- 1School of Food and Biological Engineering, Hubei University of Technology, Wuhan, China
- 2Guelph Food Research Centre, Agriculture and Agri-Food Canada, Guelph, ON, Canada
- 3State Key Laboratory of Food Science and Technology/International Joint Laboratory on Food Safety, Jiangnan University, Wuxi, China
- 4State Key Laboratory of Food Science and Technology, Nanchang University, Nanchang, China
Enterotoxigenic Escherichia coli (ETEC) infection causes the death of Caenorhabditis elegans, which can be prevented by certain Lactobacillus isolates. The host response of C. elegans to ETEC infection and its regulation by the isolates are, however, largely unclear. This study has revealed that, in agreement with the results of life-span assays, the expression of the genes encoding p38 mitogen-activated protein kinase (MAPK) pathway (nsy-1, sek-1, and pmk-1), insulin/insulin-like growth factor (DAF/IGF) pathway (daf-16), or antimicrobial peptides (lys-7, spp-1, and abf-3) and other defensing molecules (abf-2, clec-85) was upregulated significantly when the wild-type nematode (N2) was subjected to ETEC infection. This upregulation was further enhanced by the pretreatment with Lactobacillus zeae LB1, but not with L. casei CL11. Mutants defective in the cell signaling of C. elegans were either more susceptible (defective in NSY-1, SEK-1, PMK-1, or DAF16) or more resistant (defective in AGE-1, DBL-1, SKN-1, or SOD-3) to ETEC infection compared with the wild-type. Mutants defective in antimicrobial peptides (LYS-7, SPP1, or ABF-3) were also more susceptible. In addition, mutants that are defective in NSY-1, SEK-1, PMK-1, DAF16, ABF-3, LYS-7, or SPP1 showed no response to the protection from L. zeae LB1. The expression of the genes encoding antimicrobial peptides (lys-7, spp-1, and abf-3) and other defensing molecules (abf-2, clec-60, and clec-85) were almost all upregulated in AGE-1- or DBL-1-defective mutant compared with the wild-type, which was further enhanced by the pretreatment of L. zeae LB1. The expression of these genes was, however, mostly downregulated in NSY-1- or DAF-16-defective mutant. These results suggest that L. zeae LB1 regulates C. elegans signaling through the p38 MAPK and DAF/IGF pathways to control the production of antimicrobial peptides and defensing molecules to combat ETEC infection.
Introduction
Probiotics are defined as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (1) and have widely been used as a food component to promote human health, including enhancement of host immunity and control of enteric bacterial infection (2, 3). Enterotoxigenic Escherichia coli (ETEC) are a pathogen commonly causing diarrhea in humans in addition to pigs (4–6). Previous studies have shown that ETEC infection can be controlled by probiotics (7–9). However, the mechanisms underlying probiotic effects, including host responses, remain largely unknown, which are critical for effective development and application of probiotics.
Caenorhabditis elegans is a small free-living soil nematode. The short life-span, clear genetic background, easy for culturing and gene manipulation, the simple microbiota and availability of various mutants of C. elegans have made the nematode an excellent laboratory animal model for studying bacteria and host interactions (10–12). The usefulness of this laboratory animal model in elucidating the molecular mechanisms of probiotic effects has particularly been highlighted by a recent study that revealed a secreted antigen A (SagA, with peptidoglycan hydrolase activity) from Enterococcus faecium with a function to protect C. elegans against Salmonella pathogenesis by promoting pathogen tolerance in a tol-1-dependent manner (13). In the past, a number of bacterial pathogens, such as Pseudomonas aeruginosa (10), Salmonella enterica (14, 15), Staphylococcus aureus (16), and Enterococcus faecalis (11) have been assessed for their virulence with the nematode model. In addition, C. elegans has increasingly been used for preselecting probiotics for pathogen control (17–20). Interestingly, probiotic bacteria preselected by using C. elegans were reported to reduce pig diarrhea, demonstrating a correlation of probiotic effect between the two animals (18). Recently, we determined that isolate Lactobacillus zeae LB1 was able to protect C. elegans from death caused by ETEC infection, which was mediated by inhibiting ETEC enterotoxin production, rather than by interfering colonization of the pathogen in the worm gut (20). The finding was based on the following observations: (1) the expression of ETEC enterotoxin genes was significantly down-regulated during the infection of C. elegans by ETEC, which had been pretreated with isolate LB1; (2) the clone with either estA or estB (enterotoxin genes) expressed in a non-pathogenic E. coli was able to effectively kill the nematode and the killing by the clones could also be prevented by isolate LB1; (3) the same isolate only partially inhibited the gene expression of enterotoxins in ETEC or in the clones in vitro. Whether L. zeae LB1 can also modulate the host immunity to protect C. elegans is unclear and is addressed in the present study.
It is known that C. elegans immune defense mechanisms are evolutionarily conserved, including the DAF/insulin-like growth factor (DAF/IGF) pathway, p38 mitogen-activated protein kinase (p38 MAPK) pathway, and transforming growth factor-β (TGF-β) signaling pathway (21–24). The C. elegans innate immune response consists of the production of numerous antimicrobial proteins and among these proteins, many of their gene expression are inducible upon pathogen infection (25–27). Moreover, some of these putative antimicrobial genes are regulated by signaling pathways involved in the defense of nematodes and mammals against pathogen infection (22, 24, 28). By investigating the behavior of a wild-type C. elegans strain, when exposed to ETEC infection and Lactobacillus protection, and corresponding gene expression of key components in the p38 MAPK and DAF/IGF pathways and antimicrobial peptides, followed by assessing various mutants defective in particular components of the pathways or antimicrobial peptides for their behavior, we were able to determine that L. zeae LB1 could regulate C. elegans cell signaling through the p38 MAPK and DAF/IGF pathways to control the production of antimicrobial peptides and related molecules to combat ETEC infection. The results are presented herein.
Materials and Methods
C. elegans and Bacteria
Caenorhabditis elegans N2 Bristol wild-type and mutants that are defective in lys-7 (mutant ok1384), nsy-1 (mutant ag3), pmk-1 (mutant km25), sek-1 (mutant ag1), skn-1 (mutant zu67), dbl-1 (mutant nk3), spp-1 (mutant ok2703), abf-3 (mutant ok3366), daf-16 (mutant mu86), age-1 (mutant hx546), sod-3 (mutant gk235), or ced-9 (mutant n1950) were obtained from Caenorhabditis Genetics Center, University of Minnesota, Minneapolis, MN, USA. The double mutant defective in both daf-16 and pmk-1 was kindly provided by Dr. Dennis Kim (Department of biology, Massachusetts Institute of Technology, Boston). C. elegans strains were routinely maintained on nematode growth medium (NGM) plates seeded with E. coli OP50 using standard procedures (29).
K88+ ETEC strain JG280 is a hemolytic E. coli of serotype O149: K88 (F4), a porcine isolate possessing the toxin genes of elt, estA, estB, and astA, and antibiotic resistance to tetracycline, ceftiofur, ampicillin, spectinomycin, apramycin, gentamicin, neomycin, and trimethoprim/sulfonamide (6). The ETEC JG280 was cultured in Luria–Bertani (LB) medium at 37°C for 16 h. Following three washes with M9 medium, 200 µl of cell suspension (108 CFU/ml) was spread on a NGM plate (100 mm in diameter) and dried for 3 h at 22°C before beginning of the assay. Lactobacillus isolates (L. zeae LB1 and L. casei CL11) were originally obtained from the adult chicken or pig intestine (18). Either de Man Rogosa Sharpe broth or agar was used to culture Lactobacillus isolates at 37°C for 18–24 h in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI, USA) with an atmosphere of 85% N2, 10% CO2, and 5% H2. After three washes with M9 medium, 200 µl cell suspension of each Lactobacillus isolate (108 CFU/ml) was spread on a NGM plate (100 mm in diameter) and dried for 3 h at 22°C prior to the use.
Life-Span Assay of C. elegans
The life-span assays of C. elegans were performed using the methods published previously (17, 20) with some modifications. Briefly, the synchronized C. elegans were transferred to NGM agar with E. coli OP50 at 25°C for 48–60 h until they reached the L4 stage. In the assay, there were usually three types of a treatment including: (1) Control, (2) ETECT Infection only, and (3) Probiotic Pretreatment. In the control group, the nematode was treated with E. coli OP50 (food for C. elegans) only throughout the entire assay. In the group of ETECT infection only, the nematode was incubated with E. coli OP50 for 18 h followed by incubation with ETEC for up to 15 days in the absence of E. coli OP50. In the group of Probiotic Pretreatment, the nematode was incubated with a Lactobacillus isolate (either L. zeae LB1 or L. casei CL11) for 18 h followed by incubation with ETEC for up to 15 days in the absence of E. coli OP50. The incubation temperature was 25°C. Each assay was started by transferring L4 stage worms (50 worms per replicate and 3 replicates per treatment) onto the agar plates seeded with either E. coli OP50 or a Lactobacillus isolate, which was designated as day 0. After 18 h incubation, worms on each plate within the group of either ETEC Infection only or Probiotic Pretreatment were transferred to a fresh NGM plate daily that had been seeded with ETEC JG280 and was subsequently incubated at 25°C. In parallel, worms within the Control group were transferred to a fresh NGM plate daily that had been seeded with E. coli OP50 after the 18 h incubation with the same bacterium. The survival of nematode was examined at 24-h intervals for up to 15 days. To determine the survival of C. elegans, the number of live worms was recorded daily, and the percentage of surviving worms was calculated by the following formula: survival (%) = (live worms/total worms used) × 100. A worm was considered to be dead when it failed to respond to touch. In the assay where a mutant was examined, the procedure remained unchanged except that the wild-type nematode was replaced by the mutant. Each assay was repeated at least twice unless it is otherwise indicated.
RNA Extraction
To prepare the lysates for RNA extraction, approximately 150 worms from each treatment were sampled on day 2 of the life-span assay. The sampling date were selected based on the observation that the death of worms started to increase on days 3 and 4 after ETEC infection (Figure 1). Following two washes with PBS (pH 7.4), the worms were disrupted in 0.8 ml of a Lysis/Binding Buffer from the mirVana miRNA Isolation Kit (Ambion, TX, USA) by a bead-beater (PowerLzyer24, MO BIO Laboratories, Inc., Carlsbad, CA, USA). The beating was conducted at 3,500 rpm for two cycles followed by four cycles at 3,000 rpm and four cycles at 2,500 rpm. Each cycle lasted for 1.5 min and there was a 2-min interval between two cycles with the samples on ice. The total RNA of C. elegans was then extracted using the mirVana miRNA Isolation Kit according to manufacturer’s instructions. After RNA extraction, the samples were treated with DNase I (Ambion, TX, USA) at 37°C for 30 min and then verified as DNA-free by PCR assays with primers specific to act-1. RNA integrity was determined by visualization in an agarose gel. The concentration of total RNA was determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
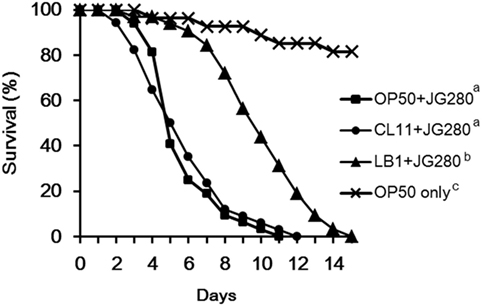
Figure 1. Effects of feeding isolates LB1 (Lactobacillus zeae) and CL11 (Lactobacillus casei) on the survival of the wild-type Caenorhabditis elegans (N2) after infection with Enterotoxigenic Escherichia coli (ETEC) JG280. The worms were first fed either with E. coli OP50 or Lactobacillus (isolate LB1, CL11) at 108 CFU/ml for 18 h and then ETEC JG280 for the remaining days. Treatments: ■, E. coli OP50 and then ETEC JG280; ●, isolate CL11 and then ETEC JG280; ▲, isolate LB1 and then ETEC JG280; and ×, treated with E. coli OP50 only. All the groups showing different letters were significantly different (P ≤ 0.01) in their survival curves.
Reverse Transcription and Real-Time QPCR Analysis
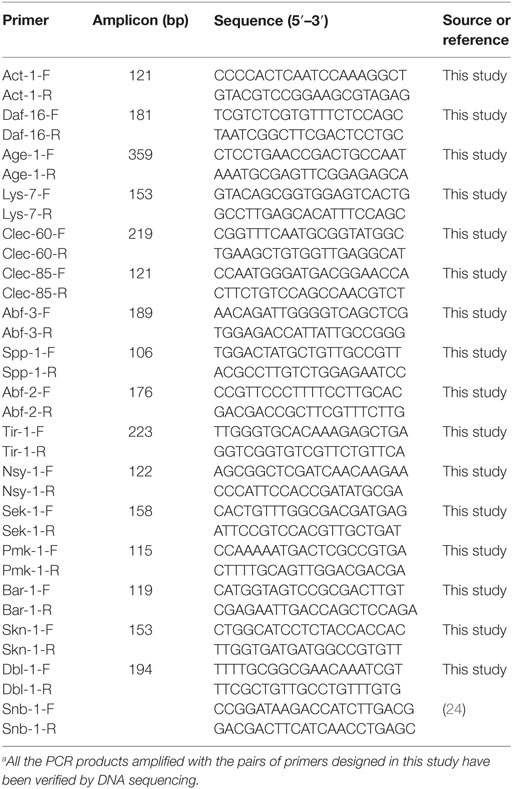
The C. elegans gene expression was determined by reverse transcription and quantitative PCR (QPCR) analysis as described previously (30) with some modifications. Briefly, a RNA sample was used for first-strand cDNA synthesis using the SuperScript first-strand synthesis system (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions. Housekeeping genes snb-1 and act-1 were used as internal controls for relative quantification of gene expression. QPCR assays were performed using 7500 Real-Time PCR System (Applied Biosystems, Foster, CA, USA) and brilliant SYBR green QPCR master mix (Bio-Rad Laboratories, Richmond, VA, USA). All the primers for PCR assays are listed in Table 1. The primers developed from the present study were designed using the primer designing tool by NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast) and verified experimentally by sequencing the amplicons from each pair of the primers (data not shown). In the QPCR assays, 1μl of each cDNA sample was included in a 24-µl reaction mixture containing 12.5 µl Master Mix, 3.75 µl each of the primers at 150 nM, and 4 µl irradiated and double autoclaved dH2O. The QPCR program included 5 min at 95°C and 40 cycles of 95°C for 30 s, 56°C for 1 min, and 72°C for 30 s. Fluorescence was measured after each annealing during the cycles.
The QPCR data were analyzed using the 2−ΔΔCt method to determine the relative abundance (fold changes) of target genes (31). The cycle threshold, Ct, is the point at which fluorescence above the background is statistically significant. Ct values were determined with the 7500 Real-Time PCR System software based on a threshold line that was manually defined above the non-informative fluorescent data. The ΔCt represents the difference between the Ct value with the primers to a target gene and the Ct value to the housekeeping genes. The ΔΔCt represents the difference between the ΔCt value of treatment group (either treated with Lactobacillus or ETEC JG280, or a mutant) and the ΔCt value of control group (treated with E. coli OP50, or the wild-type nematode). The values derived from 2−ΔΔCt represent fold changes of samples in abundance relative to the reference samples. The reference samples (either the samples treated with E. coli OP50 or the samples of wild-type nematode) had the 2−ΔΔCt value of 1.
Statistical Analysis
All statistical computation analyses were performed using the Statistical Analysis System (SAS release 9.2, SAS Institute Inc., Cary, NC, USA). Survival curves for C. elegans were compared using the Kaplan–Meier survival analysis followed by a log-rank test. One-way analysis of variance and the Tukey’s multiple comparisons were carried out to test for significant differences between the means. Means with P values ≤0.05 were considered to differ significantly.
Results
Enhancement in the Resistance of C. elegans to ETEC JG280 Infection by L. zeae LB1
In our previous study, a temperature-sensitive mutant (SS104) of C. elegans that harbors a temperature-sensitive allele of glp-4 (bn2) was used to develop a C. elegans infection and protection assay for preselection of probiotics (20). In the present study, identification of the host response of C. elegans at the molecular level to the infection of ETEC and also to the protection offered by Lactobacillus was the goal. Since there was a need to use various mutants of C. elegans, the wild-type nematode (N2) had to be used in the present study in order to compare the behavior of the wild-type and mutants in both life-span assays and gene expression experiments. Figure 1 shows the effects of isolates L. zeae LB1 and L. casei CL11 on the life-span of C. elegans infected with ETEC. The wild-type nematode (N2) pretreated with isolate LB1 had a significantly extended life-span (P ≤ 0.05) than those infected with ETEC JG280 only or pretreated with isolate CL11. No significant difference in the life-span (P > 0.05) was detected between the worms infected with ETEC JG280 only or pretreated with isolate CL11 followed by ETEC infection. These results are similar to our previously reported observations with a temperature-sensitive mutant (SS104) of C. elegans (20).
Response of Wild-Type C. elegans in Gene Expression to ETEC Pretreated With Lactobacillus Isolates
To examine the host immune response against infection, the wild-type worms on day 2 of the life-span assay were selected since the numbers of viable worms started to dramatically decrease on day 3 and day 4 (Figure 1) and the major components in the p38 MAPK (tir-1, nsy-1, sek-1, and pmk-1) and DAF/IGF (daf-16 and age-1) pathways, previously identified antimicrobial peptides (lys-7, spp-1, abf-2, clec-85, clec-60, and abf-3), and other reported defense molecules (sod-3, dbl-1, and skn-1) were used as the indicators for the host response and signaling transduction. As shown in Figure 2, when infection with ETEC the expression of almost all selected genes was upregulated (P ≤ 0.05) except for tir-1, clec-60, skn-1, and sod-3. More specifically, expression of the genes associated with the p38 MAPK pathway, such as nsy-1, sek-1, and pmk-1, or associated with the DAF/IGF pathway, such as daf-16 and age-1, or dbl-1 gene was all increased significantly (nearly two- to fourfold, P ≤ 0.05). Notably, the transcription of antimicrobial peptide, genes such as lys-7, spp-1, abf-2, clec-85, and abf-3 were all upregulated (almost 8–10-fold, P ≤ 0.05). Compared with the groups infected with ETEC only, pretreatment of the worms with L. casei CL11 showed no effect on the expression of all the selected genes. In contrast, the pretreatment with L. zeae LB1 significantly upregulated (P ≤ 0.05) the expression of all the selected genes except for tir-1, age-1, dbl-1, and skn-1.
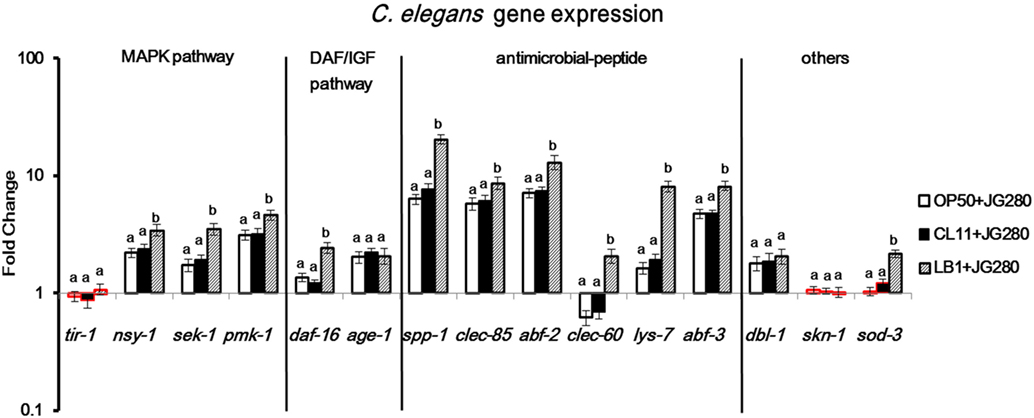
Figure 2. Gene expression of the wild-type Caenorhabditis elegans in response to the treatment of Enterotoxigenic Escherichia coli (ETEC) JG280 only, or to ETEC JG280 after pretreatment with Lactobacillus zeae LB1 or L. casei CL11. The nematode was sampled on day 2 of the life-span assays. The baseline is the level of gene expression in C. elegans treated with E. coli OP50 only. Relative expression was determined using the 2−ΔΔCt method as the ratio of transcript level of each target gene to the housekeeping genes within each group and expressed as fold changes by comparing each treatment group to the control group (E. coli OP50). The first ΔCT is the comparison between target genes and housekeeping genes. The second ΔCT represents the comparison between treatment group and control group. Data are presented as mean ± SD. Means marked with “a,” “b” were significantly different (P ≤ 0.05) for the same gene among different treatments. While the treatment groups represented by the bars with their borders in red had no significant difference (P > 0.05) between each treatment and the control group (fed E. coli OP50 only) in the gene expression level of the same gene, the remaining treatment groups represented by the bars with their borders in black differed significantly from the control group (P ≤ 0.05).
Resistance of C. elegans to ETEC Infection Involves the p38 MAPK and DAF/IGF Signaling Pathways as Well as Antimicrobial Peptides and Other Related Molecules
To determine the roles of C. elegans cell signaling in the resistance to ETEC infection, 12 different mutants were used to investigate the life-span of C. elegans after ETEC infection. The mutants were divided into four groups: (1) defective in the p38 MAPK pathway including mutants ag3 (defective in nsy-1), ag1 (sek-1), and km25 (pmk-1); (2) defective in the DAF/IGF pathway including mutants mu86 (daf-16) and hx546 (age-1); (3) defective in antimicrobial peptides including mutants ok3366 (abf-3), ok1384 (lys-7), and ok2703 (spp-1); and (4) defective in other molecules with a defense function, including mutants nk3 (dbl-1), gk235 (sod-3), n1950 (ced-9), and zu67 (skn-1). All the tested three mutants in Group 1 and mutant mu86 (defective in daf-16) in Group 2 were highly susceptible to ETEC infection (Figures 3A,B). More specifically, the life-span of these four mutants was reduced by more than 30% compared to the wild-type nematode (N2). Similar results were also observed with the mutants in Group 3 that are defective in antimicrobial peptides (Figure 3C). In contrast, mutant hx546 (defective in age-1) in Group 2 and the three mutants (nk3, gk235, and zu67) in Group 4 that are defective in dbl-1, sod-3, or skn-1 were more resistant to ETEC infection (Figures 3B,D). However, mutant n1950 (defective in ced-9) showed no changes in the life-span compared to the wild-type (Figure 3D).
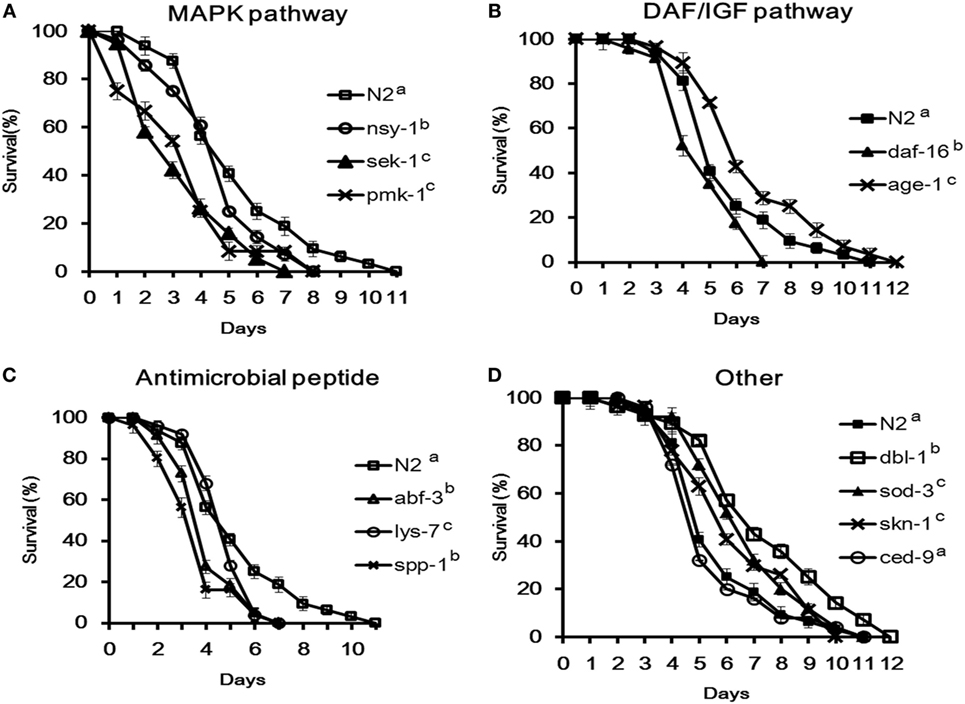
Figure 3. Survival of Caenorhabditis elegans mutants infected with Enterotoxigenic Escherichia coli (ETEC) JG280. (A) Survival curves of the mutants defective in the p38-mitogen-activated protein kinase pathway: nsy-1(ag3), sek-1(ag1), or pmk-1(km25); (B) survival curves of the mutants defective in the DAF/IGF pathway: daf-16(mu86) or age-1(hx546); (C) survival curves of the mutants defective in antimicrobial peptide genes: lys-7(ok1384), abf-3(ok3366), or spp-1(ok2703); (D) survival curves of the mutants defective in genes with other functions: dbl-1(nk3), sod-3(gk235), skn-1(zu67), or ced-9(n1950). All the groups were infected with ETEC JG280 after 18 h incubation with E. coli OP50 in the life-span assay. The wild-type (N2) served as a reference. All the groups showing different letters were significantly different (P ≤ 0.01) in their survival curves.
Immunomodulatory Activity of L. zeae LB1 Involves Some Components in the p38 MAPK and DAF/IGF Signaling Pathways as Well as Antimicrobial Peptides
To determine if L. zeae LB1 could trigger cell signaling of C. elegans to resist ETEC infection, the 12 mutants were also examined for their resistance to ETEC infection after pretreatment with the Lactobacillus isolate. Interestingly, only the four mutants that are defective in nsy-1, sek-1, pmk-1, or daf-16 and more susceptible to ETEC infection than the wild-type showed no changes to ETEC infection even they were pretreated with L. zeae LB1 (Figures 4A–C,E), indicating no protection from the isolate. Moreover, L. zeae LB1 showed no protection to the three mutants defective in antimicrobial peptide genes (lys-7, spp-1, or abf-3), i.e., no changes to ETEC infection regardless of the pretreatment with the Lactobacillus isolate (Figures 4J–L). In contrast, pretreatment with L. zeae LB1 significantly increased (P ≤ 0.05) the life-span of the five mutants (Figures 4D,F–I) that were either more resistant or no change in the resistance compared to the wild-type when treated with ETEC JG280 only.
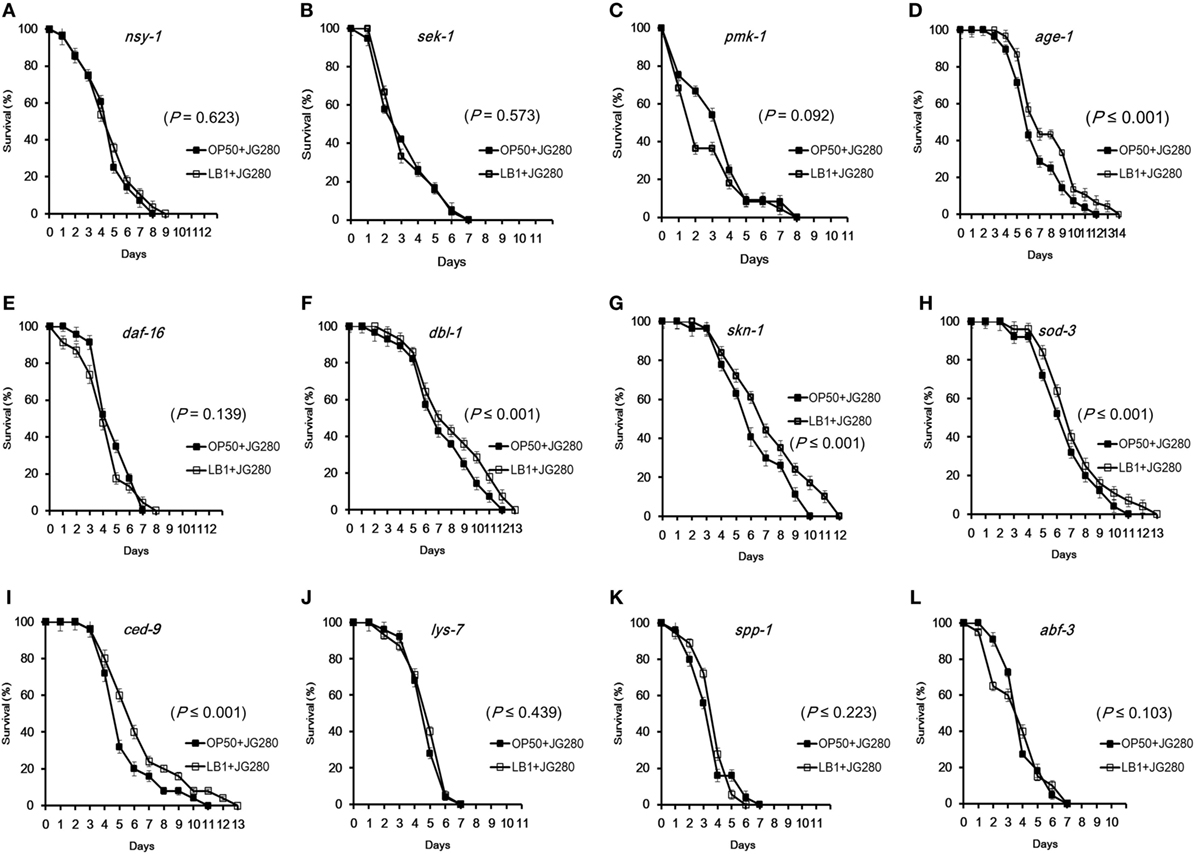
Figure 4. Effects of Lactobacillus zeae LB1 on the resistance to Enterotoxigenic Escherichia coli (ETEC) infection of Caenorhabditis elegans mutants defective in the genes for cell signaling or defense molecules and antimicrobial peptides. The mutants are defective in nsy-1 (A), sek-1 (B), pmk-1 (C), age-1 (D), daf-16 (E), dbl-1 (F), skn-1 (G), sod-3 (H), ced-9 (I), lys-7 (J), spp-1 (K), or abf-3 (L). The life-span assay was conducted in the absence or presence of L. zeae LB1 during the pretreatment. All C. elegans samples were treated with 108 CFU/ml isolate LB1 or E. coli OP50 (Control group) for 18 h and then ETEC JG280 for remaining days. Comparisons were made between the worms pre-exposed to L. zeae LB1 (LB1 + JG280) and those exposed to ETEC JG280 only (JG280). The P value for each comparison is indicated inside of each panel.
Regulation of C. elegans in Immunomodulatory Activity by L. zeae LB1 Through Cell Signaling
To elucidate the specific immune response stimulated by L. zeae LB1, pretreatment with L. zeae LB1 of two ETEC-resistant C. elegans mutants that are defective in age-1 (Figure 3B) or dbl-1 (Figure 3D), two ETEC-sensitive C. elegans mutants that are defective in nsy-1 (Figure 3A) or daf-16 (Figure 3B), and one mutant defective in both pmk-1 and daf-16 were employed to investigate the regulation of eight selected genes, including lys-7, spp-1, abf-2, clec-85, abf-3, clec-60, sek-1, and pmk-1. The expression of most tested genes, including lys-7, abf-2, clec-60, clec-85, sek-1, and pmk-1 was downregulated significantly (P ≤ 0.05) in the mutant defective in nsy-1 compared to the wild-type, regardless of the treatments (Figure 5A). Similarly, the expression of most tested genes, including lys-7, abf-2, clec-60, clec-85, and spp-1 was also downregulated (P ≤ 0.05) in the mutant defective in daf-16 compared to the wild-type, regardless of the treatments (Figure 5B). A similar trend was also observed in the mutant defective in both pmk-1 and daf-16, in which the expression of all the tested genes except for abf-3 was downregulated (P ≤ 0.05), regardless of the treatments (Figure 5E). On the contrary, the transcription of all the tested genes was all increased substantially (P ≤ 0.05) in the mutants either defective in dbl-1 or age-1, except for clec-60 in the mutant defective in dbl-1 compared to the wild-type, regardless of the treatments (Figures 5C,D). In view of the responses of all the tested mutants to the three different treatments, i.e., subjected to E. coli OP50 only, ETEC infection only, or to the pretreatment with L. zeae LB1 before the infection, the three ETEC-sensitive mutants (defective in nsy-1 or daf-16, or double defective in pmk-1 and daf-16) showed no significant differences (P > 0.05) in the gene expression within each tested gene (Figures 5A,B,E). In the two ETEC-resistant mutants (defective in dbl-1 or age-1), all the tested genes had no significant differences (P > 0.05) in the gene expression between the treatments with E. coli OP50 only or with ETEC only (Figures 5C,D). However, the expression of all the tested genes, except for clec-85 in the mutant defective in dbl-1 and clec-60 in the mutant defective age-1, was upregulated (P ≤ 0.05) by the pretreatment with L. zeae LB1.
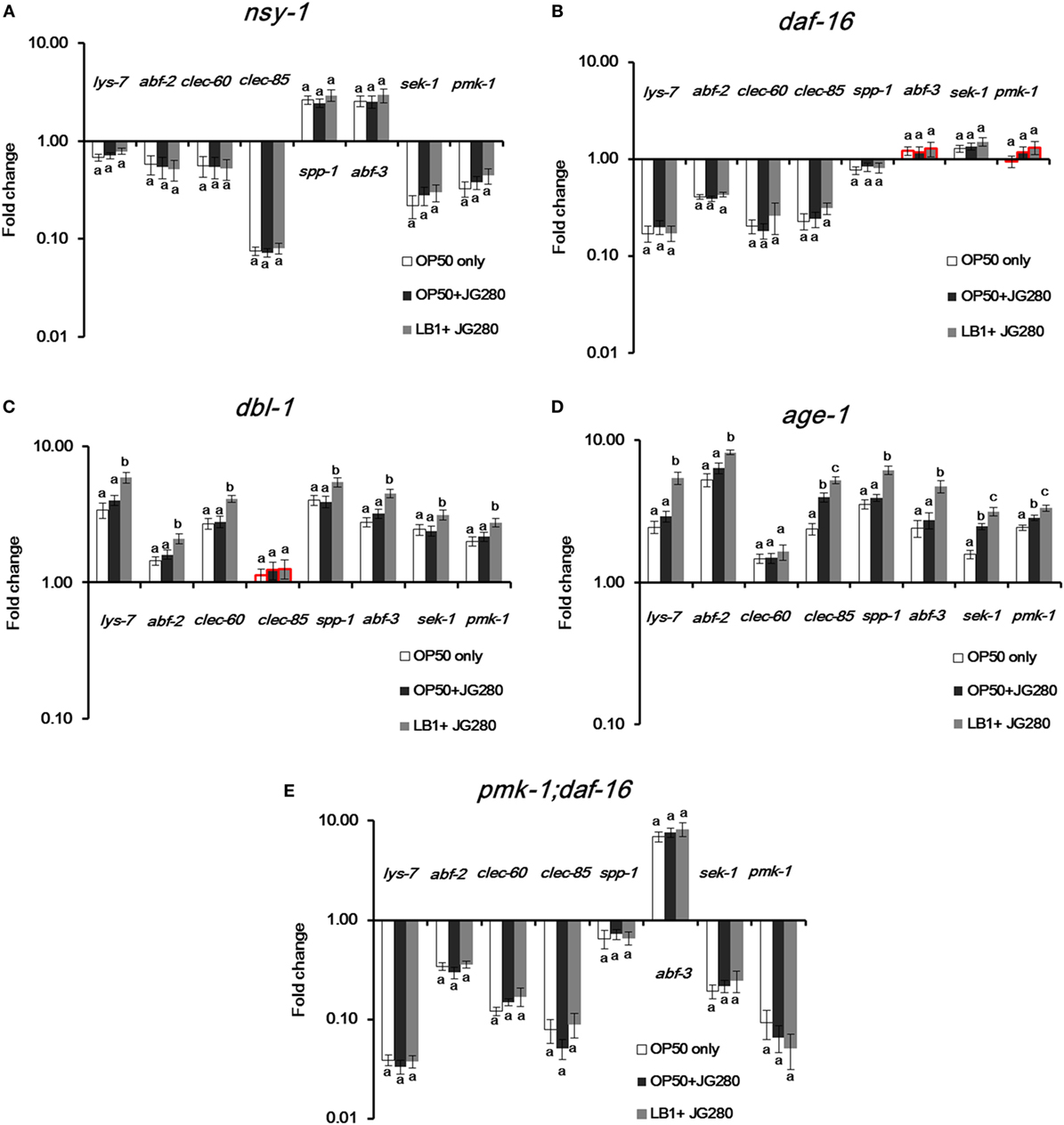
Figure 5. Gene expression of selected Caenorhabditis elegans mutants in response to the treatment of E. coli OP50, Enterotoxigenic Escherichia coli (ETEC) JG280 only, or ETEC JG280 after pretreatment with Lactobacillus zeae LB1. The nematode was sampled on day 2 of the life-span assay. The mutants are defective in dbl-1 (A), age-1 (B), nsy-1 (C), daf-16 (D), or in both dpmk-1 and daf-16 (E). The control group is the level of gene expression in C. elegans fed OP50 only (open bars). Relative expression was determined using the 2−ΔΔCt method. The first ΔCT is the comparison between target genes and housekeeping genes. The second ΔCT represents the comparison between the mutants and wild-type. The value of each gene in the wild-type is considered to be 1. Means marked with “a,” “b,” and “c” were significantly different (P ≤ 0.05) for the same gene among different treatments. While the groups represented by the bars with their borders in red had no significant difference (P > 0.05) in the gene expression level of the same gene between each mutant and the wild-type strain (N2) that had been subjected to the same treatment, the remaining groups represented by the bars with their borders in black differed significantly for the same gene within the same treatment when compared to the wild-type (P ≤ 0.05).
Discussion
Although many aspects of innate immunity are shared with higher vertebrates (24), C. elegans lacks a cell-mediated immune system and the production of antimicrobial peptides is, therefore, part of its innate immunity to combat bacterial infection (32). The lysozyme (LYS) family, Ascaris suum antibacterial factor (ABF) family, SPP (Caenopores are the saposin-like proteins) family, and C-type lectins family are some examples of the nematode innate immunity, which have been shown to play an important role in the general and more specifically induced immune responses to bacterial infection (26, 28, 33). Notably in the present study, the transcription of antimicrobial peptide genes, including spp-1, clec-85, abf-2, lys-7, and abf-3 was significantly upregulated in the wild-type of C. elegans in response to ETEC infection (Figure 2), suggesting a vital role of the antimicrobial peptides in the defense of C. elegans against ETEC infection. This notion is also supported by the fact that C. elegans with a mutation in an antimicrobial peptide gene (lys-7, spp-1, or abf-3) showed significant shorter life-span than the wild-type when subjected to ETEC infection (Figure 3C). It has been documented from previous studies that the expression of antimicrobial peptide genes in C. elegans could be induced by bacterial infection (25, 26, 34–36), which was mainly controlled by the p38 MAPK and DAF/IGF pathways (32, 37). Thus, it can be concluded that antimicrobial peptides play a crucial role in the defense system of C. elegans against ETEC infection. Further to this conclusion, there have been reports on specific regulation in the production of antimicrobial peptides by different signaling pathways, e.g., the expression of lys-1, lys-8, clec-85, and nlp-29 through the p38 MAPK pathway (32). In the present study, one new observation was the down regulation in the gene expression of both abf-2 and clec-60 in the mutants either defective in nsy-1 or daf-16 (Figures 5A,B). In contrast, the expression of spp-1 was differentially regulated in these two mutants. These results suggest that both abf-2 and clec-60 were controlled by both the p38 MAPK and DAF/IGF pathways while spp-1 was regulated by the DAF/IGF pathway only, which were reported for the first time to the best of our knowledge.
Caenorhabditis elegans possesses three major cell signaling pathways in its defense system, including the p38 MAPK, DAF/IGF, and TGF-β pathways (38). The p38 MAPK pathway is the most ancient signal transduction cascade in the nematode immunity, which is mainly associated with antimicrobial responses (39). In parallel, the DAF/IGF signaling pathway transcriptionally regulates many genes involved in the immune and stress responses that is linked to the longevity of C. elegans (21, 24). Many candidate antimicrobial genes have been identified in the genome of C. elegans; but the role of the signaling pathways in regulating these antimicrobial peptides in response to bacterial infection is yet to be fully elucidated (40–42). In the present study, four genes (tir-1, nsy-1, sek-1, and pmk-1) in the p38 MAPK pathway, two genes (daf-16 and age-1) in the DAF/IGF pathway, six genes (lys-7, spp-1, abf-2, clec-85, and clec-60, and abf-3) reported previously that encode antimicrobial peptides (27), and three other genes (sod-3, dbl-1, and skn-1) reported previously with a defense function (23, 43, 44) were initially examined for their possible involvement in the immune response of the wild-type nematode. All the tested genes except for tir-1, clec-60, and skn-1 were upregulated significantly as the results of responding to ETEC infection (Figure 2). While the upregulation suggests the involvement of antimicrobial peptides and defense molecules in the defense system of C. elegans as discussed above, it also implies a role of the p38 MAPK and DAF/IGF pathways in regulating the production of antimicrobial peptides and defense molecules. Additional evidence to support the regulatory role of these two pathways is the downregulation of tested genes coding for antimicrobial peptides and defense molecules in the mutants either defective in nsy-1 or daf-16 (Figures 5A,B) and in the mutant defective in both pmk-1 and daf-16 (Figure 5E). In agreement with these data, the life-span assay indicates that the mutants either defective in nsy-1, sek-1, or pmk-1 (the major components of p38 MAPK pathway) or defective in daf-16 (a major component of DAF/IGF pathway) were more susceptible to ETEC infection with over 30% reduction in the life-span compared to the wild-type (Figures 3A,B). In view of the data described above, it appears that the regulation of C. elegans response in the production of antimicrobial peptides and other molecules with a defense function is mediated mainly through the p38 MAPK and DAF/IGF pathways of cell signaling.
It has been noted from the present study that the regulation in the production of antimicrobial peptides and other molecules with a defense function through the cell signaling pathways could be either positive or negative. This statement is supported by the results from both study of gene expression in the mutants and the life-span assay. In the study of gene expression, tested genes encoding antimicrobial peptides or other molecules with a defense function were upregulated in the mutants either defective in dbl-1 or age-1, but downregulated in the mutants defective either in nsy-1 or daf-16 (Figure 5). In agreement with these observations, both mutants either defective in dbl-1 or age-1 were more resistant to ETEC infection, whereas the two mutants either defective in nsy-1 or daf-16 became more susceptible (Figure 3). These results suggest that genes dbl-1 and age-1 have a negative role, while genes nsy-1 and daf-16 play a positive role in regulating the production of antimicrobial peptides and defense molecules in the wild-type nematode.
Recently, Kim and Mylonakis (19) reported that the pretreatment of C. elegans with Lactobacillus acidophilus enhanced the resistance of C. elegans to the infection by Gram-positive pathogens via the p38 MAPK pathway. Moreover, Bifidobacteria have also been shown to extend the C. elegans longevity through the modulation of the p38 MAPK and DAF/IGF pathways (45). Nevertheless, these reports have not yet identified the components in the pathways that Lactobacillus or Bifidobacterium interacted with to provide protection to the nematode. Very recently, Kamaladevi and Balamurugan (37) reported that L. case triggered TLR-mediated RACK-1-dependent p38 MAPK pathway to increase host resistance and protect nematode against K. pneumoniae infection. Bifidobacterium longum strain BB68 increased the longevity of nematodes by activating the TIR-1—JNK-1—DAF-16 signaling pathway (44). In the present study, the expression of almost all the tested genes encoding antimicrobial peptides or other molecules with a defense function was enhanced significantly by the pretreatment with L. zeae LB1 in the mutants that are defective in dbl-1 or age-1 when compared to those treated with E. coli OP50 or ETEC only (Figures 5C,D). Such enhancement was, however, not detected in the mutants that are defective in nsy-1 or daf-16 (Figures 5A,B), or defective in both pmk-1 and dal-16 (Figure 5E). Interestingly, the same mutants that are defective in dbl-1 or age-1 exhibited longer life-span, whereas the mutants that are defective in nsy-1 or daf-16 had shorter life-span than the wild-type (Figure 3). Furthermore, the four mutants that are defective in nsy-1, sek-1, pmk-1, or daf-16 showed no response to the pretreatment with L. zeae LB1 and their life-span was similar to the same mutants infected with ETEC only in the life-span assay (Figures 4A–C,E). These data suggest that Nsy-1, Sek-1, Pmk-1, and Daf-16 could be the functional sites in the signaling pathways targeted by L. zeae LB1 for the protection.
In conclusion, the present study has revealed: (1) the host response of C. elegans to ETEC infection mainly involves both the p38 MAPK and DAF/IGF pathways of cell signaling that regulate the production of antimicrobial peptides and defense molecules; (2) L. zeae LB1 alters the production of antimicrobial peptides and other molecules with a defense function through the regulation of both the p38 MAPK and DAF/IGF pathways of cell signaling; (3) gene nsy-1 or daf-16 may play a positive role in the regulation, while the role of gene dbl-1 or age-1 is negative in the wild-type of C. elegans; (4) Nsy-1, Sek-1, Pmk-1, and Daf-16 appear to be the functional sites in the signaling pathways targeted by L. zeae LB1 for regulating the production of antimicrobial peptides and other molecules with a defense function. To summarize the new findings from the present study, a schematic diagram has been generated (Figure 6), which also speculates the immunomodulatory mechanism by L. zeae.
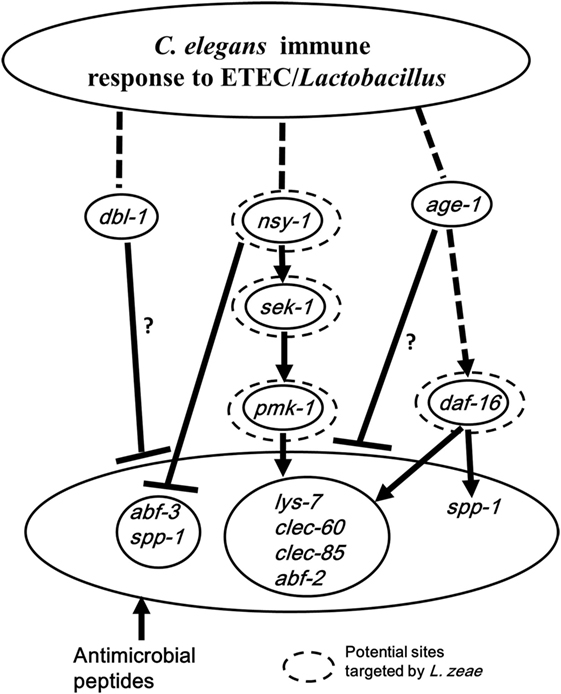
Figure 6. Schematic diagram speculating the immunomodulatory mechanism by Lactobacillus zeae. Pretreatment with L. zeae LB1 altered the production of antimicrobial peptides through the p38 mitogen-activated protein kinase (MAPK) and DAF/IGF pathways of cell signaling, therefore, changed the resistance of Caenorhabditis elegans to Enterotoxigenic Escherichia coli (ETEC) infection. The hypothesis was based on the data of both life-span assays and gene expression of various C. elegans mutants either pretreated or not treated with L. zeae LB1 before the ETEC infection. While dashed lines represent the cascades described in previous literatures, solid lines indicate the effects observed in the present study. ▾, Upregulation; ┴, downregulation. Dashed circle: potential functional sites in the cell signaling targeted by L. zeae LB1. ?, Indicating that the regulation could be direct or indirect. More specifically, the downregulation in the production of antimicrobial peptides could be controlled by dbl-1 or age-1 directly or through pmk-1 and sek-1 in the p38-MAPK pathway in the wild-type nematode (Figures 5C,D).
Author Contributions
MZ and XL performed the experiments. MZ, HY, XL, and JG analyzed the data. JG, WC, S-PN, and M-YX contributed for reagents/materials/analysis tools. MZ and JG wrote the paper. MZ, HY, XY, and JG designed the experiments. JG and WC conceived the research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The double mutant (defective in both pmk-1 and daf-16) was a gift from Dr. Dennis Kim (Department of biology, Massachusetts Institute of Technology, Boston).
Funding
This research was funded by Agriculture & Agri-Food Canada through the A-base Program (J-001391.001.02). MZ is a visiting graduate student financially supported by the China Scholarship Council through the MOE-AAFC Ph.D. Research Program and National Natural Science Foundation of China (No:31601455).
References
1. FAO/WHO. Evaluation of Health and Nutritional Properties of Powder Milk with Live Lactic Acid Bacteria. Report of FAO/WHO Expert Consultation. (2001).
2. Matsuzaki T, Chin J. Modulating immune responses with probiotic bacteria. Immunol Cell Biol (2000) 78(1):67–73. doi:10.1046/j.1440-1711.2000.00887.x
3. Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol (2010) 8(3):171–84. doi:10.1038/nrmicro2297
4. Nagy B, Wilson RA, Whittam TS. Genetic diversity among Escherichia coli isolates carrying f18 genes from pigs with porcine postweaning diarrhea and edema disease. J Clin Microbiol (1999) 37(5):1642–5.
5. Frydendahl K. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhea and edema disease in pigs and a comparison of diagnostic approaches. Vet Microbiol (2002) 85(2):169–82. doi:10.1016/S0378-1135(01)00504-1
6. Noamani BN, Fairbrother JM, Gyles CL. Virulence genes of O149 enterotoxigenic Escherichia coli from outbreaks of postweaning diarrhea in pigs. Vet Microbiol (2003) 97(1–2):87. doi:10.1016/j.vetmic.2003.08.006
7. Roselli M, Finamore A, Britti MS, Mengheri E. Probiotic bacteria Bifidobacterium animalis MB5 and Lactobacillus rhamnosus GG protect intestinal Caco-2 cells from the inflammation-associated response induced by enterotoxigenic Escherichia coli K88. Br J Nutr (2006) 95(6):1177–84. doi:10.1079/BJN20051681
8. Carey CM, Kostrzynska M, Ojha S, Thompson S. The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157:H7. J Microbiol Methods (2008) 73(2):125–32. doi:10.1016/j.mimet.2008.01.014
9. Daudelin JF, Lessard M, Beaudoin F, Nadeau É, Bissonnette N, Boutin Y, et al. Administration of probiotics influences F4 (K88)-positive enterotoxigenic Escherichia coli attachment and intestinal cytokine expression in weaned pigs. Vet Res (2011) 42(1):1–11. doi:10.1186/1297-9716-42-69
10. Tan M-W, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A (1999) 96(2):715–20. doi:10.1073/pnas.96.2.715
11. Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, et al. A simple model host for identifying gram-positive virulence factors. Proc Natl Acad Sci U S A (2001) 98(19):10892–7. doi:10.1073/pnas.191378698
12. Kwok TC, Ricker N, Fraser R, Chan AW, Burns A, Stanley EF, et al. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature (2006) 441(7089):91–5. doi:10.1038/nature04657
13. Rangan KJ, Pedicord VA, Wang YC, Kim B, Lu Y, Shaham S, et al. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science (2016) 353(6306):1434. doi:10.1126/science.aaf3552
14. Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol (2000) 10(23):1539–42. doi:10.1016/S0960-9822(00)00830-7
15. Labrousse A, Chauvet S, Couillault C, Léopold Kurz C, Ewbank JJ. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr Biol (2000) 10(23):1543–5. doi:10.1016/S0960-9822(00)00833-2
16. Sifri CD, Begun J, Ausubel FM, Calderwood SB. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect Immun (2003) 71(4):2208–17. doi:10.1128/IAI.71.4.2208-2217.2003
17. Ikeda T, Yasui C, Hoshino K, Arikawa K, Nishikawa Y. Influence of lactic acid bacteria on longevity of Caenorhabditis elegans and host defense against Salmonella enterica serovar Enteritidis. Appl Environ Microbiol (2007) 73(20):6404–9. doi:10.1128/AEM.00704-07
18. Wang C, Wang J, Gong J, Yu H, Pacan JC, Niu Z, et al. Use of Caenorhabditis elegans for preselecting Lactobacillus isolates to control Salmonella Typhimurium. J Food Prot (2011) 74(1):86–93. doi:10.4315/0362-028X.JFP-10-155
19. Kim Y, Mylonakis E. Caenorhabditis elegans immune conditioning with the probiotic bacterium Lactobacillus acidophilus strain NCFM enhances gram-positive immune responses. Infect Immun (2012) 80(7):2500–8. doi:10.1128/IAI.06350-11
20. Zhou M, Yu H, Yin X, Sabour PM, Chen W, Gong J. Lactobacillus zeae protects Caenorhabditis elegans from enterotoxigenic Escherichia coli-caused death by inhibiting enterotoxin gene expression of the pathogen. PLoS One (2014) 9(2):e89004. doi:10.1371/journal.pone.0089004
21. Kurz CL, Ewbank JJ. Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat Rev Genet (2003) 4(5):380–90. doi:10.1038/nrg1067
22. Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature (2003) 424(6946):277. doi:10.1038/nature01789
23. Zugasti O, Ewbank JJ. Neuroimmune regulation of antimicrobial peptide expression by a noncanonical TGF-β signaling pathway in Caenorhabditis elegans epidermis. Nat Immunol (2009) 10(3):249–56. doi:10.1038/ni.1700
24. Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol (2010) 10(1):47–58. doi:10.1038/nri2689
25. Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, et al. Inducible antibacterial defense system in C. elegans. Curr Biol (2002) 12(14):1209–14. doi:10.1016/S0960-9822(02)00928-4
26. Couillault C, Pujol N, Reboul J, Sabatier L, Guichou J-F, Kohara Y, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol (2004) 5(5):488–94. doi:10.1038/ni1060
27. Bogaerts A, Beets I, Schoofs L, Verleyen P. Antimicrobial peptides in Caenorhabditis elegans. Invert Surv J (2010) 7(1):45–52.
28. Huffman DL, Abrami L, Sasik R, Corbeil J, van der Goot FG, Aroian RV. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc Natl Acad Sci U S A (2004) 101(30):10995–1000. doi:10.1073/pnas.0404073101
29. Breger J, Fuchs BB, Aperis G, Moy TI, Ausubel FM, Mylonakis E. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog (2007) 3(2):e18. doi:10.1371/journal.ppat.0030018
30. Yin X, Chambers JR, Wheatcroft R, Johnson RP, Zhu J, Liu B, et al. Adherence of Escherichia coli O157: H7 mutants in vitro and in ligated pig intestines. Appl Environ Microbiol (2009) 75(15):4975–83. doi:10.1128/AEM.00297-09
31. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods (2001) 25(4):402–8. doi:10.1006/meth.2001.1262
32. Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol (2007) 27(15):5544–53. doi:10.1128/MCB.02070-06
33. Mochii M, Yoshida S, Morita K, Kohara Y, Ueno N. Identification of transforming growth factor-beta-regulated genes in Caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc Natl Acad Sci U S A (1999) 96(26):15020–25.
34. O’Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res (2006) 16(8):1005–16. doi:10.1101/gr.50823006
35. Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, Tan M-W. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci U S A (2006) 103(38):14086–91. doi:10.1073/pnas.0603424103
36. Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet (2006) 2(11):e183. doi:10.1371/journal.pgen.0020183
37. Kamaladevi A, Balamurugan K. Lactobacillus casei triggers a TLR mediated RACK-1 dependent p38 MAPK pathway in Caenorhabditis elegans to resist Klebsiella pneumoniae infection. Food Funct (2016) 7(7):3211. doi:10.1039/c6fo00510a
38. Pujol N, Zugasti O, Wong D, Couillault C, Kurz CL, Schulenburg H, et al. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog (2008) 4(7):e1000105. doi:10.1371/journal.ppat.1000105
39. Kim DH, Ausubel FM. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science (2002) 297(5581):623. doi:10.1126/science.1073759
40. Nicholas HR, Hodgkin J. Responses to infection and possible recognition strategies in the innate immune system of Caenorhabditis elegans. Mol Immunol (2004) 41(5):479–93. doi:10.1016/j.molimm.2004.03.037
41. Schulenburg H, Léopold Kurz C, Ewbank JJ. Evolution of the innate immune system: the worm perspective. Immunol Rev (2004) 198(1):36–58. doi:10.1111/j.0105-2896.2004.0125.x
42. Gravato-Nobre MJ, Hodgkin J. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell Microbiol (2005) 7(6):741–51. doi:10.1111/j.1462-5822.2005.00523.x
43. Keshet A, Mertenskötter A, Winter SA, Brinkmann V, Dölling R, Paul RJ. PMK-1 p38 MAPK promotes cadmium stress resistance, the expression of SKN-1/Nrf and DAF-16 target genes, and protein biosynthesis in Caenorhabditis elegans. Mol GenetGenom (2017) 292(6):1341–61. doi:10.1007/s00438-017-1351-z
44. Liang Z, Yang Z, Liu R, Zheng X, Min Z, Guo H, et al. The transcription factor DAF-16 is essential for increased longevity in C. elegans exposed to Bifidobacterium longum BB68. Sci Rep (2017) 7:7408. doi:10.1038/s41598-017-07974-3
Keywords: Lactobacillus, enterotoxigenic Escherichia coli, Caenorhabditis elegans, mitogen-activated protein kinase pathway, DAF/IGF pathway, antimicrobial peptides
Citation: Zhou M, Liu X, Yu H, Yin X, Nie S-P, Xie M-Y, Chen W and Gong J (2018) Cell Signaling of Caenorhabditis elegans in Response to Enterotoxigenic Escherichia coli Infection and Lactobacillus zeae Protection. Front. Immunol. 9:1745. doi: 10.3389/fimmu.2018.01745
Received: 11 January 2018; Accepted: 16 July 2018;
Published: 10 September 2018
Edited by:
Janice C. Telfer, University of Massachusetts Amherst, United StatesReviewed by:
Hai-Peng Liu, Xiamen University, ChinaKatherine Buckley, Carnegie Mellon University, United States
Copyright: © 2018 Nie, Xie, Chen and Her Majesty the Queen in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Chen, Y2hlbndlaTY2QGppYW5nbmFuLmVkdS5jbg==;
Joshua Gong, am9zaHVhLmdvbmdAYWdyLmdjLmNh
 Mengzhou Zhou
Mengzhou Zhou Xiaozhen Liu2,4
Xiaozhen Liu2,4 Wei Chen
Wei Chen Joshua Gong
Joshua Gong