
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 18 March 2020
Sec. Epigenomics and Epigenetics
Volume 11 - 2020 | https://doi.org/10.3389/fgene.2020.00205
This article is part of the Research Topic Epigenetic Variation Influences on Livestock Production and Disease Traits View all 24 articles
Somatic cell nuclear transfer (SCNT) has broad applications but is limited by low cloning efficiency. In this review, we mainly focus on SCNT-mediated epigenetic reprogramming in livestock and also describe mice data for reference. This review presents the factors contributing to low cloning efficiency, demonstrates that incomplete epigenetic reprogramming leads to the low developmental potential of cloned embryos, and further describes the regulation of epigenetic reprogramming by long non-coding RNAs, which is a new research perspective in the field of SCNT-mediated epigenetic reprogramming. In conclusion, this review provides new insights into the epigenetic regulatory mechanism during SCNT-mediated nuclear reprogramming, which could have great implications for improving cloning efficiency.
Somatic cell nuclear transfer (SCNT) is an assisted reproduction technology for the generation of cloned mammals that involves the culture of donor somatic cells and oocytes, transplantation of donor cell nuclei into enucleated oocytes, activation of reconstructed embryos, and transfer of cloned embryos into surrogates (Figure 1). SCNT enables the reprogramming of terminally differentiated cells into totipotent cells, which has revolutionized our understanding of cell fate determination and development, and has significant value for theoretical research and production applications.
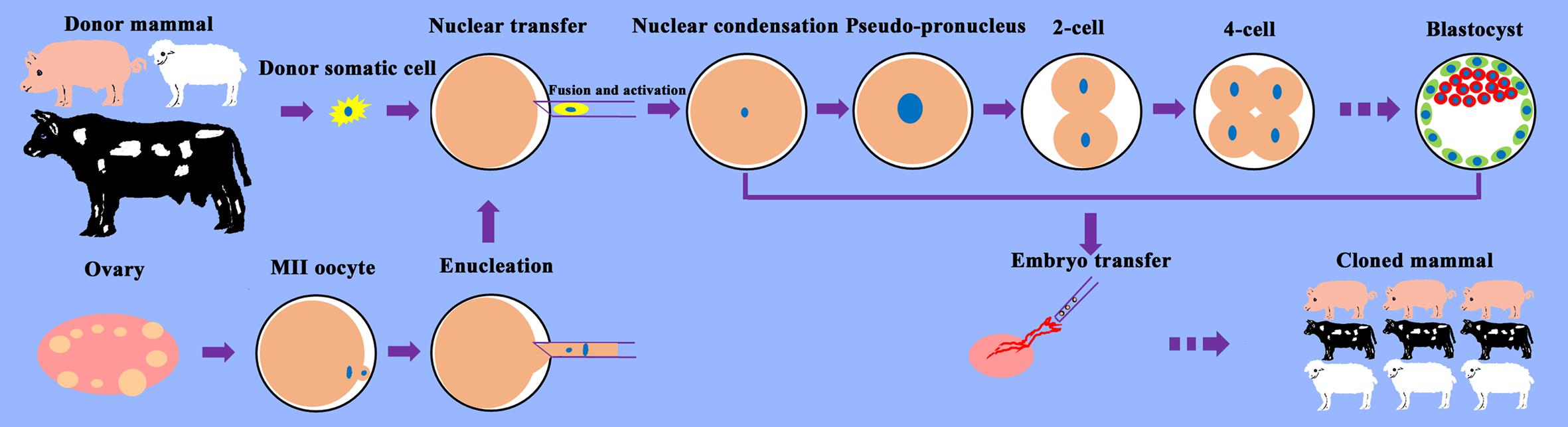
Figure 1. Schematic illustration of the SCNT process. Somatic cells from the desired donor mammal are cultured for SCNT. Oocytes are recovered from the ovaries obtained from the slaughterhouse and allowed to mature into metaphase (M)II oocytes. MII oocytes are enucleated, and donor somatic cells are transferred into the perivitelline space of oocytes. After the fusion and activation of cell–cytoplast complexes, the reconstructed cloned embryos begin to develop, undergoing nuclear condensation and nuclear swelling, followed by pseudo-pronucleus, 2-cell, 4-cell, etc. stages and form blastocysts in vitro. Cloned embryos are transferred into surrogates and develop into cloned mammals.
The first SCNT mammal was a sheep known as Dolly that was born in 1997 (Wilmut et al., 1997), and since then, SCNT has entered a new era (Table 1). A series of cloned mammals, including cow, mouse, goat, pig, and cat, have been produced with this technology (Cibelli et al., 1998; Wakayama et al., 1998; Baguisi et al., 1999; Polejaeva et al., 2000; Chesne et al., 2002; Shin et al., 2002; Galli et al., 2003; Woods et al., 2003; Zhou et al., 2003; Lee et al., 2005; Li et al., 2006; Berg et al., 2007; Shi et al., 2007; Wani et al., 2010). In 2018, the first non-human primate species, the macaque monkey, was successfully cloned by SCNT, further attracting worldwide attention on SCNT technology (Liu et al., 2018; Matoba and Zhang, 2018). Using this technology, a large number of mammals have been successfully produced, showing many potential applications (Campbell et al., 2007; Ogura et al., 2013; Telugu et al., 2017). In agriculture, SCNT can rescue endangered species, protect the genetic resources of commercially important species, and accelerate the propagation of breeding livestock, including pigs, cows, and sheep (Gomez et al., 2009; Keefer, 2015). In combination with genome-modification technologies such as the recently developed clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9-mediated genome editing, SCNT can rapidly produce cloned mammals with desirable traits including rapid growth, disease resistance, and good meat quality, thereby cultivating novel varieties, and shortening breeding cycle (Galli et al., 2012; Wells and Prather, 2017; Lee et al., 2020). In biomedicine, SCNT can create a mammary gland bioreactor to produce therapeutic proteins, establish animal models to investigate the pathogenesis of human diseases, and produce genetically modified xenograft organs for patient transplantation (Lotti et al., 2017; Niu et al., 2017; Telugu et al., 2017). SCNT can also generate blastocyst-derived stem cells, namely, nuclear transfer embryonic stem cells (ntESCs), especially human ntESCs, which are isogenic to the donor and do not cause immune rejection when transplanted, thus providing an important tool for organ regeneration (Tachibana et al., 2013). In basic research, SCNT has been used to investigate interactions between the nucleus and cytoplasm, which has enhanced our understanding of the mechanisms of cell fate determination (Long et al., 2014). Moreover, SCNT has promoted the generation and development of induced pluripotent stem cells (iPSCs), which have similar therapeutic applications to ntESCs (Takahashi and Yamanaka, 2006).
Although SCNT has been successfully used to clone many species of mammals with significant improvements in cloning efficiency in more than 20 years since the birth of Dolly, the proportion of cloned embryos that develop to full term remains very low, greatly limiting the application of SCNT technology (Czernik et al., 2019). To improve the birth of cloned mammals and cloning efficiency, researchers have investigated the effects of donor cell type, oocyte maturation stage, embryo activation method, etc. on the developmental competence of cloned embryos (Table 1), and to some extent, cloning efficiency has been shown to increase through optimizing these parameters (Blelloch et al., 2006; Campbell et al., 2007; Kurome et al., 2013). However, cloning efficiency remains low, the abortion of cloned fetus frequently occurs, and the rate of abnormality or mortality is high. Moreover, developmental defects still occur in cloned mammals even after birth (Campbell et al., 2007; Loi et al., 2016). These phenomena demonstrate that optimizing these technology parameters of SCNT cannot make significant improvements in cloning efficiency and only clarifying the theoretical molecular mechanism underlying SCNT could understand the cause of the poor and abnormal development of cloned embryos. However, SCNT-mediated nuclear reprogramming is still poorly understood, and the key factors determining the developmental potential of cloned embryos remain unclear (Matoba and Zhang, 2018). Therefore, fully and clearly revealing the molecular mechanism underlying SCNT-mediated nuclear reprogramming is needed to enhance the development of cloned embryos.
During development, totipotent embryos differentiate into pluripotent stem cells and subsequently into differentiated cells. Cell fate determination is largely achieved by activating some genes while suppressing other genes through epigenetic modification such as DNA methylation, histone modification, genomic imprinting, and X chromosome inactivation (XCI) (Reik et al., 2003). These heritable changes in gene expression without alterations in genomic DNA sequences occur during the progression from fertilized oocyte to differentiated embryo and also play a key role in embryo development following SCNT (Niemann, 2016). It is thought that the low cloning efficiency, abnormal embryo phenotype, and low viability of animals generated by SCNT are due to incomplete reprogramming of donor nuclei (Yang et al., 2007). Epigenetic changes during the SCNT process are discussed in greater detail below (Figure 2).
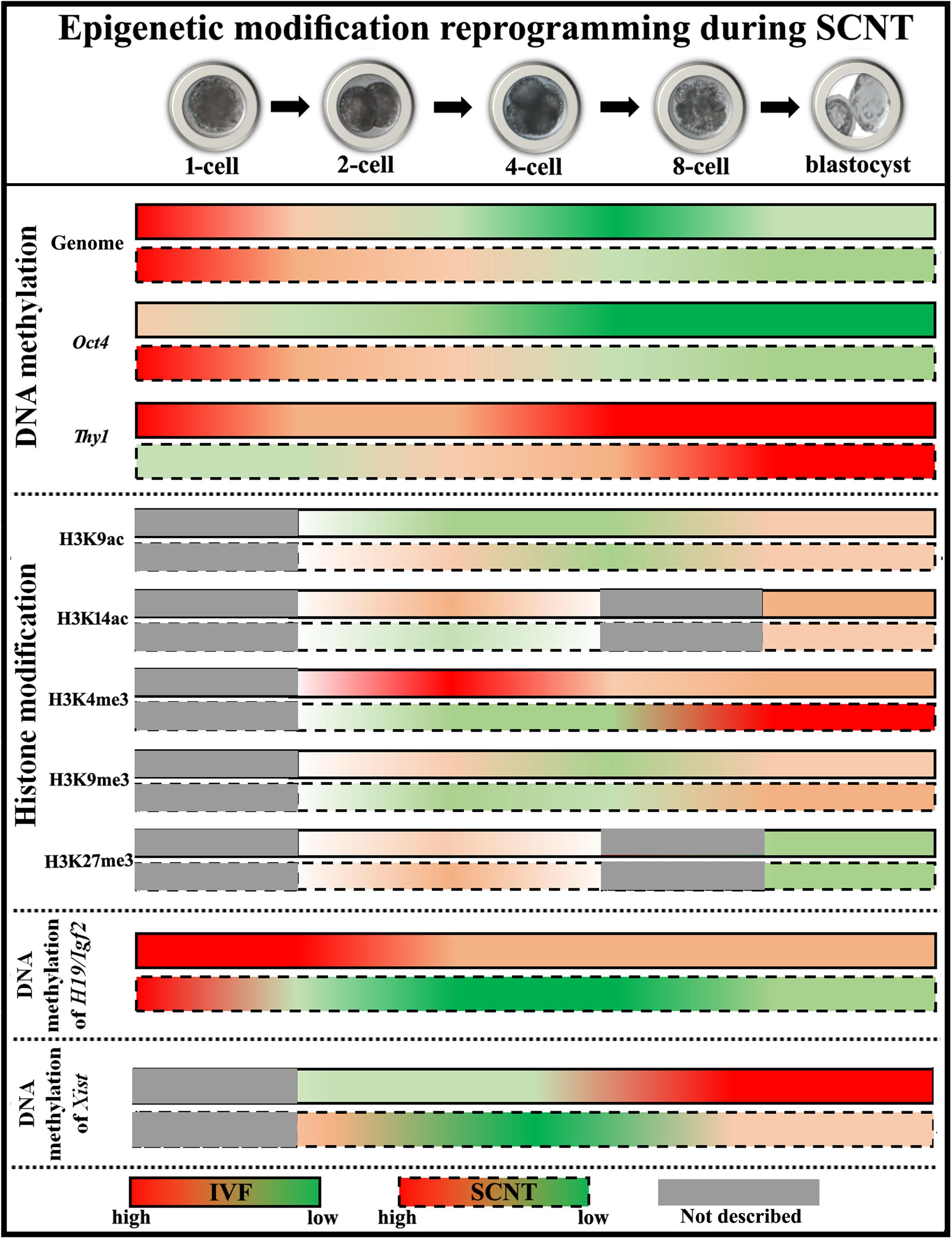
Figure 2. Diagram of epigenetic modification changes during SCNT-mediated nuclear reprogramming. The data of pig embryos are adopted to describe epigenetic modification reprogramming. For DNA methylation, cloned embryos demonstrate delayed DNA demethylation and incomplete DNA remethylation of genome (the data shown here represent DNA methylation status at centromeric repeats, which partly reflects genome DNA status), high DNA methylation status of pluripotency-related gene Oct4, and low DNA methylation levels of tissue-specific gene Thy1, respectively. For histone modifications, low levels of histone acetylation (H3K9ac at the ZGA stage and H3K14ac at the blastocyst stage) and H3K4me3, and high levels of histone methylation (H3K9me3 after ZGA and H3K27me3 at the 2-cell stage) are observed in cloned embryos. For genomic imprinting, DNA methylation of H19/Igf2 is not maintained during SCNT. For XCI, DNA methylation of Xist is not fully established in female cloned embryos.
DNA methylation occurs at cytosine residues in the CpG dinucleotide and is generally associated with transcriptional silencing (Schubeler, 2015). In the life cycle, the genome undergoes DNA methylation maintenance, DNA demethylation, and DNA remethylation, which allows organisms to activate or silence specific genes according to the requirements of organism growth and development (Li and Zhang, 2014). DNA methyltransferases (Dnmts) such as Dnmt1 and Dnmt3 (Dnmt3a, Dnmt3b, and Dnmt3l) are responsible for DNA methylation maintenance and de novo DNA methylation (Chen and Zhang, 2019). DNA demethylation occurs through the oxidation-base excision repair pathway. Oxidative DNA demethylation enzymes include ten–eleven translocation (Tet)1, Tet2, Tet3, activation-induced cytidine deaminase, and DNA glycosylases (Ito et al., 2010; Iqbal et al., 2011; Shen et al., 2013). Other pathways also contribute to active DNA demethylation during early embryonic development (Wang et al., 2014). Dnmt1 maintains methyl marks on genomic DNA and ensures that the DNA methylation pattern of offspring cells is identical to that of parental cells (Lyko, 2018). After fertilization, the genome demonstrates a combination of active and passive DNA methylation, and the paternal genomic DNA is actively demethylated while maternal genomic DNA is passively demethylated (Guo et al., 2014). When fertilized embryos develop to the blastocyst or subsequent implantation stage, genomic DNA is remethylated (Reik et al., 2001; Yang et al., 2007). In cloned embryos, the genomic DNA of donor somatic cells is highly methylated and DNA methylation reprogramming (especially DNA demethylation) is necessary for development to proceed normally. The genome also undergoes de-/remethylation during SCNT, but this is delayed and incomplete compared with normal embryos (Bourc’his et al., 2001; Dean et al., 2001; Yang et al., 2007). Tissue-specific and pluripotency-related genes in cloned embryos show low and high DNA methylation levels, respectively (Figure 2, DNA methylation) (Ng and Gurdon, 2005; Kremenskoy et al., 2006; Yamazaki et al., 2006; Huan et al., 2014, 2015a). Following zygotic genome activation (ZGA), the erroneous reconstitution of DNA methylation pattern caused by aberrant expression of genes related to DNA methylation reprogramming and, consequently, of key genes required for the normal development of cloned embryos results in low cloning efficiency and abnormalities and death in cloned animals (Bourc’his et al., 2001; Bortvin et al., 2003; Chung et al., 2003; Kiefer et al., 2016; Gao et al., 2018). Thus, a DNA methylation pattern similar to that in normal fertilized embryos is necessary for the successful development of SCNT embryos.
Chromatin structure and histone modification are key factors that regulate gene expression (Sproul et al., 2005; Yi and Kim, 2018). The basic structural unit of chromatin is the nucleosome, a histone octamer consisting of two copies each of H2A, H2B, H3, and H4 wrapped by 146 bp of DNA and H1 as a linker (Kobayashi and Kurumizaka, 2019). Gene expression depends on chromatin accessibility, which is controlled by chromatin remodeling factors and through covalent modification (e.g., acetylation, methylation, and phosphorylation) of amino acids in the histone tail (Qin et al., 2016; Kobayashi and Kurumizaka, 2019). Chromatin accessibility during the SCNT-mediated epigenetic reprogramming has not been extensively investigated as it requires a large number of embryos. Recently, progress is being made in mice owing to technologic advances such as low-input DNase I hypersensitive site (DHS) sequencing and transposase-accessible chromatin sequencing (Wu et al., 2016; Djekidel et al., 2018). DHSs, which are positively correlated with gene expression, are present in donor somatic cells and are reprogrammed in cloned embryos. However, specific DHSs of donor somatic cells fail to be reprogrammed to those of embryos, which prevents the binding of chromatin remodeling factors to regulate gene expression in cloned embryos (Djekidel et al., 2018).
Histone acetylation is regulated by histone acetyltransferase (Hat) and histone deacetylase (Hdac). Hat opens up chromatin, which allows transcription factor binding and leads to activation of gene transcription, whereas Hdac promotes gene inactivation (Sun et al., 2003). After fertilization, histone acetylation such as histone H3 acetylation occurs and allows the appropriate expression of genes related to early embryonic development (Rybouchkin et al., 2006; Ziegler-Birling et al., 2016). During SCNT, histone acetylation marks decrease and gradually disappear, for instance, Lys9 acetylation of H3 (H3K9ac) at the ZGA stage and H3K14ac at the blastocyst stage (Rybouchkin et al., 2006; Wee et al., 2006; Liu et al., 2012; Zhai et al., 2018). Histone methylation mainly occurs on lysine and arginine, and involves three methylation patterns including monomethylation, dimethylation, and trimethylation (Izzo and Schneider, 2010). Trimethyl of H3 Lys4 (H3K4me3) and H3K27me3 are the most typical modifications. H3K4me3 is regulated by the Trithorax group (TrxG) complex and is associated with gene activation, while H3K27me3 is mediated by the Polycomb group (PcG) proteins and leads to gene silencing (Liu X. et al., 2016). During SCNT-mediated nuclear reprogramming, H3K4me3 level decreases and H3K27me3 level increases (Cao et al., 2015; Xie et al., 2016; Zhai et al., 2018; Zhou et al., 2019). Another modification, H3K9me3, is catalyzed by suppressor of variegation 39H1/2 (Suv39H1/2) and removed by lysine demethylase (Kdm)4 (Kdm4a, Kdm4b, Kdm4d, and Kdm4e), and can alter chromatin conformation to inhibit gene expression (Ninova et al., 2019). H3K9me3 can be removed in donor somatic cells, but incomplete H3K9me3 demethylation in cloned embryos inhibits their development. Studies have shown that H3K9me3 is enriched in the promoters of genes against SCNT-mediated nuclear reprogramming, suggesting that incomplete H3K9me3 demethylation is an inhibitor of the development of cloned embryos (Matoba et al., 2014; Liu W. et al., 2016; Zhai et al., 2018). These disrupted histone modifications finally affect chromatin accessibility, lead to the disordered expression of genes required for the normal development of cloned embryos, and result in low cloning efficiency (Figure 2, Histone modification) (Liu et al., 2012; Xie et al., 2016; Zhai et al., 2018). Several histone variants also exhibit abnormalities such as the delayed change of H1foo (oocyte-specific H1) to somatic H1s, macroH2A expression before the endogenous activation, and the existing replacement of donor cell H3 carrying repressive modification by maternal H3.3 in cloned embryos, which contributes to incomplete SCNT-mediated nuclear reprogramming (Gao et al., 2004; Chang et al., 2010; Wen et al., 2014). Therefore, histone modification is a critical determinant in the development of cloned embryos.
Genomic imprinting, an epigenetically regulated phenomenon that shows monoallelic parent-specific gene expression, is controlled by the differentially methylated region (DMR) or specific histone modifications. The DMR is protected by DNA-binding complexes composed of Dnmt1, zinc finger protein 57, and tripartite motif-containing 28, and the H3K27me3 mark (Barlow and Bartolomei, 2014; Inoue et al., 2017a). In general, paternal and maternal gene imprinting promotes and inhibits, respectively, offspring growth and development (Barlow and Bartolomei, 2014). Therefore, parental imprinted genes compete with or complement each other, and the balance between the expression of paternal and maternal imprinted genes is important for normal developmental progression. H19/insulin-like growth factor (Igf)2 is a typical genomic imprinting locus with the DMR methylated on the paternal allele, on which H19 silencing stimulates IGF2 activity and cell growth. In contrast, H19 on the maternal allele is an inhibitory factor that has a cis-silencing effect on Igf2 expression. Inhibiting Igf2 expression leads to fetal growth retardation, whereas Igf2 overexpression or H19 transcription deficiency results in fetal overgrowth (Sasaki et al., 2000). Genomic imprinting is erased and established during gametogenesis and is maintained throughout the lifetime of an organism (Simon et al., 1999; MacDonald and Mann, 2014). Thus, the restoration of a diploid genome in fertilized embryos and mutually compensatory expression of monoallelic parent-specific imprinted genes ensure normal growth and development of early embryos. However, genomic imprinting is not effectively maintained in cloned embryos, resulting in the aberrant expression of imprinted genes that gives rise to development defects such as placental hypertrophy and fetal abortion and death (Figure 2, Genome imprinting) (Mann et al., 2003; Shi et al., 2003; Yang et al., 2007; Wei et al., 2010; Zhang et al., 2014; Huan et al., 2015b). For example, hypomethylated H19/Igf2 imprinting results in increased H19 transcription and suppresses the growth of cloned fetuses, whereas Igf2 overexpression mediated by hypermethylated H19/Igf2 imprinting leads to their overgrowth. Therefore, disrupted genome imprinting during the development of cloned embryos results in developmental abnormalities and death in cloned offspring, constraining the cloning efficiency.
Female mammals have two X chromosomes, whereas only one is present in males. In order to balance gene dosage, female mammals silence one X chromosome through the activity of the Xist gene product, a long non-coding (lnc)RNA on the inactive X chromosome that recruits transcriptional repressors such as PcG proteins (Latham, 2005; Galupa and Heard, 2015). During normal embryonic development of female mammals, both X chromosomes are active and XCI occurs at the blastocyst stage, resulting in random inactivation of the X chromosome in the inner cell mass (ICM). Meanwhile, the paternal X chromosome is inactivated in the trophoblast (Yang et al., 2007; Payer, 2016). During SCNT, the inactivated X chromosome in female donor cells is reactivated during early development of cloned embryos, with XCI occurring at the blastocyst stage. In theory, XCI should occur randomly in the ICM, with the trophoblast exhibiting XCI as in the donor cells. However, irrespective of the sex of cloned embryos, DNA methylation level of Xist is lower than that in fertilized embryos, and the consequent upregulation of Xist expression represses the transcription of numerous X-linked genes (Figure 2, XCI) (Xue et al., 2002; Nolen et al., 2005; Inoue et al., 2010; Xu et al., 2013; Yuan et al., 2014; Ruan et al., 2018). Such abnormal XCI has also been detected in the placenta and carcass of dead cloned animals and could be due to the absence of a H3K27me3 mark in the Xist promoter following SCNT, and H3K9me3 may also determine the expression level of Xist in cloned embryos (Xue et al., 2002; Inoue et al., 2010; Inoue et al., 2017b; Ruan et al., 2018). Therefore, the abnormal XCI pattern seriously affects the development of cloned fetuses and placentas.
As epigenetic modification regulates gene expression, disrupted epigenetic modification during SCNT leads to the abnormal transcription of genes related to development in cloned embryos. The persistently high expression of donor somatic cell-specific genes and failure to activate genes related to embryo development are against SCNT-mediated nuclear reprogramming (Matoba et al., 2014; Liu W. et al., 2016). Therefore, epigenetic modification status determines gene expression levels and the developmental potential of cloned embryos, further suggesting that only the full and effective reconstruction of epigenetic modifications during SCNT-mediated nuclear reprogramming can support the full-term development of cloned embryos.
Presently, the aim of SCNT-mediated nuclear reprogramming-related research is to improve epigenetic reconstruction in cloned embryos, as the degree of epigenetic reprogramming determines the developmental competence of cloned embryos (Niemann, 2016).
Improving DNA methylation reprogramming has been applied in cloned embryos (Enright et al., 2003; Huan et al., 2015a; Liao et al., 2015). One way in which this is accomplished is by recapitulating the DNA methylation pattern of normal fertilized embryos using DNA-demethylating agents or by Dnmts including Dnmt1 and Dnmt3l gene silencing. The application of DNA demethylation reagents and Dnmts knockdown have successfully ameliorated genome DNA methylation and histone modification in cloned embryos. The nucleoside analog 5-aza-2′-deoxycytidine (5-aza-dC) is incorporated into the genome during DNA replication, inhibiting DNMT1 activity and resulting in DNA hypomethylation (Enright et al., 2003). Genomic DNA hypomethylation by 5-aza-dC treatment has been shown to improve the development of cloned embryos, whereas Dnmt1 or Dnmt3l knockdown in somatic cells or cloned embryos increases gene-specific DNA methylation and histone modification reprogramming and, consequently, developmental competence (Diao et al., 2013; Huan et al., 2015a; Liao et al., 2015; Song et al., 2017b). Additionally, the expression level of Tet3 in oocytes has been shown to be positively correlated with the developmental competence, and Tet3 overexpression in donor cells restores normal DNA hypermethylation and increases the full-term development of cloned embryos (Han et al., 2018). Therefore, ameliorating DNA methylation reprogramming in cloned embryos is a feasible strategy to enhance cloning efficiency.
Modifying histone marks is another approach for increasing the development competence of cloned embryos. Hdac inhibitor treatment increases histone acetylation and opens up the chromatin structure, which facilitates the binding of transcription factors that activate genes involved in early embryonic development. Hdac inhibitors have been used to improve the developmental ability of cloned embryos. For example, trichostatin A (the class I and II Hdac inhibitor), scriptaid (a synthetic Hdac inhibitor with low toxicity), and valproic acid all increase histone acetylation levels, especially H3K9ac and H3K14ac, improve gene expression levels in cloned embryos, and thus enhance SCNT-mediated nuclear reprogramming (Enright et al., 2003; Kishigami et al., 2006; Bui et al., 2011; Costa-Borges et al., 2010; Liu et al., 2012; Zhai et al., 2018). These results suggest that histone acetylation is beneficial for the development of cloned embryos. The increased H3K4me3 has been shown to improve the epigenetic modifications and the developmental efficiency of cloned embryos (Zhai et al., 2018). H3K9me3 has been reported to be a barrier for SCNT-mediated nuclear reprogramming, and removal of H3K9me3 through injection of Kdm4 mRNA activates the appropriate expression of repressed genes and increases the developmental competence of cloned embryos (Antony et al., 2013; Matoba et al., 2014; Chung et al., 2015; Liu et al., 2018; Weng et al., 2019). Importantly, the positive effect of histone acetylation on cloning efficiency could also be mediated through H3K9me3 removal (Matoba et al., 2014). Moreover, the loss of H3K9me3 can also be realized by introducing protamines in the nuclei of donor somatic cells, holding great potential to improve cloning efficiency (Iuso et al., 2015). Additionally, blocking H3K27me3 has been shown to promote nuclear reprogramming and embryonic development following SCNT (Xie et al., 2016; Zhou et al., 2019). Therefore, improvements in histone modification can correct the expression pattern of genes required for the normal development of cloned embryos and greatly enhance cloning efficiency.
Importantly, with the enhanced development of cloned embryos induced by histone modification improvements, genome imprinting in cloned embryos, fetuses, and offspring is also effectively maintained, suggesting that epigenetic modifications form mutually regulatory networks (Cervera et al., 2009; Xu et al., 2013; Huan et al., 2015b; Inoue et al., 2017a). H19 knockdown in abnormal imprinting fetal fibroblasts has also been shown to rescue damaged imprinting and the reduced development of cloned embryos (Song et al., 2017a). Therefore, restoring normal epigenetic marks and expression of imprinted-related genes can directly or indirectly ensure genome imprinting and the normal development of cloned embryos.
Targeting XCI is another strategy for improving developmental potential in SCNT. When Xist is deleted, the pattern of X-linked gene expression is corrected in cloned embryos, and the birth rate of cloned mammals is improved (Inoue et al., 2010; Matoba et al., 2011; Ruan et al., 2018). The inhibition of Xist also results in a remarkable improvement in the development of male cloned embryos (Zeng et al., 2016; Yang et al., 2019). Therefore, Xist deletion or knockdown restores X-linked gene expression patterns in cloned embryos and increases cloning efficiency.
With the understanding of epigenetic modifications during SCNT, researchers have successfully cloned macaque monkeys (Liu et al., 2018). Therefore, the existing evidence indicates that the epigenetic status of cloned embryos is an important determinant in cloning efficiency, and improving epigenetic modifications can be a good strategy to support the successful long-term development of cloned embryos (Figure 3).
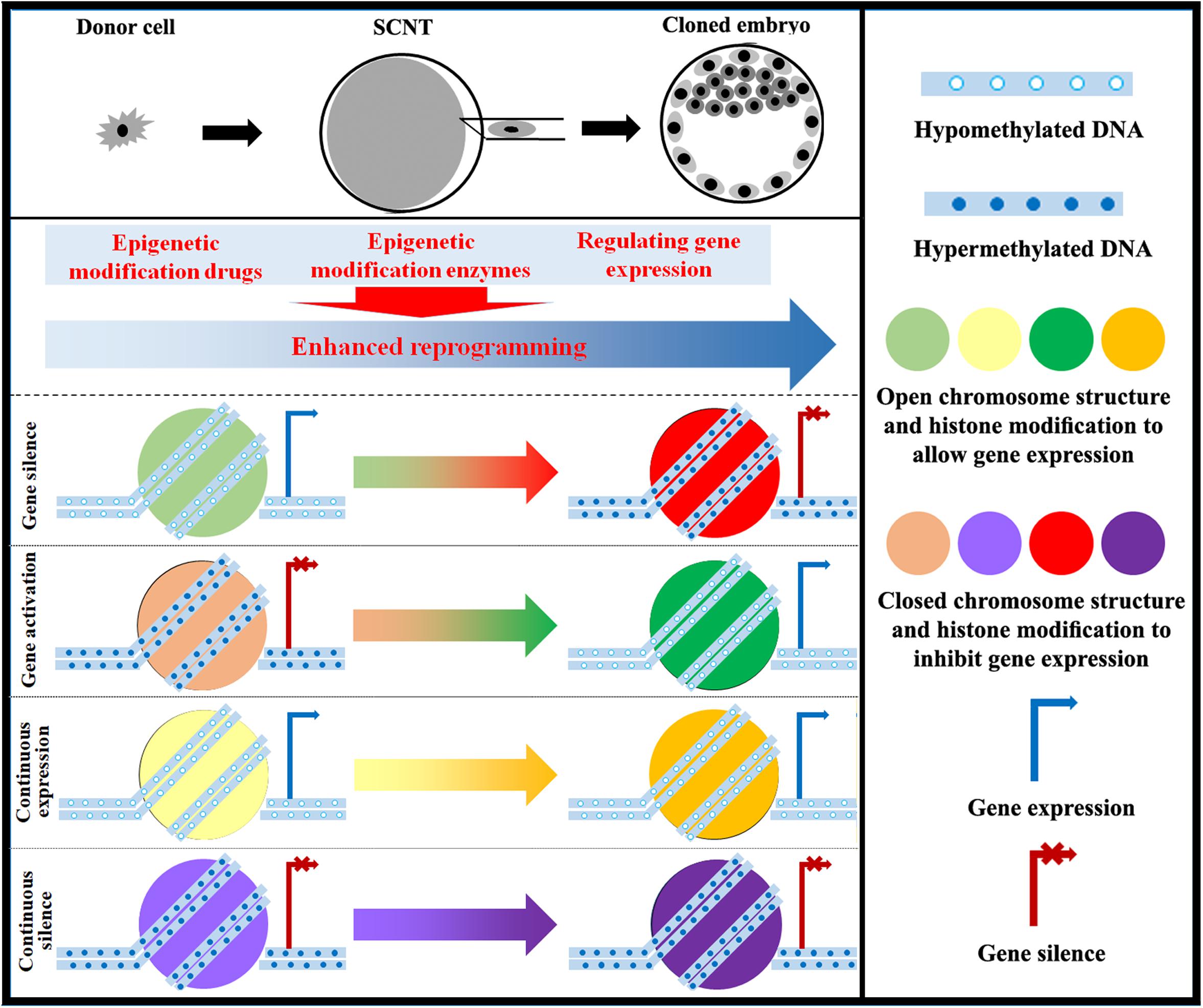
Figure 3. Strategy for improving epigenetic reprogramming during SCNT. The application of epigenetic modification drugs and the regulation of enzymes involved in epigenetic modification as well as associated genes can induce changes in DNA methylation, chromosome structure, histone modification, genomic imprinting, and XCI in cloned embryos close to those in normal fertilized embryos. Targeting epigenetic reprogramming to activate or silence genes can yield cloned embryos with high developmental competence.
An increasing number of studies have shown that the degree of epigenetic reprogramming determines the developmental potential of cloned embryos (Niemann, 2016; Matoba and Zhang, 2018). However, the molecular regulatory network involved in SCNT-mediated epigenetic reprogramming remains unclear. Therefore, exploring the molecular mechanism underlying nuclear reprogramming induced by SCNT and clarifying how highly differentiated somatic cells effectively become pluripotent cloned embryos through the epigenetic reprogramming process are areas of great interest.
LncRNAs are gene transcripts longer than 200 nucleotides that do not encode proteins but nonetheless play a critical role in gene regulation in nearly all physiologic processes, as well as in cell fate determination during development (Pauli et al., 2011; Chen and Zhang, 2016; Wang et al., 2018). A comprehensive and systematic exploration and analysis of lncRNA function has become a frontier in the field of life science (Flynn and Chang, 2014). As mentioned above, H19 and Xist have been shown to regulate the development of cloned embryos, suggesting that lncRNAs play a key role during SCNT-mediated nuclear reprogramming (Inoue et al., 2010; Song et al., 2017a). Exploring the molecular mechanism of lncRNAs in mediating epigenetic reconstruction during SCNT can provide new ideas for improving cloning efficiency.
Recent studies have shown that lncRNAs participate in many epigenetic modification processes, such as DNA methylation, histone modification, genome imprinting and XCI, and regulate the activation or silencing of genes according to cell function requirements (Mercer and Mattick, 2013; Holoch and Moazed, 2015). During DNA methylation reprogramming, lncRNAs can interact with enzymes related to DNA methylation or demethylation (Dnmts and Tets), and determine DNA methylation reconstruction to regulate gene expression. When a gene needs to be activated, lncRNAs recruit DNA demethylation-related enzymes, such as Tets, to the gene promoter and help to achieve gene DNA demethylation, and when a gene needs to be silenced, lncRNAs interact with Dnmts to establish and maintain DNA methylation of a gene promoter (Bao et al., 2015; Hamazaki et al., 2015; Wang et al., 2015; Zhou et al., 2015; Kimura et al., 2017). Therefore, lncRNAs can interact with the DNA methylation reprogramming-related enzymes to regulate DNA methylation reprogramming and regulate gene expression. LncRNAs can also alter gene expression by regulating histone modification. Studies have demonstrated that lncRNAs recruit the TrxG proteins to catalyze H3K4me3 and enhance gene transcription or PcG proteins to silence gene expression through H3K27me3 (Tsai et al., 2010; Wang et al., 2011; Liu et al., 2015). LncRNAs are also involved in the regulation of genomic imprinting, as evidenced by the finding that every genomic imprinting center contains at least one lncRNA, such as H19 and Meg3 in the H19/Igf2 and Dlk1/Meg3 imprinting regions, respectively, that regulates monoallelic parental-specific gene expression through epigenetic silencing (Barlow and Bartolomei, 2014). During XCI, Xist has been displayed to recruit PcG protein to certain gene loci, establish H3K27me3 modification and DNA methylation, and lead to gene silencing on the X chromosome (Lee, 2009; Inoue et al., 2017b). Therefore, lncRNAs can determine epigenetic modification construction.
After SCNT, epigenetic and gene expression profiles undergo substantial changes in cloned embryos. Naturally, lots of differentially expressed lncRNAs exist during the development of cloned embryos. However, research on key lncRNAs during the SCNT process is very limited. Encouragingly, the existing studies have suggested that lncRNAs can regulate the developmental competence of cloned embryos (Inoue et al., 2010; Song et al., 2017a; Ruan et al., 2018). Studies of parthenogenetic and semi-cloned mice have shown that these animals also express different levels of H19, which are associated with variable patterns of epigenetic modification and gene expression and influence developmental potential (Kono et al., 2004; Zhong and Li, 2017). Additionally, in iPSCs, lncRNAs are also shown to either promote or inhibit the reconstitution of epigenetic modifications during nuclear reprogramming (Kim et al., 2015). Therefore, lncRNAs could determine the developmental competence of cloned embryos. Given the importance of lncRNAs during SCNT-mediated epigenetic reprogramming, exploring and clarifying the underlying molecular mechanism of lncRNA-mediated epigenetic reprogramming during SCNT could lead to improvements in cloning efficiency (Figure 4).
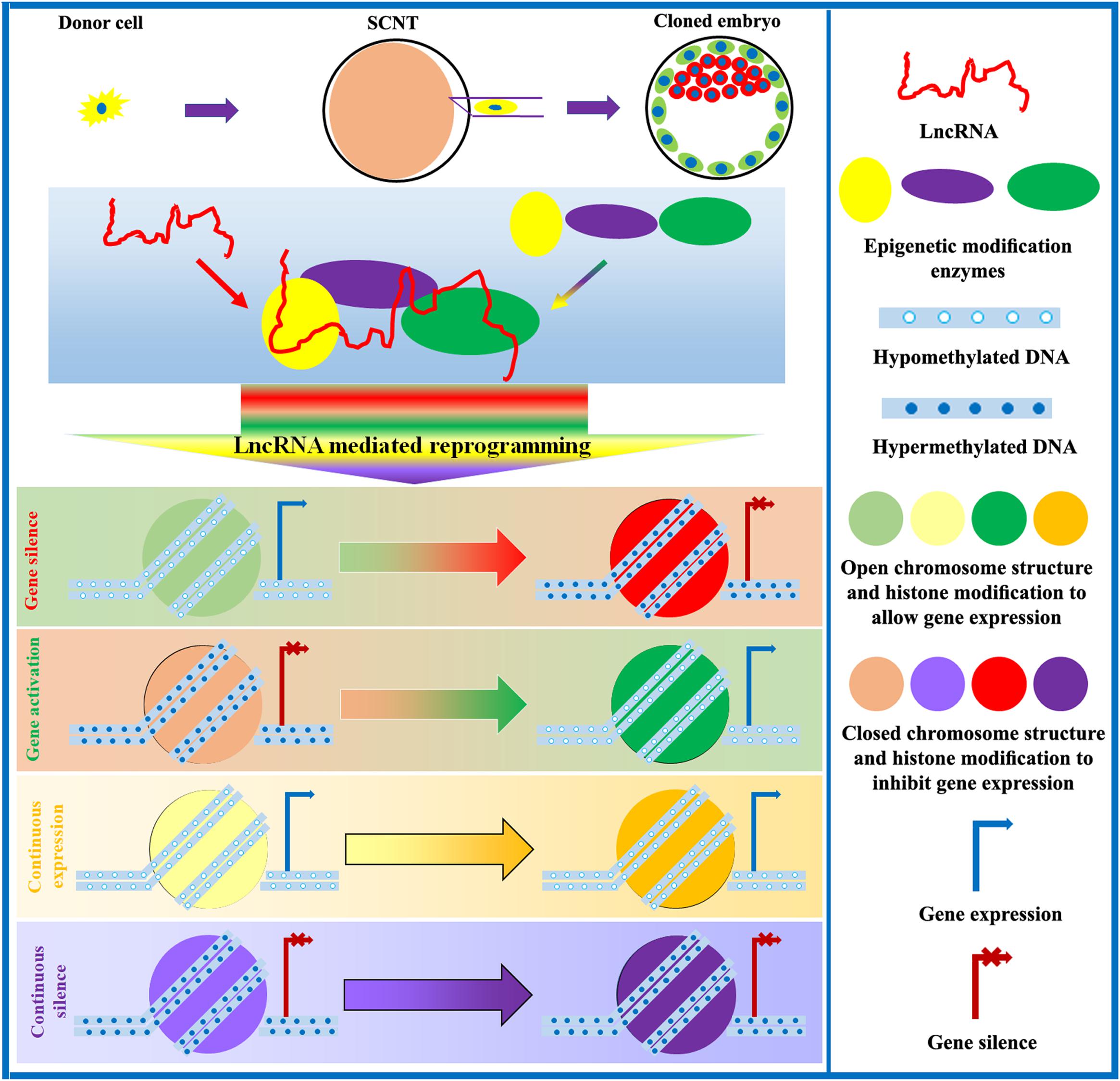
Figure 4. Role of lncRNAs in the regulation of SCNT-mediated epigenetic reprogramming. LncRNAs interact with epigenetic modification enzymes to modulate DNA methylation, chromosome structure, histone modification, genomic imprinting, and XCI, thereby controlling gene expression and promoting the development of cloned embryos.
It is known to be difficult to produce cloned embryos, and their developmental competence remains poor. Moreover, studies on the factors that regulate epigenetic reprogramming during SCNT, especially those investigating lncRNAs, are still limited. To enhance the developmental potential of cloned embryos, a systematic analysis of the factors and mechanisms involved in SCNT is required. In recent years, with technological advancements, particularly the application of single-cell transcriptome sequencing, great progress has been made in discovering and identifying the reprogramming factors related to the development of cloned embryos. To date, lots of novel genes and lncRNAs have been revealed (Bai et al., 2016; Liu X. et al., 2016; Wu et al., 2018). However, the relevant reprogramming factors, including the emerging lncRNAs that regulate the developmental potential of cloned embryos, have not yet been deeply explored. Therefore, more detailed studies are needed to elucidate the molecular mechanisms underlying SCNT-mediated epigenetic reprogramming in order to improve cloning efficiency.
Efforts to improve cloning efficiency have also promoted the application of SCNT technology. Presently, a series of agriculturally and economically important animals can be cloned, which could not only enable the protection of endangered species but also accelerate the utilization of livestock. Gland bioreactors can be created through SCNT to produce therapeutic proteins. Animal models can also be generated through SCNT to investigate the pathogenesis of human diseases. Moreover, with the CRISPR/Cas9-mediated genome editing technology, SCNT can produce desired animals or models for specific applications. In short, if cloning efficiency is greatly improved, the application of SCNT technology will be more extensive.
SCNT has important theoretical and practical research value. In this review, we present our understanding of SCNT-mediated nuclear reprogramming, especially the factors contributing to low cloning efficiency, and that incomplete epigenetic reprogramming leads to the low developmental potential of cloned embryos. We further demonstrate that the application of epigenetic modification methods can improve cloning efficiency. We also describe the regulation of epigenetic reprogramming by lncRNAs and provide a new research perspective in the field of SCNT-mediated epigenetic reprogramming. The elucidation of these mechanisms has enhanced cloning efficiency and expanded the application of SCNT technology in agriculture, regenerative medicine, and other areas. We believe that with further advances in technology, more molecular mechanisms will be revealed to enhance the development of cloned embryos, and improved cloning efficiency will promote the extensive application of SCNT technology.
YH designed the manuscript. XW, JQ, and JL wrote the manuscript. YH, HH, and ZL provided the writing guidance and revised the manuscript. All authors read and approved the final manuscript.
This work was supported by grants from the Key Research and Development Program of Shandong Province (2017NC210007 to YH), the Entrepreneurship and Innovation Leading Talent Program of Qingdao (19-3-2-2-zhc to YH), the Agriculture Variety Project of Shandong Province (2019LZGC011 to YH), the Excellent Youth Innovation Team in Shandong Colleges and Universities (2019KJF005 to YH), the National Natural Science Foundation of China (31602019 to YH), the Open Projects of Key Laboratory of Animal Cellular and Genetic Engineering of Heilongjiang Province (KF201704 to YH), and the High-level Talent Research Foundation of Qingdao Agricultural University (6631116029 to YH).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Jiang Zhu at Tongji University for the manuscript revision and English polishing.
Antony, J., Oback, F., Chamley, L. W., Oback, B., and Laible, G. (2013). Transient JMJD2B-mediated reduction of H3K9me3 levels improves reprogramming of embryonic stem cells into cloned embryos. Mol. Cell. Biol. 33, 974–983. doi: 10.1128/MCB.01014-12
Baguisi, A., Behboodi, E., Melican, D. T., Pollock, J. S., Destrempes, M. M., Cammuso, C., et al. (1999). Production of goats by somatic cell nuclear transfer. Nat. Biotechnol. 17, 456–461.
Bai, L., Li, M., Sun, J., Yang, X., Lu, Y., Lu, S., et al. (2016). RNA-Seq profiling of intact and enucleated oocyte SCNT embryos reveals the role of pig oocyte nucleus in somatic reprogramming. PLoS One 11:e0153093. doi: 10.1371/journal.pone.0153093
Bao, X., Wu, H., Zhu, X., Guo, X., Hutchins, A. P., Luo, Z., et al. (2015). The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 25, 80–92. doi: 10.1038/cr.2014.165
Barlow, D. P., and Bartolomei, M. S. (2014). Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6:a018382. doi: 10.1101/cshperspect.a018382
Berg, D. K., Li, C., Asher, G., Wells, D. N., and Oback, B. (2007). Red deer cloned from antler stem cells and their differentiated progeny. Biol. Reprod. 77, 384–394. doi: 10.1095/biolreprod.106.058172
Blelloch, R., Wang, Z., Meissner, A., Pollard, S., Smith, A., and Jaenisch, R. (2006). Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells 24, 2007–2013. doi: 10.1634/stemcells.2006-0050
Bortvin, A., Eggan, K., Skaletsky, H., Akutsu, H., Berry, D. L., Yanagimachi, R., et al. (2003). Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development 130, 1673–1680. doi: 10.1242/dev.00366
Bourc’his, D., Le Bourhis, D., Patin, D., Niveleau, A., Comizzoli, P., Renard, J. P., et al. (2001). Delayed and incomplete reprogramming of chromosome methylation patterns in bovine cloned embryos. Curr. Biol. 11, 1542–1546. doi: 10.1016/s0960-9822(01)00480-8
Bui, H. T., Seo, H. J., Park, M. R., Park, J. Y., Thuan, N. V., Wakayama, T., et al. (2011). Histone deacetylase inhibition improves activation of ribosomal RNA genes and embryonic nucleolar reprogramming in cloned mouse embryos. Biol. Reprod. 85, 1048–1056. doi: 10.1095/biolreprod.110.089474
Campbell, K. H., Fisher, P., Chen, W. C., Choi, I., Kelly, R. D., Lee, J. H., et al. (2007). Somatic cell nuclear transfer: Past, present and future perspectives. Theriogenology 68, (Suppl. 1), S214–S231.
Cao, Z., Li, Y., Chen, Z., Wang, H., Zhang, M., Zhou, N., et al. (2015). Genome-wide dynamic profiling of histone methylation during nuclear transfer-mediated porcine somatic cell reprogramming. PLoS One 10:e0144897. doi: 10.1371/journal.pone.0144897
Cervera, R. P., Marti-Gutierrez, N., Escorihuela, E., Moreno, R., and Stojkovic, M. (2009). Trichostatin A affects histone acetylation and gene expression in porcine somatic cell nucleus transfer embryos. Theriogenology 72, 1097–1110. doi: 10.1016/j.theriogenology.2009.06.030
Chang, C.-C., Gao, S., Sung, L.-Y., Corry, G. N., Ma, Y., Nagy, Z. P., et al. (2010). Rapid elimination of the histone variant MacroH2A from somatic cell heterochromatin after nuclear transfer. Cell. Reprogram. 12, 43–53. doi: 10.1089/cell.2009.0043
Chen, L., and Zhang, S. (2016). Long noncoding RNAs in cell differentiation and pluripotency. Cell Tissue Res. 366, 509–521. doi: 10.1007/s00441-016-2451-5
Chen, Z., and Zhang, Y. (2019). Role of mammalian DNA methyltransferases in development. Annu. Rev. Biochem. doi: 10.1146/annurev-biochem-103019-102815 [Epub ahead of print].
Chesne, P., Adenot, P. G., Viglietta, C., Baratte, M., Boulanger, L., and Renard, J. P. (2002). Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat. Biotechnol. 20, 366–369. doi: 10.1038/nbt0402-366
Chung, Y. G., Matoba, S., Liu, Y., Eum, J. H., Lu, F., Jiang, W., et al. (2015). Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell 17, 758–766. doi: 10.1016/j.stem.2015.10.001
Chung, Y. G., Ratnam, S., Chaillet, J. R., and Latham, K. E. (2003). Abnormal regulation of DNA methyltransferase expression in cloned mouse embryos. Biol. Reprod. 69, 146–153. doi: 10.1095/biolreprod.102.014076
Cibelli, J. B., Stice, S. L., Golueke, P. J., Kane, J. J., Jerry, J., Blackwell, C., et al. (1998). Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science 280, 1256–1258. doi: 10.1126/science.280.5367.1256
Costa-Borges, N., Santalo, J., and Ibanez, E. (2010). Comparison between the effects of valproic acid and trichostatin A on the in vitro development, blastocyst quality, and full-term development of mouse somatic cell nuclear transfer embryos. Cell. Reprogram. 12, 437–446. doi: 10.1089/cell.2009.0108
Czernik, M., Anzalone, D. A., Palazzese, L., Oikawa, M., and Loi, P. (2019). Somatic cell nuclear transfer: failures, successes and the challenges ahead. Int. J. Dev. Biol. 63, 123–130. doi: 10.1387/ijdb.180324mc
Dean, W., Santos, F., Stojkovic, M., Zakhartchenko, V., Walter, J., Wolf, E., et al. (2001). Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. U.S.A. 98, 13734–13738. doi: 10.1073/pnas.241522698
Diao, Y. F., Naruse, K. J., Han, R. X., Li, X. X., Oqani, R. K., Lin, T., et al. (2013). Treatment of fetal fibroblasts with DNA methylation inhibitors and/or histone deacetylase inhibitors improves the development of porcine nuclear transfer-derived embryos. Anim. Reprod. Sci. 141, 164–171. doi: 10.1016/j.anireprosci.2013.08.008
Djekidel, M. N., Inoue, A., Matoba, S., Suzuki, T., Zhang, C., Lu, F., et al. (2018). Reprogramming of chromatin accessibility in somatic cell nuclear transfer is DNA replication independent. Cell Rep. 23, 1939–1947. doi: 10.1016/j.celrep.2018.04.036
Enright, B. P., Kubota, C., Yang, X., and Tian, X. C. (2003). Epigenetic characteristics and development of embryos cloned from donor cells treated by trichostatin A or 5-aza-2′-deoxycytidine. Biol. Reprod. 69, 896–901. doi: 10.1095/biolreprod.103.017954
Flynn, R. A., and Chang, H. Y. (2014). Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14, 752–761. doi: 10.1016/j.stem.2014.05.014
Galli, C., Lagutina, I., Crotti, G., Colleoni, S., Turini, P., Ponderato, N., et al. (2003). Pregnancy: a cloned horse born to its dam twin. Nature 424:635. doi: 10.1038/424635a
Galli, C., Lagutina, I., Perota, A., Colleoni, S., Duchi, R., Lucchini, F., et al. (2012). Somatic cell nuclear transfer and transgenesis in large animals: current and future insights. Reprod. Domest. Anim. 47, 2–11. doi: 10.1111/j.1439-0531.2012.02045.x
Galupa, R., and Heard, E. (2015). X-chromosome inactivation: new insights into cis and trans regulation. Curr. Opin. Genet. Dev. 31, 57–66. doi: 10.1016/j.gde.2015.04.002
Gao, R., Wang, C., Gao, Y., Xiu, W., Chen, J., Kou, X., et al. (2018). Inhibition of aberrant DNA re-methylation improves post-implantation development of somatic cell nuclear transfer embryos. Cell Stem Cell 23, doi: 10.1016/j.stem.2018.07.017
Gao, S., Chung, Y. G., Parseghian, M. H., King, G. J., Adashi, E. Y., and Latham, K. E. (2004). Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Dev. Biol. 266, 62–75. doi: 10.1016/j.ydbio.2003.10.003
Gomez, M. C., Pope, C. E., Ricks, D. M., Lyons, J., Dumas, C., and Dresser, B. L. (2009). Cloning endangered felids using heterospecific donor oocytes and interspecies embryo transfer. Reprod. Fertil. Dev. 21, 76–82.
Guo, F., Li, X., Liang, D., Li, T., Zhu, P., Guo, H., et al. (2014). Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–459. doi: 10.1016/j.stem.2014.08.003
Hamazaki, N., Uesaka, M., Nakashima, K., Agata, K., and Imamura, T. (2015). Gene activation-associated long noncoding RNAs function in mouse preimplantation development. Development 142, 910–920. doi: 10.1242/dev.116996
Han, C., Deng, R., Mao, T., Luo, Y., Wei, B., Meng, P., et al. (2018). Overexpression of Tet3 in donor cells enhances goat somatic cell nuclear transfer efficiency. FEBS J. 285, 2708–2723. doi: 10.1111/febs.14515
Holoch, D., and Moazed, D. (2015). RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84. doi: 10.1038/nrg3863
Huan, Y., Wu, Z., Zhang, J., Zhu, J., Liu, Z., and Song, X. (2015a). Epigenetic modification agents improve gene-specific methylation reprogramming in porcine cloned embryos. PLoS One 10:e0129803. doi: 10.1371/journal.pone.0129803
Huan, Y., Zhu, J., Huang, B., Mu, Y., Kong, Q., and Liu, Z. (2015b). Trichostatin A rescues the disrupted imprinting induced by somatic cell nuclear transfer in pigs. PLoS One 10:e0126607. doi: 10.1371/journal.pone.0126607
Huan, Y. J., Zhu, J., Wang, H. M., Wu, Z. F., Zhang, J. G., Xie, B. T., et al. (2014). Epigenetic modification agents improve genomic methylation reprogramming in porcine cloned embryos. J. Reprod. Dev. 60, 377–382. doi: 10.1262/jrd.2014-062
Inoue, A., Jiang, L., Lu, F., Suzuki, T., and Zhang, Y. (2017a). Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424. doi: 10.1038/nature23262
Inoue, A., Jiang, L., Lu, F., and Zhang, Y. (2017b). Genomic imprinting of Xist by maternal H3K27me3. Genes Dev. 31, 1927–1932. doi: 10.1101/gad.304113.117
Inoue, K., Kohda, T., Sugimoto, M., Sado, T., Ogonuki, N., Matoba, S., et al. (2010). Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 330, 496–499. doi: 10.1126/science.1194174
Iqbal, K., Jin, S. G., Pfeifer, G. P., and Szabo, P. E. (2011). Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U.S.A. 108, 3642–3647. doi: 10.1073/pnas.1014033108
Ito, S., D’Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. doi: 10.1038/nature09303
Iuso, D., Czernik, M., Toschi, P., Fidanza, A., Zacchini, F., Feil, R., et al. (2015). Exogenous expression of human protamine 1 (hPrm1) remodels fibroblast nuclei into spermatid-like structures. Cell Rep. 13, 1765–1771. doi: 10.1016/j.celrep.2015.10.066
Izzo, A., and Schneider, R. (2010). Chatting histone modifications in mammals. Brief. Funct. Genomics 9, 429–443. doi: 10.1093/bfgp/elq024
Keefer, C. L. (2015). Artificial cloning of domestic animals. Proc. Natl. Acad. Sci. U.S.A. 112, 8874–8878. doi: 10.1073/pnas.1501718112
Kiefer, H., Jouneau, L., Campion, E., Rousseau-Ralliard, D., Larcher, T., and Martin-Magniette, M. L. (2016). Altered DNA methylation associated with an abnormal liver phenotype in a cattle model with a high incidence of perinatal pathologies. Sci. Rep. 6:38869. doi: 10.1038/srep38869
Kim, D. H., Marinov, G. K., Pepke, S., Singer, Z. S., He, P., Williams, B., et al. (2015). Single-cell transcriptome analysis reveals dynamic changes in lncRNA expression during reprogramming. Cell Stem Cell 16, 88–101. doi: 10.1016/j.stem.2014.11.005
Kimura, A. P., Yoneda, R., Kurihara, M., Mayama, S., and Matsubara, S. (2017). A long noncoding RNA, lncRNA-Amhr2, plays a role in Amhr2 gene activation in mouse ovarian granulosa cells. Endocrinology 158, 4105–4121. doi: 10.1210/en.2017-00619
Kishigami, S., Mizutani, E., Ohta, H., Hikichi, T., Thuan, N. V., Wakayama, S., et al. (2006). Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem. Biophys. Res. Commun. 340, 183–189. doi: 10.1016/j.bbrc.2005.11.164
Kobayashi, W., and Kurumizaka, H. (2019). Structural transition of the nucleosome during chromatin remodeling and transcription. Curr. Opin. Struct. Biol. 59, 107–114. doi: 10.1016/j.sbi.2019.07.011
Kono, T., Obata, Y., Wu, Q., Niwa, K., Ono, Y., Yamamoto, Y., et al. (2004). Birth of parthenogenetic mice that can develop to adulthood. Nature 428, 860–864. doi: 10.1038/nature02402
Kremenskoy, M., Kremenska, Y., Suzuki, M., Imai, K., Takahashi, S., Hashizume, K., et al. (2006). DNA methylation profiles of donor nuclei cells and tissues of cloned bovine fetuses. J. Reprod. Dev. 52, 259–266. doi: 10.1262/jrd.17098
Kurome, M., Geistlinger, L., Kessler, B., Zakhartchenko, V., Klymiuk, N., Wuensch, A., et al. (2013). Factors influencing the efficiency of generating genetically engineered pigs by nuclear transfer: multi-factorial analysis of a large data set. BMC Biotechnol. 13:43. doi: 10.1186/1472-6750-13-43
Latham, K. E. (2005). X chromosome imprinting and inactivation in preimplantation mammalian embryos. Trends Genet. 21, 120–127. doi: 10.1016/j.tig.2004.12.003
Lee, B. C., Kim, M. K., Jang, G., Oh, H. J., Yuda, F., Kim, H. J., et al. (2005). Dogs cloned from adult somatic cells. Nature 436:641. doi: 10.1038/436641a
Lee, J. T. (2009). Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 23, 1831–1842. doi: 10.1101/gad.1811209
Lee, K., Uh, K., and Farrell, K. (2020). Current progress of genome editing in livestock. Theriogenology. doi: 10.1016/j.theriogenology.2020.01.036 [Epub ahead of print].
Li, E., and Zhang, Y. (2014). DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 6:a019133. doi: 10.1101/cshperspect.a019133
Li, Z., Sun, X., Chen, J., Liu, X., Wisely, S. M., Zhou, Q., et al. (2006). Cloned ferrets produced by somatic cell nuclear transfer. Dev. Biol. 293, 439–448. doi: 10.1016/j.ydbio.2006.02.016
Liao, H. F., Mo, C. F., Wu, S. C., Cheng, D. H., Yu, C. Y., Chang, K. W., et al. (2015). Dnmt3l-knockout donor cells improve somatic cell nuclear transfer reprogramming efficiency. Reproduction 150, 245–256. doi: 10.1530/REP-15-0031
Liu, G. Y., Zhao, G. N., Chen, X. F., Hao, D. L., Zhao, X., Lv, X., et al. (2015). The long noncoding RNA Gm15055 represses Hoxa gene expression by recruiting PRC2 to the gene cluster. Nucleic Acids Res. 44, 2613–2627. doi: 10.1093/nar/gkv1315
Liu, L., Liu, Y., Gao, F., Song, G., Wen, J., Guan, J., et al. (2012). Embryonic development and gene expression of porcine SCNT embryos treated with sodium butyrate. J. Exp. Zool. Part B Mol. Dev. Evol. 318, 224–234. doi: 10.1002/jez.b.22440
Liu, W., Liu, X., Wang, C., Gao, Y., Gao, R., Kou, X., et al. (2016). Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov. 2:16010. doi: 10.1038/celldisc.2016.10
Liu, X., Wang, C., Liu, W., Li, J., Li, C., Kou, X., et al. (2016). Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562. doi: 10.1038/nature19362
Liu, Z., Cai, Y., Wang, Y., Nie, Y., Zhang, C., Xu, Y., et al. (2018). Cloning of macaque monkeys by somatic cell nuclear transfer. Cell 172, 881–887.e7. doi: 10.1016/j.cell.2018.01.020
Loi, P., Iuso, D., Czernik, M., and Ogura, A. (2016). A new dynamic Era for somatic cell nuclear transfer? Trends Biotechnol. 34, 791–797. doi: 10.1016/j.tibtech.2016.03.008
Long, C. R., Westhusin, M. E., and Golding, M. C. (2014). Reshaping the transcriptional frontier: epigenetics and somatic cell nuclear transfer. Mol. Reprod. Dev. 81, 183–193. doi: 10.1002/mrd.22271
Lotti, S. N., Polkoff, K. M., Rubessa, M., and Wheeler, M. B. (2017). Modification of the genome of domestic animals. Anim. Biotechnol. 28, 198–210. doi: 10.1080/10495398.2016.1261874
Lyko, F. (2018). The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92. doi: 10.1038/nrg.2017.80
MacDonald, W. A., and Mann, M. R. (2014). Epigenetic regulation of genomic imprinting from germ line to preimplantation. Mol. Reprod. Dev. 81, 126–140. doi: 10.1002/mrd.22220
Mann, M. R., Chung, Y. G., Nolen, L. D., Verona, R. I., Latham, K. E., and Bartolomei, M. S. (2003). Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod. 69, 902–914. doi: 10.1095/biolreprod.103.017293
Matoba, S., Inoue, K., Kohda, T., Sugimoto, M., Mizutani, E., Ogonuki, N., et al. (2011). RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc. Natl. Acad. Sci. U.S.A. 108, 20621–20626. doi: 10.1073/pnas.1112664108
Matoba, S., Liu, Y., Lu, F., Iwabuchi, K. A., Shen, L., Inoue, A., et al. (2014). Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159, 884–895. doi: 10.1016/j.cell.2014.09.055
Matoba, S., and Zhang, Y. (2018). Somatic cell nuclear transfer reprogramming: mechanisms and applications. Cell Stem Cell 23, 471–485. doi: 10.1016/j.stem.2018.06.018
Mercer, T. R., and Mattick, J. S. (2013). Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307. doi: 10.1038/nsmb.2480
Ng, R. K., and Gurdon, J. B. (2005). Maintenance of epigenetic memory in cloned embryos. Cell Cycle 4, 760–763. doi: 10.4161/cc.4.6.1743
Niemann, H. (2016). Epigenetic reprogramming in mammalian species after SCNT-based cloning. Theriogenology 86, 80–90. doi: 10.1016/j.theriogenology.2016.04.021
Ninova, M., Fejes Toth, K., and Aravin, A. A. (2019). The control of gene expression and cell identity by H3K9 trimethylation. Development 146, dev181180. doi: 10.1242/dev.181180
Niu, D., Wei, H. J., Lin, L., George, H., Wang, T., Lee, I. H., et al. (2017). Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 357, 1303–1307. doi: 10.1126/science.aan4187
Nolen, L. D., Gao, S., Han, Z., Mann, M. R., Gie Chung, Y., Otte, A. P., et al. (2005). X chromosome reactivation and regulation in cloned embryos. Dev. Biol. 279, 525–540. doi: 10.1016/j.ydbio.2005.01.016
Ogura, A., Inoue, K., and Wakayama, T. (2013). Recent advancements in cloning by somatic cell nuclear transfer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20110329. doi: 10.1098/rstb.2011.0329
Pauli, A., Rinn, J. L., and Schier, A. F. (2011). Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 12, 136–149. doi: 10.1038/nrg2904
Payer, B. (2016). Developmental regulation of X-chromosome inactivation. Semin. Cell Dev. Biol. 56, 88–99. doi: 10.1016/j.semcdb.2016.04.014
Polejaeva, I. A., Chen, S. H., Vaught, T. D., Page, R. L., Mullins, J., Ball, S., et al. (2000). Cloned pigs produced by nuclear transfer from adult somatic cells. Nature 407, 86–90. doi: 10.1038/35024082
Qin, H., Zhao, A., Zhang, C., and Fu, X. (2016). Epigenetic control of reprogramming and transdifferentiation by histone modifications. Stem Cell Rev. Rep. 12, 708–720. doi: 10.1007/s12015-016-9682-4
Reik, W., Dean, W., and Walter, J. (2001). Epigenetic reprogramming in mammalian development. Science 293, 1089–1093. doi: 10.1126/science.1063443
Reik, W., Santos, F., Mitsuya, K., Morgan, H., and Dean, W. (2003). Epigenetic asymmetry in the mammalian zygote and early embryo: relationship to lineage commitment? Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 1403–1409. doi: 10.1098/rstb.2003.1326
Ruan, D., Peng, J., Wang, X., Ouyang, Z., Zou, Q., Yang, Y., et al. (2018). XIST derepression in active X chromosome hinders pig somatic cell nuclear transfer. Stem Cell Rep. 10, 494–508. doi: 10.1016/j.stemcr.2017.12.015
Rybouchkin, A., Kato, Y., and Tsunoda, Y. (2006). Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol. Reprod. 74, 1083–1089. doi: 10.1095/biolreprod.105.047456
Sasaki, H., Ishihara, K., and Kato, R. (2000). Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J. Biochem. 127, 711–715. doi: 10.1093/oxfordjournals.jbchem.a022661
Schubeler, D. (2015). Function and information content of DNA methylation. Nature 517, 321–326. doi: 10.1038/nature14192
Shen, L., Wu, H., Diep, D., Yamaguchi, S., D’Alessio, A. C., Fung, H. L., et al. (2013). Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153, 692–706. doi: 10.1016/j.cell.2013.04.002
Shi, D., Lu, F., Wei, Y., Cui, K., Yang, S., Wei, J., et al. (2007). Buffalos (Bubalus bubalis) cloned by nuclear transfer of somatic cells1. Biol. Reprod. 77, 285–291. doi: 10.1095/biolreprod.107.060210
Shi, W., Zakhartchenko, V., and Wolf, E. (2003). Epigenetic reprogramming in mammalian nuclear transfer. Differentiation 71, 91–113. doi: 10.1046/j.1432-0436.2003.710201.x
Shin, T., Kraemer, D., Pryor, J., Liu, L., Rugila, J., Howe, L., et al. (2002). A cat cloned by nuclear transplantation. Nature 415:859. doi: 10.1038/nature723
Simon, I., Tenzen, T., Reubinoff, B. E., Hillman, D., McCarrey, J. R., and Cedar, H. (1999). Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature 401, 929–932. doi: 10.1038/44866
Song, X., Li, F., Jiang, Z., Sun, Y., Li, H., Gao, S., et al. (2017a). Imprinting disorder in donor cells is detrimental to the development of cloned embryos in pigs. Oncotarget 8, 72363–72374. doi: 10.18632/oncotarget.20390
Song, X., Liu, Z., He, H., Wang, J., Li, H., Li, J., et al. (2017b). Dnmt1s in donor cells is a barrier to SCNT-mediated DNA methylation reprogramming in pigs. Oncotarget 8, 34980–34991. doi: 10.18632/oncotarget.16507
Sproul, D., Gilbert, N., and Bickmore, W. A. (2005). The role of chromatin structure in regulating the expression of clustered genes. Nat. Rev. Genet. 6, 775–781. doi: 10.1038/nrg1688
Sun, J. M., Spencer, V. A., Chen, H. Y., Li, L., and Davie, J. R. (2003). Measurement of histone acetyltransferase and histone deacetylase activities and kinetics of histone acetylation. Methods 31, 12–23. doi: 10.1016/s1046-2023(03)00083-5
Tachibana, M., Amato, P., Sparman, M., Nuria Gutierrez, M., Tippner-Hedges, R., Ma, H., et al. (2013). Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153, 1228–1238. doi: 10.1016/j.cell.2013.05.006
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Telugu, B. P., Park, K. E., and Park, C. H. (2017). Genome editing and genetic engineering in livestock for advancing agricultural and biomedical applications. Mamm. Genome 28, 338–347. doi: 10.1007/s00335-017-9709-4
Tsai, M. C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. doi: 10.1126/science.1192002
Wakayama, T., Perry, A. C., Zuccotti, M., Johnson, K. R., and Yanagimachi, R. (1998). Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 394, 369–374. doi: 10.1038/28615
Wang, J., Wang, L., Feng, G., Wang, Y., Li, Y., Li, X., et al. (2018). Asymmetric expression of LincGET biases cell fate in two-cell mouse embryos. Cell 175, 1887–1901.e18. doi: 10.1016/j.cell.2018.11.039
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. doi: 10.1038/nature09819
Wang, K. Y., Chen, C. C., and Shen, C. K. (2014). Active DNA demethylation of the vertebrate genomes by DNA methyltransferases: deaminase, dehydroxymethylase or demethylase? Epigenomics 6, 353–363. doi: 10.2217/epi.14.21
Wang, L., Zhao, Y., Bao, X., Zhu, X., Kwok, Y. K., Sun, K., et al. (2015). LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 25, 335–350. doi: 10.1038/cr.2015.21
Wani, N. A., Wernery, U., Hassan, F. A. H., Wernery, R., and Skidmore, J. A. (2010). Production of the first cloned camel by somatic cell nuclear transfer1. Biol. Reprod. 82, 373–379. doi: 10.1095/biolreprod.109.081083
Wee, G., Koo, D. B., Song, B. S., Kim, J. S., Kang, M. J., Moon, S. J., et al. (2006). Inheritable histone H4 acetylation of somatic chromatins in cloned embryos. J. Biol. Chem. 281, 6048–6057. doi: 10.1074/jbc.m511340200
Wei, Y., Zhu, J., Huan, Y., Liu, Z., Yang, C., Zhang, X., et al. (2010). Aberrant expression and methylation status of putatively imprinted genes in placenta of cloned piglets. Cell. Reprogram. 12, 213–222. doi: 10.1089/cell.2009.0090
Wells, K. D., and Prather, R. S. (2017). Genome-editing technologies to improve research, reproduction, and production in pigs. Mol. Reprod. Dev. 84, 1012–1017. doi: 10.1002/mrd.22812
Wen, D., Banaszynski, L. A., Rosenwaks, Z., Allis, C. D., and Rafii, S. (2014). H3.3 replacement facilitates epigenetic reprogramming of donor nuclei in somatic cell nuclear transfer embryos. Nucleus 5, 369–375. doi: 10.4161/nucl.36231
Weng, X. G., Cai, M. M., Zhang, Y. T., Liu, Y., Liu, C., and Liu, Z. H. (2019). Improvement in the in vitro development of cloned pig embryos after kdm4a overexpression and an H3K9me3 methyltransferase inhibitor treatment. Theriogenology. doi: 10.1016/j.theriogenology.2019.11.027 [Epub ahead of print].
Wilmut, I., Schnieke, A. E., McWhir, J., Kind, A. J., and Campbell, K. H. S. (1997). Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813. doi: 10.1038/385810a0
Woods, G. L., White, K. L., Vanderwall, D. K., Li, G. P., Aston, K. I., Bunch, T. D., et al. (2003). A mule cloned from fetal cells by nuclear transfer. Science 301:1063. doi: 10.1126/science.1086743
Wu, F., Liu, Y., Wu, Q., Li, D., Zhang, L., Wu, X., et al. (2018). Long non-coding RNAs potentially function synergistically in the Cell. Reprogram. of SCNT embryos. BMC Genomics 19:631. doi: 10.1186/s12864-018-5021-2
Wu, J., Huang, B., Chen, H., Yin, Q., Liu, Y., Xiang, Y., et al. (2016). The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534:652. doi: 10.1038/nature18606
Xie, B., Zhang, H., Wei, R., Li, Q., Weng, X., Kong, Q., et al. (2016). Histone H3 lysine 27 trimethylation acts as an epigenetic barrier in porcine nuclear reprogramming. Reproduction 151, 9–16. doi: 10.1530/REP-15-0338
Xu, W., Li, Z., Yu, B., He, X., Shi, J., Zhou, R., et al. (2013). Effects of DNMT1 and HDAC inhibitors on gene-specific methylation reprogramming during porcine somatic cell nuclear transfer. PLoS One 8:e64705. doi: 10.1371/journal.pone.0064705
Xue, F., Tian, X. C., Du, F., Kubota, C., Taneja, M., Dinnyes, A., et al. (2002). Aberrant patterns of X chromosome inactivation in bovine clones. Nat. Genet. 31, 216–220. doi: 10.1038/ng900
Yamazaki, Y., Fujita, T. C., Low, E. W., Alarcon, V. B., Yanagimachi, R., and Marikawa, Y. (2006). Gradual DNA demethylation of the Oct4 promoter in cloned mouse embryos. Mol. Reprod. Dev. 73, 180–188. doi: 10.1002/mrd.20411
Yang, X., Smith, S. L., Tian, X. C., Lewin, H. A., Renard, J. P., and Wakayama, T. (2007). Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat. Genet. 39, 295–302. doi: 10.1038/ng1973
Yang, X., Wu, X., Yang, Y., Gu, T., Hong, L., Zheng, E., et al. (2019). Improvement of developmental competence of cloned male pig embryos by short hairpin ribonucleic acid (shRNA) vector-based but not small interfering RNA (siRNA)-mediated RNA interference (RNAi) of Xist expression. J. Reprod. Dev. 65, 533–539. doi: 10.1262/jrd.2019-070
Yi, S. J., and Kim, K. (2018). Histone tail cleavage as a novel epigenetic regulatory mechanism for gene expression. BMB Rep. 51, 211–218. doi: 10.5483/bmbrep.2018.51.5.053
Yuan, L., Wang, A., Yao, C., Huang, Y., Duan, F., Lv, Q., et al. (2014). Aberrant expression of Xist in aborted porcine fetuses derived from somatic cell nuclear transfer embryos. Int. J. Mol. Sci. 15, 21631–21643. doi: 10.3390/ijms151221631
Zeng, F., Huang, Z., Yuan, Y., Shi, J., Cai, G., Liu, D., et al. (2016). Effects of RNAi-mediated knockdown of Xist on the developmental efficiency of cloned male porcine embryos. J. Reprod. Dev. 62, 591–597. doi: 10.1262/jrd.2016-095
Zhai, Y., Zhang, Z., Yu, H., Su, L., Yao, G., Ma, X., et al. (2018). Dynamic methylation changes of DNA and H3K4 by RG108 improve epigenetic reprogramming of somatic cell nuclear transfer embryos in pigs. Cell. Physiol. Biochem. 50, 1376–1397. doi: 10.1159/000494598
Zhang, X., Wang, D., Han, Y., Duan, F., Lv, Q., and Li, Z. (2014). Altered imprinted gene expression and methylation patterns in mid-gestation aborted cloned porcine fetuses and placentas. J. Assist. Reprod. Genet. 31, 1511–1517. doi: 10.1007/s10815-014-0320-2
Zhong, C., and Li, J. (2017). Efficient generation of gene-modified mice by haploid embryonic stem cell-mediated semi-cloned technology. Methods Mol. Biol. 1498, 121–133. doi: 10.1007/978-1-4939-6472-7_8
Zhou, C., Wang, Y., Zhang, J., Su, J., An, Q., Liu, X., et al. (2019). H3K27me3 is an epigenetic barrier while KDM6A overexpression improves nuclear reprogramming efficiency. FASEB J. 33, 4638–4652. doi: 10.1096/fj.201801887R
Zhou, J., Yang, L., Zhong, T., Mueller, M., Men, Y., Zhang, N., et al. (2015). H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat. Commun. 6:10221. doi: 10.1038/ncomms10221
Zhou, Q., Renard, J. P., Le Friec, G., Brochard, V., Beaujean, N., Cherifi, Y., et al. (2003). Generation of fertile cloned rats by regulating oocyte activation. Science 302:1179. doi: 10.1126/science.1088313
Keywords: somatic cell nuclear transfer, cloning efficiency, nuclear reprogramming, epigenetic modification, long non-coding RNA
Citation: Wang X, Qu J, Li J, He H, Liu Z and Huan Y (2020) Epigenetic Reprogramming During Somatic Cell Nuclear Transfer: Recent Progress and Future Directions. Front. Genet. 11:205. doi: 10.3389/fgene.2020.00205
Received: 23 October 2019; Accepted: 21 February 2020;
Published: 18 March 2020.
Edited by:
Helene Kiefer, INRA Centre Jouy-en-Josas, FranceReviewed by:
Nathalie Beaujean, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceCopyright © 2020 Wang, Qu, Li, He, Liu and Huan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanjun Huan, aHVhbnlhbmp1bjE5ODJAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.