- 1Immunity, Inflammation, and Disease Laboratory, National Institute of Environmental Health Sciences, Durham, NC, United States
- 2Oral and Craniofacial Biomedicine Curriculum, School of Dentistry, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
The skin is a highly organized first line of defense that stretches up to 1.8 m2 and is home to more than a million commensal bacteria. The microenvironment of skin is driven by factors such as pH, temperature, moisture, sebum level, oxidative stress, diet, resident immune cells, and infectious exposure. The skin has a high turnover of cells as it continually bares itself to environmental stresses. Notwithstanding these limitations, it has devised strategies to adapt as a nutrient-scarce site. To perform its protective function efficiently, it relies on mechanisms to continuously remove dead cells without alarming the immune system, actively purging the dying/senescent cells by immunotolerant efferocytosis. Both canonical (starvation-induced, reactive oxygen species, stress, and environmental insults) and non-canonical (selective) autophagy in the skin have evolved to perform astute due-diligence and housekeeping in a quiescent fashion for survival, cellular functioning, homeostasis, and immune tolerance. The autophagic “homeostatic rheostat” works tirelessly to uphold the delicate balance in immunoregulation and tolerance. If this equilibrium is upset, the immune system can wreak havoc and initiate pathogenesis. Out of all the organs, the skin remains under-studied in the context of autophagy. Here, we touch upon some of the salient features of autophagy active in the skin.
Introduction
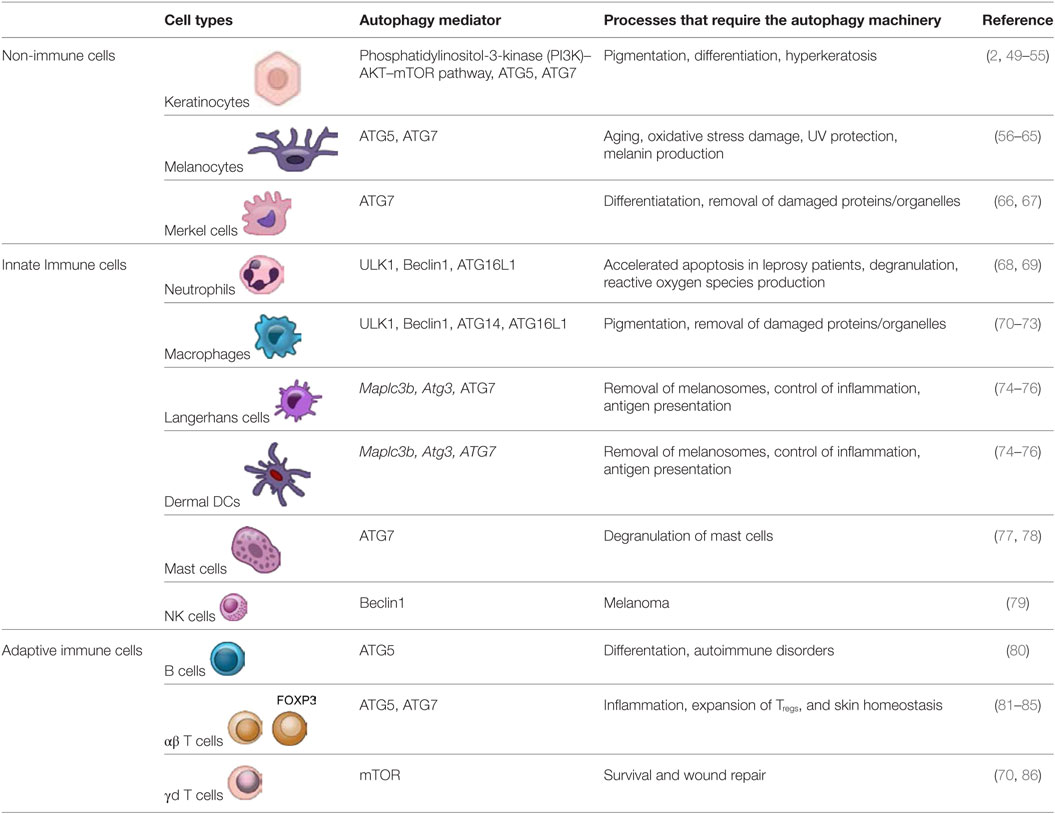
Skin architecture is designed to shield against physical as well as immunological damage by environmental assaults [such as pathogens, ultraviolet radiation (UVR), allergens, oxidative stress, and various chemical toxins like hexavalent chromium, zinc, titanium oxide, and silver nanoparticles] (1–4). The skin is a nutrient-poor environment, which exposes itself to various environmental stressors regularly and therefore, requires recycling of limited resources via the autophagy machinery to maintain homeostasis (5, 6). Nonetheless, skin has a potent arsenal of weapons at its disposal to ward off potential threats from external aggressors. The cells populating the skin have both immune and non-immune components (1, 2). The skin is comprised of the epidermis, dermis, and hypodermis (subcutaneous fat) (Figure 1A) (1, 2, 7, 8). Skin also has several appendages (adena), such as nails, sweat glands, sebaceous glands, and hair follicles, which allow sensation, lubrication, and restriction of heat loss (9). Epidermis is comprised of keratinocytes, Langerhans cells (LCs), dendritic epidermal γδ T cells (DETC), melanocytes, and merkel cells (10). Dermis is comprised of fibroblasts, immune cells [dermal DCs (dDCs), innate lymphoid cells (ILCs), NK cells, B cells, macrophages, and T cells], endothelial cells, and neurons, which build up the extracellular matrix (1, 2). The hypodermis is comprised of adipocytes, nerves, blood, and lymphatic vessels.
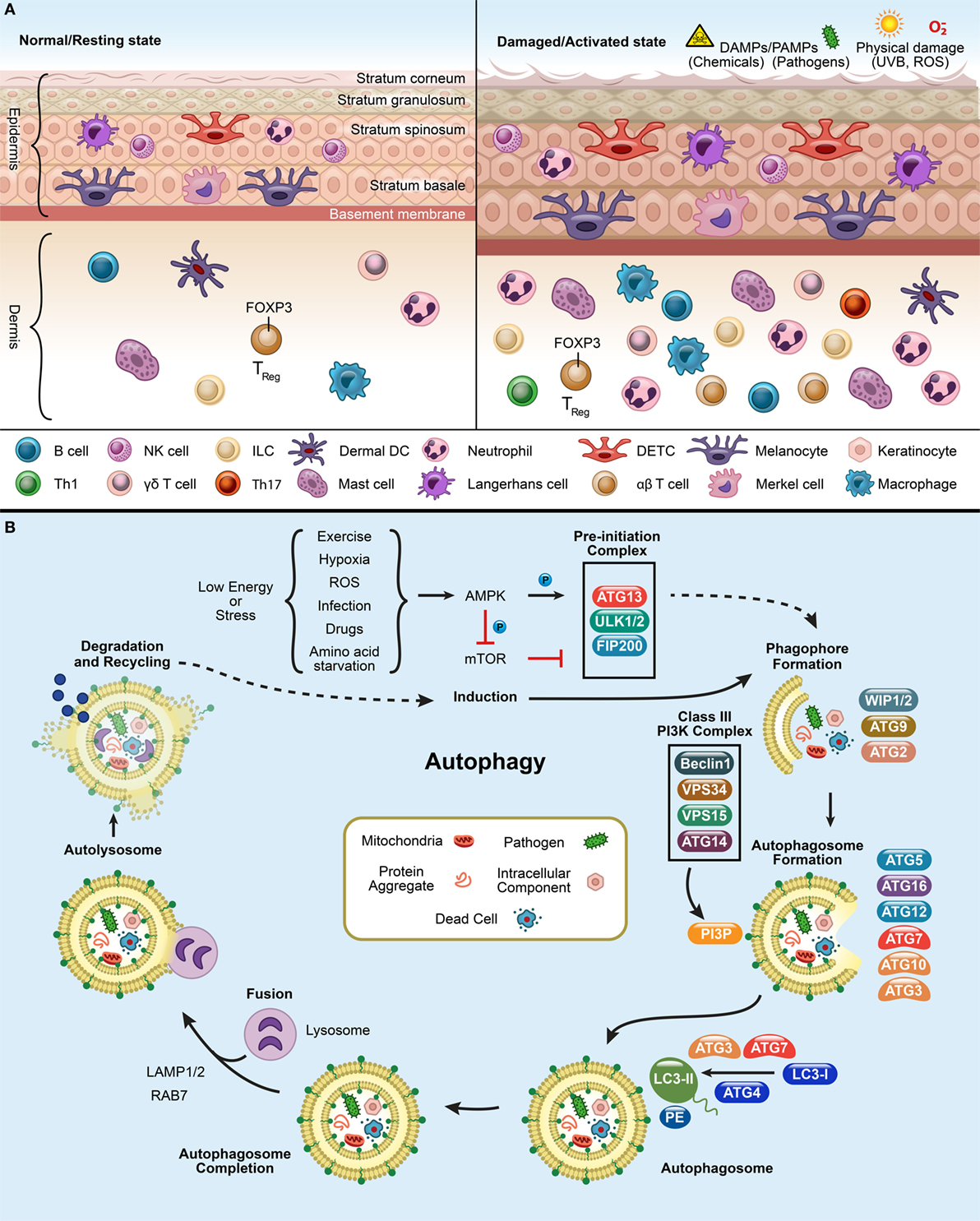
Figure 1. Panel (A) depicts the resting and activated state of the skin. In a normal or resting state, epidermis and dermis have circulating immune cells [DETCs, αβ T cells, γd T cells, macrophages, neutrophils, LCs, dermal DCs (dDCs), NK cells, B cells, innate lymphoid cells (ILCs)] and non-immune cells (melanocytes, keratinocytes, and merkel cells). Upon exposure to pathogens, chemicals, UV, or reactive oxygen species (ROS), the immune cells infiltrate at the site of infliction to defend the host and finally to resolve the inflammation after damage. Panel (B) shows the process of autophagy. mTOR inhibition triggers the activation of AMPK and initiates an autophagy-inducing signals during a low energy state such as starvation, ROS, exercise, infection, drugs, and hypoxic stress. This initiates the formation of pre-initiation complex (ULK1/2, ATG13, and FIP200) in the presence of unwanted cargo (such as, mitochondria, pathogens, protein aggregates, and intracellular components). This will, in turn activates the Class III phosphatidylinositol-3-kinase (PI3K) complex, composed of ATG14 (UVRAG)-VPS15-VPS34-Beclin1. The Class III PI3K complex completes the autophagosome formation by producing PI3P which recruits downstream ubiquitin-like conjugation systems (ATG5–12) and converts LC3-I to form LC3-PE. Finally, lysosome fuses with the autophagosome to form the autolysosome to degrade the enclosed cargo. The degraded cargo is finally assimilated and recycled.
Apart from the innate and adaptive immune cells present in the skin, the complement systems, antibodies, and antimicrobial peptides (AMPs) aid the immune system in clearing out pathogens and foreign particles. Autophagy participates in various physiological activities to ensure the smooth and quiescent operation of the immunotolerant environment and to maintain skin integrity. These activities include maintaining homeostasis, performing efferocytosis, as well as determining skin color, host defense, longevity, antigen presentation, and survival (11).
Autophagy as a Cell Survival Mechanism
Autophagy means self (auto) eating (phagy) and is a highly conserved cellular process across eukaryotes, which allows cells to recycle cytoplasmic materials via the lysosome and survive periods of nutrient deprivation (11). The term autophagy is derived from ancient Greek, but the word first garnered attention when Christian de Duve not only coined it but also won the Nobel prize in Physiology or Medicine in 1974 for his work on lysosomes (12–16). More recently Dr. Yoshinori Oshumi, described the autophagy-related genes (ATG) in yeast in 1993 and received the Nobel prize in 2016 (11, 12, 17). His pioneering work led to the discovery of other ATG genes and its human orthologs.
Autophagy pathways include macroautophagy (canonical autophagy/autophagy), microautophagy, and chaperone-mediated autophagy (CMA) (11, 13, 18). Traditionally, autophagy is orchestrated by the group of ATG proteins, which precisely control the autophagic process (11, 19). The process kickstarts the formation of the pre-initiation complex, followed by generation of the phagophore, autophagosome, and autolysosome, leading to cargo degradation (Figure 1B) (11, 19, 20). Mammalian target of rapamycin complex 1 inhibition leads to the induction of autophagy and assembles ULK1/2, ATG13, and FIP200 to form the pre-initiation complex at the phagophore (Figure 1B). Once activated, it targets the Class III phosphatidylinositol-3-kinase (PI3K) complex (Beclin1, VPS34, VPS15, and ATG14) which recruits downstream conjugation ATG proteins. During autophagosome elongation, E3(Ubiquitin)-ligase ATG7 mediates ATG5–ATG12–ATG16L1 complex formation and is recruited to the autophagosome membrane. Ubiquitin-conjugating/E2-like enzyme ATG10 mediates covalent conjugation of the ubiquitin-like ATG12–ATG5 (21). E2-like enzyme ATG3 forms ATG12–ATG3 conjugate, controls mitochondrial homeostasis (21). ATG7 can recruit ATG3 and ATG10 forming ATG7–ATG3 and ATG10–ATG3, respectively (22). Mice lacking ULK1/2, ATG3, ATG5, ATG7, ATG12, or ATG16L1 are embryonic lethal mutations (23). ATG12-conjugation is essential for the formation of preautophagosomes (24). ATG3 aids in conjugation of LC3-I with phosphatidylethanolaminie (PE) required for the formation of autophagosomes (21, 24). This facilitates the LC3 lipidation with PE and forms LC3-PE (or LC3-II). LC3-PE embeds into the mature autophagosome which finally fuses with the lysosome, wherein the cargo is degraded and recycled. The autophagy pathway is not only limited to the processes of degradation and survival during starvation but is also active in regulating other cellular functions (11). This bolsters the need for investigating autophagy’s widespread influence on different biological mechanisms.
To ensure proper scrutiny, the autophagy machinery takes on specialized roles that selectively targets and digests intracellular components and is called selective or non-canonical autophagy (25–27). Depending on the cargo engulfed, it can be classified into CMA (heat-shock cognate 70 stress protein mediated target of the substrate), aggrephagy (clearance of protein aggregates), macrolipidophagy (the degradation of lipids), pexophagy (autophagic degradation of peroxisomes), ER-phagy (endoplasmic reticulum autophagy), mitophagy (damaged mitochondria), xenophagy (intracellular pathogens), and LC3-associated phagocytosis (LAP) (efferocytosis and pathogen phagocytosis) (28–31). Selective autophagy receptors/adaptors p62/Sqstm1 (Sequestome1), OPTN (Optineurin), TAX1BP1 (T-cell leukemia virus type I binding protein 1), NDP52/CALCOCO2 (calcium binding and coiled-coil domain 2), and NBR1 (neighbor of BRCA1 gene 1) coordinate and mediate degradation of ubiquitinated cargos by delivering them to LC3-containing phagophores (Figure 1B) (11, 32–41). Mitophagy involves degradation of redundant and distressed mitochondria and normally occurs in a Parkin-PINK1-dependent manner (42, 43). After ubiquitination, autophagy adaptors, OPTN and NDP52, can recognize and deliver them to LC3-positive autophagosomes for degradation (44). Similarly, in xenophagy, cytosolic pathogens or pathogen-contained vacuole can be ubiquitinated by ubiquitin ligases (45–47). Subsequently, ubiquitinated pathogens or their substrates are recruited by autophagy receptors for autophagosomal degradation (11). However, when an extracellular pathogen is phagocytosed and it engages pathogen recognition receptor (PRR), as a result it activates a specialized autophagy process called LAP (27). The LAP pathway is also utilized for the clearance of dead cells triggered by wounds, pathogen exposure, or environmental triggers (26, 27, 48).
Autophagy in Skin Immune Cells
Skin inflammation induced by environmental irritants and pathogens requires autophagy as well as a crosstalk between immune and non-immune skin cells (listed below and in Table 1) to effectively alleviate the damage (4).
Non-Immune Components
• Melanocytes reside in the epidermis between keratinocytes, and they contain melanin or biochrome, a natural pigment found in hair, skin, and eyes (49). The process of producing melanin pigment continuously is called melanogenesis (87). Hair follicular melanogenesis (FM), unlike epidermal melanogenesis, is cyclic (88). FM starts at the anagen (active growth phase) stage of the hair cycle and is turned off by catagen (transition stage) and telogen (shedding stage) (88). Upon aging, the number of melanocytes in hair follicles is reduced and dendritic cells (including Langerhans cells) move from the upper to lower hair follicles as a response to age-related degradation of melanocytes (88).
Stress can stimulate melanocyte to activate neurotransmitters, neutropeptides, and hormones which aids in regulating skin homeostasis (89). Neuroendocrine has been shown to govern the production and secretion of l-tyrosine and l-dihydroxyphenylalanine (l-DOPA), or its derivatives, during melanogenisis (89). Melanocytes are often called “neurons of the skin” and possesses melanocytes-stimulating hormone (MSH) receptors (89, 90). Certain substrates of melanogenesis like l-tyrosine and l-DOPA can also regulate cell (melanocyte) functions and cellular metabolism through non-receptor-mediated processes (91).
Melanocytes develop into melanosomes, which can perform lysosomal degradation as seen in retinal epithelial cells (49, 56, 87). Autophagy is also involved in melanin synthesis in melanosomes (92) via FGF7/FGF7R-initiated AMPK/mTOR pathway (93–96). ATG5 and ATG7-deficiency causes premature aging and accumulation of oxidative stress-induced damage (57–60). Upon undergoing UV exposure, melanocytes can undergo photosensitization by generating superoxide radicals in cells (97). In addition, p62 is upregulated upon phototherapy (UVA radiation and light-emitting ode 585 nm) in the melanocyte (61, 62). Melanin confers protection from UV-induced DNA damage, maintains skin homeostasis, modulates the immune environment, and regulates skin color by autophagy (56, 63–65). Melanocytes can also phagocytose, present antigen via major histocompatibility complex (MHC) class II and produce cytokines like IL-1, IL-6, TNF-α, IL-3, and G-MCSF (98).
• Merkel cells populate broadly across the epidermis. They have high turnover and differentiate terminally (2, 99). Merkel cells have a longer life span than keratinocytes and require autophagy for differentiation (66). Atg7-deficient Merkel cells show accumulation of p62 (66, 67).
Keratinocytes are abundant and form the foundation of the epidermis. Out of all skin cells, they have been studied the most. Keratinocytes protect the skin cells by phagocytosing damaged melanosomes (49). Human keratinocytes also induce inflammasomes upon either UVB irradiation or viral infection (100). Autophagy in keratinocytes contributes to skin pigmentation, as it depends on the melanin from phagocytosed melanocytes engulfed by the keratinocytes (49, 92). Keratinocyte differentiation results in lysosomal enzyme activation, LC3 expression, and intracellular components degradation to form corneocytes (2, 50–52). Epidermal permeability barrier, mitophagy, and autophagy defects are observed in comparative gene identification-58 (CGI-58) (a co-activator of adipose triglyceride lipase)-deficient mice (51, 101). CGI-58 deficiency causes Chanarin–Dorfman syndrome (neutral lipid storage dysfunction) and chronic and excessive build up of keratin leading to ichthyosis (51, 101). Atg7-deficient keratinocytes are smaller, with outer root sheath thickening, acanthosis, and hyperkeratosis. They also have less keratohyalin, trichohyalin granules, and filaggrin (49, 53, 54, 102). Moreover, Atg7-deficient mice have more corneocytes (53). Atg7-deficiency leads to cellular aging and accumulation of p53 and p21 upon treatment with N,N′-dimethyl-4,4′-bipyridinium dichloride (paraquat) treatment (54). Keratinocyte growth factor (FGF7/KGF) controls human keratinocyte differentiation and induces autophagy via the PI3K–AKT–mTOR pathway (99, 103). Hence, Atg7-deficient mice demonstrate the importance of autophagy in epidermal keratinization and hair growth (53, 99, 102). Autophagy inhibition either via 3-MA treatment or Atg5-deficiency also impairs keratinocytes differentiation (49, 55).
Immune Components
• Neutrophils are a most abundant granulocytes and are considered the foot soldiers of the immune system (68). Autophagy-deficient neutrophils show impaired degranulation, NADPH-oxidase-mediated reactive oxygen species (ROS) production, and inflammatory responses (68). Neutrophils from the skin of leprosy patients release TNF-α, show increased autophagy and exhibit accelerated apoptosis in vitro (69).
• Macrophages are skin resident phagocytes surveilling the epidermis and dermis (2). They help in maintaining the immunotolerant environment of the skin (2, 92). Specialized macrophages in the skin called melanophages can engulf melanocyte fragments and melanin. These melanocyte fragments and melanin are processed by autophagy (70–72). In leprosy patients, skin-derived macrophages have significant upregulation of autophagy genes, including Beclin1 and Atg14 (73).
• Epidermal DCs, also known as Langerhans cells (LCs) are dendritic cells of the skin and are found among the keratinocytes in the epidermis (2, 104). The LCs surveil the epidermis and promote tolerance to environmental stressors (104–106). Once activated, LCs migrate to the draining lymph nodes and aid in T cell polarization via antigen presentation to define the adaptive immune environment (106). Cytosolic and endocytosed antigens are processing and presentation (or cross presentation) by DCs to either CD4+ or CD8+ T cells via MHC class II and MHC class I molecules, respectively, and this process requires the autophagy machinery (74). Macroautophagy (canonical autophagy) has been implicated in the intracellular antigen loading on MHC class II molecule, allows the autophagosomal membrane to fuse with the MHC class II-loading compartment and thus, allows efficient MHC class II presentation (75). Mintern et al. showed that CD8+ splenic DCs from ATG7-deficient mice have impaired cross presentation of antigen via MHC class I pathway but can efficiently load antigen on MHC class II molecule (74). Thus, suggesting a role of autophagy in antigen presentation and cross presentation by DCs.
Both LCs and dDCs can induce interferon (IFN) and pro-inflammatory responses (107). Type I IFN can induce autophagy during viral infection, autoimmune disorders, and certain cancers (108, 109). ATG, Map1lc3b and Atg3 have been shown to be important in the production of hapten 2,4-dinitrofluorobenzene-induced (DNFB) ROS in skin DCs (76).
• Mast cells are granulocytes crucial for allergic inflammatory reactions (77). In the skin, they are triggered by UV or ROS-inducing irritants and undergo degranulation (77, 78). In addition, the degranulation process requires ATG7 (77).
• NK cells are cytotoxic innate immune lymphocytes present in the dermis. A recent study showed that targeting autophagy by inhibiting Beclin1 can increase NK cells resulting in melanoma tumor (79). Beclin1 inhibition reduces degradation of NK-derived Granzyme B as a result of which melanoma cells can thrive even in the presence of NK cells driven by CCL5 (79).
• ILCs are a new addition to the family of innate immune cells, and they are involved in shaping the immune system (110, 111). There are three types of ILCs defined by their cytokine profile: ILC1 produces IFN-γ, ILC2 produce IL-5 and IL-13, and ILC3 produces IL-17A and IL-22 (110). In human and murine skins, ILC2 are the most prelevant. The role of ILCs in skin autophagy is not well reported.
• Human skin contains approximately 20 billion skin T cells or T lymphocytes (112). T cells define the adaptive immune responses and are differentiated from other lymphocytes by the presence of surface T cell receptor (TCR) (112). TCR engagement activates autophagy, induces ROS, and causes activation of nuclear factor of activated T-cells (NFATC1) and lysosome-associated membrane protein 2 (LAMP-2) (113). Autophagy can alter the T cell repertoire through selection and aid in cross presentation by DCs (113–116).
✓ Alpha beta (αβ) T cells (CD8+ T cells and CD4+ T cells) survive and persist in the skin long after the immune reaction is over (113, 117–123). This ensures rapid protection from future exposures to pathogens or antigens (112, 124). Skin resident-memory Treg cells mitigate inflammation and regulate immune response via autophagy (81, 82). Tregs (20–60% of CD4+ T cells) have also been shown to alleviate autoimmune disorder by suppressing DC autophagy (81, 83). Atg7-deficient Tregs cannot establish skin homeostasis (84, 85).
✓ Gamma delta (γd) T cells comprise 1–5% of the circulating T cells found in both mice and humans (125). However, in skin about 50% of γd T cells are present among the T cells (126). They populate the skin epidermis and dermis and are named dendritic epidermal γd T cells (DETCs) and dermal γd T cells, respectively (125, 127). Like αβ T cells, they possess effector functions as well as mediate tissue repair (86, 125). γd T cells are the first responders to skin damage and are responsible for maintaining homeostasis, wound repair, and production of cytokines like insulin-like growth factor 1, TNF-α, and KGF-1 (86). DETCs are in close contact with keratinocytes and aid in wound healing by releasing KGF-7 and KGF-10 (128, 129). Skin γd T cells are adaptable under immunosuppression by rapamycin (inhibits mTOR pathway) or roseotoxin B (suppresses activated T cells but does not effect naïve T cells), and they undergo autophagy to survive in the absence of cytokines (70, 86).
• B cells are a component of the adaptive immune system populating the dermal layers and produces antibodies. In B cells, ATG5 seems to play an important role in differentiation and survival (80). Autoantibodies induce autophagy and can sometimes lead to autoimmune disorders (80).
Skin Exposures and Autophagy
When the skin encounters pathogens, injury, or UVR, it deploys various defense mechanisms. Autophagy responds to these unwanted encounters to ensure inflammation resolution. Defects in autophagy can cause a hyperinflammatory skin reaction due to inflammasome activation (as seen in human keratinocytes), unpredicted ROS activation via UVR, and aberrant pro-inflammatory cytokine release (Figure 1A) (4, 113, 130–133).
Pathogens
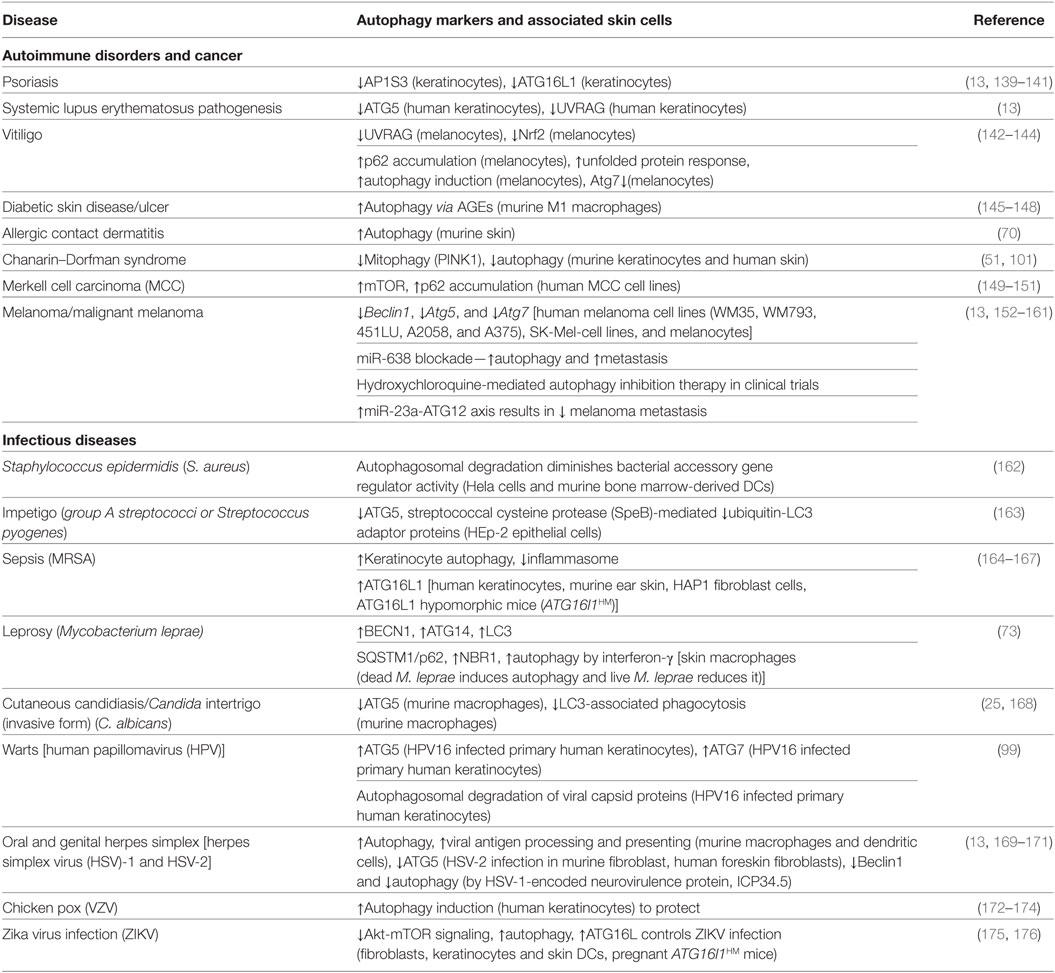
Evolutionarily, epidermal pathogenic bacteria have devised multiple evasion mechanisms to avoid autophagic clearance (134, 135). Numerous instances of the autophagy machinery engaging skin pathogens during cutaneous infection have been described (Table 2). Autophagy is critical for the clearance of group A streptococci (GAS) which evades endosomal capture (13, 136). A recent study showed that the elimination of GAS is severely reduced in Atg5-deficient cells elucidating its vital role in pathogen clearance (13, 136). Streptococcus pyogenes is responsible for causing impetigo, a common skin infection in children (137, 138). Similarly, autophagy is essential in preventing infectious skin diseases such as Staphylococcus epidermidis infection, leprosy, and sepsis (Table 1).
The skin is susceptible to fungal infections, such as cutaneous candidiasis and Candida intertrigo caused by Candida albicans (168). Studies have shown that the intracellular clearance of C. albicans depends on both autophagy (ATG5) and LAP (168, 177). Autophagy in skin also plays a unique role in anti-viral function (Table 2). Herpes simplex virus (HSV) is a double-stranded DNA virus. It is categorized into HSV-1 and HSV-2, which causes oral herpes and genital herpes (13). Autophagy aids in processing and presentation of HSV-1 antigens on MHC class I molecule for effective viral elimination (178–180). HSV-2 is more susceptible to the host ATG5 (13, 179, 181). Furthermore, skin-related viruses, including human papillomavirus, varicella zoster virus, and Zika virus can induce autophagy to degrade viral capsid proteins in the skin cells and keratinocytes (172, 182).
Wound Healing
During wound healing after an injury or pathogen invasion, skin immune responses halt the ongoing inflammation to initiate the restoration process (2). In rats, autophagy heals burnt hair follicle epithelium (183). A recent study shows that the use of mesenchymal stem cells (MSCs) in skin repair requires autophagy (184). Rapamycin-induced autophagy in MSCs causes secretion of vascular endothelial growth factor (VEGF) and improves VEGF-mediated blood circulation which in turn promotes skin wound healing and tissue regeneration (184).
Ultraviolet Radiation
Sun-generated UVR is a mixture of UVA and UVB (185). Upon exposure to UVR, basal autophagy increases in keratinocytes (28) and causes epidermal thickening or hyperkeratosis (28). This in turn, leads to epidermal hyperplasia which prevents the UVR (UVA or UVB, depending on the wavelength) to penetrate the skin (28, 186). UVR-induced cell death in the skin can promote autoimmunity due to defective clearance of apoptotic keratinocytes (5, 108). Patients with systemic lupus erythematosus, an autophagy-related autoimmune disorder, are unable to clear dead cells and suffer from severe cutaneous lesions upon exposure to UVR as a result (187–191). UVR also inhibits antigen presentation by LCs and Treg migration, as seen in cutaneous T cell-mediated dermatitis (70, 192–196).
UVA (long wavelength) irradiation penetrates the dermis, and its exposure induces autophagy to remove p62-associated protein aggregates in keratinocytes and melanocytes (5, 61, 197, 198). Chronic UVA exposure causes apoptosis of epidermal and dermal cells, photoaging, and skin pathogenesis, depending upon the presence of melanin (28, 185). Luteolin (3, 4, 5, 7-tetrahydroxyflavone), a flavonoid showing anti-cancer properties, has recently been shown to decrease UVA-induced autophagy in human skin fibroblasts by scavenging ROS (95, 191).
UVB irradiation entry is limited to the epidermis (198–200). UVB-induced autophagy involves glycogen synthase kinase signaling, which helps protect the epidermal cells from UVB-induced apoptosis (201). UVB-induced ROS downregulates mTOR in skin epidermal cells and induces autophagy (28, 187, 202). Thus, ROS inhibition prevents T cell-mediated dermatitis in mice (203). UVB-treated murine splenocytes show systemic immunosuprression by inhibiting both IFN-γ and IL-10 cytokine production 24 h post-irradiation (204).
UVB radiation also stimulates AMPs (psoriasin, RNase 7, human β-defensin [HBD 1–4]) in human keratinocytes (7, 8, 205). To protect against UVB-induced DNA damage, oxidative stress, cancer, and skin cells produce fat soluble vitamin D (206). Epidermal AMPs like cathelicidin (hBD18) expressed in keratinocytes is induced by 1,25 (OH)2 vitamin D3 from 7-dehydrocholesterol and protects the skin against pathogens (8, 206, 207). Vitamin D promotes autophagy (ATG16L1 in autoimmune disorders like inflammatory bowel disorder) and suppresses pro-inflammatory pathways (such as p38 MAPK-mediated signaling pathway, prostaglandin pathway, nuclear factor kappa B signaling pathway) (206, 208, 209).
UV stimulates the central stress response center via hypothalamic–pituitary–adrenal axis, however, the mechanism is not well understood (204). UVR-induced β-endorphin and corticotropin-releasing hormone release from the skin causes soluble neuro-endocrine-immune factors to seep into the systemic circulation (204, 210). The skin immune cells stimulated by UV act as “second messengers” allowing crosstalk between neuroendocrine system and immune system (210). Certain neuropeptides, such as calcitonin gene-related peptide (CGRP), mediate anti-inflammatory environment by UVR (211). Steriods like proopiomelanocortin produced from α-melanocyte-stimulating hormone (α-MSH) of UVR-triggered epidermal, dermal cells and macrophages induce immunosuppression (210, 211).
Skin Cancer and Autoimmune Disorders
Autoimmune disorders are the result of the adaptive immune system generating autoreactive lymphocytes (T and B cells) that target self-antigens. Skin disorders often arise as a secondary complication, as reported in diabetes, cancer, and dermatitis. The role of autophagy reported in skin-related disorders (Table 1) is limited, yet nonetheless deserves recognition.
Atg5-deficiency, Atg7-deficiency, and Beclin-1 partial deletion can spur spontaneous tumor growth commonly seen in most cancers (152, 212–214). Contrary to that, targeting Beclin-1 inhibits autophagy, overexpresses CCL5 and aids in recruit NK cells to the melanoma tumor (79). Melanoma is fatal in its aggressive form and is highly metastatic (153). Atg5-deficient melanoma cells in vitro have diminished survival (152, 154). Interestingly, in advanced stages, melanoma cells promote a tumor-suppressive environment by hijacking the autophagy machinery to ease stress induced by drugs (215). This suggests that cancer cells can manipulate the autophagy machinery to resist treatment (154, 216).
Diabetes mellitus (both type 1 and type 2) patients have defective insulin signaling in the keratinocytes and often suffer from skin lesions and foot ulcers (145–147, 217). In diabetic patients, IRF8 (an autophagy regulator) activation induces autophagy, poises the macrophages to permit inflammation and thus, impairs cutaneous wound healing (145, 217). Chronic hyperglycemia and hyperlipidemia disrupt ER homeostasis and resulted in increased unfolded protein burden (147, 217). Both autophagy and mitophagy are defective in diabetic patients and inhibit keratinocyte proliferation and migration that are requisite for wound healing (145–147).
Psoriasis is a chronic, polygenic autoimmune disease characterized by epidermal hyperplasia, defective keratinization, and infiltration of immune cells within the skin, causing dermatitis and thickened plaques formation. Genetic screening of psoriasis patients of Estonian origin reveals several single-nucleotide polymorphisms (SNPs) associated with ATG16L1, though the functional role of ATG16L1 variants in skin biology is unclear (139, 218). In pustular psoriasis cases, mutations in AP1S3, a gene encoding an autophagosome trafficking protein, result in the disruption of autophagy in keratinocytes and drive them to produce pro-inflammatory cytokines, including IL-1β, IL-8, and IL-36A (140). Furthermore, a recent study demonstrated that autophagy inhibition in vivo shows aberrations in keratinocyte differentiation, resulting in dysregulation of autophagy in psoriatic epidermis (219).
Vitiligo is a pigmentation disorder characterized by sharp demarcated white macules on skin due to the CD8+ T cell-mediated destruction of melanocytes (220, 221). This results in localized (segmental vitiligo) and/or generalized (non-segmental vitiligo) partial loss of melanin. In addition, melanocytes show increased autophagy due to misfolding of tyrosinase (tyr) and X-box binding protein 1 (Xbp1) in the ER (220, 221). Individuals suffering from vitiligo may have defective epidermal permeability barrier functions which can be alleviated by the use of topical histamine treatment (222, 223). Vitiligo patients from Chinese Han population show abnormal antioxidant Nrf2 [nuclear factor (erythroid-derived 2)-like 2] gene expressions in autophagy and associated pathways (142, 224, 225). Compared to normal melanocytes, vitiligo melanocytes in vitro show greater similarity with Atg7-deficient melanocytes (57, 60). Both exhibit impaired redox-sensitive Nrf2 activation and decreased activation of the antioxidant enzyme system in response to oxidative challenges induced by environmental stress and ROS (93, 226, 227). Multiple evidences indicate that autophagy controls melanosome degradation (92, 228). Furthermore, a Korean cohort study demonstrated an association between two UVRAG gene SNPs and non-segmental vitiligo (vitiligo vulgaris) (229). Skin autoimmune disorders are overly complicated, involve many aspects of the immune system and arise due to defects in autophagy machinery. Accumulating evidence displaying the importance of autophagy in skin disorders demonstrates the need for further research.
Concluding Remarks
Being at the forefront of the environmental defense system, a plethora of specialized cells reinforces the skin. It is a unique surface that is highly specialized in preserving immunotolerance. Therefore, autophagic clearance of senescent and damaged cells is necessary for the maintenance of specialized cellular machinery to effectively keep inflammatory triggers at bay. Several research groups have implicated the role of ATG5, ATG7, and ATG16L1 in keratinocytes, melanocytes, and immune cells in autoimmunity and cancer. Skin in the context of autophagy remains an uncharted territory and warrants further investigation. Autophagy dictates the immune response and resolution in skin cells to neutralize pathogens, clear senescent cells, or heal wounds. However, the autophagy machinery can either protect or cause autoimmune disorders. The physiological circumstances that govern this “balancing act” are not well understood. The cytokine microenvironment from dying immune cells, pathogens, or senescent cells can potentially direct the autophagic response in skin. In addition, there are evidences suggesting potential communication between autophagic machinery and skin cells (both immune and non-immune cells) in an inflammatory state that resonates in the literature; however, the mechanism is not yet reported (230). Adequate tools to examine the molecular response are lacking.
In addition, skin microbiota has been shown to shape the immune response by producing AMPs, complement, IL-1, and IL-17 as well as modulating the local T cell response (3, 231). The interaction with skin cells and microbiota may reveal underlying mechanisms to aid in conscientious operation of the autophagic machinery as seen in Crohn’s disease and colorectal cancer (232–234). Overall, further research investigating the role of autophagy in barrier function will pave the way forward for therapeutic advancements in the field of skin inflammaging and dermatology.
Author Contributions
PS and JM outlined the structure and theme of the paper. PS and S-WW performed the systemic literature search. S-WW helped with the disease section and table 2 of the manuscript. PS developed and wrote the final version manuscript. PS also performed the revisions suggested by the reviewers. JM provided valuable guidance while formulating the manuscript and supervised the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by grants from the National Institutes of Health (1ZIAES10328601).
References
1. Dainichi T, Hanakawa S, Kabashima K. Classification of inflammatory skin diseases: a proposal based on the disorders of the three-layered defense systems, barrier, innate immunity and acquired immunity. J Dermatol Sci (2014) 76(2):81–9. doi:10.1016/j.jdermsci.2014.08.010
2. Richmond JM, Harris JE. Immunology and skin in health and disease. Cold Spring Harb Perspect Med (2014) 4(12):a015339. doi:10.1101/cshperspect.a015339
3. Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science (2014) 346(6212):954–9. doi:10.1126/science.1260144
4. Chen RJ, Lee YH, Yeh YL, Wang YJ, Wang BJ. The roles of autophagy and the inflammasome during environmental stress-triggered skin inflammation. Int J Mol Sci (2016) 17(12):1–16. doi:10.3390/ijms17122063
5. Sukseree S, Eckhart L, Tschachler E, Watanapokasin R. Autophagy in epithelial homeostasis and defense. Front Biosci (Elite Ed) (2013) 5:1000–10.
6. Slominski AT, Zmijewski MA, Skobowiat C, Zbytek B, Slominski RM, Steketee JD. Sensing the environment: regulation of local and global homeostasis by the skin’s neuroendocrine system. Adv Anat Embryol Cell Biol (2012) 212:v,vii,1–115. doi:10.1007/978-3-642-19683-6_1
7. Elias PM. Skin barrier function. Curr Allergy Asthma Rep (2008) 8(4):299–305. doi:10.1007/s11882-008-0048-0
8. Elias PM. The skin barrier as an innate immune element. Semin Immunopathol (2007) 29(1):3–14. doi:10.1007/s00281-007-0060-9
9. Buechner S, Erne P, Resink TJ. T-cadherin expression in the epidermis and adnexal structures of normal skin. Dermatopathology (Basel) (2016) 3(4):68–78. doi:10.1159/000451024
10. Xu Z, Parra D, Gómez D, Salinas I, Zhang YA, von Gersdorff Jørgensen L, et al. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc Natl Acad Sci U S A (2013) 110(32):13097–102. doi:10.1073/pnas.1304319110
11. Sil P, Muse G, Martinez J. A ravenous defense: canonical and non-canonical autophagy in immunity. Curr Opin Immunol (2017) 50:21–31. doi:10.1016/j.coi.2017.10.004
12. Harnett MM, Pineda MA, Latre de Late P, Eason RJ, Besteiro S, Harnett W, et al. From Christian de Duve to Yoshinori Ohsumi: more to autophagy than just dining at home. Biomed J (2017) 40(1):9–22. doi:10.1016/j.bj.2016.12.004
13. Yu T, Zuber J, Li J. Targeting autophagy in skin diseases. J Mol Med (Berl) (2015) 93(1):31–8. doi:10.1007/s00109-014-1225-3
14. De Duve C. The significance of lysosomes in pathology and medicine. Proc Inst Med Chic (1966) 26(4):73–6.
15. De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol (1966) 28:435–92. doi:10.1146/annurev.ph.28.030166.002251
16. De Duve C. Principles of tissue fractionation. J Theor Biol (1964) 6(1):33–59. doi:10.1016/0022-5193(64)90065-7
17. Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol (1992) 119(2):301–11. doi:10.1083/jcb.119.2.301
18. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature (2008) 451(7182):1069–75. doi:10.1038/nature06639
19. Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, et al. Molecular definitions of autophagy and related processes. EMBO J (2017) 36(13):1811–36. doi:10.15252/embj.201796697
20. Shiba H, Yabu T, Sudayama M, Mano N, Arai N, Nakanishi T, et al. Sequential steps of macroautophagy and chaperone-mediated autophagy are involved in the irreversible process of posterior silk gland histolysis during metamorphosis of Bombyx mori. J Exp Biol (2016) 219(Pt 8):1146–53. doi:10.1242/jeb.130815
21. Bestebroer J, V’kovski P, Mauthe M, Reggiori F. Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic (2013) 14(10):1029–41. doi:10.1111/tra.12091
22. Kaiser SE, Mao K, Taherbhoy AM, Yu S, Olszewski JL, Duda DM, et al. Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures. Nat Struct Mol Biol (2012) 19(12):1242–9. doi:10.1038/nsmb.2415
23. Kuma A, Komatsu M, Mizushima N. Autophagy-monitoring and autophagy-deficient mice. Autophagy (2017) 13(10):1619–28. doi:10.1080/15548627.2017.1343770
24. Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol (2004) 36(12):2503–18. doi:10.1016/j.biocel.2004.05.009
25. Nicola AM, Albuquerque P, Martinez LR, Dal-Rosso RA, Saylor C, De Jesus M, et al. Macrophage autophagy in immunity to Cryptococcus neoformans and Candida albicans. Infect Immun (2012) 80(9):3065–76. doi:10.1128/IAI.00358-12
26. Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol (2015) 17(7):893–906. doi:10.1038/ncb3192
27. Wong SW, Sil P, Martinez J. Rubicon: LC3-associated phagocytosis and beyond. FEBS J (2017) 285(8):1379–88. doi:10.1111/febs.14354
28. D’Orazio J, Jarrett S, Amaro-Ortiz A, Scott T. UV radiation and the skin. Int J Mol Sci (2013) 14(6):12222–48. doi:10.3390/ijms140612222
29. Marshall RS, Vierstra RD. Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol (2018) 69:173–208. doi:10.1146/annurev-arplant-042817-040606
30. Munch C, Dikic I. Hitchhiking on selective autophagy. Nat Cell Biol (2018) 20(2):122–4. doi:10.1038/s41556-018-0036-0
31. Wu MY, Song JX, Wang SF, Cai CZ, Li M, Lu JH. Selective autophagy: the new player in the fight against neurodegenerative diseases? Brain Res Bull (2018) 137:79–90. doi:10.1016/j.brainresbull.2017.11.009
32. Whang MI, Tavares RM, Benjamin DI, Kattah MG, Advincula R, Nomura DK, et al. The ubiquitin binding protein TAX1BP1 mediates autophagasome induction and the metabolic transition of activated T cells. Immunity (2017) 46(3):405–20. doi:10.1016/j.immuni.2017.02.018
33. Sukseree S, Bergmann S, Pajdzik K, Tschachler E, Eckhart L. Suppression of autophagy perturbs turnover of sequestosome-1/p62 in Merkel cells but not in keratinocytes. J Dermatol Sci (2018) 90(2):209–11. doi:10.1016/j.jdermsci.2018.01.008
34. Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem (2008) 283(33):22847–57. doi:10.1074/jbc.M802182200
35. Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A (2016) 113(15):4039–44. doi:10.1073/pnas.1523926113
36. Shen WC, Li HY, Chen GC, Chern Y, Tu PH. Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy (2015) 11(4):685–700. doi:10.4161/auto.36098
37. Viret C, Rozieres A, Faure M. Novel insights into NDP52 autophagy receptor functioning. Trends Cell Biol (2018) 28(4):255–7. doi:10.1016/j.tcb.2018.01.003
38. Minowa-Nozawa A, Nozawa T, Okamoto-Furuta K, Kohda H, Nakagawa I. Rab35 GTPase recruits NDP52 to autophagy targets. EMBO J (2017) 36(22):3405. doi:10.15252/embj.201798293
39. Verlhac P, Viret C, Faure M. [NDP52, autophagy and pathogens: “the war then ceased for lack of combatants”]. Med Sci (Paris) (2015) 31(6–7):594–7. doi:10.1051/medsci/20153106007
40. Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, et al. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci (2013) 126(Pt 4):939–52. doi:10.1242/jcs.114819
41. Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle (2009) 8(13):1986–90. doi:10.4161/cc.8.13.8892
42. Hirota Y, Aoki Y, Kanki T. [Mitophagy: selective degradation of mitochondria by autophagy]. Seikagaku (2011) 83(2):126–30.
43. Chen L, Ma K, Han J, Chen Q, Zhu Y. Monitoring mitophagy in mammalian cells. Methods Enzymol (2017) 588:187–208. doi:10.1016/bs.mie.2016.10.038
44. Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature (2015) 524(7565):309–14. doi:10.1038/nature14893
46. Alexander DE, Leib DA. Xenophagy in herpes simplex virus replication and pathogenesis. Autophagy (2008) 4(1):101–3. doi:10.4161/auto.5222
47. Mao K, Klionsky DJ. Xenophagy: a battlefield between host and microbe, and a possible avenue for cancer treatment. Autophagy (2017) 13(2):223–4. doi:10.1080/15548627.2016.1267075
48. Kim JH, Kim TH, Lee HC, Nikapitiya C, Uddin MB, Park ME, et al. Rubicon modulates antiviral type I interferon (IFN) signaling by targeting IFN regulatory factor 3 dimerization. J Virol (2017) 91(14):e00248–17. doi:10.1128/JVI.00248-17
49. Lichtman JS, Alsentzer E, Jaffe M, Sprockett D, Masutani E, Ikwa E, et al. The effect of microbial colonization on the host proteome varies by gastrointestinal location. ISME J (2016) 10(5):1170–81. doi:10.1038/ismej.2015.187
50. Aymard E, Barruche V, Naves T, Bordes S, Closs B, Verdier M, et al. Autophagy in human keratinocytes: an early step of the differentiation? Exp Dermatol (2011) 20(3):263–8. doi:10.1111/j.1600-0625.2010.01157.x
51. Grond S, Radner FP, Eichmann TO, Kolb D, Grabner GF, Wolinski H, et al. Skin barrier development depends on CGI-58 protein expression during late-stage keratinocyte differentiation. J Invest Dermatol (2017) 137(2):403–13. doi:10.1016/j.jid.2016.09.025
52. Li Z, Hu L, Elias PM, Man MQ. Skin care products can aggravate epidermal function: studies in a murine model suggest a pathogenic role in sensitive skin. Contact Dermatitis (2018) 78(2):151–8. doi:10.1111/cod.12909
53. Rossiter H, König U, Barresi C, Buchberger M, Ghannadan M, Zhang CF, et al. Epidermal keratinocytes form a functional skin barrier in the absence of Atg7 dependent autophagy. J Dermatol Sci (2013) 71(1):67–75. doi:10.1016/j.jdermsci.2013.04.015
54. Song X, Narzt MS, Nagelreiter IM, Hohensinner P, Terlecki-Zaniewicz L, Tschachler E, et al. Autophagy deficient keratinocytes display increased DNA damage, senescence and aberrant lipid composition after oxidative stress in vitro and in vivo. Redox Biol (2017) 11:219–30. doi:10.1016/j.redox.2016.12.015
55. Chikh A, Sanzà P, Raimondi C, Akinduro O, Warnes G, Chiorino G, et al. iASPP is a novel autophagy inhibitor in keratinocytes. J Cell Sci (2014) 127(Pt 14):3079–93. doi:10.1242/jcs.144816
56. Schraermeyer U, Peters S, Thumann G, Kociok N, Heimann K. Melanin granules of retinal pigment epithelium are connected with the lysosomal degradation pathway. Exp Eye Res (1999) 68(2):237–45. doi:10.1006/exer.1998.0596
57. Zhang CF, Gruber F, Ni C, Mildner M, Koenig U, Karner S, et al. Suppression of autophagy dysregulates the antioxidant response and causes premature senescence of melanocytes. J Invest Dermatol (2015) 135(5):1348–57. doi:10.1038/jid.2014.439
58. Zheng X, Li Y, Zhao R, Yan F, Ma Y, Zhao L, et al. xCT deficiency induces autophagy via endoplasmic reticulum stress activated p38-mitogen-activated protein kinase and mTOR in sut melanocytes. Eur J Cell Biol (2016) 95(6–7):175–81. doi:10.1016/j.ejcb.2016.03.002
59. Yang F, Yang L, Wataya-Kaneda M, Hasegawa J, Yoshimori T, Tanemura A, et al. Dysregulation of autophagy in melanocytes contributes to hypopigmented macules in tuberous sclerosis complex. J Dermatol Sci (2018) 89(2):155–64. doi:10.1016/j.jdermsci.2017.11.002
60. Setaluri V. Autophagy as a melanocytic self-defense mechanism. J Invest Dermatol (2015) 135(5):1215–7. doi:10.1038/jid.2015.19
61. Sample A, Zhao B, Wu C, Qian S, Shi X, Aplin A, et al. The autophagy receptor adaptor p62 is up-regulated by UVA radiation in melanocytes and in melanoma cells. Photochem Photobiol (2017). doi:10.1111/php.12809
62. Chen L, Xu Z, Jiang M, Zhang C, Wang X, Xiang L. Light-emitting diode 585nm photomodulation inhibiting melanin synthesis and inducing autophagy in human melanocytes. J Dermatol Sci (2018) 89(1):11–8. doi:10.1016/j.jdermsci.2017.10.001
63. Abdel-Malek Z, Swope V, Smalara D, Babcock G, Dawes S, Nordlund J. Analysis of the UV-induced melanogenesis and growth arrest of human melanocytes. Pigment Cell Res (1994) 7(5):326–32. doi:10.1111/j.1600-0749.1994.tb00635.x
64. Andersson E, Rosdahl I, Törmä H, Vahlquist A. Differential effects of UV irradiation on nuclear retinoid receptor levels in cultured keratinocytes and melanocytes. Exp Dermatol (2003) 12(5):563–71. doi:10.1034/j.1600-0625.2003.00090.x
65. Slominski A, Tobin DJ, Shibahara S, Wortsman J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol Rev (2004) 84(4):1155–228. doi:10.1152/physrev.00044.2003
66. Sukseree S, Rossiter H, Tschachler E, Eckhart L. Keeping in touch with autophagy – a mouse model with Atg7-deficient Merkel cells. J Invest Dermatol (2015) 135:S66–66.
67. Sukseree S, Rossiter H, Tschachler E, Eckhart L. K14-Cre-mediated deletion of Atg7 leads to accumulation of sequestosome 1 in Merkel cells. Exp Dermatol (2016) 25(3):E6–6.
68. Bhattacharya A, Wei Q, Shin JN, Abdel Fattah E, Bonilla DL, Xiang Q, et al. Autophagy is required for neutrophil-mediated inflammation. Cell Rep (2015) 12(11):1731–9. doi:10.1016/j.celrep.2015.08.019
69. Oliveira RB, Moraes MO, Oliveira EB, Sarno EN, Nery JA, Sampaio EP. Neutrophils isolated from leprosy patients release TNF-alpha and exhibit accelerated apoptosis in vitro. J Leukoc Biol (1999) 65(3):364–71. doi:10.1002/jlb.65.3.364
70. Wang X, Hu C, Wu X, Wang S, Zhang A, Chen W, et al. Roseotoxin B improves allergic contact dermatitis through a unique anti-inflammatory mechanism involving excessive activation of autophagy in activated T lymphocytes. J Invest Dermatol (2016) 136(8):1636–46. doi:10.1016/j.jid.2016.04.017
71. Facchetti F, De Wolf-Peeters C, van den Oord JJ, Desmet VJ. Melanophages in inflammatory skin disease. J Am Acad Dermatol (1989) 21(6):1315. doi:10.1016/S0190-9622(89)80319-6
72. Busam KJ, Charles C, Lee G, Halpern AC. Morphologic features of melanocytes, pigmented keratinocytes, and melanophages by in vivo confocal scanning laser microscopy. Mod Pathol (2001) 14(9):862–8. doi:10.1038/modpathol.3880402
73. Silva BJ, Barbosa MG, Andrade PR, Ferreira H, Nery JA, Côrte-Real S, et al. Autophagy is an innate mechanism associated with leprosy polarization. PLoS Pathog (2017) 13(1):e1006103. doi:10.1371/journal.ppat.1006103
74. Mintern JD, Macri C, Chin WJ, Panozza SE, Segura E, Patterson NL, et al. Differential use of autophagy by primary dendritic cells specialized in cross-presentation. Autophagy (2015) 11(6):906–17. doi:10.1080/15548627.2015.1045178
75. Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity (2007) 26(1):79–92. doi:10.1016/j.immuni.2006.10.018
76. Luis A, Martins JD, Silva A, Ferreira I, Cruz MT, Neves BM. Oxidative stress-dependent activation of the eIF2alpha-ATF4 unfolded protein response branch by skin sensitizer 1-fluoro-2,4-dinitrobenzene modulates dendritic-like cell maturation and inflammatory status in a biphasic manner [corrected]. Free Radic Biol Med (2014) 77:217–29. doi:10.1016/j.freeradbiomed.2014.09.008
77. Nakano H, Ushio H. An unexpected role for autophagy in degranulation of mast cells. Autophagy (2011) 7(6):657–9. doi:10.4161/auto.7.6.15384
78. Moon TC, Befus AD, Kulka M. Mast cell mediators: their differential release and the secretory pathways involved. Front Immunol (2014) 5:569. doi:10.3389/fimmu.2014.00569
79. Mgrditchian T, Arakelian T, Paggetti J, Noman MZ, Viry E, Moussay E, et al. Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc Natl Acad Sci U S A (2017) 114(44):E9271–9. doi:10.1073/pnas.1703921114
80. Zhou XJ, Zhang H. Autophagy in immunity: implications in etiology of autoimmune/autoinflammatory diseases. Autophagy (2012) 8(9):1286–99. doi:10.4161/auto.21212
81. Ali N, Rosenblum MD. Regulatory T cells in skin. Immunology (2017) 152(3):372–81. doi:10.1111/imm.12791
82. Rosenblum MD, Way SS, Abbas AK. Regulatory T cell memory. Nat Rev Immunol (2016) 16(2):90–101. doi:10.1038/nri.2015.1
83. Alissafi T, Banos A, Boon L, Sparwasser T, Ghigo A, Wing K, et al. Tregs restrain dendritic cell autophagy to ameliorate autoimmunity. J Clin Invest (2017) 127(7):2789–804. doi:10.1172/JCI92079
84. Le Texier L, Lineburg KE, Cao B, McDonald-Hyman C, Leveque-El Mouttie L, Nicholls J, et al. Autophagy-dependent regulatory T cells are critical for the control of graft-versus-host disease. JCI Insight (2016) 1(15):e86850. doi:10.1172/jci.insight.86850
85. Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J Clin Invest (2011) 121(11):4503–15. doi:10.1172/JCI57456
86. Mills RE, Taylor KR, Podshivalova K, McKay DB, Jameson JM. Defects in skin gamma delta T cell function contribute to delayed wound repair in rapamycin-treated mice. J Immunol (2008) 181(6):3974–83. doi:10.4049/jimmunol.181.6.3974
87. Vavricka CJ, Christensen BM, Li J. Melanization in living organisms: a perspective of species evolution. Protein Cell (2010) 1(9):830–41. doi:10.1007/s13238-010-0109-8
88. Slominski A, Wortsman J, Plonka PM, Schallreuter KU, Paus R, Tobin DJ. Hair follicle pigmentation. J Invest Dermatol (2005) 124(1):13–21. doi:10.1111/j.0022-202X.2004.23528.x
89. Slominski A. Neuroendocrine activity of the melanocyte. Exp Dermatol (2009) 18(9):760–3. doi:10.1111/j.1600-0625.2009.00892.x
90. Pawelek JM, Chakraborty AK, Osber MP, Orlow SJ, Min KK, Rosenzweig KE, et al. Molecular cascades in UV-induced melanogenesis: a central role for melanotropins? Pigment Cell Res (1992) 5(5 Pt 2):348–56. doi:10.1111/j.1600-0749.1992.tb00561.x
91. Slominski A, Zmijewski MA, Pawelek J. L-tyrosine and L-dihydroxyphenylalanine as hormone-like regulators of melanocyte functions. Pigment Cell Melanoma Res (2012) 25(1):14–27. doi:10.1111/j.1755-148X.2011.00898.x
92. Murase D, Hachiya A, Takano K, Hicks R, Visscher MO, Kitahara T, et al. Autophagy has a significant role in determining skin color by regulating melanosome degradation in keratinocytes. J Invest Dermatol (2013) 133(10):2416–24. doi:10.1038/jid.2013.165
93. Qiao Z, Wang X, Xiang L, Zhang C. Dysfunction of autophagy: a possible mechanism involved in the pathogenesis of vitiligo by breaking the redox balance of melanocytes. Oxid Med Cell Longev (2016) 2016:3401570. doi:10.1155/2016/3401570
94. Belleudi F, Purpura V, Scrofani C, Persechino F, Leone L, Torrisi MR. Expression and signaling of the tyrosine kinase FGFR2b/KGFR regulates phagocytosis and melanosome uptake in human keratinocytes. FASEB J (2011) 25(1):170–81. doi:10.1096/fj.10-162156
95. Yan M, Liu Z, Yang H, Li C, Chen H, Liu Y, et al. Luteolin decreases the UVA-induced autophagy of human skin fibroblasts by scavenging ROS. Mol Med Rep (2016) 14(3):1986–92. doi:10.3892/mmr.2016.5517
96. Ou R, Zhang X, Cai J, Shao X, Lv M, Qiu W, et al. Downregulation of pyrroline-5-carboxylate reductase-2 induces the autophagy of melanoma cells via AMPK/mTOR pathway. Tumor Biol (2016) 37(5):6485–91. doi:10.1007/s13277-015-3927-8
97. An HT, Yoo JY, Lee MK, Shin MH, Rhie GE, Seo JY, et al. Single dose radiation is more effective for the UV-induced activation and proliferation of melanocytes than fractionated dose radiation. Photodermatol Photoimmunol Photomed (2001) 17(6):266–71. doi:10.1111/j.1600-0781.2001.170604.x
98. Le Poole IC, van den Wijngaard RM, Westerhof W, Verkruisen RP, Dutrieux RP, Dingemans KP, et al. Phagocytosis by normal human melanocytes in vitro. Exp Cell Res (1993) 205(2):388–95. doi:10.1006/excr.1993.1102
99. Li L, Chen X, Gu H. The signaling involved in autophagy machinery in keratinocytes and therapeutic approaches for skin diseases. Oncotarget (2016) 7(31):50682–97. doi:10.18632/oncotarget.9330
100. Sand J, Haertel E, Biedermann T, Contassot E, Reichmann E, French LE, et al. Expression of inflammasome proteins and inflammasome activation occurs in human, but not in murine keratinocytes. Cell Death Dis (2018) 9(2):24. doi:10.1038/s41419-017-0009-4
101. Zhang J, Xu D, Nie J, Han R, Zhai Y, Shi Y. Comparative gene identification-58 (CGI-58) promotes autophagy as a putative lysophosphatidylglycerol acyltransferase. J Biol Chem (2014) 289(47):33044–53. doi:10.1074/jbc.M114.573857
102. Yoshihara N, Ueno T, Takagi A, Oliva Trejo JA, Haruna K, Suga Y, et al. The significant role of autophagy in the granular layer in normal skin differentiation and hair growth. Arch Dermatol Res (2015) 307(2):159–69. doi:10.1007/s00403-014-1508-0
103. Belleudi F, Purpura V, Caputo S, Torrisi MR. FGF7/KGF regulates autophagy in keratinocytes: a novel dual role in the induction of both assembly and turnover of autophagosomes. Autophagy (2014) 10(5):803–21. doi:10.4161/auto.28145
104. Kaplan DH. Langerhans cells: not your average dendritic cell. Trends Immunol (2010) 31(12):437. doi:10.1016/j.it.2010.10.003
105. Green DR, Ferguson T, Zitvogel L, Kroemer G. Immunogenic and tolerogenic cell death. Nat Rev Immunol (2009) 9(5):353–63. doi:10.1038/nri2545
106. Kaplan DH. In vivo function of Langerhans cells and dermal dendritic cells. Trends Immunol (2010) 31(12):446–51. doi:10.1016/j.it.2010.08.006
107. Handfield C, Kwock J, MacLeod AS. Innate antiviral immunity in the skin. Trends Immunol (2018) 75(3):159–66. doi:10.1016/j.it.2018.02.003
108. Kemp MG. Crosstalk between apoptosis and autophagy: environmental genotoxins, infection, and innate immunity. J Cell Death (2017) 9:1179670716685085. doi:10.1177/1179670716685085
109. Schmeisser H, Bekisz J, Zoon KC. New function of type I IFN: induction of autophagy. J Interferon Cytokine Res (2014) 34(2):71–8. doi:10.1089/jir.2013.0128
110. Kim BS, Siracusa MC, Saenz SA, Noti M, Monticelli LA, Sonnenberg GF, et al. TSLP elicits IL-33-independent innate lymphoid cell responses to promote skin inflammation. Sci Transl Med (2013) 5(170):170ra16. doi:10.1126/scitranslmed.3005374
111. Villanova F, Flutter B, Tosi I, Grys K, Sreeneebus H, Perera GK, et al. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J Invest Dermatol (2014) 134(4):984–91. doi:10.1038/jid.2013.477
112. Clark RA. Skin-resident T cells: the ups and downs of on site immunity. J Invest Dermatol (2010) 130(2):362–70. doi:10.1038/jid.2009.247
113. Botbol Y, Guerrero-Ros I, Macian F. Key roles of autophagy in regulating T-cell function. Eur J Immunol (2016) 46(6):1326–34. doi:10.1002/eji.201545955
114. Klein L, Munz C, Lunemann JD. Autophagy-mediated antigen processing in CD4(+) T cell tolerance and immunity. FEBS Lett (2010) 584(7):1405–10. doi:10.1016/j.febslet.2010.01.008
115. Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature (2008) 455(7211):396–400. doi:10.1038/nature07208
116. Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med (2011) 208(13):2625–32. doi:10.1084/jem.20110640
117. Sundarasetty B, Volk V, Theobald SJ, Rittinghausen S, Schaudien D, Neuhaus V, et al. Human effector memory T helper cells engage with mouse macrophages and cause graft-versus-host-like pathology in skin of humanized mice used in a nonclinical immunization study. Am J Pathol (2017) 187(6):1380–98. doi:10.1016/j.ajpath.2017.02.015
118. Li G, Larregina AT, Domsic RT, Stolz DB, Medsger TA, Lafyatis R Jr, et al. Skin-resident effector memory CD8(+)CD28(-) T cells exhibit a profibrotic phenotype in patients with systemic sclerosis. J Invest Dermatol (2017) 137(5):1042–50. doi:10.1016/j.jid.2016.11.037
119. Clark RA, Watanabe R, Teague JE, Schlapbach C, Tawa MC, Adams N, et al. Skin effector memory T cells do not recirculate and provide immune protection in alemtuzumab-treated CTCL patients. Sci Transl Med (2012) 4(117):117ra7. doi:10.1126/scitranslmed.3003008
120. Homey B. [Psoriasis or atopic eczema: skin-infiltrating effector memory T cells make the difference]. Hautarzt (2012) 63(1):52. doi:10.1007/s00105-011-2305-8
121. Anderson BE, Tang AL, Wang Y, Froicu M, Rothstein D, McNiff JM, et al. Enhancing alloreactivity does not restore GVHD induction but augments skin graft rejection by CD4(+) effector memory T cells. Eur J Immunol (2011) 41(9):2782–92. doi:10.1002/eji.201141678
122. Pober JS, Kluger MS, Schechner JS. Human endothelial cell presentation of antigen and the homing of memory/effector T cells to skin. Ann N Y Acad Sci (2001) 941:12–25. doi:10.1111/j.1749-6632.2001.tb03706.x
123. Yawalkar N, Hunger RE, Pichler WJ, Braathen LR, Brand CU. Human afferent lymph from normal skin contains an increased number of mainly memory/effector CD4(+) T cells expressing activation, adhesion and co-stimulatory molecules. Eur J Immunol (2000) 30(2):491–7. doi:10.1002/1521-4141(200002)30:2<491::AID-IMMU491>3.0.CO;2-H
124. Mackay CR, Marston WL, Dudler L. Naive and memory T cells show distinct pathways of lymphocyte recirculation. J Exp Med (1990) 171(3):801–17. doi:10.1084/jem.171.3.801
125. Nielsen MM, Witherden DA, Havran WL. Gammadelta T cells in homeostasis and host defence of epithelial barrier tissues. Nat Rev Immunol (2017) 17(12):733–45. doi:10.1038/nri.2017.101
126. Lawand M, Dechanet-Merville J, Dieu-Nosjean MC. Key features of gamma-delta T-cell subsets in human diseases and their immunotherapeutic implications. Front Immunol (2017) 8:761. doi:10.3389/fimmu.2017.00761
127. Stetson DB, Voehringer D, Grogan JL, Xu M, Reinhardt RL, Scheu S, et al. Th2 cells: orchestrating barrier immunity. Adv Immunol (2004) 83:163–89. doi:10.1016/S0065-2776(04)83005-0
128. Jameson JM, Sharp LL, Witherden DA, Havran WL. Regulation of skin cell homeostasis by gamma delta T cells. Front Biosci (2004) 9:2640–51. doi:10.2741/1423
129. Jameson J, Ugarte K, Chen N, Yachi P, Fuchs E, Boismenu R, et al. A role for skin gammadelta T cells in wound repair. Science (2002) 296(5568):747–9. doi:10.1126/science.1069639
130. Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol (2011) 12(3):222–30. doi:10.1038/ni.1980
131. Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature (2008) 456(7219):264–8. doi:10.1038/nature07383
132. Niebler M, Qian X, Hofler D, Kogosov V, Kaewprag J, Kaufmann AM, et al. Post-translational control of IL-1beta via the human papillomavirus type 16 E6 oncoprotein: a novel mechanism of innate immune escape mediated by the E3-ubiquitin ligase E6-AP and p53. PLoS Pathog (2013) 9(8):e1003536. doi:10.1371/journal.ppat.1003536
133. Wlaschek M, Tantcheva-Poór I, Naderi L, Ma W, Schneider LA, Razi-Wolf Z, et al. Solar UV irradiation and dermal photoaging. J Photochem Photobiol B (2001) 63(1–3):41–51. doi:10.1016/S1011-1344(01)00201-9
134. Barnett TC, Liebl D, Seymour LM, Gillen CM, Lim JY, Larock CN, et al. The globally disseminated M1T1 clone of group A Streptococcus evades autophagy for intracellular replication. Cell Host Microbe (2013) 14(6):675–82. doi:10.1016/j.chom.2013.11.003
135. Singer AJ, Talan DA. Management of skin abscesses in the era of methicillin-resistant Staphylococcus aureus. N Engl J Med (2014) 370(11):1039–47. doi:10.1056/NEJMra1212788
136. Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science (2004) 306(5698):1037–40. doi:10.1126/science.1103966
137. Iwasaki A, Kawakami H, Okubo Y. Granulocyte/monocyte adsorption apheresis as a novel therapeutic approach in the treatment of an impetigo herpetiformis case. Ther Apher Dial (2018). doi:10.1111/1744-9987.12653
138. D’Cunha NM, Peterson GM, Baby KE, Thomas J. Impetigo: a need for new therapies in a world of increasing antimicrobial resistance. J Clin Pharm Ther (2018) 43(1):150–3. doi:10.1111/jcpt.12639
139. Douroudis K, Kingo K, Traks T, Reimann E, Raud K, Ratsep R, et al. Polymorphisms in the ATG16L1 gene are associated with psoriasis vulgaris. Acta Derm Venereol (2012) 92(1):85–7. doi:10.2340/00015555-1183
140. Mahil SK, Twelves S, Farkas K, Setta-Kaffetzi N, Burden AD, Gach JE, et al. AP1S3 mutations cause skin autoinflammation by disrupting keratinocyte autophagy and up-regulating IL-36 production. J Invest Dermatol (2016) 136(11):2251–9. doi:10.1016/j.jid.2016.06.618
141. Varshney P, Saini N. PI3K/AKT/mTOR activation and autophagy inhibition plays a key role in increased cholesterol during IL-17A mediated inflammatory response in psoriasis. Biochim Biophys Acta (2018) 1864(5 Pt A):1795–803. doi:10.1016/j.bbadis.2018.02.003
142. Wang P, Li Y, Nie H, Zhang X, Shao Q, Hou X, et al. The changes of gene expression profiling between segmental vitiligo, generalized vitiligo and healthy individual. J Dermatol Sci (2016) 84(1):40–9. doi:10.1016/j.jdermsci.2016.07.006
143. Ezzedine K, Lim HW, Suzuki T, Katayama I, Hamzavi I, Lan CC, et al. Revised classification/nomenclature of vitiligo and related issues: the Vitiligo Global Issues Consensus Conference. Pigment Cell Melanoma Res (2012) 25(3):E1–13. doi:10.1111/j.1755-148X.2012.00997.x
144. Nie HQ, Wang P, Zhang XY, Ding C, Liu J, Xu AE. [Relationship between autophagy of melanocytes in patients with vitiligo and clinical types]. Zhonghua Yi Xue Za Zhi (2016) 96(26):2064–9. doi:10.3760/cma.j.issn.0376-2491.2016.26.006
145. Guo Y, Lin C, Xu P, Wu S, Fu X, Xia W, et al. AGEs induced autophagy impairs cutaneous wound healing via stimulating macrophage polarization to M1 in diabetes. Sci Rep (2016) 6:36416. doi:10.1038/srep36416
146. Yang JS, Lu CC, Kuo SC, Hsu YM, Tsai SC, Chen SY, et al. Autophagy and its link to type II diabetes mellitus. Biomedicine (Taipei) (2017) 7(2):8. doi:10.1051/bmdcn/2017070201
147. Carvalho E, Burgeiro A, Leal E, Emanuelli T. Endoplasmic reticulum stress and autophagy dysfunction: implications in skin repair in a mouse model of diabetes. Diabetologia (2017) 60:S470.
148. Sun K, Wang W, Wang C, Lao G, Liu D, Mai L, et al. AGEs trigger autophagy in diabetic skin tissues and fibroblasts. Biochem Biophys Res Commun (2016) 471(3):355–60. doi:10.1016/j.bbrc.2016.02.020
149. Lin Z, McDermott A, Shao L, Kannan A, Morgan M, Stack BC Jr, et al. Chronic mTOR activation promotes cell survival in Merkel cell carcinoma. Cancer Lett (2014) 344(2):272–81. doi:10.1016/j.canlet.2013.11.005
150. Kannan A, Lin Z, Shao Q, Zhao S, Fang B, Moreno MA, et al. Dual mTOR inhibitor MLN0128 suppresses Merkel cell carcinoma (MCC) xenograft tumor growth. Oncotarget (2016) 7(6):6576–92. doi:10.18632/oncotarget.5878
151. Demerjian M, Crumrine DA, Milstone LM, Williams ML, Elias PM. Barrier dysfunction and pathogenesis of neutral lipid storage disease with ichthyosis (Chanarin-Dorfman syndrome). J Invest Dermatol (2006) 126(9):2032–8. doi:10.1038/sj.jid.5700332
152. Liu H, He Z, von Rütte T, Yousefi S, Hunger RE, Simon HU. Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma. Sci Transl Med (2013) 5(202):202ra123. doi:10.1126/scitranslmed.3005864
153. Ndoye A, Weeraratna AT. Autophagy – an emerging target for melanoma therapy. F1000Res (2016) 5(F1000 Faculty Rev):1888. doi:10.12688/f1000research.8347.1
154. Marino ML, Pellegrini P, Di Lernia G, Djavaheri-Mergny M, Brnjic S, Zhang X, et al. Autophagy is a protective mechanism for human melanoma cells under acidic stress. J Biol Chem (2012) 287(36):30664–76. doi:10.1074/jbc.M112.339127
155. Guo W, Wang H, Yang Y, Guo S, Zhang W, Liu Y, et al. Down-regulated miR-23a contributes to the metastasis of cutaneous melanoma by promoting autophagy. Theranostics (2017) 7(8):2231–49. doi:10.7150/thno.18835
156. Xie X, Koh JY, Price S, White E, Mehnert JM. Atg7 overcomes senescence and promotes growth of BrafV600E-driven melanoma. Cancer Discov (2015) 5(4):410–23. doi:10.1158/2159-8290.CD-14-1473
157. Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy (2014) 10(8):1391–402. doi:10.4161/auto.29119
158. Lazova R, Klump V, Pawelek J. Autophagy in cutaneous malignant melanoma. J Cutan Pathol (2010) 37(2):256–68. doi:10.1111/j.1600-0560.2009.01359.x
159. Liu H, He Z, Simon HU. Autophagy suppresses melanoma tumorigenesis by inducing senescence. Autophagy (2014) 10(2):372–3. doi:10.4161/auto.27163
160. Ma XH, Piao S, Wang D, McAfee QW, Nathanson KL, Lum JJ, et al. Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res (2011) 17(10):3478–89. doi:10.1158/1078-0432.CCR-10-2372
161. Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy (2014) 10(8):1369–79. doi:10.4161/auto.29118
162. O’Keeffe KM, Wilk MM, Leech JM, Murphy AG, Laabei M, Monk IR, et al. Manipulation of autophagy in phagocytes facilitates Staphylococcus aureus bloodstream infection. Infect Immun (2015) 83(9):3445–57. doi:10.1128/IAI.00358-15
163. Nakagawa I. Streptococcus pyogenes escapes from autophagy. Cell Host Microbe (2013) 14(6):604–6. doi:10.1016/j.chom.2013.11.012
164. Schnaith A, Kashkar H, Leggio SA, Addicks K, Krönke M, Krut O. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J Biol Chem (2007) 282(4):2695–706. doi:10.1074/jbc.M609784200
165. Soong G, Paulino F, Wachtel S, Parker D, Wickersham M, Zhang D, et al. Methicillin-resistant Staphylococcus aureus adaptation to human keratinocytes. MBio (2015) 6(2):e00289–15. doi:10.1128/mBio.00289-15
166. Popov LM, Marceau CD, Starkl PM, Lumb JH, Shah J, Guerrera D, et al. The adherens junctions control susceptibility to Staphylococcus aureus alpha-toxin. Proc Natl Acad Sci U S A (2015) 112(46):14337–42. doi:10.1073/pnas.1510265112
167. Maurer K, Reyes-Robles T, Alonzo F III, Durbin J, Torres VJ, Cadwell K. Autophagy mediates tolerance to Staphylococcus aureus alpha-toxin. Cell Host Microbe (2015) 17(4):429–40. doi:10.1016/j.chom.2015.03.001
168. Tam JM, Mansour MK, Acharya M, Sokolovska A, Timmons AK, Lacy-Hulbert A, et al. The role of autophagy-related proteins in Candida albicans infections. Pathogens (2016) 5(2). doi:10.3390/pathogens5020034
169. McFarlane S, Aitken J, Sutherland JS, Nicholl MJ, Preston VG, Preston CM. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus 1. J Virol (2011) 85(9):4212–21. doi:10.1128/JVI.02435-10
170. Meier JL, Grose C. Variable effects of autophagy induction by trehalose on herpesviruses depending on conditions of infection. Yale J Biol Med (2017) 90(1):25–33.
171. Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe (2007) 1(1):23–35. doi:10.1016/j.chom.2006.12.001
172. Takahashi MN, Jackson W, Laird DT, Culp TD, Grose C, Haynes JI II, et al. Varicella-zoster virus infection induces autophagy in both cultured cells and human skin vesicles. J Virol (2009) 83(11):5466–76. doi:10.1128/JVI.02670-08
173. Buckingham EM, Carpenter JE, Jackson W, Grose C. Autophagy and the effects of its inhibition on varicella-zoster virus glycoprotein biosynthesis and infectivity. J Virol (2014) 88(2):890–902. doi:10.1128/JVI.02646-13
174. Grose C, Buckingham EM, Carpenter JE, Kunkel JP. Varicella-zoster virus infectious cycle: ER stress, autophagic flux, and amphisome-mediated trafficking. Pathogens (2016) 5(4):67. doi:10.3390/pathogens5040067
175. Proenca-Modena JL, Milanez GP, Costa ML, Judice CC, Maranhão Costa FT. Zika virus: lessons learned in Brazil. Microbes Infect (2018). doi:10.1016/j.micinf.2018.02.008
176. Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, et al. Biology of Zika virus infection in human skin cells. J Virol (2015) 89(17):8880–96. doi:10.1128/JVI.00354-15
177. Tam JM, Mansour MK, Khan NS, Seward M, Puranam S, Tanne A, et al. Dectin-1-dependent LC3 recruitment to phagosomes enhances fungicidal activity in macrophages. J Infect Dis (2014) 210(11):1844–54. doi:10.1093/infdis/jiu290
178. English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol (2009) 10(5):480–7. doi:10.1038/ni.1720
179. Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity (2010) 32(2):227–39. doi:10.1016/j.immuni.2009.12.006
180. O’Connell D, Liang C. Autophagy interaction with herpes simplex virus type-1 infection. Autophagy (2016) 12(3):451–9. doi:10.1080/15548627.2016.1139262
181. Yakoub AM, Shukla D. Basal autophagy is required for herpes simplex virus-2 infection. Sci Rep (2015) 5:12985. doi:10.1038/srep12985
182. Griffin LM, Cicchini L, Pyeon D. Human papillomavirus infection is inhibited by host autophagy in primary human keratinocytes. Virology (2013) 437(1):12–9. doi:10.1016/j.virol.2012.12.004
183. Tan JQ, Zhang HH, Lei ZJ, Ren P, Deng C, Li XY, et al. The roles of autophagy and apoptosis in burn wound progression in rats. Burns (2013) 39(8):1551–6. doi:10.1016/j.burns.2013.04.018
184. An Y, Liu WJ, Xue P, Ma Y, Zhang LQ, Zhu B, et al. Autophagy promotes MSC-mediated vascularization in cutaneous wound healing via regulation of VEGF secretion. Cell Death Dis (2018) 9(2):58. doi:10.1038/s41419-017-0082-8
185. Marais TLD, Kluz T, Xu D, Zhang X, Gesumaria L, Matsui MS, et al. Transcription factors and stress response gene alterations in human keratinocytes following solar simulated ultra violet radiation. Sci Rep (2017) 7(1):13622. doi:10.1038/s41598-017-13765-7
186. Scott TL, Christian PA, Kesler MV, Donohue KM, Shelton B, Wakamatsu K, et al. Pigment-independent cAMP-mediated epidermal thickening protects against cutaneous UV injury by keratinocyte proliferation. Exp Dermatol (2012) 21(10):771–7. doi:10.1111/exd.12012
187. Bijl M, Reefman E, Limburg PC, Kallenberg CG. Inflammatory clearance of apoptotic cells after UVB challenge. Autoimmunity (2007) 40(4):244–8. doi:10.1080/08916930701357125
188. Leventhal JS, Ross MJ. LAPping up dead cells to prevent lupus nephritis: a novel role for noncanonical autophagy in autoimmunity. Kidney Int (2016) 90(2):238–9. doi:10.1016/j.kint.2016.06.001
189. Wang L, Law HK. The role of autophagy in lupus nephritis. Int J Mol Sci (2015) 16(10):25154–67. doi:10.3390/ijms161025154
190. Mistry P, Kaplan MJ. Cell death in the pathogenesis of systemic lupus erythematosus and lupus nephritis. Clin Immunol (2017) 185:59–73. doi:10.1016/j.clim.2016.08.010
191. Herrling T, Jung K, Fuchs J. Measurements of UV-generated free radicals/reactive oxygen species (ROS) in skin. Spectrochim Acta A Mol Biomol Spectrosc (2006) 63(4):840–5. doi:10.1016/j.saa.2005.10.013
192. Malina L. [Effect of ultraviolet radiation on the immune system and the effect of exogenous photochemoprotecive agents on the ultraviolet radiation induced immunosuppression]. Cas Lek Cesk (2002) 141(11):338–42.
193. Moyal DD, Fourtanier AM. Broad-spectrum sunscreens provide better protection from solar ultraviolet-simulated radiation and natural sunlight-induced immunosuppression in human beings. J Am Acad Dermatol (2008) 58(5 Suppl 2):S149–54. doi:10.1016/j.jaad.2007.04.035
194. Moyal DD, Fourtanier AM. Broad-spectrum sunscreens provide better protection from the suppression of the elicitation phase of delayed-type hypersensitivity response in humans. J Invest Dermatol (2001) 117(5):1186–92. doi:10.1046/j.0022-202x.2001.01545.x
195. Moyal DD, Fourtanier AM. Efficacy of broad-spectrum sunscreens against the suppression of elicitation of delayed-type hypersensitivity responses in humans depends on the level of ultraviolet A protection. Exp Dermatol (2003) 12(2):153–9. doi:10.1034/j.1600-0625.2003.00020.x
196. Schwarz A, Navid F, Sparwasser T, Clausen BE, Schwarz T. In vivo reprogramming of UV radiation-induced regulatory T-cell migration to inhibit the elicitation of contact hypersensitivity. J Allergy Clin Immunol (2011) 128(4):826–33. doi:10.1016/j.jaci.2011.06.005
197. Zhao Y, Zhang CF, Rossiter H, Eckhart L, König U, Karner S, et al. Autophagy is induced by UVA and promotes removal of oxidized phospholipids and protein aggregates in epidermal keratinocytes. J Invest Dermatol (2013) 133(6):1629–37. doi:10.1038/jid.2013.26
198. Strozyk E, Kulms D. The role of AKT/mTOR pathway in stress response to UV-irradiation: implication in skin carcinogenesis by regulation of apoptosis, autophagy and senescence. Int J Mol Sci (2013) 14(8):15260–85. doi:10.3390/ijms140815260
199. Wang Q, Ye Y, Liu W, Jiang S, Tashiro S, Onodera S, et al. Dual effects of silibinin treatment on autophagy-regulated dermal apoptosis retardation and epidermal apoptosis up-regulation in UVB-induced skin inflammation. J Asian Nat Prod Res (2012) 14(7):688–99. doi:10.1080/10286020.2012.685725
200. Henson PM. Cell removal: efferocytosis. Annu Rev Cell Dev Biol (2017) 33:127–44. doi:10.1146/annurev-cellbio-111315-125315
201. Yang Y, Wang H, Wang S, Xu M, Liu M, Liao M, et al. GSK3beta signaling is involved in ultraviolet B-induced activation of autophagy in epidermal cells. Int J Oncol (2012) 41(5):1782–8. doi:10.3892/ijo.2012.1620
202. Son YO, Wang X, Hitron JA, Zhang Z, Cheng S, Budhraja A, et al. Cadmium induces autophagy through ROS-dependent activation of the LKB1-AMPK signaling in skin epidermal cells. Toxicol Appl Pharmacol (2011) 255(3):287–96. doi:10.1016/j.taap.2011.06.024
203. Esser PR, Wölfle U, Dürr C, von Loewenich FD, Schempp CM, Freudenberg MA, et al. Contact sensitizers induce skin inflammation via ROS production and hyaluronic acid degradation. PLoS One (2012) 7(7):e41340. doi:10.1371/journal.pone.0041340
204. Skobowiat C, Postlethwaite AE, Slominski AT. Skin exposure to ultraviolet B rapidly activates systemic neuroendocrine and immunosuppressive responses. Photochem Photobiol (2017) 93(4):1008–15. doi:10.1111/php.12642
205. Gläser R, Navid F, Schuller W, Jantschitsch C, Harder J, Schröder JM, et al. UV-B radiation induces the expression of antimicrobial peptides in human keratinocytes in vitro and in vivo. J Allergy Clin Immunol (2009) 123(5):1117–23. doi:10.1016/j.jaci.2009.01.043
206. Jeon SM, Shin EA. Exploring vitamin D metabolism and function in cancer. Exp Mol Med (2018) 50(4):20. doi:10.1038/s12276-018-0038-9
207. Weber G, Heilborn JD, Chamorro Jimenez CI, Hammarsjo A, Torma H, Stahle M. Vitamin D induces the antimicrobial protein hCAP18 in human skin. J Invest Dermatol (2005) 124(5):1080–2. doi:10.1111/j.0022-202X.2005.23687.x
208. Berridge MJ. Vitamin D deficiency accelerates ageing and age-related diseases: a novel hypothesis. J Physiol (2017) 595(22):6825–36. doi:10.1113/JP274887
209. Sun J. VDR/vitamin D receptor regulates autophagic activity through ATG16L1. Autophagy (2016) 12(6):1057–8. doi:10.1080/15548627.2015.1072670
210. Slominski AT, Zmijewski MA, Plonka PM, Szaflarski JP, Paus R. How UV light touches the brain and endocrine system through skin, and why. Endocrinology (2018) 159(5):1992–2007. doi:10.1210/en.2017-03230
211. Seiffert K, Granstein RD. Neuropeptides and neuroendocrine hormones in ultraviolet radiation-induced immunosuppression. Methods (2002) 28(1):97–103. doi:10.1016/S1046-2023(02)00214-1
212. Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest (2003) 112(12):1809–20. doi:10.1172/JCI20039
213. Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature (1999) 402(6762):672–6. doi:10.1038/45257
214. Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev (2011) 25(8):795–800. doi:10.1101/gad.2016211
215. Corazzari M, Fimia GM, Lovat P, Piacentini M. Why is autophagy important for melanoma? Molecular mechanisms and therapeutic implications. Semin Cancer Biol (2013) 23(5):337–43. doi:10.1016/j.semcancer.2013.07.001
216. Flemming A. Cancer: autophagy presents Achilles heel in melanoma. Nat Rev Drug Discov (2011) 10(7):491. doi:10.1038/nrd3482
217. Emanuelli T, Burgeiro A, Carvalho E. Effects of insulin on the skin: possible healing benefits for diabetic foot ulcers. Arch Dermatol Res (2016) 308(10):677–94. doi:10.1007/s00403-016-1686-z
218. Douroudis K, Kingo K, Traks T, Rätsep R, Silm H, Vasar E, et al. ATG16L1 gene polymorphisms are associated with palmoplantar pustulosis. Hum Immunol (2011) 72(7):613–5. doi:10.1016/j.humimm.2011.03.009
219. Akinduro O, Sully K, Patel A, Robinson DJ, Chikh A, McPhail G, et al. Constitutive autophagy and nucleophagy during epidermal differentiation. J Invest Dermatol (2016) 136(7):1460–70. doi:10.1016/j.jid.2016.03.016
220. Iannella G, Greco A, Didona D, Didona B, Granata G, Manno A, et al. Vitiligo: pathogenesis, clinical variants and treatment approaches. Autoimmun Rev (2016) 15(4):335–43. doi:10.1016/j.autrev.2015.12.006
221. Harris JE. Cellular stress and innate inflammation in organ-specific autoimmunity: lessons learned from vitiligo. Immunol Rev (2016) 269(1):11–25. doi:10.1111/imr.12369
222. Liu J, Man WY, Lv CZ, Song SP, Shi YJ, Elias PM, et al. Epidermal permeability barrier recovery is delayed in vitiligo-involved sites. Skin Pharmacol Physiol (2010) 23(4):193–200. doi:10.1159/000288166
223. Liu J, Xu Y, Lin TK, Lv C, Elias PM, Man MQ. Topical histamine stimulates repigmentation of nonsegmental vitiligo by a receptor-dependent mechanism. Skin Pharmacol Physiol (2017) 30(3):139–45. doi:10.1159/000464335
224. Guan CP, Zhou MN, Xu AE, Kang KF, Liu JF, Wei XD, et al. The susceptibility to vitiligo is associated with NF-E2-related factor2 (Nrf2) gene polymorphisms: a study on Chinese Han population. Exp Dermatol (2008) 17(12):1059–62. doi:10.1111/j.1600-0625.2008.00752.x
225. Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol (2010) 30(13):3275–85. doi:10.1128/MCB.00248-10
226. Aristizabal AM, Caicedo LA, Martínez JM, Moreno M, Echeverri GJ. Clinical xenotransplantation, a closer reality: literature review. Cir Esp (2017) 95(2):62–72. doi:10.1016/j.ciresp.2016.12.008
227. Qiu L, Song Z, Setaluri V. Oxidative stress and vitiligo: the Nrf2-ARE signaling connection. J Invest Dermatol (2014) 134(8):2074–6. doi:10.1038/jid.2014.241
228. Kalie E, Razi M, Tooze SA. ULK1 regulates melanin levels in MNT-1 cells independently of mTORC1. PLoS One (2013) 8(9):e75313. doi:10.1371/journal.pone.0075313
229. Jeong TJ, Shin MK, Uhm YK, Kim HJ, Chung JH, Lee MH. Association of UVRAG polymorphisms with susceptibility to non-segmental vitiligo in a Korean sample. Exp Dermatol (2010) 19(8):e323–5. doi:10.1111/j.1600-0625.2009.01039.x
230. Hor JL, Heath WR, Mueller SN. Neutrophils are dispensable in the modulation of T cell immunity against cutaneous HSV-1 infection. Sci Rep (2017) 7:41091. doi:10.1038/srep41091
231. Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science (2012) 337(6098):1115–9. doi:10.1126/science.1225152
232. Levy J, Romagnolo B. Autophagy, microbiota and intestinal oncogenesis. Oncotarget (2015) 6(33):34067–8. doi:10.18632/oncotarget.5966
233. Wang C, Yu Z, Shi X, Tang X, Wang Y, Wang X, et al. Triclosan enhances the clearing of pathogenic intracellular salmonella or Candida albicans but disturbs the intestinal microbiota through mTOR-independent autophagy. Front Cell Infect Microbiol (2018) 8:49. doi:10.3389/fcimb.2018.00049
Keywords: autophagy, skin autoimmunity, selective autophagy, skin diseases, skin cancers
Citation: Sil P, Wong S-W and Martinez J (2018) More Than Skin Deep: Autophagy Is Vital for Skin Barrier Function. Front. Immunol. 9:1376. doi: 10.3389/fimmu.2018.01376
Received: 02 April 2018; Accepted: 04 June 2018;
Published: 25 June 2018
Edited by:
Herman Waldmann, University of Oxford, United KingdomReviewed by:
Andrzej T. Slominski, University of Alabama at Birmingham, United StatesJames Harris, Monash University, Australia
Copyright: © 2018 Sil, Wong and Martinez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Payel Sil, cGF5ZWwuc2lsQG5paC5nb3Y=
 Payel Sil
Payel Sil Sing-Wai Wong
Sing-Wai Wong Jennifer Martinez
Jennifer Martinez