- 1Department of Physiology, McGill University, Montreal, QC, Canada
- 2Department of Microbiology and Immunology, McGill University, Montreal, QC, Canada
- 3McGill Research Center for Complex Traits, McGill University, Montreal, QC, Canada
- 4Department of Microbiology and Immunology, Stanford University, Stanford, CA, United States
- 5Australian Animal Health Laboratory, Health and Biosecurity Business Unit, Commonwealth Scientific and Industrial Research Organisation, Geelong, VIC, Australia
A majority of viruses that have caused recent epidemics with high lethality rates in people, are zoonoses originating from wildlife. Among them are filoviruses (e.g., Marburg, Ebola), coronaviruses (e.g., SARS, MERS), henipaviruses (e.g., Hendra, Nipah) which share the common features that they are all RNA viruses, and that a dysregulated immune response is an important contributor to the tissue damage and hence pathogenicity that results from infection in humans. Intriguingly, these viruses also all originate from bat reservoirs. Bats have been shown to have a greater mean viral richness than predicted by their phylogenetic distance from humans, their geographic range, or their presence in urban areas, suggesting other traits must explain why bats harbor a greater number of zoonotic viruses than other mammals. Bats are highly unusual among mammals in other ways as well. Not only are they the only mammals capable of powered flight, they have extraordinarily long life spans, with little detectable increases in mortality or senescence until high ages. Their physiology likely impacted their history of pathogen exposure and necessitated adaptations that may have also affected immune signaling pathways. Do our life history traits make us susceptible to generating damaging immune responses to RNA viruses or does the physiology of bats make them particularly tolerant or resistant? Understanding what immune mechanisms enable bats to coexist with RNA viruses may provide critical fundamental insights into how to achieve greater resilience in humans.
Introduction
An estimated ~60% of emerging infectious diseases are caused by pathogens which originate from a non-human animal source, referred to as zoonoses (1–3). Moreover, the frequency of outbreaks caused by zoonotic pathogens has been increasing over time in the human population, with viruses being the most successful at crossing the species barrier (2–4). Given the impact of viral zoonoses on global public health, considerable resources have been invested into better understanding patterns in their emergence to improve predictions of where they might arise. One key variable in such predictions is to determine the animal reservoir populations within which these novel viruses can be maintained indefinitely (with or without disease) and which therefore act as sources for transmission to humans (5). In some instances, epidemiological associations may provide clues to identifying a reservoir host species, and the detection of natural infection through seroconversion or the virus itself provides further evidence. Recently, phylogenetic analyses have also been used to investigate viral origins—with a presence of greater diversity and of strains ancestral to those in humans being indicative of a virus circulating within a particular natural host population (6).
Once identified, viral reservoirs have historically been critical levers through which to reduce human cases (5). However, reservoir hosts may also provide us with fundamental insights into host-pathogen interactions and are a rich opportunity to examine the immunological processes that contribute to patterns governing which pathogens cross into humans, cause disease and why (7, 8). This can be particularly informative as in many instances, the zoonotic viruses that are so pathogenic in humans do not cause disease in the reservoirs with which they coexist.
Bats Are the Reservoirs for Many Human Viruses
Bats have been confirmed as reservoir hosts for many viruses, several of which are associated with fatality rates as high as 90% among diagnosed human cases. It has long been appreciated that rabies and other lyssaviruses causing lethal encephalitis can be transmitted from numerous bat species (9, 10). Live Marburg virus (MARV) has been isolated from Rousettus aegyptiacus fruit bats which, jointly with epidemiologic evidence and detection of viral RNA, strongly suggests that R. aegyptiacus is a reservoir host of this filovirus (11). The related ebolavirus (EBOV) likely also circulates in African fruit bats, with a few species having been implicated so far—the mobility of which accounts for the sudden appearance of Ebola in West Africa during the 2014 outbreak, a region where ebolavirus had not previously been detected (12, 13). The highly pathogenic henipaviruses, of which Hendra virus emerged in Australia and Nipah virus in South-east Asia via horse and pig intermediate hosts respectively, have been shown to be transmitted from Pteropus bats (14, 15). In China, horseshoe Rhinolophus bats have been identified as the reservoirs for SARS coronavirus via palm civet intermediate hosts, the cause of a large outbreak of atypical pneumonia across several countries that began in 2002 in China (16–18). More recently, MERS coronavirus that has caused lethal respiratory infections mostly in Saudi Arabia, likely transmitted via dromedary camels, was shown to be closely related to several bat coronaviruses, including those sequenced from Neoromicia capensis, Pipistrellus abramus, and Vespertilio superans bats (19, 20). Moreover, additional viruses may continue to emerge from bats, as in the single case of sosuga virus infection in a wildlife biologist collecting bats in South Sudan (21).
In addition to these emerging zoonotic viruses, bats may be the source of a number of viruses with which humans have older evolutionary associations. For instance, bats harbor viruses closely related to both mumps (rubula virus) and measles (morbilli virus) and have likely been donors of these viruses to other mammalian groups, possibly including humans (6, 22). Furthermore, both Old and New World bats carry diverse hepadnaviruses, some of which are related to hepatitis B virus and can infect human hepatocytes (23). Hepaciviruses that are related to hepatitis C virus and pegiviruses that are related to human GB viruses were detected in the sera of many different bat species, and given the basal position of these bat viruses in phylogenetic trees, may also represent strains ancestral to those found in humans (24, 25).
The preponderance of links between bat and human pathogens has led to a debate about whether bats disproportionately contribute to emerging viral infections crossing the species barrier into humans (26–30). Given the diversity of the Chiroptera order (Figure 1), we may simply see more bat viruses because there are so many (>1,300) species of bats (31). However, even when accounting for the fact that they make up ~20% of extant terrestrial mammals, bats are overrepresented as reservoir hosts of pathogens with a high potential for spilling into human populations (32, 33). In fact, no known predictors that have been described to impact the likelihood of crossing the species barrier, including reservoir host ecology, phylogenetic relatedness to humans or frequency of reservoir-human contact, explain this pattern (32). Thus, why bats are such a frequent source of pathogenic human viruses remains a tantalizing mystery.
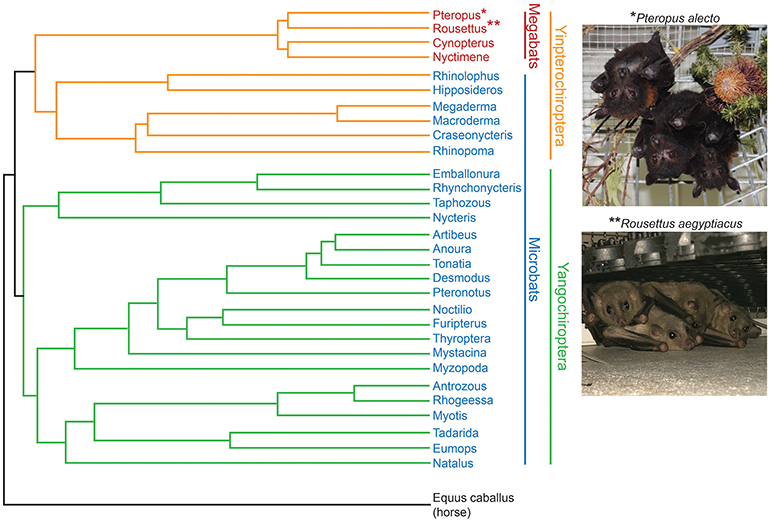
Figure 1. Phylogeny of bat genera [modified from Teeling et al. (31), note that branch lengths are not to scale] indicating the two most studied bat species so far, Pteropus alecto and Rousettus aegyptiacus. Photos provided by Susanne Wilson, CSIRO, Australia (Pteropus); Anne Balkema-Buschmann and Nils Kiley, Friedrich-Loeffler-Institut, Germany (Rousettus).
Among viruses, those that have genomes encoded by RNA generally jump across species boundaries more frequently, presumably due to their inherently greater mutation rates that facilitate the rapid adaptation to replicating within new hosts (34). Interestingly, all pathogenic viruses that have made the jump to humans for which bat species may be reservoirs share the common feature that they have single-stranded RNA genomes (with the exception of hepadnaviruses which have a DNA genome but replicate via an RNA intermediate). So far, available evidence suggests that bats remain disease-free when infected with the RNA viruses they carry—even those highly pathogenic to humans—and are able to coexist with them without detectable fitness costs using measures such as changes in temperature, loss of body weight, or overt signs of inflammation (35). Indeed, so far only one RNA virus studied which circulates in a bat population has been shown to consistently cause significant morbidity and mortality: tacaribe virus in the Jamaican fruit bat (Artibeus jamaicensis), which recent evidence suggests is not a reservoir host for this virus (36). Data from experimental rabies and lyssavirus infections suggests that rhabdoviruses may also cause disease in bats, although experimental infection outcome is very dependent on the infection route. Intracerebral infection with different strains and in different bat species invariably led to death (37, 38). In contrast, intramuscular infection led to muscle weakness, paralysis and visible histological CNS lesions in 30% of experimentally infected flying foxes (Pteropus poliocephalus) (39). Similarly, a subset of vampire bats (Desmodus rotundus) experimentally infected intramuscularly with a high dose of rabies virus remained healthy despite viral shedding in the saliva and survived (40). Naturally infected bats are thought to either die or remain healthy and seroconvert, but transmission in free-ranging populations remains incompletely understood (41).
While bats seem to be frequent hosts for RNA viruses, current available data indicates that primates and humans disproportionately harbor DNA viruses such as herpesviruses (32). Interestingly, it is these DNA viruses that can persist in an individual which can also be found in isolated, small indigenous groups—perhaps suggestive of humans having a more ancient relationship with such DNA viruses (42). It may even be the case that persistent DNA viruses in humans impact immune responses specifically to RNA viruses, but this has not yet been examined. It is likely that differences in evolutionary history of pathogen exposure between bats and humans have led to distinct adaptations in anti-viral immune responses and the ability to tolerate certain infections without disease while being susceptible to others. Importantly, bats differ in many aspects of their physiology and behavior from humans that may have direct or indirect effects on immune function.
Bat Life History Traits
Bats are a monophyletic mammalian group traditionally divided by morphological data into two suborders, the megabats and microbats, which more recent molecular data has revised into the Yinpterochiroptera and Yangochiroptera suborders (Figure 1). Bats possess a suite of traits that make them distinct from other mammals in a number of ways. These unique life history traits may play a role in understanding which pathogens bats have evolved to coexist with and why. In particular, such traits may explain the ability of bat populations to maintain particular viral pathogens indefinitely, and may have effects on immune function through specific energetic or evolutionary trade-offs we have yet to better define.
Longevity, Metabolic Rate, and Hibernation
Despite the diversity of viruses carried by bats, they are not typically known to cause mass bat die-offs or reduce bats' remarkable longevity. In this respect, bats represent a potential opportunity for long-term persistence of viruses within a population and across generations. Bats live significantly longer than similarly-sized terrestrial mammals and, despite their small size, are characterized as “slow” mammals in the slow-fast continuum (43, 44). Although their weights range from 2 grams to 2 kilograms, with respect to longevity bats group with large mammals such as humans and non-human primates (45). Aerial living has an obvious advantage in avoiding predation, but bats outlive even birds. For example, the Brandt's bat (Myotis brandtii) lives up to 41 years, compared to Selasphorus platycercus, a bird species of similar size that lives for ~14 years (45, 46). Thus, flight can only partially account for their extraordinarily long lives. Initially, the longevity of some bats was attributed to seasonal hibernation, as temperate-zone species enter continuous torpor of up to 75 days, with a dramatic drop in metabolic rate such that small fat reserves can sustain them throughout the entire hibernating season (43). However, even non-hibernating bat species live three times longer, on average, than predicted by their size, and heterothermy is not an accurate predictor of lifespan in other mammalian orders, suggesting that the driving force behind their surprising longevity is intrinsic to bats as a group (47–49).
Like other “slow” mammals, bat females typically only have one offspring per year, perhaps because the volant lifestyles of bats make it difficult to rear more than one offspring, as pregnant females and those with recent births must navigate and forage with added weight; on average, neonatal bat pups are ¼ of their mother's weight (50). The physical and energetic constraints of rearing multiple offspring may necessitate small litters, which would in turn require prolonged reproductive capability and enhanced longevity to ensure maintenance of the population over generations. Thus, in bats, the dependence of colony survival as a whole may depend upon enhanced individual survival and delayed senescence (51). Genetic analyses of several bat species have shown differences in the growth hormone (GH)/insulin-like growth factor 1 (IGF1) axis which in humans is associated with aging, resistance to diabetes and cancer (52).
The determinants of adult survival in bats have been historically difficult to identify, as this requires tracking individuals over many years, and until recently longitudinal studies of bat mortality were conducted using tagged bats, of which only a fraction were recovered (53). Recently, a 19-year study of a colony of Bechstein's bats demonstrated that unlike terrestrial mammals, survival could not be predicted by common indicators such as season, age, and body size. Instead, the only accurate predictor of mortality was a single cataclysmic weather event that affected multiple countries in north-central Europe. Additionally, even the oldest female bats were reproductively capable, indicating that bat survival is primarily affected by catastrophic natural events rather than factors that normally dictate an individual's fitness (45).
Echolocation
Molecular phylogenetic studies of bats suggest that there are massive gaps in bat fossil records. As bats are the second most diverse order of mammals, outnumbered only by rodents, the number of species unrepresented in the fossil records is staggering. Over half of microbat and nearly all of megabat fossil histories are missing (31, 54). The enormous incompleteness of the fossil records has made it difficult to identify when specific morphological traits of bats arose. As molecular phylogeny groups two echolocation-reliant microbat species with megabats (also called Old World bats or pteropodids), which do not rely on echolocation, there is some debate as to whether echolocation first arose in the common ancestor of bats and was subsequently lost in megabats, or whether it arose twice, independently (31). Pteropodids have adaptations that enhance visual acuity at night (55), and they do not require echolocation for foraging (56). There are multiple types of echolocation that can be partially delineated by species, but are more clearly categorized by the type of environment. Divergent species that inhabit the same type of environment, such as those that hunt in large, open spaces, often use the same form of echolocation, suggesting that habitat has a greater influence on echolocation than phylogeny (31). Importantly, echolocation can result in the production of droplets or small-particle aerosols of oropharyngeal fluids, mucus, or saliva, thus facilitating transmission of viruses between individuals in close proximity (57, 58). The unique navigation tactic of many bat species may inadvertently facilitate virus transmission among bats in the same habitat.
Flight
Bats are the only mammal capable of powered flight, which likely evolved ~65 million years ago alongside birds following radical ecological changes that resulted in the extinction of the dinosaurs (54, 59). During flight, bats consume approximately four times as much oxygen, and they have a markedly higher concentration of red blood cells compared to small terrestrial mammals (60). Bat flight is markedly different from that of birds and insects, whose wing surfaces are typically composed of inflexible material, such as feathers or chitin. Bat wings are constructed from live skin stretched across elongated arm and finger bones, making them extraordinarily malleable and sensitive to environmental cues (59). The plasticity of bats' wings allows them to navigate and inhabit diverse ecospheres, contributing to their extensive speciation. Moreover, the capability of powered flight can allow the efficient spread of viruses and thus the introduction of pathogens to which colonies may otherwise have remained naïve.
As flight is extremely metabolically demanding, in addition to evolving the physical mechanisms required for flight, bats have also evolved necessary underlying molecular mechanisms. The mitochondrial respiratory chain accounts for nearly all ATP required for mobility in eukaryotes, and genetic analysis of both micro- and megabat species revealed an enrichment of genes specific to the oxidative phosphorylation (OXPHOS) pathway. Specifically, 4.9% of nuclear-encoded and 23% of mitochondrial OXPHOS genes have evidence of positive selection in bats, which is markedly higher than the expected 2% of orthologous genes in previous genome-wide studies that show evidence of positive selection (61). Genomic analysis of Pteropus alecto and M. davidii suggests positive selection for the DNA damage checkpoint pathway and changes in overlapping aspects of this pathway with the innate immune system, indicating that evolutionary adaptations important for flight may have secondarily affected bat immunity (62).
Social Interaction and Communities
As a group, bats exhibit the greatest diversity of social systems in mammals. Tropical species are primarily responsible for this diversity, as temperate species are more restricted in their social behavior. Generally, however, bats are extremely social creatures that tend to form dense roosting colonies (63), and almost all temperate-zone species live in closed societies with very little infiltration of foreign bats into established roosts (63, 64). In particular, female bats form maternity colonies in which males do not take part. As bats are capable of long-distance flight, dispersal barriers cannot explain the philopatry of females. Instead, benefits such as knowledge of foraging areas and social thermoregulation likely selected for these colony types. Additionally, there is evidence that forming closed societies limits the potential invasion of new pathogens, thereby protecting colony members that would otherwise be vulnerable to infection. For example, Pseudogymnaoscus destructans has decimated North American bat populations that do not live in the type of closed societies observed elsewhere (64). DNA analysis of a closed society of Bechstein's bats revealed extraordinarily high conservation of mitochondrial DNA and relatively low conservation of nuclear DNA, suggesting stable maternal populations within colonies and gene flow between colonies via promiscuous mating with males. It is possible that the mating patterns of temperate-zone species may allow transmission of pathogens between colonies via traveling males while the more insular females may allow viruses to persist throughout generations within a colony.
Anti-Viral Immune Responses of Bats
An important commonality among pathogenic RNA viruses in humans presenting with disease is that the host response is an important contributor to the disease process, with dysregulated and excessive innate immune responses being particularly important drivers of tissue damage during infection (8). Given the general absence of clinical signs of disease in bats infected with the same viruses that are so lethal in humans or other non-natural hosts infected experimentally, a critical question has been to understand whether bats might establish effective disease tolerance, thus maintaining fitness despite pathogen replication, or whether bats are more resistant to infection through more successful control of pathogen replication and what the contribution of the immune response is (65, 66). The lack of many fundamental immunological tools enabling the probing of bat immune responses has meant that truly mechanistic studies of bat immunity have been very limited, although recently there has been some progress in establishing approaches such as flow cytometry to identify distinct bat immune cell populations (67, 68). So far, studies of bat immunity have primarily taken one of three approaches, whereby each comes with important strengths and weaknesses that have to be kept in mind: (i) comparative genome studies, (ii) in vitro cell culture assays, and (iii) experimental infections.
Comparative genome studies have confirmed that the critical components of the innate and adaptive immune system are conserved in bats at the gene level and that bats have the machinery for innate responses to pathogen-associated molecular patterns (PAMPs), the production of anti-viral effector molecules such as type I interferons (IFN), T cell responses (variable T cell receptors, MHCI and MHCII), and B cell responses [reviewed in (35)]. Interestingly, based on the 10 bat genomes sequenced so far, the only family of genes lost entirely in all of them are PYHIN genes (69). Members of the PYHIN family are DNA sensors capable of recognizing foreign DNA, including DNA viruses and damaged self DNA which can be generated by RNA viral infection. Recognition of DNA results in production of IFN through interaction with stimulator of interferon genes (STING). The PYHIN family also encode the only identified class of DNA sensors capable of activating the inflammasome. It has been hypothesized that the absence of the PYHIN family may allow bats to limit activation of the innate immune response to damaged self-DNA generated by RNA viral infection, thus avoiding excessive inflammation (69, 70). Genome comparisons highlighting contractions or expansions of specific gene families, specific genes under positive selection, or non-conserved sequence differences in critical protein domains can thus provide the basis for hypotheses worth testing further. However, it is important to note that much can be missed in absence of data on gene regulation, especially during infection when gene expression kinetics can make a critical difference to the infection outcome. Moreover, the absence of a gene or gene family does not rule out that other proteins have evolved to compensate for their loss of function. Thus, while whole genome analyses can provide a context for specific questions or be hypothesis-generating, on their own they cannot distinguish tolerance from resistance mechanisms. The repeated identification of signatures of positive selection in innate immune genes in particular, does however lend credence to the idea that bats have specific adaptations as a result of a long co-evolutionary history with viruses.
Cell culture assays with bat cell lines, or, in some instances, primary bat cells, have been used to assess whether bats are permissive for viral replication and to determine whether particular immune receptor signaling pathways are intact. As discussed below, such studies have probed the type I IFN pathway in particular, revealing some possible species-specific differences among bats (71–83). However, it is important to note that in some instances immortalized cells can behave differently from primary cells and that such cultures may miss additional differences imposed by changes in cell localization, cell recruitment or cell-cell interactions in a whole animal. Careful experiments measuring the quality, magnitude, and kinetics of immune responses in bats during infection and upon administration with defined stimuli for which we have comparative information from humans remain to be done to provide additional evidence that specific innate immune pathways are wired differently.
Experimental infections come with the enormous challenge of having to house and/or breed colonies of bats and to have biosafety-level 4 facilities in place to perform infections with viruses lethal to humans. Moreover, some trial and error is involved in determining which route and dose leads to viral replication, establishing a source of the virus (human-adapted strains tend to replicate less well in bats than strains obtained from naturally infected bats), and amplifying this viral stock without extensive tissue culture passaging. Studies to date have examined the kinetics of viral replication by quantifying the extent of viremia and dissemination to other tissues, and assessing changes in white blood cell counts, body mass, and temperature. Given the generally low levels of viral shedding and short infectious periods observed so far it remains poorly understood how transmission occurs in the wild to sufficient levels that cross-species jumps occur. Some infection experiments have also provided evidence that a particular bat species is unlikely to be a reservoir despite epidemiological evidence, for example for R. aegyptiacus and ebolavirus. Certainly, once good experimental infection models are established, such studies have the potential to be hugely informative with regard to anti-viral immune responses elicited using, for instance, comparative transcriptome analyses. One drawback may be that experimental infections do not mimic the impact of chronic stress arising from the disruption of wildlife populations, which bats are particularly sensitive to Jones et al. (84). Comparison of either cave-roosting or foliage-roosting species in areas of Malaysian Borneo designated as actively logged forest, recovering forest, or fragmented forest revealed varying impacts of habitat disturbance on stress and circulating white blood cells (85).
Overall, the limited studies of bat immunity that have been done have focused largely on 2 species: P. alecto and R. aegyptiacus. We summarize this work below, but comparisons of observations made across species suggest that although a number of species appear to be capable of avoiding the pathological effects of RNA virus infection, each bat species may have achieved this through distinct pathways, possibly involving changes to both increase pathogen replication control and to mitigate any immunopathology through decreased inflammatory responses and hence increased disease tolerance.
Pteropid Bats
The most well studied bat species with regard to antiviral immune responses is the Australian black flying fox (P. alecto). This interest has stemmed from the fact that pteropid bats have been identified as the natural reservoirs for the deadly Hendra and Nipah viruses (86), which continue to cause outbreaks [such as most recently in India in May 2018 (87)]. To date, several studies have examined the kinetics of viral infection in Pteropus bats and the nature of transmission and replication in other susceptible species (88–91). In Australia, all four species of pteropid bats (P. alecto, P. poliocephalus, P. scapulatus, and P. conspicillatus) have antibodies to Hendra virus but only P. alecto and P. conspicillatus are considered to be the primary reservoir hosts (14, 92, 93). In South East Asia, both pteropus spp. occurring in Malaysia have been found to be seropositive for Nipah virus neutralizing antibodies, and the virus has been isolated from P. hypomelanus and P. vampyrus (15, 94).
Experimental infections of pteroid bats with Hendra or Nipah virus result in sub-clinical infection with short periods of virus replication and shedding, and low antibody titres (88–91). Upon subcutaneous infection of P. poliocephalus with Hendra virus, viral antigen was detected by immunohistochemistry at 10 dpi in blood vessels of spleen, kidney and placenta (89). Similarly, oronasal Hendra virus infection of P. alecto led to the presence of viral genome in lung, spleen, liver and kidney 3 weeks later, but virus isolation was unsuccessful at this timepoint (89, 91). The Malaysian flying fox, P. vampyrus and the Australian species, P. poliocephalus demonstrate similarly short periods of viremia upon infection with Nipah virus. In subcutaneously infected P. poliocephalus, virus was isolated from the kidney and uterus of bats euthanized at 7dpi, but no virus was isolated at any of the other timepoints examined (3, 5, 10, 12, or 14 dpi) and there was no evidence of antigen in any tissue by immunohistochemistry, including tissues collected at 7 dpi. In this study, low neutralizing antibodies were detected in all bats with the exception of one individual that developed a significant neutralizing antibody titre — possibly reflecting the fact that P. poliocephalus is not the natural host for Nipah virus (90). In P. vampyrus challenged by oronasal Nipah inoculation, viral genome was detected in a throat swab at 4 dpi and a rectal swab of the same individual at 8 dpi but virus was undetectable in tissues collected at post-mortem from all individuals (49, 50, or 51dpi), consistent with a short period of viremia. Similar to previous studies, antibody titres were low in all P. vampyrus bats (91). Overall, these results are consistent with bats controlling replication rapidly, at least following experimental infections which involve higher doses of virus compared to what bats would likely be naturally exposed to in the wild. The absence of a robust antibody response also appears to be typical of all experimental Hendra and Nipah virus infections performed to date. Since antibody responses are the only immune parameter that has been measured during experimental infections of bats so far, it is difficult to speculate on the mechanisms responsible for control of viral infections in vivo.
Pteropus alecto was among the first bat species to have its genome described in detail. Genomic studies provided initial clues for possible differences in the innate immune system of bats, with evidence for selection of key innate immune genes and the expansion or contraction of specific immune gene families (62, 68, 95). The MHCI region is contracted (96), as is the type I IFN locus, which in P. alecto contains fewer IFN genes than any other mammalian species sequenced, with only three functional IFN-α loci (68). In contrast, pteropid bats have the largest and most diverse family of APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) proteins identified in any mammal (95). APOBECs interfere with the replication of retroviruses by deaminating cytosine residues in nascent retroviral DNA. This is notable, as bats are an important source of mammalian retroviruses, many of which have been transmitted to other mammals (97, 98). APOBEC diversification may therefore have occurred to counteract the effect of retroviruses and possibly other viruses, as APOBECs have been shown to restrict the replication of other virus families including hepadnaviruses, and parvoviruses (99, 100). Members of the APOBECA3 protein family exhibit direct antiviral activity through DNA cytosine deamination which results in hypermutation of the nascent retroviral DNA which is then degraded or rendered non-functional (101). The mechanism of antiviral activity against non-retroviruses remains largely unknown. For parvovirus adeno-associated virus, APOBEC meditated inhibition has been speculated to involve direct interaction with the viral DNA or the replication machinery (102). Whether the expanded family of ABOBECs in bats have evolved other mechanisms to control DNA and RNA viruses remains to be determined. As APOBECs can be induced by even low levels of type I IFN (103), one hypothesis to be tested is that bats, through their multiple APOBECs, are able to restrict viral replication without causing inflammation. Pteropus alecto is the only bat species to date in which APOBEC genes have been mapped, and whether the expansion of this gene family extends to other bat species remains to be determined.
In addition to the identification of putative immune pathways distinct in P. alecto through genome studies, differences have been identified in the activation of innate immune effectors in P. alecto from studies performed in vitro, primarily using cell lines derived from tissues including the kidney and lung. IFNs are the first line of defense following viral infection and unsurprisingly, because of this, they have been the most extensively studied group of genes in bats. Both type I (IFNA and IFNB) and III (IFNL) IFNs are detectable in bat cells. Curiously, a unique characteristic of pteropid bats is the constitutive expression of mRNA for IFNA and the signaling molecule, IFN regulatory factor 7 (IRF7) in unstimulated tissues and cells [75, 68a]. Constitutively expressed IFNA and IRF7 may allow bats to respond more rapidly to infection, thus avoiding the lag time between pathogen detection and response. Furthermore, viral infection or stimulation with synthetic ligands result in little IFNA induction in pteropid bat cells (68). The constitutive expression of IFNA has been described in two species of pteropid bats (P. alecto and Cynopterus brachyotis) and is a first for any species. IFNB and IFNL are activated following stimulation of cells from P. alecto and P. vampyrus with synthetic ligands such as polyIC (71–74). Moreover, bat IFNs demonstrate antiviral activity (68, 71–74, 104). However, viral infection of P. alecto splenocytes results in induction of IFNL but not IFNB, hinting at differences in the function of type I and III IFNs (74). In humans and mice, IFNL has recently been demonstrated to have a role not only in controlling virus replication, but also in dampening damage-inducing neutrophil functions and in modulating tissue-damaging, transcription-independent responses such as production of ROS (77, 80). A hypothesis yet to be tested is whether upregulation of IFNL rather than IFNB has a similar function in bats.
The endoplasmic reticulum (ER) membrane protein, STING, is involved in induction of type I IFN by cytosolic DNA (105). Stimulation of bat splenocytes with GMP-AMP, which is produced following sensing of cytosolic DNA by cGAS, results in little induction of IFN compared to responses observed in mouse splenocytes (83). Bat STING contains an amino acid substitution of the highly conserved and functionally important serine residue S358 which may be responsible for dampening STING-dependent IFN activation in bat cells in response to DNA. However, comparable levels of IFN induction in mouse and bat cells in response to the RNA viral mimic polyIC indicate that STING-associated inhibition of the IFN response does not extend to RNA viruses (83), thus the relevance to RNA viruses in bats remains unknown.
Downstream of the induction of IFNs, novel subsets of IFN stimulated genes (ISGs) have been detected in unstimulated and stimulated pteropid bat cells indicative of a response that is less damaging to the host. Furthermore, the ISG response is elevated for a shorter period of time in P. alecto compared to human cell lines which again may be a strategy to avoid tissue damage (78, 81). The less inflammatory profile of ISGs may be the key to the ability of bats to tolerate higher IFN expression without adverse consequences. The balance between resistance and tolerance may therefore be achieved through careful selection of the pathways that are activated and shorter periods of activation or limited activation to prevent inflammation. In this regard, studies of the regulation of IFN signaling in bats is likely to provide important additional insights.
Rousettus Bats
A second bat species whose host responses to viral infections has been studied more recently is the Egyptian fruit bat (R. aegyptiacus). Marburg virus (MARV) has been repeatedly isolated from this species with demonstrated seasonal pulses of active MARV replication in juvenile bats living in caves in Uganda (11, 106). Moreover, R. aegyptiacus were a suspected reservoir for ebolavirus (EBOV) based on epidemiological evidence and detected seroreactivity to EBOV, but no infectious virus has been isolated thus far from wild rousettus bats (107). Indeed, while cell lines from R. aegyptiacus are equally susceptible to MARV and EBOV (79, 108), experimental infections of R. aegyptiacus seem to confirm that it is a reservoir for MARV, but is unlikely to be the source of EBOV spillover to humans. Subcutaneous EBOV infection results in very low viral replication, no viremia, little dissemination to other tissues, and no viral shedding, although some animals seroconvert, suggesting that R. aegyptiacus are unlikely to perpetuate EBOV in the wild (109, 110). In contrast, experimental MARV infection of R. aegyptiacus resulted in acute viremia that peaked on days 5–6 post-infection (although generally at lower levels than in humans), oral shedding that peaked on days 7–8 post-infection, and dissemination to other tissues including spleen, liver, kidney and salivary glands (109, 111–113). Interestingly, viral replication was not associated with increases in white blood cell counts, any clinical signs of infection such as changes in body temperature or body weight, and infected tissues showed little evidence of inflammatory infiltrates (109). In all experiments, viremia was cleared by day 13 and oral shedding ceased by day 19. Intriguingly, a cohousing experiment resulted in MARV transmissions to uninfected bats 4–7 months after experimental infection, raising the question of whether persistent infection with intermittent shedding is possible or whether very long latent periods without detectable viral replication could follow exposure (114). Upon secondary challenge of previously MARV-infected bats, none showed any detectable viral replication or shedding, providing evidence that protective immunity is established (115).
Unlike for pteropus bats, no constitutive expression of type I IFNs has been detected in R. aegyptiacus (79), but type I IFNs are induced in R. aegyptiacus cell lines upon stimulation with Sendai virus as seen in other mammals (82). Furthermore, in R. aegyptiacus the type I IFN genes are expanded, again in contrast to P. alecto (82), but like for P. alecto a number of genes in the type I IFN pathway or involved in innate immune recognition of PAMPs show signs of having been under positive selection (82). Whether positive selection of genes in either bat species is associated with tolerance remains to be determined, especially given that innate immune genes in humans have also been under positive selection (116). A transcriptome study which generated 20 RNA sequencing libraries from 11 tissues taken from 1 female and 1 male R. aegyptiacus found a reduced coverage of NK cell related genes compared to other mammals, but confirmed that in these bats the predominant T cells had an αβ T cell receptor, and showed that IgE, IgG, IgM, and IgA, as well as a number of pro- and anti-inflammatory cytokines, were all detectable (117). The recently sequenced R. aegyptiacus genome revealed substantial differences in the repertoire of NK cell receptors, with this bat species entirely lacking functional killer cell immunoglobulin receptors (KIRS) and with all killer lectin-like receptors (KLRs) encoding either activating and inhibitory interaction motifs, or inhibitory interaction motifs only (82). NK cells are important immune cell players in an antiviral response but without assessment of the consequences of these genomic differences it is difficult to draw any specific conclusions with regard to viral control or the magnitude of inflammation elicited upon infection with viruses like MARV. Nonetheless, these genomic data provide some interesting hypotheses to be tested in the future.
Other Bat Species
Some additional studies probing the induction of cytokines upon stimulation of bat cells with defined innate immune stimuli provides some evidence that innate immune recognition of viruses may be altered, leading to a reduction in pro-inflammatory responses. Stimulation of kidney and myeloid cells from the big brown bat (Eptesicus fuscus) with polyinosinic-polycytidylic acid (polyI:C) resulted in only limited activation of the inflammatory cytokine, tumor necrosis factor alpha (TNFα) compared to human cells which display a robust TNFα response. Induction of TNFα is controlled by transcription factors, including the NF-kappa B (NF-κB) family which consists of five members, [RelA (p65), RelB, c-Rel, NFκB-1 (p50), and NFκB-2 (p52)] which form homo- or hetero-dimers that are bound by molecules of the inhibitor of NFκB (IκB) family and retained in the cytoplasm of the cell in an inactivated state (118). In E. fuscus, a potential repressor (c-Rel) binding motif was identified in the TNFα promoter region which may explain the difference in induction of TNFα in E. fuscus cells. Consistent with this hypothesis, partial knockdown of c-Rel transcripts significantly increased basal levels of TNFα transcripts in E. fuscus cells (104). The transcription factor, c-Rel has also undergone positive selection in the bat ancestor which may indicate that this mechanism is common to other species of bats (62). Of note, low levels of TNFα induction have also been associated with tolerance in European bank voles which are a natural reservoir for Puumala hantavirus (PUUV) (119).
Stimulation of macrophages from the greater mouse eared bat (Myotis myotis) suggested that this species may have also evolved mechanisms to avoid excessive inflammation caused by cytokines. While high levels of TNFα, IL1β, and IFNβ were produced in response to in vitro challenge with lipopolysaccharides (LPS) and PolyI:C, there was also a sustained, high-level transcription of the anti-inflammatory cytokine IL-10, which was not observed in mouse macrophages (120). Furthermore, unlike in the mouse, M. myotis macrophages did not produce the proinflammatory and cytotoxic mediator, nitric oxide, in response to LPS. The same study also showed evidence of bat specific adaptations in genes involved in antiviral and pro-inflammatory signaling pathways through comparison with other mammalian taxa, including RIG-I, IL1b, IL-18, NLRP3, STING, and CASP1, further supporting the evolution of adaptations associated with reducing inflammatory responses in bats (120).
Bat Immune Responses to Non-Viral Pathogens
Even less is known about immune responses of bats to non-viral pathogens than to viral pathogens, but it is clear that while anti-inflammatory responses may be characteristic of anti-viral responses in bats, they are susceptible to disease upon infection with particular pathogens—in some instances due to dysregulated and damaging immune responses. One particular example of this is the emerging infectious disease, white nose syndrome (WNS), that has decimated North American bat populations beginning in 2006, in what will likely rank as one of the most devastating wildlife diseases in history (121–123). For reasons that remain poorly understood, the psychrophilic fungus Pseudogymnoascus destructans (formerly Geomyces destructans) causes no mass mortality in European bats despite being abundantly detected (124, 125). Indeed, evidence suggests that a single P. destructans genotype was introduced to North American bat species from Europe (125). In North America, P. destructans infection is not specific to a particular bat genus, replicating in many different bat species during hibernation and targeting the furless skin of the wings, ears, and muzzle (126). Distinct hypotheses have been proposed for why P. destructans is so deadly in North American bats, ascribing the impaired tolerance to infection compared to European bat counterparts to either physiological or immunological factors. On the one hand, more frequent arousal, electrolyte depletion, and dehydration are thought to contribute to mortality following infection (127, 128). The destruction of wing tissue in WNS results in a marked electrolyte imbalance, as the wings play a critical role in maintaining water levels, especially during hibernation, during which bats are particularly vulnerable to dehydration (129, 130). Dehydration catalyzes arousal in hibernating bats, which is extraordinarily metabolically costly and rapidly depletes the fat reserves necessary to survive until spring (127). An alternative hypothesis posits that the restoration of the immune system following emergence from hibernation induces the fatal pathology of WNS. During hibernation, destruction of cutaneous tissue is limited and infiltrating immune cells are entirely absent, yet in the weeks following arousal, infected bats exhibit overt wing damage and corresponding neutrophilic and lymphocytic infiltration (131). Hibernation does not preclude a localized immune response to P. destructans at the site of infection and transcriptomic analysis of infected tissue showed upregulation of some acute inflammatory genes in infected tissue (132, 133). However, the observed immune responses likely occur during arousal periods, which are more common in infected bats. Ultimately, immunosuppression during torpor allows P. destructans to colonize infected bats relatively unchecked (124), and upon emergence from hibernation, the exuberant immune response may result in deadly immunopathology during WNS (131).
In addition to general studies of immune cell recruitment and transcriptional responses during WNS, body mass and white blood cell counts were examined following LPS administration in four bat species (134–137). Subcutaneous LPS challenge in of Pallas's mastiff bats (Molossus molossus) led to a loss of body mass of ~7% within the first day, but did not result in changes in circulating white blood cell counts or body temperature (135). Seba's short-tailed fruit bat (Carollia perspicillata) also showed a decrease in body mass following LPS challenge, but this was associated with increases in white blood cell counts as well as increases in derivatives of reactive oxidative metabolites (dROM) (134). Subdermal LPS challenge of fish-eating Myotis (Myotis vivesi) led to body mass decreases, increased resting metabolic rate and skin temperature (136), while intraperitoneal LPS challenge of wrinkle-lipped bats (Chaerephon plicatus) caused an increase in circulating leukocytes, but did not result in a reduction in body mass compared to controls (137). The differential responses to LPS challenge suggest that the immune response to bacterial infection varies across species. Of note, post-mortem examinations of ~500 dead bats comprising 19 species from Germany revealed inflammatory lesions, many of which had evidence of underlying bacterial or parasitic infections, particularly in the lung (138).
Conclusions
Bats have an array of unique life history characteristics that not only allow them to be particularly good reservoirs for viruses that are highly pathogenic in other species, but also appear to have shaped their immune systems. Although research on bat antiviral immunity has focused on only a few species to date, at the genomic level, selection on genes is concentrated on the innate immune system across both suborders of bats. However, while these studies have provided a rich source of hypotheses, the majority remain to be tested at the functional level and many questions remain that cannot be answered from comparative genome studies. Experimental studies to date have demonstrated some functional differences between bat species, with the common emerging theme that the overall antiviral response appears to converge on a lower inflammatory profile, with tight regulation of the cytokine and inflammatory response key to clearing viral infection without the pathological outcomes typically associated with infection. However, whether this is due to specific tolerance mechanisms that are at play or increased resistance to RNA virus replication still remains unclear. Fewer studies have examined the adaptive immune system than those probing innate immune pathways, but experimental infections with bat borne viruses have demonstrated that bats generate low or absent antibody responses which often wane rapidly. This is reminiscent of the response of another reservoir host, the sooty mangabey which is the natural reservoir for simian immunodeficiency virus (SIV) and for yellow fever virus. Sooty mangabeys given an attenuated yellow fever virus vaccine strain generate much lower, transient antibody responses as compared to humans or rhesus macaques. Changes to innate immune responses are also evident in sooty mangabeys (139). Thus, intriguingly, different reservoir hosts may have arrived at similar solutions to avoid the pathological consequences that follow viral infection in non-natural hosts.
Despite the ability of bats to avoid disease associated with viral infection, this trait does not extend to all pathogens, as evidenced by the severe consequences associated with infection of North American bats with the fungus that causes WNS. Thus, the pathways associated with the control of other pathogens have not been under the same selection pressures as those responsible for controlling infections with RNA viruses—or there are immunological trade offs involved which lead to greater susceptibilities to some pathogens than others. Overall, it is clear that studying host-pathogen interactions in reservoir hosts has considerable potential to provide novel insights into host tolerance mechanisms that eventually could assist in the treatment of diseases in humans and other susceptible hosts and may also offer solutions for the treatment of diseases that are a conservation threat to bats themselves.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a shared affiliation, though no other collaboration, with the authors JM and CS.
References
1. Taylor LH, Latham SM, Woolhouse ME. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci. (2001) 356:983–9. doi: 10.1098/rstb.2001.0888
2. Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature (2008) 451:990–3. doi: 10.1038/nature06536
3. Smith KF, Goldberg M, Rosenthal S, Carlson L, Chen J, Chen C, et al. Global rise in human infectious disease outbreaks. J R Soc Interface (2014) 11:20140950. doi: 10.1098/rsif.2014.0950
4. Morse SS, Mazet JA, Woolhouse M, Parrish CR, Carroll D, Karesh WB, et al. Prediction and prevention of the next pandemic zoonosis. Lancet (2012) 380:1956–65. doi: 10.,1016/S0140-6736(12)61684-5
5. Haydon DT, Cleaveland S, Taylor LH, Laurenson MK. Identifying reservoirs of infection: a conceptual and practical challenge. Emerg Infect Dis. (2002) 8:1468–73. doi: 10.3201/eid0812.010317
6. Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T, et al. Bats host major mammalian paramyxoviruses. Nat Commun. (2012) 3:796. doi: 10.1038/ncomms1796
7. Bean AG, Baker ML, Stewart CR, Cowled C, Deffrasnes C, Wang LF, et al. Studying immunity to zoonotic diseases in the natural host - keeping it real. Nat Rev Immunol. (2013) 13:851–61. doi: 10.1038/nri3551
8. Mandl JN, Ahmed R, Barreiro LB, Daszak P, Epstein JH, Virgin HW, et al. Reservoir host immune responses to emerging zoonotic viruses. Cell (2015) 160:20–35. doi: 10.1016/j.cell.2014.12.003
9. Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, Rupprecht CE. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science (2010) 329:676–9. doi: 10.1126/science.1188836
10. Fisher CR, Streicker DG, Schnell MJ. The spread and evolution of rabies virus: conquering new frontiers. Nat Rev Microbiol. (2018) 16:241–55. doi: 10.1038/nrmicro.2018.11
11. Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. (2009) 5:e1000536. doi: 10.1371/journal.ppat.1000536
12. Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, et al. Fruit bats as reservoirs of Ebola virus. Nature (2005) 438:575–6. doi: 10.1038/438575a
13. Hassanin A, Nesi N, Marin J, Kadjo B, Pourrut X, Leroy E, et al. Comparative phylogeography of African fruit bats (Chiroptera, Pteropodidae) provide new insights into the outbreak of Ebola virus disease in West Africa, 2014-2016. C R Biol. (2016) 339:517–28. doi: 10.1016/j.crvi.2016.09.005
14. Halpin K, Young PL, Field HE, Mackenzie JS. Isolation of hendra virus from pteropid bats: a natural reservoir of hendra virus. J Gen Virol. (2000) 81:1927–32. doi: 10.1099/0022-1317-81-8-1927
15. Chua KB, Koh CL, Hooi PS, Wee KF, Khong JH, Chua BH, et al. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. (2002) 4:145–51. doi: 10.1016/S1286-4579(01)01522-2
16. Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. (2005) 102:14040–5. doi: 10.1073/pnas.0506735102
17. Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science (2005) 310:676–9. doi: 10.1126/science.1118391
18. Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature (2013) 503:535–8. doi: 10.1038/nature12711
19. Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, Drosten C, et al. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol. (2014) 88:11297–303. doi: 10.1128/JVI.01498-14
20. Hu B, Ge X, Wang LF, Shi Z. Bat origin of human coronaviruses. Virol J. (2015) 12:221. doi: 10.1186/s12985-015-0422-1
21. Amman BR, Albarino CG, Bird BH, Nyakarahuka L, Sealy TK, Balinandi S, et al. A recently discovered pathogenic paramyxovirus, sosuga virus, is present in Rousettus aegyptiacus fruit bats at multiple locations in Uganda. J Wildl Dis. (2015) 51:774–9. doi: 10.7589/2015-02-044
22. Barr J, Smith C, Smith I, de Jong C, Todd S, Melville D, et al. Isolation of multiple novel paramyxoviruses from pteropid bat urine. J Gen Virol. (2015) 96:24–9. doi: 10.1099/vir.0.068106-0
23. Drexler JF, Geipel A, Konig A, Corman VM, van Riel D, Leijten LM, et al. Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc Natl Acad Sci USA. (2013) 110:16151–6. doi: 10.1073/pnas.1308049110
24. Epstein JH, Quan PL, Briese T, Street C, Jabado O, Conlan S, et al. Identification of GBV-D, a novel GB-like flavivirus from old world frugivorous bats (Pteropus giganteus) in Bangladesh. PLoS Pathog. (2010) 6:e1000972. doi: 10.1371/journal.ppat.1000972
25. Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ, et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci USA. (2013) 110:8194–9. doi: 10.1073/pnas.1303037110
26. Dobson AP. Virology. What links bats to emerging infectious diseases? Science (2005) 310:628–9. doi: 10.1126/science.1120872
27. Wang LF, Walker PJ, Poon LL. Mass extinctions, biodiversity and mitochondrial function: are bats ‘special’ as reservoirs for emerging viruses? Curr Opin Virol. (2011) 1:649–57. doi: 10.1016/j.coviro.2011.10.013
28. Olival K, Epstein JH, Wang LF, Field HE, Daszak P. Are bats exceptional viral reservoirs? In: Aguirre AA, Ostfeld R, Daszak P, editors. New Directions in Conservation Medicine. Oxford, UK: Oxford University Press (2012). p. 195–212.
29. Luis AD, Hayman DT, O'Shea TJ, Cryan PM, Gilbert AT, Pulliam JR, et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc Biol Sci. (2013) 280:20122753. doi: 10.1098/rspb.2012.2753
30. Brook CE, Dobson AP. Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. (2015) 23:172–80. doi: 10.1016/j.tim.2014.12.004
31. Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science (2005) 307:580–4. doi: 10.1126/science.1105113
32. Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature (2017) 546:646–50. doi: 10.1038/nature22975
33. Plourde BT, Burgess TL, Eskew EA, Roth TM, Stephenson N, Foley JE. Are disease reservoirs special? taxonomic and life history characteristics. PLoS ONE (2017) 12:e0180716. doi: 10.1371/journal.pone.0180716
34. Holmes EC. The evolutionary genetics of emerging viruses. Annu Rev Eco Evol Syst. (2009) 40:353–72. doi: 10.1146/annurev.ecolsys.110308.120248
35. Baker ML, Schountz T, Wang LF. Antiviral immune responses of bats: a review. Zoonoses Pub Health (2013) 60:104–16. doi: 10.1111/j.1863-2378.2012.01528.x
36. Malmlov A, Seetahal J, Carrington C, Ramkisson V, Foster J, Miazgowicz KL, et al. Serological evidence of arenavirus circulation among fruit bats in Trinidad. PLoS ONE (2017) 12:e0185308. doi: 10.1371/journal.pone.0185308
37. Stamm DD, Kissling RE, Eidson ME. Experimental rabies infection in insectivorous bats. J Infect Dis. (1956) 98:10–4. doi: 10.1093/infdis/98.1.10
38. Suu-Ire R, Begeman L, Banyard AC, Breed AC, Drosten C, Eggerbauer E, et al. Pathogenesis of bat rabies in a natural reservoir: comparative susceptibility of the straw-colored fruit bat (Eidolon helvum) to three strains of Lagos bat virus. PLoS Negl Trop Dis. (2018) 12:e0006311. doi: 10.1371/journal.pntd.0006311
39. McColl KA, Chamberlain T, Lunt RA, Newberry KM, Middleton D, Westbury HA. Pathogenesis studies with Australian bat lyssavirus in grey-headed flying foxes (Pteropus poliocephalus). Aust Vet J. (2002) 80:636–41. doi: 10.1111/j.1751-0813.2002.tb10973.x
40. Aguilar-Setien A, Loza-Rubio E, Salas-Rojas M, Brisseau N, Cliquet F, Pastoret PP, et al. Salivary excretion of rabies virus by healthy vampire bats. Epidemiol Infect. (2005) 133:517–22. doi: 10.1017/S0950268805003705
41. Allendorf SD, Cortez A, Heinemann MB, Harary CM, Antunes JM, Peres MG, et al. Rabies virus distribution in tissues and molecular characterization of strains from naturally infected non-hematophagous bats. Virus Res. (2012) 165:119–25. doi: 10.1016/j.virusres.2012.01.011
42. Black FL. Infectious diseases in primitive societies. Science (1975) 187:515–8. doi: 10.1126/science.163483
44. Gaillard JM, Yoccoz NG, Lebreton J, Bonenfant C, Devillard S, Loison A, et al. Generation time: a reliable metric to measure life-history variation among mammalian populations. Am Nat. (2005) 166:119–23. doi: 10.1086/430330
45. Fleischer T, Gampe J, Scheuerlein A, Kerth G. Rare catastrophic events drive population dynamics in a bat species with negligible senescence. Sci Rep. (2017) 7:7370. doi: 10.1038/s41598-017-06392-9
46. Holmes DJ, Ottinger MA. Birds as long-lived animal models for the study of aging. Exp Gerontol. (2003) 38:1365–75. doi: 10.1016/j.exger.2003.10.018
47. Austad SN, Fischer KE. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J Gerontol. (1991) 46:B47–53. doi: 10.1093/geronj/46.2.B47
48. Holmes DJ, Austad SN. Fly now, die later: life-history correlates of gliding and flying in mammals. Am Soc Mammalo. (1994) 75:224–6. doi: 10.2307/1382255
49. Brunet-Rossinni AK. Reduced free-radical production and extreme longevity in the little brown bat (Myotis lucifugus) versus two non-flying mammals. Mech Ageing Dev. (2004) 125:11–20. doi: 10.1016/j.mad.2003.09.003
50. Barclay MR. Constraints on reproduction by flying vertebrates: energy and calcium. Am Soc Nat. (1994) 144:1021–31. doi: 10.1086/285723
51. Heppell SS, Caswell H, Crowder LB. Life histories and elasticity patterns: perturbation analysis for species with minimal demographic data. Ecology (2000) 81:654–65. doi: 10.1890/0012-9658(2000)081[0654:LHAEPP]2.0.CO;2
52. Seim I, Fang X, Xiong Z, Lobanov AV, Huang Z, Ma S, et al. Genome analysis reveals insights into physiology and longevity of the Brandt's bat Myotis brandtii. Nat Commun. (2013) 4:2212. doi: 10.1038/ncomms3212
53. O'Shea TJ, Ellison LE, Stanley TR. Survival estimation in bats: historical overview, critical appraisal, and suggestions for new approaches. In: Thompson WL, editor. Sampling Rare or Elusive Species: Concepts, Designs and Techniques for Estimating Population Parameters. Washington, DC: Island Press (2004). p. 297–336.
54. Hedenström A, Johansson LC. Bat flight. Curr Biol. (2015) 25:R399–402. doi: 10.1016/j.cub.2015.04.002
55. Ollivier FJ, Samuelson DA, Brooks DE, Lewis PA, Kallberg ME, Komáromy AM. Comparative morphology of the tapetum lucidum (among selected species). Vet Opthalmol. (2004) 7:11–22. doi: 10.1111/j.1463-5224.2004.00318.x
56. Jones G, Teeling EC. The evolution of echolocation in bats. Trends Ecol Evol. (2006) 21:149–56. doi: 10.1016/j.tree.2006.01.001
57. Constantine DG, Emmons RW, Woodie JD. Rabies virus in nasal mucosa of naturally infected bats. Science (1972) 175:1255–6. doi: 10.1126/science.175.4027.1255
58. Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. (2006) 19:531–45. doi: 10.1128/CMR.00017-06
59. Hedenström A, Johansson LC. Bat flight: aerodynamics, kinematics and flight morphology. J Exp Biol. (2015) 218:653–63. doi: 10.1242/jeb.031203
60. Thomas SP, Suthers RA. The physiology and energetics of bat fligth. J Exp Biol. (1972) 57:317–35.
61. Shen YY, Liang L, Zhu ZH, Zhou WP, Irwin DM, Zhang YP. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc Natl Acad Sci USA. (2010) 107:8666–71. doi: 10.1073/pnas.0912613107
62. Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science (2013) 339:456–60. doi: 10.1126/science.1230835
63. Prat Y, Taub M, Yovel Y. Everyday bat vocalizations contain information about emitter, addressee, context, and behavior. Sci Rep. (2016) 6:39419. doi: 10.1038/srep39419
64. Kerth G, Van Schaik J. Causes and consequences of living in closed societies: lessons from a long-term socio-genetic study on Bechstein's bats. Mol Ecol. (2012) 21:633–46. doi: 10.1111/j.1365-294XX.2011.05233.x
65. Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol. (2008) 8:889–95. doi: 10.1038/nri2432
66. Ayres JS, Schneider DS. Tolerance of infections. Annu Rev Immunol. (2012) 30:271–94. doi: 10.1146/annurev-immunol-020711-075030
67. Martinez Gomez JM, Periasamy P, Dutertre CA, Irving AT, Ng JH, Crameri G, et al. Phenotypic and functional characterization of the major lymphocyte populations in the fruit-eating bat Pteropus alecto. Sci Rep. (2016) 6:37796. doi: 10.1038/srep37796
68. Zhou P, Chionh YT, Irac SE, Ahn M, Jia Ng JH, Fossum E, et al. Unlocking bat immunology: establishment of Pteropus alecto bone marrow-derived dendritic cells and macrophages. Sci Rep. (2016) 6:38597. doi: 10.1038/srep38597
69. Ahn M, Cui J, Irving AT, Wang LF. Unique loss of the PYHIN gene family in bats amongst mammals: implications for inflammasome sensing. Sci Rep. (2016) 6:21722. doi: 10.1038/srep21722
70. Li N, Parrish M, Chan TK, Yin L, Rai P, Yoshiyuki Y, et al. Influenza infection induces host DNA damage and dynamic DNA damage responses during tissue regeneration. Cell Mol Life Sci. (2015) 72:2973–88. doi: 10.1007/s00018-015-1879-1
71. Crameri G, Todd S, Grimley S, McEachern JA, Marsh GA, Smith C, et al. Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS ONE (2009) 4:e8266. doi: 10.1371/journal.pone.0008266
72. Kepler T, Sample C, Hudak K, Roach J, Haines A, Walsh A, et al. Chiropteran types I and II interferon genes inferred from genome sequencing traces by a statistical gene-family assembler. BMC Genomics (2010) 11:444. doi: 10.1186/1471-2164-11-444
73. Virtue ER, Marsh GA, Baker ML, Wang L-F. Interferon production and signaling pathways are antagonized during henipavirus infection of fruit bat cell lines. PLoS ONE (2011) 6:e22488. doi: 10.1371/journal.pone.0022488
74. Zhou P, Cowled C, Todd S, Crameri G, Virtue ER, Marsh GA, et al. Type III IFNs in pteropid bats: differential expression patterns provide evidence for distinct roles in antiviral immunity. J Immunol. (2011) 186:3138–47. doi: 10.4049/jimmunol.1003115
75. Zhou P, Cowled C, Mansell A, Monaghan P, Green D, Wu L, et al. IRF7 in the Australian black flying fox, Pteropus alecto: evidence for a unique expression pattern and functional conservation. PLoS ONE (2014) 9:e103875. doi: 10.1371/journal.pone.0103875
76. Zhou P, Tachedjian M, Wynne JW, Boyd V, Cui J, Smith I, et al. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc Nat Acad Sci USA. (2016) 113:2696–701. doi: 10.1073/pnas.1518240113
77. Broggi A, Tan Y, Granucci F, Zanoni I. IFN-λ suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat Immunol. (2017) 18:1084–93. doi: 10.1038/ni.3821
78. De La Cruz-Rivera PC, Kanchwala M, Liang H, Kumar A, Wang L-F, Xing C, et al. The IFN response in bat cells consists of canonical and non-canonical ISGs with unique temporal expression kinetics. bioRxiv. [Preprint] (2017). doi: 10.1101/167999
79. Kuzmin IV, Schwarz TM, Ilinykh PA, Jordan I, Ksiazek TG, Sachidanandam R, et al. Innate immune responses of bat and human cells to filoviruses: commonalities and distinctions. J Virol. (2017) 91:e02471–16. doi: 10.1128/JVI.02471-16
80. Zanoni I, Granucci F, Broggi A. Interferon (IFN)-λ takes the helm: immunomodulatory roles of Type III IFNs. Front Immunol. (2017) 8:1661. doi: 10.3389/fimmu.2017.01661
81. Zhang Q, Zeng L-P, Zhou P, Irving AT, Li S, Shi Z-L, et al. IFNAR2-dependent gene expression profile induced by IFN-α in Pteropus alecto bat cells and impact of IFNAR2 knockout on virus infection. PLoS ONE (2017) 12:e0182866. doi: 10.1371/journal.pone.0182866
82. Pavlovich SS, Lovett SP, Koroleva G, Guito JC, Arnold CE, Nagle ER, et al. The Egyptian rousette genome reveals unexpected features of bat antiviral immunity. Cell (2018) 173:1098–110.e18. doi: 10.1016/j.cell.2018.03.070
83. Xie J, Li Y, Shen X, Goh G, Zhu Y, Cui J, et al. Dampened STING-dependent interferon activation in bats. Cell Host Microbe. (2018) 23:297–301.e4. doi: 10.1016/j.chom.2018.01.006
84. Jones G, Jacobs DS, Kunz TH, Willig MR, Racey PA. Carpe noctem: the importance of bats as bioindicators. Endangered Species Res. (2009) 8:93–115. doi: 10.3354/esr00182
85. Seltmann A, Czirjak GA, Courtiol A, Bernard H, Struebig MJ, Voigt CC. Habitat disturbance results in chronic stress and impaired health status in forest-dwelling paleotropical bats. Conserv Physiol. (2017) 5:cox020. doi: 10.1093/conphys/cox020
86. Middleton DJ, Weingartl HM. Henipaviruses in their natural animal hosts. Curr Top Microbiol Immunol. (2012) 359:105–21. doi: 10.1007/82_2012_210
87. World Health Organization (2018). Outbreak of Nipah Virus Encephalitis in Kerala State of India. Available online at: http://www.searo.who.int/entity/emerging_diseases/links/nipah_virus/en/
88. Williamson MM, Hooper PT, Selleck PW, Gleeson LJ, Daniels PW, Westbury HA, et al. Transmission studies of Hendra virus (equine morbilli-virus) in fruit bats, horses and cats. Australian Vet J. (1998) 76:813–8. doi: 10.1111/j.1751-0813.1998.tb12335.x
89. Williamson MM, Hooper PT, Selleck PW, Westbury HA, Slocombe RF. Experimental Hendra virus infection in pregnant guinea-pigs and fruit bats (Pteropus poliocephalus). J Comp Pathol. (1999) 122:201–7. doi: 10.1053/jcpa.1999.0364
90. Middleton DJ, Morrissy CJ, van der Heide BM, Russell GM, Braun MA, Westbury HA, et al. Experimental nipah virus infection in pteropid bats (Pteropus poliocephalus). J Comp Pathol. (2007) 136:266–72. doi: 10.1016/j.jcpa.2007.03.002
91. Halpin K, Hyatt AD, Fogarty R, Middleton D, Bingham J, Epstein JH, et al. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am J Trop Med Hyg. (2011) 85:946–51. doi: 10.4269/ajtmh.2011.10-0567
92. Peter LY, Kim H, Paul WS, Hume EF, Jenny LG, Mark AK, et al. Serologic evidence for the presence in pteropus bats of a paramyxovirus related to equine morbillivirus. Emer Infect Dis. (1996) 2:239–240. doi: 10.3201/eid0203.960315
93. Field H, Jordan D, Edson D, Morris S, Melville D, Parry-Jones K, et al. Spatiotemporal aspects of hendra virus infection in pteropid bats (Flying-Foxes) in Eastern Australia. PLoS ONE (2015) 10:e0144055. doi: 10.1371/journal.pone.0144055
94. Johara Mohd Y, Hume F, Azmin Mohd R, Christopher M, Brenda van der H, Paul R, et al. Nipah virus infection in bats (Order Chiroptera) in Peninsular Malaysia. Emer Infect Dis. (2001) 7:439–41. doi: 10.3201/eid0703.017312
95. Hayward JA, Tachedjian M, Cui J, Cheng AZ, Johnson A, Baker ML, et al. Differential evolution of antiretroviral restriction factors in pteropid bats as revealed by APOBEC3 Gene complexity. Mol Biol Evol. (2018) 35:1626–37. doi: 10.1093/molbev/msy048
96. Ng JHJ, Tachedjian M, Deakin J, Wynne JW, Cui J, Haring V, et al. Evolution and comparative analysis of the bat MHC-I region. Sci Rep. (2016) 6:21256. doi: 10.1038/srep21256
97. Hayward JA, Tachedjian M, Cui J, Field H, Holmes EC, Wang LF, et al. Identification of diverse full-length endogenous betaretroviruses in megabats and microbats. Retrovirology (2013) 10:35. doi: 10.1186/1742-4690-10-35
98. Cui J, Tachedjian G, Wang L-F. Bats and rodents shape mammalian retroviral phylogeny. Sci Rep. (2015) 5:16561. doi: 10.1038/srep16561
99. Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. (2006) 16:480–5. doi: 10.1016/j.cub.2006.01.031
100. Renard M, Henry M, Guétard D, Vartanian J-P, Wain-Hobson S. APOBEC1 and APOBEC3 cytidine deaminases as restriction factors for hepadnaviral genomes in non-humans in vivo. J Mol Biol. (2010) 400:323–34. doi: 10.1016/j.jmb.2010.05.029
101. Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. (2013) 371:1–27. doi: 10.1007/978-3-642-37765-5_1
102. Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, et al. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. (2009) 5:e1000439. doi: 10.1371/journal.ppat.1000439
103. Mohanram V, Skold AE, Bachle SM, Pathak SK, Spetz AL. IFN-alpha induces APOBEC3G, F, and A in immature dendritic cells and limits HIV-1 spread to CD4+ T cells. J Immunol. (2013) 190:3346–53. doi: 10.4049/jimmunol.1201184
104. Banerjee A, Rapin N, Bollinger T, Misra V. Lack of inflammatory gene expression in bats: a unique role for a transcription repressor. Sci Rep. (2017) 7:2232. doi: 10.1038/s41598-017-01513-w
105. Ishikawa H, Barber GN. STING an endoplasmic reticulum adaptor that facilitates innate immune signaling. Nature (2008) 455:674–8. doi: 10.1038/nature07317
106. Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, Swanepoel R, et al. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. (2012) 8:e1002877. doi: 10.1371/journal.ppat.1002877
107. Pourrut X, Souris M, Towner JS, Rollin PE, Nichol ST, Gonzalez JP, et al. Large serological survey showing cocirculation of Ebola and Marburg viruses in Gabonese bat populations, and a high seroprevalence of both viruses in Rousettus aegyptiacus. BMC Infect Dis. (2009) 9:159. doi: 10.1186/1471-2334-9-159
108. Jordan I, Horn D, Oehmke S, Leendertz FH, Sandig V. Cell lines from the Egyptian fruit bat are permissive for modified vaccinia Ankara. Virus Res. (2009) 145:54–62. doi: 10.1016/j.virusres.2009.06.007
109. Jones ME, Schuh AJ, Amman BR, Sealy TK, Zaki SR, Nichol ST, et al. Experimental inoculation of egyptian rousette bats (Rousettus aegyptiacus) with viruses of the ebolavirus and marburgvirus genera. Viruses (2015) 7:3420–42. doi: 10.3390/v7072779
110. Paweska JT, Storm N, Grobbelaar AA, Markotter W, Kemp A, Jansen van Vuren P. Experimental inoculation of Egyptian fruit bats (Rousettus aegyptiacus) with ebola virus. Viruses (2016) 8:E29. doi: 10.3390/v8020029
111. Paweska JT, Jansen van Vuren P, Masumu J, Leman PA, Grobbelaar AA, Birkhead M, et al. Virological and serological findings in Rousettus aegyptiacus experimentally inoculated with vero cells-adapted hogan strain of Marburg virus. PLoS ONE (2012) 7:e45479. doi: 10.1371/journal.pone.0045479
112. Amman BR, Jones ME, Sealy TK, Uebelhoer LS, Schuh AJ, Bird BH, et al. Oral shedding of Marburg virus in experimentally infected Egyptian fruit bats (Rousettus aegyptiacus). J Wildl Dis. (2015) 51:113–24. doi: 10.7589/2014-08-198
113. Paweska JT, Jansen van Vuren P, Fenton KA, Graves K, Grobbelaar AA, Moolla N, et al. Lack of marburg virus transmission from experimentally infected to susceptible in-contact Egyptian fruit bats. J Infect Dis. (2015) 212 (Suppl. 2):S109–18. doi: 10.1093/infdis/jiv132
114. Schuh AJ, Amman BR, Jones ME, Sealy TK, Uebelhoer LS, Spengler JR, et al. Modelling filovirus maintenance in nature by experimental transmission of Marburg virus between Egyptian rousette bats. Nat Commun. (2017) 8:14446. doi: 10.1038/ncomms14446
115. Schuh AJ, Amman BR, Sealy TK, Spengler JR, Nichol ST, Towner JS. Egyptian rousette bats maintain long-term protective immunity against Marburg virus infection despite diminished antibody levels. Sci Rep. (2017) 7:8763. doi: 10.1038/s41598-017-07824-2
116. Deschamps M, Laval G, Fagny M, Itan Y, Abel L, Casanova JL, et al. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am J Hum Genet. (2016) 98:5–21. doi: 10.1016/j.ajhg.2015.11.014
117. Lee AK, Kulcsar KA, Elliott O, Khiabanian H, Nagle ER, Jones ME, et al. De novo transcriptome reconstruction and annotation of the Egyptian rousette bat. BMC Genomics (2015) 16:1033. doi: 10.1186/s12864-015-2124-x
118. Lawrence T. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harbor Persp Biol. (2009) 1:a001651. doi: 10.1101/cshperspect.a001651
119. Guivier E, Galan M, Salvador AR, Xuéreb A, Chaval Y, Olsson GE, et al. Tnf-α expression and promoter sequences reflect the balance of tolerance/resistance to Puumala hantavirus infection in European bank vole populations. Infect Genet Evol. (2010) 10:1208–17. doi: 10.1016/j.meegid.2010.07.022
120. Kacprzyk J, Hughes GM, Palsson-McDermott EM, Quinn SR, Puechmaille SJ, O'Neill LAJ, et al. A potent anti-inflammatory response in bat macrophages may be linked to extended longevity and viral tolerance. Acta Chiropterol. (2017) 19:219–28. doi: 10.3161/15081109ACC2017.19.2.001
121. Cryan PM, Meteyer CU, Boyles JG, Blehert DS. White-nose syndrome in bats: illuminating the darkness. BioMed Central Biol. (2013) 11:1–4. doi: 10.1186/1741-7007-11-47
122. Hoyt JR, Cheng TL, Langwig KE, Hee MM, Frick WF, Kilpatrick AM. Bacteria isolated from bats inhibit the growth of Pseudogymnoascus destructans, the causative agent of white-nose syndrome. PLoS ONE (2015) 10:e0121329. doi: 10.1371/journal.pone.0121329
123. United States Geological Survey (2018). United States Geological Survey. Available online at: https://www.nwhc.usgs.gov/disease_information/white-nose_syndrome/
124. Wibbelt G, Puechmaille SJ, Ohlendorf B, Muhldorfer K, Bosch T, Gorfol T, et al. Skin lesions in European hibernating bats associated with Geomyces destructans, the etiologic agent of white-nose syndrome. PLoS ONE (2013) 8:e74105. doi: 10.1371/journal.pone.0074105
125. Zukal J, Bandouchova H, Brichta J, Cmokova A, Jaron KS, Kolarik M, et al. White-nose syndrome without borders: Pseudogymnoascus destructans infection tolerated in Europe and Palearctic Asia but not in North America. Sci Rep. (2016) 6:19829. doi: 10.1038/srep19829
126. Verant M, Meteyer CU, Speakman JR, Cryan PM, Lorch JM, Blehert DS. White-nose syndrome initiates a casacde of physiological disturbances in the hibernating bat host. BioMed Central Physiol. (2014) 14:1–11. doi: 10.1186/s12899-014-0010-4
127. Reeder DM, Frank CL, Turner GG, Meteyer CU, Kurta A, Britzke ER, et al. Frequent arousal from hibernation linked to severity of infection and mortality in bats with white-nose syndrome. PLoS ONE (2012) 7:e38920. doi: 10.1371/journal.pone.0038920
128. Cryan PM, Meteyer CU, Blehert DS, Lorch JM, Reeder DM, Turner GG, et al. Electrolyte depletion in white-nose syndrome bats. J Wildl Dis. (2013) 49:398–402. doi: 10.7589/2012-04-121
129. Cryan PM, Meteyer CU, Boyles JG, Blehert DS. Wing pathology of white-nose syndrome in bats suggests life-threatening disruption of physiology. BioMed Central Biol. (2010) 8:1–8. doi: 10.1186/1741-7007-8-135
130. Willis CK, Menzies AK, Boyles JG, Wojciechowski MS. Evaporative water loss is a plausible explanation for mortality of bats from white-nose syndrome. Integr Comp Biol. (2011) 51:364–73. doi: 10.1093/icb/icr076
131. Meteyer CU, Barber D, Mandl JN. Pathology in euthermic bats with white nose syndrome suggests a natural manifestation of immune reconstitution inflammatory syndrome. Virulence (2012) 3:583–8. doi: 10.4161/viru.22330
132. Field KA, Johnson JS, Lilley TM, Reeder SM, Rogers EJ, Behr MJ, et al. The white-nose syndrome transcriptome: activation of anti-fungal host responses in wing tissue of hibernating little brown myotis. PLoS Pathog. (2015) 11:e1005168. doi: 10.1371/journal.ppat.1005168
133. Lilley TM, Prokkola JM, Johnson JS, Rogers EJ, Gronsky S, Kurta A, et al. Immune responses in hibernating little brown myotis (Myotis lucifugus) with white-nose syndrome. Proc Biol Sci. (2017) 284:20162232. doi: 10.1098/rspb.2016.2232
134. Schneeberger K, Czirjak GA, Voigt CC. Inflammatory challenge increases measures of oxidative stress in a free-ranging, long-lived mammal. J Exp Biol. (2013) 216:4514–9. doi: 10.1242/jeb.090837
135. Stockmaier S, Dechmann DK, Page RA, O'Mara MT. No fever and leucocytosis in response to a lipopolysaccharide challenge in an insectivorous bat. Biol Lett. (2015) 11:20150576. doi: 10.1098/rsbl.2015.0576
136. Otalora-Ardila A, Herrera ML, Flores-Martinez JJ, Welch KC Jr. Metabolic cost of the activation of immune response in the fish-eating myotis (Myotis vivesi): the effects of inflammation and the acute phase response. PLoS ONE (2016) 11:e0164938. doi: 10.1371/journal.pone.0164938
137. Weise P, Czirjak GA, Lindecke O, Bumrungsri S, Voigt CC. Simulated bacterial infection disrupts the circadian fluctuation of immune cells in wrinkle-lipped bats (Chaerephon plicatus). PeerJ. (2017) 5:e3570. doi: 10.7717/peerj.3570
138. Mühldorfer K, Speck S, Wibbelt G. Diseases in free-ranging bats from Germany. BioMed Central Vet Res. (2011) 7:1–11. doi: 10.1186/1746-6148-7-61
Keywords: bats (Chiroptera), viral immunology, host pathogen interaction, disease tolerance, comparative genome analyses, innate immunity
Citation: Mandl JN, Schneider C, Schneider DS and Baker ML (2018) Going to Bat(s) for Studies of Disease Tolerance. Front. Immunol. 9:2112. doi: 10.3389/fimmu.2018.02112
Received: 18 June 2018; Accepted: 28 August 2018;
Published: 20 September 2018.
Edited by:
Irah L. King, McGill University, CanadaReviewed by:
Christopher J. A. Duncan, Newcastle University, United KingdomAnna-Lena Spetz, Stockholm University, Sweden
Copyright © 2018 Mandl, Schneider, Schneider and Baker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Judith N. Mandl, anVkaXRoLm1hbmRsQG1jZ2lsbC5jYQ==
 Judith N. Mandl
Judith N. Mandl Caitlin Schneider2,3
Caitlin Schneider2,3 Michelle L. Baker
Michelle L. Baker