- 1College of Plant Protection, Hunan Agricultural University, Changsha, China
- 2Hunan Agricultural Biotechnology Research Institute, Changsha, China
- 3Hunan Academy of Agricultural Sciences (CAAS), Changsha, China
The strain B-4, isolated from a field in Changsha (China), presents strong antifungal activities, as identified by the Kirby–Bauer test, especially for pathogens that harm crops. Here, we obtained the complete genome sequence of the strain B-4 by Pacific Biosciences single-molecule real-time sequencing, making it well analyzed for understanding mechanisms and creating biological agents. Its 3,919-kb circular chromosome genome has 3,725 protein-coding genes [coding sequences (CDSs)] and 46.7% guanine–cytosine content. A comparative genome analysis of B-4 with other published strains (including Bacillus velezensis, Bacillus amyloliquefaciens, and Bacillus subtilis) revealed that the strain B-4 is a B. velezensis strain. These different strains have 2,889 CDSs in common, whereas 179 CDSs were found to be unique in the strain B-4, which is a far greater number than that in other strains. Regarding the antifungal activities of B-4, we were specifically concerned with the genes involved in the biosynthesis of secondary metabolites. In total, more than 19.56% of the genome was annotated to 12 gene clusters relating to synthesis of antimicrobial metabolites, which contained various enzyme-encoding operons for non-ribosomal peptide synthetases, polyketide synthases, and lantipeptide synthesis proteins. They were all considered to be related to the production of bacteriostatic substances or stimulation of induced systemic resistance by bacterial metabolites. These situations also present an advantage over those of other strains for biocontrol potential. We provide evidence that the biological control effect of the strain B-4, as demonstrated in antibacterial activity experiments and predicted from the complete genome sequence analysis, provides the basis for research promoting agricultural research on sustainable development, especially the contribution of biotechnology to agriculture.
Introduction
Control of crop diseases has been an important process in agriculture production. Chemical pesticides have been used to control crop disease in popular and efficient ways, but to achieve highly effective control efficiency, pesticides have always been overdosed by practitioners who work in agriculture, which has had a detrimental effect on the environment or has caused crops to become resistant (Chang, 2018). With increasing requirements for the quality of food safety, the protection of crops cannot depend on chemical pesticides (Syed Ab Rahman et al., 2018). For many years, antagonistic bacteria have been used to control crop diseases caused by fungi, bacteria, viruses, or nematodes. This method can be classified as biological control and has received unusual attention because of its safety and environmentally friendly impact on crops (Fernando et al., 2007; Beneduzi et al., 2012).
Bacillus velezensis is a species that differs from Bacillus amyloliquefaciens but was previously reported as a later heterotypic synonym. The use of B. velezensis in biological control research has become popular. Moreover, Bacillus sp. are considered to have extensive bacteriostatic properties; this species has been reported to produce a variety of lipoprotein antibacterial substances, such as fengycin (Ongena et al., 2007). Using confocal scanning laser microscopy to analyze Arabidopsis root surfaces treated with Bacillus subtilis 6051, a stable, extensive biofilm and secreted surfactin were noted to combat exogenous pathogens (Bais et al., 2004). The reason that Bacillus sp. can produce antibiotics via a secondary metabolic activity is related to genes and proteins. B. velezensis FZB42 is an environmental bacterium that suppresses competitive harmful organisms. These activities were assigned to complete polyketide synthase (PKS) gene clusters, and it was founded that the pks1 (bae) and pks3 (dif) gene clusters encode proteins necessary for the biosynthesis of the polyene antibiotics (Chen et al., 2006). Molecular biological techniques should be useful for understanding the mechanism of bacterial resistance to diseases at the gene level. A recent example, B. velezensis M75, was isolated from cotton waste; the authors used the complete genome sequence and found that the genome contained operons that are related to the biosynthesis of secondary metabolites (Kim et al., 2017). In conclusion, ample gene coding is related to the mechanism of an antimicrobial activity, and complete-genome sequencing technology could help to better study biological control. Although many research results have suggested that bacteria play a major role in biological control, which could inhibit the growth of other strains, the results concerning the mechanism of action and interpretation at the genetic level remain inconclusive, and the complete genomes of only a limited number of antagonistic bacteria have been determined.
In this study, B. velezensis B-4 was isolated from a field in Changsha, China. We identified and reported a B. velezensis B-4 antibacterial activity and complete genome sequencing results. Interestingly, the strain B-4 could be developed as a novel biocontrol agent on the basis of the actual bacteriostatic tests and gene features.
Materials and Methods
Strains, Fungi, and Culture Media
Here, the strain B-4 was newly isolated from a field, and as a bacterial strain, it was obtained with GenBank ID (CP031424.1). Sclerotinia sclerotiorum, Thanatephorus cucumeris, and Fusarium graminearum were kept in the microbiology laboratory of Hunan Agricultural Biotechnology Research Institute (Changsha, China). The Luria–Bertani (LB) medium comprised 2.5 g of yeast extract, 5 g of peptone, 5 g of NaCl, and 500 ml of distilled water (pH 7.0). The center of potato dextrose agar (PDA) medium comprised potato 200 g, glucose 20 g, and agar 15 g dissolved in 1,000 ml of distilled water. All of these media were maintained for 30 min in 121°C.
Bacterial Growth Curve and the Linear Regression Analysis
The B-4 strain [2 ml, bacterial suspension recovered from 4°C, optical density (OD) 600 = 0.322] was grown in LB medium (98 ml) on a shaker incubator (200 rpm) at 28°C, using the UV spectrophotometer measured OD (λ = 600 nm) every hour. At the same time, the number of plate colonies was determined by colony counting method. There were constructed by GraphPad Prism (version 8.3.1) that concluded the strain B-4 bacterial growth curves and the linear regression between OD and number of living bacterium (Zuluaga et al., 2009).
Evaluation of the Antifungal Activities of the Strain B-4
We evaluated whether the strain B-4 has a strong antimicrobial effect for several plant pathogenic bacteria by the Kirby–Bauer (K-B) method (Gopinath et al., 2012; Gautam et al., 2013).
The K-B method was utilized to assess the activity of the strain B-4 against plant pathogenic bacteria, including but not limited to S. sclerotiorum, T. cucumeris, and F. graminearum. The B-4 strain was grown in LB medium on a shaker incubator (200 rpm) at 28°C. After 40 h, fungus was inoculated on the center of PDA plates. An 8-mm susceptibility paper disk was placed on the PDA plates for the inoculation of the strain B-4. Ten microliters of fermentation broth (72.61 × 109 CFU/ml) of the strain B-4 was then inoculated into the paper disk. After the completion of the above process, the plates were incubated at 25°C for 2–3 days, and the zone of inhibition was observed. Meanwhile, the control group, with plates that did not receive a drop of the fermentation broth of the strain B-4 and only contained the pathogenic bacteria, was incubated in the same environment as the test group. Finally, the Fisher least significant difference method was used to analyze the significance of the difference through R software (version 3.6.3).
Complete-Genome Sequencing and Assembly
The complete genome of the strain B-4 was sequenced by Pacific Biosciences (PacBio) single-molecule real-time (SMRT) sequencing technology with an approximately 15-kb SMRTbellTM template library (MajorBio Co., Shanghai, China). Sequencing raw data were analyzed after quality inspection (Watson, 2014). To create a high-quality genomic sequence, all reads were spliced into contigs and then combined into scaffolds that did not include errors and gaps.
The genome sequences were submitted to a bioinformatic analysis to further reveal biological information. Therefore, in this study, Barrnap 0.4.2 and tRNAscan-SE v1.3.1 were used to predict the rRNA and tRNA genes, respectively (Lowe and Eddy, 1997). The prediction of the strain B-4 genes was carried out using Glimmer 3.02.
Complete-Genome Annotation and Comparative Genomic Analysis
All of the genes from the strain B-4 were annotated by BLAST 2.2.28 + program1, covering the Nr, Genes, STRING, and Gene Ontology (GO) databases (Ashburner et al., 2000; Kanehisa et al., 2016; O’Leary et al., 2016; Szklarczyk et al., 2017). In a comparative genomic analysis, firstly, all of genes were categorized into different types by the Clusters of Orthologous Groups of proteins (COGs) database, and then a private project was constructed comprising B. velezensis FZB42 (NC_009725.1), B. velezensis M75 (NZ_CP016395.1), B. amyloliquefaciens DSM7 (NC_014551.1), B. amyloliquefaciens Y2 (NC_017912.1), B. amyloliquefaciens CC178 (NC_022653.1), B. amyloliquefaciens MT45 (NZ_CP011252.1), B. subtilis 168 (NC_000964.3), and the strain B-4, with known genome sequences. To construct a phylogenetic tree by RAxML for this project, OrthoMC software was used to determine the core and specific genes of the genomes (Li et al., 2003; Stamatakis, 2006). In the following step, a comparative circular genome map was created using BRIG (BLAST Ring Image Generator, 0.95-dev.0004, Alikhan et al., 2011), and Mauve (version 2.4.0, Darling et al., 2004) was used to evaluate synteny. We obtained gene clusters for secondary metabolite biosynthesis using the antiSMASH3.0 program and using Blast with other strains (Weber et al., 2015).
Results
Bacterial Growth Pattern of the Strain B-4
Through a curve fitting analysis, we constructed bacterial growth curve (Figure 1A), which confirmed the exponential phase of the strain B-4 from 4 to 15 h. During this phase, we analyzed the linear regression equation and correlation coefficient between the OD of fermentation broth and the number of living bacterium (Figure 1B).
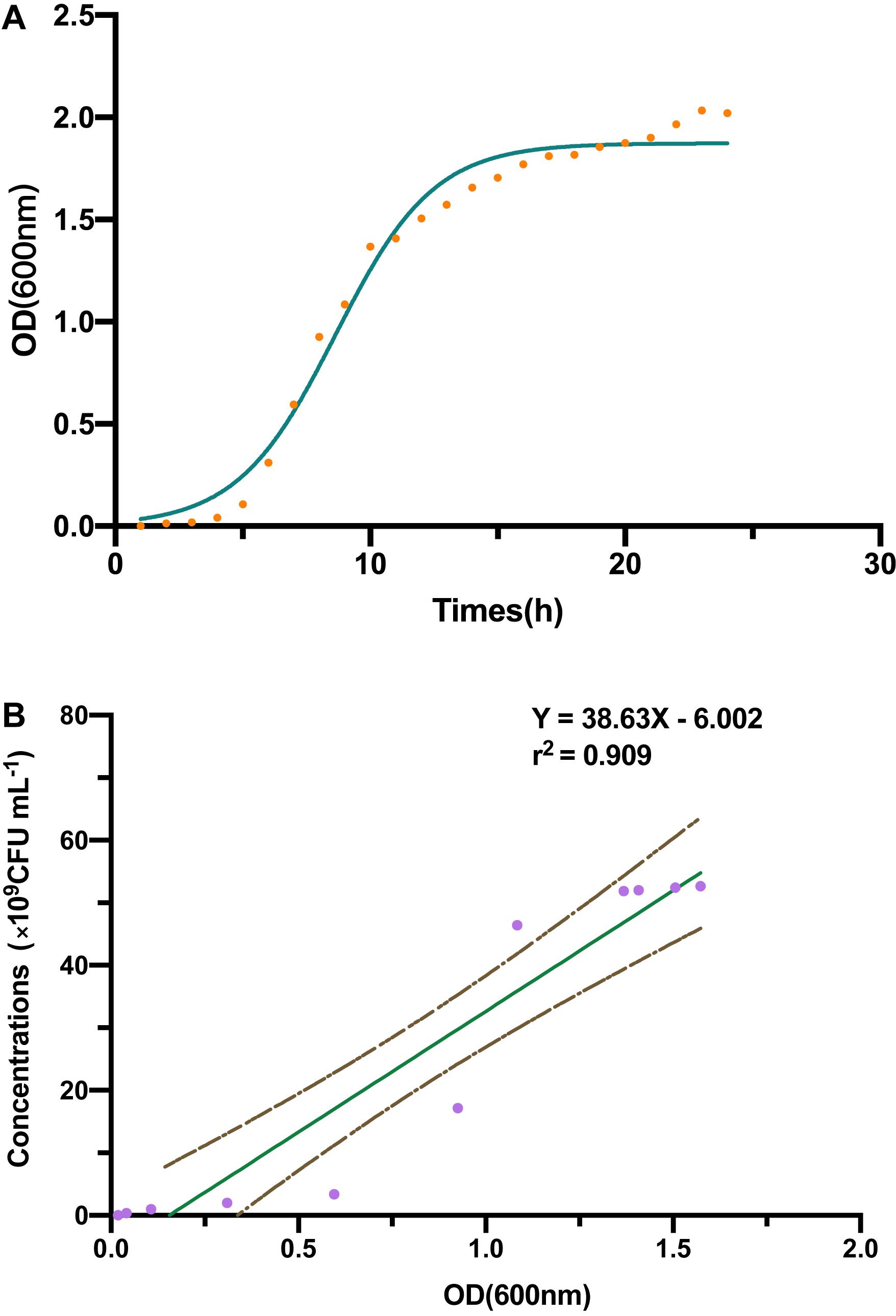
Figure 1. (A) The strain B-4 growth curve. (B) The linear regression between optical density (OD) and number of living bacterium.
Antifungal Activities of the Strain B-4
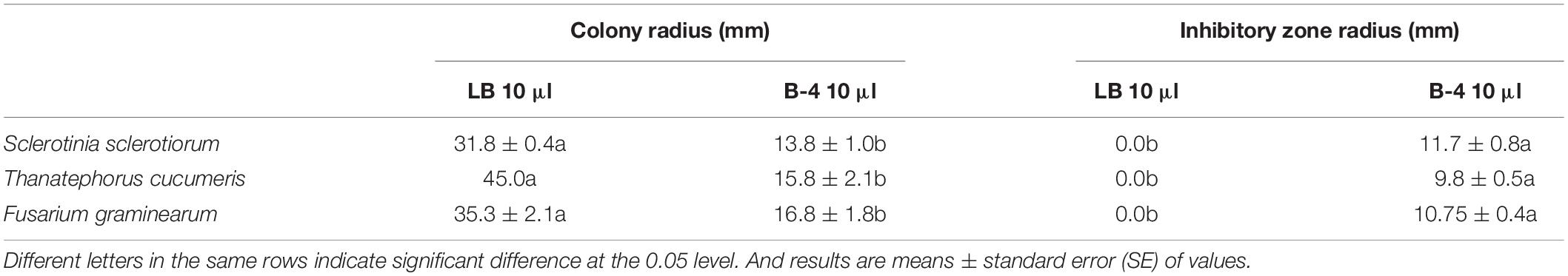
The strain B-4 showed excellent antifungal activities against pathogenic fungus. We measured the linear distance from the center of the susceptibility paper to the edge of the pathogen as the bacteriostatic radius and measured colony growth radius of the fungus. The mean inhibitory zone of the four replicates was measured, and the average amount was calculated. From Table 1, compared with the control, the strain B-4 had a significant antifungal activity. In the experimental group, the strain B-4 formed a very clear blank growth area between the disk with the bacterial suspension and the pathogenic bacteria (Figure 2). As expected, the strain B-4, which acts mainly against pathogenic bacteria, had a growth advantage in a flat confrontation test.
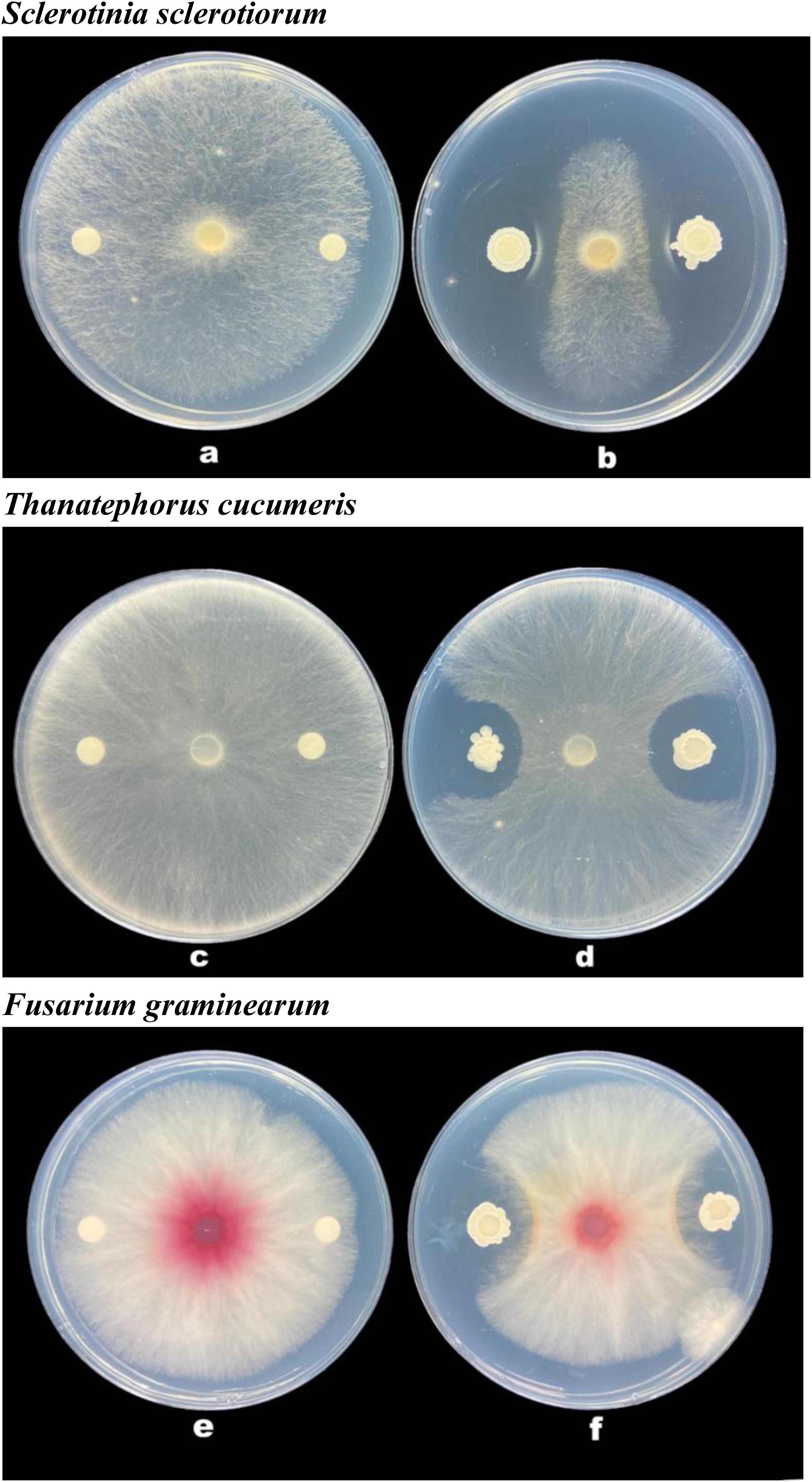
Figure 2. Antifungal activity of Bacillus velezensis B-4 against fungus. (a,c,e) Disk paper with Luria–Bertani (LB) medium added. (b,d,f) Disk paper with the strain B-4 fermentation broth.
Genome Sequencing and Phylogenomic Analysis of the Strain B-4
A total of 99,637 reads were sequenced, and the average length of reads was 5,904 bp. These reads were assembled to a circular chromosome of the strain B-4 genome with a size of 3,919,798 bp and a guanine–cytosine (GC) content of 56.67% (Table 2). The chromosome contains 4,127 predicted genes [4,014 coding sequences (CDSs), 27 rRNAs, and 86 tRNAs]. In addition, different functions of genes were classified based on a COG analysis, which revealed a large proportion of “metabolism” (43.48%); this category is considered to be related to predicted proteins for a secondary metabolism activity. Among them, 1,260 proteins were classified into eight COG functions (Figure 3), and in particular, 7.78% of proteins were related to “secondary metabolites biosynthesis, transport and catabolism.” This situation was more considerable than that in other strains and was strong evidence of the antifungal properties of the strain B-4.
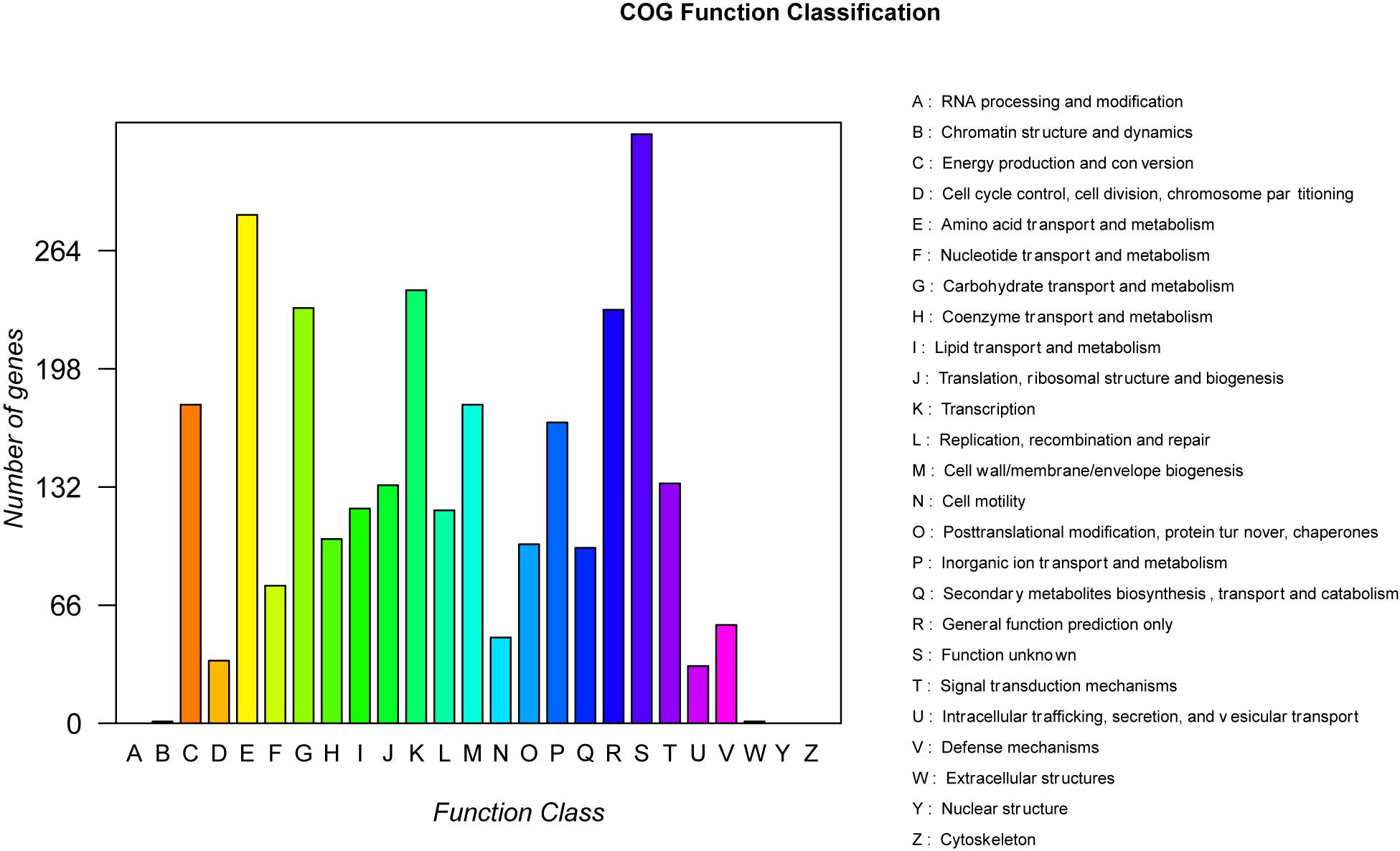
Figure 3. The statistical results of the Clusters of Orthologous Groups of proteins (COG) functional classification of genomic protein.
The genome of the strain B-4 was compared with that of other strains of the same genus (Bacillus sp.). Based on the results from OrthoMC software, the gene numbers of B-4 were found to be higher than those of the strains M75, DSM7, CC178, and MT45; and the number of core genes was 2,889, with all compared strains containing the strains M75, DSM7, CC178, MT45, Y2, and 168. Additionally, 179 CDSs of the strain B-4 were singletons. Among these genes, 33.5% were annotated to hypothetical proteins with unknown functions (Figure 4). Then, to classify the strain B-4, a phylogenomic tree of the complete genome was constructed and compared with closely related genomes. Based on the analysis results, the strain B-4 formed a close genetic relationship with the strain M75 and showed a distinct branch from the strains 168, DSM7, CC178, MT45, and Y2 (Figure 5). Therefore, the strain B-4 is a newly found antifungal strain recognized as B. velezensis.
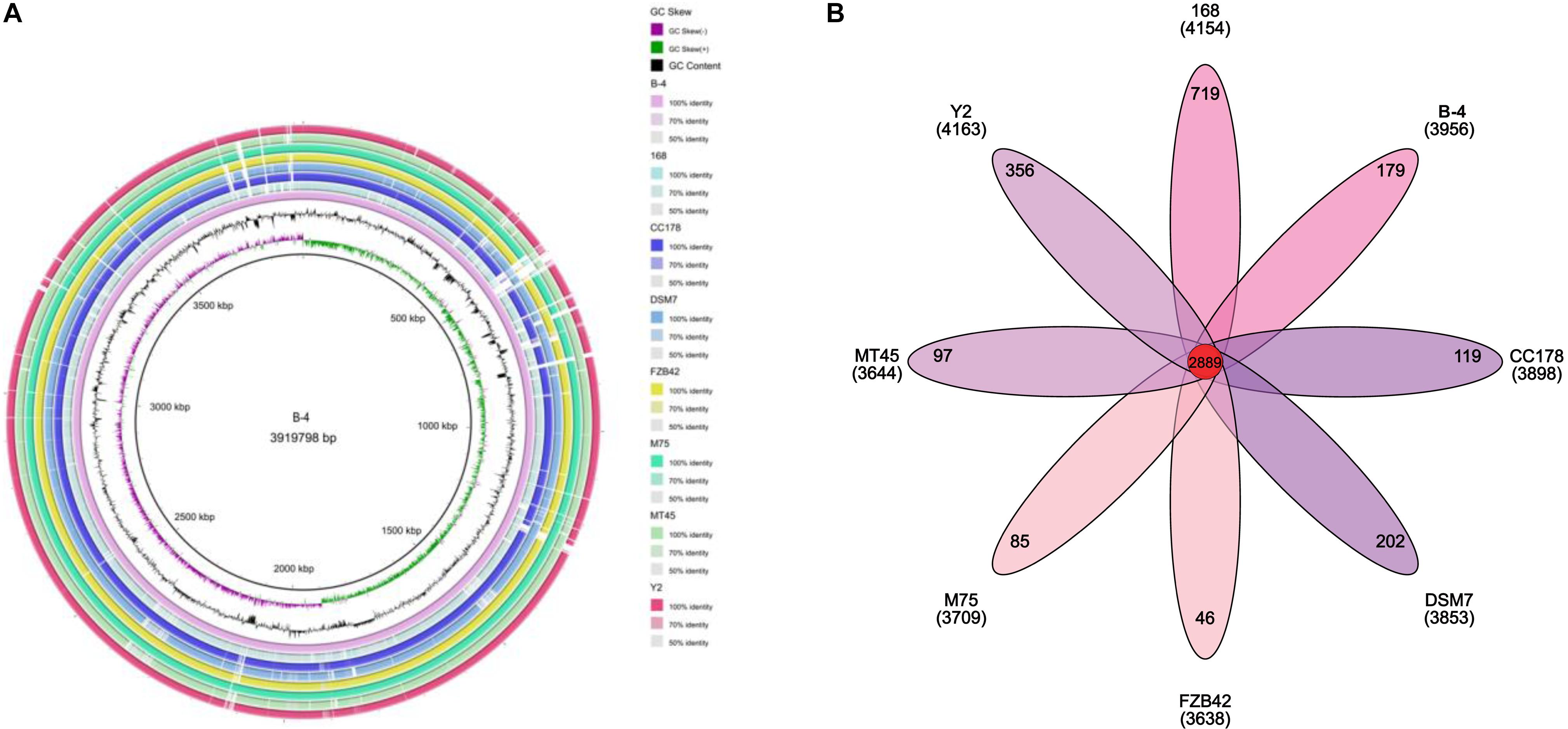
Figure 4. Comparative genome analysis. (A) Comparative circular genome map. The figure compares multiple genomes, locates genes in other genomes through known genome information according to the homology of coding order and structure, and summarizes the potential functions of each gene and the changes in the internal structure of the genome. (B) Comparative genome analysis based on protein cluster analysis shows shared and unique protein clusters.
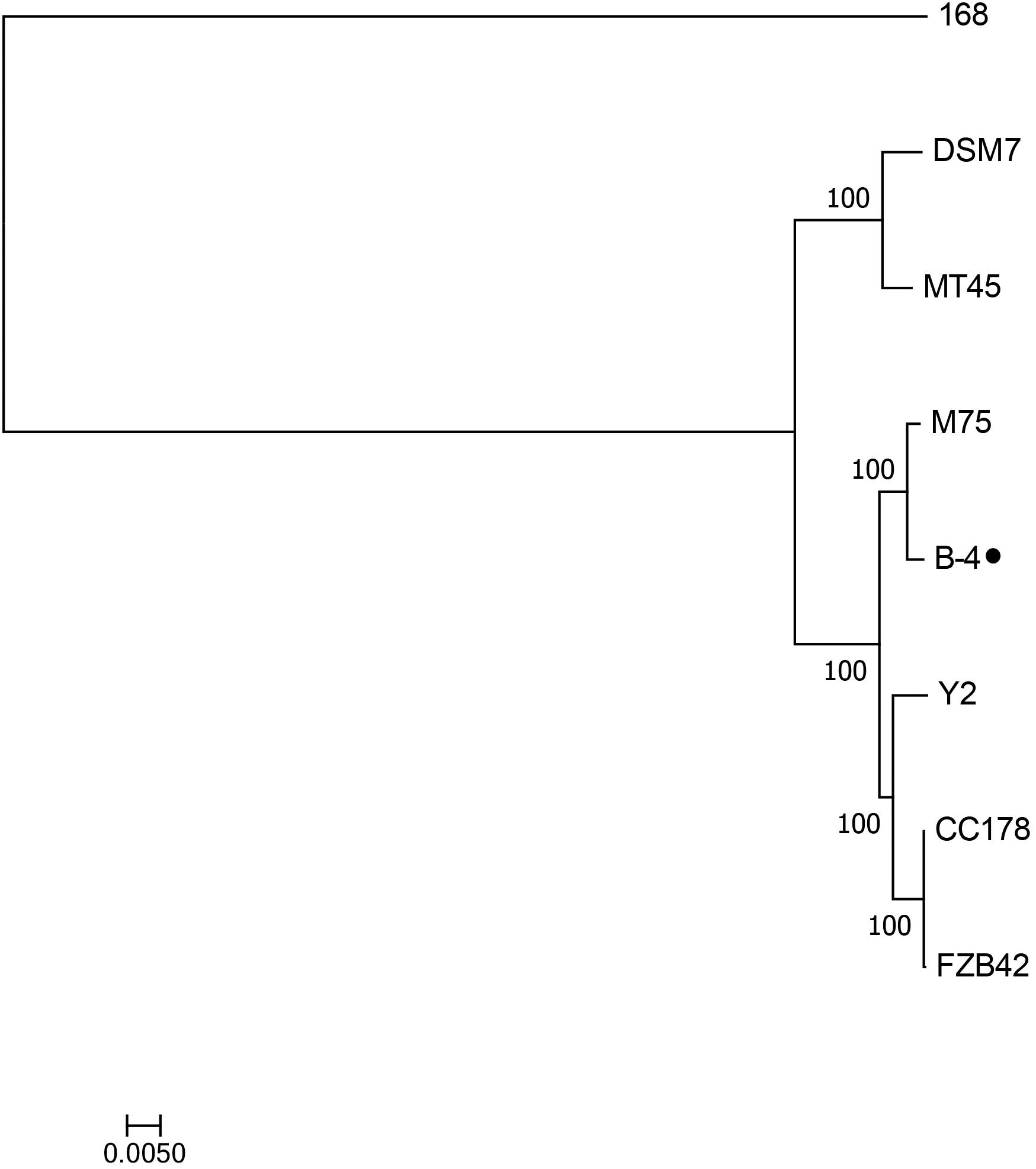
Figure 5. Phylogenetic tree based on whole genome showed the phylogenetic position of the strain B-4 (⚫) and that the strain B-4 belonged to Bacillus velezensis. And the Bacillus subtilis 168 (NC_000964.3) was used as the out group.
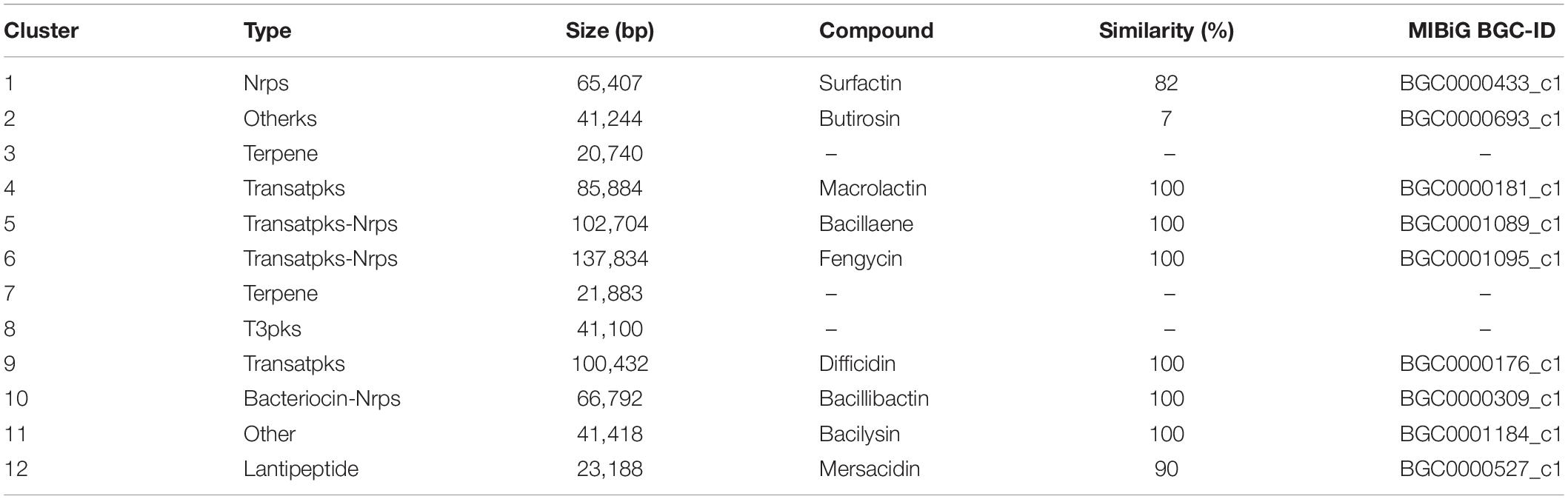
In an analysis of the gene clusters, 12 secondary metabolite gene clusters of B. velezensis B-4 were identified by the antiSMASH (version 4.0.2) database (Table 3). Those included one Nrps, one terpene, two transatpks, two transatpks-Nrps, one terpene, one T3pks, one bacteriocin-Nrps, and one lantipeptide clusters, and most of them were related to the production of non-ribosomal peptide synthetase (NRPS) and PKS systems. In comparison with other strains published, clusters 2 and 12 were unique metabolite pathways related to butirosin and mersacidin, respectively.
Discussion and Conclusion
Research on the strain of B. velezensis B-4 showed outstanding growth rate inhibition of S. sclerotiorum, which is a devastating necrotrophic plant pathogen. Simultaneously, farmers use high levels of carbendazim to control classic crop disease pathogens including S. sclerotiorum, T. cucumeris, and F. graminearum; and high resistance has been reported in China (Wang et al., 2014). High-dose applications are accompanied by excessive pesticide residues in the environment, impacting the health of humans and animals. Rape, rice, and wheat are very important crops but are affected by diseases all year round; fortunately, the effective inhibition radius of B. velezensis B-4 on S. sclerotiorum, T. cucumeris, and F. graminearum reached 11.7 ± 0.8, 9.8 ± 0.5, and 10.75 ± 0.4 mm separately, which was the same as that of Bacillus sp., which have the ability to inhibit the growth and to proliferate harmful bacteria, pathogenic bacteria, and so forth. Through bioassay experiments developed in our laboratory, B. velezensis B-4 was confirmed to be an ideal biological control agent to meet the needs of sustainable farmland development.
The complete-genome sequencing and comparative genomics analysis provided further molecular biology evidence for inhibiting pathogens with B. velezensis B-4. Microbial genome sequencing produces numerous sequences of deduced proteins, and information about these proteins can also be annotated by the COGs database, which includes a detailed prediction of the biological function(s) of representative complete genomes from all bacteria (Galperin et al., 2015). Specifically, the complete genome sequence of B. velezensis B-4 was grouped by category using the COG database. In all, 43.48% of its genome is devoted to the process of metabolism, including 7.78% participating in secondary metabolite biosynthesis, transport, and catabolism. These results further support the ideal biocontrol function of B. velezensis B-4. There are also other possible analyses. A GO database annotation makes it easier for us to understand the biological meaning behind genes (Götz et al., 2008; Pusztahelyi et al., 2015). The genome annotation of the metabolic process category had an absolute enrichment in the GO database, which may be an important factor in the inhibition of pathogens. In addition, some types of genomes with a number of beneficial traits, such as cellular process, single-organism process, and binding and catalytic activities, showed that the activity of B. velezensis B-4 was concentrated mainly in differentiation, enzyme catalysis, and metabolism. However, further investigations combining two categories of annotation methods found that a portion of the genome was not annotated. Relative to other strains, B. velezensis B-4 contains a large number of unique genes that were not found in the compared strains, which showed that B. velezensis B-4 was unique and had development potential. We suggest that these genes also have general effects on the physiological activity of the strain. For instance, 11.35% of the genome was not annotated in the COG database, which could be involved in the activity of inhibiting pathogenic bacteria. Taken together, these findings contribute to our understanding of B. velezensis B-4, which has antibacterial effects, on the basis of comparative genomics, and it contains numerous unknown genome regions that can be developed.
The comparative genomics analysis of the complete genome of B. velezensis B-4 revealed that it had excellent antibacterial effects. Moreover, recent research has shown that activities of biocontrol bacteria are closely related to secondary metabolites that have no clear function in life activities (Guo et al., 2013; Chowdhury et al., 2015). According to gene cluster research in Bacillus sp., various antifungal materials are produced by Bacillus sp. For example, a study of B. subtilis 916 demonstrated that bacillomycin Ls and fengycins produced by the strain contributed mainly to an antifungal activity (Luo et al., 2015). In addition, B. velezensis LS69 was found to have a series of gene clusters relevant to the activity against pathogenic bacteria (Liu et al., 2017, p. 916). Thus, the search for secondary metabolites in the complete genome of B. velezensis B-4 confirmed that it possessed an antagonistic activity. Here, we found four gene clusters playing a major role in non-ribosomal synthesis and three gene clusters involved in PKS, as well as a biosynthetic gene cluster for mersacidin, which is a type of lantipeptide belonging to ribosomally synthesized natural products (RNPs). In fact, secondary metabolites such as fengycins and surfactin are also the major compounds involved in the inhibitory activity. The above substances belong to lipopeptides, which constitute one of the main classes of biosurfactants. González-Jaramillo et al. (2017) evaluated the B. subtilis EA-CB0015 strain and found that iturin A and fengycin C inhibited mycelial growth and ascospore germination of Mycosphaerella fijiensis, whereas surfactin was not effective. However, we detected a great quantity of gene clusters, showing that B. velezensis B-4 had an antibacterial activity; among them, seven gene clusters were corroborated to separately produce macrolactin, bacillaene, fengycin, difficidin, bacillibactin, and bacilysin, which were 80–100% similar to those of others that are known. Four metabolites (bacilysin, bacillibactin, fengycin, and surfactin) produced via NRPS and three metabolites (bacillaene, difficidin, and macrolactin) produced via PKS were reported to be responsible for well-known antimicrobial activities. In contrast to other bacteria, such as the strain DSM7, strain FZB42, or strain M75, there was no lantipeptide-type gene cluster, which is often well known for potent antibacterial activities (Kim et al., 2017). According to alignment, the biosynthetic enzymes for the secondary metabolites bacilysin (BGC0001184_c1), bacillaene (BGC0001089_c1), bacillibactin (BGC0000309_c1), difficidin (BGC0000176_c1), fengycin (BGC0001095_c1), macrolactin (BGC0000181_c1), and mersacidin (BGC0000527_c1) were highly homologous among all the strains. The surfactin-containing gene cluster (involves four genes: srfAA, AB, AC, and ATE) had a high level of similarity to that in the others, but other components of genes showed significant differences compared with those of other strains. Interestingly, mersacidin biosynthetic gene clusters (BGC0000527_c1), which had 90% of the genes, showed similarity only in the strain B-4. In addition, mersacidin, as an aminoglycoside antibiotic metabolite, was found in the genome of the strain B-4, which showed a similarity of 7% (BGC0000693_c1). However, there were still some genetic clusters that had been detected or annotated with a function. Similarly, according to Nodwell, it is of tremendous clinical use to analyze bacterial specialized metabolites as bioactive molecules, combined with transcript elongation and other methods (Nodwell, 2017). Therefore, the secretion of secondary metabolites produced by B. velezensis B-4 against pathogenic bacteria was confirmed at the level of gene clusters and from additional information that was revealed.
Based on all the research on B. velezensis B-4, including evaluation of antifungal activities, complete-genome sequencing by Pacific Biosciences SMRT, and data analysis by comparative genomics, B. velezensis B-4 was shown to belong to the antagonist bacteria and to have an excellent antifungal activity against pathogens. Therefore, B. velezensis B-4 can be considered a promising biocontrol agent for biological control in crop production. Furthermore, the unknown genes that we found via complete-genome sequencing need to be studied. The results provide useful genomic information that may affect the development of biocontrol agents and the elucidation of functions of Bacillus sp. genome.
Data Availability Statement
The datasets generated for this study can be found in the B. velezensis (CP031424.1), B. velezensis FZB42(NC_009725.1), B. velezensis M75(NZ_CP016395.1), B. amyloliquefaciens DSM7(NC_014551.1), B. amyloliquefaciens Y2(NC_017912.1), B. amyloliquefaciens CC178(NC_022653.1), B. amyloliquefaciens MT45(NZ_CP011252.1), and B. subtilis 168(NC_000964.3).
Author Contributions
ZZ selected Bacillus velezensis B-4, wrote the manuscript, carried out experimental work, and analyzed the complete genomics sequencing data. QP and YM helped in the bioassay experiments. LB and XZ supervised the entire study. DP helped with the design of the experiments and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (31501601), the Natural Science Foundation of Hunan Province (2016JJ6059), and the Scientific-Innovative of Hunan Agricultural Sciences and Technology (2017GC01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bais, H. P., Fall, R., and Vivanco, J. M. (2004). Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134, 307–319. doi: 10.1104/pp.103.028712
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. P. (2012). Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. doi: 10.1590/S1415-47572012000600020
Chang, G.-R. (2018). Persistent organochlorine pesticides in aquatic environments and fishes in Taiwan and their risk assessment. Environ. Sci. Pollut. Res. Int. 25, 7699–7708. doi: 10.1007/s11356-017-1110-z
Chen, X.-H., Vater, J., Piel, J., Franke, P., Scholz, R., Schneider, K., et al. (2006). Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 188, 4024–4036. doi: 10.1128/JB.00052-06
Chowdhury, S. P., Hartmann, A., Gao, X., and Borriss, R. (2015). Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 - a review. Front. Microbiol. 6:780. doi: 10.3389/fmicb.2015.00780
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Fernando, W. G. D., Nakkeeran, S., Zhang, Y., and Savchuk, S. (2007). Biological control of Sclerotinia sclerotiorum (Lib.) de Bary by Pseudomonas and Bacillus species on canola petals. Crop Prot. 26, 100–107. doi: 10.1016/j.cropro.2006.04.007
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. doi: 10.1093/nar/gku1223
Gautam, V., Singhal, L., Arora, S., Jha, C., and Ray, P. (2013). Reliability of Kirby-Bauer disk diffusion method for detecting carbapenem resistance in Acinetobacter baumannii-calcoaceticus complex isolates. Antimicrob. Agents Chemother. 57, 2003–2004. doi: 10.1128/AAC.01450-12
González-Jaramillo, L. M., Aranda, F. J., Teruel, J. A., Villegas-Escobar, V., and Ortiz, A. (2017). Antimycotic activity of fengycin C biosurfactant and its interaction with phosphatidylcholine model membranes. Coll. Surf. B Biointerfaces 156, 114–122. doi: 10.1016/j.colsurfb.2017.05.021
Gopinath, V., MubarakAli, D., Priyadarshini, S., Priyadharsshini, N. M., Thajuddin, N., and Velusamy, P. (2012). Biosynthesis of silver nanoparticles from Tribulus terrestris and its antimicrobial activity: a novel biological approach. Coll. Surf. B Biointerfaces 96, 69–74. doi: 10.1016/j.colsurfb.2012.03.023
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Guo, Y.-W., Guo, H.-L., Li, X., Huang, L.-L., Zhang, B.-N., Pang, X.-B., et al. (2013). Two type III polyketide synthases from Polygonum cuspidatum: gene structure, evolutionary route and metabolites. Plant Biotechnol. Rep. 7, 371–381. doi: 10.1007/s11816-012-0271-y
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. doi: 10.1093/nar/gkv1070
Kim, S. Y., Lee, S. Y., Weon, H.-Y., Sang, M. K., and Song, J. (2017). Complete genome sequence of Bacillus velezensis M75, a biocontrol agent against fungal plant pathogens, isolated from cotton waste. J. Biotechnol. 241, 112–115. doi: 10.1016/j.jbiotec.2016.11.023
Li, L., Stoeckert, C. J., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Liu, G., Kong, Y., Fan, Y., Geng, C., Peng, D., and Sun, M. (2017). Whole-genome sequencing of Bacillus velezensis LS69, a strain with a broad inhibitory spectrum against pathogenic bacteria. J. Biotechnol. 249, 20–24. doi: 10.1016/j.jbiotec.2017.03.018
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Luo, C., Liu, X., Zhou, H., Wang, X., and Chen, Z. (2015). Nonribosomal peptide synthase gene clusters for lipopeptide biosynthesis in Bacillus subtilis 916 and their phenotypic functions. Appl. Environ. Microbiol. 81, 422–431. doi: 10.1128/AEM.02921-14
Nodwell, J. R. (2017). Antimicrobials: expressing antibiotic gene clusters. Nat. Microbiol. 2:17061. doi: 10.1038/nmicrobiol.2017.61
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Ongena, M., Jourdan, E., Adam, A., Paquot, M., Brans, A., Joris, B., et al. (2007). Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. doi: 10.1111/j.1462-2920.2006.01202.x
Pusztahelyi, T., Holb, I. J., and Pócsi, I. (2015). Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 6:573. doi: 10.3389/fpls.2015.00573
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Syed Ab Rahman, S. F., Singh, E., Pieterse, C. M. J., and Schenk, P. M. (2018). Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. Int. J. Exp. Plant Biol. 267, 102–111. doi: 10.1016/j.plantsci.2017.11.012
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi: 10.1093/nar/gkw937
Wang, Y., Hou, Y.-P., Chen, C.-J., and Zhou, M.-G. (2014). Detection of resistance in Sclerotinia sclerotiorum to carbendazim and dimethachlon in Jiangsu Province of China. Austral. Plant Pathol. 43, 307–312. doi: 10.1007/s13313-014-0271-1
Watson, M. (2014). Quality assessment and control of high-throughput sequencing data. Front. Genet. 5:235. doi: 10.3389/fgene.2014.00235
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H. U., Bruccoleri, R., et al. (2015). antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Keywords: antibacterial activity, complete genome, comparative genome, Bacillus velezensis B-4, biology control
Citation: Zhu Z, Peng Q, Man Y, Li Z, Zhou X, Bai L and Peng D (2020) Analysis of the Antifungal Properties of Bacillus velezensis B-4 Through a Bioassay and Complete-Genome Sequencing. Front. Genet. 11:703. doi: 10.3389/fgene.2020.00703
Received: 28 November 2019; Accepted: 10 June 2020;
Published: 16 July 2020.
Edited by:
Xiaochen Bo, Academy of Military Medical Sciences, ChinaReviewed by:
Néstor Walter Soria, Catholic University of Córdoba, ArgentinaWei Xu, Texas A&M University Corpus Christi, United States
Copyright © 2020 Zhu, Peng, Man, Li, Zhou, Bai and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lianyang Bai, YmFpbGlhbnlhbmcyMDA1QGFsaXl1bi5jb20=; Di Peng, c21pbGVhZGlAMTI2LmNvbQ==
 Zheyuan Zhu
Zheyuan Zhu Qiong Peng2
Qiong Peng2 Lianyang Bai
Lianyang Bai Di Peng
Di Peng