
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 31 July 2018
Sec. Nutritional Immunology
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.01765
This article is part of the Research Topic Xenobiotics and the gut microbiome in health and disease View all 15 articles
 Jiyan Su1†
Jiyan Su1† Lu Su2,3†
Lu Su2,3† Dan Li3,4
Dan Li3,4 Ou Shuai3
Ou Shuai3 Yifan Zhang1
Yifan Zhang1 Huijia Liang3
Huijia Liang3 Chunwei Jiao3
Chunwei Jiao3 Zhanchi Xu4
Zhanchi Xu4 Yong Lai2*
Yong Lai2* Yizhen Xie1,3*
Yizhen Xie1,3*
As breast cancer is the leading cause of cancer-related deaths in women population worldwide, ongoing endeavor has been made for alternative regimens with improved efficacy but fewer adverse effects. Recently, active components from the spores of Ganoderma lucidum have attracted much attention for their versatile biological activities owing to the advance in sporoderm-breaking technology. Here, anticancer potential of an extract derived from the sporoderm-breaking spores of G. lucidum (ESG) was explored in a 4T1-breast cancer xenograft mice model. Results showed that ESG was able to suppress 4T1 tumor growth in vivo rather than in vitro. Flowcytometry analysis revealed that ESG could significantly increase both cytotoxic T cell (Tc) population and the ratio of Tc to helper T cell (Th) in peripheral blood of the tumor-bearing mouse; similar promotion on Tc was also found in tumor-infiltrating lymphocyte. Moreover, ESG evidently downregulated the two immune checkpoints, programmed cell death protein-1 (PD-1, in the spleen) and cytotoxic T lymphocyte antigen-4 (CTLA-4, in the tumor), suggesting that ESG could effectively restore the T cell paradigm by recovering the exhaustion status via suppressing the co-inhibitory checkpoints. By 16S rRNA gene sequence analysis on the fecal microbiota, it was found that ESG would remodeling the overall structure of the samples from tumor-bearing mice toward that of the normal counterparts, including 18 genera in 5 phyla, together with regulations on several genes that are responsible for signaling pathways involved in metabolism, cellular processes, and environmental information processing. Collectively, this study demonstrated that ESG would serve as a natural anticancer adjuvant via a restoration on the exhausted Tc, highlighting important clinical implications for the treatment of breast cancer.
Breast cancer is one of the most frequently diagnosed cancers and leading causes of cancer-related death both worldwide (1, 2). It is a highly heterogeneous disease. Besides the multiple signaling pathways that mediate its initiation and progression, accumulating epidemiology studies have proposed several suspected risk factors for breast cancer, including exposures to cancerigenic substances, certain lifestyles (such as lack of physical exercise, intemperance), aging and family history (3). To date, surgical resection, adjuvant chemotherapy, radiotherapy, and hormone therapy represent the main treatment options for early-stage breast cancer (4), but they are still unsatisfied due to the emerging drug resistance and adverse toxic effects (5, 6). Even more disturbing, triple-negative breast cancer does not respond to hormone therapies (7).
Despite the slow progress in the above regimes, recent success of cancer immunotherapy utilizing immune checkpoint blockade has reformed the treatment algorithms in various aggressive neoplastic diseases (8, 9). Immunotherapy is based upon the “3E immunoediting theory” (Eliminate, Equilibrium, and Escape) (10). It reminds that immune surveillance stands at the very heart of the fighting against cancer, but T cells would be exhausted and have decreased effector function and proliferative capacity during cancer progression, due the overexpression of immune checkpoints, and the interaction with their ligands (11, 12). So far, immunotherapy targeting programmed cell death protein-1 (PD-1)/programmed death-ligand 1 (PD-L1) has been on it way of clinical trials (13) for breast cancer, although optimal selection of ideal candidates to the immune therapy remains a challenge. On the other hand, increasing evidence highlights the cardinal role of gut microbiota in tumour genesis (14, 15) and that in the outcomes of chemotherapy (16) and immunotherapy (17, 18), due to their intrinsic capacity of drug metabolism and the influence on host metabolizing homeostasis (19, 20).
Ganoderma lucidum (Leyss. ex Fr.) Karst. is a valuable medical macrofungi that has long been used in traditional Chinese medicine for health and longevity for thousands of years (21). In addition to numerous benefit for the treatment of allergy, cardiovascular diseases, diabetics (22, 23), ample evidence have revealed that G. lucidum exerts anticancer effects not only via direct cytotoxicity approaches, such as cell cycle arrest (24), apoptosis induction (25), and migration inhibition (26), but also, more importantly, through several ways of immune enhancement (27–29). Recently, although active components from fruiting body or mycelia are still hotspots of G. lucidum studies, those from the spores of G. lucidum (SG) have attracted much attention for their versatile biological activities owing to the advance in sporoderm-breaking technology. Studies reported that SG displayed anticancer potentials against Sarcoma 180 and HCT116 (30–32). Moreover, it has been found that polysaccharide content in SG was higher than that in fruiting body (33) and that sporoderm-breaking would potentiate the immunoregulation activity of spore (32, 34, 35). These studies suggest that SG may serve as promising anticancer agent for cancer therapy. In this study, anticancer potential of an extract from the sporoderm-breaking SG, for the first time, was investigated in a breast cancer xenograft mice model. The exploration focused on the interaction between T cell restoration and gut microbiota, to make a comprehensive interception for the anticancer activity of the sporoderm-breaking SG.
BALB/c mice (female, 18–22 g, aged 6–8 weeks) were provided by Guangdong Medical Laboratory Animal Center (Guangzhou, Guangdong, China). All animals were housed at 20 ± 2°C with a humidity of 50 ± 5% in a 12 h light/dark cycle with food and water ad libitum. The animals were acclimatized for 7 days, and the experiment was performed according to the Guidelines of Guangdong Institute of Microbiology Laboratory Animal Center, Guangdong Institute of Microbiology Laboratory Animal Ethics Committee. The experimental protocols were approved by the Guangdong Institute of Microbiology Laboratory Animal Ethics Committee.
The sporoderm-breaking SG was provided by Guangdong Yuewei Edible Fungi Technology Co. Ltd. It was extracted with boiling water (15 L/kg) for 2 h and concentrated under vacuum. The concentrated extract was subjected to two to three cycles of precipitation with anhydrous ethanol at a final ethanol percentage of 85%. The obtained precipitate was dissolved in water and dialyzed with 3,500 Da dialysis tube (MWCO). Content in the 3,500 Da dialysis tube was then dialyzed in 100 kDa dialysis tube (MWCO). The dialysate was pooled, concentrated, and lyophilized, to obtain ESG with a yield of 0.4%.
Characteristics analysis for ESG was performed as described by Qiao et al. (36) with mild modification. Results showed that sugar content of ESG was 50% (determined by the phenol–sulfuric acid method using glucose as standard), while protein was hardly detected by BCA methods with the BCA protein kit (Kang wei shiji Co. Ltd., Beijing, China), suggesting that ESG is rich in polysaccharide. Weight average molecular weight (Mw) by size-exclusive high performance liquid chromatography showed that polysaccharide in ESG was about 3.6 kDa (Table S1 and Figure S1 in Supplementary Material). Monosaccharide composition analysis by gas chromatograph showed that ESG was mainly made up of glucose (Figure S2 in Supplementary Material).
Murine metastatic breast cancer 4T1-cell line was obtained from the Cell bank of Chinese Academy of Sciences, Shanghai, China. 4T1 cells were cultured in high glucose DMEM medium (4.5 mg/mL, Gibco, NY, USA) supplemented with 10% fetal bovine serum (Gibco, NY, USA) and 1% penicillin/streptomycin (Gibco, NY, USA) and maintained in humidified incubators at 37°C under an atmosphere of 5% CO2.
4T1 cells were seeded in a 96-well plate at a density of 1.25 × 104 cells/mL (sextuple wells in each group) and treated with ESG in complete DMEM medium at multiple concentrations (12.5, 25, 50, 100, and 200 µg/mL) for 24 and 48 h. Then, the medium was replaced with 100 µL of complete DMEM medium containing 0.5 mg/mL 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) for another 4-h incubation. At last, the medium was discarded, and 150 µL of DMSO was added to dissolve the formazan. The optical density was measured at 490 nm on a microplate reader.
To induce the breast cancer xenograft model, female BALB/c mice were implanted with 4T1 tumor cells by subcutaneous injection at the right forleg armpit (0.1 mL/mouse, 2 × 105 cells/mouse), and then they were randomly divided into Model group, paclitaxel group (PTX, Hainan Quanxing Pharmaceutical Co. Ltd., Hainan, China), and the ESGH group (400 mg/kg), ESGL group (200 mg/kg), eight for each. On the same day, control animals (Normal group, eight mice) received an injection of 0.1 mL of complete DMEM medium at the similar site. Over the following 21 days, mice in the PTX group were administrated with PTX at a dose of 15 mg/kg (i.p.) twice a week; mice of the ESG groups were given different doses of ESG (i.g.) every day. Normal group and Model group received equal volume of water.
Throughout the 21-day administration, tumor volume was measured with an electronic vernier caliper every 3 days since sixth day. Tumor volume was calculated as V = a × b2/2, where a indicates the longer diameter and b indicates the shorter diameter. 24 h after the last administration, all animals were sacrificed by cervical dislocation. Tumors, spleens, and peripheral blood were harvested for further analysis.
Tumors were weighted, photographed, cut into several segments, and then stored according to different purposes once they were harvested. One segment of the tumor was fixed in 4% neutral formalin (in PBS), embedded by paraffin, and stained with hematoxylin–eosin (HE). The stained sections were observed and photographed under a light microscope (with 200× magnification).
Peripheral blood was collected from the orbital vein plexus with EDTA-Li micro-anticoagulant tube. 50 µL of blood was stained with FITC anti-mouse CD3 (0.125 μg/test), APC anti-mouse CD4 (0.0625 μg/test), PE anti-mouse CD8 (0.125 μg/test, Invitrogen, Thermo Fisher Scientific, San Diego, CA, USA) at 4°C in dark for 30 min, and then erythrocytes were lysed in ACK Lysis Buffer for 10 min. Following by two washes with pre-cold PBS, T cell subsets in the peripheral blood were enumerated with a FACS Canto II cytometer (BD, NY, USA), and the data were analyzed by Diva software (version 6.1.3).
Tumor segment kept in cold PBS was used for TIL isolation and analysis. In brief, they were minced and digested in 3 mL digestive medium, which was mainly composed of basic RPMI-160 medium supplemented with 0.1% Type IV collagenase (Invitrogen, Thermo Fisher Scientific, Grand Isle, NY, USA), 350 U/mL DNAse I (Roche, Basel, Switzerland), and 1% penicillin–streptomycin. Then, it was ground in pre-cold PBS by passing through a 70 µm strainer, washed with PBS, and resuspended in basic RPMI-160 medium. TILs from the obtained cell suspension were separated with Mouse 1× Lymphocyte Separation Medium (Dakewe Biotechnology Co. Ltd., Shenzhen, China) according to the manufacture’s instruction. TILs were stained with FITC anti-mouse CD3 (0.125 μg/test), APC anti-mouse CD4 (0.0625 μg/test), and PE anti-mouse CD8 (0.125 μg/test) at 4°C in dark for 30 min. After two washes with pre-cold PBS, T cell subsets in TIL were enumerated with a FACS Canto II cytometer, and the data were analyzed by Diva software (version 6.1.3).
About 100 mg of spleen tissue was cut into pieces, homogenized in ice-cold PBS with proteinase inhibitor cocktail (Roche, USA) for 30 s, centrifuged at 18,000 × g for 20 min at 4°C. Protein content in the supernatant was quantified with BCA protein kit (Kang wei shiji Co. Ltd., Beijing, China). PD-1 protein content in spleen was measured using the Mouse PD1 ELISA Kit (Abcam, Cambridge, UK, Cat: ab210971) according to the manufacturer’s instruction.
Immunohistochemistry for CTLA-4 was performed with the formalin-fixed, paraffin-embedded tumor segment. Briefly, the slides were deparaffinized. Antigen retrieval was carried out by incubation in EDTA buffer (pH 8.0) via microwave heating. After being washed with PBS, endogenous peroxidase in the section was blocked with 3% H2O2 in dark for 25 min. Sections were blocked with 3% normal non-immune serum and then incubated with primary antibody against mouse CTLA-4 (1:100, Lifespan Biosciences, London, UK) at 4°C overnight, and then with HRP-conjugated secondary antibodies at room temperature for 50 min. Finally, the sections were stained with DAB substrate and counterstained with hematoxylin. The mean density of positive area was calculated as ratio of integrated optical density to the total pixel of each picture (IOD/106 pixel), which was analyzed by Image Pro Plus 6.0 software (Media Cybernetics, Silver Spring, MD, USA).
Total RNAs from spleen and tumor were extracted with TRIzol according to the manufacturer’s instructions (Invitrogen, Thermo Fisher Scientific, Grand Isle, NY, USA). 0.1 μg of total RNA was reverse transcribed using the PrimeScript™ RT reagent kit (Takara Bio, Inc., Shiga, Japan) following the supplier’s protocol. The reactions were incubated at 35°C for 15 min, then at 85°C for 5 s, and the products were stored at 4°C. The PCR primer sequences are listed in Table 1. Real-time PCR reactions were performed with SYBR® Premix Ex Taq™ II (Takara Bio, Inc., Shiga, Japan), and the reaction program in an StepOnePlus Real-Time PCR system (Thermo Fisher Scientific, Grand Isle, NY, USA) was as follows: a precycling stage at 95°C for 30 s, then 40 cycles of denaturation at 95°C for 5 s and annealing at 60°C for 30 s. Fluorescence was measured at the end of each annealing step, and the melting curves were monitored to confirm the specificity of the PCR products. The 2−ΔΔCt method was used to determine the mRNA expression levels of pd1 and ctla4 relative to control gene gapdh.
Sequencing service was provided by Personal Biotechnology Co., Ltd., Shanghai, China. Total DNA was isolated from the fecal samples as previously reported with some modification (37). Briefly, OMEGA Soil DNA Kit (OMEGA, US) was used following the manufacturer’s recommendations. The bacterial 16S rRNA gene V3–4 region was amplified by PCR using the forward primer (5′-AYTGGGYD TAAAGNG-3′) and the reverse primer (5′-TACNVGGGTATCTAATCC-3′). The PCR products were separated by gel electrophoresis and purified using the AP-GX-500 DNA Gel Extraction Kit (Axygen, Corning, USA). Library was build up with the obtained products and then sequenced on a MiSeq sequencing platform (Illumina, USA) as described by Zhao et al. (38).
The trimmed and assembled sequences from each sample were aligned to the Greengene 16S rRNA database set 10 using the best hit classification option to classify the taxonomy abundance in QIIME1 (39). Bacterial operation taxonomic units (OTU) were generated using the uclust function in QIIME.2 A Venn diagram was generated to compare OTUs between groups. The following statistics were performed by R software. ACE, Chao, Simpson, and Shannon indices were calculated for α-diversity evaluation. Principal component analysis (PCA) and UniFrac distance-based Nonmetric Multidimensional Scaling (NMDS) were employed to assess β-diversity. Hierarchical clustering analysis of the OTUs presented by heatmap was performed using the heatmap package v1.0.7 running in R v3.2.1.3 Taxon-based analysis and LefSe analysis were applied to identify different taxa microbes among lines using the default parameters (40).
Microbial functions were predicted using PICRUSt (41). The OTUs were mapped to gg13.5 database at 97% similarity by QIIME’s command “pick_closed_otus.” The OTUs’ abundance was normalized automatically using 16S rRNA gene copy numbers from known bacterial genomes in Integrated Microbial Genomes. The predicted genes and their functions were aligned to Kyoto Encyclopedia of Genes and Genomes (KEGG) database, and differences among groups were compared through software STAMP4 (42). Two-side Welch’s t-test and the Benjamini–Hochberg FDR (p < 0.05) correction were used in two-group analysis.
Statistical analysis was performed with SPSS 22 (IBM Corp., NY, USA). Datasets from each experiment were subjected to normal distribution test first. If according with the normal distribution, the data was analyzed by one-way analysis of variance (ANOVA), following by pairwise comparison with different parametric test depending on test for homogeneity of variance, otherwise the data was compared by Kruskal–Wallis H Test. In ANOVA, post hoc LSD test was applied for difference analysis under homogeneity of variance, if not, a Dunnett’s test would be applied. #p < 0.05 and ##p < 0.01 as compared with Normal group; *p < 0.05 and **p < 0.01 as compared with Model group.
In Figure 1A, it was found that viability of 4T1 cells was not affected by either 24 or 48 h treatment of ESG (12.5–200 μg/mL) in vitro, implying that the antitumor activity of ESG was not mediated by a directly cytotoxicity. However, ESG exhibited a favorite antitumor activity in the xenograft model. Two mice of the PTX group died during the experiment. As shown in Figures 1B,C, tumor volume of Model group kept increasing throughout the 21-day experiment. By comparison, tumor volumes of the PTX group and ESG groups (400 and 200 mg/kg) showed remarkable reduction since the 15th day, although they also kept increasing in the earlier days. Finally, tumors of the Model group weighted 512 ± 45 mg, while those of PTX group and ESG groups (400 and 200 mg/kg) were significantly decreased to 193 ± 45 mg (p < 0.01), 387 ± 23 mg (p < 0.05), and 450 ± 41 mg, respectively, which were in accordance with the depressed volume. Moreover, histology analysis by HE staining revealed that tumors from Model group exhibited a homogeneous distribution of viable cells, while those of PTX group and ESG groups showed significant indication of necrosis, including shrinkage of the cells, nuclear condensation, and fibrosis (Figure 1D).
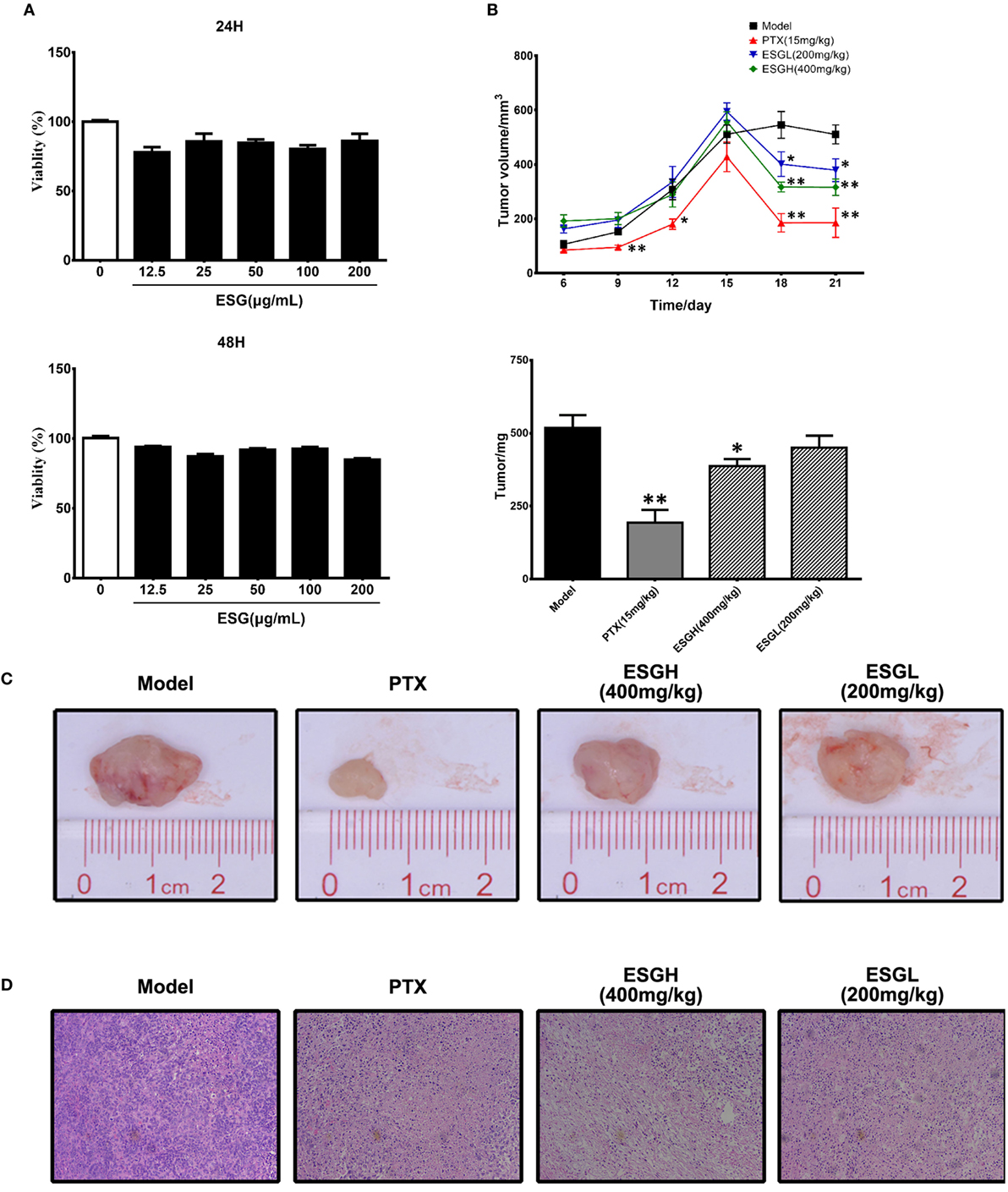
Figure 1. Effect of ESG on 4T1-xenograft model. (A) Effect of ESG on the viability of 4T1 cell in vitro (48 and 72 h). (B) Tumor volume changes and tumor mass. (C) Representative images for tumor from each group. (D) Representative hematoxylin–eosin staining images for tumor tissue from each group (200× magnification). Values were represented the means ± SEM (n = 6–8). *p < 0.05 and **p < 0.01 versus Model group.
Since the antitumor activity of ESG was independent of cytotoxicity, we analyzed the impact of oral administration of ESG on immune surveillance system by flowcytometry (FCM) as it plays a cardinal role in the suppression of tumor. Results showed that in peripheral blood (Figure 2), percentage of T cell (CD3+) of Model group was obviously lower than those of Normal group (p < 0.01), and distinct decreases were also found in the proportions of the two main subsets (p < 0.01), helper T cell (Th, CD3+CD4+) and cytotoxic T cell (Tc, CD3+CD8+). PTX made a more serious lost on T cell and Th cell when comparing with those of Model group (p < 0.01 and p < 0.05, respectively). On the other hand, although failing to lift the amount of T cell or that of the Th subset, ESG (200 mg/kg) prominently increased the percentage of Tc cell in the circulatory system (p < 0.05). Moreover, the ratio of Tc/Th was obviously elevated after ESG treatment (400 and 200 mg/kg, p < 0.01).
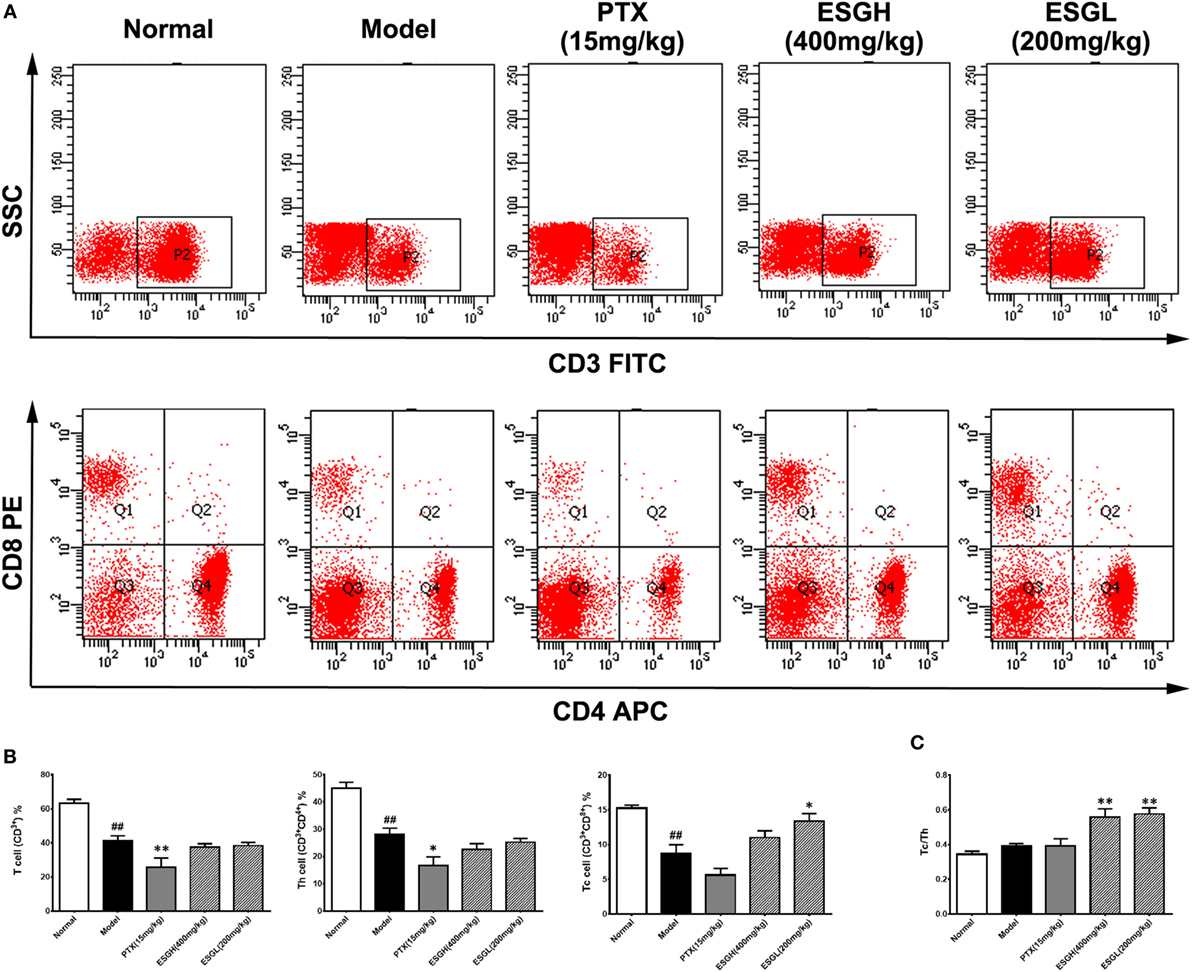
Figure 2. Effect of ESG on T cell subsets in peripheral blood. (A) Representative scatter diagram. (B) Quantitative analysis for T cell subsets in peripheral blood. (C) Ratio of Tc/Th in peripheral blood. Values were represented the means ± SEM (n = 6–8). #p < 0.05 and ##p < 0.01 versus Normal group; *p < 0.05 and **p < 0.01 versus Model group.
To make a more detail observation for the immune surveillance system in the TME, TIL was also determined by FCM. It was found that, although percentage of Th cell was decreased (p < 0.01), ESG brought out an evident increase on the Tc cell proportion in the TME (p < 0.05) accompanying with a higher Tc/Th ratio (p < 0.01), which was quite corresponding to the results of peripheral blood (Figure 3). Nevertheless, this promotion on Tc cell by ESG did not exist when ESG directly acted on splenocytes in vitro (Figure S3 in Supplementary Material).
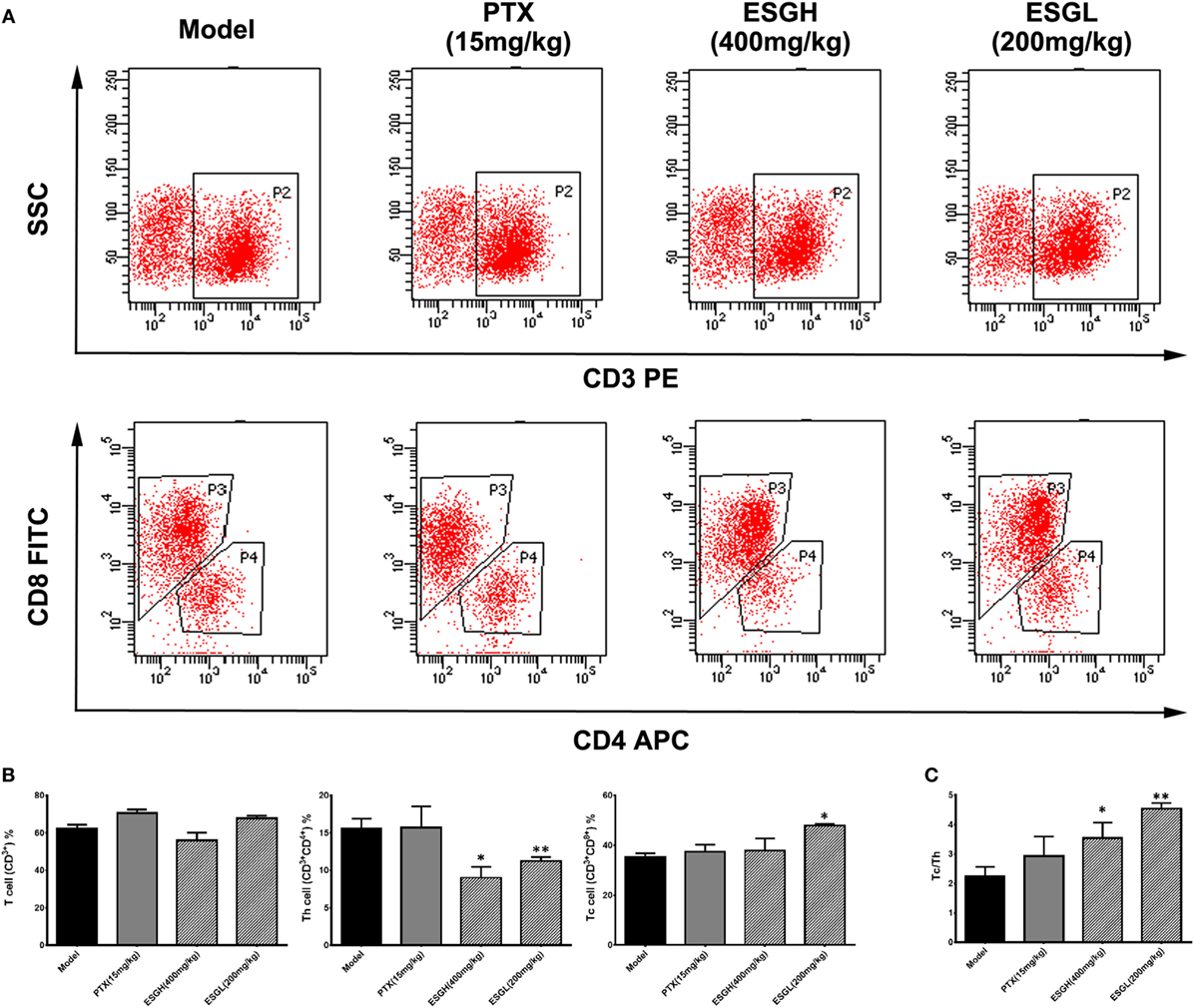
Figure 3. Effect of ESG on T cell subsets in tumor-infiltrating lymphocyte (TIL). (A) Representative scatter diagram. (B) Quantitative analysis for T cell subsets in TIL. (C) Ratio of Tc/Th in TIL. Values were represented the means ± SEM (n = 6–8). *p < 0.05 and **p < 0.01 versus Model group.
To further evaluate the enhancement of ESG on the T cell status, expressions of the two immune checkpoints, PD-1 and CTLA-4, were explored. Results from Figure 4 displayed that 4T1-cell xenograft significantly upregulated the mRNA expressions of pd1 in spleen (p < 0.01), while decreasing the protein level of PD-1 (p < 0.01). In comparison, both PTX and ESG obviously downregulated the pd1 gene expression (p < 0.01), but not affecting the protein level of PD-1. In tumor tissue, PTX evidently increased PD-1 protein expression (p < 0.05), while the mRNA level was almost the same as those from the Model group. By contrast, ESG exhibited obvious inhibition on PD-1, especially that the 400 mg/kg ESG made strong suppressions on both levels of mRNA and protein (p < 0.05 and p < 0.01, respectively).
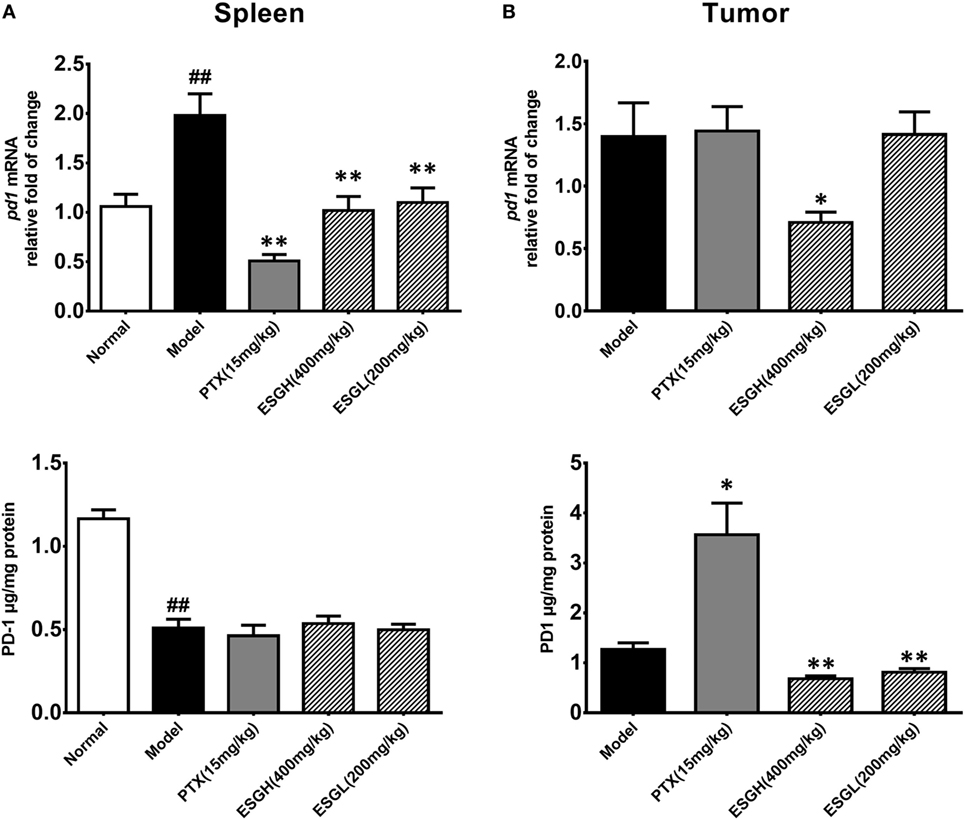
Figure 4. Effect of ESG on PD-1 in spleen (A) and tumor (B). The upper panels are results of quantitative real-time PCR for pd1. The lower panels are results of ELISA for PD-1 protein. Values were represented the means ± SEM (n = 6–8). #p < 0.05 and ##p < 0.01 versus Normal group; *p < 0.05 and **p < 0.01 versus Model group.
Regarding to CTLA-4, tumor xenograft made an evident upregulation on mRNA and protein level in spleen, while both treatments of PTX and ESG decreased them significantly (Figure 5). In tumor (Figure 6), PTX was intended to decrease mRNA level of ctla4 but to increase its protein expression. By contrast, both dosages of ESG were able to evidently downregulate ctla4 mRNA (p < 0.05), whereas not affecting the protein expression of CTLA-4 apparently.
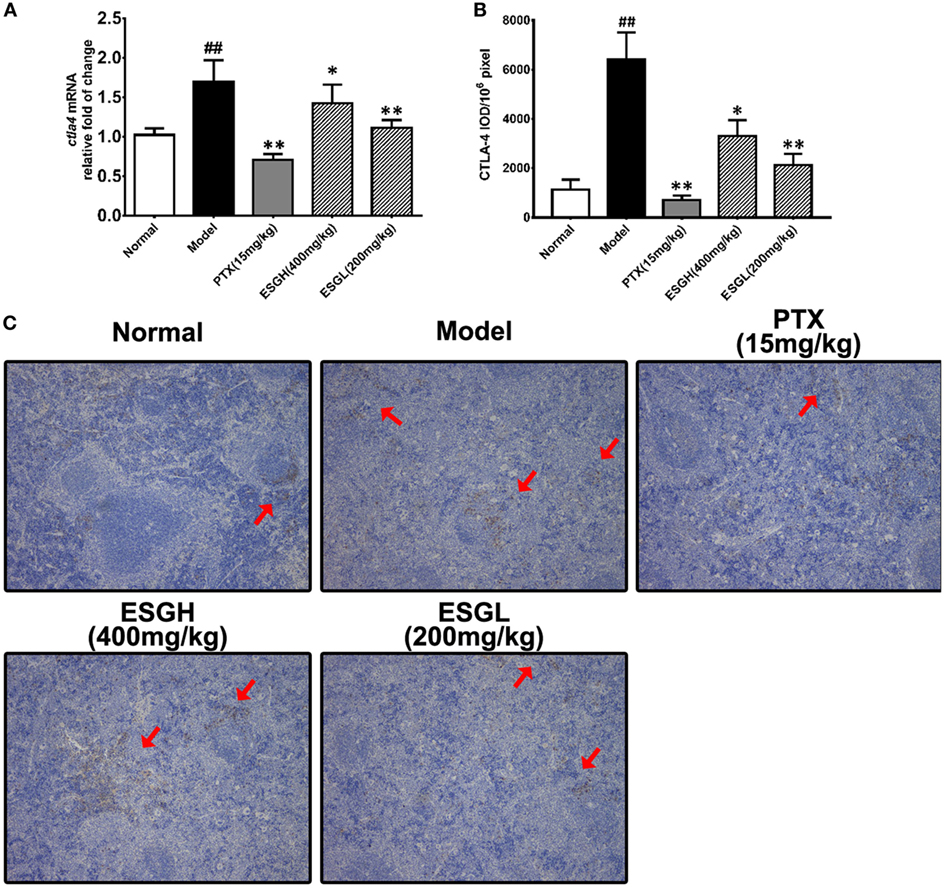
Figure 5. Effect of ESG on cytotoxic T lymphocyte antigen-4 (CTLA-4) in spleen. (A) Quantitative real-time PCR for ctla4. (B) Immunohistochemistry (IHC) for CTLA-4 in spleen. The mean density of positive area was calculated as ratio of integrated optical density to the total pixel of each picture (IOD/106 pixel). (C) Representative IHC image (400×) for CTLA-4 in spleen. Values were represented the means ± SEM (n = 6–8). #p < 0.05 and ##p < 0.01 versus Normal group; *p < 0.05 and **p < 0.01 versus Model group.
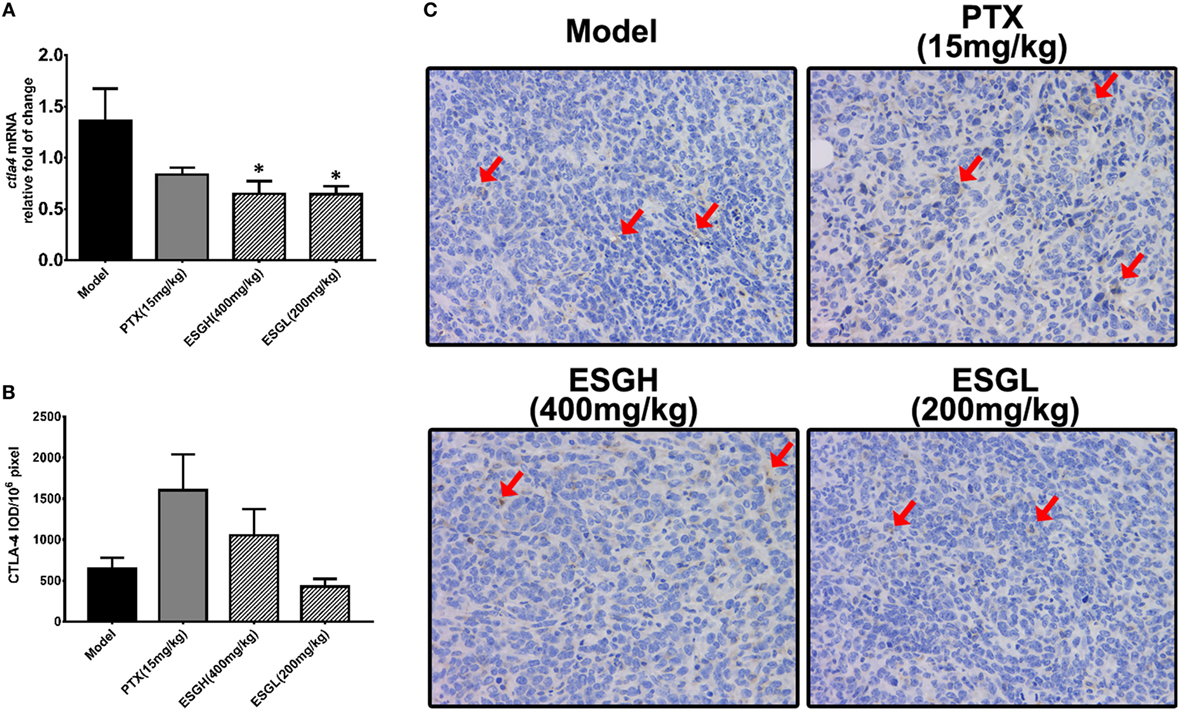
Figure 6. Effect of ESG on cytotoxic T lymphocyte antigen-4 (CTLA-4) in tumor. (A) Quantitative real-time PCR for ctla4. (B) Immunohistochemistry (IHC) for CTLA-4 in tumor. The mean density of positive area was calculated as ratio of integrated optical density to the total pixel of each picture (IOD/106 pixel). (C) Representative IHC image (400×) for CTLA-4 in tumor. Values were represented the means ± SEM (n = 6–8). *p < 0.05 and **p < 0.01 versus Model group.
Given that gut microbiota has been recognized as a pivotal assistant in chemotherapy and immunotherapy, we investigated its possible involvement in the efficacy of ESG in this part. As the above data indicated that ESGH group (400 mg/kg) exhibited better antitumor effect in the 4T1-xenograft model, gut microbiota in fecal samples from the Normal group, Model group, and ESGH group were analysis by the Illumina Miseq sequencing system. A total of 903,411 sequences were obtained from all the fecal samples, with an average of 34,962 sequences per sample (33,526–43,436 sequences). The high-quality sequences were then delineated into 25,394 OTUs at a similarity cutoff of 97% as previously reported (43). Common OTU analysis presented by Venn diagram indicated that there existed 889 unique OTUs in Normal group, 723 in Model group, and 541 in ESGH group (400 mg/kg), respectively, while 1,480 common OTUs were identified in all samples (Figure 7A).
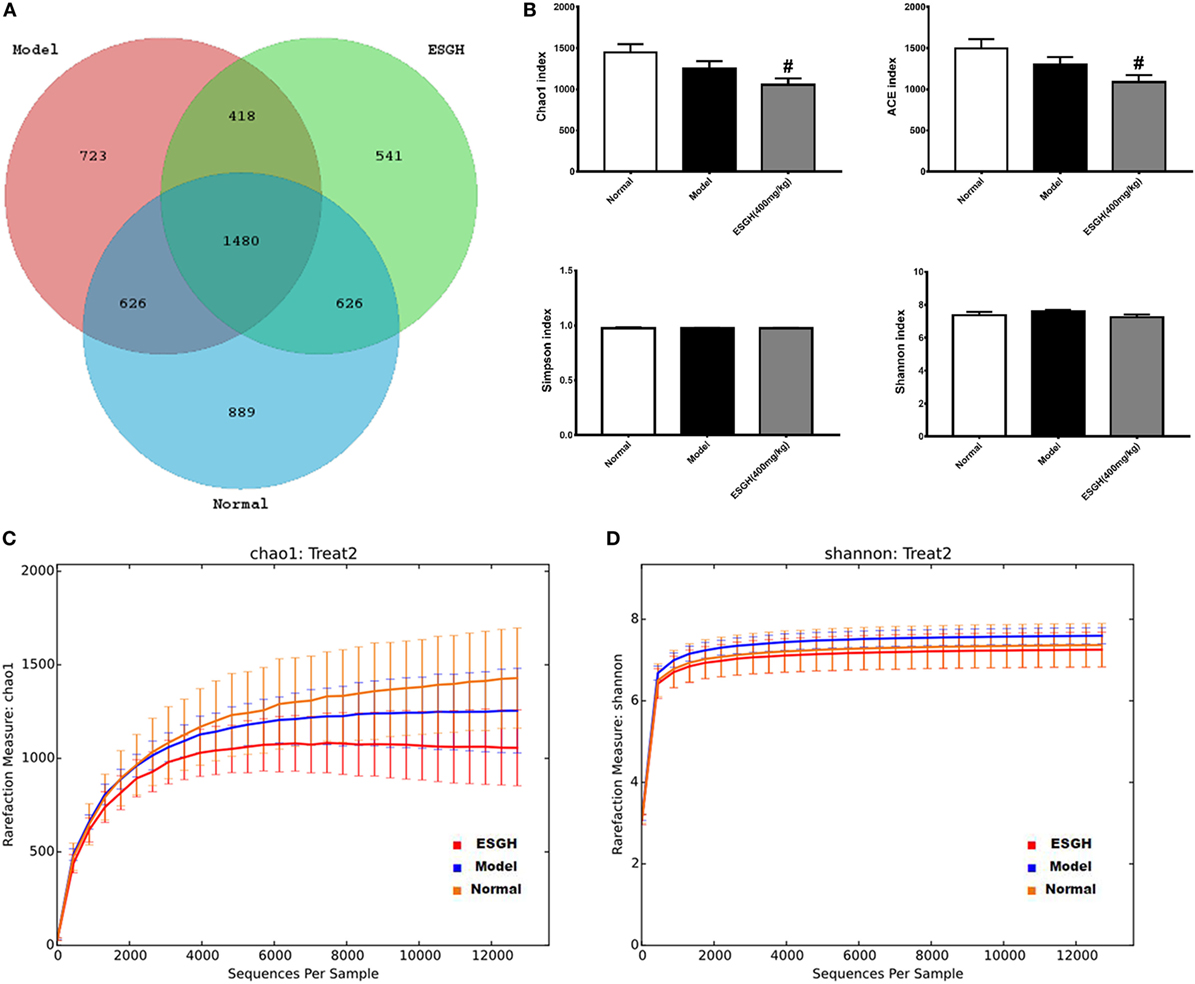
Figure 7. α-Diversity analysis on the fecal microbiota. (A) Venn diagram. (B) Alpha diversity indices comparison. (C,D) Rarefaction curve. Values were represented the means ± SEM (n = 8). #p < 0.05 and ##p < 0.01 versus Normal group; *p < 0.05 and **p < 0.01 versus Model group.
Microbiota community diversity was first evaluated by α-diversity analysis employing indices including Chao1, ACE, Shannon, and Simpson. Chao1 and ACE indices are estimators for community richness (44, 45). Shannon and Simpson indices represent community diversity and uniformity (46). As depicted by the data, gut microbiome of the ESGH group demonstrated significant reduced richness with lower Chao1 and ACE indexes (p < 0.01), compared with that of the Normal group (Figure 7B), while overall diversity was not affected. Rarefaction curve (Figures 7C–D) also presented a significant difference in the richness (Chao1), but not diversity (Shannon), among the three groups in this study.
β-Analysis was used to compare the similarity of overall community structure, which employed several unsupervised multivariate statistical assessments, including PCA and UniFrac NMDS. Both PCA (Figure 8A) and UniFrac NMDS (Figure 8B) displayed a marked structure shift in samples of Model group in contrast to those of Normal group; while after the 3-week treatment, gut microbiome from the tumor-bearing mice of ESGH group was restored to be similar with that of Normal group. UniFrac distances analysis (both weighted and unweighted, Figure 8C) made a further confirmation on the structure remodeling by ESG, as indicated by the significant intergroup difference between Normal group and Model group, as well as that between ESGH group and Model group (p < 0.01).
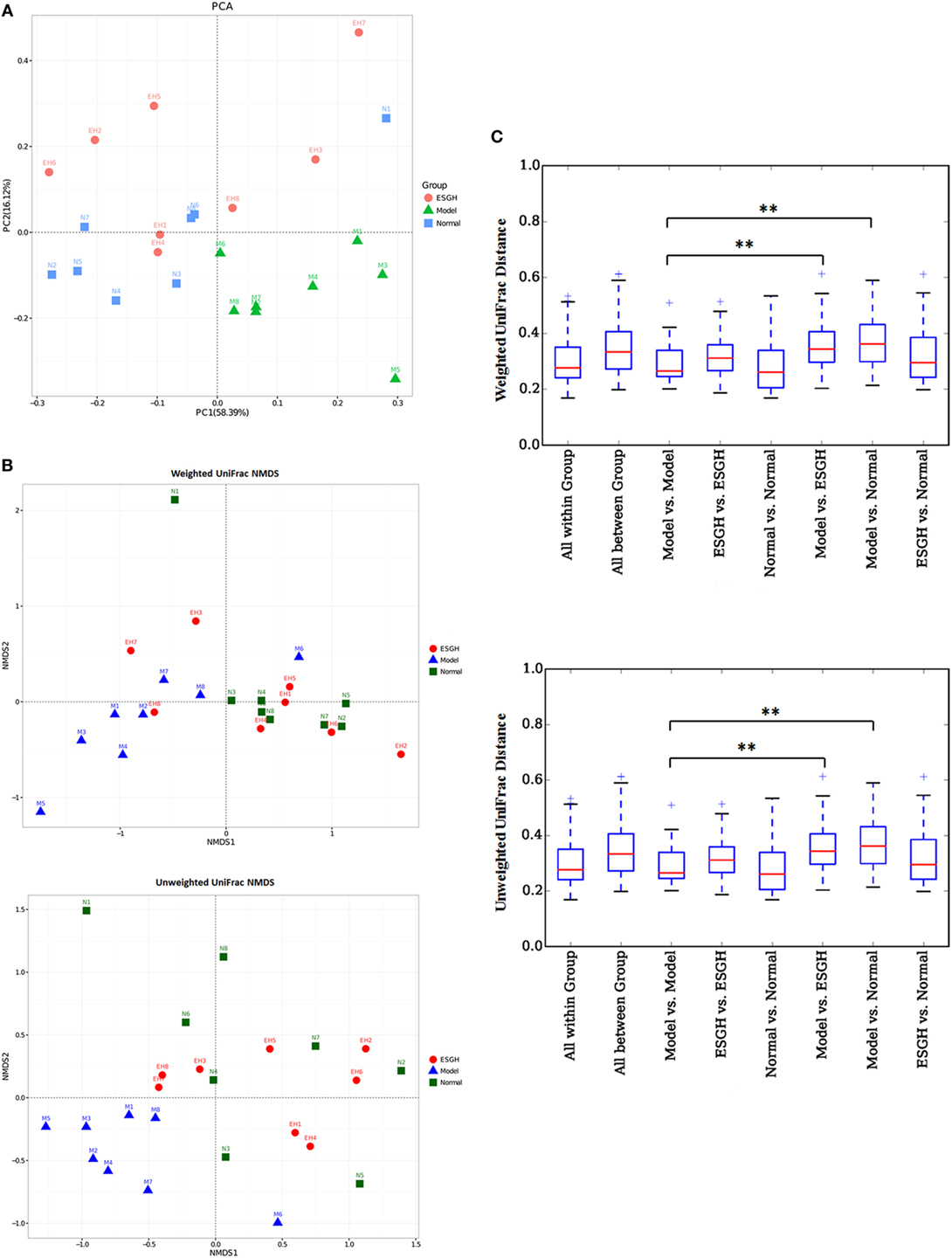
Figure 8. β-Diversity analysis on fecal microbiota. (A) Principal component analysis. The percent variation explained by each principal coordinate was indicated on the axes. (B) UniFrac distance-based Nonmetric Multidimensional Scaling (NMDS). (C) UniFrac distance analysis. Boxes are the interquartile range; median values are bands within the boxes; whiskers are 1.5 times the IQR; and open circle is an outlier value. There would be statistic difference between two groups when the intergroup distance is evidently higher than the within-group distance. *p < 0.05 and **p < 0.01 versus the distance of “Model versus Model” (n = 8).
Taxon-based analysis revealed marked differences at both phylum and genus levels among Normal, Model, and ESGH samples. Overall, a total of nine phyla were shared by samples from all groups (Figure 9A). Of them, Firmicutes and Bacteroidetes compromised over 90% of the total classified sequences. Relative abundances of Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, and Proteobacteria displayed significant differences in the three groups. In particular, ESGH treatment significantly raised the relative abundances of Firmicutes (p < 0.05) and Proteobacteria (p < 0.05) but reduced those of Actinobacteria (p < 0.01), Bacteroidetes (p < 0.01), and Cyanobacteria (p < 0.01), which apparently reversed the community shift induced by tumor xenograft (Figure 9B). At genus level, a total of 61 genera were identified from all samples (Data Sheet 2 in Supplementary Material), and hierarchical clustering analysis presented by heatmap showed that fecal microbiota from the Model group exhibited obvious community shift compared with those of Normal group and ESGH group (Figure 10A). Although LefSe analysis indicated no genus is specific for any group, there were 18 genus exhibited obvious differences among them. Briefly, three genera (Helicobacter, Rikenella, and Turicibacter) were evidently higher in both Normal group and ESGH group than those in Model group, which have been reported to be positively related to the enhanced immune response (47–50); while another 15 genera (Acinetobacter, Arthrobacter, Bacillus, Bacteroides, Blautia, Brevundimonas, Clostridium, Coprobacillus, Corynebacterium, Facklamia, Jeotgalicoccus, Parabacteroides, Prevotella, Sporosarcina, Staphylococcus, and Streptococcus) were lowered in both Normal group and ESGH group in contrast to those in Model group (Figure 10B).
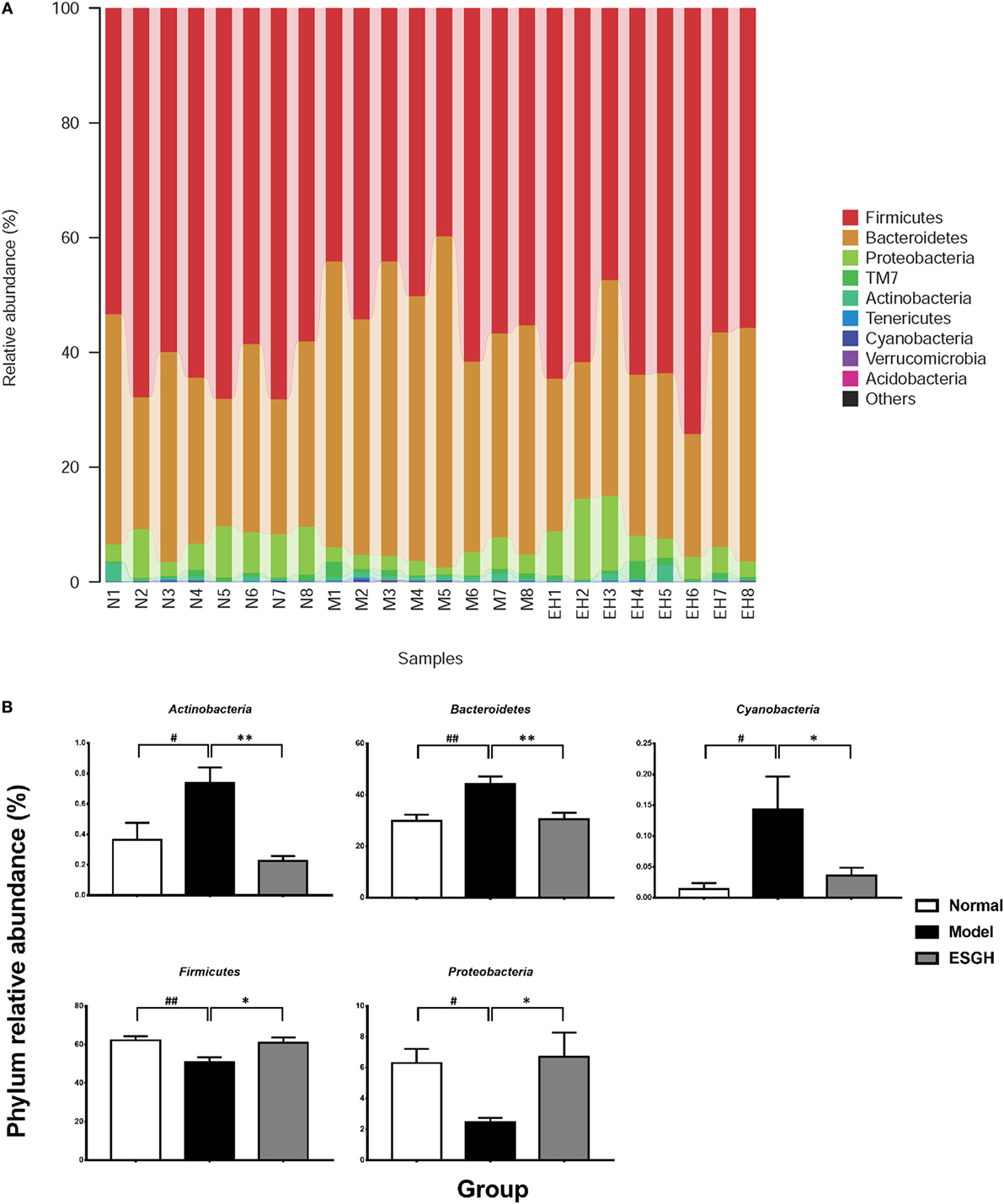
Figure 9. Taxonomy analysis on level of phyla. (A) Relative abundance of all detected phylum from each sample. (B) Significant intergroup differences were found in five phyla. Values were represented the means ± SEM (n = 8). #p < 0.05 and ##p < 0.01 versus Normal group; *p < 0.05 and **p < 0.01 versus Model group.
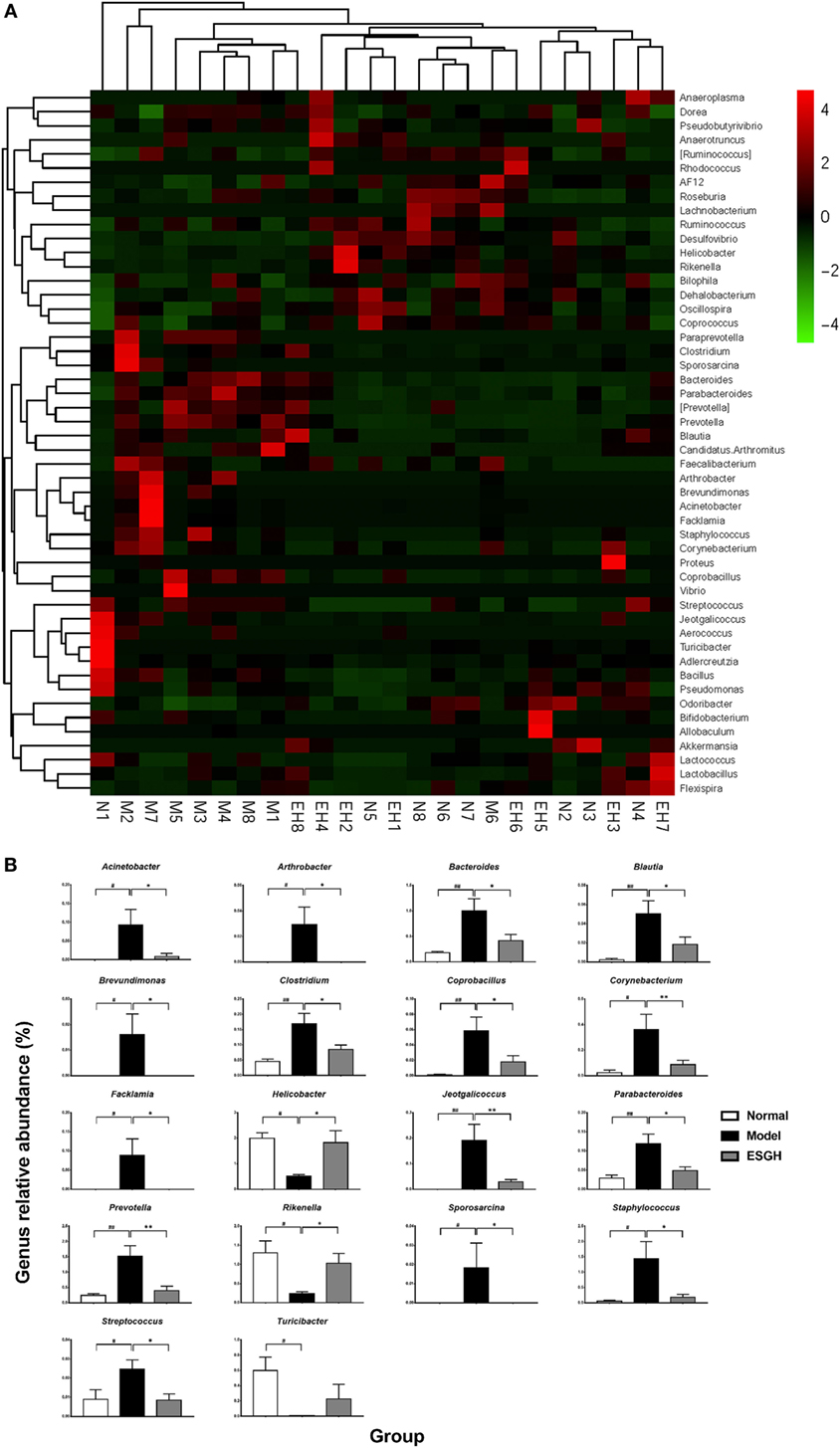
Figure 10. Taxonomy analysis on level of genera. (A) Heat map showing the relative abundance of major genera ranking top 50 from each sample. M, N, and EH denote Model group, Normal group, and ESGH groups, respectively. (B) Significant intergroup differences were found in 18 genera. Values were represented the means ± SEM (n = 8). #p < 0.05 and ##p < 0.01 versus Normal group; *p < 0.05 and **p < 0.01 versus Model group.
Via comparing the sequencing data with those collected in KEGG pathway database by PICRUSt (Figure 11), it was found that tumor xenograft significantly upregulated abundances of several genes that are responsible for five metabolism pathways, including “biosynthesis of other Secondary metabolites,” “energy metabolism,” “enzyme families,” “glycan biosynthesis and metabolism,” and “metabolism of cofactors and vitamins,” and a cellular processes pathway (“transport and catabolism”), but downregulated genes involved in the “cell mobility” and “signal transduction” pathways. After ESGH treatment, changes in the genes that referred to most of these pathways were evidently reversed, such as “transport and catabolism,” “enzyme families,” “glycan biosynthesis and metabolism,” and “biosynthesis of other Secondary metabolites,” indicating that together with the structural modulation, ESG could regulate the metabolic activities of the gut microbiota to promote the tumor immune surveillance.
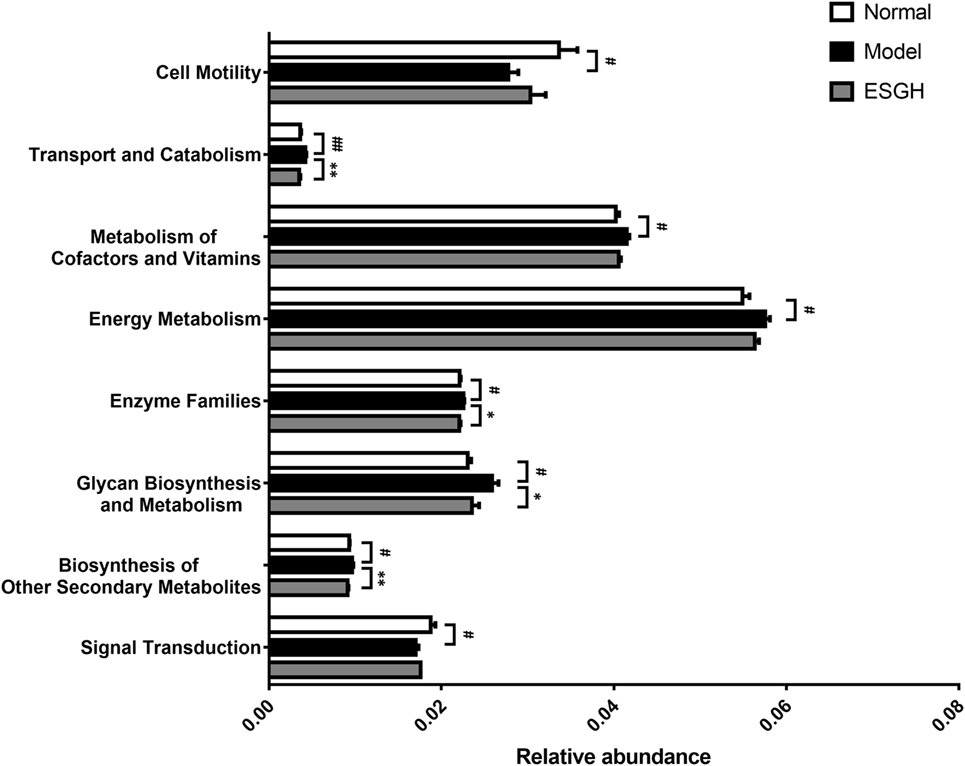
Figure 11. Microbiome function prediction according to Kyoto Encyclopedia of Genes and Genomes pathway database. Values were represented the means ± SEM (n = 8). #p < 0.05 and ##p < 0.01 versus Normal group; *p < 0.05 and **p < 0.01 versus Model group.
With the great success of paclitaxel in cancer chemotherapy, increasing attention has been paid for the natural compounds in prevention and treatment of cancer. So far, anticancer candidates derived from natural products, including alkaloids, saponins, polysaccharides, terpenoids, and flavonoids, have been extensively studied in laboratories and clinical investigations. More recently, potential of the active constituents from sporoderm-broken SG was also explored. Na et al. (30) found that the aforementioned polysaccharide exerted direct cytotoxicity on HCT116 via cell cycle arrest and apoptosis, but at a relative high concentration (1.25–7.5 mg/mL) (30); while Wang et al. (31) showed that a commercial polysaccharide from SG suppressed Sarcoma 180 only in vivo via the stimulation of NK cells, T cells, and macrophages (31).
In this study, anticancer potential of the polysaccharide-rich ESG was investigated in a murine 4T1-breast cancer xenograft model. Results showed that ESG effectively inhibited tumor growth both in terms of volumes and mass, accompanying with a significant necrosis in the tumor tissue; whereas this suppression was not mediated by a directly cytotoxicity as proved by the in vitro experiment. Numerous studies have demonstrated the importance of adaptive T cell-mediated cytotoxic responses as a dominant mechanism of host antitumor immune responses (51, 52), and TILs nesting in and around neoplastic cells have showed potential clinical implications, especially that the presence of CD8+ TIL is strongly associated with favorable prognosis in every solid human cancer studied virtually (53). Specifically, breast cancer patients with higher numbers of CD8+ TIL in the tumors are more likely to gain better outcomes concerning survival (54, 55). Tc cells (CD8+ T cells) are the most effective elements for tumor destruction (56). Spontaneously, these antitumor effector cells are first activated by the tumor cell-expressing surface molecules, such as calreticulin, tumor antigens in context of MHC class I molecules, and/or NKG2D ligands; then they lead apoptosis or inhibition on proliferation and angiogenesis to destroy the tumor tissue, mainly via increasing the cytotoxic factors within TME (including perforin, granzymes, IFN-α/β/γ, IL-1, IL-12, and TNF-α) (57). Despite not being an antitumor enforcer, Th cells (CD4+ T cells) have multiple impacts on the antitumor effect of Tc cells depending on the specific functions of various subsets, such as Th1, Th2, and regulatory Th cells (Treg). By secreting interferon-γ, Th1 cells are essential for the activation of Tc cells and have been shown to correlate with favorable survival in breast cancer (58). However, Treg cells are able to dampen the immune system so as to limit excessive immune responses that can cause collateral damage to normal tissue, which, on the other hand, resulted in a weaken on the antitumor function of Tc cells (58–61). In view of this, we hypothesized that the tumor control of ESG would be a restoration on the T cell paradigm. FCM data indicated that in the peripheral blood, percentage of Tc cells was increased by ESG treatment, and consequently, the ratio of Tc/Th was increased on account of the unchanged total T cell proportion. Intriguingly, CD8+ TILs of the tumor-bearing mice, together with the ratio of Tc/Th with TME, displayed a correspondent increase in response to ESG treatment. Moreover, it was found that ESG evidently increased the production of TNF in serum of the tumor-bearing mice but did not affect any other cytokines that are characteristic for Th1, Th2, or Th17 (Figure S4 in Supplementary Material). These collective results verified our hypothesis that by promoting the differentiation of T cell toward Tc, ESG employed Tc cells to potentiate the tumor immune surveillance, thereby effectively suppressing tumor growth. However, although ESG did not affect neither Th cell proportion in peripheral blood nor that in TME, its impact on Th cell subsets would be explored in the future due to their possible influence in tumor immune surveillance.
As depicted by the “3E immunoediting” theory, despite host antitumor immune responses stand the very heart of self-surveillance, the spontaneous tumor immunity, especially the dominant T-cell response, would not only be seized up by the tumor cell-driving “escape strategy” but also be limited in the setting of standard treatments (10, 52). Since derived from normal cells, tumor cells can develop several survival features to get immune tolerance, including reduced tumor antigens (CA15-3, CEA, PCNA, etc.), increased resistance or survival (STAT-3 or anti-apoptotic molecule Bcl2), or development of an immunosuppressive TME (releasing cytokines such as VEGF, TGF-β, or expressing immunoregulatory molecules such as IDO, PD-L1, Tim-3/galectin-9, and CTLA-4/CD80) (57). On the other hand, T-cell paradigm turns out to be anergy and exhaustion during cancer progression, due to the activation of membrane co-inhibitory signaling pathways, such as PD-1/PD-L1, and CTLA-4/B7 (11, 12), which finally results in decreased effector function and proliferative capacity. As demonstrated by our data, PTX apparently inhibited PD-1 and CTLA-4 in spleen but upregulated PD-1 in the tumor, implying a possible induction of anergy on the tumor immune surveillance. By contrast, ESG could evidently reduce the mRNA levels of pd1 and ctla4, both in tumor tissue and spleen. Although effect of ESG on the protein expressions was not exactly consistent with that on mRNA level, it was also found that protein expression of PD-1 was significantly suppressed in tumor, whereas that of CTLA-4 was only downregulated in spleen.
Indeed, growing evidence reveals the correlation between mRNA and protein abundances in the cell is notoriously poor (62). This is because there exists the translational control on the process of protein translation from mRNA, involves mRNA degradation induced by microRNA and siRNA, initiation codon scanning, ribosome assemble, and so on, and the initiation phase is often the most regulated part (63–65). Thus, translational control results in an unequal protein translation from the transcribed mature mRNA, which enables the organisms to adapt to the changed circumstances via a quick regulation on the protein biosynthesis in each cell. As showed in the data, when compared with the normal control, protein level of PD-1 in spleen was reduced in the tumor-bearing mice, although the mRNA expression of PD-1 was upregulated. By contrast, PD-1 protein of ESG groups was evidently decreased, while their mRNA levels were equal to that of normal control group. Similar poor correlation between the mRNA and protein levels of PD-1 also exhibited in tumor. These results suggested that ESG would make profound regulations on the translation control of PD-1 and CTLA-4, so as to suppress this immune checkpoints signaling. Together with the previous results, it would be rationally to speculate that ESG could effectively restore the T cell paradigm by reversing the anergy and exhaustion status via suppressing the co-inhibitory checkpoints, thereby resulting in a favorite control on breast cancer. Furthermore, impacts on the tumor immune surveillance, such as co-inhibitory signaling pathway interaction (PD-1/PD-L1 and CTLA-4/CD86) and tumoral immunological balances (such as CD28: B7 binding versus CTLA-4: B7 binding), would be explored to make a more comprehensive exploration for the antitumor activity of ESG.
One more issue to be noteworthy is that, ESG did not exhibit obvious promotion on Tc cells differentiation in vitro. As ESG was administrated orally in this work, it is highly probable that a regulation on gut microbiota by ESG promotes the restoration on the exhausted Tc cells. Besides the cardinal role in the development and efficiency of immune system, microbiota also interact with gut mucosal surfaces thereby intervene the therapeutic responses for tumors occurring outside of the intestinal tract (16, 66). Specifically in breast cancer, increasing data have shown that gut microbiome is involved with all potential related factors of breast cancer, including immune regulation, metabolism of endogenous and exogenous substances, obese status, and so forth. CD8+ TIL has been found to be positive related to a better outcomes concerning survival in breast cancer patients (54), and an optimization on the gut microbiota community brings about a promotion on CD8+ T cell-mediated immunity, thereby indicating a better prognosis and an effective outcome of immunotherapy (14, 67). As demonstrated by our data, in despite of the reduced richness in the microbiome of the tumor-bearing mice (α-diversity), mice receiving ESG treatment shared a parallel microbiome structure as those of the normal counterparts when compared with the mice of Model group (β-diversity). Furthermore, ESG was able to reverse the microbiota community shift caused by tumor xenograft, of which 5 phyla (Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, and Proteobacteria) and 18 genera were significantly affected. Among the influenced genera, Helicobacter, Rikenella, and Turicibacter were the enriched ones that accounted over 0.5%, and especially, both Helicobacter and Rikenella were restored up to about 1%. Despite as pathogenicities to certain inflammatory-related carcinoma and autoimmune disorders, it has been revealed that Helicobacter and Rikenella were positively correlated to enhanced immune response (47, 48). Turicibacter was also strongly associated with immune functions, as indicated by the fact that Turicibacter populations within the gastrointestine would be almost abolished both in innate and adaptive immunodeficiency mouse models (49, 50). On the other hand, Bacteroides and other 14 genera were declined by ESG treatment. Typically, Bacteroides took up the biggest population (1% or so) among them, and it has been considered as a beneficial commensal when retained to a proper quantity in the gut, otherwise inducing bacteremia and abscess formation in multiple body sites (68). Nevertheless, increased abundance of Bacteroides is associated with immunosuppression (69) and carcinogenesis (70). Overall, ESG made a beneficial alterations on the gut microbiome community, i.e., to enrich the immunocompetence-related genera (Helicobacter, Rikenella, and Turicibacter) and to decline certain immunosuppressive-related ones (such as Bacteroides), resulting in a restoration on the dysbiosis induced by cancer xenograft.
Indeed, gut microbiota not only experiences long-term coevolution with host (71) but also influences the host immunity (72, 73). Especially, lifestyle, including diets and medicine intake, contributes greatly to the modification on gut microbiota (71). On the other hand, microbial utilization of complex polysaccharides is a major driving force in shaping the composition of the human gut microbiota (74, 75). In our works, it was found that ESG is mainly composed of polysaccharide made up of glucose, and it restored several pathways involving metabolism (“biosynthesis of other Secondary metabolites,” “energy metabolism,” “enzyme families,” “glycan biosynthesis and metabolism,” and “metabolism of cofactors and vitamins”), cellular processes (“transport and catabolism” and “cell mobility”), and environmental information processing (“cell mobility”). So it is probable that by oral administration, ESG was able to directly affect the growth homeostasis within the microbiota via regulating its metabolism mode, and possibly, serves as selective growth promoters to certain species. Therefore, microbiota community of the ESG-treatment samples was evidently shifted with lowered community richness (with reduced Chao1 and ACE indexes) but unaltered diversity. Nevertheless, the specific organisms affected by ESG have to be figured out and confirmed via experiment on germ-free animal models. Moreover, several studies revealed that natural polysaccharides would take several weeks to influence the gut microbiota (76, 77). It is probable that the remodeling by ESG may take several days to shift the community membership and to change the metabolism characteristics. Hence, tumor growth was not suppressed until the tumor immune surveillance was powerful enough by a long-term ESG treatment. Taken together, given that the promotion by ESG on Tc-mediated tumor suppression would only achieve by oral administration, it is feasible that the modulation on gut microbiome community would contribute greatly to ESG’s tumor control by restoring the exhausted Tc cells.
Of course, there are some more affairs to be taken in account for the antitumor activity of ESG, such as oxidative stress and aerobic metabolism (also namely Warburg effect). Oxidative stress has a controversial association with cancer, which is mainly mediated by reactive oxidative species (ROS). The elevated levels of ROS not only increases gene mutations and genomic instability (78, 79) but also inactivates the phosphatases within cancer cells, so their target proteins responsible for proliferation are kept activated, resulting in an uncontrolled cell growth (80). In these view of oxidative stress, Chikara et al. have reviewed the antioxidant potential of phytochemicals in cancer chemoprevention and treatment (81). However, activation of oxidation (such as by increasing ROS) contributes to autophagy and apoptosis in cancer cells, even though it is crucial for normal-to-cancerous cell transformation and cancer development (82, 83). Therefore, whether the antioxidant effect contributes to the anticancer activity depends on the various types of compounds. For instance, it is reported that ganoderic acids induce apoptosis in human cervical cancer HeLa cells via increasing the generation of intracellular ROS (84); while a G. lucidum extract elicited antitumor effects by suppressing cell growth and inducing antioxidative/detoxification activity in human ovarian OVCAR-3 cells (85). Regarding Warburg effect, it is indeed a metabolic nature of TME shifting from oxidative phosphorylation to aerobic glycolysis (86). It is critical for cancer progress and immune escape and characterized by increased glucose uptake and accumulation of lactate, accompanying with the upregulations of transporters, glycolytic enzymes, and the responsible signaling pathway proteins (87, 88). Therefore, pathways and activities involved in Warburg effect have been considered as novel targets in cancer therapy, such as metformin that acts via reducing the expression of monocarboxylate transporter 4 on cancer-associated fibroblasts (89). Although no researches report the impact on Warburg effect, it has revealed an antioxidant effect of the spore of G. lucidum in model of ischemia/reperfusion and streptozotocin-induced neuron damage (90, 91). In the future, we would pay attention to the possible role of antioxidant effect (or Warburg effect) by ESG in its anticancer activity.
Collectively, this study revealed that a polysaccharide-rich extract from sporoderm-breaking spore of G. lucidum (ESG) would serve as natural candidate for breast cancer treatment. The underlying mechanism of ESG was probably contributed to a suppression on co-inhibitory signaling (PD-1 and/or CTLA-4) and a consequent restoration on the exhausted Tc cells, to which the gut microbiome remodeling made a great contribution. Especially, such regulation involved not only changes in microbiota structure and community membership but also alternations in several metabolism pathways within the microbiome. So far, it has been found that, there is an intricate relationship between the biology of T cells (mainly regulatory T cells and Tc cell), and the various metabolites produced by host and commensal microbes, such as vitamins and short chain fatty acids (67, 92). Hence in all probability, regulation on the metabolism of gut microbiota would be the pivotal of ESG to enhance the Tc-mediated tumor surveillance against breast cancer. On the other hand, toxicity test is demanded to make comprehensive evaluation on its safety, although no article has yet reported the adverse effect of the spore of G. lucidum, except one suggests that an extract from fruit body of G. lucidum has lethal and sub-lethal effects on zebrafish embryos (93). And rigor pharmacokinetics study, as well as multi-omics investigations, for ESG will be applied to intercept the underlying mechanism of its immune-regulation activity, as well as the potential in cancer treatment.
The experiment was performed according to the Guidelines of Guangdong Institute of Microbiology Laboratory Animal Center, Guangdong Institute of Microbiology Laboratory Animal Ethics Committee. The experimental protocols were approved by the Guangdong Institute of Microbiology Laboratory Animal Ethics Committee.
Conceived and designed the experiments; drafted and revised the manuscript: JS, YL, and YX. Performed the experiments: JS, LS, DL, OS, YZ, HL, and ZX. Analyzed the data: JS, LS, DL, and CJ.
All coauthors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Shijie Li, Ms. Cuiyun Mo, and Ms. Lixia Gan for the help in characteristics analysis for ESG. This work was financially supported by the High-level Leading Talent Introduction Program of Guangdong Academy of Science (No. 2016GDASRC-0102), the Natural Science Foundation of Guangdong Province (No. 2015A030313711), Guangdong Province Science and Technology Project (No. 2016A030303041), and Guangzhou Science and Technology Planning Project (No. 2016201604030072, No. 201707020022).
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fimmu.2018.01765/full#supplementary-material.
1. Chen Y, Zhang Y. Application of the CRISPR/Cas9 system to drug resistance in breast cancer. Adv Sci (Weinh) (2018) 5(6):1700964. doi:10.1002/advs.201700964
2. Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: burden and trends. Cancer Epidemiol Biomarkers Prev (2017) 26(4):444–57. doi:10.1158/1055-9965.EPI-16-0858
3. PDQ. Adult Treatment Editorial Board. Breast Cancer Treatment (PDQ®): Patient Version, PDQ Cancer Information Summaries. Bethesda, MD: National Cancer Institute (US) (2017).
4. Moulder S, Hortobagyi GN. Advances in the treatment of breast cancer. Clin Pharmacol Ther (2008) 83(1):26–36. doi:10.1038/sj.clpt.6100449
5. DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin (2014) 64(1):52–62. doi:10.3322/caac.21203
6. Cazzaniga M, Bonanni B. Breast cancer chemoprevention: old and new approaches. J Biomed Biotechnol (2012) 2012:985620. doi:10.1155/2012/985620
7. Reddy KB. Triple-negative breast cancers: an updated review on treatment options. Curr Oncol (2011) 18(4):e173–9. doi:10.3747/co.v18i4.738
8. Franklin C, Livingstone E, Roesch A, Schilling B, Schadendorf D. Immunotherapy in melanoma: recent advances and future directions. Eur J Surg Oncol (2017) 43(3):604–11. doi:10.1016/j.ejso.2016.07.145
9. Remon J, Pardo N, Martinez-Marti A, Cedres S, Navarro A, Martinez de Castro AM, et al. Immune-checkpoint inhibition in first-line treatment of advanced non-small cell lung cancer patients: current status and future approaches. Lung Cancer (2017) 106:70–5. doi:10.1016/j.lungcan.2017.02.002
10. Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol (2002) 3(11):991–8. doi:10.1038/ni1102-991
11. Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood (2009) 114(8):1537–44. doi:10.1182/blood-2008-12-195792
12. Thommen DS, Schumacher TN. T cell dysfunction in cancer. Cancer Cell (2018) 33(4):547–62. doi:10.1016/j.ccell.2018.03.012
13. Katz H, Alsharedi M. Immunotherapy in triple-negative breast cancer. Med Oncol (2017) 35(1):13. doi:10.1007/s12032-017-1071-6
14. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (2015) 350(6264):1084–9. doi:10.1126/science.aac4255
15. Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology (2017) 153(6):1621.e–33.e. doi:10.1053/j.gastro.2017.08.022
16. Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science (2013) 342(6161):971–6. doi:10.1126/science.1240537
17. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science (2018) 359(6371):97–103. doi:10.1126/science.aan4236
18. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science (2018) 359(6371):91–7. doi:10.1126/science.aan3706
19. Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science (2013) 341(6143):295–8. doi:10.1126/science.1235872
20. Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res (2017) 179:204–22. doi:10.1016/j.trsl.2016.08.002
21. Lin ZB. Modern Research of Ganoderma lucidum. 2nd ed. Beijing: Beijing Medical University Press (2001). p. 219–83.
22. Bishop KS, Kao CH, Xu Y, Glucina MP, Paterson RR, Ferguson LR. From 2000 years of Ganoderma lucidum to recent developments in nutraceuticals. Phytochemistry (2015) 114:56–65. doi:10.1016/j.phytochem.2015.02.015
23. Boh B, Berovic M, Zhang J, Zhi-Bin L. Ganoderma lucidum and its pharmaceutically active compounds. Biotechnol Annu Rev (2007) 13:265–301. doi:10.1016/S1387-2656(07)13010-6
24. Wu G, Qian Z, Guo J, Hu D, Bao J, Xie J, et al. Ganoderma lucidum extract induces G1 cell cycle arrest, and apoptosis in human breast cancer cells. Am J Chin Med (2012) 40(3):631–42. doi:10.1142/S0192415X12500474
25. Dai J, Miller MA, Everetts NJ, Wang X, Li P, Li Y, et al. Elimination of quiescent slow-cycling cells via reducing quiescence depth by natural compounds purified from Ganoderma lucidum. Oncotarget (2017) 8(8):13770–81. doi:10.18632/oncotarget.14634
26. Tsao SM, Hsu HY. Fucose-containing fraction of Ling-Zhi enhances lipid rafts-dependent ubiquitination of TGFbeta receptor degradation and attenuates breast cancer tumorigenesis. Sci Rep (2016) 6:36563. doi:10.1038/srep36563
27. Chang CJ, Chen YY, Lu CC, Lin CS, Martel J, Tsai SH, et al. Ganoderma lucidum stimulates NK cell cytotoxicity by inducing NKG2D/NCR activation and secretion of perforin and granulysin. Innate Immun (2014) 20(3):301–11. doi:10.1177/1753425913491789
28. Li A, Shuai X, Jia Z, Li H, Liang X, Su D, et al. Ganoderma lucidum polysaccharide extract inhibits hepatocellular carcinoma growth by downregulating regulatory T cells accumulation and function by inducing microRNA-125b. J Transl Med (2015) 13:100. doi:10.1186/s12967-015-0465-5
29. Sun LX, Li WD, Lin ZB, Duan XS, Xing EH, Jiang MM, et al. Cytokine production suppression by culture supernatant of B16F10 cells and amelioration by Ganoderma lucidum polysaccharides in activated lymphocytes. Cell Tissue Res (2015) 360(2):379–89. doi:10.1007/s00441-014-2083-6
30. Na K, Li K, Sang T, Wu K, Wang Y, Wang X. Anticarcinogenic effects of water extract of sporoderm-broken spores of Ganoderma lucidum on colorectal cancer in vitro and in vivo. Int J Oncol (2017) 50(5):1541–4. doi:10.3892/ijo.2017.3939
31. Wang PY, Zhu XL, Lin ZB. Antitumor and immunomodulatory effects of polysaccharides from broken-spore of Ganoderma lucidum. Front Pharmacol (2012) 3:135. doi:10.3389/fphar.2012.00135
32. Yue GG, Fung KP, Leung PC, Lau CB. Comparative studies on the immunomodulatory and antitumor activities of the different parts of fruiting body of Ganoderma lucidum and Ganoderma spores. Phytother Res (2008) 22(10):1282–91. doi:10.1002/ptr.2478
33. Chen JH, Wu FQ, Li WL, Li XC, Wang XR, Zhang D, et al. Comparison of contents of triterpenoids, polysaccharides and heavy metals in Ganoderma lucidum sporule powder and Ganoderma lucidum. China Pharmacy (2008) 19:2585–7.
34. Li TM, Zhu JH, Liu C, Sai Ji RH, Xin C, Li Y. Effect of extract from fruit body and spore of Ganoderma lucidum on the activity of acid phosphatase in mouse peritoneal macrophage. Acta Edulis Fungi (2004) 11(03):30–3. doi:10.16488/j.cnki.1005-9873.2004.03.006
35. Zhu JH, Li TM, Chen Y, Yang X, Cong N, Chu HW, et al. Effect of extract from fruit body and spore of Ganoderma lucidum on immune function of mice. Acta Edulis Fungi (2004) 11(04):24–7. doi:10.16488/j.cnki.1005-9873.2004.04.005
36. Qiao D, Wang L, Liu J, Sun Y, Ye H, Zeng X. Preliminary characterization of proteoglycans from Hyriopsis cumingii. Zhong Yao Cai (2013) 36(01):15–22. doi:10.13863/j.issn1001-4454.2013.01.013
37. Miller DN, Bryant JE, Madsen EL, Ghiorse WC. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol (1999) 65(11):4715–24.
38. Zhao L, Wang G, Siegel P, He C, Wang H, Zhao W, et al. Quantitative genetic background of the host influences gut microbiomes in chickens. Sci Rep (2013) 3:1163. doi:10.1038/srep01163
39. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods (2010) 7(5):335–6. doi:10.1038/nmeth.f.303
40. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol (2011) 12(6):R60. doi:10.1186/gb-2011-12-6-r60
41. Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol (2013) 31(9):814–21. doi:10.1038/nbt.2676
42. Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics (2010) 26(6):715–21. doi:10.1093/bioinformatics/btq041
43. Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One (2012) 7(8):e42529. doi:10.1371/journal.pone.0042529
44. Chao A, Hwang WH, Chen YC, Kuo CY. Estimating the number of shared species in two communities. Stat Sin (2000) 10(1):227–46.
45. Durden C, Dong Q. RICHEST – a web server for richness estimation in biological data. Bioinformation (2009) 3(7):296–8. doi:10.6026/97320630003296
47. Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest (2013) 123(2):700–11. doi:10.1172/JCI62236
48. Huang G, Khan I, Li X, Chen L, Leong W, Ho LT, et al. Ginsenosides Rb3 and Rd reduce polyps formation while reinstate the dysbiotic gut microbiota and the intestinal microenvironment in Apc(Min/+) mice. Sci Rep (2017) 7(1):12552. doi:10.1038/s41598-017-12644-5
49. Dimitriu PA, Boyce G, Samarakoon A, Hartmann M, Johnson P, Mohn WW. Temporal stability of the mouse gut microbiota in relation to innate and adaptive immunity. Environ Microbiol Rep (2013) 5(2):200–2. doi:10.1111/j.1758-2229.2012.00393.x
50. Kellermayer R, Dowd SE, Harris RA, Balasa A, Schaible TD, Wolcott RD, et al. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice. FASEB J (2011) 25(5):1449–60. doi:10.1096/fj.10-172205
51. Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature (2012) 482(7385):400–4. doi:10.1038/nature10755
52. Wick DA, Webb JR, Nielsen JS, Martin SD, Kroeger DR, Milne K, et al. Surveillance of the tumor mutanome by T cells during progression from primary to recurrent ovarian cancer. Clin Cancer Res (2014) 20(5):1125–34. doi:10.1158/1078-0432.CCR-13-2147
53. Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer (2012) 12(4):298–306. doi:10.1038/nrc3245
54. Margolis KL, Rodabough RJ, Thomson CA, Lopez A, McTiernan A; Women’s Health Initiative Research Group. Prospective study of leukocyte count as a predictor of incident breast, colorectal, endometrial, and lung cancer and mortality in postmenopausal women. Arch Intern Med (2007) 167(17):1837–44. doi:10.1001/archinte.167.17.1837
55. Ali HR, Provenzano E, Dawson SJ, Blows FM, Liu B, Shah M, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol (2014) 25(8):1536–43. doi:10.1093/annonc/mdu191
56. Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I. Nat Rev Immunol (2015) 15(7):405–14. doi:10.1038/nri3845
57. Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases – elimination, equilibrium and escape. Curr Opin Immunol (2014) 27:16–25. doi:10.1016/j.coi.2014.01.004
58. West NR, Kost SE, Martin SD, Milne K, Deleeuw RJ, Nelson BH, et al. Tumour-infiltrating FOXP3(+) lymphocytes are associated with cytotoxic immune responses and good clinical outcome in oestrogen receptor-negative breast cancer. Br J Cancer (2013) 108(1):155–62. doi:10.1038/bjc.2012.524
59. Allaoui R, Hagerling C, Desmond E, Warfvinge CF, Jirstrom K, Leandersson K. Infiltration of gammadelta T cells, IL-17+ T cells and FoxP3+ T cells in human breast cancer. Cancer Biomark (2017) 20(4):395–409. doi:10.3233/CBM-170026
60. Eftekhari R, Esmaeili R, Mirzaei R, Bidad K, de Lima S, Ajami M, et al. Study of the tumor microenvironment during breast cancer progression. Cancer Cell Int (2017) 17:123. doi:10.1186/s12935-017-0492-9
61. Taylor NA, Vick SC, Iglesia MD, Brickey WJ, Midkiff BR, McKinnon KP, et al. Treg depletion potentiates checkpoint inhibition in claudin-low breast cancer. J Clin Invest (2017) 127(9):3472–83. doi:10.1172/JCI90499
62. Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett (2009) 583(24):3966–73. doi:10.1016/j.febslet.2009.10.036
63. Kong J, Lasko P. Translational control in cellular and developmental processes. Nat Rev Genet (2012) 13(6):383–94. doi:10.1038/nrg3184
64. Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev (2006) 20(5):515–24. doi:10.1101/gad.1399806
65. Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol (2010) 11(2):113–27. doi:10.1038/nrm2838
66. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science (2013) 342(6161):967–70. doi:10.1126/science.1240527
67. Bhattacharya N, Yuan R, Prestwood TR, Penny HL, DiMaio MA, Reticker-Flynn NE, et al. Normalizing microbiota-induced retinoic acid deficiency stimulates protective CD8(+) T cell-mediated immunity in colorectal cancer. Immunity (2016) 45(3):641–55. doi:10.1016/j.immuni.2016.08.008
68. Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev (2007) 20(4):593–621. doi:10.1128/CMR.00008-07
69. Zhang Z, Liu L, Tang H, Jiao W, Zeng S, Xu Y, et al. Immunosuppressive effect of the gut microbiome altered by high-dose tacrolimus in mice. Am J Transplant (2018) 18(7):1646–56. doi:10.1111/ajt.14661
70. Zakharzhevskaya NB, Vanyushkina AA, Altukhov IA, Shavarda AL, Butenko IO, Rakitina DV, et al. Outer membrane vesicles secreted by pathogenic and nonpathogenic Bacteroides fragilis represent different metabolic activities. Sci Rep (2017) 7:5008. doi:10.1038/s41598-017-05264-6
71. Moeller AH, Li Y, Mpoudi Ngole E, Ahuka-Mundeke S, Lonsdorf EV, Pusey AE, et al. Rapid changes in the gut microbiome during human evolution. Proc Natl Acad Sci U S A (2014) 111(46):16431–5. doi:10.1073/pnas.1419136111
72. Sigal M, Meyer TF. Coevolution between the human microbiota and the epithelial immune system. Dig Dis (2016) 34(3):190–3. doi:10.1159/000443349
73. Yang L, Liu S, Ding J, Dai R, He C, Xu K, et al. Gut microbiota co-microevolution with selection for host humoral immunity. Front Microbiol (2017) 8:1243. doi:10.3389/fmicb.2017.01243
74. Tamura K, Hemsworth GR, Dejean G, Rogers TE, Pudlo NA, Urs K, et al. Molecular mechanism by which prominent human-gut Bacteroidetes utilize mixed-linkage beta-glucans, major health-promoting cereal polysaccharides. Cell Rep (2017) 21(2):417–30. doi:10.1016/j.celrep.2017.09.049
75. El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol (2013) 11(7):497–504. doi:10.1038/nrmicro3050
76. Tang C, Sun J, Zhou B, Jin C, Liu J, Kan J, et al. Effects of polysaccharides from purple sweet potatoes on immune response and gut microbiota composition in normal and cyclophosphamide treated mice. Food Funct (2018) 9(2):937–50. doi:10.1039/c7fo01302g
77. Nguyen SG, Kim J, Guevarra RB, Lee JH, Kim E, Kim SI, et al. Laminarin favorably modulates gut microbiota in mice fed a high-fat diet. Food Funct (2016) 7(10):4193–201. doi:10.1039/C6FO00929H
78. Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med (2010) 49(11):1603–16. doi:10.1016/j.freeradbiomed.2010.09.006
79. Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science (2008) 320(5876):661–4. doi:10.1126/science.1156906
80. Lee SR, Yang KS, Kwon J, Lee C, Jeong W, Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem (2002) 277(23):20336–42. doi:10.1074/jbc.M111899200
81. Chikara S, Nagaprashantha LD, Singhal J, Horne D, Awasthi S, Singhal SS. Oxidative stress and dietary phytochemicals: role in cancer chemoprevention and treatment. Cancer Lett (2018) 413:122–34. doi:10.1016/j.canlet.2017.11.002
82. Tong Y, Zhang G, Li Y, Xu J, Yuan J, Zhang B, et al. Corilagin inhibits breast cancer growth via reactive oxygen species-dependent apoptosis and autophagy. J Cell Mol Med (2018). doi:10.1111/jcmm.13647
83. Wu CL, Huang AC, Yang JS, Liao CL, Lu HF, Chou ST, et al. Benzyl isothiocyanate (BITC) and phenethyl isothiocyanate (PEITC)-mediated generation of reactive oxygen species causes cell cycle arrest and induces apoptosis via activation of caspase-3, mitochondria dysfunction and nitric oxide (NO) in human osteogenic sarcoma U-2 OS cells. J Orthop Res (2011) 29(8):1199–209. doi:10.1002/jor.21350
84. Liu RM, Li YB, Liang XF, Liu HZ, Xiao JH, Zhong JJ. Structurally related ganoderic acids induce apoptosis in human cervical cancer HeLa cells: involvement of oxidative stress and antioxidant protective system. Chem Biol Interact (2015) 240:134–44. doi:10.1016/j.cbi.2015.08.005
85. Hsieh TC, Wu JM. Suppression of proliferation and oxidative stress by extracts of Ganoderma lucidum in the ovarian cancer cell line OVCAR-3. Int J Mol Med (2011) 28(6):1065–9. doi:10.3892/ijmm.2011.788
86. Warburg O, Posener K, Negelein E. Über den Stoffwechsel der Carcinomzelle. Biochem Z (1924) 152(12):309–44.
87. Warburg O. On the origin of cancer cells. Science (1956) 123(3191):309–14. doi:10.1126/science.123.3191.309
88. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell (2012) 21(3):297–308. doi:10.1016/j.ccr.2012.02.014
89. Romero IL, Mukherjee A, Kenny HA, Litchfield LM, Lengyel E. Molecular pathways: trafficking of metabolic resources in the tumor microenvironment. Clin Cancer Res (2015) 21(4):680–6. doi:10.1158/1078-0432.CCR-14-2198
90. Levin RM, Xia L, Wei W, Schuler C, Leggett RE, Lin AD. Effects of Ganoderma lucidum shell-broken spore on oxidative stress of the rabbit urinary bladder using an in vivo model of ischemia/reperfusion. Mol Cell Biochem (2017) 435(1–2):25–35. doi:10.1007/s11010-017-3053-6
91. Zhou Y, Qu ZQ, Zeng YS, Lin YK, Li Y, Chung P, et al. Neuroprotective effect of preadministration with Ganoderma lucidum spore on rat hippocampus. Exp Toxicol Pathol (2012) 64(7–8):673–80. doi:10.1016/j.etp.2010.12.011
92. Zeng H, Chi H. Metabolic control of regulatory T cell development and function. Trends Immunol (2015) 36(1):3–12. doi:10.1016/j.it.2014.08.003
Keywords: spore of Ganoderma lucidum, cytotoxic T cells, exhaustion, immune checkpoints, gut microbiota
Citation: Su J, Su L, Li D, Shuai O, Zhang Y, Liang H, Jiao C, Xu Z, Lai Y and Xie Y (2018) Antitumor Activity of Extract From the Sporoderm-Breaking Spore of Ganoderma lucidum: Restoration on Exhausted Cytotoxic T Cell With Gut Microbiota Remodeling. Front. Immunol. 9:1765. doi: 10.3389/fimmu.2018.01765
Received: 15 November 2017; Accepted: 17 July 2018;
Published: 31 July 2018
Edited by:
Zhaoping Li, Ronald Reagan UCLA Medical Center, United StatesReviewed by:
Lei Lei, Northwest University, ChinaCopyright: © 2018 Su, Su, Li, Shuai, Zhang, Liang, Jiao, Xu, Lai and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Lai, bGFpeW9uZzg4NzlAc2luYS5jb20=;
Yizhen Xie, eGlleWl6aGVuQDEyNi5jb20=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.