- 1Department of Agronomy, Faculty of Agriculture at Kamphaeng Saen, Kasetsart University, Kamphaeng Saen Campus, Nakhon Pathom, Thailand
- 2Center of Excellence on Agricultural Biotechnology: (AG-BIO/PERDO-CHE), Bangkok, Thailand
- 3Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
Zombi pea [Vigna vexillata (L.) A. Rich] is a legume crop found in Africa. Wild zombi pea is widely distributed throughout the tropical and subtropical regions, whereas domesticated zombi pea is rarely cultivated. Plant domestication is an evolutionary process in which the phenotypes of wild species, including seed dormancy, pod shattering, organ size, and architectural and phenological characteristics, undergo changes. The molecular mechanism underlying the domestication of zombi pea is relatively unknown. In this study, the genetic basis of the following 13 domestication-related traits was investigated in an F2 population comprising 198 individuals derived from a cross between cultivated (var. macrosperma) and wild (var. vexillata) zombi pea accessions: seed dormancy, pod shattering, days-to-flowering, days-to-maturity, stem thickness, stem length, number of branches, leaf area, pod length, 100-seed weight, seed width, seed length, and seeds per pod. A genetic map containing 6,529 single nucleotide polymorphisms constructed for the F2 population was used to identify quantitative trait loci (QTLs) for these traits. A total of 62 QTLs were identified for the 13 traits, with 1–11 QTLs per trait. The major QTLs for days-to-flowering, stem length, number of branches, pod length, 100-seed weight, seed length, and seeds per pod were clustered in linkage group 5. In contrast, the major QTLs for seed dormancy and pod shattering belonged to linkage groups 3 and 11, respectively. A comparative genomic analysis with the cowpea [Vigna unguiculata (L.) Walp.] genome used as the reference sequence (i.e., the genome of the legume species most closely related to zombi pea) enabled the identification of candidate genes for the major QTLs. Thus, we revealed the genomic regions associated with domestication-related traits and the candidate genes controlling these traits in zombi pea. The data presented herein may be useful for breeding new varieties of zombi pea and other Vigna species.
Introduction
Plant domestication is a process in which wild forms of crop (cultivated) plants evolve and adapt to agricultural conditions and human needs via conscious and unconscious selections by humans and natural selection due to environmental factors. Domestication is one of the most important technological innovations in human history (Doebley et al., 2006). Although the processes and spatiotemporal requirements for domestication vary among crops, the processes generally lead to similar results regarding morphological and phenological changes to the wild forms. The changes from wild species to domesticated/cultivated species are collectively referred to as the “domestication syndrome” (Hammer, 1984). Compared with wild forms, domesticated forms exhibit a lack of or decreased seed dormancy and seed dispersal, produce larger organs (fruit, seed, leaf, and stem), flower and mature earlier, and have a more robust and firm plant architecture. During the last 20 years, plant domestication studies have become a major part of crop evolution and adaptation research. In fact, domestication is the earliest form of plant breeding (Doebley et al., 2006). The information generated in investigations of the evolutionary relationships between crops and their wild progenitors may be useful for exploiting wild plants for crop improvement (Vaughan et al., 2007). The genetic basis of the domestication syndrome of several plant species, but mainly cereals and legumes, including rice, maize, wheat, sorghum, soybean, common bean, and cowpea, has been investigated via molecular genetic analyses, especially quantitative trait locus (QTL) mapping. Several genes controlling the domestication-related traits of some of these crops have been identified and cloned (Doebley et al., 2006; Vaughan et al., 2007; Ishimaru et al., 2013).
Zombi pea [Vigna vexillata (L.) A. Rich] is an underutilized legume crop. Wild zombi pea is widely distributed in Africa, Asia, Australia, and the Americas (Dachapak et al., 2017). Because of its wide distribution, several wild forms of zombi pea have been identified and described (Maxted et al., 2004). Additionally, there are two types of cultivated zombi pea (i.e., seed and tuberous root) (Ferguson, 1954; Karuniawan et al., 2006). The cultivated seed type is grown in Africa (Ferguson, 1954), whereas the cultivated tuberous root type exists only in Bali, Indonesia, and East Timor (Karuniawan et al., 2006). The former is grown principally for its seeds, although the tuberous roots are sometimes consumed, whereas the latter is cultivated mainly for its tuberous roots. An analysis of molecular diversity based on microsatellite (i.e., simple sequence repeat) markers revealed that these two forms were domesticated independently (Dachapak et al., 2017). There exists a strong crossing barrier between the cultivated tuberous root type and the cultivated seed type/wild forms of zombi pea.
The genus Vigna includes as many as nine cultivated/domesticated species and several wild species that are used as human food, animal feed, and cover crops. Several comparative genomic analyses revealed that the genomes of Vigna species are highly conserved (Chaitieng et al., 2006; Isemura et al., 2007, 2010; Dachapak et al., 2018; Amkul et al., 2019; Yundaeng et al., 2019). We previously developed linkage maps of zombi pea and determined that its genome is highly conserved with those of cowpea [Vigna unguiculata (L.) Walp.], mung bean [Vigna radiata (L.) Wilczek], azuki bean [Vigna angularis (Willd.) Ohwi and Ohashi], and rice bean [Vigna umbellata (Thunb.) Ohwi and Ohashi] (Dachapak et al., 2018, Dachapak et al., 2019; Amkul et al., 2019). A recent study proved that the zombi pea genome and the genomes of common bean (Phaseolus vulgaris L.) and soybean [Glycine max (L.) Merr.] (Amkul et al., 2019) are also highly conserved. Additionally, QTLs controlling domestication-related traits have been identified in Vigna species, including mung bean (Isemura et al., 2012), azuki bean (Isemura et al., 2007), rice bean (Isemura et al., 2010), cowpea (Kongjaimun et al., 2012; Lo et al., 2018), and zombi pea (Dachapak et al., 2018). Although these studies indicated that the QTLs for the same traits are generally conserved among Vigna species, the major QTLs for some important traits, such as seed size and weight, vary between species.
The genetic basis of zombi pea domestication is particularly interesting because the two cultivated types were likely domesticated from the same or different wild zombi pea species via different processes (Dachapak et al., 2017). The seed-type zombi pea plant produces relatively few branches and exhibits a determinate growth habit. In contrast, the tuberous root-type zombi pea is a highly branched, vine-like plant with an indeterminate growth habit. Dachapak et al. (2018) identified QTLs for the domestication of the tuberous root form of zombi pea. However, because of the sterility of the mapping population, the QTLs of key domestication-related traits, including pod dehiscence (pod shattering) and seed dormancy, were not detected (Dachapak et al., 2018). Therefore, identifying the QTLs controlling domestication-related traits in the seed-type zombi pea may provide important insights regarding the genetic basis of the domestication of this species and other Vigna species, which may facilitate the identification of new candidate genes for crop improvement. The objectives of this study were to (i) detect the QTLs associated with domestication-related traits and (ii) identify candidate genes for these traits in the seed-type zombi pea.
Materials and Methods
Mapping Population
An F2 population comprising 198 individuals developed from a cross between TVNu 240 and TVNu 1623 was used in this study. Specifically, TVNu 240 is a cultivated zombi pea (V. vexillata var. macrosperma) accession from the Central African Republic, whereas TVNu 1623 is a wild zombi pea (V. vexillata var. vexillata) accession from Nigeria. The F2 population was previously used for developing high-density linkage maps and for the mapping of QTLs for bruchid (Callosobruchus spp.) resistance (Amkul et al., 2019). Briefly, the F2 plants and 10 plants of each parent were grown under field conditions, with 0.75 m × 0.75 m spacing, at Kasetsart University, Kamphaeng Sean Campus, Nakhon Pathom, Thailand between December 2017 and March 2018.
Measurement of Domestication-Related Traits
Thirteen domestication-related traits were assessed in the F2 and parental plants (Table 1). All of the traits, except for leaf area (LFA), were analyzed in previous studies on Vigna species, including mung bean (Isemura et al., 2012), azuki bean (Isemura et al., 2007), rice bean (Isemura et al., 2010), cowpea (Kongjaimun et al., 2012), and zombi pea (Dachapak et al., 2018). The LFA of nine fully expanded leaves was measured with the LI-3100C area meter (LI-COR Biosciences, United States) at 50 days after sowing. Stem length (STL) and stem thickness (STT) were recorded at harvest. The number of days from sowing to first flowering (FLD) and the number of days from sowing to harvesting of the first pod (PDDM) were also recorded. The mature pods and seeds of each plant were harvested separately to analyze the pod-related and seed-related traits. Specifically, pod length (PDL) was measured and the number of seeds per pod (SDNPPD) was determined based on 10 pods. Pod shattering was evaluated by counting the number of twists along the pod (PDT) of five pods maintained at room temperature. The seed length and width (SDL and SDW, respectively) were recorded as the average value of five seeds. The 100-seed weight (SD100WT) was measured twice using intact seeds. Seed dormancy was assessed by measuring seed water absorption (SDWA). Specifically, 40 intact seeds harvested from each plant were immersed in water in a plastic container and incubated at 25°C for 7 days. The number of seeds that imbibed water was recorded.
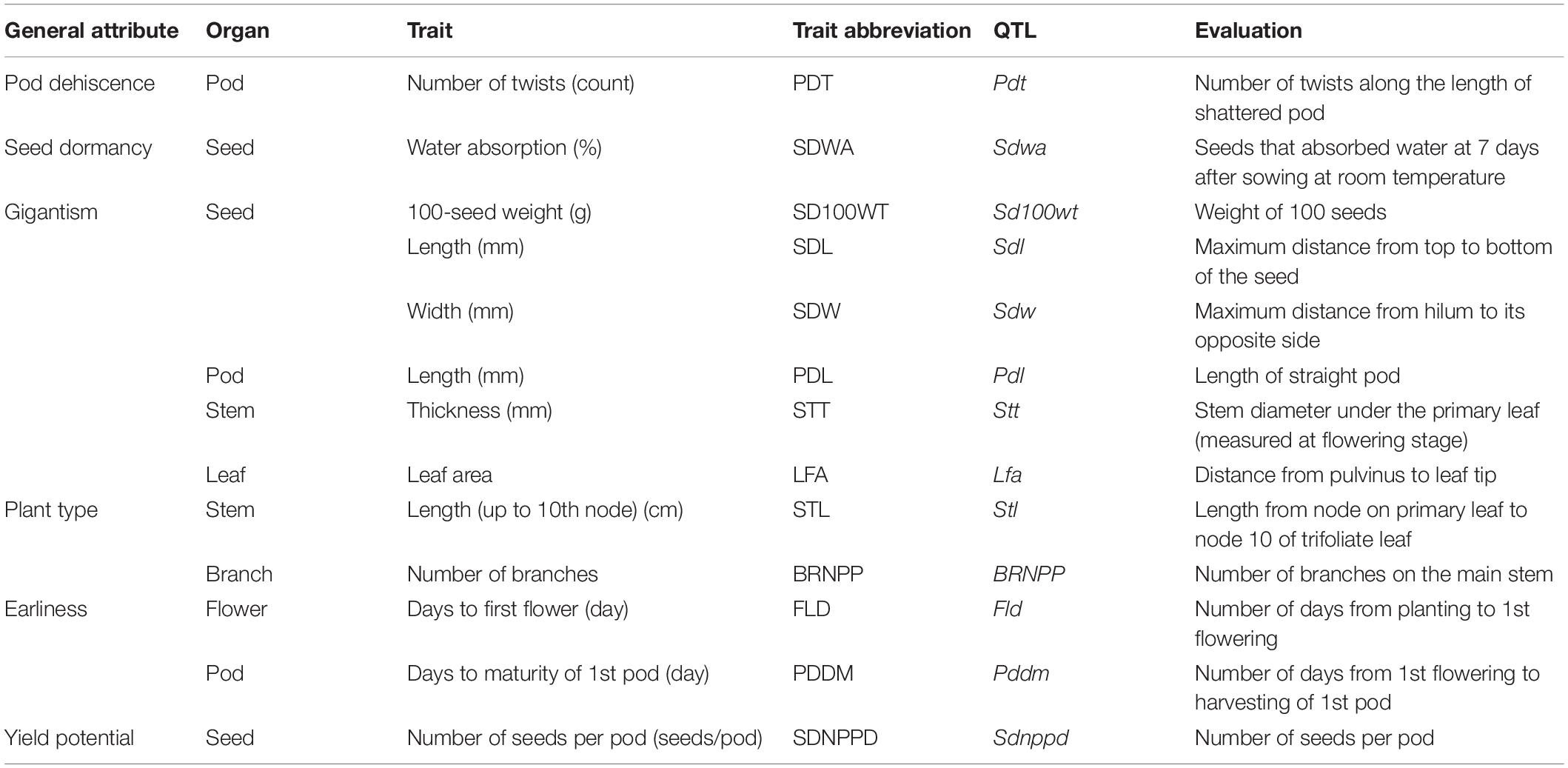
Table 1. Domestication-related traits in the F2 population derived from a cross between TVNu 240 (cultivated) and TVNu 1623 (wild).
Heritability Estimation and Correlation Analysis
The broad-sense heritability (H2) (Allard, 1999) of each trait was calculated using the following formula:
where and are the variances of the F2 population, TVNu 240, and TVNu 1623, respectively. The correlation between traits was calculated with the R program (version 10.12.0).
Quantitative Trait Loci Mapping
The F2 population was previously used for constructing a single nucleotide polymorphism (SNP)-based linkage map and for mapping QTLs for bruchid resistance (Amkul et al., 2019). The SNP markers were generated via a specific locus amplified fragment sequencing method (Sun et al., 2013). The map consisted of 11 linkage groups (LGs) and comprised 6,529 SNP markers (Amkul et al., 2019). The linkage map and genotypic (SNP marker) data were used for analyzing the QTLs of domestication-related traits in the present study.
The QTLs were analyzed according to the inclusive composite interval mapping (ICIM) method (Li et al., 2007) of the QTL IciMapping 4.2 software1, with 0.1 cM steps. A probability of stepwise regression of 0.001 was used for the ICIM. A logarithm of odds of 3.0 was used as the threshold for assessing the significance of the QTLs.
Identification of Candidate Genes for Domestication-Related Traits
Candidate genes at the major QTLs [i.e., phenotypic variance explained (PVE) ≥10%] for domestication-related traits were identified based on the nearest DNA markers flanking the QTLs on the cowpea reference genome sequence (Lonardi et al., 2019)2. All of the annotated genes between the markers were identified, after which the genes with functions potentially associated with the specific trait were selected as candidate genes.
Results
Variation and Heritability of Domestication-Related Traits
The data for the domestication-related traits of the F2 population (mean and range) and the traits of the parents (mean) are summarized in Table 2. The values for the following traits were higher for the cultivated parent (TVNu 240) than for the wild parent (TVNu 1623): germination (SDWA), organ size (SD100WT, LFA, SDL, PDL, and STL), yield (SDNPPD), and earliness (FLD and PDDM). In contrast, the STT and branching (BRNPP) values were higher for the wild parent than for the cultivated parent. The parents had very similar PDT and SDL values. The SDWA, SD100WT, LFA, SDW, STT, BRNPP, and SDNPPD mean values of the F2 population were between those of the parents (Table 2). However, the PDT and SDL mean values were similar among the F2 population and both parents. The F2 population mean values for PDL and STL were almost the same as those of the wild parent and cultivated parent, respectively, whereas the SDNPPD mean value was lower for the F2 population than for either parent (Table 2). All traits exhibited a continuous distribution (Figure 1), indicative of a quantitative trait inheritance. Specifically, SDNPPD exhibited a bimodal distribution, suggesting that this trait may be controlled by a few loci. Transgressive segregation was clearly detected for several traits, including PDT, SDWA, LFA, PDL, STT, STL, BRNPP, FLD, PDDM, and SDNPPD (Figure 1 and Table 2), implying that both parents possessed alleles that positively and negatively affected these traits. Most of the traits exhibited moderate to relatively high heritability (Table 2), with the heritability value exceeding 0.70 for seven traits.
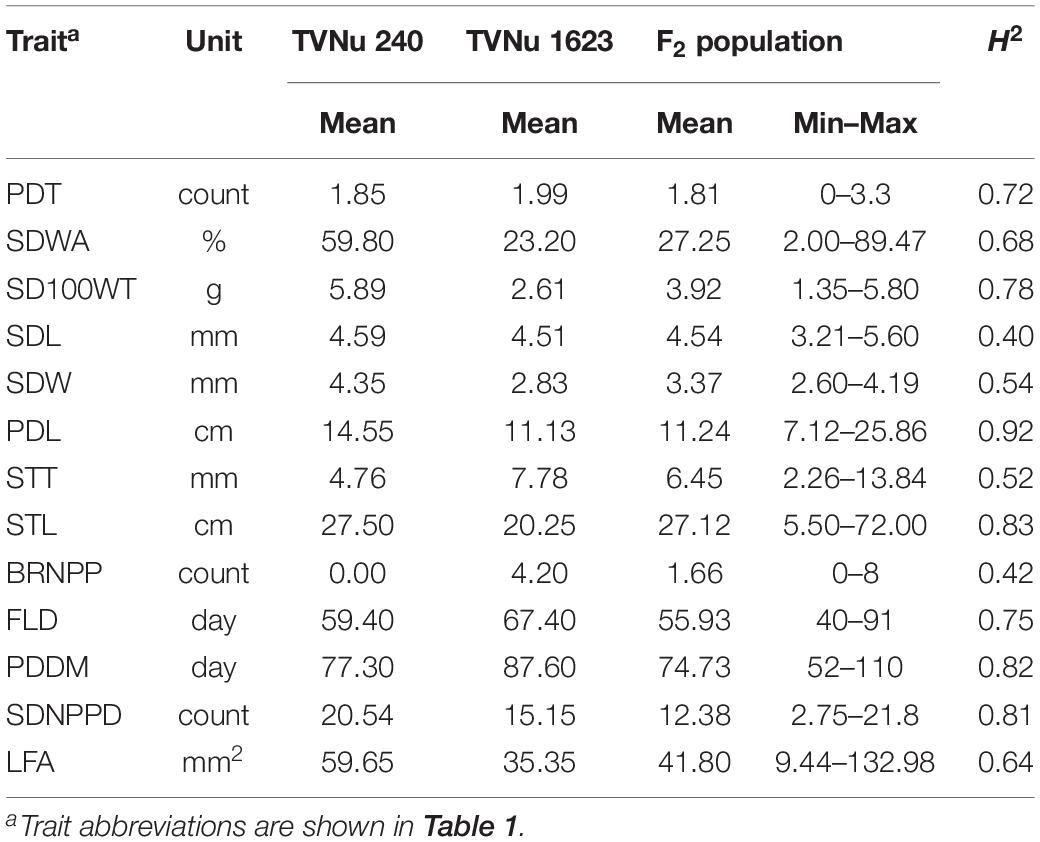
Table 2. Mean, minimum, and maximum values as well as the broad-sense heritability (H2) of domestication-related traits of the zombi pea F2 population derived from a cross between TVNu 240 (cultivated) and TVNu 1623 (wild).
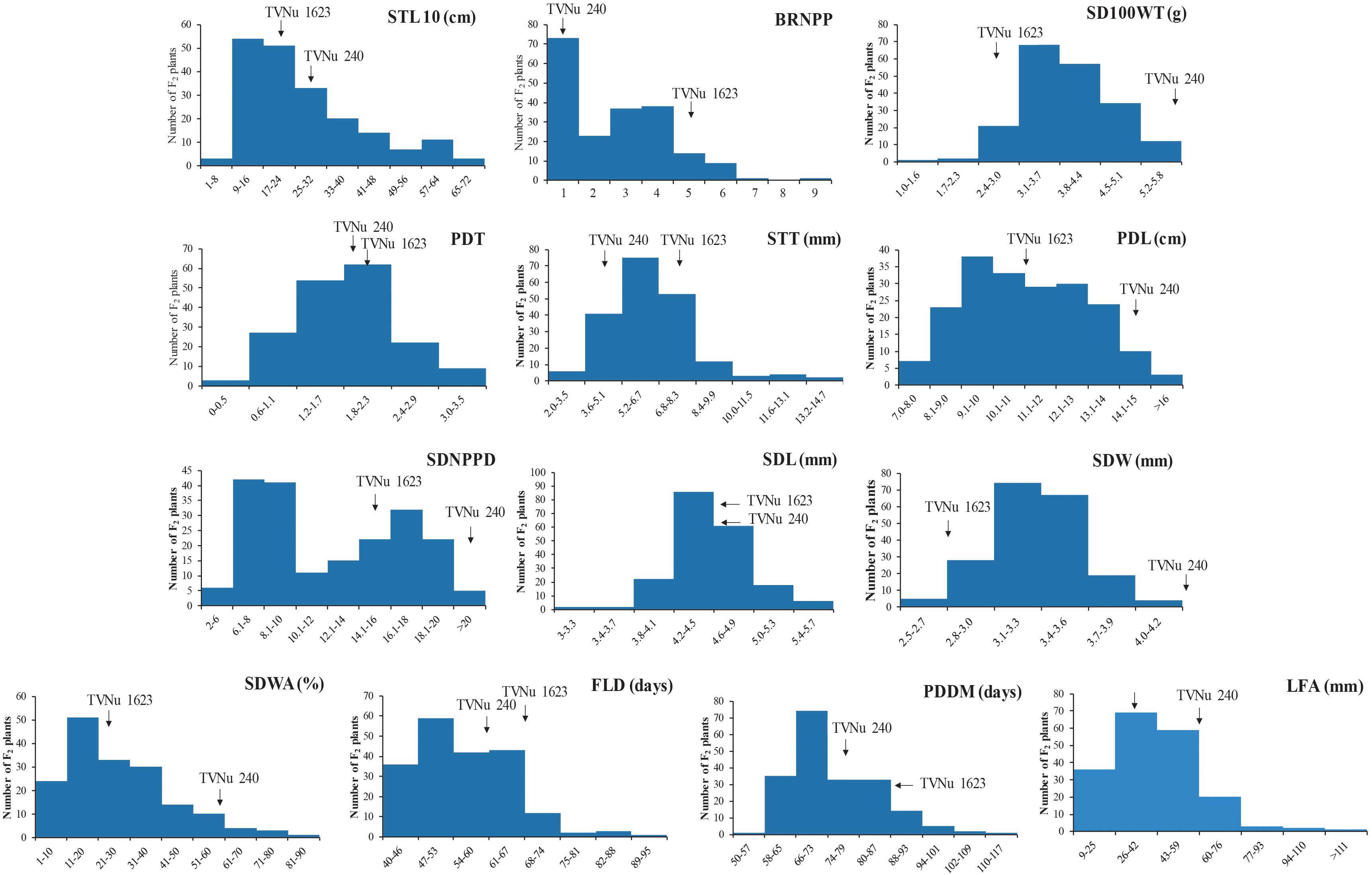
Figure 1. Frequency distribution of 13 domestication-related traits in the zombi pea F2 population developed from a cross between cultivated zombi pea (TVNu 240; V. vexillata var. macrosperma) and wild zombi pea (TVNu 1623; V. vexillata var. vexillata).
The correlations among the examined traits are listed in Supplementary Table S2. There were significant and positive correlations between related traits, including SD100WT and SDW or SDL, PDL and SDNPPD, and FLD and PDDM. Additionally, SD100WT was positively correlated with PDT, STT, STL, BRNPP, and LFA, but negatively correlated with SDNPPD. Interestingly, LFA was significantly and positively correlated with all of the other traits, with the exception of SDWA and PDT. Moreover, SDWA was significantly and negatively correlated with PDT, FLD, PDDM, and BRNPP. The data also revealed a significant correlation between BRNPP and all other analyzed traits.
QTLs and Candidate Genes for Domestication-Related Traits
A total of 62 QTLs were identified for the 13 analyzed traits, with 1–11 loci per trait (Table 3 and Figure 2). These 62 QTLs were distributed in all LGs, except for LG10. The number, location, and effect of the QTLs identified for each trait are summarized in Table 3.
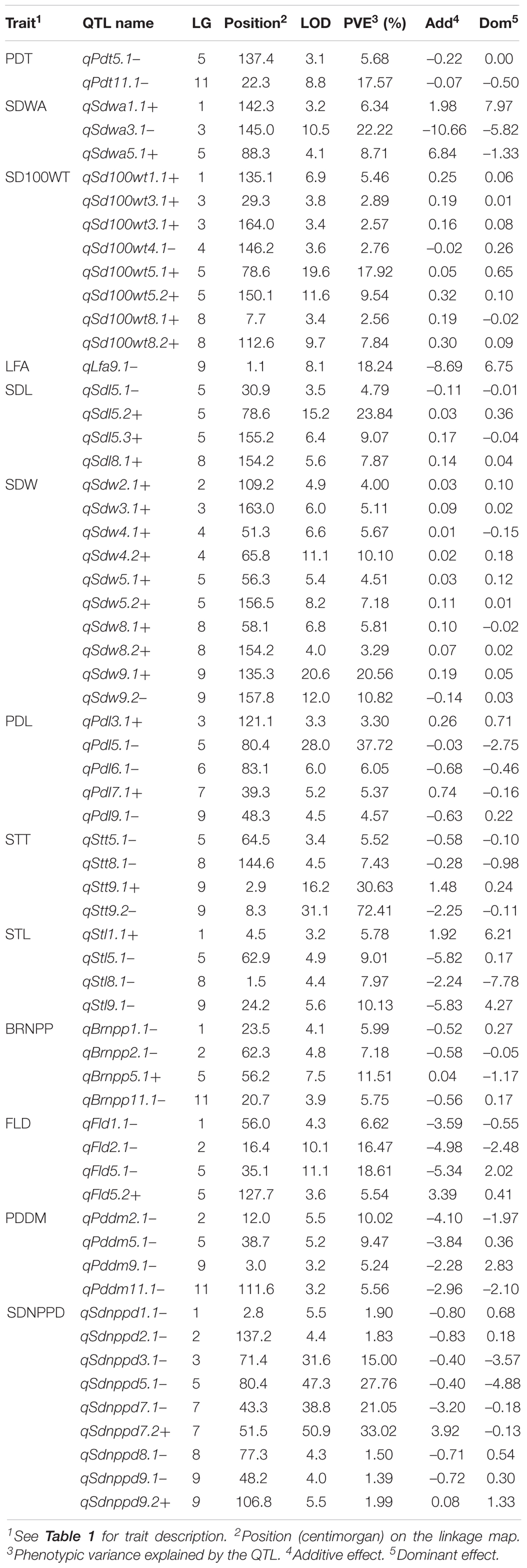
Table 3. Quantitative trait loci for 13 domestication-related traits in the zombi pea F2 population derived from a cross between TVNu 240 (female cultivated) and TVNu 1623 (male wild).
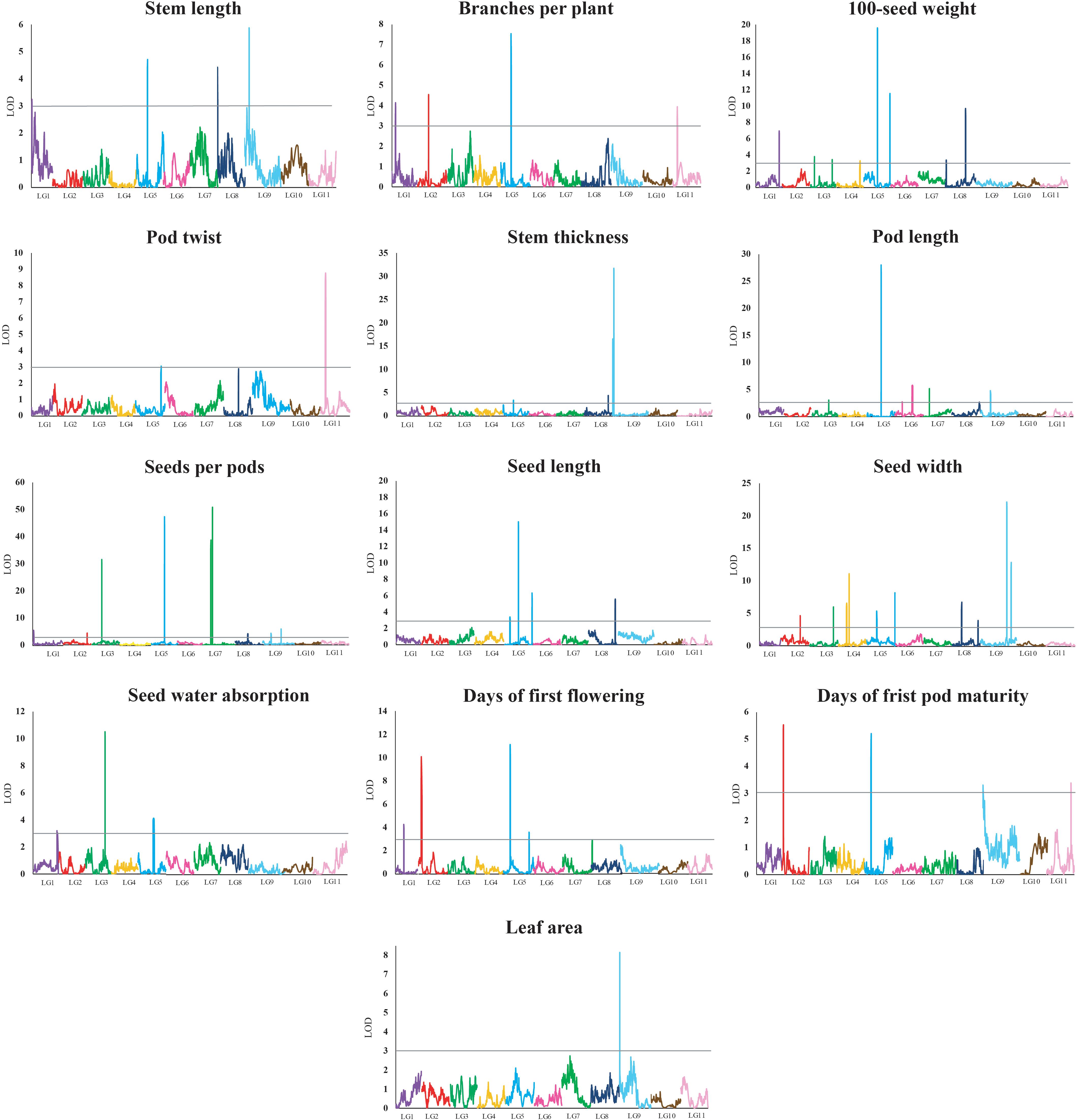
Figure 2. Quantitative trait locus plots for 13 domestication-related traits in the zombi pea F2 population developed from a cross between cultivated zombi pea (TVNu 240; V. vexillata var. macrosperma) and wild zombi pea (TVNu 1623; V. vexillata var. vexillata). The x-axis indicates the linkage groups, whereas the y-axis indicates the logarithm of odds (LOD) scores. The gray line horizontal to the y-axis indicates the LOD significance threshold.
The wild and cultivated parents differed considerably regarding SDWA (23.20 and 59.80%, respectively). The three QTLs detected for this trait, qSdwa1.1+, qSdwa3.1-, and qSdwa5.1+, were in LGs 1, 3, and 5, respectively (Table 3 and Figure 2). The QTL in LG3, qSdwa3.1-, had the largest effect (PVE of 22.2 %). Interestingly, at this QTL, the cultivated zombi pea alleles decreased the seed coat permeability (decreased germination). The qSdwa1.1+ and qSdwa5.1+ QTLs both accounted for about 9% of the total variation. At these two QTLs, the cultivated zombi pea alleles increased the seed coat permeability. The qSdwa3.1- region consisted of approximately 179.0 kb on cowpea chromosome 3 and included 16 genes (Supplementary Table S3). Of these genes, Vigun03g339300, encoding a calmodulin-binding protein-like protein, was identified as a candidate gene for this QTL. The qSdwa5.1+ region spanned about 1.74 Mb on cowpea chromosome 5 and contained 38 genes (Supplementary Table S3), with Vigun05g170600 and Vigun05g170700, both of which encode gibberellin 2-beta-dioxygenase 1, identified as candidate genes for this QTL.
The wild zombi pea exhibited extensive pod dehiscence, in contrast to the cultivated zombi pea (Table 2). Two QTLs were identified for this trait, with one each in LGs 5 and 11 (Table 3 and Figure 2). The QTL in LG11, qPdt11.1−, had a PVE of 17.6%, whereas the QTL in LG5, qPdt5.1−, had a PVE of only 5.7%. At both QTLs, the cultivated zombi pea alleles decreased pod dehiscence. The qPdt11.1− and qPdt5.1− regions spanned approximately 406 and 72.7 kb on cowpea chromosomes 11 and 5, and included eight and seven genes, respectively (Supplementary Table S3). These genes included Vigun11g053700 [encoding the bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 (UGE1)] and Vigun11g054200 [encoding glycosyltransferase family 14 (GT14)], which were considered as candidate genes for qPdt11.1−. The Vigun05g241400 gene, encoding UDP-apiose/xylose synthase, was selected as a candidate gene for qPdt5.1−.
The cultivated zombi pea seeds were about 2.5-fold larger than the wild zombi pea seeds (Table 2). Regarding the seed size-related traits (SD100WT, SDL, and SDW), 4–10 QTLs were identified, with at least one QTL in all LGs, with the exception of LGs 10 and 11 (Table 3 and Figure 2). At all QTLs, except for qSd100tw4.1− and qSdw9.1−, the cultivated zombi pea alleles were associated with increases in seed size. The QTLs with the largest effects on SD100WT and SDL were qSd100wt5.1+ (PVE of 17.9%) and qSdl5.2+ (PVE of 23.8%), respectively, both of which belonged to LG5 (Table 3). The QTL that contributed the most to SDW was qSdw9.1+ (PVE of 20.6%), which was in LG9. Additionally, LG5 and LG8 included 11 and 5 QTLs for seed size, respectively. Interestingly, qSd100wt5.1+ and qSdl5.2+ were mapped to the same position. Another large-effect QTL for SD100WT was qSd100tw5.2 + (PVE of about 10%). The qSd100wt5.1+ and qSdl5.2+ region covered 1.03 Mb on cowpea chromosome 5 and contained 38 genes (Supplementary Table S3). The identified candidate genes included Vigun05g139100, which encodes an auxin-responsive protein-related protein, and Vigun05g140300, which encodes a protein with an unknown function (DUF538). The qSd100wt5.2+ region comprised 217.9 kb on cowpea chromosome 5 and contained 30 genes (Supplementary Table S3). Among these genes, Vigun05g277600, encoding an IAA-amino acid hydrolase ILR1-like 1-related protein, was considered as a candidate gene for qSd100wt5.2+. Moreover, the qSdw9.1+ region covered a 130.0-kb region on cowpea chromosome 9 and comprised 12 genes (Supplementary Table S3). Both Vigun09g215300 (WRKY transcription factor 40-related protein) and Vigun09g215400 (WRKY DNA-binding protein) were selected as candidate genes for this QTL.
The leaves of the cultivated parent were larger than those of the wild parent. The only QTL detected for LFA, qLfaA9.1− (PVE of 18.2%), was mapped to LG9 (Table 3 and Figure 2). Moreover, the cultivated parent alleles at this QTL decreased the LFA value. The qLfa9.1− region spanned 201.1kb on cowpea chromosome 9 and comprised 17 genes (Supplementary Table S3). Among these genes, Vigun09g018300 and Vigun09g017000, encoding a WD40 repeat-containing protein and falz-related bromodomain-containing protein/transcription factor GTE1, respectively, were identified as candidate genes for this QTL.
The cultivated zombi pea pods were approximately 3.5 cm longer than the wild zombi pea pods (Table 2). Five QTLs related to PDL were detected on different LGs (Table 3 and Figure 2). At QTLs qPdl3.1+ and qPdl7.1+, the cultivated zombi pea alleles increased the PDL value. Additionally, at QTLs qPdl5.1−, qPdl6.1−, and qPdl9.1−, the wild zombi pea alleles increased the PDL value. The qPdl5.1− QTL in LG5 had a PVE of 37.7%, which was considerably higher than the PVE for the other four QTLs (<5%). The qPdl5.1− region spanned 860.4 kb on cowpea chromosome 5 and contained 41 genes (Supplementary Table S3). The Vigun05g125800 gene, encoding pectate lyase 1, was chosen as a candidate gene for qPdl5.1−.
The cultivated zombi pea plants had shorter stems (the first 10 internodes), but more branches, than the wild zombi pea plants. The four QTLs for STL belonged to different LGs (Table 3 and Figure 2). The PVE of each of the QTLs for STL was ≤10%, but was highest for qStl9.1− in LG9. The cultivated parent alleles at three of the four QTLs decreased the internode length. Similarly, four QTLs on different LGs were detected for BRNPP, and the PVE of each QTL was ≤12%, but was highest for qBrnpp5.1− in LG5 (Table 3 and Figure 2). The cultivated parent alleles at three of the four QTLs decreased the number of branches. Only one marker flanking qStl9.1−, Marker293030, matched a sequence in the cowpea reference genome. The Vigun09g037900 gene, encoding a MOB-like protein phocein (MOB1), which was detected approximately 113 kb from Marker293030, was considered to be a candidate gene for qStl9.1−. The qBrnpp5.1− region was less than 1.0 kb on cowpea chromosome 5 and included Vigun05g082200, which encodes an auxin efflux carrier family protein (Supplementary Table S3).
Domestication generally leads to cultivated crops that flower and mature earlier than their wild progenitors. The cultivated zombi pea plants flowered and matured approximately 10 and 8 days earlier, respectively, than the wild zombi pea plants (Table 2). Four QTLs on three LGs were identified for FLD (Table 3 and Figure 2). The effects of QTLs qFld2.1− in LG2 and qFld5.1− in LG5 on FLD were similar (PVE of 16.5 and 18.6%, respectively). With the exception of qFld5.2+, the cultivated parent alleles at these QTLs enhanced flowering. The qFld2.1− region spanned 391.5 kb on cowpea chromosome 2 and comprised 28 genes (Supplementary Table S3). Both Vigun02g051100 (WRKY transcription factor 57) and Vigun02g052800 (ring finger and CCH-type zinc finger domain-containing protein) were selected as candidate genes for qFld2.1−. An analysis of qFld5.1− revealed that this QTL spanned 90.5 kb on cowpea chromosome 5 and contained eight genes (Supplementary Table S3), of which Vigun05g043100, encoding a bHLH transcription factor PRE1-related protein, was identified as a candidate gene.
Similarly to the FLD analysis, four QTLs on different LGs were identified for PDDM (Table 3 and Figure 2). The qPddm2.1− (LG2) and qPddm5.1− (LG5) QTLs had the largest effects on this trait (PVE of about 10%). At all four QTLs, the cultivated parent alleles accelerated maturity. Additionally, qFld2.1− and qPddm2.1− were located close to each other, whereas qFld5.1− was mapped close to qPddm5.1−. The qPddm2.1− region covered 83.3 kb on cowpea chromosome 2 and contained seven genes, including Vigun02g050200, encoding the 14-3-3-like protein GF14 IOTA, which was chosen as a candidate gene. The qPddm5.1− region spanned only 19.0 kb on cowpea chromosome 5 and contained four genes (Supplementary Table S3). The Vigun05g050300 gene, which encodes 3-deoxy-8-phosphooctulonate synthase/phospho-2-keto-3-deoxyoctonate aldolase, was designated as a candidate gene for qPddm5.1−.
The cultivated parent produced more seeds per pod than the wild parent. Nine QTLs were detected for SDNPPD (Table 3 and Figure 2). The qSdnppd3.1−, qSdnppd5.1−, qSdnppd7.1−, and qSdnppd7.2+ QTLs had larger effects on SDNPPD (PVE of 15.0, 27.8, 21.1, and 33.0%, respectively) than the other QTLs (PVE of only about 2.0%). Regarding the qSdnppd7.2+ and qSdnppd9.2+ QTLs, the cultivated zombi pea alleles increased the seed yield. In contrast, at the other QTLs, the cultivated zombi pea alleles had the opposite effect on seed yield. The qSdnppd3.1− region covered about 221.6 kb on cowpea chromosome 3 and comprised five genes, including Vigun03g187300, which encodes abscisic acid (ABA)-insensitive 5-like protein 6. The qSdnppd5.1− and qSdnppd7.2+ regions spanned approximately 860.4 kb and 2.38 Mb on cowpea chromosomes 5 and 7 and contained 41 and 60 genes, respectively. Moreover, Vigun05g126900, encoding the MALE STERILE 5 protein, was identified as a candidate gene for qSdnppd5.1− (Supplementary Table S3). Both Vigun07g094201 and Vigun07g096300, encoding the B3-like transcription factor and the B3 DNA-binding domain protein, respectively, were selected as candidate genes for qSdnppd7.2+ (Supplementary Table S3). An analysis of qSdnppd7.1−, revealed only one flanking marker (Marker225965) on cowpea chromosome 7. Furthermore, Vigun07g074900, encoding the transcription factor TCP21, and located approximately 81.6 kb from Marker225965, was identified as a candidate gene for qSdnppd7.1−.
Discussion
Compared with wild zombi pea, cultivated zombi pea has undergone phenotypic changes due to domestication. In this study, we identified 62 QTLs controlling 13 domestication-related traits in zombi pea. The QTLs were distributed in all LGs, except for LG10. The number of QTLs identified for individual traits varied from one to ten, with an average of five QTLs (Table 3). Large-effect QTLs (PVE > 15%) were detected for all examined traits, except for STL, PDDM, and BRNPP (Table 3). These results imply that the zombi pea domestication-related traits are controlled by one or only a few major QTLs along with a small number of minor QTLs. This is consistent with the results of earlier investigations of other Vigna species, including mung bean (Isemura et al., 2012), azuki bean (Isemura et al., 2007), rice bean (Isemura et al., 2010), and cowpea (Kongjaimun et al., 2012; Lo et al., 2018). However, in a recent study, Dachapak et al. (2018) identified an average of 2.3 QTLs for zombi pea domestication-related traits, which is about twofold less than the number of QTLs detected in our study. This difference may have been due to the sterility of the mapping population used in the earlier study.
In Vigna species, the major QTLs for domestication-related traits, especially organ size, have always been mapped to the same LG (Isemura et al., 2007, 2010, 2012; Kongjaimun et al., 2012; Lo et al., 2018; Yundaeng et al., 2019). In our study, the major QTLs for the domestication-related traits of zombi pea, including SD100WT, SDL, PDL, STL, BRNPP, FLD, PDDM, and SDNPPD, were clustered in LG5, corresponding to cowpea chromosome 5. In an earlier study of cowpea, which is the legume crop most closely related to zombi pea, the major QTLs for domestication-related traits, such as pod shattering, seed dormancy, seed size (weight, width, and length), pod size (length and width), stem size (thickness and length), and leaf size (width and length), were clustered in LG7 (Kongjaimun et al., 2012), which corresponds to chromosome 5. These findings suggest that the genetics underlying the effects of domestication on the organ size (e.g., seed size, pod length, and stem length) of these two African Vigna crops may be the same. However, the genetic basis of seed dormancy and pod shattering, which are key domestication-related traits, varies between these two species.
Domestication results in a decrease in or a loss of seed dormancy, enabling the uniform germination of seeds, which is relevant for agricultural production. The seed dormancy of cultivated zombi pea is suppressed. In this study, we identified one major and two minor QTLs for the seed dormancy-related trait (Table 3). Specifically, qSdwa3.1− was detected as the major QTL, accounting for 22.2% of the phenotypic variation. Because the wild alleles at qSdwa3.1− positively affected SDWA (i.e., decreased dormancy), their introgression into cultivated zombi pea may be useful for decreasing seed dormancy. The Vigun03g339300 gene, encoding a calmodulin-binding protein-like protein, was identified at qSdwa3.1−. Calmodulin (CaM) and CaM-like (CML) proteins are the major Ca2+-binding proteins. The CaM/CML signals are involved in the ABA-induced inhibition of seed germination and seedling growth (Zhou et al., 2018). In Arabidopsis thaliana (Arabidopsis), IQM4, which is a CaM-binding protein, was recently reported to affect seed dormancy by modulating ABA biosynthesis and ABA signaling during seed maturation and germination (Zhou et al., 2018). In cowpea, Sdp1.1+ is the major QTL related to seed dormancy, explaining about 40% of the trait variation (Kongjaimun et al., 2012). The markers flanking Sdp1.1+ suggest that this QTL is located on chromosome 3 between positions 45,527,961 and 51,968,469. Thus, qSdwa3.1− and Sdp1.1+ are different loci. The minor QTL qSdwa5.1+ accounted for about 9% of the phenotypic variation, and included Vigun05g170600 and Vigun05g170700, both of which encode gibberellin 2-beta-dioxygenase 1 (GA2OX1). Gibberellin is a key plant hormone that regulates seed dormancy and germination by antagonistically suppressing ABA-triggered seed dormancy (Shu et al., 2016). Loss-of-function mutations to GA2ox2, GA2ox7, and GA2ox8, which encode GA2OX, reportedly enhance seed germination in Arabidopsis (Schomburg et al., 2003; Yamauchi et al., 2007).
Preventing or limiting pod dehiscence (pod shattering) may minimize pre-harvest yield losses, resulting in a more efficient harvest. Thus, pod indehiscence may be an advantageous trait during the harvesting of seeds, making it an important consideration during crop domestication. A major QTL in LG11 and a minor QTL in LG5 were related to pod twisting (Table 3). The major QTL, qPdt11.1+, explained 17.6% of the dehiscence variation, and included two genes, Vigun11g053700, encoding UGE1, and Vigun11g054200, encoding the glycosyltransferase GT14. Earlier studies confirmed that UGE is a nucleotide sugar interconversion enzyme that mediates the interconversion of UDP-D-glucose and UDP-D-galactose and contributes to cell wall biosynthesis (Barber et al., 2006; Rösti et al., 2007). Glycosyltransferases in plants are mainly involved in the biosynthesis of cell wall polysaccharides and glycoproteins (Hansen et al., 2012). Although the functions of plant GT14 remain uncharacterized, two GT14 genes identified in a hybrid aspen species (Populus tremula × P. tremuloides) reportedly exhibit xylem-specific expression (Aspeborg et al., 2005). Accordingly, GT14 may participate in secondary cell wall biosynthesis. In cowpea, three major QTLs for pod shattering have been identified (Kongjaimun et al., 2012; Suanum et al., 2016; Lo et al., 2018). Candidate genes for these QTLs include Vigun02g095200 (cellulose synthase), Vigun03g306000 (NAC domain transcription factor), Vigun03g302600 (C2H2 zinc finger protein), Vigun05g262100 (MYB46 transcription factor), Vigun05g273500 (MYB26 transcription factor), and Vigun05g266600 (beta glucosidase) (Suanum et al., 2016; Lo et al., 2018; Takahashi et al., 2019; Watcharatpong et al., 2020). Thus, the major QTLs/genes which control pod shattering differ between zombi pea and cowpea.
In most cases, domestication increases organ sizes, especially the harvested or consumed or exploited organs. Seed size is a key determinant of seed yield in legume crops as well as evolutionary fitness. It is a polygenic trait affected by various cellular processes. Twenty-two QTLs were identified for seed size-related traits, of which the QTLs with the largest effects on seed weight (qSd100wt5.1+) and seed length (qSdl5.1+) were mapped to the same position of LG5 (Table 3). Two genes were identified at these two QTLs, namely Vigun05g139100, encoding an auxin-responsive protein-related protein, and Vigun05g140300, encoding DUF538. Auxin is important for seed development, with specific effects on the embryo, endosperm, and seed coat, all of which influence seed size and yield (Cao et al., 2020). Genes encoding auxin-related proteins control seed size (Schruff et al., 2006; Ishimaru et al., 2013; Liu et al., 2015; Sun et al., 2017). In Arabidopsis mutant line F08314, the overexpression of RAFL06-12-J05 (encoding DUF538) results in seeds that are larger than the wild-type seeds (Akiyama et al., 2014)3. In addition to qSd100wt5.1+, qSd100wt5.2+ also substantially affects seed weight. We identified Vigun05g277600, which encodes an IAA-amino acid hydrolase ILR1-like 1-related protein, as a candidate gene for qSd100wt5.2+. Earlier studies revealed ILR1 is involved in auxin conjugation and affects auxin homeostasis (Rampey et al., 2004; and see Cao et al., 2020 for review). In rice, TGW3, encoding an IAA-glucose hydrolase, controls the rice grain weight and yield (Ishimaru et al., 2013). The QTL with the largest effect on seed length, qSdl9.1+, was included in LG9. Both Vigun09g215300 (WRKY transcription factor 40-related protein) and Vigun09g215400 (WRKY DNA-binding protein) were detected as candidate genes for qSdl9.1+. In plants, WRKY proteins have diverse biological functions related to seed development as well as developmental and hormone-controlled processes (Bakshi and Oelmüller, 2014). For example, MINISEED3, which encodes WRKY10, regulates seed size in Arabidopsis (Luo et al., 2005) and Loose Panicle1, which encodes a WRKY transcription factor, regulates seed size by increasing the seed length and width in foxtail millet [Setaria italica (L.) P. Beauv.] (Xiang et al., 2017). Additionally, SoyWRKY15a, which encodes WRKY15, is associated with changes to seed size due to the domestication of soybean [Glycine max (L.) Merr.], with expression levels that vary significantly between domesticated and wild soybeans (Gu et al., 2017).
In the current study, qLfa9.1−, with a PVE of 18.2%, was the only QTL identified for leaf size (Table 3). The Vigun09g018300 gene (WD40 repeat-containing protein) was detected as a candidate gene for this QTL. In cucumber, LITTLELEAF, which encodes a WD40 repeat domain−containing protein, regulates the size of various organs, including leaves (Yang et al., 2018). We also identified Vigun09g017000, encoding the bromodomain-containing protein GTE1, as a candidate gene. In Arabidopsis, GTE4 and GTE6 encode bromodomain-containing proteins that regulate leaf shape (and thus leaf size). Specifically, GTE4 controls the mitotic cell cycle during plant development (Airoldi et al., 2010), whereas GTE6 induces the expression of ASYMMETRIC LEAVES1, encoding a MYB-domain protein that regulates proximodistal leaf patterns (Chua et al., 2005).
Legume domestication results in increased pod length. Five QTLs were identified for pod length, but qPdl5.1− was the only major QTL, accounting for nearly 38% of the trait variation (Table 3). Pod elongation requires changes to fiber cell structures and components. The Vigun05g125800 gene, encoding pectate lyase 1, was selected as a candidate gene for qPdl5.1−. Pectin is a major polymer in the plant primary cell wall. Pectate lyase is a pectin-degrading enzyme that can degrade demethylesterified homogalacturonan (HG), which is a pectic polymer (Palusa et al., 2007). The methylesterification status of HG can regulate cellular growth and cell shape, thereby influencing plant growth and development (Wolf et al., 2009; Peaucelle et al., 2011). A previous study regarding common bean (Phaseolus vulgaris L.), which is a legume closely related to zombi pea, revealed that the major changes in cell wall polysaccharides during pod development involve pectic polymers and that the deposition of galactose-rich pectic polymers in cell walls increases in elongating pods (Stolle-Smits et al., 1999). Moreover, during pod elongation, the HG level steadily increases. Accordingly, pectate lyase may help mediate the elongation of zombi pea pods. In rice, a mutation to DEL1, which encodes a pectate lyase precursor, decreases the total pectate lyase activity, leading to an increase in the methylesterified HG content and altered cell wall compositions and structures, with implications for plant height, root length, flag leaf length, grain size, and panicle and internode lengths. In cotton, the downregulated expression of pectate lyase genes GhPEL48_Dt and GhPEL76 decreases fiber length by impeding pectin degradation (Wang et al., 2009; Sun et al., 2020).
Most domesticated legume crops exhibit greater determinate growth, with less branching, than their wild ancestors. The qStl9.1− QTL explained about 10% of the variability in the stem length (plant height) (Table 3). We identified Vigun09g037900, encoding MOB1, as a candidate gene for this QTL. Arabidopsis MOB1 proteins are critical for plant development because they promote auxin signaling (Cui et al., 2016) and regulate jasmonate accumulation (Guo et al., 2020). Mutations to the MOB1 gene in Arabidopsis negatively affect plant height (Xiong et al., 2016; Guo et al., 2020). We identified Vigun05g082200, encoding an auxin efflux carrier protein, as a candidate gene for qBrnpp5.1−, which controls the number of branches in zombi pea. The Vigun05g082200 sequence is similar to that of the PIN-LIKES 6 transporter (AT5G01990). The PIN proteins form a family of auxin efflux carriers. Auxin is well known for its ability to regulate numerous plant growth and developmental processes such as meristem formation and lateral organ formation and patterning (Wang and Jiao, 2018). For example, PIN6 contributes to auxin signaling-mediated developmental processes, including lateral/adventitious root organogenesis, primary/lateral root development and growth, and shoot apical dominance (Cazzonelli et al., 2013; Nisar et al., 2014; Simon et al., 2016). In rice, PIN1 and PIN2 are involved in tillering (Xu et al., 2005; Chen et al., 2012).
Generally, wild ancestors of domesticated crops are sensitive to short-day conditions. In this study, we identified four QTLs related to days-to-flowering in zombi pea, of which two are large-effect QTLs, qFld2.1− and qFld5.1− (PVE of 16.5 and 18.6%, respectively). We selected Vigun02g051100 (WRKY57 transcription factor) and Vigun02g052800 (RING finger and CCCH-type zinc finger protein) as candidate genes for qFLD2.1− and qFLD5.1−, respectively. The WRKY transcription factors are involved in physiological changes and responses to biotic and abiotic stresses. In Arabidopsis, WRKY12, WRKY13, WRKY71, and WRKY75 regulate flowering time (Li W. et al., 2016; Yu et al., 2016; Zhang et al., 2018). Additionally, WRKY75 interacts with DELLA proteins, which are the major components involved in gibberellic acid perception and signaling (Zhang et al., 2018). A mutation to WRKY75 reportedly delays flowering, whereas the overexpression of this gene has the opposite effect (Zhang et al., 2018). Plant zinc finger proteins comprise a large protein family and have diverse functions related to plant development and resistance to biotic and abiotic stresses. The Arabidopsis genes AtC3H17, AtZFP1, AtKHZ1, and AtKHZ2 as well as the Medicago sativa gene MsZFN, encoding CCCH zinc finger proteins, influence flowering time (Chao et al., 2014; Seok et al., 2016; Yan et al., 2017; Wang et al., 2018).
Four QTLs were detected for days-to-flowering in zombi pea, of which qPddm2.1− and qPddm5.1− had a similar PVE of about 10% (Table 3). We identified Vigun02g050200, encoding the 14-3-3-like protein GF14 IOTA, and Vigun05g050300, encoding 3-deoxy-8-phosphooctulonate synthase/phospho-2-keto-3-deoxyoctonate aldolase, as the candidate genes for qPddm2.1− and qPddm5.1−, respectively. The 14-3-3 (GF14) proteins are critical components of various cellular signaling processes and are important regulators of several physiological processes in plants. The GF14 genes are associated with fruit development and/or ripening in banana (Musa acuminata L.) (Li et al., 2012; Li M. et al., 2016) and pear (Pyrus pyrifolia Nakai.) (Shi and Zhang, 2014). Phospho-2-keto-3-deoxyoctonate aldolase is also known as 3-deoxy-d-manno-2-octulosonic acid-8-phosphate synthase (KDO8P), which has a key function related to the biosynthesis of KDO, which is a sugar present only in the rhamnogalacturonan II pectic fraction of the primary cell walls of higher plants (Delmas et al., 2003). In the common bean, which is a legume closely related to zombi pea, pod elongation and maturation are accompanied by changes to rhamnogalacturonan (Stolle-Smits et al., 1999).
The number of seeds per pod is one of the yield determinants of legumes. Some domesticated legumes have more seeds per pod than their wild ancestors. Up to nine QTLs were detected for this trait in zombi pea (Table 3). Among these QTLs, qSdnppd3.1−, qSdnppd5.1−, qSdnppd7.1−, and qSdnppd7.2+ had relatively large effects (PVE between 15 and 33%). Because the wild alleles at qSdnppd3.1−, qSdnppd5.1−, and qSdnppd7.1− were observed to increase the number of seeds per pod, they may be useful for increasing the seed yield of cultivated zombi pea. We identified Vigun03g187300 (ABA-insensitive 5-like protein 6) as a candidate gene for qSdnppd3.1−. The encoded protein is an ABA-responsive element (ABRE)-binding factor that regulates ABRE-dependent gene expression (Nakashima and Yamaguchi-Shinozaki, 2013). In Arabidopsis, an ABA-deficiency reportedly decreases the number of seeds per silique (Cheng et al., 2014). The Vigun05g126900 gene, encoding MALE STERILE 5, was selected as a candidate gene for qSdnppd5.1−. In an earlier investigation on Arabidopsis, mutations to MS5, which encodes MALE STERILE 5, resulted in the formation of “polyads” (i.e., tetrads with more than four pools of chromosomes following male meiosis) (Glover et al., 1998). Plants that are homozygous for the ms5 recessive allele reportedly exhibit stunted growth and produce empty siliques, whereas in plants that are heterozygous for MS5, silique elongation and seed set are less inhibited (Glover et al., 1998). We identified Vigun07g094201 (B3-like transcription factor) and Vigun07g096300 (B3 DNA-binding domain protein) as the candidate genes for qSdnppd7.1−. The B3 domain family proteins are associated with ovule development (Skinner and Gasser, 2009). A recent study on Arabidopsis investigated the roles of three Reproductive Meristem (REM) genes, REM34, REM35, and REM36, which encode B3 domain family transcription factors affecting female and male gametophyte development (Caselli et al., 2019). This previous study proved that the silencing of one or more of these genes via RNA interference increases the number of unfertilized ovules, thereby decreasing the number of seeds per silique. Additionally, the Vigun07g094201 sequence is highly similar to that of Arabidopsis REM39 (AT3G18990). In the current study, Vigun07g074900, which encodes the transcription factor TCP21, was selected as a candidate gene for qSdnppd7.2+. In Arabidopsis, several TCP transcription factors, including TCP21, are involved in ovule development via their interactions with SPOROCYTELESS (Chen et al., 2014; Wei et al., 2015), which is required for sporogenesis in both male and female organs (Yang et al., 1999). An earlier study involving cowpea identified a gene encoding TCP5 as a candidate gene for the number of seeds per pod (Lo et al., 2018).
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
PS and LW conceptualized the study and secured research funding. PS designed and supervised the study and revised the manuscript. KA and KL developed population and conducted data collection and analysis. PS and KA wrote the manuscript. All authors read and approved the accepted manuscript.
Funding
This research was funded by the Thailand Research Fund (Grant No. RSA5880051), China Agriculture Research System (CARS-08), and the Agricultural Science and Technology Innovation Program (ASTIP) of Chinese Academy of Agricultural Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to the Joint Legume Research Center between Chinese Academy of Agriculture Sciences and Kasetsart University for molecular lab facilities. We thank Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac) for editing the English text of a draft of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00803/full#supplementary-material
TABLE S1 | Number of markers, marker length, and distance between markers in each linkage group of the zombi pea genetic map used for detecting the QTLs of domestication-related traits in the F2 population derived from a cross between TVNu 240 and TVNu 1623. The markers are SNPs generated by specific locus amplified fragment sequencing. The map was constructed by Amkul et al. (2019).
TABLE S2 | Correlations among domestication-related traits in the zombi pea F2 population derived from a cross between TVNu 240 and TVNu 1623.
TABLE S3 | Candidate genes for each QTL.
Footnotes
- ^ http://www.isbreeding.net
- ^ https://phytozome-next.jgi.doe.gov/info/Vunguiculata_v1_2
- ^ http://rarge-v2.psc.riken.jp/cdna/RAFL06-12-J05
References
Airoldi, C. A., Rovere, F. D., Falasca, G., Marino, G., Kooiker, M., Altamura, M. M., et al. (2010). The Arabidopsis BET bromodomain factor GTE4 is involved in maintenance of the mitotic cell cycle during plant development. Plant Physiol. 152, 1320–1334. doi: 10.1104/pp.109.150631
Akiyama, K., Kurotani, A., Iida, K., Kuromori, T., Shinozaki, K., and Sakurai, T. (2014). RARGE II: an integrated phenotype database of Arabidopsis mutant traits using a controlled vocabulary. Plant Cell Physiol. 55:e4. doi: 10.1093/pcp/pct165
Amkul, K., Wang, L., Somta, P., Wang, S., and Cheng, X. (2019). Construction of a high density linkage map and genome dissection of bruchid resistance in zombi pea (Vigna vexillata (L.) A. Rich). Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-48239-5
Aspeborg, H., Schrader, J., Coutinho, P. M., Stam, M., Kallas, A., Djerbi, S., et al. (2005). Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol. 137, 983–997. doi: 10.1104/pp.104.055087
Bakshi, M., and Oelmüller, R. (2014). WRKY transcription factors: jack of many trades in plants. Plant Signal Behav. 9:e27700. doi: 10.4161/psb.27700
Barber, C., Rösti, J., Rawat, A., Findlay, K., Roberts, K., and Seifert, G. J. (2006). Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana. J. Biol. Chem 281, 17276–17285. doi: 10.1074/jbc.M512727200
Cao, J., Li, G., Qu, D., Li, X., and Wang, Y. (2020). Into the seed: auxin controls seed development and grain yield. Int. J. Mol. Sci. 21:1662. doi: 10.3390/ijms21051662
Caselli, F., Beretta, V. M., Mantegazza, O., Petrella, R., Leo, G., Guazzotti, A., et al. (2019). REM34 and REM35 control female and male gametophyte development in Arabidopsis thaliana. Front. Plant Sci. 10:1351. doi: 10.3389/fpls.2019.01351
Cazzonelli, C. I., Vanstraelen, M., Simon, S., Yin, K., Carron-Arthur, A., Nisar, N., et al. (2013). Role of the Arabidopsis PIN6 auxin transporter in auxin homeostasis and auxin-mediated development. PLoS One 8:e70069. doi: 10.1371/journal.pone.0070069
Chaitieng, B., Kaga, A., Tomooka, N., Isemura, T., Huroda, Y., and Vaughan, D. A. (2006). Development of black gram [Vigna mungo (L.) Hepper] linkage map and its comparison with an azuki bean [Vigna angularis (Wild.) Ohwi and Ohashi] linkage map. Theor Appl Genet. 13, 1261–1269. doi: 10.1007/s00122-006-0380-5
Chao, Y., Zhang, T., Yang, Q., Kang, J., Sun, Y., Gruber, M. Y., et al. (2014). Expression of the alfalfa CCCH-type zinc finger protein gene MsZFN delays flowering time in transgenic Arabidopsis thaliana. Plant Sci. 215, 92–99. doi: 10.1016/j.plantsci.2013.10.012
Chen, G. H., Sun, J. Y., Liu, M., Liu, J., and Yang, W. C. (2014). SPOROCYTELESS is a novel embryophyte-specific transcription repressor that interacts with TPL and TCP proteins in Arabidopsis. J. Genet. Genomics 41, 617–625. doi: 10.1016/j.jgg.2014.08.009
Chen, Y., Fan, X., Song, W., Zhang, Y., and Xu, G. (2012). Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 10, 139–149. doi: 10.1111/j.1467-7652.2011.00637.x
Cheng, Z. J., Zhao, X. Y., Shao, X. X., Wang, F., Zhou, C., Liu, Y. G., et al. (2014). Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26, 1053–1068. doi: 10.1105/tpc.113.121566
Chua, Y. L., Channelière, S., Mott, E., and Gray, J. C. (2005). The bromodomain protein GTE6 controls leaf development in Arabidopsis by histone acetylation at ASYMMETRIC LEAVES1. Genes Dev. 19, 2245–2254. doi: 10.1101/gad.352005
Cui, X., Guo, Z., Song, L., Wang, Y., and Cheng, Y. (2016). NCP1/AtMOB1A plays key roles in auxin-mediated Arabidopsis development. PLoS Genet. 12:e1005923. doi: 10.1371/journal.pgen.1005923
Dachapak, S., Somta, P., Naito, K., Tomooka, N., Kaga, A., and Srinives, P. (2019). Detection of quantitative trait loci for salt tolerance in zombi pea [Vigna vexillata (L.) A. Rich]. Euphytica 215:208. doi: 10.1007/s10681-019-2530-2
Dachapak, S., Somta, P., Poonchaivilaisak, S., Yimram, T., and Srinives, P. (2017). Genetic diversity and structure of the zombi pea (Vigna vexillata (L.) A. Rich) gene pool based on SSR marker analysis. Genetica 145, 189–200. doi: 10.1007/s10709-017-9957-y
Dachapak, S., Tomooka, N., Somta, P., Naito, K., Kaga, A., and Srinives, P. (2018). QTL analysis of domestication syndrome in zombi pea (Vigna vexillata), an underutilized legume crop. PLoS One 13:e0200116. doi: 10.1371/journal.pone.0200116
Delmas, F., Petit, J., Joubès, J., Séveno, M., Paccalet, T., Hernould, M., et al. (2003). The gene expression and enzyme activity of plant 3-deoxy-D-manno-2-octulosonic acid-8-phosphate synthase are preferentially associated with cell division in a cell cycle-dependent manner. Plant Physiol. 133, 348–360. doi: 10.1104/pp.103.026872
Doebley, J. F., Gaut, B. S., and Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi: 10.1016/j.cell.2006.12.006
Ferguson, H. (1954). The food crops of the SUDAN and Their relation to environment. Paper Presented at the 1953 Conference of the Philosophical Society of the Sudan on “Food and Society in the Sudan”, Philosophical Society of the Sudan Khartoum. London: McCorquodale and Co. (Sudan), Ltd.
Glover, J., Grelon, M., Craig, S., Chaudhury, A., and Dennis, E. (1998). Cloning and characterization of MS5 from Arabidopsis: a gene critical in male meiosis. Plant J. 15, 345–356. doi: 10.1046/j.1365-313x.1998.00216.x
Gu, Y., Li, W., Jiang, H., Wang, Y., Gao, H., Liu, M., et al. (2017). Differential expression of a WRKY gene between wild and cultivated soybeans correlates to seed size. J. Exp. Bot. 68, 2717–2729. doi: 10.1093/jxb/erx147
Guo, Z., Yue, X., Cui, X., Song, L., and Cheng, Y. (2020). AtMOB1 genes regulate jasmonate accumulation and plant development. Plant Physiol. 182, 1481–1493. doi: 10.1104/pp.19.01434
Hansen, S. F., Harholt, J., Oikawa, A., and Scheller, H. V. (2012). Plant glycosyltransferases beyond CAZy: a perspective on DUF families. Front. Plant Sci. 3:59. doi: 10.3389/fpls.2012.00059
Isemura, T., Kaga, A., Konishi, S., Ando, T., Tomooka, N., Han, O. K., et al. (2007). Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and their comparison with other warm season legumes. Ann. Bot. 100, 1053–1071. doi: 10.1093/aob/mcm155
Isemura, T., Kaga, A., Tabata, S., Somta, P., Srinives, P., Shimizu, T., et al. (2012). Construct of genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS One 7:e41304. doi: 10.1371/journal.pone.0041304
Isemura, T., Kaga, A., Tomooka, N., Shimizu, T., and Vaughan, D. A. (2010). The genetics of domestication of rice bean, Vigna umbellata. Ann. Bot. 106, 927–944. doi: 10.1093/aob/mcq188
Ishimaru, K., Hirotsu, N., Madoka, Y., Murakami, N., Hara, N., Onodera, H., et al. (2013). Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–711. doi: 10.1038/ng.2612
Karuniawan, A., Iswandi, A., Kale, P. R., Heinzemann, J., and Grüneberg, W. J. (2006). Vigna vexillata (L.) A. Rich. cultivated as a root crop in Bali and Timor. Genet Resour. Crop Evol. 53, 213–217. doi: 10.1007/s10722-005-1654-5
Kongjaimun, A., Kaga, A., Tomooka, N., Somta, P., Vaughan, D. A., and Srinives, P. (2012). The genetics of domestication of yardlong bean, Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. sesquipedalis. Ann. Bot. 109, 1185–2000. doi: 10.1093/aob/mcs048
Li, H., Ye, G., and Wang, J. (2007). A modified algorithm for the improvement of composite interval mapping. Genetics 175, 361–374. doi: 10.1534/genetics.106.066811
Li, M., Ren, L., Xu, B., Yang, X., Xia, Q., He, P., et al. (2016). Genome-wide identification, phylogeny, and expression analyses of the 14-3-3 family reveal their involvement in the development, ripening, and abiotic stress response in banana. Front. Plant Sci. 7:1442. doi: 10.3389/fpls.2016.01442
Li, M. Y., Xu, B. Y., Liu, J. H., Yang, X. L., Zhang, J. B., Jia, C. H., et al. (2012). Identification and expression analysis of four 14-3-3 genes during fruit ripening in banana (Musa acuminata L. AAA group, cv. Brazilian). Plant Cell Rep. 31, 369–378. doi: 10.1007/s00299-011-1172-1
Li, W., Wang, H., and Yu, D. (2016). Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol. Plant. 9, 1492–1503. doi: 10.1016/j.molp.2016.08.003
Liu, L., Tong, H., Xiao, Y., Che, R., Xu, F., Hu, B., et al. (2015). Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 11102–11107. doi: 10.1073/pnas.1512748112
Lo, S., Muñoz-Amatriaín, M., Boukar, O., Herniter, I., Cisse, N., Guo, Y.-N., et al. (2018). Identification of genetic factors controlling domestication-related traits in cowpea (Vigna unguiculata L. Walp). Sci. Rep. 19:6261. doi: 10.1038/s41598-018-24349-4
Lonardi, S., Muñoz-Amatriaín, M., Liang, Q., Shu, S., Wanamaker, S. I., Lo, S., et al. (2019). The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 98, 767–782. doi: 10.1111/tpj.14349
Luo, M., Dennis, E. S., Berger, F., Peacock, W. J., and Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 17531–17536.
Maxted, N., Mabuza-Diamini, P., Moss, H., Padulosi, S., Jarvis, A., and Guarino, L. (2004). An Ecogeographic Study: African Vigna, Systematic and Ecogeographic Studies of Crop Genepool. Rome: International Plant Genetic Resources Institutes.
Nakashima, K., and Yamaguchi-Shinozaki, K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Nisar, N., Cuttriss, A. J., Pogson, B. J., and Cazzonelli, C. I. (2014). The promoter of the Arabidopsis PIN6 auxin transporter enabled strong expression in the vasculature of roots, leaves, floral stems and reproductive organs. Plant Signal Behav. 9:e27898. doi: 10.4161/psb.27898
Palusa, S. G., Golovkin, M., Shin, S. B., Richardson, D. N., and Reddy, A. S. (2007). Organ-specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in Arabidopsis. New Phytol. 174, 537–550. doi: 10.1111/j.1469-8137.2007.02033.x
Peaucelle, A., Braybrook, S. A., Le, G. L., Bron, E., Kuhlemeier, C., and Höfte, H. (2011). Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 21, 1720–1726. doi: 10.1016/j.cub.2011.08.057
Rampey, R. A., LeClere, S., Kowalczyk, M., Ljung, K., Sandberg, G., and Bartel, B. (2004). A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135, 978–988. doi: 10.1104/pp.104.039677
Rösti, J., Barton, C. J., Albrecht, S., Dupree, P., Pauly, M., Findlay, K., et al. (2007). UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana. Plant Cell. 19, 1565–1579. doi: 10.1105/tpc.106.049619
Schomburg, F. M., Bizzell, C. M., Lee, D. L., Zeevaart, J. A. D., and Amasino, R. M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15, 151–163. doi: 10.1105/tpc.005975
Schruff, M. C., Spielman, M., Tiwari, S., Adams, S., Fenby, N., and Scott, R. J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251–261. doi: 10.1242/dev.02194
Seok, H.-Y., Woo, D.-H., Park, H.-Y., Lee, S.-Y., Tran, H. T., Lee, E.-H., et al. (2016). AtC3H17, a non-tandem CCCH zinc finger protein, functions as a nuclear transcriptional activator and has pleiotropic effects on vegetative development, flowering and seed development in Arabidopsis. Plant Cell Physiol. 57, 603–615. doi: 10.1093/pcp/pcw013
Shi, H., and Zhang, Y. (2014). Pear 14-3-3a gene (Pp14-3-3a) is regulated during fruit ripening and senescense, and involved in response to salicylic acid and ethylene signaling. J. Genet. 93, 747–753. doi: 10.1007/s12041-014-0447-z
Shu, K., Liu, X. D., Xie, Q., and He, Z. H. (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant. 9, 34–45. doi: 10.1016/j.molp.2015.08.010
Simon, S., Skùpa, P., Viaene, T., Zwiewka, M., Tejos, R., Klíma, P., et al. (2016). PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 211, 65–74. doi: 10.1111/nph.14019
Skinner, D. J., and Gasser, C. S. (2009). Expression-based discovery of candidate ovule development regulators through transcriptional profiling of ovule mutants. BMC Plant Biol. 9:29. doi: 10.1186/1471-2229-9-29
Stolle-Smits, T., Beekhuizen, J. G., Kok, M. T. C., Pijnenburg, M., Recourt, K., Derksen, J., et al. (1999). Changes in cell wall polysaccharides of green bean pods during development. Plant Physiol. 121, 363–372. doi: 10.1104/pp.121.2.363
Suanum, W., Somta, P., Kongjaimun, A., Yimram, T., Kaga, A., Tomooka, N., et al. (2016). Co-localization of QTLs for pod fiber content and pod shattering in F2 and backcross populations between yardlong bean and wild cowpea. Mol. Breed. 36:80.
Sun, H., Hao, P., Gu, L., Cheng, S., Wang, H., Wu, A., et al. (2020). Pectate lyase-like gene GhPEL76 regulates organ elongation in Arabidopsis and fiber elongation in cotton. Plant Sci. 293:110395. doi: 10.1016/j.plantsci.2019.110395
Sun, X., Liu, D., Zhang, X., Li, W., Liu, H., Hong, W., et al. (2013). SLAF-seq: an efcient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One 8:e58700. doi: 10.1371/journal.pone.0058700
Sun, Y., Wang, C., Wang, N., Jiang, X., Mao, H., Zhu, C., et al. (2017). Manipulation of auxin response factor 19 affects seed size in the woody perennial Jatropha curcas. Sci. Rep. 7:40844.
Takahashi, Y., Kongjaimun, A., Muto, C., Kobayashi, Y., Kumagai, M., Sakai, H., et al. (2019). Genetic factor for twisting legume pods identified by fine-mapping of shattering-related traits in azuki bean and yard-long bean. bioRxiv. [Preprint]. doi: 10.1101/774844
Vaughan, D. A., Balázs, E., and Heslop-Harrison, J. S. (2007). From crop domestication to super-domestication. Ann. Bot. 100, 893–901. doi: 10.1093/aob/mcm224
Wang, H., Guo, Y., Lv, F., Zhu, H., Wu, S., Jiang, Y., et al. (2009). The essential role of GhPEL gene, encoding a pectate lyase, in cell wall loosening by depolymerization of the de-esterified pectin during fiber elongation in cotton. Plant Mol. Biol. 72, 397–406. doi: 10.1007/s11103-009-9578-7
Wang, W., Zheng, H., Wang, Y., Han, G., and Sui, N. (2018). Overexpression of CCCH zinc finger protein gene delays flowering time and enhances salt tolerance in Arabidopsis by increasing fatty acid unsaturation. Acta Physiol. Plant 40:196. doi: 10.1007/s11738-018-2775-8
Wang, Y., and Jiao, Y. (2018). Auxin and above-ground meristems. J. Exp. Bot. 69, 147–154. doi: 10.1093/jxb/erx299
Watcharatpong, P., Kaga, A., Chen, X., and Somta, P. (2020). Narrowing down a major qtl region conferring pod fiber contents in yardlong bean (Vigna unguiculata), a vegetable cowpea. Genes 11:363. doi: 10.3390/genes11040363
Wei, B., Zhang, J., Pang, C., Yu, H., Guo, D., Jiang, H., et al. (2015). The molecular mechanism of sporocyteless/nozzle in controlling Arabidopsis ovule development. Cell Res. 25, 121–134. doi: 10.1038/cr.2014.145
Wolf, S., Mouille, G., and Pelloux, J. (2009). Homogalacturonan methyl-esterification and plant development. Mol. Plant. 2, 851–860. doi: 10.1093/mp/ssp066
Xiang, J., Tang, S., Zhi, H., Jia, G., Wang, H., and Diao, X. (2017). Loose Panicle1 encoding a novel WRKY transcription factor, regulates panicle development, stem elongation, and seed size in foxtail millet [Setaria italica (L.) P. Beauv.]. PLoS One 12:e0178730. doi: 10.1371/journal.pone.0178730
Xiong, J., Cui, X., Yuan, X., Yu, X., Sun, J., and Gong, Q. (2016). The Hippo/STE20 homolog SIK1 interacts with MOB1 to regulate cell proliferation and cell expansion in Arabidopsis. J. Exp. Bot. 67, 1461–1475. doi: 10.1093/jxb/erv538
Xu, M., Zhu, L., Shou, H., and Wu, P. (2005). A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46, 1674–1681. doi: 10.1093/pcp/pci183
Yamauchi, Y., Takeda-Kamiya, N., Hanada, A., Ogawa, M., Kuwahara, A., Seo, M., et al. (2007). Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 486, 555–561. doi: 10.1093/pcp/pcm023
Yan, Z., Jia, J., Yan, X., Shi, H., and Han, Y. (2017). Arabidopsis KHZ1 and KHZ2, two novel non-tandem CCCH zinc-finger and K-homolog domain proteins, have redundant roles in the regulation of flowering and senescence. Plant Mol Biol. 95, 549–565. doi: 10.1007/s11103-017-0667-8
Yang, L., Liu, H., Zhao, J., Pan, Y., Cheng, S., Lietzow, C. D., et al. (2018). LITTLELEAF (LL) encodes a WD40 repeat domain-containing protein associated with organ size variation in cucumber. Plant J. 95, 834–847. doi: 10.1111/tpj.13991
Yang, W. C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13, 2108–2117. doi: 10.1101/gad.13.16.2108
Yu, Y., Liu, Z., Wang, L., Kim, S. G., Seo, P. J., et al. (2016). WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 85, 96–106. doi: 10.1111/tpj.13092
Yundaeng, C., Somta, P., Amkul, K., Kongjaimun, A., Kaga, A., and Tomooka, N. (2019). Construction of genetic linkage map and genome dissection of domestication-related traits of moth bean (Vigna aconitifolia), a legume crop of arid areas. Mol. Genet. Genomics 294, 621–635. doi: 10.1007/s00438-019-01536-0
Zhang, L., Chen, L., and Yu, D. (2018). Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 176, 790–803. doi: 10.1104/pp.17.00657
Keywords: zombi pea, Vigna vexillata, domestication syndrome, QTL, legume
Citation: Amkul K, Somta P, Laosatit K and Wang L (2020) Identification of QTLs for Domestication-Related Traits in Zombi Pea [Vigna vexillata (L.) A. Rich], a Lost Crop of Africa. Front. Genet. 11:803. doi: 10.3389/fgene.2020.00803
Received: 29 April 2020; Accepted: 06 July 2020;
Published: 18 September 2020.
Edited by:
Gaofeng Zhou, Department of Primary Industries and Regional Development of Western Australia (DPIRD), AustraliaReviewed by:
Pei Xu, China Jiliang University, ChinaLongbiao Guo, Chinese Academy of Agricultural Sciences, China
Copyright © 2020 Amkul, Somta, Laosatit and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prakit Somta, YWdycGtzQGt1LmFjLnRo; cGtzb210YUBnbWFpbC5jb20=; Lixia Wang, d2FuZ2xpeGlhMDNAY2Fhcy5jbg==
 Kitiya Amkul1
Kitiya Amkul1 Prakit Somta
Prakit Somta Lixia Wang
Lixia Wang