- Ministry of Agriculture (MOA) Key Lab of Pest Monitoring and Green Management, College of Plant Protection, China Agricultural University, Beijing, China
Late blight caused by Phytophthora infestans, is one of the most devastating diseases of potato, and was responsible for the death of millions of people during the Irish Potato Famine in the nineteenth century. Phytophthora infestans is a heterothallic oomycete that typically requires two compatible types (mating types), A1 and A2, to complete sexual reproduction (i.e., oospore production). Oospores have critical effects on disease epidemiology because they serve as the primary inoculum in subsequent growing seasons. The sexual reproduction of Phytophthora species is regulated by α hormones. In previous studies, we proved that transformants in which selected histone deacetylase (HDAC) genes are silenced exhibit abnormal hormone production. In the current study, we compared the transcriptomes of HDAC-silenced and wild-type strains to explore the genes regulated by HDAC and the genes involved in sex hormone biosynthesis in Phytophthora species. A total of 14,423 transcripts of unigenes were identified in the wild-type strain, the HDAC family-silenced transformant (HDST), and the HDAC7-silenced transformant (H7ST). After comparing the intergroup gene expression levels, 1,612 unigenes were identified as differentially expressed among these strains. The expression levels of 16 differentially expressed genes (DEGs) were validated by quantitative real-time PCR. The functional annotation of the DEGs by gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses indicated that HDACs affect the expression of genes related to metabolic and biosynthetic processes, RNA processing, translation, ribosome biogenesis, cellular structural constituents, RNA binding, and protein binding. Moreover, HDAC7 specifically influences the transcription of genes associated with transport, methylation, mitochondria, organelle inner membranes, receptors and transporters, and hydrolase activities. We also identified 18 candidate genes related to α hormones biosynthesis, including a gene encoding the NF-Y transcription factor (PITG_10861). The overexpression of PITG_10861 increased the production of hormone α2. The results of this study revealed P. infestans genes affected by histone acetylation. The data presented herein provide useful inputs for future research on the epigenetic mechanisms and mating behaviors of Phytophthora species.
Introduction
Phytophthora species are filamentous organisms that cause severe diseases in many plant species, resulting in substantial economic losses in the global agricultural industry. Phytophthora species are oomycetes, which form a specific eukaryotic group in the kingdom Stramenopila, and are phylogenetically close to photosynthetic algae (Sogin and Silberman, 1998; Baldauf et al., 2000; Yoon et al., 2002; Adl et al., 2005). Phytophthora infestans is one of the most destructive plant pathogens. It is the causal pathogen of the Irish Potato Famine, which resulted in the death of millions of people in the middle of the nineteenth century. Additionally, it is a model heterothallic Phytophthora species, which normally requires two compatible mating types (A1 and A2) producing specific hormones to initiate selfing and hybridizations in cocultures of the same species (Judelson, 1997; Qi et al., 2005; Ko, 2007; Ojika et al., 2011).
Previous studies revealed that the sexual reproduction of Phytophthora species is regulated by two interspecies universally active, diterpene hormones, α1 and α2 (Ko, 1980; Qi et al., 2005; Ojika et al., 2011), and requires two distinct and independent processes, hormone production and hormone reception (Ko, 1978). Hormone α1, secreted by A1 strains, induces oospore production in A2 strains, whereas hormone α2, secreted by A2 strains, induces oospore production in A1 strains. The production of oospores in strains of homothallic species is stimulated by self-produced α hormones (α1, α2, or both) (Ko, 1980). Using deuterium-labeled 1H, Ojika et al. (2011) proved that A2 strains can use phytol to synthesize α2, and A1 strains can use α2 to synthesize α1. However, the hormone synthesis-related genes remain to be investigated.
Gene expression in eukaryotes can be regulated by epigenetic mechanisms, especially DNA methylation and histone modifications (Hattori and Ushijima, 2014). Histone acetylation occurs at the amino groups of lysine residues at the N-terminus of histone tails, and is regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs) (Allfrey et al., 1964; Brownell and Allis, 1996; Pflum et al., 2001). The HATs, which act as “writers,” are responsible for transferring an acetyl moiety from acetyl coenzyme A (acetyl CoA) to the ε-amino group of specific lysine residues at the histone N-terminal tails (Bertos et al., 2001; Thiagalingam et al., 2003). Consequently, RNA polymerase and other transcription factor complexes interact with DNA (Hong et al., 1993), leading to up-regulated gene expression (Mukherjee et al., 2012). In contrast, HDACs function as epigenetic “erasers” (i.e., HAT antagonists), which remove acetyl groups from histones, thereby down-regulating gene expression. The balance between “erasers” and “writers” makes chromatin regulation a dynamic process. Many studies have indicated that the expression of biosynthesis-related genes is regulated by histone acetylation. In mammals, the key regulator of cholesterol biosynthesis, sterol-regulatory element binding protein (SREBP)-2, is regulated by sirtuin (Sirt6) (Tao et al., 2013). In the endogenous cholesterol synthesis pathway, a transcription factor, CCAAT-binding factor/nuclear factor Y (CBF/NF-Y), is required for the sterol-regulated transcription of the gene encoding a key regulatory enzyme, 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) synthase (Dooley et al., 1998). In Arabidopsis thaliana, the HDACs SRT1 and SRT2 interact with EIN2 nuclear-associated protein 1 (ENAP1) to regulate the extent of the H3K9 acetylation in the ethylene signaling pathway and mediate transcriptional repression (Zhang et al., 2018). In Aspergillus flavus, the epigenetic reader SntB, which is a transcriptional regulator of the sterigmatocystin biosynthetic gene, controls the global levels of H3K9K14 acetylation and secondary metabolite synthesis (Pfannenstiel et al., 2018). The cytoplasmic effector PsAvh23 produced by the soybean pathogen Phytophthora sojae modulates HATs in plants, suppresses the activation of host defense genes by disrupting HAT complex functionality during an infection, and ultimately increases the plant susceptibility to disease (Kong et al., 2017). However, the mechanism underlying the metabolic biosynthesis regulated by histone acetylation in oomycetes remains unclear.
We previously analyzed the expression of HAT and HDAC genes during various P. infestans biological stages (Wang et al., 2016). We also generated transformants in which at least one HDAC gene was silenced, which altered compatible type. Therefore, we speculated that the biosynthesis of hormones in P. infestans might be regulated by histone acetylation. The aim of this study was to investigate the genes regulated by histone acetylation and identify the genes related to sex hormone biosynthesis. We selected the wild-type (WT) P. infestans MX5-1 strain and two HDAC-silenced transformants for RNA sequencing (RNA-seq) and comparative transcriptome analyses. We then screened for genes related to hormone biosynthesis by comparing the gene expression profiles with the phenotypes of the strains.
Materials and Methods
Experimental Design
The WT A1 strain MX5-1 as well as the HDAC family-silenced transformant HDST43 and the HDAC7-silenced transformant H7ST20 were analyzed in this study (Supplementary Figure S1). The HDAC family-silenced transformants were obtained with vectors containing a partial antisense PiHDAC7 open reading frame (ORF) sequence that is conserved in P. infestans HDAC genes. The HDAC7-silenced transformants were generated using vectors containing the 5′ and 3′ untranslated regions of PiHDAC7. The down-regulated HDAC expression of gene-silenced transformants was confirmed by quantitative real-time (qRT)-PCR analyses (Supplementary Figure S1). The two transformants grew slowly and their mating types differed from that of the WT MX5-1 (Supplementary Figure S2). These three strains have a nearly identical genetic background, but vary regarding mating behavior (phenotypes). Their gene expression profiles were compared based on RNA-seq data, which were validated by qRT-PCR (Figure 1). Furthermore, differential expression profiles were examined to screen for genes potentially associated with sex hormone biosynthesis, including genes related to diverse hormone chemical structures, genes involved in terpene biosynthesis pathways, and genes encoding regulators of sex hormone biosynthesis (e.g., transcription factors). We selected one candidate gene and evaluated its effect on hormone production. All experiments were repeated with different sets of biological samples.
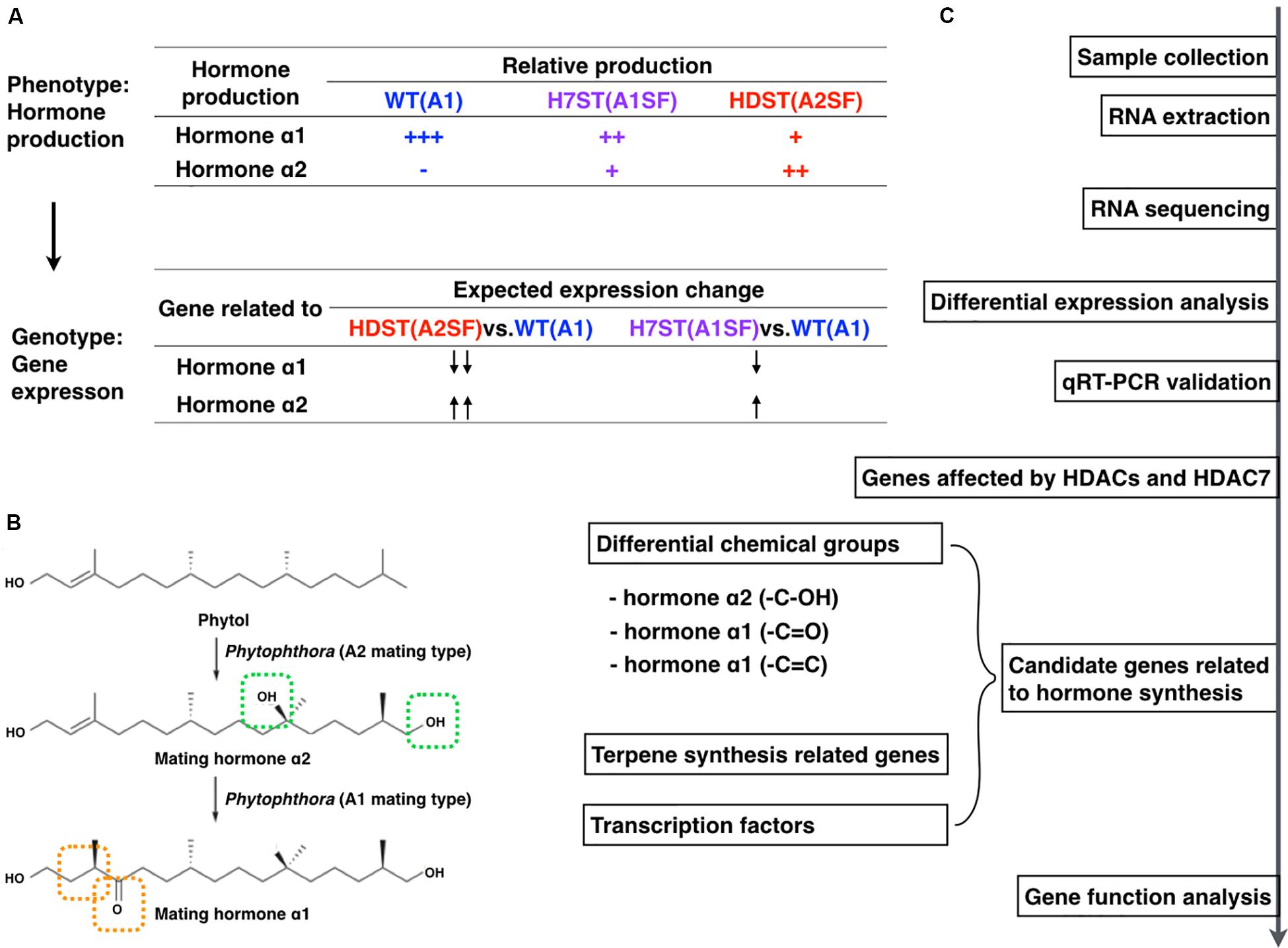
Figure 1. Experimental design and workflow. (A) Expected expression-level changes to genes associated with sex hormone biosynthesis in WT, HDST, and H7ST strains according to the differences in hormone production in these strains. (B) Chemical structures of α1 and α2 hormones and the predicted hormone biosynthesis pathway with phytol as a precursor (modified from Ojika et al., 2011). (C) Study workflow.
Sample Collection and RNA Extraction
The HDST43, H7ST20, and WT MX5-1 strains were grown on 290 tomato rye agar at 18°C in darkness for 6 days (Guo et al., 2010). All strains were analyzed with three biological replicates, each comprising five plates (60 mm). The mycelia were collected and ground in liquid nitrogen, after which total RNA was extracted with the NucleoSpin RNA Plant kit (MACHEREY-NAGEL, Düren, Germany). The purity and quality of the RNA were assessed with the Thermo NanoDrop 2000 spectrophotometer (Wilmington, United States) and by 1% agarose gel electrophoresis.
Transcriptome Profiling
After completing the RNA quality control procedures, mRNA was enriched with oligo(dT) beads and fragmented randomly in fragmentation buffer before first-strand cDNA was synthesized with random hexamers and reverse transcriptase. A custom second-strand synthesis buffer (Illumina, San Diego, CA, United States) was added with dNTPs, RNase H, and Escherichia coli polymerase I to generate the second strand via nick translation. The final cDNA library was prepared after a round of purification, terminal repair, A-tailing, ligation of sequencing adapters, size selection, and PCR enrichment. The cDNA library concentration was determined with the Qubit 2.0 fluorometer (Life Technologies), and then diluted to 1 ng/μl before checking the insert size with the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, United States) and by quantitative PCR to ensure accuracy. Libraries were sequenced with the Illumina HiSeq X Ten RNA-seq platform (Illumina, San Diego, CA, United States). The resulting raw data were transformed into sequenced reads by base calling. The raw data were recorded in a FASTQ file, which contained sequence information (reads) and the corresponding sequencing quality details. Each biological replicate was treated as an independent sample. Raw reads were then filtered to remove reads containing adapters or those of low quality. All remaining clean reads were mapped to the P. infestans reference genome (the EMBL/Genbank/DDBJ databases under the accession GCA_000142945.1) with the TopHat2 software1 (Kim et al., 2013).
Differential Expression Analysis
Gene expression levels were recorded in terms of transcript abundance, which was measured by counting the reads mapped to genes or exons. The expected number of fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) was considered for examining the sequencing depth and gene fragment lengths (Trapnell et al., 2010). The HTSeq program was used to analyze the gene expression levels. Specifically, an FPKM value of 1 was set as the threshold for determining whether a gene was expressed (Anders et al., 2015). To compare gene expression levels under different conditions, an FPKM distribution diagram and violin plot was applied. The final FPKM for each strain was the mean value of the three biological replicates. The correlation analysis between samples of each strain was used to test the reliability of the transcriptome profiles. When the correlation coefficient is larger than 0.8, the three biological samples were used as replicates in the analyses of differentially expressed genes (DEGs). The DEGs between each sample pair were completed with the DESeq R package (Robinson et al., 2010). The p-values were adjusted based on q-values. A q-value < 0.05 was set as the threshold for determining significant differences in expression. The DEGs were functionally annotated based on gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. The GO enrichment analysis was completed according to the GO-seq R package-based Wallenius non-central hypergeometric distribution (Young et al., 2010). The manually curated KEGG databases were related to genomes, biological pathways, diseases, drugs, and chemical substances. Pathway enrichment analyses of the DEGs with the KOBAS software identified significantly enriched metabolic or signal transduction pathways (Mao et al., 2005).
Quantitative Real-Time PCR Validation
We selected some genes to validate the transcriptome data by qRT-PCR, with the elongation factor gene (PiEF1) as an internal control. Regarding the cDNA synthesis, 1 μg total RNA was reverse transcribed with the oligo(dT)18 primer and Reverse Transcriptase M-MLV (TaKaRa Bio Inc., Shiga, Japan) following the manufacturer’s instructions (Supplementary Table S1). A qRT-PCR assay was performed with the ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, United States) as previously described (Wang et al., 2016). Gene expression levels were normalized against the expression of PiEF1. Each RNA sample was analyzed in triplicate. The experiments were repeated once with a different set of biological samples. The expression data were analyzed with the ABI 7500 system software.
Plasmid Construction and Generation of Phytophthora infestans Transformants
The pTOR-X vector, which is pTORmRFP4 with an added XbaI restriction site, was used to construct recombinant plasmids for transforming P. infestans. The PITG_10861 ORF was amplified with specific primers (Supplementary Table S1) and the cDNA of the WT P. infestans A2 strain 80787-94L. The PITG_10861 ORF was inserted into the pTOR-X vector in the sense orientation and was verified by DNA sequencing. The constructed recombinant plasmid was used for overexpressing PITG_10861. The 80787-94L strain was transformed as previously described (Judelson et al., 1991; Mcleod et al., 2008; Guo et al., 2017). All transformants were validated by measuring mycelial PITG_10861 transcript levels in a qRT-PCR assay with specific primers (Supplementary Table S1).
Detection of α Hormone Production
The bioassay method developed by Ko (1980) was used for analyzing hormone production and reception. Specifically, P. infestans strains MX5-1 (A1) and HCl7-7-2 (A2SF) were used as the receptors of the transformant-produced hormones α2 and α1, respectively. To evaluate the hormone production of the transformants, a culture block of the hormone receptor strain (1-day-old) was placed at the center of a Petri plate (60 mm in diameter) and covered with a polycarbonate membrane (Millipore HTTP04700, 0.22 μm, 47 mm in diameter). A transformant block (4-day-old) was placed on top. The cultures were then incubated in a humid chamber in darkness for 3 weeks. For each test, the oospores produced on each block were counted. All experiments were repeated with three replicates per treatment.
Results
Differences in the Hormone Production of Three Tested Strains
Phytophthora species mating types are determined based on the type of hormone production and reception. The silencing of HDAC genes resulted in transformants that differed from the WT strain regarding hormone production. The HDAC family-silenced transformant HDST43 was A2SF (self-fertile and produced more oospores with the A1 strain than with the A2 strain), whereas the HDAC7-silenced transformant H7ST20 was A1SF (self-fertile and produced more oospores with the A2 strain than with the A1 strain). The two transformants produced both hormones α1 and α2. However, the bioassay data revealed that HDST43 produced more hormone α2, whereas H7ST20 produced more hormone α1 (Supplementary Figure S2).
Overview of Illumina Sequencing Data
To investigate the effect of histone acetylation on gene expression as well as the molecular mechanism underlying the biosynthesis of sex hormones, we conducted next-generation sequencing of WT, HDST, and H7ST P. infestans strains, which varied in terms of sex hormone production. The sequencing data were used for a comparative transcriptomic analysis. After filtering the raw reads, more than 44 million clean reads (>6.5 Gb) were generated for each mycelial sample of these strains. Detailed sequencing results are summarized in Table 1.
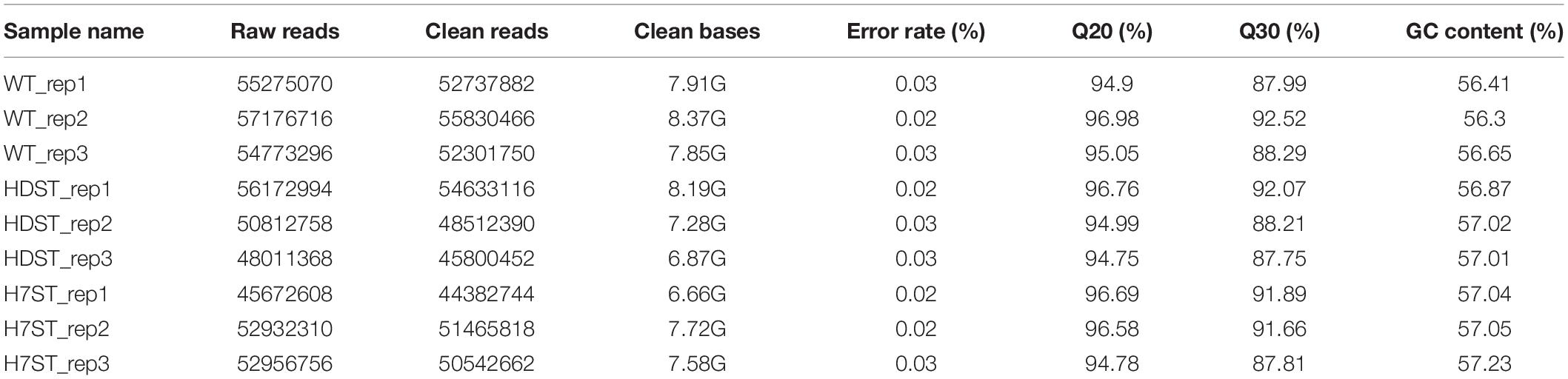
Table 1. Summary statistics of transcriptome sequencing for three different phenotypes of Phytophthora infestans.
The transcriptome assemblies obtained for three strains were pooled and used to assemble full-length transcripts with the P. infestans reference genome. After eliminating the redundant transcripts, 72.68–79.13% of the clean reads for these samples were uniquely mapped to the reference genome (Figure 2A and Supplementary Table S2). Furthermore, 91.8–95% of the reads were located within exons (Supplementary Figure S3). The transcriptome sequencing of mycelia identified a total of 27,657 unigenes (Supplementary Table S3). In this study, a gene was considered to be expressed in a strain if its transcript was detected in the cDNA library for all biological replicates. A total of 14,423 transcripts of unigenes were detected in the WT, HDST, and H7ST strains, but relatively few genes were highly expressed (Figure 2B and Supplementary Table S3). For each strain, the data were highly correlated among three biological replicates (Pearson’s r > 0.98) (Figure 2C), suggesting the transcriptome profiles were highly reproducible and reliable.
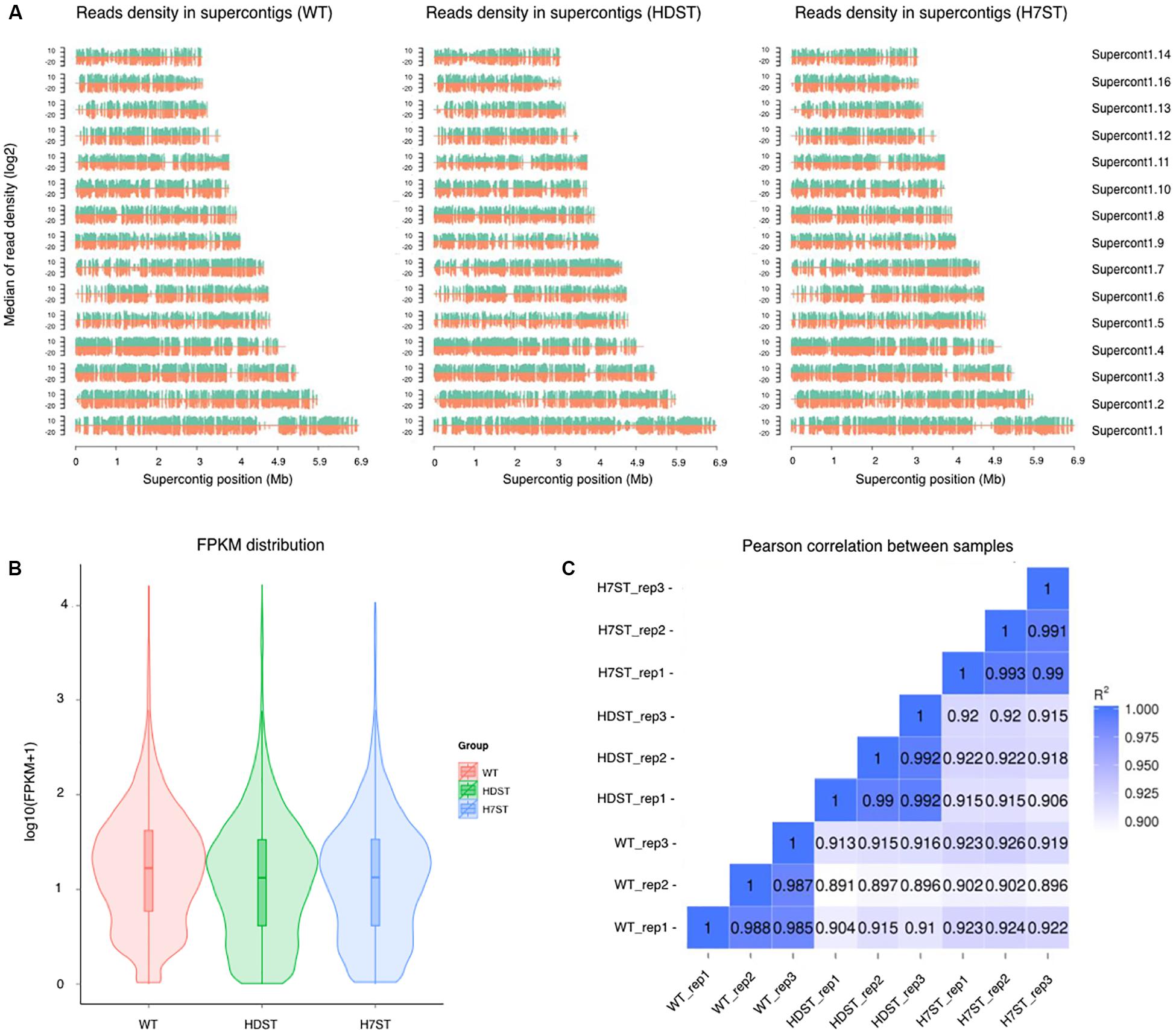
Figure 2. Analysis of the transcriptome data. (A) Distribution plot of the mapped reads in supercontigs. The x-axis indicates the chromosome length (in Mb), whereas the y-axis indicates the median log2 read density. Green and red indicate the positive and negative strands, respectively. (B) Violin plots of the FPKM values for gene expression levels in various samples. The x-axis indicates the sample names, whereas the y-axis indicates the log10(FPKM + 1) values. Each violin plot has five statistical magnitudes (maximum value, upper quartile, median, lower quartile, and minimum value). The violin width represents the gene density. (C) RNA-seq correlation analysis. Heat maps of the correlation between samples are presented. R2, the square of the Pearson correlation coefficient.
Functional Enrichment Analysis of Differentially Expressed Genes
To investigate the effect of histone acetylation on gene expression in P. infestans, we compared the three transcriptomes. We identified 7,142 DEGs between the HDST and WT strains, of which 3,446 and 3,696 DEGs exhibited up- and down-regulated expression, respectively. We also identified 6,900 DEGs between the H7ST and WT strains, including 3,441 and 3,459 DEGs with up- and down-regulated expression, respectively. Additionally, 7,001 DEGs were detected between the H7ST and HDST strains, of which 3,558 and 3,443 DEGs exhibited up- and down-regulated expression, respectively (Figure 3A). An exploration of the global transcriptional changes among samples revealed the co-expression of 4,085 DEGs between the HDST vs. WT and H7ST vs. WT comparisons as well as 1,612 DEGs among the HDST vs. WT, H7ST vs. WT, and H7ST vs. HDST comparisons (Figure 3B).
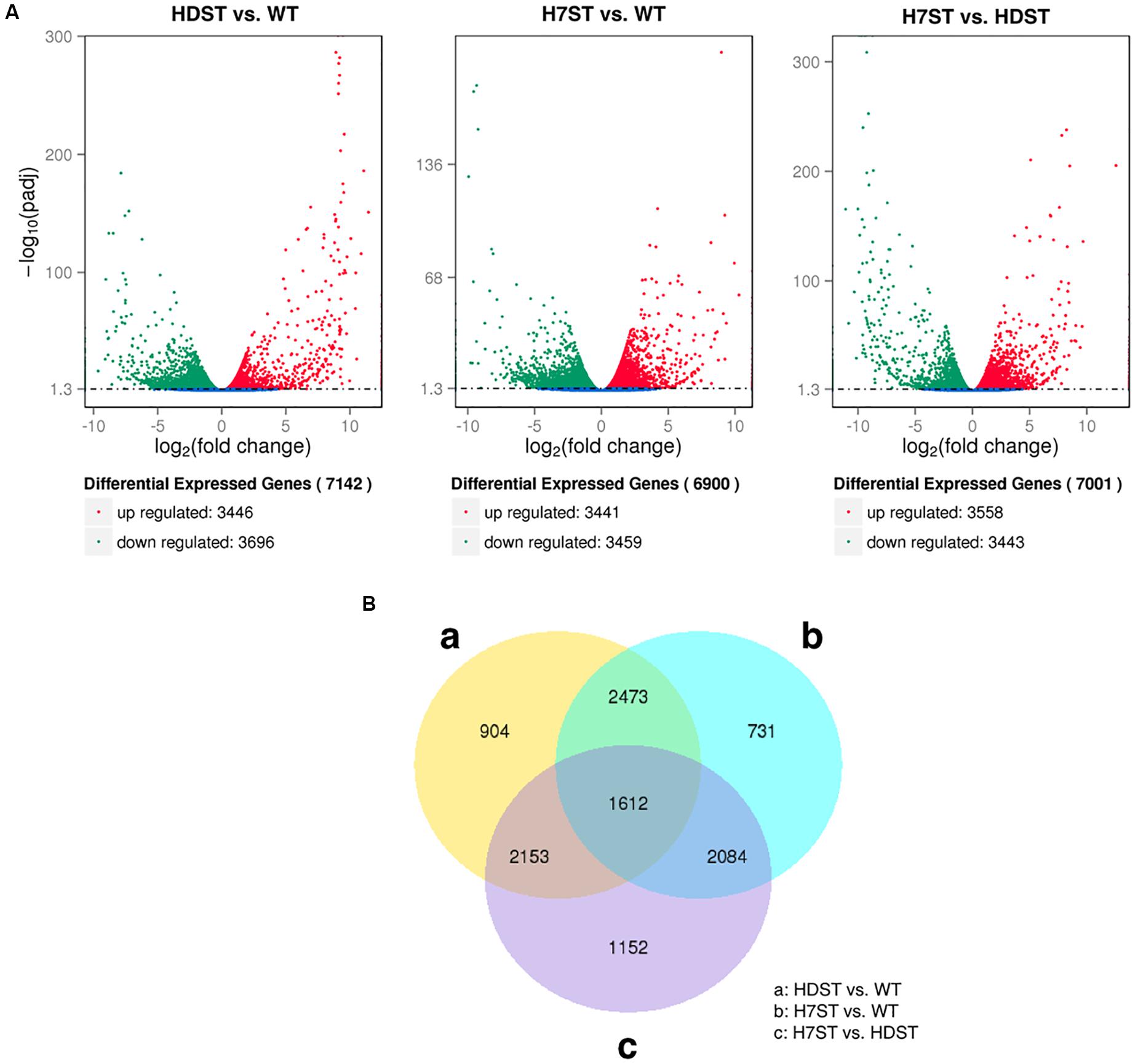
Figure 3. Differentially expressed genes (DEGs) in three phenotypically diverse strains. (A) Volcano plot for DEGs. The x-axis indicates the gene expression fold-changes between different strains, whereas the y-axis indicates the significance of the differences. Significantly up- and down-regulated genes are highlighted in red and green, respectively. The blue dots indicate the genes that were not differentially expressed between strains. (B) Venn diagram of DEGs. The sum of the numbers in each circle represents the total number of expressed genes within a comparison, whereas the numbers in the overlapping areas represent the number of expressed genes shared between groups.
To functionally characterize the DEGs, we performed a GO enrichment analysis. The 10 most significantly overrepresented GO terms were as follows: organonitrogen compound metabolic and biosynthetic process, translation, peptide metabolic and biosynthetic process, gene expression, intracellular ribonucleoprotein complex, ribosome, cytoplasm, structural constituent of ribosome, RNA binding, and protein binding (Figure 4 and Supplementary Table S4). The cellular metabolic and biosynthetic processes were the two largest subcategories within the biological process category. The most abundant cellular component subcategories were ribosome and intracellular, whereas the two largest molecular function subcategories were binding and catalytic activity. The GO analysis of the HDAC-silenced transformants and the WT strain implied that HDACs affect the expression of diverse genes, including those related to metabolic and biosynthetic processes, RNA processing, translation, ribosome biogenesis, ribosomal structural constituents, intracellular and extracellular components, the cytoplasm and organelles, RNA binding, protein binding, ion binding, and nucleic acid binding. We previously identified eight Class I and II HDACs in the P. infestans genome, with HDAC7 belonging to Class II (Wang et al., 2016). A comparison of the DEGs between H7ST vs. WT and H7ST vs. HDST indicated that HDAC7 specifically influences the expression of genes associated with transport, methylation, mitochondria, organelle inner membranes, receptors, transporters, and hydrolase activity.
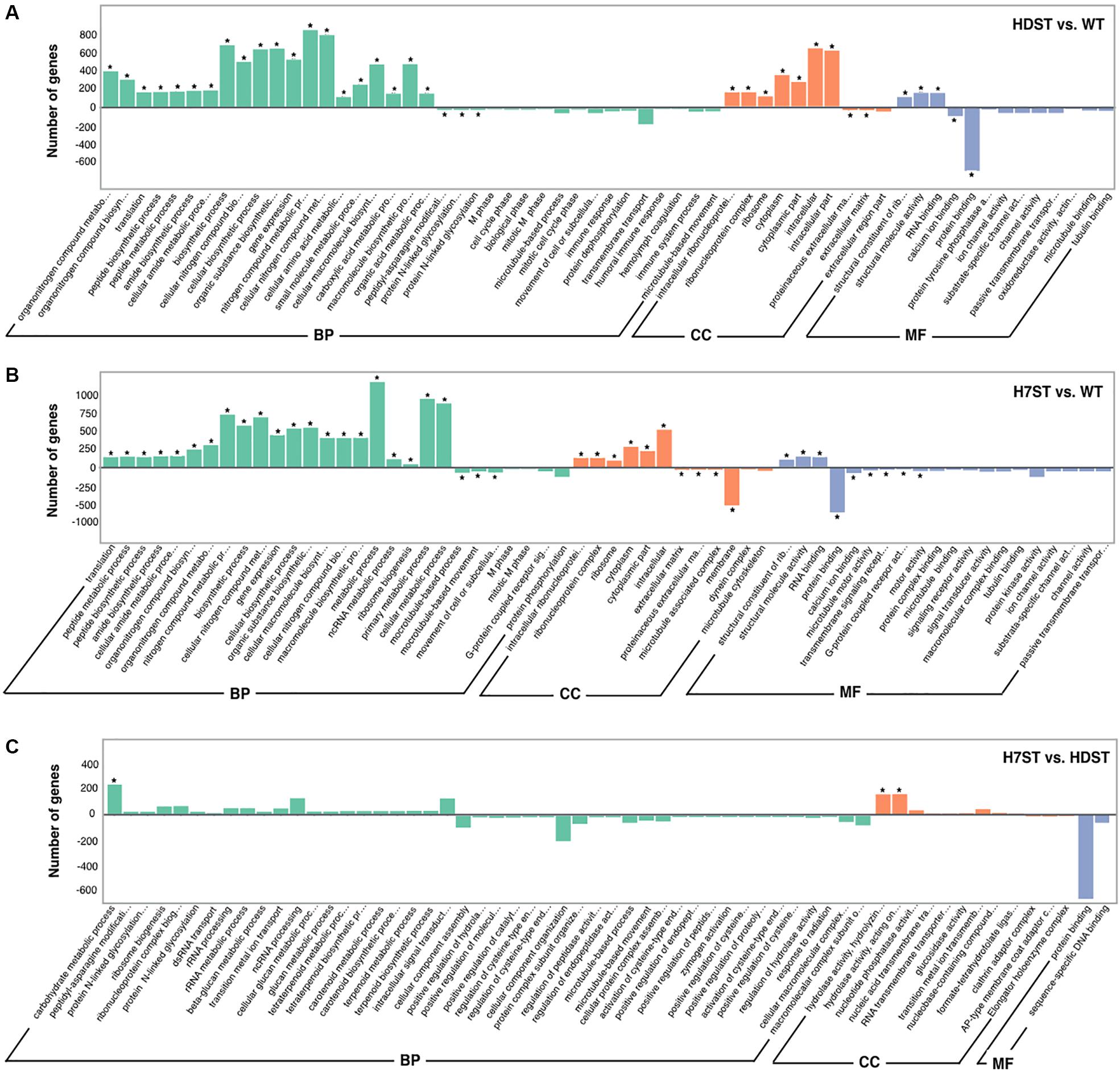
Figure 4. Gene ontology functional classification of differentially expressed genes. The x-axis indicates the enriched GO terms, whereas the y-axis indicates the number of differentially expressed genes in the HDST vs. WT (A), H7ST vs. WT (B), and H7ST vs. HDST (C) comparisons. Different colors are used to distinguish the biological process, cellular component, and molecular function GO categories. The enriched GO terms are marked with an asterisk. The ± numbers represent the up-/down-regulated genes.
The functions of the DEGs were also examined based on a KEGG pathway enrichment analysis. The DEGs detected in the HDST vs. WT, H7ST vs. WT, and H7ST vs. HDST comparisons were related to 25 pathways (Supplementary Table S5). Most of the DEGs between the HDST and WT strains appear to contribute to ribosome biogenesis, metabolic pathways, and RNA transport. The genes associated with fatty acid degradation were expressed at lower levels in the HDST strain than in the WT strain. In contrast, the expression levels of some of the DEGs involved in the biosynthesis of amino acids were up-regulated in the HDST strain. Similar trends were observed for the DEGs between the H7ST and WT strains. Additionally, some of the DEGs with lower expression levels in the H7ST strain than in the WT strain were related to endocytosis, regulation of autophagy, and tyrosine metabolism. Moreover, most of the DEGs between the H7ST and HDST strains were significantly associated with cyanoamino acid metabolism, starch and sucrose metabolism, ribosome biogenesis, and fatty acid degradation. The expression levels of DEGs involved in nicotinate and nicotinamide metabolism were down-regulated in the H7ST strain relative to the corresponding expression in the HDST strain, whereas genes related to DNA replication and other glycan degradation exhibited the opposite trend.
Validating the Differentially Expressed Genes Identified With RNA-Seq Data by Quantitative Real-Time PCR
To assess the reliability of the DEGs identified by the comparative transcriptomic analysis, we randomly selected 16 genes that were differentially expressed among strains (fold-change > 2) and expressed in at least one sample (FPKM > 1) for a qRT-PCR assay. The qRT-PCR data were highly consistent with the RNA-seq results (Pearson’s r > 0.80, p < 0.01; Figure 5).
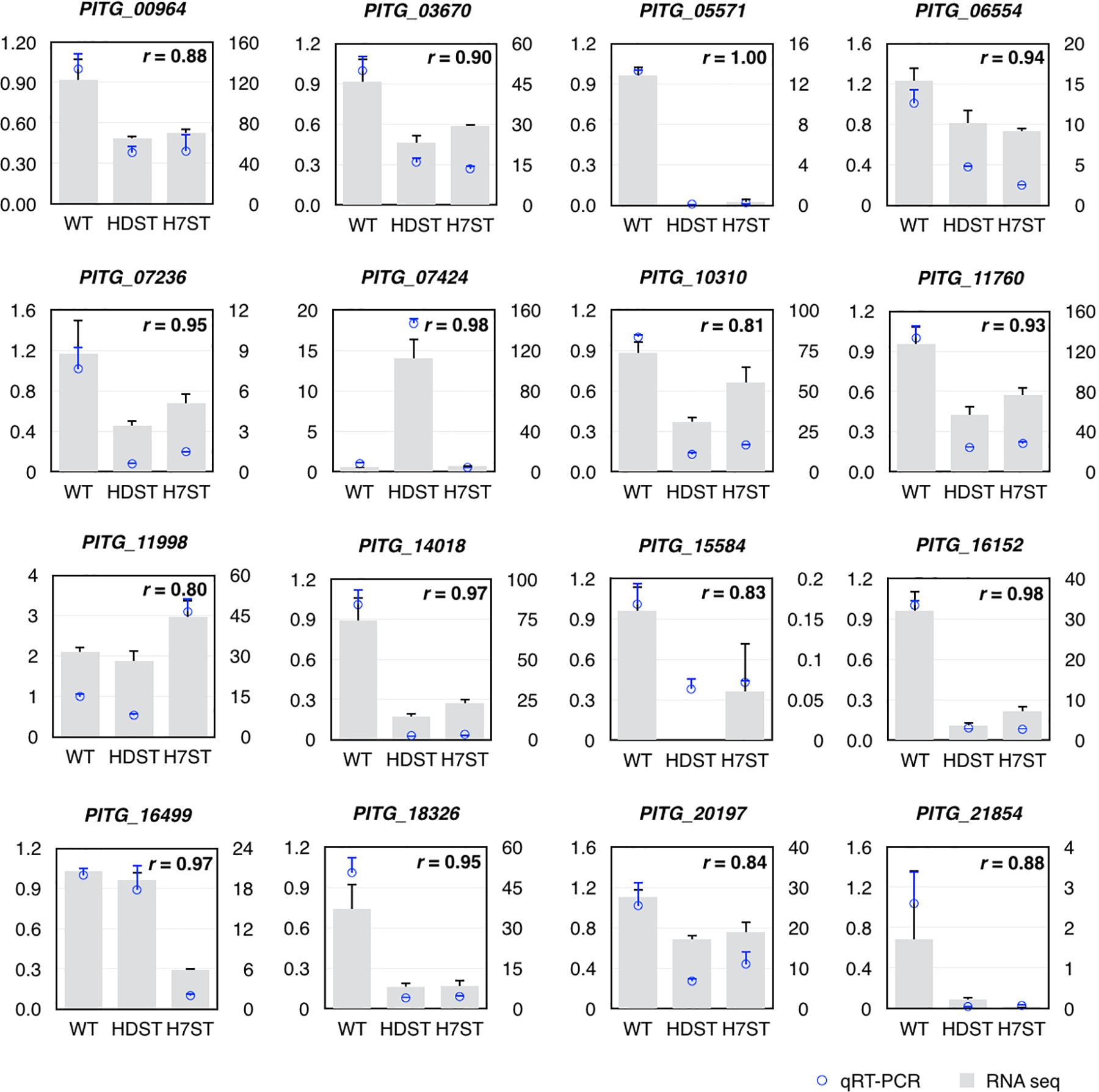
Figure 5. Validation of differentially expressed genes by qRT-PCR. The left y-axis indicates the gene expression fold-changes based on the qRT-PCR data, whereas the right y-axis indicates the FPKM values based on the RNA-seq data, which represent the relative expression of the same gene in the HDST, H7ST, and WT strains. The r value represents the Pearson correlation coefficient of two data sets.
Expression of Genes Related to Terpene Biosynthesis
In Phytophthora species, hormones α1 and α2 are straight-chain diterpenes. Because the terpene pathway in Phytophthora species has not been characterized, we screened for genes related to terpene biosynthesis and generated putative pathways according to the KEGG pathway enrichment analysis (Supplementary Figure S4). There are two main terpenoid backbone biosynthesis pathways, namely the mevalonate (MAV) pathway and the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway. After the terpenoid backbone is synthesized, there are many candidate pathways for diterpene biosynthesis. We identified some P. infestans genes related to the MAV pathway, but did not find any orthologs related to the MEP pathway. Moreover, we did not detect any orthologs of genes related to various diterpene biosynthesis pathways predicted in Phytophthora species. These results suggested that Phytophthora species either have a unique diterpene biosynthesis pathway or do not have one at all.
To investigate the terpene biosynthesis-related genes, a GO enrichment analysis was performed, resulting in the identification of 210 candidate genes annotated with 31 GO terms (Figure 6A and Supplementary Table S6). A comparison between the candidate gene expression patterns and the changes in hormone production in the analyzed strains uncovered nine genes potentially associated with hormone production. Among these genes, the following four with down-regulated expression levels in the HDST and H7ST strains might be related to hormone α1 synthesis: PITG_14532, Novel01090, PITG_15033, and PITG_01370 (p < 0.05). The other five genes (PITG_16974, PITG_14619, PITG_13614, PITG_03525, and PITG_10837), whose expression levels were up-regulated in the HDST and H7ST strains (p < 0.05; Figure 6B), might be related to hormone α2 synthesis. The molecular functions of these down-regulated genes are associated with binding during various biosynthesis-related processes, including flavin adenine dinucleotide binding, ion binding, small molecule binding, and nucleic acid binding. The up-regulated gene PITG_16974 encodes a coenzyme in cofactor metabolic processes. Two other genes with up-regulated expression, PITG_14619 and PITG_10837, encode transferases related to hexosyl groups. The proteins encoded by the remaining two up-regulated genes, PITG_13614 and PITG_03525, have hydrolase activities affecting glycosyl and ester bonds, respectively.
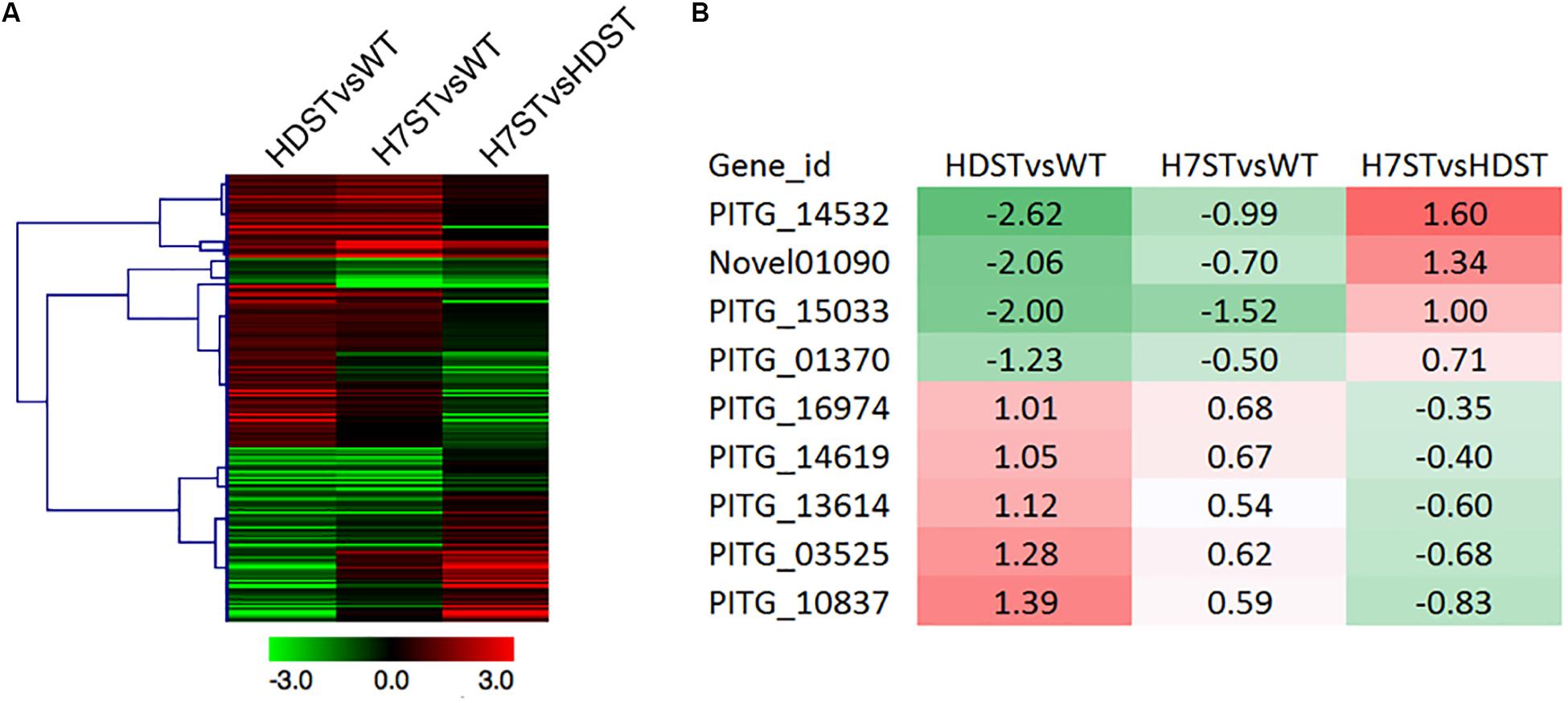
Figure 6. Expression of the genes related to terpene biosynthesis. (A) Heat map presenting the expression of 210 genes related to terpene biosynthesis. (B) Heat map presenting the expression of nine candidate genes associated with α hormone biosynthesis. Red and green denote genes with up- and down-regulated expression levels, respectively. The numbers represent the log2(fold-change) values.
Expression of Genes Related to Chemical Groups on Hormones Structures
Phytophthora species can use organic compounds with structures similar to those of diterpenes to synthesize sex hormones. Ojika et al. (2011) proved that A2 strains can use phytol to synthesize α2, whereas A1 strains can use α2 to synthesize α1. In this biosynthetic pathway, four chemical groups on the phytol undergo changes, including the addition of 11-OH and 16-OH to form hormone α2 as well as the formation of a C2–C3 double bond and an α-methyl-branching ketone at C4 on hormone α2 to synthesize hormone α1. Ojika et al. (2011) suggested that cytochrome P450s might be the enzymes catalyzing the modifications that convert phytol to hormone α2. We speculated that alcohol dehydrogenases (ADHs) might contribute to the formation of an α-methyl-branching ketone at C4 on the backbone of hormone α1 via a dehydrogenation after an OH group is added to hormone α2. Additionally, 2-coumarate reductases may help break the C2–C3 double bond on hormone α2 to form hormone α1. We then screened the candidate genes by comparing the changes in their expression levels with the changes in hormone production in the analyzed strains.
We detected 20 P450 genes in the P. infestans genome. Because the FPKM of P450 gene PITG_13866 was less than 1.0, we considered this gene to be unexpressed. The expression patterns of the other P450 genes varied (Figure 7A). The changes in the PITG_07424 and PITG_14018 expression levels (p < 0.05) were consistent with the hormone production levels in the WT, HDST, and H7ST strains (Figure 1 and Supplementary Figure S2), implying these genes are related to hormone production. The PITG_07424 expression level was significantly up-regulated in the HDST and H7ST strains, suggesting it may participate in the production of hormone α2. In contrast, PITG_14018 expression was significantly down-regulated in the HDST and H7ST strains, indicating that it may contribute to the production of hormone α1. Similarly, we identified 47 ADH genes in the P. infestans genome, of which 42 were differentially expressed among strains. Three genes with significantly down-regulated expression levels in the HDST and H7ST strains were identified (PITG_10290, PITG_11293, and PITG_11295) (p < 0.05, Figure 7B). Moreover, of the seven genes encoding 2-coumarate reductases, the expression of PITG_14479 was significantly up-regulated in the HDST and H7ST strains (p < 0.05, Figure 7C). Accordingly, this gene may mediate the biosynthesis of hormone α1.
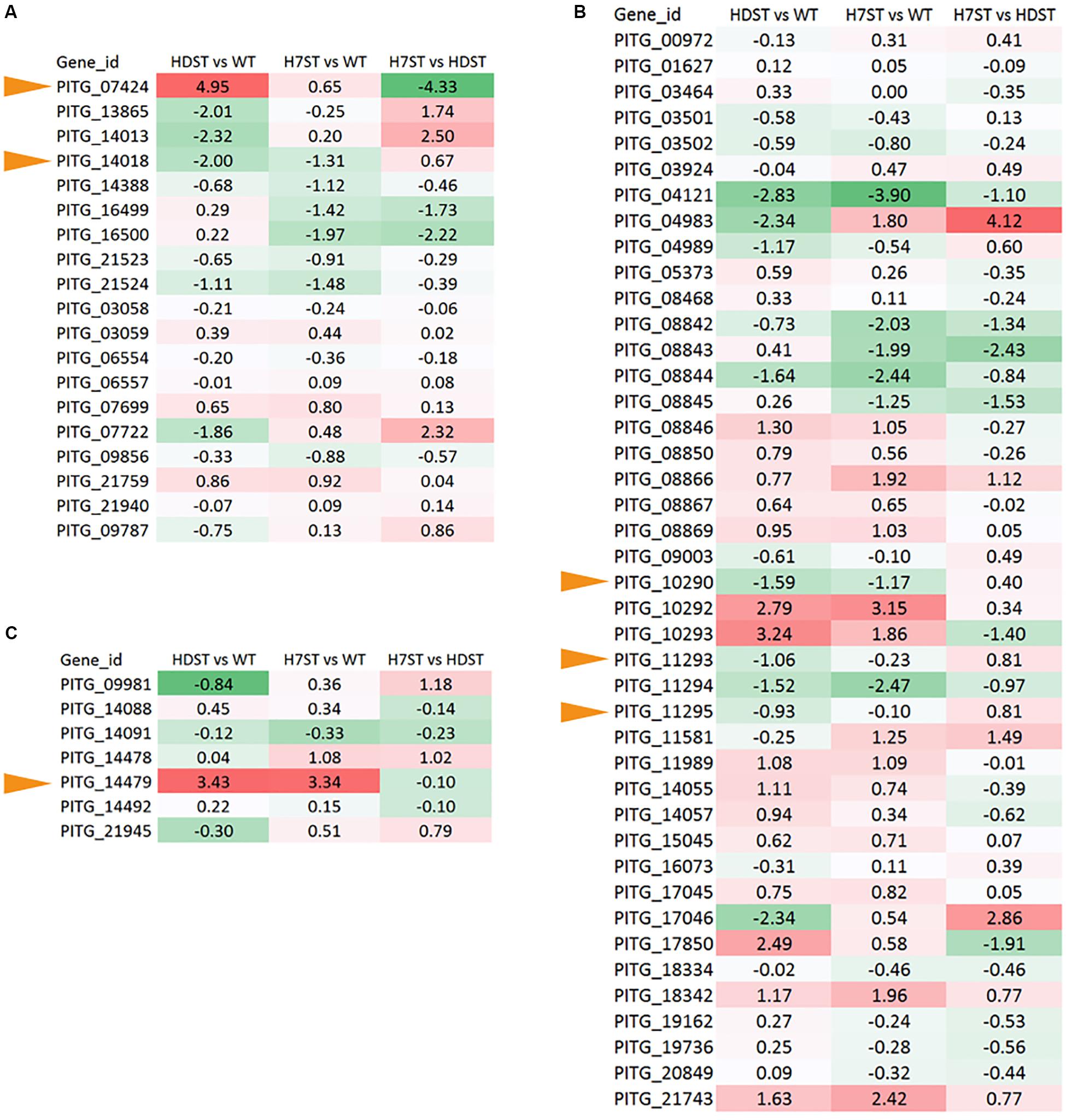
Figure 7. Expression-level differences of genes encoding (A) cytochrome P450, (B) ADH, and (C) 2-coumarate reductase, which alter hormone chemical groups. Red and green denote genes with up- and down-regulated expression levels, respectively. The numbers represent the log2(fold-change) values. Yellow arrows represent the hormone synthesis related genes.
Transcription Factors Related to Hormone Production
Transcription factors are important regulators of gene expression. A thorough analysis of the P. infestans genome uncovered 257 transcription factor genes belonging to 18 families (Supplementary Table S7). Our DEG analysis revealed a MYB family gene (PITG_16152), a homeodomain family gene (PITG_08175), and a CBF/NF-Y family gene (PITG_10861) (Figure 8). The PITG_16152 expression level was significantly down-regulated in the HDST and H7ST strains, whereas the opposite expression trend was observed for PITG_08175 and PITG_10861 (p < 0.05), suggesting these genes may differentially regulate hormone production.
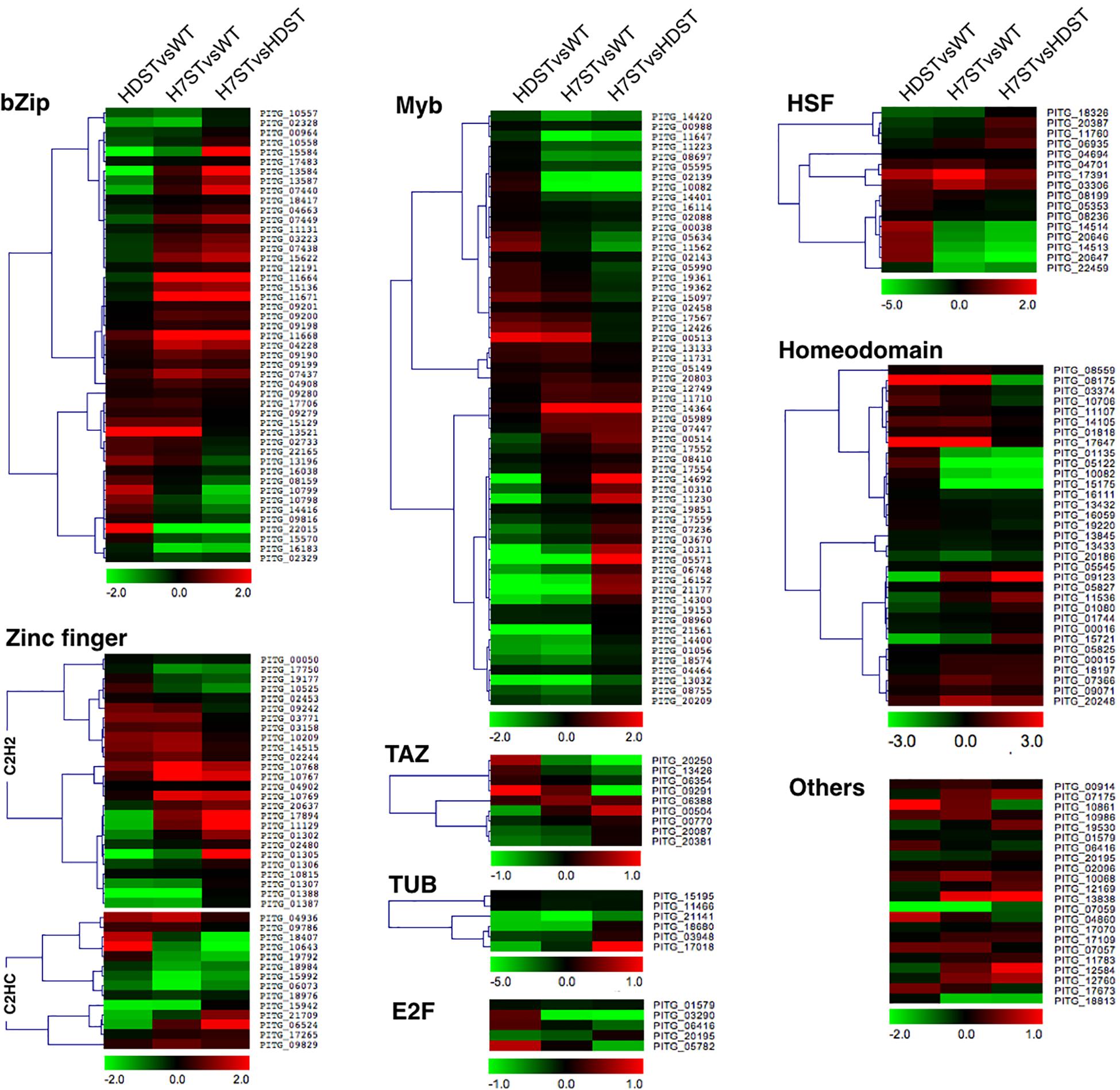
Figure 8. Expression-level differences of transcription factor genes. Red and green denote genes with up- and down-regulated expression levels, respectively. The color bar represents the log2(fold-change) value.
Effect of Transcription Factor NF-Y (PITG_10861) on Hormone Production
The expression level of a gene (PITG_10861) encoding an NF-Y transcription factor, which contains a CBF/NF-Y/archaeal histone domain, was significantly up-regulated in the HDST and H7ST strains (Figure 9A). We speculated that this transcription factor promotes the production of hormone α2. We tested this hypothesis by generating transformants overexpressing PITG_10861 (OT11, OT27, OT56, and OT57) relative to the corresponding expression levels in the WT and empty vector controls (Figure 9B). The growth rates of the PITG_10861-overexpressing transformants were similar to the WT and empty vector control growth rates (Figure 9C). We then quantified the oospore production in the A1 strain induced by these PITG_10861-overexpressing transformants as well as the WT and empty vector controls. The data revealed that the A1 strain produced more oospores with OT11, OT27, OT56, and OT57 than with the WT and empty vector controls (Figure 9D). Thus, overexpressing PITG_10861 increased the production of hormone α2 in P. infestans, which is consistent with our hypothesis. Therefore, PITG_10861 is an important regulator of hormone α2 production.
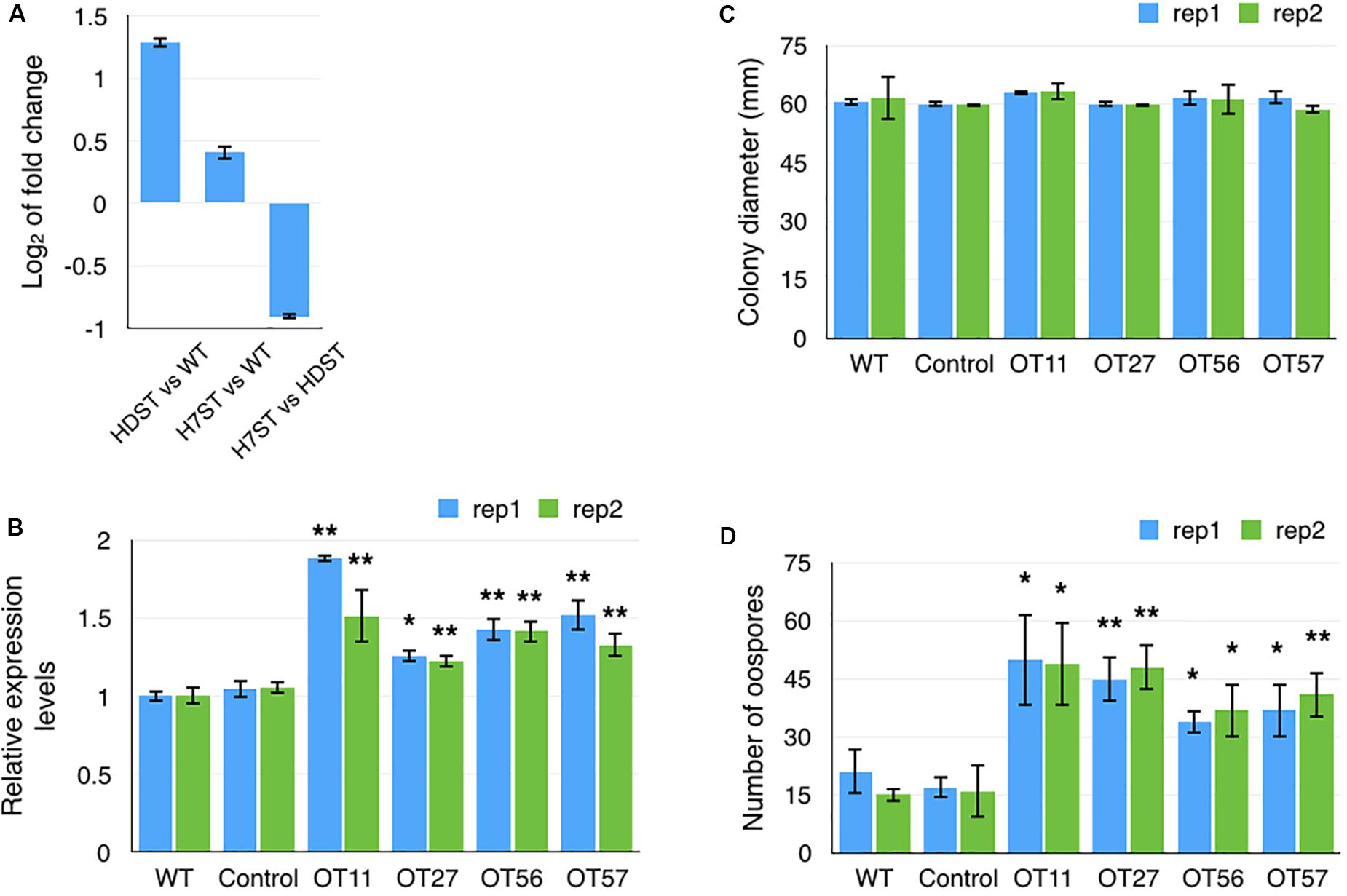
Figure 9. Transcription factor gene PITG_10861 promotes the production of hormone α2. (A) Fold-changes to PITG_10861 expression levels in the HDST, H7ST, and wild-type strains. Relative expression levels (B) and colony diameters (C) of wild-type, empty vector control, and four PITG_10861-overexpressing transformants. (D) Oospore production in the A1 strain induced by the wild-type, empty vector control, and four PITG_10861-overexpressing transformants. *p < 0.05; **p < 0.01.
Discussion
In this study, we identified many DEGs associated with histone acetylation in P. infestans based on Illumina sequencing analyses of a WT P. infestans strain, MX5-1, as well as HDAC family-silenced and HDAC7-silenced transformants. The highly correlated data among the three biological replicates for each strain are indicative of the high reproducibility and reliability of the transcriptome profiling performed in this study. The consistency between the qRT-PCR and RNA-seq data suggests the DEG analysis is accurate. In a previous study, we identified many P. infestans genes encoding HATs and HDACs that are expressed in different developmental stages as well as during infections and in response to stresses (Wang et al., 2016). The HDAC7 expression level is up-regulated during the cyst formation and sexual reproduction stages. Compared with the WT control, the HDAC-silenced transformants grew more slowly, with altered hormone production and mating types as well as defective asexual and sexual structures, indicating that HDACs have many biological roles. In this study, we used 6-day-old mycelia as samples for RNA-seq analyses because Phytophthora species regularly produce sex hormones during the mycelial growth stage. Thus, the identified DEGs were associated mainly with vegetative growth and the early sexual reproduction stage rather than with infections, the late sexual reproduction stage, or stress responses. To reveal the biological roles of HDACs related to asexual and sexual structures, infections, and stress responses, RNA-seq data for these specific biological stages will need to be generated and analyzed.
The sexual behaviors of Phytophthora species are regulated by α hormones (Ko, 1980, 2007; Qi et al., 2005). The three strains examined in this study vary regarding their ability to produce α hormones. By comparing the DEGs between the transformants and the WT control with the changes in hormone production in the transformants, we identified 18 candidate genes related to α hormone synthesis, including three transcription factor genes, two P450 genes, three ADH genes, one 2-coumarate reductase gene, and nine terpene biosynthesis-related genes. We further examined the effect of PITG_10861 on hormone production by generating transformants in which the gene was overexpressed. Subsequent analyses indicated that overexpressing PITG_10861 enhanced the production of hormone α2 in P. infestans, implying this gene encodes an important regulator of α hormone biosynthesis. This result was also suggestive of the reliability of our analysis. The genetic verification of the regulatory effects of the other genes on α hormone biosynthesis is in progress.
The PITG_10861 gene encodes the NF-Y transcription factor carrying the CBF/NF-Y/archaeal histone domain. Previous research confirmed that NF-Y is an essential transcription factor for mammalian development, from the early stages to adulthood, and in human pathogenesis (Maity, 2017). In mice, the NF-Y transcription factor interacts with the orphan nuclear receptor steroidogenic factor-1 to regulate the expression of the mouse follicle-stimulating hormone-β gene (FSHβ) (Jacobs et al., 2003). In A. thaliana, NF-YCs interact with the histone deacetylase HDA15 when exposed to light to co-target the promoters of a set of hypocotyl elongation-related genes, thereby modulating the extent of the histone H4 acetylation of the associated chromatin (Tang et al., 2017). Additionally, MYB and homeodomain transcription factors might affect α hormone biosynthesis. Previous studies revealed that MYB transcription factors regulate P. infestans sporulation (Xiang and Judelson, 2014). The AaMYB1 gene and its ortholog AtMYB61 affect terpene metabolism and trichome development in Artemisia annua and A. thaliana (Matías-Hernández et al., 2017). We are currently conducting experiments to genetically verify the regulation of α hormone biosynthesis by the MYB and homeodomain transcription factors.
Ojika et al. (2011) reported that A2 isolates can use plant phytols to synthesize the α2 hormone, whereas A1 isolates can use α2 to synthesize α1. In the associated biosynthetic pathway, four phytol chemical groups are altered by the addition of 11-OH and 16-OH to form hormone α2 and the formation of a C2–C3 double bond and an α-methyl-branching ketone at C4 to synthesize hormone α1. Ojika et al. (2011) suspected that oxidizing enzymes (cytochrome P450s) might play a role in the conversion of phytol to hormone α2. In this study, we revealed that P450 genes PITG_07424 and PITG_14018 might be involved in sex hormone biosynthesis. The PITG_07424 expression level was significantly up-regulated in the HDST and H7ST strains, suggesting it may participate in the production of hormone α2. However, PITG_14018 expression was significantly down-regulated in the HDST and H7ST strains, indicating that it may be involved in the production of hormone α1. In A. thaliana, CYP77A6 is an in-chain hydroxylase that functions after CYP86A4, the fatty acid ω-hydroxylase, during the synthesis of 10,16-dihydroxypalmitate, which is required for the synthesis of the cutin polyester at floral surfaces (Li-Beisson et al., 2009). Although the KEGG pathway analysis indicated PITG_07424 is a homolog of CYP86, we were unable to determine which P450 gene is the homolog of CYP77A6 or CYP86A4 via a BLAST search because of the low similarity between P. infestans and A. thaliana P450 genes (Supplementary Figure S5). Consequently, further experimental evidence is needed to elucidate the role of PITG_07424 related to hormone α2 synthesis. During the conversion of hormone α2 to hormone α1, alcohol dehydrogenases (ADHs) may modify the C4 site to form an α-methyl-branching ketone via a dehydrogenation after the addition of an OH group by P450s. The PITG_14018 expression profile suggested that this gene might participate in the oxidation during the biosynthesis of α-methyl-branching ketone on sex hormone α1. The genetic verification of the effects of these two cytochrome P450s on α hormone production and their roles in α hormone biosynthesis is in progress.
Considering the diterpene structures of α hormones, we also screened for genes related to terpene synthesis in Phytophthora species. Previous studies proved that MAV and MEP pathways are mainly involved in the production of the terpenoid backbone (McGarvey and Croteau, 1995; Rohmer, 1999). However, a genomic examination did not uncover orthologs related to the MEP pathway in Phytophthora species. This suggests that only the MAV pathway exists in these oomycetes. After the terpenoid backbone is formed, diterpene compounds can be formed via various pathways, including the pathway involving geranylgeraniol, which is structurally similar to the sex hormones of Phytophthora species. However, orthologs of the genes in these pathways have not been detected in Phytophthora species. Earlier research indicated that monoterpenes, diterpenes, and tetraterpenes, which are synthesized in the plastids of various organisms, are derived from the isopentenyl diphosphate produced by the MEP pathway (Lichtenthaler, 2000). However, oomycetes lost these plastids during long-term evolution (Tyler et al., 2006; Derelle et al., 2016). The ambiguities related to the terpene biosynthetic pathway are related to these contradictory observations. In the current study, we identified nine terpene biosynthesis-related genes in P. infestans. However, their roles in α hormone synthesis remain to be investigated.
This study revealed P. infestans genes affected by histone acetylation, including genes related to the biosynthesis of sex hormones and the associated regulation. The data presented herein provide useful inputs for future investigations on the epigenetic mechanisms and the regulation of mating behaviors in Phytophthora species.
Data Availability Statement
All RNA-Seq data generated in this study can be found in PRJNA608542, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA 608542.
Author Contributions
L-YG and X-WW conceived and designed the experiments and wrote the manuscript. X-WW, J-LL, and Y-RS performed the experiments. X-WW and J-LL analyzed the data. All authors have read and approved the final manuscript.
Funding
This work was funded by the Chinese Universities Scientific Fund (2017ZB001 and 88097099).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Yuan-chao Wang (Nanjing Agricultural University) and Dr. Francine Govers (Wageningen University) for providing the vector pTORmRFP, Dr. Tao Zhou (China Agricultural University) for valuable suggestions about transcriptome analysis, Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00508/full#supplementary-material
Footnotes
References
Adl, S. M., Simpson, A. G., Farmer, M. A., Andersen, R. A., Anderson, O. R., Barta, J. R., et al. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451. doi: 10.1111/j.1550-7408.2006.00123.x
Allfrey, V. G., Faulkner, R., and Mirsky, A. E. (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 51, 786–794. doi: 10.1073/pnas.51.5.786
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Baldauf, S. L., Roger, A. J., Wenk-Siefert, I., and Doolittle, W. F. (2000). A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 5493, 972–977. doi: 10.1126/science.290.5493.972
Bertos, N. R., Wang, A. H., and Yang, X. J. (2001). Class II histone deacetylases: structure, function, and regulation. Biochem. Cell Biol. 79, 243–252.
Brownell, J. E., and Allis, C. D. (1996). Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev. 6, 176–184. doi: 10.1016/s0959-437x(96)80048-7
Derelle, R., López-García, P., Timpano, H., and Moreira, D. (2016). A phylogenomic framework to study the diversity and evolution of stramenopiles (= heterokonts). Mol. Biol. Evol. 33, 2890–2898. doi: 10.1093/molbev/msw168
Dooley, K. A., Millinder, S., and Osborne, T. F. (1998). Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J. Biol. Chem. 273, 1349–1356. doi: 10.1074/jbc.273.3.1349
Guo, L. Y., Zhu, X. Q., Hu, C. H., and Ristaino, J. B. (2010). Genetic structure of Phytophthora infestans populations in China indicates multiple migration events. Phytopathology 100, 997–1006. doi: 10.1094/PHYTO-05-09-0126
Guo, T., Wang, X. W., Shan, K., Sun, W., and Guo, L. Y. (2017). The loricrin-like protein (LLP) of Phytophthora infestans is required for oospore formation and plant infection. Front. Plant Sci. 8:142. doi: 10.3389/fpls.2017.00142
Hattori, N., and Ushijima, T. (2014). Compendium of aberrant DNA methylation and histone modifications in cancers. Biochem. Bioph. Res. Co. 455, 3–9. doi: 10.1016/j.bbrc.2014.08.140
Hong, L., Schroth, G. P., Matthews, H. R., Yau, P., and Bradbury, E. M. (1993). Studies of the DNA finding properties of histone H4 amino terminus: thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J. Biol. Chem. 268, 305–314.
Jacobs, S. B., Coss, D., McGillivray, S. M., and Mellon, P. L. (2003). Nuclear factor Y and steroidogenic factor 1 physically and functionally interact to contribute to cell-specific expression of the mouse Follicle-stimulating hormone-beta gene. Mol. Endocrinol. 17, 1470–1483. doi: 10.1210/me.2002-0286
Judelson, H. S. (1997). Expression and inheritance of sexual preference and selfing potential in Phytophthora infestans. Fungal Genet. Biol. 21, 188–197.
Judelson, H. S., Tyler, B. M., and Michelmore, R. W. (1991). Transformation of the oomycete pathogen, Phytophthora infestans. Mol. Plant Microb. Interact. 4, 602–607. doi: 10.1094/mpmi-4-602
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Ko, W. H. (1978). Heterothallic Phytophthora: evidence for hormonal regulation of sexual reproduction. J. General Microbiol. 107, 15–18.
Ko, W. H. (1980). Hormonal regulation of sexual reproduction in Phytophthora. Microbiol. 116, 459–463.
Ko, W. H. (2007). Hormonal regulation of sexual reproduction in Phytophthora. Bot. Stud. 48, 365–375.
Kong, L., Qiu, X., Kang, J., Wang, Y., Chen, H., Huang, J., et al. (2017). A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Curr. Biol. 27, 981–991. doi: 10.1016/j.cub.2017.02.044
Li-Beisson, Y., Pollard, M., Sauveplane, V., Pinot, F., Ohlrogge, J., and Beisson, F. (2009). Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc. Natl. Acad. Sci. U.S.A. 106, 22008–22013. doi: 10.1073/pnas.0909090106
Lichtenthaler, H. K. (2000). Non-mevalonate isoprenoid biosynthesis: enzymes, genes and inhibitors. Biochem. Soc. Trans. 28, 785–789.
Maity, S. N. (2017). NF-Y (CBF) regulation in specific cell types and mouse models. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 598–603. doi: 10.1016/j.bbagrm.2016.10.014
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Matías-Hernández, L., Jiang, W., Yang, K., Tang, K., Brodelius, P. E., and Pelaz, S. (2017). AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. Plant J. 90, 520–534. doi: 10.1111/tpj.13509
McGarvey, D. J., and Croteau, R. (1995). Terpenoid metabolism. Plant Cell 7, 1015–1026. doi: 10.1105/tpc.7.7.1015
Mcleod, A., Fry, B. A., Zuluaga, A. P., Myers, K. L., and Fry, W. E. (2008). Toward improvements of oomycete transformation protocols. J. Eukaryot. Microbiol. 55, 103–109. doi: 10.1111/j.1550-7408.2008.00304.x
Mukherjee, K., Fischer, R., and Vilcinskas, A. (2012). Histone acetylation mediates epigenetic regulation of transcriptional reprogramming in insects during metamorphosis, wounding and infection. Front. Zool. 9:25. doi: 10.1186/1742-9994-9-25
Ojika, M., Molli, S. D., Kanazawa, H., Yajima, A., Toda, K., Nukada, T., et al. (2011). The second Phytophthora mating hormone defines interspecies biosynthetic crosstalk. Nat. Chem. Biol. 7:591. doi: 10.1038/nchembio.617
Pfannenstiel, B. T., Greco, C., Sukowaty, A. T., and Keller, N. P. (2018). The epigenetic reader SntB regulates secondary metabolism, development and global histone modifications in Aspergillus flavus. Fungal Genet. Biol. 120, 9–18. doi: 10.1016/j.fgb.2018.08.004
Pflum, M. K., Tong, J. K., Lane, W. S., and Schreiber, S. L. (2001). Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J. Biol. Chem. 276, 47733–47741. doi: 10.1074/jbc.M105590200
Qi, J., Asano, T., Jinno, M., Matsui, K., Atsumi, K., Sakagami, Y., et al. (2005). Characterization of a Phytophthora mating hormone. Science 309:1828. doi: 10.1126/science.1114756
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rohmer, M. (1999). The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 16, 565–574. doi: 10.1039/a709175c
Sogin, M. L., and Silberman, J. D. (1998). Evolution of the protists and protistan parasites from the perspective of molecular systematics. Int. J. Parasitol. 28, 11–20. doi: 10.1016/s0020-7519(97)00181-1
Tang, Y., Liu, X., Liu, X., Li, Y., Wu, K., and Hou, X. (2017). Arabidopsis NF-YCs mediate the light-controlled hypocotyl elongation via modulating histone acetylation. Mol. Plant. 10, 260–273. doi: 10.1016/j.molp.2016.11.007
Tao, R., Xiong, X., DePinho, R. A., Deng, C.-X., and Dong, X. C. (2013). Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J. Lipid Res. 54, 2745–2753. doi: 10.1194/jlr.M039339
Thiagalingam, S., Cheng, K. H., Lee, H. J., Mineva, N., Thiagalingam, A., and Ponte, J. F. (2003). Histone deacetylases: unique players in shaping the epigenetic histone code. Ann. N. Y. Acad. Sci. 983, 84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tyler, B. M., Sucheta, T., Xuemin, Z., Paramvir, D., Jiang, R. H. Y., Andrea, A., et al. (2006). Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266. doi: 10.1126/science.1128796
Wang, X. W., Guo, L. Y., Han, M., and Shan, K. (2016). Diversity, evolution and expression profiles of histone acetyltransferases and deacetylases in oomycetes. BMC Genomics 17:927. doi: 10.1186/s12864-016-3285-y
Xiang, Q., and Judelson, H. S. (2014). Myb transcription factors and light regulate sporulation in the oomycete Phytophthora infestans. PLoS One 9:e92086. doi: 10.1371/journal.pone.0092086
Yoon, H. S., Hackett, J. D., Pinto, G., and Bhattacharya, D. (2002). The single, ancient origin of chromist plastids. Proc. Natl. Acad. Sci. U.S.A. 99, 15507–15512. doi: 10.1073/pnas.242379899
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Keywords: oomycetes, epigenetic mechanisms, oospores, α hormones, P450, transcription factors, RNA-seq
Citation: Wang X-W, Lv J-L, Shi Y-R and Guo L-Y (2020) Comparative Transcriptome Analysis Revealed Genes Regulated by Histone Acetylation and Genes Related to Sex Hormone Biosynthesis in Phytophthora infestans. Front. Genet. 11:508. doi: 10.3389/fgene.2020.00508
Received: 26 February 2020; Accepted: 27 April 2020;
Published: 21 May 2020.
Edited by:
Meixiang Zhang, Nanjing Agricultural University, ChinaReviewed by:
Xiao-Ren Chen, Yangzhou University, ChinaYu Du, Northwest A&F University, China
Yonglin Wang, Beijing Forestry University, China
Copyright © 2020 Wang, Lv, Shi and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Yun Guo, cHBndW9AY2F1LmVkdS5jbg==; cHBndW9seUAxMjYuY29t
†These authors have contributed equally to this work
 Xiao-Wen Wang
Xiao-Wen Wang Jia-Lu Lv
Jia-Lu Lv Ya-Ru Shi
Ya-Ru Shi Li-Yun Guo
Li-Yun Guo