- Faculty of Medicine and Life Sciences, Vaccine Research Center, University of Tampere, Tampere, Finland
Norovirus (NoV) causes a substantial global burden of acute gastroenteritis in all age groups and the development of NoV vaccine is a high priority. There are still gaps in understanding of protective NoV-specific immunity. Antibody mediated immune responses have been widely studied, but in contrast, the research on NoV-specific human T cell-mediated immunity is very limited. We have recently reported NoV capsid VP1-specific 18-mer peptide (134SPSQVTMFPHIIVDVRQL151) to induce strong CD8+ T cell immune responses in healthy adult donors. This work extends to identify the precise NoV T cell epitope and the restricting human leucocyte antigen (HLA). Pentamer technology was used to detect HLA-A*0201-restricted T cell-mediated responses to 10-mer peptide 139TMFPHIIVDV148 of four healthy adult blood donors. Immunogenicity of the 10-mer epitope was confirmed by ELISPOT IFN-γ and intracellular cytokine staining (ICS) on flow cytometry. A population of CD3+CD8+ T lymphocytes binding to HLA-A*0201/TMFPHIIVDV pentamers was identified in two HLA-A*0201-positive donors. Recognition of the 10-mer epitope by T cells resulted in a strong IFN-γ secretion as shown by ELISPOT assay. In addition, ICS confirmed that high proportion (31 and 59%) of the TMFPHIIVDV epitope-responsive CD3+CD8+ T cells in the two donors had multifunctional phenotype, simultaneously producing IFN-γ, IL-2 and TNF-α cytokines. In the present study novel human NoV HLA-A*0201-restricted minimal 10-mer epitope 139TMFPHIIVDV148 in the capsid VP1 was identified. The HLA-peptide pentamer staining of T cells from healthy donor PBMCs and cytokine responses in ex-vivo ELISPOT and ICS assays suggest that this epitope is recognized during NoV infection and activates memory phenotype of the epitope-specific multifunctional CD8+ T cells. The importance of this epitope in protection from NoV infection remains to be determined.
Introduction
Noroviruses (NoV) are group of very contagious viruses that cause >90% of non-bacterial and approximately half of all-cause epidemic gastroenteritis worldwide. NoV gastroenteritis is a significant public health problem with high clinical and economic costs, that is estimated to cause over 200,000 deaths annually, mainly in resource-limited countries (1). Despite scientific efforts and increasing awareness of NoV burden, there is no vaccine available yet (2, 3). NoV exhibits a great genetic diversity with six currently recognized genogroups (GI-GVI), human NoVs belonging mainly to GI and GII, that include over 30 genotypes and numerous intra-genotype variants (4). GII.4 genotype has been predominant since the mid-90s, including pandemic variants US 1995/96, Farmington Hills 2002, Hunter 2004, New Orleans (NO) 2009, and most recently, Sydney 2012 (5). GII.4 variants share >95% of their VP1 capsid amino acid (aa) sequence, whereas < 85% of the capsid aa are identical between different genotypes (6). The antigenic diversity of NoVs is likely to have major impact on evading host immune responses and complicates designing of NoV vaccines. NoV research is further limited due to the lack of small animal model and efficient NoV propagation in cell culture (7, 8), but spontaneously formed NoV VP1 virus-like particles (VLPs) are successfully utilized as viral surrogates for assaying immune responses and as vaccine candidates (9, 10).
NoV infections cause acute but self-limited illness already at early age and several consecutive infections occur frequently (11–13). Protective immunity and clearance of NoV infections are not well-characterized (1, 2). Even though there is some indication of pre-existing antibodies conferring protection (14) the level of serum NoV-specific antibodies is not directly linked to protection from infection (15). The best correlate of protection from NoV infection identified so far are blocking (neutralizing) antibodies, that are able to block the binding of NoV VLPs to the putative receptors, histo-blood group antigens (HBGA's) (15). NoV-specific antibodies are cross-reactive to some extent within genogroups however, blocking antibodies are mostly genotype-specific with low protective capacity against more distinct strains (1). Although NoV-specific antibodies are extensively studied very little is published on human T cell immunity to NoV and its' role in protection (16–19). Studies using a murine norovirus (MNV)-mouse model have suggested T cell-mediated immunity to be important in clearance of MNV infection (20, 21). To this end, cytotoxic CD8+ T cell-mediated immune responses may provide substantial protection against serologically distinct viruses via recognition of cross-reactive, conserved epitopes, such as observed in influenza virus infection (22).
Using matrix peptide approach we have recently identified a NoV-specific 18-mer peptide (134SPSQVTMFPHIIVDVRQL151) containing CD8+ T cell epitope likely restricted to human leukocyte antigen (HLA)-A*0201 allele (17). Furthermore, peptide binding prediction tools have predicted 10 amino acid (aa) stretch as a minimal epitope (139TMFPHIIVDV148). In here, experiments were undertaken to experimentally confirm the recognition of the 10-mer epitope and its presentation by the HLA-A*0201 allele.
Materials and Methods
Blood Donors and Cell Isolation
Four healthy adults (age 35–45, laboratory personnel) volunteering in a study recently conducted by our laboratory were selected based on their CD8+ T cell responses to NoV GII.4 VP1-specific 18-mer peptide 134SPSQVTMFPHIIVDVRQL151 (originally named 99-20) recently described (17). The two peptide responders were HLA typed as HLA-A*02:01 carriers (17). The cells of the two donors identified as non-responders to the 18-mer peptide were used as negative controls. Peripheral blood mononuclear cells (PBMCs) of the heparin blood sample were obtained by Ficoll-Pague PLUS (GE Healthcare, Little Chalfont, United Kingdom) density gradient centrifugation. PBMCs were frozen in 10% DMSO in fetal bovine serum (FBS) using freezing container (Mr. Frosty™, Thermo Scientific,Waltham, MA, United States) with controlled rate of cooling at −80°C and transferred to liquid nitrogen. Prior to analysis, PBMCs were thawed, washed, and resuspended in culture medium (CM) containing RPMI 1640 with Glutamax® and HEPES (Gibco™ by Thermo Fisher Scientific) supplemented with 10 μg/ml Gentamicin (Gibco™) and 10% fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO, United States). The samples tested here are collected at a single bleed. Each sample aliquot was tested simultaneously by pentamer staining, enzyme-linked immunosorbent spot interferon-gamma (ELISPOT IFN-γ) assay and intracellular cytokine staining (ICS) assays, at least two times. Written informed consent was obtained from each volunteer prior to the sample collection in accordance with the Declaration of Helsinki. No approval by an ethics committee was required as per the local legislation.
Synthetic Peptides and Pentamers
To predict the optimal HLA-A*02:01 allele binding epitope within the 18-mer NoV-specific peptide 134SPSQVTMFPHIIVDVRQL151, artificial neural networks (ANN) (23) implemented at Immune Epitope Database and Analysis Resource (IEDB) was employed (24–26). A 10-mer high binding affinity sequence 139TMFPHIIVDV148 was identified (17) and the peptide was synthetized with purity >75% (Synpeptide Co. Ltd, Shanghai, China). SYFPEITHI prediction algorithm (http://www.syfpeithi.de) (27) was used for scoring the peptide affinity for HLA-A*0201 allele, as a binding score of >21 ensures synthesis of a custom pentamer by Proimmune Ltd. (Oxford, United Kingdom). HLA-A*0201/TMFPHIIVDV pentamer (Pro5® MHC Class I Pentamer) labeled with R-phycoerythrin (R-PE) was synthetized by Proimmune Ltd. In addition, HLA-A*0201 negative control pentamer conjugated to R-PE was synthetized as well. The degree of conservation of the 10-mer epitope among different NoV genotypes and genogroups was investigated using Basic Local Alignment Search Tool (BLAST) for sequence identity. Evolutionary analyses of the major capsid VP1 aa sequence of the aligned NoV genotypes were conducted in MEGA X (28). The evolutionary distances were computed using the Poisson correction method (29).
Pentamer Staining
PBMCs were treated for 10 min with Human BD Fc Block to prevent non-specific staining. To discriminate viable from non-viable cells, Horizon Fixable Viability stain 780 was used according to manufacturer's instructions. The cells (1 × 106) were incubated for 30 min on ice with either HLA-A*0201/TMFPHIIVDV pentamer or negative control pentamer, using 0.25 or 0.5 μg of the pentamers per condition. Cells were further stained with monoclonal antibodies against human CD3 (clone UCHT1, fluorescein isothiocyanate (FITC) conjugate) and CD8a (clone RPA-T8, PerCP Cy5.5 conjugate) for 30 min on ice. All reagents used for staining were purchased from BD Pharmingen (San Jose, CA, United States). After washing the cells were resuspended in 1% FBS, 2.5% formaldehyde in PBS for flow cytometry acquisition. At least 400 000 events were acquired for analysis on a 2-laser FACS CantoII flow cytometer (BD) with FACSDiva Software V 6.1.3 (Becton Dickinson, Heidelberg, Germany). Data were analyzed by FlowJo software version 10.1 (Tree Star, San Carlos, CA, United States). Lymphocytes were gated according to forward/sideward scatter (FSC/SSC) and dead cells were excluded by gating on the population negative for the viability dye. CD3+CD8+ population was further plotted on SSC-A vs. pentamer population.
ELISPOT IFN-γ Assay
PBMCs of the four donors were assayed in an ELISPOT assay for IFN-γ production as previously described (17). The cells were stimulated with increasing concentrations of 10-mer 139TMFPHIIVDV148 peptide (0.05, 0.1, 0.5, 1, 2, and 4 μg/ml final concentration), 4 μg/ml of 18-mer peptide 99-20, 4 μg/ml irrelevant 9-mer peptide (negative control) or 50 μg/ml of phytohemagglutinin (PHA, positive control). Briefly, ninety-six-well nitrocellulose filter plates (Millipore) were coated with anti-human IFN-γ capture antibody (Mabtech) and blocked with 10% FBS in CM. PBMCs (0.2 × 106 cells/well) were plated with stimulants and incubated for 20 h at +37°C and 5% CO2. Biotinylated anti-human IFN-γ antibody (Mabtech) followed by streptavidin-HRP (BD, New Jersey, United States) was used for detection. The spots were developed with Vector Nova Red substrate (Vector Labs, Burlingame, United States) and the plates were analyzed using ImmunoSpot Series II analyzer (CTL Europe, Leinfelden-Echterdingen, Germany). The results are expressed as mean spot forming cells (SFC)/106 PBMCs of the duplicate wells.
Intracellular Cytokine Staining (ICS)
An ICS assay was employed to quantify IFN-γ, TNF-α, and IL-2 producing CD3+ CD8+ T cells. PBMCs were stimulated according to the previously published protocol (17) with 4 μg/ml of 10-mer peptide 139TMFPHIIVDV148, 4 μg/ml irrelevant 9-mer peptide (negative control) or 1 μg/ml Staphylococcal enterotoxin B (SEB, Sigma) in the presence of 1 μg/ml CD28 and 1 μg/ml CD49d costimulatory antibodies (BD Biosciences, San Jose, CA, United States). PBMCs incubated in CM supplemented with the costimulatory antibodies only were used as additional control. The protein transport inhibitor brefeldin A (GolgiPlug, BD Biosciences, San Jose, CA, United States) at a concentration of 10 μg/ml was added after 2 h and the incubation was continued for 16 h at 37°C. After stimulation, the cells were treated with EDTA for 15 min and washed with FACS Stain buffer. Prior to fixation and permeabilization for ICS, PBMCs were blocked for non-specific staining, stained for viable/non-viable discrimination and surface markers CD3 and CD8a as described above for the pentamer staining. BD Fixation/Permeabilization solution was used according to the manufacturer's instructions and cells were intracellularly stained with the mixture of IFN-γ (clone 4S.B3) PE-Cy7 conjugate, IL-2 (clone MQ1-17H12) PE-conjugate, and TNF-α (clone MAb11) allophycocyanin (APC)-conjugate in 50 μl Perm/Wash buffer for 30 min on ice in the dark. Cells were resuspended in FACS Staining Buffer for acquisition and analysis using FACS CantoII flow cytometer and FACSDiva Software V 6.1.3. All reagents used for ICS were purchased from BD Pharmingen (San Jose, CA, United States). The data analysis was performed using FlowJo software version 10.1.
Results
The 10-mer Minimal Epitope Prediction and Conservation
At the outset of our study, an 18-mer NoV VP1-specific CD8+ T cell epitope (99-20, 134SPSQVTMFPHIIVDVRQL151) had been identified in two subjects, both having HLA-A*0201 allele. When analyzed by IEDB database ANN method, a 10 aa sequence 139TMFPHIIVDV148 within the 18-mer showed the highest binding affinity of IC50 21.5 nM (Table 1), whereas all other predicted sequence lengths indicated affinity >80 nM (data not shown). Peptides with IC50 < 50 nM are considered high affinity and < 500 nM intermediate affinity (30). Congruently, the same 10-mer epitope was identified to have the highest binding affinity by SMM method [data not shown (23)]. SYFPEITHI prediction algorithm confirmed a score of 27 for the peptide affinity for HLA-A*0201 allele (Table 1). When comparing the aa sequence of different NoV genotypes (Table 1) the 10-mer epitope was found to be highly conserved among different GII.4 variants and also more distant NoV genotypes belonging to GII and GI. The phylogenetic distances of the NoV genotypes compared in the Table 1 are shown in Figure 1. In addition, the 10-mer epitope sequence in all genotypes belonging to GI and GII NoV listed in the Table 1 also had good affinity binding scores for HLA-A*0201 allele regardless of up to three aa substitutions compared to the GII.4-1999 10-mer sequence. On the contrary, the 10-mer sequence in the human GIV 2010 NoV had a unique substitution 140M to 140Q which seemed to abolish its binding affinity resulting in IC50 552.6 nM and syfpeithi score 17 (Table 1). In addition, a change of the aa 148V to 148D at the c-terminus of the empirical 10-mer peptide abolished the predicted binding completely. This is not surprising as the aa at positions 140 and 148 are reported anchoring positions for HLA-A*0201 binding (31–33). In addition, deletions at the each terminus of the peptide affected the predicted binding capability of the peptide.
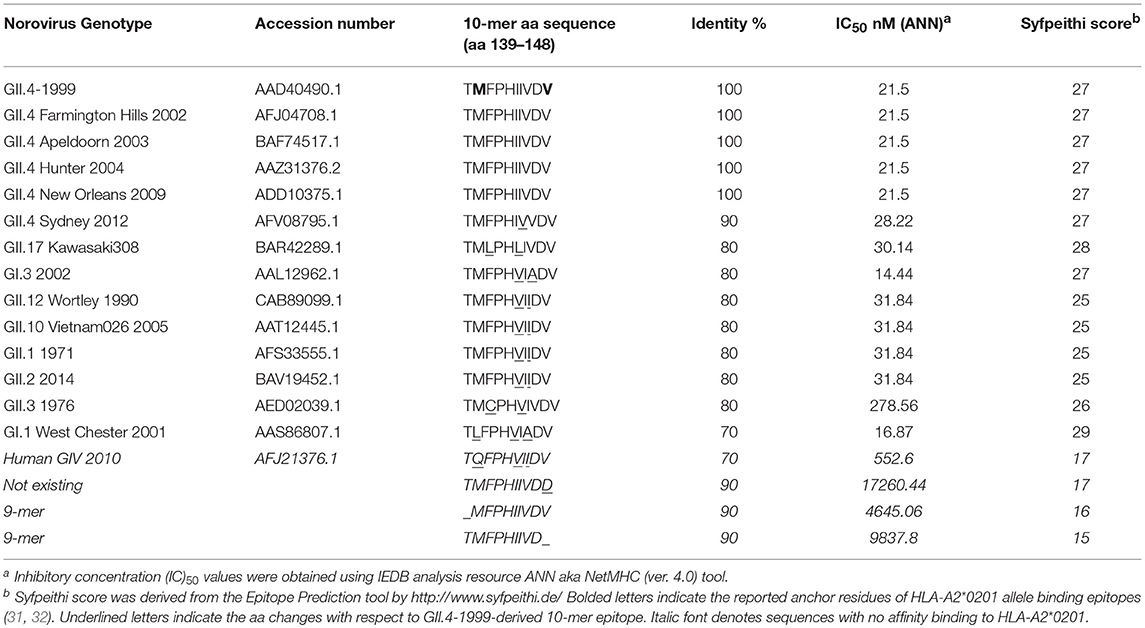
Table 1. Conservation and HLA-A2*0201 allele binding affinity prediction of the norovirus 10-mer epitope.
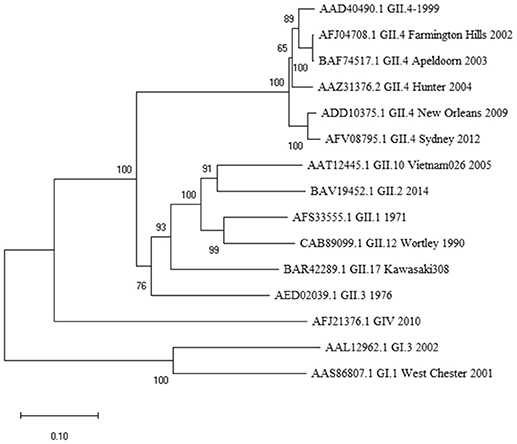
Figure 1. Phylogenetic analysis of norovirus major capsid protein VP1 amino acid sequences. The tree was inferred by Neighbor-Joining method with a bootstrap of 500 by using MegaX. The scale bar shows the genetic distance, expressed as amino acid substitutions per site.
CD8+ T Lymphocytes Bind the 10-mer Peptide 139TMFPHIIVDV148 in the Context of HLA-A*0201
The frequency of T cells specific for 139TMFPHIIVDV148 10-mer in the context of HLA-A*0201 allele was analyzed by direct binding to pentamer A*0201/TMFPHIIVDV R-PE using four-color flow cytometry. PBMCs of the two HLA-A*0201 positive donors previously responding to the 18-mer peptide 99-20 had frequency of 0.51% (Donor 1) and 1.52% (Donor 2) (Figure 2) pentamer-binding T cells of the live CD3+CD8+ gated lymphocytes. At the same time, the CD8+ T cells of the control donors non-responders to the 99-20, did not bind to A*0201/TMFPHIIVDV pentamer (Figure 2). The negative control pentamer did not show non-specific binding (Figure 2).
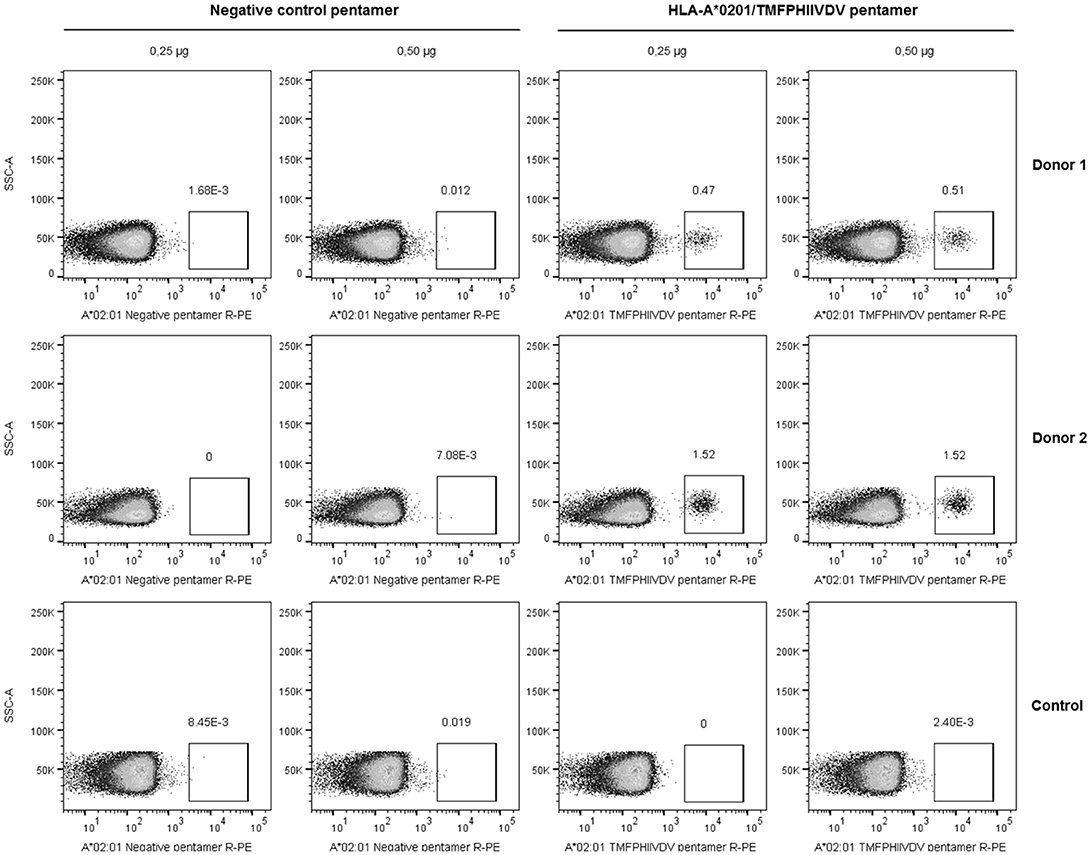
Figure 2. Pentamer staining of norovirus-derived 139TMFPHIIVDV148 epitope-specific CD8+ T cells. Peripheral blood mononuclear cells (PBMCs) from two HLA-A*0201-positive healthy adult donors and the negative control were directly stained using 0.25 and 0.5 μg HLA-A*0201/TMFPHIIVDV pentamer or negative control pentamer. The dot plots show the percentage of pentamer-positive T cells for two HLA-A*0201-positive donors and a representative negative control donor after gating on live CD3+CD8+ T lymphocyte population.
The 10-mer 139TMFPHIIVDV148 Epitope Induces Strong IFN-γ Response in HLA-A*0201 Positive Donors
IFN-γ-secreting lymphocytes specific for 139TMFPHIIVDV148 10-mer peptide were enumerated by ex-vivo ELISPOT assay with increasing concentration of the 10-mer peptide. Robust IFN-γ response to the 10-mer peptide was observed for the two HLA-A*0201 positive donors (Figure 3) but not for the two control donors (data not shown). A dose response was observed up to 1.0 μg/ml concentration of the peptide and also very low concentration (0.05 μg/ml) stimulated high IFN-γ secretion (donor 1, 260 SFC/106 cells; donor 2, 293 SFC/106 cells). IFN-γ production was not observed in the cells stimulated with the irrelevant 9-mer peptide or CM only (SFC/106 cells <50) (Figure 3A–C). All donors responded strongly to the positive control PHA (IFN-γ SFC/106 cells >2000, data not shown).
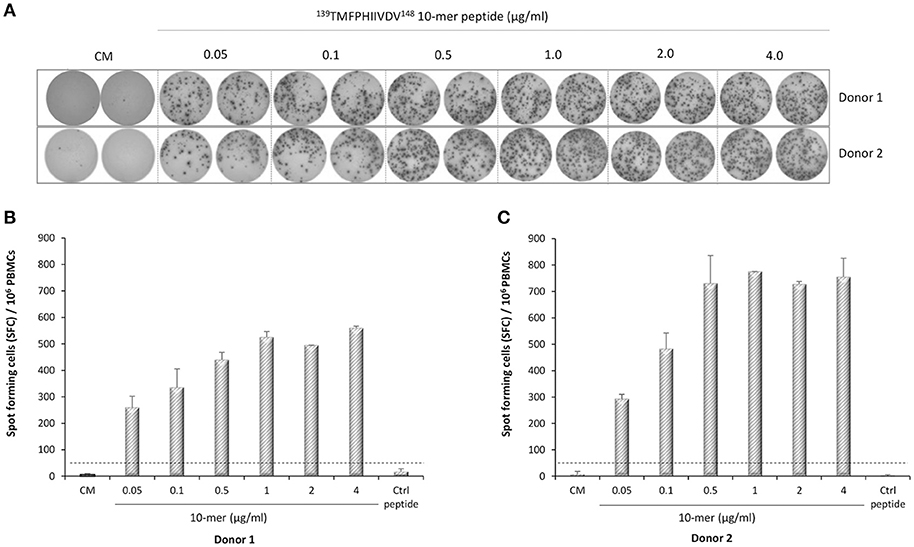
Figure 3. Norovirus epitope 139TMFPHIIVDV148 –specific enzyme-linked immunosorbent spot (ELISPOT) interferon-gamma (IFN-γ) responses. PBMCs of Donor 1 (A,B) and Donor 2 (A,C) positive for HLA-A*0201 allele were stimulated with an increasing concentration (0–4 μg/ml) of the 10-mer epitope peptide, 4 μg/ml irrelevant 9-mer peptide (Ctrl), or left unstimulated (CM, culture media). Images of the ELISPOT replicate wells with the stained spot-forming cells (SFC) are shown (A). Mean SFC/106 PBMCs of two replicate wells with the standard errors of the mean for Donor 1 (B) and Donor 2 (C). The cut-off line (dotted line) indicates the positive SFC/106 value of ≥50 SFC/106 cells and twice above the background control (CM only wells).
The 10-mer 139TMFPHIIVDV148 Epitope Induces Multifunctional CD8+ T Cells Producing IFN-γ, IL-2, and TNF-α
Along with quantification of the epitope-specific CD8+ T cells with HLA-A*0201 pentamer and IFN-γ ELISPOT, immune responses to 10-mer peptide were characterized by ICS of IFN-γ, IL-2, and TNF-α (Figure 4). The gating strategy for ICS analysis is shown in Figure 4. The viability of the lymphocytes was >98% and approximately 30% of the viable CD3+ lymphocyte population were CD8+ T cells. CD8+ T cells were further segregated into IFN-γ+ and IFN-γ− population and plotted for the expression of IL-2 and TNF-α. Robust cytokine response by CD3+CD8+ T cells toward 10-mer epitope in donor 1 (Figure 4A) and donor 2 (Figure 4B) but not in two control donors (data not shown) was observed. Both donors had CD8+ T cells producing single, double, and triple cytokines (Figures 4A,B). App. 1% of the CD8+ T cells of both donors produced IFN-γ in response to the 10-mer epitope (Figure 4) whereas irrelevant control peptide induced no IFN-γ (< 0.05%, data not shown). The 10-mer epitope induced multifunctional CD8+ T cells in both donors, donor 1 had ~30% triple cytokine (IFN-γ+, TNF-α+, and IL-2+) secreting CD3+CD8+ T cells, while donor 2 had ~60% of the triple-positive CD8+ T cells. Strong IFN-γ, IL-2, and TNF-α response was observed in both CD8+ and CD8− PBMCs against positive control SEB in all tested donors (not shown).
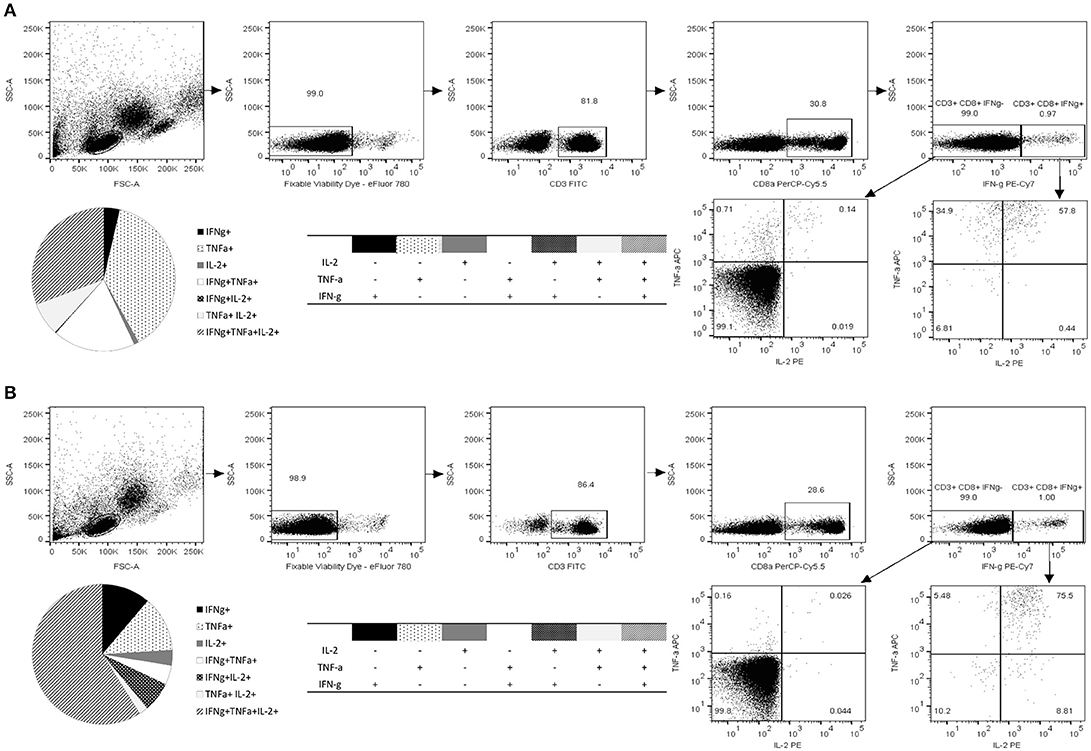
Figure 4. Functional characterization of norovirus 10-mer epitope 139TMFPHIIVDV148 –specific CD8+ T cells by intracellular cytokine staining (ICS). Production of IL-2, IFN-γ, and TNF-α measured by ICS after 16 h stimulation of the PBMC with the peptide 139TMFPHIIVDV148 (4 μg/ml). Shown are the results of the two HLA-A*0201-positive donors, Donor 1 (A) and Donor 2 (B). Live epitope-specific CD3+ CD8+ T cells were segregated into IFN-γ+ and IFN-γ− T cells. Seven distinct populations, based on production of the cytokines in any combination are depicted on the pie charts and the accompanying tables. Fractions of the cells producing one, two, or three cytokines are derived from the plotted IFN-γ+ and IFN-γ− populations.
Discussion
Despite significant efforts to define correlates of protection to NoV infection (1, 2), the role of T lymphocytes in protection and clearance of NoV infection is still largely unexplored area, with only few papers published so far (16–19, 34). We have recently tested ten healthy volunteer donors for NoV GII.4 capsid VP1-specific T cell responses using matrix peptide pools and found two CD8+ T cell responders to 18-mer NoV-specific peptide (134SPSQVTMFPHIIVDVRQL151, 99-20) (17). In here, we defined a 10-mer 139TMFPHIIVDV148 minimal epitope within this sequence and its restriction to HLA-A*0201 allele.
While the 10-mer 139TMFPHIIVDV148 epitope is derived from GII.4-1999 strain (17), the alignment with several other GII.4 variants showed epitope sequence to remain invariable until 2012 (Table 1). Interestingly, alignment with other quite distant genotypes (Figure 1) even belonging to GI viruses, indicated high degree of conservation of the epitope. Importantly, up to three aa substitutions as in the GI.1 West Chester 2001 NoV do not necessarily reduce the binding affinity to HLA-A*0201, depending on the aa position and characteristics. Aa at the position 6 and 8 of the 10-mer showed most variability among the aligned genotypes, however, these changes did not lead to considerable change in binding affinity predictions. However, a change at aa position 2 from a hydrophobic (M) to a polar aa (Q) in a human GIV 2010 NoV and at aa position 9 from V to D in an empirical sequence seemed to drastically abolish binding affinity. This is not surprising as HLA-A*0201 binding epitopes have a restricted size of 9/10 aa and hydrophobic anchor residues at positions 2 and 9/10 likewise the epitope identified in this study. The NoV 10-mer epitope contains methionine (M) at position 2 and valine (V) at position 10 which is described as a typical C-terminal anchor residue (31–33).
Direct staining of PBMCs with HLA-A*0201 pentamer loaded with 139TMFPHIIVDV148 peptide attested relatively high frequency of circulating NoV capsid-specific memory CD8+ T cells in two HLA-A*0201 positive healthy donors (0.51 and 1.52% of CD3+CD8+ gated lymphocytes, respectively). HLA-A2 is the most prevalent MHC allele family in human population and the gene frequency of HLA-A*02:01 is especially high in Caucasians (>20%), and other western ethnic groups (African Americans 12%, Hispanics 23%, North American natives 21%) (35–37), further emphasizing the importance of the identified epitope. However, it has been reported that only a small portion of the peptides with predicted and experimentally confirmed high affinity binding, are actually capable to induce T-cell responses (38). Therefore, the utilization of functional assays is essential for determining the immunogenicity of an epitope. To that end, the functionality of T cell immune responses to 10-mer epitope were determined by two functional assays, ELISPOT IFN-γ and ICS. The results of ELISPOT IFN-γ assay showed robust NoV VP1 10-mer epitope-specific CD8+ T cell immune responses in HLA-A*0201-positive individuals, whereas control donors were negative. Titration of 10-mer peptide down to 0.05 μg/ml revealed high avidity of CD8+ T cells to 10-mer peptide. These results were expanded using ICS assay to detect CD8+ T cells secreting cytokines IFN-γ, IL-2, and TNF–α. By this approach, the presence of circulating monofunctional and polyfunctional memory CD8+ T lymphocytes specific for 10-mer NoV epitope was detected in HLA-A*0201-positive healthy donors. Polyfunctional CD8+ T cells that produce more than one immune mediator, are associated with protection from viral infections, such as HIV-1 and human herpes virus (39–41). In the future we aim to utilize the pentamer technology to validate CD8+ T cell responses in a larger number of serologically positive NoV-infected HLA-A*02:01 positive subjects to strengthen the clinical significance of the finding in this study.
Pentamer staining, ELISPOT IFN-γ and ICS assays, performed simultaneously using the same sample of each donor, resulted in highly congruent data supporting high immunogenicity of the novel NoV 139TMFPHIIVDV148 CD8+ T cell epitope. Many viral infections are not contained by antibody responses alone and it is likely that T cell responses play a role in clearance of NoV infection and may have a role in protective immunity as well. Virus-specific CTL responses play a crucial role in a clearance of many other viral infections such as human immunodeficiency virus-1 (HIV-1) (40, 42, 43), influenza (22, 44) and human papilloma virus (45). Patients with chronic hepatitis B (HBV) infection typically lack effective HBV-specific T cells, whereas fully recovered patients display strong CD8+ T-cell responses (46).
To conclude, the present work describes the first human NoV CD8+ T cell epitope 139TMFPHIIVDV148 restricted to HLA-A*02:01 allele. Regarding the high conservation of the identified epitope and high frequency of HLA-A*02:01 allele in human population, it can be speculated that ~20% of the individuals exposed to divergent NoV genotypes, will develop strong CD8+ T cell responses to this 10-mer epitope. While NoV blocking antibodies are largely genotype specific, a role of T cells targeted to broadly conserved epitopes, as described in here, may be of large significance. However, the role and importance of this epitope as well as overall T cell responses in protection from NoV infection needs to be further investigated.
Author Contributions
MM sample acquisition, processing, and laboratory analysis (Pentamer staining, ELISPOT, and ICS by flow cytometry). Data acquisition, analysis, and interpretation, and writing the manuscript. TV the Head of Vaccine Research Center, revision of the manuscript text. VB the Head of the Laboratory, conception and designing the study, data interpretation, drafting, and writing the paper, critical revision of article for important intellectual content.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the technical assistance given by the laboratory personnel of the Vaccine Research Center of University of Tampere.
References
1. Lopman BA, Steele D, Kirkwood CD, Parashar UD. The vast and varied global burden of norovirus: prospects for prevention and control. PLoS Med. (2016) 13:e1001999. doi: 10.1371/journal.pmed.1001999
2. Ramani S, Estes MK, Atmar RL. Correlates of protection against norovirus infection and disease-where are we now, where do we go? PLoS Pathog. (2016) 12:e1005334. doi: 10.1371/journal.ppat.1005334
3. Vesikari T, Blazevic V. Norovirus vaccine: one step closer. J Infect Dis. (2015) 211:853–5. doi: 10.1093/infdis/jiu498
4. Parra GI, Squires RB, Karangwa CK, Johnson JA, Lepore CJ, Sosnovtsev SV, et al. Static and evolving norovirus genotypes: implications for epidemiology and immunity. PLoS Pathog. (2017) 13:e1006136. doi: 10.1371/journal.ppat.1006136
5. Eden JS, Tanaka MM, Boni MF, Rawlinson WD, White PA. Recombination within the pandemic norovirus GII.4 lineage. J Virol. (2013) 87:6270–82. doi: 10.1128/JVI.03464-12
6. Kroneman A, Vega E, Vennema H, Vinje J, White PA, Hansman G, et al. Proposal for a unified norovirus nomenclature and genotyping. Arch Virol. (2013) 158:2059–68. doi: 10.1007/s00705-013-1708-5
7. Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science (2014) 346:755–9. doi: 10.1126/science.1257147
8. Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science (2016) 353:1387–93. doi: 10.1126/science.aaf5211
9. Atmar RL, Bernstein DI, Lyon GM, Treanor JJ, Al-Ibrahim MS, Graham DY, et al. Serological correlates of protection against a GII.4 Norovirus. Clin Vaccine Immunol. (2015) 22:923–9. doi: 10.1128/CVI.00196-15
10. Blazevic V, Lappalainen S, Nurminen K, Huhti L, Vesikari T. Norovirus VLPs and rotavirus VP6 protein as combined vaccine for childhood gastroenteritis. Vaccine (2011) 29:8126–33. doi: 10.1016/j.vaccine.2011.08.026
11. Nurminen K, Blazevic V, Huhti L, Rasanen S, Koho T, Hytonen VP, et al. Prevalence of norovirus GII-4 antibodies in Finnish children. J Med Virol. (2011) 83:525–31. doi: 10.1002/jmv.21990
12. Blazevic V, Malm M, Honkanen H, Knip M, Hyoty H, Vesikari T. Development and maturation of norovirus antibodies in childhood. Microbes Infect. (2015) 18:263–9. doi: 10.1016/j.micinf.2015.12.004
13. Blazevic V, Malm M, Salminen M, Oikarinen S, Hyoty H, Veijola R, et al. Multiple consecutive norovirus infections in the first 2 years of life. Eur J Pediatr. (2015) 174:1679–83. doi: 10.1007/s00431-015-2591-8
14. Malm M, Uusi-Kerttula H, Vesikari T, Blazevic V. High serum levels of norovirus genotype-specific blocking antibodies correlate with protection from infection in children. J Infect Dis. (2014) 210:1755–62. doi: 10.1093/infdis/jiu361
15. Reeck A, Kavanagh O, Estes MK, Opekun AR, Gilger MA, Graham DY, et al. Serological correlate of protection against norovirus-induced gastroenteritis. J Infect Dis. (2010) 202:1212–8. doi: 10.1086/656364
16. Cutler AJ, Oliveira J, Ferreira RC, Challis B, Walker NM, Caddy S, et al. Capturing the systemic immune signature of a norovirus infection: an n-of-1 case study within a clinical trial. Wellcome Open Res. (2017) 2:28. doi: 10.12688/wellcomeopenres.11300.1
17. Malm M, Tamminen K, Vesikari T, Blazevic V. Norovirus-specific memory T cell responses in adult human donors. Front Microbiol. (2016) 7:1570. doi: 10.3389/fmicb.2016.01570
18. Lindesmith L, Moe C, Lependu J, Frelinger JA, Treanor J, Baric RS. Cellular and humoral immunity following Snow Mountain virus challenge. J Virol. (2005) 79:2900–9. doi: 10.1128/JVI.79.5.2900-2909.2005
19. Tacket CO, Sztein MB, Losonsky GA, Wasserman SS, Estes MK. Humoral, mucosal, and cellular immune responses to oral Norwalk virus-like particles in volunteers. Clin Immunol. (2003) 108:241–7. doi: 10.1016/S1521-6616(03)00120-7
20. Chachu KA, LoBue AD, Strong DW, Baric RS, Virgin HW. Immune mechanisms responsible for vaccination against and clearance of mucosal and lymphatic norovirus infection. PLoS Pathog. (2008) 4:e1000236. doi: 10.1371/journal.ppat.1000236
21. Tomov VT, Osborne LC, Dolfi DV, Sonnenberg GF, Monticelli LA, Mansfield K, et al. Persistent enteric murine norovirus infection is associated with functionally suboptimal virus-specific CD8 T cell responses. J Virol. (2013) 87:7015–31. doi: 10.1128/JVI.03389-12
22. Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med. (2013) 19:1305–12. doi: 10.1038/nm.3350
23. Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics (2005) 6:132. doi: 10.1186/1471-2105-6-132
24. Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. (2003) 12:1007–17. doi: 10.1110/ps.0239403
25. Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. (2008) 36:509–12. doi: 10.1093/nar/gkn202
26. Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics (2016) 32:511–7. doi: 10.1093/bioinformatics/btv639
27. Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics (1999) 50:213–9. doi: 10.1007/s002510050595
28. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
29. Zuckerkandl E, Pauling L. Evolutionary Divergence and Convergence in Proteins. New York: Academic Press (1965). p. 97. doi: 10.1016/B978-1-4832-2734-4.50017-6
30. Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast WM, et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. (1994) 153:5586–92.
31. Ruppert J, Sidney J, Celis E, Kubo RT, Grey HM, Sette A. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell (1993) 74:929–37. doi: 10.1016/0092-8674(93)90472-3
32. Falk K, Rammensee H, Jung G, Rötzschke O, Stevanovié S. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature (1991) 351:290–6. doi: 10.1038/351290a0
33. Parker KC, Bednarek MA, Hull LK, Utz U, Cunningham B, Zweerink HJ, et al. Sequence motifs important for peptide binding to the human MHC class I molecule, HLA-A2. J Immunol. (1992) 149:3580–7.
34. Lindesmith LC, Donaldson E, Leon J, Moe CL, Frelinger JA, Johnston RE, et al. Heterotypic humoral and cellular immune responses following Norwalk virus infection. J Virol. (2010) 84:1800–15. doi: 10.1128/JVI.02179-09
35. Ellis JM, Henson V, Slack R, Ng J, Hartzman RJ, Hurley CK. Frequencies of HLA-A2 alleles in five U.S. population groups Predominance of A02011 and identification of HLA-A0231. Hum Immunol. (2000) 61:334–40. doi: 10.1016/S0198-8859(99)00155-X
36. Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernández-Viña MA. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol. (2001) 62:1009–30. doi: 10.1016/S0198-8859(01)00298-1
37. Krausa P, Browning MJ. HLA-A2 polymorphism and immune functions. Eur J Immunogenet. (1996) 23:261–74. doi: 10.1111/j.1744-313X.1996.tb00122.x
38. Zhong W, Reche PA, Lai CC, Reinhold B, Reinherz EL. Genome-wide characterization of a viral cytotoxic T lymphocyte epitope repertoire. J Biol Chem. (2003) 278:45135–44. doi: 10.1074/jbc.M307417200
39. Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood (2006) 107:4781–9. doi: 10.1182/blood-2005-12-4818
40. Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. (2008) 8:247–58. doi: 10.1038/nri2274
41. Lepone L, Rappocciolo G, Knowlton E, Jais M, Piazza P, Jenkins FJ, et al. Monofunctional and polyfunctional CD8+ T cell responses to human Herpesvirus 8 lytic and latency proteins. Clin Vaccine Immunol. (2010) 17:1507–16. doi: 10.1128/CVI.00189-10
42. Blazevic V, Sahgal N, Kessler HA, Landay AL, Shearer GM. T cell responses to recall antigens, alloantigen, and mitogen of HIV-infected patients receiving long-term combined antiretroviral therapy. AIDS Res Hum Retroviruses (2000) 16:1887–93. doi: 10.1089/08892220050195847
43. Rosenberg ES, Billingsley JM, Caliendo AM, Boswell SL, Sax PE, Kalams SA, et al. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science (1997) 278:1447–50. doi: 10.1126/science.278.5342.1447
44. Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. (2012) 18:274–80. doi: 10.1038/nm.2612
45. Stanley M. Immune responses to human papillomavirus. Vaccine (2006) 24 (Suppl. 1):16. doi: 10.1016/j.vaccine.2005.09.002
Keywords: norovirus, CD8 T cell epitope, HLA-A2*0201, multifunctional T cells, pentamer, ELISPOT IFN-gamma, cellular immunity
Citation: Malm M, Vesikari T and Blazevic V (2018) Identification of a First Human Norovirus CD8+ T Cell Epitope Restricted to HLA-A*0201 Allele. Front. Immunol. 9:2782. doi: 10.3389/fimmu.2018.02782
Received: 05 July 2018; Accepted: 12 November 2018;
Published: 27 November 2018.
Edited by:
Linda F. Van Dyk, University of Colorado Denver, United StatesReviewed by:
Lijun Xin, Cincinnati Children's Hospital Medical Center, United StatesJorma Hinkula, Linköping University, Sweden
Copyright © 2018 Malm, Vesikari and Blazevic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vesna Blazevic, vesna.blazevic@uta.fi
 Maria Malm
Maria Malm Timo Vesikari
Timo Vesikari Vesna Blazevic
Vesna Blazevic