
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Genet. , 28 April 2020
Sec. Evolutionary and Genomic Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00438
This article is part of the Research Topic New Advances in Genetic Studies to Understand Yeast Adaptation to Extreme and Fermentative Environments View all 13 articles
Overexpression of MSN2, which is the transcription factor gene in response to stress, is well-known to increase the tolerance of the yeast Saccharomyces cerevisiae cells to a wide variety of environmental stresses. Recent studies have found that the Msn2 is a feasible potential mediator of proline homeostasis in yeast. This result is based on the finding that overexpression of the MSN2 gene exacerbates the cytotoxicity of yeast to various amino acid analogs whose uptake is increased by the active amino acid permeases localized on the plasma membrane as a result of a dysfunctional deubiquitination process. Increased understanding of the cellular responses induced by the Msn2-mediated proline incorporation will provide better comprehension of how cells respond to and counteract to different kinds of stimuli and will also contribute to the breeding of industrial yeast strains with increased productivity.
Cells of microorganisms are frequently exposed to a variety of stresses in their environment, such as prolonged nutrient starvation, free radicals and toxic molecules, imbalances in osmotic pressure and pH level, and non-optimal growth temperatures (Mager and De Kruijff, 1995; Ruis and Schüller, 1995). To survive in the face of such threatening and detrimental surroundings, eukaryotic cells have evolved a series of defense mechanisms at the transcriptional, protein, as well as metabolic levels (Boy-Marcotte et al., 1998; Toone and Jones, 1998; Estruch, 2000; Gasch et al., 2000; Kandror et al., 2004). External stimuli are perceived and transduced via the signal transduction pathways to cause global remodeling of gene expression, which is governed by transcriptional activators and repressors. In general, cellular stresses severely affect both transcription and translation activities, resulting in the inhibition of de novo protein synthesis. Moreover, environmental fluctuations may cause protein-related damages, such as the inhibition of enzyme activities, destabilization of cellular structures, and instability of chemical gradients, which eventually result in cell disruption. Thus, protein quality control and protein homeostasis are essential prerequisites for stress responses. Under harsh stresses, cells also undergo the systematic downregulation of energy-producing and energy-consuming processes in order to enter into a quiescent state, often accompanied by a dynamic shift in the central metabolic pathways that convert nutrients into energy and biomass. Cells possess tight and precise regulation systems to coordinate all the changes that are interconnected at those different levels.
In recent years, extensive research advances have been made in the field of stress responses using a eukaryotic model organism, the budding yeast Saccharomyces cerevisiae (Causton et al., 2001; Gasch, 2003). Earlier studies revealed the importance of the highly conserved stress-responsive transcription factors. Heat-shock factor 1 (Hsf1) was identified as a transcription activator that governs the expression of heat-shock proteins in response to elevated temperature (Sorger, 1990; Smith and Yaffe, 1991). The basic leucine-zipper transcription factor Yap1 is required for the induction of stress-responsive genes under oxidative stress conditions (Harshman et al., 1988; Moye-Rowley et al., 1989). Notably, S. cerevisiae cells have also developed species-specific transcription factors, namely Msn2 and Msn4 (Msn2/4) (Estruch and Carlson, 1993; Martínez-Pastor et al., 1996; Görner et al., 1998). Msn2/4 play pivotal roles in stress responses through the activation of hundreds of stress-related genes as a consequence to various stress conditions (Estruch, 2000; Gasch et al., 2000; Hasan et al., 2002; Berry and Gasch, 2008).
S. cerevisiae cells are also equipped with stress response mechanisms at the protein level to ensure protein quality at different subcellular locations, such as the cytosol (Hiraishi et al., 2009; Nillegoda et al., 2010; Theodoraki et al., 2012), endoplasmic reticulum (Brodsky, 2012; Thibault and Ng, 2012; Gardner et al., 2013; Wu et al., 2014), nucleus (Gardner et al., 2005; Rosenbaum and Gardner, 2011), mitochondria (Haynes and Ron, 2010; Baker and Haynes, 2011), and plasma membrane (Zhao et al., 2013; MacGurn, 2014; Shiga et al., 2014). The protein quality control includes all processes that ensure proper protein folding and thus prevent the toxic consequences of protein misfolding (Goldberg, 2003; Turcu et al., 2009). Irreversibly damaged proteins are selectively and effectively removed through proteasomal and/or vacuolar degradation systems, both of which consist of multiple fine-tuned steps including protein ubiquitination and deubiquitination (Finley et al., 2012).
Intracellular metabolism is dynamically changed in response to various stresses in S. cerevisiae, as well as in many other organisms. When the nutrient levels (e.g., carbon and nitrogen sources) are reduced, yeast cells reprogram the modes of energy metabolism from fermentation to respiration, a process termed a diauxic shift, in order to maximize the efficiency of energy production (Gray et al., 2004). Simultaneously, cells accumulate storage carbohydrates, such as trehalose and glycogen, which improve their survival rates under stress conditions and extend their life span (Fontana et al., 2010). In addition to carbon metabolites, nitrogen metabolites are consumed and produced in response to external stimuli. Recent studies have reported the significant importance of amino acids not only as building blocks of proteins but also in the control of cellular physiology (Sharma and Dietz, 2006; Takagi, 2008; Zhang et al., 2017). For instance, proline is accumulated and acts as an osmoprotectant in many plant and bacterial cells in response to osmotic stress (Csonka and Hanson, 1991; Kavi Kishor and Sreenivasulu, 2014). Besides that, glutamine plays a substantial role in mammalian cell growth where it facilitates the transport of other amino acids such as leucine into the cells and subsequently activates mTORC1 (Gonzalez and Hall, 2017). Collectively, these multiple regulatory mechanisms at the transcriptional, protein, and metabolic levels constitute a network that protects yeast cells from harmful conditions and allows them to adapt to new environments.
The environmental stress response (ESR) controlled by the yeast-specific transcription factors Msn2 and Msn4 (Msn2/4) includes responses to various stresses, such as oxidative stress, osmotic shock, glucose starvation, high ethanol concentrations, temperature upshift, and freezing stress (Gasch et al., 2000; Izawa et al., 2007; Sadeh et al., 2011, 2012; Sasano et al., 2012a,b); together, these constituent responses of the ESR are required for both acute stress responses and cell survival during prolonged stress (Reiter et al., 2013). Although the Msn2/4 proteins were first reported to be 41% identical to each other and functionally redundant (Estruch and Carlson, 1993), subsequent studies demonstrated that Msn2 and Msn4 individually can induce the expression of different set of genes under certain stress conditions (Gasch, 2003; Watanabe et al., 2007; Berry and Gasch, 2008). Additionally, while the MSN2 gene is constitutively expressed, transcription of the MSN4 gene is induced by stress in an Msn2/4-dependent manner (Gasch et al., 2000). Thus, the roles of Msn2/4 are mostly overlapped but can be distinguished in part. Several studies suggest that Msn2 plays a role in transcriptional repression as well. The repression likely occurs via gene expression for transcription repressors or growth inhibitors. Msn2 activates the transcription of the DOT1 gene, which encodes a repressor of the ribosome biogenesis gene (Elfving et al., 2014). Transcription of the XBP1 gene, which encodes a repressor of cell-cycle associated genes, is also Msn2-dependent (Miles et al., 2013).
Under non-stress growth conditions, Msn2/4 are phosphorylated by cAMP-dependent protein kinase A (PKA) and reside in the cytoplasm. Once yeast cells are challenged by environmental perturbations, Msn2/4 are rapidly dephosphorylated and translocated into the nucleus (Görner et al., 1998; Beck and Hall, 1999). They then bind to the stress-response element sequence (STRE; AGGGG) in the promoter region of the target genes and subsequently activate the transcription (Boy-Marcotte et al., 1998, 1999; Gasch et al., 2000; Causton et al., 2001). Previous studies identified functional domains of Msn2, which include the C-terminal zinc finger DNA-binding domain (DBD) (Marchler et al., 1993; Martínez-Pastor et al., 1996; Schmitt and McEntee, 1996; Moskvina et al., 1998), the nuclear localization signal (NLS) region (Görner et al., 1998, 2002), the nuclear export signal (NES) region (Görner et al., 1998), and the imperative transcriptional activating domain (TAD) at the N terminus (Boy-Marcotte et al., 2006). In addition to phosphorylation by PKA, multiple upstream pathways are involved in the regulation of Msn2 and/or Msn4: the target-of-rapamycin (TOR) signaling-dependent cytoplasmic localization (Beck and Hall, 1999), the karyopherin Msn5-dependent nuclear export (Chi et al., 2001; Görner et al., 2002), proteasome-mediated degradation (Durchschlag et al., 2004), the ubiquitin ligase Rsp5-dependent nuclear export of mRNA (Haitani and Takagi, 2008), and the protein kinase Rim15-dependent phosphorylation (Lee et al., 2013).
To understand how Msn2/4 contribute to stress responses, the downstream target genes of Msn2/4 have been comprehensively investigated. First, Msn2/4 directly induce the expression of the genes encoding antioxidant enzymes, such as CTT1 (for catalase), SOD1 and SOD2 (for superoxide dismutases), and PRX1 and TSA2 (for thiol peroxidases) (Hasan et al., 2002; Drakulic et al., 2005; Sadeh et al., 2011). Since various kinds of stresses lead to the imbalanced generation of reactive oxygen species (ROS) causing cell death, the elimination of ROS by the antioxidant enzymes is an important stress response. Second, Msn2/4 are essential to the induction of the genes involved in protein quality control, such as the molecular chaperone gene HSP12, the sHSP-family genes HSP26 and HSP42, the HSP70-family genes SSA1 and SSA4, and the HSP90-family genes HSP82 and HSP104 (Kandror et al., 2004; Eastmond and Nelson, 2006). Under stress conditions, the expression of the polyubiquitin precursor gene UBI4 is also upregulated to mark proteins for selective degradation via the ubiquitin-proteasome system (Simon et al., 1999). Third, Msn2/4 triggers metabolic reprogramming in response to stress by inducing the expression of the mitochondrial respiratory genes COX5b, COX17, and COX20, the pentose phosphate pathway genes SOL4, GND2, and TKL2, the trehalose synthetic genes TPS1, TPS2, TPS3, and TSL1, and the glycogen synthetic genes GSY1, GSY2, and GLC3 (Estruch, 2000; Gasch et al., 2000; Causton et al., 2001; Sadeh et al., 2011).
Overexpression of MSN2 or MSN4 has been a promising approach for the construction of industrial yeast strains to improve their stress resistance as well as fermentation ability (Cardona et al., 2007; Watanabe et al., 2009; Sasano et al., 2012a,d). For instance, baker's yeast cells are challenged by a variety of baking-associated stresses during dough fermentation, including freeze-thaw, air-drying, and a high sugar content, which trigger the oxidative stress due to the accumulation of intracellular ROS caused by protein misfolding and mitochondrial damage (Kitagaki and Takagi, 2014). Our previous study reported that baker's yeast cells that overexpressed MSN2 have shown a higher tolerance to freezing stress and enhanced the ability of yeast cells to ferment productively in frozen dough (Sasano et al., 2012a). Furthermore, in second-generation bioethanol production with lignocellulosic biomass, several growth/fermentation inhibitors such as furfural and 5-hydroxymethylfurfural are generated and they are known to produce ROS (Allen et al., 2010). The overexpression of MSN2 in bioethanol yeast strains that were grown in the presence of furfural upregulates the transcription of antioxidant gene, which in turn increases the cell resistance and leads to recovery of cell growth (Sasano et al., 2012d). Our recent study also showed that the overexpression of MSN2 shortens the replicative lifespan of yeast cells by reducing the intracellular proline levels (Mukai et al., 2019).
Among major carbon and nitrogen metabolites, Takagi's laboratory has focused on amino acids as hallmarks and mediators of S. cerevisiae stress responses (Takagi et al., 2000; Morita et al., 2002; Matsuura and Takagi, 2005; Kaino et al., 2008; Takagi, 2008; Nishimura et al., 2010). Amino acids essentially serve as a nitrogen source and the building blocks of proteins in yeast. Furthermore, they also contribute to the proliferation and durability of yeast cells following exposure to stresses. For instance, proline and charged amino acids such as arginine and glutamate were suggested to be pivotal for cell resistance in yeast and Escherichia coli cells, as these amino acids can inhibit the denaturation of proteins (Takagi et al., 1997; Morita et al., 2002; Shiraki et al., 2002; Chattopadhyay et al., 2004; Golovanov et al., 2004).
In terms of stress-resistance activity, proline is one of the most studied among the 20 naturally occurring amino acids. It has a cryoprotective activity in S. cerevisiae cells, as well as in many other kinds of cells (Sleator and Hill, 2001; Maggio et al., 2002; Krishnan et al., 2008; Liang et al., 2013). Although proline synthesis is not induced in response to stress, intracellular proline accumulation via the engineering or modification of the enzymes involved in the proline synthesis and degradation pathway in industrial baker's yeast strains elevates the resistance of freeze-thaw stress, leading to an enhanced fermentation ability in frozen dough (Kaino et al., 2008; Sasano et al., 2012c; Tsolmonbaatar et al., 2016). In addition to the freeze-thaw stress tolerance, proline confers tolerance to high osmolality, desiccation, high concentrations of ethanol, and weak acids (Takagi et al., 2000, 2016; Sasano et al., 2012b). Although the involvement of proline carries considerable importance in general stress responses, the roles played by Msn2/4 that link to the proline homeostasis is poorly understood.
The concerted processes of biosynthesis, degradation, and transport of proline administer the cellular proline homeostasis. Proline, which is incorporated into proteins, is synthesized from glutamate in three enzymatically catalyzed steps. First, the γ-glutamyl kinase Pro1 catalyzes the conversion of glutamate to glutamate-5-phosphate (Brandriss, 1979). Then, the unstable glutamate-5-phosphate is converted to glutamate semialdehyde by the γ-glutamyl phosphate reductase Pro2 (Tomenchok and Brandriss, 1987). Glutamate semialdehyde, then spontaneously cyclizes to form Δ1-pyrroline-5-carboxylate (P5C), which is converted to proline by the P5C reductase Pro3 (Brandriss and Falvey, 1992). Pro1 is sensitive to proline feedback inhibition, and thus, several known amino acid changes, such as Ile150Thr and Asp154Asn, in Pro1 alleviate feedback inhibition and elevate the level of intracellular proline (Morita et al., 2003; Sekine et al., 2007). At the transcriptional level, only the expression of the PRO2 gene is under the general amino acid control system (Natarajan et al., 2001), and it is still unknown whether the proline-synthetic pathway genes are coordinately transcribed by a certain external stimulus. To assimilate proline as a nitrogen source, it is degraded into glutamate via the proline oxidase Put1 and the P5C dehydrogenase Put2, both of which are mitochondrial enzymes (Brandriss, 1979). Loss of the Put1 function contributes to an increase of the intracellular proline content (Takagi et al., 2000). Nitrogen catabolite repression (NCR) transcriptionally represses both the PUT1 and PUT2 genes (Hofman-Bang, 1999; Georis et al., 2009), which are positively regulated by the transcription activator Put3 (Ann et al., 1996). NCR prevents the utilization of proline as a nitrogen source when rich nitrogen compounds, such as ammonia and glutamine, are present.
In S. cerevisiae, the amino-acid-polyamine-organocation (APC) superfamily consists of 24 permease proteins whose function is to transport amino acids and other amines into the cells (Nelissen et al., 1997; Jack et al., 2000). Four of them, namely Gap1, Put4, Agp1, and Gnp1 are responsible to incorporate proline (Andréasson et al., 2004). Gap1 encodes a high capacity transporter for all naturally occurring amino acids and is regulated by the quality of the nitrogen source present in the growth medium (Grenson et al., 1970; Chen and Kaiser, 2002). Put4 is required for the high-affinity transport of proline and is regulated at the transcriptional level by NCR (Xu et al., 1995; Ter Schure et al., 2000). On the other hand, Agp1 and Gnp1 encode permeases with broad substrate specificity and a high affinity for glutamine, respectively (Zhu et al., 1996; Iraqui et al., 1999). The AGP1 and GNP1 genes are induced by the regulation of the Ssy1-Ptr3-Ssy5 amino acid sensor complex (Didion et al., 1998; Iraqui et al., 1999; Forsberg et al., 2001; Ljungdahl, 2009).
Structural analogs of amino acids have been widely used to analyze amino acid homeostasis. L-azetidine-2-carboxylic acid (AZC), a toxic analog for proline, is used in both fundamental and applied researches, as it has been proven to be beneficial to study the cellular metabolism and the production of macromolecules in both prokaryotes and eukaryotes (Bach and Takagi, 2013). AZC is a non-protein amino acid originally found in plants and has a heterocyclic structure with a four-membered nitrogen ring and a carboxylic acid group on one of the ring carbon atoms. The main difference between AZC and proline is that the former has a four-ring member while the latter has a five-ring member. AZC, as well as many other amino acid analogs, is thought to be toxic to cells, because it is carried into the cells through proline permeases, and competes with proline during incorporation into nascent proteins, which consequently causes protein misfolding and cell death (Trotter et al., 2001; Weids and Grant, 2014).
Our recent study found that overexpression of MSN2 increased the sensitivity of yeast cells to several toxic amino acid analogs, namely AZC, o-fluoro-DL-phenylalanine (OFP), and L-canavanine (Can), which are known to be the analogs of proline, phenylalanine, and arginine, respectively (Mat Nanyan et al., 2019a). This suggests that MSN2 overexpression negatively controls the growth and survival of yeast cells in media containing those toxic compounds, indicating that overexpression of MSN2 is involved in the incorporation of amino acids into the cells (Mat Nanyan et al., 2019a). Further investigation showed that the increased AZC sensitivity in MSN2-overexpressing (MSN2-OE) cells could be due to the increased incorporation of AZC, since higher AZC levels were detected in MSN2-OE cells than that observed in wild-type cells (Mat Nanyan et al., 2019a). Not only AZC, but also the overexpression of MSN2 increased proline levels shortly after the addition of proline into the culture (Mat Nanyan et al., 2019a). Our study also found that quadruple disruption of proline permease genes (GAP1, PUT4, AGP1, and GNP1) in MSN2-OE cells with the strain CAY29 background conferred higher resistance to AZC (Mat Nanyan et al., 2019a), similar to that observed in wild-type cells with the same quadruple disruption (Andréasson et al., 2004). Moreover, a single disruption of GNP1 showed the most striking effect among single deletions of the permease genes, where Δgnp1 cells showed a higher resistance against AZC, highlighting a predominant role played by Gnp1 in the AZC incorporation as compared to the other three permeases, which suggest that they may have redundant roles (Mat Nanyan et al., 2019a). Consistently, the overexpression or deletion of GNP1 has no significant effect on the growth of yeast when grown in the media containing OFP or Can because Gnp1 does not incorporate phenylalanine (transported by Gap1, Agp1, and Bap2) or arginine (transported by Can1; Ljungdahl and Daignan-Fornier, 2012). Intriguingly, although the transcription of the typical Msn2-targeted gene CTT1 was highly upregulated under MSN2 overexpression, none of the levels of proline permease gene mRNA transcripts were significantly upregulated in MSN2-OE cells compared to that of in the wild-type cells, which signifies that GNP1 or other proline permease genes are not transcriptionally activated by Msn2 (Mat Nanyan et al., 2019a). More importantly, the Gnp1-GFP signal is highly detected in MSN2-OE cells, which is largely distributed on the plasma membrane rather than other parts of the cells (Mat Nanyan et al., 2019a). Thus, these results indicate that the endocytic degradation of Gnp1 was defective under MSN2 overexpression, which highlights the newly discovered network between the ESR which is mediated by Msn2 and proline homeostasis in yeast. It would be intriguing to further examine whether proline permeases and intracellular proline homeostasis under different stress conditions are enhanced via the blocking of endocytosis.
Ubiquitination and deubiquitination are involved in the control of endocytic degradation of amino acid permeases localized on the plasma membrane (Springael and André, 1998; Kimura et al., 2009; Kimura and Tanaka, 2010; Jones et al., 2012; MacGurn, 2014). Since the E3 ubiquitin ligase Rsp5 is responsible for regulating the localization of Gnp1 (Sasaki and Takagi, 2013), MSN2 overexpression might negatively control the protein ubiquitination mediated by Rsp5. The expression of the kinase gene NPR1 or other associated inhibitor genes for Rsp5-dependent protein ubiquitination might be induced as a result of MSN2 overexpression, and could thereby inhibit the endocytosis of Gnp1 (Figure 1). Although it is still hard to predict the target of Msn2 that is responsible for the endocytosis of Gnp1, it is crucial to explore and determine the target of many regulators, including the TORC1 pathway, Rsp5, and endocytosis. Interestingly, our recent study demonstrated that yeast cells overexpressing MSN2 exhibited a higher level of ubiquitinated proteins and a shortage of free ubiquitin content (Mat Nanyan et al., 2019b), similar to what is observed in Δubp6 cells, where the absence of UBP6 gene encoding one of the yeast deubiquitinating enzymes (DUBs) and is classified into ubiquitin-specific-protease (USP) family caused depletion of free cellular ubiquitin (Chernova et al., 2003; Hanna et al., 2003). These results suggest that the excess level of Msn2 reduces or impairs the activity of DUBs, which could result in the inhibition of endocytosis of Gnp1 (Mat Nanyan et al., 2019b). Further investigations showed that the transcription of UBP6 and other DUB genes was not significantly changed in MSN2-OE cells, indicating that Msn2 does not repress UBP6 and other DUB genes transcriptionally (Mat Nanyan et al., 2019b). The disruption of UBP6 in yeast cells makes them more susceptible to the toxicity of translational inhibitors such as cycloheximide and the toxic arginine analog, Can, which are caused by a deficiency of free ubiquitin (Chernova et al., 2003; Hanna et al., 2003). Intriguingly, deletion of UBP6, UBP3 as well as OTU1 genes aggravates the growth inhibitory effect of toxic amino acid analogs in yeast cells (Mat Nanyan et al., 2019b). In particular, disruption of UBP6 led to the most remarkable sensitivity toward AZC, OFP, and Can (Mat Nanyan et al., 2019b). These phenomena were also observed in MSN2-OE cells, suggesting that the excess level of Msn2 and Ubp6 deficiency confer a common phenotype of defective resistance against toxic amino acid analogs. The combination of the MSN2 overexpression and UBP6 disruption resulted in growth similar to that seen with Δubp6 cells in the presence of low concentrations of AZC (Mat Nanyan et al., 2019b). Moreover, the co-overexpression of MSN2 and UBP6 increased the resistance to AZC, OFP, and Can (Mat Nanyan et al., 2019b). Further investigations should be carried out to clarify whether Msn2 and Ubp6 affect AZC resistance via the same mechanism or whether Ubp6 functions independently to counteract the toxicity of amino acid analogs (Figure 1). Intriguingly, the Gnp1-GFP signals were elevated in Δubp6 cells and were mostly localized on the plasma membrane, similar to what was observed in MSN2-OE cells (Mat Nanyan et al., 2019b). The impaired deubiquitination mediated by an excess of Msn2 might cause the inhibition of the endocytic degradation of proline permeases by unknown mechanisms. We propose here a novel role of Msn2 in the control of intracellular uptake of proline (Figure 1). Msn2 is known for its global effect in the control of various regulatory networks in the face of stress conditions such as regulating the antioxidant enzyme genes and molecular chaperones, as well as reprogramming the carbon metabolism. In addition to that, the novel link suggested between Msn2 and proline uptake might further contribute to a deeper comprehension of global stress responses in S. cerevisiae in order to withstand various fluctuating growth conditions.
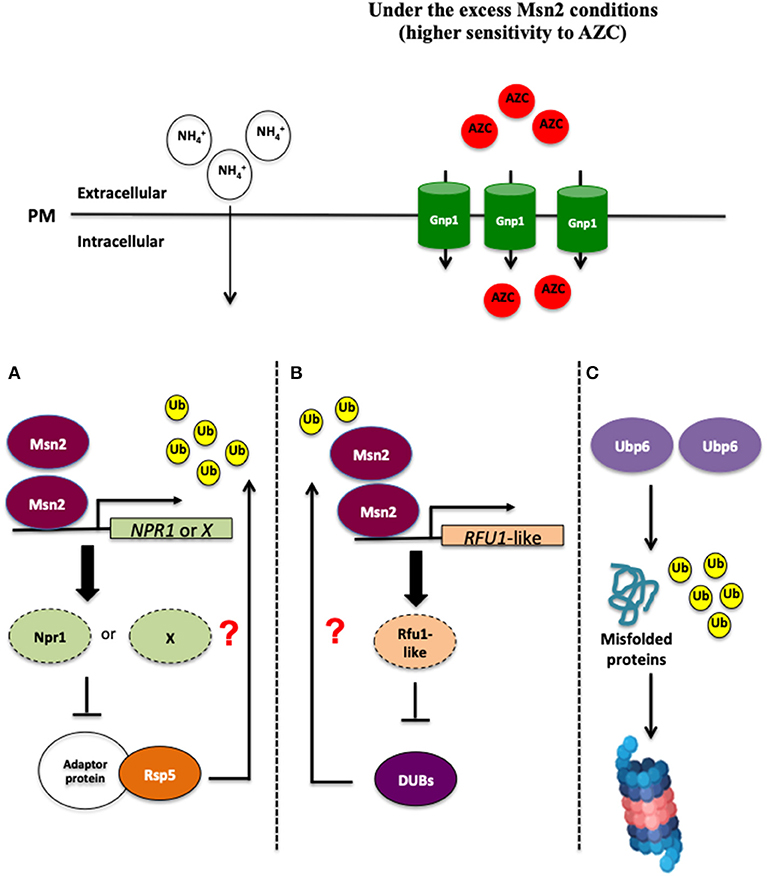
Figure 1. An excess of Msn2 inhibits the endocytic degradation of Gnp1 by unknown mechanisms. There are at least three plausible mechanisms, which could explain this phenomenon. (A) Overexpression of MSN2 might induce the expression of NPR1 or the associated inhibitor genes for Rsp5-independent protein ubiquitination, therefore Gnp1 could not be ubiquitinated and endocytosed. (B) Secondly, a high level of Msn2 may activate the gene expression of DUB repressors such as Rfu1-like protein(s), leading to a reduced or loss function of DUBs. (C) In yeast cells overexpressing UBP6, an excess of Ubp6 may enhance the ubiquitination and accelerate the degradation of misfolded proteins by unknown mechanisms, for example, the proteasome-mediated mechanism in the MSN2-independent manner, leading to the tolerance toward proteotoxic stress caused by intracelullar accumulation of the misfolded proteins.
In this mini-review, we discuss the current understanding of the stress-responsive transcription factor Msn2 and the stress response in yeast. In particular, the overexpression of MSN2 has been shown to increase the tolerance of yeast cells for various kinds of stress conditions, such as oxidative and freezing stresses. In addition, we shed light on a potentially important link, namely that the inhibition of endocytic degradation mediated by Msn2 plays an essential role in regulating the proline homeostasis under stress conditions, evidently caused by the loss of function of DUBs. Further investigations will elucidate the profound relationship among Msn2, DUBs, and Gnp1 in the regulation of proline homeostasis, which may serve as a foundation to engineer more robust industrial yeast strains, which might be advantageous in fermentation industries.
NM and HT substantially and intellectually contribute to this work, and reviewed and approved the final version of manuscript.
The study was partially supported by a grant from the Project of the NARO Bio-oriented Technology Research Advancement Institution (Research program on development of innovative technology) to HT under grant number 30017B.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Allen, S. A., Clark, W., McCaffery, J. M., Cai, Z., Lanctot, A., Slininger, P. J., et al. (2010). Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol. Biofuels. 3:2. doi: 10.1186/1754-6834-3-2
Andréasson, C., Neve, E. P. A., and Ljungdahl, P. O. (2004). Four permeases import proline and the toxic proline analogue azetidine-2-carboxylate into yeast. Yeast 21, 193–199. doi: 10.1002/yea.1052
Ann, S., Etages, G., Fahey, D. A., Reecet, R. J., and Brandriss, M. C. (1996). Functional analysis of the PUT3 transcriptional activator of the proline utilization pathway in Saccharomyces cerevisiae. Genetics 142, 1069–1082. doi: 10.1002/zamm.19660460126
Bach, T. M. H., and Takagi, H., (2013). Properties, metabolisms, and applications of L-proline analogues. Appl. Microbiol. Biotechnol. 97, 6623–6634. doi: 10.1007/s00253-013-5022-7
Baker, B. M., and Haynes, C. M. (2011). Mitochondrial protein quality control during biogenesis and aging. Trends Biochem. Sci. 36, 254–261. doi: 10.1016/j.tibs.2011.01.004
Beck, T., and Hall, M. N. (1999). The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402, 689–692. doi: 10.1038/45287
Berry, D. B., and Gasch, A. P. (2008). Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol. Biol. Cell. 19, 4580–4587. doi: 10.1091/mbc.E07
Boy-Marcotte, E., Garmendia, C., Garreau, H., Lallet, S., Mallet, L., and Jacquet, M. (2006). The transcriptional activation region of Msn2p, in Saccharomyces cerevisiae, is regulated by stress but is insensitive to the cAMP signalling pathway. Mol. Genet. Genomics. 275, 277–287. doi: 10.1007/s00438-005-0017-4
Boy-Marcotte, E., Lagniel, G., Perrot, M., Bussereau, F., Boudsocq, A., Jacquet, M., et al. (1999). The heat shock response in yeast: Differential regulations and contributions of the Msn2p/Msn4p and Hsf1 regulons. Mol. Microbiol. 33, 274–283. doi: 10.1046/j.1365-2958.1999.01467.x
Boy-Marcotte, E., Perrot, M., Bussereau, F., Boucherie, H., and Jacquet, M. (1998). Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180, 1044–1052.
Brandriss, M. C. (1979). Isolation and preliminary characterization of Saccharomyces cerevisiae proline auxotrophs. J. Bacteriol. 138, 816–822.
Brandriss, M. C., and Falvey, D. A. (1992). Proline biosynthesis in Saccharomyces cerevisiae: molecular analysis of the PRO1 gene, which encodes γ-glutamyl kinase. J. Bacteriol. 174, 4148–4156. doi: 10.1128/jb.174.12.4148-4156.1992
Brodsky, J. L. (2012). Cleaning up : ER-associated degradation to the rescue. Cell 151, 1163–1167. doi: 10.1016/j.cell.2012.11.012
Cardona, F., Carrasco, P., Pérez-Ortín, J. E., Olmo, M. I., and Aranda, A. (2007). A novel approach for the improvement of stress resisitance in wine yeasts. Int. J. Food Microbiol. 114, 83–91. doi: 10.1016./j.ijfoodmicro.2006.10.043
Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E., Jennings, E. G., et al. (2001). Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 12, 323–337. doi: 10.1091/mbc.12.2.323
Chattopadhyay, M. K., Kern, R., Mistou, M. Y., Dandekar, A. M., Uratsu, S. L., and Richarme, G. (2004). The chemical chaperone proline relieves the thermosensitivity of a dnaK deletion mutant at 42°C. J. Bacteriol. 186, 8149–8152. doi: 10.1128/JB.186.23.8149-8152.2004
Chen, E. J., and Kaiser, C. A. (2002). Amino acids regulate the intracellular trafficking of the general amino acid permease of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 99, 14837–14842. doi: 10.1073/pnas.232591899
Chernova, T. A., Allen, K. D., Wesoloski, L. M., Shanks, J. R., Chernoff, Y. O., and Wilkinson, K. D. (2003). Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J. Biol. Chem. 278, 52102–52115. doi: 10.1074/jbc.M310283200
Chi, Y., Huddleston, M. J., Zhang, X., Young, R. A., Annan, R. S., Carr, S. A., et al. (2001). Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15, 1078–1092. doi: 10.1101/gad.867501
Csonka, L. N., and Hanson, A. D. (1991). Prokaryotic osmoregulation: genetics and physiology. Ann. Rev. Microbiol. 45, 569–606. doi: 10.1146/annurev.mi.45.100191.003033
Didion, T., Regenberg, B., Jørgensen, M. U., Kielland-Brandt, M. C., and Andersen, H. A. (1998). The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27, 643–650. doi: 10.1046/j.1365-2958.1998.00714.x
Drakulic, T., Temple, M. D., Guido, R., Jarolim, S., Breitenbach, M., Attfield, P. V., et al. (2005). Involvement of oxidative stress response genes in redox homeostasis, the level of reactive oxygen species, and ageing in Saccharomyces cerevisiae. FEMS Yeast Res. 5, 1215–1228. doi: 10.1016/j.femsyr.2005.06.001
Durchschlag, E., Reiter, W., Ammerer, G., and Schüller, C. (2004). Nuclear localization destabilizes the stress-regulated transcription factor Msn2. J. Biol. Chem. 279, 55425–55432. doi: 10.1074/jbc.M407264200
Eastmond, D. L., and Nelson, H. C. M. (2006). Genome-wide analysis reveals new roles for the activation domains of the Saccharomyces cerevisiae heat shock transcription factor (Hsf1) during the transient heat shock response. J. Biol. Chem. 281, 32909–32921. doi: 10.1074/jbc.M602454200
Elfving, N., Chereji, R. V., Bharatula, V., Bjorklund, S., Morozov, A. V., and Broach, J. R. (2014). A dynamic interplay of nucleosome and Msn2 binding regulates kinetics of gene activation and repression following stress. Nucleic Acids Res. 42, 5468–5482. doi: 10.1093/nar/gku176
Estruch, F. (2000). Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24, 469–486. doi: 10.1111/j.1574-6976.2000.tb00551.x
Estruch, F., and Carlson, M. (1993). Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol. Cell Biol. 13, 3872–3881. doi: 10.1128/MCB.13.7.3872
Finley, D., Ulrich, H. D., Sommer, T., and Kaiser, P. (2012). The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192, 319–360. doi: 10.1534/genetics.112.140467
Fontana, L., Partridge, L., and Longo, V. D. (2010). Extending healthy life span-from yeast to humans. Science 328, 321–326. doi: 10.1126/science.1172539
Forsberg, H., Gilstring, C. F., Zargari, A., Martínez, P., and Ljungdahl, P. O. (2001). The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol. Microbiol. 42, 215–228. doi: 10.1046/j.1365-2958.2001.02627.x
Gardner, B. M., Pincus, D., Gotthardt, K., Gallagher, C. M., and Walter, P. (2013). Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 5:a013169. doi: 10.1101/cshperspect.a013169
Gardner, R. G., Nelson, Z. W., and Gottschling, D. E. (2005). Degradation-mediated protein quality control in the nucleus. Cell 120, 803–815. doi: 10.1016/j.cell.2005.01.016
Gasch, A. P. (2003). “The environmental stress response : a common yeast response to diverse environmental stresses,” in Yeast Stress Response, eds S. Hohmann and W. H. Mager (Berlin: Springer-Verlag), 11–70.
Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., et al. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 11, 4241–4257. doi: 10.1091/mbc.11.12.4241
Georis, I., Feller, A., Tate, J. J., Cooper, T. G., and Dubois, E. (2009). Nitrogen catabolite repression-sensitive transcription as a readout of Tor pathway regulation: the genetic background, reporter gene and GATA factor assayed determine the outcomes. Genetics 181, 861–874. doi: 10.1534/genetics.108.099051
Goldberg, A. L. (2003). Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899. doi: 10.1038/nature02263
Golovanov, A. P., Hautbergue, G. M., Wilson, S. A., and Lian, L. Y. (2004). A simple method for improving protein solubility and long-term stability. J. Am. Chem. Soc. 126, 8933–8939. doi: 10.1021/ja049297h
Gonzalez, A., and Hall, M. N. (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 36, 397–408. doi: 10.15252/embj.201696010
Görner, W., Durchschlag, E., Martinez-Pastor, M. T., Estruch, F., Ammerer, G., Hamilton, B., et al. (1998). Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12, 586–597. doi: 10.1101/gad.12.4.586
Görner, W., Durchschlag, E., Wolf, J., Brown, E. L., Ammerer, G., Ruis, H., et al. (2002). Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21, 135–144. doi: 10.1093/emboj/21.1.135
Gray, J. V., Petsko, G. A., Johnston, G. C., Ringe, D., Singer, R. A., and Werner-Washburne, M. (2004). “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68:187206. doi: 10.1128/MMBR.68.2.187
Grenson, M., Hou, C., and Crabeel, M. (1970). Multiplicity of the amino acid permeases in Saccharomyces cerevisiae IV. Evidence for a general amino acid permease. J. Bacteriol. 103, 770–777.
Haitani, Y., and Takagi, H. (2008). Rsp5 is required for the nuclear export of mRNA of HSF1 and MSN2/4 under stress conditions in Saccharomyces cerevisiae. Genes Cells 13, 105–116. doi: 10.1111/j.1365-2443.2007.01154.x
Hanna, J., Leggett, D. S., and Finley, D. (2003). Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol. Cell. Biol. 23, 9251–9261. doi: 10.1128/MCB.23.24.9251
Harshman, K. D., Moye-Rowley, W. S., and Parker, C. S. (1988). Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell 53, 321–330. doi: 10.1016/0092-8674(88)90393-5
Hasan, R., Leroy, C., Isnard, A.-D., Labarre, J., Boy-Marcotte, E., and Toledano, M. B. (2002). The control of the yeast H2O2 response by the Msn2/4 transcription factors. Mol. Microbiol. 45, 233–241. doi: 10.1046/j.1365-2958.2002.03011.x
Haynes, C. M., and Ron, D. (2010). The mitochondrial UPR–protecting organelle protein homeostasis. J. Cell Sci. 123, 3849–3855. doi: 10.1242/jcs.075119
Hiraishi, H., Shimada, T., Ohtsu, I., Sato, T., and Takagi, H. (2009). The yeast ubiquitin ligase Rsp5 downregulates the alpha subunit of nascent polypeptide-associated complex Egd2 under stress conditions. FEBS J. 276, 5287–5297. doi: 10.1111/j.1742-4658.2009.07226.x
Hofman-Bang, J. (1999). Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol. Biotechnol. 12, 35–73. doi: 10.1385/MB:12:1:35
Iraqui, L., Vissers, S., Bernard, F., Craene, J. D. E., Boles, E., Urrestarazu, A., et al. (1999). Amino acid signaling in Saccharomyces cerevisiae : a permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19, 989–1001. doi: 10.1128/MCB.19.2.989
Izawa, S., Ikeda, K., Ohdate, T., and Inoue, Y. (2007). Msn2p/Msn4p-activation is essential for the recovery from freezing stress in yeast. Biochem. Biophys. Res. Commun. 352, 750–755. doi: 10.1016/j.bbrc.2006.11.100
Jack, D. L., Paulsen, I. T., and Saier, J. (2000). The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146, 1797–1814. doi: 10.1099/00221287-146-8-1797
Jones, C. B., Ott, E. M., Keener, J. M., Curtiss, M., Sandrin, V., and Babst, M. (2012). Regulation of membrane protein degradation by starvation-response pathways. Traffic 13, 468–482. doi: 10.1111/j.1600-0854.2011.01314.x
Kaino, T., Tateiwa, T., Mizukami-Murata, S., Shima, J., and Takagi, H. (2008). Self-cloning baker's yeasts that accumulate proline enhance freeze tolerance in doughs. Appl. Environ. Microbiol. 74, 5845–5849. doi: 10.1128/AEM.00998-08
Kandror, O., Bretschneider, N., Kreydin, E., Cavalieri, D., and Goldberg, A. L. (2004). Yeast adapt to near-freezing temperatures by STRE/Msn2,4-dependent induction of trehalose synthesis and certain molecular chaperones. Mol. Cell 13, 771–781. doi: 10.1016/S1097-2765(04)00148-0
Kavi Kishor, P. B., and Sreenivasulu, N. (2014). Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ. 37, 300–311. doi: 10.1111/pce.12157
Kimura, Y., and Tanaka, K. (2010). Regulatory mechanisms involved in the control of ubiquitin homeostasis. J. Biochem. 147, 793–798. doi: 10.1093/jb/mvq044
Kimura, Y., Yashiroda, H., Kudo, T., Koitabashi, S., Murata, S., Kakizuka, A., et al. (2009). An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell 137, 549–559. doi: 10.1016/j.cell.2009.02.028
Kitagaki, H., and Takagi, H. (2014). Mitochondrial metabolism and stress response of yeast: application in fermentation technologies. J. Biosci. Bioeng. 117:383–393. doi: 10.1016/j/jbiosc.2013.09.011
Krishnan, N., Dickman, M. B., and Becker, D. F. (2008). Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic. Biol. Med. 44, 671–681. doi: 10.1016/j.freeradbiomed.2007.10.054
Lee, P., Kim, M. S., Paik, S.-M., Choi, S.-H., Cho, B.-R., and Hahn, J.-S. (2013). Rim15-dependent activation of Hsf1 and Msn2/4 transcription factors by direct phosphorylation in Saccharomyces cerevisiae. FEBS Lett. 587, 3648–3655. doi: 10.1016/j.febslet.2013.10.004
Liang, X., Zhang, L., Natarajan, S. K., and Becker, D. F. (2013). Proline mechanisms of stress survival. Antioxid. Redox Signal. 19, 998–1011. doi: 10.1089/ars.2012.5074
Ljungdahl, P. O. (2009). Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem. Soc. Trans. 37, 242–247. doi: 10.1042/BST0370242
Ljungdahl, P. O., and Daignan-Fornier, B. (2012). Regulation of amino acid, nucleotide, and phosphate metabolisn in Saccharomyces cerevisiae. Genetics 190, 885–929. doi: 10.1534/genetics.111.133306
MacGurn, J. A. (2014). Garbage on, garbage off: new insights into plasma membrane protein quality control. Curr. Opin. Cell Biol. 29, 92–98. doi: 10.1016/j.ceb.2014.05.001
Mager, W. H., and De Kruijff, A. J. J. (1995). Stress-induced transcriptional activation. Microbiol. Rev. 59, 506–531.
Maggio, A., Miyazaki, S., Veronese, P., Fujita, T., Ibeas, J. I., Damsz, B., et al. (2002). Does proline accumulation play an active role in stress-induced growth reduction? Plant J. 31, 699–712. doi: 10.1046/j.1365-313X.2002.01389.x
Marchler, G., Schüller, C., Adam, G., and Ruis, H. (1993). A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 12, 1997–2003. doi: 10.1002/jsfa.2740301103
Martínez-Pastor, M. T., Marchler, G., Schüller, C., Marchler-Bauer, A., Ruis, H., and Estruch, F. (1996). The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15, 2227–2235. doi: 10.1002/j.1460-2075.1996.tb00576.x
Mat Nanyan, N. S., Watanabe, D., Sugimoto, Y., and Takagi, H. (2019a). Involvement of the stress-responsive transcription factor gene MSN2 in the control of amino acid uptake in Saccharomyces cerevisiae. FEMS Yeast Res. 19:foz052. doi: 10.1093/femsyr/foz052
Mat Nanyan, N. S., Watanabe, D., Sugimoto, Y., and Takagi, H. (2019b). Effect of the deubiquitination enzyme gene UBP6 on the stress-responsive transcription factor Msn2-mediated control of the amino acid permease Gnp1 in yeast. J. Biosci. Bioeng. 129, 423–427. doi: 10.1016/j.jbiosc.2019.10.002
Matsuura, K., and Takagi, H. (2005). Vacuolar functions are involved in stress-protective effect of intracellular proline in Saccharomyces cerevisiae. J. Biosci. Bioeng. 100, 538–544. doi: 10.1263/jbb.100.538
Miles, S., Li, L., Davison, J., and Breeden, L. L. (2013). Xbp1 directs global repression of budding yeast transcription during the transition to quiescence and is important for the longevity and reversibility of the quiescent state. PLoS Genet. 9:e1003854. doi: 10.1371/journal.pgen.1003854
Morita, Y., Nakamori, S., and Takagi, H. (2002). Effect of proline and arginine metabolism on freezing stress of Saccharomyces cerevisiae. J. Biosci. Bioeng. 94, 390–394. doi: 10.1016/S1389-1723(02)80214-6
Morita, Y., Nakamori, S., and Takagi, H. (2003). L-proline accumulation and freeze tolerance in Saccharomyces cerevisiae are caused by a mutation in the PRO1 gene encoding γ-glutamyl kinase. Appl. Environ. Microbiol. 69, 212–219. doi: 10.1128/AEM.69.1.212-219.2003
Moskvina, E., Schüller, C., Maurer, C. T. C., Mager, W. H., and Ruis, H. (1998). A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast 14, 1041–1050. doi: 10.1002/(SICI)1097-0061(199808)14:11<1041::AID-YEA296>3.0.CO;2-4
Moye-Rowley, W. S., Harshman, K. D., and Parker, C. S. (1989). Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 3, 283–292. doi: 10.1101/gad.3.3.283
Mukai, Y., Kamei, Y., Liu, X., Jiang, S., Sugimoto, Y., Mat Nanyan, N., et al. (2019). Proline metabolism regulates replicative lifespan in the yeast Saccharomyces cerevisiae. Microb. Cell. 6, 482–490. doi: 10.15698/mic2019.10.694
Natarajan, K., Meyer, M. R., Jackson, B. M., Slade, D., Roberts, C., Hinnebusch, A. G., et al. (2001). Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21, 4347–4368. doi: 10.1128/MCB.21.13.4347
Nelissen, B., De Wachter, R., and Goffeau, A. (1997). Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 21, 113–134. doi: 10.1016/S0168-6445(97)00053-3
Nillegoda, N. B., Theodoraki, M. A., Mandal, A. K., Mayo, K. J., Ren, H. Y., Sultana, R., et al. (2010). Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol. Biol. Cell. 21, 2102–2116. doi: 10.1091/mbc.E10
Nishimura, A., Kotani, T., Sasano, Y., and Takagi, H. (2010). An antioxidative mechanism mediated by the yeast N-acetyltransferase Mpr1: oxidative stress-induced arginine synthesis and its physiological role. FEMS Yeast Res. 10, 687–698. doi: 10.1111/j.1567-1364.2010.00650.x
Reiter, W., Klopf, E., De Wever, V., Anrather, D., Petryshyn, A., Roetzer, A., et al. (2013). Yeast protein phosphatase 2A-Cdc55 regulates the transcriptional response to hyperosmolarity stress by regulating Msn2 and Msn4 chromatin recruitment. Mol. Cell. Biol. 33, 1057–1072. doi: 10.1128/MCB.00834-12
Rosenbaum, J. C., and Gardner, R. G. (2011). How a disordered ubiquitin ligase maintains order in nuclear protein homeostasis. Nucleus 2, 264–270. doi: 10.4161/nucl.2.4.16118
Ruis, H., and Schüller, C. (1995). Stress signaling in yeast. BioEssays 17, 959–965. doi: 10.1002/bies.950171109
Sadeh, A., Baran, D., Volokh, M., and Aharoni, A. (2012). Conserved motifs in the Msn2-activating domain are important for Msn2-mediated yeast stress response. J. Cell Sci. 125, 3333–3342. doi: 10.1242/jcs.096446
Sadeh, A., Movshovich, N., Volokh, M., Gheber, L., and Aharoni, A. (2011). Fine-tuning of the Msn2/4-mediated yeast stress responses as revealed by systematic deletion of Msn2/4 partners. Mol. Biol. Cell 22, 3127–3138. doi: 10.1091/mbc.E10-12-1007
Sasaki, T., and Takagi, H. (2013). Phosphorylation of a conserved Thr357 in yeast Nedd4-like ubiquitin ligase Rsp5 is involved in down-regulation of the general amino acid permease Gap1. Genes Cells 18, 459–475. doi: 10.1111/gtc.12049
Sasano, Y., Haitani, Y., Hashida, K., Ohtsu, I., Shima, J., and Takagi, H. (2012a). Overexpression of the transcription activator Msn2 enhances the fermentation ability of industrial baker's yeast in frozen dough. Biosci. Biotechnol. Biochem. 76, 624–627. doi: 10.1271/bbb.110959
Sasano, Y., Haitani, Y., Hashida, K., Ohtsu, I., Shima, J., and Takagi, H. (2012b). Simultaneous accumulation of proline and trehalose in industrial baker's yeast enhances fermentation ability in frozen dough. J. Biosci. Bioeng. 113, 592–595. doi: 10.1016/j.jbiosc.2011.12.018
Sasano, Y., Haitani, Y., Ohtsu, I., Shima, J., and Takagi, H. (2012c). Proline accumulation in baker's yeast enhances high-sucrose stress tolerance and fermentation ability in sweet dough. Int. J. Food Microbiol. 152, 40–43. doi: 10.1016/j.ijfoodmicro.2011.10.004
Sasano, Y., Watanabe, D., Ukibe, K., Inai, T., Ohtsu, I., Shimoi, H., et al. (2012d). Overexpression of the yeast transcription activator Msn2 confers furfural resistance and increases the initial fermentation rate in ethanol production. J. Biosci. Bioeng. 113, 451–455. doi: 10.1016/j.jbiosc.2011.11.017
Schmitt, A. P., and McEntee, K. (1996). Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Biochemistry 93, 5777–5782. doi: 10.1073/pnas.93.12.5777
Sekine, T., Kawaguchi, A., Hamano, Y., and Takagi, H. (2007). Desensitization of feedback inhibition of the Saccharomyces cerevisiae γ-glutamyl kinase enhances proline accumulation and freezing tolerance. Appl. Environ. Microbiol. 73, 4011–4019. doi: 10.1128/AEM.00730-07
Sharma, S. S., and Dietz, K. J. (2006). The significance of amino acids and amino acid-derived molecules in plant responses and adaptation to heavy metal stress. J. Exp. Bot. 57, 711–726. doi: 10.1093/jxb/erj073
Shiga, T., Yoshida, N., Shimizu, Y., Suzuki, E., Sasaki, T., Watanabe, D., et al. (2014). Quality control of plasma membrane proteins by Saccharomyces cerevisiae Nedd4-like ubiquitin ligase Rsp5p under environmental stress conditions. Eukaryot. Cell 13, 1191–1199. doi: 10.1128/EC.00104-14
Shiraki, K., Kudou, M., Fujiwara, S., Imanaka, T., and Takagi, M. (2002). Biophysical effect of amino acids on the prevention of protein aggregation. J. Biochem. 132, 591–595. doi: 10.1093/oxfordjournals.jbchem.a003261
Simon, J. R., Treger, J. M., and McEntee, K. (1999). Multiple independent regulatory pathways control UBI4 expression after heat shock in Saccharomyces cerevisiae. Mol. Cell. Biol. 31, 823–832. doi: 10.1046/j.1365-2958.1999.01220.x
Sleator, R., and Hill, C. (2001). Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 25, 49–71. doi: 10.1016/S0168-6445(01)00071-7
Smith, B. J., and Yaffe, M. P. (1991). A mutation in the yeast heat-shock factor gene causes temperature-sensitive defects in both mitochondrial protein import and the cell cycle. Mol. Cell. Biol. 11, 2647–2655. doi: 10.1016/0962-8924(91)90074-J
Sorger, P. K. (1990). Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell 62, 793–805. doi: 10.1016/0092-8674(90)90123-V
Springael, J. Y., and André, B. (1998). Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol. Biol. Cell. 9, 1253–1263. doi: 10.1091/mbc.9.6.1253
Takagi, H. (2008). Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications. Appl. Microbiol. Biotechnol. 81, 211–223. doi: 10.1007/s00253-008-1698-5
Takagi, H., Iwamoto, F., and Nakamori, S. (1997). Isolation of freeze-tolerant laboratory strains of Saccharomyces cerevisiae from proline-analogue-resistant mutants. Appl. Microbiol. Biotechnol. 47, 405–411. doi: 10.1007/s002530050948
Takagi, H., Sakai, K., Morida, K., and Nakamori, S. (2000). Proline accumulation by mutation or disruption of the proline oxidase gene improves resistance to freezing and desiccation stresses in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 184, 103–108. doi: 10.1111/j.1574-6968.2000.tb08998.x
Takagi, H., Taguchi, J., and Kaino, T. (2016). Proline accumulation protects Saccharomyces cerevisiae cells in the stationary phase from ethanol stress by reducing reactive oxygen species levels. Yeast 33, 355–363. doi: 10.1002/yea.3154
Ter Schure, E. G., Van Riel, N. A. W., and Verrips, C. T. (2000). The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 24, 67–83. doi: 10.1016/S0168-6445(99)00030-3
Theodoraki, M. A., Nillegoda, N. B., Saini, J., and Caplan, A. J. (2012). A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast. J. Biol. Chem. 287, 23911–23922. doi: 10.1074/jbc.M112.341164
Thibault, G., and Ng, D. T. W. (2012). The endoplasmic reticulum-associated degradation pathways of budding yeast. Cold Spring Harb. Perspect. Biol. 4:a013193. doi: 10.1101/cshperspect.a013193
Tomenchok, D. M., and Brandriss, M. C. (1987). Gene-enzyme relationships in the proline biosynthetic pathway of Saccharomyces cerevisiae. J. Bacteriol. 169, 5364–5372. doi: 10.1128/jb.169.12.5364-5372.1987
Toone, W. M., and Jones, N. (1998). Stress-activated signalling pathways in yeast. Genes Cells 3, 485–498. doi: 10.1046/j.1365-2443.1998.00211.x
Trotter, E. W., Berenfeld, L., Krause, S. A., Petsko, G. A., and Gray, J. V. (2001). Protein misfolding and temperature up-shift cause G1 arrest via a common mechanism dependent on heat shock factor in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 98, 7313–7318. doi: 10.1073./pnas.121172998
Tsolmonbaatar, A., Hashida, K., Sugimoto, Y., Watanabe, D., Furukawa, S., and Takagi, H. (2016). Isolation of baker's yeast mutants with proline accumulation that showed enhanced tolerance to baking-associated stresses. Int. J. Food Microbiol. 238:233–240. doi: 10.1016/j.ijfoodmicro.2016.09.015
Turcu, F. E. R., Ventii, K. H., and Wilkinson, K. D. (2009). Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397. doi: 10.1146/annurev.biochem.78.082307.091526
Watanabe, M., Tamura, K., Magbanua, J. P., Takano, K., Kitamoto, K., Kitagaki, H., et al. (2007). Elevated expression of genes under the control of stress response element (STRE) and Msn2p in an ethanol-tolerance sake yeast Kyokai no. 11. J. Biosci. Bioeng. 104, 163–170. doi: 10.1263/jbb.104.163
Watanabe, M., Watanabe, D., Akao, T., and Shimoi, H. (2009). Overexpression of MSN2 in a sake yeast strain promotes ethanol tolerance and increases ethanol production in sake brewing. J. Biosci. Bioeng. 107, 516–518. doi: 10.1016/j.jbiosc.2009.01.006
Weids, A. J., and Grant, C. M. (2014). The yeast peroxiredoxin Tsa1 protects against protein-aggregate-induced oxidative stress. J. Cell. Sci. 127, 1327–1335. doi: 10.1242/jcs.144022
Wu, H., Ng, B. S. H., and Thibault, G. (2014). Endoplasmic reticulum stress response in yeast and humans. Biosci. Rep. 34, 321–330. doi: 10.1042/BSR20140058
Xu, S., Falvey, D. A., and Brandriss, M. C. (1995). Roles of URE2 and GLN3 in the proline utilization pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 2321–2330. doi: 10.1128/MCB.15.4.2321
Zhang, J., Pavlova, N. N., and Thompson, C. B. (2017). Cancer cell metabolism: the essential role of the non-essential amino acid, glutamine. EMBO J. 36, 1302–1315. doi: 10.15252/embj.201696151
Zhao, Y., MacGurn, J. A., Liu, M., and Emr, S. (2013). The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. ELife. 2, 1–18. doi: 10.7554/eLife.00459
Keywords: Saccharomyces cerevisiae, stress-responsive transcription factor, Msn2, proline homeostasis, proline permease, Gnp1, deubiquitinating enzymes, Ubp6
Citation: Mat Nanyan NSb and Takagi H (2020) Proline Homeostasis in Saccharomyces cerevisiae: How Does the Stress-Responsive Transcription Factor Msn2 Play a Role? Front. Genet. 11:438. doi: 10.3389/fgene.2020.00438
Received: 13 February 2020; Accepted: 09 April 2020;
Published: 28 April 2020.
Edited by:
Amparo Querol, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Johan M. Thevelein, KU Leuven, BelgiumCopyright © 2020 Mat Nanyan and Takagi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroshi Takagi, aGlyb0Bicy5uYWlzdC5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.