- 1College of Life Sciences, Institute for Conservation and Utilization of Agro-Bioresources in Dabie Mountains, Xinyang Normal University, Xinyang, China
- 2College of Horticulture and Forestry, Huazhong Agricultural University, Key Laboratory of Horticultural Plant Biology, Ministry of Education, Wuhan, China
- 3College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
Brassica juncea is an important dietary vegetable cultivated and consumed in China for its edible stalks and leaves. The purple leaf mustard, which is rich in anthocyanins, is eye-catching and delivers valuable nutrition. However, the molecular mechanism involved in anthocyanin biosynthesis has not been well studied in B. juncea. Here, histological and transcriptome analyses were used to characterize the purple leaf color and gene expression profiles. Free-hand section analysis showed that the anthocyanin was mainly accumulated in the adaxial epidermal leaf cells. The anthocyanin content in the purple leaves was significantly higher than that in the green leaves. To investigate the critical genes and pathways involved in anthocyanin biosynthesis and accumulation, the transcriptome analysis was used to identify the differentially expressed genes (DEGs) between the purple and green leaves from the backcrossed BC3 segregation population in B. juncea. A total of 2,286 different expressed genes were identified between the purple and green leaves. Among them, 1,593 DEGs were up-regulated and 693 DEGs were down-regulated. There were 213 differently expressed transcription factors among them. The MYB and bHLH transcription factors, which may regulate anthocyanin biosynthesis, were up-regulated in the purple leaves. Interestingly, most of the genes involved in plant–pathogen interaction pathway were also up-regulated in the purple leaves. The late biosynthetic genes involved in anthocyanin biosynthesis were highly up-regulated in the purple leaves of B. juncea. The up regulation of BjTT8 and BjMYC2 and anthocyanin biosynthetic genes (BjC4H, BjDFR, and BjANS) may activate the purple leaf formation in B. juncea. This study may help to understand the transcriptional regulation of anthocyanin biosynthesis in B. juncea.
Introduction
Anthocyanin, which includes numbers of natural water-soluble pigments, colored the fruit and flowers of many plants (Glover and Martin, 2012). Anthocyanin could produce various colors, such as red, purple, blue, yellow, and orange, in different plant organs and is beneficial to health (Castañeda-Ovando et al., 2009). High-anthocyanin food consumption could help to stimulate the immune system and reduce cancer risks (Thomasset et al., 2009). Dietary anthocyanins, which are important health-promoting antioxidants, make major contribution to the quality of fruits or leaves. Furthermore, dietary anthocyanin accumulation is also recognized as a visible biomarker of plants that have suffered from environmental stresses in many studies (Xie Y. et al., 2016; Zheng et al., 2019; Sun et al., 2020). Anthocyanin biosynthesis and regulation pathway has been well studied in Cruciferae plants including Arabidopsis, Brassica rapa, Brassica oleracea, Brassica napus, etc. (Guo et al., 2014; Xie Y. et al., 2016; Goswami et al., 2018; Yan et al., 2019).
In Arabidopsis, anthocyanin biosynthesis genes can be divided into early biosynthesis genes (EBGs) and late biosynthesis genes (LBGs). The ternary complex of MYB-bHLH-WD40 influences anthocyanin biosynthesis pathway through regulating the expression intensity and pattern of the structural genes by binding to their promoter regions (Petroni and Tonelli, 2011). Anthocyanin accumulation is also induced by different environmental stress (Xie Y. et al., 2016). In Arabidopsis, overexpression of the R2R3-MYB transcription factors (PAP1, PAP2, MYB113, and MYB114) increased anthocyanin accumulation (Gonzalez et al., 2008). Overexpression of MYB75 increased anthocyanin and flavonol levels which enhance plant resistance to a specialist caterpillar (Pieris brassicae) in Arabidopsis (Onkokesung et al., 2014). The up-regulation of BoMYB2 and BoTT8 leads to the anthocyanin accumulation in red cabbage and purple cauliflower (Yuan et al., 2009; Chiu et al., 2010). Transcriptional regulation of BrTT8 promotes anthocyanin biosynthesis in purple bok choy (B. rapa var. chinensis) (Zhang et al., 2014). And the transcript level of BjTT8 is associated with up-regulation of most anthocyanin biosynthetic genes (Xie et al., 2014). The loss expression of BoMYBL2-1 gene resulted in the establishment of purple B. oleracea (Song et al., 2018). In Arabidopsis, the TT8 is required for expression of flavonoid late biosynthetic genes, DFR and BAN (Nesi et al., 2000). The expression profiling of genes involved in transcriptional regulation and structure genes of anthocyanin biosynthesis were identified in the high-anthocyanin resynthesized B. napus (Goswami et al., 2018). It was reported that BrEGL3.1 and BrEGL3.2, which encode bHLH transcription factors, are two candidate genes for anthocyanin accumulation in zicaitai (Guo et al., 2015). These studies provide insight into the mechanisms of coloration in Cruciferae crops.
In recent years, RNA-seq has been used to identify differentially expressed genes (DEGs) involved in anthocyanin biosynthesis. The metabolism combined with transcriptome methods had been used to identify novel genes involved in flavonoid biosynthesis in Arabidopsis (Tohge et al., 2005). Transcriptome analysis showed that most of the anthocyanin biosynthetic genes were highly up-regulated in purple leaves investigated in Brassica (Mushtaq et al., 2016). Transcriptome analysis has been used for comprehensive understanding of the molecular mechanisms of stem development in B. juncea (Sun et al., 2012). Transcriptome had also been used to identify genes for the biosynthesis of flavonoids of B. juncea seed coat (Liu et al., 2013). Transcriptomic and metabolomic study revealed that the main genes involved in flavonoid biosynthesis were up-regulated in red kale than green kale (Jeon et al., 2018a). RNA-seq analysis was employed to seek the candidate genes involved in dark-purple leaves from B. rapa (Xie L. et al., 2016). Mustard (B. juncea) is widely cultivated as a typical condiment, cruciferous vegetables, and oilseed crops worldwide (Chen et al., 2013). The cultivated mustard varieties have been widely used as a fresh and pickled vegetable in the southwest of China (Fan et al., 2017). The water-soluble anthocyanins are widely distributed in the purple leaf mustard. The anthocyanin-rich mustard is beneficial to heath due to its high antioxidant activity and colorful germplasm resource for health vegetables breeding. A previous study has identified 67 anthocyanins, 102 flavonol glycosides, and 40 hydroxycinnamic acid derivatives from the red mustard greens (B. juncea Coss variety) (Lin et al., 2011). And more than 20 different types of anthocyanins were separated and identified from the purple tumorous stem mustard by high performance liquid chromatography electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) (Xie et al., 2014). The genome sequence of B. juncea has facilitated us to select anthocyanin biosynthesis genes in it (Yang et al., 2016). Although a number of genes involved in anthocyanin biosynthesis and regulation pathway have been studied in Brassica species, only few study mining candidate genes involved in anthocyanin formation in mustard by using RNA sequencing. Thus, comprehensive understanding of anthocyanin biosynthesis and regulation will promote the utilization of the anthocyanin in mustard. To better understand the molecular mechanism of anthocyanin biosynthesis, RNA-seq technology was employed to analyze the expression profile of different expressed genes between purple leaves and green leaves. These results may facilitate to promote molecular function analysis of the key genes involved in biosynthesis of anthocyanin and elucidate the molecular mechanism about it in B. juncea.
Materials and Methods
Plant Materials
The purple leaf (ZiYi) line and the green leaf (LvYi) line were planted in the experimental field at Xinyang Normal University (Xinyang, China). The ZiYi and LvYi plants were crossed to produce the F1 generation. The F1 plants were backcrossed with the recessive LvYi plants to construct the BC1 population in 2016. Then, the purple plants from the BC2 population planted in 2017 were backcrossed with the recessive LvYi parent line to construct the BC3 population in year 2018 (Supplementary Figure S1). The leaves from the purple and green plants in the BC3 population were frozen in liquid nitrogen and stored at −80°C until use for RNA extraction in Dec 2018.
Extraction and Measurement of Anthocyanin Content
A total of anthocyanins was extracted from the purple and green plants in the BC3 population with three replicates, respectively. The purple and green leaves from the BC3 population were extracted into a mixed liquid with 1 ml of 95% ethanol and 1.5 M HCl (4:1, v/v) for 24 h at 4°C in the dark. Then, the extracts were centrifuged at 12,000 g for 10 min at 4°C. The absorption of the supernatant extracts at 530 nm was determined using a Thermo ScientificTM NanoDropTM One. Total anthocyanin content was measured by the methods described previously (Li et al., 2016). The data were analyzed by independent samples T test and significant differences with P < 0.01. Error bars indicated the standard error of the average anthocyanin contents.
RNA Extraction and RNA-Seq
Total RNA was extracted from the ZiYi and LvYi plants in the BC3 population with three replications each. TaKaRa MiniBEST Plant RNA Extraction Kit (Code No: 9769) was used to extract total RNA following the manufacturer’s protocols. Agilent 2100 RNA Nano 6000 Assay Kit (Agilent Technologies, Santa Clara, CA, United States) was used to detect the integrity and concentration of RNA samples. RNA seq was performed by using the Illumina Hiseq X Ten platform.
Bioinformatics Analysis of Differential Expression Genes
Raw data were processed with the adopted filtering criteria as follows: remove the adaptor-polluted reads, remove the low-quality reads, and remove reads with number of N bases accounting for more than 5%. The obtained clean data will be carried out on statistics to analyze its quantity and quality, including Q30, data quantity and base content statistics, etc. Bowtie/Bowtie2 was used for building the genome index files. Clean Data was mapped to the B. juncea genome1 by using tophat v2.0.12. Fragments count for each gene in each sample was counted by HTSeq v0.6.0, and FPKM (fragments per kilobase per million mapped fragments) was calculated to estimate the expression level of genes in each sample. DESeq/DESeq2 was used for DEGs between ZiYi and LvYi with three biological replicates using a model based on the negative binomial distribution. The P-value could be assigned to each gene and adjusted by the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with a fold change >2 and a P-value < 0.05 were identified as DEGs. GO2 (Gene Ontology) enrichment of DEGs was implemented by the hypergeometric test, in which P-value is calculated and adjusted as q-value, and data background is genes in the whole genome. GO terms with q < 0.05 were considered to be significantly enriched. GO enrichment analysis could exhibit the biological functions of the DEGs. KEGG3 (Kyoto Encyclopedia of Genes and Genomes) is a database resource containing a collection of manually drawn pathway maps representing our knowledge on the molecular interaction and reaction networks. The KEGG enrichment of DEGs was implemented by the hypergeometric test, in which P-value was adjusted by multiple comparisons as q-value. KEGG terms with q < 0.05 were considered to be significantly enriched.
Quantitative Real-Time PCR
The cDNA from different samples were synthesized from total RNA (3 μg) by using a PrimeScriptTM 1st Strand cDNA Synthesis Kit (Code No: 6110A). Then, they were diluted to 80 times. Each qPCR included 8.4 μl of the diluted cDNA template, 0.8 μl of each primer (10 μM), and 10 μl of SYBR Green PCR master mix (ABI, United States). Reactions were carried out on the ABI PRISM 7300 Real-Time PCR System (Applied Biosystems). Quantitative real-time PCR (qRT-PCR) was performed as follows: 95°C for 5 min; 40 cycles at 95°C for 10 s, 60°C for 15 s, and 72°C for 30 s; and a final extension step at 72°C for 5 min. Results were analyzed using the CFX Manager software according to the 2–Δ Δ CT method (Livak and Schmittgen, 2001). Three biological replicates were carried for gene expression analysis. The primers used for quantitative real time PCR (qRT-PCR) analysis were designed by Primer express software (Applied Biosystems, United States). The primers used in our study are listed in Supplementary Table S1. The BjActin gene was used as an internal control gene for mRNA expression (Heng et al., 2018).
Results
The Inheritance and Different Anthocyanin Content in the Purple Leaves in B. juncea
Purple leaf mustard showed higher accumulation of anthocyanin in leaves than that of the green leaf mustard (Figures 1A–D). With the leaf develops, the anthocyanin was mainly accumulated on both adaxial and abaxial epidermis and subepidermal cell layers (Figure 1D). Only in the middle vein (Figure 1B), the anthocyanin appeared to be not accumulated on the abaxial side. The plants from the segregation population were further used to investigate the genetic pattern of the purple leaf color. All the F1 plants showed purple leaf color. The F2 segregation population showed a normal Mendelian segregation ratio of 3:1 (purple vs. green leaves). The backcrossed segregation population showed a normal Mendelian segregation ratio of 1:1. Our result showed that a dominant gene controlled the purple leaf trait in B. juncea (Supplementary Table S2). To accurately determine the anthocyanin content in ZiYi, total anthocyanin was extracted from purple leaf and green leaf mustard. The total anthocyanin content from ZiYi was 4.4 times higher in purple leaves when compared to the green leaves in mustard (Figure 1E).
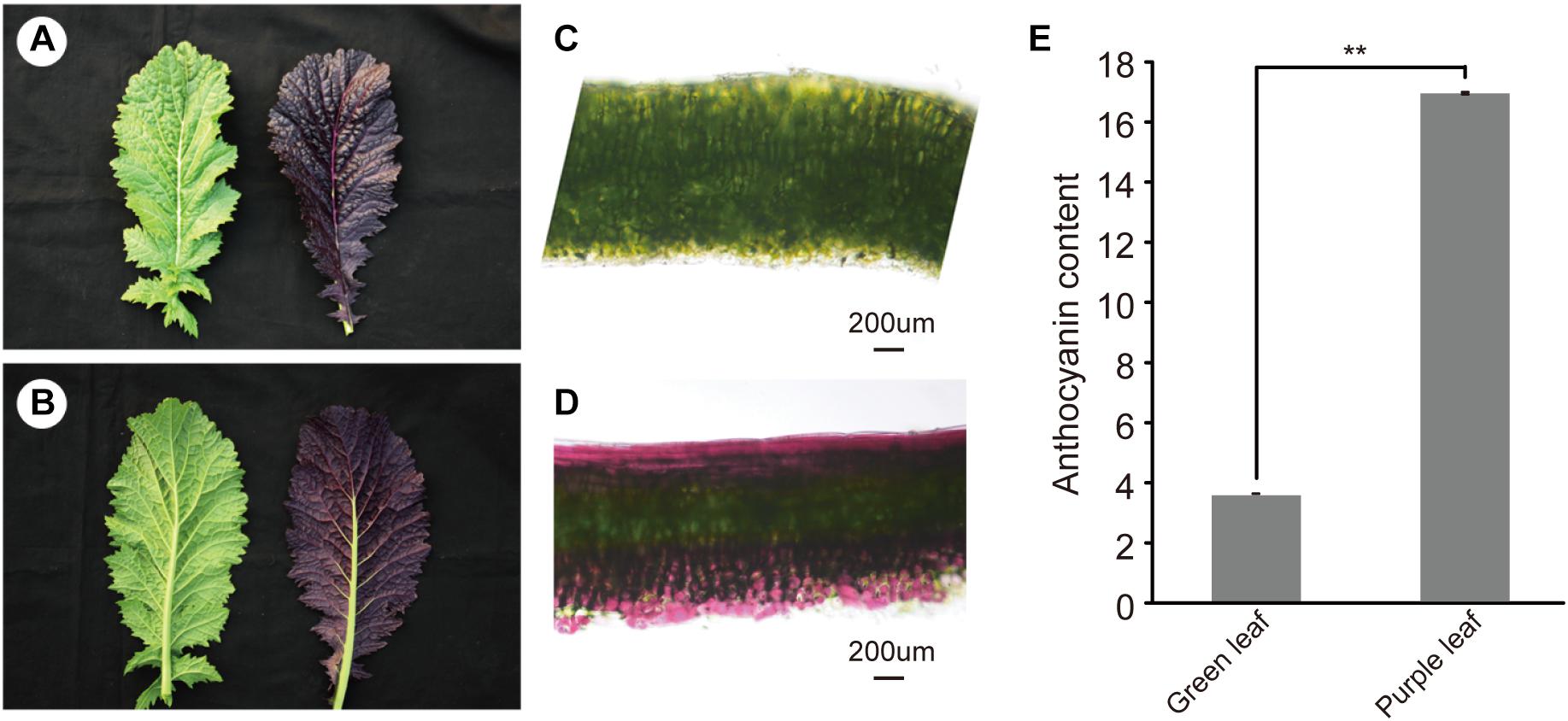
Figure 1. Morphology of purple leaf and anthocyanin content in B. juncea. (A) Upper epidermis of green (LvYi) and purple (ZiYi) leaf. (B) Lower epidermis of green and purple leaf. (C) Transverse section of a green leaf. Bars = 200 μm. (D) Transverse section of a purple leaf. Bars = 200 μm. (E) Total anthocyanin contents of green and purple leaves. **Signifcantly diferent at P < 0.01.
De novo Sequencing, Assembly, and Identification of Differentially Expressed Genes
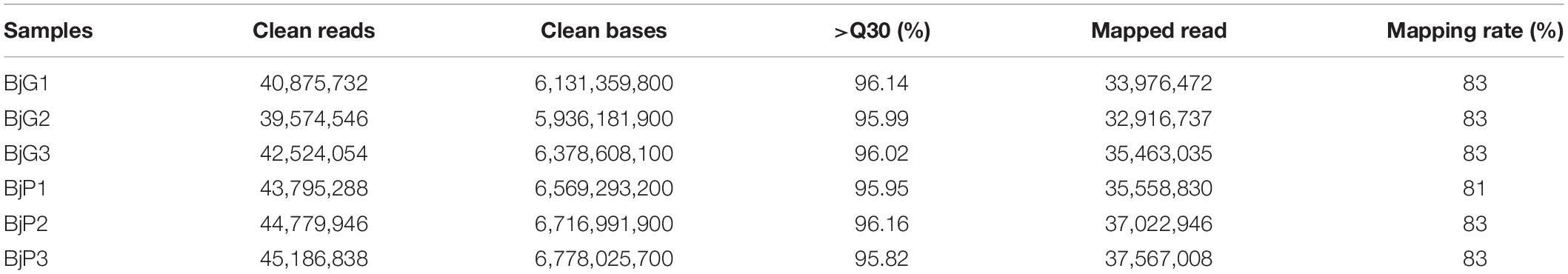
To obtain a genome-wide gene expression profile for purple leaf color formation, six sequencing libraries were constructed from purple leaf and green leaf mustard with three replicates each. A total of 150.01 million and 136.57 million clean reads were obtained by Solexa/Illumina sequencing of samples. About 256.73 million clean reads were selected after the initial screening with clean read ratios more than 95.82% (Table 1). As a result, we obtained 1.91–5.24 × 106 mapped reads for the six libraries. Then, these sequences were assembled into the B. juncea genome from Brassica database (BRAD). In total, 81–83% reads for each individual library could be aligned against the B. juncea genome. The gene expression levels of purple leaf and green leaf mustard were estimated by calculating the FPKM (fragments per kilobase per million mapped fragments) values (log2 Fold change ≥ 1 and FDR ≤ 0.01). A total of 2,286 DEGs (Figure 2A) were identified between the purple leaves and green leaves (Supplementary Table S3). Among them, 1,593 genes were found to be up-regulated and 693 genes were significantly down-regulated in the purple leaves (Figure 2B).
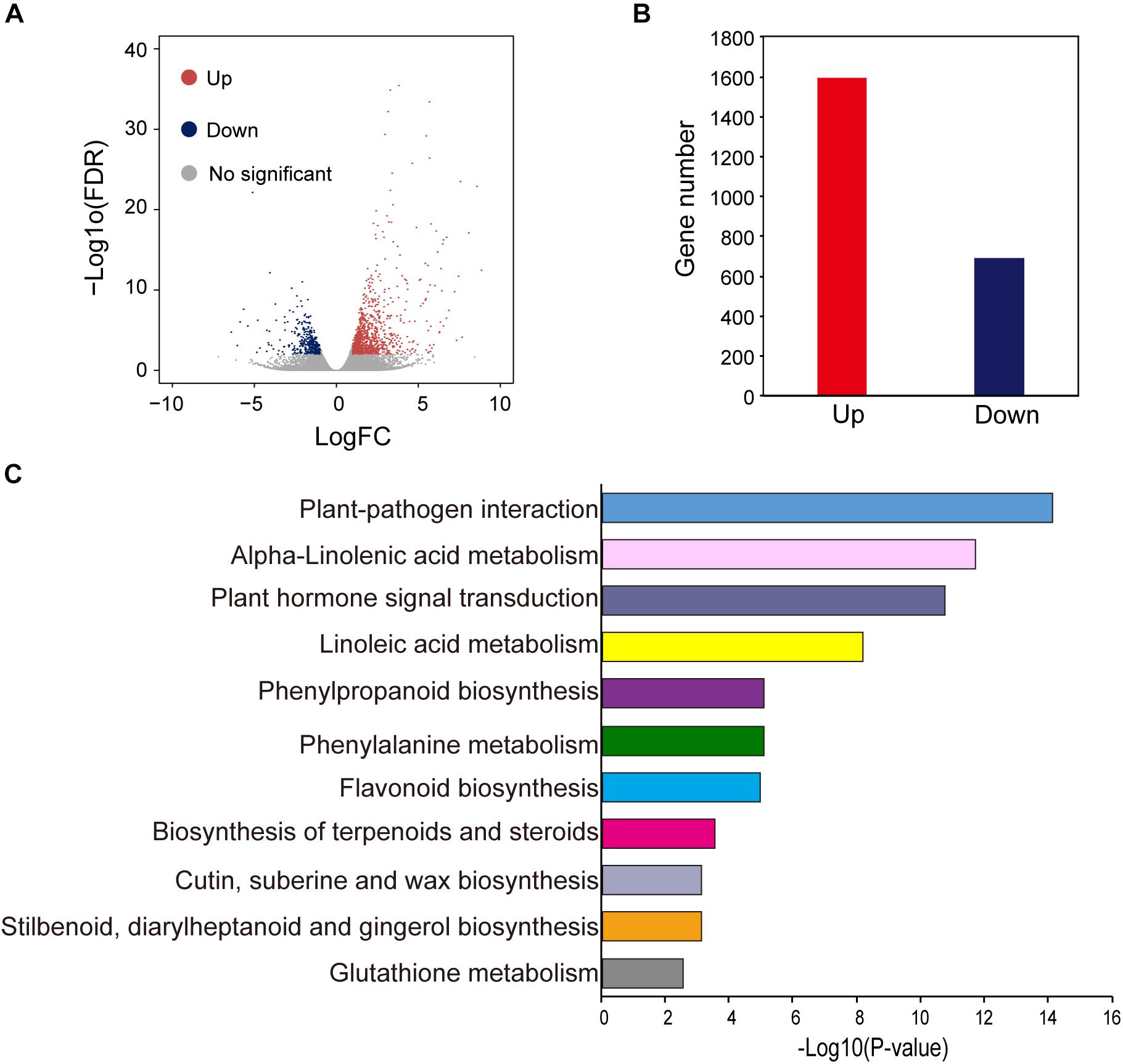
Figure 2. Different expressed genes between purple and green leaf. (A) The volcano map of DEGs between ZiYi and LvYi. Each dot represents a gene. (B) The number of up- and down-regulated genes in ZiYi. (C) KEGG metabolism pathway analysis of the DEGs between ZiYi and LvYi. Histograms represent the metabolism pathway distribution; the X axial represents the number of genes that have been annotated to the metabolism pathway.
Different Biological Process and Pathways Between Purple and Green Leaves
Gene ontology terms were used to analyze the functional classification of the DEGs in purple and green leaves, respectively. GO analysis divided these DEGs based on biological process (BP), cellular component (CC), and molecular function (MF) (Supplementary Figure S2). The largest BP subcategory for the DEGs in BjP vs. BjG was involved in cellular processes, metabolic process, single-organism process, and response to stimulus. The largest CC subcategory for the DEGs in BjP vs. BjG was related to cell part, organelle, and membrane. And the largest MF subcategory for the DEGs in BjP vs. BjG was binding, catalytic, and nucleic acid binding transcription factor. The number of up-regulated genes involved in different processes of BP, CC, and MF were much higher than the number of down-regulated genes. These results suggested that the purple leaf may undergo more complex metabolic activities involved in anthocyanin biosynthesis.
To further explore the biological pathways involved in anthocyanin biosynthesis, DEGs were annotated in the Kyoto Encyclopedia of Genes and Genomes (KEGG) by using KeggArray software. Among them, the plant-pathogen interaction (62 members), alpha-linolenic acid metabolism (22 members), plant hormone signal transduction (72 members), and linoleic acid metabolism (12 members) were the most significant changed typical pathways between the purple and green leaves (Figure 2C). In particular, pathways enriched in phenylpropanoid biosynthesis (25 members), phenylalanine metabolism (30 members), and flavonoid biosynthesis (12 members), which are hallmarks of metabolic plasticity that provide plant color and adapt to biotic and abiotic stress and promote health benefits, were significantly affected during purple leaf color formation. Most of these DEGs involved in the typical pathways mentioned above were up-regulated (Figure 2C).
The Expression Profiles of Different Transcription Factor Families
To better understand the gene regulatory networks involved in anthocyanin biosynthesis, the different expressed transcription factors were further identified. Blast analysis the genome sequence of B. juncea revealed that there were 5,258 transcription factors in it. And a total of 213 differentially expressed transcription factors (Supplementary Table S4) was also found between the purple leaves and green leaves (Figure 3A). They were divided into more than 22 different types (Figure 3B). The ERF, MYB, and bHLH transcription factors are the three most TFs among them. Among the 35 differentially expressed ERF transcription factors, more than 60% of them were up-regulated in the purple leaf mustard. Cluster analysis found that these TF families could be classified into three clusters with distinctive expression patterns (Figure 3C). The number of each TF family members showed substantial diversity (Figure 3D). Blast analysis suggested that these ERF transcription factors were mainly involved in abiotic stress. It is known that numbers of MYB and bHLH transcription factors had been reported in anthocyanin biosynthesis (Albert et al., 2014). These differently expressed TFs may be involved in regulating anthocyanin biosynthesis genes in B. juncea. And BjuB004115 encoding a bHLH regulation factor, which is homologous to AtTT8 that regulate proanthocyanidin and anthocyanin biosynthesis (Nesi et al., 2000), is up-regulated in purple leaf mustard. BjMYC2 (BjuO001663), which encodes a basic helix-loop-helix (bHLH) DNA-binding family protein (MYC2), was also up-regulated in purple leaf mustard. The upregulation of BjTT8 and BjMYC2 may contribute to the anthocyanin accumulation in purple leaf.
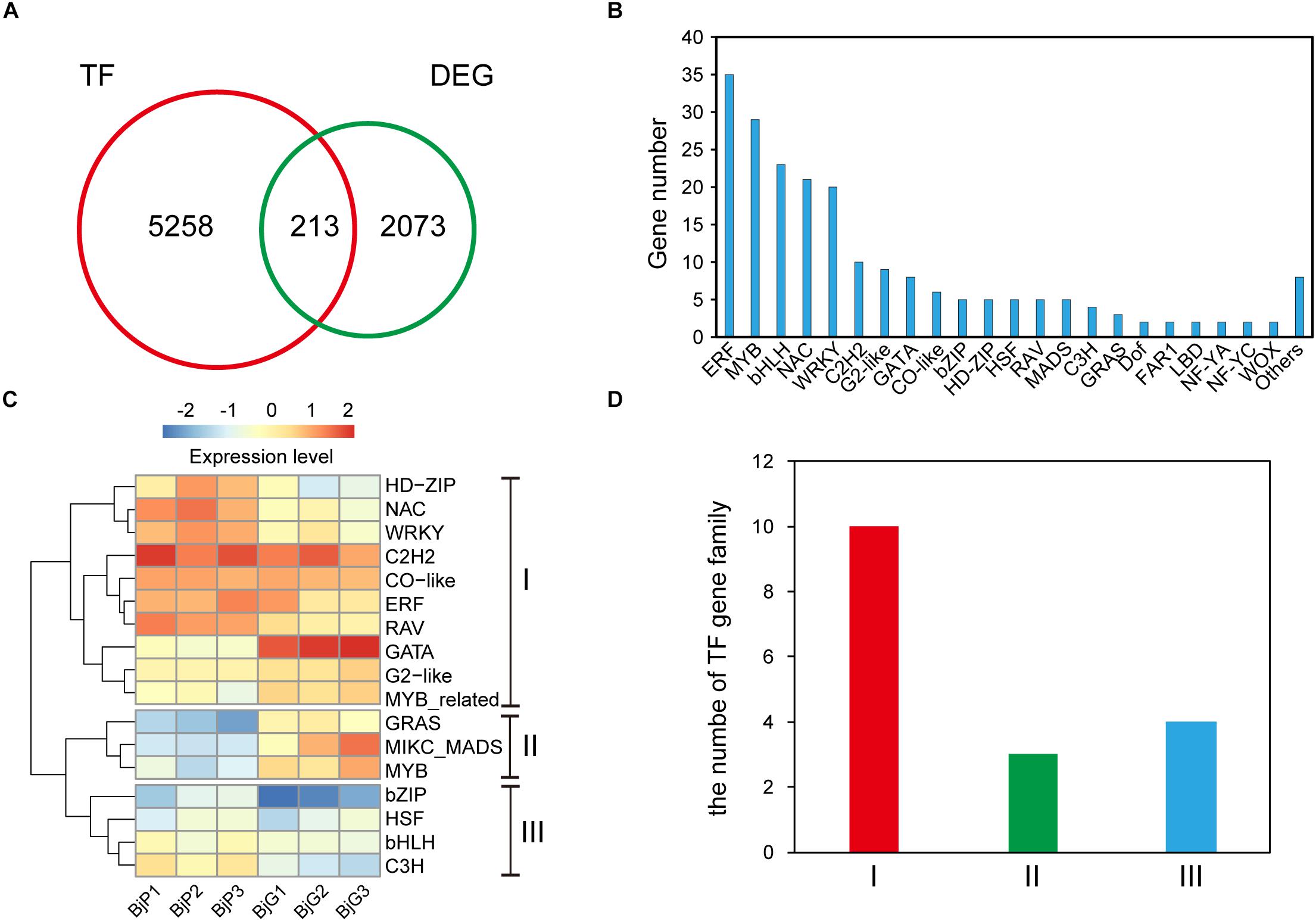
Figure 3. Distinct expression patterns of transcription factors. (A) The number of different expressed transcription factors between ZiYi and LvYi. (B) Gene numbers of different type transcription factors among the DEGs between ZiYi and LvYi. (C) Heatmaps representing the distinct expression patterns of transcription factors between ZiYi and LvYi. BjP1, BjP2, and BjP3 representative three biological replication in ZiYi. BjG1, BjG2, and BjG3 representative three biological replication in LvYi. (D) Comparison of the clusters of TF families.
Analysis of Differentially Expressed Enzymes Involved in Anthocyanin Biosynthesis
To further investigate the key genes involved in anthocyanin biosynthesis and their regulation pathway, the genes involved in anthocyanin biosynthesis in B. juncea were blasted against their orthologous genes from Arabidopsis (Supplementary Table S5). The DEGs involved in anthocyanin biosynthesis were screened from 2,286 DEGs between purple and green mustard. The anthocyanin is biosynthesized through the flavonoid pathway. The key enzymes such as phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumaryol CoA ligase (4CL) were involved in the phenylpropanoid pathway. And anthocyanin synthesis, which includes chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), and flavanone 3′-hydroxylase (F3′H), was involved in the early anthocyanin biosynthesis pathway. And dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and UDP-glucose flavonoid 3-O-glucosyl transferase (UFGT) were involved in the late anthocyanin biosynthesis pathway (Figure 4A). In our study, the transcript expression levels of most of the main enzymes involved in anthocyanins biosynthesis were up-regulated in purple leaves when compared with green leaves (Figure 4B). Among them, at least one copy of each key enzyme was up-regulated in purple leaves. It was found that three of the PAL genes were up-regulated in purple leaves. And four of the ANS genes were up-regulated in purple leaves. These up-regulated enzymes may promote the anthocyanins biosynthesis in purple leaf mustard. Apart from them, the 4CL was found to be down-regulated in purple leaves. The F3′H, which encodes flavanone 3′-hydroxylase, was not up-regulated in purple leaves significantly. And the F3′5 ′H gene, which encodes flavanone 3′, 5′-hydroxylase, was not found in our transcriptome data. These results suggested that the pelargonidin-3-glucoside biosynthesis pathway is the main pathway involved in anthocyanin biosynthesis, which contributes to purple leaf color formation in B. juncea.
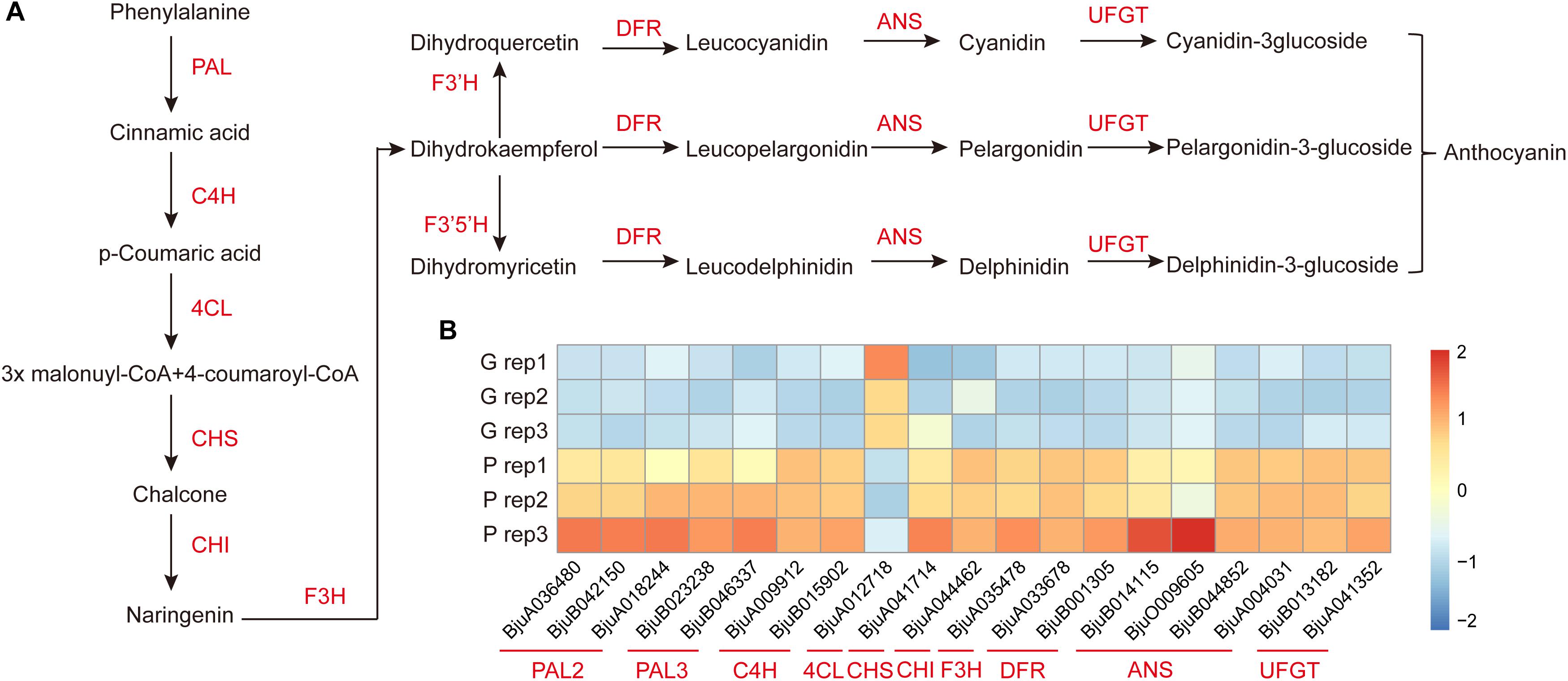
Figure 4. Diagram and heatmaps of genes involved the anthocyanin biosynthesis pathway. (A) Schematic diagram of genes involved in the anthocyanin biosynthesis pathway. (B) Heatmaps of DEGs involved in the accumulation of anthocyanin in ZiYi B. juncea.
qRT-PCR Analysis of DEGs Involved in Anthocyanin Biosynthesis
To confirm the transcript expression patterns of DEGs involved in anthocyanin biosynthesis, the key genes involved in anthocyanin biosynthesis-related enzymes were further analyzed through qRT-PCR. Nine DEGs related to anthocyanin biosynthesis pathways were selected for qRT-PCR analysis. The expression pattern of all these DEGs was consistent with the transcriptome data (Figure 5). Among them, phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), dihydroflavonol 4-reductase (DFR), and anthocyanidin synthase (ANS) were significantly higher expressed in the purple leaf. But the 4-coumaryol CoA ligase (4CL) gene, which was involved in the phenylpropanoid pathway, was down-regulated in purple leaf. These results further confirmed the validity of our transcriptome data. And the high transcription level of BjC4H, BjDFR, and BjANS may be a direct cause of the purple leaf formation in B. juncea.
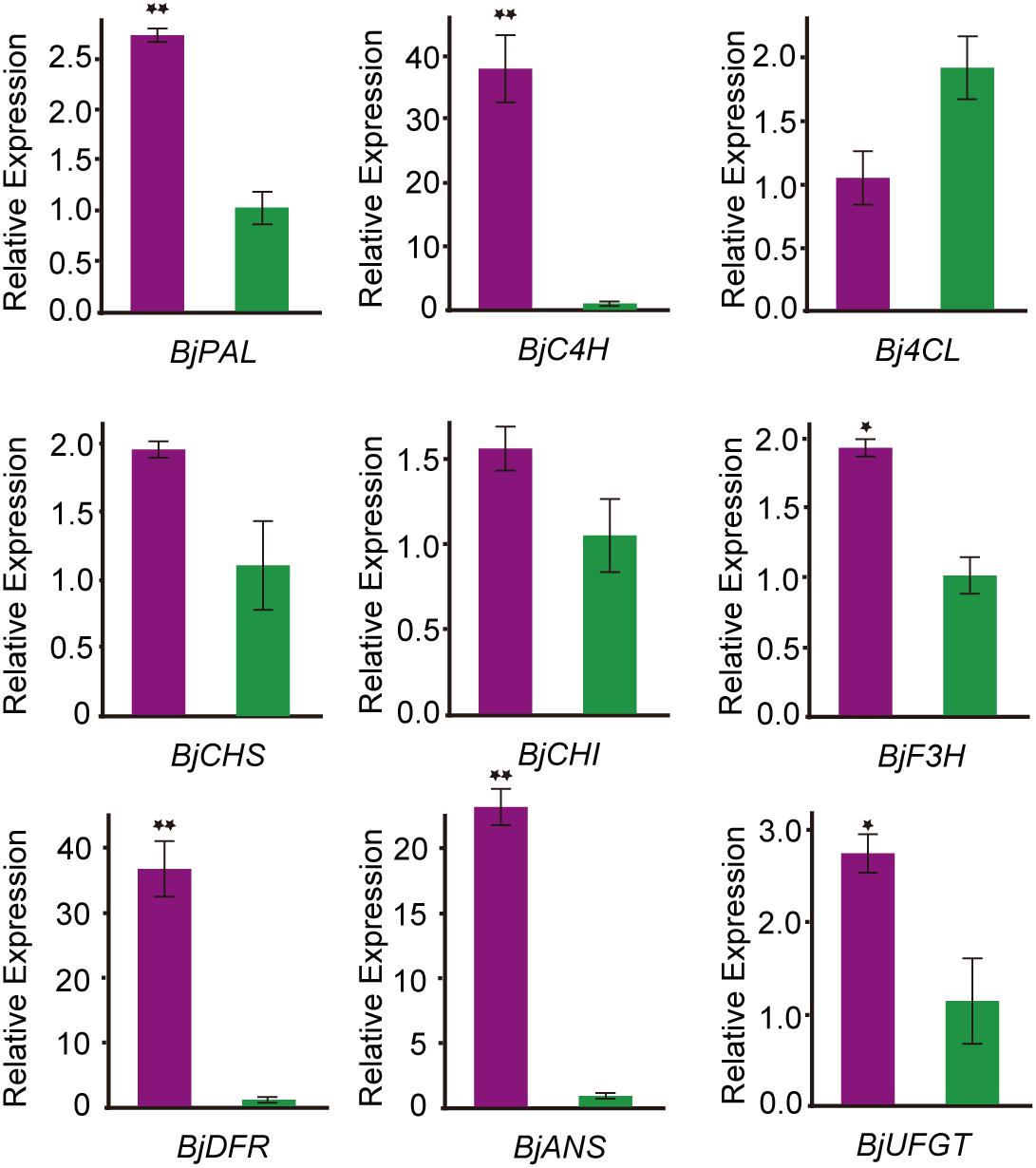
Figure 5. qRT-PCR analysis the expression pattern of genes involved in anthocyanin biosynthesis B. juncea. The purple color represents transcript expression level of genes in ZiYi B. juncea with purple leaf. The green color represents transcript expression level of genes in LvYi B. juncea with green leaf. *Signifcantly diferent at P < 0.05; **Signifcantly diferent at P < 0.01.
Discussion
It has been known that anthocyanins could promote human health with high antioxidant activities (Khoo et al., 2017). Thus, studying the molecular mechanism of the anthocyanin biosynthesis and cultivating anthocyanin-rich plants is necessary for healthy food production. Transcriptome sequencing has been used as a powerful tool to study genome-wide gene activities and helps to understand the molecular mechanisms underlying different biological process. However, knowledge of the transcriptome analysis of purple leaf formation in B. juncea is still lacking. Since the anthocyanin contents of purple leaf B. juncea greatly increased, this study was initiated with the aim to explore differently expressed anthocyanin regulate and biosynthesis genes in purple leaf B. juncea.
Many structural and regulatory genes participating in anthocyanin biosynthesis have been described in Arabidopsis and other Brassica. It has been reported that there was a single dominant gene control purple leaf color inheritance in B. juncea, Chinese Ziye mustard (Luo et al., 2011). A spontaneous mutant 1280-1 with purple leaves was controlled by a dominant gene (BjPl1), and it was mapped to a 225-kb interval on linkage group B2 of B. juncea (Zhao et al., 2017). The phenotype of purple leaf in B. juncea “HunanQianyang” was also controlled by a single dominant gene inheritance and an R2R3-MYB transcription factor enhanced anthocyanin over accumulation in the B. rapa introgression line within the darker-purple (Xie L. et al., 2016). Genetic study indicated that the purple leaf B. juncea in our study was also controlled by a single dominant gene. The genetic pattern suggested that these purple leaf inbred lines may be controlled by the same causal gene. However, the purple leaves in B. napus were controlled by a candidate incomplete dominant gene BnAPR2, which encodes adenosine 5′-phosphosulfate reductase, localized at the end of chromosome A03 (Li et al., 2016). And the leaf color is apparently different from those reported in B. juncea. The different models of genetic inheritance suggested that the purple leaf gene involved in the molecular mechanism of the anthocyanin biosynthesis may be different.
It has been widely known that the anthocyanin biosynthesis is mainly controlled by a ternary complex of MYB-bHLH-WD40. In different plant tissues, the bHLH transcription factors could bind DNA either alone or interact with MYB transcription factors to regulate anthocyanin and PA biosynthesis (Heim et al., 2003). Up-regulated the regulatory gene BjTT8 and all biosynthetic genes may contribute to accumulation of anthocyanin in the purple Tumorous Stem Mustard (Xie et al., 2014). Transcriptome analysis showed that TT8 was highly up-regulated in all purple leaves from B. rapa, B. napus, B. juncea, B. oleracea, and B. carinata (Mushtaq et al., 2016). The BjTT8 was also up-regulated in purple leaf of B. juncea. In our study, another bHLH factor BjMYC2 was also up-regulated in the purple leaves. In Citrus sinensis, the highest expression of CsMYC2 was correlated with the purple leaf formation in the “Poros” lemon (Cultrone et al., 2009). The upregulation of BjTT8 and BjMYC2 may contribute to the anthocyanin accumulation in purple leaf of B. juncea. The ERF TFs have also been reported to be important in response to abiotic stresses, such as high salt, drought, hypoxia, and low temperature, etc. The anthocyanins are involved in plant defensive mechanisms against abiotic and biotic stresses (Asem et al., 2015). Most of the ERF TFs up-regulated in the purple leaves may help to defend against environmental stress.
In addition, based on RNA-seq data sets related to the purple leaf formation, numbers of genes involved in phenylpropanoid biosynthesis, phenylalanine metabolism, and flavonoid biosynthesis were found to be different expressed between purple and green leaves. The DEGs revealed that most of the anthocyanin biosynthesis genes were up-regulated in purple leaves. In purple-fleshed sweet potato, a root-preferential expressed gene, which encodes dihydrokaempferol reductase (IbDHKR), contributes to the anthocyanin biosynthesis in tuber roots (Liu et al., 2017). The MBW complex could activate the transcription of late anthocyanin biosynthesis pathway genes including DFR, ANS, etc. (Patra et al., 2013; Xu et al., 2015). In our research, BjDFR and BjANS were significantly higher expressed in purple leaves. This results well explained that these up-regulated anthocyanin biosynthesis genes in our study synergistically affect the purple leaf color. But the 4-coumaryol CoA ligase (4CL) gene, which catalyzes p-Coumaric acid to form 4-coumaroyl-CoA, was down-regulated in purple leaf. However, the transcript expression level of Br4CL1 from purple pakchoi cultivar “8389” was up-regulated (Jeon et al., 2018b). It was reported that 4CL5, which participated in phenylalanine metabolism, was expressed at very low level in different Brassica species. And apart from B. oleracea, almost no difference in transcript expression level of 4CL5 was found between purple and green leaves (Mushtaq et al., 2016). The 4CL gene played an important role in the pathways of lignin biosynthesis (Jiang et al., 2019). The down-regulated 4CL in our study may decrease the lignin biosynthesis and increase anthocyanin biosynthesis. Surprisingly, the most significant typical pathways between the purple and green leaves were the plant-pathogen interaction, alpha-linolenic acid metabolism, plant hormone signal transduction, and linoleic acid metabolism. Up expressed genes involved in typical pathways may be helpful to the adaptability to various environmental stresses. In summary, these data indicated the different response mechanism occurred between the purple leaves and green leaves. A better understanding of the anthocyanin biosynthesis and regulate network in Brassica is a prerequisite to develop anthocyanin-rich vegetables.
Conclusion
In conclusion, we identified different anthocyanin contents and transcriptome profile between purple and green leaves in B. juncea. Histological analysis showed that different leaf color was mainly caused by the accumulation of anthocyanin on the surface of the leaf. The differences in the transcriptome profile of the BC3 population were consistent with the different leaf color observed. The upregulation of BjTT8 and BjMYC2 may activate the anthocyanin biosynthesis in purple leaf.
Data Availability Statement
RNA seq data were deposited in the NCBI Sequence Read Archive (SRA) under accession number: PRJNA544908.
Author Contributions
SH, ZW, JS, and TF contributed to the design of the research. SH, HH, XY, CG, MC, ML, and QL performed the sample collection, RNA isolation, and gene expression analysis. SH and LW performed the bioinformatics analysis. SH wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the Key Scientific Research Projects of colleges and universities in Henan Province (20B210021), the Science and Technology Research Key Project of Henan Province (202102110001), the China Agriculture Research System (CARS-24-A-06), and Nanhu Scholars Program for Young Scholars of XYNU.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the reviewers for their constructive comments for revisions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00322/full#supplementary-material
FIGURE S1 | Technology roadmap of construction of the BC3 population.
FIGURE S2 | Gene ontology (GO) classification of different expressed genes between the purple and green leaves. The x-axis represents the enrichment of DEGs in each category, and the y-axis represents the enrichment ratio and numbers of each GO term. The jacinth bars represent the percentages of up-regulated genes in each category/the total number of up-regulated genes in each GO term. The light green bars represent the percentages of down-regulated genes in each category/the total number of down-regulated genes in each GO term.
TABLE S1 | The primers used for qPCR in this study.
TABLE S2 | Genetic analyses of the purple leaves in F2 and backcrossed segregating populations.
TABLE S3 | Detail of DEGs identified between the purple leaves and green leaves.
TABLE S4 | Differentially expressed transcription factors between the purple leaves and green leaves.
TABLE S5 | Orthologous genes involved in anthocyanin biosynthesis in B. juncea.
Footnotes
References
Albert, N. W., Davies, K. M., Lewis, D. H., Zhang, H., Montefiori, M., Brendolise, C., et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962–980. doi: 10.1105/tpc.113.122069
Asem, I. D., Imotomba, R. K., Mazumder, P. B., and Laishram, J. M. (2015). Anthocyanin content in the black scented rice (Chakhao): its impact on human health and plant defense. Symbiosis 66, 47–54. doi: 10.1007/s13199-015-0329-z
Castañeda-Ovando, A., Pacheco-Hernández, M. D. L., Páez-Hernández, M. E., Rodríguez, J. A., and Galán-Vidal, C. A. (2009). Chemical studies of anthocyanins: a review. Food Chem. 113, 859–871. doi: 10.1016/j.foodchem.2008.09.001
Chen, S., Wan, Z., Nelson, M. N., Chauhan, J. S., Redden, R., Burton, W. A., et al. (2013). Evidence from genome-wide simple sequence repeat markers for a polyphyletic origin and secondary centers of genetic diversity of Brassica juncea in China and India. J. Hered. 104, 416–427. doi: 10.1093/jhered/est015
Chiu, L. W., Zhou, X., Burke, S., Wu, X., Prior, R. L., and Li, L. (2010). The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol. 154, 1470–1480. doi: 10.1104/pp.110.164160
Cultrone, A., Cotroneo, P. S., and Reforgiato Recupero, G. (2009). Cloning and molecular characterization of R2R3-MYB and bHLH-MYC transcription factors from Citrus sinensis. Tree Genet. Genom. 6, 101–112. doi: 10.1007/s11295-009-0232-y
Fan, Y., Shen, J., and Dong, D. (2017). Development status and research prospect of mustard vegetables industry. Agricult. Sci. Technol. 18, 556–564.
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313x.2007.03373.x
Goswami, G., Nath, U. K., Park, J. I., Hossain, M. R., Biswas, M. K., Kim, H. T., et al. (2018). Transcriptional regulation of anthocyanin biosynthesis in a high-anthocyanin resynthesized Brassica napus cultivar. J. Biol. Res. (Thessalon) 25:19.
Guo, N., Wu, J., Zheng, S., Cheng, F., Liu, B., Liang, J., et al. (2015). Anthocyanin profile characterization and quantitative trait locus mapping in zicaitai (Brassica rapa L. ssp. chinensis var. purpurea). Mol. Breed. 35:113
Guo, N., Cheng, F., Wu, J., Liu, B., Zheng, S., and Liang, J. (2014). Anthocyanin biosynthetic genes inBrassica rapa. BMC Genomics 15:426. doi: 10.1186/1471-2164-15-426
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P. C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Heng, S., Gao, J., Wei, C., Chen, F., Li, X., Wen, J., et al. (2018). Transcript levels of orf288 are associated with the hau cytoplasmic male sterility system and altered nuclear gene expression in Brassica juncea. J. Exp. Bot. 69, 455–466. doi: 10.1093/jxb/erx443
Jeon, J., Kim, J. K., Kim, H., Kim, Y. J., Park, Y. J., Kim, S. J., et al. (2018a). Transcriptome analysis and metabolic profiling of green and red kale (Brassica oleracea var. acephala) seedlings. Food Chem. 241, 7–13. doi: 10.1016/j.foodchem.2017.08.067
Jeon, J., Lim, C. J., Kim, J. K., and Park, S. U. (2018b). Comparative metabolic profiling of green and purple pakchoi (Brassica Rapa Subsp. Chinensis). Molecules 23:1613. doi: 10.3390/molecules23071613
Jiang, J., Zhu, S., Yuan, Y., Wang, Y., Zeng, L., Batley, J., et al. (2019). Transcriptomic comparison between developing seeds of yellow- and black-seeded Brassica napus reveals that genes influence seed quality. BMC Plant Biol. 19:203.
Khoo, H. E., Azlan, A., Tang, S. T., and Lim, S. M. (2017). Anthocyanidins and anthocyanins: colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 61:1361779. doi: 10.1080/16546628.2017.1361779
Li, H., Zhu, L., Yuan, G., Heng, S., Yi, B., Ma, C., et al. (2016). Fine mapping and candidate gene analysis of an anthocyanin-rich gene, BnaA.PL1, conferring purple leaves in Brassica napus L. Mol. Genet. Genom. 291, 1523–1534. doi: 10.1007/s00438-016-1199-7
Lin, L. Z., Sun, J., Chen, P., and Harnly, J. (2011). UHPLC-PDA-ESI/HRMS/MS(n) analysis of anthocyanins, flavonol glycosides, and hydroxycinnamic acid derivatives in red mustard greens (Brassica juncea coss variety). J. Agric. Food Chem. 59, 12059–12072. doi: 10.1021/jf202556p
Liu, X. J., Lu, Y., Yuan, Y. H., Liu, S. Y., Guan, C. Y., Chen, S. Y., et al. (2013). De novo transcriptome of brassica juncea seed coat and identification of genes for the biosynthesis of flavonoids. PLoS One 8:e71110. doi: 10.1371/journal.pone.0071110
Liu, X., Min, X., Yufang, F., Chunxian, Y., Lingjiang, Z., Qitang, Z., et al. (2017). Root-preferential DFR-like gene encoding dihydrokaempferol reductase involved in anthocyanin biosynthesis of purple-fleshed sweet potato. Front. Plant Sci. 8:279. doi: 10.3389/fpls.2017.00279
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, Y. X., Du, D. Z., Fu, G., Xu, L., Li, X. P., Xing, X. R., et al. (2011). Inheritance of leaf color and sequence-related amplified polymorphic (SRAP) molecular markers linked to the leaf color gene in Brassica juncea. Afr. J. Biotechnol. 10. 14724–14730.
Mushtaq, M. A., Pan, Q., Chen, D., Zhang, Q., Ge, X., and Li, Z. (2016). Comparative leaves transcriptome analysis emphasizing on accumulation of anthocyanins in brassica: molecular regulation and potential interaction with photosynthesis. Front. Plant Sci. 7:311.
Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. doi: 10.1105/tpc.12.10.1863
Onkokesung, N., Reichelt, M., Van Doorn, A., Schuurink, R. C., Van Loon, J. J., and Dicke, M. (2014). Modulation of flavonoid metabolites in Arabidopsis thaliana through overexpression of the MYB75 transcription factor: role of kaempferol-3,7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J. Exp. Bot. 65, 2203–2217. doi: 10.1093/jxb/eru096
Patra, B., Schluttenhofer, C., Wu, Y., Pattanaik, S., and Yuan, L. (2013). Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta Gene Regul. Mech. 1829, 1236–1247. doi: 10.1016/j.bbagrm.2013.09.006
Petroni, K., and Tonelli, C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181, 219–229. doi: 10.1016/j.plantsci.2011.05.009
Song, H., Yi, H., Lee, M., Han, C. T., Lee, J., Kim, H., et al. (2018). Purple Brassica oleracea var. capitata F. rubra is due to the loss of BoMYBL2-1 expression. BMC Plant Biol. 18:82.
Sun, C., Deng, L., Du, M., Zhao, J., Chen, Q., Huang, T., et al. (2020). A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol. Plant 13, 42–58. doi: 10.1016/j.molp.2019.10.010
Sun, Q., Zhou, G., Cai, Y., Fan, Y., Zhu, X., Liu, Y., et al. (2012). Transcriptome analysis of stem development in the tumourous stem mustard Brassica juncea var. tumida Tsen et Lee by RNA sequencing. BMC Plant Biol. 12:53. doi: 10.1186/1471-2229-12-53
Thomasset, S., Teller, N., Cai, H., Marko, D., Berry, D. P., Steward, W. P., et al. (2009). Do anthocyanins and anthocyanidins, cancer chemopreventive pigments in the diet, merit development as potential drugs? Cancer Chemother. Pharmacol. 64, 201–211. doi: 10.1007/s00280-009-0976-y
Tohge, T., Nishiyama, Y., Hirai, M. Y., Yano, M., Nakajima, J.-I., Awazuhara, M., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42, 218–235. doi: 10.1111/j.1365-313x.2005.02371.x
Xie, L., Li, F., Zhang, S., Zhang, H., Qian, W., Li, P., et al. (2016). Mining for candidate genes in an introgression line by using RNA sequencing: the anthocyanin overaccumulation phenotype in brassica. Front. Plant Sci. 7:1245.
Xie, Q., Hu, Z., Zhang, Y., Tian, S., Wang, Z., Zhao, Z., et al. (2014). Accumulation and molecular regulation of anthocyanin in purple tumorous stem mustard (Brassica juncea var. tumida Tsen et Lee). J. Agric. Food Chem. 62, 7813–7821. doi: 10.1021/jf501790a
Xie, Y., Tan, H., Ma, Z., and Huang, J. (2016). DELLA Proteins Promote Anthocyanin Biosynthesis via Sequestering MYBL2 and JAZ Suppressors of the MYB/bHLH/WD40 Complex in Arabidopsis thaliana. Mol Plant 9, 711–721. doi: 10.1016/j.molp.2016.01.014
Xu, W., Dubos, C., and Lepiniec, L. C. (2015). Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Yan, C., An, G., Zhu, T., Zhang, W., Zhang, L., Peng, L., et al. (2019). Independent activation of the BoMYB2 gene leading to purple traits in Brassica oleracea. Theoret. Appl. Genet. 132, 895–906. doi: 10.1007/s00122-018-3245-9
Yang, J., Liu, D., Wang, X., Ji, C., Feng, C., Liu, B., et al. (2016). The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nature Genetics. 48, 1225–1232 doi: 10.1038/ng.3657
Yuan, Y., Chiu, L. W., and Li, L. (2009). Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta 230, 1141–1153. doi: 10.1007/s00425-009-1013-4
Zhang, Y. J., Chen, G. P., Dong, T. T., Pan, Y., Zhao, Z. P., Tian, S. B., et al. (2014). Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple bok-choy (Brassica rapa var. chinensis). J. Agricult. Food Chem. 62, 12366–12376. doi: 10.1021/jf503453e
Zhao, Z., Xiao, L., Xu, L., Xing, X., Tang, G., and Du, D. (2017). Fine mapping the BjPl1 gene for purple leaf color in B2 of Brassica juncea L. through comparative mapping and whole-genome re-sequencing. Euphytica 213:80
Keywords: anthocyanin, purple leaves, Brassica juncea, transcriptome, gene expression
Citation: Heng S, Wang L, Yang X, Huang H, Chen G, Cui M, Liu M, Lv Q, Wan Z, Shen J and Fu T (2020) Genetic and Comparative Transcriptome Analysis Revealed DEGs Involved in the Purple Leaf Formation in Brassica juncea. Front. Genet. 11:322. doi: 10.3389/fgene.2020.00322
Received: 05 December 2019; Accepted: 18 March 2020;
Published: 24 April 2020.
Edited by:
Luis Herrera-Estrella, Texas Tech University, United StatesReviewed by:
Cheng Qin, Zunyi Vocational and Technical College, ChinaFei Zhang, Yale University, United States
Copyright © 2020 Heng, Wang, Yang, Huang, Chen, Cui, Liu, Lv, Wan, Shen and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuangping Heng, c2h1YW5ncGluZ2hlbmdAMTI2LmNvbQ==
 Shuangping Heng
Shuangping Heng Lei Wang
Lei Wang Xi Yang1
Xi Yang1 Jinxiong Shen
Jinxiong Shen