
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 22 July 2020
Sec. Evolutionary and Population Genetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00743
This article is part of the Research Topic Time Domains of Hypoxia Adaptation: Evolutionary Insights and Applications View all 24 articles
Over millions of years, vertebrate species populated vast environments spanning the globe. Among the most challenging habitats encountered were those with limited availability of oxygen, yet many animal and human populations inhabit and perform life cycle functions and/or daily activities in varying degrees of hypoxia today. Of particular interest are species that inhabit high-altitude niches, which experience chronic hypobaric hypoxia throughout their lives. Physiological and molecular aspects of adaptation to hypoxia have long been the focus of high-altitude populations and, within the past decade, genomic information has become increasingly accessible. These data provide an opportunity to search for common genetic signatures of selection across uniquely informative populations and thereby augment our understanding of the mechanisms underlying adaptations to hypoxia. In this review, we synthesize the available genomic findings across hypoxia-tolerant species to provide a comprehensive view of putatively hypoxia-adaptive genes and pathways. In many cases, adaptive signatures across species converge on the same genetic pathways or on genes themselves [i.e., the hypoxia inducible factor (HIF) pathway). However, specific variants thought to underlie function are distinct between species and populations, and, in most cases, the precise functional role of these genomic differences remains unknown. Efforts to standardize these findings and explore relationships between genotype and phenotype will provide important clues into the evolutionary and mechanistic bases of physiological adaptations to environmental hypoxia.
Some of the most challenging environments for terrestrial life are those in which oxygen availability is limited. For terrestrial vertebrates, such hypoxic environments occur in high-altitude niches, where ambient oxygen levels are reduced relative to sea level due to lower barometric pressure, and in underground burrows, wherein animal respiration combined with poor gas diffusion through the surrounding soils results in decreased oxygen availability. A key distinction between these environments is that hypoxia is constant at high-altitude, whereas in underground burrows, hypoxia varies both in its intensity and consistency. Despite the challenges that hypoxia imposes on physiology, humans have inhabited hypoxic high-altitude regions for 10, 000 of years, while other mammals have evolved to adapt to life under conditions of hypoxia over millions of years or were brought to high altitudes via human intervention (e.g., domesticated animals) within a few hundred to several 1000 years ago (Qi et al., 2013; Witt and Huerta-Sanchez, 2019). Therefore, it is possible to explore common physiological and genetic adaptations to hypoxia across varied and distinct time spans and environmental conditions in both human and animal populations.
Limited oxygen availability is one of the strongest drivers of evolutionary adaptation and has resulted in the appearance of a wide variety of adaptive strategies. Over the past several decades, much effort has gone into characterizing physiological and molecular adaptations to hypoxia in various species adapted to life at high altitude or in underground burrows, and this work has been reviewed elsewhere (Hochachka, 1986; Hochachka et al., 1996; Bickler and Buck, 2007; Ramirez et al., 2007; Pamenter, 2014, 2016; Dzal et al., 2015; Pamenter and Powell, 2016; McClelland and Scott, 2019). These adaptations mitigate the impact of reduced environmental oxygen availability by enhancing its delivery through the body (i.e., increased supply) or, in situations where enhanced delivery is not sufficient, by reducing the need for oxygen at the tissue level to generate cellular energy (i.e., by decreasing metabolism; decreased demand) (Dzal et al., 2015; McClelland and Scott, 2019). The primary sites of adaptation in terms of oxygen supply reside within the cardio-respiratory system, whose primary function is to extract oxygen from the atmosphere and deliver it to the mitochondria of cells through four primary steps: ventilation, diffusion of oxygen from the air into the blood, circulation, and diffusion of oxygen from the blood into the cells (Figure 1). Common adaptations to hypoxia that have been observed in this oxygen transport cascade include changes in the sensitivity of chemosensory cells and organs to hypoxia, alterations in the anatomy, mechanics, and neural/cellular control of ventilation and cardiac function, biochemical changes to the oxygen carrying capacity of the blood, and anatomical adjustments that alter the diffusion distance across which gases must travel between the blood and tissues (and vice versa) (Dzal et al., 2015).
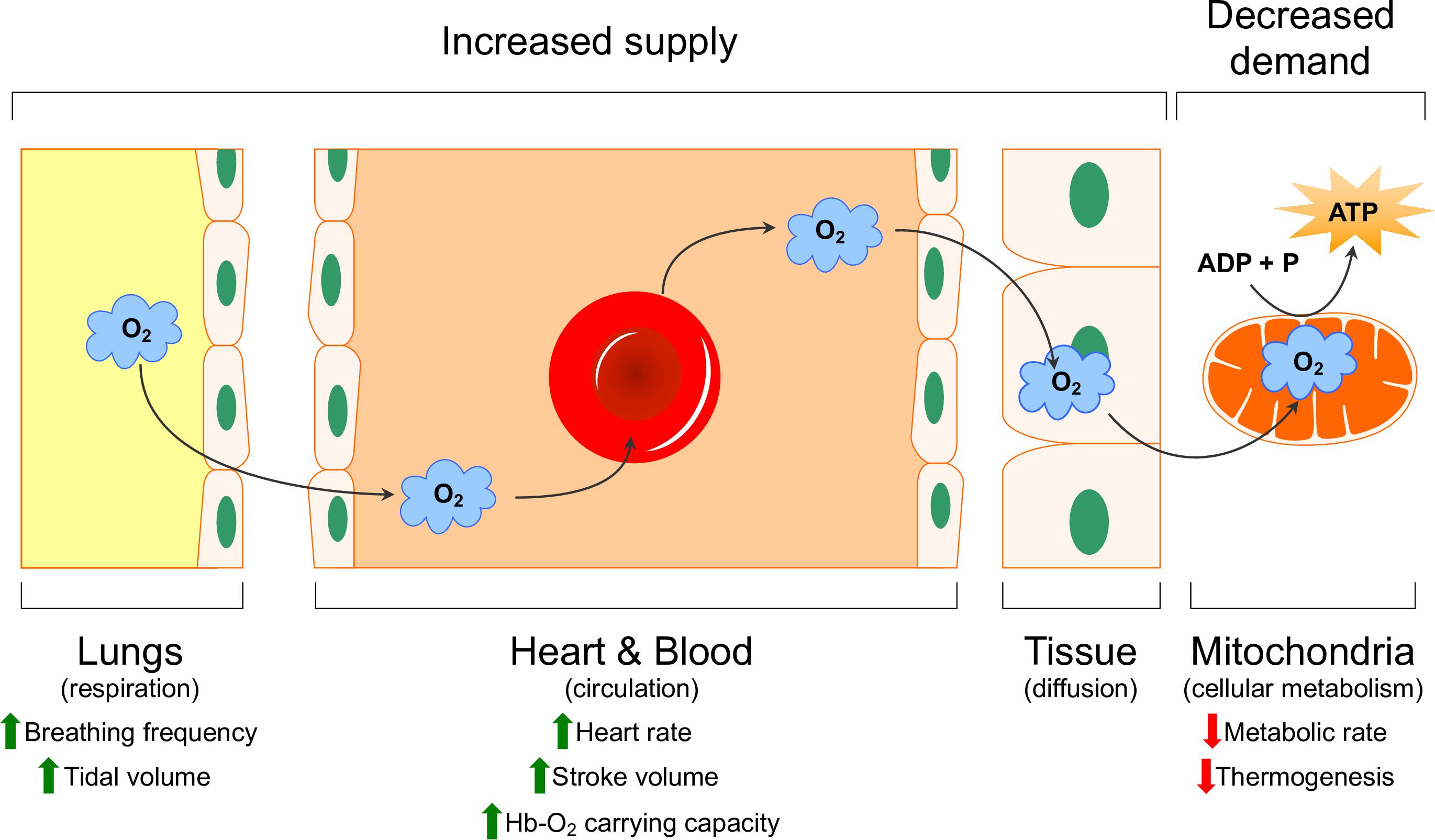
Figure 1. Schematic representation of the oxygen transport cascade. Physiological mechanisms that increase oxygen supply (green arrows) and decreased demand (red arrows) are indicated. Modified with permission from Dzal et al. (2015).
On the demand side of the hypoxia/energy balance reside systemic and cellular adaptations that minimize energy demand, either through reduced behaviors (i.e., torpor or hibernation), shutting down non-essential tissues to preserve energy for oxygen-sensitive organs, or by reducing energy demand at the cellular level by turning off specific processes (e.g., protein synthesis, thermogenesis, etc.) (Hochachka, 1986; Hochachka et al., 1996). Examining differences in the function and composition of the various components of the oxygen transport cascade and also cellular consumers of energy, and the control of these responses between species adapted to environments with variable oxygen availability has provided important insight into the evolution and function of oxygen-dependent processes.
Until somewhat recently, most studies have relied exclusively upon physiological and/or molecular approaches. While these studies have provided remarkable insight into the scope and variability of physiological responses and cellular adaptations to hypoxia, it is only with more recent genomic analysis of hypoxia-adapted human and animal populations that we have begun to understand the genetic underpinnings of these adaptations, their evolutionary origins, and, in a very few cases, links between genetic mutations and specific physiological and molecular characteristics.
In this review, we focus on the genomic aspects of hypoxia-adapted animals and humans from whom such information is available (regardless of the availability of matching physiological data) and identify overlap in common genes reported as top candidates for selection. The most commonly reported adaptation-associated genes are those related to the control and function of the hypoxia-inducible factor (HIF) pathway and/or downstream genes of this family of transcriptional regulators. This commonality has received considerable attention in comparative analysis of hypoxia-mediated genetic adaptation; however, many other genes have been identified across species, suggesting that there are multiple genetic avenues by which molecular and physiological adaptations to hypoxia might have evolved (Simonson, 2015). Adaptations in mammalian, bird, and ectothermic species that are native to high-altitude environments or that have been brought to high altitude as domesticated animals by humans, as well as species that experience hypoxia at low altitudes (e.g., mammals that live in densely populated underground burrows) are included for comparative purposes. There are many hypoxia-tolerant species in which physiological mechanisms of hypoxia-tolerance have been extensively described but for which genetic information is not available; discussion of these adaptations is outside the scope of the present review. However, many excellent reviews have been published recently that describe physiological adaptations to hypoxia and the interested reader is encouraged to consult these for further information (e.g., Bickler and Buck, 2007; Gilbert-Kawai et al., 2014; Dzal et al., 2015; Simonson, 2015; Moore, 2017).
Hypoxia inducible factors (HIFs) are the master regulators of cellular response to hypoxia. These transcriptional factors are involved in many processes such as cellular metabolism, angiogenesis, erythropoiesis, regulation of bone and connective tissue development, and fetal development (Bigham and Lee, 2014). The HIF family of transcriptional factors is comprised of four protein orthologs, HIF-1α, HIF-2α, HIF-3α, and HIF-1β (Semenza, 2020). HIF-1α and HIF-1β are present and highly conserved among metazoans (Loenarz et al., 2011) and are expressed in all mammalian tissues (Wiener et al., 1996). HIF-2α and HIF-3α are found in vertebrates and are expressed in a cell-type specific manner (Semenza, 2020). HIF-1α has been shown to be essential to embryonic development (Cerychova and Pavlinkova, 2018), and HIF-2α is noted for regulation of EPO transcription, to produce erythropoietin (EPO) protein postnatally in the kidney (Gruber et al., 2007; Haase, 2013). While these transcription factors regulate thousands of genes, the roles of HIF-3α remain to be extensively characterized (Yang S. L. et al., 2015; Duan, 2016). The genes encoding the HIF proteins, HIF1A (HIF-1α), EPAS1 (HIF-2α), and HIF3A (HIF-3α), each encode a number of protein isoforms via alternative splicing adding additional functional capabilities (Drutel et al., 2000; Gothié et al., 2000; Maynard et al., 2003; Dales et al., 2010; Pasanen et al., 2010; Heikkilä et al., 2011; Yang S. L. et al., 2015).
HIFs 1-3 share a conserved domain structure consisting of an N-terminal basic helix-loop-helix (bHLH) DNA binding domain, two Per-Arnt-Sim (PAS) domains, an oxygen-dependent degradation domain, and two transactivation domains (Figure 2; Beaudry et al., 2016). HIF-1β, also known as the aryl hydrocarbon nuclear translocator (ARNT) protein, shares some homology to HIFs 1-3 but lacks an oxygen-dependent degradation domain. The bHLH and PAS-A domains interact with DNA while the PAS domains are key for heterodimerization (Jiang et al., 1996; Wu et al., 2015). The oxygen-dependent degradation domain interacts with a number of regulatory proteins which modulate molecule stability and target the molecule for degradation (Metzen and Ratcliffe, 2004), and the transactivation domains recruit coactivators p300 and CREB-binding protein in the nucleus (Arany et al., 1996).
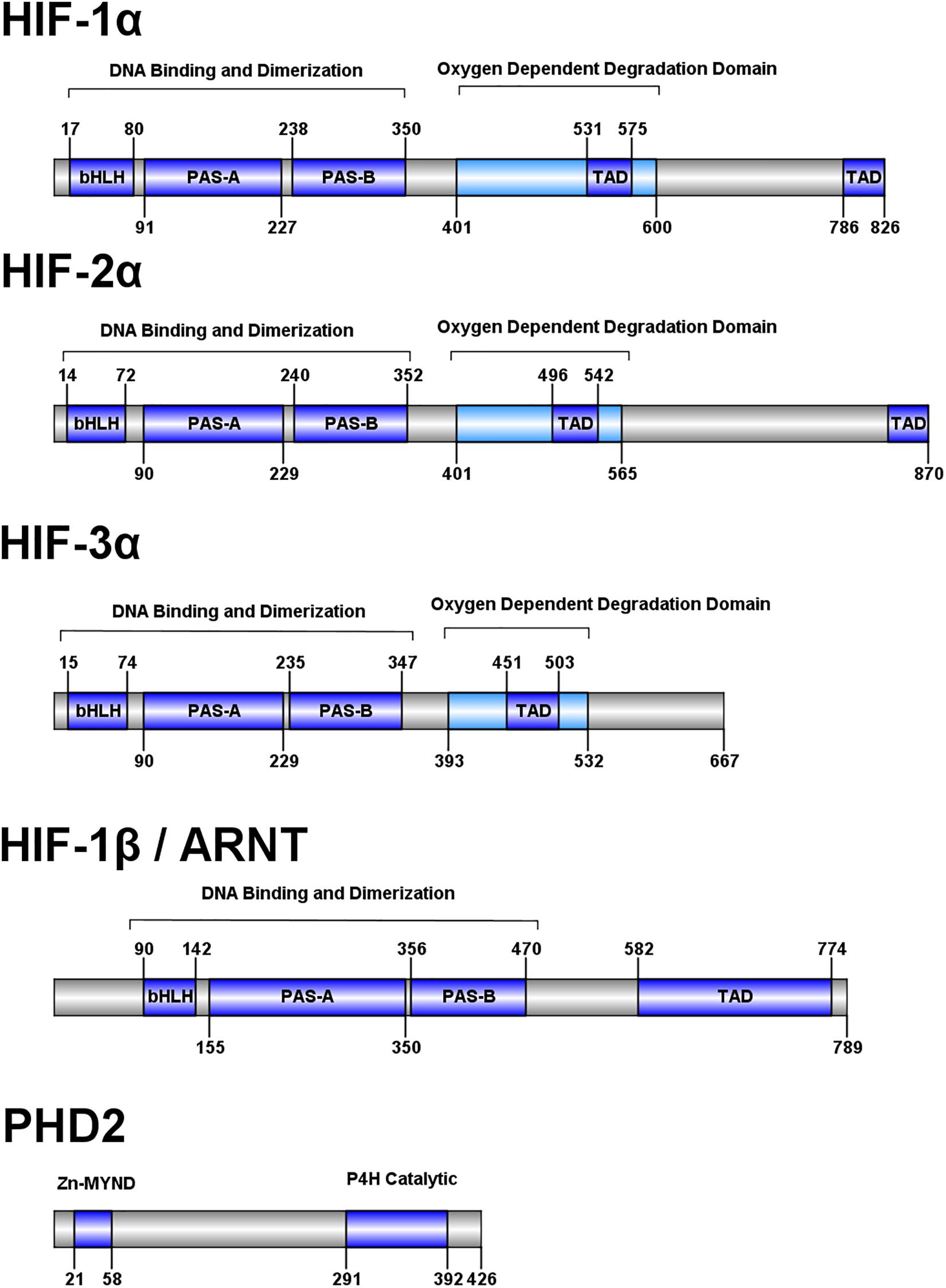
Figure 2. Domain structure of human HIF pathway proteins. Domain structure for HIF-1α, HIF-2α, HIF-3α, HIF-1β, and PHD2 are shown. The overall domain layout for the HIF proteins are similar with an N-terminal bHLH domain, followed by two PAS domains and an oxygen dependent degradation domain (absent in HIF-1β). HIF-1α and HIF-2α contain two transactivation domains in the C-terminal portion of the protein, while HIF-3α and HIF-1β have one. PHD2 has a domain layout containing an N-terminal MYND-type (myeloid, Nervy, and DEAF-1) zinc finger domain and a C-terminal prolyl-4-hydroxylase catalytic domain.
Under normoxic conditions, HIF-1α and HIF-2α are regulated post-translationally via hydroxylation of specific proline residues within the oxygen-dependent degradation domain (Jaakkola et al., 2001; Figure 3). Hydroxylation is carried out by prolyl hydrolase domain (PHD) proteins PHD1-3 (Appelhoff et al., 2004). Hydroxylated HIF-α is recognized by von Hippel-Lindau (VHL) protein, which targets HIFs for degradation through the ubiquitin–proteasome pathway (Maxwell et al., 1999; Ivan et al., 2001). Under hypoxic conditions, this oxygen-dependent modification is arrested, leading to stabilization and accumulation of HIF-1α and/or HIF-2α within the cell (Taylor et al., 2016). HIF-1α and/or HIF-2α localize to the nucleus and dimerize with HIF-1β, which is required for DNA binding and transcriptional modulation (Ruas and Poellinger, 2005; Taylor et al., 2016). HIF-1 and HIF-2 complexes (HIF-1α/2α and HIF-1β dimer) recognize hypoxia response elements (HREs), which contain short conserved DNA sequences (5′-RCGTG-3′) (Semenza et al., 1996; Wu et al., 2015). HREs activate the expression of hypoxia-associated genes [e.g., EPO, VEGF (angiogenesis), ALDOA, ENO1, and LDHA (glycolytic enzymes) (Semenza et al., 1996)]. In addition to gene activation, HIF complexes can activate transcriptional repressors via the HIF target gene RE1-silencing transcription factor (REST) (Cavadas et al., 2016).
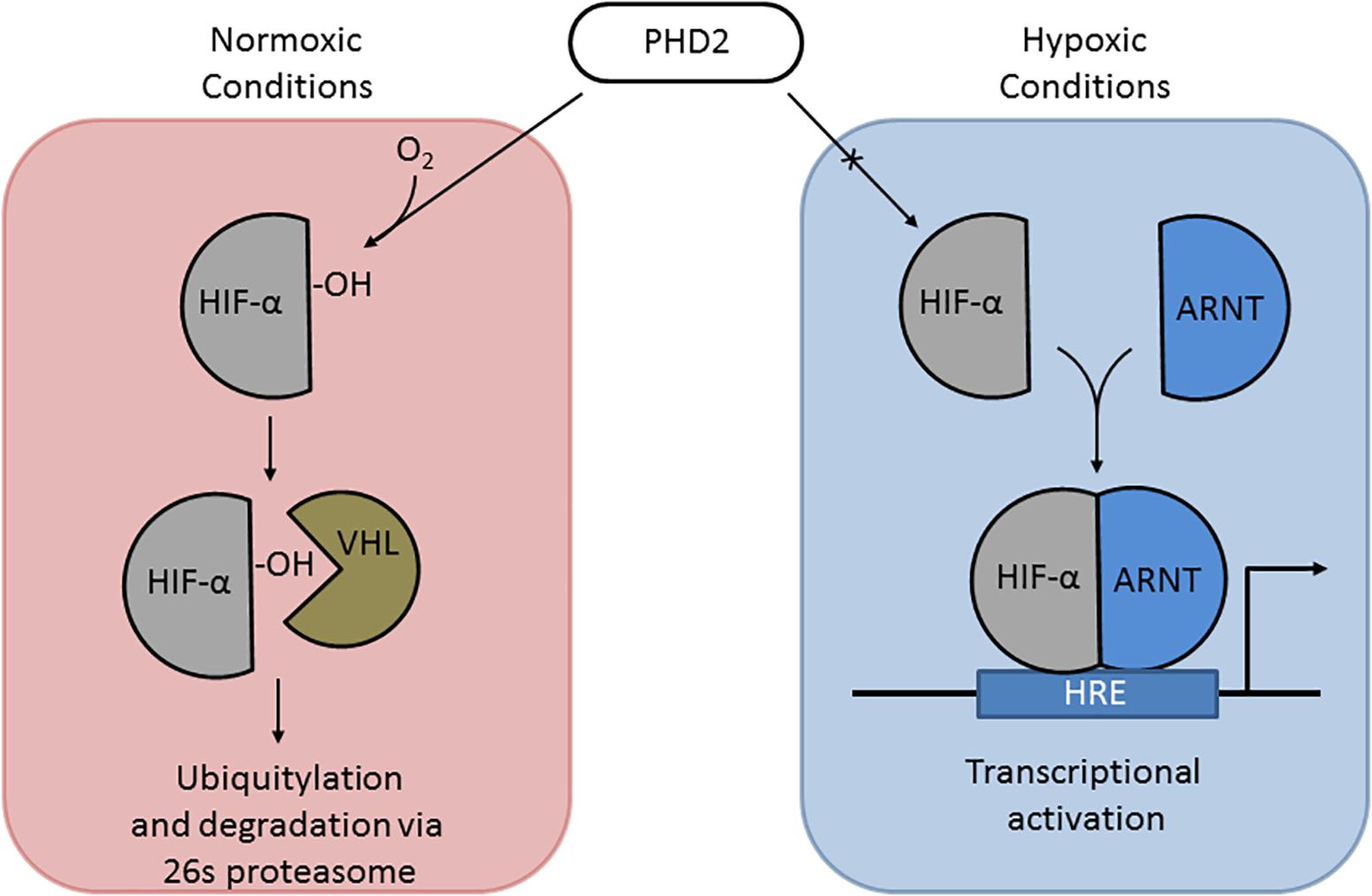
Figure 3. The HIF pathway. (Left) Under normoxic conditions, PHD2 hydroxylates proline 531 of HIF-2α, which can then be recognized by VHL, targeting it for ubiquitylation and degradation. (Right) Under hypoxic conditions, PHD2 cannot hydroxylate HIF α subunits, allowing it to be stabilized and translocate to the nucleus to dimerize with ARNT; the HIF-α-ARNT complex in the nucleus triggers transcriptional activation at hypoxia response elements (HREs).
Genomic studies in search of adaptive signatures to high altitude indicate many HIF pathway genes and their downstream targets are top candidates for positive selection in humans (e.g., EPAS1, the gene the encodes HIF-2α and EGLN1, the gene that encodes PHD2) (reviewed in Bigham and Lee, 2014; Simonson, 2015). Studies in various high-altitude species have focused on these candidates and report specific variants, many of which are missense mutations and appear to be deleterious. It is interesting to note that while HIF-1α and HIF-2α have many overlapping and distinct targets, HIF1A does not repeatedly appear in selective sweeps like EPAS1. This may be due to the tendency of HIF mutations to be deleterious, HIF-1α playing a key role in embryonic development, and HIF-2α playing a major role in other processes with more plasticity.
Numerous variants have been identified within HIF-2α which have been associated with high-altitude adapted species and disease states (Table 1). With a number of crystal structures available for HIF-2α, PHD2, and VHL, structural analysis and insights into how alterations in protein sequence translate to perturbation of HIF pathway signaling are under investigation. The structure of the DNA binding and dimerization domain of HIF-2α (Residues 8-360) in complex with ARNT (Wu et al., 2015) is shown in Figure 4. Post-translational modification sites in HIF-2α that have been identified in humans cluster along the “top” of this domain while the majority of variants among species cluster along dimerization interfaces on the opposite side of the PAS domains. While little is known about how these mutations affect protein structure and function, predictive analysis and co-immunoprecipitation studies suggest mutations along the bHLH and PAS domains are typically disruptive in nature, leading to loss-of-function (Wu et al., 2015; VonHoldf et al., 2017). Understanding how these variants affect HIF-2α structure, domain dynamics, and protein−protein interactions with binding partners (e.g., ARNT and PHD2) are key for understanding the role HIF-2α structure plays in high-altitude adaptation and hypoxic signaling within the cell (Hall et al., 2020).
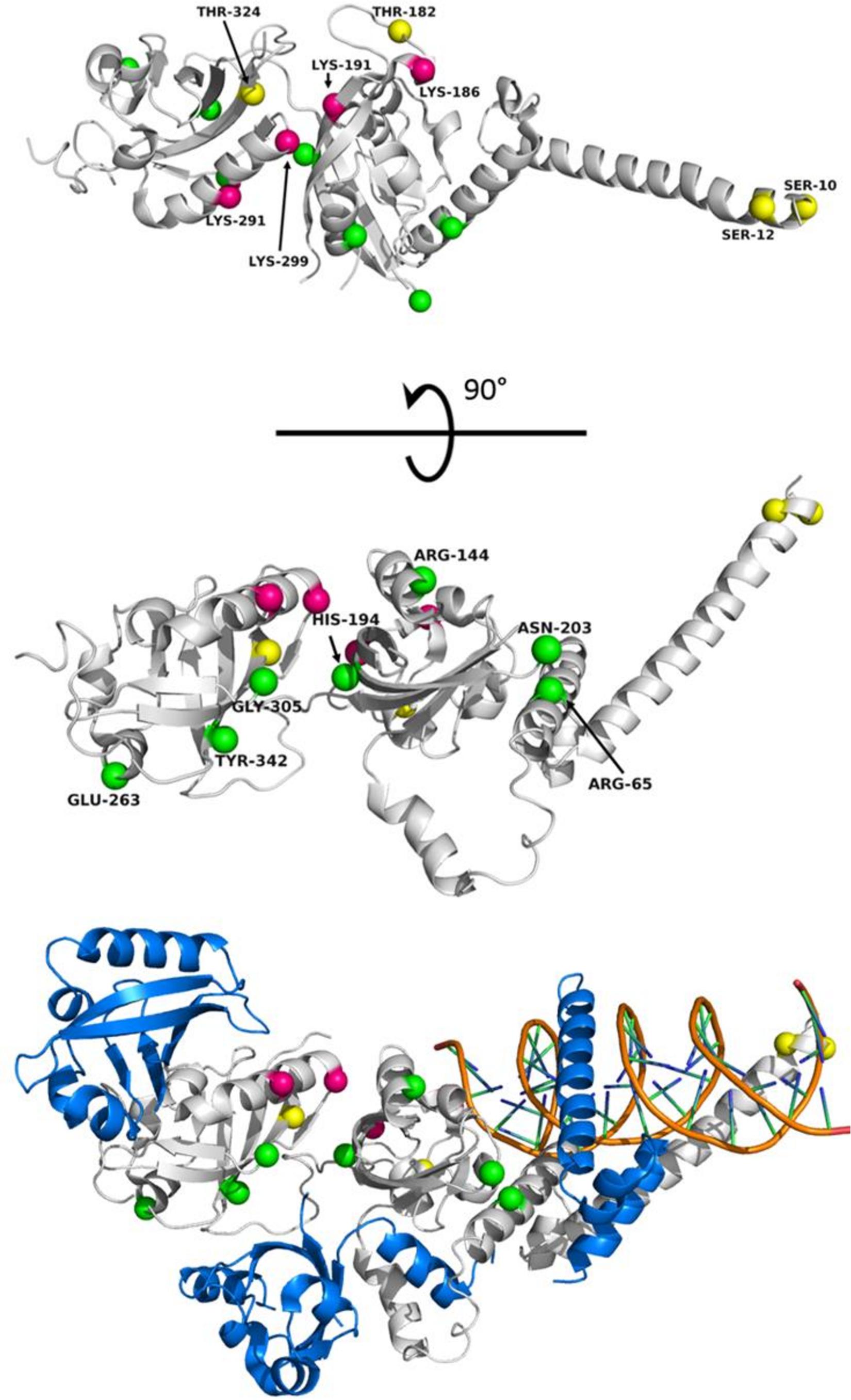
Figure 4. Known EPAS1 variants mapped to the structure of the DNA binding and dimerization domain of HIF-2α in various species. (Top) “Top” view of HIF-2α structure (PDB ID 4ZPK). (Middle) “Side” view of HIF-2α. Spheres representing locations of interest are mapped onto HIF-2α with magenta representing sites of ubiquitylation in humans (K186, K191, K291, and K299) (Akimov et al., 2018), yellow representing phosphorylation sites (S10, S12, T182, and T324), and green representing the locations of identified variants from genetic studies. The species, variation, and publication are listed in Table 1. (Bottom) “Side” view of HIF-2α bound to ARNT and DNA (PDB ID 4ZPK). HIF-2α is shown in gray. ARNT is shown in blue. The DNA double helix is shown in Orange. Note: Tyr333 in chickens maps to residue Tyr342 in humans.
Of the variants listed in Table 1, only a few have been functionally investigated in vivo to establish the effects upon hypoxic signaling. Within the PAS domain of HIF-2α, R144C has been identified as a gain-of-function adaptation in Tibetan horses (Liu et al., 2019), G305S has been identified in high-altitude dogs (Gou et al., 2014), and N203H has been associated with patent ductus arteriosus in Tibetan humans (Pan et al., 2018). The R144C variant was functionally investigated in human lung adenocarcinoma A549 cells, and this variation is reported to stabilize the PAS domain leading to increased ARNT binding. In the case of high-altitude dogs, G305 is highly conserved and invariant among other animals. Homology modeling and prediction of functional effects has suggested that the mutation of G305S may affect thermostability of the PAS domain and is most likely causal mutation for the EPAS1 selective sweep in high-altitude dogs (Gou et al., 2014; Wang et al., 2014). The N203H variant has been linked to non-syndromic congenital heart disease in Tibetans, and functional studies suggest that this particular mutation not only shows decreased transcriptional activity but also results in enhanced protein−protein interactions with both PHD2 and VHL (Pan et al., 2018). Outside of the PAS domains, other identified mutations identified as gain-of-function in the HIF-2α gene are reported in patients with erythrocytosis (Gardie et al., 2014; Pan et al., 2018). These variations are found in the oxygen-dependent degradation domain, clustering around Proline 531, which can disrupt residue hydroxylation and/or recognition of HIF-2α by VHL (Furlow et al., 2009), leading to increased cellular levels of HIF-2α. Aside from general perturbation of protein structure and stability, these variations within the HIF-2α PAS domain highlight how amino acid substitutions can affect protein−protein interactions, post-translational modification, and transcriptional activation, leading to a myriad of adaptive or maladaptive results. These effects can further be perturbed through additional variation within PHD2 and VHL, which have been identified in disease states associated with erythrocytosis and several cancers (Gardie et al., 2014).
Recently, H194R, located at an intramolecular domain interface within HIF-2α that is also part of a binding groove for ARNT binding (Figure 4), was identified as a missense mutation in high-altitude Andeans (Eichstaedt et al., 2017). The location of the H194R mutation may play a role in not only the stability of the intramolecular domain interface of HIF-2α but also in ARNT binding. In addition to structural effects, H194R is adjacent to K191, which has been identified in proteomic studies as a ubiquitylation site in humans (Akimov et al., 2018). The proximity of a bulky positive residue next to this ubiquitylation site may interfere with ubiquitylation (Radivojac et al., 2010) via the VHL-associated E3 ubiquitin ligase complex, which would in turn lead to elevated levels of HIF-2α within the cell. While more functional investigation for variants within HIF-2α and PHD2 need to be conducted to determine specific contributions of the individual variations on hypoxic signaling, the information currently available on the locations and specific variations in EPAS1 among different species showcase how this gene, and others, can provide many different avenues for affecting transcriptional regulation and physiological adaptation toward hypoxia.
Human populations have resided for hundreds of generations in the Tibetan, Andean and Ethiopian highlands in Central Asia, South America, and East Africa, respectively. Over several millennia and despite challenges imposed by environmental hypoxia at altitudes greater than 3500 m, these populations have continuously inhabited and successfully passed down heritable traits necessary for survival. The Tibetan highlands have been continuously inhabited the longest, with a human presence noted 30, 000−40, 000 years before present (Zhang et al., 2018). Initial settlements of high-altitude regions in the Andes date to approximately the Terminal Pleistocene (∼ 12,000 years before present) shortly after the arrival of modern humans in South America (Rademaker et al., 2014), while populations have migrated in and out of the Ethiopian highlands for possibly the past 70, 000 years (Hassen, 1990). The unique sets of traits exhibited by highlanders today suggest distinct population histories have shaped the adaptive and maladaptive milieu in each population, although there is notable within-population variation in several key traits.
The key physiological traits exhibited by each continental population have been recently summarized (Gilbert-Kawai et al., 2014; Simonson, 2015; Moore, 2017; Bhandari and Cavalleri, 2019). On average, Tibetan and Ethiopian highlanders tend to exhibit hemoglobin concentrations that are lower relative to acclimatized sojourners and Andeans at high altitude (Beall, 2007). There is, however, substantial variation within these groups, and Andeans have increased incidence of excessive erythrocytosis and chronic mountain sickness (CMS) (Monge et al., 1989; León-Velarde et al., 2005; Corante et al., 2018). Many Andeans, relative to Tibetans, also exhibit a decreased hypoxic ventilatory response and minute ventilation (Beall et al., 1997) and experience further hypoxia challenges during sleep (Pham et al., 2017, Heinrich et al., 2020), which may exacerbate mal-adaptations in this population. In addition to these hallmark differences, there are notable distinctions across developmental stages and various physiological systems. A detailed list of gene names, protein names, and functions mentioned in this review can be found in Table 2. Efforts to examine the functional roles of this variation are necessary to unravel the complexities of adaptive and maladaptive physiological responses (Hall et al., 2020).
Advances in high-throughput genomic technologies have provided unprecedented insight into population-specific patterns of variation, including signals indicative of genetic adaptations. The power of this approach was first illustrated in human data, whereby adaptive regions of the genome may be revealed in as few as 30 individuals (Pickrell et al., 2007). The extent of distinct and shared genetic adaptations and how these factors relate to the distinct suite of physiological traits exhibited within and across populations is an active area of research (Wuren et al., 2014; Jha et al., 2016; Cole et al., 2017).
The Tibetan Plateau is one of the largest and longest continuously inhabited high-altitude regions in the world, and its inhabitants are the most extensively studied human populations in terms of genetic adaptation to hypoxia. Some of the first genomic studies of Tibetans, revealed adaptive candidate genes involved in hypoxia sensing and response pathways (Beall et al., 2010; Bigham et al., 2010; Simonson et al., 2010; Yi Z. et al., 2010), including EPAS1 and EGLN1 and PPARA (Peroxisome Proliferator Activated Receptor Alpha); these genes were further associated with relatively lower hemoglobin concentration in Tibetans (reviewed in Simonson, 2015). Subsequent studies identified associations between the adaptive PPARA candidate gene and metabolic parameters suggesting reduced fatty acid oxidation (Ge et al., 2012), in addition to greater oxygen utilization and protection from oxidative stress in skeletal muscle (Horscroft et al., 2017; O’Brien et al., 2019). In addition to Tibetan populations, various studies across Sherpa and Central Asian populations provide further support of key adaptive signatures in these populations (Foll et al., 2014; Jeong et al., 2014; Arciero et al., 2018; Gnecchi-Ruscone et al., 2018).
Numerous studies, conducted across various locations within the Qinghai-Tibetan Plateau and Central Asia, corroborate many of the initial findings regarding key genetic adaptations (Bigham et al., 2010; Peng et al., 2011; Xu et al., 2011; Jeong et al., 2014; Wuren et al., 2014). In addition to EPAS1 and EGLN1 HIF-related genes, several non-HIF pathway targets have been reported in more than one independent study, including the hemoglobin gene cluster HBB/HBG2, HFE, PKLR, CYP17A1, and HMOX2 (Simonson, 2015). A recent study across Himalayan populations provides additional gene candidates, including HLa-DQB1/HLA-DPB1, ANKH, RPaa-384F7.2, AC068633.1, ZNF532, and COL4A4 (Yang et al., 2016a; Arciero et al., 2018). Despite tremendous progress on the genomics front, the precise functional variants that provide functional benefits in Tibetans and other populations remain largely unknown (Hall et al., 2020). One exception is at the EGLN1 gene, whereby variants in the first exon found at high frequency in Tibetans (Asp4Glu; Cys127Ser) are reported to result in a gain of PHD2 function, which leads to increased HIF degradation under hypoxic conditions and erythroid progenitor disruption (Lorenzo et al., 2014). Other reports suggest these variants underlie a loss of PHD2 function via defective binding of co-chaperone p23 that would lead to increased HIF activity (Aggarwal et al., 2010; Song et al., 2014) and possibly ventilatory responses (Song et al., 2020).
Whole-genome sequencing of DNA from archaic populations, including Neanderthal and Denisovan individuals, has provided the opportunity to search for genetic admixture within the genomes of modern human populations. Some of the exchanged genetic material has proven adaptive over many generations and provide examples of what is termed adaptive introgression. One notable example of adaptive introgression occurred in Tibetans at the EPAS1 gene locus, which exhibits one of the strongest adaptive signatures in Tibetans and is most similar to Denisovan DNA compared to DNA from other human populations (Huerta-Sánchez et al., 2014; Hu et al., 2017). Therefore, archaic genetic admixture provided variation that putatively helped Tibetans adapt to the high-altitude environment. This finding highlights the importance of understanding distinct population histories and unique genetic backgrounds in studies of genetic adaptation to high altitude. Interestingly, EPAS1 is one of the genes reported most frequently as a candidate gene for high-altitude adaptation in other high-altitude species (Supplementary Table S1), and is further noted to exhibit a signature of adaptive introgression across species.
The first analysis of genome-wide adaptation in two Andean populations, the Quechua and Aymara, and subsequent comparison with Tibetans, concluded the genetic basis for altitude adaptation was dissimilar in the two continental populations, although an exception was noted at the EGLN1 gene region (Bigham et al., 2010). Of the reported Tibetan variants in the first exon of EGLN1, c.12C > G (Asp4Glu) is absent and 380G > C (Cys127Ser) is found at low frequency in Andeans from Cerro de Pasco, Peru (Heinrich et al., 2019), further supporting the occurrence of different adaptive mechanisms despite an overlap in the adaptive genetic signal. Preliminary epigenetic investigation of specific sites in the EGLN1 region in Andeans suggests distinct levels of methylation between Andeans with and without excessive erythrocytosis (Julian, 2017; Julian and Moore, 2019), and recent studies show variants at this locus are associated with exercise capacity in Andeans (Brutsaert et al., 2019). More than 30 other genes were reported by Bigham et al. (2010), including EDNRA and PRKAA1, which were both associated with birthweight and the latter with metabolic homeostasis in subsequent genotype-phenotype analysis (Bigham et al., 2014). Several additional genes reported in these studies have since been reported as adaptive in other Andean populations or identified in other highland populations (e.g., the beta hemoglobin (HBB) cluster region and EDNRA) (Simonson, 2015; Supplementary Table S1).
Additional studies of Andean adaptation suggest genes related to cardiac function, rather than hypoxia responses, are essential for survival in this population. Cardiac-related genes VEGFB and ELTD1 were identified within the strongest regions of selection in the Colla population from the Argentinian Andes (Eichstaedt et al., 2014). Analysis of low-coverage whole-genome sequence highlighted a distinct set of genes related to cardiovascular function (BRINP3, NOS2, and TBX5) and suggested these genes, rather than those in hypoxia-response pathways, are the strongest targets of selection in Andeans (Crawford et al., 2017).
Another whole-genome analysis based on analyses of Andeans with and without CMS reported SENP1 and ANP32D as top targets for adaptation (Zhou et al., 2013), and gene expression in fibroblasts is lower in cells derived from individuals without compared to those with CMS (Zhou et al., 2013). Additional studies based on single nucleotide polymorphism (SNP) microarray analysis indicate FAM213A and SFTPD (Valverde et al., 2015), are associated with oxidative stress and respiration and innate defenses, respectively, in Quechua and Aymara populations.
Using a new statistical approach to detect very recent positive selection, Eichstaedt et al. (2017) compared whole-genome sequences from high-altitude Argentinians and lowland Native Americans and identified a missense variant in the EPAS1 gene, one of the major adaptive genes reported in Tibetans, and another in GPR126, which was associated with lung function (Eichstaedt et al., 2017). This is the first study to report EPAS1 as adaptive in Andean populations and, like EGLN1, suggests different variants play pivotal roles in each population’s adaptive genetic profile.
Highland Ethiopians also exhibit distinct physiological and genetic adaptations to high altitude, although they have been less studied than their Tibetan and Andean counterparts. The population history of Ethiopian highlanders is complex, due to repeated migrations into and out of the Ethiopian highlands spanning the past 70, 000 years (Hassen, 1990). Several genomic studies published thus far provide important insights into convergence of the HIF as well as distinct adaptive pathways (Scheinfeldt et al., 2012; Bigham and Lee, 2014; Petousi and Robbins, 2014; Simonson, 2015; Moore, 2017).
Putatively adaptive copies of the THRB gene region as well as PPARA and EPAS1 identified in Tibetans (Simonson et al., 2010) show relationships with hemoglobin concentration in Amhara Ethiopians (Scheinfeldt et al., 2012). EDNRB (endothelial receptor B), previously reported as a top selection candidate in Andeans (Bigham et al., 2010), is also reported in Amhara Ethiopians, and knockdown of this gene increases hypoxia tolerance in mice (Udpa et al., 2014). The gene family member EDNRA is also a top candidate gene in Tibetans (Simonson et al., 2010). BHLHE41, although not associated with hemoglobin, is a key HIF pathway gene and top selection candidate in Amhara, Oromo, and Tigray Ethiopians (Huerta-Sánchez et al., 2013). In addition to these hypoxia-associated genes, three others (VAV3, which encodes vav guanine nucleotide exchange factor 3, and RORA that encodes the RAR-related orphan receptor A), are reported as top candidates in Amhara Ethiopians (Scheinfeldt and Tishkoff, 2013). Whole-genome sequence analyses indicate three genes contained within the same region of chromosome 19 identified as adaptive targets in Oromo and Simen Ethiopians, CIC, LIPE, and PAFAH1B3 (that encode capicua transcriptional repressor, lipase E hormone-sensitive type, and platelet-activating factor acetylhydrolase 1b catalytic subunit 2, respectively) have orthologs in Drosophila that afford tolerance to hypoxia (Udpa et al., 2014). Comparison of genome-wide epigenetic profiles from saliva samples collected in high and low altitude Oromo Ethiopians indicated differences at several genes (Alkorta-Aranburu et al., 2012).
While various candidate genes are highlighted due to replication and/or association with phenotype, hundreds of distinct putatively adaptive gene regions have been identified and may or may not be reported in individual studies to date. Therefore, inconsistencies among studies may reflect differences in analytical approaches and reporting, variation within a continental region, and/or the stage and degree of adaptation. Additional efforts to standardize analysis within and across continental populations are needed to fully understand the extent of overlap among humans.
Many genes originally highlighted as adaptive targets in human high-altitude studies have also emerged as top candidates for genetic adaptation in other species under various conditions of environmental hypoxia. For example, as in high-altitude human populations, adaptations involving hematological values are found in most hypoxia-adapted high-altitude animal populations. Specifically, high-O2 affinity hemoglobin is found in many highland animals, including alpacas (Piccinini et al., 1990), Andean and bar-headed geese (Jessen et al., 1991; Zhang et al., 1996; Liang et al., 2001), deer mice (Storz et al., 2007, 2009), and yaks (Weber et al., 1988) (among others), and also in many lowland animals that are adapted to hypoxia, such as naked mole-rats (Johansen et al., 1976). Despite major overlap in genetic pathways (i.e., the hemoglobin gene cluster and associations with blood-O2 affinity and the hypoxia HIF pathway genes, e.g., EPAS1 and EGLN1), different variants with putatively distinct functions are reported as targets of selection among highland human and other animal populations.
More recent analyses of whole genome sequences (e.g., in Andeans, Zhou et al., 2013; in Tibetans, Hu et al., 2017) indicate that non-protein coding variants, including those in heterochromatic or DNA methylated portions of the genome, are crucial for adaptation. In such cases, the effects of increased or decreased gene expression could vary across tissues and/or stages of development (in contrast to protein-coding variants that result in uniform alterations across all cells). An understanding of these fine-tuned, context-specific changes could provide much needed insight into molecular mechanisms of adaptation that influence genetic pathways in ways that are similar or not to other hypoxia-adapted species.
As humans spread across the globe and to altitude, they brought a variety of domesticated mammals for companionship, labor, defense, and food. As a result, these animals have undergone strong selection for the tasks they perform for their human masters (e.g., for size, etc.), and so analysis of genetic adaptations to altitude in these populations must take this into account. Nonetheless, the study of domesticated high-altitude mammal populations provides an opportunity to contrast adaptive strategies to hypoxia between relatively short- and long-term (on a generational scale) exposures to low oxygen environments (Witt and Huerta-Sanchez, 2019).
One good example of such anthropocentric-driven short-term adaptation to chronic hypoxia in domesticated mammals can be found in feral horses of the Andean Paramo. This region of the Andes is a challenging environment with large temperature, humidity, and rainfall variance, high radiation, and minimal food availability. As a result, ungulates that had naturally evolved to live in this niche have largely shifted their range to more temperate regions. Conversely, populations of feral horses, which were originally introduced by Spanish conquistadores in the 1500’s, have thrived in this niche and the success of these introduced horses over the past ∼200 generations provides an important opportunity to evaluate genetic changes driven by this relatively short-term population-history exposure to hypoxia at altitude. Importantly, genes in Andean feral horse populations can be easily compared to those of their lowland Iberian ancestors (Hendrickson, 2013). A recent genomic analysis between these two populations revealed a mutation in EPAS1 in the high-altitude population. Specifically, BIEC2– 310909 (rs69041973) is highly different between the lowland and highland species, although this SNP is intronic and has no known function. Unfortunately, little is known regarding physiological adaptations to hypoxia in this species and so it is difficult to draw direct comparisons between human studies in this case; however, the commonality of the occurrence of mutations in HIF-related genes in high-altitude populations is important to note and will be a recurring theme in this section.
Another useful study model are dogs, whose range has expanded in step with their human companions since the paleolithic era (Germonpre et al., 2009). On the Tibetan plateau, Tibetan mastiffs, which are derived from the native lowland Chinese Native dog, have undergone human-driven selection for life at high altitude (Li and Zhang, 2012). As a result, these mastiffs have lower hemoglobin levels than do Chinese native dogs (Wen and Yuan, 1998), and genome analysis revealed a mutation in EPAS1 and also in SIRT7, which negatively regulates HIF1 and HIF2 (Li et al., 2014). The frequency of specific variants within these genes scale with altitude, as demonstrated by a second study that compared the genomes of five dog species whose range spans continuous altitudes along the Tea Horse Road in the Tibetan Plateau and a European dog species as an out-group (Gou et al., 2014). In this analysis, four novel non-synonymous mutations in EPAS1 were found between the high-altitude dog population and both the mid- and low-altitude populations studied. Furthermore, all remaining genes that varied in this comparison are regulated by the HIF pathways, including the HBB cluster region, similar to Tibetan and Andean human populations. These researchers also examined hematological properties in these species and found that the high-altitude populations had a lower peripheral vascular resistance, which would enhance the flow of blood otherwise made more viscous by higher hemoglobin concentrations at altitude.
Intriguingly, and as we see in high-altitude human populations, this adaptation in high-altitude adapted dogs may be due to accelerated adaptations through admixture: i.e., the spread of beneficial alleles between closely related species. In this case, there is evidence that mutations to EPAS1 in high-altitude dogs is due to admixture from interbreeding with Tibetan wolves (VonHoldf et al., 2017). The gray wolf is the most widely distributed terrestrial mammal with as many as 32 sub-species. One of these is the Tibetan gray wolf (Canis lupis chanco), which is endemic to the Tibetan plateau (Aggarwal et al., 2003). Genetic comparison between the Tibetan gray wolf and low-altitude wolf populations in China revealed positive selection of hypoxia-related genes in HIF-signaling pathways, including three SNPs unique to the highland populations in EPAS1, two in RYR2, and one in ANGPT1 (Zhang W. P. et al., 2014). Along with EPAS1, ANGPT1 functions in the HIF pathway and can increase vascularization and thereby oxygen delivery (Prabhakar and Semenza, 2012). ANGPTL4 functions in this same gene pathway is reported as an adaptive gene candidate in Tibetans (Simonson et al., 2010). A recent study examined the degree of admixture between Tibetan mastiffs and wolves at the EPAS1 gene and reported an excess of highland gray wolf ancestry at the EPAS1 locus in the highland domestic dogs (VonHoldf et al., 2017). This finding suggests that an adaptive variant of EPAS1 was transferred from wolves to dogs through interbreeding, thereby accelerating the adaptation to high altitude in the later species through adaptive introgression.
A similar adaptive history is apparent in analysis of Tibetan sheep (Ovis aries), which were also brought to altitude as a domesticated animal by humans [∼ 3, 100 years ago (Hu et al., 2019)]. Contrary to in dogs, higher hemoglobin concentration is observed in high altitude-resident populations of Tibetan sheep relative to lowland populations, along with higher hematocrit (Wei et al., 2016) (although peripheral vascular resistance has not been examined in this species and adaptations here may compensate for increased blood viscosity). These adaptations are associated with shorter-term life histories at altitude. However, positive selection has occurred in the form of 12 mutations to the EPAS1 gene in Tibetan sheep (Wei et al., 2016). Importantly, this study also examined the effect of several splice variants of EPAS1 on hematological parameters and determined that at least one mutation was associated with a gain of hemoglobin concentration. This divergent physiological phenotype, despite evolutionary pressure on the same gene, highlights the complexity of relating physiological and genetic studies of adaptation to high altitude. Interestingly, as in dogs, the changes in the sheep genome may be due to adaptive introgression from local species, in this case from argali (Ovis ammon), which may have accelerated the adoption of mutations to commonly targeted genes (Hu et al., 2019), similar to reports of high-altitude dog populations.
Another interesting model, and one of the largest resident mammals on the Tibetan Plateau, are yaks (Bos grunniens). Yaks currently exist in both domesticated and wild populations on the plateau, but which were initially brought to altitude ∼ 4, 500 years ago by humans (Meyer et al., 2009; Qi et al., 2013; Qiu et al., 2015). Relative to other high-altitude mammals, the physiology of yaks has been reasonably well-characterized, and adaptations to hypoxia in this species include an increased pulmonary surface area, reduced gas diffusion barriers, lower hemoglobin, and larger lungs, than are found in lowland cattle (Qi et al., 2019). Yaks at sea level and at altitudes of 2260−4500 m exhibit lower pulmonary artery pressures (Ishizaki et al., 2005). Genomic analysis of yaks indicates several HIF-specific mutations have occurred in high vs. low altitude populations, including to two important regulators of HIF (ADAM17 and ARG2) (Qiu et al., 2012, 2015; Guang-Xin et al., 2019). Furthermore, transcriptomic analysis of yaks endemic to an altitudinal gradient demonstrated that EPAS1 expression increases with altitude (Qi et al., 2019). Intriguingly, genomic analysis of Tibetan cattle also suggests introgression of adaptive mutations driven by life in hypoxia due to hybridization with yaks (Wu et al., 2018). Clearly, this is a common route of accelerating adaptive mutations in domesticated species brought to high-altitude by humans.
At the other end of this spectrum are species that have lived at high altitude for hundreds of thousands of years in the wild (and/or that have been domesticated by high altitude human populations but not transported to high altitude). One example species are Tibetan wild boars, which live at ∼ 4300m. A recent genome comparison between this species and the European domestic Duroc pig revealed 13 positively selected genes in the “response to hypoxia” category (Li et al., 2013), of which several are downstream targets of HIF pathways, including FIGF, PGF, and VEFPC. A similar phenotype is found in Tibetan pigs, which are native to the Tibetan Plateau and inhabit a niche ranging in altitude from 2, 900 to 4, 300 m. Intriguingly, and similarly to adapted Tibetan human populations, Tibetan pigs have a blunted erythropoietic response to high-altitude hypoxia and have lower hemoglobin concentrations relative to lowland pigs at sea level or acclimated to high-altitude (Ai et al., 2014). At the genetic level, genome analysis revealed that Tibetan pig populations (after considering various sub populations and back flow of genes from low altitude populations) exhibit a loss of function mutation in EPAS1 (Ai et al., 2013, Ai et al., 2014; Li et al., 2013). In addition, another study demonstrated that mutations to an intron 5′-CGTG-3′ sequence of TMPRSS6 increase with increasing residence altitude in Tibetan pigs (Kong et al., 2019). TMPRSS6 is a downstream gene of the HIF pathway that is associated with serum iron concentration and hemoglobin levels (Chambers et al., 2009; Tanaka et al., 2010). Hemoglobin also scaled with altitude in this study (Kong et al., 2019). The 5′-CGTG-3′ intron sequence is associated with binding to HREs (Camenisch et al., 2001), suggesting that HIF modulation of blood properties is under Darwinian selection with increasing altitude in this species. This mutation thus likely contributes to blunted erythropoietic response to hypoxia in these pigs.
An opposing hematological phenotype is found in Tibetan cashmere goats (Capra hircus), which are one of the more ancient domesticated species in Tibet and have the broadest altitudinal range of Chinese herbivores, spanning from sea level to the top of the Tibetan Plateau (∼ 0 – 5,000 m) (Wang et al., 2011). Unlike many large high-altitude mammals, some physiological data is available regarding their adaptations to altitude relative to lowland populations: highland populations have a higher hemoglobin concentration and a lower resting heart rate (Huang, 1980; Renzheng and Ciren, 1992). This is in striking contrast to previous studies in Tibetan human and pig populations, wherein hemoglobin concentrations are lower relative to non-adapted human populations but blood flow is higher. Here, genetic analysis revealed a missense mutation in EPAS1 which results in an amino acid substitution in HIF-2α at a site adjacent to its dimerization interface with HIF-1β (Wang et al., 2011; Song et al., 2016). This is intriguing and suggests that mutations to EPAS1 may result in either an up or down-regulation of HIF-related signaling pathways. This divergence in gene-driven physiological function, which is dependent upon the site of mutation, may be a key determinant of adaptation vs. maladaptation to high altitude in various animal and human populations.
Another ungulate, the Tibetan antelope (Pantholops hodgsonii), is a particularly athletic species found at elevations of ∼4, 000−5, 000 m. A recent genomic study examined conserved genetic changes between this species and two species of high-altitude pika: the American Pika (Ochotona princeps), and the Tibetan Pika (O. curzoniae) (Ge et al., 2013). This study identified seven genes (ADORA2A, CCL2, ENG, PIK3C2A, PKLR, ATP12A, and NOS3) as being under positive selection in all three high-altitude species. Of particular interest is NOS3, which influences nitric oxide production and thereby blood vessel diameter. In addition, PKLR is a top candidate of selection in Tibetans (Simonson et al., 2010, 2012; Yi X. et al., 2010).
One of the best studied high-altitude mammals is also one of the smallest. The plateau zokor (Myospalax baileyi) inhabits the Tibetan plateau and dwells in underground nests, in which the ambient oxygen level is likely even lower than that at the surface of the plateau (Zhou and Dou, 1990). Relative to lowland rodents, plateau zokors have high hemoglobin and hematocrit, and a low heart rate (Wei and Ma, 2001; Wei et al., 2006). A genetic comparison between this species and the Norwegian rat (Rattus norvegicus) revealed positively selected genes in the blood vessel development category, including ANGPTL3, PRCP, SEC24B, EPHA1, MCAM, KAT6A, MYH9, CYSLTR2, MYLK, SPINT1, PPAP2B, and STRA6 (Shao et al., 2015). Most of these genes are under regulation by the HIF pathway. In addition, evaluation of parallel evolution between the plateau zokor and the naked mole-rat (see below) revealed several genes related to hypoxia that were similarly modified in both hypoxia-tolerant species, including EPAS1, SLC29A1, MYO5B, AJUBA, TGFBR3, VASN, ACAA2, PINK1, SIRT1, SIRT2, SOD2, and NARFL. Of particular interest is that EPAS1 mutations in both naked mole-rats and plateau zokors are identical. AJUBA is also a key regulator of HIF function (Bridge and Sharp, 2012) and is similarly mutated in both species (Shao et al., 2015). Furthermore, transcriptome analysis between different populations of Zokors endemic to increasing elevations indicated that mutations to EPAS1, COX1, and EGLN1 all scaled with increasing altitude (Cai et al., 2018).
Many avian species live in high-altitude regions and/or transverse high-altitude regions in their annual migrations (Altshuler, 2006; Bishop et al., 2015). In addition to the hypoxic and hypothermic challenges faced by other species at altitude, birds face additional challenges during active flight related to low air density and cold air (Sun et al., 2016). Some birds that are resident to high-altitude environments compensate for these challenges with increases in wing size and stroke amplitude (Altshuler et al., 2004; Sun et al., 2016); however, this comes with the added cost of greater body mass and a higher metabolic rate, which in turn enhances energetic challenges in hypobaric hypoxia. Thus, the lifestyles of different bird species likely have a large effect on the evolution of adaptations to life at high-altitude.
Perhaps the best example of an avian species endemic to high altitude with high exercise requirements are apex avian predators, such as raptors. On the Tibetan Plateau, this is the saker falcon (Falco cherrug), which colonized this region as recently as 2000 years ago (Pan et al., 2017). Little is known regarding physiological adaptations to hypoxia in this species, but a recent study compared the genomes of falcon populations across Eurasia at three elevations (lowland, steppe, and Tibetan Plateau) and found a strong positive exonic selection signal on the EPAS1 gene (Pan et al., 2017), despite this very short colonization time at altitude. Hemoglobin was also altered in this comparison, and transcriptomic analyses revealed that 50% of the transcripts (6/12) that were upregulated in the saker falcon were related to hypoxia response or hematopoiesis. Here, transcripts that are regulated by EPAS1 transcription were also elevated in the saker falcon and transcripts whose expression levels correlated with that of EPAS1 were almost uniformly elevated (21/24 transcripts).
Another avian population of interest are Andean ducks, of which there are many species that include both low and high-altitude populations, making these an excellent model for comparison. A recent study examined convergent evolution at the HIF pathway between high and low elevation populations of the yellow-billed pintail (Anas georgica), the cinnamon teal (A. cyanoptera), and the speckled teal (A. flavirostris flavirostris/A. f. oxyptera) (Graham and McCracken, 2019). This study revealed strong support for convergent evolution on the HIF pathway, with common mutations found in the high-altitude populations on the exonic regions of EPAS1 and the intronic regions of ELGN1.
An interesting alternative model are flightless birds. For example, the Tibetan ground tit (Parus humilis), which is a largely ground-bound bird that is endemic to the Tibetan Plateau and is only found above the treeline (i.e., above 3, 300 m) (Qu et al., 2013). Genetic analysis of the Tibetan ground tit revealed that the majority of gene ontology (GO) categories modified in this species [relative to the closely related great tit (P. major) and yellow-cheeked tit (P. spilonotus)], were associated with increased energy metabolism (Qu et al., 2013), presumably to support heat production in this cold climate. Furthermore, and similar to Tibetan human populations, some genes related to hypoxia are also under positive selection in the ground tit, including the HIF-1 alpha subunit inhibitor (HIF1AN), ANGP4 (ANGPTL4), which is involved in angiogenesis, and ADAM9 (ADAMTS9), which is an important regulator of HIF1 (Qiu et al., 2012; Qu et al., 2013). Each of these is reported within the top 2% of selection candidate genes in Tibetans (Simonson et al., 2010). Interestingly, EPAS1 was not under selection in this avian species, at least in this comparison.
Similarly, the Tibetan chicken (Gallus gallus), which is an aboriginal breed found on the Tibetan plateau, is flightless and adapted to life at high altitude. Compared to low altitude chickens, the Tibetan chicken is smaller, and has higher hematocrit and hemoglobin concentration and also a higher hemoglobin-oxygen affinity (Zhang et al., 2007). A recent study sequenced and compared SNPs of Tibetan and lowland chickens to test for potential variations in EPAS1 in these species (Li et al., 2017), and this analysis revealed two SNPs in G. gallus EPAS1 that correlate with high-altitude adaptation.
A key consideration in evaluating genetic changes in animals and human populations that are resident to high altitude is that such environments also incorporate numerous powerful evolutionary drivers in addition to hypoxia. For example, reduced atmospheric protection at altitude results in increased UV exposure and radiation (Blumthaler et al., 1997); annual and daily temperature and humidity swings are also more pronounced at high altitude than at lower altitudes. As a result, it can be difficult to filter out genetic changes that underlie adaptations to life in hypoxia from genetic changes that are due to other selective pressures. This limitation can be partially overcome by studying ectothermic species that live at high altitude, in which endogenous thermoregulatory complications endemic to life at elevation may be removed from consideration. Toward this aim, the genomes of three Tibetan hot-spring snakes (Thermophis) were recently sequenced and compared to those of lowland ectotherms (Li J. T. et al., 2018). Not surprisingly, this study identified three shared amino acid replacement mutations in the EPAS1 gene of hot-spring snakes that are consistent with a downregulation in function relative to that of the lowland species. One of these mutations was in the DNA-binding domain of EPAS1, and this mutation limited erythropoietin expression in human 293T cells when transfected using a plasmid approach. This study indicates that common mutations in the HIF-signaling architecture have occurred in response to the adaptive pressure of high-altitude hypoxia in both resident endothermic and ectothermic species and that, similar to the other studies discussed above, point mutations in EPAS1 may have up or down-regulating effects on physiological adaptations.
Another ectothermic species of interest is the toad-headed sand lizard (Phrynocephalus erythrurus), which typically live at altitudes of >4, 500 m and are generally considered to be the most high-altitude-adapted lizard known (Yang Y. Z. et al., 2015). A comparative analysis between the genomes of this species and that of a lower-altitude adapted congener (P. putjatia) revealed that UBE2D1 was positively selected for in the high-altitude population (Yang Y. Z. et al., 2015). This gene enables the ubiquitination of HIF1 alpha. Not surprisingly, the hemoglobin concentration and capillary density of P. erythrurus is greater than in the lowland cousin (He et al., 2013). Another study comparing lowland and highland populations of toad-headed sand lizards (P. przewalskii and P. vlangalii, respectively) found similar results (Yang et al., 2014). Specifically, ADAM17 was positively selected in the high-altitude population, and so was MB, which encodes for myoglobin. Once again, in this comparison the high-altitude population expresses higher hemoglobin concentration and hematocrit than the lowland population (He et al., 2013). Interestingly, a similar analysis of a pair of ranid frog species, including the high-altitude dwelling Rana kukunoris and the low-altitude dwelling R. chensinensis similarly revealed positive selection on UBE2D1, suggesting convergent evolution at this site in ectotherms adapted to hypobaric hypoxia (Yang Y. Z. et al., 2015).
As is clear from the studies reviewed above, the study of genomic adaptations to life at high altitude has largely focused on endothermic species and a small number of ectothermic vertebrates. However, there are also numerous aquatic species that are native to highland bodies of water and fish in these niches are beginning to draw attention as model systems in which to study adaptations to life at altitude. To date, no study has examined genomic adaptation in a highland fish but the genomes of two fish species that are endemic to the Tibetan plateau – Glyptosternon maculatum and Trilophysa tibetana – have been recently sequenced with an eye toward future use of these genomes to evaluate mutations associated with life at high altitude (Liu et al., 2018; Yang et al., 2019).
It is clearly apparent that life in hypobaric hypoxia at altitude has driven genetic and physiological adaptations related to hematological parameters in almost all species studied to date. Similarly, this evolutionary pressure appears to have modified angiogenesis, which is largely initiated by the HIF pathway (Rey and Semenza, 2010), suggesting that vascular changes are a crucial response to life in chronic hypoxia. Indeed, candidate genes that contribute to angiogenesis and/or vasculogenesis have been detected in numerous species living at high altitudes, although there is little overlap in specific genes among species. For example, positive selection of SRF, TXNRD2, and WNT7B were detected in the ground tit (Qu et al., 2013), NOS3 was detected in the Tibetan antelope (Ge et al., 2013), and ADAM17 was detected in both the pig (Ai et al., 2013), toad-headed lizard (Yang et al., 2014), yak (Qiu et al., 2012), and humans (Simonson et al., 2010), and all modify vascularity. These findings suggest the potential occurrence of convergent evolution along multiple genetic pathways toward a common physiological phenotype in high-altitude adapted populations, although additional studies are required to link genotypes to phenotypes. As we will see in the next section, hypoxia in lowland niches has driven similar adaptations. An extensive table of genes associated with high-altitude adaptation shared among humans and other hypoxia-adapted species, along with references, can be found in Supplementary Table S1.
Beyond studies in high-altitude resident ectotherms, studies in species that are native to low altitude hypoxic environments are perhaps more useful in teasing out genetic adaptations that commonly arise as a result of hypoxia specifically, from those related to damage repair (e.g., following radiation) or thermoregulation. Indeed, hypoxia and hypothermia often have opposing effects on development, with the former favoring small body size and low metabolic rates and the later favoring the opposite. Therefore, physiological and genetic studies of high-altitude populations may be confounded by these opposing evolutionary drivers, clouding the impact of hypoxia-alone on genetic mutations and physiological adaptations. Fortunately, hypoxic environments are commonly found beyond high altitude niches. For example, underground burrows and frozen lakes and ponds and are lowland niches in which many hypoxia-tolerant species are found, across the animal kingdom. A key caveat in this comparison, however, is that lowland hypoxic environments are likely not consistently or uniformly hypoxia; denizens of these niches likely experience gradients of hypoxia and even periods of normoxia in their day-to-day activities. This periodic intermittent hypoxic exposure results in divergent stresses to which these animals must adapt (e.g., reoxygenation injury, hypercapnia) and therefore likely exerts a different selection pressure than life in hypobaric hypoxia; the effects of normobaric and hypobaric hypoxia further remain unclear and require additional standardized analyses (Coppel et al., 2015). However, genomic analysis where available has revealed genetic mutations that in many cases are common in their target to those found in high-altitude adapted animal populations.
Of particular interest are naked mole-rats (Hetercephalus glaber), which are among the most hypoxia-tolerant mammal presently identified and tolerate minutes of complete anoxia, hours at 3% O2, and days to weeks at 8% O2 (Pamenter et al., 2015; Chung et al., 2016; Park et al., 2017). Naked mole-rats are endemic to north eastern Africa and are not found at high-altitudes. However, they live in large colony groups and putatively experience severe hypoxia in their crowded nest chambers due to the combination of a large number of animals breathing in small space and poor gas diffusion through the surrounding soil. A recent genome study of this species revealed mutations in the VHL binding domain (Kim et al., 2011), which results in a high endogenous expression of HIF protein in this species. Perhaps not surprisingly, naked mole-rats exhibit a hemoglobin-oxygen affinity that is similar to that of neonatal rodents (Johansen et al., 1976). These observations highlight that chronic hypoxia may not be required to drive the physiological and genomic mutations that are commonly found in high-altitude vertebrate populations.
As mentioned above, genes in the HIF pathway have been targets of selection across multiple species. How these variants relate to phenotypes within and across species will provide import insight into the primary and secondary effects of adaptations and maladaptation to hypoxia (Storz and Scott, 2019) and require detailed functional assessments (Hall et al., 2020).
From studies of high-altitude adapted human and animal populations it has become abundantly clear that life in hypobaric hypoxia has driven the convergent evolution of specific genetic pathways across very disparate animals. In species that have lived at altitude for shorter periods of time, admixture from other species that have lived at altitude for longer has played a key role in accelerating these adjustments. Intriguingly, the occurrence of hematological adaptations that enhance blood flow seems to be a common theme at both the physiological and genetic levels, which are easily studied across species. In recent years, genetic analyses have in some ways outpaced physiological studies as high-throughput population-wide approaches become more widely utilized. However, the physiological impact of many of the mutations identified in high-altitude populations and species are unknown, and it is highly likely that adaptations beyond hematological function will prove to be important in tolerating hypoxia at altitude. Indeed, it is very likely that adaptations at the metabolic and cellular levels are critical to tolerating life in hypoxia. Conversely, numerous studies have examined physiological adaptations to hypoxia in a systemic context in a wide variety of comparative animal species; however, genetic information is lacking for many of these species. As such, the next major step in this field will be to marry genetic, omic, and physiological knowledge to fully elucidate beneficial physiological adaptations to hypoxia and their evolutionary origins.
MP and TS designed the report. All authors produced figures and drafted and contributed comments to the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00743/full#supplementary-material
Adachi, K., Kim, J., Asakura, T., and Schwartz, E. (1990). Characterization of two types of fetal hemoglobin: alpha 2G gamma 2 and alpha 2A gamma 2. Blood 75, 2070–2075. doi: 10.1182/blood.v75.10.2070.2070
Aggarwal, R. K., Ramadevi, J., and Singh, L. (2003). Ancient origin and evolution of the Indian wolf: evidence from mitochondrial DNA typing of wolves from Trans-Himalayan region and Pennisular India. Genome Biol. 4:6.
Aggarwal, S., Negi, S., Jha, P., Singh, P. K., Stobdan, T., Pasha, M. Q., et al. (2010). EGLN1 involvement in high-altitude adaptation revealed through genetic analysis of extreme constitution types defined in Ayurveda. Proc. Natl. Acad. Sci. U.S.A. 107, 18961–18966.
Ahmed, S. I. Y., Ibrahim, M. E., and Khalil, E. A. G. (2017). High altitude and pre-eclampsia: adaptation or protection. Med. Hypotheses 104, 128–132. doi: 10.1016/j.mehy.2017.05.007
Ai, H. S., Huang, L. S., and Ren, J. (2013). Genetic diversity, linkage disequilibrium and selection signatures in Chinese and Western Pigs revealed by genome-wide SNP markers. PLoS One 8:e56001. doi: 10.1371/journal.pone.0056001
Ai, H. S., Yang, B., Li, J., Xie, X. H., Chen, H., and Ren, J. (2014). Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genomics 15:834. doi: 10.1186/1471-2164-15-834
Akimov, V., Barrio-Hernandez, I., Hansen, S. V. F., Hallenborg, P., Pedersen, A.-K., Bekker-Jensen, D. B., et al. (2018). UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 25, 631–640. doi: 10.1038/s41594-018-0084-y
Alam, H., Weck, J., Maizels, E., Park, Y., Lee, E. J., Ashcroft, M., et al. (2009). Role of the phosphatidylinositol-3-kinase and extracellular regulated kinase pathways in the induction of hypoxia-inducible factor (HIF)-1 activity and the HIF-1 target vascular endothelial growth factor in ovarian granulosa cells in response to follicle-stimulating hormone. Endocrinology 150, 915–928. doi: 10.1210/en.2008-0850
Alekseyenko, A. A., Walsh, E. M., Zee, B. M., Pakozdi, T., Hsi, P., Lemieux, M. E., et al. (2017). Ectopic protein interactions within BRD4–chromatin complexes drive oncogenic megadomain formation in NUT midline carcinoma. Proc. Natl. Acad. Sci. U.S.A. 114, E4184–E4192.
Alkorta-Aranburu, G., Beall, C. M., Witonsky, D. B., Gebremedhin, A., Pritchard, J. K., and Di Rienzo, A. (2012). The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 8:e1003110. doi: 10.1371/journal.pgen.1003110
Altshuler, D. L. (2006). Flight performance and competitive displacement of hummingbirds across elevational gradients. Am. Nat. 167, 216–229. doi: 10.1086/498622
Altshuler, D. L., Dudley, R., and Mcguire, J. A. (2004). Resolution of a paradox: hummingbird flight at high elevation does not come without a cost. Proc. Natl. Acad. Sci. U.S.A. 101, 17731–17736. doi: 10.1073/pnas.0405260101
Appelhoff, R. J., Tian, Y. M., Raval, R. R., Turley, H., Harris, A. L., Pugh, C. W., et al. (2004). Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 279, 38458–38465. doi: 10.1074/jbc.m406026200
Arany, Z., Huang, L. E., Eckner, R., Bhattacharya, S., Jiang, C., Goldberg, M. A., et al. (1996). An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 93, 12969–12973. doi: 10.1073/pnas.93.23.12969
Arciero, E., Kraaijenbrink, T., Haber, M., Mezzavilla, M., Ayub, Q., Wang, W., et al. (2018). Demographic history and genetic adaptation in the Himalayan region inferred from genome-wide SNP genotypes of 49 populations. Mol. Biol. Evol. 35, 1916–1933. doi: 10.1093/molbev/msy094
Azmi, S., Sun, H., Ozog, A., and Taneja, R. (2003). mSharp-1/DEC2, a basic helix-loop-helix protein functions as a transcriptional repressor of E box activity Andstra13expression. J. Biol. Chem. 278, 20098–20109. doi: 10.1074/jbc.m210427200
Baay-Guzman, G. J., Bebenek, I. G., Zeidler, M., Hernandez-Pando, R., Vega, M. I., Garcia-Zepeda, E. A., et al. (2012). HIF-1 expression is associated with CCL2 chemokine expression in airway inflammatory cells: implications in allergic airway inflammation. Respir. Res. 13:60. doi: 10.1186/1465-9921-13-60
Beall, C. M. (2007). Two routes to functional adaptation: tibetan and Andean high-altitude natives. Proc. Natl. Acad. Sci. U.S.A. 104, 8655–8660. doi: 10.1073/pnas.0701985104
Beall, C. M., Cavalleri, G. L., Deng, L., Elston, R. C., Gao, Y., Knight, J., et al. (2010). Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. U.S.A. 107, 11459–11464. doi: 10.1073/pnas.1002443107
Beall, C. M., Strohl, K. P., Blangero, J., Williams−Blangero, S., Almasy, L. A., Decker, M. J., et al. (1997). Ventilation and hypoxic ventilatory response of Tibetan and Aymara high altitude natives. Am. J. Phys. Anthropol. 104, 427–447. doi: 10.1002/(sici)1096-8644(199712)104:4<427::aid-ajpa1>3.0.co;2-p
Beaudry, M., Hidalgo, M., Launay, T., Bello, V., and Darribère, T. (2016). Regulation of myogenesis by environmental hypoxia. J. Cell Sci. 129:2887. doi: 10.1242/jcs.188904
Bhandari, S., and Cavalleri, G. L. (2019). Population history and altitude-related adaptation in the sherpa. Front. Physiol. 10:1116. doi: 10.3389/fphys.2019.01116
Bickler, P. E., and Buck, L. T. (2007). Hypoxia tolerance in reptiles, amphibians, and fishes: life with variable oxygen availability. Annu. Rev. Physiol. 69, 145–170. doi: 10.1146/annurev.physiol.69.031905.162529
Bigham, A., Bauchet, M., Pinto, D., Mao, X., Akey, J. M., Mei, R., et al. (2010). Identifying signatures of natural selection in tibetan and andean populations using dense genome scan data. PLoS Genet. 6:e1001116. doi: 10.1371/journal.pgen.1001116
Bigham, A. W., Julian, C. G., Wilson, M. J., Vargas, E., Browne, V. A., Shriver, M. D., et al. (2014). Maternal PRKAA1 and EDNRA genotypes are associated with birth weight, and PRKAA1 with uterine artery diameter and metabolic homeostasis at high altitude. Physiol. Genomics 46, 687–697. doi: 10.1152/physiolgenomics.00063.2014
Bigham, A. W., and Lee, F. S. (2014). Human high-altitude adaptation: forward genetics meets the HIF pathway. Genes Dev. 28, 2189–2204. doi: 10.1101/gad.250167.114
Bigham, A. W., Mao, X., Mei, R., Brutsaert, T., Wilson, M. J., Julian, C. G., et al. (2009). Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum. Genomics 4, 79–90.
Bishop, C. M., Spivey, R. J., Hawkes, L. A., Batbayar, N., Chua, B., Frappell, P. B., et al. (2015). The roller coaster flight strategy of bar-headed geese conserves energy during Himalayan migrations. Science 347, 250–254. doi: 10.1126/science.1258732
Blumthaler, M., Ambach, W., and Ellinger, R. (1997). Increase in solar UV radiation with altitude. J. Photochem. Photobiol. B Biol. 39, 130–134. doi: 10.1016/s1011-1344(96)00018-8
Briançon-Marjollet, A., Monneret, D., Henri, M., Hazane-Puch, F., Pepin, J.-L., Faure, P., et al. (2016). Endothelin regulates intermittent hypoxia-induced lipolytic remodelling of adipose tissue and phosphorylation of hormone-sensitive lipase. J. Physiol. 594, 1727–1740. doi: 10.1113/jp271321
Bridge, K. S., and Sharp, T. V. (2012). Regulators of the hypoxic response: a growing family. Future Oncol. 8, 491–493. doi: 10.2217/fon.12.46
Brutsaert, T. D., Kiyamu, M., Revollendo, G. E., Isherwood, J. L., Lee, F. S., Rivera-Ch, M., et al. (2019). Association of EGLN1 gene with high aerobic capacity of Peruvian Quechua at high altitude. Proc. Natl. Acad. Sci. U.S.A. 116, 24006–24011. doi: 10.1073/pnas.1906171116
Cai, Z. Y., Wang, L. Y., Song, X. Y., Tagore, S., Li, X. F., Wang, H. H., et al. (2018). Adaptive transcriptome profiling of subterranean zokor, myospalax baileyi, to high- altitude stresses in Tibet. Sci. Rep. 8:4671.
Camenisch, G., Stroka, D. M., Gassmann, M., and Wenger, R. H. (2001). Attenuation of HIF-1 DNA-binding activity limits hypoxia-inducible endothelin-1 expression. Pflugers Arch. Eur. J. Physiol. 443, 240–249. doi: 10.1007/s004240100679
Carbone, C., Piro, G., Merz, V., Simionato, F., Santoro, R., Zecchetto, C., et al. (2018). Angiopoietin-like proteins in angiogenesis, inflammation and cancer. Int. J. Mol. Sci. 19:431. doi: 10.3390/ijms19020431
Cavadas, M. A. S., Mesnieres, M., Crifo, B., Manresa, M. C., Selfridge, A. C., Keogh, C. E., et al. (2016). REST is a hypoxia-responsive transcriptional repressor. Sci. Rep. 6:31355.
Cerychova, R., and Pavlinkova, G. (2018). HIF-1, metabolism, and diabetes in the embryonic and adult heart. Front. Endocrinol. 9:460. doi: 10.3389/fendo.2018.00460
Chambers, J. C., Zhang, W. H., Li, Y., Sehmi, J., Wass, M. N., Zabaneh, D., et al. (2009). Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. Nat. Genet. 41, 1170–1172. doi: 10.1038/ng.462
Chen, F., Welker, F., Shen, C.-C., Bailey, S. E., Bergmann, I., Davis, S., et al. (2019). A late middle pleistocene denisovan mandible from the Tibetan Plateau. Nature 569, 409–412. doi: 10.1038/s41586-019-1139-x
Chen, J.-Y., Lin, C.-H., and Chen, B.-C. (2017). Hypoxia-induced ADAM 17 expression is mediated by RSK1-dependent C/EBPβ activation in human lung fibroblasts. Mol. Immunol. 88, 155–163. doi: 10.1016/j.molimm.2017.06.029
Chen, R., Dioum, E. M., Hogg, R. T., Gerard, R. D., and Garcia, J. A. (2011). Hypoxia increases sirtuin 1 expression in a hypoxia-inducible factor-dependent manner. J. Biol. Chem. 286, 13869–13878. doi: 10.1074/jbc.m110.175414
Cheng, J., Kang, X., Zhang, S., and Yeh, E. T. H. (2007). SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 131, 584–595. doi: 10.1016/j.cell.2007.08.045
Choksi, S., Lin, Y., Pobezinskaya, Y., Chen, L., Park, C., Morgan, M., et al. (2011). A HIF-1 target, ATIA, protects cells from apoptosis by modulating the mitochondrial thioredoxin, TRX2. Mol. Cell 42, 597–609. doi: 10.1016/j.molcel.2011.03.030
Chu, W., Li, X., Li, C., Wan, L., Shi, H., Song, X., et al. (2011). TGFBR3, a potential negative regulator of TGF-β signaling, protects cardiac fibroblasts from hypoxia-induced apoptosis. J. Cell. Physiol. 226, 2586–2594. doi: 10.1002/jcp.22604
Chung, D., Dzal, Y. A., Seow, A., Milsom, W. K., and Pamenter, M. E. (2016). Naked mole rats exhibit metabolic but not ventilatory plasticity following chronic sustained hypoxia. Proc. Biol. Sci. 283:20160216. doi: 10.1098/rspb.2016.0216
Cole, A. M., Cox, S., Jeong, C., Petousi, N., Aryal, D. R., Droma, Y., et al. (2017). Genetic structure in the Sherpa and neighboring Nepalese populations. BMC Genomics 18:102. doi: 10.1186/s12864-016-3469-5
Coppel, J., Hennis, P., Gilbert-Kawai, E., and Grocott, M. P. (2015). The physiological effects of hypobaric hypoxia versus normobaric hypoxia: a systematic review of crossover trials. Extr. Physiol. Med. 4:2.
Corante, N., Anza-Ramírez, C., Figueroa-Mujica, R., Macarlupú, J. L., Vizcardo-Galindo, G., Bilo, G., et al. (2018). Excessive erythrocytosis and cardiovascular risk in andean highlanders. High Altitude Med. Biol. 19, 221–231. doi: 10.1089/ham.2017.0123
Crawford, J. E., Amaru, R., Song, J., Julian, C. G., Racimo, F., Cheng, J. Y., et al. (2017). Natural selection on genes related to cardiovascular health in high-altitude adapted Andeans. Am. J. Hum. Genet. 101, 752–767. doi: 10.1016/j.ajhg.2017.09.023
Cui, H., Wang, Y., Huang, H., Yu, W., Bai, M., Zhang, L., et al. (2014). GPR126 protein regulates developmental and pathological angiogenesis through modulation of VEGFR2 receptor signaling. J. Biol. Chem. 289, 34871–34885. doi: 10.1074/jbc.m114.571000
Dales, J.-P., Beaufils, N., Silvy, M., Picard, C., Pauly, V., Pradel, V., et al. (2010). Hypoxia inducible factor 1α gene (HIF-1α) splice variants: potential prognostic biomarkers in breast cancer. BMC Med. 8:44. doi: 10.1186/1741-7015-8-44
David, Y., Ziv, T., Admon, A., and Navon, A. (2010). The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J. Biol. Chem. 285, 8595–8604. doi: 10.1074/jbc.m109.089003
Drutel, G., Kathmann, M., Héron, A., Gros, C., Macé, S., Schwartz, J. C., et al. (2000). Two splice variants of the hypoxia−inducible factor HIF−1α as potential dimerization partners of ARNT2 in neurons. Eur. J. Neurosci. 12, 3701–3708. doi: 10.1046/j.1460-9568.2000.00266.x
Duan, C. (2016). Hypoxia-inducible factor 3 biology: complexities and emerging themes. Am. J. Physiol. Cell Physiol. 310, C260–C269.
Dzal, Y. A., Jenkin, S. E., Lague, S. L., Reichert, M. N., York, J. M., and Pamenter, M. E. (2015). Oxygen in demand: how oxygen has shaped vertebrate physiology. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 186, 4–26. doi: 10.1016/j.cbpa.2014.10.029
Eichstaedt, C. A., Antao, T., Pagani, L., Cardona, A., Kivisild, T., and Mormina, M. (2014). The Andean adaptive toolkit to counteract high altitude maladaptation: genome-wide and phenotypic analysis of the Collas. PLoS One 9:e93314. doi: 10.1371/journal.pone.0093314
Eichstaedt, C. A., Pagani, L., Antao, T., Inchley, C. E., Cardona, A., Mörseburg, A., et al. (2017). Evidence of early-stage selection on EPAS1 and GPR126 genes in Andean high altitude populations. Sci. Rep. 7:13042.
Ekambaram, P., and Parasuraman, P. (2017). Differential expression of sirtuin 2 and adipocyte maturation restriction: an adaptation process during hypoxia in fish. Biol. Open 6, 1375–1382. doi: 10.1242/bio.027334
Eltzschig, H. K., Abdulla, P., Hoffman, E., Hamilton, K. E., Daniels, D., Schönfeld, C., et al. (2005). HIF-1–dependent repression of equilibrative nucleoside transporter (ENT) in hypoxia. J. Exp. Med. 202, 1493–1505. doi: 10.1084/jem.20050177
Enns, C. A. (2006). Possible roles of the hereditary hemochromatosis protein, HFE, in regulating cellular iron homeostasis. Biol. Res. 39, 105–111.
Foll, M., Gaggiotti, O. E., Daub, J. T., Vatsiou, A., and Excoffier, L. (2014). Widespread signals of convergent adaptation to high altitude in Asia and america. Am. J. Hum. Genet. 95, 394–407. doi: 10.1016/j.ajhg.2014.09.002
Furlow, P. W., Percy, M. J., Sutherland, S., Bierl, C., Mcmullin, M. F., Master, S. R., et al. (2009). Erythrocytosis-associated HIF-2alpha mutations demonstrate a critical role for residues C-terminal to the hydroxylacceptor proline. J. Biol. Chem. 284, 9050–9058. doi: 10.1074/jbc.m808737200
Gale, N. W., and Yancopoulos, G. D. (1999). Growth factors acting via endothelial cell-specific receptor tyrosine kinases: VEGFs, angiopoietins, and ephrins in vascular development. Genes Dev. 13, 1055–1066. doi: 10.1101/gad.13.9.1055
Gardie, B., Percy, M. J., Hoogewijs, D., Chowdhury, R., Bento, C., Arsenault, P. R., et al. (2014). The role of PHD2 mutations in the pathogenesis of erythrocytosis. Hypoxia 2, 71–90.
Gautier-Veyret, E., Arnaud, C., Bäck, M., Pépin, J.-L., Petri, M. H., Baguet, J.-P., et al. (2013). Intermittent hypoxia-activated cyclooxygenase pathway: role in atherosclerosis. Eur. Respir. J. 42, 404–413. doi: 10.1183/09031936.00096512
Ge, R. L., Cai, Q. L., Shen, Y. Y., San, A., Ma, L., Zhang, Y., et al. (2013). Draft genome sequence of the Tibetan antelope. Nat. Commun. 4:1858.
Ge, R.-L., Simonson, T. S., Cooksey, R. C., Tanna, U., Qin, G., Huff, C. D., et al. (2012). Metabolic insight into mechanisms of high-altitude adaptation in Tibetans. Mol. Genet. Metab. 106, 244–247. doi: 10.1016/j.ymgme.2012.03.003
Germonpre, M., Sablin, M. V., Stevens, R. E., Hedges, R. E. M., Hofreiter, M., Stiller, M., et al. (2009). Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes. J. Archaeol. Sci. 36, 473–490. doi: 10.1016/j.jas.2008.09.033
Gilbert-Kawai, E. T., Milledge, J. S., Grocott, M. P., and Martin, D. S. (2014). King of the mountains: Tibetan and Sherpa physiological adaptations for life at high altitude. Physiology (Bethesda) 29, 388–402. doi: 10.1152/physiol.00018.2014
Gnecchi-Ruscone, G. A., Abondio, P., De Fanti, S., Sarno, S., Sherpa, M. G., Sherpa, P. T., et al. (2018). Evidence of polygenic adaptation to high altitude from Tibetan and Sherpa genomes. Genome Biol. Evol. 10, 2919–2930.
Gothié, E., Richard, D. E., Berra, E., Pagès, G., and Pouysségur, J. (2000). Identification of alternative spliced variants of human hypoxia-inducible factor-1α. J. Biol. Chem. 275, 6922–6927. doi: 10.1074/jbc.275.10.6922
Gou, X., Wang, Z., Li, N., Qiu, F., Xu, Z., Yan, D. W., et al. (2014). Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 24, 1308–1315. doi: 10.1101/gr.171876.113
Graham, A. M., and McCracken, K. G. (2019). Convergent evolution on the hypoxia-inducible factor (HIF) pathway genes EGLN1 and EPAS1 in high-altitude ducks. Heredity (Edinb) 122, 819–832. doi: 10.1038/s41437-018-0173-z
Gruber, M., Hu, C. J., Johnson, R. S., Brown, E. J., Keith, B., and Simon, M. C. (2007). Acute postnatal ablation of Hif-2alpha results in anemia. Proc. Natl. Acad. Sci. U.S.A. 104, 2301–2306. doi: 10.1073/pnas.0608382104
Guang-Xin, E., Basang, W. D., and Zhu, Y. B. (2019). Whole-genome analysis identifying candidate genes of altitude adaptive ecological thresholds in yak populations. J. Anim. Breed. Genet. 136, 371–377. doi: 10.1111/jbg.12403
Haase, V. H. (2013). Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev. 27, 41–53. doi: 10.1016/j.blre.2012.12.003
Hagedorn, M. (2011). PRCP: a key to blood vessel homeostasis. Blood 117, 3705–3706. doi: 10.1182/blood-2011-02-335992
Hall, J. E., Lawrence, E. S., Simonson, T. S., and Fox, K. (2020). Seq-ing higher ground: functional investigation of adaptive variation associated with high-altitude adaptation. Front. Genet. 11:471. doi: 10.3389/fgene.2020.00471
Hankinson, O. (2002). “Aryl hydrocarbon receptor nuclear translocator (ARNT) (hypoxia inducible factor 1β [HIF-1β]),” in Wiley Encyclopedia of Molecular Medicine (New York, NY: John Wiley & Sons), 1–5.
He, J. Z., Xiu, M. H., Tang, X. L., Yue, F., Wang, N. B., Yang, S. B., et al. (2013). The different mechanisms of hypoxic acclimatization and adaptation in lizard Phrynocephalus vlangalii living on qinghai-Tibet Plateau. J. Exp. Zool. Part A Ecol. Genet. Physiol. 319a, 117–123. doi: 10.1002/jez.1776
Heaton, M. P., Smith, T. P. L., Carnahan, J. K., Basnayake, V., Qiu, J., Simpson, B., et al. (2016). Using diverse U.S. beef cattle genomes to identify missense mutations in EPAS1, a gene associated with pulmonary hypertension. F1000Res. 5, 2003. doi: 10.12688/f1000research.9254.1
Heikkilä, M., Pasanen, A., Kivirikko, K. I., and Myllyharju, J. (2011). Roles of the human hypoxia-inducible factor (HIF)-3α variants in the hypoxia response. Cell. Mol. Life Sci. 68, 3885–3901. doi: 10.1007/s00018-011-0679-5
Heinrich, E. C., Orr, J. E., Gilbertson, D., Anza-Ramirez, C., Deyoung, P. N., Djokic, M. A., et al. (2020). Relationships between chemoreflex responses, sleep quality, and hematocrit in andean men and women. Front. Physiol. 11:437. doi: 10.3389/fphys.2020.00437
Heinrich, E. C., Wu, L., Lawrence, E. S., Cole, A. M., Anza-Ramirez, C., Villafuerte, F. C., et al. (2019). Genetic variants at the EGLN1 locus associated with high-altitude adaptation in Tibetan are absent or found at low frequency in highland Andean. Ann. Hum. Genet. 83, 171–176. doi: 10.1111/ahg.12299
Hendrickson, S. L. (2013). A genome wide study of genetic adaptation to high altitude in feral Andean Horses of the paramo. BMC Evol. Biol. 13:273. doi: 10.1186/1471-2148-13-273
Hochachka, P. W. (1986). Defense strategies against hypoxia and hypothermia. Science 231, 234–241. doi: 10.1126/science.2417316
Hochachka, P. W., Buck, L. T., Doll, C. J., and Land, S. C. (1996). Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc. Natl. Acad. Sci. U.S.A. 93, 9493–9498. doi: 10.1073/pnas.93.18.9493
Horscroft, J. A., Kotwica, A. O., Laner, V., West, J. A., Hennis, P. J., Levett, D. Z., et al. (2017). Metabolic basis to Sherpa altitude adaptation. Proc. Natl. Acad. Sci. U.S.A. 114, 6382–6387.
Hu, H., Petousi, N., Glusman, G., Yu, Y., Bohlender, R., Tashi, T., et al. (2017). Evolutionary history of Tibetans inferred from whole-genome sequencing. PLoS Genet. 13:e1006675. doi: 10.1371/journal.pgen.1006675
Hu, X. J., Yang, J., Xie, X. L., Lv, F. H., Cao, Y. H., Li, W. R., et al. (2019). The genome landscape of Tibetan sheep reveals adaptive introgression from argali and the history of early human settlements on the Qinghai-Tibetan Plateau. Mol. Biol. Evol. 36, 283–303. doi: 10.1093/molbev/msy208
Huang, W. (1980). “Some physiological parameters of domestic animals at various altitudes in Xizang,” in Proceedings of symposium on Qinghai-Xizang (Tibet) Plateau (Beijing: CAB International), 182–183.
Hubbi, M. E., Hu, H., Kshitiz, Gilkes, D. M., and Semenza, G. L. (2013). Sirtuin-7 inhibits the activity of hypoxia-inducible factors. J. Biol. Chem. 288, 20768–20775. doi: 10.1074/jbc.m113.476903
Huerta-Sánchez, E., Degiorgio, M., Pagani, L., Tarekegn, A., Ekong, R., Antao, T., et al. (2013). Genetic signatures reveal high-altitude adaptation in a set of Ethiopian populations. Mol. Biol. Evol. 30, 1877–1888. doi: 10.1093/molbev/mst089
Huerta-Sánchez, E., Jin, X., Asan, Bianba, Z., Peter, B. M., Vinckenbosch, N., et al. (2014). Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197. doi: 10.1038/nature13408
Ishizaki, T., Koizumi, T., Ruan, Z., Wang, Z., Chen, Q., and Sakai, A. (2005). Nitric oxide inhibitor altitude-dependently elevates pulmonary arterial pressure in high-altitude adapted yaks. Respir. Physiol. Neurobiol. 146, 225–230. doi: 10.1016/j.resp.2004.12.002
Ito, Y., Ahmad, A., Kewley, E., and Mason, R. J. (2011). Hypoxia-inducible factor regulates expression of surfactant protein in alveolar type II cells in vitro. Am. J. Respir. Cell Mol. Biol. 45, 938–945. doi: 10.1165/rcmb.2011-0052oc
Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., et al. (2001). HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468. doi: 10.1126/science.1059817
Jaakkola, P., Mole, D. R., Tian, Y.-M., Wilson, M. I., Gielbert, J., Gaskell, S. J., et al. (2001). Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472. doi: 10.1126/science.1059796
Jarrar, Y., Zihlif, M., Al Bawab, A. Q., and Sharab, A. (2020). Effects of intermittent hypoxia on expression of glucose metabolism genes in MCF7 breast cancer cell line. Curr. Cancer Drug Targets 20, 216–222. doi: 10.2174/1568009619666191116095847
Jeong, C., Alkorta-Aranburu, G., Basnyat, B., Neupane, M., Witonsky, D. B., Pritchard, J. K., et al. (2014). Admixture facilitates genetic adaptations to high altitude in Tibet. Nat. Commun. 5:3281.
Jessen, T. H., Weber, R. E., Fermi, G., Tame, J., and Braunitzer, G. (1991). Adaptation of bird hemoglobins to high-altitudes – demonstration of molecular mechanism by protein engineering. Proc. Natl. Acad. Sci. U.S.A. 88, 6519–6522. doi: 10.1073/pnas.88.15.6519
Jha, A. R., Zhou, D., Brown, C. D., Kreitman, M., Haddad, G. G., and White, K. P. (2016). Shared genetic signals of hypoxia adaptation in Drosophila and in high-altitude human populations. Mol. Biol. Evol. 33, 501–517. doi: 10.1093/molbev/msv248
Jiang, B.-H., Rue, E., Wang, G. L., Roe, R., and Semenza, G. L. (1996). Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 271, 17771–17778. doi: 10.1074/jbc.271.30.17771
Jiang, W., Sun, B., Song, X., Zheng, Y., Wang, L., Wang, T., et al. (2015). Arginase inhibition protects against hypoxia-induced pulmonary arterial hypertension. Mol. Med. Rep. 12, 4743–4749. doi: 10.3892/mmr.2015.3994
Johansen, K., Lykkeboe, G., Weber, R. E., and Maloiy, G. M. (1976). Blood respiratory properties in the naked mole rat Heterocephalus glaber, a mammal of low body temperature. Respir. Physiol. 28, 303–314. doi: 10.1016/0034-5687(76)90025-6
Jones, E. Y., Fugger, L., Strominger, J. L., and Siebold, C. (2006). MHC class II proteins and disease: a structural perspective. Nat. Rev. Immunol. 6, 271–282. doi: 10.1038/nri1805
Julian, C. G. (2017). Epigenomics and human adaptation to high altitude. J. Appl. Physiol. 123, 1362–1370. doi: 10.1152/japplphysiol.00351.2017
Julian, C. G., and Moore, L. G. (2019). Human genetic adaptation to high altitude: evidence from the andes. Genes 10:150. doi: 10.3390/genes10020150
Kim, E. B., Fang, X., Fushan, A. A., Huang, Z., Lobanov, A. V., Han, L., et al. (2011). Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479, 223–227. doi: 10.1038/nature10533
Kong, X., Dong, X., Yang, S., Qian, J., Yang, J., Jiang, Q., et al. (2019). Natural selection on TMPRSS6 associated with the blunted erythropoiesis and improved blood viscosity in Tibetan pigs. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 233, 11–22. doi: 10.1016/j.cbpb.2019.03.003
Krishnan, J., Ahuja, P., Bodenmann, S., Knapik, D., Perriard, E., Krek, W., et al. (2008). Essential role of developmentally activated hypoxia-inducible factor 1alpha for cardiac morphogenesis and function. Circ. Res. 103, 1139–1146. doi: 10.1161/01.res.0000338613.89841.c1
Lacombe, C., and Mayeux, P. (2008). “Biology of EPO and EPO-receptor,” in Recombinant Human Erythropoietin (rhEPO) in Clinical Oncology, ed. M. R. Nowrousian (Vienna: Springer), 67–80. doi: 10.1007/978-3-211-69459-6_2
León-Velarde, F., Maggiorini, M., Reeves, J. T., Aldashev, A., Asmus, I., Bernardi, L., et al. (2005). Consensus statement on chronic and subacute high altitude diseases. High Altitude Med. Biol. 6, 147–157. doi: 10.1089/ham.2005.6.147
Lerman, M. I. (2002). “VHL (von Hippel-Lindau) Protein,” in Wiley Encyclopedia of Molecular Medicine (New York, NY: John Wiley & Sons), 1–5.
Li, H., Zhou, L., and Dai, J. (2018). Retinoic acid receptor-related orphan receptor RORα regulates differentiation and survival of keratinocytes during hypoxia. J. Cell. Physiol. 233, 641–650. doi: 10.1002/jcp.25924
Li, J. T., Gao, Y. D., Xie, L., Deng, C., Shi, P., Guan, M. L., et al. (2018). Comparative genomic investigation of high-elevation adaptation in ectothermic snakes. Proc. Natl. Acad. Sci. U.S.A. 115, 8406–8411. doi: 10.1073/pnas.1805348115
Li, M. Z., Tian, S. L., Jin, L., Zhou, G. Y., Li, Y., Zhang, Y., et al. (2013). Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 45, 1431–U1180.
Li, S. C., Li, D. Y., Zhao, X. L., Wang, Y., Yin, H. D., Zhou, L. Y., et al. (2017). A non-synonymous SNP with the allele frequency correlated with the altitude may contribute to the hypoxia adaptation of Tibetan chicken. PLoS One 12:e0172211. doi: 10.1371/journal.pone.0172211
Li, X. (2001). Novel VEGF family members: VEGF-B, VEGF-C and VEGF-D. Int. J. Biochem. Cell Biol. 33, 421–426. doi: 10.1016/s1357-2725(01)00027-9
Li, Y., Wu, D. D., Boyko, A. R., Wang, G. D., Wu, S. F., Irwin, D. M., et al. (2014). Population variation revealed high-altitude adaptation of Tibetan Mastiffs. Mol. Biol. Evol. 31, 1200–1205. doi: 10.1093/molbev/msu070
Li, Y., and Zhang, Y. P. (2012). High genetic diversity of Tibetan Mastiffs revealed by mtDNA sequences. Chin. Sci. Bull. 57, 1483–1487. doi: 10.1007/s11434-012-4995-4
Liang, Y. H., Hua, Z. Q., Liang, X., Xu, Q., and Lu, G. Y. (2001). The crystal structure of bar-headed goose hemoglobin in deoxy form: the allosteric mechanism of a hemoglobin species with high oxygen affinity. J. Mol. Biol. 313, 123–137. doi: 10.1006/jmbi.2001.5028
Liao, B., Zheng, Y.-M., Yadav, V. R., Korde, A. S., and Wang, Y.-X. (2011). Hypoxia induces intracellular Ca2+ release by causing reactive oxygen species-mediated dissociation of FK506-binding protein 12.6 from ryanodine receptor 2 in pulmonary artery myocytes. Antioxid. Redox Signal. 14, 37–47. doi: 10.1089/ars.2009.3047
Lin, W., Wadlington, N. L., Chen, L., Zhuang, X., Brorson, J. R., and Kang, U. J. (2014). Loss of PINK1 attenuates HIF-1α induction by preventing 4E-BP1-dependent switch in protein translation under hypoxia. J. Neurosci. 34, 3079–3089. doi: 10.1523/jneurosci.2286-13.2014
Liu, B., Cheng, S., Xing, W., Pourteymoor, S., and Mohan, S. (2015). RE1-silencing transcription factor (Rest) is a novel regulator of osteoblast differentiation. J. Cell. Biochem. 116, 1932–1938. doi: 10.1002/jcb.25148
Liu, H., Liu, Q., Chen, Z., Liu, Y., Zhou, C., Liang, Q., et al. (2018). Draft genome of Glyptosternon maculatum, an endemic fish from Tibet Plateau. Gigascience 7:giy104.
Liu, H., Tang, F., Su, J., Ma, J., Qin, Y., Ji, L., et al. (2020). EPAS1 regulates proliferation of erythroblasts in chronic mountain sickness. Blood Cells Mol. Dis. 84:102446. doi: 10.1016/j.bcmd.2020.102446
Liu, H., and Xia, Y. (2015). Beneficial and detrimental role of adenosine signaling in diseases and therapy. J. Appl. Physiol. 119, 1173–1182. doi: 10.1152/japplphysiol.00350.2015
Liu, X., Zhang, Y., Li, Y., Pan, J., Wang, D., Chen, W., et al. (2019). EPAS1 gain-of-function mutation contributes to high-altitude adaptation in Tibetan horses. Mol. Biol. Evol. 36, 2591–2603. doi: 10.1093/molbev/msz158
Liu, X.-B., Jiang, J., Gui, C., Hu, X.-Y., Xiang, M.-X., and Wang, J.-A. (2008). Angiopoietin-1 protects mesenchymal stem cells against serum deprivation and hypoxia-induced apoptosis through the PI3K/Akt pathway. Acta Pharmacol. Sin. 29, 815–822. doi: 10.1111/j.1745-7254.2008.00811.x
Loenarz, C., Coleman, M. L., Boleininger, A., Schierwater, B., Holland, P. W. H., Ratcliffe, P. J., et al. (2011). The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep. 12, 63–70. doi: 10.1038/embor.2010.170
Lorenzo, F. R., Huff, C., Myllymäki, M., Olenchock, B., Swierczek, S., Tashi, T., et al. (2014). A genetic mechanism for Tibetan high-altitude adaptation. Nat. Genet. 46, 951–956.
Maguire, J. J., and Davenport, A. P. (2015). Endothelin receptors and their antagonists. Semin. Nephrol. 35, 125–136. doi: 10.1016/j.semnephrol.2015.02.002
Mariyama, M., Leinonen, A., Mochizuki, T., Tryggvason, K., and Reeders, S. T. (1994). Complete primary structure of the human alpha 3(IV) collagen chain. Coexpression of the alpha 3(IV) and alpha 4(IV) collagen chains in human tissues. J. Biol. Chem. 269, 23013–23017.
Maurer, E., Gütschow, M., and Stirnberg, M. (2012). Matriptase-2 (TMPRSS6) is directly up-regulated by hypoxia inducible factor-1: identification of a hypoxia-responsive element in the TMPRSS6 promoter region. Biol. Chem. 393, 535–540. doi: 10.1515/hsz-2011-0221
Maxwell, P. H., Wiesener, M. S., Chang, G.-W., Clifford, S. C., Vaux, E. C., Cockman, M. E., et al. (1999). The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275. doi: 10.1038/20459
Maynard, M. A., Qi, H., Chung, J., Lee, E. H., Kondo, Y., Hara, S., et al. (2003). Multiple splice variants of the human HIF-3α locus are targets of the von Hippel-Lindau E3 ubiquitin ligase complex. J. Biol. Chem. 278, 11032–11040. doi: 10.1074/jbc.m208681200
McClelland, G. B., and Scott, G. R. (2019). Evolved mechanisms of aerobic performance and hypoxia resistance in high-altitude natives. Annu. Rev. Physiol. 81, 561–583. doi: 10.1146/annurev-physiol-021317-121527
Metzen, E., and Ratcliffe, P. J. (2004). HIF hydroxylation and cellular oxygen sensing. Biol. Chem. 385, 223–230.
Meyer, M. C., Hofmann, C. C., Gemmell, A. M. D., Haslinger, E., Hausler, H., and Wangda, D. (2009). Holocene glacier fluctuations and migration of Neolithic yak pastoralists into the high valleys of northwest Bhutan. Quat. Sci. Rev. 28, 1217–1237. doi: 10.1016/j.quascirev.2008.12.025
Monge, C., Leon-Velarde, F., and Arregui, A. (1989). Increasing prevalence of excessive erythrocytosis with age among healthy high-altitude miners. New Engl. J. Med. 321:1271. doi: 10.1056/nejm198911023211817
Moore, L. G. (2017). Measuring high-altitude adaptation. J. Appl. Physiol. 123, 1371–1385. doi: 10.1152/japplphysiol.00321.2017
Morfoisse, F., Renaud, E., Hantelys, F., Prats, A.-C., and Garmy-Susini, B. (2015). Role of hypoxia and vascular endothelial growth factors in lymphangiogenesis. Mol. Cell Oncol. 2:e1024821. doi: 10.1080/23723556.2015.1024821
Nechiporuk, T., Urness, L. D., and Keating, M. T. (2001). ETL, a novel seven-transmembrane receptor that is developmentally regulated in the heart. ETL is a member of the secretin family and belongs to the epidermal growth factor-seven-transmembrane subfamily. J. Biol. Chem. 276, 4150–4157. doi: 10.1074/jbc.m004814200
O’Brien, K. A., Horscroft, J. A., Devaux, J., Lindsay, R. T., Steel, A. S., Clark, A. D., et al. (2019). PPARα-independent effects of nitrate supplementation on skeletal muscle metabolism in hypoxia. Biochim. Biophys. Acta 1865, 844–853. doi: 10.1016/j.bbadis.2018.07.027
Olson, E. N., and Nordheim, A. (2010). Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11, 353–365. doi: 10.1038/nrm2890
Otto, T., and Fandrey, J. (2008). Thyroid hormone induces hypoxia-inducible factor 1alpha gene expression through thyroid hormone receptor beta/retinoid x receptor alpha-dependent activation of hepatic leukemia factor. Endocrinology 149, 2241–2250. doi: 10.1210/en.2007-1238
Pamenter, M. E. (2014). Mitochondria: a multimodal hub of hypoxia tolerance. Can. J. Zool. 92, 569–589. doi: 10.1139/cjz-2013-0247
Pamenter, M. E. (2016). Comparative insights into mitochondrial adaptations to anoxia in brain. Neural Regen. Res. 11, 723–724.
Pamenter, M. E., Dzal, Y. A., and Milsom, W. K. (2015). Adenosine receptors mediate the hypoxic ventilatory response but not the hypoxic metabolic response in the naked mole rat during acute hypoxia. Proc. Biol. Sci. 282:20141722. doi: 10.1098/rspb.2014.1722
Pamenter, M. E., and Powell, F. L. (2016). Time domains of the hypoxic ventilatory response and their molecular basis. Compr. Physiol. 6, 1345–1385. doi: 10.1002/cphy.c150026
Pan, H., Chen, Q., Qi, S., Li, T., Liu, B., Liu, S., et al. (2018). Mutations in EPAS1 in congenital heart disease in Tibetans. Biosci. Rep. 38:BSR20181389.
Pan, S. K., Zhang, T. Z., Rong, Z. Q., Hu, L., Gu, Z. R., Wu, Q., et al. (2017). Population transcriptomes reveal synergistic responses of DNA polymorphism and RNA expression to extreme environments on the Qinghai-Tibetan Plateau in a predatory bird. Mol. Ecol. 26, 2993–3010. doi: 10.1111/mec.14090
Pandey, P., Mohammad, G., Singh, Y., and Qadar Pasha, M. A. (2015). ROCK2 and MYLK variants under hypobaric hypoxic environment of high altitude associate with high altitude pulmonary edema and adaptation. Appl. Clin. Genet. 8, 257–267.
Park, H. S., Kim, J. H., Sun, B. K., Song, S. U., Suh, W., and Sung, J.-H. (2016). Hypoxia induces glucose uptake and metabolism of adipose-derived stem cells. Mol. Med. Rep. 14, 4706–4714. doi: 10.3892/mmr.2016.5796
Park, T. J., Reznick, J., Peterson, B. L., Blass, G., Omerbasic, D., Bennett, N. C., et al. (2017). Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science 356, 305–308.
Pasanen, A., Heikkilä, M., Rautavuoma, K., Hirsilä, M., Kivirikko, K. I., and Myllyharju, J. (2010). Hypoxia-inducible factor (HIF)-3α is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int. J. Biochem. Cell Biol. 42, 1189–1200. doi: 10.1016/j.biocel.2010.04.008
Peng, Y., Yang, Z., Zhang, H., Cui, C., Qi, X., Luo, X., et al. (2011). Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol. Biol. Evol. 28, 1075–1081. doi: 10.1093/molbev/msq290
Petousi, N., and Robbins, P. A. (2014). Human adaptation to the hypoxia of high altitude: the Tibetan paradigm from the pregenomic to the postgenomic era. J. Appl. Physiol. 116, 875–884. doi: 10.1152/japplphysiol.00605.2013
Pham, L. V., Meinzen, C., Arias, R. S., Schwartz, N. G., Rattner, A., Miele, C. H., et al. (2017). Cross-sectional comparison of sleep-disordered breathing in native Peruvian highlanders and lowlanders. High Alt. Med. Biol. 18, 11–19. doi: 10.1089/ham.2016.0102
Piccinini, M., Kleinschmidt, T., Jurgens, K. D., and Braunitzer, G. (1990). Primary structure and oxygen-binding properties of the hemoglobin from guanaco (Lama guanacoë, Tylopoda). Biol. Chem. Hoppe Seyler 371, 641–648. doi: 10.1515/bchm3.1990.371.2.641
Pickrell, J., Clerget−Darpoux, F., and Bourgain, C. (2007). Power of genome−wide association studies in the presence of interacting loci. Genet. Epidemiol. 31, 748–762. doi: 10.1002/gepi.20238
Popeijus, H. E. (2018). “Peroxisome proliferator-activated receptor alpha (PPAR-Alpha),” in Encyclopedia of Signaling Molecules, ed. S. Choi (New York, NY: Springer), 3890–3895. doi: 10.1007/978-3-319-67199-4_101552
Prabhakar, N. R., and Semenza, G. L. (2012). Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol. Rev. 92, 967–1003. doi: 10.1152/physrev.00030.2011
Qi, X. B., Cui, C. Y., Peng, Y., Zhang, X. M., Yang, Z. H., Zhong, H., et al. (2013). Genetic evidence of paleolithic colonization and neolithic expansion of modern humans on the Tibetan Plateau. Mol. Biol. Evol. 30, 1761–1778. doi: 10.1093/molbev/mst093
Qi, X. B., Zhang, Q., He, Y. X., Yang, L. X., Zhang, X. M., Shi, P., et al. (2019). The transcriptomic landscape of Yaks reveals molecular pathways for high altitude adaptation. Genome Biol. Evol. 11, 72–85.
Qiu, Q., Wang, L. Z., Wang, K., Yang, Y. Z., Ma, T., Wang, Z. F., et al. (2015). Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat. Commun. 6:10283.
Qiu, Q., Zhang, G., Ma, T., Qian, W., Wang, J., Ye, Z., et al. (2012). The yak genome and adaptation to life at high altitude. Nat. Genet. 44, 946–949.
Qu, Y. H., Zhao, H. W., Han, N. J., Zhou, G. Y., Song, G., Gao, B., et al. (2013). Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat. Commun. 4:2071.
Rademaker, K., Hodgins, G., Moore, K., Zarrillo, S., Miller, C., Bromley, G. R. M., et al. (2014). Paleoindian settlement of the high-altitude Peruvian Andes. Science 346:466. doi: 10.1126/science.1258260
Radivojac, P., Vacic, V., Haynes, C., Cocklin, R. R., Mohan, A., Heyen, J. W., et al. (2010). Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78, 365–380. doi: 10.1002/prot.22555
Ramirez, J. M., Folkow, L. P., and Blix, A. S. (2007). Hypoxia tolerance in mammals and birds: from the wilderness to the clinic. Annu. Rev. Physiol. 69, 113–143. doi: 10.1146/annurev.physiol.69.031905.163111
Ren, H., Panchatcharam, M., Mueller, P., Escalante-Alcalde, D., Morris, A. J., and Smyth, S. S. (2013). Lipid phosphate phosphatase (LPP3) and vascular development. Biochim. Biophys. Acta 1831, 126–132. doi: 10.1016/j.bbalip.2012.07.012
Renzheng, J. J., and Ciren, B. (1992). Comparison on several hematologic value of goat in Tibet Plateau at different altitude, Southwest China. J. Agr. Sci. 1:014.
Rey, S., and Semenza, G. L. (2010). Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodelling. Cardiovasc. Res. 86, 236–242. doi: 10.1093/cvr/cvq045
Rodríguez-Manzaneque, J. C., Fernández-Rodríguez, R., Rodríguez-Baena, F. J., and Luisa Iruela-Arispe, M. (2015). ADAMTS proteases in vascular biology. Matrix Biol. 44-46, 38–45. doi: 10.1016/j.matbio.2015.02.004
Ruas, J. L., and Poellinger, L. (2005). Hypoxia-dependent activation of HIF into a transcriptional regulator. Semin. Cell Dev. Biol. 16, 514–522. doi: 10.1016/j.semcdb.2005.04.001
Scheinfeldt, L. B., Soi, S., Thompson, S., Ranciaro, A., Woldemeskel, D., Beggs, W., et al. (2012). Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 13:R1.
Scheinfeldt, L. B., and Tishkoff, S. A. (2013). Recent human adaptation: genomic approaches, interpretation and insights. Nat. Rev. Genet. 14, 692–702. doi: 10.1038/nrg3604
Schofield, C. J., and Ratcliffe, P. J. (2004). Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5, 343–354. doi: 10.1038/nrm1366
Semenza, G. L. (2020). The genomics and genetics of oxygen homeostasis. Annu. Rev. Genomics Hum. Genet. 7, [Epub ahead of print].
Semenza, G. L., Jiang, B. H., Leung, S. W., Passantino, R., Concordet, J. P., Maire, P., et al. (1996). Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537. doi: 10.1074/jbc.271.51.32529
Seza, E. G., Güderer, I., Ermis, Ç, and Banerjee, S. (2019). PRKAA1 (protein kinase AMP-activated catalytic subunit alpha 1). Atlas Genet. Cytogenet. Oncol. Haematol. 23, 105–111.
Shao, Y., Li, J. X., Ge, R. L., Zhong, L., Irwin, D. M., Murphy, R. W., et al. (2015). Genetic adaptations of the plateau zokor in high-elevation burrows. Sci. Rep. 5:17262.
Shih, I. M., and Kurman, R. J. (1996). Expression of melanoma cell adhesion molecule in intermediate trophoblast. Lab. Invest. 75, 377–388.
Shukla, S., Levine, C., Sripathi, R. P., Elson, G., Lutz, C. S., and Leibovich, S. J. (2018). The kat in the HAT: the histone acetyl transferase Kat6b (MYST4) is downregulated in murine macrophages in response to LPS. Mediat. Inflamm. 2018:7852742.
Simonson, T. S. (2015). Altitude adaptation: a glimpse through various lenses. High Alt. Med. Biol. 16, 125–137. doi: 10.1089/ham.2015.0033
Simonson, T. S., Mcclain, D. A., Jorde, L. B., and Prchal, J. T. (2012). Genetic determinants of Tibetan high-altitude adaptation. Hum. Genet. 131, 527–533. doi: 10.1007/s00439-011-1109-3
Simonson, T. S., Yang, Y., Huff, C. D., Yun, H., Qin, G., Witherspoon, D. J., et al. (2010). Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75.
Song, D., Li, L.-S., Arsenault, P. R., Tan, Q., Bigham, A. W., Heaton-Johnson, K. J., et al. (2014). Defective Tibetan PHD2 binding to p23 links high altitude adaption to altered oxygen sensing. J. Biol. Chem. 289, 14656–14665. doi: 10.1074/jbc.m113.541227
Song, D., Navalsky, B. E., Guan, W., Ingersoll, C., Wang, T., Loro, E., et al. (2020). Tibetan PHD2, an allele with loss-of-function properties. Proc. Natl. Acad. Sci. U.S.A. 117, 12230–12238.
Song, S., Yao, N., Yang, M., Liu, X. X., Dong, K. Z., Zhao, Q. J., et al. (2016). Exome sequencing reveals genetic differentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra hircus). BMC Genomics 17:122. doi: 10.1186/s12864-016-2449-0
Stobdan, T., Zhou, D., Williams, A. T., Cabrales, P., and Haddad, G. G. (2018). Cardiac-specific knockout and pharmacological inhibition of Endothelin receptor type B lead to cardiac resistance to extreme hypoxia. J. Mol. Med. 96, 975–982. doi: 10.1007/s00109-018-1673-2
Storz, J. F., Runck, A. M., Sabatino, S. J., Kelly, J. K., Ferrand, N., Moriyama, H., et al. (2009). Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc. Natl. Acad. Sci. U.S.A. 106, 14450–14455. doi: 10.1073/pnas.0905224106
Storz, J. F., Sabatino, S. J., Hoffmann, F. G., Gering, E. J., Moriyama, H., Ferrand, N., et al. (2007). The molecular basis of high-altitude adaptation in deer mice. PLoS Genetics 3:e45. doi: 10.1371/journal.pgen.0030045
Storz, J. F., and Scott, G. R. (2019). Life ascending: mechanism and process in physiological adaptation to high-altitude hypoxia. Annu. Rev. Ecol. Evol. Syst. 50, 503–526. doi: 10.1146/annurev-ecolsys-110218-025014
Sun, Y. F., Ren, Z. P., Wu, Y. F., Lei, F. M., Dudley, R., and Li, D. M. (2016). Flying high: limits to flight performance by sparrows on the Qinghai-Tibet Plateau. J. Exp. Biol. 219, 3642–3648. doi: 10.1242/jeb.142216
Susen, R. M., Bauer, R., Olesch, C., Fuhrmann, D. C., Fink, A. F., Dehne, N., et al. (2019). Macrophage HIF-2α regulates tumor-suppressive Spint1 in the tumor microenvironment. Mol. Carcinog. 58, 2127–2138. doi: 10.1002/mc.23103
Tan, T., Yu, R. M. K., Wu, R. S. S., and Kong, R. Y. C. (2017). Overexpression and knockdown of hypoxia-inducible factor 1 disrupt the expression of steroidogenic enzyme genes and early embryonic development in Zebrafish. Gene Regul. Syst. Bio. 11:1177625017713193.
Tanaka, T., Roy, C. N., Yao, W. L., Matteini, A., Semba, R. D., Arking, D., et al. (2010). A genome-wide association analysis of serum iron concentrations. Blood 115, 94–96. doi: 10.1182/blood-2009-07-232496
Taylor, S. E., Bagnall, J., Mason, D., Levy, R., Fernig, D. G., and See, V. (2016). Differential sub-nuclear distribution of hypoxia-inducible factors (HIF)-1 and -2 alpha impacts on their stability and mobility. Open Biol. 6:160195. doi: 10.1098/rsob.160195
Tian, F., Zhou, A.-X., Smits, A. M., Larsson, E., Goumans, M.-J., Heldin, C.-H., et al. (2010). Endothelial cells are activated during hypoxia via endoglin/ALK-1/SMAD1/5 signaling in vivo and in vitro. Biochem. Biophys. Res. Commun. 392, 283–288. doi: 10.1016/j.bbrc.2009.12.170
Tjoelker, L. W., and Stafforini, D. M. (2000). Platelet-activating factor acetylhydrolases in health and disease. Biochim. Biophys. Acta 1488, 102–123.
Tudisco, L., Orlandi, A., Tarallo, V., and De Falco, S. (2017). Hypoxia activates placental growth factor expression in lymphatic endothelial cells. Oncotarget 8, 32873–32883. doi: 10.18632/oncotarget.15861
Udpa, N., Ronen, R., Zhou, D., Liang, J., Stobdan, T., Appenzeller, O., et al. (2014). Whole genome sequencing of Ethiopian highlanders reveals conserved hypoxia tolerance genes. Genome Biol. 15:R36.
Ulc, A., Gottschling, C., Schäfer, I., Wegrzyn, D., Van Leeuwen, S., Luft, V., et al. (2017). Involvement of the guanine nucleotide exchange factor Vav3 in central nervous system development and plasticity. Biol. Chem. 398, 663–675. doi: 10.1515/hsz-2016-0275
Valverde, G., Zhou, H., Lippold, S., De Filippo, C., Tang, K., Herráez, D. L., et al. (2015). A novel candidate region for genetic adaptation to high altitude in Andean populations. PLoS One 10:e0125444. doi: 10.1371/journal.pone.0125444
van Patot, M. C. T., and Gassmann, M. (2011). Hypoxia: adapting to high altitude by mutating EPAS-1, the gene encoding HIF-2α. High Alt. Med. Biol. 12, 157–167. doi: 10.1089/ham.2010.1099
Verma, P., Sharma, A., Sodhi, M., Thakur, K., Kataria, R. S., Niranjan, S. K., et al. (2018). Transcriptome analysis of circulating PBMCs to understand mechanism of high altitude adaptation in native cattle of ladakh region. Sci. Rep. 8:7681.
VonHoldf, B., Fan, Z. X., Vecchyo, D. O. D., and Wayne, R. K. (2017). EPAS1 variants in high altitude Tibetan wolves were selectively introgressed into highland dogs. PeerJ 5:e3522. doi: 10.7717/peerj.3522
Wang, G.-D., Fan, R.-X., Zhai, W., Liu, F., Wang, L., Zhong, L., et al. (2014). Genetic convergence in the adaptation of dogs and humans to the high-altitude environment of the Tibetan Plateau. Genome Biol. Evol. 6, 2122–2128. doi: 10.1093/gbe/evu162
Wang, G. L., Jiang, B. H., Rue, E. A., and Semenza, G. L. (1995). Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514. doi: 10.1073/pnas.92.12.5510
Wang, Y., Liu, S., Zhang, Y., and Yang, J. (2019). Myosin heavy chain 9: oncogene or tumor suppressor gene? Med. Sci. Monit. 25, 888–892. doi: 10.12659/msm.912320
Wang, Y., Wang, J., Zi, X. D., Huatai, C. R., Ouyang, X., and Liu, L. S. (2011). Genetic diversity of Tibetan goats of Plateau type using microsatellite markers. Arch. Fur Tierzucht 54, 188–197. doi: 10.5194/aab-54-188-2011
Weber, R. E., Lalthantluanga, R., and Braunitzer, G. (1988). Functional-characterization of fetal and adult yak hemoglobins – an oxygen binding cascade and its molecular-basis. Arch. Biochem. Biophys. 263, 199–203. doi: 10.1016/0003-9861(88)90628-5
Wei, C. H., Wang, H. H., Liu, G., Zhao, F. P., Kijas, J. W., Ma, Y. J., et al. (2016). Genome-wide analysis reveals adaptation to high altitudes in Tibetan sheep. Sci. Rep. 6:26770.
Wei, D. B., and Ma, J. B. (2001). Comparison of the content of myoglobin and lactate dehydrogenase in cardiac and skeleton muscle of plateau zorkor and mouse. J. Qinghai Univ 19, 20–21.
Wei, D. B., Wei, L., Zhang, J. M., and Yu, H. Y. (2006). Blood-gas properties of plateau zokor (Myospalax baileyi). Comp. Biochem. Physiol. 145, 372–375. doi: 10.1016/j.cbpa.2006.07.011
Wen, J., and Yuan, N. (1998). Analysis on the parameters of blood in healthy Tibetan Mastiffs. J. Qinghai Med. College 20, 7–9.
Wiener, C. M., Booth, G., and Semenza, G. L. (1996). In vivoexpression of mRNAs encoding hypoxia-inducible factor 1. Biochem. Biophys. Res. Commun. 225, 485–488. doi: 10.1006/bbrc.1996.1199
Witt, K. E., and Huerta-Sanchez, E. (2019). Convergent evolution in human and domesticate adaptation to high-altitude environments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374:20180235. doi: 10.1098/rstb.2018.0235
Wu, D., Potluri, N., Lu, J., Kim, Y., and Rastinejad, F. (2015). Structural integration in hypoxia-inducible factors. Nature 524, 303–308. doi: 10.1038/nature14883
Wu, D. D., Ding, X. D., Wang, S., Wojcik, J. M., Zhang, Y., Tokarska, M., et al. (2018). Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nat. Ecol. Evol. 2, 1139–1145. doi: 10.1038/s41559-018-0562-y
Wuren, T., Simonson, T. S., Qin, G., Xing, J., Huff, C. D., Witherspoon, D. J., et al. (2014). Shared and unique signals of high-altitude adaptation in geographically distinct Tibetan populations. PLoS One 9:e88252. doi: 10.1371/journal.pone.0088252
Xu, S., Li, S., Yang, Y., Tan, J., Lou, H., Jin, W., et al. (2011). A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol. Biol. Evol. 28, 1003–1011. doi: 10.1093/molbev/msq277
Yang, D., Peng, Y., Cui, C., Wang, L., Xiang, K., He, Y., et al. (2016a). HMOX2 functions as a modifier gene for high−altitude adaptation in Tibetans. Hum. Mutat. 37, 216–223. doi: 10.1002/humu.22935
Yang, D., Peng, Y., Ouzhuluobu, Bianbazhuoma, Cui, C., Bianba, Wang, L., Xiang, K., He, Y., Zhang, H., Zhang, X., Liu, J., Shi, H., Pan, Y., Duojizhuoma, Dejiquzong, Cirenyangji, Baimakangzhuo, Gonggalanzi, Liu, S., Gengdeng, Wu, T., Chen, H., Qi, X., and Su, B. (2016b). HMOX2 Functions as a Modifier Gene for High-Altitude Adaptation in Tibetans. Hum. Mutat. 37, 216–223.
Yang, S. L., Wu, C., Xiong, Z. F., and Fang, X. (2015). Progress on hypoxia-inducible factor-3: its structure, gene regulation and biological function. Mol. Med. Rep. 12, 2411–2416. doi: 10.3892/mmr.2015.3689
Yang, W. Z., Qi, Y., and Fu, J. Z. (2014). Exploring the genetic basis of adaptation to high elevations in reptiles: a comparative transcriptome analysis of two toad-headed agamas (Genus Phrynocephalus). PLoS One 9:e112218. doi: 10.1371/journal.pone.0112218
Yang, X., Liu, H., Ma, Z., Zou, Y., Zou, M., Mao, Y., et al. (2019). Chromosome-level genome assembly of Triplophysa tibetana, a fish adapted to the harsh high-altitude environment of the Tibetan Plateau. Mol. Ecol. Resour. 19, 1027–1036. doi: 10.1111/1755-0998.13021
Yang, Y. Z., Wang, L. Z., Han, J., Tang, X. L., Ma, M., Wang, K., et al. (2015). Comparative transcriptomic analysis revealed adaptation mechanism of Phrynocephalus erythrurus, the highest altitude Lizard living in the Qinghai-Tibet Plateau. BMC Evol. Biol. 15:101. doi: 10.1186/s12862-015-0371-8
Yi, X., Liang, Y., Huerta-Sanchez, E., Jin, X., Cuo, Z. X. P., Pool, J. E., et al. (2010). Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78.
Yi, Z., Jun, W., Xingbo, S., Xiaojun, L., Liu, D., and Binwu, Y. (2010). Genetic analysis of 17 Y-chromosomal STRs haplotypes of Chinese Tibetan ethnic minority group. Leg. Med. 12, 108–111. doi: 10.1016/j.legalmed.2009.11.003
Yoshioka, J. (2015). Thioredoxin reductase 2 (Txnrd2) regulates mitochondrial integrity in the progression of age-related heart failure. J. Am. Heart Assoc. 4:e002278.
Zaka, R., and Williams, C. J. (2006). Role of the progressive ankylosis gene in cartilage mineralization. Curr. Opin. Rheumatol. 18, 181–186. doi: 10.1097/01.bor.0000209432.36355.6e
Zhang, H., Wu, C. X., Chamba, Y., and Ling, Y. (2007). Blood characteristics for high altitude adaptation in Tibetan chickens. Poult. Sci. 86, 1384–1389. doi: 10.1093/ps/86.7.1384
Zhang, J., Hua, Z. Q., Tame, J. R. H., Lu, G. Y., Zhang, R. J., and Gu, X. C. (1996). The crystal structure of a high oxygen affinity species of haemoglobin (bar-headed goose haemoglobin in the oxy form). J. Mol. Biol. 255, 484–493. doi: 10.1006/jmbi.1996.0040
Zhang, P., Yao, Q., Lu, L., Li, Y., Chen, P.-J., and Duan, C. (2014). Hypoxia-inducible factor 3 is an oxygen-dependent transcription activator and regulates a distinct transcriptional response to hypoxia. Cell Rep. 6, 1110–1121. doi: 10.1016/j.celrep.2014.02.011
Zhang, W. P., Fan, Z. X., Han, E. J., Hou, R., Zhang, L., Galaverni, M., et al. (2014). Hypoxia adaptations in the Grey Wolf (Canis lupus chanco) from Qinghai-Tibet Plateau. PLoS Genet. 10:e1004466. doi: 10.1371/journal.pgen.1004466
Zhang, X. L., Ha, B. B., Wang, S. J., Chen, Z. J., Ge, J. Y., Long, H., et al. (2018). The earliest human occupation of the high-altitude Tibetan Plateau 40 thousand to 30 thousand years ago. Science 362, 1049. doi: 10.1126/science.aat8824
Zhou, D., Udpa, N., Ronen, R., Stobdan, T., Liang, J., Appenzeller, O., et al. (2013). Whole-genome sequencing uncovers the genetic basis of chronic mountain sickness in Andean highlanders. Am. J. Hum. Genet. 93, 452–462. doi: 10.1016/j.ajhg.2013.07.011
Zhou, W. Y., and Dou, F. M. (1990). Studies on activity and home range of plateau zokor. Acta Theriol Sinica 10, 31–39.
Keywords: hypoxia, high-altitude adaption, Tibetan, Andean, Ethiopian, EPAS1, HIF pathway, genomic adaptations
Citation: Pamenter ME, Hall JE, Tanabe Y and Simonson TS (2020) Cross-Species Insights Into Genomic Adaptations to Hypoxia. Front. Genet. 11:743. doi: 10.3389/fgene.2020.00743
Received: 16 February 2020; Accepted: 22 June 2020;
Published: 22 July 2020.
Edited by:
José M. Álvarez-Castro, University of Santiago de Compostela, SpainReviewed by:
Niroj Kumar Sethy, Defence Institute of Physiology and Allied Sciences (DRDO), IndiaCopyright © 2020 Pamenter, Hall, Tanabe and Simonson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tatum S. Simonson, dHNpbW9uc29uQGhlYWx0aC51Y3NkLmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.