- Division of Cell Signaling and Immunology, School of Life Sciences, University of Dundee, Dundee, United Kingdom
Cytokines comprise a large family of secreted ligands that are critical for the regulation of immune homeostasis. Cytokines initiate signaling via dimerization or oligomerization of the cognate receptor subunits, triggering the activation of the Janus Kinases (JAKs)/ signal transducer and activator of transcription (STATs) pathway and the induction of specific gene expression programs and bioactivities. Deregulation of cytokines or their downstream signaling pathways are at the root of many human disorders including autoimmunity and cancer. Identifying and understanding the mechanistic principles that govern cytokine signaling will, therefore, be highly important in order to harness the therapeutic potential of cytokines. In this review, we will analyze how biophysical (ligand-receptor binding geometry and affinity) and cellular (receptor trafficking and intracellular abundance of signaling molecules) parameters shape the cytokine signalosome and cytokine functional pleiotropy; from the initial cytokine binding to its receptor to the degradation of the cytokine receptor complex in the proteasome and/or lysosome. We will also discuss how combining advanced protein engineering with detailed signaling and functional studies has opened promising avenues to tackle complex questions in the cytokine signaling field.
Introduction
Cytokines comprise a large family of soluble factors, which control virtually every aspect of mammalian physiology (1–5). Deregulation of cytokines or cytokine-related pathways can result in human diseases such as asthma, severe combined immunodeficiency (SCID) and certain cancers (6–13), making this family highly relevant to human health. A poor mechanistic understanding of how cytokine signaling is initiated and regulated in space and time, however, has hindered the translation of these ligands to the clinic.
The cytokine signaling paradigm encompasses the binding of a cytokine to its surface receptors, followed by the activation of receptor associated tyrosine kinases of the Janus Kinases family (JAKs) (1, 14). JAKs in turn phosphorylate tyrosines in the cytokine receptor intracellular domains (ICD), generating docking sites for the signal transducer and activator of transcription (STAT) factors (2, 3, 15, 16). Upon receptor binding, STATs are phosphorylated by JAKs, forming homo- and hetero-dimers, which translocate to the nucleus, bind specific promoter sequences and induce defined gene expression programs and bioactivities (17–19). In recent years however, a series of biophysical and protein engineering studies have provided new evidence which highlights the large complexity of signaling triggered by a cytokine-cytokine receptor complex. This complexity allows cytokines to produce a wide range of biological responses despite using a very minimal set of surface receptors and effector signaling molecules. In this review, we will focus on cytokines that engage the JAK/STAT signaling pathway and on how the engineering of agonistic surrogate cytokines has expanded our understanding of cytokine signaling and biology; in addition we will discuss future directions in the context of cytokine-based therapies.
Stoichiometry of the Cytokine-Cytokine Receptor Complex in the Plasma Membrane
One of the most debated questions in the cytokine field concerns the stoichiometry of the cytokine receptor complex in the absence of ligand (20, 21). At first glance, this question seems to be unimportant, given that all models agree that ligand binding is the initial step for activating cytokine receptors. However, how cytokine receptors are activated by cytokine binding has clear functional implications—in particular for targeted engineering of desired cytokine properties. Two opposing models have emerged in the past years. The first model postulates that cytokine receptors exist as preformed inactive dimers in the plasma membrane that become active upon cytokine binding through a conformational/structural rearrangement. Evidence supporting this model is found primarily in homo-dimeric systems such as erythropoietin (Epo) (22–26), thrombopoietin (Tpo) (27, 28), and Growth Hormone (GH) (29–31), although some reports in hetero-dimeric systems have also being reported (32–35). The erythropoietin receptor (EpoR) was found to exist as a dimer in crystals that did not include Epo (23), and at the cell surface by immunofluorescence (26). Similar observations were made for the GH receptor (GH-R) via co-immunoprecipitation of differentially tagged receptors or fluorescence and bioluminescence resonance energy transfer techniques (29, 30).
A second model postulates that cytokine receptors diffuse freely in the plasma membrane as monomers and only upon cytokine binding are recruited into a complex to trigger signaling. According to this model, cytokine receptor assembly is driven by affinities, interaction rate constants and the respective concentrations of all involved reactants. This leads to dynamic equilibria between monomeric and assembled receptors subunits, which can be tuned by affinities and receptor concentrations according to the law of mass action. There are also several lines of evidence supporting this model: (a) this model predicts a step-wise formation of cytokine receptor complexes. Indeed, all cytokines described to date bind one of the receptor chains with significantly higher affinity than the other one and step-wise complex formation has been shown for several cytokines both in in vitro and in vivo studies, including Epo and IFN systems (36–38). (b) cytokine receptor chimeras where the extracellular domains and/or transmembrane domains are swapped by those of any other receptor still trigger signaling in a ligand dependent manner (39–45). (c) surrogate cytokine ligands, e.g., antibodies (46–49) can trigger signaling, arguing against precise conformational changes required for signal activation. (d) Single particle fluorescence imaging studies in several cytokine systems has shown that receptor subunits exist as monomers on the surface of live cells at physiologically relevant cell surface densities, and only form dimers upon ligand binding (48, 50–55). Additionally, cytokines with mutations in the low-affinity chain binding site (“site 2”) fail to induce receptor dimers in agreement with the classical two-step binding mode.
A point to consider from all these studies is that in many instances modified/tagged receptor constructs that are ectopically expressed are used. Thus, the possibility that these modifications inhibit or induce receptor assembly on their own cannot be formally excluded, making it difficult to decide which model is true for a given cytokine receptor system. Nonetheless, due to the strong evidences supporting either model, it is plausible that both models are correct to some extent and that their relative contribution to cytokine signaling could vary depending on cellular context, i.e., receptor and signaling molecules abundance, as has been reported for the Epo system.
Cytokine-Cytokine Receptor Complex Stability vs. Activity
A key factor contributing to signaling and bioactivity potency and specificity by cytokines is the stability of the cytokine-cytokine receptor interactions. Type I Interferons (IFNs) have been used as a model system to study how receptor complex stability influences signaling. The type I IFN family comprises more than 15 different subtypes, all binding to the same receptor complex formed by IFNAR1 and IFNAR2 subunits and activating to the same extent the same JAK/STAT pathway (36, 56–58). Yet, different IFN subtypes induce anti-viral and anti-cancer responses with very different potencies (59–64). While all IFNs exhibit a comparable antiviral activity, only IFNβ has an exceptional antiproliferative activity, which is linked to its anti-cancer potential. A series of biophysical, structural and engineering studies has started to address this apparent lack of correlation between signaling and activity output in this family. Early studies elegantly showed that complex stability critically contributed to the differential activities exhibited by IFNs (59, 65–69). Indeed, an IFNα2 variant, engineered to mimic the properties of IFNβ by enhancing IFNAR1 binding affinity acquired potent antiproliferative activity (59, 66, 67). More recently, structural and engineering studies have shown that the topologies of the IFN receptor complexes formed by the different IFNs are very similar and that their differential activities likely result from different receptor binding kinetics and signal activation (64, 70). Indeed, this differential kinetics of STAT activation by type I IFNs result in the induction of two sets of genes: robust genes that drive the antiviral response and only require short pulses of IFN at low concentrations, and tuneable genes that require sustained activation with higher doses of IFNs and are linked to the anti-cancer responses. Induction of robust genes is not very sensitive to changes in complex stability, while the induction of tuneable genes is (71–73).
The ability of cytokine receptors to translate binding stability into biological output potency is not restricted to type I IFNs and can be found in other cytokine systems. IL-4 and IL-13 are two important immunomodulatory cytokines that bind the same receptor complex comprised of IL-4Rα and IL-13Rα1 and activate STAT6 (8, 74). Yet, their activities are not completely overlapping, with each cytokine exhibiting pockets of specificity (75–77). Biophysical and structural studies have shown that the kinetics of complex formation by these two cytokines is at the root of their differences (5, 78, 79). While IL-4 binds first IL-4Rα with high affinity and then recruits IL-13Rα1, IL-13 first binds IL-13Rα1 and then recruits IL-4Rα. Importantly, in most immune cells IL-4Rα appears to be the limiting factor (78, 80–83). As a consequence, IL-4 by recruiting IL-4Rα with higher efficiency can activate signaling more efficiently than IL-13 and overall elicit more potent biological responses (77). Despite this, IL-13 can elicit specific biological responses not induced by IL-4, for which we still lack a mechanistic understanding.
Viruses have taken advantage of the functional plasticity exhibited by the cytokine system. Viruses often encode open-reading frames that share sequence identity with known human cytokines and mimic their biological properties (56, 84–86). A classic example of this is viral IL-10 (vIL-10) (87). IL-10 is a key immune-modulatory cytokine that controls the extent and potency of the immune response by engaging a surface receptor formed by IL-10Rα and IL-10Rβ receptor subunits to activate STAT3 (11, 56, 88, 89). Interestingly two viruses, cytomegalovirus (CMV) and Epstein-Barr (EBV), encode in their genomes homologs of this cytokine (87). Of particular interest is the ebvIL-10, which it is better known as vIL-10. Despite sharing a high degree of sequence and structural homology with human IL-10 (hIL-10), vIL-10 only engages the anti-inflammatory responses elicited by hIL-10 (90–92). vIL-10 inhibits the expression of MHC class II in monocytes and macrophages and the proliferation of T cells (93), but fails to promote other hIL-10 activities such as induction of thymocytes and mast cell proliferation or upregulation of MHC class II by B cells (94–96). This differential effect could be traced to the different complex stabilities elicited by the two ligands, with vIL-10 binding more weakly to IL-10Rα than hIL-10 (91). Overall, these studies describe an intricate relationship between ligand-receptor complex stability and signaling and biological outcomes by cytokines, which could act as a source of functional heterogeneity and potentially be exploited for therapeutic purposes.
Additional Factors Contributing to Cytokine-Cytokine Receptor Complex Stability
Signal activation by cytokines is a very efficient process where cytokines exhibiting a wide range of binding affinities activate signaling to a similar extent. This suggests that other factors beyond the sole affinity of the ligand for its receptor contribute to form and stabilize the cytokine receptor complex (Figure 1). Here we will highlight three cellular determinants that have been the focus of attention in recent studies:
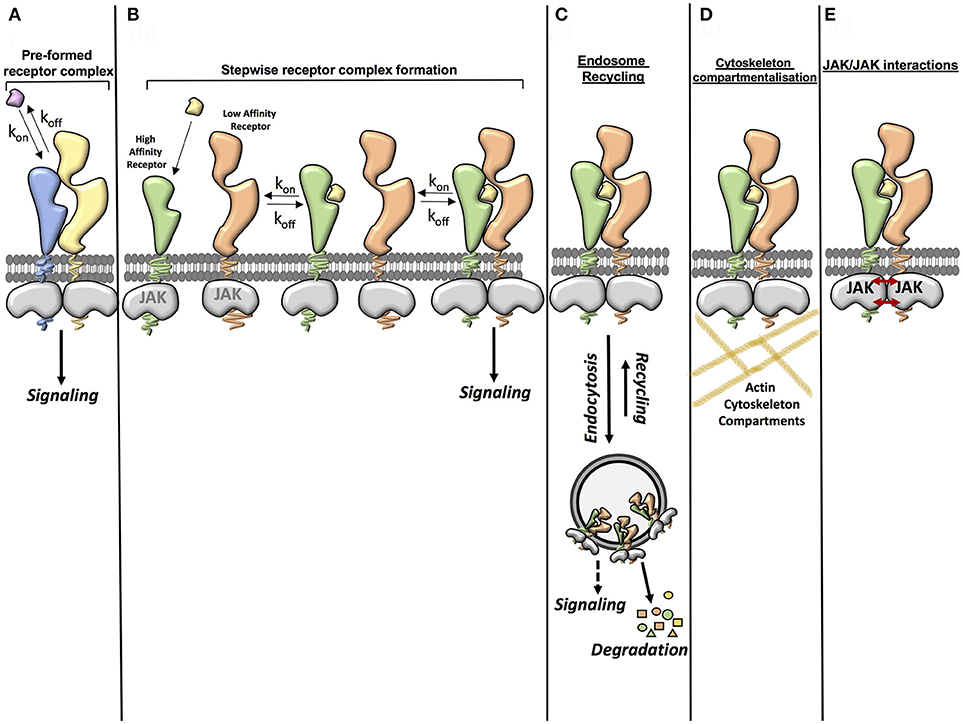
Figure 1. Factors contributing to cytokine-cytokine receptor complex stability. (A) Ligand affinity for the receptor (pre-formed receptor complex). The affinity of the ligand for the pre-formed receptor can enhance the stability of the ligand-receptor complex. (B) Ligand affinity for the receptor (stepwise receptor complex formation). The ligand first binds to the high affinity receptor chain. This then allows recruitment of the second receptor chain. The affinity of the ligand for the high affinity receptor chain and the affinity of the ligand-high affinity receptor complex for the second receptor chain can influence the stability of the complete complex. (C) Endosome recycling. Endocytosis of the ligand-receptor complex from the cell surface to intracellular vesicles depletes ligands and receptors from the plasma membrane, negatively regulating signaling. However some studies have suggested that there is a possibility that some ligand-receptor complexes may also be stabilized in the endosome, leading to a stable complex being formed. Under certain circumstances the recycling of receptor components and ligands from the endosome back to the plasma membrane also contributes to the formation of a stable complex. (D) Cytoskeleton compartmentalization. Cytoskeleton components such as actin can contribute to the stability of a receptor complex by confining the movement of the receptor and thus increasing the opportunity for rebinding of the ligand. (E) JAK/JAK interactions. Some studies have shown that interactions between JAKs can aid dimerization of the receptor complex and that JAKs can act in trans in certain receptor systems.
The role of the endosomal compartment in cytokine signaling initiation and diversification has been proposed but not formally proven (97–102). Early work in the EGF system showed that EGF mutants with impaired EGFR binding affinity paradoxically elicited more potent signaling responses (103). Through a series of studies the authors showed that receptor complexes formed by these mutants did not survive the endosomal acidic pH leading to dissociation and recycling of the ligands and receptor to the membrane, contributing to more sustained signal activation by these mutants (103). More recently, studies utilizing TIRF microscopy have revealed that the endosomal compartment contributes to the formation and stabilization of the cytokine-cytokine receptor complex, thus ensuring signaling fitness at a wide range of environmental conditions (50, 104). Whether endosomes serve as signaling platforms where cytokine receptors encounter alternative signaling molecules to fine-tune their activities, however, still remains an open question. Some evidence of this can be found in a study which showed that phosphorylated JAK1 and Tyk2 could be found in EEA1 positive endosomes upon IFN stimulation (105). Additionally, mutations in the G-CSF receptor that altered its intracellular traffic differentially affected the signaling output and bioactivities engaged by this receptor (106, 107). However, to date no direct evidence demonstrating that endosomes function as signaling hubs for cytokine receptors has been described. This dearth of knowledge originates from the technical challenge that following and blocking cytokine receptor complexes to intracellular compartments represents. Future studies combining biochemistry and imaging methodologies will be required to address this long-standing question.
Another factor contributing to cytokine-cytokine receptor complex formation and stabilization is the actin cytoskeleton. Two recent studies in the IFN and IL-4 systems have shown that cytokine receptors are confined to cytoskeletal microcompartments at the plasma membrane, which allows quick reassembly of the cytokine-cytokine receptor complex after dissociation (52, 55). Manipulation of these actin compartments with small molecule inhibitors altered signaling downstream of the IFN or IL-4 complexes (52, 55).
Yet another factor contributing to cytokine-cytokine receptor complex stability are the JAK kinases associated to the receptors intracellular domains. Early studies with type I IFN showed that mutations in JAKs which did not affect receptor surface expression decreased the number of high affinity IFN binding sites in cells, suggesting an inside-out communication between JAKs and the IFN receptor (108). More recent studies have confirmed this initial observation and provided mechanistic insight into this JAK-receptor communication. A first study showed that two JAK2 molecules could interact in trans via their kinase/pseudokinase domains when bound to the GH-R homodimer, contributing to signaling initiation and propagation (31, 109). A follow up study showed that a productive JAK1-Tyk2 interaction was required to obtain maximal dimerization efficiency in the IFN system. Indeed, lack of JAK1 resulted in a reduction of the number of complexes formed by IFNα2, which could not be assigned to lower levels of IFNAR1 or IFNAR2 on the surface (51).
Overall these studies suggest that the cytokine system has developed a series of check points to ensure that the cytokine-cytokine receptor complex is formed and activates signaling. The next topic we will address is whether we can exploit these different factors contributing to cytokine receptor complex formation to fine-tune cytokine signaling and responses.
Exploiting Cytokine Engineering to Discover New Cytokine Biology
Manipulation of cytokine binding properties via protein engineering is a valuable tool with which we can better understand cytokine biology and to fine-tune cytokine responses. Above we have already introduced some examples focused on the IFN system that help to better understand IFN biology. Next, we will describe additional examples in other cytokine systems which highlight the potential of cytokine engineering to address complex biological problems.
IL-2 plays a critical role in regulating T cell responses, making it an attractive target to treat autoimmune diseases and cancer (5, 110–113). However, its use in the clinic is limited due to severe toxicity resulting from its functional pleiotropy (114–117). IL-2 can engage two types of receptor complexes on the surface of responsive cells: the high affinity receptor complex comprised of IL-2Rα, IL-2Rβ, and γc receptor subunits and the intermediate affinity complex formed by IL-2Rβ and γc subunits (5). Thus, T cells control their sensitivity to IL-2 by modulating their levels of the alpha receptor (113). Many attempts to improve the clinical efficacy of IL-2 by fine tuning its receptor binding properties have been carried out over the years. One of the first studies was performed by Shanafelt and colleagues, who proposed that IL-2-derived toxicity resulted from engagement of the intermediate receptor present on NK cells, which are believed to be the major source of the cytokines and inflammatory mediators causing most of the toxicity associated with high-dose IL-2 therapy (118). In order to specifically target IL-2 to T cells and thus decrease its toxicity they used site directed mutagenesis to reduce the binding affinity of IL-2 to IL-2Rβ (119). This IL-2 mutant could not engage the intermediate affinity receptor, but still could activate signaling in the context of the high affinity receptor, leading to a more than 3,000-fold specificity for T cells over NK cells (119). In an experimental lung metastasis model, sensitive to IL-2 therapy, this IL-2 mutant showed similar levels of tumor inhibition to IL-2 but elicited lower levels of morbidity as scored by general health examination (119). However, in a later phase I trial this mutant did not show advantage over wt IL-2 in anti-tumor responses or toxicity, highlighting the complexity of this cytokine in an in vivo setting (120). Another example of IL-2 engineering is found in studies by the Wittrup lab. Using yeast surface display, the authors engineered an IL-2 variant with high affinity for IL-2Rα. This variant induced T cell proliferation more potently than wt IL-2, suggesting that it could be a better alternative than wt for cancer immunotherapy since lower doses of the variant would be required to show efficacy which could result in lower toxicity (121–123).
More recently, studies by Garcia and collaborators have provided a series of IL-2 variants that have furthered our understanding of IL-2 biology. Using yeast surface display, Levin and colleagues engineered an IL-2 variant (Super-2) binding 200-fold tighter to IL-2Rβ than wt (124). Super-2 can signal through the intermediate affinity receptor as potently as through the high affinity receptor, thus negating the regulatory role of the alpha subunit. This in turn resulted in a stronger anti-tumor response by Super-2 with a significantly lower toxicity when compared to wt IL-2 (124). In a second study, Suman and colleagues used Super-2 as a backbone to engineer a series of Super-2-based cytokines where binding to γc chain was reduced (125). Strikingly, the authors observed that rather than a complete loss in response, these new variants activated signaling with different amplitudes ranging from 100% activity to 50 and 10% in accordance with their binding affinity (125). Interestingly, the IL-2 mutant activating 50% activity could induce proliferation of activated T cells, but not of naïve T cells, suggesting different signaling thresholds required for proliferation in different T cell differentiation stages (125).
In addition, a recent study has shown that IL-2 receptor binding specificity can also be altered in a mutation-independent manner by introducing PEG molecules in the IL-2 region interacting with IL-2Rα. This new IL-2 variant, named NKTR-214, has shown promising anti-tumor responses and decreased toxicity and it is now finding its way to the clinic (126).
The IL-4/IL-13 system has also been the subject of protein engineering studies. As described above, IL-4 binds two surface receptor complexes: The type I receptor, consisting of the IL-4Rα and γc subunits, which is found exclusively on hematopoietic cells; and the type II receptor, composed of the IL-4Rα and IL-13Rα1 chains, which is also shared by IL-13 (5, 78). A recent work by Junttila and collaborators shed some light onto the differential activities elicited by the two IL-4 complexes. Using yeast surface display, the authors engineered two IL-4 variants exhibiting high specificity for either the type I or the type II IL-4 receptors (127). Detailed functional characterization of these variants revealed that while T cell responses were exclusively dependent on the type I IL-4 complex, in agreement with the specific expression of this receptor in T cells, dendritic cell maturation was dependent on the IL-4 type II complex (127). These results agreed with previously published observations and revealed functional dichotomy between the Type I and Type II IL-4 receptors (128, 129).
The impact of complex formation kinetics and stability on signaling and activities by the IL-4/IL-13 complex was further explored in a recent work (50). In this study, we engineered a range of IL-13 variants exhibiting different binding affinities for the IL-13Rα1 receptor subunit. When we functionally characterized these variants, we observed that large decreases in binding affinity were required to marginally alter signaling efficiency. Further increases in binding affinities, however, did not improve signaling by IL-13. Through a series of modeling simulations and experiments we concluded that transition of the cytokine-cytokine receptor complex to the endosomal compartment was the limiting rate factor for signaling potency in the IL-13 system. Cytokine-cytokine receptor complexes capable of undergoing endocytosis would be stabilized due to the high local cytokine receptor concentration achieved in the limited area of endosomes. Further stabilization of the complex beyond that required to transit to the endosomes will have minimal influence on signaling (50). Indeed, our data agreed with a recent study highlighting the role of the endosomal compartment in the formation of the IL-4/IL-13 complexes (104, 130). Interestingly, this disconnect between binding affinity and signaling output was not found when more complex biological responses, e.g., TF-1 cell proliferation and dendritic cell differentiation, were analyzed, which in turn directly correlated with the stability of the IL-13 complex (50). A possible explanation for this apparent lack of correlation between signaling and activity could be found in the different times used to study these processes. While signaling is measured during the first few hours of cytokine stimulation, biological responses often take days to be observed. Surface receptors, signaling molecules and ligand concentrations could be altered with time, leading to functional diversification from an apparently similar starting point. An example of this can be found in the type I IFN system, where IFN stimulation leads to the upregulation of negative regulators that preferentially inhibit short-lived IFN complexes (51, 131, 132).
Conclusions and Remarks
In this mini-review, we have summarized recent studies that have underlined the intricate interplay of cytokine-receptor complex stability and signaling and biological responses. Additionally we have discussed recent findings that support a scaffolding role for the JAK kinases in complex formation, as well as interesting observations regarding the contribution of the actin cytoskeleton and the endosomal compartment to signaling robustness by cytokine-receptor complexes. However, key standing questions remains in the field such as how binding of a cytokine to its receptor triggers signaling, how signaling specificity is generated, are endosomes contributing to fine tune cytokine signaling and biology and how cytokine functional pleiotropy is generated. In order to answer these questions, which would allow us to rationally manipulate cytokine responses and harness their full therapeutic potential, future studies will need to take advantage of recent advances in cryo-EM and membrane protein structural biology to fully understand the complex interconnectivity of the cytokine/cytokine receptor/JAK/STAT complex. Additionally, advance microscopy studies combined with proximity labeling methodologies such as bioID could provide us with new insights into the role that the endosomal compartment plays in shaping cytokine signaling and responses.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by Wellcome Trust (grant # 202323/Z/16/Z to IM), European Research Council (grant # 714680 to IM, JM-F); SW was funded by the EMBO (ALTF 454-2017); CG was funded by Wellcome Trust 4 year Ph.D. (203752/Z/16/Z).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Haan C, Kreis S, Margue C, Behrmann I. Jaks and cytokine receptors–an intimate relationship. Biochem Pharmacol. (2006) 72:1538–46. doi: 10.1016/j.bcp.2006.04.013
2. Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. (2007) 178:2623–9. doi: 10.4049/jimmunol.178.5.2623
3. Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. (2007) 282:20059–63. doi: 10.1074/jbc.R700016200
4. Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. (2009) 9:480–90. doi: 10.1038/nri2580
5. Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. (2009) 27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616
6. Foster PS, Martinez-Moczygemba M, Huston DP, Corry DB. Interleukins-4,−5, and−13: emerging therapeutic targets in allergic disease. Pharmacol Ther. (2002) 94:253–64. doi: 10.1016/S0163-7258(02)00220-6
7. Kovanen PE, Leonard WJ. Cytokines and immunodeficiency diseases: critical roles of the gamma(c)-dependent cytokines interleukins 2, 4, 7, 9, 15, and 21, and their signaling pathways. Immunol Rev. (2004) 202:67–83. doi: 10.1111/j.0105-2896.2004.00203.x
8. Wills-Karp M. Interleukin-13 in asthma pathogenesis. Curr Allergy Asthma Rep. (2004) 4:123–31. doi: 10.1007/s11882-004-0057-6
9. Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. (2006) 6:595–601. doi: 10.1038/nri1901
10. Boyman O, Purton JF, Surh CD, Sprent J. Cytokines and T-cell homeostasis. Curr Opin Immunol. (2007) 19:320–6. doi: 10.1016/j.coi.2007.04.015
11. Mosser DM, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol Rev. (2008) 226:205–18. doi: 10.1111/j.1600-065X.2008.00706.x
12. Romo de Vivar Chavez A, de Vera ME, Liang X, Lotze MT. The biology of interleukin-2 efficacy in the treatment of patients with renal cell carcinoma. Med Oncol. (2009) 26(Suppl. 1):3–12. doi: 10.1007/s12032-008-9162-z
13. Steel JC, Waldmann TA, Morris JC. Interleukin-15 biology and its therapeutic implications in cancer. Trends Pharmacol Sci. (2012) 33:35–41. doi: 10.1016/j.tips.2011.09.004
14. Yamaoka K, Saharinen P, Pesu M, Holt VE III, Silvennoinen O, O'Shea JJ. The Janus kinases (Jaks). Genome Biol. (2004) 5:253. doi: 10.1186/gb-2004-5-12-253
15. Leonard WJ, O'Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. (1998) 16:293–322. doi: 10.1146/annurev.immunol.16.1.293
16. Heim MH. The Jak-STAT pathway: cytokine signalling from the receptor to the nucleus. J Recept Signal Transduct Res. (1999) 19:75–120. doi: 10.3109/10799899909036638
17. Ramana CV, Chatterjee-Kishore M, Nguyen H, Stark GR. Complex roles of Stat1 in regulating gene expression. Oncogene (2000) 19:2619–27. doi: 10.1038/sj.onc.1203525
18. Ihle JN. The Stat family in cytokine signaling. Curr Opin Cell Biol. (2001) 13:211–7. doi: 10.1016/S0955-0674(00)00199-X
19. Levy DE, Darnell JE Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. (2002) 3:651–62. doi: 10.1038/nrm909
21. Atanasova M, Whitty A. Understanding cytokine and growth factor receptor activation mechanisms. Crit Rev Biochem Mol Biol. (2012) 47:502–30. doi: 10.3109/10409238.2012.729561
22. Constantinescu SN, Ghaffari S, Lodish HF. The erythropoietin receptor: structure, activation and intracellular signal transduction. Trends Endocrinol Metab. (1999) 10:18–23. doi: 10.1016/S1043-2760(98)00101-5
23. Livnah O, Stura EA, Middleton SA, Johnson DL, Jolliffe LK, Wilson IA. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science (1999) 283:987–90. doi: 10.1126/science.283.5404.987
24. Remy I, Wilson IA, Michnick SW. Erythropoietin receptor activation by a ligand-induced conformation change. Science (1999) 283:990–3. doi: 10.1126/science.283.5404.990
25. Constantinescu SN, Huang LJ, Nam H, Lodish HF. The erythropoietin receptor cytosolic juxtamembrane domain contains an essential, precisely oriented, hydrophobic motif. Mol Cell (2001) 7:377–85. doi: 10.1016/S1097-2765(01)00185-X
26. Constantinescu SN, Keren T, Socolovsky M, Nam H, Henis YI, Lodish HF. Ligand-independent oligomerization of cell-surface erythropoietin receptor is mediated by the transmembrane domain. Proc Natl Acad Sci USA. (2001) 98:4379–84. doi: 10.1073/pnas.081069198
27. Staerk J, Defour JP, Pecquet C, Leroy E, Antoine-Poirel H, Brett I, et al. Orientation-specific signalling by thrombopoietin receptor dimers. EMBO J. (2011) 30 4398–413. doi: 10.1038/emboj.2011.315
28. Matthews EE, Thevenin D, Rogers JM, Gotow L, Lira PD, Reiter LA, et al. Thrombopoietin receptor activation: transmembrane helix dimerization, rotation, and allosteric modulation. FASEB J. (2011) 25:2234–44. doi: 10.1096/fj.10-178673
29. Gent J, van Kerkhof P, Roza M, Bu G, Strous GJ. Ligand-independent growth hormone receptor dimerization occurs in the endoplasmic reticulum and is required for ubiquitin system-dependent endocytosis. Proc Natl Acad Sci USA. (2002) 99:9858–63. doi: 10.1073/pnas.152294299
30. Brown RJ, Adams JJ, Pelekanos RA, Wan Y, McKinstry WJ, Palethorpe K, et al. Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat Struct Mol Biol. (2005) 12:814–21. doi: 10.1038/nsmb977
31. Brooks AJ, Dai W, O'Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science (2014) 344:1249783. doi: 10.1126/science.1249783
32. Pillet AH, Lavergne V, Pasquier V, Gesbert F, Theze J, Rose T. IL-2 induces conformational changes in its preassembled receptor core, which then migrates in lipid raft and binds to the cytoskeleton meshwork. J Mol Biol. (2010) 403:671–92. doi: 10.1016/j.jmb.2010.08.056
33. Tenhumberg S, Schuster B, Zhu L, Kovaleva M, Scheller J, Kallen KJ. gp130 dimerization in the absence of ligand: preformed cytokine receptor complexes. Biochem Biophys Res Commun. (2006) 346:649–57. doi: 10.1016/j.bbrc.2006.05.173
34. Malka Y, Hornakova T, Royer Y, Knoops L, Renauld JC, Constantinescu SN, et al. Ligand-independent homomeric and heteromeric complexes between interleukin-2 or−9 receptor subunits and the gamma chain. J Biol Chem. (2008) 283:33569–77. doi: 10.1074/jbc.M803125200
35. Krause CD, Digioia G, Izotova LS, Xie J, Kim Y, Schwartz BJ, et al. Ligand-independent interaction of the type I interferon receptor complex is necessary to observe its biological activity. Cytokine (2013) 64:286–97. doi: 10.1016/j.cyto.2013.06.309
36. Piehler J, Thomas C, Garcia KC, Schreiber G. Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol Rev. (2012) 250:317–34. doi: 10.1111/imr.12001
37. Moraga I, Spangler J, Mendoza JL, Garcia KC. Multifarious determinants of cytokine receptor signaling specificity. Adv Immunol. (2014) 121:1–39. doi: 10.1016/B978-0-12-800100-4.00001-5
38. Spangler JB, Moraga I, Mendoza JL, Garcia KC. Insights into cytokine-receptor interactions from cytokine engineering. Annu Rev Immunol. (2015) 33:139–67. doi: 10.1146/annurev-immunol-032713-120211
39. Ohashi H, Maruyama K, Liu YC, Yoshimura A. Ligand-induced activation of chimeric receptors between the erythropoietin receptor and receptor tyrosine kinases. Proc Natl Acad Sci USA. (1994) 91:158–62. doi: 10.1073/pnas.91.1.158
40. Fujiwara H, Hanissian SH, Tsytsykova A, Geha RS. Homodimerization of the human interleukin 4 receptor alpha chain induces Cepsilon germline transcripts in B cells in the absence of the interleukin 2 receptor gamma chain. Proc Natl Acad Sci USA. (1997) 94:5866–71. doi: 10.1073/pnas.94.11.5866
41. Pattyn E, Van Ostade X, Schauvliege L, Verhee A, Kalai M, Vandekerckhove J, et al. Dimerization of the interferon type I receptor IFNaR2–2 is sufficient for induction of interferon effector genes but not for full antiviral activity. J Biol Chem. (1999) 274:34838–45. doi: 10.1074/jbc.274.49.34838
42. Schaeffer M, Schneiderbauer M, Weidler S, Tavares R, Warmuth M, de Vos G, et al. Signaling through a novel domain of gp130 mediates cell proliferation and activation of Hck and Erk kinases. Mol Cell Biol. (2001) 21:8068–81. doi: 10.1128/MCB.21.23.8068-8081.2001
43. Kawahara M, Ogo Y, Tsumoto K, Kumagai I, Ueda H, Nagamune T. Growth control of hybridoma cells with an artificially induced EpoR-gp130 heterodimer. Cytotechnology (2006) 52:171–9. doi: 10.1007/s10616-006-9035-2
44. Heller NM, Qi X, Gesbert F, Keegan AD. The extracellular and transmembrane domains of the gammaC and interleukin (IL)-13 receptor alpha1 chains, not their cytoplasmic domains, dictate the nature of signaling responses to IL-4 and IL-13. J Biol Chem. (2012) 287:31948–61. doi: 10.1074/jbc.M112.348896
45. Moraga I, Spangler JB, Mendoza JL, Gakovic M, Wehrman TS, Krutzik P, et al. Synthekines are surrogate cytokine and growth factor agonists that compel signaling through non-natural receptor dimers. Elife (2017). 6:e22882. doi: 10.7554/eLife.22882
46. Cwirla SE, Balasubramanian P, Duffin DJ, Wagstrom CR, Gates CM, Singer SC, et al. Peptide agonist of the thrombopoietin receptor as potent as the natural cytokine. Science (1997) 276:1696–9. doi: 10.1126/science.276.5319.1696
47. Elliott S, Lorenzini T, Yanagihara D, Chang D, Elliott G. Activation of the erythropoietin (EPO) receptor by bivalent anti-EPO receptor antibodies. J Biol Chem. (1996) 271:24691–7. doi: 10.1074/jbc.271.40.24691
48. Moraga I, Wernig G, Wilmes S, Gryshkova V, Richter CP, Hong WJ, et al. Tuning cytokine receptor signaling by re-orienting dimer geometry with surrogate ligands. Cell (2015) 160:1196–208. doi: 10.1016/j.cell.2015.02.011
49. Rui H, Lebrun JJ, Kirken RA, Kelly PA, Farrar WL. JAK2 activation and cell proliferation induced by antibody-mediated prolactin receptor dimerization. Endocrinology (1994) 135:1299–306. doi: 10.1210/endo.135.4.7925093
50. Moraga I, Richter D, Wilmes S, Winkelmann H, Jude K, Thomas C, et al. Instructive roles for cytokine-receptor binding parameters in determining signaling and functional potency. Sci Signal. (2015) 8:ra114. doi: 10.1126/scisignal.aab2677
51. Wilmes S, Beutel O, Li Z, Francois-Newton V, Richter CP, Janning D, et al. Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. J Cell Biol. (2015) 209:579–93. doi: 10.1083/jcb.201412049
52. You C, Marquez-Lago TT, Richter CP, Wilmes S, Moraga I, Garcia KC, et al. Receptor dimer stabilization by hierarchical plasma membrane microcompartments regulates cytokine signaling. Sci Adv. (2016) 2:e1600452. doi: 10.1126/sciadv.1600452
53. Ho CCM, Chhabra A, Starkl P, Schnorr PJ, Wilmes S, Moraga I, et al. Decoupling the functional pleiotropy of stem cell factor by tuning c-kit signaling. Cell (2017) 168:1041–1052 e18. doi: 10.1016/j.cell.2017.02.011
54. Kim AR, Ulirsch JC, Wilmes S, Unal E, Moraga I, Karakukcu M, et al. Functional Selectivity in Cytokine Signaling Revealed Through a Pathogenic EPO Mutation. Cell (2017) 168:1053–1064 e15. doi: 10.1016/j.cell.2017.02.026
55. Richter D, Moraga I, Winkelmann H, Birkholz O, Wilmes S, Schulte M, et al. Ligand-induced type II interleukin-4 receptor dimers are sustained by rapid re-association within plasma membrane microcompartments. Nat Commun. (2017) 8:15976. doi: 10.1038/ncomms15976
56. Walter MR. Structural analysis of IL-10 and Type I interferon family members and their complexes with receptor. Adv Protein Chem. (2004) 68:171–223. doi: 10.1016/S0065-3233(04)68006-5
57. de Weerd NA, Samarajiwa SA, Hertzog PJ. Type I interferon receptors: biochemistry and biological functions. J Biol Chem. (2007) 282:20053–7. doi: 10.1074/jbc.R700006200
58. Pestka S. The interferons: 50 years after their discovery, there is much more to learn. J Biol Chem. (2007) 282:20047–51. doi: 10.1074/jbc.R700004200
59. Jaitin DA, Roisman LC, Jaks E, Gavutis M, Piehler J, Van der Heyden J, et al. Inquiring into the differential action of interferons (IFNs): an IFN-alpha2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-beta. Mol Cell Biol. (2006) 26:1888–97. doi: 10.1128/MCB.26.5.1888-1897.2006
60. Jaks E, Gavutis M, Uze G, Martal J, Piehler J. Differential receptor subunit affinities of type I interferons govern differential signal activation. J Mol Biol. (2007) 366:525–39. doi: 10.1016/j.jmb.2006.11.053
61. Marijanovic Z, Ragimbeau J, van der Heyden J, Uze G, Pellegrini S. Comparable potency of IFNalpha2 and IFNbeta on immediate JAK/STAT activation but differential down-regulation of IFNAR2. Biochem J. (2007) 407:141–51. doi: 10.1042/BJ20070605
62. Moraga I, Harari D, Schreiber G, Uze G, Pellegrini S. Receptor density is key to the alpha2/beta interferon differential activities. Mol Cell Biol. (2009) 29:4778–87. doi: 10.1128/MCB.01808-08
63. Subramaniam PS, Khan SA, Pontzer CH, Johnson HM. Differential recognition of the type I interferon receptor by interferons tau and alpha is responsible for their disparate cytotoxicities. Proc Natl Acad Sci USA. (1995) 92:12270–4. doi: 10.1073/pnas.92.26.12270
64. Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A, et al. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146:621–32. doi: 10.1016/j.cell.2011.06.048
65. Chill JH, Quadt SR, Levy R, Schreiber G, Anglister J. The human type I interferon receptor: NMR structure reveals the molecular basis of ligand binding. Structure (2003) 11:791–802. doi: 10.1016/S0969-2126(03)00120-5
66. Kalie E, Jaitin DA, Abramovich R, Schreiber G. An interferon alpha2 mutant optimized by phage display for IFNAR1 binding confers specifically enhanced antitumor activities. J Biol Chem. (2007) 282:11602–11. doi: 10.1074/jbc.M610115200
67. Kalie E, Jaitin DA, Podoplelova Y, Piehler J, Schreiber G. The stability of the ternary interferon-receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J Biol Chem. (2008) 283:32925–36. doi: 10.1074/jbc.M806019200
68. Roisman LC, Jaitin DA, Baker DP, Schreiber G. Mutational analysis of the IFNAR1 binding site on IFNalpha2 reveals the architecture of a weak ligand-receptor binding-site. J Mol Biol. (2005) 353:271–81. doi: 10.1016/j.jmb.2005.08.042
69. Li H, Sharma N, General IJ, Schreiber G, Bahar I. Dynamic modulation of binding affinity as a mechanism for regulating interferon signaling. J Mol Biol. (2017) 429:2571–89. doi: 10.1016/j.jmb.2017.06.011
70. Mendoza JL, Schneider WM, Hoffmann HH, Vercauteren K, Jude KM, Xiong A, et al. The IFN-lambda-IFN-lambdaR1-IL-10Rbeta complex reveals structural features underlying type III IFN functional plasticity. Immunity (2017) 46:379–92. doi: 10.1016/j.immuni.2017.02.017
71. Schreiber G. The molecular basis for differential type I interferon signaling. J Biol Chem. (2017) 292:7285–94. doi: 10.1074/jbc.R116.774562
72. Urin V, Levin D, Sharma N, Harari D, Schreiber G. Fine Tuning of a type 1 interferon antagonist. PLoS ONE (2015) 10:e0130797. doi: 10.1371/journal.pone.0130797
73. Levin D, Schneider WM, Hoffmann HH, Yarden G, Busetto AG, Manor O, et al. Multifaceted activities of type I interferon are revealed by a receptor antagonist. Sci Signal. (2014) 7:ra50. doi: 10.1126/scisignal.2004998
74. Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. (2010) 28:445–89. doi: 10.1146/annurev-immunol-030409-101212
75. Chomarat P, Banchereau J. Interleukin-4 and interleukin-13: their similarities and discrepancies. Int Rev Immunol. (1998) 17:1–52. doi: 10.3109/08830189809084486
76. Liang HE, Reinhardt RL, Bando JK, Sullivan BM, Ho IC, Locksley RM. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat Immunol. (2012) 13:58–66. doi: 10.1038/ni.2182
77. Junttila IS, Mizukami K, Dickensheets H, Meier-Schellersheim M, Yamane H, Donnelly RP, et al. Tuning sensitivity to IL-4 and IL-13: differential expression of IL-4Ralpha, IL-13Ralpha1, and gammac regulates relative cytokine sensitivity. J Exp Med. (2008) 205:2595–608. doi: 10.1084/jem.20080452
78. LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, et al. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell (2008) 132:259–72. doi: 10.1016/j.cell.2007.12.030
79. Kelly-Welch AE, Hanson EM, Boothby MR, Keegan AD. Interleukin-4 and interleukin-13 signaling connections maps. Science (2003) 300:1527–8. doi: 10.1126/science.1085458
80. Zhang JL, Buehner M, Sebald W. Functional epitope of common gamma chain for interleukin-4 binding. Eur J Biochem. (2002) 269:1490–9. doi: 10.1046/j.1432-1033.2002.02796.x
81. Zhang JL, Simeonowa I, Wang Y, Sebald W. The high-affinity interaction of human IL-4 and the receptor alpha chain is constituted by two independent binding clusters. J Mol Biol. (2002) 315:399–407. doi: 10.1006/jmbi.2001.5243
82. Andrews AL, Holloway JW, Puddicombe SM, Holgate ST, Davies DE. Kinetic analysis of the interleukin-13 receptor complex. J Biol Chem. (2002) 277:46073–8. doi: 10.1074/jbc.M209560200
83. Andrews AL, Holloway JW, Holgate ST, Davies DE. IL-4 receptor alpha is an important modulator of IL-4 and IL-13 receptor binding: implications for the development of therapeutic targets. J Immunol. (2006) 176:7456–61. doi: 10.4049/jimmunol.176.12.7456
84. Boulanger MJ, Chow D, Brevnova E, Martick M, Sandford G, Nicholas J, et al. Molecular mechanisms for viral mimicry of a human cytokine: activation of gp130 by HHV-8 interleukin-6. J Mol Biol. (2004) 335:641–54. doi: 10.1016/j.jmb.2003.10.070
85. Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat Rev Immunol. (2003) 3:36–50. doi: 10.1038/nri980
86. Chatterjee M, Osborne J, Bestetti G, Chang Y, Moore PS. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science (2002) 298:1432–5. doi: 10.1126/science.1074883
87. Redpath S, Ghazal P, Gascoigne NR. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. (2001) 9:86–92. doi: 10.1016/S0966-842X(00)01919-3
88. Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol. (2004) 22:929–79. doi: 10.1146/annurev.immunol.22.012703.104622
89. Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. (2001) 19:683–765. doi: 10.1146/annurev.immunol.19.1.683
90. Schreiber G, Walter MR. Cytokine-receptor interactions as drug targets. Curr Opin Chem Biol. (2010) 14:511–9. doi: 10.1016/j.cbpa.2010.06.165
91. Yoon SI, Jones BC, Logsdon NJ, Walter MR. Same structure, different function crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain. Structure (2005) 13:551–64. doi: 10.1016/j.str.2005.01.016
92. Jones BC, Logsdon NJ, Josephson K, Cook J, Barry PA, Walter MR. Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proc Natl Acad Sci USA. (2002) 99:9404–9. doi: 10.1073/pnas.152147499
93. Liu Y, de Waal Malefyt R, Briere F, Parham C, Bridon JM, Banchereau J, et al. The EBV IL-10 homologue is a selective agonist with impaired binding to the IL-10 receptor. J Immunol. (1997) 158:604–13.
94. Vieira P, de Waal-Malefyt R, Dang MN, Johnson KE, Kastelein R, Fiorentino DF, et al. Isolation and expression of human cytokine synthesis inhibitory factor cDNA clones: homology to Epstein-Barr virus open reading frame BCRFI. Proc Natl Acad Sci USA. (1991) 88:1172–6. doi: 10.1073/pnas.88.4.1172
95. MacNeil IA, Suda T, Moore KW, Mosmann TR, Zlotnik A. IL-10, a novel growth cofactor for mature and immature T cells. J Immunol. (1990) 145:4167–73.
96. Go NF, Castle BE, Barrett R, Kastelein R, Dang W, Mosmann TR, et al. Interleukin 10, a novel B cell stimulatory factor: unresponsiveness of X chromosome-linked immunodeficiency B cells. J Exp Med. (1990) 172:1625–31. doi: 10.1084/jem.172.6.1625
97. Becker V, Schilling M, Bachmann J, Baumann U, Raue A, Maiwald T, et al. Covering a broad dynamic range: information processing at the erythropoietin receptor. Science (2010) 328:1404–8. doi: 10.1126/science.1184913
98. Bulut GB, Sulahian R, Ma Y, Chi NW, Huang LJ. Ubiquitination regulates the internalization, endolysosomal sorting, and signaling of the erythropoietin receptor. J Biol Chem. 286 6449–57. doi: 10.1074/jbc.M110.186890
99. Keeler C, Jablonski EM, Albert YB, Taylor BD, Myszka DG, Clevenger CV, et al. The kinetics of binding human prolactin, but not growth hormone, to the prolactin receptor vary over a physiologic pH range. Biochemistry (2007) 46:2398–410. doi: 10.1021/bi061958v
100. Claudinon J, Monier MN, Lamaze C. Interfering with interferon receptor sorting and trafficking: impact on signaling. Biochimie (2007) 89:735–43. doi: 10.1016/j.biochi.2007.03.014
101. Shah M, Patel K, Mukhopadhyay S, Xu F, Guo G, Sehgal PB. Membrane-associated STAT3 and PY-STAT3 in the cytoplasm. J Biol Chem. (2006) 281:7302–8. doi: 10.1074/jbc.M508527200
102. German CL, Sauer BM, Howe CL. The STAT3 beacon: IL-6 recurrently activates STAT 3 from endosomal structures. Exp Cell Res. (2010) 317:1955–69. doi: 10.1016/j.yexcr.2011.05.009
103. Reddy CC, Niyogi SK, Wells A, Wiley HS, Lauffenburger DA. Engineering epidermal growth factor for enhanced mitogenic potency. Nat Biotechnol. (1996) 14:1696–9. doi: 10.1038/nbt1296-1696
104. Gandhi H, Worch R, Kurgonaite K, Hintersteiner M, Schwille P, Bokel C, et al. Dynamics and interaction of interleukin-4 receptor subunits in living cells. Biophys J. (2014) 107:2515–27. doi: 10.1016/j.bpj.2014.07.077
105. Payelle-Brogard B, Pellegrini S. Biochemical monitoring of the early endocytic traffic of the type I interferon receptor. J Interferon Cytokine Res. (2010) 30:89–98. doi: 10.1089/jir.2009.0044
106. Avalos BR, Gasson JC, Hedvat C, Quan SG, Baldwin GC, Weisbart RH, et al. Human granulocyte colony-stimulating factor: biologic activities and receptor characterization on hematopoietic cells and small cell lung cancer cell lines. Blood (1990) 75:851–7.
107. Irandoust MI, Aarts LH, Roovers O, Gits J, Erkeland SJ, Touw IP. Suppressor of cytokine signaling 3 controls lysosomal routing of G-CSF receptor. EMBO J. (2007) 26:1782–93. doi: 10.1038/sj.emboj.7601640
108. Gauzzi MC, Barbieri G, Richter MF, Uze G, Ling L, Fellous M, et al. The amino-terminal region of Tyk2 sustains the level of interferon alpha receptor 1, a component of the interferon alpha/beta receptor. Proc Natl Acad Sci USA. (1997) 94:11839–44. doi: 10.1073/pnas.94.22.11839
109. Waters MJ, Brooks AJ, Chhabra Y. A new mechanism for growth hormone receptor activation of JAK2, and implications for related cytokine receptors. JAKSTAT (2014) 3:e29569. doi: 10.4161/jkst.29569
110. Boyman O, Cho JH, Sprent J. The role of interleukin-2 in memory CD8 cell differentiation. Adv Exp Med Biol. (2010) 684:28–41. doi: 10.1007/978-1-4419-6451-9_3
111. Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. (2011) 12:551–9. doi: 10.1038/ni.2030
112. Boyman O, Krieg C, Homann D, Sprent J. Homeostatic maintenance of T cells and natural killer cells. Cell Mol Life Sci. (2012) 69:1597–608. doi: 10.1007/s00018-012-0968-7
113. Ross SH, Cantrell DA. Signaling and function of interleukin-2 in T lymphocytes. Annu Rev Immunol. (2018) 36:411–433. doi: 10.1146/annurev-immunol-042617-053352
114. Pellegrini M, Mak TW, Ohashi PS. Fighting cancers from within: augmenting tumor immunity with cytokine therapy. Trends Pharmacol Sci. (2010) 31:356–63. doi: 10.1016/j.tips.2010.05.003
115. Smith FO, Downey SG, Klapper JA, Yang JC, Sherry RM, Royal RE, et al. Treatment of metastatic melanoma using interleukin-2 alone or in conjunction with vaccines. Clin Cancer Res. (2008) 14:5610–8. doi: 10.1158/1078-0432.CCR-08-0116
116. Klapper JA, Downey SG, Smith FO, Yang JC, Hughes MS, Kammula US, et al. High-dose interleukin-2 for the treatment of metastatic renal cell carcinoma : a retrospective analysis of response and survival in patients treated in the surgery branch at the National Cancer Institute between 1986 and 2006. Cancer (2008) 113:293–301. doi: 10.1002/cncr.23552
117. Boyman O, Surh CD, Sprent J. Potential use of IL-2/anti-IL-2 antibody immune complexes for the treatment of cancer and autoimmune disease. Expert Opin Biol Ther. (2006) 6:1323–31. doi: 10.1517/14712598.6.12.1323
118. Smith KA. Interleukin-2: inception, impact, and implications. Science (1988) 240:1169–76. doi: 10.1126/science.3131876
119. Shanafelt AB, Lin Y, Shanafelt MC, Forte CP, Dubois-Stringfellow N, Carter C, et al. A T-cell-selective interleukin 2 mutein exhibits potent antitumor activity and is well tolerated in vivo. Nat Biotechnol. (2000) 18:1197–202. doi: 10.1038/81199
120. Margolin K, Atkins MB, Dutcher JP, Ernstoff MS, Smith JW, 2nd, Clark JI, et al. Phase I trial of BAY 50–4798, an interleukin-2-specific agonist in advanced melanoma and renal cancer. Clin Cancer Res. (2007) 13:3312–9. doi: 10.1158/1078-0432.CCR-06-1341
121. Rao BM, Driver I, Lauffenburger DA, Wittrup KD. High-affinity CD25-binding IL-2 mutants potently stimulate persistent T cell growth. Biochemistry (2005) 44:10696–701. doi: 10.1021/bi050436x
122. Rao BM, Driver I, Lauffenburger DA, Wittrup KD. Interleukin 2 (IL-2) variants engineered for increased IL-2 receptor alpha-subunit affinity exhibit increased potency arising from a cell surface ligand reservoir effect. Mol Pharmacol. (2004) 66:864–9. doi: 10.1124/mol.66.4
123. Rao BM, Girvin AT, Ciardelli T, Lauffenburger DA, Wittrup KD. Interleukin-2 mutants with enhanced alpha-receptor subunit binding affinity. Protein Eng. (2003) 16:1081–7. doi: 10.1093/protein/gzg111
124. Levin AM, Bates DL, Ring AM, Krieg C, Lin JT, Su L, et al. Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine'. Nature (2012) 484:529–33. doi: 10.1038/nature10975
125. Mitra S, Ring AM, Amarnath S, Spangler JB, Li P, Ju W, et al. Interleukin-2 activity can be fine tuned with engineered receptor signaling clamps. Immunity (2015) 42:826–38. doi: 10.1016/j.immuni.2015.04.018
126. Charych DH, Hoch U, Langowski JL, Lee SR, Addepalli MK, Kirk PB, et al. NKTR-214, an Engineered cytokine with biased IL2 receptor binding, increased tumor exposure, and marked efficacy in mouse tumor models. Clin Cancer Res. (2016) 22:680–90. doi: 10.1158/1078-0432.CCR-15-1631
127. Junttila IS, Creusot RJ, Moraga I, Bates DL, Wong MT, Alonso MN, et al. Redirecting cell-type specific cytokine responses with engineered interleukin-4 superkines. Nat Chem Biol. (2012) 8:990–8. doi: 10.1038/nchembio.1096
128. Lutz MB, Schnare M, Menges M, Rossner S, Rollinghoff M, Schuler G, et al. Differential functions of IL-4 receptor types I and II for dendritic cell maturation and IL-12 production and their dependency on GM-CSF. J Immunol. (2002) 169:3574–80. doi: 10.4049/jimmunol.169.7.3574
129. Shanafelt AB, Forte CP, Kasper JJ, Sanchez-Pescador L, Wetzel M, Gundel R, et al. An immune cell-selective interleukin 4 agonist. Proc Natl Acad Sci USA. (1998) 95:9454–8. doi: 10.1073/pnas.95.16.9454
130. Kurgonaite K, Gandhi H, Kurth T, Pautot S, Schwille P, Weidemann T, et al. Essential role of endocytosis for interleukin-4-receptor-mediated JAK/STAT signalling. J Cell Sci. (2015) 128:3781–95. doi: 10.1242/jcs.170969
131. Francois-Newton V, Magno de Freitas Almeida G, Payelle-Brogard B, Monneron D, Pichard-Garcia L, Piehler J, et al. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon alpha response. PLoS ONE (2011) 6:e22200. doi: 10.1371/journal.pone.0022200
Keywords: cytokine signaling, protein engineering, JAK/STAT signaling pathway, endosomal trafficking, endosomes signaling
Citation: Gorby C, Martinez-Fabregas J, Wilmes S and Moraga I (2018) Mapping Determinants of Cytokine Signaling via Protein Engineering. Front. Immunol. 9:2143. doi: 10.3389/fimmu.2018.02143
Received: 30 May 2018; Accepted: 30 August 2018;
Published: 27 September 2018.
Edited by:
Jorge Bernardino De La Serna, United Kingdom Research and Innovation, United KingdomReviewed by:
Mark Walter, University of Alabama at Birmingham, United StatesJan Tavernier, Ghent University, Belgium
Copyright © 2018 Gorby, Martinez-Fabregas, Wilmes and Moraga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ignacio Moraga, imoragagonzalez@dundee.ac.uk
 Claire Gorby
Claire Gorby Ignacio Moraga
Ignacio Moraga