- 1Division of Veterinary Biotechnology, ICAR-Indian Veterinary Research Institute (IVRI), Bareilly, India
- 2DBT-National Institute of Animal Biotechnology, Hyderabad, India
- 3The Ohio State University, Columbus, Ohio, OH, United States
- 4Division of Biological Products, ICAR-Indian Veterinary Research Institute (IVRI), Bareilly, India
- 5Division of Virology, ICAR-Indian Veterinary Research Institute (IVRI), Mukteswar, India
- 6ICAR- Directorate of Foot and Mouth Disease, Mukteswar, India
- 7ARIS Cell, ICAR-Indian Veterinary Research Institute (IVRI), Bareilly, India
In this study, the miRNAome and proteome of virulent Peste des petits ruminants virus (PPRV) infected goat peripheral blood mononuclear cells (PBMCs) were analyzed. The identified differentially expressed miRNAs (DEmiRNAs) were found to govern genes that modulate immune response based on the proteome data. The top 10 significantly enriched immune response processes were found to be governed by 98 genes. The top 10 DEmiRNAs governing these 98 genes were identified based on the number of genes governed by them. Out of these 10 DEmiRNAs, 7 were upregulated, and 3 were downregulated. These include miR-664, miR-2311, miR-2897, miR-484, miR-2440, miR-3533, miR-574, miR-210, miR-21-5p, and miR-30. miR-664 and miR-484 with proviral and antiviral activities, respectively, were upregulated in PPRV infected PBMCs. miR-210 that inhibits apoptosis was downregulated. miR-21-5p that decreases the sensitivity of cells to the antiviral activity of IFNs and miR-30b that inhibits antigen processing and presentation by primary macrophages were downregulated, indicative of a strong host response to PPRV infection. miR-21-5p was found to be inhibited on IPA upstream regulatory analysis of RNA-sequencing data. This miRNA that was also highly downregulated and was found to govern 16 immune response genes in the proteome data was selected for functional validation vis-a-vis TGFBR2 (TGF-beta receptor type-2). TGFBR2 that regulates cell differentiation and is involved in several immune response pathways was found to be governed by most of the identified immune modulating DEmiRNAs. The decreased luciferase activity in Dual Luciferase Reporter Assay indicated specific binding of miR-21-5p and miR-484 to their target thus establishing specific binding of the miRNAs to their targets.This is the first report on the miRNAome and proteome of virulent PPRV infected goat PBMCs.
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs (22 nucleotides) found to regulate the expression of genes post-transcriptionally in animals, plants, and some viruses (1). They regulate different cellular processes, including reproduction, development, pathogenesis, and apoptosis (2–4). miRNAs are also effective in regulating immune response and cellular differentiation (5–7). The regulation process generally takes place by binding of miRNA at its seed sequence (2–8 nucleotides from 5′-end) to the 3′ untranslated region (3′UTR) of specific mRNAs of the genes that govern the biological processes. However, several instances of miRNAs binding to 5' UTR or coding regions in the regulation process have also been reported (8, 9).
Viral pathogenesis is greatly influenced by cellular miRNAs (10–12). Several cellular miRNAs have been demonstrated to play a regulatory role in the host-virus interaction networks (13, 14). Cellular miRNA expression profile is profoundly influenced by viral infections and vice-versa (15). For example, miR-122 is reported to enhance replication of Hepatitis C virus (16) and miR-142 suppresses replication of Eastern Equine Encephalitis virus (17). The HIV-1 virus has been found to increase the expression of various host miRNAs, including miR-370, miR-122, miR-297, and miR-373, and suppress the expression of miR-17-92 cluster (18). With the advent of deep sequencing technology, it has become possible to explore changes in miRNA expression in the host, in response to various viral infections like enterovirus 71, avian influenza, PPRV, Japanese Encephalitis virus and hepatitis C virus (19–23).
Peste des petits ruminants (PPR) characterized by fever, sore mouth, conjunctivitis, gastroenteritis, and pneumonia, is an acute, highly contagious viral disease of sheep and goats. However, a more severe form of the clinical disease has been reported in goats than in sheep, since goats are more susceptible (24–29). The recovery is also slower in goats than in sheep (27). However, regions having large sheep populations have reported severe outbreaks of PPR (27, 30, 31). Our earlier in-vitro transcriptome analysis studies to evaluate host response of goat PBMCs to PPR live attenuated vaccine virus uncovered several transcription factors that modulate immune response (32, 33). Also, dysregulation in the host miRNAome in lung and spleen of experimentally infected goats and sheep by virulent PPRV suggests a strong host immune response in sheep and goats (21). However, the host miRNAome of PPRV infected goat and sheep PBMCs has not been explored to date. Lymphocytes are the primary targets of PPRV infection from where it reaches different tissues by piggybacking on PBMCs (34). A higher viral load is reported to be at 9 days post-infection (dpi), which coincides with the peak clinical signs of the disease (22, 35). In the present study, control (0 day) and PPRV infected PBMCs (9 dpi) of goats were isolated and subjected to microRNA sequencing (miRNA-seq) and proteome profiling. DEmiRNAs were identified from miRNA-seq data and correlated with the proteome data to identify the miRNAs that govern the immune processes. Among the miRNAs, miR-21-5p was found to be highly downregulated in miRNA-seq data, inhibited in RNA sequencing (RNA-seq) data (unpublished) and involved in regulation of various immune response genes. This miR-21-5p was selected for annotation and functional validation. Additionally, one more miR, miR-484 was randomly chosen from top 10 immunoregulating DEmiRNAs for functional validation.
Materials and Methods
Ethics Statement and Animal Experiment
The study is a part of vaccine potency testing experiment conducted at ICAR-Indian Veterinary Research Institute Mukteshwar Campus as per the guidelines of Indian pharmacopeia-2014. The permission to conduct the experiment was sought from Indian Veterinary Research Institute–Institutional Animal Ethics Committee (IVRI–IAEC) under the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), India and was approved vide letter no 387/CPCSEA. The animals that were apparently healthy and negative for the presence of PPRV antibody by competitive ELISA and serum neutralization test (SNT) were used in this study. Virulent PPRV [accession number KR140086.1 (36)], a lineage IV isolate and strain Izatnagar/94, was used as a challenge virus and infection was confirmed in goats by RT–PCR, qRT-PCR, and sandwich ELISA. PBMCs were isolated from the blood collected from PPRV (Izatnagar/94) infected goats at 9 dpi (The animals succumbed to the disease at 10 dpi). The PBMCs isolated from blood collected from apparently healthy animals (0 day) acted as a control. PBMCs were isolated using Histopaque-1077 (Sigma), USA.
MicroRNA Sequencing
Total RNA from the PBMCs of goats was isolated using the RNeasy Mini kit (Qiagen GmbH, Germany) following the manufacturer's protocol. To access integrity and quality of the RNA, RNA integrity number (RIN) value of each sample was measured on Bioanalyzer (Agilient Technologies, Inc.). The RIN value was found to be >8, which is considered suitable for further processing (37). The library was prepared using NEBNext Multiplex Small RNA Library Prep Kit (New England Biolabs Inc.) as per the manufacturer's protocol. Hundred nanogram of total RNA from each sample was used for small RNA library preparation. The quality of the libraries was assessed on Bioanalyzer. Libraries were quantified using Qubit 2.0 Fluorometer (Life Technologies) and by qPCR (38).The high-throughput sequencing was performed on Illumina–NextSeq500 platform to generate 75 bp single-end reads as per manufacturer's protocol. The data was submitted to the GEO database with accession number GSE109799.
Processing miRNA-seq Data
The miRNA reads trimming and preprocessing was performed with CLC Genomic workbench v6.0 (CLC bio, Denmark) to remove adaptor sequences and low quality reads using default parameters. Since the cattle genome (mirBase–Release 21) is relatively better annotated and as the miRNAs are conserved across species, cattle genome was used to map these clean reads. The map files for the infected and control samples were created independently. From the toolbox of CLC workbench, an experiment was created, the read count was quantitatively normalized and the expression values were obtained. Proportion based statistics–Kal's test was used to identify differentially expressed miRNAs at 9dpi PPRV infected PBMCs of goat.
Proteomics Data Generation and Analysis
The proteomic data was generated from control and PPRV infected 9 dpi goats PBMCs and analyzed following the standard procedure as described in the previous study (21). Briefly, proteome from goat PBMCs was quantitatively analyzed using trypsin in conjugation with C18 Nano-LC column separation, followed by analysis on the Waters Synapt G2 Q-TOF instrument for MS. The raw data was processed by MassLynx 4.1 WATERS, and MSMS spectra were matched to the database sequence using PLGS software. The identified proteins in the three runs of each sample were compared with each other as control (healthy) and infected samples. Quantification was done using expression analysis package of the PLGS software. The ion counts matching with the peptides of a specific protein corresponding between the two samples in the three runs were averaged and the ratio was calculated for the whole protein.
Functional Annotation of Differentially Expressed miRNAs
To explore the regulatory role of the differentially expressed miRNAs, the target genes governed by each of the DEmiRNAs were obtained using TargetScan tool (39)1 All these target genes were pooled up and compared to the dysregulated proteins identified in proteomics data. The genes common to both data were selected for functional annotation via ClueGo (ver. 2.3.3) and CluePedia (ver. 1.3.3) (40) in Cytoscape (ver. 3.2.1) (41). The genes involved in top 10 significantly enriched immune response processes were identified. Out of these immune response genes, the genes governed by each of the DEmiRNAs were identified and top 10 DEmiRNAs were selected.
Processing RNA-seq Data
From IPA analysis (Ingenuity Pathway Analysis)2 of the RNA-sequencing data generated in our lab (unpublished), the upstream miRNA regulators governing the differentially expressed genes (data not shown) were identified. The overview of the entire analysis is given in Figure 1.
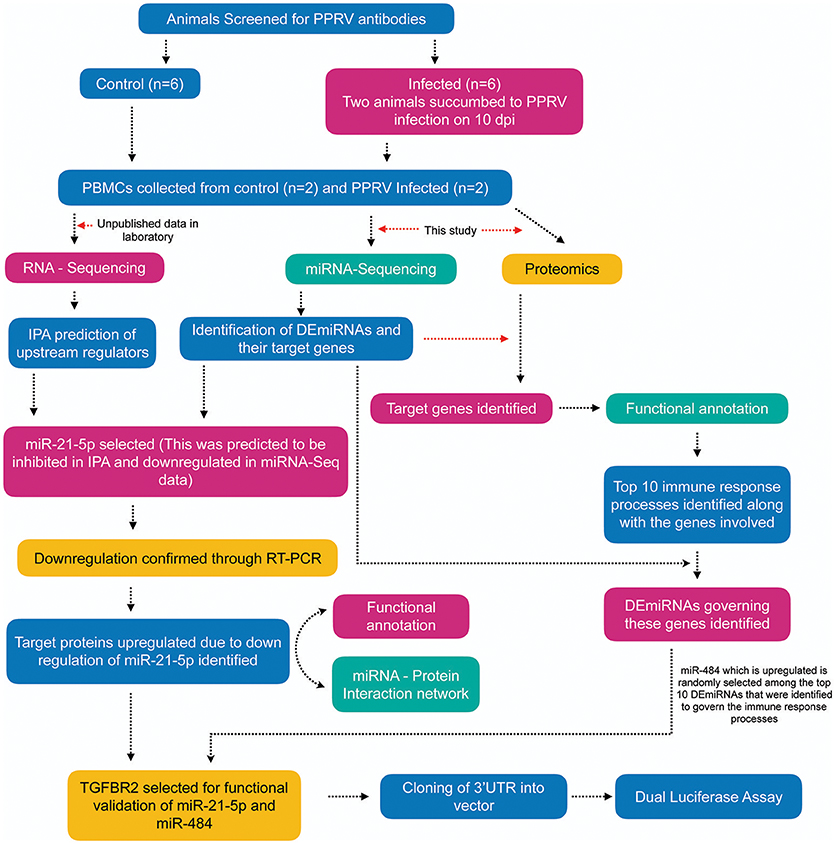
Figure 1. Overview of the experiment. Apparently healthy goats (n = 6) that were negative for the presence of PPRV antibody by competitive ELISA and serum neutralization test (SNT) were infected with virulent PPRV strain Izatnagar/94. These animals acted as control in the vaccine potency experiment. Two animals in the group succumbed to disease on 10 dpi. PBMCs from blood collected at 9dpi from these two succumbed animals and PBMCs isolated from blood collected on 0 day (control) were sent for miRNA-Sequencing, RNA-Sequencing (unpublished data in the lab) and Proteome profiling. Differentially expressed miRNAs (DEmiRNAs) and proteins were identified from the miRNA-Seq and proteome data, repectively. The DEmiRNAs were functionally annotated w.r.t the proteins governed in the proteome data. The top 10 DEmiRNAs governing the top 10 immune response processes were identified. miR-21-5p which was found to be inhibited on IPA upstream regulatory analysis of RNA-Sequencing data, highly downregulated in miRNA-Seq data (validated by qRT-PCR) and found to govern 16 immune response genes in the proteome data was selected for functional validation vis-a-vis TGFBR2 (TGF-beta receptor type-2). TGFBR2 regulates cell differentiation and is involved in several immune response pathways was found to be governed by most of the identified immune modulating DEmiRNAs. Dual luciferease assay was done using mimics of miR-21-5p and miR-484 and 3′UTR of the target gene TGFBR2 to establish the specificity of binding.
Downregulation of miR-21-5p in qRT-PCR
The expression of the miR-21-5p in PPRV infected PBMCs was validated by qRT-PCR. Total RNA, including small RNA from the PBMCs of control and infected goats, was isolated using mirVana™ miRNA isolation kit (Invitrogen). Reverse transcriptase reactions were performed using RT specific primers of miR-21-5p and U6snRNA by TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, USA, #4366596). Total RNA from the PBMCs was diluted to a concentration of 10 ng/μl and 1 μl of RNA was added to the reaction mix containing 0.15 μl 100 mM dNTPs, 1 μl of RT enzyme (50 U/μl), 1.5 μl 10 × RT buffer, 0.19 μl RNase inhibitor (20 U/μl), 3 μl 5 × RT specific-primer and 8.16 μl DEPC-treated water to obtain a final volume of 15 μl. The reaction conditions were 16°C for 30 min followed by 42°C at 30 min and 85°C for 5 min to stop the reaction. The cDNA was then used for the Real-time PCR. Real-time PCR was performed using a standard TaqMan PCR kit protocol on Applied Biosystems 7,500 fast Sequence Detection System. The 10 μl PCR included 5 μl of 2 × Taqman Gene Expression Mastermix (Thermo Fisher Scientific Inc., Wilmington, DE, USA), 0.5 ul of 20 × Taqman probe (Assay ID 005982-mat), 2 μl (0.134 ng) of RT product and 2.5 μl of NFW. The reactions were incubated in a 96-well plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All reactions were run in triplicate. The expression of miRNA-21-5p was assayed taking the expression of U6snRNA as an internal control. The relative expression of miR-21-5p was calculated using the 2−ΔΔCT method with the control group as calibrator (42). Student's t-test was done in JMP9 (SAS Institute Inc., Cary, USA) to test the significance of difference and difference between groups was considered significant at P ≤ 0.05.
Prediction of miR-21-5p Target Genes and Functional Annotation
The target genes governed by miRNA-21-5p were obtained using TargetScan tool to explore its regulatory role (39). The target genes obtained were compared to the upregulated proteins from proteome data to identify the proteins that are upregulated because of downregulation of miR-21-5p. The miRNA-protein-network was created based on the expression profile of target genes and miR-21-5p using Cytoscape (ver. 3.2.1). Functional annotation of these genes was performed by ClueGo (ver. 2.3.3) and CluePedia (ver. 1.3.3) (40) in Cytoscape (ver. 3.2.1) (41).
Functional Validation of miR-21-5p and miR-484
Prediction of miR Target site in 3′ UTR
TGFBR2 that is governed by miR-21-5p and miR-484, and found connected to most significantly enriched GO terms, was selected for further validation of the miRNAs. The 3'UTR target site of this gene and mature miRNA were extracted from NCBI and analyzed in miRanda (43) tool to evaluate the strength of interaction using the parameters ΔG and total score value.
Design of Wild Type and Mutant Type miR Target Sites and miRNA Mimic and Control
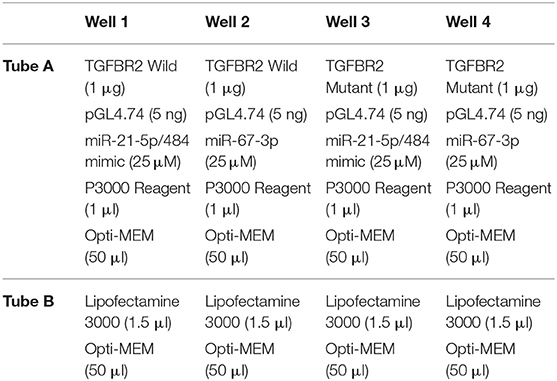
While two wild-type oligonucleotides (62 bp) were constructed from 3′UTR of TGFBR2 mRNA flanking the miR-21-5p and miR-484 target sites, respectively, the mutant of both was created by replacing the target site either with poly A or poly T sequence. pGL4.13 vector (Promega) was used to clone the oligonucleotide sequences (wild-type and mutant-type separately) at XbaI RE site of this vector. pGL4.74 was used as a control vector for the normalization of the transfection efficiency. Likewise, mimics of miR-21-5p and miR-484 were chemically synthesized. miR-67-3p was used as control for it is reported to have least sequence identity with known miRNAs in humans, rat, and mouse (44).
Co-Transfection Strategy for Carrying Out Dual-Luciferase Reporter Assay
HEK293 cells were used for the co-transfection of the vectors, pGL4.13, and pGL4.74. The cells were maintained in MEM medium with 10% FBS, antibiotic and antimycotic (Himedia), and placed in an incubator at 37°C with 5% CO2. The experiment was performed in triplicates. Briefly, the co-transfection complex for each vector was prepared in 2 tubes, one containing Opti-MEM + Lipofectamine and the other tube containing Opti-MEM + Vectors + Mimic. The complex was formulated as shown in Table 1. Contents of tube A and B were mixed and incubated for 15 min at room temperature to allow the formation of transfection complex. After the formation of the complex, Opti-MEM was added to the complex to make the volume up to 400 μl. The medium from the 24 well plate was removed and the co-transfection complexes were gently loaded into each well of the plate. The plate was kept in an incubator for 4 h. After 4 h, the medium with the complex was removed from the wells and fresh 1% MEM (500 μl) was added to the wells. The cells were lysed 48 h post co-transfection and the luciferase activity was measured using Dual Luciferase Assay kit (Promega) according to the manufacturer's protocol. The assay results were represented as relative luciferase activity. Student's t-test was done in JMP9 (SAS Institute Inc, Cary, USA) to test the significance of difference, and differences between groups were considered significant at P ≤ 0.05.
Results
Confirmation of PPRV Infection
Viral infection in the PBMCs of goats infected with PPRV was confirmed by RT-PCR amplification of 351 bp N gene fragment (Figure 2). The viral infection was further confirmed by sandwich ELISA and qRT-PCR (data not shown).
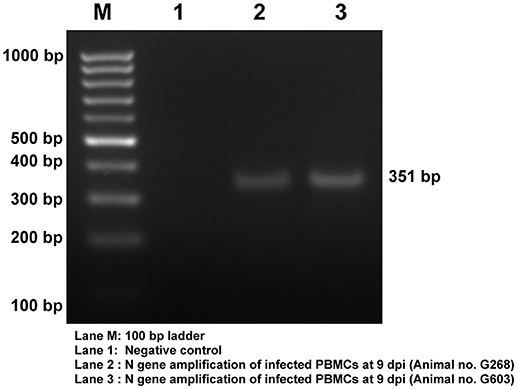
Figure 2. Confirmation of PPRV infection in PBMCs of goats. Amplification of 351 bp N gene by RT- PCR. Lane M, 100 bp ladder; Lane 1, NTC; Lane 2, Infected Goat PBMCs (Animal no: G268) Lane 3, Infected Goat PBMCs (Animal No: G603).
miRNAs Governing the Immune Processes Were Identified
In goat PBMCs infected with PPRV, a total of 68 miRNAs were significantly (P < 0.05) differentially expressed (42 down-regulated and 26 up-regulated) (Table S1). From the proteomics data (Table S2) generated from PPRV infected goat PBMCs, 1,965 and 3,509 proteins were identified to be downregulated and upregulated, respectively. From the TargetScan data, 15,341 genes were found to be governed by the 68 DEmiRNAs, out of which 4,027 proteins were found to be dysregulated in the proteomics data. On ClueGo analysis of these genes, the top 10 significantly enriched immunological processes included immunoglobulin mediated immune response, NK T cell differentiation lymphocyte mediated immunity, adaptive immune response, positive regulation of gamma-delta T cell activation, T cell differentiation, regulation of leukocyte differentiation, positive regulation of NK T cell differentiation, positive regulation of lymphocyte differentiation, positive regulation of innate immune response (Figure 3). The genes (from the proteomics data) enriched under these GO terms are given in Table S3. A total of 98 genes were found to be enriched in these top 10 significant immune response processes governed by 42 DEmiRNAs (18 upregulated and 24 downregulated) (Tables S4, S5). The top 10 DEmiRNAs based on the number of immune response genes governed are given in Table 2. On comparing the DEmiRNAs of PPRV infected PBMCs with DEmiRNAs of the PPRV infected lung and spleen, reported in our earlier study (21), we found there are 3 DEmiRNAs common among the lung, spleen and PBMCs and 9 DEmiRNAs common between the lung and PBMCs. However, there was no DEmiRNA exclusively common between PBMCs and spleen (Table S6). Of the 9 DEmiRNAs common between lung and PBMCs, miR-378b, miR-342, miR-30f, miR-339a were found to be downregulated while miR-1246 and miR-2440 were upregulated in both. In addition, miR-181a-1, miR-181a-2 and miR-7-1 were found upregulated in lung but downregulated in PBMCs. Of the 3 DEmiRNAs common to each, miR-574 was found upregulated. miR-21-5p was found upregulated in spleen and lung but downregulated in PBMCs and vice-versa in case of miR-744.
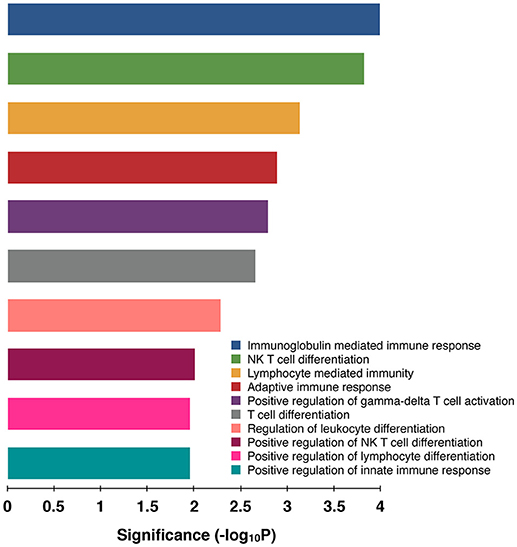
Figure 3. Top 10 Significant (P < 0.05) Immunological processes involved in virulent PPRV goats PBMCs. Color differentiates the immune processes.
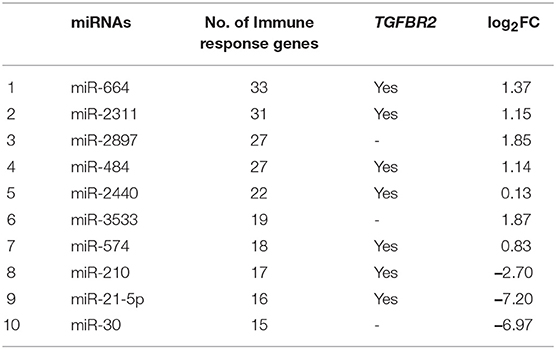
Table 2. Ten DEmiRNAs based on the number of immune response genes, involvement of TGFBR2 and fold-change values.
miR-21-5p Was Selected for Functional Annotation and Validation
On analyzing RNA-seq data (from the lab) of the 9 dpi PBMCs, 5,150 differentially expressed genes (DEGs) were identified in PPRV infected goats. These genes were further subjected to IPA using various modules based on knowledge database to predict the biological function of DEGs, the role of the molecules in various disease processes, upstream regulators (transcription factors, miRNAs, and drugs) that regulate the function of the downstream target genes and possible interactions among them. In the present study, we concentrated only on those miRNAs, which act as upstream regulators for DEGs. Twenty-seven miRNAs were identified regulating these DEGs. Of the 27 identified miRNAs, 26 were inhibited (z score < −2) and only 1 was activated (z score >2) (Figure 4). Further, out of these 27, only four miRNAs viz miR-129, miR-21-5p, Let-7a, and miR-200 were found be differentially expressed in the miRNA-seq data. miR-21-5p was found highly downregulated in miRNA-seq data (Table 2), inhibited in RNA-seq data and involved in regulation of various immune response genes. This miR-21-5p was further selected for annotation and functional validation.
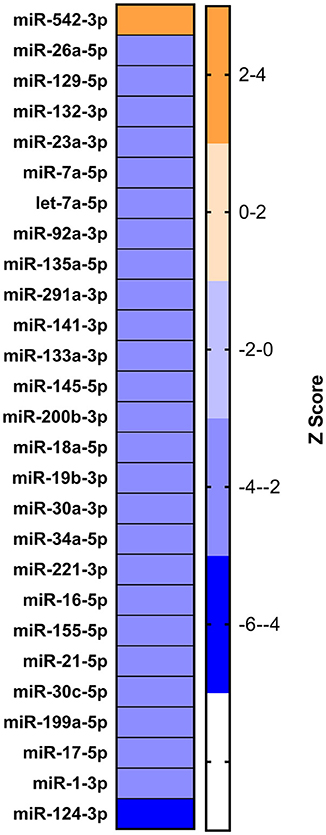
Figure 4. Identification of dysregulated miRNA from RNA sequencing data. IPA analysis to identify the upstream regulators governing the differentially expressed genes yielded 27 miRNAs.
Validation of miR-21-5p by qRT-PCR
To confirm the downregulation of miR-21-5p, qRT-PCR was used to validate its expression in PPRV infected goat PBMCs. This miRNA was found to be downregulated and was in concordance with the miRNA-seq results (Figure 5).
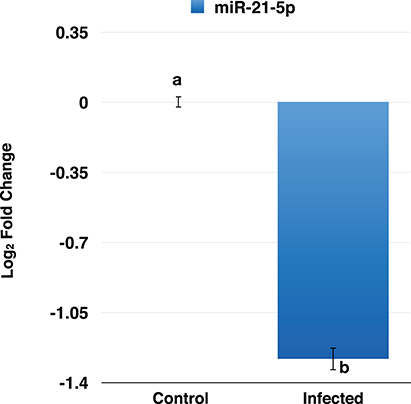
Figure 5. RT-qPCR analysis of miR-21-5p in PPRV infected goats PBMCs. The miR-21-5p was found to be downregulated in PPRV infected goat PBMCs.
Prediction of miRNA-21-5p Targets, Gene Ontology Analysis, and Target Selection for Functional Validation
From the TargetScan data, 356 genes were found to be governed by miR-21-5p, out of which 66 proteins were found to be upregulated (since miR-21-5p was downregulated, the study was concentrated only on upregulated proteins) in the proteomics data. These target proteins on GO analysis were enriched in Wnt signaling pathway, cell surface receptor signaling pathway, pathway-restricted SMAD phosphorylation, morphogenesis processes, positive regulation of cellular processes, multicellular organismal development, etc., (Figure 6). TGFBR2 gene, which is connected to most of the GO terms viz pathway-restricted SMAD phosphorylation, activin receptor signaling pathway, Wnt signaling pathway, morphogenesis of lung and heart and blood vessels, and osteoclast differentiation was selected as the target of miR-21-5p. TGFBR2 was also found to be governed by 21 identified immune regulating DEmiRNAs (Table 2 and Table S7).
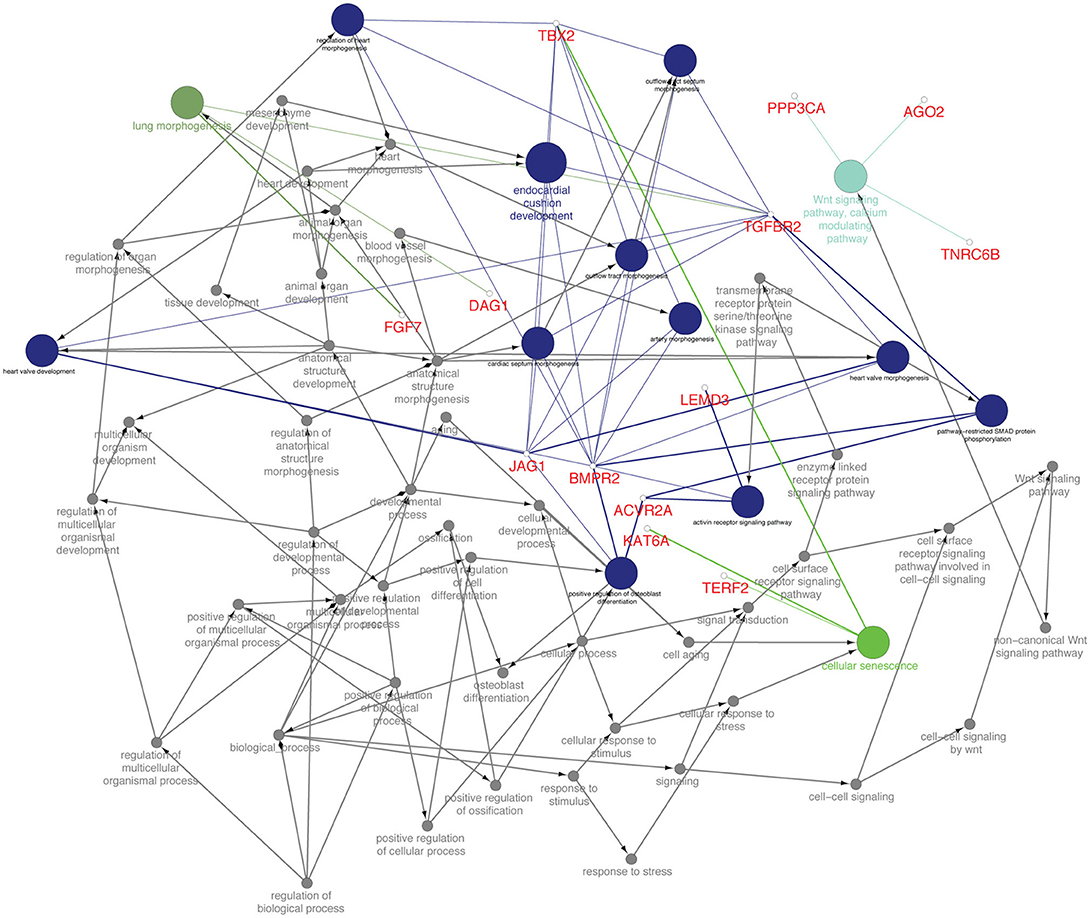
Figure 6. Functionally annotation of miR-21-5p based on the upregulated target genes (45). Gene ontology was visualized in these genes visualized in ClueGo (ver. 2.3.3) + CluePedia (ver. 1.3.3) plugin of Cytoscape (ver. 3.2.1).
miRNA-Protein Regulatory Network of miR-21-5p
The miR-21-5p and 66 upregulated proteins interacting with it are represented in a network (Figure 7). Among the 66, 15 proteins (ACBD5, ADNP, CD97, CDH6, CREBRF, CYSLTR1, DNAJC16, FBXO11, HIPK3, JAG1, KAT6A, PAG1, PJA2, RAD21, TGFBR2) were found to be involved in immune response processes (Table 3). This suggested the involvement of miR-21-5p in the regulation of immune response in PPRV infected PBMCs.
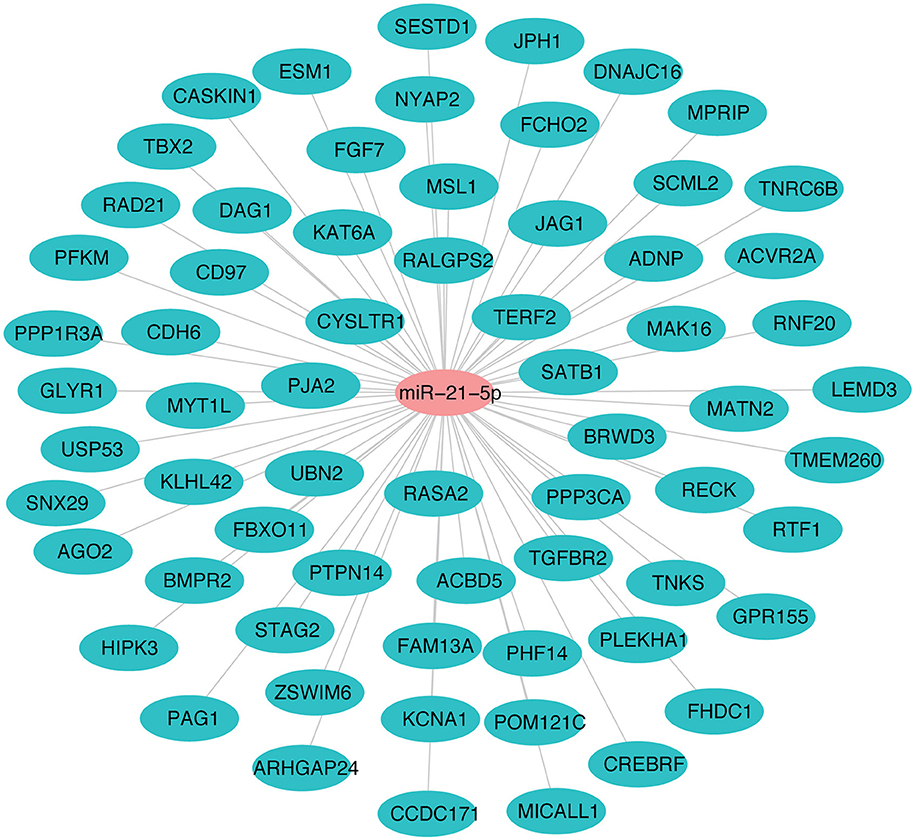
Figure 7. miRNA-protein interaction network of target proteins upregulated due to downregulation of miR-21-5p. Student's t-test was done in JMP9 (SAS Institute Inc, Cary, USA) to test the significance of difference and difference between groups was considered significant at P ≤ 0.05.
miR-21-5p and miR-484 Were Functionally Validated Using Dual-Luciferase Reporter Assay
The miR-21-5p and miR-484 sequences were found to be complementary to sequences from 329-349 and 613-634 at 3′ UTR of TGFBR2 gene, respectively. Further, the strength of interaction between the target site on TGFBR2 for miR-21-5p as evaluated on the basis of ΔG value and total score value was−18.70 kCal/Mol and 152, respectively. The parameters of the interaction for the miR-484 and its target site at 3′ UTR of TGFBR2 were−18.16 kCal/Mol and 148 (Figure 8). The wild-type and mutated type target sequences of TGFBR2 were cloned into XbaI site of the vector pGL4.13. The transfection results showed complementary binding between miRNA mimics and their specific target sites expressed in the form of decreased expression of the luciferase gene in case of wild-type construct and increased expression in case of mutant type (Figure 9). The overview of abstract form of results is given in Data Sheet 1.
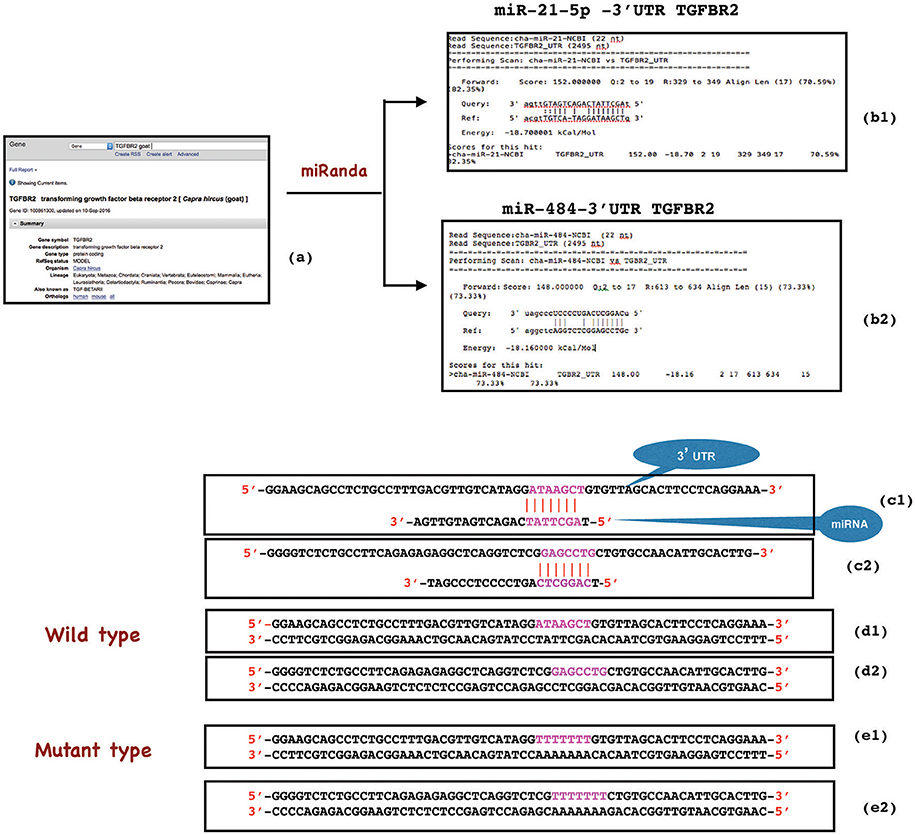
Figure 8. Construction of wild type and mutant type sequences flanking the miRNA target sequence. 3'UTR target site of TGFBR2 gene was retrieved from NCBI (a) and analyzed in miRanda (b1,b2). The complementary binding (in red), ΔG value (−18.70 and−18.16) and total score value (152 and 148) indicated the strength of interaction between miR-21-5p and TGFBR2 (329-349 3'UTR), and miR-484 and TGFBR2 (613-634 3'UTR) respectively (b1, c1, b2, c2). While wild type oligonucleotide (62 bp) was constructed from 3'UTR of TGFBR2 mRNA flanking the miRNA target site (d1, d2), the mutant was created by replacing the target site either with poly A or poly T sequence (red) (e1, e2).
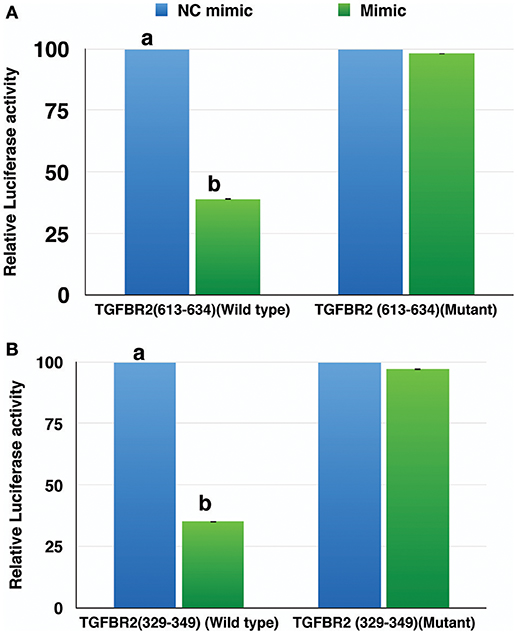
Figure 9. Relative luciferase activity in wild type and mutant of TGFBR2 gene. The decrease in luciferase activity in the wild type indicated specific binding of miR-484 (A) and miR-21-5p (B) to their target site. Student's t-test was done in JMP9 (SAS Institute Inc, Cary, USA) to test the significance of difference and differences between groups were considered significant at P ≤ 0.05.
Discussion
In an attempt to explore the role of microRNAs in modulating the host immune response against PPRV infection, we studied the differential expression of miRNAs in PPRV infected PBMCs, evaluated the influence of these DEmiRNAs on immune response processes from the proteomics data and functionally validated two miRNAs through Dual-Luciferase Reporter Assay.
The miRNA-seq data was analyzed in CLC genomics3 The analysis tools in CLC Genomics Workbench are designed to facilitate trimming of reads, and counting and annotation of the resulting tags using miRBase in general and microRNA of reference organism in particular. Functional analyses of miRNAs or miRNA high-throughput datasets commonly use the Gene Ontology annotations associated with the genes or gene products that the miRNAs are predicted to regulate. Therefore, it is critical to identify targets for understanding their biological function and molecular mechanism (62). TargetScan, an online tool allows the user to extract target genes against broadly conserved or poorly conserved miRNA families across several species or target miRNAs against a particular gene. Thus, it is imperative to identify proteins that are regulated by miRNAs.
The miRNA-protein network analysis suggests that one miRNA could participate in several biological processes by targeting different mRNAs, and one biological process could be influenced by multiple miRNAs (2). The DEmiRNAs identified were found to govern 98 genes that regulate several immune response pathways. miR-664, miR-2311, miR-484, miR-2440, miR-574, miR-210, miR-21-5p, miR-2897, miR-3533, and miR-30 were the top 10 miRNAs governing the immune response processes. miR-664 has been demonstrated to be upregulated during influenza A infection of A549 cells and was found to have proviral activity (63). Similarly, overexpression of miR-484 has been found to inhibit Dengue viral infection in-vitro (64). These miRNAs—miR-664 and miR-484 were found to be upregulated in our study indicating their possible role in the immune response in PPRV infected PBMCs. miR-210 has been reported to inhibit apoptosis (65). This miR-210 was downregulated in PPRV infected PBMCs thus promoting apoptosis in animals that succumbed to the disease. PPRV has been reported to cause apoptosis in PBMCs (66). miR-21-5p has been found to decrease the sensitivity of the cell to the antiviral activity of IFNs, decrease the production of the Th1 cytokine IFN γ and inactivate the T-cell (67), thus facilitating viral replication. In our previous study, IFN γ has been found to increase in PBMCs infected with PPRV infection (35). So, the downregulation of miR-21-5p could be a contributing factor for the increase in IFN γ level during PPRV infection. miR-30b plays an inhibitory role in antigen processing and presentation by primary macrophages and dendritic cells (68) and suppresses TLR/MyD88 activation and cytokine expression in THP-1 cells during MTB H37Rv infection (69). The downregulation of miR-21-5p and miR-30 in our study is indicative of strong host response to PPRV infection.
To investigate the role of immune regulating DEmiRNAs in regulating immune response genes in goat, we choose to confirm the binding between miRNAs and a common gene to evaluate the effect of the interaction. Here, we selected miR-21-5p for functional validation vis-à-vis TGFBR2 as a target gene. TGFBR2 along with TGFBR1 transduces signals of cytokines like TGFB1, TGFB2, and TGFB3 from the cell surface to cytoplasm (70). TGFB signaling pathway plays an important role not only in tissue development and morphogenesis (71, 72) but also in wound healing (73). In this study, TGFBR2 was found to be regulated by 21 identified DEmiRNAs including most of the top 10 immune regulating miRNAs. We identified TGFBR2 to be connected to various development processes like lung morphogenesis and cardiovascular system in GO analysis, suggesting its role in restoring host physiology under PPRV infection. Further, the development, differentiation, and tolerance of T cells and homeostasis of T and B cells are regulated separately by TGFB signaling pathway and Wnt signaling pathway (74–76). Activin receptor signaling pathway shares the SMAD proteins with TGFB signaling pathway (77, 78) and plays a crucial role in inflammation (79). The involvement of TGFBR2 in TGFB signaling pathway and its direct association with Activin receptor signaling pathway and indirect association with Wnt signaling pathway, as predicted in the GO analysis in this study, highlights how TGFBR2 regulates immune response under PPRV infection. Moreover, miR-484 was selected at random from the top 10 immune regulating DEmiRNAs for functional validation against TGFBR2 to provide further evidence of interaction between miRs and genes.
Genetic reporter systems provide efficient means of studying the regulation of eukaryotic gene expression by exploiting the “biochemical requirements” for luminescence of reporter molecules (80). In Dual Luciferase Reporter (DLR) Assay, the target sites of the TGFBR2 for miR-21-5p and for miR-484 were cloned into pGL4.13 vector having luciferase reporter. The interaction between the target mRNA and the specific miRNA mimic lead to an inevitable knockdown in expression of the target mRNA. The ligation of the target site (3'UTR) of TGFBR2 to the luciferase reporter gene in the vector pGL4.13 prevent the luciferase gene from getting translated whenever miRNA-21-5p and miR-484 mimics were cotransfected with the vector. Hence, the significant reduction in expression of luciferase activity in wild-type TGFBR2 in comparison to mutant type TGFBR2 was observed.
In this study, we identified miRNAs that are instrumental in regulating immune response to PPRV infection following integrative analysis of miRNA-seq data and proteome profiling data.
Author Contributions
RS, BPM, and RG conceived and designed the research. KR and DM performed the vaccine potency experiment. YS and RG maintained the server for analysis. AK, ARS, SW, and SS conducted the wet lab work. AK, ARS, RK, AP, and RG analyzed the data. RK, WM, RG, APS, PM, and BM helped in manuscript drafting and editing. RS, BPM, and RG proofread the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported in part by Centre for Agricultural Bioinformatics (ICAR-IASRI) and SubDIC (BTISnet), ICAR-IVRI. We also thank Department of Biotechnology for providing fellowship and contingency for students - Amit Ranjan Sahu (DBT Fellow No. DBT/2014/IVRI/170) and Raja Ishaq Nabi Khan (DBT Fellow No. DBT/2017/IVRI/768).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2018.02631/full#supplementary-material
Table S1. Summary of miRNA sequencing data.
Table S2. Proteome data.
Table S3. Gene Symbol of proteins identified in GO terms pertinent to immune response.
Table S4. Total immune response genes governed by various upregulated miRNAs.
Table S5. Total immune response genes governed by various downregulated miRNAs.
Table S6. Comaprsion of DEmiRNA from PPRV infected PBMCs with DEmiRNA from PPRV infected lung and spleen.
Table S7. DEmiRNAs that regulates the expression of TGFBR2.
Data Sheet 1. Overview of results.
Footnotes
1. ^http://www.targetscan.org/vert_71/.
2. ^https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/
3. ^https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/.
References
1. Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome (2006) 17:189–202. doi: 10.1007/s00335-005-0066-3
2. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
3. Sahu AR. Host-virus interaction: role of miRNA and bioinformatics tools for miRNA target prediction. Adv Animal Vet Sci. (2015) 3:30–6. doi: 10.14737/journal.aavs/2015/3.4s.30.36
4. Ambros V. The functions of animal microRNAs Nature 431: 350–355. Proc Natl Acad Sci USA. (2004) 103:3687–92. doi: 10.1038/nature02871
5. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science (2007) 316:608–11. doi: 10.1126/science.1139253
6. Thai T-H, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by microRNA-155. Science (2007) 316:604–8. doi: 10.1126/science.1141229
7. Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature (2008) 451:1125–9. doi: 10.1038/nature06607
8. Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. (2011) 39:7716–29. doi: 10.1093/nar/gkr426
9. Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature (2008) 455:1124–8. doi: 10.1038/nature07299
10. Guo XK, Zhang Q, Gao L, Li N, Chen XX, Feng WH. Increasing expression of microRNA 181 inhibits porcine reproductive and respiratory syndrome virus replication and has implications for controlling virus infection. J Virol. (2013) 87:1159–71. doi: 10.1128/JVI.02386-12
11. Li S, Duan X, Li Y, Liu B, McGilvray I, Chen L. MicroRNA-130a inhibits HCV replication by restoring the innate immune response. J Viral Hepat. (2014) 21:121–8. doi: 10.1111/jvh.12131
12. Mizuguchi Y, Takizawa T, Uchida E. Host cellular microRNA involvement in the control of hepatitis B virus gene expression and replication. World J Hepatol. (2015) 7:696–702. doi: 10.4254/wjh.v7.i4.696
13. Grassmann R, Jeang KT. The roles of microRNAs in mammalian virus infection. Biochim Biophys Acta (2008) 1779:706–11. doi: 10.1016/j.bbagrm.2008.05.005
14. Scaria V, Hariharan M, Maiti S, Pillai B, Brahmachari SK. Host-virus interaction: a new role for microRNAs. Retrovirology (2006) 3:68. doi: 10.1186/1742-4690-3-68
15. Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. (2010) 64:123–41. doi: 10.1146/annurev.micro.112408.134243
16. Chang J, Guo JT, Jiang D, Guo H, Taylor JM, Block TM. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol. (2008) 82:8215–23. doi: 10.1128/JVI.02575-07
17. Trobaugh DW, Gardner CL, Sun C, Haddow AD, Wang E, Chapnik E, et al. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature (2014) 506:245–8. doi: 10.1038/nature12869
18. Roberts AP, Lewis AP, Jopling CL. The role of microRNAs in viral infection. Progr Molecul Biol Transl Sci. (2011). 102.101–39. doi: 10.1016/B978-0-12-415795-8.00002-7
19. Cui L, Guo X, Qi Y, Qi X, Ge Y, Shi Z, et al. Identification of microRNAs involved in the host response to enterovirus 71 infection by a deep sequencing approach. J Biomed Biotechnol. (2010) 2010:425939. doi: 10.1155/2010/425939
20. Wang Y, Brahmakshatriya V, Zhu H, Lupiani B, Reddy SM, Yoon BJ, et al. Identification of differentially expressed miRNAs in chicken lung and trachea with avian influenza virus infection by a deep sequencing approach. BMC Genomics (2009) 10:512. doi: 10.1186/1471-2164-10-512
21. Pandey A, Sahu AR, Wani SA, Saxena S, Kanchan S, Sah V, et al. Modulation of Host miRNAs transcriptome in lung and spleen of peste des petits ruminants virus infected sheep and goats. Front Microbiol. (2017) 8:1146. doi: 10.3389/fmicb.2017.01146
22. Patel A, Rajak KK, Balamurugan V, Sen A, Sudhakar SB, Bhanuprakash V, et al. Cytokines expression profile and kinetics of Peste des petits ruminants virus antigen and antibody in infected and vaccinated goats. Virologica Sinica (2012) 27:265–71. doi: 10.1007/s12250-012-3240-2
23. Kumari B, Jain P, Das S, Ghosal S, Hazra B, Trivedi AC, et al. Dynamic changes in global microRNAome and transcriptome reveal complex miRNA-mRNA regulated host response to Japanese Encephalitis Virus in microglial cells. Sci Rep. (2016) 6:20263. doi: 10.1038/srep20263
25. Nanda Y, Chatterjee A, Purohit A, Diallo A, Innui K, Sharma R, et al. The isolation of peste des petits ruminants virus from Northern India. Vet Microbiol. (1996) 51:207–16.
26. Dhar P, Sreenivasa B, Barrett T, Corteyn M, Singh R, Bandyopadhyay S. Recent epidemiology of peste des petits ruminants virus (PPRV). Vet Microbiol. (2002) 88:153–9. doi: 10.1016/S0378-1135(02)00102-5
27. Singh R, Saravanan P, Sreenivasa B, Singh R, Bandyopadhyay S. Prevalence and distribution of peste des petits ruminants virus infection in small ruminants in India. Rev Sci Tech. (2004) 23:807–19. doi: 10.20506/rst.23.3.1522
28. Delil F, Asfaw Y, Gebreegziabher B. Prevalence of antibodies to peste des petits ruminants virus before and during outbreaks of the disease in Awash Fentale district, Afar, Ethiopia. Trop Anim Health Produc. (2012) 44:1329–30. doi: 10.1007/s11250-012-0110-8
29. Truong T, Boshra H, Embury-Hyatt C, Nfon C, Gerdts V, Tikoo S, et al. Peste des petits ruminants virus tissue tropism and pathogenesis in sheep and goats following experimental infection. PLoS ONE (2014) 9:e87145. doi: 10.1371/journal.pone.0087145
30. Raghavendra A, Gajendragad M, Sengupta P, Patil S, Tiwari C, Balumahendiran M, et al. Seroepidemiology of peste des petits ruminants in sheep and goats of southern peninsular India. Revue Sci et Tech. (2008) 27:861. doi: 10.1007/2Fs13337-013-0177-5
31. Maganga GD, Verrier D, Zerbinati RM, Drosten C, Drexler JF, Leroy EM. Molecular typing of PPRV strains detected during an outbreak in sheep and goats in south-eastern Gabon in 2011. Virol J. (2013) 10:82. doi: 10.1186/1743-422X-10-82
32. Manjunath S, Mishra BP, Mishra B, Sahoo AP, Tiwari AK, Rajak KK, et al. Comparative and temporal transcriptome analysis of peste des petits ruminants virus infected goat peripheral blood mononuclear cells. Virus Res. (2017) 229:28–40. doi: 10.1016/j.virusres.2016.12.014
33. Manjunath S, Kumar GR, Mishra BP, Mishra B, Sahoo AP, Joshi CG, et al. Genomic analysis of host - Peste des petits ruminants vaccine viral transcriptome uncovers transcription factors modulating immune regulatory pathways. Vet Res. (2015) 46:15. doi: 10.1186/s13567-015-0153-8
34. Pope RA, Parida S, Bailey D, Brownlie J, Barrett T, Banyard AC. Early events following experimental infection with peste-des-petits ruminants virus suggest immune cell targeting. PLoS ONE (2013) 8:e55830. doi: 10.1371/journal.pone.0055830
35. Wani SA, Sahu AR, Saxena S, Rajak KK, Saminathan M, Sahoo AP, et al. Expression kinetics of ISG15, IRF3, IFNγ, IL10, IL2 and IL4 genes vis-a-vis virus shedding, tissue tropism and antibody dynamics in PPRV vaccinated, challenged, infected sheep and goats. Microb Pathog. (2018) 117:206–18. doi: 10.1016/j.micpath.2018.02.027
36. Sahu AR, Wani SA, Saminathan M, Rajak KK, Sahoo AP, Pandey A, et al. Genome sequencing of an Indian peste des petits ruminants virus isolate, Izatnagar/94, and its implications for virus diversity, divergence and phylogeography. Arch Virol. (2017) 162:1677–93. doi: 10.1007/s00705-017-3288-2
37. Kukurba KR, Montgomery SB. RNA Sequencing and Analysis. Cold Spring Harb Protoc. (2015) 2015:951–69. doi: 10.1101/pdb.top084970
38. Robin JD, Ludlow AT, LaRanger R, Wright WE, Shay JW. Comparison of DNA quantification methods for next generation sequencing. Sci Rep. (2016) 6:24067. doi: 10.1038/srep24067
39. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife (2015) 12:4. doi: 10.7554/eLife.05005
40. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics (2009) 25:1091–3. doi: 10.1093/bioinformatics/btp101
41. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303
42. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C T method. Nature Protoc. (2008) 3:1101. doi: 10.1038/nprot.2008.73
43. Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. (2003) 5:R1. doi: 10.1186/gb-2003-5-1-r1
44. Jin Y, Chen Z, Liu X, Zhou X. Evaluating the microRNA targeting sites by luciferase reporter gene assay. Methods Mol Biol. (2013) 936:117–27. doi: 10.1007/978-1-62703-083-0_10
45. Mandel S, Rechavi G, Gozes I. Activity-dependent neuroprotective protein (ADNP) differentially interacts with chromatin to regulate genes essential for embryogenesis. Dev Biol. (2007) 303:814–24. doi: 10.1016/j.ydbio.2006.11.039
46. Nazarko TY, Ozeki K, Till A, Ramakrishnan G, Lotfi P, Yan M, et al. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J Cell Biol. (2014) 204:541–57. doi: 10.1083/jcb.201307050
47. Punzo F, Mientjes E, Rohe C, Scianguetta S, Amendola G, Oostra B, et al. A mutation in the acyl-coenzyme A binding domain-containing protein 5 gene (ACBD5) identified in autosomal dominant thrombocytopenia. J Thrombosis Haemostasis (2010) 8:2085–7. doi: 10.1111/j.1538-7836.2010.03979.x
48. Safaee M, Clark AJ, Ivan ME, Oh MC, Bloch O, Sun MZ, et al. CD97 is a multifunctional leukocyte receptor with distinct roles in human cancers. Intl J Oncol. (2013) 43:1343–50. doi: 10.3892/ijo.2013.2075
49. Leemans JC, te Velde AA, Florquin S, Bennink RJ, de Bruin K, van Lier RA, et al. The epidermal growth factor-seven transmembrane (EGF-TM7) receptor CD97 is required for neutrophil migration and host defense. J Immunol. (2004) 172:1125–31. doi: 10.4049/jimmunol.172.2.1125
50. Shimoyama Y, Gotoh M, Terasaki T, Kitajima M, Hirohashi S. Isolation and sequence analysis of human cadherin-6 complementary DNA for the full coding sequence and its expression in human carcinoma cells. Cancer Res. (1995) 55:2206–11.
51. Yang Y, Jin Y, Martyn AC, Lin P, Song Y, Chen F, et al. Expression pattern implicates a potential role for LRF in the process of implantation in uteri and development of preimplantation embryos in mice. J Reprod Dev. (2012) 59:245–51. doi: 10.1262/2Fjrd.2012-137
52. Audas TE, Hardy-Smith PW, Penney J, Taylor T, Lu R. Characterization of nuclear foci-targeting of Luman/CREB3 recruitment factor (LRF/CREBRF) and its potential role in inhibition of herpes simplex virus-1 replication. Eur J Cell Biol. (2016) 95:611–22. doi: 10.1016/j.ejcb.2016.10.006
53. Theron AJ, Steel HC, Tintinger GR, Gravett CM, Anderson R, Feldman C. Cysteinyl leukotriene receptor-1 antagonists as modulators of innate immune cell function. J Immunol Res. (2014) 2014:608930. doi: 10.1155/2014/608930
54. Qiu X-B, Shao Y-M, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Molecul Life Sci. (2006) 63:2560–70. doi: 10.1007/s00018-006-6192-6
55. Abbas T, Mueller AC, Shibata E, Keaton M, Rossi M, Dutta A. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Molecul Cell (2013) 49:1147–58. doi: 10.1016/j.molcel.2013.02.003
56. Curtin JF, Cotter TG. JNK regulates HIPK3 expression and promotes resistance to Fas-mediated apoptosis in DU 145 prostate carcinoma cells. J Biol Chem. (2004) 279:17090–100. doi: 10.1074/jbc.M307629200
57. Le Friec G, Sheppard D, Whiteman P, Karsten CM, Shamoun SA-T, Laing A, et al. The CD46-Jagged1 interaction is critical for human T H 1 immunity. Nat Immunol. (2012) 13:1213. doi: 10.1038/ni.2454
58. Newman DM, Sakaguchi S, Lun A, Preston S, Pellegrini M, Khamina K, et al. Acetylation of the Cd8 locus by KAT6A determines memory T cell diversity. Cell Rep. (2016) 16:3311–21. doi: 10.1016/j.celrep.2016.08.056
59. Brdicka T, Pavlistová D, Leo A, Bruyns E, Korínek V, Angelisová P, et al. Phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG), a novel ubiquitously expressed transmembrane adaptor protein, binds the protein tyrosine kinase csk and is involved in regulation of T cell activation. J Exp Med. (2000) 191:1591–604. doi: 10.1084/jem.191.9.1591
60. Faust TB, Li Y, Jang GM, Johnson JR, Yang S, Weiss A, et al. PJA2 ubiquitinates the HIV-1 Tat protein with atypical chain linkages to activate viral transcription. Sci Rep. (2017) 7:45394. doi: 10.1038/srep45394
61. Pati D, Zhang N, Plon SE. Linking sister chromatid cohesion and apoptosis: role of Rad21. Molecul Cell Biol. (2002) 22:8267–77. doi: 10.1128/2FMCB.22.23.8267-8277.2002
62. Tang Z, Yang Y, Wang Z, Zhao S, Mu Y, Li K. Integrated analysis of miRNA and mRNA paired expression profiling of prenatal skeletal muscle development in three genotype pigs. Sci Rep. (2015) 5:15544. doi: 10.1038/srep15544
63. Wolf S, Wu W, Jones C, Perwitasari O, Mahalingam S, Tripp RA. MicroRNA regulation of human genes essential for influenza A (H7N9) replication. PLoS ONE (2016) 11:e0155104. doi: 10.1371/journal.pone.0155104
64. Castrillón-Betancur JC, Urcuqui-Inchima S. Overexpression of miR-484 and miR-744 in Vero cells alters Dengue virus replication. Memórias do Instituto Oswaldo Cruz (2017) 112:281–91. doi: 10.1590/2F0074-02760160404
65. Hu S, Huang M, Li Z, Jia F, Ghosh Z, Lijkwan MA, et al. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation (2010) 122(Suppl. 11):S124–31. doi: 10.1161/CIRCULATIONAHA.109.928424
66. Mondal B, Sreenivasa B, Dhar P, Singh R, Bandyopadhyay S. Apoptosis induced by peste des petits ruminants virus in goat peripheral blood mononuclear cells. Virus Res. (2001) 73:113–9. doi: 10.1016/S0168-1702(00)00214-8
67. Yang CH, Li K, Pfeffer SR, Pfeffer LM. The type I IFN-induced miRNA, miR-21. Pharmaceuticals (2015) 8:836–47. doi: 10.3390/2Fph8040836
68. Naqvi AR, Fordham JB, Ganesh B, Nares S. miR-24, miR-30b and miR-142-3p interfere with antigen processing and presentation by primary macrophages and dendritic cells. Sci Rep. (2016) 6:32925. doi: 10.1038/2Fsrep32925
69. Wu Y, Sun Q, Dai L. Immune regulation of miR-30 on the Mycobacterium tuberculosis-induced TLR/MyD88 signaling pathway in THP-1 cells. Exp Therapeutic Med. (2017) 14:3299–303. doi: 10.3892/etm.2017.4872
70. Wieser R, Wrana J, Massague J. GS domain mutations that constitutively activate T beta R-I, the downstream signaling component in the TGF-beta receptor complex. EMBO J. (1995) 14:2199–208. doi: 10.1002/j.1460-2075.1995.tb07214.x
71. Ogata S, Morokuma J, Hayata T, Kolle G, Niehrs C, Ueno N, et al. TGF-β signaling-mediated morphogenesis: modulation of cell adhesion via cadherin endocytosis. Genes Dev. (2007) 21:1817–31. doi: 10.1101/gad.1541807
72. Buschke S, Stark H-J, Cerezo A, Prätzel-Wunder S, Boehnke K, Kollar J, et al. A decisive function of transforming growth factor-β/Smad signaling in tissue morphogenesis and differentiation of human HaCaT keratinocytes. Molecul Biol Cell (2011) 22:782–94. doi: 10.1091/mbc.E10-11-0879
73. Finnson KW, McLean S, Di Guglielmo GM, Philip A. Dynamics of transforming growth factor beta signaling in wound healing and scarring. Adv Wound Care (2013) 2:195–214. doi: 10.1089/wound.2013.0429
74. Spender L, O'Brien D, Simpson D, Dutt D, Gregory C, Allday M, et al. TGF-β induces apoptosis in human B cells by transcriptional regulation of BIK and BCL-X L. Cell Death Diff. (2009) 16:593. doi: 10.1038/cdd.2008.183
75. Roes J, Choi BK, Cazac BB. Redirection of B cell responsiveness by transforming growth factor β receptor. Proc Natl Acad Sci. USA. (2003) 100:7241–6. doi: 10.1073/pnas.0731875100
76. Reya T, O'Riordan M, Okamura R, Devaney E, Willert K, Nusse R, et al. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity (2000) 13:15–24. doi: 10.1016/S1074-7613(00)00004-2
77. Lebrun J-J, Takabe K, Chen Y, Vale W. Roles of pathway-specific and inhibitory Smads in activin receptor signaling. Molecul Endocrinol. (1999) 13:15–23.
78. Deacu E, Mori Y, Sato F, Yin J, Olaru A, Sterian A, et al. Activin type II receptor restoration in ACVR2-deficient colon cancer cells induces transforming growth factor-β response pathway genes. Cancer Res. (2004) 64:7690–6. doi: 10.1158/0008-5472.CAN-04-2082
79. Palin NK, Savikko J, Pasternack A, Rintala JM, Kalra B, Mistry S, et al. Activin inhibition limits early innate immune response in rat kidney allografts-a pilot study. Transpl Int. (2017) 30:96–107. doi: 10.1111/tri.12876
Keywords: miRNAome, proteome, PPR, goats, host-pathogen interaction, immunopathogenesis
Citation: Khanduri A, Sahu AR, Wani SA, Khan RIN, Pandey A, Saxena S, Malla WA, Mondal P, Rajak KK, Muthuchelvan D, Mishra B, Sahoo AP, Singh YP, Singh RK, Gandham RK and Mishra BP (2018) Dysregulated miRNAome and Proteome of PPRV Infected Goat PBMCs Reveal a Coordinated Immune Response. Front. Immunol. 9:2631. doi: 10.3389/fimmu.2018.02631
Received: 02 April 2018; Accepted: 25 October 2018;
Published: 21 November 2018.
Edited by:
Ju-Tao Guo, Baruch S. Blumberg Institute, United StatesReviewed by:
Jianzhong Zhu, Yangzhou University, ChinaNoemi Sevilla, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), Spain
Copyright © 2018 Khanduri, Sahu, Wani, Khan, Pandey, Saxena, Malla, Mondal, Rajak, Muthuchelvan, Mishra, Sahoo, Singh, Singh, Gandham and Mishra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bishnu Prasad Mishra, YnBtaXNocmFfMUBob3RtYWlsLmNvbQ==
Ravi Kumar Gandham, Z2FuZGhhbTcxQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Alok Khanduri
Alok Khanduri Amit Ranjan Sahu
Amit Ranjan Sahu Sajad Ahmad Wani
Sajad Ahmad Wani Raja Ishaq Nabi Khan
Raja Ishaq Nabi Khan Aruna Pandey
Aruna Pandey Shikha Saxena
Shikha Saxena Waseem Akram Malla
Waseem Akram Malla Piyali Mondal
Piyali Mondal Kaushal Kishor Rajak
Kaushal Kishor Rajak D. Muthuchelvan5
D. Muthuchelvan5 Aditya P. Sahoo
Aditya P. Sahoo Ravi Kumar Gandham
Ravi Kumar Gandham