
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 27 November 2018
Sec. Comparative Immunology
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.02758
Macrophages in teleosts are less sensitive to lipopolysaccharide (LPS) compared to mammals. The functional equivalent of the mammalian LPS surface receptor in teleost macrophages for the pro-inflammatory response is either non-existent or replaced by negative regulation. LPS signaling in teleost macrophages remains unclear. Here, we found a scavenger receptor class B 2a (PaSRB2a) that played a crucial role in LPS signaling in teleost macrophages. The internalization of LPS and subsequent pro-inflammatory responses in macrophages were mediated by PaSRB2a, which is a novel isoform of the mammalian SRB2 gene. LPS internalization by PaSRB2a is dependent on its C-terminal intracellular domain. Following LPS internalization, it interacts with the ayu intracellular receptors nucleotide-binding oligomerization domain protein 1 (PaNOD1) and PaNOD2. Moreover, LPS pre-stimulation with sub-threshold concentrations reduced the effect of secondary LPS treatment on pro-inflammatory responses that were mediated by PaSRB2a. The pro-inflammatory responses in LPS-treated ayu were down-regulated upon PaSRB2a knockdown by lentivirus siRNA delivery. In grass carp and spotted green pufferfish, SRB2a also mediated LPS internalization and pro-inflammatory responses. Our work identifies a novel LPS signaling pathway in teleosts that differs from those in mammals, and contributes to our understanding of the evolution of pathogen recognition in vertebrates.
Lipopolysaccharides (LPS), a major component of the outer membrane of gram-negative bacteria, are sensed by immune receptors (1). Since LPS commonly exists in pathogenic gram-negative bacteria, it has been identified as a major endotoxin in infectious diseases in vertebrates (2, 3). Macrophages are the main target cells of LPS (4, 5). After LPS recognition, macrophages are activated to produce pro-inflammatory cytokines (6). It has long been known that leukocytes of teleosts are less sensitive to LPS compared with those of mammals (7–9). This suggests that the mechanisms of LPS recognition differ between teleosts and mammals.
Extracellular LPS recognition plays an important role in pro-inflammatory responses (10, 11). Toll-like receptors (TLRs) participate in innate immune recognition as pattern recognition receptors (12–14). Among the TLRs, TLR4 was the first to be identified and is involved in the recognition of LPS, which is implicated in the etiology of LPS-induced septic shock (15). The TLR4 signaling pathway mediated by extracellular LPS recognition also includes myeloid differentiation protein 2 (MD2) and CD14 (16). In TLR4 knockout mouse macrophages, even 100 μg/ml LPS does not induce up-regulation of TNF, suggesting the critical role of TLR4 in LPS recognition of mammalian macrophages (17). Moreover, human TLR2 is also identified as a LPS receptor (18). In teleosts, TLR4 is lacking in some species and has been identified as a negative regulator of TLR signaling in zebrafish (19). The CD14 and MD2 genes of the TLR4 signaling pathway are not found in teleost genomes (19, 20). TLR2 genes have been identified in several teleosts species (zebrafish, Japanese flounder, fugu, and catfish). The precise mechanisms underlying extracellular LPS recognition in teleosts are unclear.
In addition to surface receptor interaction, LPS can also activate pro-inflammatory responses by binding with intracellular receptors via internalization by electroporation (21–23). In the inflammasome, intracellular sensor protein activation regulates the down-stream nuclear factor-κB (NF-κB), which participates in the pro-inflammatory response (24, 25). Nucleotide-binding oligomerization domain protein 1 (NOD1) has been implicated in the recognition of intracellular bacteria and LPS not only in mammals (26), but also in teleosts (27, 28), suggesting that the intracellular LPS recognition pattern is similar in mammals and teleosts. In mammals, outer membrane vesicles of bacteria are internalized by macrophages and release LPS in the cytoplasm (23, 29). It is still unclear how LPS enters the cytoplasm of macrophages in teleosts.
Teleost macrophage studies have revealed notable similarities and differences in the molecular strategies by which fish and higher vertebrates control their pro-inflammatory responses (6, 30). Here, we have found a novel pathway for LPS entrance into the cytoplasm of the teleost macrophages, via binding to a scavenger receptor. The ayu scavenger receptor B 2a (PaSRB2a) mediated the pro-inflammatory response in macrophages by interacting with LPS and internalizing it, activating NOD1 and NOD2. SRB2a expression was further confirmed to be dramatically down-regulated after LPS treatment in ayu macrophages, which partially explains the LPS tolerance of teleosts. Furthermore, SRB2a, a novel gene which is common in teleosts but not in mammals, was also shown to play an important role in LPS internalization and pro-inflammatory responses in grass carp and spotted green pufferfish.
Ayu (Plecoglossus altivelis), weighing 40 ± 5 g each, were used in all experiments. This strain has undergone seven successive generations of mass selection (31). The fish were kept at 20–22°C in a recirculating system using filtered water. They were held in the laboratory for two weeks before use in experiments. Grass carp, with a body weight of 100−120 g, were obtained from a commercial farm (Hunan, China). The fish were kept in freshwater tanks at 20–22°C in a recirculating system. One-year old wild-type spotted green pufferfish, weighing 3–6 g, were kept in recirculating water at 26–28°C. They were held in the laboratory for at least 2 weeks, with healthy appearance and normal activity, prior to use in experiments. For cell cultures, 7–8 fish were used in each experiment. The experimental conditions and procedures were approved by the Ningbo University Institutional Animal Care and Use Committee and were carried out in compliance with the National Institutes of Health's Guide for the Care and Use of Laboratory Animals.
LPS was extracted from Aeromonas hydrophila (ATCC 7966) and Vibrio anguillarum (Ayu-H080701) with the Lipopolysaccharide Extraction Kit (iNtRON Biotechnology, Inc., Burlington, MA, USA) according to the manufacturer's instructions. LPS extract was dissolved and treated with 2 mg/ml proteinase K at 37°C overnight, then collected by ethanol precipitation. After washing with 70% ethanol, pellets were dried, weighed, and reconstituted at a concentration of 1 mg/ml in distilled water. LPS purity was confirmed by its inability to induce IL-6 production in macrophages isolated from the LPS-unresponsive C3H/HeJ mouse strain (32). LPS from Escherichia coli O55:B5 and O111:B4 was purchased from Sigma Aldrich (St. Louis, MO, USA). Purpald assay was employed to quantify LPS. To each of the duplicated wells, we added 50 μl of the sample and 50 μl of 32 mm NaIO4 before incubating at room temperature for 25 min in the dark. After this, purpald reagent was added and the plate was incubated for 20 min in the dark. After NaIO4 was added, the absorbance was read at 550 nm.
The cDNA sequence of the PaSRB2a and PaSRB2b genes were obtained by the transcriptome analysis of ayu macrophages, and transcriptomic data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE40221 (33). PCR, cloning, and sequencing were used to confirm the authenticity of these sequences. The partial cDNA sequence of PaTLR2a, PaNOD1, and PaNOD2 genes were also obtained by transcriptome analysis of ayu macrophages. The full-length cDNA sequence of PaTLR2a, PaNOD1, and PaNOD2 were subsequently obtained by using the rapid amplification of cDNA ends (RACE) method and the sequence specific primers. First strand cDNA was synthesized using the SMART RACE kit (Clontech, Palo Alto, CA, USA). Gene-specific primers for PaNOD1, PaNOD2, and PaTLR2a were designed based on the corresponding gene sequences (Table 1). The cDNA sequences of PaTLR2a, PaSRB2a, PaSRB2b, PaNOD1, and PaNOD2 were deposited in GenBank under accession numbers MG674831, MH699855, JP736791, MG674829, and MG674830, respectively.
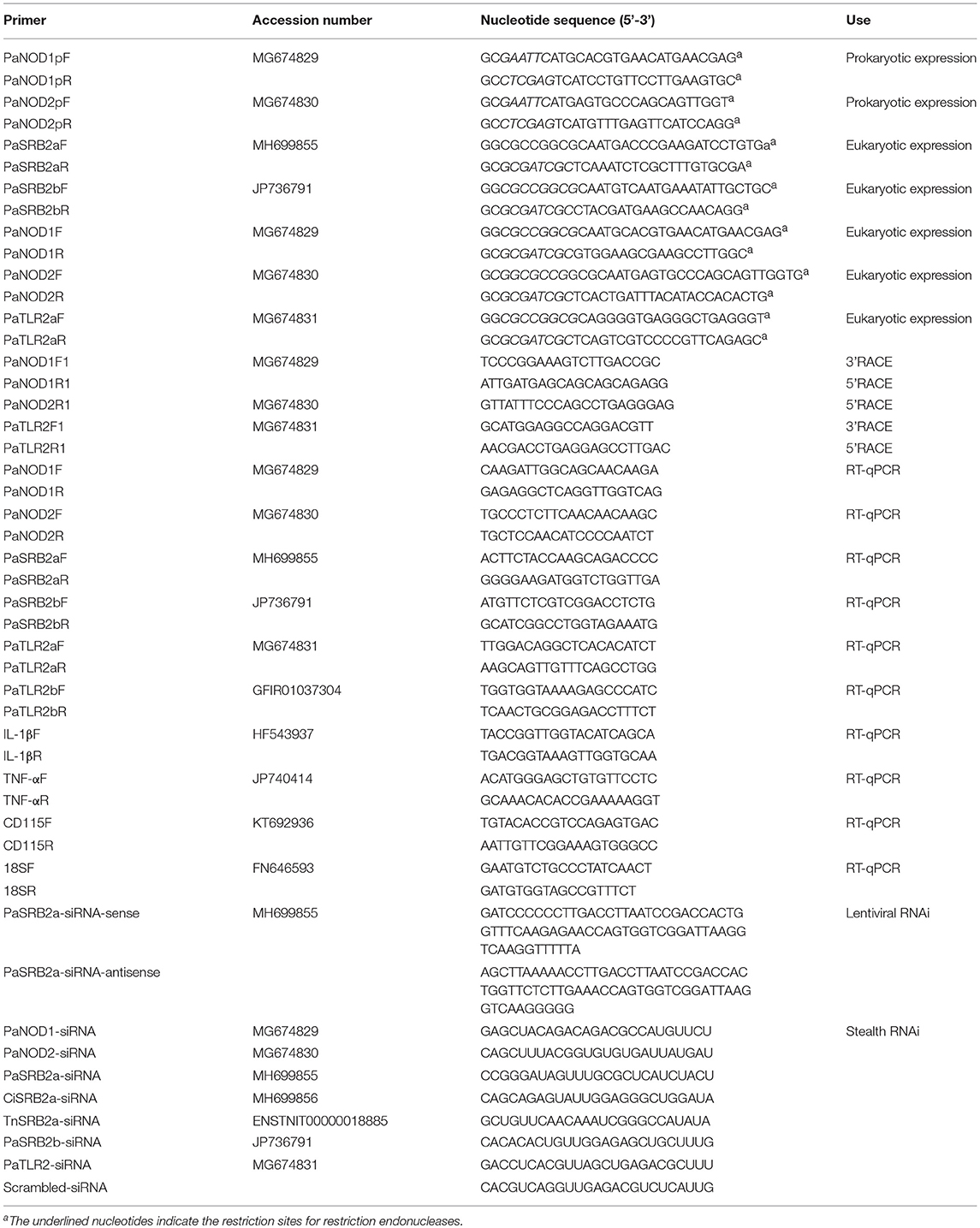
Table 1. Oligonucleotide primers used for amplifying cDNAs, expressing constructs, and gene expression analysis.
The similarity between the obtained sequences and other known sequences were analyzed using the basic local alignment search tool (https://blast.ncbi.nlm.nih.gov/). Phylogenetic and molecular evolutionary analyses were conducted using the molecular evolutionary genetics analysis (MEGA) tool (version 5). The accession numbers of TLR genes are listed in Table 2. The accession numbers of SRB genes are listed Table 3.
After ayu were sacrificed, the head kidney was rapidly removed to isolate the macrophages. Ayu head kidney was washed in RPMI 1,640 medium supplemented with 2% fetal bovine serum (FBS; Invitrogen, Shanghai, China), penicillin (100 U/ml), streptomycin (100 μg/ml), and heparin (20 U/ml). The buffy coat cells were separated by using Ficoll Hypaque PREMIUM (1.077 ± 0.001 g/ml; Invitrogen) in combination with centrifugation according to the manufacturer's instructions. The cells were then seeded in 35 mm dishes at a density of 2 × 107/ml. Non-adherent cells were washed off and the attached cells were incubated in complete medium (RPMI 1640, 5% ayu serum, 5% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin) at 24°C with 5% CO2. According to Giemsa staining results, over 96% of adherent cells were macrophages.
To inhibit clathrin-mediated endocytosis, sucrose hypertonic treatment was performed by incubating cells at 37°C with a 0.45 M sucrose solution; potassium (K+) depletion was performed by washing the cells at 37°C with PBS twice, followed by 5 min with hypotonic PBS (1:3 v/v with water), and five quick washes followed by 10 min incubation in K+-depletion solution (150 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 2 mM HEPES pH7.4, 0.5% DMSO, 0.5% BSA) (34). To inhibiting clathrin-independent endocytosis, we employed 5-(N-ethyl-N-isopropyl) amiloride (EIPA, 25 μm) and IPA-3 (IPA, 20 μm) to treat the cells (35).
Total RNA was extracted from ayu tissues or macrophages using RNAiso reagents (TaKaRa, Dalian, China) for RT-qPCR. Each RNA sample (5 μg) was incubated with 1 U DNase I (Thermo Fisher Scientific, Grand Island, NY, USA) for 30 min at 37°C to remove residual genomic DNA. The first-strand cDNA was then synthesized using reverse transcriptase M-MLV (TaKaRa). Gene-specific primers for PaTLR2a, PaSRB2a, PaSRB2b, PaNOD1, PaNOD2, CD115, TNF, IL-1β, and housekeeping gene 18S rRNA are listed in Table 1. Gene 18S rRNA was utilized as an internal reference for normalization. The RT-qPCR reaction was performed using SYBR premix Ex Taq (Perfect Real Time, TaKaRa). The reaction mixture was incubated for 300 s at 95°C, followed by 35 amplification cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C, on an ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Ct values of PaTLR2a, PaSRB2a, PaSRB2b, PaNOD1, PaNOD2, TNF, and IL-1β for all samples were normalized to 18S rRNA using the 2−ΔΔCt method.
LPS was diluted to the appropriate concentrations in PBS to infect ayu macrophages for different times and culture supernatant was collected for ELISA. Microplates (Nunc, Roskilde, Denmark) were coated overnight using concentrated cell supernatant. Plates were precoated using Poly-L-Lysine (Sigma Aldrich) to increase protein binding. Blocking of unbound binding sites was performed using 5% teleost gelatin (Sigma Aldrich). The anti-IL-1β and anti-TNF antibodies come from our previous studies (31, 34). One hundred microliters of 5 μg/ml anti-IL-1β or anti-TNF antibody was added to each well, incubated for 1 h, and washed three times. One hundred microliter of a 1:2500 dilution in PBS of a horseradish peroxidase (HRP)-labeled secondary antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) in PBS was added to each well for 1 h and washed three times. Finally, alkaline phosphatase yellow liquid substrate system for ELISA (Sigma Aldrich) was used and optical density read at 405 nm.
The target gene siRNA and scrambled siRNA of PaTLR2a,PaSRB2a, PaSRB2b, PaNOD1, and PaNOD2 for RNAi were designed and synthesized by Invitrogen, respectively. The siRNA sequences of target genes are listed in Table 1. Transfection of cells with siRNA was performed using the Lipofectamine 2,000 transfection reagent (Invitrogen) according to the manufacturer's protocol. Briefly, 5 μl of Lipofectamine 2,000 in 250 μl of Opti-MEM (Invitrogen) was mixed with either 100 pmol siRNA or 100 pmol scrambled siRNA in 250 μl of Opti-MEM. The mixture was then incubated for 20 min at room temperature before adding to macrophages with a final siRNA concentration of 40 nm. After a 5.5-h incubation, media were changed to complete media, and cells were cultured for another 48, 72, or 96 h before collection for expression analysis of PaTLR2a, PaSRB2a, PaSRB2b, PaNOD1, and PaNOD2. RT-qPCR and western blot analyses confirmed the knockdown of PaTLR2a, PaSRB2a, PaSRB2b, PaNOD1, and PaNOD2 mRNA and protein, respectively.
Ayu macrophages were washed twice in PBS and lysed in a buffer [20 mM HEPES [pH7.4], 1.5 mM MgCl2, 0.2 mM EDTA, 100 mM NaCl, 0.2 mM DTT, 0.5 mM sodium orthovanadate, and 0.4 mM PMSF] containing phosphatase inhibitor (Phosphatase Inhibitor Cocktail, Sigma Aldrich). The protein concentration of the supernatant was measured in each soluble fraction using the Bradford method, and samples were subjected to SDS-PAGE (12% acrylamide gel) and transferred to PVDF (Pall, New York, NY, USA). Membranes were blocked for 1 h in a 10% non-fat dry milk solution in TBS-Tween. After a 1.5-h incubation with antibodies against PaTLR2a, PaSRB2a, PaSRB2b, PaNOD1, or PaNOD2, membranes were washed and incubated for 1 h with HRP-labeled secondary antibody (1:5000, Santa Cruz Biotechnology). Proteins were visualized by enhanced chemiluminescence (Santa Cruz Biotechnology). Ayu β-actin was used as the control. The intensity of each band obtained by western blot was analyzed using the NIH ImageJ program.
The antibodies of PaTLR2a, PaSRB2a, and PaSRB2b were prepared using peptides derived from these proteins (PaTLR2a: 123-139 aa, PaSRB2a: 243-259 aa, and PaSRB2b: 336-352 aa, GL Biochem, Shanghai, China). These peptides were synthesized to generate the respective monoclonal antibodies (GL Biochem; 1 mg/ml). The antibodies were validated using flow cytometry and Western blot (Supplemental Figures 1A–C). For flow cytometry, the antibodies to PaTLR2a, PaSRB2a, and PaSRB2b were used at dilutions of 1:200, 1:300, and 1:200, respectively. For western blotting, antibodies to PaTLR2a, PaSRB2a, and PaSRB2b were used at dilutions of 1:400, 1:600, and 1:400, respectively. Antibodies to PaNOD1 and PaNOD2 were prepared using recombinant proteins. The recombinant plasmids were constructed and transformed into E. coli BL21 (DE3) for over-expression. PaNOD1 and PaNOD2 were expressed as inclusion bodies and were purified using a Ni-NTA column (QIAGEN, Shanghai, China), according to the manufacturer's instructions. Endotoxin in the recombinant proteins was < 0.1 EU/mg after processing with an endotoxin-removal column (Pierce, Rockford, IL). The concentration of the PaNOD1 and PaNOD2 antibodies were both 2.5 mg/ml quantified by the Bradford method. The primer sequences for the target genes are listed in Table 1. The antibodies were prepared from the rabbit against PaNOD1 and PaNOD2. The specificity of anti-PaNOD1 and anti-PaNOD2 antibodies was validated using ELISA and Western blot (Supplemental Figures 1D,E). For ELISA, the antibodies to PaNOD1 and PaNOD2 were used at dilutions of 1:300 and 1:400, respectively. For Western blot, antibodies to PaNOD1 and PaNOD2 were used at dilutions of 1:600 and 1:800, respectively.
Macrophages or HEK293T cells were incubated at 24°C with FITC-LPS (Sigma Aldrich). After incubation, cells were washed with PBS and further treated with 250 μg/ml proteinase K (Sigma Aldrich) for 30 min to remove cell surface-bound LPS. The remaining LPS were considered to be intracellular. FITC-LPS in macrophages was measured by a Gallios flow cytometer (Beckman Coulter, Miami, FL, USA). To evaluate the participation of scavenger receptors in LPS uptake, cells were pretreated with oxLDL and polyG for 30 min before measurement of FITC-LPS uptake.
For confocal microscopy analysis, cells were washed three times with PBS and fixed with 200 μl of 4% paraformaldehyde in PBS for 30 min after LPS internalization, then air-dried. After three washes with PBS, cells were probed with rhodamine phalloidin for 30 min (66 nm, Invitrogen) to indicate filamentous cell membrane (red). DAPI (10 μg/ml, Sigma-Aldrich, Shanghai, China) was used to stain the cell nucleus for 5 min after washing with PBS. Coverslips were viewed using an IX81-FV1000 microscope (Olympus, Tokyo, Japan).
Chimeric DNA constructs of PaSRB2a and PaSRB2b were created using a standard overlap extension PCR technique. Chimeric PCR products were cloned into the pcDNA3.1 vector. Sequencing was performed for all DNA constructs, which were used to transfect HEK293T cells. For PaSRB2a overexpression in macrophages, PaSRB2a was prepared using a PCR-based method. For expression, a lentiviral vector carrying cDNA of PaSRB2a was constructed using the ViraPower lentiviral expression system (Invitrogen). Viral particles were produced in HEK293T cells according to the manufacturer's instructions. Cells were treated with blasticidin for selecting cells with successful integration of the plasmid. Lentiviral vectors were transduced into ayu macrophages at a multiplicity of infection of three.
HEK293T cells were co-transfected with PaSRB2a, PaNOD1, or PaNOD2 luciferase reporter vector, NF-κB luciferase reporter plasmid (or IFNβ luciferase reporter plasmid), and Renilla luciferase plasmid. After 24 or 48 h, the cells were collected and assayed for reporter activity using the Dual-Luciferase reporter system following the manufacturer's instructions (Promega, Shanghai, China). The relative luciferase activity was achieved against the Renilla luciferase control.
The polysaccharide moiety of LPS was biotin-labeled using biotin hydrazide (Thermo Fisher Scientific) according to the manufacturer's instructions. HEK293T cells were transfected with PaSRB2a, PaNOD1, or PaNOD2. Cells were washed and lysed for immunoprecipitations. The cell lysates were incubated with biotinylated LPS at 4°C overnight. Biotinylated LPS protein complexes were collected by addition of 20 μl of packed streptavidin beads. The presence of PaSRB2a, PaNOD1, or PaNOD2 in the LPS complexes was assessed by western blotting.
For gene knockdown in vivo by lentivirus delivery, the small interfering RNAs (siRNAs) against PaSRB2a were predicted by BLOCK-iT RNAi Designer (Invitrogen). Short hairpin RNAs containing the selected siRNA sequences, as well as scrambled siRNA, were designed and listed in Table 1. DNA oligonucleotides for short hairpin RNA expression were synthesized by Invitrogen, annealed, and constructed into pSUPER vector (Oligoengine, Seattle, WA, USA) downstream of the H1 promoter, as previously described (36). The generated constructs with different siRNAs (1 μg) or control pSUPER (1 μg) along with overexpression plasmid pcDNA3.1-PaSRB2a (1 μg) were co-transfected into HEK293T cells in 12-well plates. The efficiency of siRNAs against PaSRB2a was determined by RT-qPCR.
The U6 promoter cassette in lentiviral vector pLB was replaced by the H1-siRNA cassette excised from the most effective siRNA constructs to produce lentiviral vectors. Lentiviruses were produced by transient transfection of packaging cell line HEK293T cells. In brief, Lentiviral vectors (20 μg) were co-transfected with pCMV-dR8.2 dvpr (15 μg) and pCMV-VSV-G (6 μg) packaging vectors into HEK293T cells using FuGENE 6 Transfection Reagent (Promega). The lentiviral supernatant was harvested at 48–72 h post transfection, concentrated via ultracentrifugation at 25,000 rpm for 90 min at 4°C (SW-41Ti rotor) to dissolve in 100 μl PBS, and then purified and concentrated using the Fast-Trap Lentivirus Purification and Concentration Kit (Millipore, Bedford, MA, USA). Lentivirus titers were determined by transduction analysis of GFP expression in HEK293T cells. Infected cells were examined under a fluorescent microscope investigation (Nikon, Tokyo, Japan).
The silencing efficiency of the constructed lentivirus was determined both in vitro and in vivo. In vitro, HEK293T cells were transfected with pcDNA3.1-PaSRB2a. Concentrated lentivirus (10 μl) was added to the culture medium and 4 μg/ml of polybrene (Sigma Aldrich) was simultaneously added to increase the infection efficiency. The silencing efficiency of lentivirus was detected by RT-qPCR. For in vivo assay, the lentiviruses (1 × 108 TU/fish/day) was repeatedly delivered into fish by injection once a day for 3 d. Total RNA from liver and spleen were isolated and reverse transcribed into cDNA. RT-qPCR was conducted to evaluate the efficiency of in vivo suppression of PaSRB2a.
Survival rate assay was performed as previously described (37). Briefly, fish were divided into different treatment groups, each containing 30 fish. The fish in the experimental groups received ip. injections of LPS with 0.2 mg/g body weight, while the control group received PBS. Morbidity was monitored for 7 days after challenge, and the results were recorded every 24 h.
Clondronate-liposome suspension has been given to deplete tissue macrophages in rodents and some kinds of teleosts (37, 38). Ayu macrophage depletion was performed by ip injection with clodronate-liposomes or PBS-liposomes on days −4 and −2. The clodronate-liposomes were given ip at a dose of 0.25 ml/100 g. CD115 has been identified as a marker of monocytes/macrophages in mammal and teleosts (39). The depletion of monocytes/macrophages was confirmed by flow cytometry analysis of CD115-positive cells in the blood and RT-qPCR analysis of CD115 mRNA in peripheral blood mononuclear cells 4 days after first injection of liposomes (37). Peripheral blood mononuclear cells were isolated from freshly collected blood by Ficoll (Invitrogen) density gradient centrifugation, washed, and resuspended in PBS. Antibodies to ayu CD115 were prepared at 1 mg/ml in previous work (37) and used at a dilution of 1:300 for flow cytometry.
Data are presented as mean ± SEM. The animals used in the experiments were randomly chosen. Animal experiments were performed by an observer blinded to the experimental conditions. The survival curves were analyzed using the Kaplan-Meier method. All other data were analyzed by one-way ANOVA. When variances were significantly different (p < 0.05), logarithmic transformation was used to stabilize the variance. *, **, and *** represent p-values, 0.05, 0.01, and 0.001, respectively.
It has been reported that high concentrations of LPS are required to activate leukocytes from several types of teleosts in vitro (7, 40). We measured TNF and IL-1β mRNA expression in ayu macrophages at different time points after LPS treatment at 10 μg/ml. TNF and IL-1β were both up-regulated after LPS O55:B5 treatment (Supplemental Figures 2A,B). We further measured TNF and IL-1β mRNA expression in macrophages at different LPS concentrations. LPS treatment induced a dramatic up-regulation of TNF and IL-1β mRNA expression at 10 and 100 μg/ml (Supplemental Figures 2C,D). There was no up-regulation of TNF and IL-1β observed in LPS-treated macrophages with concentrations ≤ 1 μg/ml. Up-regulation of TNF and IL-1β proteins were found at 4, 8, and 16 h after LPS treatment at 10 μg/ml (Supplemental Figures 2E,F). These data suggest that LPS initiates inflammatory responses in teleost macrophages at high concentrations.
TLR4 activation by LPS does not lead to a pro-inflammatory reaction in zebrafish (7). However, it is still unclear whether TLR2 mediates LPS signaling in teleost macrophages. We first cloned the PaTLR2a and PaTLR2b genes from ayu. PaTLR2a and PaTLR2b associated with the TLR2 genes from other animals to form a cluster in a phylogenetic tree (Supplemental Figure 3). PaTLR2a was expressed in ayu macrophages (Supplemental Figures 4A,B). However, PaTLR2b could not been detect in ayu macrophages by RT-qPCR. Moreover, PaTLR2b is non-existent in the transcriptome of ayu macrophages (35). We further investigate whether PaTLR2a mediates LPS effect in ayu macrophages. We knocked down PaTLR2a gene in ayu macrophages by RNAi (Supplemental Figures 4A,B). The PaTLR2a mRNA was down-regulated at 48, 72, and 96 h after PaTLR2a siRNA treatment in ayu macrophages (Supplemental Figure 4A). Western blot results confirmed that PaTLR2a protein was down-regulated at 96 h after PaTLR2a siRNA treatment in macrophages (Supplemental Figure 4B). We further investigated the effect of PaTLR2a on TNF and IL-1β expression. PaTLR2a siRNA treatment did not affect TNF and IL-1β expression in macrophages treated with LPS (Supplemental Figures 4C,D).
Since TLR2 also does not mediate the effect of LPS on the pro-inflammatory reaction, we further asked whether LPS can enter ayu macrophages to induce pro-inflammatory responses. First, we measured whether LPS can enter HEK293T cells. FITC-LPS were incubated with HEK293T cells for 4 h, and proteinase K was used to remove extracellular LPS. No FITC-LPS signaling was detected by flow cytometry in HEK293T cells (Figure 1A). We further collected ayu macrophages at different time points after FITC-LPS treatment. Specific internalization of LPS was detected by flow cytometry (Figure 1B). Since high concentrations of LPS are required to activate teleost leukocytes, we further measured LPS internalization in macrophages with different LPS concentrations. The MFI of internalization of LPS at 1 μg/ml was 1.7-fold that at 103 ng/ml, while the MFI of internalization of LPS at 10 μg/ml was 7.8-fold that at 104 ng/ml (Figure 1C). Furthermore, confocal microscopy indicated that LPS was internalized within the cell cytoplasm 30 min after incubation (Figure 1D).
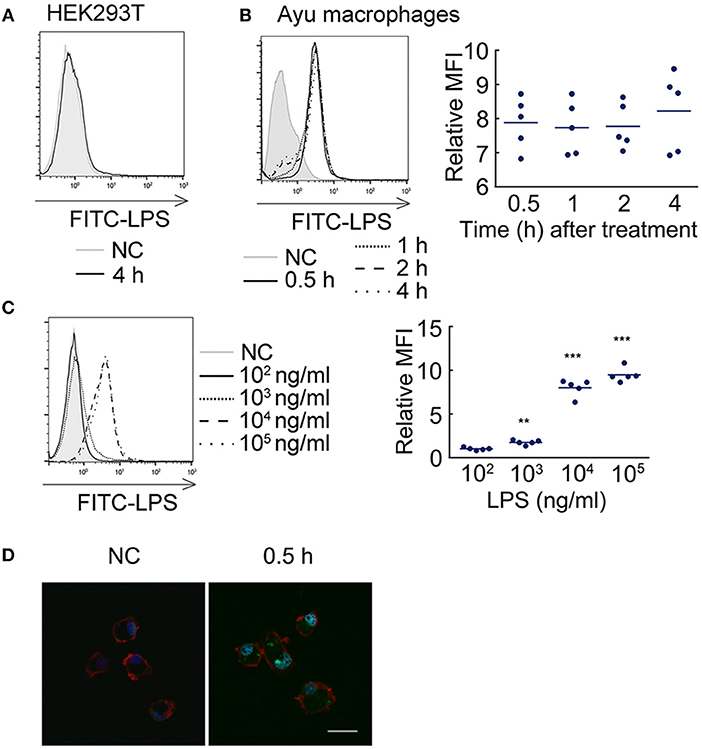
Figure 1. LPS internalization in ayu macrophages. (A) LPS did not enter HEK293T cells after 4 h incubation with 10 μg/ml LPS. (B) LPS detection in ayu macrophages after LPS treatment at different time points. The LPS concentration was 10 μg/ml. (C) LPS detection in ayu macrophages after LPS treatment for 4 h at different LPS concentrations. (D) Internalization of LPS in ayu macrophages was observed by confocal microscopy. Green color: FITC-LPS, red color: rhodamine-phalloidin, blue color: 4', 6'-diamidino-2-phenylindole (DAPI). Bar, 20 μm. Each bar represents the mean ± SE. n = 5. Data are representative of two (A–C) and three (D) independent experiments. **P < 0.01, ***P < 0.001.
It is well-known that scavenger receptors mediate the recognition and internalization of endotoxin, not only in mammals (41), but also in teleosts (42). We further investigated what types of scavenger receptors mediated LPS internalization. First, we employed antagonists of scavenger receptor B (SRB, oxLDL) and scavenger receptor A (SRA, polyG) to measure their effect on LPS internalization. OxLDL reduced the MFI of FITC-LPS in ayu macrophages (MFI: 8.1 at 0 μg/ml, 3.2 at 10 μg/ml, 1.8 at 50 μg/ml, respectively). PolyG did not affect LPS internalization in macrophages at 1 and 5 mg/ml. These data suggest that SRB may participate in LPS internalization in macrophages. We then cloned two SRB genes from ayu macrophages, defined as PaSRB2a and PaSRB2b. The receptor internalization of PaSRB2a and PaSRB2b was measured after LPS treatment by flow cytometry and western blot. LPS treatment at different time points led to the internalization of PaSRB2a, but not PaSRB2b (Figures 2A,B). PaSRB2a and LPS internalization was not affected after sucrose hypertonic treatment and K+ depletion, which both inhibit clathrin-mediated endocytosis (Figures 2C,D). However, PaSRB2a and LPS internalization was prevented after treatment with 5-(N-ethyl-N-isopropyl) amiloride (EIPA) or IPA-3 (IPA), which inhibit clathrin-independent endocytosis (Figures 2C,D). These data suggest that PaSRB2a may play an important role in LPS internalization by ayu macrophages.
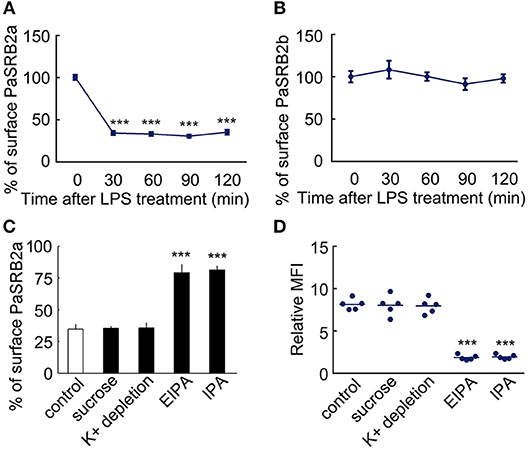
Figure 2. LPS internalization accompanied PaSRB2a endocytosis in ayu macrophages. (A) Surface levels of PaSRB2a were measured by flow cytometry at different time points after treatment with 10 μg/ml LPS. (B) Surface levels of PaSRB2b were measured by flow cytometry at different time points. (C) Surface levels of PaSRB2a after treatment with chemical clathrin-mediated endocytosis inhibitors (sucrose and K+ depletion) or clathrin-independent endocytosis inhibitors (EIPA, 5-(N-ethyl-N-isopropyl) amiloride and IPA, IPA-3). (D) LPS internalization after treatment with membrane endocytosis inhibitors. Each bar represents the mean ± SE. n = 5. Data are representative of three independent experiments. ***P < 0.001.
To confirm the effect of PaSRB2a on LPS internalization, we knocked down PaSRB2a in ayu macrophages by siRNA. The PaSRB2a mRNA was down-regulated at all tested time points after PaSRB2a siRNA treatment in ayu macrophages (Figure 3A). Western blot results confirmed that PaSRB2a protein was down-regulated at 96 h after PaSRB2a siRNA treatment in macrophages (Figure 3B). PaSRB2a siRNA treatment reduced LPS internalization in macrophages (Figure 3C). Furthermore, PaSRB2b siRNA treatment also knocked down the expression of PaSRB2b in macrophages (Figure 3D). Western blot results confirmed that PaSRB2b protein was down-regulated at 96 h after PaSRB2b siRNA treatment in macrophages (Figure 3E). However, there was no change in LPS internalization observed in macrophages after PaSRB2b siRNA treatment (Figure 3F). We further measured whether the internalization of LPS mediated by PaSRB2a affects TNF and IL-1β production. TNF and IL-1β protein levels at both 4 and 8 h were lower in macrophages treated with PaSRB2a siRNA than in those treated with scrambled siRNA (Figures 3G,H). Moreover, PaSRB2a was transfected into HEK293T cells with no LPS internalization before transfection. LPS internalization was then observed in HEK293T cells after transfection with PaSRB2a (Figures 3I, J). Our results demonstrate that PaSRB2a mediates TNF and IL-1β production after LPS induction in ayu macrophages by regulating LPS internalization.
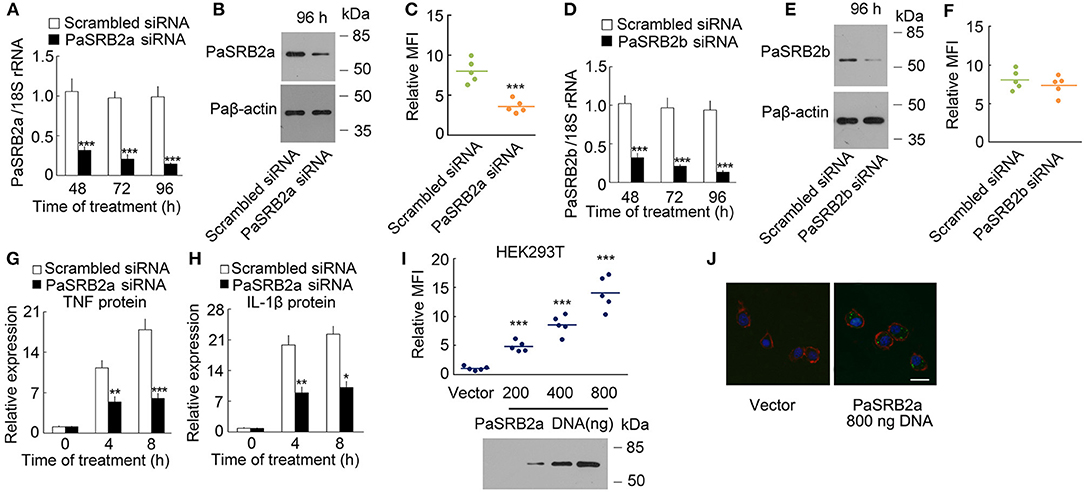
Figure 3. PaSRB2a mediated LPS internalization and pro-inflammatory response. (A) PaSRB2a mRNA levels in ayu macrophages treated with PaSRB2a siRNA. (B) PaSRB2a protein levels in ayu macrophages treated with PaSRB2a siRNA. Representative blots of three independent experiments are shown. (C) Effect of PaSRB2a siRNA on LPS internalization in ayu macrophages. (D) PaSRB2b mRNA levels in ayu macrophages treated with PaSRB2b siRNA. (E) PaSRB2b protein levels in ayu macrophages treated with PaSRB2b siRNA. Representative blots of three independent experiments are shown. (F) Effect of PaSRB2b siRNA on LPS internalization in ayu macrophages. (G) PaSRB2a effect on TNF expression in LPS-treated ayu macrophages. (H) PaSRB2a effect on IL-1β expression in LPS-treated ayu macrophages. (I) LPS internalization in HEK293T cells transfected with a PaSRB2a expression plasmid by flow cytometry. Representative blots of three independent experiments are shown. (J) Internalization of LPS in HEK293T was observed by confocal microscopy. Green color: FITC-LPS, red color: rhodamine-phalloidin, blue color: 4', 6'-diamidino-2-phenylindole (DAPI). Bar, 20 μm. Each bar represents the mean ± SE. n = 5. Data are representative of two (A,C,D,F–I) and three (B,E,J) independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
We then determined the presence of an intracellular receptor for LPS. NOD-like receptors play an important role in host defense as intracellular receptors for pathogen-associated molecular patterns (28, 43). We cloned the nucleotide-binding oligomerization domain 1 (PaNOD1) and PaNOD2 genes from ayu macrophages, and employed RNAi to knockdown the expression of NOD1 and NOD2 in ayu macrophages. Both PaNOD1 and PaNOD2 siRNAs reduced the RNA levels of PaNOD1 and PaNOD2 in ayu macrophages at 48, 72, and 96 h after siRNA treatment (Figures 4A,B). Western blot results confirmed that PaNOD1 and PaNOD2 protein levels were down-regulated at 96 h after PaNOD1 and PaNOD2 siRNA treatment in macrophages (Figures 4A,B). TNF and IL-1β were measured in LPS-treated macrophages after siRNA incubation. Both PaNOD1 and PaNOD2 siRNA incubation resulted in the down-regulation of TNF and IL-1β protein expression in macrophages at 4 and 8 h after LPS treatment (Figures 4C,D).
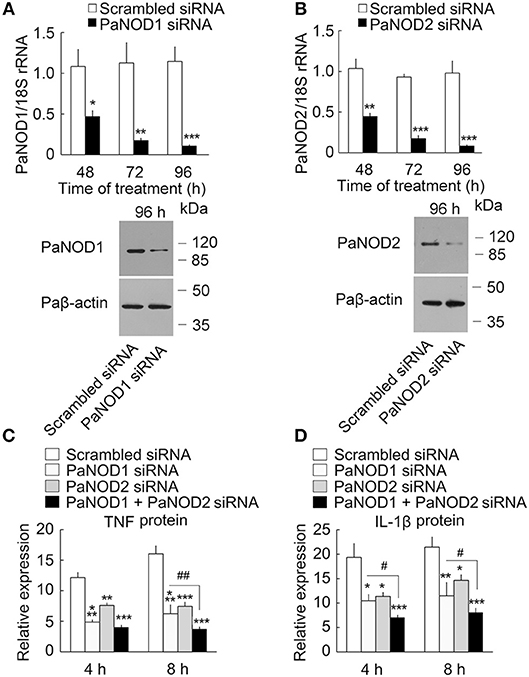
Figure 4. Effect of PaNOD1 and PaNOD2 on the production of TNF and IL-1β in ayu macrophages. (A) PaNOD1 mRNA and protein expression in macrophages treated with PaNOD1 siRNA. Representative blots of three independent experiments are shown. (B) PaNOD2 mRNA and protein expression in macrophages treated with PaNOD2 siRNA. Representative blots of three independent experiments are shown. (C) TNF production in macrophages after PaNOD1 siRNA, PaNOD2 siRNA, or PaNOD1 plus PaNOD2 siRNA treatment. (D) IL-1β expression in macrophages after PaNOD1 siRNA, PaNOD2 siRNA, or PaNOD1 plus PaNOD2 siRNA treatment. Each bar represents the mean ± SE. n = 5. Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. #P < 0.05, ##P < 0.01 vs. PaNOD1 siRNA.
We found that ayu PaSRB2a and PaSRB2b were clustered together in a phylogenetic tree built with the SRBS genes from several other animals (Figure 5A). PaSRB2a and PaSRB2b share a high degree of sequence similarity. However, PaSRB2a, but not PaSRB2b, mediated LPS internalization in ayu macrophages. Thus, we expressed a chimeric PaSRB2a and PaSRB2b receptor to determine the domains of PaSRB2a responsible for LPS internalization (Figure 5B). When we expressed the chimeric receptor consisting of the N-terminal intracellular and extracellular domains of PaSRB2b and the C-terminal intracellular domain of PaSRB2a, we detected a increase in LPS internalization (Figure 5C), suggesting that the C-terminal intracellular domain of PaSRB2a plays an important role in LPS internalization.
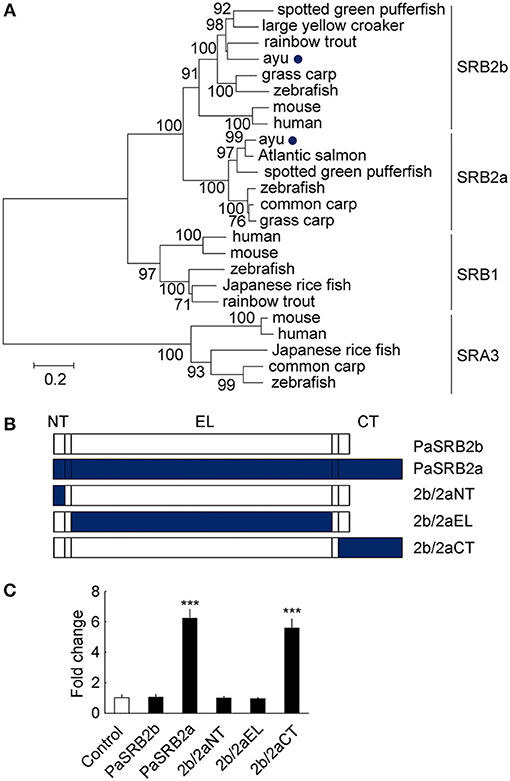
Figure 5. The critical domains for PaSRB2a uptake of LPS. (A) Phylogenetic (neighbor-joining) analysis of SRB amino acid sequences using MEGA 5.0. Node values represent the percentage bootstrap confidence derived from 1,000 replicates. The accession numbers of scavenger receptor genes are listed in Table 3. (B) Organization of cDNAs encoding PaSRB2a and PaSRB2b chimeras. PaSRB2a and PaSRB2b are predicted to have an intracellular N terminus, large extracellular domain, and intracellular C terminus. (C) LPS internalization in HEK293T cells transfected with PaSRB2a-PaSRB2b chimeras. 2b/2aNT: PaSRB2b-PaSRB2a N-terminal domain chimera, 2b/2aEL: PaSRB2b-PaSRB2a extracellular loop domain chimera, 2b/2aCT: PaSRB2b-PaSRB2a C-terminal chimera. Data are expressed as fold-change compared to the control. ***P < 0.001.
Our results illustrate that PaSRB2a, PaNOD1, and PaNOD2 were important in the LPS-mediated pro-inflammatory reaction in ayu macrophages. We then attempted to determine whether PaNOD1 and PaNOD2 act downstream of PaSRB2a. HEK293T cells were transfected with increasing amounts of PaNOD1 to measure the effect of LPS on NF-κB activation. The relative luciferase activity of NF-κB was not affected in PaNOD1-transfected HEK293T cells after LPS incubation without electroporation (Figure 6A). The relative luciferase activity was dramatically up-regulated after LPS electroporation (Figure 6A). PaNOD2 transfection also led to the up-regulation of relative luciferase activity of NF-κB in HEK293T cells with LPS electroporation, but not in HEK293T cells without LPS electroporation (Figure 6B). The relative luciferase activity of NF-κB was not affected by PaSRB2a transfection in LPS-treated HEK293T cells with or without electroporation (Figure 6C). Furthermore, both PaNOD1 and PaNOD2 increased the relative luciferase activity of NF-κB after LPS incubation without electroporation in PaSRB2a transfected HEK293T cells (Figure 6D). The relative luciferase activity of IFNβ was not changed after LPS incubation in PaSRB2a-, PaNOD1-, and PaNOD2-transfected HEK293T cells (Supplemental Figure 5). We further investigated if PaSRB2a, PaNOD1, and PaNOD2 directly bind LPS. LPS precipitation assay illustrated that LPS interacted physically with PaSRB2a, PaNOD1, and PaNOD2 (Figure 6E). These data suggest that PaSRB2a is necessary for LPS internalization to activate inflammatory responses.
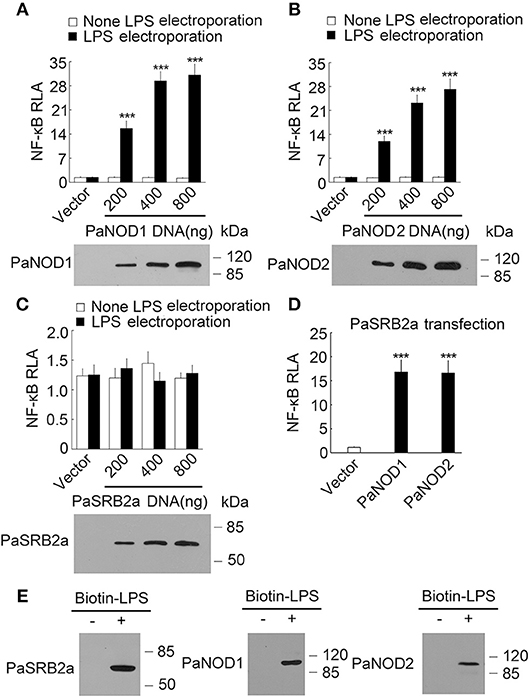
Figure 6. LPS activated NF-κB via PaNOD1, PaNOD2, and PaSRB2a. (A) PaNOD1 effect on NF-κB activation in LPS-treated HEK293T cells. (B) PaNOD2 effect on NF-κB activation in LPS-treated HEK293T cells. (C) PaSRB2a effect on NF-κB activation in LPS-treated HEK293T cells. (D) The effect of PaNOD1 and PaNOD2 in HEK293T cells with PaSRB2a transfection after LPS treatment. Bottom panels present western blots of cell lysates to monitor protein expression. (E) Physical interaction between LPS and PaSRB2a, PaNOD1, PaNOD2. Each lane represents the cell lysate incubated with biotinylated LPS (Biotin-LPS) and avidin beads. Each bar represents the mean ± SE. Representative blots of three independent experiments are shown. n = 5. Data are representative of two (A–D) and three (E) independent experiments. ***P < 0.001.
We further investigated PaSRB2a function in vivo by delivering lentivirus into ayu tissues. Using HEK293T cells, we identified the most effective siRNA (85.5%) for pSUPER-PaSRB2a (Figure 7A). The interference efficiency of this system for PaSRB2a reached 77.1% in liver and 91.5% in spleen (Figure 7B). The siRNA expression efficiency of the lentiviruses detected in HEK293T cells reached > 90% using the expression of GFP as an indicator for PaSRB2a (Figure 7C). The survival rates of pSUPER-PaSRB2a-treated ayu were up-regulated at 96 h after LPS administration compared with pSUPER empty vector (Figure 7D). In pSUPER-PaSRB2a-transfected ayu, the TNF and IL-1β mRNA levels in liver were down-regulated compared with those in pSUPER empty vector transfected ayu (Figures 7E, F). Both TNF and IL-1β were also down-regulated in the spleen in pSUPER-PaSRB2a-transfected ayu compared with those in pSUPER-transfected ayu (Figures 7G,H).
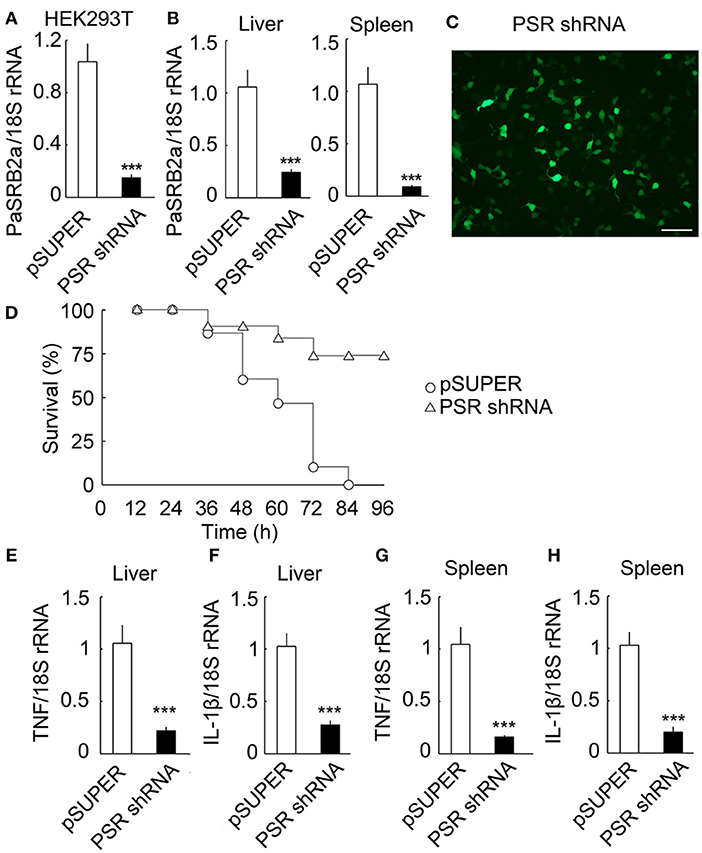
Figure 7. Effect of PaSRB2a on outcome of LPS-treated ayu. (A) PaSRB2a shRNA decreased the expression of PaSRB2a in HEK293T cells. (B) PaSRB2a mRNA expression in liver and spleen after delivery of lentiviruses into ayu. (C) shRNA expression efficiency of lentiviruses. Bar: 100 μm. GFP fluorescence was detected in HEK293T cells by microscopy. (D) Survival in ayu treated with pSUPER and pSUPER-PaSRB2a shRNA. n = 30 per group. RT-qPCR analysis of TNF (E) and IL-1β (F) expression in ayu liver. RT-qPCR analysis of TNF (G) and IL-1β (H) expression in ayu spleen. PSR shRNA: PaSRB2a shRNA. Each bar represents the mean ± SE. n = 5. Data are representative of two (A,B, E–H) and three (C,D) independent experiments. ***P < 0.001.
It has long been known that sub-threshold stimulatory concentrations of LPS for 6 h result in the enhancement of pro-inflammatory responses after secondary LPS stimulation in mammals (44). We investigated the effect of secondary LPS stimulation on sub-threshold LPS pre-stimulated ayu macrophages. In 10 μg/ml LPS-stimulated ayu macrophages, the protein levels of TNF and IL-1β were down-regulated at 10, 100, and 1,000 μg/ml LPS pre-stimulated macrophages compared to those without LPS pre-stimulation (Figures 8A,B). We then investigated whether PaSRB2a mediated the down-regulation of pro-inflammatory responses in LPS pre-stimulated macrophages. First, we found that PaSRB2a was down-regulated in LPS pre-stimulated macrophages (Figures 8C,D). We delivered lentivirus to ayu macrophages to increase PaSRB2a expression (Figure 8E). PaSRB2a lentivirus restored the induction of pro-inflammatory responses by LPS in LPS pre-stimulated macrophages (Figures 8F,G). These data demonstrate that sub-threshold stimulatory concentrations of LPS result in the down-regulation of pro-inflammatory responses after secondary LPS stimulation via reducing PaSRB2a expression in ayu.
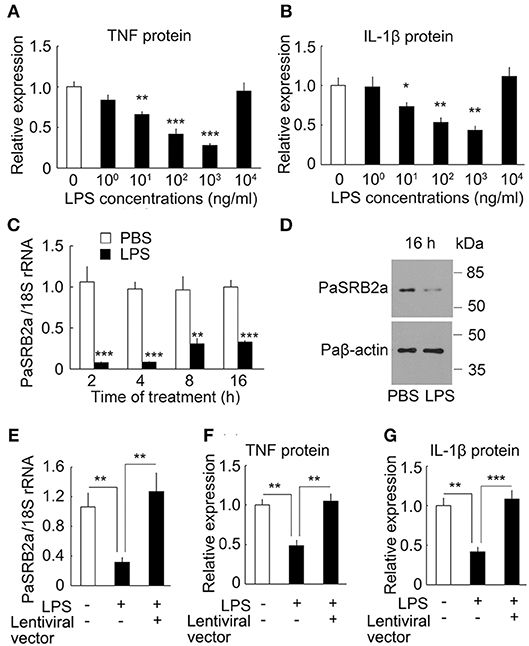
Figure 8. PaSRB2a mediated the effect LPS pre-stimulation on the expression of pro-inflammatory cytokines in ayu macrophages. Effect of LPS pre-stimulation on the expression of TNF (A) and IL-1β (B). Protein levels of pro-inflammatory cytokines were determined by ELISA. Different LPS concentrations were incubated for 6 h followed by 10 μg/ml LPS treatment for an additional 8 h. (C) LPS treatment induced the down-regulation of PaSRB2a mRNA. Macrophage cultures were treated with 1 μg/ml LPS. (D) PaSRB2a protein levels at 16 h after LPS treatment. Representative blots of three independent experiments are shown. (E) Over-expression of PaSRB2a by lentiviral vector in LPS-treated macrophages. Protein levels of TNF (F) and IL-1β (G) in PaSRB2a over-expressed macrophages after LPS treatment. Each bar represents the mean ± SE n = 5. Data are representative of two (A–C, E–G) and three (D) independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
For the depletion of macrophage, we employed CD115 antibodies to identify ayu monocytes/macrophages in blood by flow cytometry and RT-qPCR. In fish injected with clodronate-liposomes alone, the number of CD115+ monocytes/macrophages was significantly decreased in blood compared with that in PBS-liposomes treated ayu (Supplemental Figure 6A). Furthermore, the mRNA expression of CD115+ was also down-regulation in blood of cllodronateliposomes treated ayu (Supplemental Figure 6B). Then we employed macrophage-depleted ayu to investigate the effect of PaSRB2a on pro-inflammatory responses in LPS-treated ayu. PaSRB2a shRNA treatment resulted in up-regulation of survival rate in PBS-liposomes treated ayu (Figure 9A), while the survival rate was not altered in the PaSRB2a shRNA-treated group compared with the pSUPER control in macrophage-depleted fish (Figure 9A). Furthermore, PaSRB2a shRNA treatment did not change TNF and IL-1β expression in the liver of macrophage-depleted fish (Figures 9B,C). In the spleen, PaSRB2a shRNA treatment did not change TNF and IL-1β expression in macrophage-depleted fish (Figures 9D,E). These data suggest that macrophages play an important role in PaSRB2a-mediated pro-inflammatory responses to LPS in vivo.
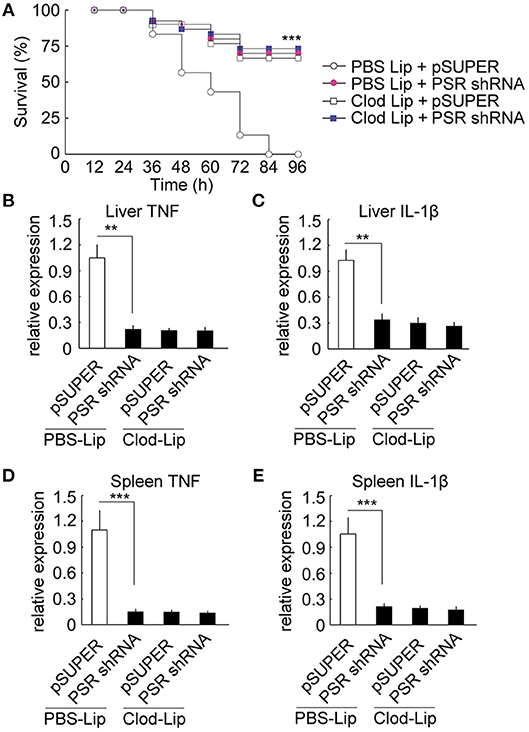
Figure 9. Macrophages mediated PaSRB2a inducement of the pro-inflammatory effect of LPS in vivo. (A) Survival in ayu treated with pSUPER-PaSRB2a after macrophage depletion. Following macrophage depletion, ayu were treated with LPS after lentivirus delivery. n = 30 per group. RT-qPCR analysis of TNF (B) and IL-1β (C) expression in ayu liver. RT-qPCR analysis of TNF (D) and IL-1β (E) expression in ayu spleen. Cytokine genes were normalized to 18S rRNA. Each bar represents the mean ± SE n = 5. Data are representative of two (A) and three (B–E) independent experiments. **P < 0.01, ***P < 0.001. PBS Lip: PBS liposome, Clod Lip: clodronate liposome, PSR shRNA: PaSRB2a shRNA.
We further compare the internalization of LPS from Escherichia coli O55:B5, E. coli O111:B4, Vibrio anguillarum, and Aeromonas hydrophila (O55:B5-LPS, O111:B4-LPS, Va-LPS, and Ah-LPS). All four types of LPS were observed to enter ayu macrophages 4 h after treatment (Figure 10A). PaSRB2a siRNA treatment reduced internalization of all 4 types of LPS (Figure 10A). LPS precipitation assay illustrated that PaSRB2a could interact with all types of LPS (Figure 10B). O55:B5-LPS, O111:B4-LPS, Va-LPS, and Ah-LPS antagonized the internalization of LPS from other bacteria, respectively (Figure 10C), suggesting that LPS from different bacteria may employ similar pathways in ayu macrophages. Furthermore, PaSRB2a siRNA treatment reduced TNF and IL-1β expression in ayu macrophages treated with O55:B5-LPS, O111:B4-LPS, Va-LPS, or Ah-LPS, respectively (Figure 10D). These data suggest that PaSRB2a plays an important role in the pro-inflammatory responses of various LPS derived from different bacteria.
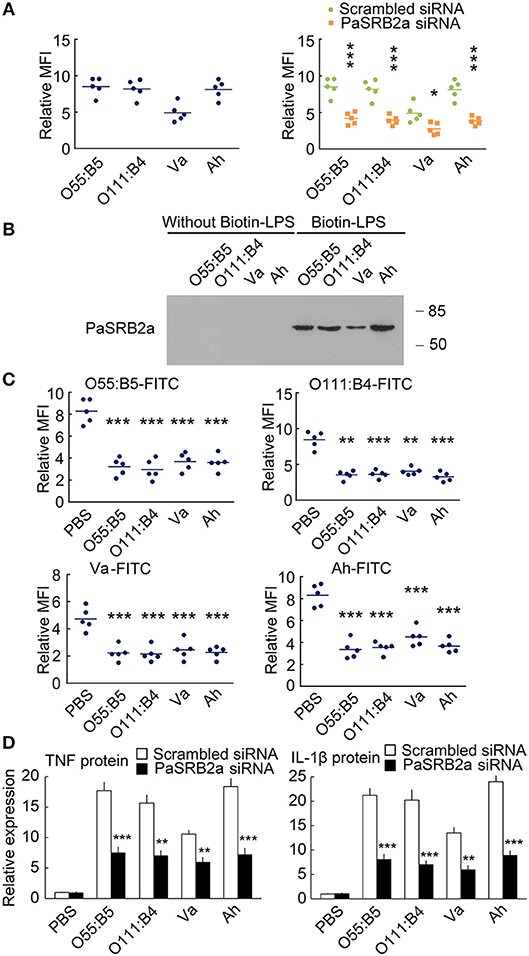
Figure 10. PaSRB2a mediated the inflammatory effect of LPS derived from various bacteria. (A) FITC fluorescence was detected in ayu macrophages after treatment with LPS from E. coli O55:B5, E. coli O111:B4, V. anguillarum, or A. hydrophila. (B) Physical interaction between PaSRB2a and LPS from various bacteria. Representative blots of three independent experiments are shown. (C) LPS-FITC was internalized by ayu macrophages after treatment with LPS from various bacteria. (D) Protein levels of TNF and IL-1β in ayu macrophages after treatment with LPS from various bacteria. n = 5. Data are representative of two (A, C,D) and three (B) independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Since SRB2a also exist in other teleosts, we determined if SRB2a genes in other teleosts mediate LPS internalization and pro-inflammatory responses in macrophages. In grass carp, SRB2a siRNA treatment led to the down-regulation of SRB2a mRNA expression at 48, 72, and 96 h (Figure 11A). SRB2a siRNA treatment reduced LPS internalization in grass carp macrophages (Figure 11B). SRB2a siRNA treatment also decreased TNF and IL-1β mRNA expression in grass carp macrophages (Figure 11C). In spotted green pufferfish, SRB2a siRNA treatment also decreased LPS internalization and pro-inflammatory responses in macrophages (Figures 11D–F). Hence, there may be a common pathway of SRB2a-mediated LPS internalization and pro-inflammatory responses in teleosts.
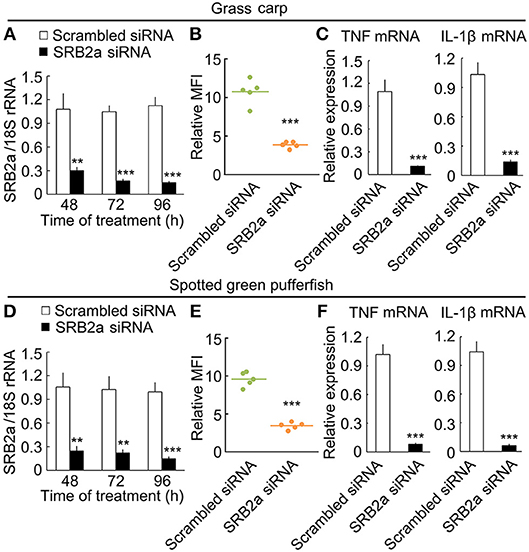
Figure 11. SRB2a mediated LPS internalization and pro-inflammatory effect in the macrophages of grass carp and spotted green pufferfish. (A) mRNA levels of SRB2a in macrophages treated with SRB2a siRNA in grass carp. (B) LPS detection in macrophages 4 h after 10 μg/ml LPS treatment in grass carp. (C) RT-qPCR analysis of TNF and IL-1β expression in grass carp macrophages. (D) mRNA levels of SRB2a in macrophages treated with SRB2a siRNA in spotted green pufferfish. (E) LPS detection in macrophages 4 h after 10 μg/ml LPS treatment in spotted green pufferfish. (F) RT-qPCR analysis of TNF and IL-1β expression in spotted green pufferfish macrophages. n = 5. Data are representative of three independent experiments. **P < 0.01, ***P < 0.001.
Although a relatively high concentration of LPS is necessary to activate the immune response of macrophages in teleosts compared to mammals (7), LPS is still an important pathogen-associated molecule involved in the first line of host defense in teleosts (45, 46). Here, we found that LPS internalization was necessary for the pro-inflammatory response in teleost macrophages. SRB2a, a novel gene in teleosts that does not exist in mammals, mediated LPS internalization and pro-inflammatory responses in macrophages. We further identified that SRB2a mediated intracellular activation of NOD1, NOD2, and NF-κB by interacting and internalizing LPS. The C-terminal intracellular domain of SRB2a played an important role in LPS internalization. Moreover, LPS treatment led to the dramatic down-regulation of SRB2a expression in macrophages, which partly explains the LPS tolerance observed in teleosts. Although LPS is an influential pathogen-associated molecule in the teleost immune system (47), this is the first study to characterize the mechanism of LPS internalization to activate pro-inflammatory responses in teleost macrophages.
TLR2 is also a candidate for the LPS signaling receptor in mammals (17, 18). Although several TLR2 genes have been cloned and characterized in teleosts (8, 48), prior to our study it was unclear whether teleost TLR2 is the receptor for LPS. It has been found that two TLR2 genes existed in several fish such as, common carp, large yellow croaker, and rainbow trout (49). Furthermore, the expression of TLR2a is relatively higher in macrophages of common carp compared with TLR2b, suggesting that TLR2a and TLR2b in teleost may play different roles in immune system (49). In ayu, we also found two TLR2 genes, PaTLR2a, and PaTLR2b. PaTLR2a is expressed in ayu macrophages, while PaTLR2b mRNA could not be detected. We also did not find PaTLR2b expression in the transcriptome of ayu macrophages (33). Since PaTLR2b is not expressed in macrophages we further investigated whether PaTLR2a is the LPS receptor in the macrophages of teleosts. We found that PaTLR2a knockdown did not change the effect of LPS on teleost macrophages, suggesting that PaTLR2a is not the teleost LPS receptor. Although further investigation is necessary to demonstrate whether a TLR is the LPS receptor in teleosts, our results support the concept that LPS recognition of teleost macrophages is completely different from that in mammals.
Since the surface receptor for the LPS signaling pathway in teleosts is unclear, it is possible that LPS can enter the cytoplasm to directly activate pro-inflammatory responses in teleosts. In mammals, when the LPS receptor TLR4 in cell surface is knocked out, even 100 μg/ml LPS cannot activate macrophages in mammals (17), suggesting that extracellular LPS itself does not enter macrophages to activate pro-inflammatory responses. In fact, intracellular LPS in mammals is recognized by the outer membrane of gram-negative bacteria-mediated delivery in macrophages (29). In teleost macrophages, 10 μg/ml LPS activated the production of pro-inflammatory cytokines, suggesting that LPS can enter the cytoplasm of teleost macrophages via by a surface receptor.
Mammalian macrophages express SRB, which does not participate in LPS-induced pro-inflammatory responses in mammalian macrophages (17, 50). In teleosts, a scavenger receptor in teleosts has been found to negatively regulate NF-κB activation (51). Here, we found that SRB but not SRA participates in LPS internalization. There are two SRB2 genes in teleosts: SRB2a, which is a novel gene not found in mammals, while SRB2b is the homolog of mammalian SRB2. According to previous studies, redundant genes may regulate the crucial functions of the teleost immune system (36, 52–54). Here, we found that LPS treatment of macrophage culture led to LPS internalization and a pro-inflammatory response mediated by SRB2a. SRB2b knockdown did not affect LPS internalization, and SRB2a knockdown reduced LPS internalization by 55% in teleost macrophages, suggesting that SRB2a is the main receptor in teleost macrophages that mediate LPS internalization. Hence, we have determined that the novel surface receptor SRB2a mediates the pro-inflammatory response in LPS treated teleost macrophages, which differs from CD14- and TLR-mediated LPS responses in mammals. Although mammalian SRB2 in macrophages does not participate in LPS-induced pro-inflammatory responses, SRB2 does indeed mediates bacterial adhesion and uptake in mammalian macrophages (55). Moreover, SRB2 also participates in cytokine production following bacterial infection in mammals (56). These results suggest that mammalian SRB2 also recognize bacterial components to mediate pro-inflammatory response in macrophages, but the LPS may not play a crucial role in the SRB2 effect on mammalian macrophages. Since teleosts lack the LPS recognition signaling proteins, CD14 and MD2, it may be a complement signaling pathway through which teleost SRB2a interacts with LPS. These results also illustrate that the signaling pathways of LPS recognition vary in vertebrates. Further investigation will focus on how teleosts develop different LPS signaling pathway in response to their water environment.
We further investigated the downstream signaling of SRB2a in teleost macrophages. We first found that the C-terminal intracellular domain but not the extracellular domain of SRB2a mediated LPS internalization. Teleost SRB2a transfection alone could not activate NF-κB in HEK293T cells, while the mammalian TLR4 surface receptor complex can activate NF-κB in HEK293T cells (57, 58). These results suggest that PaSRB2a is important for LPS signaling in teleost macrophages, but it may not be the receptor that directly activates the pro-inflammatory responses to LPS. Moreover, we found two important NOD-like receptors expressed in teleost macrophages, PaNOD1 and PaNOD2 (33). PaNOD1 and PaNOD2 knockdown led to stronger down-regulation of the pro-inflammatory responses in teleost macrophages. These data suggest that NOD1 and NOD2 are the main intracellular receptors of LPS in teleost macrophages following LPS internalization mediated by SRB2a.
Since SRB2a downstream signaling in teleosts differs from TLR4 in mammals, we further investigated whether the unique LPS recognition mechanism of SRB2a affects LPS priming and tolerance. Although LPS tolerance is observed after LPS pre-stimulation for 24 h (59), LPS pre-stimulation with sub-threshold concentrations for 6 h increase the pro-inflammatory responses in mammalian macrophages (44). Here, we found that LPS pre-stimulation with sub-threshold concentrations for 6 h reduced the pro-inflammatory responses in teleost macrophages. Moreover, PaSRB2a was down-regulated in teleost macrophages treated with a sub-threshold LPS concentration. Over expression of PaSRB2a restored the secondary LPS treatment-induced pro-inflammatory responses. Hence, the down-regulation of SRB2a expression after LPS induction suggests a negative feedback regulation of LPS in teleost macrophages. In mammalian macrophages, the expression of LPS receptor TLR4 is not down-regulated after LPS treatment (60). Firstly, the TLR4 surface receptor complex in mammals can activate NF-κB, but SRB2a in teleosts cannot. Secondly, SRB2a in teleosts is down-regulated after LPS treatment, but TLR4 in mammals is not. These results may partly explain the LPS-tolerant mechanism in teleosts. Our results also support the concept that differences in immune responses between teleosts and mammals contribute to the understanding of the evolution of the immune system in vertebrates (61). The interaction of LPS with its receptor is critical for immune responses after bacterial infections, and its dysregulation is thought to promote aberrant cytokine production during bacterial sepsis (62), when LPS release during infection is enhanced when bacterial biomass increases (63). Hence, LPS detection is a crucial process when bacterial numbers increases in vivo during infection. Pre-treatment with low doses of LPS leads to increased pro-inflammatory responses in mammalian macrophages in the early stages of infection, while the decreased pro-inflammatory responses seen in teleost macrophages may contribute to their LPS tolerance. In mammals, knockout of the LPS receptor TLR4 confers resistance to bacteria induced sepsis (64), suggesting that dramatic LPS-induced pro-inflammatory responses are detrimental in sepsis. LPS tolerance in teleosts may therefore be beneficial to severe infection. Further investigation is necessary to illustrate the differential mechanisms underlying the recognition of pathogen-associated molecular patterns between teleosts and mammals.
LPS, a major component of the outer membrane of gram-negative bacteria, is important to the host defense when vertebrates are infected with bacteria. Although teleosts are less sensitive to LPS compared with mammals, LPS is lethal at high doses (65). In both teleosts and mammals, LPS is thought to be an important bacterial component, inducing cytokine expression and death (66). Here, we found that the survival rate was up-regulated and the expression of pro-inflammatory cytokines was down-regulated in LPS-treated ayu that had been administered with lentivirus containing specific small hairpin RNAs, targeting teleost SRB2a. Furthermore, SRB2a shRNA treatment did not change survival rates or the expression of pro-inflammatory cytokines in macrophage-depleted ayu. This further suggests that teleost SRB2a in macrophages regulates pro-inflammatory responses in vivo.
In summary, our study characterized a novel pathway for LPS signaling in the macrophages of vertebrates (Figure 12). Although we do not exclude the existence of other LPS signals in teleost immune cells, to our knowledge, this is the first study to reveal an LPS receptor on the cell surface that mediates pro-inflammatory responses in teleosts. This study illustrates that the LPS signaling in teleosts are different from that in mammals, contributing to our understanding of the evolution of pathogen recognition of vertebrates.
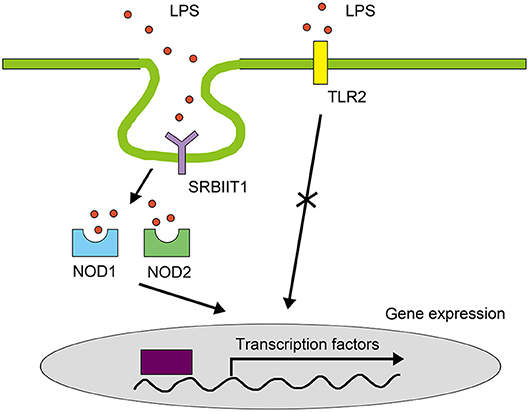
Figure 12. LPS signaling pathway in teleost macrophages. In teleosts, SRB2a mediates LPS internalization for interaction with NOD1 and NOD2 to initiate NF-κB in macrophages. TLRs do not mediate LPS signals in the pro-inflammatory response of teleost macrophages.
The sequences presented in this article have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers MG674831 (PaTLR2), MH699855 (PaSRB2a), JP736791 (PaSRB2b), MG674829 (PaNOD1), and MG674830 (PaNOD2).
X-JL, Y-JN, HL, and LN performed the experiments and data analysis. X-JL and JC designed the in vivo and in vitro experiments. X-JL and JC wrote the manuscript and JC directed the study.
This study was supported by the Program for the National Natural Science Foundation of China (31772876, 41776151), Zhejiang Provincial Natural Science Foundation of China (LZ18C190001; LR18C040001), the Scientific Innovation Team Project of Ningbo (2015C110018), Young Talent Cultivation Project of Zhejiang Association for Science and Technology (Grant No. 2016YCGC003), and the KC Wong Magna Fund of Ningbo University, Ningbo, China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2018.02758/full#supplementary-material
1. Granucci F. The family of LPS signal transducers increases: the arrival of chanzymes. Immunity (2018) 48:4–6. doi: 10.1016/j.immuni.2017.12.016
2. Delano MJ. and Ward PA. Sepsis-induced immune dysfunction: can immune therapies reduce mortality? J Clin Invest. (2016) 126:23–31. doi: 10.1172/jci82224
3. Zhang N, Zhang XJ, Song YL, Lu XB, Chen DD, Xia XQ, et al. Preferential combination between the light and heavy chain isotypes of fish immunoglobulins. Dev Comp Immunol. (2016) 61:169–79. doi: 10.1016/j.dci.2016.04.001
4. Morris MC, Gilliam EA, Li L. Innate immune programing by endotoxin and its pathological consequences. Front Immunol. (2015) 5:680. doi: 10.3389/fimmu.2014.00680
5. Wiegertjes GF, Wentzel AS, Spaink HP, Elks PM, Fink IR. Polarization of immune responses in fish: the 'macrophages first' point of view. Mol Immunol. (2016) 69:146–56. doi: 10.1016/j.molimm.2015.09.026
6. Zou J, Secombes C. The function of fish cytokines. Biology (2016) 5:23. doi: 10.3390/biology5020023
7. Sepulcre MP, Alcaraz-Pérez F, López-Muñoz A, Roca FJ, Meseguer J, Cayuela ML, et al. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-κB activation. J Immunol. (2009) 182:1836–45. doi: 10.4049/jimmunol.0801755
8. Ribeiro CMS, Hermsen T, Taverne-Thiele AJ, Savelkoul HFJ, Wiegertjes GF. Evolution of recognition of ligands from gram-positive bacteria: similarities and differences in the TLR2-mediated response between mammalian vertebrates and teleost fish. J Immunol. (2010) 184:2355–68. doi: 10.4049/jimmunol.0900990
9. Álvarez CA, Ramírez-Cepeda F, Santana P, Torres E, Cortés J, Guzmán F, et al. Insights into the diversity of NOD-like receptors: identification and expression analysis of NLRC3, NLRC5 and NLRX1 in rainbow trout. Mol Immunol. (2017) 87:102–13. doi: 10.1016/j.molimm.2017.03.010
10. Triantafilou M, Triantafilou K. Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol. (2002) 23:301–4. doi: 10.1016/S1471-4906(02)02233-0
11. Rosadini CV, Kagan JC. Early innate immune responses to bacterial LPS. Curr Opin Immunol. (2017) 44:14–9. doi: 10.1016/j.coi.2016.10.005
12. Cao X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat Rev Immunol. (2016) 16:35–50. doi: 10.1038/nri.2015.8
13. Li S, Gong P, Tai L, Li X, Wang X, Zhao C, et al. Extracellular vesicles secreted by neospora caninum are recognized by toll-like receptor 2 and modulate host cell innate immunity through the MAPK signaling pathway. Front Immunol. (2018) 9:1633. doi: 10.3389/fimmu.2018.01633
14. Nie L, Cai SY, Shao JZ, Chen J. Toll-Like receptors, associated biological roles, and signaling networks in non-mammals. Front Immunol. (2018) 9:1523. doi: 10.3389/fimmu.2018.01523
15. Kuzmich N, Sivak K, Chubarev V, Porozov Y, Savateeva-Lyubimova T, Peri F. TLR4 signaling pathway modulators as potential therapeutics in inflammation and sepsis. Vaccines (2017) 5:34. doi: 10.3390/vaccines5040034
16. Neyen C, Lemaitre B. Sensing gram-negative bacteria: a phylogenetic perspective. Curr Opin Immunol. (2016) 38:8–17. doi: 10.1016/j.coi.2015.10.007
17. Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity (1999) 11:443–51. doi: 10.1016/S1074-7613(00)80119-3
18. Yang RB, Mark MR, Gurney AL, Godowski PJ. Signaling events induced by lipopolysaccharide-activated toll-like receptor 2. J Immunol. (1999) 163:639–43.
19. Iliev DB, Roach JC, Mackenzie S, Planas JV, Goetz FW. Endotoxin recognition: in fish or not in fish? FEBS Lett. (2005) 579:6519–28. doi: 10.1016/j.febslet.2005.10.061
20. Pietretti D, Spaink HP, Falco A, Forlenza M, Wiegertjes GF. Accessory molecules for Toll-like receptors in Teleost fish. Identification of TLR4 interactor with leucine-rich repeats (TRIL). Mol Immunol. (2013) 56:745–56. doi: 10.1016/j.molimm.2013.07.012
21. Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science (2013) 341:1246–9. doi: 10.1126/science.1240248
22. Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature (2014) 514:187–92. doi: 10.1038/nature13683
23. Santos JC, Dick MS, Lagrange B, Degrandi D, Pfeffer K, Yamamoto M, et al. LPS targets host guanylate-binding proteins to the bacterial outer membrane for non-canonical inflammasome activation. EMBO J. (2018) 37:e98089. doi: 10.15252/embj.201798089
24. Lechtenberg BC, Mace PD, Riedl SJ. Structural mechanisms in NLR inflammasome signaling. Curr Opin Struct Biol. (2014) 29:17–25. doi: 10.1016/j.sbi.2014.08.011
25. Candel S, Tyrkalska SD, Garcia-Moreno D, Meseguer J, Mulero V. Identification of evolutionarily conserved Md1 splice variants that regulate innate immunity through differential induction of NF-small ka, CyrillicB. J Immunol. (2016) 197:1379–88. doi: 10.4049/jimmunol.1502052
26. Inohara N, Ogura Y, Chen FF, Muto A, Nuñez G. Human nod1 confers responsiveness to bacterial lipopolysaccharides. J Biol Chem. (2001) 276:2551–4. doi: 10.1074/jbc.M009728200
27. Li S, Chen X, Hao G, Geng X, Zhan W, Sun J. Identification and characterization of a novel NOD-like receptor family CARD domain containing 3 gene in response to extracellular ATP stimulation and its role in regulating LPS-induced innate immune response in Japanese flounder (Paralichthys olivaceus) head kidney macrophages. Fish Shellfish Immunol. (2016) 50:79–90. doi: 10.1016/j.fsi.2016.01.029
28. Bi D, Wang Y, Gao Y, Li X, Chu Q, Cui J, et al. Recognition of lipopolysaccharide and activation of NF-κB by cytosolic sensor NOD1 in teleost fish. Front Immunol. (2018) 9:1413. doi: 10.3389/fimmu.2018.01413
29. Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD, et al. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell (2016) 165:1106–19. doi: 10.1016/j.cell.2016.04.015
30. Grayfer L, Kerimoglu B, Yaparla A, Hodgkinson JW, Xie J, Belosevic M. Mechanisms of fish macrophage antimicrobial immunity. Front Immunol. (2018) 9:1105. doi: 10.3389/fimmu.2018.01105
31. Lu XJ, Chen Q, Rong YJ, Chen J. Mobilisation and dysfunction of haematopoietic stem/progenitor cells after Listonella anguillarum infection in ayu, Plecoglossus altivelis. Sci Rep. (2016) 6:28082. doi: 10.1038/srep28082
32. Yeo SJ, Yoon JG, Yi AK. Myeloid differentiation factor 88-dependent post-transcriptional regulation of cyclooxygenase-2 expression by CpG DNA: tumor necrosis factor-alpha receptor-associated factor 6, a diverging point in the Toll-like receptor 9-signaling. J Biol Chem. (2003) 278:40590–600. doi: 10.1074/jbc.M306280200
33. Lu XJ, Hang XY, Yin L, He YQ, Chen J, Shi YH, et al. Sequencing of the first ayu (Plecoglossus altivelis) macrophage transcriptome and microarray development for investigation the effect of LECT2 on macrophages. Fish Shellfish Immunol. (2013) 34:497–504. doi: 10.1016/j.fsi.2012.11.047
34. Lu XJ, Chen J, He YQ, Shi YH. Molecular characterization of an IL-1beta gene from ayu, Plecoglossus altivelis. Fish Shellfish Immunol. (2013) 34:1253–9. doi: 10.1016/j.fsi.2013.02.009
35. Boucrot E, Ferreira AP, Almeida-Souza L, Debard S, Vallis Y, Howard G, et al. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature (2015) 517:460–5. doi: 10.1038/nature14067
36. Zhu LY, Lin AF, Shao T, Nie L, Dong WR, Xiang LX, et al. B cells in teleost fish act as pivotal initiating APCs in priming adaptive immunity: an evolutionary perspective on the origin of the B-1 cell subset and B7 molecules. J Immunol. (2014) 192:2699–714. doi: 10.4049/jimmunol.1301312
37. Chen J, Chen Q, Lu XJ, Li CH. LECT2 improves the outcomes in ayu with Vibrio anguillarum infection via monocytes/macrophages. Fish Shellfish Immunol. (2014) 41:586–92. doi: 10.1016/j.fsi.2014.10.012
38. Van Rooijen N, Sanders A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J Immunol Methods (1994) 174:83–93. doi: 10.1016/0022-1759(94)90012-4
39. Katzenback BA, Belosevic M. Colony-stimulating factor-1 receptor protein expression is a specific marker for goldfish (Carassius auratus L.) macrophage progenitors and their differentiated cell types. Fish Shellfish Immunol. (2012) 32:434–45. doi: 10.1016/j.fsi.2011.12.003
40. Johansson P, Wang T, Collet B, Corripio-Miyar Y, Monte MM, Secombes CJ, et al. Identification and expression modulation of a C-type lectin domain family 4 homologue that is highly expressed in monocytes/macrophages in rainbow trout (Oncorhynchus mykiss). Dev Comp Immunol. (2016) 54:55–65. doi: 10.1016/j.dci.2015.08.005
41. Hampton RY, Golenbock DT, Penman M, Krieger M, Raetz CR. Recognition and plasma clearance of endotoxin by scavenger receptors. Nature (1991) 352:342–4. doi: 10.1038/352342a0
42. Seternes T, Dalmo RA, Hoffman J, Bogwald J, Zykova S, Smedsrod B. Scavenger-receptor-mediated endocytosis of lipopolysaccharide in Atlantic cod (Gadus morhua L.). J Exp Biol. (2001) 204:4055–64. Available online at: http://jeb.biologists.org/content/204/23/4055.long
43. Meunier E, Broz P. Evolutionary convergence and divergence in NLR function and structure. Trends Immunol. (2017) 38:744–57. doi: 10.1016/j.it.2017.04.005
44. Zhang X, Morrison DC. Lipopolysaccharide-induced selective priming effects on tumor necrosis factor alpha and nitric oxide production in mouse peritoneal macrophages. J Exp Med. (1993) 177:511–6. doi: 10.1084/jem.177.2.511
45. Abós B, Wang T, Castro R, Granja AG, Leal E, Havixbeck J, et al. Distinct differentiation programs triggered by IL-6 and LPS in teleost IgM+ B cells in the absence of germinal centers. Sci Rep. (2016) 6:30004. doi: 10.1038/srep30004
46. Mottaz H, Schönenberger R, Fischer S, Eggen RIL, Schirmer K, Groh KJ. Dose-dependent effects of morphine on lipopolysaccharide (LPS)-induced inflammation, and involvement of multixenobiotic resistance (MXR) transporters in LPS efflux in teleost fish. Environ Pollut. (2017) 221:105–15. doi: 10.1016/j.envpol.2016.11.046
47. Hsu AY, Gurol T, Sobreira TJP, Zhang S, Moore N, Cai C, et al. Development and characterization of an endotoxemia model in zebra fish. Front Immunol. (2018) 9:607. doi: 10.3389/fimmu.2018.00607
48. Palti, Y. Toll-like receptors in bony fish: from genomics to function. Dev Comp Immunol. (2011) 35:1263–72. doi: 10.1016/j.dci.2011.03.006
49. Fink IR, Pietretti D, Voogdt CGP, Westphal AH, Savelkoul HFJ, Forlenza M, et al. Molecular and functional characterization of Toll-like receptor (Tlr)1 and Tlr2 in common carp (Cyprinus carpio). Fish Shellfish Immunol. (2016) 56:70–83. doi: 10.1016/j.fsi.2016.06.049
50. Baranova IN, Vishnyakova TG, Bocharov AV, Leelahavanichkul A, Kurlander R, Chen, et al . Class B scavenger receptor types I and II and CD36 mediate bacterial recognition and proinflammatory signaling induced by Escherichia coli, lipopolysaccharide, and cytosolic chaperonin 60. J Immunol. (2012) 188:1371–80. doi: 10.4049/jimmunol.1100350
51. Meng Z, Zhang XY, Guo J, Xiang LX, Shao JZ. Scavenger receptor in fish is a lipopolysaccharide recognition molecule involved in negative regulation of NF-κB activation by competing with TNF receptor-associated factor 2 recruitment into the TNF-α signaling pathway. J Immunol. (2012) 189:4024–39. doi: 10.4049/jimmunol.1201244
52. Hong S, Li R, Xu Q, Secombes CJ, Wang T. Two types of TNF-α exist in teleost fish: phylogeny, expression, and bioactivity analysis of Type-II TNF-α3 in rainbow trout Oncorhynchus mykiss. J Immunol. (2013) 191:5959–72. doi: 10.4049/jimmunol.1301584
53. Zhang XJ, Zhang XY, Zhang N, Guo X, Peng KS, Wu H, et al. Distinctive structural hallmarks and biological activities of the multiple cathelicidin antimicrobial peptides in a primitive teleost fish. J Immunol. (2015) 194:4974–87. doi: 10.4049/jimmunol.1500182
54. Piazzon MC, Wentzel AS, Wiegertjes GF, Forlenza M. Carp Il10a and Il10b exert identical biological activities in vitro, but are differentially regulated in vivo. Dev Comp Immunol. (2017) 67:350–60. doi: 10.1016/j.dci.2016.08.016
55. Vishnyakova TG, Kurlander R, Bocharov AV, Baranova IN, Chen Z, Abu-Asab MS, et al. CLA-1 and its splicing variant CLA-2 mediate bacterial adhesion and cytosolic bacterial invasion in mammalian cells. Proc Natl Acad Sci USA. (2006) 103:16888–93. doi: 10.1073/pnas.0602126103
56. Leelahavanichkul A, Bocharov AV, Kurlander R, Baranova IN, Vishnyakova TG, Souza AC, et al. Class B scavenger receptor types I and II and CD36 targeting improves sepsis survival and acute outcomes in mice. J Immunol. (2012) 188:2749–58. doi: 10.4049/jimmunol.1003445
57. Youn HS, Kim YS, Park ZY, Kim SY, Choi NY, Joung SM, et al. Sulforaphane suppresses oligomerization of TLR4 in a thiol-Dependent manner. J Immunol. (2009) 184:411–9. doi: 10.4049/jimmunol.0803988
58. Rallabhandi P, Phillips RL, Boukhvalova MS, Pletneva LM, Shirey KA, Gioannini TL, et al. Respiratory syncytial virus fusion protein-induced toll-like receptor 4 (TLR4) signaling is inhibited by the TLR4 antagonists rhodobacter sphaeroides lipopolysaccharide and eritoran (E5564) and requires direct interaction with MD-2. mBio (2012) 3:e00218–12. doi: 10.1128/mBio.00218-12
59. Rajaiah R, Perkins DJ, Polumuri SK, Zhao A, Keegan AD, Vogel SN. Dissociation of endotoxin tolerance and differentiation of alternatively activated macrophages. J Immunol. (2013) 190:4763–72. doi: 10.4049/jimmunol.1202407
60. Matsuguchi T, Musikacharoen T, Ogawa T, Yoshikai Y. Gene expressions of toll-like receptor 2, but not toll-like receptor 4, is induced by LPS and inflammatory cytokines in mouse macrophages. J Immunol. (2000) 165:5767–72. doi: 10.4049/jimmunol.165.10.5767
61. Sunyer JO. Fishing for mammalian paradigms in the teleost immune system. Nat Immunol. (2013) 14:320–6. doi: 10.1038/ni.2549
62. Tan Y, Kagan JC. A cross-disciplinary perspective on the innate immune responses to bacterial lipopolysaccharide. Mol Cell (2014) 54:212–23. doi: 10.1016/j.molcel.2014.03.012
63. Van Den Berg C, de Neeling AJ, Schot CS, Hustinx WN, Wemer J, de Wildt DJ. Delayed antibiotic-induced lysis of Escherichia coli in vitro is correlated with enhancement of LPS release. Scand J Infect Dis. (1992) 24:619–27.
64. Roger T, Froidevaux C, Le Roy D, Reymond MK, Chanson AL, Mauri D, et al. Protection from lethal gram-negative bacterial sepsis by targeting Toll-like receptor 4. Proc Natl Acad Sci USA. (2009) 106:2348–52. doi: 10.1073/pnas.0808146106
65. Novoa B, Bowman TV, Zon L, Figueras A. LPS response and tolerance in the zebrafish (Danio rerio). Fish Shellfish Immunol. (2009) 26:326–31. doi: 10.1016/j.fsi.2008.12.004
Keywords: teleost, macrophages, lipopolysaccharide, scavenger receptor class B2, inflammation
Citation: Lu X-J, Ning Y-J, Liu H, Nie L and Chen J (2018) A Novel Lipopolysaccharide Recognition Mechanism Mediated by Internalization in Teleost Macrophages. Front. Immunol. 9:2758. doi: 10.3389/fimmu.2018.02758
Received: 27 August 2018; Accepted: 09 November 2018;
Published: 27 November 2018.
Edited by:
Geert Wiegertjes, Wageningen University & Research, NetherlandsReviewed by:
Magdalena Chadzinska, Jagiellonian University, PolandCopyright © 2018 Lu, Ning, Liu, Nie and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Jiang Lu, bHhqNzExMDQzQDE2My5jb20=
Jiong Chen, amNoZW4xOTc1QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.