- State Key Laboratory of Cotton Biology, Key Laboratory of Plant Stress Biology, School of Life Sciences, Henan University, Kaifeng, China
Respiratory burst oxidase homologs (Rbohs) play a predominant role in reactive oxygen species (ROS) production, which is crucial in plant growth, differentiation, as well as their responses to biotic and abiotic stresses. To date, however, there is little knowledge about the function of cotton Rboh genes. Here, we identified a total of 87 Rbohs from five sequenced Gossypium species (the diploids Gossypium arboreum, Gossypium raimondii, and Gossypium australe, and the allotetraploids Gossypium hirsutum and Gossypium barbadense) via BLAST searching their genomes. Phylogenetic analysis of the putative 87 cotton Rbohs revealed that they were divided into seven clades. All members within the same clade are generally similar to each other in terms of gene structure and conserved domain arrangement. In G. barbadense, the expression levels of GbRbohs in the CladeD were induced in response to a fungal pathogen and to hormones (i.e., jasmonic acid and abscisic acid), based upon which the main functional member in CladeD was discerned to be GbRboh5/18. Further functional and physiological analyses showed that the knock-down of GbRboh5/18 expression attenuates plant resistance to Verticillium dahliae infection. Combined with the molecular and biochemical analyses, we found less ROS accumulation in GbRboh5/18-VIGS plants than in control plants after V. dahliae infection. Overexpression of GbRboh5/18 in G. barbadense resulted in more ROS accumulation than in control plants. These results suggest that GbRboh5/18 enhances the cotton plants’ resistance against V. dahliae by elevating the levels of ROS accumulation. By integrating phylogenetic, molecular, and biochemical approaches, this comprehensive study provides a detailed overview of the number, phylogeny, and evolution of the Rboh gene family from five sequenced Gossypium species, as well as elucidating the function of GbRboh5/18 for plant resistance against V. dahliae. This study sheds fresh light on the molecular evolutionary properties and function of Rboh genes in cotton, and provides a reference for improving cotton’s responses to the pathogen V. dahliae.
Introduction
Reactive oxygen species (ROS) play dual roles in plant growth, and development, and in their responses to biotic and abiotic stresses (Torres and Dangl, 2005; Suzuki et al., 2011). Hydrogen peroxide (H2O2), the most stable species of ROS, can serve as a secondary signaling molecule for transduction in the cytoplasmic at a low basal level. However, because of the damage they cause to plant cell’s DNA, proteins, and lipids, ROS species are toxic when the accumulation exceeds a certain threshold, which can occur for the superoxide anion radical (O2–), hydroxyl radical (⋅OH), and H2O2 (Baxter et al., 2014).
Respiratory burst oxidase homologs (Rbohs), which are plant-specific NADPH oxidases, are key enzymes that move electrons from cytoplasmic NADPH to O2 and thus foster the generation of O2–, followed by the dismutation of O2– into H2O2 (Torres et al., 1998; Sagi and Fluhr, 2006). The first plant Rboh gene was found in rice, named OsRbohA, for which the mammalian homolog is gp91phox/Nox2 (Keller et al., 1998). Then more Rbohs from plants were successively identified from model plants (including Arabidopsis and tobacco) and crops, such as tomato, potato, and cucumber (Torres et al., 1998; Yoshioka et al., 2003; Sagi et al., 2004; Kobayashi et al., 2006; Xia et al., 2009; Yu et al., 2019). Generally, plant Rboh proteins are best described as integral plasma membrane proteins containing six transmembrane (TM) domains. The heme groups usually bind two pairs of histidine residues, respectively located in third and fifth TMs, to facilitate electron transport. Two Ca2+-binding EF-hand motifs and phosphorylation sites in the cytological N-terminal region participate in regulating enzymatic activity. Similar to the mammalian gp91phox, there are flavin adenine dinucleotide (FAD) and NADPH-binding domains on the C-terminus of Rboh proteins in plants (Wong et al., 2007).
Ten and nine Rbohs have been identified from Arabidopsis and rice, referred to as AtRbohA–AtRbohJ and OsRbohA–OsRbohI, respectively (Torres et al., 1998). Previous studies suggest they have distinct spatio-temporal expression, enzyme activity, and functional diversification (Lin et al., 2009; Kaur and Pati, 2018; Kaur et al., 2018; Kaya et al., 2019). AtrbohB fulfills important roles in the germination of Arabidopsis seeds through variant splicing transcripts (Muller et al., 2009). AtRbohD and AtRbohF present a different spatial expression pattern either in shoot or in root during Arabidopsis development (Jiao et al., 2013; Morales et al., 2016). Finally, AtRbohC, -D, -F, -H, and -J are synergistically activated by Ca2+-binding and then phosphorylated to produce ROS (Ogasawara et al., 2008; Han et al., 2019; Kaya et al., 2019).
Previous studies have proven that Rbohs figure predominantly in ROS production during pathogen-associated molecular pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) responses in Arabidopsis (Torres and Dangl, 2005; Nuhse et al., 2007; Kadota et al., 2014; Li et al., 2014b). Perhaps the best example is RbohD (and RbohF in some cases), a key player not only in intracellular oxidative stresses but also in plant responses and resistance to pathogens (Yun et al., 2011; Chaouch et al., 2012; Ma et al., 2016; Angelos and Brandizzi, 2018). AtRbohD showed the highest expression of the 10 AtRbohs: it functions via ROS production in abscisic acid-induced stomatal closure, flagellin-induced immune responses (Pogany et al., 2009), resistance to Botrytis cinereal (Galletti et al., 2008), as well as phosphorylation mediated by CRK2 in plant defense against the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Kimura et al., 2020). AtRbohD-dependent H2O2 production was positively regulated by H2Bub1 in the responses against Vd-toxins in Arabidopsis (Zhao et al., 2020). DORN-mediated phosphorylation on the N-terminus of RbohD led to ROS accumulation and stomatal closure of Arabidopsis in biotic stresses (Chen et al., 2017). NbrbohB participates in H2O2 accumulation, resulting in elicitor-induced stomatal closure, but it does not function in elicitor-induced HR in Nicotiana benthamiana (Yoshioka et al., 2003; Kobayashi et al., 2007; Zhang et al., 2009). Ethylene signaling can regulate ROS production and enhance disease resistance, in which both are mediated by OsEIL2 binding to the promoter of OsrbohA and OsrbohB in rice (Yang et al., 2017).
Cotton (Gossypium spp.) is a globally important fiber crop that is the largest source of natural renewable textile fibers. The allotetraploids Gossypium hirsutum and Gossypium barbadense are cultivated worldwide because of their high yield and superior fiber quality, respectively (Wendel, 1989; Paterson et al., 2012; Wang et al., 2019). Both G. hirsutum and G. barbadense originated from two diploid progenitor species: Gossypium arboreum (A-genome donor) and Gossypium raimondii (D-genome donor) ca. 1–2 million years ago (Wendel, 1989). Because of their tetraploidy and repeat sequences, G. hirsutum and G. barbadense have large genomes; hence, their study from a genetics and genomics perspective is quite difficult (Paterson et al., 2012; Li et al., 2014a, 2015; Zhang et al., 2015). Moreover, the wild diploid cotton species Gossypium australe possesses excellent traits for plant resistance to disease, and its genome was fully sequenced and assembled last year (Cai et al., 2019). Nevertheless, with recent advances in sequencing technology and high-quality genome assemblies and annotations, it is much easier now to pursue detailed analyses of plant genomics (Wang et al., 2019).
Verticillium wilt, caused by the soil-born fungus Verticillium dahliae Kleb., is a serious widespread disease that can severely affect plant growth and fiber quality in cotton production fields (Shaban et al., 2018; Wang et al., 2018). The disease symptoms of V. dahliae infection consists of necrotic areas on leaves, yellowing of leaves, wilting and defoliation, and discoloration of vascular tissues (Li et al., 2017). Currently, verticillium wilt has yet to be controlled effectively by either genetic means or fungicide applications, so analyzing the resistance mechanisms of cotton against verticillium wilt should help to resolve it. Although some studies have investigated the function of Rbohs in biotic stress defense responses of plants, detailed genome-wide phylogenetic and functional analyses of the Rboh genes family in G. barbadense are still lacking.
To better understand the dynamics of Rboh genes in Gossypium plants and to foster future research on this important enzyme family, here we aimed to provide a detailed overview of their number, phylogeny, and function in defense against V. dahliae, by investigating five sequenced Gossypium species. From these, we identified 87 Rbohs. We determined that not only did GbRboh5/18 in the CladeD respond to V. dahliae infection and hormones, but it was also able to enhance plant resistance against verticillium wilt through elevated ROS accumulation in G. barbadense.
Materials and Methods
Sequence Retrieval and Identification of Cotton Rboh Genes
The motif FAD_binding_8 (PF08022) and EF-hand 2 (PF09068) were each downloaded from the Pfam database1. The whole genome sequence from the ancestral diploids G. arboreum (A2) (Li et al., 2014a; Du et al., 2018) and G. raimondii (D5) (Paterson et al., 2012), as well as those of their descendant tetraploids, G. barbadense (AD2) (Wang et al., 2019), and G. hirsutum (AD1) (Li et al., 2015; Zhang et al., 2015; Wang et al., 2019) were downloaded from the CottonGen database2, while that of G. australe (G) (Cai et al., 2019) was downloaded from NCBI3 (accession number: SMMG00000000), to carry out a genome-wide search for Rboh genes in the Gossypium genus. Splice variants were excluded and only the first variant was retained for further analysis. The molecular weights (kDa) and isoelectric points (pI) of Rboh proteins were determined using ProtParam4, and the subcellular localization of each protein was predicted by SherLoc25.
Phylogenetic, Gene Structure and Conserved Domain Analysis
A total of 10 Arabidopsis Rboh gene sequences were downloaded from TAIR 106. Multiple sequence alignments of all identified Rbohs from cotton and Arabidopsis were performed in ClustalX 2.0. The phylogenetic tree of deduced amino acid sequences was constructed by applying the neighbor-joining (NJ) method, with default parameters and 1000 bootstrap replicates, in MEGA7.07. To analyze the exon-intron distributions of the Rboh gene, the gene structure display server (GSDS)8 was utilized. Conserved motifs were then predicted using the MEME9 tool, under these parameters: number of repetitions, “any”; maximum number of motifs, “20”; and 60≥ “widths” ≥5, and between 6 and 300 residues. Only those motifs associated with an E-value < e–5 were retained.
Chromosomal Localization
The positional information of a given cotton Rboh gene was obtained from parsed general feature format (GFF) files, downloaded from the CottonGen website. The Rbohs in the five Gossypium species genomes were all mapped onto the chromosome. A shared identity of genes between different Gossypium spp. above 91%, or an identity of genes between the A and D subgenomes on tetraploid Gossypium spp. higher than 98% were connected by different-colored lines and depicted with the Circos genome visualization tool10.
Plant Materials, Growth Condition, and Treatments
Unless otherwise indicated, seeds of G. barbadense cultivar (“Xinhai15”) were inoculated in ddH2O for 24 h and then germinated for another 24 h under 25°C. After they had absorbed water, two seeds were sown per pot containing vermiculite. The pots were kept on a tray covered with a plastic dome, in a growth chamber at 25°C, with 120 μE m–2 s–1 light illumination under a 12-h-light/12-h-dark photoperiod. The domes were removed when two cotyledons emerged on all seedlings. When the first two true leaves fully expanded and the third true leaf emerged, the cotton plants could be treated with V. dahliae or hormones. For the former, the V. dahliae suspension was injected into the vermiculite nearby root; for the latter, plants were sprayed with jasmonic acid (JA; 100 μM concentration) or with abscisic acid (ABA; 50 μM concentration).
RNA Extraction, Reverse Transcription and Quantitative Real-Time PCR Analyses
Total RNA was extracted from cotton leaves using the RNAprep Pure Plant Plus Kit (DP441, Tiangen). Reverse transcription was performed with the HiScript II Q Select RT SuperMix (R233, Vazyme Biotech, Nanjing, China) following the manufacturer’s instructions. The quantitative PCR reactions employed the ChamQ SYBR Color qPCR Master Mix (Q411-02, Vazyme Biotech) and were run on the LightCycler480II Real-time PCR system (Roche) (every Q-PCR reaction was carried in triplicate for each sample). The cotton UBQ7 gene served as an internal reference gene for the normalization of the data. The sequences of all primer used can be found in Supplementary Table S2. For a given sample, the gene’s Ct values were standardized and its relative changes analyzed using the 2–ΔΔCT method.
Vector Construction
The TRV-VIGS vectors were constructed as described previously (Liu et al., 2002). The CDS fragment of GbRboh5/18 (Gbar_A05G025370/Gbar_D05G026620) and the fragment of 297 bp for virus-induced gene silencing (VIGS) with BamHI/KpnI at the 5′ and 3′ ends, were amplified from G. barbadense L. “Xinhai15” cDNA by PCR with gene-specific primers (listed in Supplementary Table S2) and cloned into the pMD18-T vector. The GbRboh5/18-VIGS fragment was inserted into the pTRV2 vector by BamHI/KpnI. All plasmids (including empty vector pTRV1, pTRV2, and recombinant vectors pTRV2- GbRboh5/18, pTRV2-CLA as the positive control) were respectively electroporated into the Agrobacterium tumefaciens strain GV3101, which was used to inject cotton cotyledons for VIGS.
The full length CDS fragment of GbRboh5/18 was amplified from G. barbadense L. “Xinhai15” cDNA by PCR with specific primers (listed in Supplementary Table S2). The overexpression vector was constructed using the Gateway Technology (Invitrogen). The PCR product was first conducted in BP reaction with pDONOR221 vector. Then, the positive plasmid pDONOR221-GbRboh5/18 underwent an LR reaction with the pK7WG2.0 vector to generate the p35S::GbRboh5/18 construct. The positive plasmid pK7WG2.0-GbRboh5/18 was electroporated into A. tumefaciens strain LBA4404, which was used to transform cotton plants for transient overexpression.
Virus-Induced Gene Silencing of GbRboh5/18 in Cotton
Virus-induced gene silencing assays were performed as described previously (Li et al., 2016) with minor modifications. The cotton plants with two fully expanded cotyledons were selected for infiltration for the VIGS assay. The A. tumefaciens strain GV3101 cells carrying the corresponding plasmid were inoculated into an LB liquid medium (containing 50 μg mL–1 kanamycin, 50 μg mL–1 rifampicin, and 25 μg mL–1 gentamicin). Grown overnight at 28°C under continuous agitation (200 rpm), A. tumefaciens cells were collected until OD600 = 0.8–1.2 by centrifugation and resuspended in the infiltration buffer (10 mM 2-(N-morpholino) ethanesulfonic acid [MES], 10 mM MgCl2, 200 μM AS). The density of A. tumefaciens cells suspension was set to OD600 = 0.6–0.8. Next, the A. tumefaciens cells containing pTRV1, pTRV2, or its derivatives, were mixed in a 1:1 ratio and these A. tumefaciens suspensions were incubated at room temperature for 3 h. Finally, the A. tumefaciens suspension was infiltrated fully on the underside of cotton cotyledons.
V. dahliae Culturing and Infection
Verticillium dahliae culturing and infection was performed as described (Zhao et al., 2017) with minor modifications. V. dahliae strain Vd991 was streak cultivated onto fresh PDA plates from glycerol stocks and incubated for 1 week at 25°C. The mycelia of one plate (90 mm) was transferred by a sterilized blade into a 200 mL Czapek-Dox medium and cultivated for 1 week with shaking at 200 rpm at 25°C in the dark to collect conidia. After filtrating the fungal cultures with 4 layers of sterilized gauze to remove mycelia, the conidial suspension was collected. The concentration of conidia in suspension was measured using a hemocytometer and was adjusted to approximately 1 × 107 conidia mL–1 for infection.
After two weeks of A. tumefaciens infiltration, cotton seedlings were inoculated with root-dipping into V. dahliae suspension, and then replanted into pots. After growing for another two weeks, the third true leaves of each plant were removed for staining experiments. Pretreated leaves were treated in 10 μM diphenyleneiodonium chloride (DPI, NADPH oxidase inhibitor) for 3 h before ROS staining or trypan blue staining.
Evaluation of Resistance to Verticillium Wilt
Verticillium wilt disease severity was performed as described (Meng et al., 2018) with minor modifications. The disease index of cotton plants was assessed 21 days and 25 days post-inoculation (dpi) with Vd991. The grade of disease severity was recorded and the disease index (DI) was calculated. The verticillium wilt disease severity level was recorded as five grades: Grade 0 (normal); Grade 1 (disease leaves less than 25%); Grade 2 (disease leaves between 25 and 50%); Grade 3 (disease leaves between 50 and 75%); and Grade 4 (disease leaves over 75%, almost all the dead plants). Disease index was scored with at least 30 plants per treatment and each treatment was repeated at least three times. Data were analyzed by Student’s t-test: ∗, p < 0.05; ∗∗, p < 0.01.
The lesion area of the transverse section of the cotton stem at 25 dpi with Vd991 was photographed and the browning degree was calculated using imageJ as described (Long et al., 2020). Fungal recovery was performed as described (Zhao et al., 2017).
Transient Overexpression of GbRboh5/18 in G. barbadense
Transient overexpression was performed as described (Li et al., 2018) with minor modifications. Overnight culturing of A. tumefaciens LBA4404 cells containing the pK7WG2.0-GbRboh5/18 plasmid at 28°C until the OD600 = 1.2–1.5 A. tumefaciens cells were collected and suspended in the infection solution [1/2 MS liquid medium,165 μM acetosyringone (AS), 0.01% Tween-20], with this mixture adjusted to OD600 = 0.8–1.0 to infect the cotton seedlings. After seeds of G. barbadense L. “Xinhai15” germinated, they were cultured in Hoagland’s solution until their cotyledons had fully expanded and the first true leaf emerged. These seedlings were washed with sterile water and 70% alcohol, and then sterilized in 0.01% HgCl2 for 2 min. Seedlings were washed for 2 min with sterile water, 5 times, then placed in a sterile jar to which the infection buffer was added, without Tween-20, for 2 h. After vacuuming for 15 min, the buffer was discarded. The seedlings were incubated in the A. tumefaciens suspensions at 28°C for 5 h (at 150 rpm) after seedlings were washed several times with sterile water to reduce the risk of subsequent bacterial contamination. Finally, the transformed seedlings were cultured in 1/2 MS solid medium and grown for 5 days; these seedlings were used to detect the expression levels or for H2DCFDA staining.
ROS Staining, H2DCFDA Staining and Trypan Blue Staining
The H2O2 content and cell death were detected by DAB staining and trypan blue staining, respectively. DAB staining and trypan blue staining were performed as described (Kumar et al., 2014) with minor modifications. Treated or control cotton true leaves were inoculated in 1 mg mL–1 of DAB staining solution in 10 mM Tris-acetic acid (pH 5.0) or in 0.4% (W/V) trypan blue staining buffer, and then vacuumed for 10 min. Tissue staining proceeded overnight (DAB staining) or for 3 h (trypan blue staining) at room temperature. Then the staining solution was drained off and absolute ethanol was added before tissue immersion in a boiling water-bath for 10 min to remove chlorophyll. The H2O2 was visualized as a reddish-brown color, while cell death appeared in blue. The H2DCFDA staining was used to detect ROS accumulation in cells. Overexpression cotton true leaves (vector as the negative control) were inoculated in 10 μM H2DCFDA solution in PBS buffer (pH 7.5), for 30 min at 37°C, after which the staining solution was drained off and the leaves washed four times with PBS buffer (pH 7.5). Fluorescence was visualized under a Zeiss LSM710 microscope with an excitation light of 488nm.
Results
Identification of Rbohs in Five Gossypium Species
To identify all the Rboh genes in the five sequenced Gossypium species, we performed a BLASTP search in the diploids G. arboreum (A2), G. raimondii (D5), and G. australe (G), as well as the tetraploids G. hirsutum (AD1) and G. barbadense (AD2). The protein database was queried by PF08022 and PF09068, which are conserved in the AtRbohs. Overall, we found 87 Rbohs on the cotton genomes: 13 Rbohs in A2, 13 Rbohs in D5, 10 Rbohs in G, 24 Rbohs in AD1, and 27 Rbohs in AD2 (Table 1). The number of Rboh genes in the At sub-genome of tetraploid Gossypium species was the same as that in the diploid G. arboretum (A2), while the Rboh genes in the Dt sub-genome of tetraploid Gossypium species was less than that in the diploid G. raimondii (D5), leaving the remainder 2 GbRbohs anchored to unmapped scaffolds. The genome harboring the fewest Rboh genes was that of G. australe (G).
According to the chromosomal locations of the corresponding genes, we renamed all Rbohs consecutively, as GaRboh1–GaRboh13, GrRboh1–GrRboh13, GauRboh1–GauRboh10, GhRboh1–GhRboh24, and GbRboh1–GbRboh27. For the 87 Rboh genes found in cotton, their encoded proteins consisted of 743–1015 amino acids. The predicted molecular weight of the Rboh proteins in the five Gossypium species ranged from 84.2 to 115.47 kDa, with a theoretical pI between 8.65 and 9.35. Their predicted subcellular localization showed that 71 of the 87 Rbohs were located in the cytoplasm, except for GaRboh9, GrRboh5, GhRboh2/15, and GbRboh2/15 that were restricted to mitochondria (Supplementary Table S1).
Phylogenetic Analysis of Rbohs in Five Gossypium Species
To understand their evolutionary relationships, a phylogenetic analysis of the putative 87 Rbohs from the five Gossypium species and 10 AtRbohs was carried out. Based on the homologous relationship in Arabidopsis, the 87 Rbohs in cotton were divided into seven clades (named CladeA, -B, -D, -E, -F, -H, and -I according to their homologs in Arabidopsis) (Figures 1, 2A). There were 20 members in CladeA, -D, and -F, 7 members in CladeB, -E, and -H, 6 members in CladeI, respectively. The cotton Rboh members within each clade displayed a high similarity to their orthologs in Arabidopsis. These results suggested the Rbohs in the five Gossypium species are highly conserved.
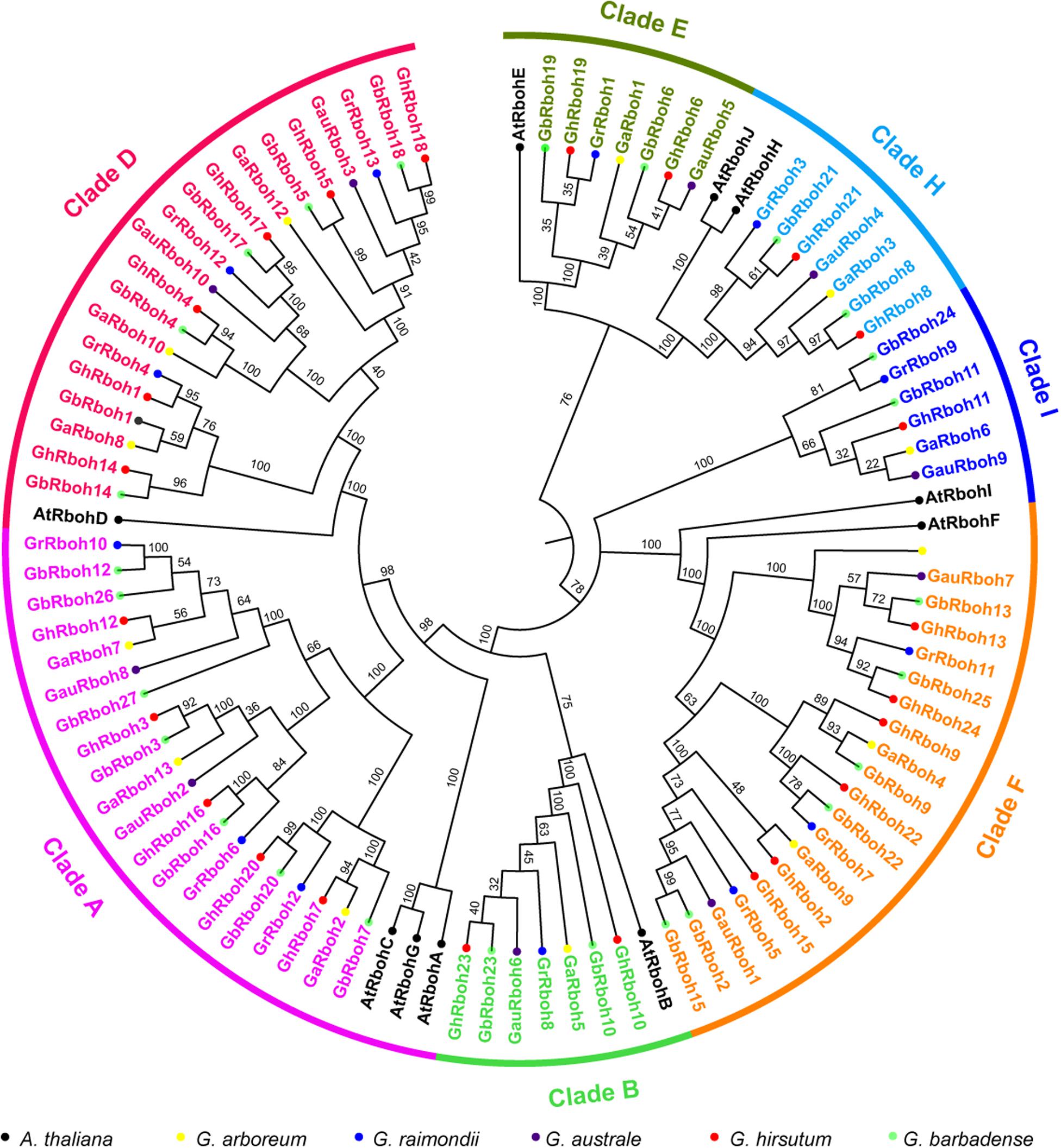
Figure 1. Phylogenetic analysis of Rbohs in cotton and Arabidopsis. A phylogenetic tree of Rboh proteins from G. arboreum, G. raimondii, G. australe, G. hirsutum, G. barbadense, and A. thaliana. The full-length amino acid sequences of the Rboh proteins were aligned using ClustalX in MEGA7.0. The unrooted tree was generated by the neighbor-joining (NJ) method (n = 1000 bootstraps). Clades are distinguished by different colors. Cotton Rbohs are colored clade specifically and the Arabidopsis Rbohs are in black. Different-colored solid circles indicate genes in different Gossypium species or Arabidopsis.
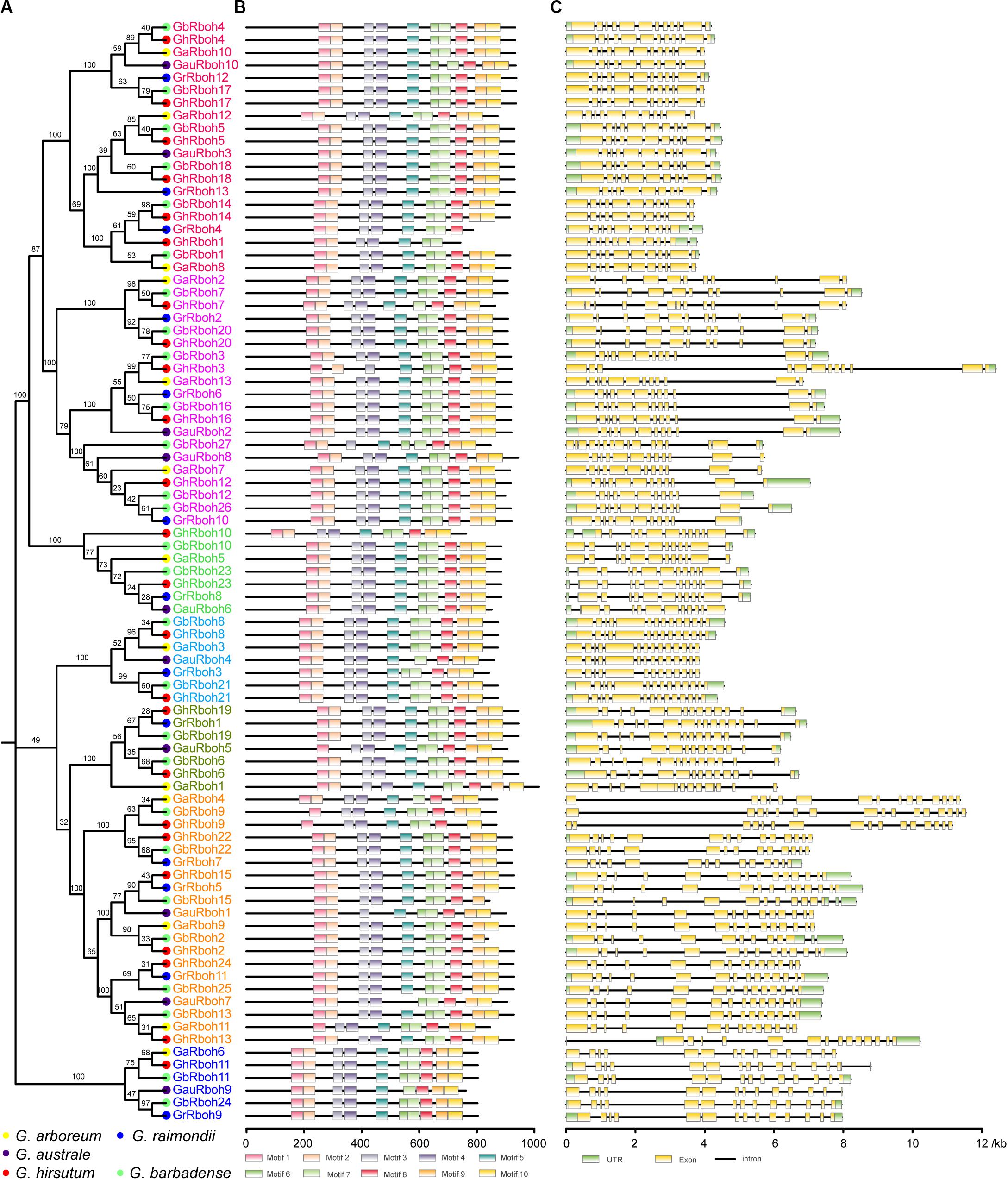
Figure 2. Characterization of the 87 Rbohs in five Gossypium species. The characteristics include the homologous relationship, conserved domain location, and exon-intron structure. (A) Phylogenetic relationships among the Rbohs in five Gossypium species. Rbohs in same clade are indicated by the same color. Different-colored solid circles indicate genes in different Gossypium species. (B) Conserved domain architecture of cotton Rboh proteins. Motifs were identified through a MEME analysis (http://meme-suite.org/) and protein length was estimated via the scale at the bottom. (C) Exon-intron organization analyses of cotton Rboh genes. The gene length was estimated by the scale at the bottom through gene structure analyses (GSDS: http://gsds.cbi.pku.edu.cn/). Green boxes indicate 5′- or 3′-untranslated regions. Exons and introns are distinguished in yellow boxes and in black lines, respectively.
The conserved motif analysis revealed that all these Rboh proteins contained at least 10 conserved motifs: two EF-hands in the N-terminus, six TM domains, and a NADPH-binding motif and a FAD-binding motif in the C-terminal region (Figure 2B). The number and arrangement of conserved domains within the same clade were similar. Comparing their gene structures, the exon-intron organizations revealed that Rboh genes in both CladeD and CladeH possessed densely arranged exons, with shorter introns, whereas longer introns characterized the rest of the cotton Rboh genes (Figure 2C). These results suggested a similar gene structure and conserved motif arrangement within the same clade.
Localization of Rboh Genes on Five Gossypium Chromosomes
To better understand their evolutionary relationships, we further analyzed the orthologous and paralogous Rboh genes in the five Gossypium species. Many gene blocks were found among the chromosomes of two sub-genomes of G. hirsutum and G. barbadense, marked by similarities to those in the sequenced diploid cotton genomes (A2, D5, and G). Compared to G. raimondii, G. arboreum, and G. australe, there, respectively were 22, 26, and 18 pairs of putative outparalogous genes in G. barbadense (Figure 3A). In G. hirsutum, there were 18, 20, and 14 pairs of putative orthologous Rbohs genes when compared to G. raimondii, G. arboretum, and G. australe, respectively (Figure 3B).
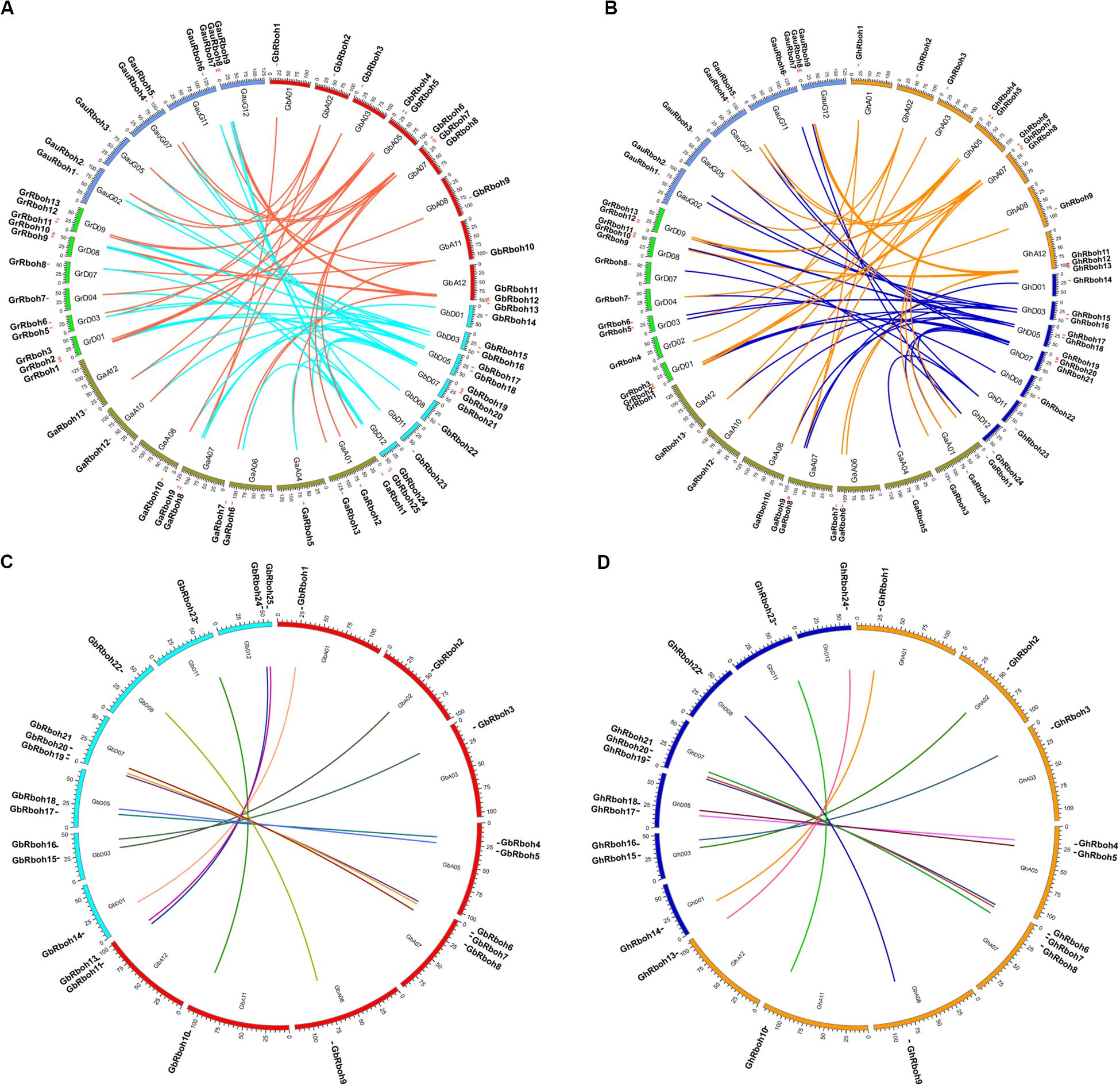
Figure 3. Localization of cotton Rbohs on chromosomes. The genomic (At and Dt) analysis of Rboh genes in panels (A) G. barbadense and (B) G. hirsutum vis-à-vis the diploid Gossypium spp. Different-colored links indicate translocation from the A or D sub-genome of an allotetraploid cotton to the diploid cotton. Lined gene coupling represents a shared identity between them that exceeded 91%. The inter-genomic (At and Dt) analyses of Rboh genes in panels (C) G. barbadense and (D) G. hirsutum between the A and D sub-genomes. Different-colored lines indicate gene couples whose identity is over 98%, depicted using the Circos genome visualization tool (http://www.circos.ca/).
Further analysis of the paralogous Rboh genes, between the A and D subgenomes in the two tetraploid cotton species, showed that 24 of 27 could be one-to-one paired in G. barbadense between the A and D sub-genomes, in addition to GbRboh12, GbRboh26, and GbRboh27 (Figure 3C). There were 11 syntenic gene blocks in G. hirsutum, with the exception of GhRboh11 and GhRboh12 (Figure 3D); these paired genes may exist as alleles on chromosomes between the A and D sub-genomes. These results implied that Rbohs are generally highly conserved between A and D sub-genomes during the evolution of tetraploid cotton plants.
CladeD (mainly GbRboh5/18) Response to Pathogen and Hormones in G. barbadense
To detect which Rbohs may contribute to plant resistance against verticillium wilt, or which can respond to hormones in G. barbadense, we designed specific primers for the conserved regions to detect the post-treatment level of transcripts for each clade’s genes. Within 24 h of treatment with V. dahliae, JA, or ABA, the transcripts of CladeA, -B, -D, and -F showed dramatic or slight changes, whereas the transcripts of GbRbohs in the other remaining three clades went undetected (Figure 4). Among the four responsive clades, CladeD underwent the starkest fold-change during the treatment with V. dahliae (6-fold at 24 h), JA (3.2-fold at 4 h), and ABA (9.6-fold at 4 h).
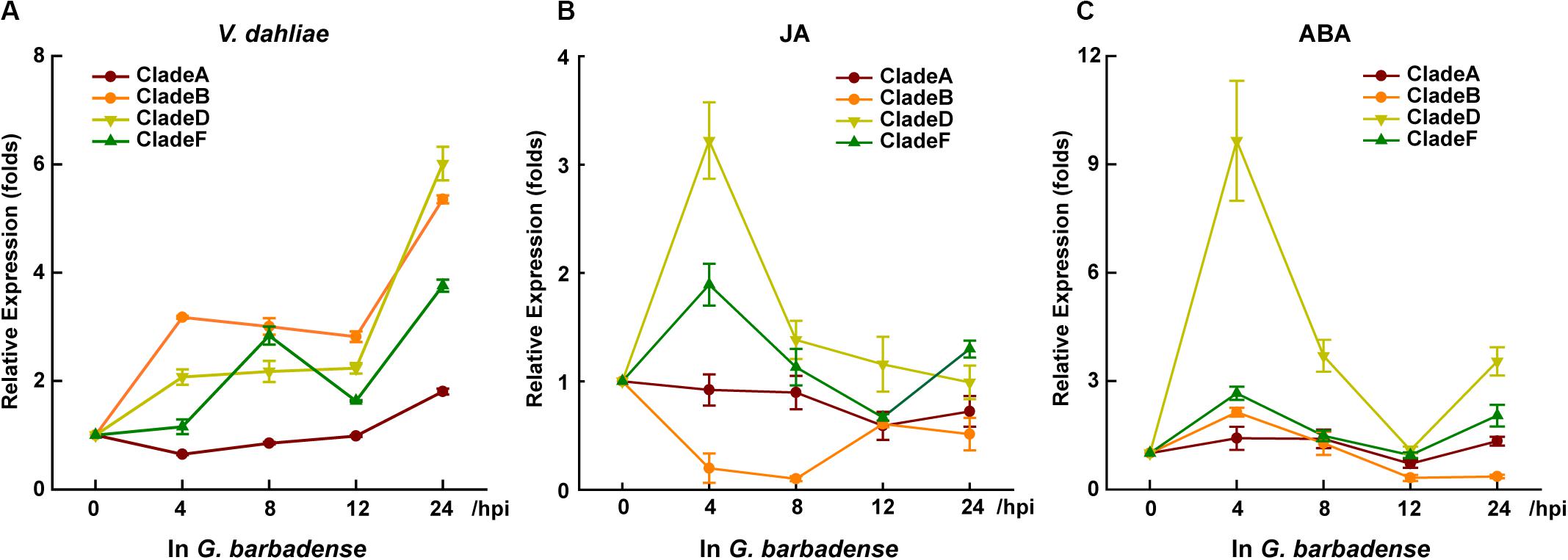
Figure 4. GbRbohs in the CladeD respond to V. dahliae infection and hormones in G. barbadense. Quantitative analyses of each clade GbRbohs’ expression levels in “Xinhai15” cotton plant leaves treated with (A) V. dahliae, (B) jasmonic acid, and (C) abscisic acid. Total RNAs were extracted from leaves of 3-week-old seedlings at 0, 4, 8, 12, and 24 h post-inoculation. The values are the mean ± SD for three technical replicates. Relative expression levels of all clade genes were determined after normalizing to the expression level at 0 h, which was set to 1.0. hpi, hours post-inoculation. The experiments were repeated at least three times, with similar results.
In G. barbadense, CladeD included three gene couples: GbRboh1/14, GbRboh4/17, and GbRboh5/18. We further detected their respective transcript levels by Q-PCR to identify which members contributed chiefly. The transcript of GbRboh5/18 was clearly induced by V. dahliae, or the JA, or ABA treatments (Figure 5), whereas the transcripts of GbRboh1/14 and GbRboh4/17 were not detected. Furthermore, the trend in the variation of GbRboh5/18’s transcript level was consistent with CladeD; hence we deduced that GbRboh5/18 is the cardinal member of CladeD in G. barbadense.
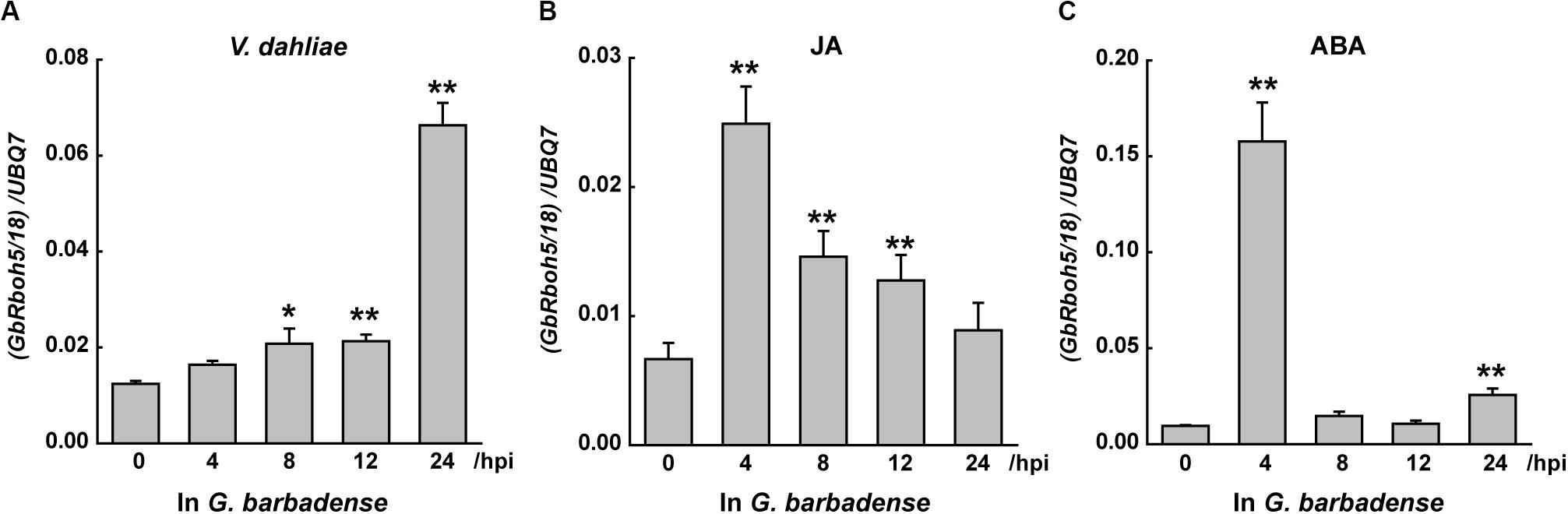
Figure 5. GbRboh5/18 are the main functional genes of CladeD in G. barbadense. Quantitative analyses of GbRboh5/18 expression levels in “Xinhai15” cotton plant leaves inoculated with (A) V. dahliae, (B) jasmonic acid, and (C) abscisic acid. Total RNAs were extracted from leaves of 3-week-old seedlings at 0, 4, 8, 12, and 24 h post-inoculation. The experiments were repeated three times, with similar results. The values are the means ± SD for three technical replicates. Transcript levels of each gene couple were first normalized to UBQ7. Asterisks indicate significant differences compared with 0 h under same treatment (*p < 0.05; **p < 0.01, Student’s t-test).
Knock-Down of GbRboh5/18 Attenuates Resistance to V. dahliae in G. barbadense
To verify the function of GbRbohD in cotton plant resistance to V. dahliae, we utilized VIGS, a powerful technique, to achieve knock-down in the transcript activity of GbRboh5/18, the gene mainly responsible in CladeD. When “Xinhai15” (cultivar of G. barbadense) seedlings had grown for 10 days, we infiltrated cotyledons with TRV: 00 (as negative control) or TRV: GbRboh5/18. After 2 weeks of treatment with V. dahliae, the GbRboh5/18-VIGS plants displayed more severe disease symptoms than the control plants, such as wilted leaves and dark vascular bundles (Figure 6A). The successful knock-down of the GbRboh5/18 transcript was confirmed by the Q-PCR analysis (Figure 6B), and the disease index was consistent with the plant phenotype (Figure 6C). Further, a lesion area analysis and fungal recovery assay closely resembled the phenotype and disease index (Figures 6D,E). These results suggested that the knock-down of GbRboh5/18 transcription is able to attenuate plant resistance to V. dahliae infection.
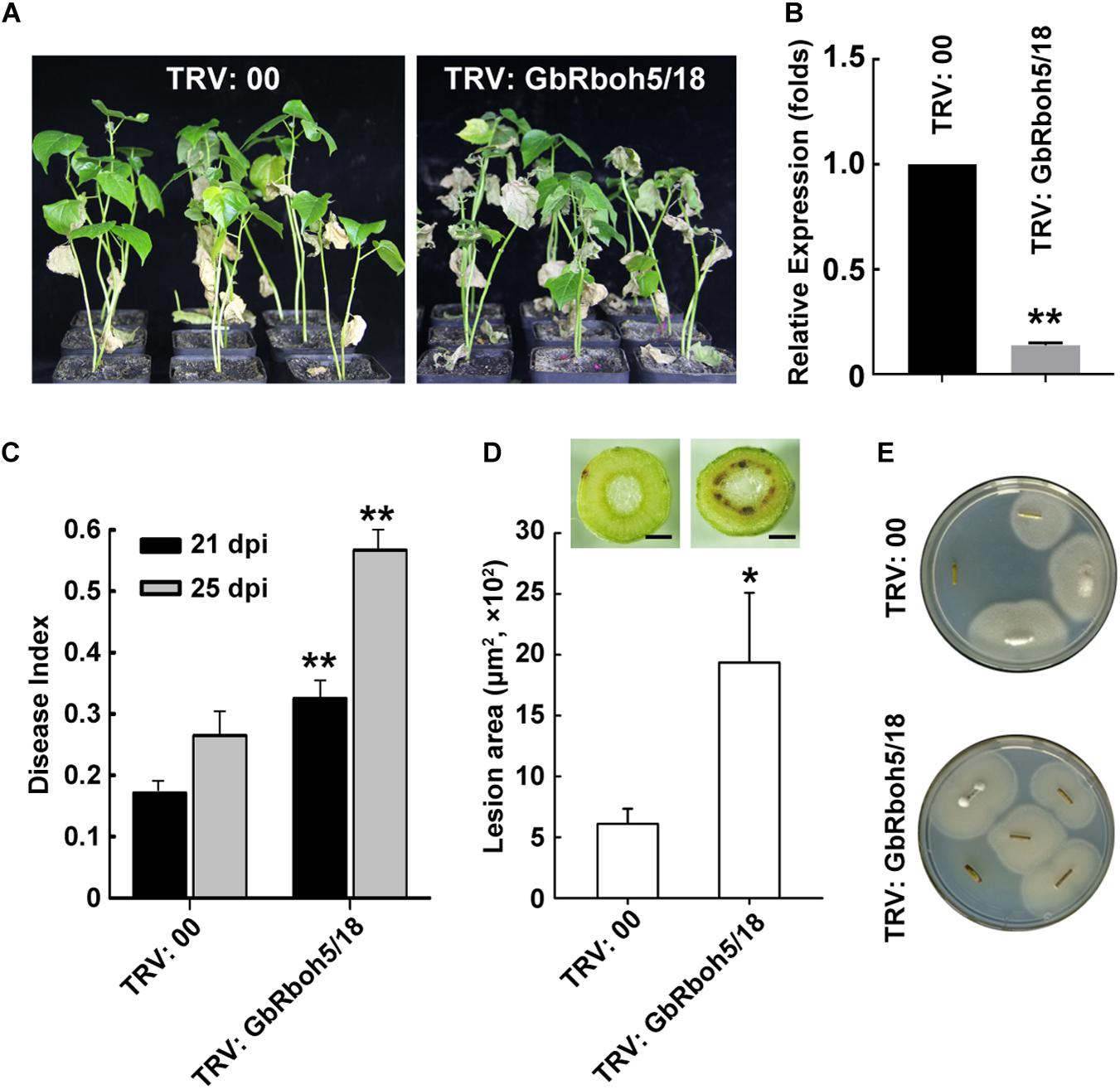
Figure 6. Knock-down of GbRboh5/18 attenuates plant resistance to V. dahliae. (A) Disease phenotype of cotton plants (“Xinhai15”) at 25 days post-infection with V. dahliae strain Vd991. Seedlings were inoculated with V. dahliae two weeks after VIGS and photographed 25 days later. (B) Expression of GbRboh5/18 in GbRboh5/18-VIGS and control plants. UBQ7 was the internal reference gene (**p < 0.01; Student’s t-tests). (C) Disease index of GbRboh5/18-VIGS and control plants at 21 and 25 dpi with V. dahliae. Asterisks indicate significant differences compared with TRV: 00 at same time (*p < 0.05; **p < 0.01, Student’s t-test). (D) Lesion area of GbRboh5/18-VIGS and control plants’ stems at 25 dpi with V. dahliae. Transverse section of stems were photographed with an OLYMPUS CX31 microscope. The values are the mean ± SD, n ≥ 30. Bar = 0.5 mm. (E) Fungal recovery assay. Sterile stems approximately 1 cm from same position of plants was plated on PDA plates at 25°C to allow V. dahliae recovery; photographs were taken 3 days post-inoculation. All experiments were repeated at least three times, with similar results.
GbRboh5/18 Promotes ROS Accumulation in G. barbadense
To clarify the function of GbRboh5/18, we detected cell death in the GbRboh5/18-VIGS plants and control plants, via trypan blue staining. There was no significant difference between GbRboh5/18-VIGS and control leaves, but more of the blue stain appeared in GbRboh5/18-VIGS leaves than control leaves after infection with V. dahliae. Pretreatment with DPI, an inhibitor of the NADPH oxidase, before V. dahliae infection, resulted in almost no staining in all leaves (Figure 7A). Further, the DAB staining showed less ROS accumulation in GbRboh5/18-VIGS leaves than in control leaves after V. dahliae infection, yet this did not occur in both groups leaves when they were pretreated with DPI (Figure 7B). These results indicated that the knock-down of GbRboh5/18 caused more cell death mediated by less ROS accumulation when plants were infected with the V. dahliae pathogen.
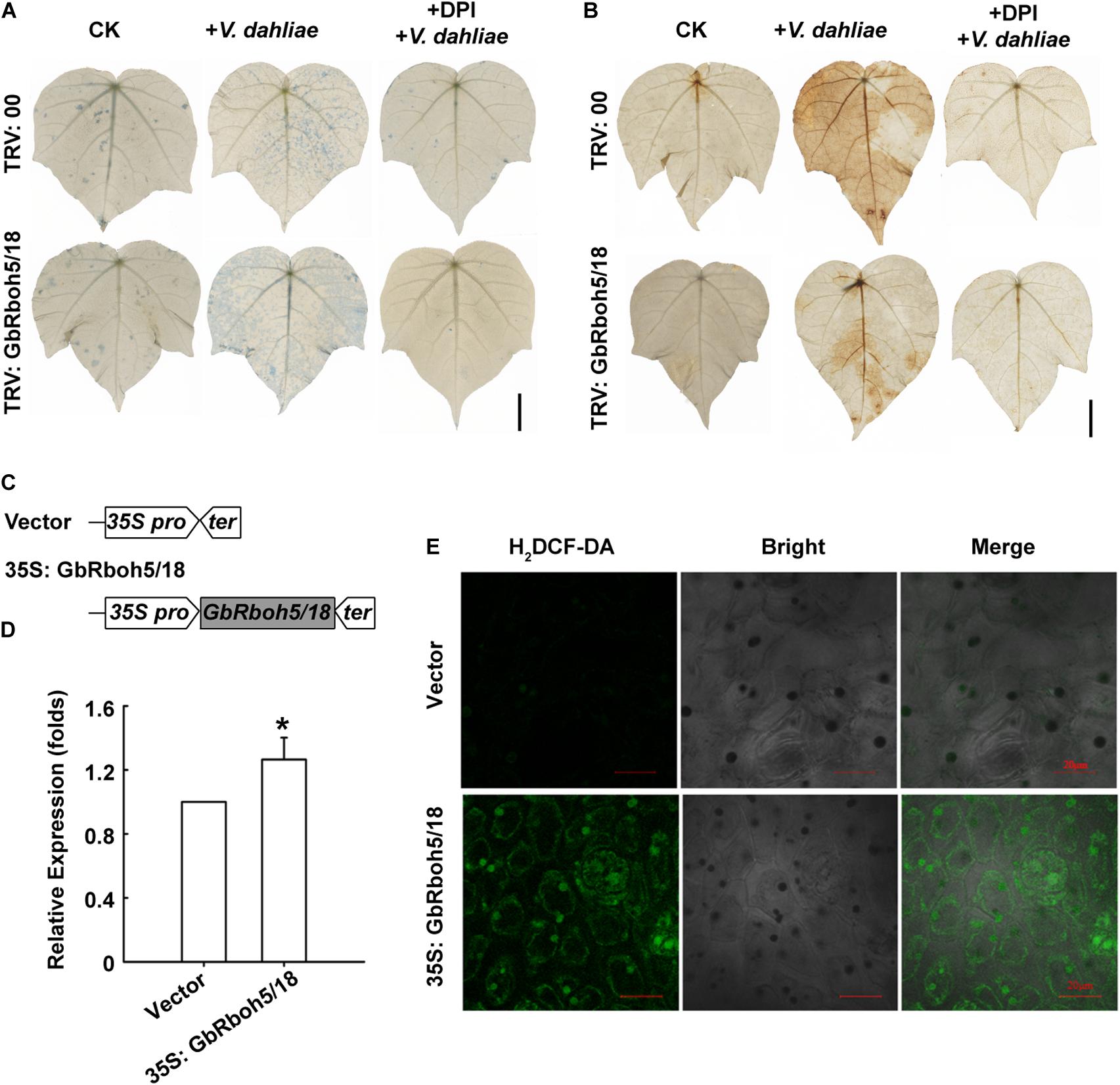
Figure 7. GbRboh5/18 promotes ROS accumulation in G. barbadense. (A) Trypan blue staining of cell death and (B) DAB staining of ROS in GbRboh5/18-VIGS and control leaves. Leaves were stained at 48 h post-inoculation with or without V. dahliae. Leaves were pretreated with or without 10 μM DPI before trypan blue staining or DAB staining. Bar = 1 cm. (C) Schematic diagram showing the structure of overexpression vector. Gray-filled squares indicate ORF of GbRboh5/18; the left pentagon and right pentagon are the 35S promoter and 35S terminator, respectively. (D) Expression of GbRboh5/18 in GbRboh5/18-OE and vector plant leaves. UBQ7 was the internal reference gene. (*p < 0.05; Student’s t-tests). (E) H2DCF-DA fluorescence probe of ROS in GbRboh5/18-OE and vector leaves cells. Bar = 20 μm. After 5 days of growth, the first true leaves were used to Q-PCR or H2DCF-DA staining. All experiments were repeated at least three times with similar results.
Nonetheless, we constructed the GbRboh5/18-OE vector to verify the functional mechanism of GbRboh5/18 (Figure 7C). We utilized a transient overexpression strategy to detect ROS accumulation in cotton seedlings. Overexpression of GbRboh5/18 was confirmed by the Q-PCR analysis (Figure 7D). The H2DCF-DA fluorescent assay was used to observe ROS accumulation in GbRboh5/18-OE leaves, which revealed stronger fluorescent signals in GbRboh5/18-OE than in the control (Figure 7E). These results showed that GbRboh5/18 can increase the ROS accumulation.
Discussion
Rbohs, a group of plant-specific NADPH oxidases, have been phylogenetically and functionally characterized for a variety of plant species (Torres et al., 1998; Xia et al., 2009; Kaur and Pati, 2018), but not yet clearly for cotton (Gossypium spp.). Five Gossypium genomes were sequenced and assembled over the past few years (Li et al., 2014a, 2015; Zhang et al., 2015; Du et al., 2018; Ma et al., 2018, 2019; Cai et al., 2019; Hu et al., 2019; Wang et al., 2019). With the development of sequencing and assembly technology, information of genomes is becoming more and more complete. Utilizing bioinformatics, Q-PCR, VIGS, and plant-pathogen biology, in this study we systematically reported on the Rboh gene family in five sequenced Gossypium species. This integrated approach included building a phylogenetic tree of the Rboh genes in cotton and Arabidopsis; analyses of gene structure and conserved domains, and transcript levels’ response to V. dahliae; and verifying the function of GbRboh5/18 in conferring resistance to V. dahliae. Collectively, this provides a basic understanding of cotton Rboh genes and highlights the key role of GbRboh5/18 in responses to V. dahliae.
We systematically identified 87 Rboh genes in these representative five sequenced Gossypium species: G. arboretum, G. raimondii, G. australe, G. hirsutum, and G. barbadense. Both G. arboretum and G. raimondii harbored 13 Rboh genes, a result that agrees with the findings of Zhang et al. (2020). The main difference with that work is that we found 24 and 27 Rboh genes in G. hirsutum and G. barbadense, while they found 26 and 19, respectively (Wang et al., 2020). For G. hirsutum, 4 of 26 GhRbohs were anchored to unmapped scaffolds in the NAU-NBI v1.1 assembly genome (assembled in 2015), whereas we found that all 24 GhRbohs in the HAU v1.0 assembly genome (assembled in 2018) mapped onto the At or Dt sub-genome chromosomes. For G. barbadense, we found more GbRbohs in the HAU v2.0 assembly genome (assembled in 2018) than in the NAU assembly genome (Table 1, Supplementary Table S1). This disparity may be due to recent advances in assembly technology that have improved the quantity and quality of genomic information, resulted in a more accurate genome. In addition, this study uncovered 10 GauRbohs in the G. austral genome, making it now possible to further explore the function of the Rboh gene family in this species.
The number of Rboh genes in G. hirsutum was less than its sum in the two diploids, suggesting that gene loss events happened in the Rboh gene family during the polyploidization of G. hirsutum. By contrast, the number of Rboh genes in G. barbadense was more than the sum of the Rboh genes’ number in the A2 and D5 genomes, suggesting gene expansion occurred in the Rboh gene family during the polyploidization of G. barbadense. These results are consistent with previous work showing that many gene loss and duplication events have occurred in allotetraploid cotton plants (Li et al., 2015; Zhang et al., 2015; Wang et al., 2019; Yang et al., 2019). According to their phylogenetic relationships with Arabidopsis orthologs, the Rbohs were assigned to seven clades. In addition to CladeA and -I, there were twice as many genes of other clades in the tetraploids as in the diploid ancestors (G. arboretum and G. raimondii). This suggests that the Rboh gene family was conserved during the evolution of cotton.
Rbohs play pivotal roles in ROS production during the PTI and ETI responses in Arabidopsis and rice (Pogany et al., 2009; Chaouch et al., 2012; Dubiella et al., 2013). Verticillium wilt, a “cancer” of cotton production, is a vascular bundle disease caused by the fungus V. dahliae (Fradin et al., 2009). Overexpression of polyamine oxidase GhPAO contributes to the resistance of plants against V. dahliae by elevating the level of H2O2 accumulation (Mo et al., 2015). These previous findings contended that ROS content influences plant resistance to V. dahliae. Yet in our study we did not detect the transcripts of CladeE (GbRboh6/19), CladeH (GbRboh8/21), or CladeI (GbRboh11/24) when plants were treated with either the pathogen or hormones. Accordingly, these results lead us to speculate that CladeE, CladeH, and CladeI may participate in certain physiological developmental processes, but not in the cotton plant defense response against this pathogen.
AtRbohD maintains hydrogen peroxide (H2O2) homeostasis in Arabidopsis plants under different stresses (Kobayashi et al., 2012; Morales et al., 2016; Yao et al., 2017). The latest findings suggest that RbohD-dependent H2O2 production is regulated by the PTP-MPK3/6-WRKY pathway in the response against Vd-toxins in Arabidopsis (Zhao et al., 2020). Since GbRboh5/18 mainly contributes to CladeD, we also determined the function of GbRboh5/18, via VIGS and transient overexpression experiments. Knock-down of GbRboh5/18 resulted in a lowered ROS content and reduced resistance to V. dahliae, whereas overexpressing GbRboh5/18 resulted in higher ROS accumulations (Figures 6, 7). So we may conclude that GbRboh5/18 enhances cotton plant resistance by increasing its ROS accumulation.
In conclusion, we showed that the Rboh gene family and GbRboh5/18, in particular, contributed to resistance against verticillium wilt in cotton. The molecular functional mechanism of GbRboh5/18 in the regulation of H2O2 production now awaits investigation.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.cottongen.org/ [BGI-CGB v2.0 assembly genome for G. arboreum (A2)], [JGI v2.0 assembly data for G. raimondii (D5)], [HAU v1.0 assembly genome for G. hirsutum (AD1)], [HAU v2.0 assembly genome for G. barbadense (AD2)]; https://www.ncbi.nlm.nih.gov/ [assembly CCRI_HENU_Gaus_1.1 for G. australe (G)].
Author Contributions
YC and BL designed the experiments. BL and SG analyzed the genes’ data. QS and RG performed the Q-PCR, VIGS, and the ROS detection. BL and YZ carried out the overexpression of cotton seedlings. JL constructed the phylogenetic tree. YC and YFC wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the National Key Research and Development Program of China (grant numbers 2016YFD0101902 and 2018YFD0100304), the Natural Science Foundation of China (grant numbers U1704104 and 31571724), and the Innovation and Talent Introduction Program on Crop Stress Biology (111 Project).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00788/full#supplementary-material
Footnotes
- ^ http://pfam.xfam.org/
- ^ https://www.cottongen.org/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://web.expasy.org/protparam
- ^ https://abi-services.informatik.uni-tuebingen.de/sherloc2/webloc.cgi
- ^ http://www.arabidopsis.org
- ^ https://www.megasoftware.net
- ^ http://gsds.cbi.pku.edu.cn/
- ^ http://meme-suite.org/tools/meme
- ^ http://www.circos.ca/
References
Angelos, E., and Brandizzi, F. (2018). NADPH oxidase activity is required for ER stress survival in plants. Plant J. 96, 1106–1120. doi: 10.1111/tpj.14091
Baxter, A., Mittler, R., and Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Cai, Y., Cai, X., Wang, Q., Wang, P., Zhang, Y., Cai, C., et al. (2019). Genome sequencing of the Australian wild diploid species Gossypium austral highlights disease resistance and delayed gland morphogenesis. Plant Biotechnol. J. 18, 814–828 doi: 10.1111/pbi.13249
Chaouch, S., Queval, G., and Noctor, G. (2012). AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 69, 613–627. doi: 10.1111/j.1365-313X.2011.04816.x
Chen, D., Cao, Y., Li, H., Kim, D., Ahsan, N., Thelen, J., et al. (2017). Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat. Commun. 8, 2265–2278. doi: 10.1038/s41467-017-023402343
Du, X., Huang, G., He, S., Yang, Z., Sun, G., Ma, X., et al. (2018). Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 50, 796–802. doi: 10.1038/s41588-018-0116-x
Dubiella, U., Seybold, H., Durian, G., Komander, E., Lassig, R., Witte, C. P., et al. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. U S A 110, 8744–8749. doi: 10.1073/pnas.1221294110
Fradin, E. F., Zhang, Z., Juarez Ayala, J. C., Castroverde, C. D., Nazar, R. N., Robb, J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. doi: 10.1104/pp.109.136762
Galletti, R., Denoux, C., Gambetta, S., Dewdney, J., Ausubel, F. M., De Lorenzo, G., et al. (2008). The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 148, 1695–1706. doi: 10.1104/pp.108.127845
Han, J. P., Koster, P., Drerup, M. M., Scholz, M., Li, S., Edel, K. H., et al. (2019). Fine-tuning of RBOHF activity is achieved by differential phosphorylation and Ca2 + binding. New. Phytol. 221, 1935–1949. doi: 10.1111/nph.15543
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. doi: 10.1038/s41588-019-0371375
Jiao, Y., Sun, L., Song, Y., Wang, L., Liu, L., Zhang, L., et al. (2013). AtrbohD and AtrbohF positively regulate abscisic acid-inhibited primary root growth by affecting Ca2 + signalling and auxin response of roots in Arabidopsis. J. Exp. Bot. 64, 4183–4192. doi: 10.1093/jxb/ert228
Kadota, Y., Sklenar, J., Derbyshire, P., Stransfeld, L., Asai, S., Ntoukakis, V., et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell. 54, 43–55. doi: 10.1016/j.molcel.2014.02.021
Kaur, G., Guruprasad, K., Temple, B. R. S., Shirvanyants, D. G., Dokholyan, N. V., and Pati, P. K. (2018). Structural complexity and functional diversity of plant NADPH oxidases. Amino. Acids 50, 79–94. doi: 10.1007/s00726-017-24912495
Kaur, G., and Pati, P. K. (2018). In silico insights on diverse interacting partners and phosphorylation sites of respiratory burst oxidase homolog (Rbohs) gene families from Arabidopsis and rice. BMC Plant Biol. 18:161. doi: 10.1186/s12870-018-1378-2
Kaya, H., Takeda, S., Kobayashi, M. J., Kimura, S., Iizuka, A., Imai, A., et al. (2019). Comparative analysis of the reactive oxygen species-producing enzymatic activity of Arabidopsis NADPH oxidases. Plant J. 98, 291–300. doi: 10.1111/tpj.14212
Keller, T., Damude, H. G., Werner, D., Doerner, P., Dixon, R. A., and Lamb, C. (1998). A Plant Homolog of the Neutrophil NADPH Oxidase gp91phox Subunit Gene Encodes a Plasma Membrane Protein with Ca2 + Binding Motifs. Plant Cell 10, 255–266. doi: 10.1105/tpc.10.2.255
Kimura, S., Hunter, K., Vaahtera, L., Tran, H. C., Citterico, M., Vaattovaara, A., et al. (2020). CRK2 and C-terminal Phosphorylation of NADPH Oxidase RBOHD Regulate Reactive Oxygen Species Production in Arabidopsis. Plant Cell. 32, 1063–1080. doi: 10.1105/tpc.19.00525
Kobayashi, M., Kawakita, K., Maeshima, M., Doke, N., and Yoshioka, H. (2006). Subcellular localization of Strboh proteins and NADPH-dependent -generating activity in potato tuber tissues. J. Exp. Bot. 57, 1373–1379. doi: 10.1093/jxb/erj113
Kobayashi, M., Ohura, I., Kawakita, K., Yokota, N., Fujiwara, M., Shimamoto, K., et al. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19, 1065–1080. doi: 10.1105/tpc.106.048884
Kobayashi, M., Yoshioka, M., Asai, S., Nomura, H., Kuchimura, K., Mori, H., et al. (2012). StCDPK5 confers resistance to late blight pathogen but increases susceptibility to early blight pathogen in potato via reactive oxygen species burst. New. Phytol. 196, 223–237. doi: 10.1111/j.1469-8137.2012.04226.x
Kumar, D., Yusuf, M. A., Singh, P., Sardar, M., and Sarin, N. B. (2014). Histochemical Detection of Superoxide and H2O2 Accumulation in Brassica juncea Seedlings. Bio. Protocol. 4:e1108.
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014a). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Li, L., Li, M., Yu, L., Zhou, Z., Liang, X., Liu, Z., et al. (2014b). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host. Microbe. 15, 329–338. doi: 10.1016/j.chom.2014.02.009
Li, H., Li, K., Guo, Y., Guo, J., Miao, K., Botella, J. R., et al. (2018). A transient transformation system for gene characterization in upland cotton (Gossypium hirsutum). Plant Methods 14:50. doi: 10.1186/s13007-018-0319312
Li, T., Ma, X., Li, N., Zhou, L., Liu, Z., Han, H., et al. (2017). Genome-wide association study discovered candidate genes of Verticillium wilt resistance in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 15, 1520–1532. doi: 10.1111/pbi.12734
Li, Y. B., Han, L. B., Wang, H. Y., Zhang, J., Sun, S. T., Feng, D. Q., et al. (2016). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 170, 2392–2406. doi: 10.1104/pp.15.01930
Lin, F., Zhang, Y., and Jiang, M. Y. (2009). Alternative Splicing and Differential Expression of Two Transcripts of Nicotine Adenine Dinucleotide Phosphate Oxidase B Gene from Zea mays. J. Integr. Plant Biol. 51, 287–298. doi: 10.1111/j.1744-7909.2008.00808.x
Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313x.2002.01297.x
Long, L., Xu, F.-C., Zhao, J., Li, B., Xu, L., and Gao, W. (2020). GbMPK3 overexpression increases cotton sensitivity to Verticillium dahliae by regulating salicylic acid signaling. Plant Sci. 292:110374. doi: 10.1016/j.plantsci.2019.110374
Ma, X., Wang, W., Bittner, F., Schmidt, N., Berkey, R., Zhang, L., et al. (2016). Dual and Opposing Roles of Xanthine Dehydrogenase in Defense-Associated Reactive Oxygen Species Metabolism in Arabidopsis. Plant Cell 28, 1108–1126. doi: 10.1105/tpc.15.00880
Ma, X., Wang, Z., Li, W., Zhang, Y., Zhou, X., Liu, Y., et al. (2019). Resequencing core accessions of a pedigree identifies derivation of genomic segments and key agronomic trait loci during cotton improvement. Plant Biotechnol. J. 17, 762–775. doi: 10.1111/pbi.13013
Ma, Z., He, S., Wang, X., Sun, J., Zhang, Y., Zhang, G., et al. (2018). Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 50, 803–813. doi: 10.1038/s41588-018-0119117
Meng, J., Gao, H., Zhai, W., Shi, J., Zhang, M., Zhang, W., et al. (2018). Subtle regulation of cotton resistance to Verticillium wilt mediated by MAPKK family members. Plant Sci. 272, 235–242. doi: 10.1016/j.plantsci.2018.05.003
Mo, H., Wang, X., Zhang, Y., Zhang, G., Zhang, J., and Ma, Z. (2015). Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae. Plant J. 83, 962–975. doi: 10.1111/tpj.12941
Morales, J., Kadota, Y., Zipfel, C., Molina, A., and Torres, M. A. (2016). The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 67, 1663–1676. doi: 10.1093/jxb/erv558
Muller, K., Carstens, A. C., Linkies, A., Torres, M. A., and Leubner-Metzger, G. (2009). The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol. 184, 885–897. doi: 10.1111/j.1469-8137.2009.03005.x
Nuhse, T. S., Bottrill, A. R., Jones, A. M., and Peck, S. C. (2007). Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51, 931–940. doi: 10.1111/j.1365-313X.2007.03192.x
Ogasawara, Y., Kaya, H., Hiraoka, G., Yumoto, F., Kimura, S., Kadota, Y., et al. (2008). Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2 + and phosphorylation. J. Biol. Chem. 283, 8885–8892. doi: 10.1074/jbc.M708106200
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Pogany, M., von Rad, U., Grun, S., Dongo, A., Pintye, A., Simoneau, P., et al. (2009). Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol. 151, 1459–1475. doi: 10.1104/pp.109.141994
Sagi, M., Davydov, O., Orazova, S., Yesbergenova, Z., Ophir, R., Stratmann, J. W., et al. (2004). Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum. Plant Cell 16, 616–628. doi: 10.1105/tpc.019398
Sagi, M., and Fluhr, R. (2006). Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 141, 336–340. doi: 10.1104/pp.106.078089
Shaban, M., Miao, Y., Ullah, A., Khan, A. Q., Menghwar, H., Khan, A. H., et al. (2018). Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 125, 193–204. doi: 10.1016/j.plaphy.2018.02.011
Suzuki, N., Miller, G., Morales, J., Shulaev, V., Torres, M. A., and Mittler, R. (2011). Respiratory burst oxidases: the engines of ROS signaling. Curr. Opin. Plant Biol. 14, 691–699. doi: 10.1016/j.pbi.2011.07.014
Torres, M. A., and Dangl, J. L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin. Plant Biol. 8, 397–403. doi: 10.1016/j.pbi.2005.05.014
Torres, M. A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K. E., and Jones, J. D. G. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14, 365–370. doi: 10.1046/j.1365-313x.1998.00136.x
Wang, J., Tian, L., Zhang, D. D., Short, D. P. G., Zhou, L., Song, S. S., et al. (2018). SNARE-encoding genes VdSec22 and VdSso1 mediate protein secretion required for full virulence in Verticillium dahliae. Mol. Plant Microbe. Interact. 31, 651–664. doi: 10.1094/MPMI-12-17-0289-R
Wang, M., Tu, L., Yuan, D., Zhu, D., Shen, C., Li, J., et al. (2019). Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 51, 224–229. doi: 10.1038/s41588-018-0282-x
Wang, W., Chen, D., Liu, D., Cheng, Y., Zhang, X., Song, L., et al. (2020). Comprehensive analysis of the Gossypium hirsutum L. respiratory burst oxidase homolog (Ghrboh) gene family. BMC Genomics 21:91. doi: 10.1186/s12864-020-65036506
Wendel, J. F. (1989). New World tetraploid cottons contain Old World cytoplasm. Proc. Natl. Acad, Sci, U S A 86, 4132–4136. doi: 10.1073/pnas.86.11.4132
Wong, H. L., Pinontoan, R., Hayashi, K., Tabata, R., Yaeno, T., Hasegawa, K., et al. (2007). Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19, 4022–4034. doi: 10.1105/tpc.107.055624
Xia, X. J., Wang, Y. J., Zhou, Y. H., Tao, Y., Mao, W. H., Shi, K., et al. (2009). Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 150, 801–814. doi: 10.1104/pp.109.138230
Yang, C., Li, W., Cao, J., Meng, F., Yu, Y., Huang, J., et al. (2017). Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 89, 338–353. doi: 10.1111/tpj.13388
Yang, Z., Ge, X., Yang, Z., Qin, W., Sun, G., Wang, Z., et al. (2019). Extensive intraspecific gene order and gene structural variations in upland cotton cultivars. Nat. Commun. 10:2989. doi: 10.1038/s41467-019-10820-x
Yao, Y., He, R. J., Xie, Q. L., Zhao, X. H., Deng, X. M., He, J. B., et al. (2017). ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)-dependent mechanism in response to different stresses in Arabidopsis. New Phytol. 213, 1667–1681. doi: 10.1111/nph.14278
Yoshioka, H., Numata, N., Nakajima, K., Katou, S., Kawakita, K., Rowland, O., et al. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15, 706–718. doi: 10.1105/tpc.008680
Yu, S., Kakar, K. U., Yang, Z., Nawaz, Z., Lin, S., Guo, Y., et al. (2019). Systematic study of the stress-responsive Rboh gene family in Nicotiana tabacum: Genome-wide identification, evolution and role in disease resistance. Genomics 19, S0888–S7543. doi: 10.1016/j.ygeno.2019.08.010
Yun, B.-W., Feechan, A., Yin, M., Saidi, N. B. B., Le Bihan, T., Yu, M., et al. (2011). S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268. doi: 10.1038/nature10427
Zhang, G., Yue, C., Lu, T., Sun, L., and Hao, F. (2020). Genome-wide identification and expression analysis of NADPH oxidase genes in response to ABA and abiotic stresses, and in fibre formation in Gossypium. PeerJ. 8:e8404. doi: 10.7717/peerj.8404
Zhang, H., Fang, Q., Zhang, Z., Wang, Y., and Zheng, X. (2009). The role of respiratory burst oxidase homologues in elicitor-induced stomatal closure and hypersensitive response in Nicotiana benthamiana. J. Exp. Bot. 60, 3109–3122. doi: 10.1093/jxb/erp146
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhao, J., Chen, Q., Zhou, S., Sun, Y., Li, X., and Li, Y. (2020). H2Bub1 Regulates RbohD-Dependent Hydrogen Peroxide Signal Pathway in the Defense Responses to Verticillium dahliae Toxins. Plant Physiol. 182, 640–657. doi: 10.1104/pp.19.00913
Keywords: respiratory burst oxidase homologs (Rbohs), reactive oxygen species (ROS), Gossypium, V. dahliae, GbRboh5/18
Citation: Chang Y, Li B, Shi Q, Geng R, Geng S, Liu J, Zhang Y and Cai Y (2020) Comprehensive Analysis of Respiratory Burst Oxidase Homologs (Rboh) Gene Family and Function of GbRboh5/18 on Verticillium Wilt Resistance in Gossypium barbadense. Front. Genet. 11:788. doi: 10.3389/fgene.2020.00788
Received: 19 April 2020; Accepted: 02 July 2020;
Published: 11 September 2020.
Edited by:
Hongjian Wan, ZheJiang Academy of Agricultural Sciences, ChinaReviewed by:
Huiming Guo, Chinese Academy of Agricultural Sciences (CAAS), ChinaZhikun Li, Agricultural University of Hebei, China
Copyright © 2020 Chang, Li, Shi, Geng, Geng, Liu, Zhang and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingfan Cai, eWluZ2ZhbmNhaUBvdXRsb29rLmNvbQ==
†These authors have contributed equally to this work
 Ying Chang
Ying Chang Bo Li
Bo Li Qian Shi
Qian Shi Shuaipeng Geng
Shuaipeng Geng Jinlei Liu
Jinlei Liu Yingfan Cai
Yingfan Cai