- 1Department of Immunology and Infectious Disease, The John Curtin School of Medical Research, The Australian National University, Canberra, ACT, Australia
- 2Centre for Research in Mathematics and Data Science, School of Computer, Data and Mathematical Sciences, Western Sydney University, Parramatta, NSW, Australia
- 3Australian Center for Blood Diseases, Faculty of Medicine, Nursing and Health Sciences, Monash University, Melbourne, VIC, Australia
- 4Institute of Health and Biomedical Innovation, School of Biomedical Sciences, Queensland University of Technology, Brisbane, QLD, Australia
- 5Translational Research Institute, Brisbane, QLD, Australia
- 6Centre for Chronic Disease, Faculty of Health, The University of Queensland, Brisbane, QLD, Australia
- 7Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, University of Melbourne, Melbourne, VIC, Australia
- 8Menzies School of Health Research, Darwin, NT, Australia
The F2RL3 gene encoding protease activated receptor 4 (PAR4) contains a single nucleotide variant, rs773902, that is functional. The resulting PAR4 variants, Thr120, and Ala120, are known to differently affect platelet reactivity to thrombin. Significant population differences in the frequency of the allele indicate it may be an important determinant in the ethnic differences that exist in thrombosis and hemostasis, and for patient outcomes to PAR antagonist anti-platelet therapies. Here we determined the frequency of rs773902 in an Indigenous Australian group comprising 467 individuals from the Tiwi Islands. These people experience high rates of renal disease that may be related to platelet and PAR4 function and are potential recipients of PAR-antagonist treatments. The rs773902 minor allele frequency (Thr120) in the Tiwi Islanders was 0.32, which is similar to European and Asian groups and substantially lower than Melanesians and some African groups. Logistic regression and allele distortion testing revealed no significant associations between the variant and several markers of renal function, as well as blood glucose and blood pressure. These findings suggest that rs773902 is not an important determinant for renal disease in this Indigenous Australian group. However, the relationships between rs773902 genotype and platelet and drug responsiveness in the Tiwi, and the allele frequency in other Indigenous Australian groups should be evaluated.
Introduction
Platelet activation by thrombin during hemostasis and thrombosis is mediated through two therapeutically tractable protease-activated receptors, PAR1 and PAR4. The gene encoding PAR4 (F2RL3) contains a functionally important single nucleotide polymorphism (rs773902) expressing either an Ala120 or Thr120 variant receptor. The Thr120 variant is associated with higher aggregation and Ca2+ flux induced by PAR4 agonists in isolated platelets (Edelstein et al., 2014; Whitley et al., 2018), and greater ex vivo thrombosis formation (Tourdot et al., 2018). The allele frequencies for this variant differ according to ethnicity, with the Thr120 variant relatively common in some African populations (up to 68%) and less common in Asians and Europeans (19–29%; Edelstein et al., 2014; Heenkenda et al., 2018). Previous studies comparing PAR4-dependent platelet functions in African and “white” Americans indicated distinct differences (Edelstein et al., 2013; Tourdot et al., 2014), which in other studies have been directly associated with Ala120/Thr120 variant status (Edelstein et al., 2014). The variant may also be clinically important. One study has shown a greater risk of thrombosis amongst carriers of the Thr120 in a general American population (Whitley et al., 2018). In addition, the relative benefits and risks of using the PAR1 antagonist vorapaxar may vary according to the PAR4 sequence variant, with a suggestion of lower bleeding rates in Thr120-expressing individuals treated with vorapaxar than in Ala120-expressing individuals (Tricoci et al., 2018). Furthermore, PAR4 antagonists are in clinical development (French and Hamilton, 2017; Wong et al., 2017) and the impact of the rs773902 PAR4 SNP on the efficacy and safety of these agents remains unexplored.
Indigenous Australians, who represent a distinct population with ancestral and genetic histories that predate all cultures outside of Africa, suffer very high burdens of cardiovascular disease, stroke, diabetes, and chronic kidney disease (CKD; Australian Institute of Health and Welfare, 2016). For example, diabetes deaths in Indigenous Australians are increased 15-fold, cardiovascular deaths by 3–6 fold (Hoy, 1996), and rates of treated end-stage renal disease are 20 times more than in non-Indigenous Australians (Spencer et al., 1998). Studies in these people have also reported familial aggregation amongst cases of CKD (Van Buynder et al., 1993), and significant genetic heritability and genetic associations with markers of renal function (Duffy et al., 2016; Thomson et al., 2019). Both platelets and PAR4 have been implicated in kidney function and CKD pathogenesis (Lambert, 2016; Madhusudhan et al., 2016; Palygin et al., 2016), and renal disease patients are prone to bleeding and thrombotic events (Saheb Sharif-Askari et al., 2017). However, the allele frequencies of rs773902 have not been investigated in Indigenous Australians. Therefore, we determined the genotype and allele frequencies of rs773902 in an Indigenous Australian group native to The Tiwi Islands and analyzed the associations between this variant and markers of renal disease.
Methods
Study Participants
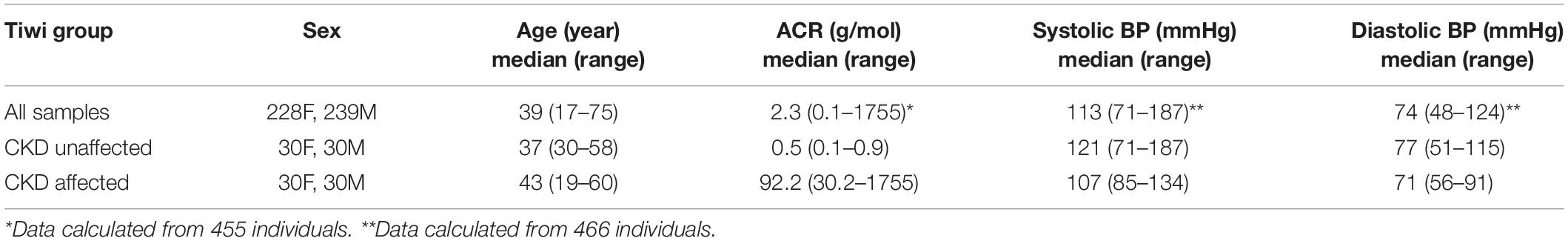
The study participants all lived on The Tiwi Islands, located off the northern coast of Australia in the Arafura Sea. Tiwi Islanders exhibit a distinct genetic ancestry compared to other ethnicities (Thomson et al., 2019), and are considered most closely related to mainland Indigenous Australians (Tiwi Land Council, 2018). The current total population is approximately 2,500 individuals (ABS, 2016). All the participants were self-identifying Tiwi Islanders. They were recruited as clinic outpatients and consented to collection of blood and urine samples, primarily for characterization of renal function and DNA for genetic studies. The numbers of participants, their age ranges and the respective clinical measures used in the study are shown in Table 1. None of the participants were experiencing renal failure or receiving renal replacement therapy at the time the study was performed. A previous analysis of a separate cohort of self-identifying Tiwi Islanders (n = 73 individuals) indicated low admixture with other populations, including Europeans (Thomson et al., 2019). The study received the full support of The Tiwi Island Land Council and was approved by the human research ethics committees of The Northern Territory Department of Health (2012–1767), The Australian National University (2014–663), The University of Queensland (2012001146), and The University of Tasmania (H0012832).
Genotyping and Allele Frequency Analysis
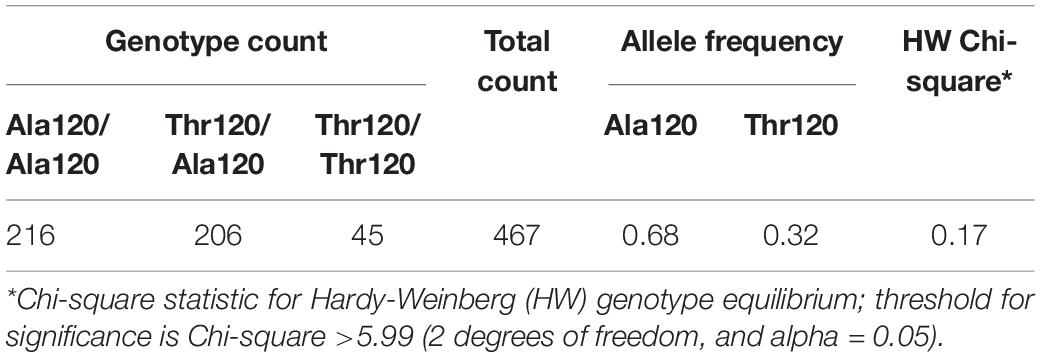
A total of 467 participants were successfully genotyped for the rs773902 allele. Genotyping was performed on 2.5 ng of genomic DNA prepared from blood samples using a TaqMan assay and 7900HT Fast Real-Time PCR System (Applied Biosystems), and standard protocols. A subset of randomly chosen samples (n = 70) subjected to replicate genotyping were in 100% agreement. Chi-square test for genotype frequency distortions from Hardy Weinberg equilibrium were performed using the WPcalc online tool (Wow-Company).
Clinical Measures
Blood and urine specimens were non-fasting and collected without regard for time of day. Serum was separated immediately after clotting, within 20 min in all cases. All specimens were then refrigerated until transfer to the pathology laboratory by air, within 24 h. Albumin (mass) and creatinine (molar) concentrations were analyzed in the spot urine sample and an albumin/creatinine ratio (ACR) calculated (g/mol). The estimated glomerular filtration rate (eGFR, mL/min/1.73 m2) was determined from serum creatinine levels using the MDRD Study equation (Levey et al., 1999). Blood glucose levels were determined using standard diagnostic laboratory assay. Blood and leukocyte positive urine samples were identified using urine test strips (Bayer Multistix 10 SG Reagent Urinalysis Test Strips), defined as small, moderate or large, and ≥1+, respectively. Systolic and diastolic blood pressure (BP) were measured by a single observer using a mercury sphygmomanometer. The median and range of the ACR, systolic and diastolic BP measures amongst the study participants are provided in Table 1.
Association Studies
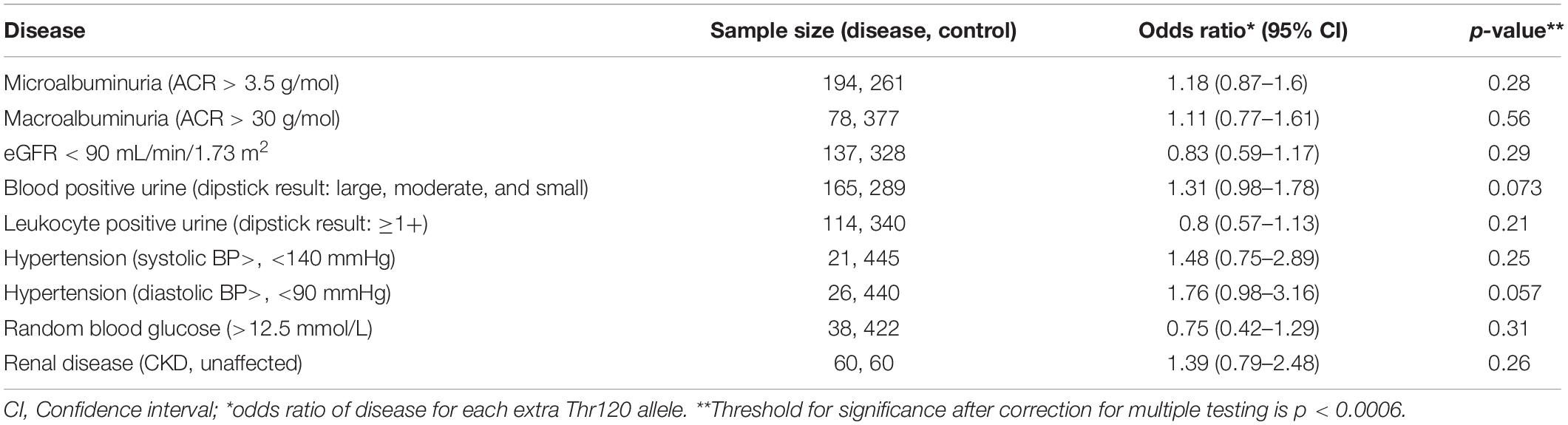
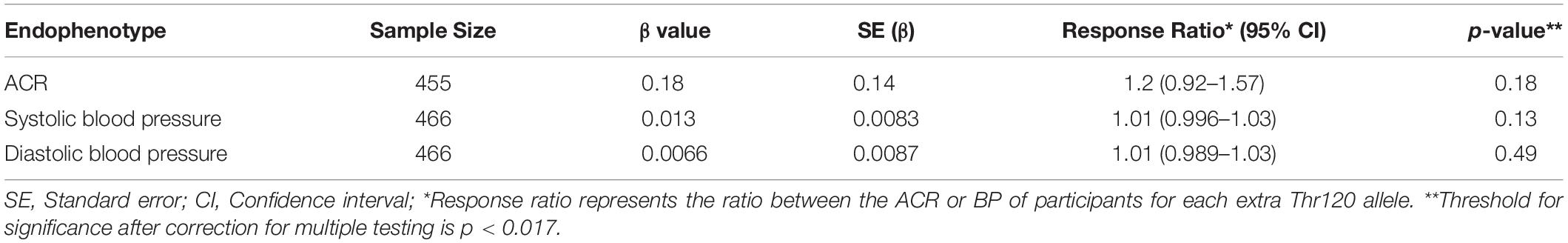
Logistic regression was used to estimate the odds ratio between disease markers and the rs773902 Thr120 allele using a dose dependent model, and adjustments for age and sex. The markers used were micro- and macroalbuminuria (ACR > 3.5 g/mol and ACR > 30 g/mol; available for 455 participants), eGFR < 90 mL/min/1.73 m2 (467 participants), blood positive urine (dipstick result: large, moderate and small; 454 participants), leukocyte positive urine (dipstick result: ≥1+; 454 participants), hypertension (systolic blood BP > 140 and diastolic BP > 90; 466 participants), and random blood glucose (>12.5 mmol/L; 460 participants). The association between renal disease and the rs773902 Thr120 allele was also examined with a logistic regression model using a subset of 60 CKD affected and 60 unaffected individuals (Table 1). The 60 CKD affected individuals were the youngest 30 males and 30 females of the cohort with ACR ≥ 34 g/mol and any eGFR value. The 60 unaffected individuals were the oldest 30 males and 30 females of the cohort with ACR ≤ 3.4 g/mol and eGFR ≥ 90 mL/min/1.73 m2. These groupings were assigned in consultation with renal physician Prof. Matthew Jose, University of Tasmania. Selection according to age was important because of the high frequency of early-onset renal disease in the Tiwi Islander population (Hoy et al., 1998), and was applied to minimize inclusion of healthy younger aged individuals in the unaffected group (who may go to develop CKD at a later age) and maximize inclusion of those with early-onset disease in the affected group. The age distributions of each group are presented in the Supplementary Figure S1. Linear regression modeling was used to examine the relationship between the rs773902 Thr120 allele and the response variables, ACR, systolic and diastolic BP. The response variable was log transformed for all models and the beta coefficients were exponentiated to provide response ratios. All models were adjusted for age and sex, and p-value thresholds of significance were adjusted for multiple testing using the Bonferroni method.
Results and Discussion
The allele frequencies for rs773902 determined for the entire cohort satisfied Hardy-Weinberg equilibrium and the minor allele frequency (MAF), encoding Thr120, was 0.32 (Table 2). Logistic regression analyses indicated no significant relationships between the Thr120 allele and indicators of abnormal renal function, including microalbuminuria, macroalbuminuria, eGFR < 90 mL/min/1.73 m2, and blood-positive and leukocyte-positive urine samples, or hypertension and random blood glucose concentration. A separate analysis of the allele frequencies in individuals categorized as CKD affected versus unaffected (n = 60/60) also revealed no significant relationship (Table 3). Analyses using linear regression modeling likewise did not detect any significant associations with ACR or BP within the whole study cohort (Table 4). The small sample sizes in our study may be a limiting factor for detecting statistically significant differences; by comparison, the seminal study reporting significant associations between rs773902 and stroke risk included more than 6000 individuals (Whitley et al., 2018). Due to an absence of data in our cohort, it was not possible to directly test if cardiovascular disease or thrombosis were associated with rs773902.
The MAF for rs773902 measured in the Tiwi Islanders (0.32) was slightly higher than the 0.19–0.29 range observed in various Asian and European groups, but substantially lower than the 0.57–0.63 range reported in some African groups [(Edelstein et al., 2014; Heenkenda et al., 2018); ExAc and gnomAD data sets]. Notably, the Tiwi Islander frequency was also lower than Melanesian and Papua New Guinean frequencies (0.55, Human Genome Diversity Project data set), who are considered to have a close genetic relationship to Indigenous Australians (McEvoy et al., 2010) – although it should also be noted the Melanesian allele data are from only 35 individuals. The method of participant recruitment for this study was not randomized across the population, however, the relatively large sample size (representing approximately 20% of the entire Tiwi Island population) and the similarities in disease burdens to other community-wide based surveys of the Tiwi (Hoy et al., 1998; Hoy et al., 2017) indicate the rs773902 allele frequencies reported here are an accurate reflection of the Tiwi Islander people.
The high burden of diseases related to platelet function in Indigenous Australians and the concomitant use of anti-platelet therapies should motivate further investigation to understand the functional effects of the variant in these populations. It will also be of interest to determine if the PAR4 rs773902 allele frequency found in the Tiwi is representative of other Indigenous Australian groups since many have existed in relative isolation from each other for several 1000 years.
Data Availability Statement
The datasets for this article are not publicly available due to privacy concerns for the study participants and the indigenous community involved. Requests to access the datasets should be directed to the corresponding author and/or the Tiwi Land Council, YWRtaW5AdGl3aWxhbmRjb3VuY2lsLmNvbQ==.
Ethics Statement
The studies involving human participants were reviewed and approved by the human research ethics committees of The Northern Territory Department of Health (2012-1767), The Australian National University (2014-663), The University of Queensland (2012001146), and The University of Tasmania (H0012832). The patients/participants provided their written informed consent to participate in this study. The study received the full support of The Tiwi Island Land Council.
Author Contributions
BM, RT, and JH wrote the manuscript. BM, EG, and JH conceived the study. BM, DN, and VT carried out the genotyping. RT and SN carried out the statistical analyses. JM, WH, and SF founded the Tiwi kidney disease and genetic studies, were instrumental in working with the Tiwi community and Tiwi Land Council, and contributed clinical knowledge. All authors read and approved the final manuscript.
Funding
The study was supported by grants from National Health and Medical Research Council of Australia (APP490040, APP511081, APP951250, APP951342, and APP1024207). The Colonial Foundation of Australia and The Australian Kidney Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge the following people: Barry Ullungurra for his help as the key contact person with the Tiwi Islanders; Bev Mcleod and Ceri Flowers for their project management and sample and data collection; Maria Scarlett for her considerable advice and guidance on the ethics of this project; and Beverley Hayhurst for the original sample collection and most notably the study participants and the Tiwi Land Council for their time and ongoing support for this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00432/full#supplementary-material
References
Australian Institute of Health and Welfare (2016). Australian Burden of Disease Study: Impact and Causes of Illness and Death in Aboriginal and Torres Strait Islander People 2011, ed. Australian A Institute of Health and Welfare. Canberra: Australian Government.
Duffy, D. L., McDonald, S. P., Hayhurst, B., Panagiotopoulos, S., Smith, T. J., Wang, X. L., et al. (2016). Familial aggregation of albuminuria and arterial hypertension in an Aboriginal Australian community and the contribution of variants in ACE and TP53. BMC Nephrol. 17:183. doi: 10.1186/s12882-016-0396-2
Edelstein, L. C., Simon, L. M., Lindsay, C. R., Kong, X., Teruel-Montoya, R., Tourdot, B. E., et al. (2014). Common variants in the human platelet PAR4 thrombin receptor alter platelet function and differ by race. Blood 124, 3450–3458. doi: 10.1182/blood-2014-04-572479
Edelstein, L. C., Simon, L. M., Montoya, R. T., Holinstat, M., Chen, E. S., Bergeron, A., et al. (2013). Racial differences in human platelet PAR4 reactivity reflect expression of PCTP and miR-376c. Nat. Med. 19, 1609–1616. doi: 10.1038/nm.3385
French, S. L., and Hamilton, J. R. (2017). Drugs targeting protease-activated receptor-4 improve the anti-thrombotic therapeutic window. Ann. Transl. Med. 5:464. doi: 10.21037/atm.2017.11.31
Heenkenda, M. K., Lindahl, T. L., and Osman, A. (2018). Frequency of PAR4 Ala120Thr variant associated with platelet reactivity significantly varies across sub-Saharan African populations. Blood 132, 2103–2106. doi: 10.1182/blood-2018-05-852335
Hoy, W. E., Mathews, J. D., McCredie, D. A., Pugsley, D. J., Hayhurst, B. G., Rees, M., et al. (1998). The multidimensional nature of renal disease: rates and associations of albuminuria in an Australian Aboriginal community. Kidney Int. 54, 1296–1304. doi: 10.1046/j.1523-1755.1998.00099.x
Hoy, W. E., Mott, S. A., and McLeod, B. J. (2017). Transformation of mortality in a remote Australian Aboriginal community: a retrospective observational study. BMJ Open 7:e016094. doi: 10.1136/bmjopen-2017-016094
Lambert, M. P. (2016). Platelets in liver and renal disease. Hematol. Am. Soc. Hematol. Educ. Program 2016, 251–255.
Levey, A. S., Bosch, J. P., Lewis, J. B., Greene, T., Rogers, N., and Roth, D. (1999). A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann. Int. Med. 130, 461–470. doi: 10.7326/0003-4819-130-6-199903160-00002
Madhusudhan, T., Kerlin, B. A., and Isermann, B. (2016). The emerging role of coagulation proteases in kidney disease. Nat. Rev. Nephrol. 12, 94–109. doi: 10.1038/nrneph.2015.177
McEvoy, B. P., Lind, J. M., Wang, E. T., Moyzis, R. K., Visscher, P. M., van Holst Pellekaan, S. M., et al. (2010). Whole-genome genetic diversity in a sample of Australians with deep Aboriginal ancestry. Am. J. Hum. Genet. 87, 297–305. doi: 10.1016/j.ajhg.2010.07.008
Palygin, O., Ilatovskaya, D. V., and Staruschenko, A. (2016). Protease-activated receptors in kidney disease progression. Am. J. Physiol. Renal Physiol. 311, F1140–F1144. doi: 10.1152/ajprenal.00460.2016
Saheb Sharif-Askari, F., Syed Sulaiman, S. A., and Saheb Sharif-Askari, N. (2017). Anticoagulation therapy in patients with chronic kidney disease. Adv. Exp. Med. Biol. 906, 101–114. doi: 10.1097/MD.0000000000017628
Spencer, J. L., Silva, D. T., Snelling, P., and Hoy, W. E. (1998). An epidemic of renal failure among Australian Aboriginals. Med. J. Aust. 168, 537–541.
Thomson, R. J., McMorran, B., Hoy, W., Jose, M., Whittock, L., Thornton, T., et al. (2019). New genetic loci associated with chronic kidney disease in an indigenous Australian population. Front. Genet. 10:330. doi: 10.3389/fgene.2019.00330
Tiwi Land Council (2018). Timeline: Dreamtime to 1978. Available online at: http://tiwilandcouncil.com (accessed April 5).
Tourdot, B. E., Conaway, S., Niisuke, K., Edelstein, L. C., Bray, P. F., and Holinstat, M. (2014). Mechanism of race-dependent platelet activation through the protease-activated receptor-4 and Gq signaling axis. Arterioscler Thromb. Vasc. Biol. 34, 2644–2650. doi: 10.1161/ATVBAHA.114.304249
Tourdot, B. E., Stoveken, H., Trumbo, D., Yeung, J., Kanthi, Y., Edelstein, L. C., et al. (2018). Genetic variant in human PAR (Protease-Activated Receptor) 4 Enhances Thrombus Formation Resulting in Resistance to Antiplatelet Therapeutics. Arterioscler Thromb Vasc. Biol. 38, 1632–1643. doi: 10.1161/ATVBAHA.118.311112
Tricoci, P., Neely, M., Whitley, M. J., Edelstein, L. C., Simon, L. M., Shaw, C., et al. (2018). Effects of genetic variation in protease activated receptor 4 after an acute coronary syndrome: analysis from the TRACER trial. Blood Cells Mol. Dis. 72, 37–43. doi: 10.1016/j.bcmd.2018.07.004
Van Buynder, P. G., Gaggin, J. A., and Mathews, J. D. (1993). Renal disease patterns in aboriginal Australians. A family-based study in a high incidence community. Med. J. Aust. 159, 82–87.
Whitley, M. J., Henke, D. M., Ghazi, A., Nieman, M., Stoller, M., Simon, L. M., et al. (2018). The protease-activated receptor 4 Ala120Thr variant alters platelet responsiveness to low-dose thrombin and protease-activated receptor 4 desensitization, and is blocked by non-competitive P2Y12 inhibition. J. Thromb. Haemost 16, 2501–2514. doi: 10.1111/jth.14318
Keywords: protease activated receptor 4, rs773902, renal disease, Australian Aboriginal and Torres Strait Islanders, indigenous genetics
Citation: Ningtyas D, Thomson RJ, Tarlac V, Nagaraj SH, Hoy W, Mathews JD, Foote SJ, Gardiner EE, Hamilton JR and McMorran BJ (2020) Analysis of the F2LR3 (PAR4) Single Nucleotide Polymorphism (rs773902) in an Indigenous Australian Population. Front. Genet. 11:432. doi: 10.3389/fgene.2020.00432
Received: 02 September 2019; Accepted: 07 April 2020;
Published: 30 April 2020.
Edited by:
Cheryl Ann Winkler, Frederick National Laboratory for Cancer Research (NIH), United StatesReviewed by:
Yanina Timasheva, Institute of Biochemistry and Genetics of Ufa Scientific Centre (RAS), RussiaTugce Karaderi, University of Copenhagen, Denmark
Marvin T. Nieman, Case Western Reserve University, United States
Copyright © 2020 Ningtyas, Thomson, Tarlac, Nagaraj, Hoy, Mathews, Foote, Gardiner, Hamilton and McMorran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brendan J. McMorran, YnJlbmRhbi5tY21vcnJhbkBhbnUuZWR1LmF1
 Dian Ningtyas
Dian Ningtyas Russell J. Thomson
Russell J. Thomson Volga Tarlac
Volga Tarlac Shivashankar H. Nagaraj4,5
Shivashankar H. Nagaraj4,5 Wendy Hoy
Wendy Hoy Elizabeth E. Gardiner
Elizabeth E. Gardiner Justin R. Hamilton
Justin R. Hamilton Brendan J. McMorran
Brendan J. McMorran