
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 03 July 2020
Sec. RNA
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00716
 Junyu Zhai1,2,3
Junyu Zhai1,2,3 Shang Li2,3
Shang Li2,3 Sushmita Sen1
Sushmita Sen1 Jessica Opoku-Anane1
Jessica Opoku-Anane1 Yanzhi Du2,3
Yanzhi Du2,3 Zi-Jiang Chen2,3
Zi-Jiang Chen2,3 Linda C. Giudice1*
Linda C. Giudice1*Adenomyosis is a prevalent, estrogen-dependent uterine disorder wherein endometrial cells are abnormally present in the myometrium and are surrounded by hyperplastic/hypertrophic smooth muscle. Its etiology is unclear, although endometrial cell invasion into the myometrium has been postulated. RNA methylation, particularly N6-methyladenosine (m6A), plays an important role in regulating various physiological processes and invasive disorders. The goal of this in silico and lab-based experimental study was to explore a possible role for m6A in adenomyosis. Gene expression profiles of both the endometrium and myometrium of women with adenomyosis (cases) and without disease (controls) were obtained from the publicly available Gene Expression Omnibus (GEO) database. In the endometrium, STRING database analysis revealed that METTL3 functions as a “hub” gene of m6A RNA methylation regulators, and the genes involved in m6A regulation, including METTL3, FTO, ZC3H13, and YTHDC1 expression, were significantly decreased in cases versus controls. Functional, co-expression, and correlational analyses of endometrium from cases versus controls revealed decreased total m6A levels, induced by METTL3, and the downstream elevated insulin−like growth factor−1(IGF1) and D-Dopachrome Tautomerase (DDT), with the latter two having known functions in epithelial proliferation and cell migration, which are important processes in the pathogenesis of adenomyosis in endometrium. m6A RNA methylation regulators, including RBM15/15B, ALKBH5, FTO, YTHDF1/2, KIAA1429, HNRNPC, METTL3, ZC3H13, and YTHDC2, were also differentially expressed in the myometrium from cases versus controls. We validated decreased total m6A levels and differential expression of m6A RNA methylation regulators in the myometrium of patients with adenomyosis using qRT-PCR, immunohistochemistry and tissues available from our biorepository. Possible target genes, including cadherin 3(CDH3), sodium channelβ-subunit 4 (SCN4B), and placenta-specific protein 8 (PLAC8), which are involved in cell adhesion, muscle contraction and immune response in the myometrium of adenomyosis patients were also validated. Thus, through extensive public database mining and validation of select genes, this study, for the first time, implicates m6A and its methylation regulators in the pathogenesis of adenomyosis. Follow on functional studies are anticipated to elucidate mechanisms involving m6A and its regulators and down-stream effectors in the pathogenesis of this enigmatic reproductive disorder and potentially identify druggable targets to control its associated symptoms.
Adenomyosis is a common disease of the uterus in which endometrial epithelial cells and stromal fibroblasts abnormally are found in the myometrium, wherein they elicit hyperplasia and hypertrophy of surrounding smooth muscle cells (Bird et al., 1972). It occurs in 8–27% of reproductive age women (Kissler et al., 2008) and results in a diffusely enlarged uterus, pelvic pain, heavy menstrual bleeding and infertility in those affected (Benagiano et al., 2012). Historically, definitive diagnosis was based on histological examination of hysterectomy specimens. Currently, decreased echogenicity or signal intensity on ultrasound and magnetic resonance imaging (MRI), respectively, is commonly used to diagnose adenomyosis which can occur in a diffuse pattern, as discrete adenomyomas, or cystic lesions in the uterine smooth muscle layer (Reinhold et al., 1998). As mechanisms underlying the pathogenesis and pathophysiology of adenomyosis are not well understood, therapies are inadequate to control symptoms or to facilitate successful pregnancy (Li et al., 2018).
Disruption of the “inner myometrium,” i.e., the normal boundary between the endometrial basal layer and the myometrium, has been postulated to underlie adenomyosis pathogenesis, with subsequent endometrial tissue and cells migrating into the adjacent smooth muscle compartment (Vercellini et al., 2006). As adenomyosis is more prevalent in women with previous cesarean section, it has been postulated that the endometrium invades a predisposed myometrium or a traumatized endometrial-myometrial interface during periods of regeneration, healing, and reepithelization (Curtis et al., 2002). In addition, there also evidence of familial predisposition in which genetic, immunological and other factors are involved (Arnold et al., 1995). Abnormal Müllerian and mesenchymal interactions during uterine development also may contribute to its pathogenesis, and tissue injury typically activates adult stem cells, which may establish endometrial lineage cells through disruption of endometrial stem/progenitor cell niches (Gargett et al., 2016). Thus, both compartments (endometrium and myometrium) have been implicated in the pathogenesis of adenomyosis, although more research is required to understand, mechanistically, the initiation and progression of the disease. Herein, we focus on RNA methylation in both compartments.
Epigenetic modifications play an important role in regulation of human physiology and invasive diseases, among which DNA and RNA methylation are involved. While most studies have focused on the role of DNA methylation in the female reproductive system, less data are available regarding RNA methylation, especially in adenomyosis. N6-methyladenosine (m6A) is the most abundant modification on mRNAs (Zaccara et al., 2019). m6A provides dynamic regulation of the nucleation, splicing, translation and stability of mRNA molecules (Wang et al., 2014; Roundtree and He, 2016; Deng et al., 2018), thereby influencing fundamental biological and pathological processes such as proliferation, differentiation, cellular response to stress and tumorigenesis (Peer et al., 2017). The m6A modification regulators are classified into the three groups: writers, erasers and readers (Wu et al., 2020). “Writers” include the m6A methyltransferases that promote methylation of m6A. They are mainly composed of methyltransferase−like 3 (METTL3) and 14 (METTL14) and Wilms’ tumor 1−associating protein (WTAP). Moreover, KIAA1429, zinc finger CCCH−type containing 13 (ZC3H13), METTL16 and RNA binding motif protein 15/15B (RBM15/15B) are also contribute to the RNA methylation (Wen et al., 2018; Yue et al., 2018). “Erasers” are demethylases which consist of fat mass and obesity−associated protein (FTO) and AlkB homolog 5 (ALKBH5). “Readers” are proteins that bind to the m6A site and promoting the function of m6A. They are predominantly in the YT521−B homology (YTH) protein family [YTH domain family 1/2/3 (YTHDF1/2/3) and YTH domain containing 1/2 (YTHDC1/2)], nuclear heterogeneous protein HNRNP family and IGF2BP protein family (Liao et al., 2018).
Previous studies indicated that m6A RNA methylation mediates cell proliferation and apoptosis in different cell types (Gu et al., 2018). Notably, increased proliferation and inhibition of cellular apoptosis are characteristics of the endometrium in women with adenomyosis (Li et al., 2019). m6A also contributes to the epithelial-to-mesenchymal transition (EMT) of cancer cells (Li et al., 2020) and EMT is considered to be a possible mechanism for the transfer of epithelial cell into the myometrium in adenomyosis patients (Hu et al., 2020). Moreover, m6A and its methylation regulators also regulate T cell activity (Winkler et al., 2019) and vascular development, which is involved in endometrial dysfunction of adenomyosis patients (Ota et al., 1998; Benagiano and Brosens, 2012). Thus, m6A RNA methylation may also play a role in the pathogenesis of adenomyosis.
m6A and its methylation regulators can also play roles in endometrial function in other settings. Liu et al. reported that about 70% of endometrial tumors exhibit reductions in m6A RNA methylation due to reduced METTL3 expression. Moreover, m6A mRNA methylation is regarded as an oncogenic mechanism in endometrial cancer through regulation of AKT signaling (Liu et al., 2018). A previous study indicated that adenomyosis and type I endometrial cancer are linked to sex steroid action and exhibit gene expression profiling supporting a relationship between endometrial cancer and adenomyosis (Inoue et al., 2019), and women with adenomyosis are at higher risks of endometrial cancer (Yeh et al., 2018). The PI3K-AKT pathway, BCL2 apoptosis regulator and other factors are implicated in both adenomyosis and endometrial cancer (Roddy and Chapman, 2017). Thus, m6A RNA methylation may also contribute to endometrial dysfunction in women with adenomyosis.
Herein, we have investigated expression of m6A RNA methylation regulators in both endometrium and myometrium of women with versus without adenomyosis, providing a novel perspective and laying the foundation to elucidate underlying mechanisms of adenomyosis pathogenesis and pathophysiology.
We searched the associated gene expression profiles of the eutopic endometrium of adenomyosis patients in Gene Expression Omnibus (GEO) database1, using the keywords “adenomyosis”, “eutopic endometrium”, and “Homo sapiens.” We chose GSE78851 (Herndon et al., 2016) for analysis (5 control and 3 adenomyosis). All the eight samples of eutopic endometrium are in proliferative phase and we retained gene expression datasets from the Affymetrix Human Gene 1.0 ST Array (HuGene-1_0-st) and detected gene expression changes in the eutopic endometrium between three patients with adenomyosis and 5 healthy women (control).
We further searched the gene expression profiles of the myometrium of women with adenomyosis in GEO database using “adenomyosis,” “myometrium” and “Homo sapiens” and chose GSE7307 to investigate the mechanism of adenomyosis from the view of myometrium (10 women with adenomyosis versus 40 without adenomyosis). The gene expression was got from Affymetrix Human Genome U133 Plus 2.0 Array (HG-U133_Plus_2). We detected gene expression of myometrium between 10 women with adenomyosis (cases) and 40 without adenomyosis (controls).
After downloading the GSE78851 and GSE7307 from GEO, the “impute” package of R software was used to impute the missing expression data while “limma” package was used to normalize the gene expression and identify the differentially expressed genes separately. The significant difference was defined as log FC > 1 and P < 0.05.
We first assembled a list of eighteen m6A RNA methylation regulators from published literature and review (Wu et al., 2020), and then we restricted the list to sixteen genes with available RNA expression data separately from the GSE78851 and GSE7307 in GEO dataset. This yielded a total of sixteen m6A RNA methylation regulators. Then, we systematically compared the expression of these sixteen m6A RNA methylation regulators in the eutopic endometrium and myometrium of women with and without adenomyosis separately using Wilcox test in R software (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Interactions among m6A RNA methylation regulators were analyzed using the STRING2. Moreover, the correlation between m6A RNA methylation regulators and highly enriched Gene ontology (GO) terms related DEGs were identified using Spearman correlation in the “Corrplot” package of R software. p < 0.001 was considered as significantly correlated to each other.
To verify the potential relationship and the co-expression genes of m6A RNA methylation regulators that were differentially expressed in the women with and without adenomyosis, we used another method to analyze the DEGs in the eutopic endometrium and myometrium separately. WGCNA assigns a connection weight to each gene pair in the network, being more meaningful compared to traditional methods that use binary information. Thus, WGCNA can be used to find modules of highly correlated genes and identify candidate target genes (Langfelder and Horvath, 2008). We used WGCNA to analyze the DEGs and identified the relationship between m6A RNA methylation regulators and the potential target genes of m6A RNA methylation regulators in the eutopic endometrium and myometrium separately.
GO analysis was used to identify the possible molecular function and visualize the potential biological meaning behind DEGs, whereas Kyoto Encyclopedia of Genes and Genomes (KEGG) was to analyze the potential functions of these genes. The gene ID was set using the “org.Hs.eg.db” of R software, and then GO and KEGG pathway enrichment analyses were performed with “clusterProfiler.” p < 0.05 was identified as significant.
Clinical symptoms and histologic evaluation of hysterectomy specimens identified samples from cases and controls (latter had hysterectomies due to uterine fibroids or dysmenorrhea). Full thickness uterine specimens (including endometrium, inner myometrium and outer myometrium) were collected and stored in OCT at −80°C. All participants (n = 9 cases; n = 9 controls) were in the proliferative phase of the cycle, confirmed by endometrial histology (Noyes et al., 1950) and serum estrogen (E2), progesterone levels (P4). All participants were documented to be not pregnant and had not received hormonal or gonadotropin-releasing hormone agonist (GnRHa) therapies for at least 3 months prior to tissue sampling. 6 cases and 6 controls were used for the detection of the percentage of m6A in total RNA and validation of real-time polymerase chain reaction (qRT-PCR) while the rest of the cases and controls samples (3 for each group) were used for immunohistochemistry (IHC) and m6A quantification of only mRNA. The clinical samples were collected from the Endometrial Tissue and DNA Bank at the University of California, San Francisco under an approved human subjects protection protocol (IRB # 10-02786), after written informed consent of all participants.
The full thickness uterine tissue was embedded in OCT and stored at −80°C. Five μm-thick tissue frozen sections were prepared and then OCT blocks were returned to −80°C and stored at this temperature until further use. Slides were brought to room temperature and left for at least an hour prior to fixing the tissue in 100% ethanol for 10 min. Then, the slides were stained in hematoxylin for 7 min and differentiated by hydrochloric acid for 30 s. Finally, the sections were incubated in eosin for 1 min before covering the slide and visualizing using a microscope (Zeiss, Oberkochen, Germany).
The endometrium and myometrium of OCT blocks were separated on ice according to H&E stain. We chose the myometrium close to the interface between the endometrium and myometrium, so that most of myometrial samples were mainly inner myometrium. Total RNA from endometrium and myometrium was extracted, separately, using an RNA isolation kit (Macherey-Nagel, Bethlehem, PA, United States) and reversely transcribed into cDNA (TAKARA, Dalian, China). The mRNA expression of target genes was detected using qRT-PCR. Results were analyzed by ΔΔCt method. The ratio of the target gene over β-Actin was calculated as the target mRNA level. The primer sequences used for targeting genes are shown in Supplementary Table.
The m6A RNA Methylation Assay Kit (Abcam, Cambridge, United Kingdom) was used to evaluate the content of m6A in total RNA as protocol. Total RNA was bound to the strip wells using an RNA high binding solution provided by the manufacturer. Briefly, 200 ng total RNA were coated on each assay well, followed by specific capture with N6-methyladenosine antibody and the detection antibody. Then the detected signal was enhanced and quantified colorimetrically by reading the absorbance in a microplate spectrophotometer at a wavelength of 450 nm. The amount of m6A is proportional to the OD intensity measured. Finally, calculations using OD450 values were performed based on the standard curve to get the final content of m6A level in total RNA.
Five μm-thick tissue frozen sections were prepared using OCT blocks of full thickness uterine tissue. Slides were rehydrated and then blocked using blocking buffer for 1 h at room temperature. Heat-mediated antigen retrieval was carried out with 10 mM sodium citrate, 0.05% Tween 20, pH 6, and then slides were incubated in anti-METTL3 antibody (1:200 dilution; Proteintech, Wuhan, China) overnight at 4°C. After being washed with PBS, the slides were processed with the secondary antibody (1:400) for 1 h at room temperature, and then the color reaction was visualized by exposure to diaminobenzidine (DAB). Slides were counterstained with hematoxylin and dehydrated through graded alcohols and xylene before visualizing using a microscope (Zeiss). Staining was assessed using Image J.
RNA m6A quantification by LC-MS/MS was performed as described previously (Liu et al., 2014). In brief, total RNA from the endometrium and myometrium of women with and without adenomyosis was isolated using TRIzol reagent (Invitrogen, CA, United States), and polyadenylated RNAs were extracted by oligo d(T)25 magnetic beads (NEB, Ipswich, MA, United States), followed by removal of rRNA with RiboMinus Eukaryote Kit (Ambion, Austin, TX, United States). 200 ng mRNA were digested by nuclease P1 (1 U, Sigma-Aldrich, St. Louis, MO, United States) in 20 μL buffer which contained 25 mM NaCl, 2.5 mM ZnCl2 for 2 h at 37°C. After an additional incubation at 37°C for 2 h, the solution was centrifuged at 13000 rpm for 10 min at 4°C, and 10 μL of the solution was injected into LC-MS/MS. Quantification was performed by comparison with the standard curve obtained from pure nucleoside standards. The ratio of m6A to A in polyadenylated RNAs was calculated based on the calculated concentrations.
Results are presented as mean ± SEM. The m6A level and qRT-PCR quantification of target genes between women with and without adenomyosis were analyzed in unpaired Student’s t-test with SPSS software (IBM, New York, NY, United States). Statistical significance is shown as ∗P < 0.05, ∗∗P < 0.01, or ∗∗∗P < 0.001.
From the analysis of the gene expression profile of GSE78851, we found that METTL3, ZC3H13, FTO, and YTHDC1 were all significantly decreased in the eutopic endometrium of adenomyosis patients versus controls (Figure 1A). METTL3 is a “writer,” and ZC3H13 aids in the RNA methylation process; while FTO belongs to the “erasers,” and YTHDC1 is a “reader” (Wu et al., 2020). Thus, we pursued whether m6A levels are regulated by the above-mentioned factors in cases versus controls. Specifically, we examined m6A modification-related interactions and correlations among the 16 m6A RNA methylation regulators. STRING database analysis suggested that METTL3 and WTAP are “hub” genes of the m6A RNA methylation regulators, as both interact with 12 m6A RNA methylation regulators (interaction score ≥ 0.7) (Figures 1B,C). Moreover, the expression of METTL3 was also significantly correlated to expression of all other differentially expressed m6A RNA methylation regulators in endometrium of cases versus controls, including YTHDC1, FTO, and ZC3H13 (Figure 1D, Spearman R) without any change in the expression of WATP. Therefore, METTL3 is a prime candidate as the “hub” gene of m6A RNA methylation regulators involved in endometrial dysfunction in the setting of adenomyosis.
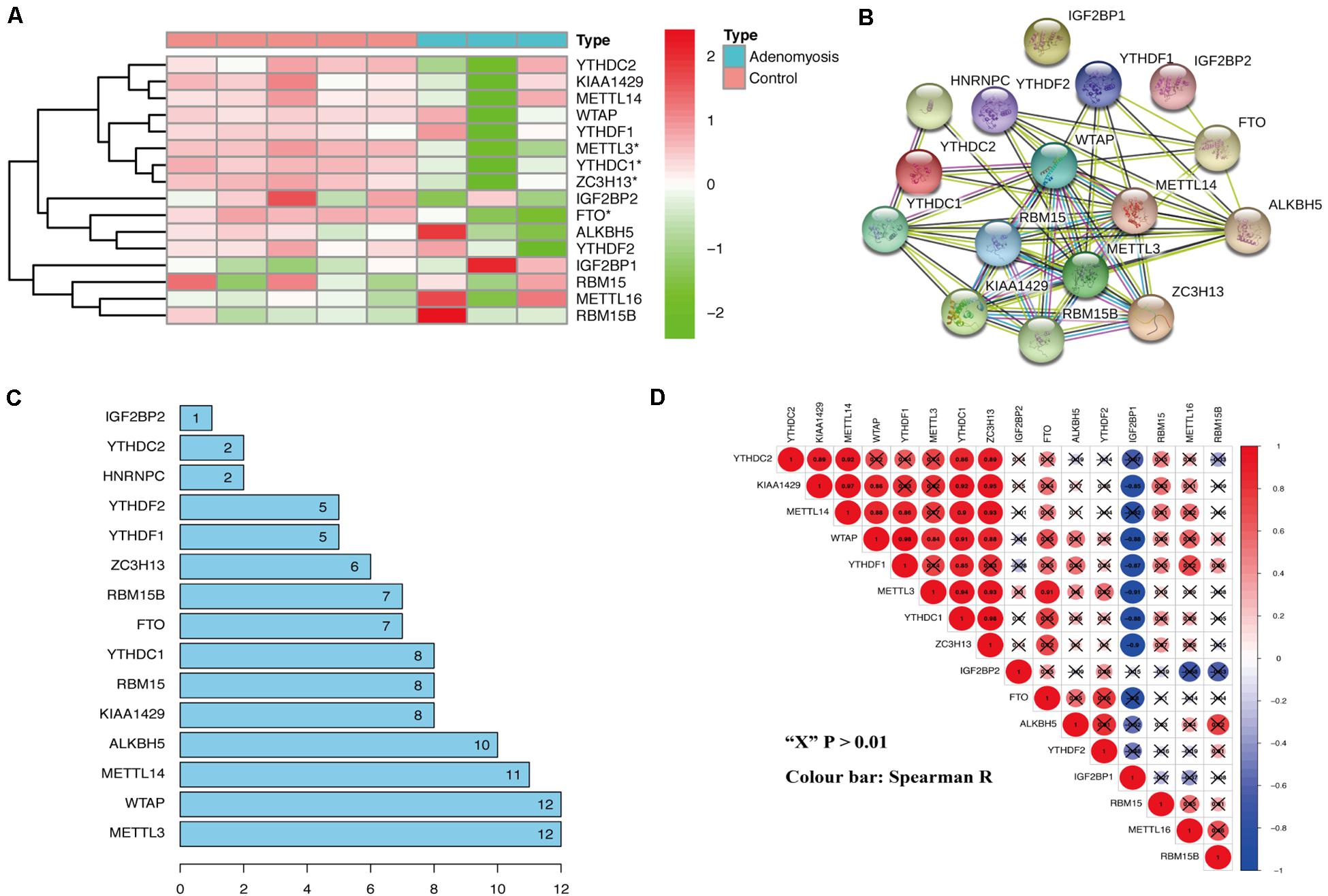
Figure 1. The expression and interaction of m6A RNA methylation regulators in the eutopic endometrium of women with and without adenomyosis. (A) The expression levels of sixteen m6A RNA methylation regulators in the eutopic endometrium. (B) The m6A modification-related interactions among the 16 m6A RNA methylation regulators. (C) The interaction counts of 16 m6A RNA methylation regulators. (D) Spearman correlation analysis of the 16 m6A modification regulators.
Using “limma” package, we analyzed GSE78851 and found 791 transcripts differentially expressed (191 up regulated and 600 down regulated) in endometrium of cases versus controls (Figure 2A). Biological functions of these DEGs were identified using GO and KEGG enrichment. With GO, enrichment was noted in “epithelial cell proliferation,” “regulation of fibroblast proliferation,” “regulation of cell migration,” “cell-substrate adherens junction,” “regulation of Wnt signaling pathway” and other biological processes were highly enriched in DEGs [FDR (adjust P) < 0.05]. In addition, other significantly enriched biological processes were related to m6A function, including “RNA splicing,” “regulation of mRNA stability,” “translation preinitiation complex” and “structural constituent of ribosome” (Figure 2B).
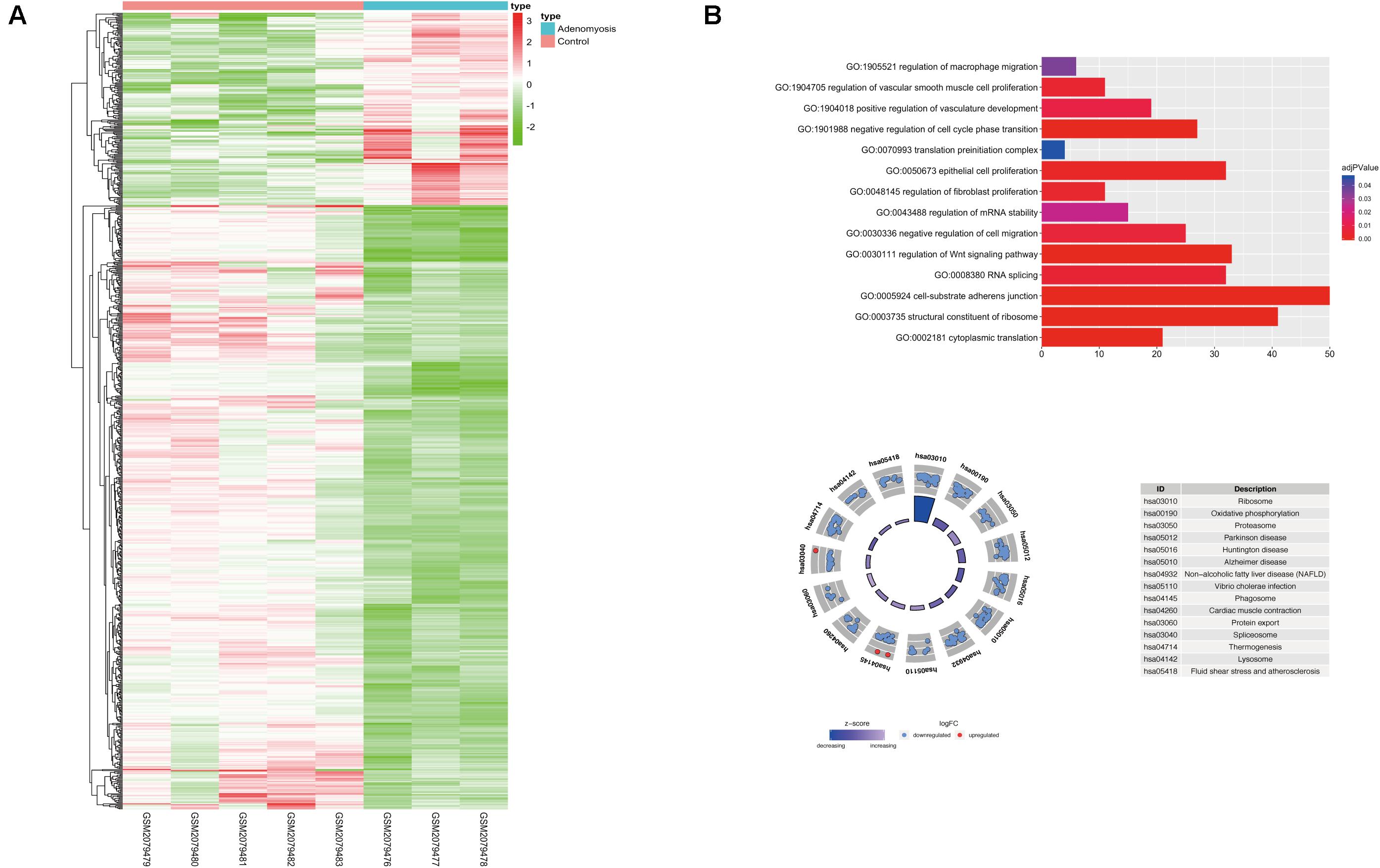
Figure 2. The expression, functional enrichment of DEGs in the eutopic endometrium of women with and without adenomyosis. (A) The differentially expressed transcripts in the eutopic endometrium of adenomyosis patients versus controls (3 adenomyosis patients and 5 controls). (B) Functional annotation of the DEGs of eutopic endometrium using GO terms of biological processes (upper) and KEGG pathway (lower).
As previously described, METTL3 is considered as the “hub” m6A RNA methylation regulator. Accordingly, further exploration of the correlation between the 217 DEGs from these highly enriched GO terms and the expression of METTL3 was performed to clarify a possible role of m6A RNA methylation regulators in the dysfunction of endometrium of women with adenomyosis (Figure 3A). We found 67 genes were significantly correlated to the expression of METTL3. Nine of these were involved in “epithelial cell proliferation,” 2 were involved in “negative regulation of cell migration” and 2 contributed to “negative regulation of cell cycle phase transition.”
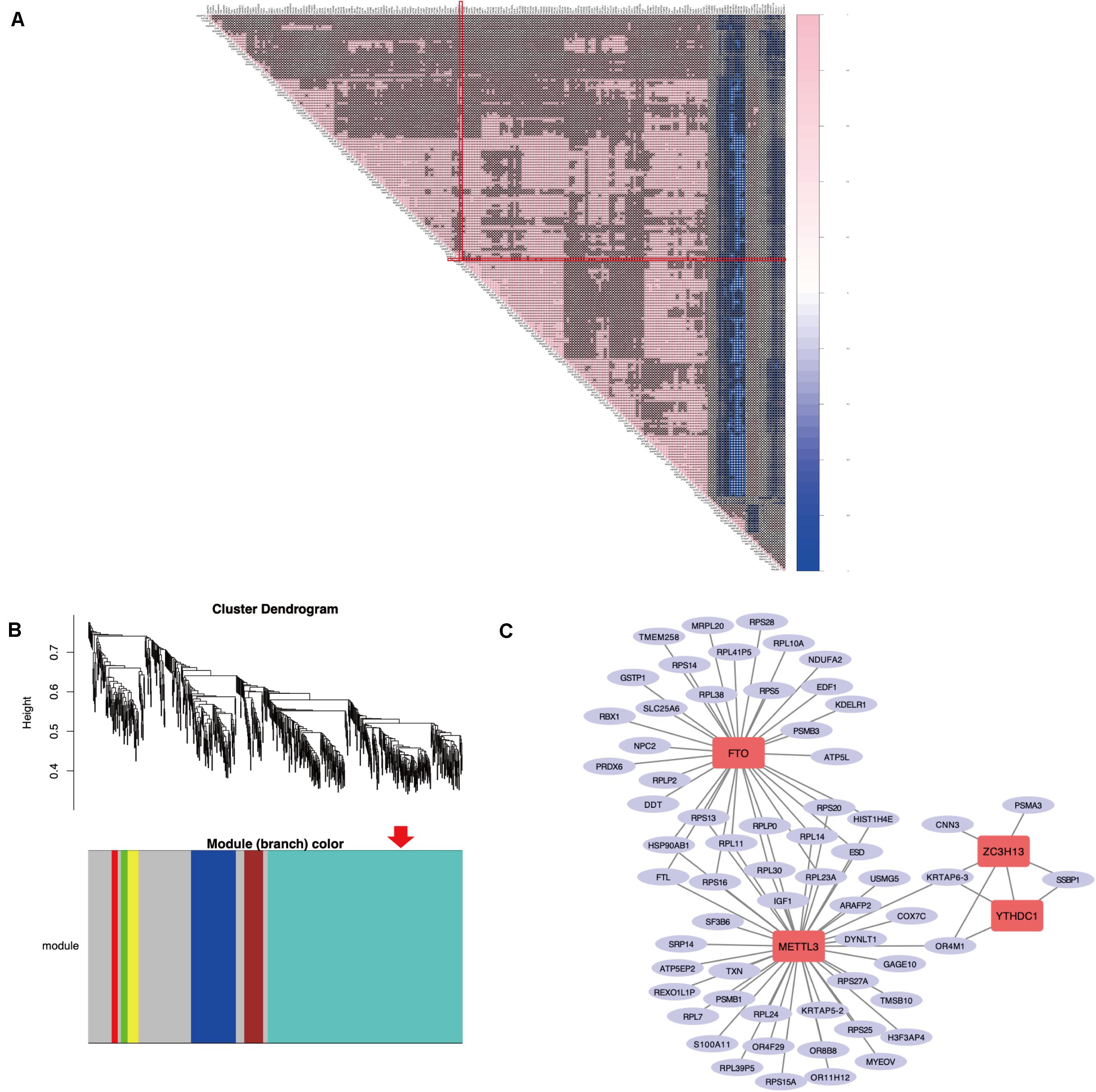
Figure 3. The correlation analysis and WGCNA of DEGs in the eutopic endometrium. (A) The Spearman correlation analysis of the m6A modification regulators and the DEGs enriched in the GO terms that related to eutopic endometrium dysfunction of adenomyosis. “X” p > 0.001, Red line clarified that correlation between METTL3 and other DEGs. (B) WGCNA of DEGs in the eutopic endometrium of women with adenomyosis versus without. Red arrow revealed the module that the m6A RNA methylation regulators belong to. (C) The co-expressed genes of four differential expressed m6A RNA methylation regulators using WGCNA analysis (threshold = 0.8).
WGCNA was used to further identify target genes of differentially expressed m6A RNA methylation regulators in endometrium of cases versus controls. Firstly, we found all differentially expressed m6A RNA methylation regulators (METTL3, FTO, ZC3H13, and YTHDC1) belong to the “turquoise” module (Figure 3B), demonstrating their close relationship to each other. Secondly, genes that were significantly co-expressed or correlated to the four differentially expressed m6A RNA methylation regulators were identified, and the network is shown in Figure 3C. Combining the Spearman correlation between METTL3 and DEGs and WGCNA results, a total of 19 co-expressed genes were found, which may be target genes of m6A RNA methylation regulators in eutopic endometrium of cases versus controls (threshold = 0.8) (Table 1).
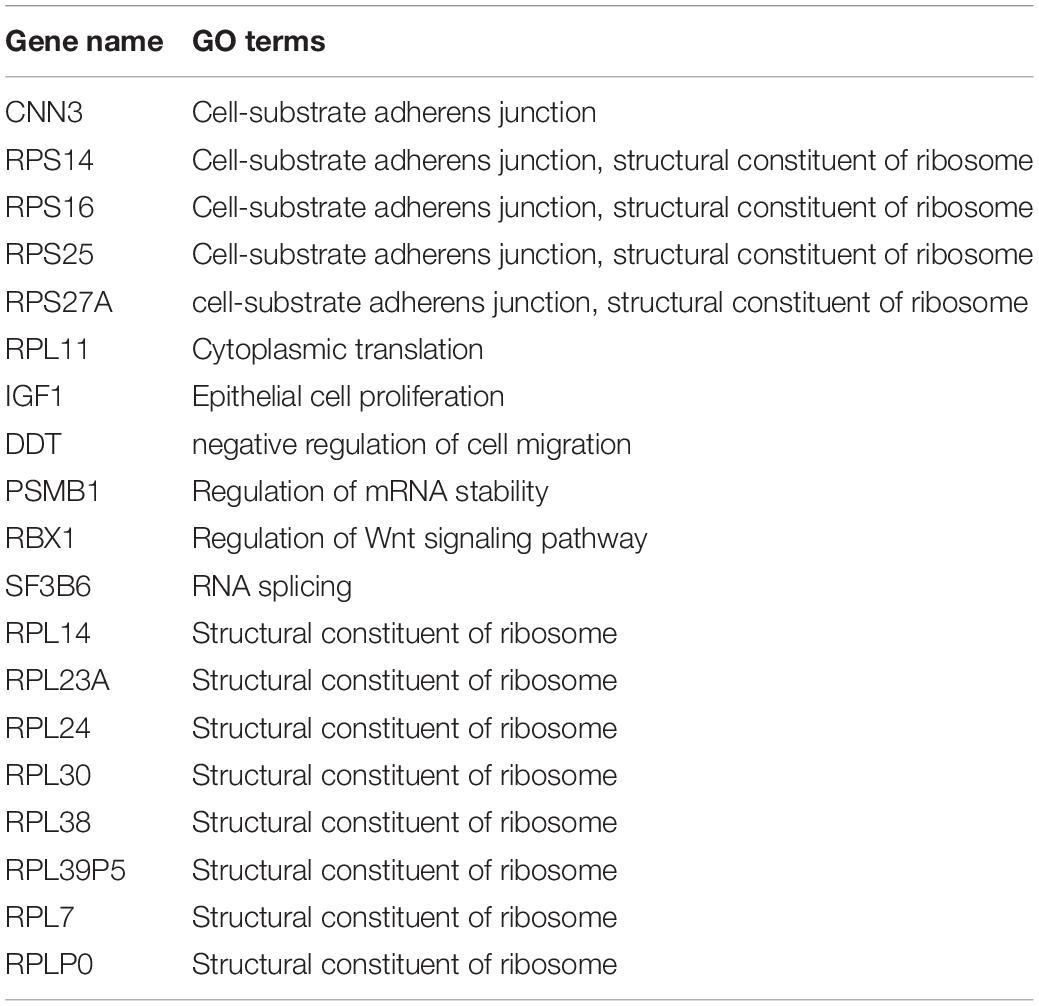
Table 1. The potential target genes of differential expressed m6A RNA methylation regulators in the eutopic endometrium.
After combing the results from the bioinformatics analyses, m6A levels and METTL3 were considered as likely functional regulators in endometrium of women with adenomyosis. To validate this hypothesis, we studied total m6A levels and the relative expression of m6A RNA methylation regulators in endometrium from a cohort of women with adenomyosis (n = 6) and controls (n = 6) – all in the proliferative phase of the menstrual cycle. The percentage of m6A content in total RNA of endometrium was significantly reduced (Figure 4A), while a trend for lower amounts of m6A was detected in polyadenylated RNA of cases versus controls (Supplementary Figure 1A), similar to endometrial cancer (Liu et al., 2018). Moreover, METTL3, and YTHDC1 mRNA were also decreased (Figure 4B). We also detected METTL3 protein in endometrium via IHC. We found high expression of METTL3 protein both in the glands and stroma of control endometrium (Supplementary Figure 2A). The protein level of METTL3 was significantly decreased in the endometrium of adenomyosis patients when compared with control (p = 0.034), consistent with its mRNA level.
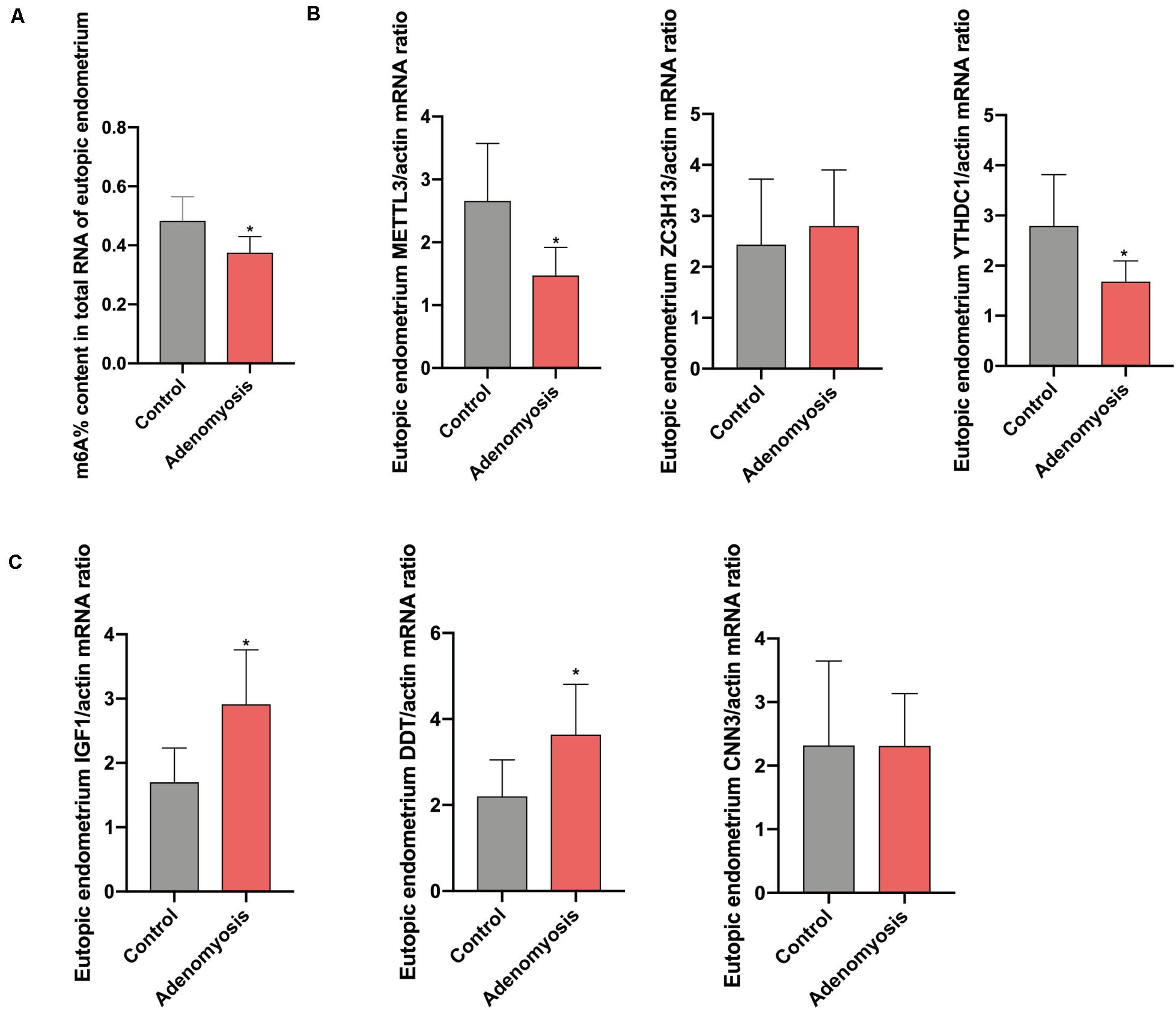
Figure 4. The total m6A level and qRT-PCR validation of bioinformatics data in eutopic endometrium of women with and without adenomyosis. (A) m6A% content in total RNA in the eutopic endometrium of women with and without adenomyosis. (B) The expression of METTL3, ZC3H13, YTHDC1 mRNA level in the eutopic endometrium. (C) The expression of IGF1, DDT, and CNN3 mRNA level in the eutopic endometrium. n = 6 for each group. *p < 0.05.
Furthermore, we verified the potential target genes of differentially expressed m6A RNA methylation regulators identified above. Messenger RNA for insulin−like growth factor−1 (IGF1), the key regulator of the epithelial proliferation and the AKT pathway (Stitt et al., 2004; Merritt et al., 2016), was significantly increased in cases versus controls. Furthermore, D-dopachrome tautomerase (DDT), works as a gene responsible for cell migration and cell proliferation (Merk et al., 2012), were also significantly highly expressed in cases versus controls, without any change in Calponin 3 (CNN3) (Figure 4C).
Gene expression profiles of myometrium of women with and without adenomyosis were mined from GSE7307. Gene expression of the myometrium of adenomyosis group (n = 10) was compared to the control group (n = 40) comprised of myometrial samples without adenomyosis, endometriosis and/or cancer. The comparison identified 563 DEGs, of which 278 genes were down regulated, and 285 genes were upregulated from cases versus controls. Expression of 16 m6A RNA methylation regulators is shown in Figure 5A, and 11 of them were significantly differentially expressed in the myometrium of adenomyosis patients including RBM15/15B, ALKBH5, FTO, YTHDF1/2, KIAA1429, HNRNPC, METTL3, ZC3H13, and YTHDC2 (Figure 5A).
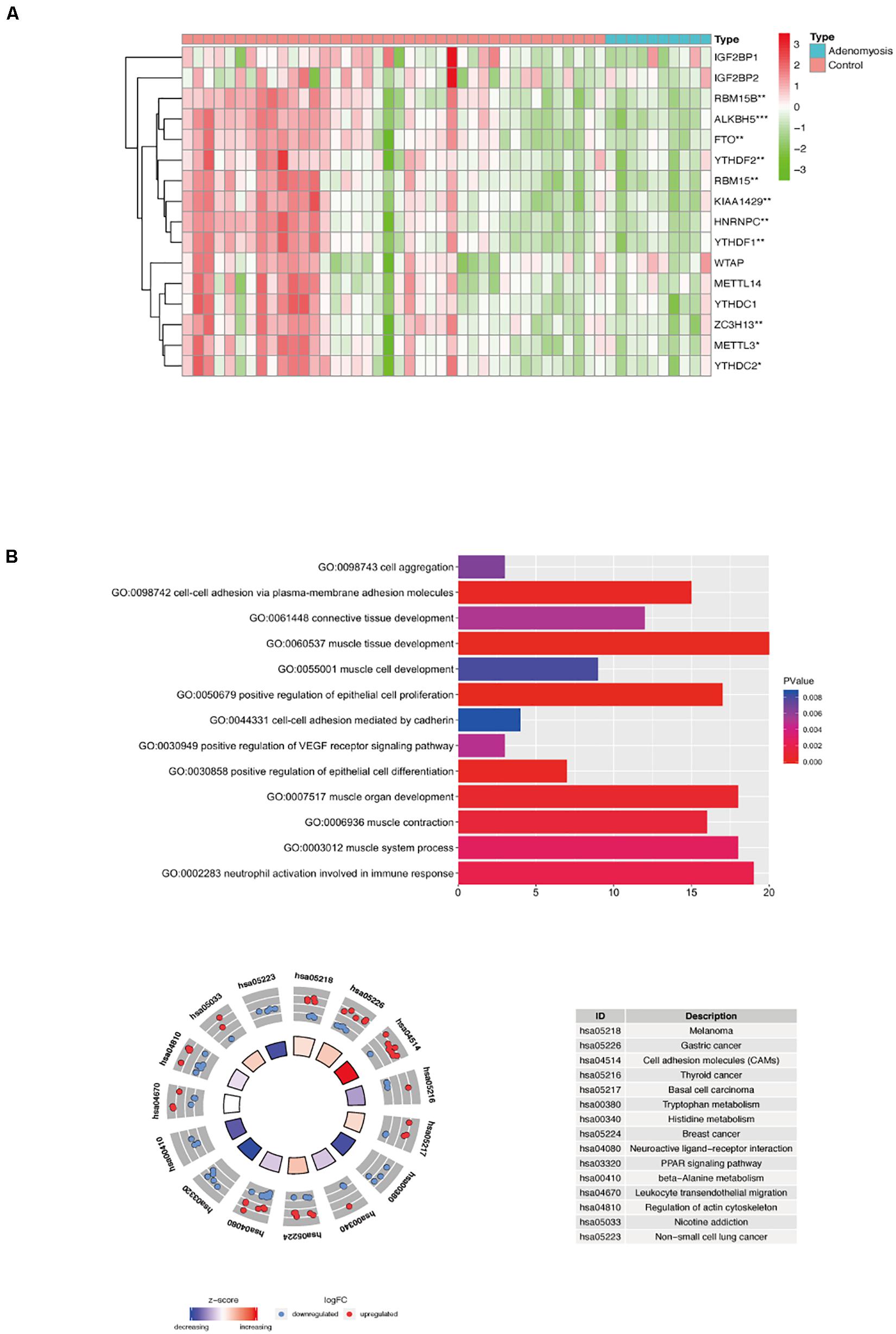
Figure 5. The expression of m6A RNA methylation regulators and the functional enrichment of DEGs in the myometrium of women with and without adenomyosis. (A) The expression levels of sixteen m6A RNA methylation regulators in the myometrium (10 women with adenomyosis and 40 without adenomyosis). (B) Functional enrichment of the DEGs of myometrium using GO terms of biological processes (upper) and KEGG pathway (lower).
Functional analysis revealed that “muscle tissue development,” “cell-cell adhesion via plasma-membrane adhesion molecules,” “positive regulation of epithelial cell differentiation,” “muscle contraction,” “neutrophil activation involved in immune response,” “connective tissue development” and other biological processes were highly enriched in DEGs in myometrium of cases versus controls (Figure 5B).
Different from endometrium, 13 of 16 m6A RNA methylation regulators in myometrium were significantly correlated to each other, and most of them were down regulated in adenomyosis patients using Spearman correlation (Figures 5A, 6). Additionally, the 13 m6A RNA methylation regulators were all attributed to the “blue” module using WGCNA (Supplementary Figure 3). Thus, the 13 m6A RNA methylation regulators (RBM15/15B, YTHDF1, WTAP, KIAA1429, ZC3H13, YTHDC2, METTL3, METTL14, YTHDC1, ALKBH5, and FTO) were identified as a “cluster” of m6A methylation regulators in the myometrium of women.

Figure 6. The Spearman correlation between m6A RNA methylation regulators and the DEGs of myometrium enriched in the GO terms that related to myometrium dysfunction of adenomyosis patients. “X” p > 0.001, Red line clarified that correlation between the 13 m6A regulation “cluster” and other DEGs.
We analyzed the correlation between the 107 DEGs in these highly enriched biological processes in myometrium of women with adenomyosis and controls and m6A regulator “cluster” to clarify potential mechanisms underlying regulation of m6A RNA methylation regulators in the setting of disease. Genes that significantly correlated to the expression of the “m6A regulator cluster” were identified as potential target genes of m6A RNA methylation regulators in the myometrium. Fifteen genes were identified, 4 of which were involved in “muscle contraction”, 3 with “cell-cell adhesion” and 2 genes were involved in “neutrophil activation involved in immune response” (Table 2; Figure 6).
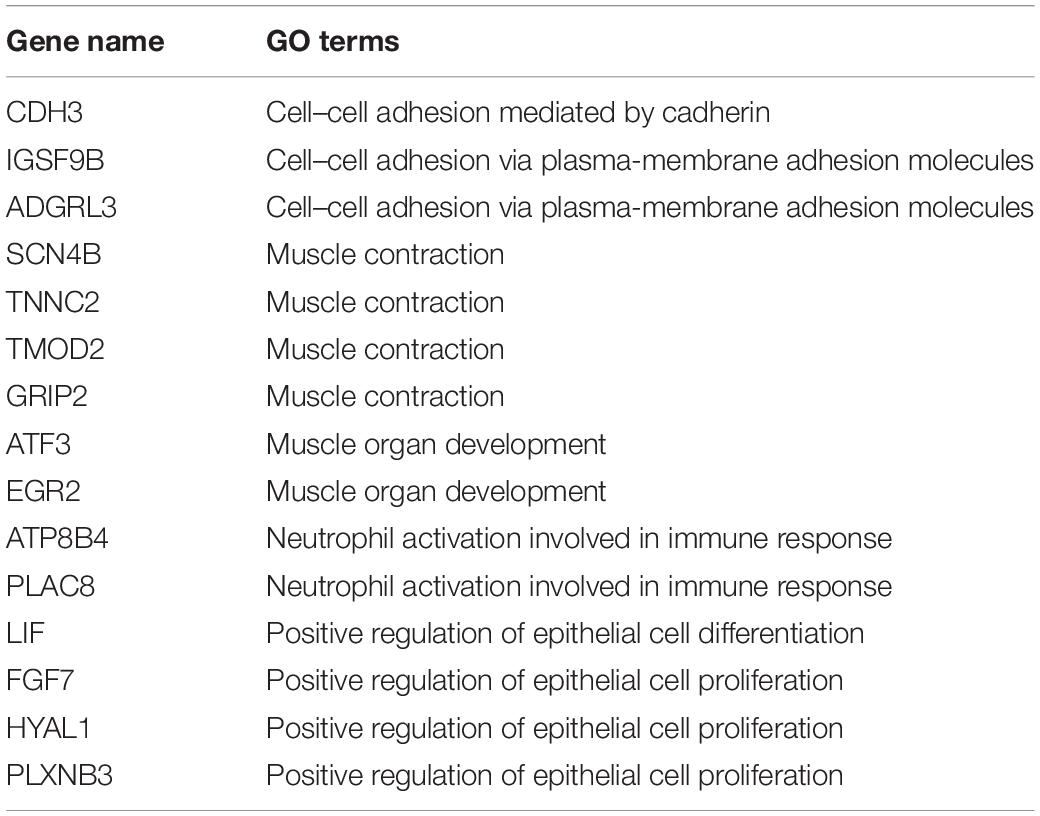
Table 2. The DEGs from high enriched GO terms that are significantly correlated to m6A RNA methylation regulators cluster in the myometrium.
Equivalent to the eutopic endometrium, the m6A% content in total RNA of myometrium was also significantly reduced (Figure 7A) while a decreased tendency of m6A% was detected in polyadenylated RNA of women with adenomyosis compared with controls (Supplementary Figure 1B). In addition, expression of METTL3 and FTO mRNA was decreased without changes in METTL14 and ALKBH5 (Figure 7B). The protein level of METTL3 appeared lower in myometrium of women with adenomyosis compared with controls (p = 0.198) (Supplementary Figure 2B). Expression of identified genes that were significantly correlated with the m6A regulator “cluster” in the myometrium was also verified. As a key molecule in cell-cell adhesion and EMT (Sousa et al., 2019), cadherin 3 (CDH3) mRNA was significantly increased in adenomyosis patients. Additionally, sodium channelβ-subunit 4 (SCN4B) mRNA was decreased, suggesting possible regulation of m6A RNA methylation regulators to cell adhesion in adenomyosis. Finally, placenta-specific protein 8 (PLAC8) mRNA was also increased in the myometrium of adenomyosis patients, indicating the role of immune response in the myometrium of women with adenomyosis (Johnson et al., 2012; Figure 7C).
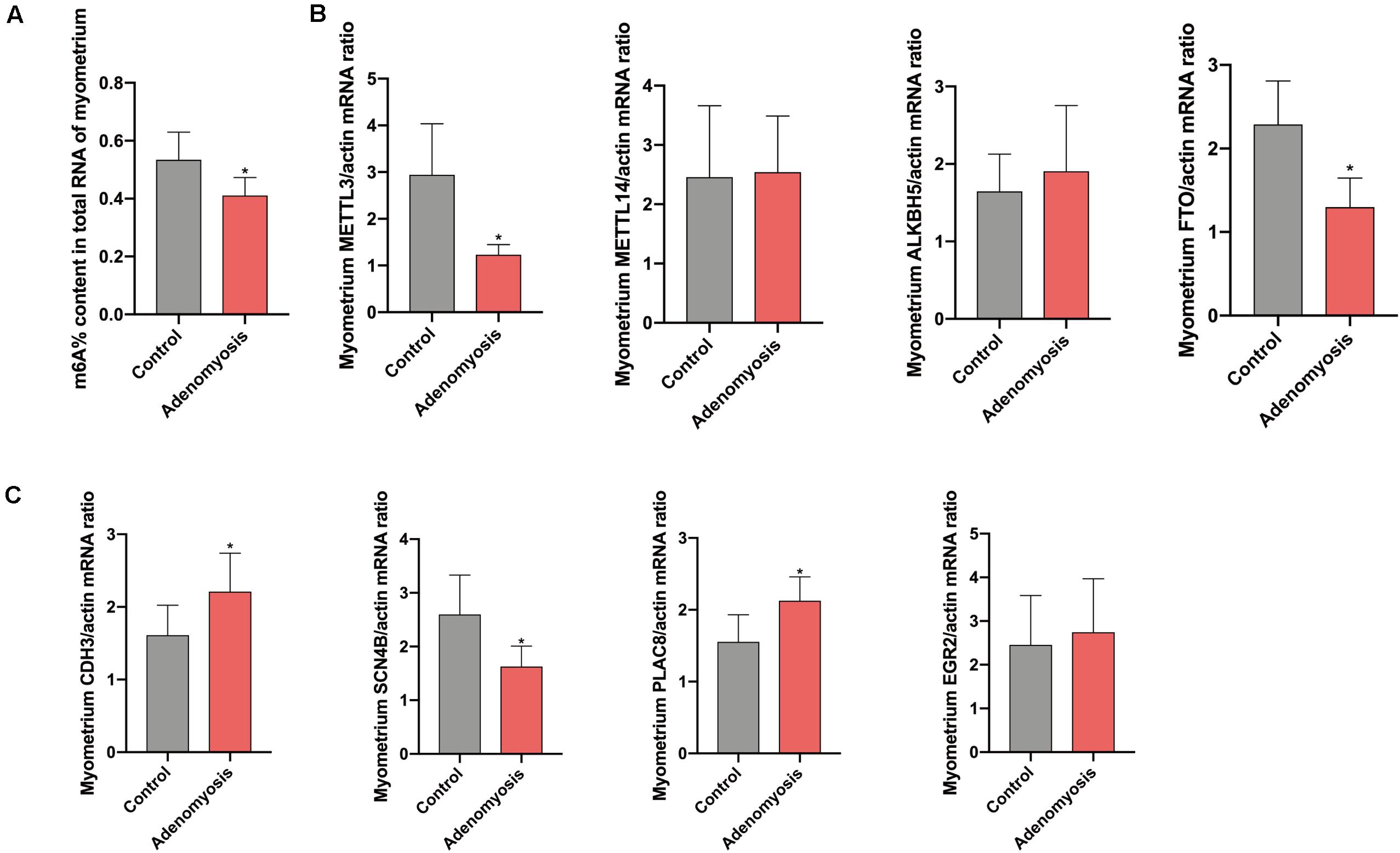
Figure 7. The total m6A level and qRT-PCR validation of bioinformation data in myometrium of women with and without adenomyosis. (A) m6A% content in total RNA in the myometrium of adenomyosis and control patients. (B) The expression of METTL3, METTL14, ALKBH5, FTO mRNA level in the myometrium. (C) The expression of CDH3, SCN4B, PLAC8, and EGR2 mRNA level in the myometrium. n = 6 for each group. *p < 0.05.
Adenomyosis is a disease with unknown pathogenesis, and diagnostics rely on the utilization of imaging techniques based on differences in the appearance of smooth muscle, particularly the inner myometrium (Kissler et al., 2008). One hypothesis of the pathogenesis of adenomyosis is that the adenomyosis lesions originate from invaginating endometrium basalis, due to the similarities between these tissues. Thus, the disruption of the normal boundary (inner myometrium) may result in invasion of endometrial cells into the myometrium, inducing myocyte hypertrophy (Vercellini et al., 2006). At the same time, in vitro studies have demonstrated that myocytes from adenomyosis enhance invasion of endometrial stromal cells, compared to normal myocytes. Moreover, some studies have reported misexpression of estrogen receptor (ER) and progesterone receptor (PR) in the inner myometrium, including increased ER-β and decreased PR-A and PR-B (Mehasseb et al., 2011b). While myometrial dysfunction in the pathogenesis of adenomyosis has been proposed (Mehasseb et al., 2010b), other studies demonstrate that disruption of the myometrium does not necessarily result in adenomyosis (Mehasseb et al., 2010a). With regard to the endometrium, there is increased invasiveness of the E-cadherin negative epithelial cells (Gaetje et al., 1997; Benagiano and Brosens, 2012; Brosens et al., 2012) and abnormal estrogen. Thus, adenomyosis appears to be a disease of both the myometrial and endometrial compartments, although further research is needed to understand mechanisms contributing to the pathogenesis and pathophysiology of this disorder.
RNA methylation, especially m6A, contributes to biological processes such as cell proliferation, immunology (Winkler et al., 2019; Zhang et al., 2019), tumorigenesis (Deng et al., 2018) and tissue development (Klungland and Dahl, 2014; Wu et al., 2018; Heck and Wilusz, 2019), all of which may be involved in the pathogenesis of adenomyosis. Moreover, reduced METTL3 and m6A level have been detected in endometrial cancer, which shares some characteristics with adenomyosis.
METTL3 expression was significantly decreased in the endometrium of adenomyosis patients with reduced total m6A levels, similar to those in endometrial cancer (Liu et al., 2018). As a result, reduced m6A may contribute to the endometrial dysfunction in the setting of adenomyosis via different functional pathways. We found that DEGs were involved in epithelial cell proliferation, vasculature development, cell migration and macrophage migration, processes consistent with recent RNA-seq data of the endometrium in women with adenomyosis (Xiang et al., 2019). Moreover, Benagiano and Brosens have also suggested that some of enriched biological processes (e.g., increase angiogenesis, proliferation of endometrium and Wnt signaling pathway) are involved in the pathogenesis of adenomyosis (Benagiano and Brosens, 2012). We further investigated the correlation and co-expression among the DEGs in these biological processes and METTL3, the “hub” gene of m6A RNA methylation regulators in the endometrium, suggesting the possible target genes of METTL3 and m6A regulation.
IGF1 is an effective growth factor in disease progression and plays an important role in cellular growth, proliferation, invasion, and angiogenesis of several tissues. Our study revealed that IGF1 was differentially expressed in the microarray analysis and was significantly correlated with expression of METTL3 in the endometrium of adenomyosis patients. Previous study demonstrated that the decreased METTL3 and m6A RNA methylation level can promote the cell proliferation through the AKT pathway in the endometrium, while IGF1 contributes to the regulation of AKT (Liu et al., 2018). Thus, combining the bioinformatic analyses herein with data from the literature, we propose that METTL3 regulates m6A and contributes to increased expression of IGF1, which further promotes cell proliferation and invasion of endometrial cells into the myometrium via AKT pathway. However, the mechanism underlying the regulation of METTL3 and m6A to IGF1 and AKT pathway still need to be further investigated.
EMT, induced in the basal endometrium by high levels of estrogen, resulted in invagination of endometrium into myometrium, thereby playing an important role in the pathogenesis of adenomyosis (Hu et al., 2020). Herein, in our data analysis we found the decreased expression of cadherin 1(E-cadherin), a marker of the epithelial cell and EMT. Wnt pathway activation induces EMT in several tissues (Teeuwssen and Fodde, 2019); we found that WNT5A mRNA was differentially expressed in the endometrium of adenomyosis patients. WNT5A may contribute to the pathogenesis of adenomyosis through proliferation of epithelial and fibroblast cells and regulation of vasculature development. However, expression of WNT5A was not correlated to METTL3 in endometrium, suggesting it is not under the control of m6A RNA methylation regulators. Beside Wnt signaling, previous studies demonstrated that METTL3 and m6A levels contribute to EMT in lung cancer (Wanna-Udom et al., 2020). IGF1 upregulates components of the Wnt signaling pathway and promotes EMT, and it is co-expressed with METTL3 (Verras and Sun, 2005). Thus, increased IGF1 in eutopic endometrium may mediate the regulation of METTL3 and contribute to the endometrial dysfunction in adenomyosis women through EMT.
Besides of EMT, multipotent stem cells that travel from the endometrium to the myometrium and subsequently differentiate into lineage epithelial and/or stromal fibroblasts provide another possibility for adenomyosis pathogenesis. METTL3 and m6A can also contribute to the differentiation of stem cells in different tissues, indicating another possible role of m6A and METTL3 in adenomyosis (Lee et al., 2019).
DDT is the a member of the macrophage migration inhibitory factor (MIF) protein superfamily (Merk et al., 2012) and is involved in regulation of cell migration and other biological processes in various tumors. Previous studies found that DDT and MIF are involved in proliferation, migration, and invasion of cervical cancer (Wang et al., 2017). Moreover, MIF contributes to development of endometriosis, and MIF expression is increased in the endometrium of adenomyosis women (Rakhila et al., 2014). In our data analysis results, we also found MIF to be significantly differentially expressed in the endometrium of women with adenomyosis. However, its correlation to METTL3 and other m6A RNA methylation regulators was not significant. Thus, MIF may be involved in endometrial dysfunction of women with adenomyosis, but not under the regulation of m6A RNA methylation regulators. DDT is significantly co-expressed with m6A RNA methylation regulators and binds to MIF cell surface receptor, inducing similar cell signaling and effector functions. Thus, the increased DDT mRNA in the endometrium of adenomyosis patients could mediate regulation of m6A RNA methylation regulators relevant to cell migration of endometrium in the setting of adenomyosis.
Our study also revealed decreased m6A content and 11 differentially expressed m6A RNA methylation regulators in myometrium of adenomyosis patients, providing a possible mechanism for myometrial dysfunction of adenomyosis. Moreover, downstream factors of m6A regulation may participate.
While immune activation is mainly observed in endometrium of women with adenomyosis, e.g., increased endometrial macrophages and autoantibodies such as anti-phosphatidylinositol IgG, anti-phosphatidylglycerol IgG (Ota et al., 1998), immune dysregulation may also occur in the myometrium of affected women. m6A has been shown to be involved in immune regulation (Wang et al., 2019), and our functional enrichment analysis of DEGs in myometrium herein highlights neutrophil activation in cases versus controls. Additionally, PLAC8, a multi-faceted protein involved in various cellular physical processes (such as the regulation of immunity, cell differentiation and apoptosis) (Li et al., 2014), was also increased in the myometrium in adenomyosis patients and was closely related to m6A RNA methylation regulator “cluster.” Thus, the myometrium has processes in place for immune cell response in the setting of adenomyosis, associated with m6A RNA methylation regulators.
SCN4B is the β-subunit of voltage-gated sodium channels (VGSCs), required for generation of action potentials in excitable cells, and it also functions in cell–cell adhesion (Shimizu et al., 2017). Expression of SCN4B has been detected in the longitudinal smooth muscle layer of rat myometrium (Seda et al., 2007), suggesting its role in myometrial homeostasis and contractility. Functional enrichment analysis of myometrial DEGs in our study identified SCN4B associated with “muscle contraction” and was also significantly correlated with expression of the m6A regulator “cluster.” Moreover, expression of SCN4B mRNA was decreased in myometrium of adenomyosis patients, as were METTL3 and FTO. Thus, m6A RNA methylation regulators may regulate cell adhesion through SCN4B, further contributing to adenomyosis development.
In the myometrium, the GO enrichment of DEGs revealed that WNT5A was decreased and was involved in “the connective tissue development,” “muscle development” and other processes. However, it was also not correlated to the m6A RNA methylation regulators “cluster” herein. Thus, WNT5A may contribute to the pathogenesis of adenomyosis but is not regulated through the regulation of m6A RNA methylation regulators.
The strengths of this study include mining publicly available databases with abundant gene expression on human endometrium and myometrium separately from women with and without adenomyosis. Also, the endometrial and myometrial specimens used for validation were obtained using standard operating procedures (SOPs) from the UCSF NIH Human Endometrial Tissue and DNA Bank with well annotated clinical data in our RedCap Database (Sheldon et al., 20113). Moreover, our study is the first to propose possible involvement of RNA methylation in the pathogenesis of adenomyosis. It thus provides a novel paradigm needing subsequent mechanistic validation. It also potentially opens new avenues for novel targeted therapeutic approaches for symptoms associated with this disorder.
The limitations of our study were that the roles of the m6A RNA methylation regulators and their downstream factors in adenomyosis was deduced from gene expression profile analysis. Mechanisms underlying a role for m6A RNA methylation regulation still needs to be demonstrated through animal and in vitro experiments. In addition, although we did the validation using clinical samples, the sample size was limited, and the protein levels of target genes still needed to be detected. Other limitations include: the type of adenomyosis lesions (diffuse, adenomyoma, or cystic), clinical data, and cycle phase of subjects of the myometrium samples used in the data reported in the GEO database are not known. Notably, our validation approach used myometrium solely from the proliferative phase of the cycle, and previous studies have demonstrated expression of some genes in myometrium in adenomyosis patients do not vary with cycle phase (Mehasseb et al., 2011a), giving some mitigation to this limitation.
Herein, we investigated gene expression and the interactome of m6A RNA methylation regulators and total m6A levels in adenomyosis patients. Decreased METTL3 and total m6A levels in endometrium of adenomyosis patients may contribute to cell proliferation and invasion through IGF1 and DDT. The RNA methylation levels of specific and target genes such as IGF1, DDT, PLAC8, and SCN4B remain to be investigated using methods such as methylated RNA immunoprecipitation sequencing (MeRIP-seq) and MeRIP-qPCR. Furthermore, in the myometrium, m6A RNA methylation regulators work as a cluster and play roles in cell adhesion, muscle contraction and immune response. In conclusion, m6A RNA methylation regulators may be involved the pathogenesis of adenomyosis through aberrant expression and actions in both the uterine endometrium and myometrium.
The two public gene expression datasets GSE78851 and GSE7303 can be downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). All other data presented in this study are included in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the Committee on Human Research (CHR) at UCSF. The patients/participants provided their written informed consent to participate in this study.
JZ, SL, JO-A, and LG contributed to the design of the experiments, collection of samples, acquisition of data, analysis and interpretation of data. JZ, SL, YD, and Z-JC analyzed the data and made the figures. JZ, SS, and LG finished drafting the manuscript or revising it critically for important intellectual content. LG is responsible for the final approval of the version to be published. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was supported by the NIH Eunice Kennedy Shriver National Institute for Child Health and Human Development P50 HD055764-12, National Centers for Translational Research in Reproduction and Infertility Program (LG), the Kerfuffle Foundation (LG), and the National Natural Science Foundation (81901549).
We gratefully acknowledge all research volunteers for participating in the study. We also acknowledge to the UCSF NIH Human Endometrial Tissue and DNA Bank and P50 NIH grant Project 4.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00716/full#supplementary-material
Arnold, L. L., Meck, J. M., and Simon, J. A. (1995). Adenomyosis: evidence for genetic cause. Am. J. Med. Genet. 55, 505–506. doi: 10.1002/ajmg.1320550423
Benagiano, G., and Brosens, I. (2012). The endometrium in adenomyosis. Womens Health 8, 301–312. doi: 10.2217/whe.12.8
Benagiano, G., Habiba, M., and Brosens, I. (2012). The pathophysiology of uterine adenomyosis: an update. Fertil. Steril. 98, 572–579. doi: 10.1016/j.fertnstert.2012.06.044
Bird, C. C., Mcelin, T. W., and Manalo-Estrella, P. (1972). The elusive adenomyosis of the uterus–revisited. Am. J. Obstet. Gynecol. 112, 583–593. doi: 10.1016/0002-9378(72)90781-8
Brosens, I., Brosens, J. J., and Benagiano, G. (2012). The eutopic endometrium in endometriosis: are the changes of clinical significance? Reprod. Biomed. Online 24, 496–502. doi: 10.1016/j.rbmo.2012.01.022
Curtis, K. M., Hillis, S. D., Marchbanks, P. A., and Peterson, H. B. (2002). Disruption of the endometrial-myometrial border during pregnancy as a risk factor for adenomyosis. Am. J. Obstet. Gynecol. 187, 543–544. doi: 10.1067/mob.2002.124285
Deng, X., Su, R., Feng, X., Wei, M., and Chen, J. (2018). Role of N(6)-methyladenosine modification in cancer. Curr. Opin. Genet. Dev. 48, 1–7. doi: 10.1016/j.gde.2017.10.005
Gaetje, R., Kotzian, S., Herrmann, G., Baumann, R., and Starzinski-Powitz, A. (1997). Nonmalignant epithelial cells, potentially invasive in human endometriosis, lack the tumor suppressor molecule E-cadherin. Am. J. Pathol. 150, 461–467.
Gargett, C. E., Schwab, K. E., and Deane, J. A. (2016). Endometrial stem/progenitor cells: the first 10 years. Hum. Reprod. Update 22, 137–163.
Gu, S., Sun, D., Dai, H., and Zhang, Z. (2018). N(6)-methyladenosine mediates the cellular proliferation and apoptosis via microRNAs in arsenite-transformed cells. Toxicol. Lett. 292, 1–11. doi: 10.1016/j.toxlet.2018.04.018
Heck, A. M., and Wilusz, C. J. (2019). Small changes, big implications: the impact of m(6)A RNA methylation on gene expression in pluripotency and development. Biochim. Biophys. Acta Gene Regul. Mech. 1862:194402. doi: 10.1016/j.bbagrm.2019.07.003
Herndon, C. N., Aghajanova, L., Balayan, S., Erikson, D., Barragan, F., Goldfien, G., et al. (2016). Global transcriptome abnormalities of the eutopic endometrium from women with adenomyosis. Reprod. Sci. 23, 1289–1303. doi: 10.1177/1933719116650758
Hu, R., Peng, G. Q., Ban, D. Y., Zhang, C., Zhang, X. Q., and Li, Y. P. (2020). High-expression of neuropilin 1 correlates to estrogen-induced epithelial-mesenchymal transition of endometrial cells in adenomyosis. Reprod. Sci. 27, 395–403. doi: 10.1007/s43032-019-00035-2
Inoue, S., Hirota, Y., Ueno, T., Fukui, Y., Yoshida, E., Hayashi, T., et al. (2019). Uterine adenomyosis is an oligoclonal disorder associated with KRAS mutations. Nat. Commun. 10:5785.
Johnson, R. M., Kerr, M. S., and Slaven, J. E. (2012). Plac8-dependent and inducible NO synthase-dependent mechanisms clear Chlamydia muridarum infections from the genital tract. J. Immunol. 188, 1896–1904. doi: 10.4049/jimmunol.1102764
Kissler, S., Zangos, S., Kohl, J., Wiegratz, I., Rody, A., Gatje, R., et al. (2008). Duration of dysmenorrhoea and extent of adenomyosis visualised by magnetic resonance imaging. Eur. J. Obstet. Gynecol. Reprod. Biol. 137, 204–209. doi: 10.1016/j.ejogrb.2007.01.015
Klungland, A., and Dahl, J. A. (2014). Dynamic RNA modifications in disease. Curr. Opin. Genet. Dev. 26, 47–52. doi: 10.1016/j.gde.2014.05.006
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Lee, H., Bao, S., Qian, Y., Geula, S., Leslie, J., Zhang, C., et al. (2019). Stage-specific requirement for Mettl3-dependent m(6)A mRNA methylation during haematopoietic stem cell differentiation. Nat. Cell. Biol. 21, 700–709. doi: 10.1038/s41556-019-0318-1
Li, C., Ma, H., Wang, Y., Cao, Z., Graves-Deal, R., Powell, A. E., et al. (2014). Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon cancer. J. Clin. Invest. 124, 2172–2187. doi: 10.1172/jci71103
Li, J., Chen, F., Peng, Y., Lv, Z., Lin, X., Chen, Z., et al. (2020). N6-methyladenosine regulates the expression and secretion of tgfbeta1 to affect the epithelial-mesenchymal transition of cancer cells. Cells 9:296. doi: 10.3390/cells9020296
Li, J., Yanyan, M., Mu, L., Chen, X., and Zheng, W. (2019). The expression of Bcl-2 in adenomyosis and its effect on proliferation, migration, and apoptosis of endometrial stromal cells. Pathol. Res. Pract. 215:152477. doi: 10.1016/j.prp.2019.152477
Li, J. J., Chung, J. P. W., Wang, S., Li, T. C., and Duan, H. (2018). The investigation and management of adenomyosis in women who wish to improve or preserve fertility. Biomed. Res. Int. 2018:6832685.
Liao, S., Sun, H., and Xu, C. (2018). YTH domain: a family of N(6)-methyladenosine (m(6)A) readers. Genomics Proteomics Bioinformatics 16, 99–107. doi: 10.1016/j.gpb.2018.04.002
Liu, J., Eckert, M. A., Harada, B. T., Liu, S. M., Lu, Z., Yu, K., et al. (2018). m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell. Biol. 20, 1074–1083. doi: 10.1038/s41556-018-0174-4
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Mehasseb, M. K., Bell, S. C., Brown, L., Pringle, J. H., and Habiba, M. (2011a). Phenotypic characterisation of the inner and outer myometrium in normal and adenomyotic uteri. Gynecol. Obstet. Invest. 71, 217–224. doi: 10.1159/000318205
Mehasseb, M. K., Bell, S. C., and Habiba, M. A. (2010a). Neonatal administration of tamoxifen causes disruption of myometrial development but not adenomyosis in the C57/BL6J mouse. Reproduction 139, 1067–1075. doi: 10.1530/rep-09-0443
Mehasseb, M. K., Panchal, R., Taylor, A. H., Brown, L., Bell, S. C., and Habiba, M. (2011b). Estrogen and progesterone receptor isoform distribution through the menstrual cycle in uteri with and without adenomyosis. Fertil. Steril. 95, 2228.e1–2235.e1.
Mehasseb, M. K., Taylor, A. H., Pringle, J. H., Bell, S. C., and Habiba, M. (2010b). Enhanced invasion of stromal cells from adenomyosis in a three-dimensional coculture model is augmented by the presence of myocytes from affected uteri. Fertil. Steril. 94, 2547–2551. doi: 10.1016/j.fertnstert.2010.04.016
Merk, M., Mitchell, R. A., Endres, S., and Bucala, R. (2012). D-dopachrome tautomerase (D-DT or MIF-2): doubling the MIF cytokine family. Cytokine 59, 10–17. doi: 10.1016/j.cyto.2012.03.014
Merritt, M. A., Strickler, H. D., Einstein, M. H., Yang, H. P., Sherman, M. E., Wentzensen, N., et al. (2016). Insulin/IGF and sex hormone axes in human endometrium and associations with endometrial cancer risk factors. Cancer Causes Control 27, 737–748. doi: 10.1007/s10552-016-0751-4
Noyes, R. W., Hertig, A. T., and Rock, J. (1950). Dating the endometrial biopsy. Fertil. Steril. 1, 3–25.
Ota, H., Igarashi, S., Hatazawa, J., and Tanaka, T. (1998). Is adenomyosis an immune disease? Hum. Reprod. Update 4, 360–367. doi: 10.1093/humupd/4.4.360
Peer, E., Rechavi, G., and Dominissini, D. (2017). Epitranscriptomics: regulation of mRNA metabolism through modifications. Curr. Opin. Chem. Biol. 41, 93–98. doi: 10.1016/j.cbpa.2017.10.008
Rakhila, H., Girard, K., Leboeuf, M., Lemyre, M., and Akoum, A. (2014). Macrophage migration inhibitory factor is involved in ectopic endometrial tissue growth and peritoneal-endometrial tissue interaction in vivo: a plausible link to endometriosis development. PLoS One 9:e110434. doi: 10.1371/journal.pone.0110434
Reinhold, C., Tafazoli, F., and Wang, L. (1998). Imaging features of adenomyosis. Hum. Reprod. Update 4, 337–349. doi: 10.1093/humupd/4.4.337
Roddy, E., and Chapman, J. (2017). Genomic insights in gynecologic cancer. Curr. Probl. Cancer 41, 8–36. doi: 10.1016/j.currproblcancer.2016.11.001
Roundtree, I. A., and He, C. (2016). Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Trends Genet. 32, 320–321. doi: 10.1016/j.tig.2016.03.006
Seda, M., Pinto, F. M., Wray, S., Cintado, C. G., Noheda, P., Buschmann, H., et al. (2007). Functional and molecular characterization of voltage-gated sodium channels in uteri from nonpregnant rats. Biol. Reprod. 77, 855–863. doi: 10.1095/biolreprod.107.063016
Sheldon, E., Vo, K. C., Mcintire, R. A., Aghajanova, L., Zelenko, Z., Irwin, J. C., et al. (2011). Biobanking human endometrial tissue and blood specimens: standard operating procedure and importance to reproductive biology research and diagnostic development. Fertil. Steril. 95, 2120–2122.
Shimizu, H., Tosaki, A., Ohsawa, N., Ishizuka-Katsura, Y., Shoji, S., Miyazaki, H., et al. (2017). Parallel homodimer structures of the extracellular domains of the voltage-gated sodium channel beta4 subunit explain its role in cell-cell adhesion. J. Biol. Chem. 292, 13428–13440. doi: 10.1074/jbc.m117.786509
Sousa, B., Pereira, J., and Paredes, J. (2019). The crosstalk between cell adhesion and cancer metabolism. Int. J. Mol. Sci. 20:1933. doi: 10.3390/ijms20081933
Stitt, T. N., Drujan, D., Clarke, B. A., Panaro, F., Timofeyva, Y., Kline, W. O., et al. (2004). The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell. 14, 395–403. doi: 10.1016/s1097-2765(04)00211-4
Teeuwssen, M., and Fodde, R. (2019). Wnt signaling in ovarian cancer stemness, EMT, and therapy resistance. J. Clin. Med. 8:1658. doi: 10.3390/jcm8101658
Vercellini, P., Vigano, P., Somigliana, E., Daguati, R., Abbiati, A., and Fedele, L. (2006). Adenomyosis: epidemiological factors. Best Pract. Res. Clin. Obstet. Gynaecol. 20, 465–477. doi: 10.1016/j.bpobgyn.2006.01.017
Verras, M., and Sun, Z. (2005). Beta-catenin is involved in insulin-like growth factor 1-mediated transactivation of the androgen receptor. Mol. Endocrinol. 19, 391–398. doi: 10.1210/me.2004-0208
Wang, H., Hu, X., Huang, M., Liu, J., Gu, Y., Ma, L., et al. (2019). Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat. Commun. 10:1898.
Wang, Q., Wei, Y., and Zhang, J. (2017). Combined knockdown of D-dopachrome tautomerase and migration inhibitory factor inhibits the proliferation, migration, and invasion in human cervical cancer. Int. J. Gynecol. Cancer 27, 634–642. doi: 10.1097/igc.0000000000000951
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wanna-Udom, S., Terashima, M., Lyu, H., Ishimura, A., Takino, T., Sakari, M., et al. (2020). The m6A methyltransferase METTL3 contributes to Transforming Growth Factor-beta-induced epithelial-mesenchymal transition of lung cancer cells through the regulation of JUNB. Biochem. Biophys. Res. Commun. 524, 150–155. doi: 10.1016/j.bbrc.2020.01.042
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)a methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 69, 1028.e6–1038.e6.
Winkler, R., Gillis, E., Lasman, L., Safra, M., Geula, S., Soyris, C., et al. (2019). m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20, 173–182. doi: 10.1038/s41590-018-0275-z
Wu, J., Frazier, K., Zhang, J., Gan, Z., Wang, T., and Zhong, X. (2020). Emerging role of m(6) A RNA methylation in nutritional physiology and metabolism. Obes. Rev. 21:e12942.
Wu, R., Liu, Y., Yao, Y., Zhao, Y., Bi, Z., Jiang, Q., et al. (2018). FTO regulates adipogenesis by controlling cell cycle progression via m(6)A-YTHDF2 dependent mechanism. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1863, 1323–1330. doi: 10.1016/j.bbalip.2018.08.008
Xiang, Y., Sun, Y., Yang, B., Yang, Y., Zhang, Y., Yu, T., et al. (2019). Transcriptome sequencing of adenomyosis eutopic endometrium: a new insight into its pathophysiology. J. Cell. Mol. Med. 23, 8381–8391. doi: 10.1111/jcmm.14718
Yeh, C. C., Su, F. H., Tzeng, C. R., Muo, C. H., and Wang, W. C. (2018). Women with adenomyosis are at higher risks of endometrial and thyroid cancers: a population-based historical cohort study. PLoS One 13:e0194011. doi: 10.1371/journal.pone.0194011
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4:10.
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell. Biol. 20, 608–624. doi: 10.1038/s41580-019-0168-5
Keywords: adenomyosis, m6A, METTL3, endometrium, myometrium, in silico
Citation: Zhai J, Li S, Sen S, Opoku-Anane J, Du Y, Chen Z-J and Giudice LC (2020) m6A RNA Methylation Regulators Contribute to Eutopic Endometrium and Myometrium Dysfunction in Adenomyosis. Front. Genet. 11:716. doi: 10.3389/fgene.2020.00716
Received: 25 March 2020; Accepted: 12 June 2020;
Published: 03 July 2020.
Edited by:
Graziano Pesole, National Research Council, ItalyReviewed by:
Ernesto Picardi, University of Bari Aldo Moro, ItalyCopyright © 2020 Zhai, Li, Sen, Opoku-Anane, Du, Chen and Giudice. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linda C. Giudice, bGluZGEuZ2l1ZGljZUB1Y3NmLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.