- College of Animal Science and Veterinary Medicine, Henan Agricultural University, Zhengzhou, China
Liver is the main organ of lipid metabolism in chicken, especially for laying hens. To explore the molecular mechanism of lipid metabolism in chicken, five novel genes discovered in chicken liver tissue were systematically studied. Bioinformatic analysis was used to analyze the gene characteristics. The expression patterns and regulatory molecular mechanism of the five genes were examined. Our results showed that all five novel genes contain a common NADP-binding site that belongs to the NADB-Rossmann superfamily, and the genes were designated NADB-LER1-5. Phylogenetic tree of the NADB-LERs gene family in different species suggested these five genes originated from the same ancestor. Tissue distributions showed that NADB-LER1-4 genes were highly expressed in lipid metabolism organs, including liver, kidney and duodenum, and that the NADB-LER5 gene was highly expressed in liver and kidney. The spatiotemporal expression indicated that the expression levels of NADB-LER1-5 genes in liver tissue were significantly greater in sexually mature hens than that of immature pullets (P-value ≤ 0.05). The expression levels of NADB-LER1-5 were significantly induced by 17β-estradiol in primary cultured chicken embryo hepatocytes (P-value ≤ 0.05), and 17β-estradiol regulated the expression of NADB-LER1-5 mediated by ERα. Individual assays verified that under induction of 17β-estradiol, the five novel genes were significantly upregulated, with subsequent alteration in serum TG, TC, and VLDLs in 10-week-old pullets. This study proved NADB-LERs family mainly expressed in liver, kidney, and duodenum tissues. 17β-estradiol induces the expression of NADB-LER1-5 genes predominantly mediated via ERα. They likely involved in lipid metabolism in the liver of chicken.
Introduction
In chicken, liver is the most important and central site for lipid synthesis, responsible for more than 90% of total fatty acid de novo synthesis (O’Hea and Leveille, 1969). The hormonal control of lipid metabolism is a remarkable paradigm in laying hen. Studies have shown that estrogen could directly regulates the gene expression of chicken liver for yolk precursor synthesis (Bergink et al., 1974; Kirchgessner et al., 1987). Moreover, estrogen also is likely involved in regulating lipolytic activities and lipid transfer processes, which play important roles in the deposition of yolk into the fast-growing oocytes and in subsequent embryonic yolk utilization (Speake et al., 1998).
Especially during the egg-laying stage, under the influence of estrogen, many lipids including triacylglycerols, cholesteryl esters, and free fatty acids, are largely synthesized by the liver tissue. Then they are assembled into the egg-yolk precursors very low density lipoprotein (VLDL) and vitellogenin particles (Deeley et al., 1977), and transferred through blood circulation and deposited in the fast-growing oocyte (Brady et al., 1976; Wiskocil et al., 1980; Walzem et al., 1999; Nikolay et al., 2013). Therefore, lipid metabolism physiology is highly relevant to chicken embryonic growth development and the female reproductive system.
The lipid metabolism regulation process in poultry is highly similar to that in mammalian species, while recent work found that the functions of some genes and their products in poultry are different from mammals (Wiskocil et al., 1980; Kirchgessner et al., 1987; Hermier et al., 1996; Mason, 1999). Moreover, a recent study had demonstrated that some of the genes involved in lipid metabolism (e.g., resistin, TNFα, and SERPINE1) in poultry species, compared with mammals, may have been lost during the evolutionary process (Daković et al., 2014). Therefore, the hepatic lipid metabolism regulation mechanism of chicken needs to be elucidated.
Compared to other gene expression profile technology, RNA-seq is a superior method in analyzing gene expression at different development periods or in different tissues in a single assay, and it can help in the discovery of novel genes and splice variants (Trapnell et al., 2012). Previous study on liver tissue of chickens at the pre- and peak-laying stages by RNA-seq revealed the differentially expressed genes mainly involved in regulating lipid metabolism-related processes, and may do the same of the novel discovered protein-coding genes (Li et al., 2015).
A novel uncharacterized protein-coding gene family with five members was differentially expressed in the liver of hens between these two physiological stages (Li et al., 2015). According to bioinformatic analysis, the five members contain the same highly conserved NADP(H)-binding motif and the same substrate specificity site, which belong to NADB-Rossmann superfamily (Hanukoglu, 2015). Our subsequent study showed that the five NADB members with relatively high abundant expressed in chicken liver tissue were regulated by 17β-estradiol and were named NADB-LER1, NADB-LER2, NADB-LER3, NADB-LER4, and NADB-LER5. In addition, a β-ketoacyl acyl carrier protein reductase (FabG) component and an adh_short domain were also discovered among these novel genes. FabG, an NADPH-dependent β-ketoacyl-ACP reductase (Hoang et al., 2002), is an essential and universally expressed component of type II fatty acid biosynthesis (Zhang et al., 2003). To understand the biological functions of these genes, the cDNA sequences of the five mRNAs were cloned, and the phylogenetic relationship among different species was analyzed. Their tissue expression pattern and expression regulation mechanism were systematically investigated at in vivo and in vitro levels, respectively.
This report provides the first identification and expression profiling of NADB-LERs in chicken. The findings suggested that these newly identified proteins function in processes controlling lipid metabolism in the liver of chicken.
Materials and Methods
Ethics Statement
All the animal experiments were performed according to the Institutional Animal Care and Use Committee guidelines following protocols approved by the Henan Agricultural University (Permit Number: 11-0085).
Animals and Treatments
All the birds used in this study were provided by the Center of Poultry Germplasm Resource of Henan Agricultural University. They were raised in the same environmental conditions with ad libitum access to food and water.
The Lushi blue-shelled-egg chickens were used as the study animals. Hens at the pre-laying (20 week) stage without start laying and peak-laying (30 week) stage with an egg laying rate of over 70% were used (Li et al., 2015). Six hens at each stage were slaughtered and 11 tissues including the heart, liver, spleen, lung, kidney, duodenum, pectoral, abdominal fat, glandular stomach, pancreas and ovary tissues were collected, snap-frozen in liquid nitrogen and stored at −80°C until use. In addition, six female birds from the flock hatched in the same batch were slaughtered at each of the five different time points (1-day-old, 1-, 10-, 15-, and 35-week-old), and 11 tissues were collected and stored as mentioned above.
Sixty-four healthy female Lushi blue-shelled-egg chickens at the age of 10 weeks with similar body weights were divided into 4 groups with 16 birds in each group. The birds in the control group received an intramuscular injection of (specify volume) olive oil. The birds in the three experimental groups received the intramuscular injection of 0.5, 1.0, and 2.0 mg/kg body weight of 17β-estradiol (Sigma, St. Louis, MO, United States) (dissolved in olive oil), respectively. After injection for 12 h, eight birds from each group were randomly selected, and 3–5 mL blood sample of each bird was collected from wing vein. Then, the chickens were humanely slaughtered. Subsequently, the livers and kidneys were collected and stored as described above. The remaining eight birds in each group were continuously fed, and humanely killed until 24 h after injection. The blood samples, livers and kidneys were collected and stored as mentioned above.
Blood Physiological and Biochemical Index Measurement
The serum samples were collected from the fresh whole blood at room temperature for 1 h, then centrifuged at 2000 g for 15 min and stored at −70°C until further analysis. Blood physiological indexes of serum samples, including the low-density lipoprotein cholesterol (LDL-c) (Jiancheng, Corp., Nanjing, China) and high-density lipoprotein cholesterol (HDL-c) (Jiancheng), total cholesterol (TC) (Jiancheng), and triglyceride (TG) (Jiancheng), were determined using a blood analyzer (Hitachi 7100), while the very low-density lipoprotein cholesterol (VLDL-c) could instead be determined by the cholesterol contained in the VLDL and was indirectly determined by the Friedewald formula:VLDL-c = TC-HDL-c-LDL-c (Friedewald et al., 1972; Delong et al., 1986).
Cell Culture and Stimulation
The specific-pathogen-free (SPF) fertilized eggs purchased from Beijing Meiliyaweitong Experimental Animal Technology, Co., Ltd. (Beijing, China) were incubated in a sterile environment under suitable conditions (Ma et al., 2017). When the fertilized eggs were incubated for 18-day-old subjects, the embryonic primary hepatocytes were isolated from the liver tissue of the embryos according to a modified protocol based on the method put forward by Fischer and Marks (1976). In brief, the prepared hepatocytes were seeded in six-well plates and cultured in William’s E medium (Sigma, St. Louis, MO, United States) with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin. Hepatocytes were incubated at 37°C under a water-saturated atmosphere containing 95% air humidity and 5% CO2 overnight.
When the hepatocytes were grown to 70–80% confluence and serum-starved for 12 h, cells were exposed to 0, 25, 50, and 100 nM 17β-estradiol dissolved in olive oil, with six replicates for each treatment concentration. After stimulation for 12 h, the cells were collected and stored at −80°C for further experiments. This assay was repeated independently three times.
Different estrogen receptor antagonists, including methyl-piperidino-pyrazole (MPP), an estrogen receptor alpha (ERα)-selective antagonist, and ICI 182,780 and tamoxifen (Sigma-Aldrich), which are antagonists of both ERα and ERβ and may also act as the agonists of GPR30 on certain genes in some species (Guo et al., 2006; Pang et al., 2008), were dissolved in dimethyl sulfoxide (DMSO). When the cells were prepared in six-well plates for experiments, 17β-estradiol and 17β-estradiol combined with 1 μM MPP, 1 μM ICI 182,780, or 1 μM tamoxifen were used to treat the hepatocytes. Each treatment group had six replicates. The control group suffered from olive oil and DMSO at a final concentration of 0.1% each. After treatment for 24 h, cells were collected and stored at −80°C until use. The experiment was repeated independently three times.
Gene Cloning and Sequencing
Total RNA was extracted from cells and tissues using TRIzol reagent according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, United States). Transcription of 1 μg of RNA into cDNA was performed using the Thermo ScientificTM Revert Aid First Strand cDNA Synthesis kit (Thermo Scientific) according to the manufacturer’s instructions. The cDNA samples were stored at −20°C for further analysis.
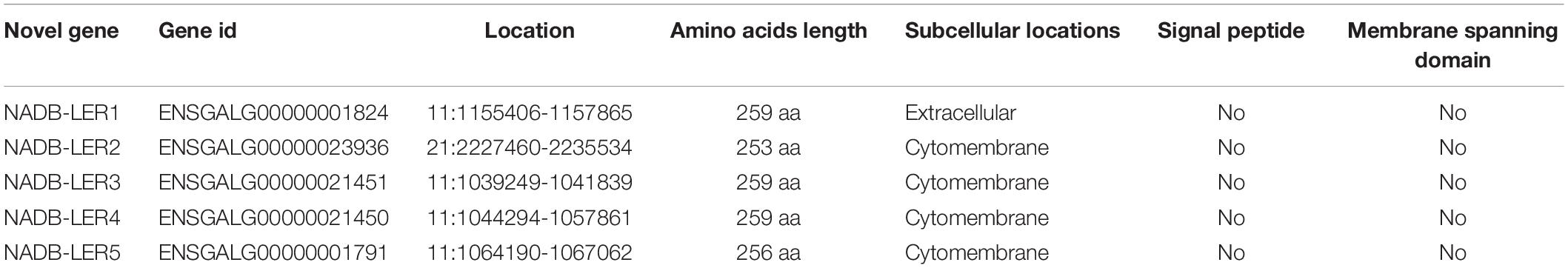
To understand the structures and potential biological functions of novel genes available from previous RNA-seq data (Li et al., 2015), bioinformatic analysis was performed to analyze the protein conserved domains of 91 up-regulated expressed novel genes. Five of these differentially expressed genes (ENSGALG00000001824, ENSGALG00000023936, ENSGALG00000021451, ENSGALG00000021450, and ENSGALG00000001791) after named NADB-LER1-5 were further studied.
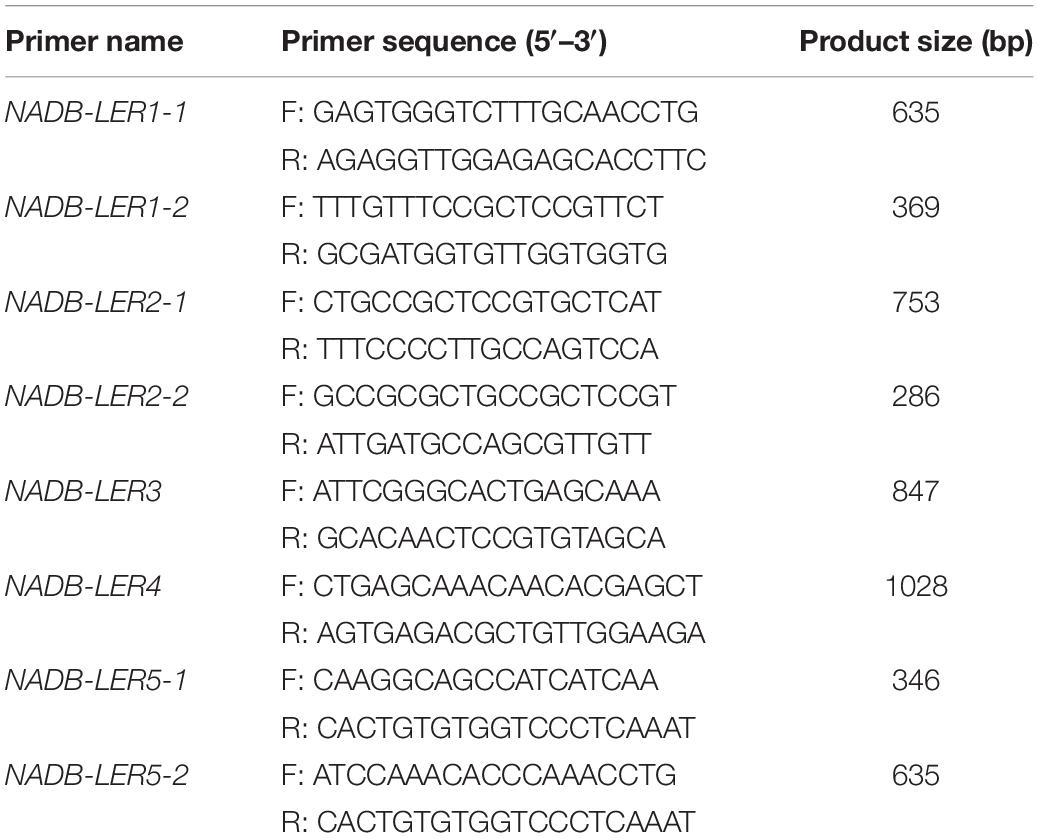
According to the predicted chicken NADB-LER1-5 sequences published in the Ensemble databank, several primer pairs were designed to produce overlapping fragments, and finally, the eight pairs of specific PCR primers in Table 1 were used for cloning the complete coding sequences (CDs) of the five mRNAs. Primers were designed using the Primer 5.0 software. PCRs were carried out in a total reaction volume of 25 μL, and the reaction component consisted of 1.0 μL of cDNA sample, 12.5 μL of 2 × pfu MasterMix (Tiangen Biotech, Co., Ltd., Beijing, China), 1.0 μL each of the forward and reverse primers (10 μM), and 9.5 μL of RNase-free water. The amplification reaction procedure was the following: an initial denaturation at 95°C for 5 min; followed by 35 cycles, with each cycle consisting of 30 s at 95°C, 30 s at 60°C and 30 s at 72°C; followed by a final elongation step at 72°C for 10 min. PCR products detected with the right fragment size were sequenced by Sangon Biotech, Co., Ltd. (Shanghai, China). Each sequence was sequenced on both strands.
Bioinformatic Analysis
To predict the protein structure and potential function of the five members, online databanks and software were used. The NCBI1 was used to predict the conserved domains. PSORT2 was used to analyze the subcellular localization. SignalP3 was used to predict the signal peptides. TMHMM4 was used to predict the membrane-spanning domains. ProtFun5 was used to predict the protein functions.
The complete CD sequences of the five members were obtained and used for phylogenetic analysis. The NADB-LER 1-5 nucleotide sequence alignments were performed using Clustal X2 (Thompson et al., 1994). Phylogenetic analyses of the five members among different species was constructed using the neighbor-joining method. Amino acid alignments were generated using Clustal V in the Molecular Evolutionary Genetics Analysis version 6.0 (MEGA 6) software (Tamura et al., 2013). The motif of the five genes were also predicted using online software MEME5.0.5.6 The evolutionary model was set to the Tamura 3-parameter model with a proportion of invariant sites and discrete gamma model implemented in PAUP∗ software (version 4.0b10) (Swofford, 2002). Evaluation of statistical confidence in nodes was based on 2000 bootstrap replicates. All branches with a bootstrap value equal to or greater than 50% are shown, while others were collapsed.
Quantitative Real-Time PCR (qPCR)
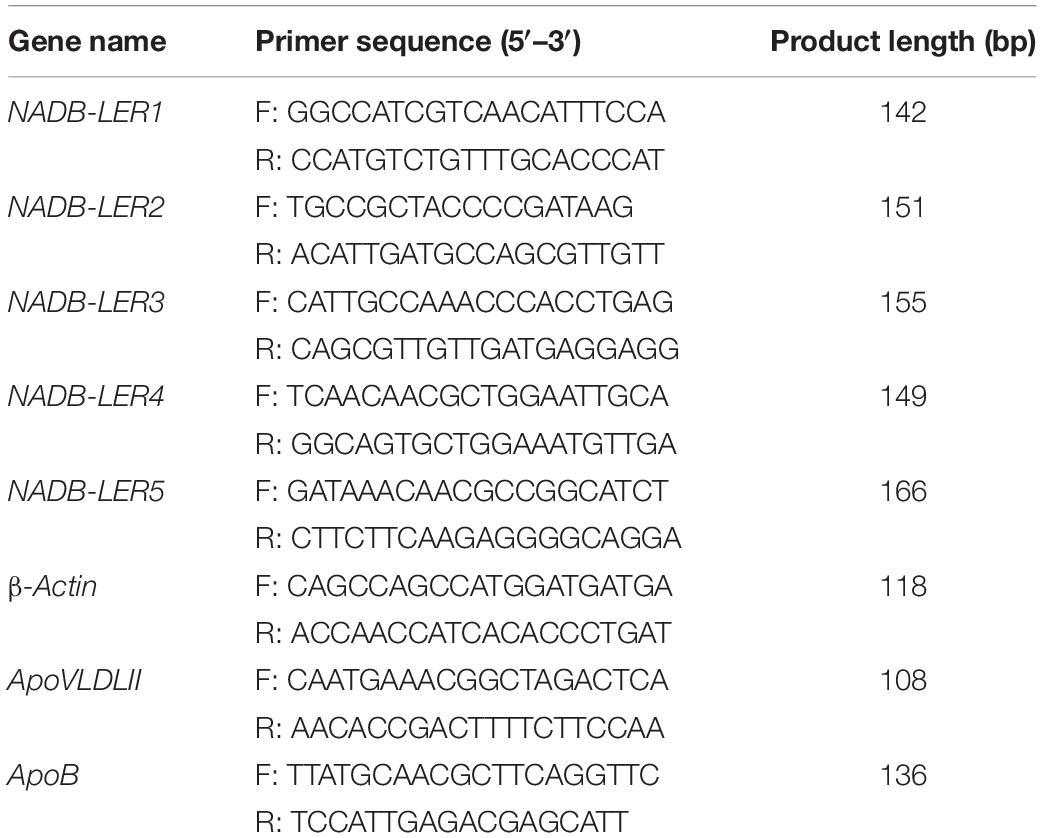
The oligonucleotide primers used for qPCR are shown in Table 2. All the primers were designed using the principle of span exons to avoid genomic DNA contamination. The qPCR was carried out on the LightCycler96 real-time PCR system (Roche Applied Science, Penzberg, Germany). Each reaction was performed in a final reaction volume of 10 μL containing 1 μL of cDNA, 5 μL of 2 × SYBR Green PCR Master Mix (Takara), 0.5 μL each of forward and reverse primer (10 μM) and 3 μL of RNase-free water. The qPCR amplification procedure was as follows: preincubation at 95°C for 5 min, and followed by 40 cycles of 30 s at 95°C, 30 s at 60°C and 30 s at 72°C. β-Actin mRNA was used as the housekeeping gene. ApoVLDL II, with the canonical estrogen response element (ERE), is an estrogen-dependent marker gene, is liver specific (Ryan et al., 1994) and was used as the positive control. All samples were analyzed in triplicate, and the average value of the triplicates was used for quantification.
Statistical Analysis
Gene expression levels were normalized to β-actin mRNAs and analyzed using the 2–ΔΔct method (Schmittgen and Livak, 2008). The One-Way Analysis of Variance followed by the Student’s t-test of the GraphPad Prism, version 5 (Graph Pad Software, Inc., San Diego, CA, United States) was used to determine the statistical significance. Due to the cDNA sample of each tissue used for expression distribution is a mixture at an equal volume of six individuals, therefore the data without significant analysis. Besides, all the rest values are presented as the mean ± SE, and differences with P-value ≤ 0.05 were considered statistically significant.
Results
Identification and Sequencing of Novel Genes in Chicken
To understand the structures and potential biological functions of NADB-LER1-5, bioinformatic analysis was performed. The five genes had the same NADP-binding motif and a substrate specific binding site shared by the NADB-Rossmann superfamily (Zhong et al., 2014). The sequence similarity of NADP-binding motifs ranged from 49.21 to 88.54%. In addition, a specific β-ketoacyl acyl carrier protein reductase (FabG) component, an Adh_short domain and a DItE domain were also discovered among the five novel genes (Supplementary Figure S1). According to the qPCR analysis, the five genes are significantly up-regulated in the liver tissue of 30 week hens relative to 20 week hens, which is in according with the RNA-seq data (Figure 1).
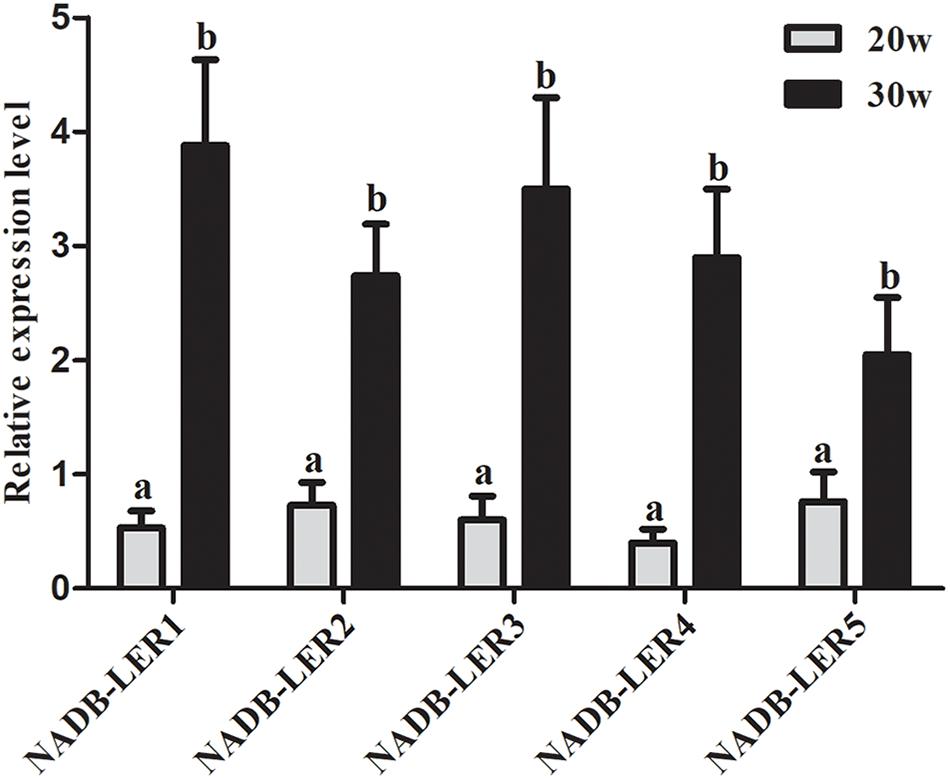
Figure 1. The relative expression levels of NADB-LERs mRNAs in the livers of chickens at the age of 20- and 30-week-old. a, b different letters indicate a significant difference between groups (P-value ≤ 0.05) (n = 6).
To identify novel chicken NADB-LERs, the draft chicken genomic database was queried using the predicted sequences published in the Ensemble databank. The PCR was performed to amplify the identified sequences using chicken liver tissue cDNA as template. The resulting DNA fragments were cloned and sequenced. By this approach, the full-length cDNA cloned from the chicken genes corresponding to NADB-LER1, NADB-LER2, NADB-LER3, NADB-LER4, and NADB-LER5 were obtained, and they are the same as the predicted sequences published in the Ensemble databank. They were found on chicken chromosomes 11 (NADB-LER1, -3, -4, and -5) and 21 (NADB-LER2). The subcellular localization analysis showed that NADB-LER1 is located extracellularly and the other four members are located on the cell membrane. In addition, none of them has a signal peptide or a membrane-spanning domain (Table 3). The amino acid alignment of the CDSs of the five chicken NADB-LERs is shown in Figure 2. The sequence identity is relatively high, and the N-myristoylation, N-glycosylation, and protein kinase C phosphorylation sites are well-conserved among the members.

Figure 2. Amino acid sequence alignment of chicken NADB-LERs by Clustal X2. The relatively conserved N-myristoylation, N-glycosylation, and protein kinase C phosphorylation sites are marked by rectangles, triangles and rhombuses, respectively. Amino acids identical in all five proteins are marked with an asterisk (∗), conservative substitutions with a colon (:), and semiconservative substitutions with a period (.).
Phylogenetic Analysis
To investigate the molecular function evolution and the relationships among the homologs, the NADB-LER amino acid sequences from chicken were aligned using phylogenetic methods. A BLAST search found that the novel genes only were presented in avian and other lower vertebrates. Multiple alignments of NADB-LER amino acid sequences from different species, including chicken and seven other representative species from avian, batrachians, reptiles and fish, were obtained by Clustal X2. The Ensemble IDs for each gene from this range of species are listed in Supplementary Table S1. The evolution analysis of these genes from different species showed that the phylogeny tree was divided into three clusters by NADB-LER2, NADB-LER5 and the other three members, and they belong to paralogs of the other three genes, while NADB-LER1, NADB-LER 3, and NADB-LER 4 are relative low homologous among different species, and divided into two small branches. In each cluster, they are almost own the similarity motifs (Figure 3).
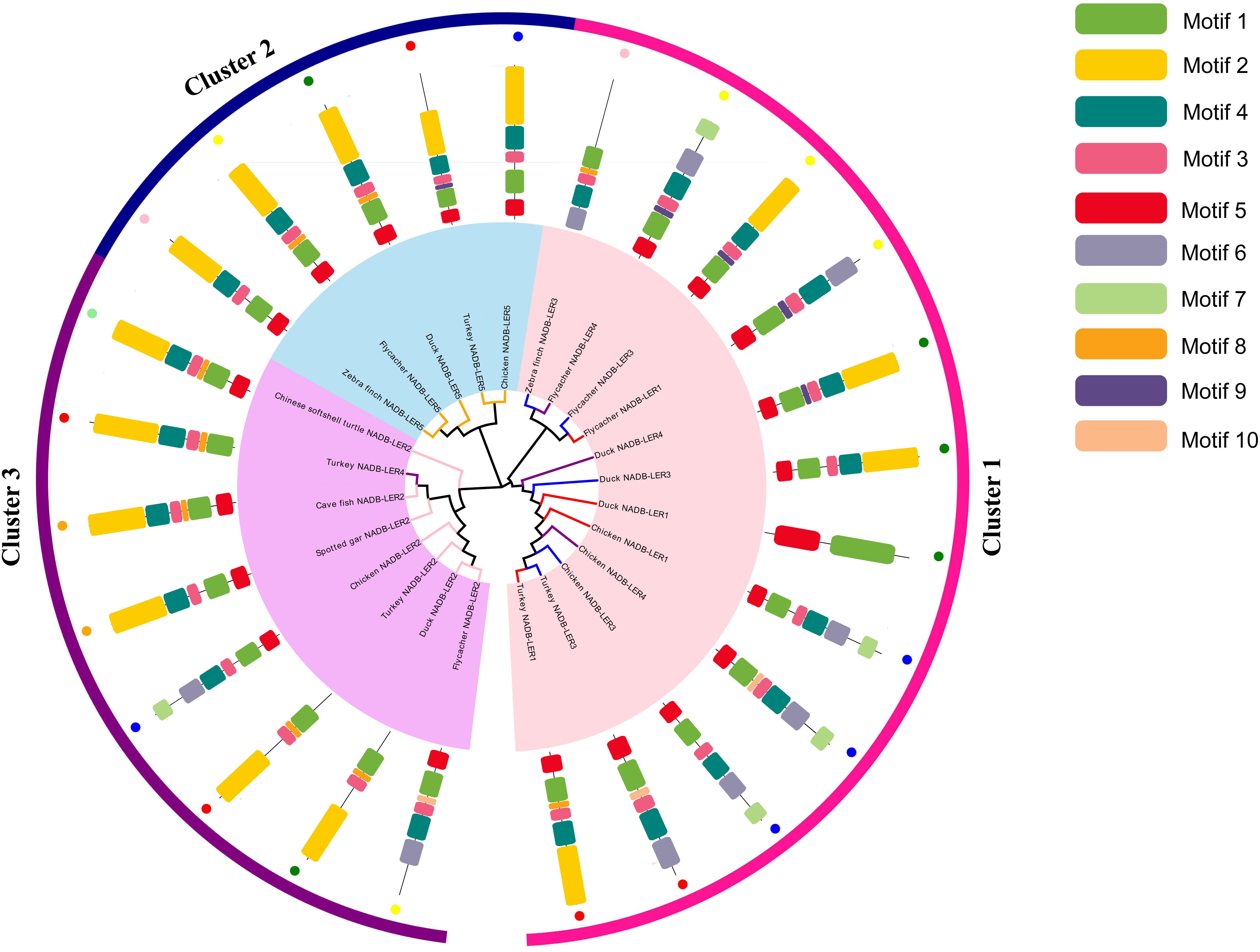
Figure 3. Consensus phylogenetic tree of NADB-LERs among different species. Different small color dots mean different species; the different color branches mean different members, except black.
Expression Profiles
To analyze the tissue expression profiles of the above-described NADB-LERs, the mRNA relative expression levels in 11 chicken tissues by qPCR were tested (Figure 4). The cDNA pool for each tissue mixed with six individuals was used as the temple. The results showed the five genes are expressed predominantly in liver tissue and expressed to a greater degree in 30-week-old hens than in 10-week-old hens. Relatively high expression levels of these genes were also measured in kidney at 10 weeks. The expression patterns of NADB-LER1 and NADB-LER4 resembled each other in different tissues. Duodenum expressed relatively high levels of NADB-LER1, -LER2, -LER3, and -LER4 and almost presented the same levels in 10- and 30-week-old hens. Lung expressed relatively high levels of NADB-LER1, -LER2, -LER4, and LER5, while the relative level is higher in 30-week-old than in 10-week-old hens. In contrast, heart, muscle and pancreas expressed relatively high levels of NADB-LER5 in 30-week-old hens.
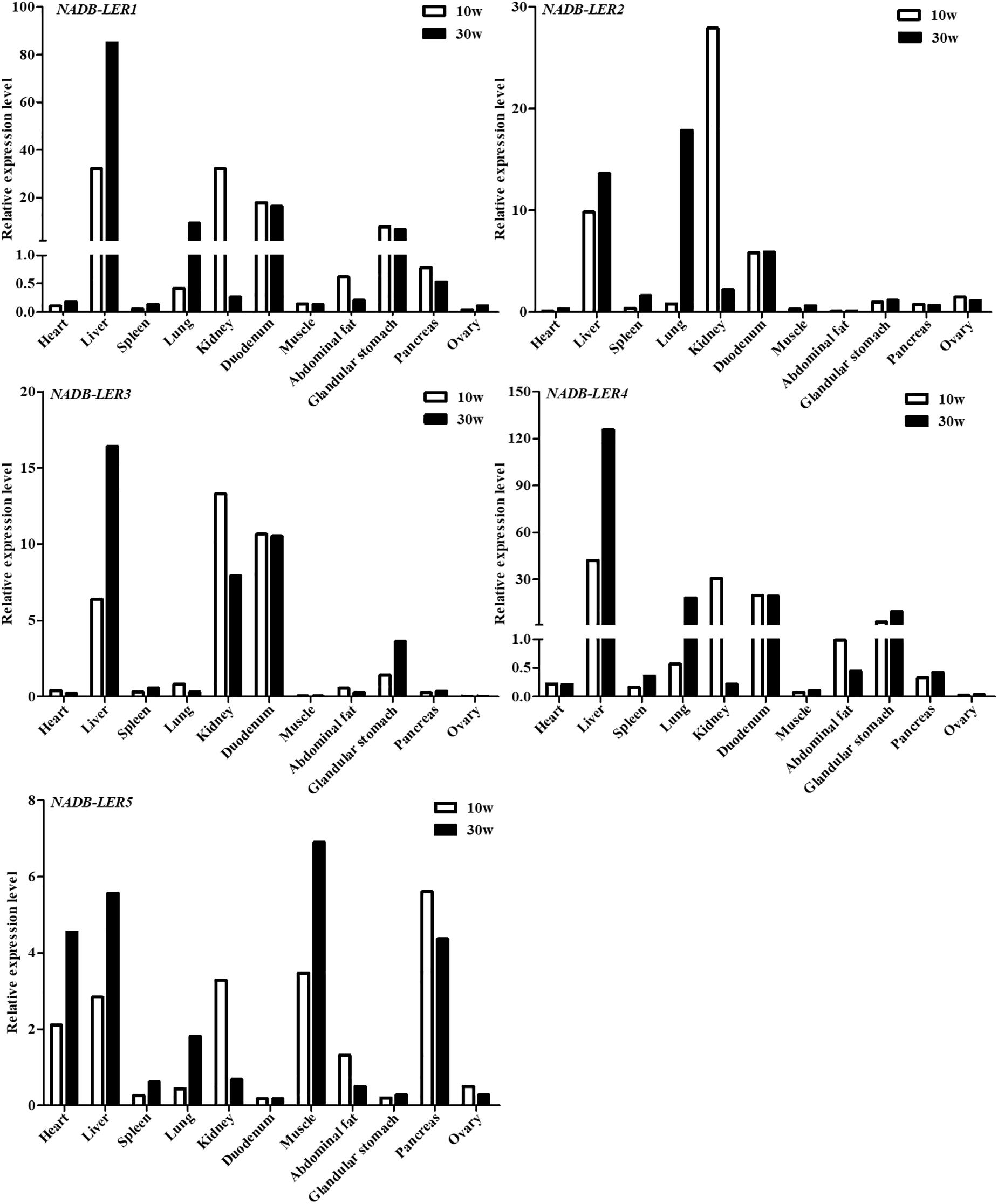
Figure 4. The relative expression levels of NADB-LERs in different tissues of 10- and 30-week-old chickens. n = 1, a cDNA pool of six individuals for each tissue sample was used as temple.
To gain insight into the spatiotemporal expression of NADB-LERs in the liver tissue of egg-laying chickens, their relative mRNA expression levels at seven different growth development stages were analyzed (Figure 5). Unexpectedly, the expression patterns of NADB-LERs resembled each other. In greater detail, the mRNA expression of the NADB-LERs at the peak-laying stages (30 and 35 week) were significantly higher than those of pre-laying stages (P-value ≤ 0.05), and there was no significant difference between the expression levels in 30- and 35-week-old hens (P-value > 0.05). Significant differences in NADB-LER expression levels were also not found among 1 day, 1-, 10-, 15- and 20-week-old hens (P-value > 0.05).
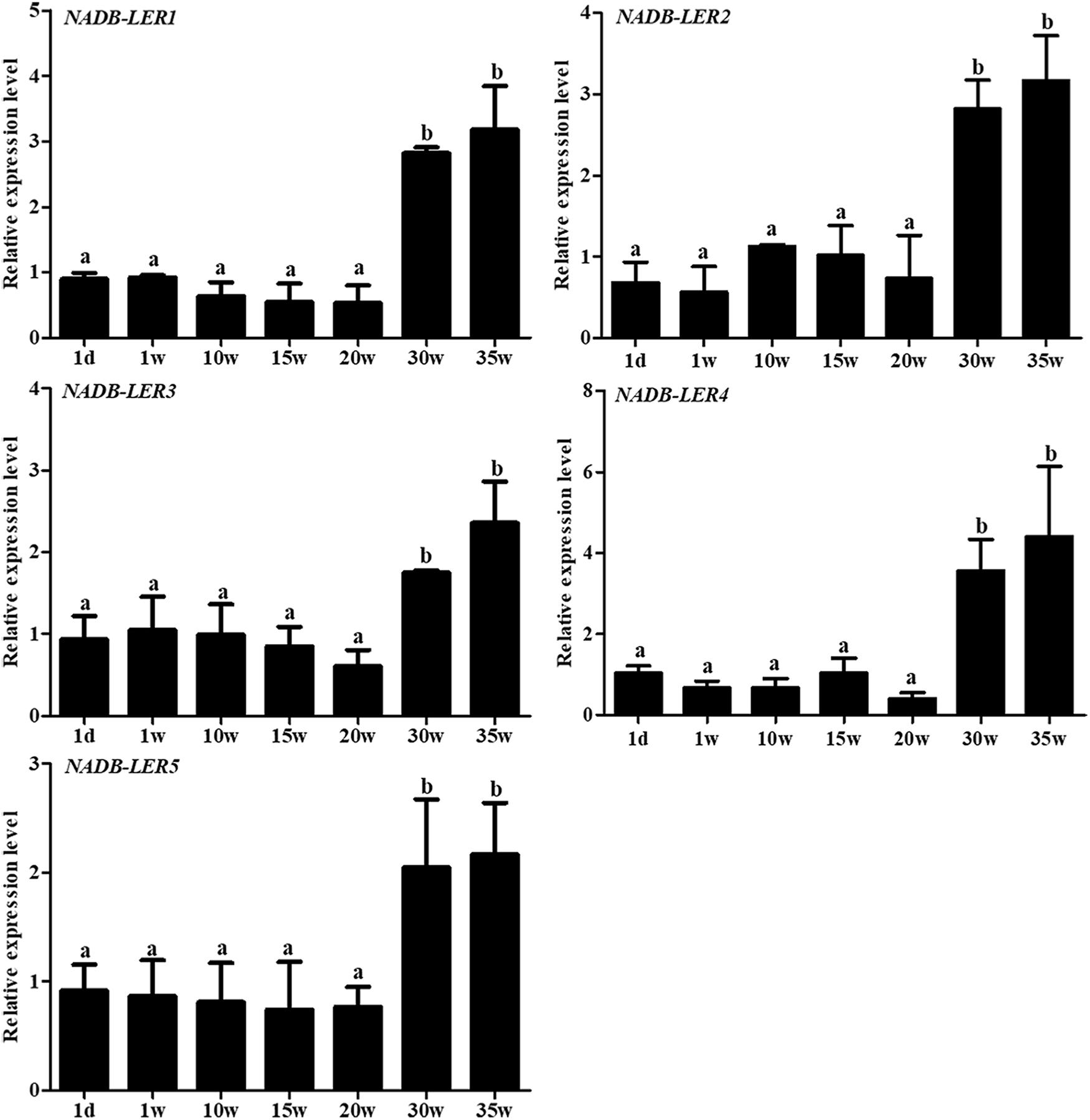
Figure 5. The relative expression levels of NADB-LERs genes in liver of chicken at different development stages. Different letters indicate a significant difference between groups (P-value ≤ 0.05). For each treatment, the data are expressed means ± SE (n = 6).
Regulation of Novel Chicken Genes by Estradiol in vitro
To understand the effect of 17β-estradiol on the expression of NADB-LERs in vitro, all the mRNA expression levels of the family members were analyzed in chicken embryonic primary hepatocytes stimulated with 17β-estradiol. The results showed that compared with their levels in the control group, both ApoVLDL II and ApoB genes were significantly upregulated by different concentrations of estradiol, and they presented dose-dependent expression (P-value ≤ 0.05) in chicken hepatocytes (Figure 6). Compared with their levels in the control, the mRNA expression levels of NADB-LER1-4 in chicken hepatocytes were significantly increased under induction by estradiol at 50 and 100 nM concentrations (P-value ≤ 0.05), while there was no significant difference between the two treatment groups (P-value > 0.05). Different from the levels of the other members, the NADB-LER5 mRNA expression level was significantly upregulated at 25 and 50 nM concentrations (P-value ≤ 0.05) but returned to the control level when cells were treated with 100 nM 17β-estradiol (P-value > 0.05) (Figure 7). In addition, 50 nM was the optimal concentration that induced the expression of NADB-LERs.
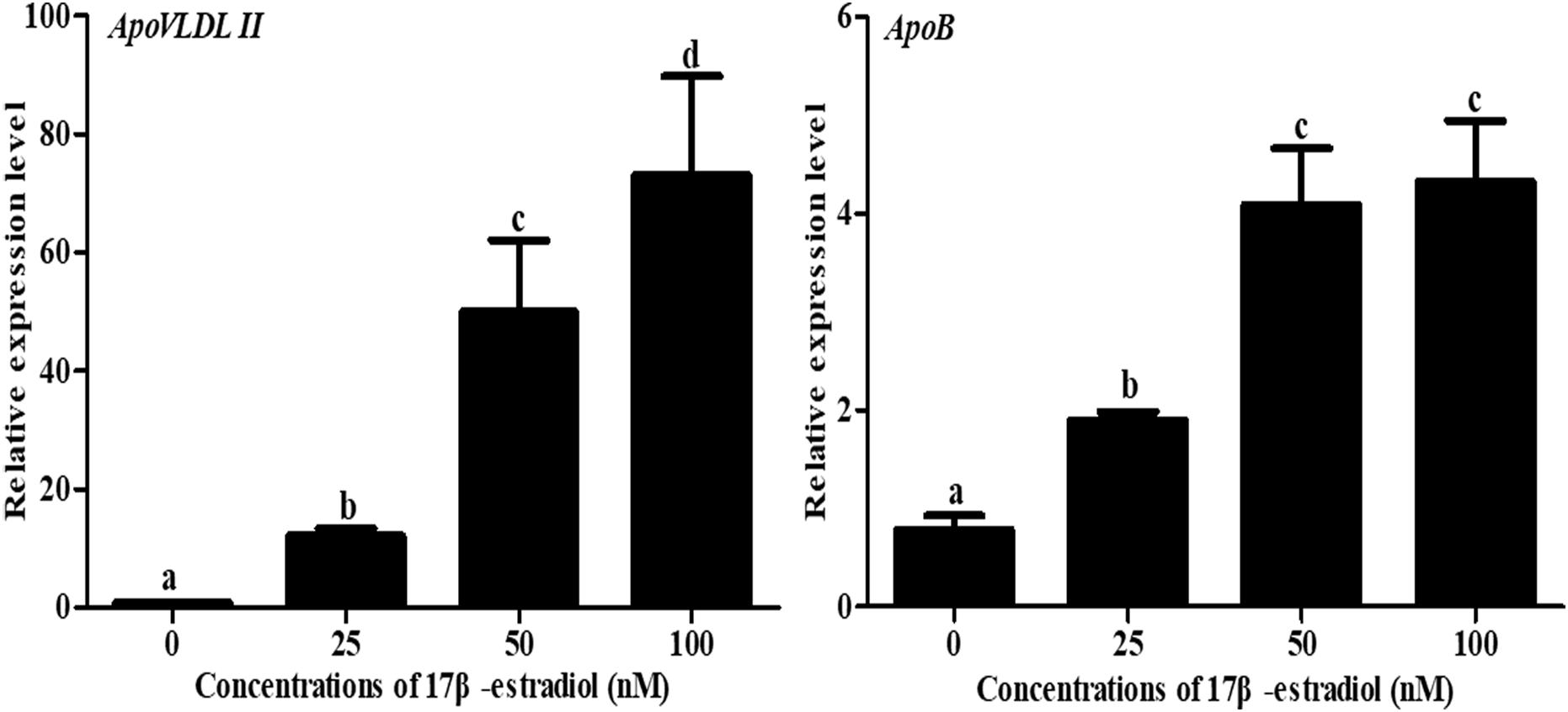
Figure 6. Effects of different doses of 17β-estradiol on the expression of ApoB and ApoVLDL II in primary hepatocytes after treatment for 12 h. Different letters indicate a significant difference (P-value ≤ 0.05). For each treatment, the data are expressed means ± SE (n = 6).
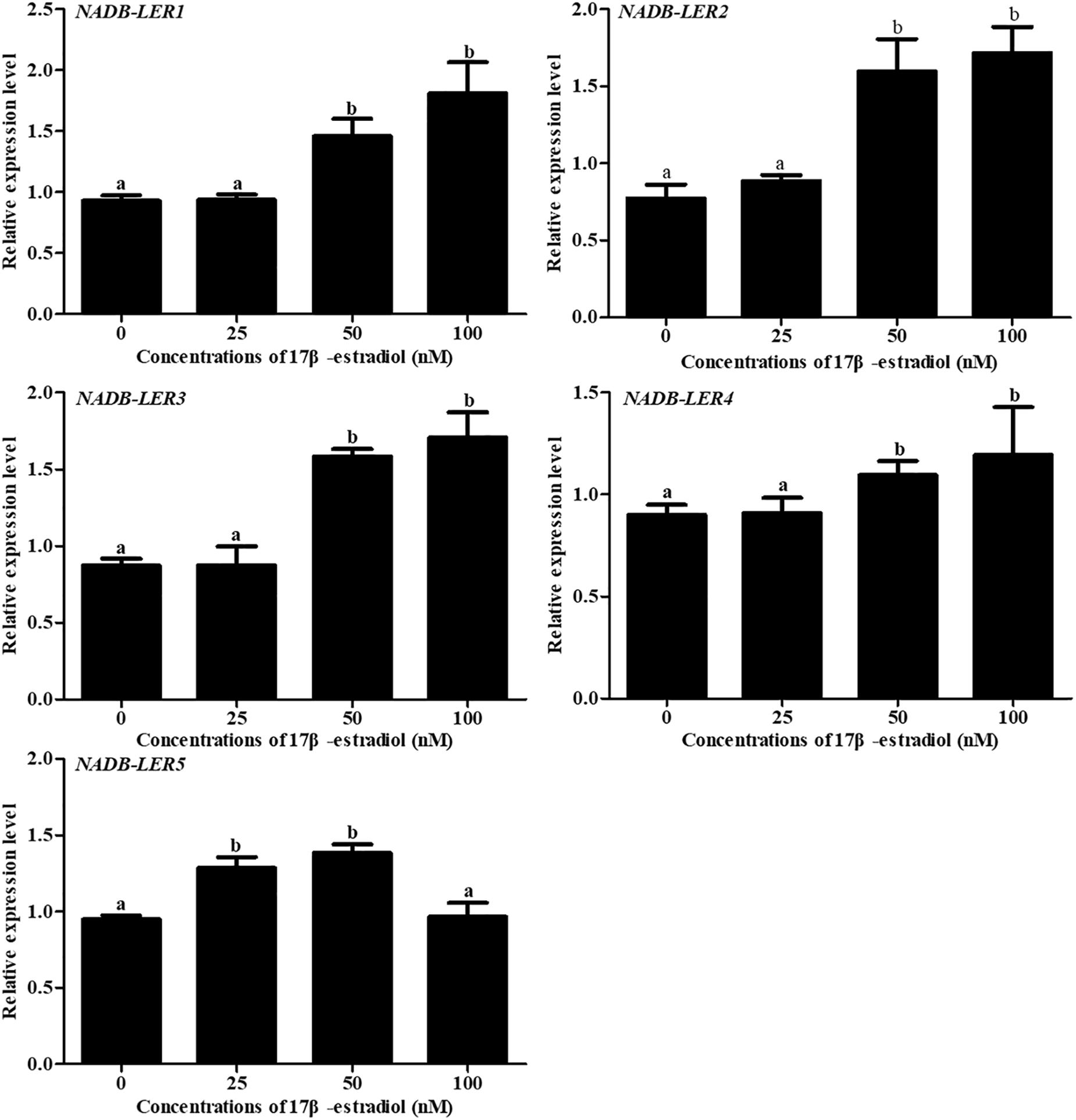
Figure 7. The effects of different doses of 17β-estradiol on the expression of NADB-LERs in primary hepatocytes after treatment for 12 h. Different letters indicate a significant difference (P-value ≤ 0.05). For each treatment, the data are expressed means ± SE (n = 6).
To further analyze the estrogen receptor (ER) subtype that mediates the expression of NADB-LER genes, chicken embryo primary hepatocytes were treated with 17β-estradiol alone or combined with different ER-subtype antagonists (Figure 8). As a positive control, compared with the 50 nM 17β-estradiol treated group, the expression of ApoVLDL II was significantly down-regulated when the hepatocytes were treated with 50 nM 17 β-estradiol combined with 1 μM MPP (P-value ≤ 0.05), and there was no significant difference among the groups treated with 50 nM 17 β-estradiol combined with either 1 μM MPP, or 1 μM tamoxifen or 1 μM ICI 182,780 (P-value > 0.05). Similarly, the expression levels of NADB-LERs were significantly decreased (P-value ≤ 0.05) when the hepatocytes were treated with 50 nM 17 β-estradiol combined with 1 μM MPP or 50 nM 17 β-estradiol combined with 1 μM tamoxifen and 1 μM ICI 182,780 (P-value ≤ 0.05). The expression levels of the genes returned to the control level in the three ER subtype antagonist-treated groups (P-value > 0.05). The results revealed that estrogen effects on the expression of chicken hepatic NADB-LERs were mediated by ERα.
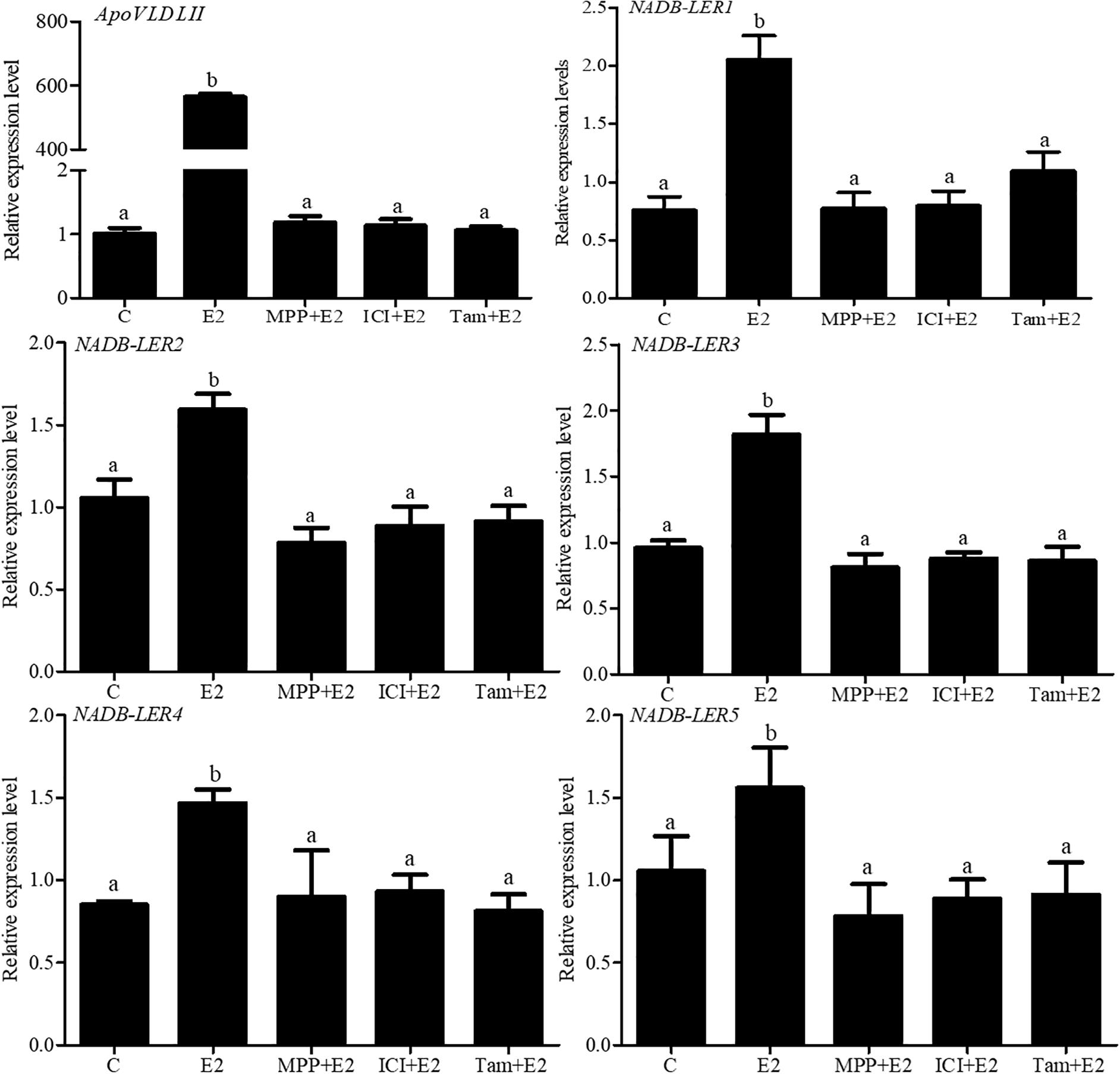
Figure 8. Effects of ER antagonists on the expression of NADB-LER1-5 in chicken embryo hepatocytes. C: the control group; E2:17β-estradiol (50 nM); ICI: ICI182,780 (1 μM), TAX: tamoxifen (1 μM), and MPP: (1,3-bis (4-hydroxyphenyl) -4-methyl-5-[4-(2-piperidinylethoxy) phenol]-1H-pyrazoledihydrochloride) (1 μM). Different letters indicate significant difference among the different treatments (P-value ≤ 0.05). For each treatment, the data are expressed means ± SE (n = 6).
Estradiol Regulation in vivo
To further validate whether estrogen influenced the expression of chicken NADB-LERs in individuals, female pullets at the age of 10 weeks were treated with different doses of 17β-estradiol for 12 h and 24 h. The blood biochemical index of the chickens was measured, and the results showed estrogen could significantly increase the concentrations of TG and TC when applied for 12 h (P-value ≤ 0.05), while the concentration of VLDLs was not changed (P-value > 0.05). Twenty-four hours of 17β-estradiol treatment at concentrations of 1 and 2 mg/kg dramatically increased the concentrations of TG, TC, and VLDLs (P-value ≤ 0.05) (Figure 9). Therefore, the optimal response time to 17β-estradiol for chicken was 24 h. In addition, both the expression of ApoVLDL II and ApoB genes were significantly increased in a dosage-dependent manner after treatment with 17β-estradiol for 24 h (P-value ≤ 0.05) (Figure 10).
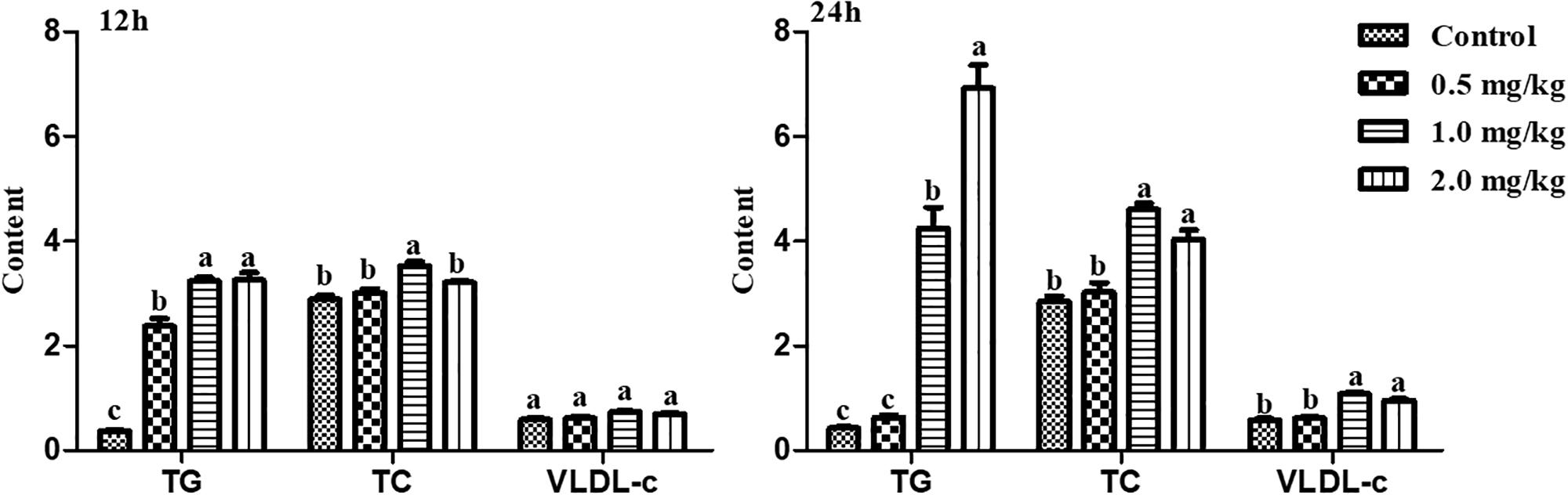
Figure 9. The differences of TG, TC, and VLDL-c contents in chicken serum treated with different concentrations of 17β-estradiol for 12 and 24 h, respectively. Different letters indicate a significant difference (P-value ≤ 0.05). For each treatment, the data are expressed as the means ± SE (n = 8).
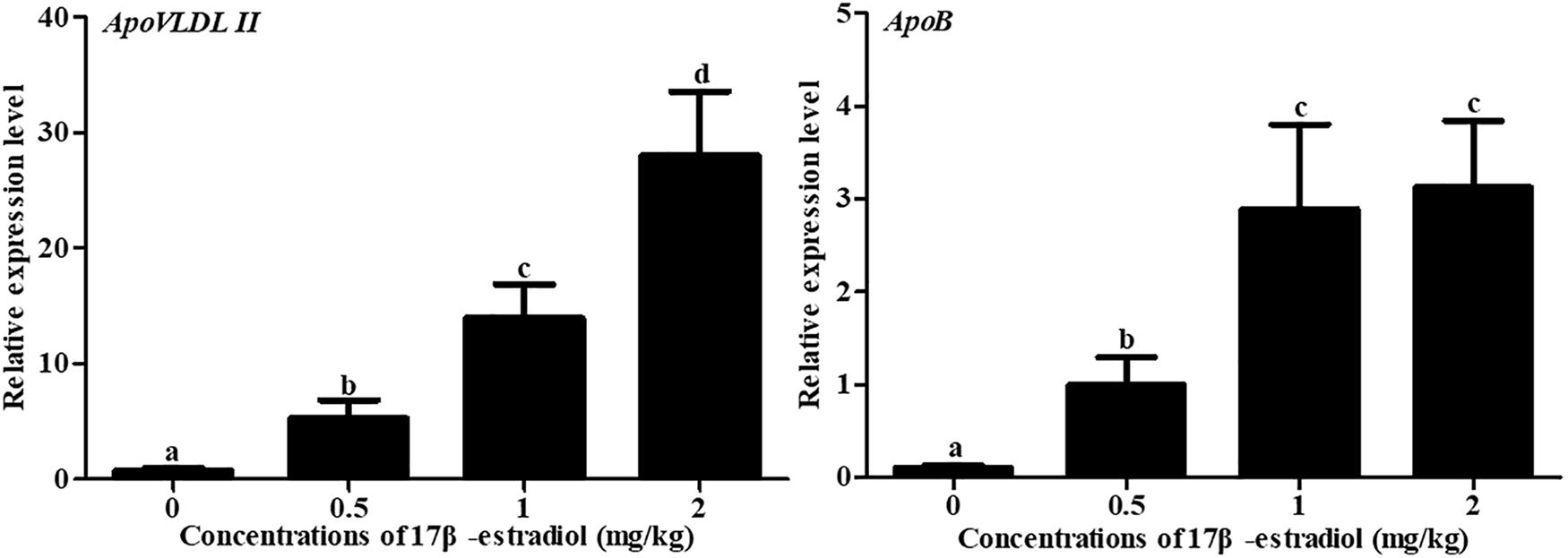
Figure 10. Effects of different doses of 17β-estradiol on the expression of ApoB and ApoVLDL II in the livers of 10-week-old pullets after treatment for 24 h. Different letters indicate a significant difference (P-value ≤ 0.05). For each treatment, the data are expressed as the means ± SE (n = 8).
To analyze the effect of 17β-estradiol on NADB-LERs in chicken liver tissue, the mRNA expression levels of NADB-LERs were analyzed in the liver of chicken treated with 17β-estradiol for 24 h. Compared to the levels in the control group, the relative expression levels of the five genes were significantly up-regulated at treatment concentrations of 1 and 2 mg/kg (P-value ≤ 0.05), while no significant difference was found between the two groups (P-value > 0.05); the expression levels of the five genes at a treatment concentration of 0.5 mg/kg were not changed from their levels in the controls (P-value > 0.05) (Figure 11).
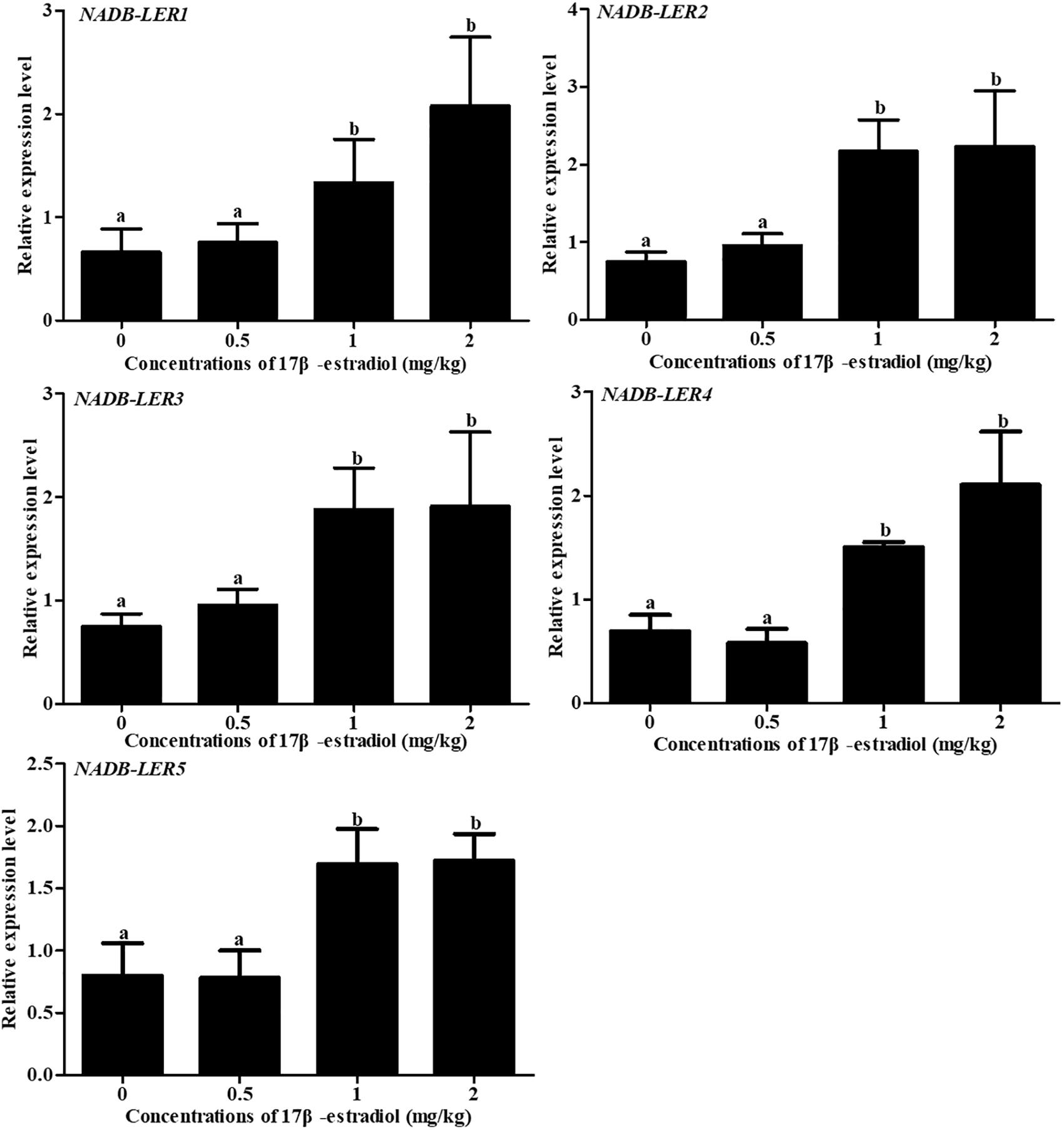
Figure 11. Effects of different doses of 17β-estradiol on the expression of NADB-LERs in the livers of 10-week-old pullets after treatment for 24 h. Different letters in each figure indicate a significant difference (P-value ≤ 0.05). For each treatment, the data are expressed as the means ± SE (n = 8).
To understand the effect of estrogen on NADB-LERs in chicken kidney, the mRNA expression levels of NADB-LERs were analyzed in the kidney tissue of chicken treated with estrogen for 24 h. Meanwhile, the expression of the ApoVLDL II gene was also analyzed. Compared to its level in the control group, ApoVLDL II was significantly up-regulated by different concentrations of 17β-estradiol and presented dose-dependent expression (P-value ≤ 0.05). In contrast, the relative expression levels of the NADB-LERs were significantly down-regulated from their control levels at different treatment concentrations (P-value ≤ 0.05), while no significant difference was found among the three treatment groups (P-value ≤ 0.05) (Figure 12).
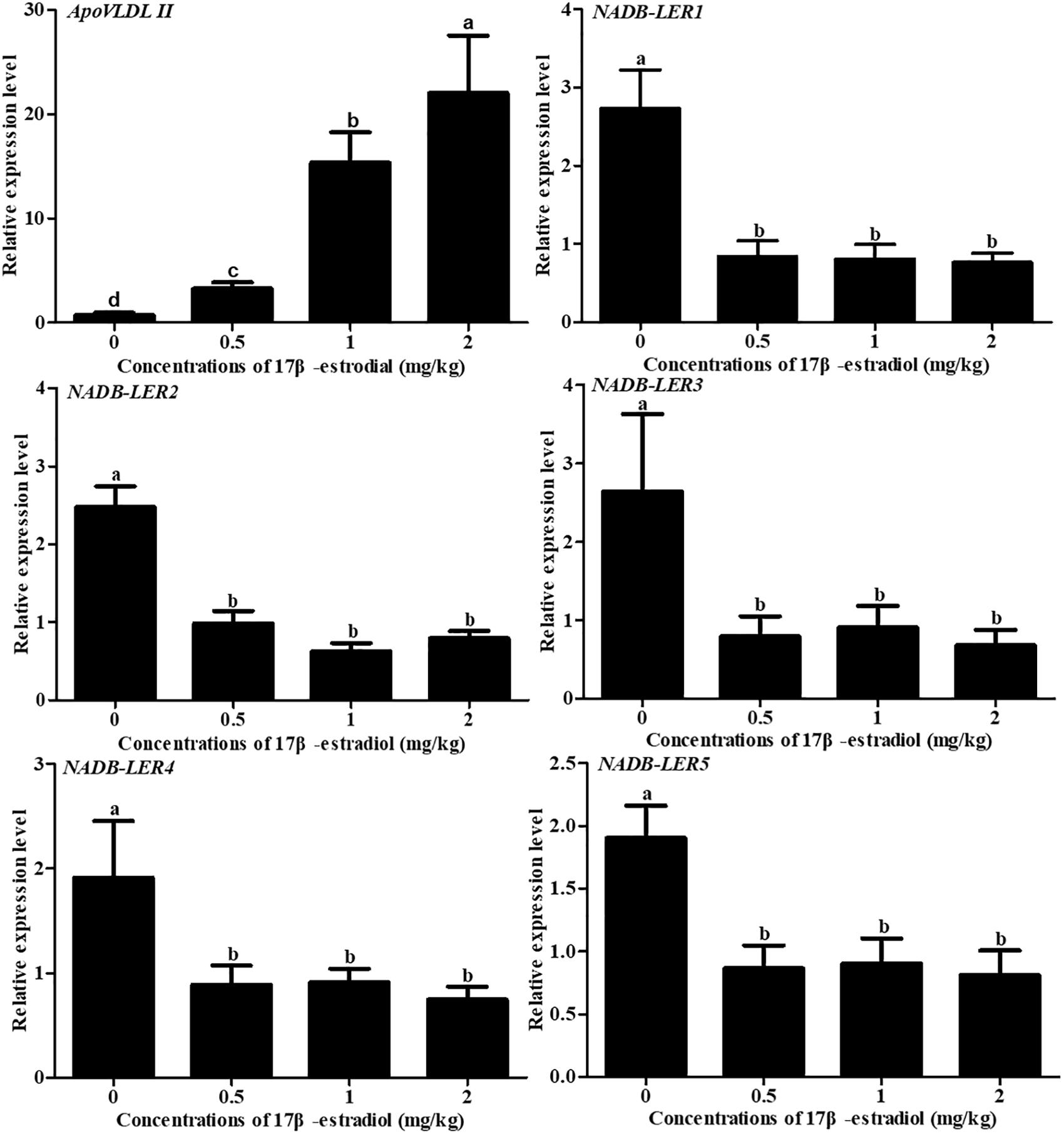
Figure 12. Effects of different doses of 17β-estradiol on the expression of ApoVLDL II and NADB-LERs in the kidney of 10-week-old pullets after being treated for 24 h. Different letters in each figure indicate a significant difference (P-value ≤ 0.05). For each treatment, the data are expressed as the means ± SE (n = 8).
Discussion
In this study, five differentially expressed novel genes called NADB-LERs were found basing on a previous RNA-seq experiment (Li et al., 2015). Our study first systemically explored the characteristics of the NADB_Rossmann superfamily in chicken. The NADB-LER members were identified to contain the same conserved NADB-Rossmann structural domain and special component FabG using bioinformatics. Phylogenetic tree analysis suggested that the NADB-LER1-5 genes originated from the same ancestor. Therefore, the NADB-LERs may have been generated through gene duplication, and different functions were produced in the process of evolution.
FabG, an essential and universally expressed component of type II fatty acid biosynthesis, appeared in NADB-LERs. FabG enzymes are members of the short-chain alcohol dehydrogenase/reductase (SDR) family (Nanson and Forwood, 2015). In addition, SDR enzymes play critical roles in lipid, amino acid, carbohydrate, cofactor, hormone, and xenobiotic metabolism and in redox sensor mechanisms (Mayoral et al., 2013). Five NADB-LER members were significantly up-regulated in the liver of peak-laying hens compared to pre-laying female chickens (Li et al., 2015). The most significant physiological difference between 20- and 30-week-old layer hens is whether they lay eggs or not; during the egg-laying period, to supply sufficient nutrients for embryo growth and development, neutral fatty acids, mainly triacylglycerols, are synthesized in the liver of hen under promotion by estrogen (Bergink et al., 1974). It was suggested that the five novel genes are related to lipid metabolism.
The five genes were extensively expressed in various tissues, while the high levels of NADB-LER1, -2, -3, and -4 in the liver, kidney and duodenum of chicken may indicate tissue-specific role(s) of these genes. Liver, kidney and duodenum, as the main lipid metabolism organs in chicken, undertake different metabolism roles. VLDLy, intermediate-density lipoprotein (IDL) and low-density lipoprotein (LDL) are mainly synthesized and secreted in liver, chyle particles (CM) are mainly synthesized in small intestine, and VLDL and high-density lipoprotein (HDL) are mainly synthesized in and secreted from kidney (Bensadoun and Rothfeld, 1972; Tarugi et al., 1998). Moreover, in chicken, most of the yolk protein, including apolipoproteins and vitellogenins, are synthesized by the liver tissue (Deeley et al., 1977). The relative mRNA levels of NADB-LER1, -2, -3, -4, and -5 were clearly more highly expressed in the liver of peak-laying hens than in that of juvenile chickens and were clearly less expressed in the kidney of peak-laying hens than in that of juvenile chickens, but no dramatic difference in their levels was found in the duodenum between the two stages. In birds, fat synthesis is extremely active in the liver at the stage of egg-laying (Macwhirter, 1999). The spatiotemporal expression of NADB-LERs in chicken liver showed that their expression was significantly up-regulated in the peak-laying stages relative to unmatured hens. It was suggested that NADB-LER1, -2, -3, -4, and -5 may be strongly related to chicken hepatic lipid metabolism and play different roles in different tissues.
Estrogen, as a class of steroid sex hormone, is responsible for the development and regulation of the female reproductive system (Mohamadi et al., 2018). Generically, lipid metabolism is tightly regulated by estrogen in chicken with the onset of an egg-laying cycle (Hermier et al., 1996; Ivessa et al., 2013). Studies demonstrated that in estrogen-stimulated groups, the expression levels of ApoVLDL II and ApoB genes presented a dose-dependent increase (Miller and Lane, 1984; Hermier et al., 1989). Consistent with previous reports, our in vitro assay results indicated that 17β-estradiol significantly promoted the expression of ApoVLDL II and ApoB mRNA in chicken embryonic primary hepatocytes, suggesting that 17β-estradiol played a role in the hepatocytes. Meanwhile, NADB-LER1, -2, -3, -4, and -5 were significantly up-regulated in 50 and 100 nM 17β-estradiol-treated groups. This result demonstrated that the expression of five novel genes could be controlled by 17β-estradiol in hepatocytes. Estrogen-inducible transcription regulation can be mediated via estrogen receptors (ERα and ERβ) or via intracellular G protein-coupled receptor 30 (GPR30) (Fietz et al., 2014), but the exact participation of the individual receptor is not clear. Our in vitro studies demonstrated that in chicken embryo hepatocytes, as a positive control, estrogen stimulates the expression of ApoVLDL II mRNAs predominantly through ER-α in the chicken liver, which were consist with previous report (Li et al., 2014). 17β-estradiol could significantly increase the expression of NADB-LER1-5, while the combined treatment of highly selective ERα antagonist MPP with 17β-estradiol almost fully blocked the effect.
In comparison with the MPP and 17β-estradiol-treated group, the combinations of ERα and ERβ antagonists ICI182,780 and tamoxifen with 17β-estradiol treatments failed to change the expression of NADB-LERs. It has been widely proven that tamoxifen, as a synthetic estrogen antagonist, represses target genes via binding to ERs at the transcriptional level (Lerner and Jordan, 1990) and that ICI 182,780 is a high-affinity ER antagonist (Turner et al., 2000). From these results, we concluded that estrogen activated the expression of NADB-LERs in chicken embryo hepatocytes mediated by ERα. The ER-mediated regulation of gene expression involves the direct binding of dimeric ER to DNA sequences known as estrogen response elements (EREs) (Klinge, 2001), or indirectly associate with promoters through protein-protein interactions with other DNA-binding transcription factors (Safe, 2001; Marino et al., 2006). To further sight into how estrogen regulated the expression of NADB-LER1-5 genes through ERα, the ERα specific EREs within the promoter region, 2.0 kb upstream the transcription start sites were predicated by using online software MEME Suite 5.1.0. The results showed that two putative ERα binding EREs occurred in the promoter regions of the NADB-LER1 and NADB-LER 3 genes, one occurred in NADB-LER4, and no one was found in NADB-LER2 and NADB-LER5. Therefore, it suggested that the ERα dimer might activate the transcription of NADB-LER1, -3 and -4 genes by directly binding to EREs within their promoter, but activate the transcription of NADB-LER2 and -5 genes by binding to other transcriptional activators which indirectly regulated the genes’ transcription. However, we could not rule out the possibility that there were some ERα specific EREs existed in the promoter regions of NADB-LER2 and -5 genes further upstream.
During the chicken egg-laying stage, the increased production of VLDLs is accompanied by the upregulation of lipid synthesis and protein components, including ApoVLDL II and ApoB (Chan et al., 1976; Capony and Williams, 1980). The formation of hepatic VLDLs in avian is tightly regulated by estrogen, and therefore genes involved in the processes would be steered by estrogen. Previous reports demonstrated that estrogen administration to immature pullets (Heald and McLachlan, 1964) and roosters (Greengard et al., 1964) could lead to plasma changes similar to those occurring in the sexually mature female. In our study, the concentrations of TG, TC, and VLDL in the blood of 10 week pullets stimulated by 17β-estradiol hormone were dramatically increased, which is in accordance with previous references. In addition, the hormone caused massive synthesis of ApoVLDL II and ApoB in the liver tissue of chicken (Kirchgessner et al., 1987; Evans et al., 1988). Moreover, the NADB-LER1-5 genes were significantly up-regulated by 17β-estradiol in the liver tissue of chicken, which further proved that NADB-LER1-5 genes are likely involved in hepatic lipid metabolism under induction by 17β-estradiol in chicken.
Meanwhile, NADB-LER1-5 genes were significantly inhibited in the kidney tissues of pullets under stimulation by 17β-estradiol. It is likely that chicken kidney could produce apoB-lipoprotein particles, but whether they are typical TG-rich particles or secreted into plasma is unknown (Kirchgessner et al., 1987). It was reported that bird kidney secretes generic VLDLs in some proximal tubule cells (Walzem et al., 1999). While the true biological role of NADB-LER1-5 in chicken kidney is unclear, further studies will be needed to prove the biological function of the NADB-LER genes in chicken kidney.
Conclusion
A novel gene family consisted of five novel genes NADB-LER1-5 was identified in chicken genome. All of the genes in the family contain a common NADP-binding site that belongs to the NADB-Rossmann superfamily. The NADB-LER genes were mainly expressed in liver, kidney and duodenum and were significantly up-regulated in liver of hens in the peak-laying stage at the age of 30 and 35 weeks. The expression of the NADB-LER genes was predominantly controlled by17β-estradiol via ERα.
Taken together, our study suggested that the gene family may be a novel regulation factor involved in chicken hepatic lipid metabolism.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
This study was carried out in accordance with the recommendations of Animal Care and Use Guideline of Institutional Animal Care and Use Committee of Henan Agricultural University. The protocol was approved by the Institutional Animal Care and Use Committee of Henan Agricultural University (Permit Number: 11-0085).
Author Contributions
HL performed the data analysis and drafted the manuscript. YL carried out the experiments. LY, DZ, and ZiL participated in the experiment. YW, RH, GL, and ZhL helped to analyze the data. YT and XK participated in the design of the experiments and the discussion. XL conceived the study, participated in the experimental design and helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. U1704233), the China Postdoctoral Science Foundation (No. 2017M620301); the Henan Province Association for Science and Technology Young Talents Support Project; and the Key Science and Technology Research Project of Henan Province (No. 151100110800).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00271/full#supplementary-material
FIGURE S1 | Analysis of the conserved domains in the amino acid sequences of NADB-LER1-5. (A) NADB-LER1; (B) NADB-LER2; (C) NADB-LER3; (D) NADB-LER4; (E) NADB-LER5.
TABLE S1 | Species and sequences selected for constructing the phylogenetic tree.
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/BLAST
- ^ https://www.genscript.com/psort.html
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ http://www.cbs.dtu.dk/services/TMHMM-2.0
- ^ http://www.cbs.dtu.dk/services/ProtFun
- ^ http://meme-suite.org/tools/meme
References
Bensadoun, A., and Rothfeld, A. (1972). The form of absorption of lipids in the chicken, Gallus domesticus. Proc. Soc. Exp. Biol. Med. Soc. Exp. Biol. Med. 141, 814–817. doi: 10.3181/00379727-141-36878
Bergink, E. W., Wallace, R. A., Gruber, M., and Geert, A. B. (1974). Estrogen-Induced synthesis of yolk proteins in roosters. Am. Zool. 14, 1177–1193. doi: 10.1093/icb/14.4.1177
Brady, L., Romsos, D. R., and Leveille, G. A. (1976). In vivo estimation of fatty acid synthesis in the chicken (Gallus domesticus) utilizing 3H2O. Comp. Biochem. Physiol. B 54, 403–407. doi: 10.1016/0305-0491(76)90265-0
Capony, F., and Williams, D. L. (1980). Apolipoprotein B of avian very low density lipoprotein: characteristics of its regulation in nonstimulated and estrogen-stimulated rooster. Biochemistry 19, 2219–2226. doi: 10.1021/bi00551a035
Chan, L., Jackson, R. L., O’Malley, B. W., and Means, A. R. (1976). Synthesis of very low density lipoproteins in the cockerel: effects of estrogen. J. Clin. Investig. 58, 368–379. doi: 10.1172/JCI108481
Daković, N., Térézol, M., Pitel, F., Maillard, V., Elis, S., Leroux, S., et al. (2014). The loss of adipokine genes in the chicken genome and implications for insulin metabolism. Mol. Biol. Evol. 31, 2637–2646. doi: 10.1093/molbev/msu208
Deeley, R. G., Gordon, J. I., Burns, A. T., Mullinix, K. P., Binastein, M., and Goldberg, R. F. (1977). Primary activation of the vitellogenin gene in the rooster. J. Biol. Chem. 252, 8310–8319.
Delong, D. M., Delong, E. R., Wood, P. D., Lippel, K., and Rifkind, B. M. (1986). A comparison of methods for the estimation of plasma low- and very low-density lipoprotein cholesterol: the lipid research clinics prevalence study. JAMA 256, 2372–2377. doi: 10.1001/jama.1986.03380170088024
Evans, M. I., Silva, R., and Burch, J. B. (1988). Isolation of chicken vitellogenin I and III cDNAs and the developmental regulation of five estrogen-responsive genes in the embryonic liver. Genes Dev. 2, 116–124. doi: 10.1101/gad.2.1.116
Fietz, D., Ratzenböck, C., Hartmann, K., Raabe, O., Kliesch, S., Weidner, W., et al. (2014). Expression pattern of estrogen receptors α and β and G-protein-coupled estrogen receptor I in the human testis. Histochem. Cell Biol. 142, 421–432. doi: 10.1007/s00418-014-1216-z
Fischer, P. W. F., and Marks, G. S. (1976). Chick embryo liver cells maintained in serum-free way mouth MD 705/1 medium. Methods Cell Sci. 2, 449–452. doi: 10.1007/BF00918339
Friedewald, W. T., Levy, R. I., and Fredrickson, D. S. (1972). Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentifuge. Clin. Chem. 18, 499–502. doi: 10.1093/clinchem/18.6.499
Greengard, O., Gordon, M., Smith, M. A., and Acs, G. (1964). Studies on the mechanism of diethylstilbestrol-induced formation of phosphoprotein in male chickens. J. Biol. Chem. 239, 2079–2082. doi: 10.2222/jsv1951.2.6
Guo, R. X., Wei, L. H., Zhao, D., Wang, J. L., and Li, X. P. (2006). Effects of ICI182780 (Faslodex) on proliferation and apoptosis induced by 17β-estradiol in endometrial carcinoma cells. J. Peking Univ. 38, 470–474.
Hanukoglu, I. (2015). Proteopedia: Rossmann fold: a beta-alpha-beta fold at dinucleotide biding sites. Biochem. Mol. Biol. Edu. 43, 206–209. doi: 10.1002/bmb.20849
Heald, P. J., and McLachlan, P. M. (1964). The isolation of phosvitin from the plasma of the oestrogen-treated immature pullet. Biochem. J. 92, 51–55. doi: 10.1042/bj0920051
Hermier, D., Catheline, D., and Legrand, P. (1996). Relationship between hepatic fatty acid desaturation and lipid secretion in the estrogenized chicken. Comp. Biochem. Physiol. A 115, 259–264. doi: 10.1016/0300-9629(96)00057-6
Hermier, D., Forgez, P., Williams, J., and Chapman, M. J. (1989). Alterations in plasma 602 lipoproteins and apolipoproteins associated with estrogen−induced hyperlipidemi in the laying hen. Eur. J. Biochem. 184, 109–118. doi: 10.1111/j.1432-1033.1989.tb14996.x
Hoang, T. T., Sullivan, S. A., Cusick, J. K., and Schweizer, H. P. (2002). β-ketoacyl acyl carrier protein reductase (FabG) activity of the fatty acid biosynthetic pathway is a determining factor of 3-oxo-homoserine lactone acyl chain lengths. Microbiology 148, 3849–3856. doi: 10.1128/MMBR.66.4.739.2002
Ivessa, N. E., Rehberg, E., Kienzle, B., Seif, F., Hermann, R., Hermann, M., et al. (2013). Molecular cloning, expression, and hormonal regulation of the chicken microsomal triglyceride transfer protein. Gene 523, 1–9. doi: 10.1016/j.gene.2013.03.102
Kirchgessner, T. G., Heinzmann, C., Svenson, K. L., Gordon, D. A., Nicosia, M., Lebherz, H. G., et al. (1987). Regulation of chicken apolipoprotein B: cloning, tissue distribution, and estrogen induction of mRNA. Gene 59, 241–251. doi: 10.1016/0378-1119(87)90332-5
Klinge, C. M. (2001). Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 29, 2905–2919. doi: 10.1093/nar/29.14.2905
Lerner, L. J., and Jordan, V. C. (1990). Development of antiestrogens and their use in breast cancer: eighth Cain memorial award lecture. Cancer Res. 50, 4177–4189.
Li, H., Wang, T., Xu, C., Wang, D., Ren, J., Li, Y., et al. (2015). Transcriptome profile of liver at different physiological stages reveals potential mode for lipid metabolism in laying hens. BMC Genomics 16:763. doi: 10.1186/s12864-015-1943-0
Li, J., Leghari, I. H., He, B., Zeng, W. D., Mi, Y. L., and Zhang, C. Q. (2014). Estrogen stimulates expression of chicken hepatic vitellogenin II and very low-density apolipoprotein II through ER-α. Theriogenology 82, 517–524. doi: 10.1016/j.theriogenology.2014.05.003
Ma, Z., Li, H., Zheng, H., Jiang, K., Jia, L., Yan, F., et al. (2017). MicroRNA-101-2-5p targets the ApoB gene in liver of chicken (Gallus gallus). Genome 60, 673–678. doi: 10.1139/gen-2017-0020
Macwhirter, P. (1999). Comparative avian nutrition. Aust. Vet. J. 77, 55–55. doi: 10.1111/j.1751-0813.1999.tb12440.x
Marino, M., Galluzzo, P., and Ascenzi, P. (2006). Estrogen signaling multiple pathways to impact gene transcription. Curr. Genomics 7, 497–508. doi: 10.2174/138920206779315737
Mason, T. M. (1999). The role of factors that regulate the synthesis and secretion of very-low-density lipoprotein by hepatocytes. Crit. Rev. Clin. Lab. Sci. 35, 461–487. doi: 10.1080/10408369891234246
Mayoral, J. G., Leonard, K. T., Nouzova, M., Noriega, F. G., Defelipe, L. A., and Turjanski, A. G. (2013). Functional analysis of a mosquito short-chain dehydrogenase cluster. Arch. Insect. Biochem. Physiol. 82, 96–115. doi: 10.1002/arch.21078
Miller, K. W., and Lane, M. D. (1984). Estradiol-induced alteration of very-low-density lipoprotein assembly. Possible competition among apoproteins for incorporation into nascent very-low-density lipoprotein. J. Biol. Chem. 259, 15277–15286.
Mohamadi, A., Aghaei, M., and Panjehpour, M. (2018). Estrogen stimulates adenosine receptor expression subtypes in human breast cancer MCF-7 cell line. Res. Pharm. Sci. 13, 57–64. doi: 10.4103/1735-5362.220968
Nanson, J. D., and Forwood, J. K. (2015). Structural characterisation of FabG from Yersinia pestis, a key component of bacterial fatty acid synthesis. PLoS One 10:e0141543. doi: 10.1371/journal.pone.0141543
Nikolay, B., Plieschnig, J. A., Subik, D., Schneider, J. D., Schneider, W. J., and Hermann, M. (2013). A novel estrogen-regulated avian apolipoprotein. Biochimie 95, 2445–2453. doi: 10.1016/j.biochi.2013.09.005
O’Hea, E. K., and Leveille, G. A. (1969). Lipid biosynthesis and transport in the domestic chick (Gallus domesticus). Comp. Biochem. Physiol. 30, 149–159. doi: 10.1016/0010-406X(69)91309-7
Pang, Y., Dong, J., and Thomas, P. (2008). Estrogen signaling characteristics of Atlantic croaker G protein-coupled receptor 30 (GPR30) and evidence it is involved in maintenance of oocyte meiotic arrest. Endocrinology 149, 3410–3426. doi: 10.1210/en.2007-1663
Ryan, A. K., Schrader, T. J., Wright, R. B., Buchanan, L., and Deeley, R. G. (1994). Characterization of protein interactions with positive and negative elements regulating the apoVLDLII gene. DNA Cell Biol. 13, 987–999. doi: 10.1089/dna.1994.13.987
Safe, S. (2001). Transcriptional activation of genes by 17β-estradiol through estrogen receptor-Sp1 interactions. Vitam. Horm. 62, 231–252. doi: 10.1016/S0083-6729(01)62006-5
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Speake, B. K., Murray, A. M. B., and Noble, R. C. (1998). Transport and transformations of yolk lipid during development of the avian embryo. Prog. Lipid Res. 37, 1–32. doi: 10.1177/016555159702300202
Swofford, D. L. (2002). PAUP∗: Phylogenetic Analysis Using Parsimony (∗ and Other Methods), Version 4.0 b10. Sunderland, MA: Sinauer Associates.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tarugi, P., Ballarini, G., Pinotti, B., Franchini, A., Ottaviani, E., and Calandra, S. (1998). Secretion of apoB- and apoA-I-containing lipoproteins by chick kidney. J. Lipid Res. 39, 731–743. doi: 10.1089/jir.1998.18.273
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Turner, R. T., Evans, G. L., and Dobnig, H. (2000). The high-affinity estrogen receptor antagonist ICI 182,780 has no effect on bone growth in young male rats. Calcif. Tissue Int. 66, 461–464. doi: 10.1007/s002230010092
Walzem, R. L., Hansen, R. J., Williams, D. L., and Hamilton, R. L. (1999). Estrogen induction of VLDLy assembly in egg-laying hens. J. Nutr. 129, 467S–472S. doi: 10.1046/j.1365-277x.1999.00131.x
Wiskocil, R., Bensky, P., Dower, W., Goldberger, R. F., Gordon, J. I., and Deeley, R. G. (1980). Coordinate regulation of two estrogen-dependent genes in avian liver. Proc. Natl. Acad. Sci. U.S.A. 77, 4474–4478. doi: 10.1073/pnas.77.8.4474
Zhang, Y. M., Wu, B., Zheng, J., and Rock, C. O. (2003). Key residues responsible for acyl carrier protein and beta-ketoacyl-acyl carrier protein reductase (FabG) interaction. J. Biol. Chem. 278, 52935–52943. doi: 10.1074/jbc.M309874200
Keywords: chicken, hepatocytes, lipid, expression regulation, estrogen, liver
Citation: Li H, Li Y, Yang L, Zhang D, Liu Z, Wang Y, Han R, Li G, Li Z, Tian Y, Kang X and Liu X (2020) Identification of a Novel Lipid Metabolism-Associated Hepatic Gene Family Induced by Estrogen via ERα in Chicken (Gallus gallus). Front. Genet. 11:271. doi: 10.3389/fgene.2020.00271
Received: 17 February 2019; Accepted: 06 March 2020;
Published: 31 March 2020.
Edited by:
ChunLei Zhang, Jiangsu Normal University, ChinaReviewed by:
Hai Lin, Shandong Agricultural University, ChinaHui Li, Northeast Agricultural University, China
Copyright © 2020 Li, Li, Yang, Zhang, Liu, Wang, Han, Li, Li, Tian, Kang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Liu, eGpsaXUyMDA4QGhvdG1haWwuY29t
 Hong Li
Hong Li Yanmin Li
Yanmin Li Guoxi Li
Guoxi Li Xiangtao Kang
Xiangtao Kang Xiaojun Liu
Xiaojun Liu