- 1Department of Health Sciences, University of Catanzaro “Magna Graecia”, Catanzaro, Italy
- 2Colorectal Unit, Hospital Universitario y Politécnico La Fe, Valencia, Spain
- 3Department of Medical, Surgical, Neurological, Metabolic and Ageing Sciences, Università della Campania “Luigi Vanvitelli”, Naples, Italy
- 4Rheumatology Institute of Lucania (IReL) – Rheumatology Department of Lucania, “San Carlo” Hospital of Potenza and “Madonna delle Grazie” Hospital of Matera, Potenza, Italy
- 5Basilicata Ricerca Biomedica (BRB) Foundation, Potenza, Italy
- 6Department of Biomedicine and Prevention, University Tor Vergata, Rome, Italy
- 7Department of Nuclear Medicine, IRCCS Neuromed, Pozzilli, Italy
- 8Department of Medical Sciences, Clinica Medica Unit, University of Ferrara, Ferrara, Italy
Metformin (dimethyl biguanide) is a synthetic derivative of guanidine, isolated from the extracts of Galega officinalis, a plant with a prominent antidiabetic effect. Since its discovery more than 50 years ago, metformin represents a worldwide milestone in treatment of patients with type 2 diabetes (T2D). Recent evidence in humans indicates novel pleiotropic actions of metformin which span from its consolidated role in T2D management up to various regulatory properties, including cardio- and nephro-protection, as well as antiproliferative, antifibrotic, and antioxidant effects. These findings, together with ground-breaking studies demonstrating its ability to prolong healthspan and lifespan in mice, provided the basis for defining metformin as a potential antiaging molecule. Moreover, emerging in vivo and in vitro evidence support the novel hypothesis that metformin can exhibit immune-modulatory features. Studies suggest that metformin interferes with key immunopathological mechanisms involved in systemic autoimmune diseases, such as the T helper 17/regulatory T cell balance, germinal centers formation, autoantibodies production, macrophage polarization, cytokine synthesis, neutrophil extracellular traps release, and bone or extracellular matrix remodeling. These effects may represent a powerful contributor to antiaging and anticancer properties exerted by metformin and, from another standpoint, may open the way to assess whether metformin can be a candidate molecule for clinical trials involving patients with immune-mediated diseases. In this article, we will review the available preclinical and clinical evidence regarding the effect of metformin on individual cells of the immune system, with emphasis on immunological mechanisms related to the development and maintenance of autoimmunity and its potential relevance in treatment of autoimmune diseases.
Introduction
Since its discovery more than 50 years ago, metformin represents a worldwide milestone in treatment of patients with type 2 diabetes (T2D). Metformin (dimethyl biguanide) is a synthetic derivative of guanidine, isolated from the extracts of Galega officinalis, a plant with a prominent antidiabetic effect (1). Indeed, metformin lowers both fasting and post-prandial glucose levels by inhibiting hepatic glucose production, reducing intestinal glucose absorption, and improving glucose uptake and utilization by peripheral tissues. However, the exact pharmacodynamic properties have been elusive for many years still remaining a matter of debate (2). Recent evidence in humans indicates novel pleiotropic actions of metformin which span from its consolidated role in T2D management up to various regulatory properties, including cardio- and nephro-protection, as well as antiproliferative, antifibrotic, and antioxidant effects (3). These findings, together with ground-breaking studies demonstrating its ability to prolong healthspan and lifespan in mice (4), provided the basis for defining metformin as a potential antiaging molecule (5). This fascinating hypothesis is currently being tested through the ongoing Targeting Aging with Metformin study, a placebo-controlled trial (5) aimed at investigating the potential role of metformin in delaying the onset of aging-associated diseases.
Notably, emerging in vivo and in vitro evidences suggest that metformin can exhibit immune-modulatory features (6, 7). These effects may represent a powerful contributor to antiaging and anticancer properties exerted by metformin.
In addition to recent evaluation of the metabolic and immunological roles of metformin (7), this article will appraise current preclinical and clinical data regarding the effect of metformin on individual cells of the immune system. Furthermore, the immunological mechanisms exerted by metformin will be explored as well as its potential relevance in the treatment of autoimmune diseases.
Pharmacokinetics and Mechanism of Action of Metformin
Pharmacokinetics
Upon oral administration in humans (typical therapeutic doses range from 1,000 to 3,000 mg/day), approximately 70% of metformin is absorbed from the small intestine (8), with the remaining component passing into the colon before being excreted in feces (9). The intestinal absorption is primarily mediated by the plasma membrane monoamine transporter, which is expressed on the luminal side of enterocytes; however, the organic cation transporters 1 and 3 may also contribute (10). Following absorption, metformin is mainly distributed in the intestine, liver, and kidneys and finally excreted unchanged in urine by means of active tubular secretion, with half-life of ~5 h (8).
Mechanism of Action
Consistent data show that mitochondria are the main subcellular targets of metformin. Indeed, the drug accumulates selectively in mitochondria reaching up to 1,000-fold higher concentrations than those observed in the extracellular medium (11). Recent studies demonstrate that metformin transiently inhibits NADH:ubiquinone oxidoreductase (also referred to as “Complex I” of the mitochondrial electron transport chain), an entry enzyme of oxidative phosphorylation located in the inner mitochondrial membrane. This inhibitory effect evokes a drop in the energetic state of the cell, thereby leading to a reduced ATP production and increased AMP:ATP ratio (2). The resultant metabolic shift leads to the activation of the energy sensor 5′-AMP-activated protein kinase (AMPK). Upon activation, AMPK coordinates different signaling networks in the attempt to restore a physiologic energy balance by switching on ATP-generating catabolic pathways and, at the same time, shutting down ATP-consuming anabolic mechanisms. The most of the well-characterized glucose-lowering effects of metformin relies on AMPK activation, although AMPK-independent mechanisms have been recently disclosed (2).
Downstream consequences of AMPK activation may account for many of the effects of metformin on immune homeostasis. Indeed, immunological functions are associated with specific metabolic pathways and targeting immunocytes. Upon activation, these cells, even those with prominent anti-inflammatory properties, undergo metabolic reprogramming that is essential for disease development and maintenance (12, 13). Pro-inflammatory cell subset, i.e., neutrophils, M1 macrophages, and effector T cells preferentially produce ATP through glycolysis, whereas cells with an anti-inflammatory lineage, i.e., memory and regulatory T cells (Tregs) and M2 macrophages, favor mitochondrial ATP generation (13). In this context, AMPK activation promotes the oxidation of substrates in mitochondria, thereby limiting the glycolytic capacity of cells (14).
The mammalian target of rapamycin (mTOR) is a downstream target of AMPK involved in regulating energy homeostasis by modulating cellular processes, such as protein synthesis and autophagy implicated in cell proliferation and tumorigenesis in cancer (15). With regard to immune function, mTOR is crucial for fine tuning T cells activation and differentiation (16) through its interaction with the signal transducer and activator of transcription (STAT) pathway (17). Metformin has been demonstrated to downregulate mTOR signaling through either AMPK-dependent (18) or -independent mechanisms (19, 20). Moreover, metformin can regulate other pathways relevant to autoimmunity, including the nuclear factor kappaB (NF-kB) (21) and mitogen-activated protein kinase (MAPK)/c-Jun NH2-terminal kinase (JNK) (22).
Effects of Metformin on Immune Cells Involved in Autoimmune Diseases
Several studies investigated the effect of metformin on different cells involved in the induction and maintenance of autoimmunity and inflammation. Herein, we will review the available evidence regarding the effect of metformin on main cell types and related pathways (summarized in Figure 1).
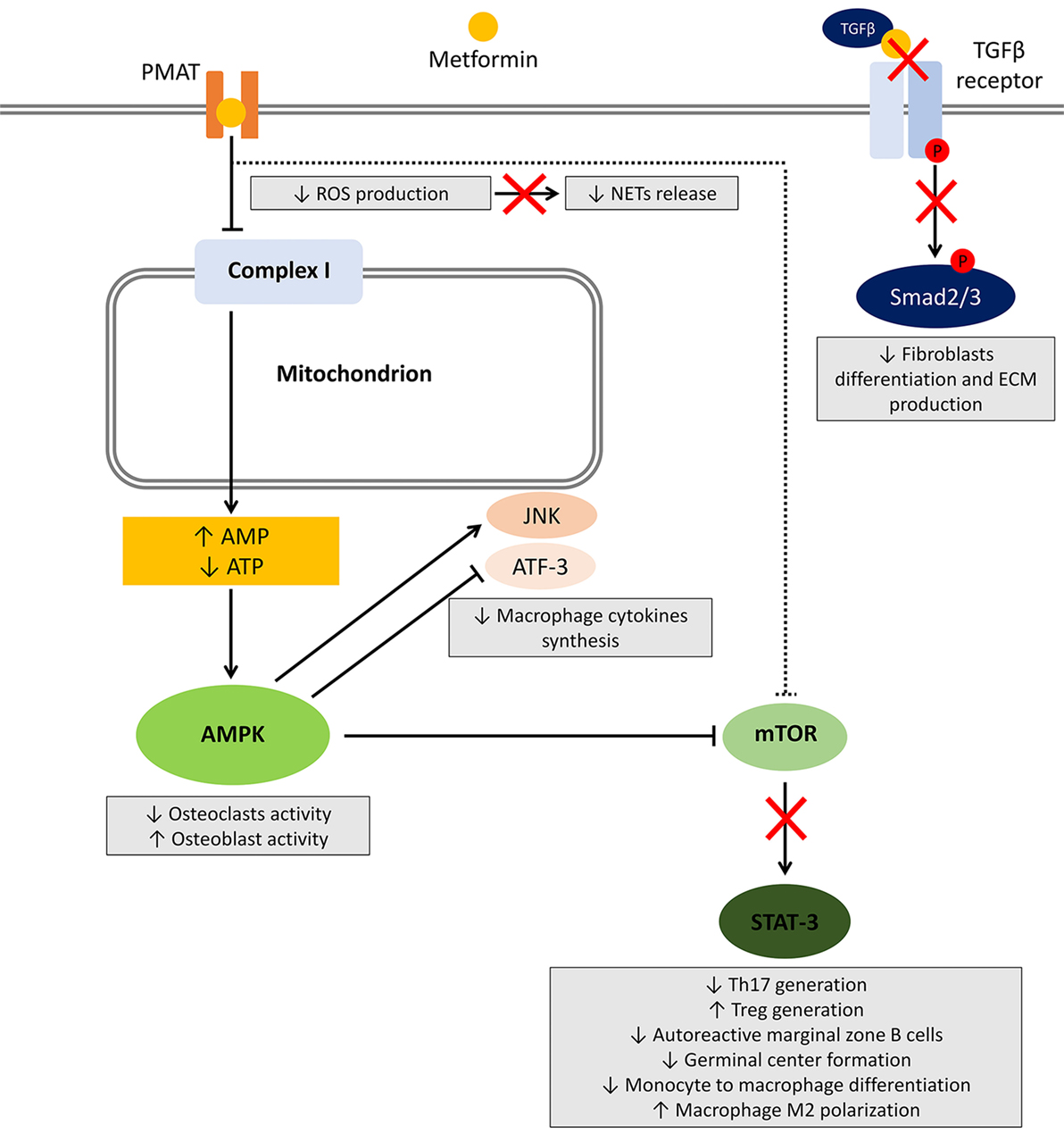
Figure 1. Effects of metformin on immune cells. After entering the cell, metformin transiently inhibits NADH:ubiquinone oxidoreductase (complex I) located in the inner mitochondrial membrane leading to a reduced ATP production and increased AMP:ATP ratio. The resultant metabolic shift stimulates the activation of the energy sensor 5'-AMP-activated protein kinase (AMPK). Among several targets, AMPK activation inhibits mammalian target of rapamycin (mTOR), responsible for most of the effects on the immune system. Other potential mechanisms include AMPK-independent inhibition of mTOR, reduced reactive oxygen species (ROS) production, and block of the TGF-β receptor/Smad interplay. The effects on immune cells are summarized in gray boxes.
Effects of Metformin on T Cells
T cells are key master players in the delicate equilibrium leading to immune tolerance breakdown and autoimmunity (23). The traditional dichotomy between Th1- and Th2-mediated diseases has been overwhelmed in the last years by the identification of novel T cell subsets, including T helper 17 (Th17) and Treg populations. Th17 cells are characterized by the production of the pro-inflammatory cytokine IL-17 and the expression of the transcriptional factor retinoic-acid-receptor-related orphan nuclear receptor gamma (RORγt) and exert pro-inflammatory effects relevant to the pathophysiology of several autoimmune diseases (24). Conversely, Tregs are characterized by the expression of the transcription factor forkhead box P3 and act as negative regulators of immune-mediated inflammation (25). Globally, their balance represents the subtle edge on which autoimmunity develops and maintains chronically (25, 26). T cell metabolic derangement has been largely demonstrated in autoimmune diseases such as systemic lupus erythematosus (SLE), with a shift toward oxidation, mitochondrial abnormalities, activation of mTORC1, and increased glucose flux (27).
In this context, the mTOR pathway—a key downstream molecule of AMPK signaling—and its crosstalk with STAT-mediated signaling, plays a crucial role in regulating of T cell subset differentiation (17, 28). mTOR-deficient T cells fail to differentiate into Th1, Th2, or Th17 effector cells, while maintaining their ability to differentiate into Tregs (29). Specularly, the addition of rapamycin (mTOR inhibitor) to CD25-cell cultures selectively induces the Treg phenotype and function (30). Finally, treatment with mTOR-targeting agents, such as sirolimus (31) or N-acetylcysteine (32), improves disease activity in patients with SLE.
Nath et al. (33) evaluated the effect of oral metformin on experimental autoimmune encephalomyelitis (EAE), a T-cell-mediated mouse model of multiple sclerosis (MS). In this study, treatment with metformin resulted in a slower disease progression, reduced infiltration of inflammatory cells in the central nervous system and expression of inflammatory cytokines IFN-γ, TNF-α, IL-17, IL-1β, and IL-6. Notably, T cells isolated from mice treated with metformin showed reduced expression of IFN-γ and IL-17 along with the two transcription factors T-box transcription factor and RORγt suggestive of Th1 and Th17 differentiation, respectively. A similar effect was obtained when metformin was associated with lovastatin (34). Complementary with the reduction of Th17, metformin has been also demonstrated to increase Tregs in EAE through the inhibition of mTOR pathway (35).
Similarly, Kang et al. evaluated the effect of metformin on collagen antibody-induced arthritis (CAIA), a well-established animal model of rheumatoid arthritis (RA) (36). In CAIA mice, metformin treatment resulted in a significant improvement of arthritis score, with reduced bone destruction, inflammatory cytokines production, and RORγt-expressing T cells associated with the AMPK/mTOR-mediated inhibition of STAT3 signaling.
The beneficial AMPK/mTOR/STAT3-dependent effect of metformin on Th17-mediated inflammation was further confirmed in collagen-induced arthritis (CIA) (37–39), dextran sulfate sodium-induced colitis (reminiscent of human inflammatory bowel disease, IBD) (40), Roquinsan/san model of SLE (41), and acute graft-versus-host disease (42).
In addition to the AMPK/mTOR/STAT3 pathway, other T cell pathogenic mechanisms may be targeted by metformin. Several other metabolic pathways regulate the function of T cells in health and diseased conditions (43, 44). Mitochondrial oxidative metabolism has been shown to be the main ATP synthesis pathway in experimental SLE (27, 45). Using the triple congenic B6.Sle1.Sle2.Sle3 lupus-prone mouse model, Yin et al. (46) demonstrated that metformin can restore T cell metabolism and reduce IFN-γ production. Moreover, the simultaneous treatment of mice with metformin and 2-deoxy-d-glucose—a glycolysis inhibitor—significantly reduced anti-nuclear antibodies and anti-double-stranded DNA (dsDNA) antibodies production and improved the severity of associated nephritis. Similar findings were obtained from the same group in B6.lpr mice, another model of spontaneous SLE (47).
Taken together, data from animal models of T cell-mediated autoimmunity strongly support an immune-modulatory effect of metformin with the ability to restore the balance between pathogenic and tolerogenic T cell populations by acting on the AMPK/mTOR/STAT3 and on the normalization of T cell mitochondrial metabolism.
Despite the promising results obtained with experimental models, data in humans are still scarce. Yin et al. (46) demonstrated that CD4+ T cells from SLE patients exhibit elevated cellular metabolism when compared with healthy controls, a feature that correlates with T cell activation and subset distribution. Moreover, their excessive IFNγ production was significantly reduced by adding metformin in vitro. Furthermore, immune function is known to be impaired in T2D, a feature that explains an increased susceptibility to infection and autoimmune diseases (48). Dworacki et al. (49, 50) showed that T2D is characterized by impaired thymic output, as documented by reduced number of circulating recent thymic emigrants (RTEs) and CD127+ CD132+ naïve T cells, mainly attributable to the conversion of RTE in terminally differentiated memory cells. Among diabetic patients, however, those treated with metformin had the highest thymic output of a similar magnitude to that observed in nondiabetic subjects (50). IL-17 and IFN-γ levels were higher in patients with poor glucoregulation at baseline (defined as a glycated hemoglobin >7%); nonetheless, a 12-week treatment with metformin could reduce IL-17 in diabetic patients. Finally, in patients with polycystic ovary syndrome (PCOS), the administration of metformin combined with drospirenone/ethinylestradiol has been shown to reduce the frequency of CD4+CD28null T cells (51), a subset of CD4+ Th1 cells lacking the co-stimulatory receptor CD28 thought to be involved in autoimmune diseases such as RA (52).
Only limited direct evidence of the potential therapeutic effect of metformin in patients with autoimmune diseases are available to date. In a cohort study by Negrotto et al. (53), MS patients with comorbid metabolic syndrome were treated with metformin (850–1,500 mg/day) or pioglitazone resulting in a significant decrease in enlarging T2-weighted and gadolinium-enhanced lesions (as a measure of disease burden) on brain magnetic resonance imaging compared with matched controls. This result was already evident after 6 months of therapy and maintained up to 24 months. Moreover, in peripheral blood mononuclear cell isolated from patients treated with metformin, the authors observed an enhanced AMPK expression, a reduced prevalence of IFN-γ- and IL-17-producing cells, and an increase of Treg cell percentage. In a proof-of-concept trial by Wang et al. (54), 113 SLE patients were randomly assigned to receive add-on (500–1,500 mg/day) metformin or conventional treatment alone. Metformin treatment resulted in a 51% reduction of risk of disease flares and significantly less corticosteroid exposure.
Effects of Metformin on B Cells
In the network of mutual interactions between immune cells, the interplay between T and B cells represents a crucial component for the development of adaptive immune responses (55). Despite many autoimmune conditions have been considered T cell-driven diseases for a long time, several B cell-dependent mechanisms are emerging in recent years extending beyond their traditional role as autoantibody-producing cells (56, 57).
The stronger support for a potential role of metformin on B cells biology arises from studies investigating its effect on hematological malignancies. Several lines of evidence indicate that aberrant activation of the mTOR pathway is common in both Hodgkin lymphomas and many types of B-cell non-Hodgkin lymphomas (58), thereby contributing to tumorigenesis and cell autophagy during response to anticancer agents. Notably, defects in autophagy mechanisms has recently been proposed to play a role in lymphoma progression, thus implying that autophagy represents a promising target for novel lymphoma therapeutics (59). Notably, mTOR-driven autophagy is also an important mechanism in autoimmune diseases (60, 61).
Shi et al. (62) reported the antitumor action of metformin-mediated AMPK activation in lymphoma. Metformin treatment induced a dose-dependent suppression of lymphoma cells proliferation through negative control of the mTOR pathway. Importantly, lymphoma cells sensitivity to anticancer agents was enhanced by concomitant treatment of metformin via induction of autophagy. Furthermore, metformin use has been associated with an improved response rate and progression-free survival in diabetic diffuse large B-cell lymphoma patients (63).
Recent studies suggested that mTOR activity plays a critical role in B cells during autoimmune diseases. Conditional deletion of the mTOR genes in B cells markedly impairs B cell proliferation and germinal center (GC) differentiation (64). Using the Roquesan/san model of SLE, Lee et al. (41) demonstrated that oral metformin can attenuate signs of autoimmunity including anti-dsDNA antibodies production and kidney and liver inflammation. This clinical improvement was accompanied by a significant reduction in autoreactive marginal zone B cells and reduced GC formation, the main site of differentiation of B cells in long-lived autoreactive plasma cells. At the molecular level, this was characterized by an increased activation of AMPK with subsequent decrease in phosphorylation of mTOR and STAT3 (41). Conversely, protective antibody response may be even boosted by metformin. Diaz et al. (65) evaluated the antibody response to influenza vaccination in T2D patients’ naïve and on metformin treatment, respectively. The drug enhanced vaccine-specific antibody titers in vivo and in vitro, and this effect was accompanied by an increase in switched memory B cell and a decrease in late/exhausted B cells, known to impair antibody responses. These mechanisms were associated with an increased AMPK phosphorylation and reduced intrinsic B cell inflammation upon exposure to metformin, as demonstrated by a reduction in TNF-α, miR-155, and miR-16 expression, known to affect the ability of B cells to respond to antigenic stimulation (41).
Among several B-cell subpopulations so far identified (66), the innate-like B-1a cells represent a distinct subpopulation that can significantly contribute to generate circulating IgM natural antibodies, mucosal immunity, and immunoregulation (67, 68). Furthermore, two distinct phenotypes have been identified in B-1a cells. These are distinguished by the relative expression of the plasma cell alloantigen 1 (PC1, also known as ENPP1—ectonucleotide pyrophosphatase/phosphodiesterase 1) and are named PC-1low and PC-1high, with the latter subsets exhibiting a more pronounced regulatory function (69, 70). The same glycoprotein PC-1 has been demonstrated to have a role in insulin resistance associated with T2D (71). Stefanovic et al. (72) demonstrated an increased PC-1 expression in T2D lymphocytes, which was reverted by a 3-month metformin treatment and combined with a significant improvement of insulin sensitivity. Several lines of evidence support a potential role of B1a cells in the development of autoimmunity (73); thus, the modulation of PC-1 by metformin may represent an additional immune-regulatory mechanism deserving further studies.
Effects of Metformin on Monocytes/Macrophages
Macrophages represent the main tissue-resident immune cells in many organs providing a quick first-line response against pathogens. Upon activation, macrophages can polarize in two major phenotypes, i.e., the pro-inflammatory “classically” activated (M1) and the “alternatively” activated (M2, further subclassified as M2a, b, or c), mainly associated with resolution of inflammation and tissue repair processes (74, 75). Inflammatory infiltrates in autoimmune diseases are characterized by a predominance of a distinct macrophage lineage: active RA synovial tissue shows abundance of M1 macrophages, whereas in spondyloarthritis patients, the M2 phenotype predominates (76). Consistently with their respective functions, the cytokine portfolio is significantly diversified, with M1 macrophages producing mainly pro-inflammatory cytokines relevant to the pathogenesis of autoimmune diseases (i.e., TNF-α, IL-1, IL-6, IL-12, IL-23, and MCP-1), as opposed to M2 macrophages releasing cytokines with anti-inflammatory properties, e.g., IL-10 and TGF-β (77).
In vitro, metformin demonstrated anti-inflammatory properties on macrophages via AMP-dependent and -independent mechanisms. In THP-1 acute monocytic leukemia cell line, metformin treatment abrogates phorbol 12-myristate 13-acetate-induced monocyte-to-macrophage differentiation and IL-1β, TNF-α, and MCP-1 production (78). This effect is primarily mediated by AMPK activation resulting in a decrease in JNK1 phosphorylation (influencing inflammatory cytokine synthesis and release) and the negative regulating STAT3 phosphorylation (modulating the effects on differentiation). In primary murine peritoneal macrophages, metformin treatment dose-dependently suppresses lipopolysaccharide (LPS)-induced TNF-α and IL-6 expression. This effect was attributable, at least in part, to the AMPK-dependent ATF-3 induction via competition with NF-κB for binding to TNF-α and IL-6 promoters (79). Similarly, Kelly et al. (80) demonstrated that metformin dose-dependently inhibits LPS-induced pro-IL-1β while boosting IL-10 expression in murine bone marrow-derived macrophages. In this case, the effect on pro-IL1β was independent of AMPK activation and associated with reduced reactive oxygen species (ROS) production as a direct consequence of metformin-induced mitochondrial complex I suppression. In high-fat diet (HFD) mice, a classical model of obesity-induced T2D and chronic inflammation, treatment with metformin resulted in reduced serum levels of IL-6 and TNF-α and in the AMPK-mediated modulation of macrophage polarization with a shift toward an anti-inflammatory M2 phenotype (81).
Similarly, in human macrophages, metformin was able to suppress the LPS-induced expression of TNF-α and MCP-1 and ROS production in an AMPK-dependent manner. This effect was associated with a reduced NF-kB and MAPK activity (82). In impaired glucose tolerance patients treated with fenofibrate, addition of metformin for 12 weeks was able to reduce LPS-induced production of TNF-α and IL-6 (83) by peripheral blood monocytes. Likewise, in impaired fasting glucose patients treated with simvastatin, addition of metformin was able to reduce LPS-induced TNF-α, IL-1β, IL-6, IL-8, and MCP-1 (84). Using cultures of human adipose tissue, Bruun et al. (85) demonstrated that metformin reduces the release of MCP-1 by resident macrophages.
Macrophage migration inhibitory factor (MIF) is a pleiotropic cytokine, acting as potent M1-polarizing factor (86, 87). Evidence for a role for MIF in autoimmunity has been provided by studies showing that MIF is expressed at increasing levels in different experimental models of disease. Immunoneutralization or genetic deletion of MIF confers protection from pathologic progression (88–91). In addition, both the circulating levels and the tissue expression of MIF are elevated in patients with autoimmune inflammatory disorders, and high-expression MIF alleles have been associated with more severe end-organ damage in RA (92, 93), SLE (94), and scleroderma (95). As for the macrophage polarization, synovial fluids in RA patients contain high levels of M1 macrophage-derived mediators, along with low levels of M2 macrophage-derived mediators (96). Adoptive transplantation of M2, but not M1, macrophages significantly reduced SLE severity in lymphocyte-derived DNA (ALD-DNA) induced lupus mice (97). Dandona et al. (98) demonstrated that plasma MIF concentrations and MIF mRNA expression in the mononuclear cells are elevated in obese patients, and oral metformin treatment for 6 weeks suppresses MIF levels. Further studies are eagerly awaited to further define the role of MIF in autoimmune diseases.
Effects of Metformin on Neutrophils
Neutrophils are the most numerous circulating leukocytes in humans, providing a powerful first-line defense against bacterial and fungal pathogens. Despite these cells densely infiltrate various tissues in autoimmune diseases, their exact role was puzzling until the last years (99), when a number of studies demonstrated the involvement of neutrophils in different phases of autoimmune diseases pathogenesis (100).
From a clinical standpoint, the neutrophil count and neutrophil-to-lymphocyte ratio (NLR, normal range: 0.78–3.53) emerged in the last years as an accessible, inexpensive measure of systemic inflammation (101). NLR has been extensively studied as a prognostic marker in cancer patients (102); moreover, it correlates with disease activity in SLE (103), RA (104), MS (105), and IBD (106). In a large cohort of diabetic patients, Cameron et al. (107) demonstrated that treatment with metformin, but not with sulfonylureas, significantly reduces NLR after 8–16 months. Also, metformin has been shown to reduce neutrophil count in young women with PCOS (108) and in girls born small for gestational age with exaggerated adrenarche and precocious pubarche (108), two conditions characterized by a pronounced systemic inflammatory state. Finally, in diabetic patients undergoing endarterectomy for carotid artery stenosis, Eilenberg et al. (109) demonstrated that metformin treatment was associated with reduced expression of neutrophil gelatinase-associated lipocalin—a protein released by activated neutrophils and associated with atherosclerotic plaques vulnerability—thus contributing to lower cerebral embolization events.
More recently, a novel, exciting feature of neutrophils has been disclosed, namely, the production of neutrophil extracellular traps (NETs) (110). NETs are DNA structures released upon chromatin decondensation and spreading in the extracellular space. Several proteins adhere to the DNA scaffold, including histones and components of primary and secondary granules, such as elastase, myeloperoxidase, cathepsin G, lactoferrin, pentraxin 3, gelatinase, proteinase 3, LL37, and peptidoglycan-binding proteins. Exaggerated NETosis is increasingly being recognized as a contributing mechanism in induction and maintenance of autoimmunity and a major source of autoantibodies generation in SLE (111) and RA (112). Furthermore, Wang et al. demonstrated that mitochondrial DNA, in addition to nuclear DNA, can be found in NETs obtained by stimulating neutrophils from SLE patients and anti-mitochondrial DNA antibody response associates with disease activity even better than anti-dsDNA. Moreover, treatment with metformin in SLE patients resulted in a reduction of disease flares and corticosteroid use (54). NETosis can be experimentally induced in vitro following exposure of human peripheral blood white cells to high glucose concentration (113); similarly, exaggerated NETosis has been clearly demonstrated in patients with T2D (113, 114), and this process can be restored by treatment with metformin (115). Finally, in patients with pre-diabetes, metformin treatment reduces the concentration of NET components independently from glycemic control (116).
Effects of Metformin on Other Cells Involved in Autoimmune Diseases
Effects of Metformin on Fibroblasts
Fibroblasts actively participate in the pathophysiology of several autoimmune conditions (117), but their role is emphasized in systemic sclerosis (SSc)—the prototypical systemic fibrosing disease—in which activated myofibroblasts drive uncontrolled extracellular matrix accumulation in the skin and internal organs. At a subcellular level, TGF-β is the key cytokine regulating fibroblasts aggressive behavior in SSc (118). TGF-β signals through cell-surface serine/threonine kinase receptors to the intracellular Smad proteins, which in turn accumulate in the nucleus to regulate gene expression (119).
Extensive preclinical data suggest a potent antifibrotic effect of metformin. In cardiac fibroblasts, metformin treatment has been shown to inhibit fibrosis and collagen synthesis via the TGF-β/Smad3 signaling pathway (120) and to impair the differentiation into myofibroblasts (121). A similar effect was observed in other cellular models, including nasal polyp-derived fibroblasts (122), renal fibroblasts (123), hepatic stellate cells (124), and in fibroblasts harvested from capsular ligaments of ankylosing spondylitis patients (125). In lungs, metformin attenuates gefitinib-induced exacerbation of pulmonary fibrosis by inhibiting the TGF-β/SMAD2/3 signaling pathway (126). Finally, our group demonstrated that metformin can ameliorate skin fibrosis in the bleomycin-induced murine model of scleroderma (127), as indicated by reduced dermal thickness, collagen accumulation, and number of lesional myofibroblasts. The mechanisms leading to metformin-mediated downregulation of the TGF-β axis have been elusive until recently, when an elegant study demonstrated that metformin is able to directly bind to TGF-β, thereby preventing the interaction with its receptor (128).
Effects of Metformin on Osteoblasts/Osteoclasts
Under physiological conditions, bone homeostasis is maintained through a harmonic balance between bone formation and resorption, orchestrated by osteoblasts and osteoclasts, respectively (129). During RA and other autoimmune diseases, pro-inflammatory cytokines such as IL-1 and TNF-α alter this subtle equilibrium in favor of bone-resorptive mechanisms, ultimately leading to local (bony erosions) (130) and systemic (osteoporosis) consequences (129). AMPK plays an important role in regulating bone turnover by suppressing osteoclasts (131) while concurrently stimulating osteoblasts (132). Consequently, metformin exhibited protective effects in animal models of osteoporosis (133, 134). Furthermore, Son et al. (37) demonstrated that improvement in bone erosion and cartilage destruction in CIA mice following exposure to metformin is associated with reduced osteoclastogenesis via an AMPK/mTOR/STAT3-dependent mechanism. In humans, it is well accepted that T2D is associated with an increased risk of osteoporosis (135). While some antidiabetic medications, such as thiazolidinediones, demonstrated clear pro-osteoporotic effects (136), metformin treatment has been associated with reduced risk of fractures in a large, case–control study (137). So far, there are no data on the effect of metformin on bone homeostasis in patients with rheumatic disease.
Metformin and Gut Microbiota
Growing evidence indicates that the gut microbiota—i.e., the approximately 100 trillion germs located into the gastrointestinal lumen, especially in the distal segments—plays a role in human body homeostasis and health state (138). Alterations in the gut microbiota composition, a condition referred to as dysbiosis, has been associated with the development of several autoimmune diseases, including RA (139), SLE (140), MS (139), and Behcet’s disease (141).
Several factors and agents have been shown to interfere or even modulate the microbiota; however, little is known about the effects exerted by metformin on human gut and vice versa. Cabreiro et al. (142) investigated the impact of metformin on life cycle of the nematode Caenorhabditis elegans. Following on previous studies showing that metformin extended the lifespan of C. elegans (143), the authors aimed at investigating the mechanisms underlying these effects and demonstrated that metformin slowed aging of C. elegans rather than reducing the risk of death. Furthermore, using Escherichia coli co-cultures, they found that metformin inhibits bacterial folate and methionine metabolism, both mechanisms thought to contribute significantly the therapeutic efficacy of metformin. The conclusions support the C. elegans/E. coli theory of evolution (144), which implies that the animal or plant, with all associated microorganisms, are considered a “unit” of selection during evolution (“the holobiont”). Under this light, studies aiming at understating the mechanisms mediating the effects of metformin need to be conducted on adequately designed models and will require accurate assessment of the gut microbiota.
Further evidence of an axis connecting the gut microorganisms with metformin mechanism of action is provided by an experimental study in which normal diet or HFD mice were treated with metformin for 6 weeks. Compared to untreated (control) HFD mice, those receiving metformin improved the glycemic profile, and this effect was associated with higher abundance of Akkermansia muciniphila, a mucin-degrading bacterium. Furthermore, oral administration of A. muciniphila reduced visceral adipose tissue inflammation and enhanced glucose tolerance in HFD mice (145). Similar findings were confirmed at some extent by Lee and Ko (146) who showed that the abundance of A. muciniphila (12.44 ± 5.26%) and Clostridium cocleatum (0.10 ± 0.09%) significantly increased after metformin treatment in HFD mice. The same authors also addressed the role of Akkermansia and other microbiota genera in obese, aged mice treated with metformin (147). HFD mice treated with metformin had significantly increased abundance of Akkermansia, Bacteroides, Butyricimonas, and Parabacteroides, and this correlated with a reduced expression of inflammatory cytokines (IL-1β and IL-6) in the adipose tissue. It has been suggested that metformin treatment could restore glucose sensing by regulating the expression of small intestinal sodium glucose cotransporter-1 in rats, which is reduced by HFD. Upper small intestine treatment with metformin has been shown to change the microbiota by increasing, at least in part, the abundance of Lactobacillus (148). Finally, Wu et al. assessed metformin–microbiota interactions in a gut simulator and confirmed that metformin alters the biological functions of different microbial phyla, including the regulation of genes encoding for metalloproteins or metal transporters (149).
Several clinical studies investigating the relationship between gut microbiota and metformin treatment have been performed in subjects with T2D. Forslund et al. (150) analyzed 784 human metagenomes and observed a shift in the microbiota during metformin treatment with a depletion of butyrate-producing taxa. They proposed the potential role of a microbial influence on the effects of metformin, which could be related with short-chain fatty acids production (151). In keeping with this finding and with those observed with A. muciniphila in experimental models (145–147), a study recruiting 28 patients with T2D (14 on metformin) and 84 matched controls confirmed an association between glucose tolerance and metformin-modulated gut microbiota. In details, in addition to A. muciniphila, other gut microbes were more abundant in individuals receiving metformin, namely Butyrivibrio, Bifidobacterium bifidum, and Megasphaera, which are known to produce short-chain fatty acids. Conversely, Clostridiaceae 02d06 were more abundant in patients with T2D not taking metformin (150).
In an exploratory, unblinded study (NCT01357876), Napolitano et al. recruited 12 T2D patients who were receiving a stable dose of metformin (≥1,000 mg/day) for more than 3 months. They collected post-prandial blood samples, stool samples, upper small intestine bile, and fasting plasma samples for metformin concentrations at scheduled intervals. They observed that abundance of the Firmicutes in the microbiota was positively correlated with changes in cholic acid and conjugates, while Bacteroidetes abundance was negatively correlated with those of bile acids. Both were also correlated with levels of serum peptide tyrosine–tyrosine, further highlighting a complex gut-based pharmacology underlying the mechanism of action of metformin (152).
Indeed, modulating the gut microbiota by increasing favorable phyla, such as Akkermansia spp., might enhance the antidiabetic effects exerted by metformin. On the other side, microbiota modifications induced by metformin may affect immune function. In NOD mice, A. muciniphila has been demonstrated to protect from the development of islet cell autoimmunity (153) and restore intestinal immunity and homeostasis in experimental models of IBD (154, 155).
The relationship between metformin and microbiota is likely bi-directional, with a pronounced microbial effect on the mechanisms of action and efficacy of the drug, and with metformin affecting functions and abundance of specific phyla. Further data on this complex drug–microbiota interplay may extend our knowledge on metformin and identify novel targets for tailored treatments based on gut microbiota manipulation exploitable in autoimmune diseases.
Conclusion
Metformin is a safe, inexpensive medication with a history of more than 50 years of clinical experience in treating patients with T2D. In the last decade, several preclinical and clinical studies highlighted pleiotropic beneficial effects of this molecule on other clinical domains, including cancer susceptibility and cardiovascular disease risk. More recently, different in vitro studies demonstrated that metformin can regulate the function of many cell types involved in autoimmunity development and maintenance. Concurrently, metformin has been shown to be able to restore immune homeostasis and improve disease severity in animal models of autoimmune diseases. Based on this preclinical background and because of a well-established safety profile, metformin should be reconsidered in clinical trials designed to prove its efficacy in patients with autoimmune diseases. Largely available retrospective data (i.e., by analyzing RA patients treated with metformin for comorbid T2D) may contribute to provide further support to longitudinal studies. We are now going to test this hypothesis in humans through a double-blind placebo-controlled clinical trial (Metformin Treatment in Systemic Sclerosis, METSS—EudraCT number: 2018-000733-12) that recently received funding from Italian Ministry of Health. Time will tell us whether a well known and relatively safe drug, will be a new “bullet” available to physicians dealing with patients with autoimmune/rheumatological conditions.
Author Contributions
FU, ER, and RD conceived the review idea. FU, ER, GP, SD, AC, GD, RM, and RD contributed to prepare the first draft of the manuscript. GD, RM, and RD critically revised the final draft. All the authors discussed the results and contributed to the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Italian Ministry of Health for granting funding to the project GR-2016-02361286 aimed at investigating the efficacy of metformin in systemic sclerosis.
Funding
This work was supported by Fondi Ateneo Ricerca (FAR) 2014—Prof. R. Manfredini.
References
1. Bailey CJ, Day C. Metformin: its botanical background. Pract Diabetes Int (2004) 21(3):115–7. doi:10.1002/pdi.606
2. Rena G, Hardie DG, Pearson ER. The mechanisms of action of metformin. Diabetologia (2017) 60(9):1577–85. doi:10.1007/s00125-017-4342-z
3. Anabtawi A, Miles JM. Metformin: nonglycemic effects and potential novel indications. Endocr Pract (2016) 22(8):999–1007. doi:10.4158/EP151145.RA
4. Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, et al. Metformin improves healthspan and lifespan in mice. Nat Commun (2013) 4:2192. doi:10.1038/ncomms3192
5. Barzilai N, Crandall JP, Kritchevsky SB, Espeland MA. Metformin as a tool to target aging. Cell Metab (2016) 23(6):1060–5. doi:10.1016/j.cmet.2016.05.011
6. Pollak M. The effects of metformin on gut microbiota and the immune system as research frontiers. Diabetologia (2017) 60(9):1662–7. doi:10.1007/s00125-017-4352-x
7. Tomczynska M, Bijak M, Saluk JM. Metformin - The Drug for the Treatment of Autoimmune Diseases; A New Use of a Known Anti‐Diabetic Drug. Curr Top Med Chem (2016) 16(19):2223–30.
8. Zhang L, Dresser MJ, Gray AT, Yost SC, Terashita S, Giacomini KM. Cloning and functional expression of a human liver organic cation transporter. Mol Pharmacol (1997) 51(6):913–21. doi:10.1124/mol.51.6.913
9. Graham GG, Punt J, Arora M, Day RO, Doogue MP, Duong JK, et al. Clinical pharmacokinetics of metformin. Clin Pharmacokinet (2011) 50(2):81–98. doi:10.2165/11534750-000000000-00000
10. Gong L, Goswami S, Giacomini KM, Altman RB, Klein TE. Metformin pathways: pharmacokinetics and pharmacodynamics. Pharmacogenet Genomics (2012) 22(11):820–7. doi:10.1097/FPC.0b013e32834f94cb
11. Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J (2000) 348(Pt 3):607–14. doi:10.1042/0264-6021:3480607
12. Rhoads JP, Major AS, Rathmell JC. Fine tuning of immunometabolism for the treatment of rheumatic diseases. Nat Rev Rheumatol (2017) 13(5):313–20. doi:10.1038/nrrheum.2017.54
13. Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity (2013) 38(4):633–43. doi:10.1016/j.immuni.2013.04.005
14. O’Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature (2013) 493(7432):346–55. doi:10.1038/nature11862
15. Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature Rev Mol Cell Biol (2011) 12(1):21–35. doi:10.1038/nrm3025
16. Liu Y, Zhang DT, Liu XG. mTOR signaling in T cell immunity and autoimmunity. Int Rev Immunol (2015) 34(1):50–66. doi:10.3109/08830185.2014.933957
17. Saleiro D, Platanias LC. Intersection of mTOR and STAT signaling in immunity. Trends Immunol (2015) 36(1):21–9. doi:10.1016/j.it.2014.10.006
18. Dowling RJ, Zakikhani M, Fantus IG, Pollak M, Sonenberg N. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res (2007) 67(22):10804–12. doi:10.1158/0008-5472.CAN-07-2310
19. Kalender A, Selvaraj A, Kim SY, Gulati P, Brule S, Viollet B, et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab (2010) 11(5):390–401. doi:10.1016/j.cmet.2010.03.014
20. Ben Sahra I, Regazzetti C, Robert G, Laurent K, Le Marchand-Brustel Y, Auberger P, et al. Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res (2011) 71(13):4366–72. doi:10.1158/0008-5472.CAN-10-1769
21. Chaudhary SC, Kurundkar D, Elmets CA, Kopelovich L, Athar M. Metformin, an antidiabetic agent reduces growth of cutaneous squamous cell carcinoma by targeting mTOR signaling pathway. Photochem Photobiol (2012) 88(5):1149–56. doi:10.1111/j.1751-1097.2012.01165.x
22. Wu N, Gu C, Gu H, Hu H, Han Y, Li Q. Metformin induces apoptosis of lung cancer cells through activating JNK/p38 MAPK pathway and GADD153. Neoplasma (2011) 58(6):482–90. doi:10.4149/neo_2011_06_482
23. Skapenko A, Leipe J, Lipsky PE, Schulze-Koops H. The role of the T cell in autoimmune inflammation. Arthritis Res Ther (2005) 7(Suppl 2):S4–14. doi:10.1186/ar1509
24. Singh RP, Hasan S, Sharma S, Nagra S, Yamaguchi DT, Wong DT, et al. Th17 cells in inflammation and autoimmunity. Autoimmun Rev (2014) 13(12):1174–81. doi:10.1016/j.autrev.2014.08.019
25. Grant CR, Liberal R, Mieli-Vergani G, Vergani D, Longhi MS. Regulatory T-cells in autoimmune diseases: challenges, controversies and – yet – unanswered questions. Autoimmun Rev (2015) 14(2):105–16. doi:10.1016/j.autrev.2015.05.007
26. Noack M, Miossec P. Th17 and regulatory T cell balance in autoimmune and inflammatory diseases. Autoimmun Rev (2014) 13(6):668–77. doi:10.1016/j.autrev.2013.12.004
27. Morel L. Immunometabolism in systemic lupus erythematosus. Nat Rev Rheumatol (2017) 13(5):280–90. doi:10.1038/nrrheum.2017.43
28. Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol (2011) 12(4):295–303. doi:10.1038/ni.2005
29. Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity (2009) 30(6):832–44. doi:10.1016/j.immuni.2009.04.014
30. Keever-Taylor CA, Browning MB, Johnson BD, Truitt RL, Bredeson CN, Behn B, et al. Rapamycin enriches for CD4(+) CD25(+) CD27(+) Foxp3(+) regulatory T cells in ex vivo-expanded CD25-enriched products from healthy donors and patients with multiple sclerosis. Cytotherapy (2007) 9(2):144–57. doi:10.1080/14653240601145223
31. Lai ZW, Kelly R, Winans T, Marchena I, Shadakshari A, Yu J, et al. Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: a single-arm, open-label, phase 1/2 trial. Lancet (2018) 391(10126):1186–96. doi:10.1016/S0140-6736(18)30485-9
32. Lai ZW, Hanczko R, Bonilla E, Caza TN, Clair B, Bartos A, et al. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: a randomized, double-blind, placebo-controlled trial. Arthritis Rheum (2012) 64(9):2937–46. doi:10.1002/art.34502
33. Nath N, Khan M, Paintlia MK, Singh I, Hoda MN, Giri S. Metformin attenuated the autoimmune disease of the central nervous system in animal models of multiple sclerosis. J Immunol (2009) 182(12):8005–14. doi:10.4049/jimmunol.0803563
34. Paintlia AS, Mohan S, Singh I. Combinatorial effect of metformin and lovastatin impedes T-cell autoimmunity and neurodegeneration in experimental autoimmune encephalomyelitis. J Clin Cell Immunol (2013) 4. doi:10.4172/2155-9899.1000149
35. Sun Y, Tian T, Gao J, Liu X, Hou H, Cao R, et al. Metformin ameliorates the development of experimental autoimmune encephalomyelitis by regulating T helper 17 and regulatory T cells in mice. J Neuroimmunol (2016) 292:58–67. doi:10.1016/j.jneuroim.2016.01.014
36. Kang KY, Kim YK, Yi H, Kim J, Jung HR, Kim IJ, et al. Metformin downregulates Th17 cells differentiation and attenuates murine autoimmune arthritis. Int Immunopharmacol (2013) 16(1):85–92. doi:10.1016/j.intimp.2013.03.020
37. Son HJ, Lee J, Lee SY, Kim EK, Park MJ, Kim KW, et al. Metformin attenuates experimental autoimmune arthritis through reciprocal regulation of Th17/Treg balance and osteoclastogenesis. Mediators Inflamm (2014) 2014:973986. doi:10.1155/2014/973986
38. Jhun J, Lee S, Kim SY, Na HS, Kim EK, Kim JK, et al. Combination therapy with metformin and coenzyme Q10 in murine experimental autoimmune arthritis. Immunopharmacol Immunotoxicol (2016) 38(2):103–12. doi:10.3109/08923973.2015.1122619
39. Kim EK, Lee SH, Lee SY, Kim JK, Jhun JY, Na HS, et al. Metformin ameliorates experimental-obesity-associated autoimmune arthritis by inducing FGF21 expression and brown adipocyte differentiation. Exp Mol Med (2018) 50(1):e432. doi:10.1038/emm.2017.245
40. Lee SY, Lee SH, Yang EJ, Kim EK, Kim JK, Shin DY, et al. Metformin ameliorates inflammatory bowel disease by suppression of the STAT3 signaling pathway and regulation of the between Th17/Treg balance. PLoS One (2015) 10(9):e0135858. doi:10.1371/journal.pone.0135858
41. Lee SY, Moon SJ, Kim EK, Seo HB, Yang EJ, Son HJ, et al. Metformin suppresses systemic autoimmunity in roquin(san/san) mice through inhibiting B cell differentiation into plasma cells via regulation of AMPK/mTOR/STAT3. J Immunol (2017) 198(7):2661–70. doi:10.4049/jimmunol.1403088
42. Park MJ, Lee SY, Moon SJ, Son HJ, Lee SH, Kim EK, et al. Metformin attenuates graft-versus-host disease via restricting mammalian target of rapamycin/signal transducer and activator of transcription 3 and promoting adenosine monophosphate-activated protein kinase-autophagy for the balance between T helper 17 and Tregs. Transl Res (2016) 173:115–30. doi:10.1016/j.trsl.2016.03.006
43. Fernandez D, Perl A. Metabolic control of T cell activation and death in SLE. Autoimmun Rev (2009) 8(3):184–9. doi:10.1016/j.autrev.2008.07.041
44. Yang Z, Matteson EL, Goronzy JJ, Weyand CM. T-cell metabolism in autoimmune disease. Arthritis Res Ther (2015) 17:29. doi:10.1186/s13075-015-0542-4
45. Wahl DR, Petersen B, Warner R, Richardson BC, Glick GD, Opipari AW. Characterization of the metabolic phenotype of chronically activated lymphocytes. Lupus (2010) 19(13):1492–501. doi:10.1177/0961203310373109
46. Yin Y, Choi SC, Xu Z, Perry DJ, Seay H, Croker BP, et al. Normalization of CD4+ T cell metabolism reverses lupus. Sci Transl Med (2015) 7(274):274ra18. doi:10.1126/scitranslmed.aaa0835
47. Yin Y, Choi SC, Xu Z, Zeumer L, Kanda N, Croker BP, et al. Glucose oxidation is critical for CD4+ T cell activation in a mouse model of systemic lupus erythematosus. J Immunol (2016) 196(1):80–90. doi:10.4049/jimmunol.1501537
48. Lu MC, Yan ST, Yin WY, Koo M, Lai NS. Risk of rheumatoid arthritis in patients with type 2 diabetes: a nationwide population-based case-control study. PLoS One (2014) 9(7):e101528. doi:10.1371/journal.pone.0101528
49. Sumarac-Dumanovic M, Jeremic D, Pantovic A, Janjetovic K, Stamenkovic-Pejkovic D, Cvijovic G, et al. Therapeutic improvement of glucoregulation in newly diagnosed type 2 diabetes patients is associated with a reduction of IL-17 levels. Immunobiology (2013) 218(8):1113–8. doi:10.1016/j.imbio.2013.03.002
50. Dworacki G, Urazayev O, Bekmukhambetov Y, Iskakova S, Frycz BA, Jagodzinski PP, et al. Thymic emigration patterns in patients with type 2 diabetes treated with metformin. Immunology (2015) 146(3):456–69. doi:10.1111/imm.12522
51. Moro F, Morciano A, Tropea A, Sagnella F, Palla C, Scarinci E, et al. Effects of drospirenone-ethinylestradiol and/or metformin on CD4(+)CD28(null) T lymphocytes frequency in women with hyperinsulinemia having polycystic ovary syndrome: a randomized clinical trial. Reprod Sci (2013) 20(12):1508–17. doi:10.1177/1933719113488444
52. Dumitriu IE. The life (and death) of CD4+ CD28(null) T cells in inflammatory diseases. Immunology (2015) 146(2):185–93. doi:10.1111/imm.12506
53. Negrotto L, Farez MF, Correale J. Immunologic effects of metformin and pioglitazone treatment on metabolic syndrome and multiple sclerosis. JAMA Neurol (2016) 73(5):520–8. doi:10.1001/jamaneurol.2015.4807
54. Wang H, Li T, Chen S, Gu Y, Ye S. Neutrophil extracellular trap mitochondrial DNA and its autoantibody in systemic lupus erythematosus and a proof-of-concept trial of metformin. Arthritis Rheumatol (2015) 67(12):3190–200. doi:10.1002/art.39296
55. Bonilla FA, Oettgen HC. Adaptive immunity. J Allergy Clin Immunol (2010) 125(2 Suppl 2):S33–40. doi:10.1016/j.jaci.2009.09.017
56. Pillai S, Mattoo H, Cariappa A. B cells and autoimmunity. Curr Opin Immunol (2011) 23(6):721–31. doi:10.1016/j.coi.2011.10.007
57. Sakkas LI, Bogdanos DP. Systemic sclerosis: new evidence re-enforces the role of B cells. Autoimmun Rev (2016) 15(2):155–61. doi:10.1016/j.autrev.2015.10.005
58. Argyriou P, Economopoulou P, Papageorgiou S. The role of mTOR inhibitors for the treatment of B-cell lymphomas. Adv Hematol (2012) 2012:435342. doi:10.1155/2012/435342
59. Gayle S, Landrette S, Beeharry N, Conrad C, Hernandez M, Beckett P, et al. Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood (2017) 129(13):1768–78. doi:10.1182/blood-2016-09-736892
60. Bhattacharya A, Eissa NT. Autophagy and autoimmunity crosstalks. Front Immunol (2013) 4:88. doi:10.3389/fimmu.2013.00088
61. Gianchecchi E, Delfino DV, Fierabracci A. Recent insights on the putative role of autophagy in autoimmune diseases. Autoimmun Rev (2014) 13(3):231–41. doi:10.1016/j.autrev.2013.10.007
62. Shi WY, Xiao D, Wang L, Dong LH, Yan ZX, Shen ZX, et al. Therapeutic metformin/AMPK activation blocked lymphoma cell growth via inhibition of mTOR pathway and induction of autophagy. Cell Death Dis (2012) 3:e275. doi:10.1038/cddis.2012.13
63. Alkhatib Y, Abdel Rahman Z, Kuriakose P. Clinical impact of metformin in diabetic diffuse large B-cell lymphoma patients: a case-control study. Leuk Lymphoma (2017) 58(5):1130–4. doi:10.1080/10428194.2016.1239822
64. Zhang S, Pruitt M, Tran D, Du Bois W, Zhang K, Patel R, et al. B cell-specific deficiencies in mTOR limit humoral immune responses. J Immunol (2013) 191(4):1692–703. doi:10.4049/jimmunol.1201767
65. Diaz A, Romero M, Vazquez T, Lechner S, Blomberg BB, Frasca D. Metformin improves in vivo and in vitro B cell function in individuals with obesity and type-2 diabetes. Vaccine (2017) 35(20):2694–700. doi:10.1016/j.vaccine.2017.03.078
66. Leandro MJ. B-cell subpopulations in humans and their differential susceptibility to depletion with anti-CD20 monoclonal antibodies. Arthritis Res Ther (2013) 15(Suppl 1):S3. doi:10.1186/ar3908
67. Le Pottier L, Devauchelle V, Pers JO, Jamin C, Youinou P. The mosaic of B-cell subsets (with special emphasis on primary Sjogren’s syndrome). Autoimmun Rev (2007) 6(3):149–54. doi:10.1016/j.autrev.2006.09.011
68. Rothstein TL, Griffin DO, Holodick NE, Quach TD, Kaku H. Human B-1 cells take the stage. Ann N Y Acad Sci (2013) 1285:97–114. doi:10.1111/nyas.12137
69. Wang H, Shin DM, Abbasi S, Jain S, Kovalchuk AL, Beaty N, et al. Expression of plasma cell alloantigen 1 defines layered development of B-1a B-cell subsets with distinct innate-like functions. Proc Natl Acad Sci U S A (2012) 109(49):20077–82. doi:10.1073/pnas.1212428109
70. Yoon J, Wang H, Kim YC, Yoshimoto M, Abbasi S, Morse Iii HC. Plasma cell alloantigen ENPP1 is expressed by a subset of human B cells with potential regulatory functions. Immunol Cell Biol (2016) 94(8):719–28. doi:10.1038/icb.2016.31
71. Maddux BA, Sbraccia P, Kumakura S, Sasson S, Youngren J, Fisher A, et al. Membrane glycoprotein PC-1 and insulin resistance in non-insulin-dependent diabetes mellitus. Nature (1995) 373(6513):448–51. doi:10.1038/373448a0
72. Stefanovic V, Antic S, Mitic-Zlatkovic M, Vlahovic P. Reversal of increased lymphocyte PC-1 activity in patients with type 2 diabetes treated with metformin. Diabetes Metab Res Rev (1999) 15(6):400–4. doi:10.1002/(SICI)1520-7560(199911/12)15:6<400::AID-DMRR66>3.0.CO;2-4
73. Deng J, Wang X, Chen Q, Sun X, Xiao F, Ko KH, et al. B1a cells play a pathogenic role in the development of autoimmune arthritis. Oncotarget (2016) 7(15):19299–311. doi:10.18632/oncotarget.8244
74. Murray PJ. Macrophage polarization. Annu Rev Physiol (2017) 79:541–66. doi:10.1146/annurev-physiol-022516-034339
75. Recalcati S, Locati M, Gammella E, Invernizzi P, Cairo G. Iron levels in polarized macrophages: regulation of immunity and autoimmunity. Autoimmun Rev (2012) 11(12):883–9. doi:10.1016/j.autrev.2012.03.003
76. Laria A, Lurati A, Marrazza M, Mazzocchi D, Re KA, Scarpellini M. The macrophages in rheumatic diseases. J Inflamm Res (2016) 9:1–11. doi:10.2147/JIR.S82320
77. Arango Duque G, Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol (2014) 5:491. doi:10.3389/fimmu.2014.00491
78. Vasamsetti SB, Karnewar S, Kanugula AK, Thatipalli AR, Kumar JM, Kotamraju S. Metformin inhibits monocyte-to-macrophage differentiation via AMPK-mediated inhibition of STAT3 activation: potential role in atherosclerosis. Diabetes (2015) 64(6):2028–41. doi:10.2337/db14-1225
79. Kim J, Kwak HJ, Cha JY, Jeong YS, Rhee SD, Kim KR, et al. Metformin suppresses lipopolysaccharide (LPS)-induced inflammatory response in murine macrophages via activating transcription factor-3 (ATF-3) induction. J Biol Chem (2014) 289(33):23246–55. doi:10.1074/jbc.M114.577908
80. Kelly B, Tannahill GM, Murphy MP, O’Neill LA. Metformin inhibits the production of reactive oxygen species from NADH:ubiquinone oxidoreductase to limit induction of interleukin-1beta (IL-1beta) and boosts interleukin-10 (IL-10) in lipopolysaccharide (LPS)-activated macrophages. J Biol Chem (2015) 290(33):20348–59. doi:10.1074/jbc.M115.662114
81. Jing Y, Wu F, Li D, Yang L, Li Q, Li R. Metformin improves obesity-associated inflammation by altering macrophages polarization. Mol Cell Endocrinol (2018) 461:256–64. doi:10.1016/j.mce.2017.09.025
82. Buldak L, Machnik G, Buldak RJ, Labuzek K, Boldys A, Okopien B. Exenatide and metformin express their anti-inflammatory effects on human monocytes/macrophages by the attenuation of MAPKs and NFkappaB signaling. Naunyn Schmiedebergs Arch Pharmacol (2016) 389(10):1103–15. doi:10.1007/s00210-016-1277-8
83. Krysiak R, Gdula-Dymek A, Okopien B. Monocyte-suppressing effect of high-dose metformin in fenofibrate-treated patients with impaired glucose tolerance. Pharmacol Rep (2013) 65(5):1311–6. doi:10.1016/S1734-1140(13)71489-0
84. Krysiak R, Okopien B. The effect of metformin on monocyte secretory function in simvastatin-treated patients with impaired fasting glucose. Metabolism (2013) 62(1):39–43. doi:10.1016/j.metabol.2012.06.009
85. Bruun JM, Lihn AS, Pedersen SB, Richelsen B. Monocyte chemoattractant protein-1 release is higher in visceral than subcutaneous human adipose tissue (AT): implication of macrophages resident in the AT. J Clin Endocrinol Metab (2005) 90(4):2282–9. doi:10.1210/jc.2004-1696
86. Stosic-Grujicic S, Stojanovic I, Nicoletti F. MIF in autoimmunity and novel therapeutic approaches. Autoimmun Rev (2009) 8(3):244–9. doi:10.1016/j.autrev.2008.07.037
87. Kim BS, Pallua N, Bernhagen J, Bucala R. The macrophage migration inhibitory factor protein superfamily in obesity and wound repair. Exp Mol Med (2015) 47:e161. doi:10.1038/emm.2015.26
88. Mikulowska A, Metz CN, Bucala R, Holmdahl R. Macrophage migration inhibitory factor is involved in the pathogenesis of collagen type II-induced arthritis in mice. J Immunol (1997) 158(11):5514–7.
89. de Jong YP, Abadia-Molina AC, Satoskar AR, Clarke K, Rietdijk ST, Faubion WA, et al. Development of chronic colitis is dependent on the cytokine MIF. Nat Immunol (2001) 2(11):1061–6. doi:10.1038/ni720
90. Denkinger CM, Denkinger M, Kort JJ, Metz C, Forsthuber TG. In vivo blockade of macrophage migration inhibitory factor ameliorates acute experimental autoimmune encephalomyelitis by impairing the homing of encephalitogenic T cells to the central nervous system. J Immunol (2003) 170(3):1274–82. doi:10.4049/jimmunol.170.3.1274
91. Leng L, Chen L, Fan J, Greven D, Arjona A, Du X, et al. A small-molecule macrophage migration inhibitory factor antagonist protects against glomerulonephritis in lupus-prone NZB/NZW F1 and MRL/lpr mice. J Immunol (2011) 186(1):527–38. doi:10.4049/jimmunol.1001767
92. Baugh JA, Chitnis S, Donnelly SC, Monteiro J, Lin X, Plant BJ, et al. A functional promoter polymorphism in the macrophage migration inhibitory factor (MIF) gene associated with disease severity in rheumatoid arthritis. Genes Immun (2002) 3(3):170–6. doi:10.1038/sj.gene.6363867
93. Radstake TR, Sweep FC, Welsing P, Franke B, Vermeulen SH, Geurts-Moespot A, et al. Correlation of rheumatoid arthritis severity with the genetic functional variants and circulating levels of macrophage migration inhibitory factor. Arthritis Rheum (2005) 52(10):3020–9. doi:10.1002/art.21285
94. Sanchez E, Gomez LM, Lopez-Nevot MA, Gonzalez-Gay MA, Sabio JM, Ortego-Centeno N, et al. Evidence of association of macrophage migration inhibitory factor gene polymorphisms with systemic lupus erythematosus. Genes Immun (2006) 7(5):433–6. doi:10.1038/sj.gene.6364310
95. Wu SP, Leng L, Feng Z, Liu N, Zhao H, McDonald C, et al. Macrophage migration inhibitory factor promoter polymorphisms and the clinical expression of scleroderma. Arthritis Rheum (2006) 54(11):3661–9. doi:10.1002/art.22179
96. Park SY, Lee SW, Lee SY, Hong KW, Bae SS, Kim K, et al. SIRT1/adenosine monophosphate-activated protein kinase alpha signaling enhances macrophage polarization to an anti-inflammatory phenotype in rheumatoid arthritis. Front Immunol (2017) 8:1135. doi:10.3389/fimmu.2017.01135
97. Li F, Yang Y, Zhu X, Huang L, Xu J. Macrophage polarization modulates development of systemic lupus erythematosus. Cell Physiol Biochem (2015) 37(4):1279–88. doi:10.1159/000430251
98. Dandona P, Aljada A, Ghanim H, Mohanty P, Tripathy C, Hofmeyer D, et al. Increased plasma concentration of macrophage migration inhibitory factor (MIF) and MIF mRNA in mononuclear cells in the obese and the suppressive action of metformin. J Clin Endocrinol Metab (2004) 89(10):5043–7. doi:10.1210/jc.2004-0436
99. Nemeth T, Mocsai A. The role of neutrophils in autoimmune diseases. Immunol Lett (2012) 143(1):9–19. doi:10.1016/j.imlet.2012.01.013
100. Kaplan MJ. Role of neutrophils in systemic autoimmune diseases. Arthritis Res Ther (2013) 15(5):219. doi:10.1186/ar4325
101. Forget P, Khalifa C, Defour JP, Latinne D, Van Pel MC, De Kock M. What is the normal value of the neutrophil-to-lymphocyte ratio? BMC Res Notes (2017) 10(1):12. doi:10.1186/s13104-016-2335-5
102. Templeton AJ, McNamara MG, Seruga B, Vera-Badillo FE, Aneja P, Ocana A, et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: a systematic review and meta-analysis. J Natl Cancer Inst (2014) 106(6):dju124. doi:10.1093/jnci/dju124
103. Wu Y, Chen Y, Yang X, Chen L, Yang Y. Neutrophil-to-lymphocyte ratio (NLR) and platelet-to-lymphocyte ratio (PLR) were associated with disease activity in patients with systemic lupus erythematosus. Int Immunopharmacol (2016) 36:94–9. doi:10.1016/j.intimp.2016.04.006
104. Chandrashekara S, Mukhtar Ahmad M, Renuka P, Anupama KR, Renuka K. Characterization of neutrophil-to-lymphocyte ratio as a measure of inflammation in rheumatoid arthritis. Int J Rheum Dis (2017) 20(10):1457–67. doi:10.1111/1756-185X.13157
105. Bisgaard AK, Pihl-Jensen G, Frederiksen JL. The neutrophil-to-lymphocyte ratio as disease activity marker in multiple sclerosis and optic neuritis. Mult Scler Relat Disord (2017) 18:213–7. doi:10.1016/j.msard.2017.10.009
106. Nishida Y, Hosomi S, Yamagami H, Yukawa T, Otani K, Nagami Y, et al. Neutrophil-to-lymphocyte ratio for predicting loss of response to infliximab in ulcerative colitis. PLoS One (2017) 12(1):e0169845. doi:10.1371/journal.pone.0169845
107. Cameron AR, Morrison VL, Levin D, Mohan M, Forteath C, Beall C, et al. Anti-inflammatory effects of metformin irrespective of diabetes status. Circ Res (2016) 119(5):652–65. doi:10.1161/CIRCRESAHA.116.308445
108. Ibanez L, Jaramillo AM, Ferrer A, de Zegher F. High neutrophil count in girls and women with hyperinsulinaemic hyperandrogenism: normalization with metformin and flutamide overcomes the aggravation by oral contraception. Hum Reprod (2005) 20(9):2457–62. doi:10.1093/humrep/dei072
109. Eilenberg W, Stojkovic S, Piechota-Polanczyk A, Kaider A, Kozakowski N, Weninger WJ, et al. Neutrophil gelatinase associated lipocalin (NGAL) is elevated in type 2 diabetics with carotid artery stenosis and reduced under metformin treatment. Cardiovasc Diabetol (2017) 16(1):98. doi:10.1186/s12933-017-0579-6
110. Yang H, Biermann MH, Brauner JM, Liu Y, Zhao Y, Herrmann M. New insights into neutrophil extracellular traps: mechanisms of formation and role in inflammation. Front Immunol (2016) 7:302. doi:10.3389/fimmu.2016.00302
111. Yu Y, Su K. Neutrophil extracellular traps and systemic lupus erythematosus. J Clin Cell Immunol (2013) 4:139. doi:10.4172/2155-9899.1000139
112. Corsiero E, Pratesi F, Prediletto E, Bombardieri M, Migliorini P. NETosis as source of autoantigens in rheumatoid arthritis. Front Immunol (2016) 7:485. doi:10.3389/fimmu.2016.00485
113. Menegazzo L, Ciciliot S, Poncina N, Mazzucato M, Persano M, Bonora B, et al. NETosis is induced by high glucose and associated with type 2 diabetes. Acta Diabetol (2015) 52(3):497–503. doi:10.1007/s00592-014-0676-x
114. Wong SL, Demers M, Martinod K, Gallant M, Wang Y, Goldfine AB, et al. Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat Med (2015) 21(7):815–9. doi:10.1038/nm.3887
115. Carestia A, Frechtel G, Cerrone G, Linari MA, Gonzalez CD, Casais P, et al. NETosis before and after hyperglycemic control in type 2 diabetes mellitus patients. PLoS One (2016) 11(12):e0168647. doi:10.1371/journal.pone.0168647
116. Menegazzo L, Scattolini V, Cappellari R, Bonora BM, Albiero M, Bortolozzi M, et al. The antidiabetic drug metformin blunts NETosis in vitro and reduces circulating NETosis biomarkers in vivo. Acta Diabetol (2018) 55(6):593–601. doi:10.1007/s00592-018-1129-8
117. Smith TJ. Insights into the role of fibroblasts in human autoimmune diseases. Clin Exp Immunol (2005) 141(3):388–97. doi:10.1111/j.1365-2249.2005.02824.x
118. Leask A. Scar wars: is TGFbeta the phantom menace in scleroderma? Arthritis Res Ther (2006) 8(4):213. doi:10.1186/ar1976
119. Weiss A, Attisano L. The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol (2013) 2(1):47–63. doi:10.1002/wdev.86
120. Xiao H, Ma X, Feng W, Fu Y, Lu Z, Xu M, et al. Metformin attenuates cardiac fibrosis by inhibiting the TGFbeta1-Smad3 signalling pathway. Cardiovasc Res (2010) 87(3):504–13. doi:10.1093/cvr/cvq066
121. Bai J, Zhang N, Hua Y, Wang B, Ling L, Ferro A, et al. Metformin inhibits angiotensin II-induced differentiation of cardiac fibroblasts into myofibroblasts. PLoS One (2013) 8(9):e72120. doi:10.1371/journal.pone.0072120
122. Park IH, Um JY, Hong SM, Cho JS, Lee SH, Lee SH, et al. Metformin reduces TGF-beta1-induced extracellular matrix production in nasal polyp-derived fibroblasts. Otolaryngol Head Neck Surg (2014) 150(1):148–53. doi:10.1177/0194599813513880
123. Lu J, Shi J, Li M, Gui B, Fu R, Yao G, et al. Activation of AMPK by metformin inhibits TGF-beta-induced collagen production in mouse renal fibroblasts. Life Sci (2015) 127:59–65. doi:10.1016/j.lfs.2015.01.042
124. Lim JY, Oh MA, Kim WH, Sohn HY, Park SI. AMP-activated protein kinase inhibits TGF-beta-induced fibrogenic responses of hepatic stellate cells by targeting transcriptional coactivator p300. J Cell Physiol (2012) 227(3):1081–9. doi:10.1002/jcp.22824
125. Qin X, Jiang T, Liu S, Tan J, Wu H, Zheng L, et al. Effect of metformin on ossification and inflammation of fibroblasts in ankylosing spondylitis: an in vitro study. J Cell Biochem (2018) 119(1):1074–82. doi:10.1002/jcb.26275
126. Li L, Huang W, Li K, Zhang K, Lin C, Han R, et al. Metformin attenuates gefitinib-induced exacerbation of pulmonary fibrosis by inhibition of TGF-beta signaling pathway. Oncotarget (2015) 6(41):43605–19. doi:10.18632/oncotarget.6186
127. Ursini F, Grembiale RD, D’Antona L, Gallo E, D’Angelo S, Citraro R, et al. Oral metformin ameliorates bleomycin-induced skin fibrosis. J Invest Dermatol (2016) 136(9):1892–4. doi:10.1016/j.jid.2016.05.097
128. Xiao H, Zhang J, Xu Z, Feng Y, Zhang M, Liu J, et al. Metformin is a novel suppressor for transforming growth factor (TGF)-beta1. Sci Rep (2016) 6:28597. doi:10.1038/srep28597
129. Tanaka Y, Nakayamada S, Okada Y. Osteoblasts and osteoclasts in bone remodeling and inflammation. Curr Drug Targets Inflamm Allergy (2005) 4(3):325–8. doi:10.2174/1568010054022015
130. Schett G, Gravallese E. Bone erosion in rheumatoid arthritis: mechanisms, diagnosis and treatment. Nat Rev Rheumatol (2012) 8(11):656–64. doi:10.1038/nrrheum.2012.153
131. Lee YS, Kim YS, Lee SY, Kim GH, Kim BJ, Lee SH, et al. AMP kinase acts as a negative regulator of RANKL in the differentiation of osteoclasts. Bone (2010) 47(5):926–37. doi:10.1016/j.bone.2010.08.001
132. Kasai T, Bandow K, Suzuki H, Chiba N, Kakimoto K, Ohnishi T, et al. Osteoblast differentiation is functionally associated with decreased AMP kinase activity. J Cell Physiol (2009) 221(3):740–9. doi:10.1002/jcp.21917
133. Mai QG, Zhang ZM, Xu S, Lu M, Zhou RP, Zhao L, et al. Metformin stimulates osteoprotegerin and reduces RANKL expression in osteoblasts and ovariectomized rats. J Cell Biochem (2011) 112(10):2902–9. doi:10.1002/jcb.23206
134. Jang WG, Kim EJ, Bae IH, Lee KN, Kim YD, Kim DK, et al. Metformin induces osteoblast differentiation via orphan nuclear receptor SHP-mediated transactivation of Runx2. Bone (2011) 48(4):885–93. doi:10.1016/j.bone.2010.12.003
135. Jackuliak P, Payer J. Osteoporosis, fractures, and diabetes. Int J Endocrinol (2014) 2014:820615. doi:10.1155/2014/820615
136. Lecka-Czernik B. Bone loss in diabetes: use of antidiabetic thiazolidinediones and secondary osteoporosis. Curr Osteoporos Rep (2010) 8(4):178–84. doi:10.1007/s11914-010-0027-y
137. Vestergaard P, Rejnmark L, Mosekilde L. Relative fracture risk in patients with diabetes mellitus, and the impact of insulin and oral antidiabetic medication on relative fracture risk. Diabetologia (2005) 48(7):1292–9. doi:10.1007/s00125-005-1786-3
138. Shamriz O, Mizrahi H, Werbner M, Shoenfeld Y, Avni O, Koren O. Microbiota at the crossroads of autoimmunity. Autoimmun Rev (2016) 15(9):859–69. doi:10.1016/j.autrev.2016.07.012
139. Yeoh N, Burton JP, Suppiah P, Reid G, Stebbings S. The role of the microbiome in rheumatic diseases. Curr Rheumatol Rep (2013) 15(3):314. doi:10.1007/s11926-012-0314-y
140. Lopez P, de Paz B, Rodriguez-Carrio J, Hevia A, Sanchez B, Margolles A, et al. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci Rep (2016) 6:24072. doi:10.1038/srep24072
141. Consolandi C, Turroni S, Emmi G, Severgnini M, Fiori J, Peano C, et al. Behcet’s syndrome patients exhibit specific microbiome signature. Autoimmun Rev (2015) 14(4):269–76. doi:10.1016/j.autrev.2014.11.009
142. Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell (2013) 153(1):228–39. doi:10.1016/j.cell.2013.02.035
143. Onken B, Driscoll M. Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans healthspan via AMPK, LKB1, and SKN-1. PLoS One (2010) 5(1):e8758. doi:10.1371/journal.pone.0008758
144. Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev (2008) 32(5):723–35. doi:10.1111/j.1574-6976.2008.00123.x
145. Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut (2014) 63(5):727–35. doi:10.1136/gutjnl-2012-303839
146. Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Appl Environ Microbiol (2014) 80(19):5935–43. doi:10.1128/AEM.01357-14
147. Lee H, Lee Y, Kim J, An J, Lee S, Kong H, et al. Modulation of the gut microbiota by metformin improves metabolic profiles in aged obese mice. Gut Microbes (2017). doi:10.1080/19490976.2017.1405209
148. Bauer PV, Duca FA, Waise TMZ, Rasmussen BA, Abraham MA, Dranse HJ, et al. Metformin alters upper small intestinal microbiota that impact a glucose-SGLT1-sensing glucoregulatory pathway. Cell Metab (2018) 27(1):101–17.e5. doi:10.1016/j.cmet.2017.09.019
149. Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med (2017) 23(7):850–8. doi:10.1038/nm.4345
150. de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM, et al. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care (2017) 40(1):54–62. doi:10.2337/dc16-1324
151. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature (2015) 528(7581):262–6. doi:10.1038/nature15766
152. Napolitano A, Miller S, Nicholls AW, Baker D, Van Horn S, Thomas E, et al. Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS One (2014) 9(7):e100778. doi:10.1371/journal.pone.0100778
153. Hanninen A, Toivonen R, Poysti S, Belzer C, Plovier H, Ouwerkerk JP, et al. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut (2017). doi:10.1136/gutjnl-2017-314508
154. Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One (2013) 8(10):e76520. doi:10.1371/journal.pone.0076520
Keywords: metformin, autoimmunity, autoimmune diseases, T cell, B cell, macrophage, neutrophil, fibroblast
Citation: Ursini F, Russo E, Pellino G, D’Angelo S, Chiaravalloti A, De Sarro G, Manfredini R and De Giorgio R (2018) Metformin and Autoimmunity: A “New Deal” of an Old Drug. Front. Immunol. 9:1236. doi: 10.3389/fimmu.2018.01236
Received: 17 April 2018; Accepted: 17 May 2018;
Published: 04 June 2018
Edited by:
Laurence Morel, University of Florida, United StatesReviewed by:
Xiangyu Teng, Shanghai JiaoTong University School of Medicine, ChinaAndras Perl, Upstate Medical University, United States
Mi-La Cho, Catholic University of Korea, South Korea
Copyright: © 2018 Ursini, Russo, Pellino, D’Angelo, Chiaravalloti, De Sarro, Manfredini and De Giorgio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Ursini, ZnJhbmNlc2NvLnVyc2luaSYjeDAwMDQwO3lhaG9vLml0
 Francesco Ursini
Francesco Ursini Emilio Russo
Emilio Russo Gianluca Pellino2,3
Gianluca Pellino2,3 Agostino Chiaravalloti
Agostino Chiaravalloti Giovambattista De Sarro
Giovambattista De Sarro Roberto Manfredini
Roberto Manfredini