- 1Biochemistry, Molecular Biology, and Biophysics Graduate Program, University of Minnesota, St. Paul, MN, United States
- 2Department of Veterinary and Biomedical Sciences, University of Minnesota, St. Paul, MN, United States
Hemorrhagic fevers (HF) resulting from pathogenic arenaviral infections have traditionally been neglected as tropical diseases primarily affecting African and South American regions. There are currently no FDA-approved vaccines for arenaviruses, and treatments have been limited to supportive therapy and use of non-specific nucleoside analogs, such as Ribavirin. Outbreaks of arenaviral infections have been limited to certain geographic areas that are endemic but known cases of exportation of arenaviruses from endemic regions and socioeconomic challenges for local control of rodent reservoirs raise serious concerns about the potential for larger outbreaks in the future. This review synthesizes current knowledge about arenaviral evolution, ecology, transmission patterns, life cycle, modulation of host immunity, disease pathogenesis, as well as discusses recent development of preventative and therapeutic pursuits against this group of deadly viral pathogens.
Introduction
Significant progress has been made in recent years to understand the role of modern means of travel in inadvertently exporting deadly arenaviruses from endemic regions, the basic biology of these viruses, their genomic evolution and modes of transmission and immune suppression, and the disease pathogenesis for which they are responsible. A detailed level of understanding of the viral life cycle, evolution, and interactions with the host's immune signaling pathways is necessary in order to design effective therapeutic and preventative measures against this group of deadly human pathogens.
Arenaviral Classification, Evolution, and Ecology
The family Arenaviridae is divided into 3 genera based on their natural hosts—Mammarenaviridae, Reptarenaviridae, and Hartmaniviridae that include viruses infecting mammals, reptiles, and fish, respectively (1, 2). Mammarenaviridae are further classified into the regions of their origins, such as the Old World (OW) viruses found in West Africa (3–9) and the New World (NW) viruses found in South America (5, 10–17), which are believed to have originated <23,000 and 41,000 years ago in those continents, respectively (18) (Figure 1). Additionally, several NW arenaviral strains have been discovered in the United States, which are suspected to potentially cause human disease (19, 20). The OW and NW subgroups are polyphyletic and contain both human-pathogenic and non-pathogenic viral strains, with 10 strains in total known to cause human diseases (21). Unlike other hemorrhagic fever viruses, such as the Ebola virus (EBOV), arenaviral transmissions to humans have been found primarily as a result of human interactions with the rodents as the natural reservoirs of these viruses, as has been directly observed as recently as the 2017–2018 Lassa virus outbreak in Nigeria (9). However, human to human transmission may play a larger role in certain viral outbreaks, such as a 2014 outbreak where strains across larger geographical areas were found to cluster closely together (22).
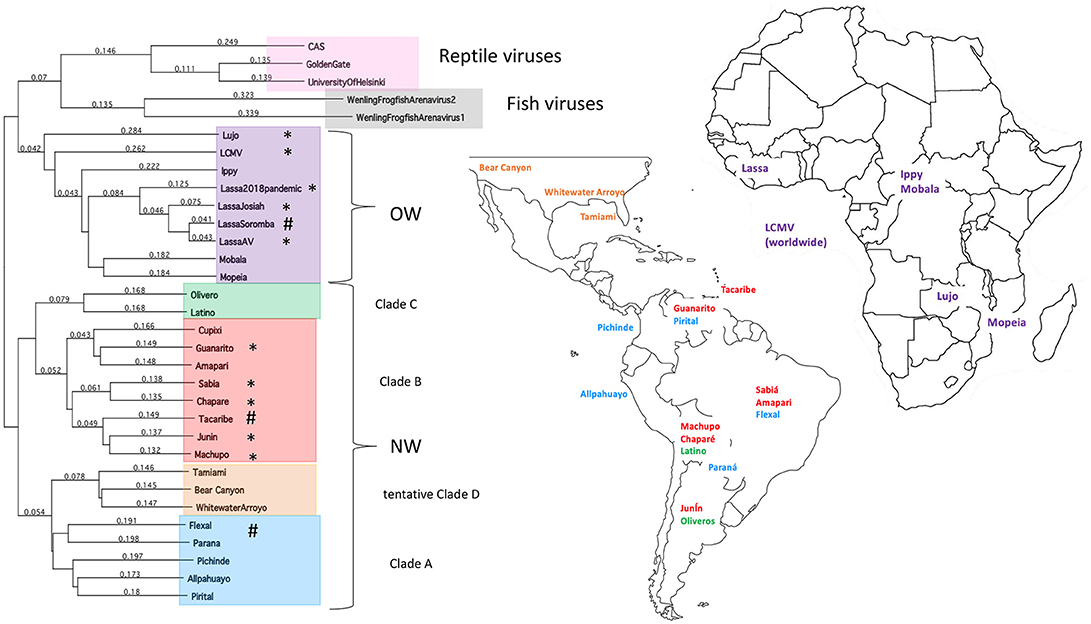
Figure 1. Taxonomy and location of arenaviruses. The phylogenetic tree for OW and NW arenaviral strains and their geographic locations. Tree was generated from full-length genomic sequences for the L polymerase protein aligned by Clustalw analysis. Asterisks designate strains that cause natural human diseases, whereas hashtags designate strains that can cause laboratory-acquired diseases in animals.
This transmission model presents an interesting challenge for determining the evolutionary history of arenaviruses. As with many zoonotic viruses that follow a co-speciation pattern to allow for infection of new hosts, arenaviruses have been previously thought to have originated in Asia along with the earliest rodents and later spread to Europe, Africa, and the Americas alongside the spread of the rodents (23, 24). However, arenaviral and rodent host phylogenetic trees almost never perfectly match (25) and some models result in rodent hosts that are randomly integrated into the arenaviral phylogeny (26). Additionally, only the Lymphocytic Choriomeningitis Virus (LCMV) has been found to circulate among European rodents (18, 27–30), but titers against LCMV have been found in human subjects worldwide (31, 32), suggesting that rodents and potentially other hosts for LCMV are more widespread than previously thought. Recent studies have revealed that LCMV can be isolated from ticks in the Ukraine (33) and in China (34) (though the low numbers of positively infected sample specimens and unknown capacity of LCMV to infect insect cells raise some doubts about ticks being a true reservoir rather than an intermittent viral carrier). Arenaviral evolution, therefore, may be more reflective of their adaptation to the available hosts based on geographic constraints (9, 18).
Local host adaption of arenaviruses is further evidenced by recent insights into diversity of arenaviruses in reptilian and marine hosts. The full genomic sequences have been determined for three reptarenaviruses isolated from boa constrictors and annulated tree boas: the CAS virus (35), Golden Gate virus (35) and University of Helsinki virus (36), necessitating the addition of the genus Reptarenaviridae into the Arenaviridae family in 2015 (1). These three reptarenaviruses have been found to be causative agents of Inclusion Body Disease (IBD), a fatal condition in snakes characterized by neurological abnormalities (37, 38), large eosinophilic aggregations inside infected cells (37–40) consisting of a 68 kDa protein (39) thought to be reptarenaviral nucleoproteins (NPs) (36, 41) and the primary source of lethality being secondary infections (38–40). Preliminary evidence has indicated that reptarenaviruses might have specifically adapted to boa constrictors, with viral replication being optimal at the reptilian 30°C and attenuated when grown at the mammalian 37°C condition (41), and infected boa constrictors having decreased rates of IBD and increased chances of becoming asymptomatic viral carriers than other snake species despite a high viral load (37, 38, 42). Like rodent reservoirs, reptarenaviruses have also been found to transmit vertically (43). Whereas, the genome of mammarenaviruses are bi-segmented with a small (S) segment and a large (L) segment, those of the reptarenaviruses are potentially more complicated, with some infected snakes carrying up to 4 different S segments and 11 different L segments per individual (44). Further adding to arenaviral genome diversity is the recent discovery of two new arenaviruses infecting ray-finned fish (2) with tri-segmented genomes, making them a potential intermediate between arenaviruses and their close “cousin” virus, the tri-segmented Bunyaviridae family found widely in arthropods (2, 45) that can also cause severe and lethal hemorrhagic infections in humans.
The focus of this article is on the OW and NW mammarenaviruses. Phylogenetically, changes in the viral polymerase of the mammarenaviruses may be more associated with older adaptive events dating to the speciation of NW arenaviruses (18), while more current adaptation, at least with the OW Lassa virus (LASV), appears to more prominently associate with the viral surface glycoproteins (46). This observation is recapitulated when comparing the genetic similarity of the four arenaviral proteins (Figure 2). The arenaviral polymerases have less homology between OW and NW viruses as well as among OW and NW strains compared to the other 3 arenaviral proteins, suggesting an earlier genetic divergence. A recent report further supports this observation by noting that a reassorted virus containing the S genomic segment that encodes the viral glycoprotein (GP) and nucleoprotein (NP) from the known virulent LASV Josiah and the L genomic segment that encodes the viral polymerase (L) and matrix protein (Z) from the known less virulent 2015/Liberia LASV strain retained the disease pathogenicity in guinea pigs (Figure 3). In contrast, another reassorted virus carrying the viral GP and NP from the 2015 Liberian strain was severely attenuated (47). Taken together, this suggests that earlier adaption to a geographically available host required fundamental changes to the internal viral proteins, such as the L polymerase, while more recent evolution is driven by optimizing viral entry efficiency and immune evasion mediated by viral GP and NP, respectively. The concept of viral proteins mediating host immune evasion will be discussed further below.
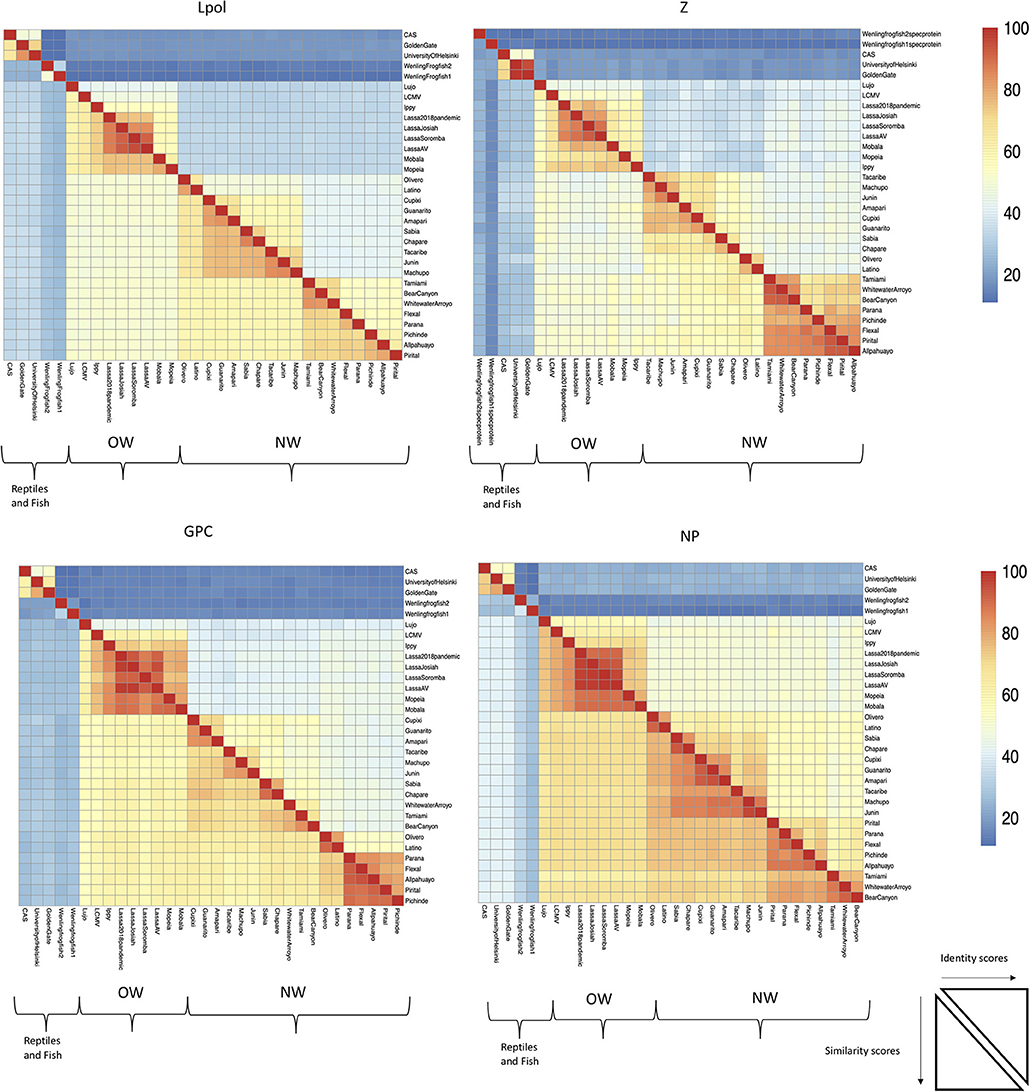
Figure 2. The L RdRp polymerase exhibits the least conservation among the arenaviral proteins. Full-length protein coding sequences from arenaviral strains were aligned by Clustlw analysis, and the matrix for pair-wise score similarity (# of shared amino acid residues/alignment length*100) was converted into a heatmap by the pheatmap module for R. The GenBank accession ID's used for alignments in Figures 1, 2 are as follows: NC_010249 (Allpahuayo L), NC_010253 (Allpahuayo S), NC_010251.1 (Amapari L), NC_010247 (Amapari S), NC_010255 (Bear Canyon L), NC_010256 (Bear Canyon S), JQ717261 (CAS L), JQ717262 (CAS S), NC_010563 (Chapare L), NC_010562 (Chapare S), NC_010252 (Cupixi L), NC_010254 (Cupixi S), NC_010759 (Flexal L), NC_010757 (Flexal S), JQ717263 (Golden Gate L), JQ717264 (Golden Gate S), NC_005082 (Guanarito L), NC_005077 (Guanarito S), NC_007906 (Ippy L), NC_007905 (Ippy S), NC_005080 (Junin L), NC_005081 (Junin S), HQ688674 (Lassa Josiah L), HQ688672 (Lassa Josiah S), MH888008 (Lassa 2018 pandemic L), MH887896, (Lassa 2018 pandemic S), FR832710 (Lassa AV L), FR832711 (Lassa AV S), KF478762 (Lassa Soromba L), KF478765 (Lassa Soromba S), NC_010760 (Latino L), NC_010758 (Latino S), AY847351 (LCMV L), AY847350 (LCMV S), NC_012777 (Lujo L), NC_012776 (Lujo S), NC_005079 (Machupo L), NC_005078 (Machupo S), NC_007904 (Mobala L), NC_007903 (Mobala S), NC_006574 (Mopeia L), NC_006575 (Mopeia S), NC_010250 (Olivero L), NC_010248 (Olivero S), NC_010761 (Parana L), NC_010756 (Parana S), NC_006439 (Pichinde L), NC_006447 (Pichinde S), NC_005897 (Pirital L), NC_005894 (Pirital S), NC_006313 (Sabia L), NC_006317 (Sabia S), NC_004292 (Tacaribe L), NC_004293 (Tacaribe S), NC_010702 (Tamiami L), NC_010701 (Tamiami S), KF297880 (University of Helsinki S), KF297881 (University of Helsinki L), MG599863 (Wenling frogfish 1 L), MG599864 (Wenling frogfish 1 S), MG59986 (Wenling frogfish 1 M), MG599866 (Wenling frogfish 2 L), MG599867 (Wenling frogfish 2 S), MG599868 (Wenling frogfish 2 M), NC_010703 (Whitewater Arroyo L), NC_010700 (Whitewater Arroyo S).
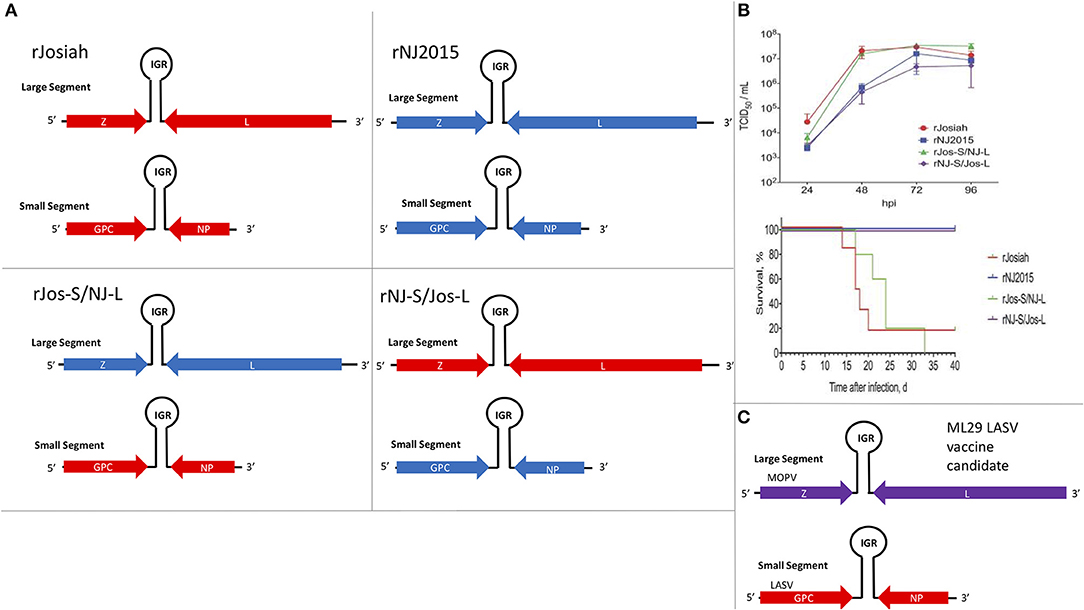
Figure 3. The S segment of the Lassa virus genome is sufficient to maintain pathogenicity. (A) Using reverse genetics strategy to produce recombinant wildtype (WT) and reassorted Lassa viruses carrying different large (L) and small (S) segments from different strains of the virus (Josiah vs. NJ2015). Viral genes: matrix Z, L RdRp, glycoprotein (GP), and nucleoprotein (NP); IGR, intergenic region. (B) Growth kinetics of viruses listed in 2A in A549 cells and the survival curve of strain 13/N guinea pigs subcutaneously infected with these viruses. Figure redrawn from Welch et al. (47). (C) Illustration of the genomic content of the ML29 LASV candidate vaccine carrying the L segment from the Mopeia virus (MOPV) and the S segment from LASV.
Disease Transmission and Social Implications
Mammarenaviruses enter their hosts by inhalation of air-borne viral particles or by eating and/or drinking virus contaminated food or water, respectively. The social structures of arenavirus endemic regions present a particular challenge for viral containment with this transmission model. Homes are often open-air spaces and contain small spaces for rats to cohabitate with humans when built from locally available materials. As a result, rodent reservoirs move easily from home to home (48) and previous surveys have found significant numbers of homes containing rodent reservoirs and instances of contact with rodent or rodent waste products, particularly at night when the activity of rodents is at its highest level (49). Rice is often the stable crop in endemic areas (particularly in western Africa), which is typically grown in fields or low-lying swamps that encourage rodent habitation (50). The challenges presented by the educational and communication infrastructure in endemic regions also prevent optimal disease control, with a recent survey finding that 76% of residents in a Nigerian urban town had inadequate knowledge of Lassa fever and 51% had poor control practices (51). Practices such as hunting rodents for food and for use in sacrificial ceremonies have also been documented to increase risks for infection (50). Future measures of disease prevention will not only need to focus on enforcing individual habits of rodent control, but also on encouraging larger political and incentive policies conducive to foster sensible habits (52, 53) and infrastructural building, as diagnostic laboratory capable of carrying out viral genomic PCR to detect arenaviral infections is often not available in endemic regions (54–56). Computational modeling of regions at risk of arenaviral pandemics will also continue to be necessary as predictive/preventive measures (53, 57–60).
Several modeling studies have indicated that several African regions in close proximity to current endemic regions are at serious risk for arenavirus spread (53, 57, 58), and new reservoirs for human pathogenic arenaviruses are continuing to be found, adding to the potential for interregional spread (61). However, the biggest factor in restricting arenaviral pandemics appears to be a strong inter-species host transmission barrier (62). Current models suggest about 10% of rodents in endemic areas are seropositive for local arenaviruses, and that rodents clear the virus in a time period significantly shorter than their life span to produce antibodies (63, 64) with some preliminary evidence suggesting that anti-arenaviral antibodies may have a small correlation with decreased survival and increased rodent capture (65).
There is increasing concern about the potential for arenaviruses to spread across regions and initiate worldwide pandemics. LASV remains the only documented arenavirus to be imported by travelers who have visited endemic regions for a variety of reasons (Supplemental Table 1). While most documented cases occurred prior to 2000 (66), more recent cases of Lassa fever importation have been reported in the United States [Pennsylvania (67), Minnesota (68), and Georgia (69)], Ghana (70), Sweden (71), and Germany (72, 73). A common theme among these cases is that, while no secondary infections as of yet have been documented, an intermittent period of remission between initial treatment and reemergence of symptoms presents a potential risk for protracted disease and transmission (74). A salient fact that arenaviruses also have the potential to be used as biological weapons (75, 76) is of particular concern for endemic regions that are engaged in constant political and military conflicts (77). Human-to-human transmissions, although are rare occurrences, have been documented through the use of contaminated medical instruments in standard and specialized medical procedures, such as organ transplantations (78). As a whole, the threat of arenaviral spread and exportation from endemic regions should be of concern for public health considerations despite natural barriers that currently help curb major outbreaks.
Arenaviral Life Cycle
Entry Mechanisms by NW and OW Arenaviruses
Arenaviruses are enveloped, ambisense single-stranded RNA viruses. Their structure consists of a membrane envelope containing the surface glycoprotein (GP) subunits surrounding a capsid, which consists of the Z matrix protein (Figure 4A). Inside the capsid are the L RNA-dependent RNA polymerase (L RdRp) and the viral bi-segmented genome encapsulated by the nucleoprotein (NP). Cell entry is mediated by GP, which is first expressed as the glycoprotein precursor complex (GPC). The host subtilase SK1-S1P proteolytically cleaves GPC into its three final subunits (79–81): GP1, GP2, and SSP (Stable Signal Peptide), which form heterotrimers on the cell membrane surface (82–84). GP1 forms spikes protruding from the viral envelope and is responsible for interacting with entry receptors, while GP2 is a class 1 fusion transmembrane protein by virtue of its 6 helix domain (85, 86). SSP (87) is an unusually small, hydrophobic and long-lived (88) signaling protein implicated in viral fusion (89, 90), transport of the GPC complex through the SK1-S1P containing Golgi (91) and as a GPC folding chaperone (92, 93). While recombinant arenaviruses expressing a SSP from a different strain are viable, recombinant viruses expressing a non-arenaviral signal peptide are not (94), indicating the specific adaptation of SSP for arenaviral proteins. This observation is strengthened by work from our laboratory, which shows that certain SSP residues attenuate viral growth independently of cell entry efficiency, illustrating the multiple roles of SSP in the viral life cycle that remain to be fully elucidated (95). GP1 and GP2 also contain N-glycosylation residues at multiple sites (96), while SSP is myristoylated (89). While the purpose of these post-translational modifications has yet to be described in detail, GP1/2 glycosylation has been linked to viral protein transport (96) and SSP myristoylation has been found to be vital for directing membrane anchor and viral fusion (89, 90).
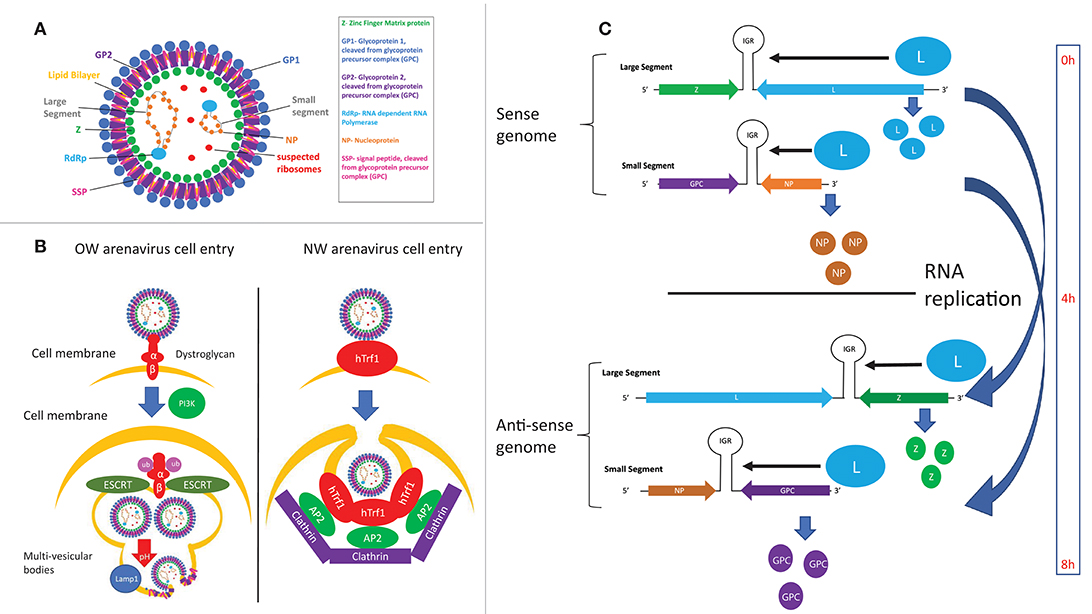
Figure 4. (A) Structure of arenaviral particles. (B) Comparison of the entry mechanism for OW and NW arenaviruses. (C) Timing of protein expression and RNA replication during the arenavirus lifecycle as determined by the genomic structure.
LASV and LCMV were first discovered to utilize α-dystroglycan as their cellular entry receptor (97), which was later confirmed to be the case for all OW and clade C NW arenaviruses (98) (Figure 4B). Glycosylation of α-dystroglycan by the glycosyltransferase LARGE is necessary for recognition by arenaviruses (99–101), which mimics the interaction between α-dystroglycan and its natural ligand laminin (102). Regulation of α-dystroglycan appears to be a driving genetic force in arenaviral endemic regions, as wide-scale genomic studies have found evidence of positive selection of polymorphisms of LARGE and other α-dystroglycan regulatory proteins in Nigerian populations (103–105). The C-terminal GP1 subunit directly interacts with α-dystroglycan (97, 106–108), while the C-terminal GP2 subunit has been found to be critical for stabilizing the α-dystroglycan-GP complex (109, 110). Residue 260 on GP1 was the first residue found to be important for interaction with α-dystroglycan (98, 111–113). X-ray crystallography analysis later identified the loop 1 structure, and specifically residues 153, 155, and 190 of GP1, to be responsible for directly interacting with α-dystroglycan (107). Residues 260 and 136 were also found to be critical for cell entry, but these residues were both located up to 25 Angstroms away from loop 1. Given that full-length GP is required for cellular entry, it is theorized that these extra residues may facilitate cell entry by complex and unknown mechanisms involving the full length protein (107). Supporting this idea, residues at the trimeric interface of the GP were recently found to also be critical for cell entry (108), highlighting the importance of the full GP for efficient cell entry.
The endosomal cellular protein LAMP1 has been identified as a critical host factor for mediating a “pH-switch” mechanism of virus-cellular membranes fusion and endocytosis of the virion particle, which can partly explain previous observations that chicken cell lines are among the few not able to be infected by LASV as they lack LAMP1 expression (106, 114). Conserved histidine residues on the GP1 subunit among OW arenaviruses have been found to mediate recognition by LAMP1 (108), and structural analysis has found a shift in GP1 conformation following its interaction with LAMP1, possibly serving as an immune cloaking mechanism (115). Another interesting mechanism has also been proposed for LAMP1-mediated LASV endocytosis, where LAMP1 raises the pH threshold for acidic endocytosis, thereby increasing virus yield by preventing deactivation of the viruses by the acidic environment inside the endosomes (116). Additional host factors and possible alternative entry receptors have also been identified for entry of arenaviruses that might involve the sodium hydrogen exchangers (117) and the phosphatidylserine receptors Axl (118) and TIM-1 (119), which need further analyses (120). A recent study has also found that the cellular NRP2 factor and CD63 appear to serve as substitutes for α-dystroglycan and LAMP1 to mediate entry of the Lujo virus (LUJV) (121), implicating the conservation of similar mechanisms in other viruses as well as the importance of conserving these mechanisms during viral evolution.
NW arenaviruses use different cellular entry receptors depending on their clade. Clade B NW arenaviruses that are pathogenic in humans have been found to use human transferrin receptor 1 (hTrf1) (122, 123) while non-pathogenic clade B viruses use hTrf1 orthologs (124, 125). A number of other viral receptors and host factors have also been found to allow Junin virus (JUNV) entry, with hTrf1 being the major candidate cellular receptor (123), and DC-SIGN/L-SIGN (126), voltage-gated calcium channels (127), and proteins involved in the clathrin-mediated pathways such as the dyn2/Eps15 endocytic pathway (128) being possible co-factors. TRIM2 has recently been identified in a siRNA screen as an inhibitor of NW arenaviral cell entry by a yet undetermined mechanism (129). The receptors for the other clades of NW arenaviruses are still unknown, though it has been shown that the tentative clade D viruses do not use hTrf1 (130).
Viral particles enter cells by clathrin-mediated pathways for α-dystroglycan-independent NW arenaviruses (131), and a clathrin-independent mechanism that involves PI3K-mediated formation of multivesicular bodies during late endocytosis and the ESCRT sorting pathway for α-dystroglycan-dependent viruses (132) (Figure 4B). GP1 disassociates from the viral particles upon exposure to acidic pH, exposing GP2 residues prior to viral fusion (133, 134). Viral fusion then ensues, of which the hydrophobic regions in GP2 (109, 135) as well as the cytosolic tail (110) have been found to play a critical role.
Viral Genome Replication, Gene Expression, and Assembly
Replication of the viral genome occurs in the cytosol following its release from the endosome. The viral NP and L proteins are the minimum known components required for viral genome replication and transcription (136–138), and the viral Z protein is an inhibitor of the viral polymerase's transcriptional initiation function (137–140).
The viral genomic RNA structural elements play important roles in the regulatory mechanisms of viral genome replication, transcription, and gene expression. The 5′ and 3′ untranslated regions (UTRs) of arenaviral genomes contain complementary sequences that are predicted to form panhandle structures and are required for effective viral RNA replication and transcription (141, 142). Specifically, 19 nucleotides located at the terminal 3′ UTR of the LCMV genome were found to serve as a minimally required promoter element and that the sequences located at the complementary 5′ UTR to form the panhandle structure appear to be required for efficient genome synthesis (142). Foscaldi and colleagues have reported that the 5′ UTR of the Tacaribe virus (TCRV) appears to contain translational stimulatory signal, whereas some sequences within the 3′ UTR can down-regulate viral gene expression (142, 143). The intergenic regions (IGRs) fold into hairpin secondary structures that are thought to help terminate transcription (12, 144, 145) and to protect the unique non-polyadenylated viral mRNA transcripts from degradation by the cellular exoribonucleases (145). These sequences appear to have a large tolerance for sequence variations that can be exploited for the development of attenuated vaccine candidates (21, 146, 147). It is noteworthy that during viral genome replication, genomic primers slip backwards from their initial binding site, resulting in a non-templated 5′ ppGpp residue (148) that is thought to act as a viral RNA decoy by competitively inhibiting the viral RNA sensing by the cellular innate-immune machinery (149). The molecular mechanisms of viral RNA sensing and innate immune evasion strategies by arenaviruses as well as on efforts to develop vaccines will be discussed in detail below.
The localization of arenaviral RNAs has also been found to be important for replication control, with both sense and anti-sense RNAs associating with cytosolic compartments containing viral NP and host factors involved in RNA metabolism (150). In this regard, recent advances in antibody-mediated dsRNA visualization (151) as well as specific probing for genomic and anti-genomic arenaviral RNA (152) show much promise for elucidating the dynamics of arenaviral RNA during the infectious life cycle. Preliminary evidence suggests that the timing of viral RNA replication is under regimented control mechanisms, as NP and L genes being transcribed and translated first and directly from the negative-sense viral genome, whereas GPC and Z are expressed several hours later after the viral genomic RNA segments encoding these genes have been replicated and then transcribed (152, 153) (Figure 4C). Additionally, virally infected cells appear to amplify viral genes in cyclical waves of expression and viral clearance, the molecular mechanism of which is unclear (152). Future studies in this area will include method development and optimization in order to image RNA species at lower copy numbers and to expand the RNA repertoire available for targeted imaging.
After the viral RNAs are replicated, transcribed, and translated, virion assembly ensues and is mediated by both viral and host cellular proteins. The C-terminal domains of the viral Z matrix protein is a central player for viral assembly, budding, and release from the infected cells. Z-L interactions (140, 154), Z-NP interactions (155), and Z-GP interactions (156, 157) have been shown to ensure co-localization of viral proteins for assembly. Z then interacts with the cellular Tsg-101 ESCRT pathway proteins to allow viral budding process to begin (158, 159), which is aided by a myristoylation residue at the N-terminus of Z to allow for interaction with the cellular membrane (160, 161). Z proteins has also been found to be capable of self-budding and the production of the viral like particles (VLPs) in the absence of the viral RNA or other proteins indicates that Z is both necessary and sufficient for budding (162–165). A recent report suggests that the late domains located in the C-terminal part of the Z protein may also be required for the release of the so-called defective interfering (DI) virion particles from the infected cells but not necessarily for the release of infectious virions, a process that may be regulated by phosphorylation of certain residues in the viral Z protein (163). DI particles are produced by many viruses, including arenaviruses, during infection and are similar to infectious virion particles in appearance and protein contents but they cannot produce productive infection (166) as their genomes contain large and deleterious deletions (167). Such deletions have been observed in the 3'UTR regions of the LCMV genome and potentially elsewhere in the viral genome, but their specific functions in inhibiting viral replication have yet to be studied in detail (168). Arenaviruses have been found to produce high levels of DI particles in cell culture (169) and in animal infection models (170), and DI particles have been theorized to contribute to viral persistence (166, 171, 172).
Disease Pathogenesis and Host Immune Suppression
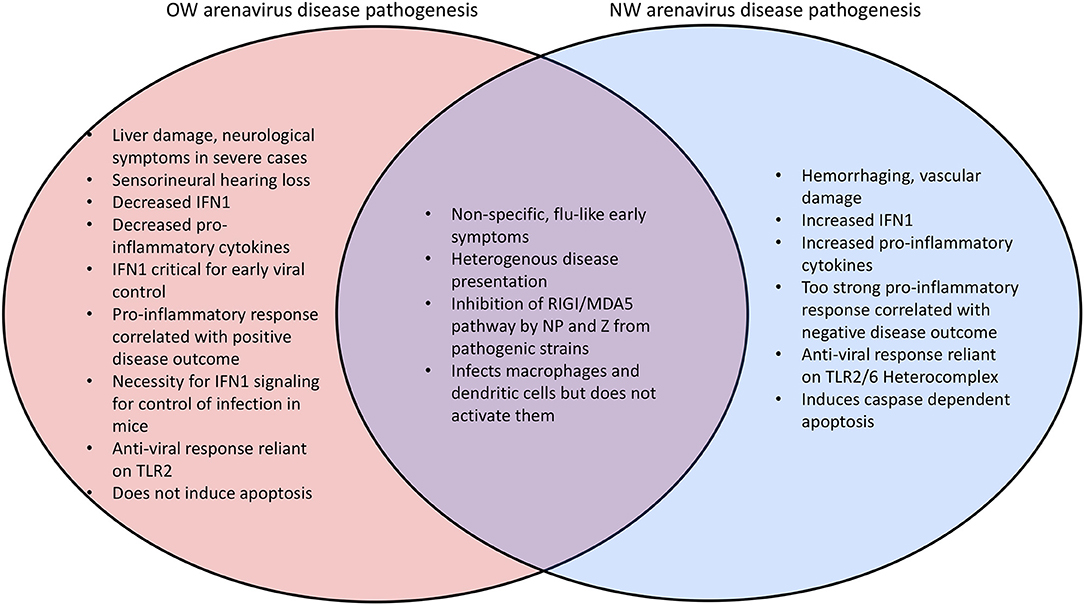
Mammarenaviral disease profiles tend to be heterogenous (173–176). Lassa fever, the most severe arenaviral disease, is estimated to cause up to 300,000 cases and 5,000 deaths per year in endemic regions of West Africa (5, 173). A recent outbreak occurred in Nigeria has resulted in 431 laboratory confirmed cases that include 37 health care workers and with a 25% overall fatality rate (177–179). Previous outbreaks have recorded lower mortality rate in hospitalized patients (180). However, the fatality rate can be as high as 87% among infected women during the third trimester of pregnancy (181), and maternal-fetal transmission of arenaviruses has been demonstrated in rodent models (64) as well as in one recent reported case in humans (182). Many other complications arise from severe arenaviral infections including liver (183–186) and vascular (185, 187, 188) damage, both of which have been recapitulated in a guinea pig arenaviral animal model (189–191) as well as in immunocompromised mice (192, 193) and a novel hamster NW arenavirus model (194). While liver damage tends to be highly prominent in infections by OW arenaviruses with platelet factors and heme breakdown products as significant biomarkers of LASV disease, hemorrhaging, and vascular damage occur more frequently in NW arenavirus infections (195) (Figure 5). Sensorineural hearing loss (SNHL) also occurs in 29% of the survivors of Lassa fever (196–199) which may be attributed to a pathogenic inflammatory response in the auditory nervous system (200, 201). Other neurological complications have also been reported for arenaviral infection (202, 203).
Monocytic cells such as alveolar macrophages and dendritic cells are the earliest target for arenaviral cell entry. However, these cells are not activated upon viral entry, as evidenced by the lack of increase in the levels of activating markers, such as CD80, CD86, CD40, CD54, and HLAs as well as cytokines TNFα, IL1β, IL6, and IL12 (204, 205). For this reason, monocytic cells are thought to act as a viral “reservoir” early in infection, where the virus can easily enter, and virus later spreads when these cells enter draining lymph nodes (206, 207). The failure of these cells to become activated is consistent with the observation of the generalized immunodepression in severely and fatally infected individuals (208, 209). Virus replication has also been directly observed in a number of cell types, including respiratory epithelial cells following infection in human (210) and animal models (211).
Studies on human and rodent survivors of mammarenaviral infection have all indicated that proper functioning of the immune responses (innate and T-cell mediated immunity) are critical to minimizing viral growth rates, presentation of symptoms, and mortality rates (209, 212). The innate immune response is a compilation of non-specific defense mechanisms against foreign antigens that is critical for early detection and inhibition of pathogen growth before the adaptive immune response has time to produce proper cell-mediated immunity, such as the development of antibodies and cytotoxic T-lymphocyte responses (CTL) against the invading pathogen and/or the pathogen-infected cells (212). Innate immunity is enacted within a few hours of host recognition of a pathogen-associated molecular pattern (PAMP). The most common viral PAMPs recognized by immune cells are unique molecular features of the viral genome, such as 5′ triphosphorylated RNA and double stranded RNA (Figure 6). Pattern recognition receptor proteins (PRRs), such as the RIG-I-like receptors (RLRs) RIG-I and MDA5, are activated by PAMPs and result in cytokine expression and the activation and recruitment of innate immune cells such as macrophages, neutrophils, and dendritic cells (213, 214).
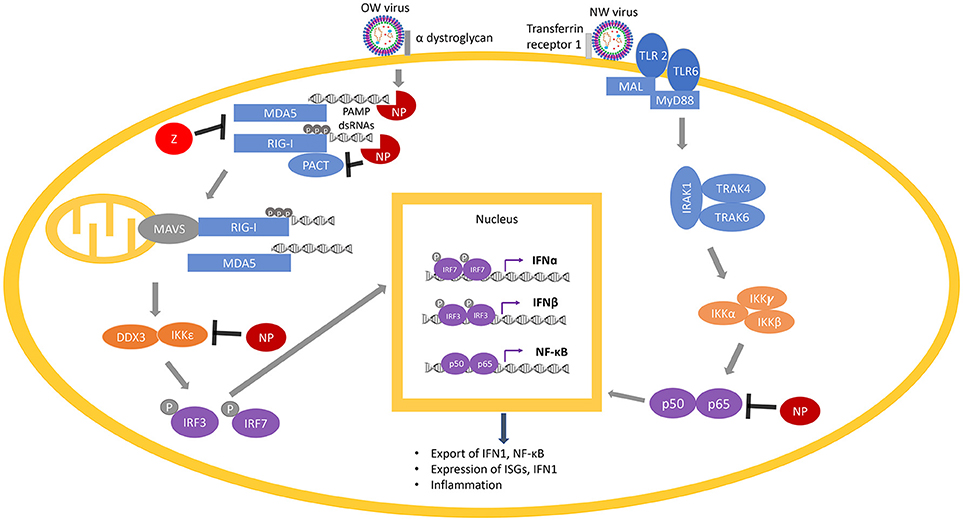
Figure 6. Arenaviral proteins (NP and Z) inhibit the RIG-I/MDA5 and NF-κB pathways. RIG-I and MDA5 are activated by PAMP dsRNAs during virus replication and are potentiated by PACT. Following the activation of MAVS (on the mitochondria) by RIG-I/MDA5, a molecular cascade involves the interaction of IKKε and DDX3, which is followed by phosphorylation of the transcription factors IRF3 and IRF7 to translocate them into the nucleus, where they dimerize and bind to transcription factor binding sites of the IFNα and IFNβ genes to activate their transcriptions. Expression and exportation of these gene products into the cellular milieu trigger the IFN1 signaling cascade in an autocrine or paracrine fashion to induce expression of hundreds of interferon-stimulated genes (ISGs) and inflammatory genes to confer virus resistance. The NF-κB pathway is activated by recognition of certain features of the virus particles by the membrane receptors TLR 2 or TLR 6. This initiates a molecular cascade resulting in the translocation of the two functional NF-κB units (p50 and p65) into the nucleus, resulting in more production of NF-κB. Arenaviral proteins (NP or Z shown in red) are known to inhibit different steps of the RIG-I/MDA5 and NF-κB pathways by either degrading the PAMP dsRNAs (through the NP RNase function) or directly inhibiting the normal function of different cellular proteins (RIG-I, MDA5, PACT, IKKε, or p50/p65).
RIG-I and MDA5, upon binding to PAMP dsRNAs in the cytosol during the process of viral replication, change their conformations from a closed to an open and activated state (215). Activated RLRs (RIG-I and MDA5) initiate several complex molecular cascades, including activation of the mitochondrial anti-viral signaling protein (MAVS), which eventually results in the translocation of transcription factors such as interferon-regulatory factors (IRF) IRF3 and IRF7 as well as nuclear factor kappa B (NF-kB) into the nucleus of the cells to activate the expression of the type 1 interferons (IFN1) that include IFNα and IFNβ, and a wide variety of interferon stimulated genes (ISG) (216, 217). A recent study has suggested that IFNα may be primarily involved in inhibiting viral growth in early stages of viral infection, while IFNβ may control viral growth in the later stages of infection (218). IFN1 is then secreted and bound by their receptors (IFNAR) on the surface of either the same cell or the neighboring cells, which activates the IFN signaling cascade in a positive feedback loop to produce more of the IFN1 and the antiviral gene products in order to confer virus resistance (219, 220).
OW arenaviral infection (e.g., LASV) in patients with moderate to severe symptoms is associated with inhibition of the innate immune response with decreased levels of IFN1 and pro-inflammatory cytokines (184, 221, 222) (Figure 5). The importance for IFN1 signaling for control of arenaviruses appears to be conserved in mammals, as wildtype (WT) mice do not exhibit symptoms during arenaviral infection but IFN receptor (IFNAR) knockout (KO) mice have been found to succumb to disease (223, 224). Additionally, in vivo infection of non-human primates (NPHs) and guinea pigs with the LASV Soromba strain isolated from local rodents in Mali, Africa, shows a significant increase in cytokine signaling and decrease in mortality compared to other LASV strains (8). Mice infected with the OW LCMV, which is capable of maintaining persistent infection with low levels of IFN1 signaling (225, 226), display an IFN1 burst 6–48 h after infection and 2–4 days before a peak in viral titers, suggesting that IFN1 signaling is most important for inhibiting viral proliferation in the earliest stages of infection (209, 227). The difference between the immune responses in rodents and humans is one of the key aspects of active investigations. Rodent reservoirs generally do not experience severe symptoms from OW arenaviral infections (65, 228, 229), and mice must be immunodeficient to experience significant symptoms. LASV infected mice also experience an early upregulation of cell adhesion molecules in the peripheral blood mononuclear cells (PBMCs) consistent with immune cells being recruited to the site of infection (230). NW arenaviruses differ from OW strains in that they tend to produce increased levels of IFN1 and cytokines, and a pro-inflammatory response is favored for inhibiting viral proliferation (222, 231–234) (Figure 5). Virus-infected A549 cell cultures (235, 236) as well as fatally infected patients (237–239) have been shown to have consistently up-regulated levels of IFN1 and TNFα. However, macrophages infected in vitro with NW arenaviruses fail to show this upregulation (240).
The arenaviral NPs have been found to inhibit the RIG-I/MDA5 pathway. The crystal structure of the LASV NP and subsequent functional studies have revealed a 3′-5′ exoribonuclease domain located within its C-terminal domain (241–243) that preferentially degrades dsRNA (242, 244). The N-terminus of NP contains a unique domain that binds ssRNA (245) and has been proposed to bind to the m7GpppN mRNA cap structure (241). NP directly interacts with RIG-I and MDA5 (246), and recent work from our lab has demonstrated that NP prevents potentiation of RIG-I function by the cellular protein PACT and that the exoribonuclease domain of NP is required for this inhibition (247), the molecular mechanism of which will be discussed in detail in the next section. The exoribonuclease domain is also required for NP inhibition of the cellular protein kinase IKKε (248) and other cellular proteins in the IRF pathway (249, 250). Abrogation of the exoribonuclease domain has allowed for IFN1 expression in normally inactivated macrophages and dendritic cells (251, 252) and subsequent activation of natural killer (NK) cells resulting in antigen-presentation (253). The exception to NP-mediated IFN1 inhibition appears to be Tacaribe virus (TCRV) NP (254), which was initially thought to be due to variance in 4 exoribonuclease residues found in this viral protein (255). However, both structural and functional studies have later demonstrated that TCRV NP can effectively degrade dsRNA and inhibit the IFN1 pathway (244, 255). These studies collectively indicate the importance of PAMP dsRNA degradation by arenaviral NPs for inhibition of the IFN1 pathway.
One important consideration in determining the larger role of NP in arenaviral pathogenesis is the fact that IFN1 inhibition by NPs is also present in strains that are not known to be pathogenic to humans. For example, NP from the non-pathogenic Pichinde virus (PICV) has been found to inhibit the IFN1 pathway in human cells (254). Additionally, work from our laboratory has demonstrated that each of the 5 exoribonuclease (RNase) catalytic residues is necessary for IFN1 inhibition by Pichinde NP as well as for optimal PICV growth in vitro (256). RNase domain-mutant viruses have also been found to exhibit diminished viral load and pathogenesis in vivo in guinea pigs (which normally experience high lethality, develop human hemorrhagic fever-like symptoms and support a high viral load when infected by the P18 PICV strain), and even result in generation of WT revertant viruses (256), implicating the important role of NP RNase function for optimal viral replication. Altogether, the ability of NP to inhibit IFN1 expression by degrading PAMP dsRNA is likely conserved across known arenaviruses. As evidenced by the tight control of gene expression and viral RNA localization within cells, it is clear that regulation of viral RNA levels and localization is a critical mechanism of the virus life cycle to allow for maximum replication without being quenched early in the cycle by the innate immune response, which may explain the absolute conservation of IFN1 inhibition by arenaviral NP proteins.
Recent studies have also been focused on the role of the viral Z protein in inhibiting the IFN1 pathway. Originally, the Z proteins from 4 tested NW arenaviral strains [JUNV, Machupo virus (MACV), TCRV, and Sabia virus (SABV)] but not from the OW LASV or LCMV strains were found to directly interact with RIG-I and inhibit IFN1 expression (257). More recently, our laboratory has demonstrated that the Z proteins from both OW and NW arenaviral strains that are known to be pathogenic in humans can directly interact with RIG-I to inhibit IFN1 expression, but Z proteins from non-pathogenic arenaviral viruses cannot (258). These results have been recapitulated in primary human macrophages infected with WT and chimeric PICVs to show that the N-terminus of LCMV Z was sufficient to inhibit macrophage activation when expressed as a chimeric Z protein with the non-pathogenic PICV (259). As a whole, the Z protein is emerging as another key target in investigating the mechanism of innate immune suppression by arenaviruses, and an encouraging potential contributor to disease pathogenesis (i.e., a potential virulence factor) that will need further characterization (105, 260).
One area of study with increasing attention is the role of innate immune cells and cytokines in initiating the adaptive immune response against mammarenaviral infection. A key pattern that has emerged from preliminary studies is that an arenavirus's ability to suppress the innate immune response appears to be positively correlated with its ability to suppress any subsequent adaptive immune response. The correlation of an inhibited innate immune response with an attenuated and delayed T cell response in those succumbing to fatal arenaviral infection has been demonstrated in humans (210, 261, 262), non-human primates (221), guinea pigs (47, 190, 211, 263–265), immunodeficient mice (223, 224, 266) as well as in immunodeficient mice transplanted with cells from WT mouse bone-marrow (193). Accordingly, individuals that survive arenaviral infections manage to keep viral loads low, peaking 10–20 days after infection for LASV, while individuals that succumb have viral loads that continually increase exponentially (205, 221, 261, 262, 267–269). In WT mice, LCMV induces robust expression of key cytokines such as the type I and II IFNs as well as IL-18, the production of type II IFN being dependent on CD8 T cells (270). This not only illustrates the close inter-connectedness of the innate and adaptive immune responses, but also further illustrates how an active immune response is key to the natural resistance against arenaviral infection in mice. The importance of the innate-adaptive interface was also recapitulated in a recent co-infection model of LCMV and E. Coli, demonstrating that an LPS-induced inhibitory mechanism of NK cells significantly attenuated the LCMV specific CD8 T cell response and the vitality of the infected mice (271).
CD8 T cells have been found to be indispensable for clearing arenaviruses (221, 272–279), while CD4 T cell (277, 280) and antibody (184) responses have been found to be dispensable. In fact, a potential contributing factor toward arenaviral pathogenicity as well as the failure of most previous arenaviral vaccine candidates in protecting against pathogenic strains may be a skewed CD4+ T cell response. STAT1 KO mice were only able to survive LCMV infection when CD4+ T cells were depleted (266), suggesting that a lack of IFN signaling could somehow overwhelmingly favor lethal CD4+ T cell proliferation. The determination of the mechanism of CD4+ T cell lethality, whether it is simply due to less CD8 T cells being activated or through some novel mechanism of CD4 T cells, has yet to be determined.
Myeloid dendritic cells (mDCs) are vital constituents in the innate-adaptive immune response interface due in combination to their large population sizes (281), their robust activation of CD8 T-cells (282), and their production of IFN1 as a CD8 T-cell activation enhancer under certain conditions, such as during LCMV infection (283). Dendritic cells (DCs) are a key target of infection by LASV (204, 205), but like macrophages, they are not activated by the infection (204, 205). However, this feature isn't consistent among arenaviral strains, as demonstrated by the well-documented comparative model of mammarenaviral immunogenicity of LASV and the Mopeia virus (MOPV). Both are OW arenaviruses and share 75% amino acid identity and the Mastomys natalensis African rat as a viral reservoir (23). However, MOPV is non-pathogenic in humans and has been considered as a potential LASV vaccine candidate in non-human primates (284), and no human cases of MOPV infection have ever been recorded (285). Initial studies found that MOPV infection of monocyte-derived dendritic cells (MoDCs) induced moderate activation and significant CD8 T-cell proliferation in the MoDC/T-cell co-culture system (286, 287), whereas LASV infected MoDCs induced low activation and weak and delayed T-cell responses (204, 287).
The same group of researchers have more recently demonstrated that both LASV and MOPV could not replicate well in myeloid dendritic cells (mDCs) isolated directly from PBMCs, and as a result, induced very low levels of IFN1 production, and that LASV-infected cells could not activate T-cells in the mDC/T-cell co-culture system (288). However, MOPV was able to induce higher levels of IFN1 and secreted cytokines known for inducing T-cell activation than in LASV-infected cells in the co-culture system, though the involvement of monocytes and potentially other cell types present in the co-culture system as a result of impurity could not be completely ruled out. Regardless, the observation seems to corroborate earlier findings indicating that DCs are dispensable for T-cell activation during LCMV infection (289). Like LASV, LCMV is known to inhibit innate immune response as demonstrated in another study (290).
Using chimeric viruses carrying different genes from either LASV or MOPV on either the LASV reverse genetics (RG) or MOPV RG backbone, the same group of investigators have shown that LASV NP is the main determinant in suppressing IFN1 production by mDCs (288), which is consistent with the known ability of all known arenaviral NPs to degrade PAMP RNA via their encoded RNase domain and thereby suppressing IFN1 production (105, 241, 244, 247, 252–256, 291). Due to the fact that mDCs did not appear to support productive replication of LASV and MOPV, it was difficult to ascertain whether the IFN1 suppressive effect was due to the expression of viral proteins in the infected cells by the different chimeric viruses. Moreover, the data could not explain the differentiation of mDC phenotype in the mDC/T-cell co-culture system between LASV and MOPV chimeric virus infections. In an attempt to provide a potential explanation for this observation, the authors have posited that various efficiencies of the NP RNase domains of different arenaviruses to suppress IFN1 production may explain differences among arenaviruses to induce DC-mediated T-cell activation. However, this hypothesis needs to be formally tested.
A chimeric LASV with a substitution of its GP with that of MOPV has been found to significantly increase IFN1 production (288), which may reflect different mechanisms of viral detection based on the different innate immune receptors (106). As an example, the Toll-like Receptor TLR2/TLR6 heterocomplex has been found to be responsible for innate immune activation by recognizing the GP of the NW JUNV upon virus infection for robust induction of the innate (292) and adaptive (293) immune responses (Figure 6), while only TLR2 has been implicated in the anti-viral activity of OW arenaviruses (294, 295). JUNV and other NW arenaviruses have also been found to initiate caspase-dependent apoptosis (233, 296–298), while OW arenaviruses fail to induce apoptosis (204, 298) or induce significantly delayed apoptosis (299) (Figure 5). The relative contributions of pathways directly mediated by the specific cellular receptor and other viral signaling mechanisms needs to be further characterized in future studies.
Contrary to earlier findings (258, 259), the Z protein was not found in the LASV/MOPV chimeric virus model to exhibit a significant effect on immune suppression (288), which might be attributed to different infectious models being utilized. It is worth noting that the experiments with the chimeric viruses did not take into consideration the potential differential gene expression levels of the viral proteins in virus-infected cells under the context of the chimeric virus backbones. Additionally, as mDCs did not appear to support productive viral replication, it raised a question of whether any of the viral proteins were synthesized in the virus-infected mDC model (288). It is therefore difficult to definitively discern the roles of the individual viral protein in modulating antiviral immunity in this unique in vitro viral infection model (288).
Modulation of Innate-Immune Protein Functions by Arenaviruses and Other Viruses
Another mammarenaviral-host interaction that has an impact on IFN1 production is that with the cellular protein PACT (Figure 6). PACT is a 313-aa protein that contains 3 conserved dsRNA binding motifs (dsRBMs) with dsRBM1 and 2 binding to dsRNA and dsRBM3 mediating activation of the dsRNA-dependent kinase PKR (300). PACT has been shown to activate RIG-I via interaction with the C-terminal repression domain of RIG-I and activates its ATPase function to potently enhance IFN1 production upon viral infection (301, 302). Even though PACT has initially been identified as a cellular binding partner and protein activator of PKR (303), activation of RIG-I by PACT does not require PKR (302). Likewise, while PACT has also been shown to interact with and stimulate the cellular Dicer protein (303, 304), Dicer does not appear to play an important role in PACT-mediated regulation of antiviral response (302).
While PACT and RIG-I pull down together in co-immunoprecipitation assays, it is less clear whether this interaction occurs strictly through protein-protein interactions or by both proteins associating with viral and/or cellular RNAs or some combination of both. The potential role of RNAs in activation of the cellular innate immune proteins (RIG-I and PACT) has been strongly implicated in recent findings that arenaviral NPs require an intact dsRNA binding domain as well as its 3′-5′ exoribonuclease activity to block PACT enhancement of RIG-I function (189–191) (Figure 6). A number of other viral nucleoproteins can block PACT enhancement of RIG-I function, which also requires an intact dsRNA binding domain (305–307), or in the case of herpes simplex virus 1 (HSV1), is independent of the RNA binding capacity of the nucleoprotein (308). Recently, our laboratory has shown that arenaviral NPs block RIG-I potentiation by PACT, and that the exoribonuclease domain is required (247). However, NP does not appear to negatively impact PACT/RIG-I direct interaction, suggesting that NP may inhibit PACT indirectly by degrading dsRNA that may be associated with the complex and is essential for its proper function. Taken together, it appears that most if not all known viral protein partners of PACT (as well as PACT itself and its cellular protein partner RIG-I) have RNA-binding properties, yet it still isn't entirely clear whether dsRNA binding is an absolute requirement to inhibit PACT-induced RIG-I activation. These studies illustrate the current lack of detail understanding of the roles of the different viral proteins, including arenaviral NPs, to inhibit PACT-mediated RIG-I activation, and the nature of the dsRNAs (viral, cellular, or both) that may intimately be involved in this process. Future studies into the exact role and molecular mechanism (s) of virus-host interactions via the PACT/RIG-I pathway can serve as an appealing pan-antiviral therapeutic target for development. Additionally, antivirals targeting PACT can influence PACT-PKR mediated NF-κB and p53 stress signaling (300, 309–316) to impact not only antiviral control but also other known cellular processes involved in cancer and other physiological and metabolic conditions.
Development of Vaccines for and Antivirals Against Arenavirus Infections
Currently, the only clinically successful anti-arenaviral vaccine is the anti-JUNV Candid #1 strain, which is currently manufactured by the Argentinian government but is not being considered for large-scale use due to its limited target population (222) as only Argentina is endemic for JUNV infection. To generate Candid #1, a human viral isolate (317) was used to passage twice in guinea pigs followed by additional passages in suckling mice and cell cultures (318–323). A number of unique mutations in Candid #1 were originally thought to attenuate Candid #1 compared to its parental WT strain (323, 324), and one such mutation (F427I in the transmembrane domain of the GP2 subunit) was consistently found to be sufficient for attenuation (325, 326). While the mechanism for this attenuation has yet to be elucidated, it is thought that the mutation may affect viral fusion or maturation efficiency (325). The equivalent mutation (F438I) in Machupo virus (MACV) was also found to be attenuated in mice (327), suggesting that this attenuation mechanism may be highly conserved. Additionally, the presence of a mutation in the SSP subunit along with the F427I mutation in GP2 is thought to prevent the virus from reverting to its wild type sequence (326). It is tempting to assume that similar attenuation techniques can be applied toward the development of other arenaviral vaccines. However, they may be limited to the JUNV vaccine for several reasons. While the IFN1 pathway (particularly IFNβ) pathway and subsequent T-cell response have been found to be critical in controlling arenaviral infection in mice (193, 209, 223, 224, 231, 266, 270, 328), arenaviral-associated immunosuppression results in limited T-cell responses (184, 204, 221, 222, 279, 287). Furthermore, the JUNV GP has been found to contain fewer glycans than the GPs from other mammarenaviruses (320, 329), and glycan residues on the glycan-rich LASV GP has been shown to promote neutralization antibody evasion (330). This is strengthened by recent observations that anti-JUNV antibodies from infected patients can neutralize other JUNV strains but cannot offer neutralization against other NW arenaviruses (331).
Partly due to the aforementioned reasons, the development of effective vaccines against other pathogenic mammarenaviruses (besides JUNV) has proven to be much more challenging. An early attempt was to use gamma-irradiated LASV particles, which produced considerable humoral responses in NHPs but failed to protect against fatal LASV challenge (332). A similar theme has been noted across subsequent studies where adaptive immunity [and even cross-protective immunity (333)] has been induced in vaccinated subjects, but has failed to protect against lethal viral challenge. Novel reverse genetic strategies are being explored to overcome some of these obstacles, such as codon optimization to increase the expression of viral protein antigens (334), tri-segmented arenaviral vectors on LCMV (335–339) or PICV (340, 341) backbones to increase viral attenuation, and single-cycle arenaviral vectors (342, 343) to minimize immunosuppression through exponential virus replication.
Another method to produce candidate vaccine in development is based on a reassorted virus known as ML29, which contains the L genomic segment of MOPV and the S genomic segment of LASV (269, 291, 344–346, 418–420) (Figure 3C). It was determined that the LASV NP and GP antigen expressions from this reassorted virus were essential to provide strong protective immunity in rodents (e.g., guinea pigs) and NHPs (e.g., marmosets and Rhesus macaques) against lethal disease caused by LASV infection (344–347). DI particles produced by persistently infected cells with ML29 have also been found to interfere with the replication of LASV, MOPV and LCMV, and to induce strong cell-mediated immunity in STAT1 KO mice (348), implicating the potential for formulation of ML29 with its DIs toward the development of a pan-arenaviral vaccine. Other LASV candidate vaccines include those expressing LASV GP or NP antigens from the genome of different recombinant viruses, such as Vesicular Stomatitis Virus (VSV) (349, 350), vaccinia virus (351, 352), MOPV (291), Yellow Fever Virus 17D (353, 354), alphavirus replicons (355, 356), and inactivated rabies virus (357), all of which have been shown to exhibit various protective efficacies against LASV infection in different animal models. Other LASV vaccine development efforts, which are being supported by a non-profit international organization called Coalition for Epidemic Preparedness Innovations (CEPI), include those that are based on new vaccine platforms, such as a recombinant measles-virus-based vaccine (358) and DNA-based vaccine (359, 360). CEPI has also supported two other LASV vaccine candidates that are based on recombinant rVSV-based vectors. The first is a rVSV construct expressing the LASV GP antigen based on the same platform applied to express the EBOV GP as a vaccine antigen that was used in a ring vaccination trial in Guinea, Africa (361, 362). The second is another rVSV-LASV GP construct with a translocated VSV-N gene and a truncated VSV-G cytoplasmic tail designed for increased vaccine safety. Similar attenuated rVSV vectors expressing single or multiple EBOV GPs have been shown to be immunogenic in NHPs (363) and successfully protected NHPs against EBOV and Marburg virus challenge (364–366), but there are currently no published pre-clinical data to support the development of a Lassa vaccine based on this platform. CEPI has also recently awarded a contract for pre-clinical development of yet another LASV-GP construct on the backbone of a non-replicating simian adenovirus, but again no pre-clinical information is available about the suitability of this viral platform for Lassa vaccine development (367). It remains to be seen how well the CEPI-supported vaccines will perform in the coming years. A formidable challenge of Lassa fever vaccine development is the requirement for a development of predominantly T-cell-mediated mechanism of protection, as strong memory CD4+ T-cell responses to LASV NP and GP2 of different strains of LASV have been demonstrated in some survivors of LASV infection in Guinea (368, 369). This is consistent with observations that, while CD4+ T-cells can provide a partial protection of ML29-vaccinated animals against lethal LASV challenge, depletion of CD8+ T-cells completely abolishes protection (370). Additionally, NP-specific CD8+ T-cells play a major protective role in mice infected with LCMV (279).
In addition to vaccine development as a preventative measure, the discovery of anti-viral drugs is a burgeoning area of arenaviral research. Currently, the only anti-viral treatment clinically in use that is specific to mammarenaviruses is convalescent plasma therapy against JUNV (175). Standard anti-viral nucleoside analogs, such as Ribavirin and Favipiravir (69, 371–374) have seen moderate clinical success, but are only effective when given in the earliest stages of infection when symptoms are primarily non-specific (173). Peptide-conjugated morpholino oligomers have also been tested as alternative nucleoside analogs, reducing the titers of several arenaviruses in cell culture and LCMV-infected mice (375). Nevertheless, recovery has been documented with symptom management in place of nucleoside analogs due to a delay in diagnosis being reached after the resolution of symptoms (376), limiting the desire for using nucleoside analogs as anti-viral compounds.
Recent advances in anti-arenaviral drug discovery have focused on identifying compounds that specifically target viral proteins or that modulate the activity of host proteins (377). The bulk of arenaviral drug discovery has focused on small molecule inhibitors of the mammarenaviral glycoprotein and cell entry, which has seen some successes with several different approaches. Amphipathic DNA polymers have been found to block LCMV GP-α-dystroglycan interaction by the virtue of its hydrophobicity rather than nucleotide sequence (378). Clotrimazole derivatives, which traditionally target the calcium-activated potassium channel KCa3.1, have also recently been found to inhibit arenaviral membrane fusion (379). However, its mechanism of action was found to be independent of KCa3.1, and thus currently unclear.
Conversely, a number of other compounds have been found to stabilize the GP-α-dystroglycan prefusion complex formation, thereby blocking pH-mediated endocytosis (380–384). One of these compounds (ST-193) has been found to reduce LASV titers in a guinea pig infection model (385). Another small molecule (LHF-535) has recently been found to inhibit a wide variety of arenaviruses with the exception of strains that contained a V434I mutation, which corresponds to the mutation responsible for attenuation in Candid #1, suggesting that it could also affect the prefusion stability complex (386). Finally, it has also been shown that a small molecule inhibitor of the cellular site 1 protease (PF-429292) can block GP processing (387, 388) as well as can serve as a general antiviral by inhibiting lipid and cholesterol synthesis needed for virus replication (389–392).
Some pharmaceutical compounds have also been found to disrupt the normal functions of mammarenaviral proteins. Given that myristoylation is necessary for Z-mediated arenavirus budding (160, 161), compounds that inhibit myristoylation enzymes (161, 393) have been found to change the cellular localization of Z and therefore inhibit virion budding from the infected cells. Aromatic compounds targeting the zinc-finger motifs of Z have also been found to inhibit arenaviral proliferation (394, 395). A variety of mechanistic effects have been characterized for one of these compounds. For example, the NSC20625 compound has been found to induce Z to unfold and accumulate in oligomeric structures (394) and thereby blocking the interaction of Z with the host PML protein and allowing nuclear bodies to form (396). The arenaviral L polymerase and NP have arguably experienced the least progress as drug targets. For the former case, one appealing potential pharmaceutical target is the cap-snatching mechanism of arenaviral polymerases. A similar drug has already been demonstrated to reduce titers of the West Nile virus (397). The specificity of viral cap snatching mechanisms (398) and the suspicion that arenaviral cap snatching mechanisms may be conserved across species (399, 400) make cap snatching a promising therapeutic target for anti-viral drug development (401). Some metal chelators have indeed been found to potently inhibit the LCMV L endonuclease domain and its subsequent cap snatching activity, demonstrating the potential for future antivirals targeting this virus feature (402). Lethal viral mutagenesis has also been identified as a possible mechanism for viral control (403, 404). While few studies have found drugs that target arenaviral NPs, one recent paper has identified a compound from a pyridine library that can reduce LCMV titers post-cell entry and is thought to affect NP and Z by unknown mechanisms (405).
Drugs that target host proteins are also being explored as potential pharmaceutical therapeutics. The tyrosine kinase inhibitor genistein is currently one of the most supported compounds, as ATF-2 phosphorylation has been found to be critical for viability of the NW PICV, which is susceptible to genistein in vitro (406, 407) and in vivo (408). Larger scale drug screens seem to implicate cellular kinase inhibition as a promising anti-viral target in general, with compounds affecting arenaviruses at multiple stages of the virus life cycle (409–411). Inhibitors of nucleoside production have also been tested in a variety of in vitro contexts (412–414) and may have additive effects when used in conjunction with nucleoside analogs (412). While these compounds didn't appear to negatively affect the vitality of cell cultures, they should be cautiously explored due to possible adverse effects on some rapidly growing cells in vivo. Finally, it may be possible to target immune signaling proteins as anti-viral control, with one anti-TLR inhibitor showing limiting LCMV-initiated cytokine responses in cells and viral replication in mice (294). For all potential anti-viral drugs, severe side effects (e.g., in neurological and embryonic development) due to known cytotoxicity of these compounds can be a great impediment to the widespread application of these compounds in treating arenavirus infections. Future anti-arenaviral drug and vaccine developments are still urgently needed to combat these deadly human pathogens.
Summary and Future Directions
It is important to reiterate the important roles of the IFN1 pathway and T-cell-mediated immunity in controlling mammarenavirus infections (209). These pathways are also known to be critical for both the innate and the adaptive immune responses to other RNA virus infection models (415, 416) as well as during vaccination (417), further illustrating the importance for future studies of each of these mechanisms separately and in concert. In addition, continuation of ecological efforts to characterize the phylogeny and spread of arenaviruses (2, 9, 18, 21, 46), socioeconomic efforts to increase the public's awareness and capacity to contain disease (49–53), and vaccine and therapeutic developments are needed in order to optimize preventative and control measures for mammarenaviruses.
Author Contributions
MB and HL contributed to the literature review and writing of the manuscript. MB prepared all Figures and Table.
Funding
This work was supported in part by NIH NIAID grant R01 AI131586 to HL and by a pre-doctoral NIH fellowship T32 DA007097 to MB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Dr. Y. Liang for her helpful comments and suggestions about the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00372/full#supplementary-material
Supplemental Table 1. Summary of documented cases of international importation of Lassa fever.
References
1. Radoshitzky SR, Bao Y, Buchmeier MJ, Charrel R, Clawson AN, Clegg CS, et al. Past, present, and future of arenavirus taxonomy. Arch Virol. (2015) 160:1851–74. doi: 10.1007/s00705-015-2418-y
2. Shi M, Lin X-D, Chen X, Tian J-H, Chen L-J, Li K, et al. The evolutionary history of vertebrate RNA viruses. Nature. (2018) 556:197–202. doi: 10.1038/s41586-018-0012-7
3. Grande-Pérez A, Gómez-Mariano G, Lowenstein PR, Domingo E. Mutagenesis-induced, large fitness variations with an invariant arenavirus consensus genomic nucleotide sequence. J Virol. (2005) 79:10451–9. doi: 10.1128/JVI.79.16.10451-10459.2005
4. Emonet S, Lemasson J-J, Gonzalez J-P, de Lamballerie X, Charrel RN. Phylogeny and evolution of old world arenaviruses. Virology. (2006) 350:251–7. doi: 10.1016/J.VIROL.2006.01.026
5. Charrel RN, de Lamballerie X, Emonet S. Phylogeny of the genus Arenavirus. Curr Opin Microbiol. (2008) 11:362–8. doi: 10.1016/j.mib.2008.06.001
6. Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. (2009) 5:e1000455. doi: 10.1371/journal.ppat.1000455
7. Albariño CG, Bird BH, Chakrabarti AK, Dodd KA, Erickson BR, Nichol ST. Efficient rescue of recombinant Lassa virus reveals the influence of S segment noncoding regions on virus replication and virulence. J Virol. (2011) 85:4020–4. doi: 10.1128/JVI.02556-10
8. Safronetz D, Strong JE, Feldmann F, Haddock E, Sogoba N, Brining D, et al. A recently isolated lassa virus from mali demonstrates atypical clinical disease manifestations and decreased virulence in cynomolgus macaques. J Infect Dis. (2013) 207:1316–27. doi: 10.1093/infdis/jit004
9. Siddle KJ, Eromon P, Barnes KG, Mehta S, Oguzie JU, Odia I, et al. Genomic analysis of lassa virus during an increase in cases in Nigeria in 2018. N Engl J Med. (2018) 379: 1745–53. doi: 10.1056/NEJMoa1804498
10. Franze-Fernandez M-T, Zetina C, Iapalucci SI, Lucero MA, Bouissou C, Lopez R, et al. Molecular structure and early events in the replication of Tacaribe arenavirus S RNA (1987). Virus Res. 7:309–24. doi: 10.1016/0168-1702(87)90045-1
11. Iapalucci S, Lopez R, Rey O, Lopez N, Franze-Fernandez MT, Cohen GN, et al. Tacaribe virus L gene encodes a protein of 2210 amino acid residues. Virology. (1989) 170:40–7.
12. Iapalucci S, López N, Franze-Fernández MT. The 3′ end termini of the tacaribe arenavirus subgenomic RNAs. Virology. (1991) 182:269–78. doi: 10.1016/0042-6822(91)90670-7
13. Zhang L, Marriott KA, Harnish DG, Aronson JF. Reassortant analysis of guinea pig virulence of pichinde virus variants. Virology. (2001) 290:30–8. doi: 10.1006/VIRO.2001.1127
14. Charrel RN, Lemasson J-J, Garbutt M, Khelifa R, De Micco P, Feldmann H, et al. New insights into the evolutionary relationships between arenaviruses provided by comparative analysis of small and large segment sequences. Virology. (2003) 317:191–6. doi: 10.1016/J.VIROL.2003.08.016
15. Cajimat MN, Fulhorst CF. Phylogeny of the Venezuelan arenaviruses. Virus Res. (2004) 102:199–206. doi: 10.1016/j.virusres.2004.01.032
16. Cajimat MNB, Milazzo ML, Hess BD, Rood MP, Fulhorst CF. Principal host relationships and evolutionary history of the North American arenaviruses. Virology. (2007) 367:235–43. doi: 10.1016/j.virol.2007.05.031
17. Delgado S, Erickson BR, Agudo R, Blair PJ, Vallejo E, Albariño CG, et al. Chapare virus, a newly discovered arenavirus isolated from a fatal hemorrhagic fever case in Bolivia. PLoS Pathog. (2008) 4:e1000047. doi: 10.1371/journal.ppat.1000047
18. Forni D, Pontremoli C, Pozzoli U, Clerici M, Cagliani R, Sironi M. Ancient evolution of mammarenaviruses: adaptation via changes in the L protein and no evidence for host-virus codivergence. Genome Biol Evol. (2018) 10:863–74. doi: 10.1093/gbe/evy050
19. Centers for Disease Control and Prevention (CDC). Fatal illnesses associated with a new world arenavirus—California, 1999–2000. MMWR Morb Mortal Wkly Rep. (2000) 49:709–11. Available online at: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm4931a1.htm
20. Milazzo ML, Campbell GL, Fulhorst CF. Novel arenavirus infection in humans, United States. Emerg Infect Dis. (2011) 17:1417–20. doi: 10.3201/eid1708.110285
21. Zapata J, Salvato M. Arenavirus variations due to host-specific adaptation. Viruses. (2013) 5:241–78. doi: 10.3390/v5010241
22. Oloniniyi OK, Unigwe US, Okada S, Kimura M, Koyano S, Miyazaki Y, et al. Genetic characterization of Lassa virus strains isolated from 2012 to 2016 in southeastern Nigeria. PLoS Negl Trop Dis. (2018) 12:e0006971. doi: 10.1371/journal.pntd.0006971
23. Bowen MD, Peters CJ, Nichol ST. Phylogenetic analysis of the arenaviridae: patterns of virus evolution and evidence for cospeciation between arenaviruses and their rodent hosts. Mol Phylogenet Evol. (1997) 8:301–16. doi: 10.1006/MPEV.1997.0436
24. Gonzalez JP, Emonet S, de Lamballerie X, Charrel R. Arenaviruses. Curr Top Microbiol Immunol. (2007) 315:253–88. doi: 10.1007/978-3-540-70962-6_11
25. Emonet SF, de la Torre JC, Domingo E, Sevilla N. Arenavirus genetic diversity and its biological implications. Infect Genet Evol. (2009) 9:417–29. doi: 10.1016/J.MEEGID.2009.03.005
26. Irwin NR, Bayerlová M, Missa O, Martínková N. Complex patterns of host switching in New World arenaviruses. Mol Ecol. (2012) 21:4137–50. doi: 10.1111/j.1365-294X.2012.05663.x
27. Blasdell KR, Becker SD, Hurst J, Begon M, Bennett M. Host range and genetic diversity of arenaviruses in rodents, United Kingdom. Emerg Infect Dis. (2008) 14:1455–8. doi: 10.3201/eid1409.080209
28. Ledesma J, Fedele CG, Carro F, Lledó L, Sánchez-Seco MP, Tenorio A, et al. Independent lineage of lymphocytic choriomeningitis virus in wood mice (Apodemus sylvaticus), Spain. Emerg Infect Dis. (2009) 15:1677–80. doi: 10.3201/eid1510.090563
29. Yama IN, Cazaux B, Britton-Davidian J, Moureau G, Thirion L, de Lamballerie X, et al. Isolation and characterization of a new strain of lymphocytic choriomeningitis virus from Rodents in Southwestern France. Vector-Borne Zoonotic Dis. (2012) 12:893–903. doi: 10.1089/vbz.2011.0892
30. Forbes KM, Voutilainen L, Jääskeläinen A, Sironen T, Kinnunen PM, Stuart P, et al. Serological survey of rodent-borne viruses in Finnish field voles. Vector Borne Zoonotic Dis. (2014) 14:278–83. doi: 10.1089/vbz.2013.1526
31. Stephensen CB, Blount SR, Lanford RE, Holmes KV, Montali RJ, Fleenor ME, et al. Prevalence of serum antibodies against lymphocytic choriomeningitis virus in selected populations from two U.S. cities. J Med Virol. (1992) 38:27–31. doi: 10.1002/jmv.1890380107
32. Riera L, Castillo E, Del Carmen Saavedra M, Priotto J, Sottosanti J, Polop J, et al. Serological study of the lymphochoriomeningitis virus (LCMV) in an inner city of Argentina. J Med Virol. (2005) 76:285–9. doi: 10.1002/jmv.20357
33. Hladkii AP. [Study of the strain of lymphocytic choriomeningitis virus isolated from IX. ricinus ticks gathered in the western regions of the Ukrainian SSR]. Mikrobiol Zh. (1965) 27:10–5.
34. Zhang L, Li S, Huang S-J, Wang Z-D, Wei F, Feng X-M, et al. Isolation and genomic characterization of lymphocytic choriomeningitis virus in ticks from northeastern China. Transbound Emerg Dis. (2018) 65:1733–9. doi: 10.1111/tbed.12946
35. Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, Reavill DR, et al. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio. (2012) 3:12. doi: 10.1128/mBio.00180-12
36. Hetzel U, Sironen T, Laurinmäki P, Liljeroos L, Patjas A, Henttonen H, et al. Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease. J Virol. (2013) 87:13. doi: 10.1128/JVI.01123-13
37. Vancraeynest D, Pasmans F, Martel A, Chiers K, Meulemans G, Mast J, et al. Inclusion body disease in snakes: a review and description of three cases in boa constrictors in Belgium. Vet Rec. (2006) 158:757–60. doi: 10.1136/VR.158.22.757
38. Chang L-W, Jacobson ER. Inclusion body disease, a worldwide infectious disease of boid snakes: a review. J Exot Pet Med. (2010) 19:216–25. doi: 10.1053/J.JEPM.2010.07.014
39. Wozniak E, McBride J, DeNardo D, Tarara R, Wong V, Osburn B. Isolation and characterization of an antigenically distinct 68- kd protein from nonviral intracytoplasmic inclusions in boa constrictors chronically infected with the inclusion body disease virus (IBDV: Retroviridae). Vet Pathol. (2000) 37:449–59. doi: 10.1354/vp.37-5-449
40. Schilliger L, Selleri P, Frye FL. Lymphoblastic lymphoma and leukemic blood profile in a red-tail boa (boa constrictor constrictor) with concurrent inclusion body disease. J Vet Diagnostic Investig. (2011) 23:159–62. doi: 10.1177/104063871102300131
41. Hepojoki J, Kipar A, Korzyukov Y, Bell-Sakyi L, Vapalahti O, Hetzel U. Replication of boid inclusion body disease-associated arenaviruses is temperature sensitive in both boid and mammalian cells. J Virol. (2015) 89:1119–28. doi: 10.1128/JVI.03119-14
42. Stenglein MD, Sanchez-Migallon Guzman D, Garcia VE, Layton ML, Hoon-Hanks LL, Boback SM, et al. Differential disease susceptibilities in experimentally reptarenavirus-infected boa constrictors and ball pythons. J Virol. (2017) 91:17. doi: 10.1128/JVI.00451-17
43. Keller S, Hetzel U, Sironen T, Korzyukov Y, Vapalahti O, Kipar A, et al. Co-infecting reptarenaviruses can be vertically transmitted in boa constrictor. PLoS Pathog. (2017) 13:e1006179. doi: 10.1371/journal.ppat.1006179
44. Stenglein MD, Jacobson ER, Chang LW, Sanders C, Hawkins MG, Guzman DSM, et al. Widespread recombination, reassortment, and transmission of unbalanced compound viral genotypes in natural arenavirus infections. PLoS Pathog. (2015) 11:e1004900. doi: 10.1371/journal.ppat.1004900
45. Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J, et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. (2015) 4:05378. doi: 10.7554/eLife.05378
46. Andersen KG, Shapiro BJ, Matranga CB, Sealfon R, Lin AE, Moses LM, et al. Clinical sequencing uncovers origins and evolution of lassa virus. Cell. (2015) 162:738–750. doi: 10.1016/j.cell.2015.07.020
47. Welch SR, Scholte FEM, Albariño CG, Kainulainen MH, Coleman-McCray JD, Guerrero LW, et al. The S genome segment is sufficient to maintain pathogenicity in intra-clade lassa virus reassortants in a guinea pig model. Front Cell Infect Microbiol. (2018) 8:240. doi: 10.3389/fcimb.2018.00240
48. Mariën J, Kourouma F, Magassouba N, Leirs H, Fichet-Calvet E. Movement patterns of small rodents in lassa fever-endemic villages in Guinea. Ecohealth. (2018) 15:348–59. doi: 10.1007/s10393-018-1331-8
49. Bonwitt J, Sáez AM, Lamin J, Ansumana R, Dawson M, Buanie J, et al. At home with mastomys and rattus: human-rodent interactions and potential for primary transmission of lassa virus in domestic spaces. Am J Trop Med Hyg. (2017) 96:16-0675. doi: 10.4269/ajtmh.16-0675
50. Dzingirai V, Bett B, Bukachi S, Lawson E, Mangwanya L, Scoones I, et al. Zoonotic diseases: who gets sick, and why? Explorations from Africa. Crit Public Health. (2017) 27:97–110. doi: 10.1080/09581596.2016.1187260
51. Olowookere SA, Adegbenro CA, Idowu A, Omisore AG, Shabi OM, Ikem UR, et al. Knowledge attitude and practices toward lassa fever control and prevention among residents of Ile-Ife, Southwest Nigeria. Int Q Community Health Edu. (2017) 37:107–12. doi: 10.1177/0272684X17701261
52. Gibb R, Moses LM, Redding DW, Jones KE. Understanding the cryptic nature of Lassa fever in West Africa. Pathog Glob Health. (2017) 111:276–88. doi: 10.1080/20477724.2017.1369643
53. Pigott DM, Deshpande A, Letourneau I, Morozoff C, Reiner RC, Kraemer MUG, et al. Local, national, and regional viral haemorrhagic fever pandemic potential in Africa: a multistage analysis. Lancet. (2017) 390:2662–72. doi: 10.1016/S0140-6736(17)32092-5
54. Iroezindu MO, Unigwe US, Okwara CC, Ozoh GA, Ndu AC, Ohanu ME, et al. Lessons learnt from the management of a case of Lassa fever and follow-up of nosocomial primary contacts in Nigeria during Ebola virus disease outbreak in West Africa. Trop Med Int Health. (2015) 20:1424–30. doi: 10.1111/tmi.12565
55. Akhuemokhan OC, Ewah-Odiase RO, Akpede N, Ehimuan J, Adomeh DI, Odia I, et al. Prevalence of Lassa Virus Disease (LVD) in Nigerian children with fever or fever and convulsions in an endemic area. PLoS Negl Trop Dis. (2017) 11:e0005711. doi: 10.1371/journal.pntd.0005711
56. Hambilon E, Raftery P, Wendland A, Dweh E, Williams G, George R, et al. The challenges of detecting and responding to a Lassa fever outbreak in an Ebola-affected setting. Int J Infect Dis. (2018) 66:65–73. doi: 10.1016/j.ijid.2017.11.007
57. Peterson AT, Moses LM, Bausch DG, Grossman S, Happi C. Mapping transmission risk of lassa fever in West Africa: the importance of quality control, sampling bias, and error weighting. PLoS ONE. (2014) 9:e100711. doi: 10.1371/journal.pone.0100711
58. Mylne AQN, Pigott DM, Longbottom J, Shearer F, Duda KA, Messina JP, et al. Mapping the zoonotic niche of Lassa fever in Africa. Trans R Soc Trop Med Hyg. (2015) 109:483–92. doi: 10.1093/trstmh/trv047
59. Lo Iacono G, Cunningham AA, Fichet-Calvet E, Garry RF, Grant DS, Khan SH, et al. Using modelling to disentangle the relative contributions of zoonotic and anthroponotic transmission: the case of lassa fever. PLoS Negl Trop Dis. (2015) 9:e3398. doi: 10.1371/journal.pntd.0003398
60. Olugasa BO, Odigie EA, Lawani M, Ojo JF. Development of a time-trend model for analyzing and predicting case-pattern of Lassa fever epidemics in Liberia, 2013–2017. Ann Afr Med. (2015) 14:89–96. doi: 10.4103/1596-3519.149892
61. Olayemi A, Cadar D, Magassouba N, Obadare A, Kourouma F, Oyeyiola A, et al. New hosts of the Lassa virus. Sci Rep. (2016) 6:25280. doi: 10.1038/srep25280
62. Gryseels S, Baird SJE, Borremans B, Makundi R, Leirs H, Goüy de Bellocq J. When viruses don't go viral: the importance of host phylogeographic structure in the spatial spread of arenaviruses. PLoS Pathog. (2017) 13:e1006073. doi: 10.1371/journal.ppat.1006073
63. Fichet-Calvet E, Becker-Ziaja B, Koivogui L, Günther S. Lassa serology in natural populations of rodents and horizontal transmission. Vector-Borne Zoonot Dis. (2014) 14:665–74. doi: 10.1089/vbz.2013.1484
64. Borremans B, Vossen R, Becker-Ziaja B, Gryseels S, Hughes N, Van Gestel M, et al. Shedding dynamics of Morogoro virus, an African arenavirus closely related to Lassa virus, in its natural reservoir host Mastomys natalensis. Sci Rep. (2015) 5:10445. doi: 10.1038/srep10445
65. Mariën J, Sluydts V, Borremans B, Gryseels S, Vanden Broecke B, Sabuni CA, et al. Arenavirus infection correlates with lower survival of its natural rodent host in a long-term capture-mark-recapture study. Parasit Vectors. (2018) 11:90. doi: 10.1186/s13071-018-2674-2
66. Macher AM, Wolfe MS. Historical Lassa fever reports and 30-year clinical update. Emerg Infect Dis. (2006) 12:835–7. doi: 10.3201/eid1205.050052
67. Amorosa V, MacNeil A, McConnell R, Patel A, Dillon KE, Hamilton K, et al. Imported Lassa fever, Pennsylvania, USA, 2010. Emerg Infect Dis. (2010) 16:1598–600. doi: 10.3201/EID1610.100774
68. Choi MJ, Worku S, Knust B, Vang A, Lynfield R, Mount MR, et al. A case of Lassa fever diagnosed at a Community Hospital-Minnesota 2014. Open forum Infect Dis. (2018) 5:ofy131. doi: 10.1093/ofid/ofy131
69. Raabe VN, Kann G, Ribner BS, Morales A, Varkey JB, Mehta AK, et al. Favipiravir and Ribavirin treatment of epidemiologically linked cases of Lassa fever. Clin Infect Dis. (2017) 65:855–9. doi: 10.1093/cid/cix406.Favipiravir
70. Kyei NNA, Abilba MM, Kwawu FK, Agbenohevi PG, Bonney JHK, Agbemaple TK, et al. Imported Lassa fever: a report of 2 cases in Ghana. BMC Infect Dis. (2015) 15:217. doi: 10.1186/s12879-015-0956-2
71. Grahn A, Bråve A, Lagging M, Dotevall L, Ekqvist D, Hammarström H, et al. Imported case of Lassa fever in Sweden with encephalopathy and sensorineural hearing deficit. Open Forum Infect Dis. (2016) 3:ofw198. doi: 10.1093/ofid/ofw198
72. Ehlkes L, George M, Samosny G, Burckhardt F, Vogt M, Bent S, et al. Management of a Lassa fever outbreak, Rhineland-Palatinate, Germany, 2016. Eurosurveillance. (2017) 22:16-00728. doi: 10.2807/1560-7917.ES.2017.22.39.16-00728
73. Lehmann C, Kochanek M, Abdulla D, Becker S, Böll B, Bunte A, et al. Control measures following a case of imported Lassa fever from Togo, North Rhine Westphalia, Germany, 2016. Eurosurveillance. (2017) 22:88. doi: 10.2807/1560-7917.ES.2017.22.39.17-00088
74. Kofman A, Choi MJ, Rollin PE. Lassa fever in travelers from West Africa, 1969–2016. Emerg Infect Dis. (2019) 25:245–8. doi: 10.3201/eid2502.180836
75. Bronze MS, Huycke MM, Machado LJ, Voskuhl GW, Greenfield RA. Viral agents as biological weapons and agents of bioterrorism. Am J Med Sci. (2002) 323:316–25. doi: 10.1097/00000441-200206000-00004
76. Hartnett JN, Boisen ML, Oottamasathien D, Jones AB, Millett MM, Nelson DS, et al. Current and emerging strategies for the diagnosis, prevention and treatment of Lassa fever. Future Virol. (2015) 10:559–84. doi: 10.2217/fvl.15.41
77. Shaffer JG, Grant DS, Schieffelin JS, Boisen ML, Goba A, Hartnett JN, et al. Lassa fever in post-conflict sierra leone. PLoS Negl Trop Dis. (2014) 8:e2748. doi: 10.1371/journal.pntd.0002748
78. Fischer SA, Graham MB, Kuehnert MJ, Kotton CN, Srinivasan A, Marty FM, et al. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N Engl J Med. (2006) 354:2235–49. doi: 10.1056/NEJMoa053240
79. Lenz O, ter Meulen J, Klenk HD, Seidah NG, Garten W. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc Natl Acad Sci USA. (2001) 98:12701–5. doi: 10.1073/pnas.221447598
80. Beyer WR, Pöpplau D, Garten W, von Laer D, Lenz O. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J Virol. (2003) 52:897–904. doi: 10.1128/jvi.77.5.2866-2872.2003
81. Rojek JM, Lee AM, Nguyen N, Spiropoulou CF, Kunz S. Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junin, Machupo, and Guanarito. J Virol. (2008) 82:6045–51. doi: 10.1128/JVI.02392-07
82. Schlie K, Maisa A, Lennartz F, Ströher U, Garten W, Strecker T. Characterization of Lassa virus glycoprotein oligomerization and influence of cholesterol on virus replication. J Virol. (2010) 84:983–92. doi: 10.1128/JVI.02039-09
83. Burri DJ, Pasquato A, da Palma JR, Igonet S, Oldstone MBA, Kunz S. The role of proteolytic processing and the stable signal peptide in expression of the Old World arenavirus envelope glycoprotein ectodomain. Virology. (2013) 436:127–33. doi: 10.1016/j.virol.2012.10.038
84. Hastie KM, Zandonatti MA, Kleinfelter LM, Heinrich ML, Rowland MM, Chandran K, et al. Structural basis for antibody-mediated neutralization of Lassa virus. Science. (2017) 356:923–8. doi: 10.1126/science.aam7260
85. Gallaher WR, DiSimone C, Buchmeier MJ. The viral transmembrane superfamily: possible divergence of Arenavirus and Filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol. (2001) 1:1. doi: 10.1186/1471-2180-1-1
86. Eschli B, Quirin K, Wepf A, Weber J, Zinkernagel R, Hengartner H. Identification of an N-terminal trimeric coiled-coil core within arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J Virol. (2006) 80:5897–907. doi: 10.1128/JVI.00008-06
87. Nunberg JH, York J. The curious case of arenavirus entry, and its inhibition. Viruses. (2012) 4:83–101. doi: 10.3390/v4010083
88. Froeschke M, Basler M, Groettrup M, Dobberstein B. Long-lived signal peptide of lymphocytic choriomeningitis virus glycoprotein pGP-C. J Biol Chem. (2003) 278:41914–20. doi: 10.1074/jbc.M302343200
89. York J, Romanowski V, Lu M, Nunberg JH. The signal peptide of the Junín arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J Virol. (2004) 78:10783–92. doi: 10.1128/JVI.78.19.10783-10792.2004
90. York J, Nunberg JH. Myristoylation of the arenavirus envelope glycoprotein stable signal peptide is critical for membrane fusion but dispensable for virion morphogenesis. J Virol. (2016) 90:8341–50. doi: 10.1128/JVI.01124-16
91. Agnihothram SS, York J, Nunberg JH. Role of the stable signal peptide and cytoplasmic domain of G2 in regulating intracellular transport of the Junín virus envelope glycoprotein complex. J Virol. (2006) 80:5189–98. doi: 10.1128/JVI.00208-06
92. Eichler R, Lenz O, Strecker T, Eickmann M, Klenk H-D, Garten W. Identification of Lassa virus glycoprotein signal peptide as a trans-acting maturation factor. EMBO Rep. (2003) 4:1084–8. doi: 10.1038/sj.embor.embor7400002
93. Schlie K, Strecker T, Garten W. Maturation cleavage within the ectodomain of Lassa virus glycoprotein relies on stabilization by the cytoplasmic tail. FEBS Lett. (2010) 584:4379–82. doi: 10.1016/j.febslet.2010.09.032
94. Messina EL, York J, Nunberg JH. Dissection of the role of the stable signal peptide of the arenavirus envelope glycoprotein in membrane fusion. J Virol. (2012) 86:6138–45. doi: 10.1128/JVI.07241-11
95. Shao J, Liu X, Ly H, Liang Y. Characterization of the glycoprotein stable signal peptide in mediating pichinde virus replication and virulence. J Virol. (2016) 90:10390–7. doi: 10.1128/JVI.01154-16
96. Eichler R, Lenz O, Garten W, Strecker T. The role of single N-glycans in proteolytic processing and cell surface transport of the Lassa virus glycoprotein GP-C. Virol J. (2006) 3:41. doi: 10.1186/1743-422X-3-41
97. Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV, et al. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. (1998) 282:2079–81. doi: 10.1126/science.282.5396.2079
98. Spiropoulou CF, Kunz S, Rollin PE, Campbell KP, Oldstone MBA. New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor. J Virol. (2002) 76:5140–6. doi: 10.1128/JVI.76.10.5140-5146.2002
99. Kanagawa M, Saito F, Kunz S, Yoshida-Moriguchi T, Barresi R, Kobayashi YM, et al. Molecular recognition by LARGE is essential for expression of functional dystroglycan. Cell. (2004) 117:953–64. doi: 10.1016/j.cell.2004.06.003
100. Kunz S, Rojek JM, Kanagawa M, Spiropoulou CF, Barresi R, Campbell KP, et al. Posttranslational modification of alpha-dystroglycan, the cellular receptor for arenaviruses, by the glycosyltransferase LARGE is critical for virus binding. J Virol. (2005) 79:14282–96. doi: 10.1128/JVI.79.22.14282-14296.2005
101. Inamori K, Yoshida-Moriguchi T, Hara Y, Anderson ME, Yu L, Campbell KP. Dystroglycan function requires xylosyl- and glucuronyltransferase activities of LARGE. Science. (2012) 335:93–6. doi: 10.1126/science.1214115
102. Rojek JM, Spiropoulou CF, Campbell KP, Kunz S. Old World and Clade C New World arenaviruses mimic the molecular mechanism of receptor recognition used by-dystroglycan's host-derived ligands. J Virol. (2007) 81:5685–95. doi: 10.1128/JVI.02574-06
103. Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. (2007) 449:913–8. doi: 10.1038/nature06250
104. Andersen KG, Shylakhter I, Tabrizi S, Grossman SR, Happi CT, Sabeti PC. Genome-wide scans provide evidence for positive selection of genes implicated in Lassa fever. Philos Trans R Soc Lond B Biol Sci. (2012) 367:868–77. doi: 10.1098/rstb.2011.0299
105. Shao J, Liang Y, Ly H. Human hemorrhagic Fever causing arenaviruses: molecular mechanisms contributing to virus virulence and disease pathogenesis. Pathogens. (2015) 4:283–306. doi: 10.3390/pathogens4020283
106. Jae LT, Raaben M, Herbert AS, Kuehne AI, Wirchnianski AS, Soh TK, et al. Lassa virus entry requires a trigger-induced receptor switch. Science. (2014) 344:1506–10. doi: 10.1126/science.1252480
107. Hastie KM, Igonet S, Sullivan BM, Legrand P, Zandonatti MA, Robinson JE, et al. Crystal structure of the prefusion surface glycoprotein of the prototypic arenavirus LCMV. Nat Struct Mol Biol. (2016) 23:513–21. doi: 10.1038/nsmb.3210
108. Acciani M, Alston JT, Zhao G, Reynolds H, Ali AM, Xu B, et al. Mutational analysis of lassa virus glycoprotein highlights regions required for alpha-dystroglycan utilization. J Virol. (2017) 91:e00574-17. doi: 10.1128/JVI.00574-17
109. Klewitz C, Klenk H-D, ter Meulen J. Amino acids from both N-terminal hydrophobic regions of the Lassa virus envelope glycoprotein GP-2 are critical for pH-dependent membrane fusion and infectivity. J Gen Virol. (2007) 88:2320–8. doi: 10.1099/vir.0.82950-0
110. Sommerstein R, Ramos da Palma J, Olschläger S, Bergthaler A, Barba L, Lee BP-L, et al. Evolution of recombinant lymphocytic choriomeningitis virus/Lassa virus in vivo highlights the importance of the GPC cytosolic tail in viral fitness. J Virol. (2014) 88:8340–8. doi: 10.1128/JVI.00236-14
111. Sevilla N, Kunz S, Holz A, Lewicki H, Homann D, Yamada H, et al. Immunosuppression and resultant viral persistence by specific viral targeting of dendritic cells. J Exp Med. (2000) 192:1249–60. doi: 10.1084/jem.192.9.1249
112. Kunz S, Sevilla N, McGavern DB, Campbell KP, Oldstone MB. Molecular analysis of the interaction of LCMV with its cellular receptor [alpha]-dystroglycan. J Cell Biol. (2001) 155:301–10. doi: 10.1083/jcb.200104103
113. Smelt SC, Borrow P, Kunz S, Cao W, Tishon A, Lewicki H, et al. Differences in affinity of binding of lymphocytic choriomeningitis virus strains to the cellular receptor alpha-dystroglycan correlate with viral tropism and disease kinetics. J Virol. (2001) 75:448–57. doi: 10.1128/JVI.75.1.448-457.2001
114. Jae LT, Brummelkamp TR. Emerging intracellular receptors for hemorrhagic fever viruses. Trends Microbiol. (2015) 23:6. doi: 10.1016/j.tim.2015.04.006
115. Cohen-Dvashi H, Cohen N, Israeli H, Diskin R. Molecular mechanism for LAMP1 recognition by Lassa virus. J Virol. (2015) 89:7584–92. doi: 10.1128/JVI.00651-15
116. Hulseberg CE, Fénéant L, Szymanska KM, White JM. Lamp1 increases the efficiency of lassa virus infection by promoting fusion in less acidic endosomal compartments. mBio. (2018) 9:17. doi: 10.1128/mBio.01818-17
117. Iwasaki M, Ngo N, de la Torre JC. Sodium hydrogen exchangers contribute to arenavirus cell entry. J Virol. (2014) 88:643–54. doi: 10.1128/JVI.02110-13
118. Fedeli C, Torriani G, Galan-Navarro C, Moraz M-L, Moreno H, Gerold G, et al. Axl can serve as entry factor for Lassa virus depending on the functional glycosylation. J Virol. (2017). doi: 10.1128/JVI.01613-17
119. Brouillette RB, Phillips EK, Patel R, Mahauad-Fernandez W, Moller-Tank S, Rogers KJ, et al. TIM-1 Mediates Dystroglycan-Independent Entry of Lassa Virus. J Virol. (2018) 92:e00093–18. doi: 10.1128/JVI.00093-18
120. Loureiro M, D'Antuono A, López N, Loureiro ME, D'Antuono A, López N. Virus-host interactions involved in lassa virus entry and genome replication. Pathogens. (2019) 8:17. doi: 10.3390/pathogens8010017
121. Raaben M, Jae LT, Herbert AS, Kuehne AI, Stubbs SH, Chou Y, et al. NRP2 and CD63 are host factors for lujo virus cell entry. Cell Host Microbe. (2017) 22:688–96.e5. doi: 10.1016/J.CHOM.2017.10.002
122. Radoshitzky SR, Abraham J, Spiropoulou CF, Kuhn JH, Nguyen D, Li W, et al. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature. (2007) 446:92–6. doi: 10.1038/nature05539
123. Zong M, Fofana I, Choe H. Human and host species transferrin receptor 1 use by North American arenaviruses. J Virol. (2014) 88:9418–28. doi: 10.1128/JVI.01112-14
124. Radoshitzky SR, Kuhn JH, Spiropoulou CF, Albariño CG, Nguyen DP, Salazar-Bravo J, et al. Receptor determinants of zoonotic transmission of New World hemorrhagic fever arenaviruses. Proc Natl Acad Sci USA. (2008) 105:2664–9. doi: 10.1073/pnas.0709254105
125. Burri DJ, da Palma JR, Kunz S, Pasquato A. Envelope glycoprotein of arenaviruses. Viruses. (2012) 4:2162–81. doi: 10.3390/v4102162
126. Martinez MG, Bialecki MA, Belouzard S, Cordo SM, Candurra NA, Whittaker GR. Utilization of human DC-SIGN and L-SIGN for entry and infection of host cells by the New World arenavirus, Junín virus. Biochem Biophys Res Commun. (2013) 441:106. doi: 10.1016/j.bbrc.2013.10.106
127. Lavanya M, Cuevas CD, Thomas M, Cherry S, Ross SR. siRNA screen for genes that affect Junín virus entry uncovers voltage-gated calcium channels as a therapeutic target. Sci Transl Med. (2013) 5:204ra131. doi: 10.1126/scitranslmed.3006827
128. Martinez MG, Forlenza MB, Candurra NA. Involvement of cellular proteins in Junin arenavirus entry. Biotechnol J. (2009) 4:866–70. doi: 10.1002/biot.200800357
129. Sarute N, Ibrahim N, Medegan Fagla B, Lavanya M, Cuevas C, Stavrou S, et al. TRIM2, a novel member of the antiviral family, limits New World arenavirus entry. PLoS Biol. (2019) 17:e3000137. doi: 10.1371/journal.pbio.3000137
130. Reignier T, Oldenburg J, Flanagan ML, Hamilton GA, Martin VK, Cannon PM. Receptor use by the Whitewater Arroyo virus glycoprotein. Virology. (2008) 371:439–46. doi: 10.1016/j.virol.2007.10.004
131. Rojek JM, Sanchez AB, Nguyen NT, de la Torre J-C, Kunz S. Different mechanisms of cell entry by human-pathogenic Old World and New World arenaviruses. J Virol. (2008) 82:7677–87. doi: 10.1128/JVI.00560-08
132. Pasqual G, Rojek JM, Masin M, Chatton J-Y, Kunz S. Old World arenaviruses enter the host cell via the multivesicular body and depend on the endosomal sorting complex required for transport. PLoS Pathog. (2011) 7:e1002232. doi: 10.1371/journal.ppat.1002232
133. Di Simone C, Zandonatti MA, Buchmeier MJ. Acidic pH triggers LCMV membrane fusion activity and conformational change in the glycoprotein spike. Virology. (1994) 198:455–65. doi: 10.1006/VIRO.1994.1057
134. Di Simone C, Buchmeier MJ. Kinetics and pH dependence of acid-induced structural changes in the lymphocytic choriomeningitis virus glycoprotein complex. Virology. (1995) 209:3–9. doi: 10.1006/VIRO.1995.1225
135. Willard K, Alston J, Acciani M, Brindley M, Willard KA, Alston JT, et al. Identification of residues in lassa virus glycoprotein subunit 2 that are critical for protein function. Pathogens. (2018) 8:1. doi: 10.3390/pathogens8010001
136. Lee KJ, Novella IS, Teng MN, Oldstone MB, de La Torre JC. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol. (2000) 74:3470–7. doi: 10.1128/jvi.74.8.3470-3477.2000
137. López N, Jácamo R, Franze-Fernández MT. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J Virol. (2001) 75:12241–51. doi: 10.1128/JVI.75.24.12241-12251.2001
138. Hass M, Gölnitz U, Müller S, Becker-Ziaja B, Günther S. Replicon system for Lassa virus. J Virol. (2004) 78:13793–803. doi: 10.1128/JVI.78.24.13793-13803.2004
139. Iwasaki M, de la Torre JC. A highly conserved leucine in mammarenavirus matrix Z protein is required for Z interaction with the virus L polymerase and Z stability in cells harboring an active viral ribonucleoprotein. J Virol. (2018) 92:e02256–17. doi: 10.1128/JVI.02256-17
140. Kranzusch PJ, Whelan SPJ. Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proc Natl Acad Sci. (2011) 108:19743–8. doi: 10.1073/pnas.1112742108
141. Salvato MS, Shimomaye EM. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology. (1989) 173:1–10.
142. Perez M, de la Torre JC. Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. (2003) 77:1184–94. doi: 10.1128/JVI.77.2.1184-1194.2003
143. Foscaldi S, D'Antuono A, Noval MG, de Prat Gay G, Scolaro L, Lopez N. Regulation of tacaribe mammarenavirus translation: positive 5′ and negative 3′ elements and role of key cellular factors. J Virol. (2017) 91:e00084–17. doi: 10.1128/JVI.00084-17
144. Tortorici M, Albariño C, Posik D, Ghiringhelli P, Lozano M, Rivera Pomar R, et al. Arenavirus nucleocapsid protein displays a transcriptional antitermination activity in vivo. Virus Res. (2001) 73:41–55. doi: 10.1016/S0168-1702(00)00222-7
145. López N, Franze-Fernández MT. A single stem-loop structure in Tacaribe arenavirus intergenic region is essential for transcription termination but is not required for a correct initiation of transcription and replication. Virus Res. (2007) 124:237–44. doi: 10.1016/J.VIRUSRES.2006.10.007
146. Iwasaki M, Ngo N, Cubitt B, Teijaro JR, de la Torre JC. General molecular strategy for development of arenavirus live-attenuated vaccines. J Virol. (2015) 89:12166–77. doi: 10.1128/JVI.02075-15
147. Iwasaki M, Cubitt B, Sullivan BM, de la Torre JC. The high degree of sequence plasticity of the arenavirus noncoding intergenic region (IGR) enables the use of a nonviral universal synthetic IGR to attenuate arenaviruses. J Virol. (2016) 90:3187–97. doi: 10.1128/JVI.03145-15
148. Garcin D, Kolakofsky D. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J Virol. (1992) 66:1370–6.
149. Marq J-B, Hausmann S, Veillard N, Kolakofsky D, Garcin D. Short double-stranded RNAs with an overhanging 5' ppp-nucleotide, as found in arenavirus genomes, act as RIG-I decoys. J Biol Chem. (2011) 286:6108–16. doi: 10.1074/jbc.M110.186262
150. Baird NL, York J, Nunberg JH. Arenavirus infection induces discrete cytosolic structures for RNA replication. J Virol. (2012) 86:11301–10. doi: 10.1128/JVI.01635-12
151. Mateer EJ, Paessler S, Huang C. Visualization of double-stranded RNA colocalizing with pattern recognition receptors in arenavirus infected cells. Front Cell Infect Microbiol. (2018) 8:251. doi: 10.3389/fcimb.2018.00251
152. King BR, Samacoits A, Eisenhauer PL, Ziegler CM, Bruce EA, Zenklusen D, et al. Visualization of arenavirus RNA species in individual cells by single-molecule fluorescence in situ hybridization (smFISH) suggests a model of cyclical infection and clearance during persistence. J Virol. (2018) 92:e02241–17. doi: 10.1128/JVI.02241-17
153. Meyer BJ, De La Torre JC, Southern PJ. Arenaviruses: genomic RNAs, transcription, and replication. Curr Top Microbiol Immunol. (2002) 262:139–57. doi: 10.1007/978-3-642-56029-3_6
154. Casabona JC, Levingston Macleod JM, Loureiro ME, Gomez GA, Lopez N. The RING domain and the L79 residue of Z protein are involved in both the rescue of nucleocapsids and the incorporation of glycoproteins into infectious chimeric arenavirus-like particles. J Virol. (2009) 83:7029–39. doi: 10.1128/JVI.00329-09
155. Shtanko O, Imai M, Goto H, Lukashevich IS, Neumann G, Watanabe T, et al. A role for the C terminus of Mopeia virus nucleoprotein in its incorporation into Z protein-induced virus-like particles. J Virol. (2010) 84:5415–22. doi: 10.1128/JVI.02417-09
156. Capul AA, Perez M, Burke E, Kunz S, Buchmeier MJ, de la Torre JC. Arenavirus Z-glycoprotein association requires Z myristoylation but not functional RING or late domains. J Virol. (2007) 81:9451–60. doi: 10.1128/JVI.00499-07
157. Urata S, Yasuda J. Cis- and cell-type-dependent trans-requirements for Lassa virus-like particle production. J Gen Virol. (2015) 96:1626–35. doi: 10.1099/vir.0.000105
158. Wilda M, Lopez N, Casabona JC, Franze-Fernandez MT. Mapping of the tacaribe arenavirus Z-protein binding sites on the L protein identified both amino acids within the putative polymerase domain and a region at the N terminus of L that are critically involved in binding. J Virol. (2008) 82:11454–60. doi: 10.1128/JVI.01533-08
159. Urata S, Noda T, Kawaoka Y, Yokosawa H, Yasuda J. Cellular factors required for Lassa virus budding. J Virol. (2006) 80:4191–5. doi: 10.1128/JVI.80.8.4191-4195.2006
160. Perez M, Greenwald DL, de La Torre JC. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J Virol. (2004) 78:11443–8. doi: 10.1128/JVI.78.20.11443-11448.2004
161. Strecker T, Maisa A, Daffis S, Eichler R, Lenz O, Garten W. The role of myristoylation in the membrane association of the Lassa virus matrix protein Z. Virol J. (2006) 3:93. doi: 10.1186/1743-422X-3-93
162. Hoenen T, Kolesnikova L, Becker S. Recent advances in filovirus- and arenavirus-like particles. Future Virol. (2007) 2:193–203. doi: 10.2217/17460794.2.2.193
163. Ziegler CM, Eisenhauer P, Bruce EA, Weir ME, King BR, Klaus JP, et al. The lymphocytic choriomeningitis virus matrix protein PPXY late domain drives the production of defective interfering particles. PLoS Pathog. (2016) 12:e1005501. doi: 10.1371/journal.ppat.1005501
164. Strecker T, Eichler R, ter Meulen J, Weissenhorn W, Dieter Klenk H, Garten W, et al. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles. J Virol. (2003) 77:10700–5. doi: 10.1128/JVI.77.19.10700
165. Perez M, Craven RC, de la Torre JC. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci USA. (2003) 100:12978–83. doi: 10.1073/pnas.2133782100
166. Huang AS, Baltimore D. Defective viral particles and viral disease processes. Nature. (1970) 226:325–7. doi: 10.1038/226325a0
167. Huang AS. Defective interfering viruses. Annu Rev Microbiol. (1973) 27:101–18. doi: 10.1146/annurev.mi.27.100173.000533
168. Meyer BJ, Southern PJ. A novel type of defective viral genome suggests a unique strategy to establish and maintain persistent lymphocytic choriomeningitis virus infections. J Virol. (1997) 71:6757–64.
169. Welsh RM, Pfau CJ. Determinants of lymphocytic choriomeningitis interference. J Gen Virol. (1972) 14:177–87. doi: 10.1099/0022-1317-14-2-177
170. Popescu M, Lehmann-Grube F. Defective interfering particles in mice infected with lymphocytic choriomengingitis virus. Virology. (1977) 77:78–83. doi: 10.1016/0042-6822(77)90407-X
171. Burns JW, Buchmeier MJ. Glycoproteins of the arenaviruses. In: Salvato, MS, editor. The Arenaviridae. Boston, MA: Springer, 17–35.
172. Oldstone MB. Viral persistence: mechanisms and consequences. Curr Opin Microbiol. (1998) 1:436–41. doi: 10.1016/S1369-5274(98)80062-3
173. Günther S, Lenz O. Lassa virus. Crit Rev Clin Lab Sci. (2004) 41:339–90. doi: 10.1080/10408360490497456
174. Damonte EB, Coto CE. Treatment of arenavirus infections: from basic studies to the challenge of antiviral therapy. Adv Virus Res. (2002) 58:125–55. doi: 10.1016/S0065-3527(02)58004-0
175. Maiztegui J, Fernandez N, De Damilano A. Efficiacy of immune plasma in treatment of Argentine haemorrhagic fever and association between treatment and a late neurological syndrome. Lancet. (1979) 314:1216–7. doi: 10.1016/S0140-6736(79)92335-3
176. Koma T, Huang C, Kolokoltsova OA, Brasier AR, Paessler S. Innate immune response to arenaviral infection: a focus on the highly pathogenic new world hemorrhagic arenaviruses. J Mol Biol. (2013) 425:4893–903. doi: 10.1016/j.jmb.2013.09.028
177. Nigeria|Relief Web. Nigeria—Lassa Fever Epidemic Outbreak (DG ECHO, NCDC/Government of Nigeria, WHO) (ECHO Daily Flash of 14 February 2018). (2018). Available online at: https://erccportal.jrc.ec.europa.eu/ECHO-Flash/ECHO-Flash-List/yy/2018/mm/2. (Accessed October 24, 2018).
178. Nigeria Centre for Disease Control. An Update of Lassa Fever Outbreak in Nigeria. (2018). Available online at: https://www.ncdc.gov.ng/diseases/sitreps/?cat=5&name=An_update_of_Lassa_fever_outbreak_in_Nigeria
179. Roberts L. Nigeria hit by unprecedented Lassa fever outbreak. Science. (2018) 359:1201–2. doi: 10.1126/science.359.6381.1201
180. McCormick JB, Webb PA, Krebs JW, Johnson KM, Smith ES. A prospective study of the epidemiology and ecology of Lassa fever. J Infect Dis. (1987) 155:437–44.
181. Price ME, Fisher-Hoch SP, Craven RB, McCormick JB. A prospective study of maternal and fetal outcome in acute Lassa fever infection during pregnancy. BMJ. (1988) 297:584–7.
182. Delaine M, Weingertner A-S, Nougairede A, Lepiller Q, Fafi-Kremer S, Favre R, et al. Congenital lymphocytic choriomeningitis virus: a case of prenatal diagnosis. J Ultrasound Med. (2018) 37:804–6. doi: 10.1002/jum.14372
183. Armstrong C. Experimental lymphocytic choriomeningitis of monkeys and mice produced by a virus encountered in studies of the 1933 St. Louis epidemic. Public Heal Rep. (1934) 49:e4581290. doi: 10.2307/4581290
184. Johnson KM, McCormick JB, Webb PA, Smith ES, Elliott LH, King IJ. Clinical virology of Lassa fever in hospitalized patients. J Infect Dis. (1987) 155:456–64. doi: 10.1093/infdis/155.3.456
185. Sewlall NH, Richards G, Duse A, Swanepoel R, Paweska J, Blumberg L, et al. Clinical features and patient management of Lujo hemorrhagic fever. PLoS Negl Trop Dis. (2014) 8:e3233. doi: 10.1371/journal.pntd.0003233
186. Yun NE, Walker DH. Pathogenesis of Lassa fever. Viruses. (2012) 4:2031–48. doi: 10.3390/v4102031
187. Kunz S. The role of the vascular endothelium in arenavirus haemorrhagic fevers. Thromb Haemost. (2009) 102:1024–9. doi: 10.1160/TH09-06-0357
188. Bell TM, Shaia CI, Bunton TE, Robinson CG, Wilkinson ER, Hensley LE, et al. Pathology of experimental machupo virus infection, chicava strain, in cynomolgus macaques (Macaca fascicularis) by intramuscular and aerosol exposure. Vet Pathol. (2015) 52:26–37. doi: 10.1177/0300985814540544
189. Liu DX, Perry DL, Evans DeWald L, Cai Y, Hagen KR, Cooper TK, et al. Persistence of lassa virus associated with severe systemic arteritis in convalescing guinea pigs (Cavia porcellus). J Infect Dis. (2018). doi: 10.1093/infdis/jiy641. [Epub ahead of print].
190. Jahrling PB, Hesse RA, Rhoderick JB, Elwell MA, Moe JB. Pathogenesis of a pichinde virus strain adapted to produce lethal infections in guinea pigs. Infect Immun. (1981) 32:872–80.
191. Bell TM, Shaia CI, Bearss JJ, Mattix ME, Koistinen KA, Honnold SP, et al. Temporal progression of lesions in guinea pigs infected with Lassa virus. Vet Pathol. (2017) 54:549–62. doi: 10.1177/0300985816677153
192. Lang E, Pozdeev VI, Shinde PV, Xu HC, Sundaram B, Zhuang Y, et al. Cholestasis induced liver pathology results in dysfunctional immune responses after arenavirus infection. Sci Rep. (2018) 8:12179. doi: 10.1038/s41598-018-30627-y
193. Oestereich L, Lüdtke A, Ruibal P, Pallasch E, Kerber R, Rieger T, et al. Chimeric mice with competent hematopoietic immunity reproduce key features of severe Lassa fever. PLoS Pathog. (2016) 12:e1005656. doi: 10.1371/journal.ppat.1005656
194. Campbell C, Phillips A, Rico A, McGuire A, Aboellail T, Quackenbush S, et al. Involvement of pro-inflammatory macrophages in liver pathology of pirital virus-infected syrian hamsters. Viruses. (2018) 10:232. doi: 10.3390/v10050232
195. Gale TV, Horton TM, Grant DS, Garry RF. Metabolomics analyses identify platelet activating factors and heme breakdown products as Lassa fever biomarkers. PLoS Negl Trop Dis. (2017) 11:e0005943. doi: 10.1371/journal.pntd.0005943
196. McCormick JB, King IJ, Webb PA. A case-control study of the clinical diagnosis and course of Lassa fever. J Infect Dis. (1987) 155:445–55.
197. Cummins D, McCormick JB, Bennett D, Samba JA, Farrar B, Machin SJ, et al. Acute sensorineural deafness in Lassa fever. JAMA J Am Med Assoc. (1990) 264:2093. doi: 10.1001/jama.1990.03450160063030
198. Ibekwe TS, Okokhere PO, Asogun D, Blackie FF, Nwegbu MM, Wahab KW, et al. Early-onset sensorineural hearing loss in Lassa fever. Eur Arch Oto-Rhino-Laryngol. (2011) 268:197–201. doi: 10.1007/s00405-010-1370-4
199. Mateer EJ, Huang C, Shehu NY, Paessler S. Lassa fever-induced sensorineural hearing loss: a neglected public health and social burden. PLoS Negl Trop Dis. (2018) 12:e0006187. doi: 10.1371/journal.pntd.0006187
200. Yun NE, Ronca S, Tamura A, Koma T, Seregin AV, Dineley KT, et al. Animal model of sensorineural hearing loss associated with Lassa virus infection. J Virol. (2015) 90:2920–7. doi: 10.1128/JVI.02948-15
201. Cashman KA, Wilkinson ER, Zeng X, Cardile AP, Facemire PR, Bell TM, et al. Immune-mediated systemic vasculitis as the proposed cause of sudden-onset sensorineural hearing loss following lassa virus exposure in cynomolgus macaques. mBio. (2018) 9:e01896–18. doi: 10.1128/mBio.01896-18
202. Wilson MR, Peters CJ. Diseases of the central nervous system caused by lymphocytic choriomeningitis virus and other arenaviruses. Handb Clin Neurol. 123:671–81. doi: 10.1016/B978-0-444-53488-0.00033-X
203. Bonthius DJ. Lymphocytic choriomeningitis virus: an underrecognized cause of neurologic disease in the fetus, child, and adult. Semin Pediatr Neurol. (2012) 19:89–95. doi: 10.1016/j.spen.2012.02.002
204. Baize S, Kaplon J, Faure C, Pannetier D, Georges-Courbot M-C, Deubel V. Lassa virus infection of human dendritic cells and macrophages is productive but fails to activate cells. J Immunol. (2004) 172:2861–9. doi: 10.4049/JIMMUNOL.172.5.2861
205. Hensley LE, Smith MA, Geisbert JB, Fritz EA, Daddario-DiCaprio KM, Larsen T, et al. Pathogenesis of Lassa fever in cynomolgus macaques. Virol J. (2011) 8:205. doi: 10.1186/1743-422X-8-205
206. González PH, Cossio PM, Arana R, Maiztegui JI, Laguens RP. Lymphatic tissue in Argentine hemorrhagic fever. Pathologic features. Arch Pathol Lab Med. (1980) 104:250–4.
207. Buchmeier MJ, de la Torre J-C, Peters CJ. Arenaviridae. In: Knipe DM, Holey P, editors. Fields Virology. Philadelphia, PA: Lippincott Williams &Wilkins (2018). p. 1283–303.
208. Geisbert TW, Jahrling PB. Exotic emerging viral diseases: progress and challenges. Nat Med. (2004) 10:S110–21. doi: 10.1038/nm1142
209. Meyer B, Ly H. Inhibition of innate immune responses is key to pathogenesis by arenaviruses. J Virol. (2016) 90:3810–8. doi: 10.1128/JVI.03049-15
210. Walker DH, McCormick JB, Johnson KM, Webb PA, Komba-Kono G, Elliott LH, et al. Pathologic and virologic study of fatal Lassa fever in man. Am J Pathol. (1982) 107:349–56.
211. Hall WC, Geisbert TW, Huggins JW, Jahrling PB. Experimental guinea pigs with venezuelan henorrhagic fever virus (Guanarito): a model of human disease. Am J Trop Med Hyg. (1996) 55:81–8.
212. Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. (2008) 89:1–47. doi: 10.1099/vir.0.83391-0
213. Marcus PI, Sekellick MJ. Defective interfering particles with covalently linked [+/-]RNA induce interferon. Nature. (1977) 266:815–9. doi: 10.1038/266815a0
214. Marcus PI, Svitlik C, Sekellick MJ. Interferon induction by viruses. X. A model for interferon induction by Newcastle disease virus. J Gen Virol. (1983) 64:2419–31. doi: 10.1099/0022-1317-64-11-2419
215. Gack MU. Mechanisms of RIG-I-like receptor activation and manipulation by viral pathogens. J Virol. (2014) 88:5213–6. doi: 10.1128/JVI.03370-13
216. Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. (2009) 1:34. doi: 10.1101/cshperspect.a000034
217. Ikushima H, Negishi H, Taniguchi T. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb Symp Quant Biol. (2013) 78:105–16. doi: 10.1101/sqb.2013.78.020321
218. Ng CT, Sullivan BM, Teijaro JR, Lee AM, Welch M, Rice S, et al. Blockade of interferon beta, but not interferon alpha, signaling controls persistent viral infection. Cell Host Microbe. (2015) 17:653–61. doi: 10.1016/j.chom.2015.04.005
219. Haller O, Kochs G, Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. (2006) 344:119–30. doi: 10.1016/j.virol.2005.09.024
220. Marie I, Durbin JE, Levy D. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. (1998) 17:6660–9. doi: 10.1093/emboj/17.22.6660
221. Baize S, Marianneau P, Loth P, Reynard S, Journeaux A, Chevallier M, et al. Early and strong immune responses are associated with control of viral replication and recovery in lassa virus-infected cynomolgus monkeys. J Virol. (2009) 83:5890–903. doi: 10.1128/JVI.01948-08
222. McLay L, Liang Y, Ly H. Comparative analysis of disease pathogenesis and molecular mechanisms of New World and Old World arenavirus infections. J Gen Virol. (2014) 95:1–15. doi: 10.1099/vir.0.057000-0
223. Yun NE, Poussard AL, Seregin AV, Walker AG, Smith JK, Aronson JF, et al. Functional interferon system is required for clearance of Lassa virus. J Virol. (2012) 86:3389–92. doi: 10.1128/JVI.06284-11
224. Rieger T, Merkler D, Günther S. Infection of type i interferon receptor-deficient mice with various old world arenaviruses: a model for studying virulence and host species barriers. PLoS ONE. (2013) 8:e72290. doi: 10.1371/journal.pone.0072290
225. Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KCF, Welch M, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. (2013) 340:207–11. doi: 10.1126/science.1235214
226. Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. (2013) 340:202–7. doi: 10.1126/science.1235208
227. Zuniga EI, Liou LY, Mack L, Mendoza M, Oldstone MBA. Persistent virus infection inhibits type I interferon production by plasmacytoid dendritic cells to facilitate opportunistic infections. Cell Host Microbe. (2008) 4:374–86. doi: 10.1016/j.chom.2008.08.016
228. Mariën J, Borremans B, Gryseels S, Soropogui B, De Bruyn L, Bongo GN, et al. No measurable adverse effects of Lassa, Morogoro and Gairo arenaviruses on their rodent reservoir host in natural conditions Joachim. Parasit Vectors. (2017) 10:210. doi: 10.1186/s13071-017-2146-0
229. Mandl JNN, Ahmed R, Barreiro LBB, Daszak P, Epstein JHH, Virgin HWW, et al. Reservoir host immune responses to emerging zoonotic viruses. Cell. (2015) 160:20–35. doi: 10.1016/j.cell.2014.12.003
230. Caballero IS, Yen JY, Hensley LE, Honko AN, Goff AJ, Connor JH. Lassa and Marburg viruses elicit distinct host transcriptional responses early after infection. BMC Genomics. (2014) 15:960. doi: 10.1186/1471-2164-15-960
231. Kolokoltsova OA, Yun NE, Poussard AL, Smith JK, Smith JN, Salazar M, et al. Mice lacking alpha/beta and gamma interferon receptors are susceptible to junin virus infection. J Virol. (2010) 84:13063–7. doi: 10.1128/JVI.01389-10
232. Huang C, Walker AG, Grant AM, Kolokoltsova OA, Yun NE, Seregin AV, et al. Potent inhibition of Junín virus infection by interferon in murine cells. PLoS Negl Trop Dis. (2014) 8:e2933. doi: 10.1371/journal.pntd.0002933
233. Kolokoltsova OA, Grant AM, Huang C, Smith JK, Poussard AL, Tian B, et al. RIG-I enhanced interferon independent apoptosis upon Junin virus infection. PLoS ONE. (2014) 9:e99610. doi: 10.1371/journal.pone.0099610
234. Peña Cárcamo JR Morell ML, Vázquez CA, Vatansever S, Upadhyay AS, Överby AK, et al. The interplay between viperin antiviral activity, lipid droplets and Junín mammarenavirus multiplication. Virology. (2018) 514:216–29. doi: 10.1016/j.virol.2017.10.012
235. Huang C, Kolokoltsova OA, Yun NE, Seregin AV, Poussard AL, Walker AG, et al. Junín virus infection activates the type I interferon pathway in a RIG-I-dependent manner. PLoS Negl Trop Dis. (2012) 6:e1659. doi: 10.1371/journal.pntd.0001659
236. Huang C, Kolokoltsova OA, Yun NE, Seregin AV, Ronca S, Koma T, et al. Highly pathogenic New World and Old World human arenaviruses induce distinct interferon responses in human cells. J Virol. (2015) 89:7079–88. doi: 10.1128/JVI.00526-15
237. Heller MV, Saavedra MC, Falcoff R, Maiztegui JI, Molinas FC. Increased tumor necrosis factor-alpha levels in Argentine hemorrhagic fever. J Infect Dis. (1992) 166:1203–4.
238. Levis SC, Saavedra MC, Ceccoli C, Feuillade MR, Enria DA, Maiztegui JI, et al. Correlation between endogenous interferon and the clinical evolution of patients with Argentine hemorrhagic fever. J Interferon Res. (1985) 5:383–9. doi: 10.1089/jir.1985.5.383
239. Marta RF, Montero VS, Hack CE, Sturk A, Maiztegui JI, Molinas FC. Proinflammatory cytokines and elastase-alpha-1-antitrypsin in Argentine hemorrhagic fever. Am J Trop Med Hyg. (1999) 60:85–9.
240. Groseth A, Hoenen T, Weber M, Wolff S, Herwig A, Kaufmann A, et al. Tacaribe virus but not junin virus infection induces cytokine release from primary human monocytes and macrophages. PLoS Negl Trop Dis. (2011) 5:e1137. doi: 10.1371/journal.pntd.0001137
241. Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, et al. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature. (2010) 468:779–83. doi: 10.1038/nature09605
242. Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci USA. (2011) 108:2396–401. doi: 10.1073/pnas.1016404108
243. West BR, Hastie KM, Saphire EO. Structure of the LCMV nucleoprotein provides a template for understanding arenavirus replication and immunosuppression. Acta Crystallogr D Biol Crystallogr. (2014) 70:1764–9. doi: 10.1107/S1399004714007883
244. Jiang X, Huang Q, Wang W, Dong H, Ly H, Liang Y, et al. Structures of arenaviral nucleoproteins with triphosphate dsRNA reveal a unique mechanism of immune suppression. J Biol Chem. (2013) 288:16949–59. doi: 10.1074/jbc.M112.420521
245. Hastie KM, Liu T, Li S, King LB, Ngo N, Zandonatti MA, et al. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci USA. (2011) 108:19365–70. doi: 10.1073/pnas.1108515108
246. Zhou S, Cerny AM, Zacharia A, Fitzgerald KA, Kurt-Jones EA, Finberg RW. Induction and inhibition of type I interferon responses by distinct components of lymphocytic choriomeningitis virus. J Virol. (2010) 84:9452–62. doi: 10.1128/JVI.00155-10
247. Shao J, Huang Q, Liu X, Di D, Liang Y, Ly H. Arenaviral nucleoproteins suppress PACT-induced augmentation of RIG-I function to inhibit type I interferon production. J Virol. (2018) 92:e00482–18. doi: 10.1128/jvi.00482-18
248. Pythoud C, Rodrigo WWSI, Pasqual G, Rothenberger S, Martinez-Sobrido L, de la Torre JC, et al. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKK. J Virol. (2012) 86:7728–38. doi: 10.1128/JVI.00187-12
249. Hastie KM, King LB, Zandonatti MA, Ollmann Saphire E. Structural basis for the dsRNA specificity of the lassa virus NP exonuclease. PLoS ONE. (2012) 7:e44211. doi: 10.1371/journal.pone.0044211
250. Rodrigo WWSI, Ortiz-Riaño E, Pythoud C, Kunz S, de la Torre JC, Martínez-Sobrido L. Arenavirus nucleoproteins prevent activation of nuclear factor kappa B. J Virol. (2012) 86:8185–97. doi: 10.1128/JVI.07240-11
251. Carnec X, Baize S, Reynard S, Diancourt L, Caro V, Tordo N, et al. Lassa virus nucleoprotein mutants generated by reverse genetics induce a robust type I interferon response in human dendritic cells and macrophages. J Virol. (2011) 85:12093–7. doi: 10.1128/JVI.00429-11
252. Reynard S, Russier M, Fizet A, Carnec X, Baize S. Exonuclease domain of the Lassa virus nucleoprotein is critical to avoid RIG-I signaling and to inhibit the innate immune response. J Virol. (2014) 88:13923–7. doi: 10.1128/JVI.01923-14
253. Russier M, Reynard S, Carnec X, Baize S. The exonuclease domain of Lassa virus nucleoprotein is involved in antigen-presenting-cell-mediated NK cell responses. J Virol. (2014) 88:13811–20. doi: 10.1128/JVI.01908-14
254. Martínez-Sobrido L, Giannakas P, Cubitt B, García-Sastre A, de la Torre JC. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol. (2007) 81:12696–703. doi: 10.1128/JVI.00882-07
255. Harmon B, Kozina C, Maar D, Carpenter TS, Branda CS, Negrete OA, et al. Identification of critical amino acids within the nucleoprotein of tacaribe virus important for anti-interferon activity. J Biol Chem. (2013) 288:8702–11. doi: 10.1074/jbc.M112.444760
256. Huang Q, Shao J, Lan S, Zhou Y, Xing J, Dong C, et al. In vitro and in vivo characterizations of pichinde viral nucleoprotein exoribonuclease functions. J Virol. (2015) 89:6595–607. doi: 10.1128/JVI.00009-15
257. Fan L, Briese T, Lipkin WI. Z Proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J Virol. (2010) 84:1785–91. doi: 10.1128/JVI.01362-09
258. Xing J, Ly H, Liang Y. The Z proteins of pathogenic but not nonpathogenic arenaviruses inhibit RIG-i-like receptor-dependent interferon production. J Virol. (2015) 89:2944–55. doi: 10.1128/JVI.03349-14
259. Xing J, Chai Z, Ly H, Liang Y. Differential inhibition of macrophage activation by lymphocytic choriomeningitis virus and pichinde virus is mediated by the Z protein N-terminal domain. J Virol. (2015) 89:12513–7. doi: 10.1128/JVI.01674-15
260. Ly H. Differential immune responses to new world and old world mammalian arenaviruses. Int J Mol Sci. (2017) 18:E1040. doi: 10.3390/ijms18051040
261. Edington GM, White HA. The pathology of lassa fever. Trans R Soc Trop Med Hyg. (1972) 66:381–9. doi: 10.1016/0035-9203(72)90268-4
262. Fisher-Hoch SP, McCormick JB, Sasso D, Craven RB. Hematologic dysfunction in Lassa fever. J Med Virol. (1988) 26:127–35. doi: 10.1002/jmv.1890260204
263. Marriott K. Comparative pathogenesis of passaged Pichinde virus variants in outbred guinea pigs. FASEB J. (1997) 11:669.
264. Scott EP, Aronson JF. Cytokine patterns in a comparative model of arenavirus haemorrhagic fever in guinea pigs. J Gen Virol. (2008) 89:2569–79. doi: 10.1099/vir.0.2008/002048-0
265. Bell TM, Bunton TE, Shaia CI, Raymond JW, Honnold SP, Donnelly GC, et al. Pathogenesis of bolivian hemorrhagic fever in guinea pigs. Vet Pathol. (2016) 53:190–9. doi: 10.1177/0300985815588609
266. Hofer MJ, Li W, Manders P, Terry R, Lim SL, King NJC, et al. Mice deficient in STAT1 but not STAT2 or IRF9 develop a lethal CD4+ T-cell-mediated disease following infection with lymphocytic choriomeningitis virus. J Virol. (2012) 86:6932–46. doi: 10.1128/JVI.07147-11
267. McCormick JB, Fisher-Hoch SP. Lassa fever. Curr Top Microbiol Immunol. (2002) 262:75–109. doi: 10.1007/978-3-642-56029-3_4
268. Oldstone MBA, Campbell KP. Decoding arenavirus pathogenesis: essential roles for alpha-dystroglycan-virus interactions and the immune response. Virology. (2011) 411:170–9. doi: 10.1016/j.virol.2010.11.023
269. Zapata J, Medina-Moreno S, Guzmán-Cardozo C, Salvato M, Zapata JC. Improving the breadth of the host's immune response to Lassa virus. Pathogens. (2018) 7:84. doi: 10.3390/pathogens7040084
270. Pien GC, Nguyen KB, Malmgaard L, Satoskar AR, Biron CA. A unique mechanism for innate cytokine promotion of T cell responses to viral infections. J Immunol. (2002) 156:1138–42. doi: 10.4049/jimmunol.169.10.5827
271. Straub T, Freudenberg MA, Schleicher U, Bogdan C, Gasteiger G, Pircher H. Bacterial coinfection restrains antiviral CD8 T-cell response via LPS-induced inhibitory NK cells. Nat Commun. (2018) 9:4117. doi: 10.1038/s41467-018-06609-z
272. Baenziger J, Hengartner H, Zinkernagel RM, Cole GA. Induction or prevention of immunopathological disease by cloned cytotoxic T cell lines specific for lymphocytic choriomeningitis virus. Eur J Immunol. (1986) 16:387–93. doi: 10.1002/eji.1830160413
273. Dixon JE, Allan JE, Doherty PC. The acute inflammatory process in murine lymphocytic choriomeningitis is dependent on Lyt-2+ immune T cells. Cell Immunol. (1987) 107:8–14. doi: 10.1016/0008-8749(87)90260-7
274. Doherty PC, Ceredig R, Allan JE. Immunogenetic analysis of cellular interactions governing the recruitment of T lymphocytes and monocytes in lymphocytic choriomeningitis virus-induced immunopathology. Clin Immunol Immunopathol. (1988) 47:19–26.
275. Doherty PC, Allan JE, Lynch F, Ceredig R. Dissection of an inflammatory process induced by CD8+ T cells. Immunol Today. (1990) 11:55–9. doi: 10.1016/0167-5699(90)90019-6
276. Fung-Leung WP, Kündig TM, Zinkernagel RM, Mak TW. Immune response against lymphocytic choriomeningitis virus infection in mice without CD8 expression. J Exp Med. (1991) 174:1425–9. doi: 10.1084/JEM.174.6.1425
277. Christensen JP, Marker O, Thomsen AR. The role of CD4+ T cells in cell-mediated immunity to LCM V: studies in MHC class I and class II deficient mice. Scand J Immunol. (1994) 40:373–82. doi: 10.1111/j.1365-3083.1994.tb03477.x
278. Oldstone MBA, von Herrath M, Lewicki H, Hudrisier D, Whitton JL, Gairin JE. Use of a high-affinity peptide that aborts mhc-restricted cytotoxic T lymphocyte activity against multiple viruses in vitro and virus-induced immunopathologic disease in vivo. Virology. (1999) 256:246–57. doi: 10.1006/VIRO.1998.9593
279. Schildknecht A, Welti S, Geuking MB, Hangartner L, van den Broek M. Absence of CTL responses to early viral antigens facilitates viral persistence. J Immunol. (2008) 180:3113–21. doi: 10.4049/JIMMUNOL.180.5.3113
280. Leist TP, Cobbold SP, Waldmann H, Aguet M, Zinkernagel RM. Functional analysis of T lymphocyte subsets in antiviral host defense. J Immunol. (1987) 138:2278–81.
281. Asselin-Paturel C, Brizard G, Pin J-J, Brière F, Trinchieri G. Mouse strain differences in plasmacytoid dendritic cell frequency and function revealed by a novel monoclonal antibody. J Immunol. (2003) 171:6466–77. doi: 10.4049/JIMMUNOL.171.12.6466
282. Collin M, McGovern N, Haniffa M. Human dendritic cell subsets. Immunology. (2013) 140:22–30. doi: 10.1111/imm.12117
283. Hervas-Stubbs S, Riezu-Boj J-I, Mancheño U, Rueda P, Lopez L, Alignani D, et al. Conventional but not plasmacytoid dendritic cells foster the systemic virus-induced type I IFN response needed for efficient CD8 T cell priming. J Immunol. (2014) 193:1151–61. doi: 10.4049/jimmunol.1301440
284. Kiley M, Lange JV, Johnson KM. Protection of rhesus monkeys from Lassa virus by immunisation with closely related arenavirus. Lancet. (1979) 314:738. doi: 10.1016/S0140-6736(79)90659-7
285. Wulff H, McIntosh BM, Hamner DB, Johnson KM. Isolation of an arenavirus closely related to Lassa virus from Mastomys natalensis in South-East Africa. Bull World Health Organ. (1977) 55:441–4.
286. Pannetier D, Faure C, Georges-Courbot M-C, Deubel V, Baize S. Human macrophages, but not dendritic cells, are activated and produce alpha/beta interferons in response to Mopeia virus infection. J Virol. (2004) 78:10516–24. doi: 10.1128/JVI.78.19.10516-10524.2004
287. Pannetier D, Reynard S, Russier M, Journeaux A, Tordo N, Deubel V, et al. Human dendritic cells infected with the nonpathogenic Mopeia virus induce stronger T-cell responses than those infected with Lassa virus. J Virol. (2011) 85:8293–306. doi: 10.1128/JVI.02120-10
288. Schaeffer J, Carnec X, Reynard S, Mateo M, Picard C, Pietrosemoli N, et al. Lassa virus activates myeloid dendritic cells but suppresses their ability to stimulate T cells. PLoS Pathog. (2018) 14:e1007430. doi: 10.1371/journal.ppat.1007430
289. Hilpert C, Sitte S, Matthies A, Voehringer D. Dendritic cells are dispensable for T cell priming and control of acute lymphocytic choriomeningitis virus infection. J Immunol. (2016) 197:2780–6. doi: 10.4049/jimmunol.1502582
290. Tsau JS, Huang X, Lai C-Y, Hedrick SM. The effects of dendritic cell hypersensitivity on persistent viral infection. J Immunol. (2018) 200:1335–46. doi: 10.4049/jimmunol.1601870
291. Carnec X, Mateo M, Page A, Reynard S, Hortion J, Picard C, et al. A vaccine platform against arenaviruses based on a recombinant hyperattenuated mopeia virus expressing heterologous glycoproteins. J Virol. (2018) 92:JVI.02230-17. doi: 10.1128/JVI.02230-17
292. Cuevas CD, Lavanya M, Wang E, Ross SR. Junin virus infects mouse cells and induces innate immune responses. J Virol. (2011) 85:11058–68. doi: 10.1128/JVI.05304-11
293. Cuevas CD, Ross SR. Toll-like receptor 2-mediated innate immune responses against Junín virus in mice lead to antiviral adaptive immune responses during systemic infection and do not affect viral replication in the brain. J Virol. (2014) 88:7703–14. doi: 10.1128/JVI.00050-14
294. Zhou S, Cerny AM, Bowen G, Chan M, Knipe DM, Kurt-Jones EA, et al. Discovery of a novel TLR2 signaling inhibitor with anti-viral activity. Antiviral Res. (2010) 87:295–306. doi: 10.1016/j.antiviral.2010.06.011
295. Hayes MW, Carrion R, Nunneley J, Medvedev AE, Salvato MS, Lukashevich IS. Pathogenic old world arenaviruses inhibit TLR2/Mal-dependent proinflammatory cytokines in vitro. J Virol. (2012) 86:7216–26. doi: 10.1128/JVI.06508-11
296. Wolff S, Becker S, Groseth A. Cleavage of the Junin virus nucleoprotein serves a decoy function to inhibit the induction of apoptosis during infection. J Virol. (2013) 87:224–33. doi: 10.1128/JVI.01929-12
297. Meyer B, Becker S, Jackson D, Groseth A, Kaufmann A, Wolff S, et al. The New World arenavirus Tacaribe virus induces caspase-dependent apoptosis in infected cells. J Gen Virol. (2016) 97:855–66. doi: 10.1099/jgv.0.000403
298. Meyer B, Groseth A. Apoptosis during arenavirus infection: mechanisms and evasion strategies. Microbes Infect. (2017) 20:65–80. doi: 10.1016/J.MICINF.2017.10.002
299. Rasmussen AL, Tchitchek N, Safronetz D, Carter VS, Williams CM, Haddock E, et al. Delayed inflammatory and cell death responses are associated with reduced pathogenicity in Lujo virus-infected cynomolgus macaques. J Virol. (2015) 89:2543–52. doi: 10.1128/JVI.02246-14
300. Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. (1998) 17:4379–90. doi: 10.1093/emboj/17.15.4379
301. Iwamura T, Yoneyama M, Koizumi N, Okabe Y, Namiki H, Samuel CE, et al. PACT, a double-stranded RNA binding protein acts as a positive regulator for type I interferon gene induced by newcastle disease virus. Biochem Biophys Res Commun. (2001) 282:515–23. doi: 10.1006/BBRC.2001.4606
302. Kok K-HH, Lui P-YY, Ng M-HJHJ, Siu K-LL, Au SWN, Jin D-YY. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe. (2011) 9:299–309. doi: 10.1016/J.CHOM.2011.03.007
303. Kok KH, Ng M-HJ, Ching Y-P, Jin D-Y. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J Biol Chem. (2007) 282:17649–57. doi: 10.1074/jbc.M611768200
304. Schlee M, Hartmann E, Coch C, Wimmenauer V, Janke M, Barchet W, et al. Approaching the RNA ligand for RIG-I? Immunol Rev. (2009) 227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x
305. Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, et al. Mutual antagonism between the ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe. (2013) 14:74–84. doi: 10.1016/j.chom.2013.06.010
306. Siu K-L, Yeung ML, Kok K-H, Yuen K-S, Kew C, Lui P-Y, et al. Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol. (2014) 88:4866–76. doi: 10.1128/JVI.03649-13
307. Tawaratsumida K, Phan V, Hrincius ER, High AA, Webby R, Redecke V, et al. Quantitative proteomic analysis of the influenza A virus nonstructural proteins NS1 and NS2 during natural cell infection identifies PACT as an NS1 target protein and antiviral host factor. J Virol. (2014) 88:9038–48. doi: 10.1128/JVI.00830-14
308. Kew C, Lui P-Y, Chan C-P, Liu X, Au SWN, Mohr I, et al. Suppression of PACT-induced type I interferon production by herpes simplex virus 1 Us11 protein. J Virol. (2013) 87:13141–9. doi: 10.1128/JVI.02564-13
309. Patel CV, Handy I, Goldsmith T, Patel RC. PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J Biol Chem. (2000) 275:37993–8. doi: 10.1074/jbc.M004762200
310. Peters GA, Hartmann R, Qin J, Sen GC. Modular structure of PACT: distinct domains for binding and activating PKR. Mol Cell Biol. (2001) 21:1908–20. doi: 10.1128/MCB.21.6.1908-1920.2001
311. Li S, Peters GA, Ding K, Zhang X, Qin J, Sen GC. Molecular basis for PKR activation by PACT or dsRNA. Proc Natl Acad Sci USA. (2006) 103:10005–10. doi: 10.1073/pnas.0602317103
312. Marques JT, White CL, Peters GA, Williams BRG, Sen GC. The role of PACT in mediating gene induction, PKR activation, and apoptosis in response to diverse stimuli. J Interferon Cytokine Res. (2008) 28:469–76. doi: 10.1089/jir.2007.0006
313. Daher A, Laraki G, Singh M, Melendez-Peña CE, Bannwarth S, Peters AHFM, et al. TRBP control of PACT-induced phosphorylation of protein kinase R is reversed by stress. Mol Cell Biol. (2009) 29:254–65. doi: 10.1128/MCB.01030-08
314. Singh M, Castillo D, Patel CV, Patel RC. Stress-induced phosphorylation of PACT reduces its interaction with TRBP and leads to PKR activation. Biochemistry. (2011) 50:4550–60. doi: 10.1021/bi200104h
315. Singh M, Patel RC. Increased interaction between PACT molecules in response to stress signals is required for PKR activation. J Cell Biochem. (2012) 113:2754–64. doi: 10.1002/jcb.24152
316. Chukwurah E, Patel RC. Stress-induced TRBP phosphorylation enhances its interaction with PKR to regulate cellular survival. Sci Rep. (2018) 8:1020. doi: 10.1038/s41598-018-19360-8
317. Parodi AS, Greenway DJ, Rugiero HR, Frigerio M, de la Barrera JM, Mettler N, et al. [Concerning the epidemic outbreak in Junin]. Dia Med. (1958) 30:2300–1.
318. Ghiringhelli PD, Rivera-Pomar RV, Lozano ME, Grau O, Romanowski V. Molecular organization of Junin virus S RNA: complete nucleotide sequence, relationship with other members of the Arenaviridae and unusual secondary structures. J Gen Virol. (1991) 72:2129–41. doi: 10.1099/0022-1317-72-9-2129
319. Piboul M, Albarino CG, Ghiringhelli PD, Romanowski V. The glycoprotein precursor gene of the attenuated junin virus vaccine strain (Candid #1). Am J Trop Med Hyg. (1997) 56:216–25. doi: 10.4269/ajtmh.1997.56.216
320. Maiztegui JI, McKee KT, Barrera Oro JG, Harrison LH, Gibbs PH, Feuillade MR, et al. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. AHF Study Group. J Infect Dis. (1998) 177:277–83.
321. Enria D, Barrera Oro J. Junin virus vaccines. Curr Top Microbiol Immunol. (2002) 263:239–61. doi: 10.1007/978-3-642-56055-2_12
322. Ambrosio AM, Riera LM, Saavedra MDC, Sabattini MS. Immune response to vaccination against argentine hemorrhagic fever in an area where different arenaviruses coexist. Viral Immunol. (2006) 19:196–201. doi: 10.1089/vim.2006.19.196
323. Goñi SE, Iserte JA, Ambrosio AM, Romanowski V, Ghiringhelli PD, Lozano ME. Genomic features of attenuated junín virus vaccine strain candidate. Virus Genes. (2006) 32:37–41. doi: 10.1007/s11262-005-5843-2
324. Goñi SE, Iserte JA, Stephan BI, Borio CS, Ghiringhelli PD, Lozano ME. Molecular analysis of the virulence attenuation process in Junín virus vaccine genealogy. Virus Genes. (2010) 40:320–8. doi: 10.1007/s11262-010-0450-2
325. Albariño CG, Bird BH, Chakrabarti AK, Dodd KA, Flint M, Bergeron E, et al. The major determinant of attenuation in mice of the Candid1 vaccine for Argentine hemorrhagic fever is located in the G2 glycoprotein transmembrane domain. J Virol. (2011) 85:10404–8. doi: 10.1128/JVI.00856-11
326. Seregin AV, Yun NE, Miller M, Aronson J, Smith JK, Walker AG, et al. The glycoprotein precursor gene of Junin virus determines the virulence of the Romero strain and the attenuation of the Candid #1 strain in a representative animal model of Argentine hemorrhagic fever. J Virol. (2015) 89:5949–56. doi: 10.1128/JVI.00104-15
327. Patterson M, Koma T, Seregin A, Huang C, Miller M, Smith J, et al. A substitution in the transmembrane region of the glycoprotein leads to an unstable attenuation of Machupo virus. J Virol. (2014) 88:10995–9. doi: 10.1128/JVI.01007-14
328. Li W, Hofer MJ, Noçon AL, Manders P, Campbell IL. Interferon regulatory factor 7 (IRF7) is required for the optimal initial control but not subsequent clearance of lymphocytic choriomeningitis virus infection in mice. Virology. (2013) 439:152–62. doi: 10.1016/J.VIROL.2013.02.015
329. Grant A, Seregin A, Huang C, Kolokoltsova O, Brasier A, Peters C, et al. Junín virus pathogenesis and virus replication. Viruses. (2012) 4:2317–39. doi: 10.3390/v4102317
330. Sommerstein R, Flatz L, Remy MM, Malinge P, Magistrelli G, Fischer N, et al. Arenavirus glycan shield promotes neutralizing antibody evasion and protracted infection. PLoS Pathog. (2015) 11:e1005276. doi: 10.1371/journal.ppat.1005276
331. Leske A, Waßmann I, Schnepel K, Shifflett K, Holzerland J, Bostedt L, et al. Assessing cross-reactivity of Junín virus-directed neutralizing antibodies. Antiviral Res. (2019) 163:106–16. doi: 10.1016/J.ANTIVIRAL.2019.01.006
332. McCormick JB, Mitchell SW, Kiley MP, Ruo S, Fisher-Hoch SP. Inactivated Lassa virus elicits a non protective immune response in rhesus monkeys. J Med Virol. (1992) 37:1–7.
333. Kotturi MF, Botten J, Maybeno M, Sidney J, Glenn J, Bui H-H, et al. Polyfunctional CD4+ T cell responses to a set of pathogenic arenaviruses provide broad population coverage. Immunome Res. (2010) 6:4. doi: 10.1186/1745-7580-6-4
334. Cheng BYH, Ortiz-Riaño E, Nogales A, de la Torre J-C, Martinez-Sobrido L. Development of live-attenuated arenavirus vaccines based on codon deoptimization of the viral glycoprotein. Virology. (2017) 501:35–46. doi: 10.1016/J.VIROL.2016.11.001
335. Popkin D, Teijaro JR, Lee AM, Lewicki H, Emonet S, de la Torre J-C, et al. Expanded potential for recombinant trisegmented lymphocytic choriomeningitis viruses: protein production, antibody production, and in vivo assessment of biological function of genes of interest. J Virol. (2011) 85:7928–32. doi: 10.1128/jvi.00486-11
336. Ortiz-Riaño E, Cheng BYH, de la Torre JC, Martínez-Sobrido L. Arenavirus reverse genetics for vaccine development. J Gen Virol. (2013) 94:1175–88. doi: 10.1099/vir.0.051102-0
337. Cheng BYH, Ortiz-Riaño E, de la Torre JC, Martínez-Sobrido L. Generation of recombinant arenavirus for vaccine development in FDA-approved Vero cells. J Vis Exp. (2013) 78:e50662. doi: 10.3791/50662
338. Cheng BYH, Ortiz-Riaño E, de la Torre JC, Martínez-Sobrido L. Arenavirus genome rearrangement for the development of live attenuated vaccines. J Virol. (2015) 89:7373–84. doi: 10.1128/JVI.00307-15
339. Martínez-Sobrido L, Cheng BYH, de la Torre JC. Reverse genetics approaches to control arenavirus. Methods Mol Biol. (2016) 1403:313–51. doi: 10.1007/978-1-4939-3387-7_17
340. Dhanwani R, Zhou Y, Huang Q, Verma V, Dileepan M, Ly H, et al. A novel live pichinde virus-based vaccine vector induces enhanced humoral and cellular immunity after a booster dose. J Virol. (2015) 90:2551–60. doi: 10.1128/JVI.02705-15
341. Dhanwani R, Ly H, Liang Y. Recombinant tri-segmented pichinde virus as a novel live viral vaccine platform. Methods Mol Biol. 1581:169–79. doi: 10.1007/978-1-4939-6869-5_10
342. Flatz L, Hegazy AN, Bergthaler A, Verschoor A, Claus C, Fernandez M, et al. Development of replication-defective lymphocytic choriomeningitis virus vectors for the induction of potent CD8+ T cell immunity. Nat Med. (2010) 16:339–45. doi: 10.1038/nm.2104
343. Rodrigo WWSI, de la Torre JC, Martínez-Sobrido L. Use of single-cycle infectious lymphocytic choriomeningitis virus to study hemorrhagic fever arenaviruses. J Virol. (2011) 85:1684–95. doi: 10.1128/JVI.02229-10
344. Lukashevich IS, Patterson J, Carrion R, Moshkoff D, Ticer A, Zapata J, et al. A live attenuated vaccine for Lassa fever made by reassortment of Lassa and Mopeia viruses. J Virol. (2005) 79:13934–42. doi: 10.1128/JVI.79.22.13934-13942.2005
345. Zapata JC, Pauza CD, Djavani MM, Rodas JD, Moshkoff D, Bryant J, et al. Lymphocytic choriomeningitis virus (LCMV) infection of macaques: a model for Lassa fever. Antiviral Res. (2011) 92:125–38. doi: 10.1016/J.ANTIVIRAL.2011.07.015
346. Zapata JC, Poonia B, Bryant J, Davis H, Ateh E, George L, et al. An attenuated Lassa vaccine in SIV-infected rhesus macaques does not persist or cause arenavirus disease but does elicit Lassa virus-specific immunity. Virol J. (2013) 10:52. doi: 10.1186/1743-422X-10-52
347. Carrion R, Patterson JL, Johnson C, Gonzales M, Moreira CR, Ticer A, et al. A ML29 reassortant virus protects guinea pigs against a distantly related Nigerian strain of Lassa virus and can provide sterilizing immunity. Vaccine. (2007) 25:4093–102. doi: 10.1016/j.vaccine.2007.02.038
348. Johnson D, Jokinen J, Lukashevich I. Attenuated replication of lassa virus vaccine candidate ML29 in STAT-1-/- mice. Pathogens. (2019) 8:9. doi: 10.3390/pathogens8010009
349. Garbutt M, Liebscher R, Wahl-Jensen V, Jones S, Möller P, Wagner R, et al. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J Virol. (2004) 78:5458–65. doi: 10.1128/JVI.78.10.5458-5465.2004
350. Geisbert TW, Jones S, Fritz EA, Shurtleff AC, Geisbert JB, Liebscher R, et al. Development of a new vaccine for the prevention of Lassa fever. PLoS Med. (2005) 2:e183. doi: 10.1371/JOURNAL.PMED.0020183
351. Clegg JCS, Lloyd G. Vaccinia recombinant expressing lassa-virus internal nucleocapsid protein protects guinea pigs against lassa fever. Lancet. (1987) 330:186–8. doi: 10.1016/S0140-6736(87)90767-7
352. Auperin DD, Esposito JJ, Lange JV, Bauer SP, Knight J, Sasso DR, et al. Construction of a recombinant vaccinia virus expressing the Lassa virus glycoprotein gene and protection of guinea pigs from a lethal Lassa virus infection. Virus Res. (1988) 9:233–48. doi: 10.1016/0168-1702(88)90033-0
353. Bredenbeek PJ, Molenkamp R, Spaan WJM, Deubel V, Marianneau P, Salvato MS, et al. A recombinant Yellow Fever 17D vaccine expressing Lassa virus glycoproteins. Virology. (2006) 345:299–304. doi: 10.1016/J.VIROL.2005.12.001
354. Jiang X, Dalebout TJ, Bredenbeek PJ, Carrion R, Brasky K, Patterson J, et al. Yellow fever 17D-vectored vaccines expressing Lassa virus GP1 and GP2 glycoproteins provide protection against fatal disease in guinea pigs. Vaccine. (2011) 29:1248–57. doi: 10.1016/j.vaccine.2010.11.079
355. Pushko P, Geisbert J, Parker M, Jahrling P, Smith J. Individual and bivalent vaccines based on alphavirus replicons protect guinea pigs against infection with Lassa and Ebola viruses. J Virol. (2001) 75:11677–85. doi: 10.1128/JVI.75.23.11677-11685.2001
356. Wang M, Jokinen J, Tretyakova I, Pushko P, Lukashevich IS. Alphavirus vector-based replicon particles expressing multivalent cross-protective Lassa virus glycoproteins. Vaccine. (2018) 36:683–90. doi: 10.1016/J.VACCINE.2017.12.046
357. Abreu-Mota T, Hagen KR, Cooper K, Jahrling PB, Tan G, Wirblich C, et al. Non-neutralizing antibodies elicited by recombinant Lassa-Rabies vaccine are critical for protection against Lassa fever. Nat Commun. (2018) 9:4223. doi: 10.1038/s41467-018-06741-w
358. Ramsauer K, Schwameis M, Firbas C, Müllner M, Putnak RJ, Thomas SJ, et al. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based chikungunya vaccine: a randomised, double-blind, placebo-controlled, active-comparator, first-in-man trial. Lancet Infect Dis. (2015) 15:519–27. doi: 10.1016/S1473-3099(15)70043-5
359. Rodriguez-Carreno MP, Nelson MS, Botten J, Smith-Nixon K, Buchmeier MJ, Whitton JL. Evaluating the immunogenicity and protective efficacy of a DNA vaccine encoding Lassa virus nucleoprotein. Virology. (2005) 335:87–98. doi: 10.1016/J.VIROL.2005.01.019
360. Cashman KA, Wilkinson ER, Shaia CI, Facemire PR, Bell TM, Bearss JJ, et al. A DNA vaccine delivered by dermal electroporation fully protects cynomolgus macaques against Lassa fever. Hum Vaccin Immunother. (2017) 13:2902–11. doi: 10.1080/21645515.2017.1356500
361. Henao-Restrepo AM, Longini IM, Egger M, Dean NE, Edmunds WJ, Camacho A, et al. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet. (2015) 386:857–66. doi: 10.1016/S0140-6736(15)61117-5
362. Henao-Restrepo AM, Camacho A, Longini IM, Watson CH, Edmunds WJ, Egger M, et al. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!). Lancet. (2017) 389:505–18. doi: 10.1016/S0140-6736(16)32621-6
363. Clarke DK, Nasar F, Chong S, Johnson JE, Coleman JW, Lee M, et al. Neurovirulence and immunogenicity of attenuated recombinant vesicular stomatitis viruses in nonhuman primates. J Virol. (2014) 88:6690–701. doi: 10.1128/JVI.03441-13
364. Matassov D, Marzi A, Latham T, Xu R, Ota-Setlik A, Feldmann F, et al. Vaccination with a highly attenuated recombinant vesicular stomatitis virus vector protects against challenge with a lethal dose of ebola virus. J Infect Dis. (2015) 212:S443–51. doi: 10.1093/infdis/jiv316
365. Mire CE, Matassov D, Geisbert JB, Latham TE, Agans KN, Xu R, et al. Single-dose attenuated Vesiculovax vaccines protect primates against Ebola Makona virus. Nature. (2015) 520:688–91. doi: 10.1038/nature14428
366. Matassov D, Mire CE, Latham T, Geisbert JB, Xu R, Ota-Setlik A, et al. Single dose trivalent vesiculovax vaccine protects macaques from lethal ebolavirus and marburgvirus challenge. J Virol. (2018) 92:e01190–17. doi: 10.1128/JVI.01190-17
367. CEPI. CEPI Awards Contract Worth Up To USD$19 Million to Oxford University and Janssen Vaccines to Develop MERS, Lassa, and Nipah Vaccines. (2018). Available online at: https://cepi.net/news_cepi/cepi-awards-contract-worth-up-to-usd19-million-to-oxford-university-and-janssen-vaccines-to-develop-mers-lassa-and-nipah-vaccines/ (Accessed February 11, 2019).
368. ter Meulen J, Badusche M, Kuhnt K, Doetze A, Satoguina J, Marti T, et al. Characterization of human CD4+ T-cell clones recognizing conserved and variable epitopes of the Lassa virus nucleoprotein. J Virol. (2000) 74:2186–92. doi: 10.1128/jvi.74.5.2186-2192.2000
369. ter Meulen J, Badusche M, Satoguina J, Strecker T, Lenz O, Loeliger C, et al. Old and New World arenaviruses share a highly conserved epitope in the fusion domain of the glycoprotein 2, which is recognized by Lassa virus-specific human CD4+ T-cell clones. Virology. (2004) 321:134–43. doi: 10.1016/J.VIROL.2003.12.013
370. Goicochea MA, Zapata JC, Bryant J, Davis H, Salvato MS, Lukashevich IS. Evaluation of Lassa virus vaccine immunogenicity in a CBA/J-ML29 mouse model. Vaccine. (2012) 30:1445–52. doi: 10.1016/j.vaccine.2011.12.134
371. Mendenhall M, Russell A, Juelich T, Messina EL, Smee DF, Freiberg AN, et al. T-705 (favipiravir) inhibition of arenavirus replication in cell culture. Antimicrob Agents Chemother. (2011) 55:782–7. doi: 10.1128/AAC.01219-10
372. Mendenhall M, Russell A, Smee DF, Hall JO, Skirpstunas R, Furuta Y, et al. Effective oral favipiravir (T-705) therapy initiated after the onset of clinical disease in a model of arenavirus hemorrhagic Fever. PLoS Negl Trop Dis. (2011) 5:e1342. doi: 10.1371/journal.pntd.0001342
373. Madelain V, Guedj J, Mentré F, Nguyen THT, Jacquot F, Oestereich L, et al. Favipiravir pharmacokinetics in nonhuman primates and insights for future efficacy studies of hemorrhagic fever viruses. Antimicrob Agents Chemother. (2017) 61:e01305–16. doi: 10.1128/AAC.01305-16
374. Delang L, Abdelnabi R, Neyts J. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res. (2018) 153:85–94. doi: 10.1016/J.ANTIVIRAL.2018.03.003
375. Neuman BW, Bederka LH, Stein DA, Ting JPC, Moulton HM, Buchmeier MJ. Development of peptide-conjugated morpholino oligomers as pan-arenavirus inhibitors. Antimicrob Agents Chemother. (2011) 55:4631–8. doi: 10.1128/AAC.00650-11
376. Ajayi N, Ukwaja K, Ifebunandu N, Nnabu R, Onwe F, Asogun D. Lassa fever—full recovery without ribavarin treatment: a case report. Afr Health Sci. (2015) 14:1074. doi: 10.4314/ahs.v14i4.40
377. McLay L, Ansari A, Liang Y, Ly H. Targeting virulence mechanisms for the prevention and therapy of arenaviral hemorrhagic fever. Antiviral Res. (2013) 97:81–92. doi: 10.1016/j.antiviral.2012.12.003
378. Lee AM, Rojek JM, Gundersen A, Ströher U, Juteau J-M, Vaillant A, et al. Inhibition of cellular entry of lymphocytic choriomeningitis virus by amphipathic DNA polymers. Virology. (2008) 372:107–17. doi: 10.1016/j.virol.2007.10.016
379. Torriani G, Trofimenko E, Mayor J, Fedeli C, Moreno H, Michel S, et al. Identification of clotrimazole-derivatives as specific inhibitors of Arenavirus fusion. J Virol. (2019). doi: 10.1128/JVI.01744-18. [Epub ahead of print].
380. Lee AM, Rojek JM, Spiropoulou CF, Gundersen AT, Jin W, Shaginian A, et al. Unique small molecule entry inhibitors of hemorrhagic fever arenaviruses. J Biol Chem. (2008) 283:18734–42. doi: 10.1074/jbc.M802089200
381. York J, Dai D, Amberg SM, Nunberg JH. pH-induced activation of arenavirus membrane fusion is antagonized by small-molecule inhibitors. J Virol. (2008) 82:10932–9. doi: 10.1128/JVI.01140-08
382. Larson RA, Dai D, Hosack VT, Tan Y, Bolken TC, Hruby DE, Amberg SM. Identification of a broad-spectrum arenavirus entry inhibitor. J Virol. (2008) 82:10768–75. doi: 10.1128/JVI.00941-08
383. Ngo N, Cubitt B, Iwasaki M, de la Torre JC. Identification and mechanism of action of a novel small-molecule inhibitor of arenavirus multiplication. J Virol. (2015) 89:10924–33. doi: 10.1128/JVI.01587-15
384. Wang P, Liu Y, Zhang G, Wang S, Guo J, Cao J, et al. Screening and identification of lassa virus entry inhibitors from an FDA-approved running title: drug repurposing for LASV entry. J Virol. (2018) 92:e00954–18. doi: 10.1128/JVI.00954-18
385. Cashman KA, Smith MA, Twenhafel NA, Larson RA, Jones KF, Allen RD, et al. Evaluation of Lassa antiviral compound ST-193 in a guinea pig model. Antiviral Res. (2011) 90:70–9. doi: 10.1016/j.antiviral.2011.02.012
386. Madu IG, Files M, Gharaibeh DN, Moore AL, Jung K-H, Gowen BB, et al. A potent Lassa virus antiviral targets an arenavirus virulence determinant. PLoS Pathog. (2018) 14:e1007439. doi: 10.1371/journal.ppat.1007439
387. Urata S, Yun N, Pasquato A, Paessler S, Kunz S, de la Torre JC. Antiviral activity of a small-molecule inhibitor of arenavirus glycoprotein processing by the cellular site 1 protease. J Virol. (2011) 85:795–803. doi: 10.1128/JVI.02019-10
388. Pasquato A, Rochat C, Burri DJ, Pasqual G, de la Torre JC, Kunz S. Evaluation of the anti-arenaviral activity of the subtilisin kexin isozyme-1/site-1 protease inhibitor PF-429242. Virology. (2012) 423:14–22. doi: 10.1016/j.virol.2011.11.008
389. Olmstead AD, Knecht W, Lazarov I, Dixit SB, Jean F. Human subtilase SKI-1/S1P is a master regulator of the HCV lifecycle and a potential host cell target for developing indirect-acting antiviral agents. PLoS Pathog. (2012) 8:e1002468. doi: 10.1371/journal.ppat.1002468
390. Blanchet M, Seidah NG, Labonté P. SKI-1/S1P inhibition: a promising surrogate to statins to block Hepatitis C virus replication. Antiviral Res. (2012) 95:159–66. doi: 10.1016/J.ANTIVIRAL.2012.05.006
391. Blanchet M, Sureau C, Guévin C, Seidah NG, Labonté P. SKI-1/S1P inhibitor PF-429242 impairs the onset of HCV infection. Antiviral Res. (2015) 115:94–104. doi: 10.1016/J.ANTIVIRAL.2014.12.017
392. Uchida L, Urata S, Ulanday GEL, Takamatsu Y, Yasuda J, Morita K, et al. Suppressive effects of the site 1 protease (S1P) inhibitor, PF-429242, on dengue virus propagation. Viruses. (2016) 8:46. doi: 10.3390/v8020046
393. Cordo SM, Candurra NA, Damonte EB. Myristic acid analogs are inhibitors of Junin virus replication. Microbes Infect. (1999) 1:609–14. doi: 10.1016/S1286-4579(99)80060-4
394. García CC, Djavani M, Topisirovic I, Borden KLB, Salvato MS, Damonte EB. Arenavirus Z protein as an antiviral target: virus inactivation and protein oligomerization by zinc finger-reactive compounds. J Gen Virol. (2006) 87:1217–28. doi: 10.1099/vir.0.81667-0
395. Sepúlveda CS, García CC, Damonte EB. Inhibition of arenavirus infection by thiuram and aromatic disulfides. Antiviral Res. (2010) 87:329–37. doi: 10.1016/J.ANTIVIRAL.2010.06.005
396. García CC, Topisirovic I, Djavani M, Borden KLB, Damonte EB, Salvato MS. An antiviral disulfide compound blocks interaction between arenavirus Z protein and cellular promyelocytic leukemia protein. Biochem Biophys Res Commun. (2010) 393:625–30. doi: 10.1016/j.bbrc.2010.02.040
397. Dong H, Ren S, Zhang B, Zhou Y, Puig-Basagoiti F, Li H, et al. West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J Virol. (2008) 82:4295–307. doi: 10.1128/JVI.02202-07
398. Reguera J, Gerlach P, Rosenthal M, Gaudon S, Coscia F, Günther S, et al. Comparative structural and functional analysis of bunyavirus and arenavirus cap-snatching endonucleases. PLOS Pathog. (2016) 12:e1005636. doi: 10.1371/journal.ppat.1005636
399. Lehmann M, Pahlmann M, Jérôme H, Busch C, Lelke M, Günther S. Role of the C terminus of Lassa virus L protein in viral mRNA synthesis. J Virol. (2014) 88:8713–7. doi: 10.1128/JVI.00652-14
400. Rosenthal M, Gogrefe N, Vogel D, Reguera J, Rauschenberger B, Cusack S, et al. Structural insights into reptarenavirus cap-snatching machinery. PLOS Pathog. (2017) 13:e1006400. doi: 10.1371/journal.ppat.1006400
401. Ferron F, Decroly E, Selisko B, Canard B. The viral RNA capping machinery as a target for antiviral drugs. Antiviral Res. (2012) 96:21–31. doi: 10.1016/J.ANTIVIRAL.2012.07.007
402. Saez-Ayala M, Laban Yekwa E, Mondielli C, Roux L, Hernández S, Bailly F, et al. Metal chelators for the inhibition of the lymphocytic choriomeningitis virus endonuclease domain. Antiviral Res. (2019) 162:79–89. doi: 10.1016/J.ANTIVIRAL.2018.12.008
403. de la Torre JC. Arenavirus extinction through lethal mutagenesis. Virus Res. (2005) 107:207–14. doi: 10.1016/J.VIRUSRES.2004.11.010
404. Moreno H, Grande-Pérez A, Domingo E, Martín V. Arenaviruses and lethal mutagenesis. Prospects for new Ribavirin-based interventions. Viruses. (2012) 4:2786–805. doi: 10.3390/v4112786
405. Miranda PO, Cubitt B, Jacob NT, Janda KD, Carlos de la Torre J. Mining a kröhnke pyridine library for anti-arenavirus activity. ACS Infect Dis. (2018) 4:815–24. doi: 10.1021/acsinfecdis.7b00236
406. Vela EM, Bowick GC, Herzog NK, Aronson JF. Genistein treatment of cells inhibits arenavirus infection. Antiviral Res. (2008) 77:153–6. doi: 10.1016/j.antiviral.2007.09.005
407. Kolokoltsov AA, Adhikary S, Garver J, Johnson L, Davey RA, Vela EM. Inhibition of Lassa virus and Ebola virus infection in host cells treated with the kinase inhibitors genistein and tyrphostin. Arch Virol. (2012) 157:121–7. doi: 10.1007/s00705-011-1115-8
408. Vela EM, Knostman KA, Mott JM, Warren RL, Garver JN, Vela LJ, et al. Genistein, a general kinase inhibitor, as a potential antiviral for arenaviral hemorrhagic fever as described in the Pirital virus-Syrian golden hamster model. Antiviral Res. (2010) 87:318–28. doi: 10.1016/J.ANTIVIRAL.2010.06.007
409. Linero FN, Scolaro LA. Participation of the phosphatidylinositol 3-kinase/Akt pathway in Junín virus replication in vitro. Virus Res. (2009) 145:166–70. doi: 10.1016/J.VIRUSRES.2009.07.004
410. Urata S, Ngo N, de la Torre JC. The PI3K/Akt pathway contributes to arenavirus budding. J Virol. (2012) 86:4578–85. doi: 10.1128/JVI.06604-11
411. Mohr EL, McMullan LK, Lo MK, Spengler JR, Bergeron É, Albariño CG, et al. Inhibitors of cellular kinases with broad-spectrum antiviral activity for hemorrhagic fever viruses. Antiviral Res. (2015) 120:40–7. doi: 10.1016/j.antiviral.2015.05.003
412. Ortiz-Riaño E, Ngo N, Devito S, Eggink D, Munger J, Shaw ML, et al. Inhibition of arenavirus by A3, a pyrimidine biosynthesis inhibitor. J Virol. (2014) 88:878–89. doi: 10.1128/JVI.02275-13
413. Dunham EC, Leske A, Shifflett K, Watt A, Feldmann H, Hoenen T, et al. Lifecycle modelling systems support inosine monophosphate dehydrogenase (IMPDH) as a pro-viral factor and antiviral target for New World arenaviruses. Antiviral Res. (2018) 157:140–50. doi: 10.1016/j.antiviral.2018.07.009
414. Sepúlveda CS, García CC, Damonte EB. Antiviral activity of A771726, the active metabolite of leflunomide, against Junín virus. J Med Virol. (2018) 90:819–27. doi: 10.1002/jmv.25024
415. Gracias DT, Stelekati E, Hope JL, Boesteanu AC, Doering TA, Norton J, et al. The microRNA miR-155 controls CD8+ T cell responses by regulating interferon signaling. Nat Immunol. (2013) 14:593–602. doi: 10.1038/ni.2576
416. Sprokholt JK, Kaptein TM, van Hamme JL, Overmars RJ, Gringhuis SI, Geijtenbeek TBH. RIG-I-like receptor activation by dengue virus drives follicular T helper cell formation and antibody production. PLoS Pathog. (2017) 13:e1006738. doi: 10.1371/journal.ppat.1006738
417. Fisher DG, Coppock GM, López CB. Virus-derived immunostimulatory RNA induces type I IFN-dependent antibodies and T-cell responses during vaccination. Vaccine. (2018) 36:4039–45. doi: 10.1016/J.VACCINE.2018.05.100
418. Lukashevich IS, Carrion R, Salvato MS, Mansfield K, Brasky K, Zapata J, et al. Safety, immunogenicity, and efficacy of the ML29 reassortant vaccine for Lassa fever in small non-human primates. Vaccine. (2008) 26:5246–54. doi: 10.1016/j.vaccine.2008.07.057
419. Lukashevich IS, Pushko P. Vaccine platforms to control Lassa fever. Expert Rev Vaccines. (2016) 15:1135–50. doi: 10.1080/14760584.2016.1184575
Keywords: arenaviruses, Lassa fever, host-virus interactions, innate and adaptive immunity, viral immunology, viral pathogenesis, host defense
Citation: Brisse ME and Ly H (2019) Hemorrhagic Fever-Causing Arenaviruses: Lethal Pathogens and Potent Immune Suppressors. Front. Immunol. 10:372. doi: 10.3389/fimmu.2019.00372
Received: 10 January 2019; Accepted: 14 February 2019;
Published: 13 March 2019.
Edited by:
Constantinos Petrovas, Vaccine Research Center (NIAID), United StatesReviewed by:
Kartika Padhan, National Institutes of Health (NIH), United StatesTakaaki Koma, Tokushima University, Japan
Copyright © 2019 Brisse and Ly. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hinh Ly, aGx5QHVtbi5lZHU=
 Morgan E. Brisse
Morgan E. Brisse Hinh Ly
Hinh Ly