Allergic and autoimmune diseases such as asthma, psoriasis, rheumatoid arthritis, and inflammatory bowel disease vary in prevalence within human populations (1, 2). The hygiene hypothesis and more recently the “old friends” hypothesis have been quoted (3) to explain this disparity of prevalence, particularly between nations in the developed and developing world. Proposed etiological mechanisms include the absence of once-common childhood infections, as well as a reduction in exposure to a variety of commensal microorganisms in childhood (3). Exposure to such organisms is proposed to educate the immune system to appropriately respond to either innocuous and dangerous stimuli. Aberrant response to innocuous stimuli results in the development of allergic and autoimmune disease (3). However, these hypotheses alone do not fully account for the variability in prevalence of allergic and autoimmune disease (2, 4, 5). Certain populations exhibit an elevated risk to autoimmune and allergic disease above the background rate of individual human populations.
The advent of Genome Wide Association Studies (GWAS) have enabled identification of predisposing genetic variants to allergic and autoimmune disease (4, 5). They have also confirmed the association of previous loci identified in linkage studies. Genetic predisposition is a “sine qua non” for the development of allergic or autoimmune disease (4, 5) due to the ongoing evolutionary battle of protection against infectious disease whilst maintaining an acceptable risk of autoimmune disease which may impact upon reproductive capacity. In the identification of allergy and autoimmune disease-associated loci, multiple overlapping loci have been identified with pathogen-induced positive selection loci (6–12) (Table 1) (In this context, positive selection refers to genomic or Darwinian selection). A commonly known example of pathogen-induced positive selection is the increased prevalence of hemoglobin variants (HbS) causing sickle cell disease in populations with high previous malaria incidence, giving partial protection to severe malarial infection (10). In autoimmune and inflammatory disease, identified overlapping genes involve the activation and function of T cells, monocytes, NK cells, and dendritic cells as well as aspects of the major histocompatibility complex and transcription factors (13–17) (Table 1). Infectious agents associated with identified genes include M. Leprae, M. Tuberculosis, Y. Pestis, diarrheal illness, and Plasmodium sp. (4, 5, 8, 9, 11, 12) (Table 1). The presence of gene variants was associated with partial protection against the development of severe disease. Existing data is skewed by ascertainment bias with the majority of allergy and autoimmune GWAS have been undertaken in European populations. Also, GWAS for infectious disease require cohorts experiencing active infection, hence past endemic organisms with a high likelihood of producing positive selection (i.e., smallpox) are unable to be evaluated. Additionally, infectious agents with near saturation and multiple strains (i.e., M. Tuberculosis) indicate strong selective pressure (10), but due to the lack of comparison cohorts, positively selected genes are difficult to identify.
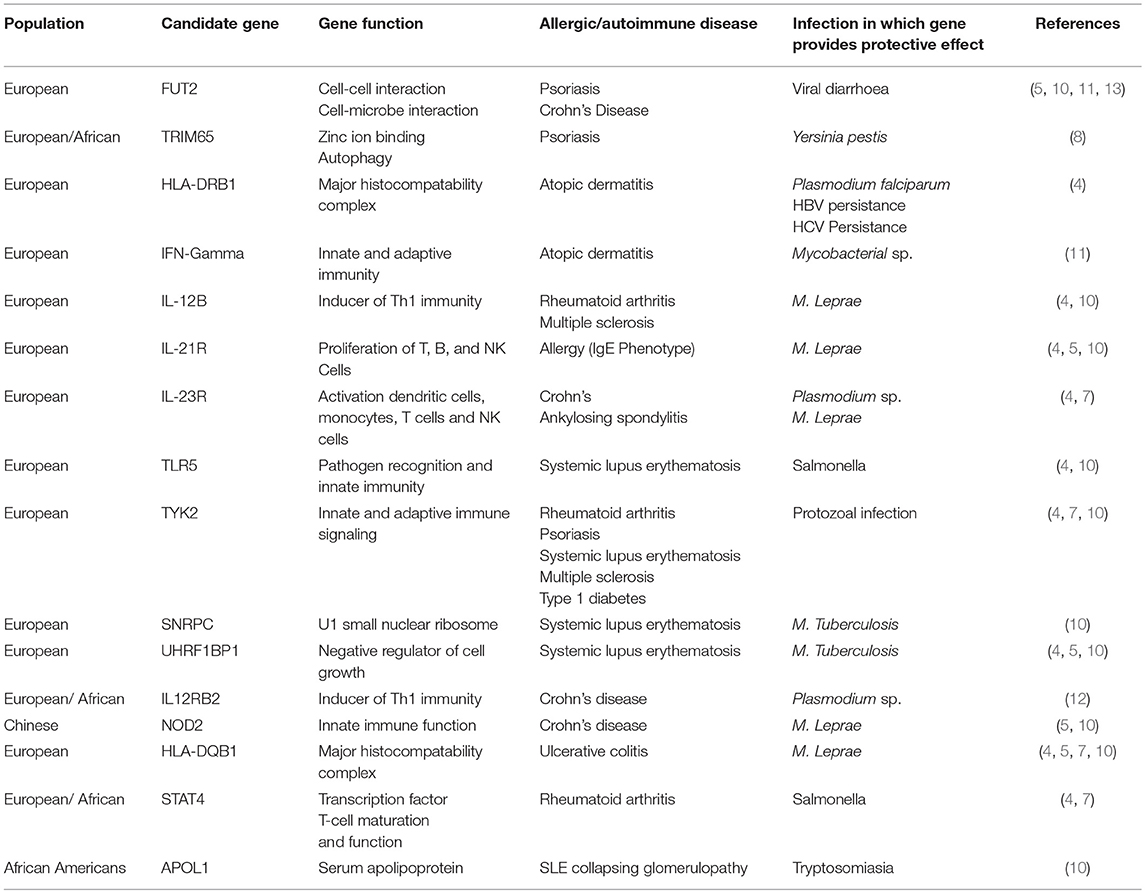
Table 1. List of identified genes identified in Autoimmune/Allergic Disease by GWAS overlapping with implicated pathogen-associated positive selection loci.
The corollary therefore is that for specific human population groups, historical exposure to infectious pathogens have positively selected for protective variants to improve survival and reproduction (6). One could hypothesize that, in the absence of infectious disease, these variants predispose to aberrant immune activation which, in the setting of appropriate environmental stimuli (such as a loss of “old friends” as well as smoking, metals, particulates, etc.) may manifest as allergy and autoimmune disease.
The implications of this correlation extend beyond population genetics to pharmacogenetics and emerging infectious diseases. The efficacy of monoclonal antibodies for control of autoimmune and allergic disease has associations with patient genetic variants (18) which have differing prevalence in various human populations. This could lead to targeted pharmacogenomic screening prior to treatment initiation. In the future this may become pertinent in emerging economies of East Asia [given the known high prevalence of NOD2 in Han Chinese, and the impact of NOD2 variants on therapy in Crohn's Disease (19)] and the Americas [in a similar vein to G6PD deficiency in Latin America (20)]. The re-emergence of vector-borne infectious diseases (such as malaria) secondary to climate change (21), may place individuals with autoimmune or allergic disease, who are being actively treated with immunomodulating therapies, at risk of infection. This risk may be greatest in those individuals where the therapy actively suppresses an inflammatory pathway known to be protective against the infectious agent. A relevant modern-day corollary is the risk of M. Tuberculosis reactivation during psoriasis treatment with TNF-alpha inhibitors in individuals of European ancestry [given the proposed mechanism of ancestral Mycobacterium sp. positively selecting for European psoriasis-associated genetic variants (22)]. This causal hypothesis requires validation in epidemiologic and functional studies.
In summary, the presence of overlapping gene associations identified by GWAS, as well as the evidence of pathogen-specific positive selection is an extension of the “hygiene/old friends” hypothesis which integrates findings from population genetics to explain disparate rates of autoimmune and allergic disease in different human populations. It also suggests avenues for further research in pharmacogenomics and susceptibility to emerging pathogens.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Supported in part by grant # UL1 TR001866 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program.
References
1. Rees F, Doherty M, Grainge MJ, Lanyon P, Zhang W. The worldwide incidence and prevalence of systemic lupus erythematosus: a systematic review of epidemiological studies. Rheumatol. (2017) 56:1945–61. doi: 10.1093/rheumatology/kex260
2. Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimoi EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. (2017) 390:2769–78. doi: 10.1016/S0140-6736(17)32448-0
3. Daley D. The evolution of the hygiene hypothesis: the role of early-life exposures to viruses and microbes and their relationship to asthma and allergic diseases. Curr Opin Allergy Clin Immunol. (2014) 14:390–6. doi: 10.1097/ACI.0000000000000101
4. Ramos PS. Population genetics and natural selection in rheumatic disease. Rheum Dis Clin North Am. (2017) 43:313–26. doi: 10.1016/j.rdc.2017.04.001
5. Raj T, Kuchroo M, Replogle JM, Raychaudhuri S, Stranger BE, De Jager PL. Common risk alleles for inflammatory diseases are targets of recent positive selection. Am J Hum Genet. (2013) 92:517–29. doi: 10.1016/j.ajhg.2013.03.001
6. Hill AVS. Evolution, revolution and heresy in the genetics of infectious disease susceptibility. Phil Trans R Soc B. (2012) 367:840–9. doi: 10.1098/rstb.2011.0275
7. Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. (2016) 48:510–8. doi: 10.1038/ng.3528
8. Corona E, Wang L, Ko D, Patel CJ. Systematic detection of positive selection in the human genome. PLoS ONE. (2018) 13:e0196676. doi: 10.1371/journal.pone.0196676
9. Withrock IC, Anderson SJ, Jefferson MA, McCormack GR, Mlynarczyk GSA, Nakama A, et al. Genetic diseases conferring resistance to infectious diseases. Genes Dis. (2015) 2:247–54. doi: 10.1016/j.gendis.2015.02.008
10. Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and Infectious diseases in human populations. Nat Rev Genet. (2014) 15:379–93. doi: 10.1038/nrg3734
11. Casanova JL, Abel L. Human genetics of infectious disease: a unified theory. EMBO J. (2007) 26:915–22. doi: 10.1038/sj.emboj.7601558
12. Frodsham AJ, Hill AVS. Genetics of infectious disease. Hum Mol Genet. (2004) 13:R187–94. doi: 10.1093/hmg/ddh225
13. Bustamante M, Standl M, Bassat Q, Vilor-Tejedor N, Medina-Gomez C, Bonilla C, et al. A genome-wide association meta-analysis of diarrhoeal disease in young children identifies FUT2 locus and provides plausible biological pathways. Hum Mol Genet. (2016) 25:4127–42. doi: 10.1093/hmg/ddw264
14. Tsoi LC, Stuart PC, Tian C, Gudjonsson JE, Das S, Zawistowski M, et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat Commun. (2017) 8:15382. doi: 10.1038/ncomms15382
15. Romagnani S. Immunological influences on allergy and the TH1/TH2 balance. J Allerg Clin Immunol. (2004) 113:395–400. doi: 10.1016/j.jaci.2003.11.025
16. Vicente CT, Revez JA, Ferreira MAR. Lessions from 10 years of genome-wide association studies of asthma. Clin Transl Immunol. (2017) 6:e165. doi: 10.1038/cti.2017.54
17. Lin YH, Machner MP. Exploitation of the host cell ubiquitin machinery by microbial effector proteins. J Cell Sci. (2017) 130:1985–96. doi: 10.1242/jcs.188482
18. Hueber W, Sands BE, Lewitzky S, Vandemeulebroecke M, Reinisch W, Higgins PD, et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of randomized, double placebo-controlled trial. Gut. (2012) 61:1693–700. doi: 10.1136/gutjnl-2011-301668
19. Kennedy NA, Lamb CA, Berry SH. The impact of NOD2 variants on fecal microbiota in Crohn's disease and controls without gastrointestinal disease. Inflamm Bowel Dis. (2018) 24:583–92. doi: 10.1093/ibd/izx061
20. Valencia SH, Ocampo ID, Arce-Plata MI. Glucose-6-Phosphate dehydrogenase deficiency prevalence and genetic variants in malaria endemic areas of Columbia. Malar J. (2016) 15:291. doi: 10.1186/s12936-016-1343-1
21. Medlock JM, Leach SA. Effect of climate change on vector-borne disease risk in the UK. Lancet Infect Dis. (2015) 15:721–30. doi: 10.1016/S1473-3099(15)70091-5
Keywords: autoimmunity, infection—immunology, genetics, allegy, monoclonal antibodies (immunology)
Citation: Frew JW (2019) The Hygiene Hypothesis, Old Friends, and New Genes. Front. Immunol. 10:388. doi: 10.3389/fimmu.2019.00388
Received: 05 November 2018; Accepted: 14 February 2019;
Published: 06 March 2019.
Edited by:
James J. Pestka, Michigan State University, United StatesReviewed by:
Kenneth Michael Pollard, The Scripps Research Institute, United StatesCopyright © 2019 Frew. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John W. Frew, jfrew@rockefeller.edu
orcid.org/0000-0001-5042-3632
 John W. Frew
John W. Frew