact us | MalariaGEN. Available online at: https://www.malariagen.net. (Accessed: November 13, 2018).
135. Scally SW, Murugan R, Bosch A, Triller G, Costa G, Mordmüller B, et al. Rare PfCSP C-terminal antibodies induced by live sporozoite vaccination are ineffective against malaria infection. J Exp Med. (2018) 215:63–75. doi: 10.1084/jem.20170869
136. Schwenk R, Lumsden JM, Rein LE, Juompan L, Kester KE, Heppner DG, et al. Immunization with the RTS,S/AS malaria vaccine induces IFN-γ +CD4 T cells that recognize only discrete regions of the circumsporozoite protein and these specificities are maintained following booster immunizations and challenge. Vaccine. (2011) 29:8847–54. doi: 10.1016/j.vaccine.2011.09.098
137. Sedegah M, Kim Y, Ganeshan H, Huang J, Belmonte M, Abot E, et al. Identification of minimal human MHC-restricted CD8+ T-cell epitopes within the Plasmodium falciparum circumsporozoite protein (CSP). Malar J. (2013) 12:185. doi: 10.1186/1475-2875-12-185
138. Tucker RP. The thrombospondin type 1 repeat superfamily. Int J Biochem Cell Biol. (2004) 36:969–74. doi: 10.1016/j.biocel.2003.12.011
139. Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell. (2002) 13:3369–87. doi: 10.1091/mbc.e02-05-0259
140. Pihlajamaa T, Kajander T, Knuuti J, Horkka K, Sharma A, Permi P. Structure of Plasmodium falciparum TRAP (thrombospondin-related anonymous protein) A domain highlights distinct features in apicomplexan von Willebrand factor A homologues. Biochem J. (2013) 450:469–76. doi: 10.1042/BJ20121058
141. Gonzalez JM. HLA-A*0201 restricted CD8 ‡ T-lymphocyte responses to malaria : identification of new Plasmodium falciparum epitopes by IFN-g ELISPOT. Parasite Immunol. (2000) 22:501–14. doi: 10.1046/j.1365-3024.2000.00331.x
142. Sztein MB. Is a human CD8 T-cell vaccine possible, and if so, what would it take? Cold Spring Harb Perspect Biol. (2017) 2017:a029546. doi: 10.1101/cshperspect.a029546
143. Carralot J-P, Lemmel C, Stevanovic S, Pascolo S. Mass spectrometric identification of an HLA-A*0201 epitope from Plasmodium falciparum MSP-1. Int Immunol. (2008) 20:1451–6. doi: 10.1093/intimm/dxn102
144. Wang R, Epstein J, Baraceros FM, Gorak EJ, Charoenvit Y, Carucci DJ, et al. Induction of CD4+ T cell-dependent CD8+ type 1 responses in humans by a malaria DNA vaccine. Proc Natl Acad Sci USA. (2001) 98:10817–822. doi: 10.1073/pnas.181123498
145. Wang R, Richie TL, Baraceros MF, Rahardjo N, Gay T, Banania JG, et al. Boosting of DNA vaccine-elicited gamma interferon responses in humans by exposure to malaria parasites boosting of DNA vaccine-elicited gamma interferon responses in humans by exposure to malaria parasites. Infect Immun. (2005) 73:2863–72. doi: 10.1128/IAI.73.5.2863-2872.2005
146. Wizel B, Houghten R, Church P, Tine JA, Lanar DE, Gordon DM, et al. HLA-A2-restricted cytotoxic T lymphocyte responses to multiple Plasmodium falciparum sporozoite surface protein 2 epitopes in sporozoite-immunized volunteers. J Immunol. (1995) 155:766–75.
147. Wizel B, Houghten RA, Parker KC, Coligan JE, Church P, Gordon DM, et al. Irradiated sporozoite vaccine induces HLA-B8-restricted cytotoxic T lymphocyte responses against two overlapping epitopes of the Plasmodium falciparum sporozoite surface protein 2. J Exp Med. (1995) 182:1435–45. doi: 10.1084/jem.182.5.1435
148. Potocnjak P, Zavala F, Nussenzweig R, Nussenzweig V. Inhibition of idiotype–anti-idiotype interaction for detection of a parasite antigen: a new immunoassay. Science. (1982) 215:1637–9. doi: 10.1126/science.6122269
149. Triller G, Scally SW, Costa G, Pissarev M, Kreschel C, Bosch A, et al. Natural parasite exposure induces protective human anti-malarial antibodies. Immunity. (2017) 47:1197–209.e10. doi: 10.1016/j.immuni.2017.11.007
150. Ménard R, Sultan AA, Cortes C, Altszuler R, van Dijk MR, Janse CJ, et al. Circumsporozoite protein is required for development of malaria sporozoites in mosquitoes. Nature. (1997) 385:336–40. doi: 10.1038/385336a0
151. Thathy V, Fujioka H, Gantt S, Nussenzweig R, Nussenzweig V, Ménard R. Levels of circumsporozoite protein in the Plasmodium oocyst determine sporozoite morphology. EMBO J. (2002) 21:1586–96. doi: 10.1093/emboj/21.7.1586
152. Cerami C, Frevert U, Sinnis P, Takacs B, Clavijo P, Santos MJ, et al. The basolateral domain of the hepatocyte plasma membrane bears receptors for the circumsporozoite protein of Plasmodium falciparum sporozoites. Cell. (1992) 70:1021–33. doi: 10.1016/0092-8674(92)90251-7
153. Doud MB, Koksal AC, Mi LZ, Song G, Lu C, Springer TA. Unexpected fold in the circumsporozoite protein target of malaria vaccines. Proc Natl Acad Sci USA. (2012) 109:7817–22. doi: 10.1073/pnas.1205737109
154. Vargas-Serrato E, Corredor V, Galinski MR. Phylogenetic analysis of CSP and MSP-9 gene sequences demonstrates the close relationship of Plasmodium coatneyi to Plasmodium knowlesi. Infect Genet Evol. (2003) 3:67–73. doi: 10.1016/S1567-1348(03)00007-8
155. Rathore D, Sacci JB, de la Vega P, McCutchan TF. Binding and invasion of liver cells by Plasmodium falciparum sporozoites. J Biol Chem. (2002) 277:7092–8. doi: 10.1074/jbc.M106862200
156. Coppi A, Pinzon-Ortiz C, Hutter C, Sinnis P. The Plasmodium circumsporozoite protein is proteolytically processed during cell invasion. J Exp Med. (2005) 201:27–33. doi: 10.1084/jem.20040989
157. Coppi A, Natarajan R, Pradel G, Bennett BL, James ER, Roggero MA, et al. The malaria circumsporozoite protein has two functional domains, each with distinct roles as sporozoites journey from mosquito to mammalian host. J Exp Med. (2011) 208:341–56. doi: 10.1084/jem.20101488
158. Espinosa DA, Gutierrez GM, Rojas-López M, Noe AR, Shi L, Tse SW, et al. Proteolytic cleavage of the Plasmodium falciparum circumsporozoite protein is a target of protective antibodies. J Infect Dis. (2015) 212:1111–9. doi: 10.1093/infdis/jiv154
159. Blum-Tirouvanziam U, Servis C, Habluetzel A, Valmori D, Men Y, Esposito F, et al. Localization of HLA-A2. 1-restricted T cell epitopes in the circumsporozoite protein of Plasmodium falciparum. J Immunol. (1995) 154:3922–31.
160. Godson GN, Ellis J, Svec P, Schlesinger DH, Nussenzweig V. Identification and chemical synthesis of a tandemly repeated immunogenic region of Plasmodium knowlesi circumsporozoite protein. Nature. (1983) 305:29–33. doi: 10.1038/305029a0
161. Zavala F, Cochrane AH, Nardin EH, Nussenzweig RS, Nussenzweig V. Circumsporozoite proteins of malaria parasites contain a single immunodominant region with two or more identical epitopes. J Exp Med. (1983) 157:1947–57. doi: 10.1084/jem.157.6.1947
162. Nardin H. Conserved repetitive epitope recognized by CD4+ clones from a malaria-immunized volunteer. Science. (1989) 246:1603–6. doi: 10.1126/science.2480642
163. Esposito. In vitro immune recognition of synthetic peptides from the Plasmodium falciparum CS protein by individuals naturally exposed to different sporozoite challenge. Immunol Lett. (1992) 33:187–99. doi: 10.1016/0165-2478(92)90046-Q
164. Nardin EH, Nussenzweig RS, McGregor IA, Bryan JH, Aikawa M. Antibodies to sporozoites: their frequent occurrence in individuals living in an area of hyperendemic malaria. Science. (1979) 206:597–9. doi: 10.1126/science.386511
165. Nardin EH. T cell responses in a sporozoite-immunized human volunteer and a chimpanzee. Immunol Lett. (1990) 25:43–7. doi: 10.1016/0165-2478(90)90089-9
166. Chougnet C, Deloron P, Lepers JP, Tallet S, Rason MD, Astagneau P, et al. Humoral and cell-mediated immune responses to the Plasmodium falciparum antigens PF155/RESA and CS protein: seasonal variations in a population recently reexposed to endemic malaria. Am J Trop Med Hyg. (1990) 43:234–42. doi: 10.4269/ajtmh.1990.43.234
167. McCutchan TF, Kissinger JC, Touray MG, Rogers MJ, Li J, Sullivan M, et al. Comparison of circumsporozoite proteins from avian and mammalian malarias: biological and phylogenetic implications. Proc Natl Acad Sci USA. (1996) 93:11889–94. doi: 10.1073/pnas.93.21.11889
168. Ying P, Shakibaei M, Patankar MS, Clavijo P, Beavis RC, Clark GF, et al. The Malaria circumsporozoite protein: interaction of the conserved regions I and II-plus with heparin-like oligosaccharides in heparan sulfate. Exp Parasitol. (1997) 85:168–82. doi: 10.1006/expr.1996.4134
169. Kappe SH, Buscaglia CA, Nussenzweig V. Plasmodium sporozoite molecular cell biology. Annu Rev Cell Dev Biol. (2004) 20:29–59. doi: 10.1146/annurev.cellbio.20.011603.150935
170. Wang Q, Fujioka H, Nussenzweig V. Mutational analysis of the GPI-anchor addition sequence from the circumsporozoite protein of Plasmodium. Cell Microbiol. (2005) 7:1616–26. doi: 10.1111/j.1462-5822.2005.00579.x
171. Flanagan KL, Lee EA, Gravenor MB, Reece WH, Urban BC, Doherty T, et al. Unique T cell effector functions elicited by Plasmodium falciparum epitopes in malaria-exposed Africans tested by three T cell assays. J Immunol. (2001) 167:4729–37. doi: 10.4049/jimmunol.167.8.4729
172. Sedegah M, Kim Y, Peters B, McGrath S, Ganeshan H, Lejano J, et al. Identification and localization of minimal MHC-restricted CD8+ T cell epitopes within the Plasmodium falciparum AMA1 protein. Malar J. (2010) 9:241. doi: 10.1186/1475-2875-9-241
173. Sette A, Sidney J. HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr Opin Immunol. (1998) 10:478–82. doi: 10.1016/S0952-7915(98)80124-6
174. Kumar A, Kumar S, Le TP, Southwood S, Sidney J, Cohen J, et al. -A*01-restricted cytotoxic T-lymphocyte epitope from the Plasmodium falciparum circumsporozoite protein. Infect Immun. (2001) 69:2766–71. doi: 10.1128/IAI.69.4.2766-2771.2001
175. López JA, Weilenman C, Audran R, Roggero MA, Bonelo A, Tiercy JM, et al. A synthetic malaria vaccine elicits a potent CD8+ and CD4+ T lymphocyte immune response in humans. Implications for vaccination strategies. Eur J Immunol. (2001) 31:1989–98. doi: 10.1002/1521-4141(200107)31:7<1989::AID-IMMU1989>3.0.CO;2-M
176. Plebanski M, Lee EA, Hannan CM, Flanagan KL, Gilbert SC, Gravenor MB, et al. Altered peptide ligands narrow the repertoire of cellular immune responses by interfering with T-cell priming. Nat Med. (1999) 5:565–71. doi: 10.1038/8444
177. Arnot DE, Barnwell JW, Tam JP, Nussenzweig V, Nussenzweig RS, Enea V. Circumsporozoite protein of Plasmodium vivax: gene cloning and characterization of the immunodominant epitope. Science. (1985) 230:815–8. doi: 10.1126/science.2414847
178. Sedegah M, Hollingdale MR, Farooq F, Ganeshan H, Belmonte M, Kim Y, et al. Sterile immunity to malaria after DNA prime/adenovirus boost immunization is associated with effector memory CD8+T cells targeting ama1 class I epitopes. PLoS ONE. (2014) 9:e106241. doi: 10.1371/journal.pone.0106241
179. Doolan DL. Plasmodium immunomics. Int J Parasitol. (2011) 41:3–20. doi: 10.1016/j.ijpara.2010.08.002
180. Tossavainen H, Pihlajamaa T, Huttunen TK, Raulo E, Rauvala H, Permi P, et al. The layered fold of the TSR domain of P. falciparum TRAP contains a heparin binding site. Protein Sci. (2006) 15:1760–8. doi: 10.1110/ps.052068506
181. Matuschewski K, Nunes AC, Nussenzweig V, Ménard R. Plasmodium sporozoite invasion into insect and mammalian cells is directed by the same dual binding system. EMBO J. (2002) 21:1597–606. doi: 10.1093/emboj/21.7.1597
182. Akhouri RR, Bhattacharyya A, Pattnaik P, Malhotra P, Sharma A. Structural and functional dissection of the adhesive domains of Plasmodium falciparum thrombospondin-related anonymous protein (TRAP). Biochem. J. (2004) 379(Pt 3):815–22. doi: 10.1042/bj20031500
183. Robson KJ, Frevert U, Reckmann I, Cowan G, Beier J, Scragg IG, et al. Thrombospondin-related adhesive protein (TRAP) of Plasmodium falciparum: expression during sporozoite ontogeny and binding to human hepatocytes. Wiley Online Libr. (1995) 14:3883–94.
184. McCormick CJ, Tuckwell DS, Crisanti A, Humphries MJ, Hollingdale MR. Identification of heparin as a ligand for the A-domain of Plasmodium falciparum thrombospondin-related adhesion protein. Mol Biochem Parasitol. (1999) 100:111–24. doi: 10.1016/S0166-6851(99)00052-3
185. King CA. Cell motility of sporozoan protozoa. Parasitol Today. (1988) 4:315–9. doi: 10.1016/0169-4758(88)90113-5
186. Kappe SH, Buscaglia CA, Bergman LW, Coppens I, Nussenzweig V. Apicomplexan gliding motility and host cell invasion: overhauling the motor model. Trends Parasitol. (2004) 20:13–6. doi: 10.1016/j.pt.2003.10.011
187. Gilbert SC, Plebanski M, Harris SJ, Allsopp CE, Thomas R, Layton GT, et al. A protein particle vaccine containing multiple malaria epitopes. Nat Biotechnol. (1997) 15:1280–4. doi: 10.1038/nbt1197-1280
188. Healer J, Crawford S, Ralph S, McFadden G, Cowman AF. Independent translocation of two micronemal proteins in developing Plasmodium falciparum merozoites. Infect Immun. (2002) 70:5751–8. doi: 10.1128/IAI.70.10.5751-5758.2002
189. Narum DL, Thomas AW. Differential localization of full-length and processed forms of PF83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Mol Biochem Parasitol. (1994) 67:59–68. doi: 10.1016/0166-6851(94)90096-5
190. Aikawa M, Miller LH, Johnson J, Rabbege J. Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J Cell Biol. (1978) 77:72–82. doi: 10.1083/jcb.77.1.72
191. Baum J, Richard D, Healer J, Rug M, Krnajski Z, Gilberger TW, et al. A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. J Biol Chem. (2006) 281:5197–208. doi: 10.1074/jbc.M509807200
192. Cowman AF, Baldi DL, Duraisingh M, Healer J, Mills KE, O'Donnell RA, et al. Functional analysis of Plasmodium falciparum merozoite antigens: implications for erythrocyte invasion and vaccine development. Philos Trans R Soc Lond B Biol Sci. (2002) 357:25–33. doi: 10.1098/rstb.2001.1010
193. Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. (2006) 124:755–66. doi: 10.1016/j.cell.2006.02.006
194. Dvorak JA, Miller LH, Whitehouse WC, Shiroishi T. Invasion of erythrocytes by malaria merozoites. Science. (1975) 187:748–50. doi: 10.1126/science.803712
195. Howell SA, Withers-Martinez C, Kocken CH, Thomas AW, Blackman MJ. Proteolytic processing and primary structure of Plasmodium falciparum apical membrane antigen-1. J Biol Chem. (2001) 276:31311–20. doi: 10.1074/jbc.M103076200
196. Hodder AN, Crewther PE, Matthew ML, Reid GE, Moritz RL, Simpson RJ, et al. The disulfide bond structure of Plasmodium apical membrane antigen-1. J Biol Chem. (1996) 271:29446–52. doi: 10.1074/jbc.271.46.29446
197. MacRaild CA, Anders RF, Foley M, Norton RS. Apical membrane antigen 1 as an anti-malarial drug target. Curr Top Med Chem. (2011) 11:2039–47. doi: 10.2174/156802611796575885
198. Coley AM, Gupta A, Murphy VJ, Bai T, Kim H, Foley M, et al. Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody. PLoS Pathog. (2007) 3:e138. doi: 10.1371/journal.ppat.0030138
199. Bateman. UniProt: the universal protein knowledgebase. Nucleic Acids Res. (2017) 45:D158–69. doi: 10.1093/nar/gkw1099
200. Guerin-Marchand C, Druilhe P, Galey B, Londono A, Patarapotikul J, Beaudoin RL, et al. A liver-stage-specific antigen of Plasmodium falciparum characterized by gene cloning. Nature. (1987) 329:164–7. doi: 10.1038/329164a0
201. Lingelbach K, Joiner KA. The parasitophorous vacuole membrane surrounding Plasmodium and Toxoplasma: an unusual compartment in infected cells. J Cell Sci. (1998) 111(Pt 11):1467–75.
202. Spielmann T, Montagna GN, Hecht L, Matuschewski K. Molecular make-up of the Plasmodium parasitophorous vacuolar membrane. Int J Med Microbiol. (2012) 302:179–86. doi: 10.1016/j.ijmm.2012.07.011
203. Spielmann T, Gardiner DL, Beck HP, Trenholme KR, Kemp DJ. Organization of ETRAMPs and EXP-1 at the parasite-host cell interface of malaria parasites. Mol Microbiol. (2006) 59:779–94. doi: 10.1111/j.1365-2958.2005.04983.x
204. Simmons D, Woollett G, Bergin-Cartwright M, Kay D, Scaife J. A malaria protein exported into a new compartment within the host erythrocyte. EMBO J. (1987) 6:485–91. doi: 10.1002/j.1460-2075.1987.tb04779.x
205. Kara UA, Stenzel DJ, Ingram LT, Bushell GR, Lopez JA, Kidson C. Inhibitory monoclonal antibody against a (myristylated) small-molecular-weight antigen from Plasmodium falciparum associated with the parasitophorous vacuole membrane. Infect Immun. (1988) 56:903–9.
206. Iriko H, Ishino T, Otsuki H, Ito D, Tachibana M, Torii M, et al. Plasmodium falciparum Exported Protein 1 is localized to dense granules in merozoites. Parasitol Int. (2018) 67:637–9. doi: 10.1016/j.parint.2018.06.001
207. Günther K, Tümmler M, Arnold HH, Ridley R, Goman M, Scaife JG, et al. An exported protein of Plasmodium falciparum is synthesized as an integral membrane protein. Mol Biochem Parasitol. (1991) 46:149–57. doi: 10.1016/0166-6851(91)90208-N
208. Lisewski M, Quiros JP, Ng CL, Adikesavan AK, Miura K, Putluri N, et al. Supergenomic network compression and the discovery of EXP1 as a glutathione transferase inhibited by artesunate. Cell. (2014) 158:916–28. doi: 10.1016/j.cell.2014.07.011
209. Le Roch G, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. (2004) 14:2308–18. doi: 10.1101/gr.2523904
210. Bozdech Z, Llinás M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. (2003) 1:e5. doi: 10.1371/journal.pbio.0000005
211. Sá E Cunha C, Nyboer B, Heiss K, Sanches-Vaz M, Fontinha D, Wiedtke E, et al. Plasmodium berghei EXP-1 interacts with host Apolipoprotein H during Plasmodium liver-stage development. Proc Natl Acad Sci USA. (2017) 114:E1138–47. doi: 10.1073/pnas.1606419114
212. Gilson PR, Nebl T, Vukcevic D, Moritz RL, Sargeant T, Speed TP, et al. Identification and stoichiometry of glycosylphosphatidylinositol-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics. (2006) 5:1286–99. doi: 10.1074/mcp.M600035-MCP200
213. Woehlbier U, Epp C, Kauth CW, Lutz R, Long CA, Coulibaly B, et al. Analysis of antibodies directed against merozoite surface protein 1 of the human malaria parasite Plasmodium falciparum. Infect Immun. (2006) 74:1313–22. doi: 10.1128/IAI.74.2.1313-1322.2006
214. O'Donnell RA, de Koning-Ward TF, Burt RA, Bockarie M, Reeder JC, Cowman AF, et al. Antibodies against merozoite surface protein (MSP)-1(19) are a major component of the invasion-inhibitory response in individuals immune to malaria. J Exp Med. (2001) 193:1403–12. doi: 10.1084/jem.193.12.1403
215. Gardiner DL, Trenholme KR. Plasmodium falciparum gametocytes: playing hide and seek. Ann Transl Med. (2015) 3:45. doi: 10.3978/j.issn.2305-5839.2015.01.23
216. Lanfrancotti A, Bertuccini L, Silvestrini F, Alano P. Plasmodium falciparum: mRNA co-expression and protein co-localisation of two gene products upregulated in early gametocytes. Exp Parasitol. (2007) 116:497–503. doi: 10.1016/j.exppara.2007.01.021
217. Silvestrini F, Bozdech Z, Lanfrancotti A, Di Giulio E, Bultrini E, Picci L, et al. Genome-wide identification of genes upregulated at the onset of gametocytogenesis in Plasmodium falciparum. Mol Biochem Parasitol. (2005) 143:100–10. doi: 10.1016/j.molbiopara.2005.04.015
218. Lobo CA, Konings RN, Kumar N. Expression of early gametocyte-stage antigens Pfg27 and Pfs16 in synchronized gametocytes and non-gametocyte producing clones of Plasmodium falciparum. Mol Biochem Parasitol. (1994) 68:151–4. doi: 10.1016/0166-6851(94)00155-3
219. Bruce MC, Carter RN, Nakamura K, Aikawa M, Carter R. Cellular location and temporal expression of the Plasmodium falciparum sexual stage antigen Pfs16. Mol Biochem Parasitol. (1994) 65:11–22. doi: 10.1016/0166-6851(94)90111-2
220. Fidock DA, Bottius E, Brahimi K, Moelans II, Aikawa M, Konings RN, et al. Cloning and characterization of a novel Plasmodium falciparum sporozoite surface antigen, STARP. Mol Biochem Parasitol. (1994) 64:219–32. doi: 10.1016/0166-6851(94)00012-3
221. Fidock DA, Pasquetto V, Gras H, Badell E, Eling W, Ballou WR, et al. Plasmodium falciparum sporozoite invasion is inhibited by naturally acquired or experimentally induced polyclonal antibodies to the STARP antigen. Eur J Immunol. (1997) 27:2502–13. doi: 10.1002/eji.1830271007
222. Ganeshan H, Kusi KA, Anum D, Hollingdale MR, Peters B, Kim Y, et al. Measurement of ex vivo ELISpot interferon-gamma recall responses to Plasmodium falciparum AMA1 and CSP in Ghanaian adults with natural exposure to malaria. Malar J. (2016) 15:55. doi: 10.1186/s12936-016-1098-8
223. Gilbert SC, Plebanski M, Hill AVS. Association of malaria parasite population structure, HLA, and immunological antagonism (human leukocyte antigen) Association of malaria parasite population structure, HLA, and immunological antagonism (human leukocyte antigen). Adv Sci. (1998) 1173:1–8.
224. Rabinovich RN, Drakeley C, Djimde AA, Hall BF, Hay SI, Hemingway J, et al. malERA: an updated research agenda for malaria elimination and eradication. PLoS Med. (2017) 14:e1002456. doi: 10.1371/journal.pmed.1002456
225. Miyakoda M, Kimura D, Yuda M, Chinzei Y, Shibata Y, Honma K, et al. Malaria-specific and nonspecific activation of CD8+ T cells during blood stage of Plasmodium berghei infection. J Immunol. (2008) 181:1420–8. doi: 10.4049/jimmunol.181.2.1420
226. Chandele A, Mukerjee P, Das G, Ahmed R, Chauhan VS. Phenotypic and functional profiling of malaria-induced CD8 and CD4 T cells during blood-stage infection with Plasmodium yoelii. Immunology. (2011) 132:273–86. doi: 10.1111/j.1365-2567.2010.03363.x
227. Anthony DD, Lehmann PV. T-cell epitope mapping using the ELISPOT approach. Methods. (2003) 29:260–9. doi: 10.1016/S1046-2023(02)00348-1
228. Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. (2008) 8:247–58. doi: 10.1038/nri2274
229. Betts R, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. (2006) 107:4781–9. doi: 10.1182/blood-2005-12-4818
230. Schussek S, Trieu A, Apte SH, Sidney J, Sette A, Doolan DL. Novel Plasmodium antigens identified via genome-based antibody screen induce protection associated with polyfunctional T cell responses. Sci Rep. (2017) 7:15053. doi: 10.1038/s41598-017-15354-0
231. Flyer DC, Ramakrishna V, Miller C, Myers H, McDaniel M, Root K, et al. Identification by mass spectrometry of CD8(+)-T-cell Mycobacterium tuberculosis epitopes within the Rv0341 gene product. Infect Immun. (2002) 70:2926–32. doi: 10.1128/IAI.70.6.2926-2932.2002
232. Cockburn JA, Tse SW, Radtke AJ, Srinivasan P, Chen YC, Sinnis P, et al. Dendritic cells and hepatocytes use distinct pathways to process protective antigen from plasmodium in vivo. PLoS Pathog. (2011) 7:e1001318. doi: 10.1371/journal.ppat.1001318
233. Doll KL, Pewe LL, Kurup SP, Harty JT. Discriminating protective from nonprotective plasmodium-specific CD8+ T cell responses. J Immunol. (2016) 196:4253–62. doi: 10.4049/jimmunol.1600155
234. Crowe SR, Miller SC, Woodland DL. Identification of protective and non-protective T cell epitopes in influenza. Vaccine. (2006) 24:452–6. doi: 10.1016/j.vaccine.2005.07.090
235. Gilbert SC. T-cell-inducing vaccines - what's the future. Immunology. (2012) 135:19–26. doi: 10.1111/j.1365-2567.2011.03517.x
236. Ishizuka A, Lyke K, DeZure A, Berry AT. R.-Medicine N, and U. 2016. Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nat Med. (2016) 22:614–23. doi: 10.1038/nm.4110
237. Gray GE, Laher F, Lazarus E, Ensoli B, Corey L. Approaches to preventative and therapeutic HIV vaccines. Curr Opin Virol. (2016) 17:104–9. doi: 10.1016/j.coviro.2016.02.010
238. Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. (2003) 421:852–6. doi: 10.1038/nature01441
Keywords: malaria, Plasmodium falciparum, CD8+, T cell epitope, HLA, restriction, cytotoxic T cells
Citation: Heide J, Vaughan KC, Sette A, Jacobs T and Schulze zur Wiesch J (2019) Comprehensive Review of Human Plasmodium falciparum-Specific CD8+ T Cell Epitopes. Front. Immunol. 10:397. doi: 10.3389/fimmu.2019.00397
Received: 20 November 2018; Accepted: 15 February 2019;
Published: 21 March 2019.
Edited by:
Noah Butler, The University of Iowa, United StatesReviewed by:
Urszula Krzych, Walter Reed Army Institute of Research, United StatesIan Andrew Cockburn, Australian National University, Australia
Copyright © 2019 Heide, Vaughan, Sette, Jacobs and Schulze zur Wiesch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julian Schulze zur Wiesch, anVsaWFuc3p3QGdtYWlsLmNvbQ==
 Janna Heide
Janna Heide Kerrie C. Vaughan
Kerrie C. Vaughan Alessandro Sette
Alessandro Sette Thomas Jacobs5
Thomas Jacobs5 Julian Schulze zur Wiesch
Julian Schulze zur Wiesch