- 1Department of Laboratory Medicine, MD Anderson Cancer Center, Houston, TX, United States
- 2Stem Cell Transplantation and Cellular Therapy, MD Anderson Cancer Center, Houston, TX, United States
Cytomegalovirus (CMV) remains a major cause of morbidity following allogeneic hematopoietic stem cell transplant. Natural killer cells expressing NKG2C have been shown to play a role in the immune surveillance of human CMV. We studied NKG2C copy number in the donor graft and the risk of CMV reactivation after double umbilical cord blood transplantation (DUCBT) in 100 CMV seropositive DUCBT recipients and their corresponding cord blood (CB) grafts (n = 200). In the setting of DUCBT, the combined graft may contain 0–4 functional copies of NKG2C gene. Sixteen patients received a combined graft with 1 or 2 NKG2C copies and 84 patients were recipients of a combined graft with 3 or 4 NKG2C copies. The 6-month cumulative incidence of CMV reactivation for the two groups was 93.7 and 58.4%, respectively (p = 0.0003). In multivariate analysis, low NKG2C copies in the graft was an independent predictor of CMV reactivation (HR = 2.72, CI = 1.59–4.64; p < 0.0001). Our study points to an important role for donor NKG2C for protection against CMV reactivation after DUCBT. These novel findings may help identify patients at a higher risk of CMV reactivation after DUCBT. Donor NKG2C genotype may be used as a potential criterion in the algorithm for graft selection for DUCBT.
Key Points
NKG2C copy number in the CB graft is an independent risk factor for CMV reactivation after double CB transplantation.
Introduction
Cytomegalovirus (CMV) is a member of the herpesviridae family and remains latent in the host cells after primary infection. The virus reactivates in the immunosuppressed host and is a leading cause of morbidity following allogeneic hematopoietic stem cell transplant (allo-HSCT) (1–3). Despite available therapeutic agents and preemptive approaches, CMV associated complications and end-organ damage remain a major cause for concern following allo-HSCT (4, 5).
Natural killer (NK) cells are a subset of effector lymphocytes involved in innate immunity. Unlike T cells, NK cells do not need prior antigen sensitization to kill their target cells and hence provide the first-line of defense against viruses (6). Natural killer (NK) cells are also known to be the first subset of lymphocytes to reconstitute after HSCT and hence play an important role in the early control of viral infections including CMV (7). Evidence for an NK cell response against CMV comes from murine studies (8, 9), and human studies reporting preferential expansion of NKG2C-expressing NK cells in CMV-infected individuals (10, 11). In the first year following allo-HSCT and cord blood (CB) transplantation, CMV reactivation induces expansion of a subset of mature “adaptive” NK cells, characterized as CD56dimCD16−NKG2C+ (12–14). Taken together, these data point to a potential role for NKG2C in CMV recognition by NK cells.
NKG2C is an activating receptor that recognizes HLA-E in complex with leader sequence peptides (15). The gene encoding NKG2C (killer-cell lectin-like receptor C2, KLRC2) is present at different copy numbers in the genomes of different individuals (16, 17). A homozygous deletion of the NKG2C gene (del/del) has been reported in up to 4% of the population, while up to 34% of the European and Japanese population have a heterozygous deletion (wt/del) (16, 18). NKG2C deletion has been reported as a risk factor for CMV (19) and HIV (20), but current data on the influence of NKG2C gene-copy number variations in the donor graft on the risk of CMV reactivation after allo-HSCT is limited. Following CBT, where the T-cell compartment is functionally naïve, the NKG2C genotype may have an even more pronounced impact on the risk of CMV reactivation. Thus, we studied NKG2C copy numbers in the donor graft and the risk of CMV reactivation in recipients of double umbilical cord blood transplantation (DUCBT).
Methods
Study Design
This retrospective study was designed to test the hypothesis that lower NKG2C copy number in the CB graft is associated with a higher risk of CMV reactivation after DUCBT. NKG2C genotyping was performed on genomic DNA collected from 200 CB donor grafts that were infused into 100 DUCBT recipients. The NKG2C gene copy number was assessed by PCR-SSP (17) as described below, based on the availability of specimen, and without preference given to patients with particular clinical characteristics.
Patients
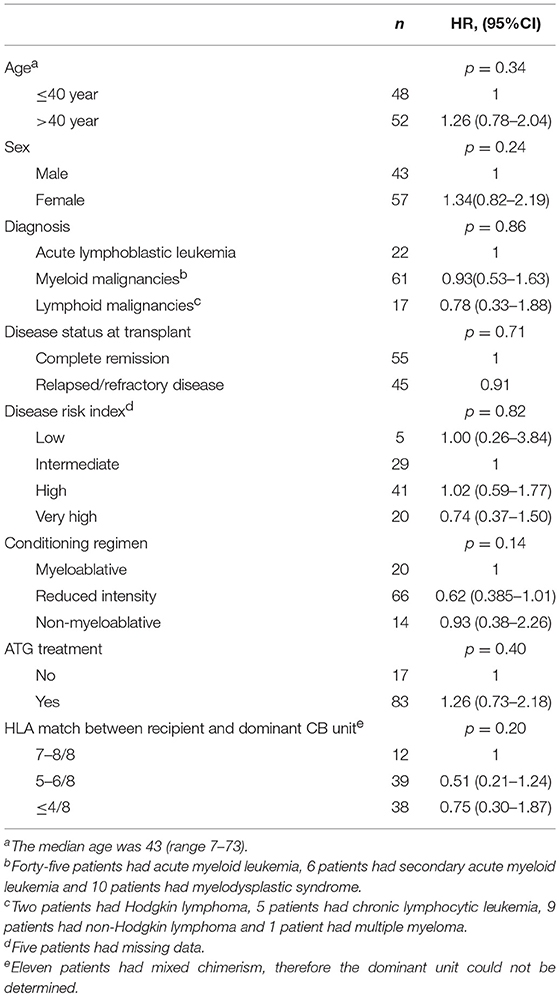
All CMV-seropositive patients who received a DUCBT for the treatment of hematologic malignancies at MD Anderson Cancer Center between 7/2005 and 12/2012 were included. All patients consented to the study in accord with the Declaration of Helsinki, and local ethics approval was obtained before sample collection. Patient characteristics are described in Table 1.
Cord unit dominance, achieved by most patients, was defined as the unit with >90% chimerism in the total DNA fraction at the time the assay was performed.
Peripheral blood was collected twice a week for CMV monitoring. The median time to CMV reactivation was 1.4-months and the 6-month cumulative incidence of CMV reactivation was 63.3%.
NKG2C Genotyping by PCR Amplification With Sequence-Specific Primers (PCR-SSP)
NKG2C gene copy numbers were assessed by PCR-SSP (17). Two pairs of primers were used to detect the NKG2C genotypes. Primer NKG2C/F (5′-CAGTGTGGATCTTCAATG-3′) and NKG2C/R (5′-TTTAGTAATTGTGTGCATCCTA-3′) amplify a 201 bp fragment from NKG2C wild type (wt) carrier. Primer NKG2Cdel/F (5′-ACTCGGATTTCTATTTGATGC-3′) and NKG2Cdel/R (5′-ACAAGTGATGTATAAGAAAAAG-3′) amplify a 411 bp fragment from NKG2C deletion (del) carrier. A single-tube PCR-SSP genotyping strategy combining the two sets of primers was validated and modified from the method by Moraru et al. (17). Briefly, genomic DNA sample was mixed with the two sets of primers at a final concentration of 1 μM for NKGC2/F and NKGC2/R, and 0.5 μM for NKG2Cdel/F, and NKG2Cdel/R, dNTPs, PCR buffer, Taq polymerase, and PCR amplified under the following thermal cycling conditions: 1 cycle of 2 min at 95°C, then 10 cycles of 20 s at 95°C, 30 s at 60°C, and 40 s at 72°C, and 20 cycles of 20 s at 95°C, 30 s at 56°C, and 40 s at 72°C. The last extension cycle of 3 min at 72°C was followed. The PCR product was visualized using gel electrophoresis and UV exposure (Supplementary Figure 1).
Statistical Methods
The probability of CMV reactivation was calculated using the cumulative incidence method. Univariate analysis was performed with standard statistical methodology. Variables found to be significant at the p < 0.15 level were included in the multivariate Fine-Gray regression analysis. Hazard ratios (HR) are reported with 95% confidence intervals (CI). All p-values are two-sided.
Results
NKG2C Copy Number in the CB Grafts Predicts for CMV Reactivation After DUCBT
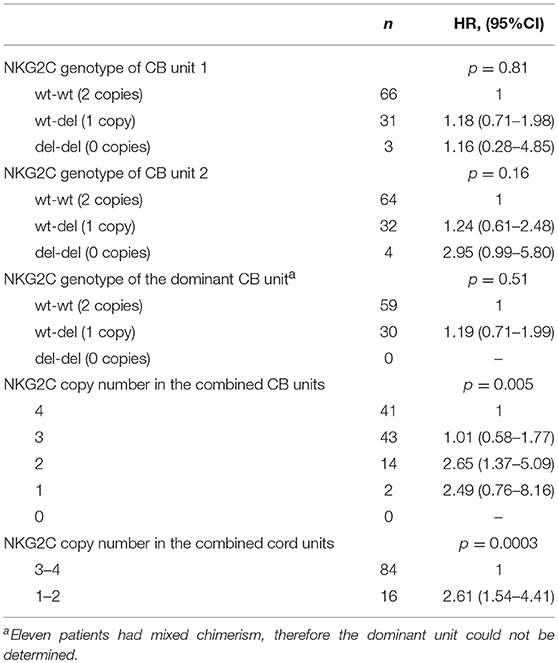
Individuals may inherit different copy numbers of the NKG2C genes (0, 1, or 2). In our cohort, ~2/3 of the CB units had both copies of the gene (wt/wt), 1/3 had only one copy (wt/del), and only a minority of units had 0 copies with both alleles deleted (del/del, Table 2). Since all patients received a DUCBT, the combined graft could contain from 0 to 4 functional copies of the NKG2C gene. Patients whose combined grafts contained only 1 or 2 NKG2C gene copies (i.e., wt/del and wt/del, or del/del and wt/wt, or wt/del and del/del) had a significantly higher probability of CMV reactivation than patients whose combined grafts had 3 or 4 NKG2C copies (i.e., wt/wt and wt/del or wt/wt and wt/wt). Namely, the 6-month cumulative incidence of CMV reactivation for the four groups was 100, 92.9, 60.5, and 55.9%, respectively (p = 0.005). No patient received a graft with zero gene copies.
For the rest of the analysis we divided the patients into two groups, namely the 84 patients who received two CB grafts with 3 or 4 NKG2C copies and the 16 patients who received two units with 1 or 2 NKG2C copies. Notably, the clinical characteristics (as defined in Table 1) for the two groups of patients were similar (data not shown). The 6-month cumulative incidence of CMV reactivation for the two groups was 58.4 and 93.7%, respectively (p = 0.0003, Figure 1). Interestingly, the NKG2C copy number of the dominant cord was not predictive for CMV reactivation, suggesting that both cord units contribute to the antiviral response early post-DUCBT (Table 2).
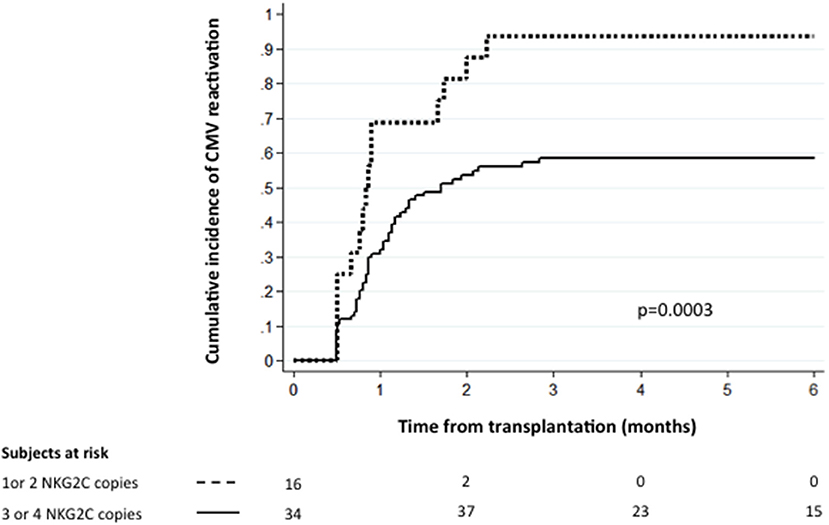
Figure 1. The cumulative incidence of CMV reactivation following DUCBT stratified by NKG2C copy number. Graft NKG2C copy number and the conditioning regimen intensity are the only independent predictors for CMV reactivation after DUCBT.
We performed univariate and multivariate analysis that included NKG2C copy number and the variables shown in Table 1. Low NKG2C copy number (1 or 2) in the combined CB graft (HR = 2.72, CI = 1.59–4.64; p < 0.0001) and reduced intensity conditioning (RIC; HR = 0.59, CI = 0.35–0.99, p = 0.046) were the only independent predictors for CMV reactivation. We also examined the influence of acute graft vs. host disease (aGVHD) on the risk of CMV reactivation using a time-dependent regression model. The development of grade II–IV or III–IV aGVHD did not significantly impact the probability of CMV reactivation (data not shown). This could be explained by the fact that most patients developed CMV reactivation prior to aGVHD onset. The 2-month cumulative incidence of CMV reactivation was 57.0% while the 2-month cumulative incidence of grade III-IV aGVHD was 7.0%. A number of studies have reported a protective effect of CMV reactivation on relapse (21, 22). However, we found no significant impact of graft NKG2C copy number on the risk of relapse after DUCBT (data not shown).
Discussion
Conventionally NKG2C-expressing NK cells have been linked to efficient recognition and elimination of CMV-infected cells (12, 13). This observation, combined with studies reporting that the frequencies and regulation of NKG2C receptor expression is dependent on the gene copy number (19, 23), led us to hypothesize that the risk of CMV reactivation after DUCBT could be directly influenced by the NKG2C copy number of the CB grafts.
To our knowledge our study is the first to assess the influence of NKG2C genotype in both CB units on CMV reactivation in the setting of DUCBT. We extensively analyzed all possible genotype combinations including dominance of engraftments looking at the distinct incidence rates of CMV reactivation, which had not previously been done.
Although the number of patients analyzed in this study is limited, our data point to an association between NKG2C gene copy number in the CB grafts and the risk of CMV reactivation after DUCBT. We show that recipients of two grafts with a wt/del and wt/del, or wt/wt and del/del genotype are inherently more susceptible to CMV reactivation, with the vast majority of patients developing CMV reactivation in the first 3-months post-CBT. This effect is likely related to quantitative differences in NKG2C expression on reconstituting NK cells after CBT. Although our study cohort did not include a case with both grafts carrying the del/del genotype, our results are in agreement with a previous study reporting CMV reactivation in three patients receiving a single CB graft with the NKG2C del/del genotype (24). Our multivariate analysis also shows that the only two independent predictors of CMV reactivation after DUCBT are low NKG2C copy number in the combined graft and reduced intensity conditioning regimen. The latter is in keeping with previous reports of better outcomes following RIC for DUCBT (25–27).
In summary, our results point to an important role for NKG2C in protection against CMV after CBT. Further studies are warranted to understand the precise mechanistic role of NKG2C+ NK cells in CMV recognition. If confirmed in larger numbers of CBT recipients, NKG2C genotyping of the CB graft may be a useful biomarker for predicting the risk of CMV infection after CBT, thus, guiding the intensity of CMV prophylaxis for individual patients. Moreover, it may provide a compelling rationale for considering NKG2C genotype in the algorithm of CB selection.
Author Contributions
KC performed experiments, designed, interpreted, analyzed, and commented on the manuscript. DM interpreted, analyzed, and wrote the manuscript. TS, WZ, NS, MD, QW, RB, and LL performed experiments, analyzed, and commented on the manuscript. GR, RS, RP, UP, CH, AO, BO, RM, RC, and ES collected clinical data, interpreted, analyzed, and commented on the manuscript. KR designed and directed the study and wrote the manuscript.
Funding
The authors' research efforts are funded in part by grants from the NIH (R01 CA211044-01), the Cancer Prevention Research Institute of Texas (RSG-15-218-01-LIB) and the M. D. Anderson's Cancer Center Support Grant CA016672.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the patients who agreed to participate in this study; the teams of nurses, pharmacists, midlevel practitioners, and physicians for their patient care.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2018.02444/full#supplementary-material
References
1. Chan ST, Logan AC. The clinical impact of cytomegalovirus infection following allogeneic hematopoietic cell transplantation: why the quest for meaningful prophylaxis still matters. Blood Rev. (2017) 31:173–83. doi: 10.1016/j.blre.2017.01.002
2. Locatelli F, Bertaina A, Bertaina V, Merli P. Cytomegalovirus in hematopoietic stem cell transplant recipients - management of infection. Expert Rev Hematol. (2016) 9:1093–105. doi: 10.1080/17474086.2016.1242406
3. Styczynski J. Who is the patient at risk of CMV recurrence: a review of the current scientific evidence with a focus on hematopoietic cell transplantation. Infect Dis Ther. (2018) 7:1–16. doi: 10.1007/s40121-017-0180-z
4. Teira P, Battiwalla M, Ramanathan M, Barrett AJ, Ahn KW, Chen M, et al. Early cytomegalovirus reactivation remains associated with increased transplant-related mortality in the current era: a CIBMTR analysis. Blood (2016) 127:2427–38. doi: 10.1182/blood-2015-11-679639
5. Maffini E, Giaccone L, Festuccia M, Brunello L, Busca A, Bruno B. Treatment of CMV infection after allogeneic hematopoietic stem cell transplantation. Expert Rev Hematol. (2016) 9:585–96. doi: 10.1080/17474086.2016.1174571
6. Lam VC, Lanier LL. NK cells in host responses to viral infections. Curr Opin Immunol. (2017) 44:43–51. doi: 10.1016/j.coi.2016.11.003
7. Mehta RS, Rezvani K. Immune reconstitution post allogeneic transplant and the impact of immune recovery on the risk of infection. Virulence (2016) 7:901–16. doi: 10.1080/21505594.2016.1208866
8. Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science (2002) 296:1323–6. doi: 10.1126/science.1070884
9. Nabekura T, Lanier LL. Tracking the fate of antigen-specific versus cytokine-activated natural killer cells after cytomegalovirus infection. J Exp Med. (2016) 213:2745–58. doi: 10.1084/jem.20160726
10. Muntasell A, Vilches C, Angulo A, Lopez-Botet M. Adaptive reconfiguration of the human NK-cell compartment in response to cytomegalovirus: a different perspective of the host-pathogen interaction. Eur J Immunol. (2013) 43:1133–41. doi: 10.1002/eji.201243117
11. Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, et al. Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci USA. (2011) 108:14725–32. doi: 10.1073/pnas.1110900108
12. Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood (2012) 119:2665–74. doi: 10.1182/blood-2011-10-386995
13. Lopez-Botet M, Muntasell A, Vilches C. The CD94/NKG2C+ NK-cell subset on the edge of innate and adaptive immunity to human cytomegalovirus infection. Semin Immunol. (2014) 26:145–51. doi: 10.1016/j.smim.2014.03.002
14. Rolle A, Brodin P. Immune adaptation to environmental influence: the case of NK cells and HCMV. Trends Immunol. (2016) 37:233–43. doi: 10.1016/j.it.2016.01.005
15. Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature (1998) 391:795–9. doi: 10.1038/35869
16. Miyashita R, Tsuchiya N, Hikami K, Kuroki K, Fukazawa T, Bijl M, et al. Molecular genetic analyses of human NKG2C (KLRC2) gene deletion. Int Immunol. (2004) 16:163–8. doi: 10.1093/intimm/dxh013
17. Moraru M, Canizares M, Muntasell A, de Pablo R, Lopez-Botet M, Vilches C. Assessment of copy-number variation in the NKG2C receptor gene in a single-tube and characterization of a reference cell panel, using standard polymerase chain reaction. Tissue Antigens (2012) 80:184–7. doi: 10.1111/j.1399-0039.2012.01911.x
18. Hikami K, Tsuchiya N, Yabe T, Tokunaga K. Variations of human killer cell lectin-like receptors: common occurrence of NKG2-C deletion in the general population. Genes Immun. (2003) 4:160–7. doi: 10.1038/sj.gene.6363940
19. Muntasell A, Lopez-Montanes M, Vera A, Heredia G, Romo N, Penafiel J, et al. NKG2C zygosity influences CD94/NKG2C receptor function and the NK-cell compartment redistribution in response to human cytomegalovirus. Eur J Immunol. (2013) 43:3268–78. doi: 10.1002/eji.201343773
20. Thomas R, Low HZ, Kniesch K, Jacobs R, Schmidt RE, Witte T. NKG2C deletion is a risk factor of HIV infection. AIDS Res Hum Retroviruses (2012) 28:844–51. doi: 10.1089/AID.2011.0253
21. Green ML, Leisenring WM, Xie H, Walter RB, Mielcarek M, Sandmaier BM, et al. CMV reactivation after allogeneic HCT and relapse risk: evidence for early protection in acute myeloid leukemia. Blood (2013) 122:1316–24. doi: 10.1182/blood-2013-02-487074
22. Manjappa S, Bhamidipati PK, Stokerl-Goldstein KE, DiPersio JF, Uy GL, Westervelt P, et al. Protective effect of cytomegalovirus reactivation on relapse after allogeneic hematopoietic cell transplantation in acute myeloid leukemia patients is influenced by conditioning regimen. Biol Blood Marrow Transplant. (2014) 20:46–52. doi: 10.1016/j.bbmt.2013.10.003
23. Muntasell A, Pupuleku A, Cisneros E, Vera A, Moraru M, Vilches C, et al. Relationship of NKG2C copy number with the distribution of distinct cytomegalovirus-induced adaptive NK cell subsets. J Immunol. (2016) 196:3818–27. doi: 10.4049/jimmunol.1502438
24. Della Chiesa M, Falco M, Bertaina A, Muccio L, Alicata C, Frassoni F, et al. Human cytomegalovirus infection promotes rapid maturation of NK cells expressing activating killer Ig-like receptor in patients transplanted with NKG2C-/- umbilical cord blood. J Immunol. (2014) 192:1471–9. doi: 10.4049/jimmunol.1302053
25. Barker JN, Weisdorf DJ, DeFor TE, Blazar BR, McGlave PB, Miller JS, et al. Transplantation of 2 partially HLA-matched umbilical cord blood units to enhance engraftment in adults with hematologic malignancy. Blood (2005) 105:1343–7. doi: 10.1182/blood-2004-07-2717
26. Barker JN, Weisdorf DJ, DeFor TE, Blazar BR, Miller JS, Wagner JE. Rapid and complete donor chimerism in adult recipients of unrelated donor umbilical cord blood transplantation after reduced-intensity conditioning. Blood (2003) 102:1915–9. doi: 10.1182/blood-2002-11-3337
Keywords: NK cells, CBT, NKG2C, CMV reactivation, graft selection
Citation: Cao K, Marin D, Sekine T, Rondon G, Zhao W, Smith NT, Daher M, Wang Q, Li L, Saliba RM, Pingali R, Popat U, Hosing C, Olson A, Oran B, Basar R, Mehta RS, Champlin R, Shpall EJ and Rezvani K (2018) Donor NKG2C Copy Number: An Independent Predictor for CMV Reactivation After Double Cord Blood Transplantation. Front. Immunol. 9:2444. doi: 10.3389/fimmu.2018.02444
Received: 29 August 2018; Accepted: 03 October 2018;
Published: 23 October 2018.
Edited by:
Agnes Yong, University of Adelaide, AustraliaReviewed by:
Subramaniam Malarkannan, Medical College of Wisconsin, United StatesWilliam K. Decker, Baylor College of Medicine, United States
Copyright © 2018 Cao, Marin, Sekine, Rondon, Zhao, Smith, Daher, Wang, Li, Saliba, Pingali, Popat, Hosing, Olson, Oran, Basar, Mehta, Champlin, Shpall and Rezvani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katayoun Rezvani, S1JlenZhbmlAbWRhbmRlcnNvbi5vcmc=
†These authors have contributed equally to this work
 Kai Cao1†
Kai Cao1† Takuye Sekine
Takuye Sekine May Daher
May Daher Rima M. Saliba
Rima M. Saliba Rafet Basar
Rafet Basar Rohtesh S. Mehta
Rohtesh S. Mehta Katayoun Rezvani
Katayoun Rezvani