- National Engineering Research Centre for Breeding Swine Industry, Guangdong Provincial Key Laboratory of Agro-Animal Genomics and Molecular Breeding, College of Animal Science, South China Agricultural University, Guangzhou, China
Being the center of the hypothalamus-pituitary-ovary (HPO) axis, the pituitary plays a key role in the onset of puberty. Recent studies show that circular RNAs (circRNAs) can perform as miRNA sponges to regulate development in animals. However, the function of pituitary-derived circRNAs in first estrus remains unclear in pigs. In this study, we performed a genome-wide identification and characterization of circRNAs using pituitaries from Landrace × Yorkshire crossbred pigs at three stages: pre-, in-, and post-puberty, to describe such pituitary-derived circRNAs in pigs. A total of 5148 circRNAs were found in the gilts' pituitaries, averaging 18 682 bp in genomic distance, which consisted of approximately 91% exonic, 6% intergenic, and 3% intronic circRNAs. Furthermore, 158 novel circRNAs were identified for the first time and classified as putative pituitary-specific circRNAs. Their expression levels during the onset of puberty, significantly exceeded those of the other circRNAs, and the parental genes of these putative pituitary-specific circRNAs were enriched in “ssc04917: prolactin signaling pathway,” “ssc04080: neuroactive ligand-receptor interaction,” and “ssc04728: dopaminergic synapse” pathways, all of which were consistent with pituitary functioning. Additionally, 17 differentially regulated circRNAs were found and investigated for their potential interaction with miRNAs, along with genes, by constructing a circRNA-targeted miRNA-gene network. Taken together, these results provide new insight into the circRNA-mediated timing of puberty in gilts at the pituitary level.
Introduction
Puberty can usually be defined as the first estrus of gilts, and the initiation of puberty implies the acquired capacity for sexual reproduction in pigs (Nonneman et al., 2016). An early onset of puberty can shorten the generation interval of livestock, and further accelerate the genetic breeding process (Rosales Nieto et al., 2014; Luo et al., 2017). Yet, surprisingly little is known about the molecular regulation of puberty's timing in pigs. Previous research has uncovered endocrinological differences across pubertal onset mainly driven by the hypothalamus-pituitary-ovary (HPO) axis (Angold et al., 1999; Blakemore et al., 2010). As the center bridging the hypothalamus and ovary in the HPO axis, the pituitary is an extremely important mediator for controlling the synthesis of hormones. During estrous cycling, an increase in the pulsatile release of gonadotropin-releasing hormone (GnRH) from the hypothalamus elicits an increased release of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) from the pituitary (Root, 1980; Coe et al., 1981). Additionally, gonadotropin hormones released from the pituitary have been shown to be directly related to animals' reproductive associated traits (Barb et al., 2012).
Circular RNAs (circRNAs) are a novel type of circular RNA molecules lacking 5′–3′ polarities and polyadenylated tails (Chen and Yang, 2015), making them more structurally stable than linear RNAs (Qu et al., 2015). Most circRNAs consist of multiple exons, as well as introns of protein-coding genes, and are conserved among different animal species (Szabo and Salzman, 2016). With the advances made in next-generation sequencing technology, much research on circRNAs has been carried out using high-throughput RNA sequencing (RNA-seq). In pigs, recent work has demonstrated circRNAs' involvement in various organismal processes. For example, through a comprehensive analysis of porcine cardiac and skeletal muscles, Chen et al. (2018) showed that circRNAs contribute to differences in aging. Moreover, circRNAs were defined as a new biomarker in metabolism-related diseases based on a study of circRNAs occurring in the subcutaneous adipose tissues of two pig breeds (Li et al., 2018). For their role in estrus, Li et al. (2018) investigated the expression of circRNAs in the sheep pituitary, finding that circRNAs there participated in the regulation of estrus. By contrast, no puberty- or even estrus-associated study has yet been performed that has tried to identify circRNAs in pigs.
Generally, since gilts have an earlier age at first estrus, they may have a longer productive life, thus farrowing multiple litters and giving birth to more piglets (Patterson et al., 2010; Saito et al., 2011). To reveal the relationships between circRNAs and puberty in the pituitary, here we conducted RNA-seq analyses using pituitaries from Landrace × Yorkshire crossbred pigs at three stages: pre-, in-, and post-puberty, to identify circRNAs and then assemble a circRNA-targeted miRNA-gene network. To our best knowledge, this study is the first to investigate the potential regulatory roles of circRNAs during the onset of puberty in gilts, and so it should provide new insight into this key developmental process at the molecular level.
Materials and Methods
Ethics Statement
Animal care and experiments were conducted following the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China; revised in June 2004) and were approved by the Animal Care and Use Committee of the South China Agricultural University, Guangzhou, China (permit number: SCAU#2013-10).
Preparation of Animals and Samples
Three stages during the onset of puberty were used: pre-, in-, and post-puberty. The onset of puberty was identified by the standing reflex with the back-pressure test and boar contact (Patterson et al., 2002). A total of nine Landrace × Yorkshire crossbred gilts were used: three gilts of 160 days in age without any pubertal signs were selected as pre-puberty gilts (weight = 81.38 ± 2.40 kg); three gilts exhibiting first pubertal signs served as the in-pubertal gilts (weight = 110.00 ± 2.00 kg); three gilts 14 days beyond the pubertal phase were designated as the post-pubertal gilts (weight = 122.82 ± 9.11 kg). After euthanizing the gilts, their brains were removed immediately and excess tissues were removed. The anterior pituitaries were carefully dissected and frozen immediately in liquid nitrogen, then stored at –80°C until further use.
RNA Sequencing and Quality Control, and the Transcriptome Assembly
Pre-, in-, and post-pubertal gilts' pituitaries were homogenized separately in liquid nitrogen. The total RNAs were extracted from porcine pituitaries with the Trizol agent (Invitrogen, Carlsbad, CA, USA), followed by quality testing of the total RNAs using the Agilent Bioanalyzer 2100 system (Agilent, Palo Alto, CA, USA). Only those RNA samples with RNA Integrity Number value > 7.0 were deemed eligible. Then, the rRNA from the eligible total RNAs was removed using an Epicentre Ribo-zero rRNA removal kit (Epicentre, Madison, WI, USA). The rRNA-depleted RNAs were used to synthesize double-stranded cDNA via the mRNA-Seq Sample Preparation Kit (Illumina, SanDiego, CA, USA), for which a total of 5 μg cDNA per sample was sequenced using a HiSeq 2500 Sequencer according to the manufacturer's instructions, and 150 bp paired-end reads were generated. These raw reads were processed by 3′ adaptor-trimming and removal of low-quality reads—having > 10% unknown bases or > 50% low-mass bases—using Cutadapt software (Martin, 2011). The reads remaining after quality control were defined as the clean reads for further analysis. These acquired clean reads were then mapped onto pig reference genome Sus scrofa11.1, using BWA software (Li and Durbin, 2010).
CircRNA identification
CIRI software (Gao et al., 2015) was applied to obtain the back-spliced junction (BSJ) reads for circRNA prediction based on the annotation file downloaded from the Ensembl genome browser (ftp://ftp.ensembl.org/pub/release-94/gtf/sus_scrofa). Then the number of circRNAs' exons and the length of circRNAs were detected by CIRI-AS module in CIRI software. The expression levels of circRNAs were quantified as the number of reads spanning the BSJ reads in terms of RPM (i.e., mapped BSJ reads per million mapped reads), by using the EBSeq package (Leng et al., 2013). The differential expression of circRNAs was determined according to these criteria: false discovery rate (FDR) < 0.05, log2|fold_change| ≥ 1, and circRNA junction reads ≥ 5. Further graphical representations of results were performed in the R platform (R Foundation for Statistical Computing, 2018). Stage-specific circRNAs were defined here as those circRNAs only expressed in one pubertal stage. Known circRNAs of pig were downloaded from the circAtlas 2.0 datasets (Ji et al., 2019), an integrated resource of circRNAs in vertebrates (http://circatlas.biols.ac.cn/). In the circAtlas 2.0 database are tens of thousands of known circRNAs identified from nine porcine tissue types: brain, heart, kidney, liver, lung, skeletal muscle, spleen, testis, and retina. The circRNAs identified in the current study were matched with the database via both starting and ending genomic positions of circRNAs, and the novel circRNAs were regarded as the putative tissue-specific circRNAs. Significant differences between any two pubertal pig groups were tested with the Welch two-sample t-test.
Functional Enrichment Analysis
The circRNAs–miRNAs interactions were predicted with miRanda software (John et al., 2004). These were filtered for predictions with a maximum binding-free energy of less than –20 kcal/mol and a miRanda match score ≥ 150. Next, targeted mRNAs of each selected miRNA were predicted by Targetscan software (Witkos et al., 2011). The competing endogenous RNAs networks among the circRNAs, miRNAs, and mRNAs were built and visualized with Cytoscape software (Su et al., 2014). Functional enrichment analysis was performed using the DAVID bioinformatics resource (Huang et al., 2007). Finally, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) terms with Benjamini-Hochberg method-adjusted P < 0.05 were identified.
qRT-PCR Analysis
Quantitative real-time reverse transcription-PCR (qRT-PCR) was carried out using the PrimeScript RT Reagent Kit (TaKaRa, Osaka, Japan) in a Mx3005P real-time PCR System (Stratagene, La Jolla, CA, USA) with SYBR Green, according to the manufacturer's protocol. Divergent primers of 5 circRNAs were designed to further test the accuracy of the RNA-seq, namely Circ 1:14408861|14457143, Circ 9:28120503|28122017, Circ 2:88184110|88206327, Circ 9:75284452|75290025, and Circ 15:74631515|74643464. GAPDH served as an internal reference to normalize the expression of circRNAs (Table S1). The PCR conditions were 94 °C denaturation for 5 min, 40 cycles at 94 °C for 10 s, 52 to 62 °C for 15 s, and 72 °C for 30 s. The 2-∆∆Ct method was used to analyze the qRT-PCR results. The Student's t test was used to assess differences in means of any two pubertal pig groups, for which a P < 0.05 was considered statistically significant.
Results
Identification of Pituitary-Derived circRNAs During the Onset of Puberty
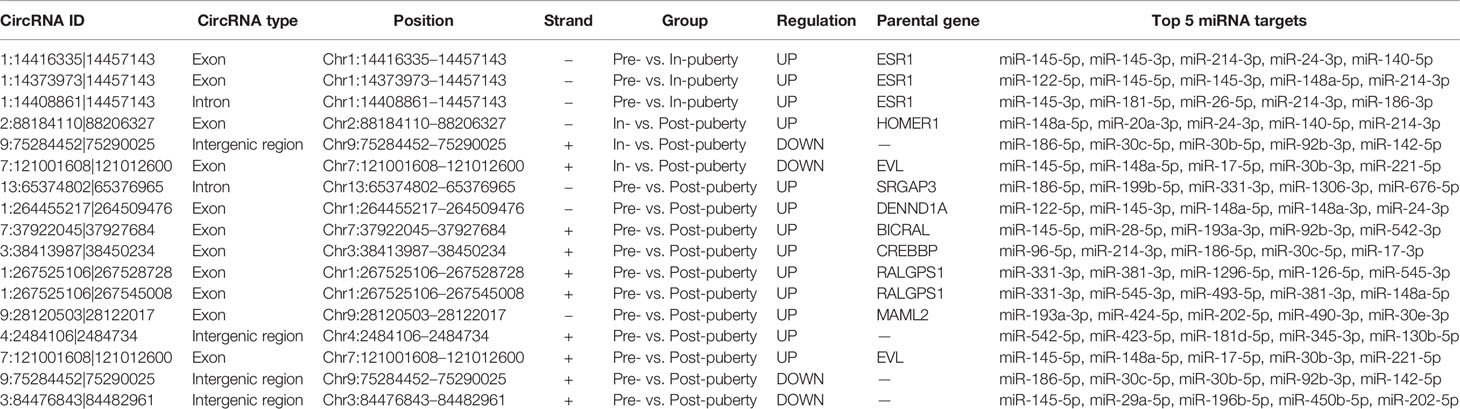
A total of 5148 circRNAs were detected in all three pubertal stages: 2779, 4062, and 3167 circRNAs respectively in the pre-, in-, and post-puberty stages of pigs (Figure 1A). The average expression level of circRNAs were dynamically changing during the onset of puberty (Figure 1B). Sus scrofa chromosome (SSC) 1 harbored the most circRNAs, while the SSC10 had the highest density of circRNAs (Figure 1C). The average genomic distance of all circRNAs found was 18 682 bp, with 92% of the circRNAs shorter than 50 000 bp, and the number of circRNAs decreased as their size lengthened (Figure 1C). The most circRNAs were made up of two exons, and the length of most circRNAs was about 200 to 300 bp (Figure 1D). After annotation with the pig genome, the found circRNAs consisted of approximately 91% exonic, 6% intergenic, and 3% intronic circRNAs, respectively (Figure 1E).
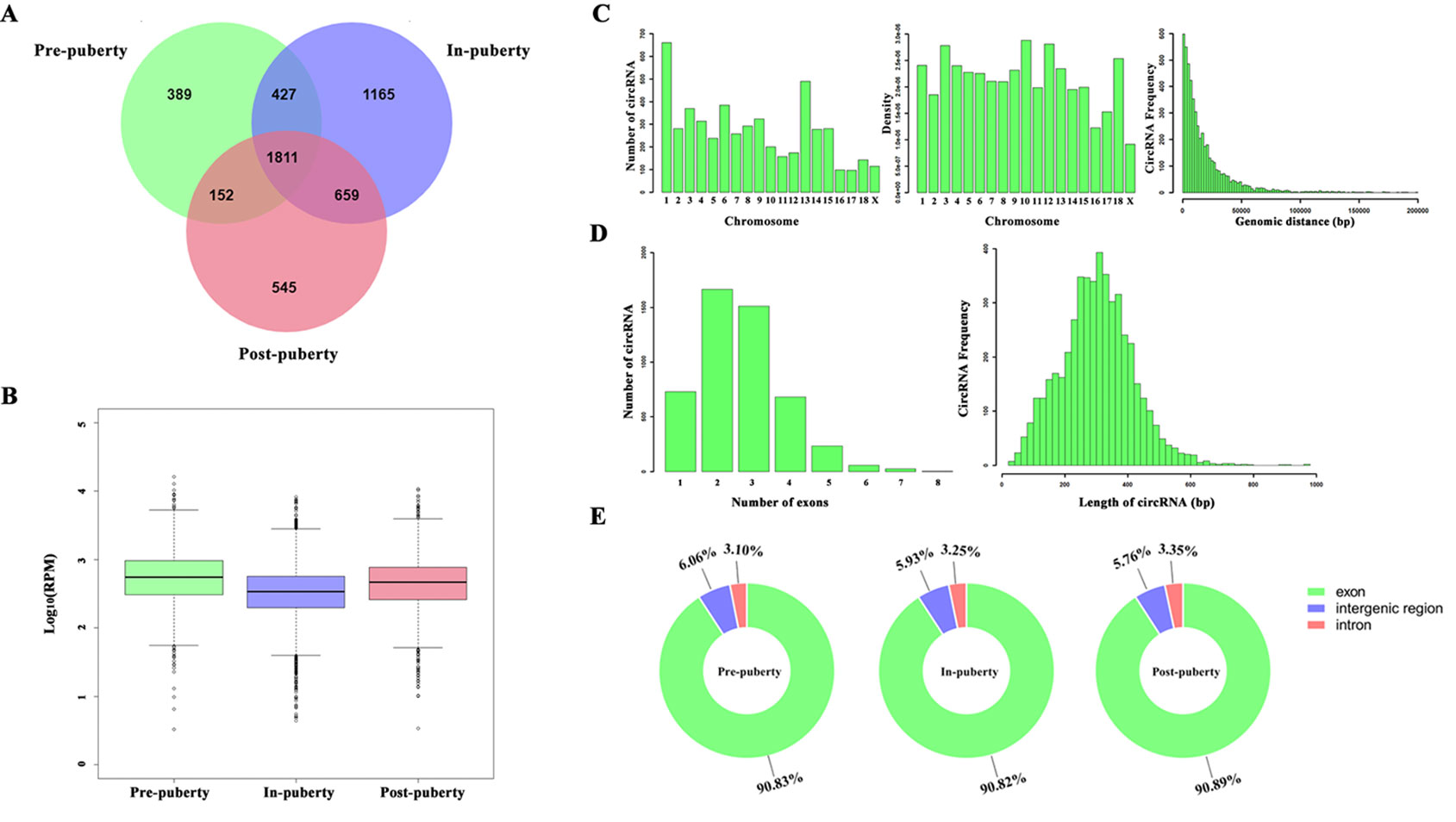
Figure 1 Identification of pituitary-derived circRNAs during the onset of puberty in pigs. (A) The Venn diagram of circRNAs detected in pre-, in-, and postpuberty; (B) Distribution and genomic distance of the circRNAs; (C) Proportion of circRNAs that originated from the exon, intergenic, and intronic regions; (D) The exon number of the circRNAs; (E) Distribution and transcript length of the circRNAs.
Putative Stage-Specific circRNAs in Gilts During the Onset of Puberty
A total of 389, 1165, and 545 circRNAs were identified as putative stage-specific circRNAs from the pre-, in-, and post-puberty groups, respectively (Figure 1A), and their pair-wise comparisons did not reveal any significant difference in bp length (t-test, P > 0.05). Further, the expression levels of pre-puberty specific circRNAs significantly exceeded those of post-puberty specific circRNAs (t-test, P < 2.20E−16), with the latter being significantly higher than the expression levels of in-puberty specific circRNAs (t-test, P < 2.20E−16) (Figure 2A). The KEGG pathways enriched using the parental genes of stage-specific circRNAs are listed in Table S2, of which the top five are shown in Figure 2B–D.
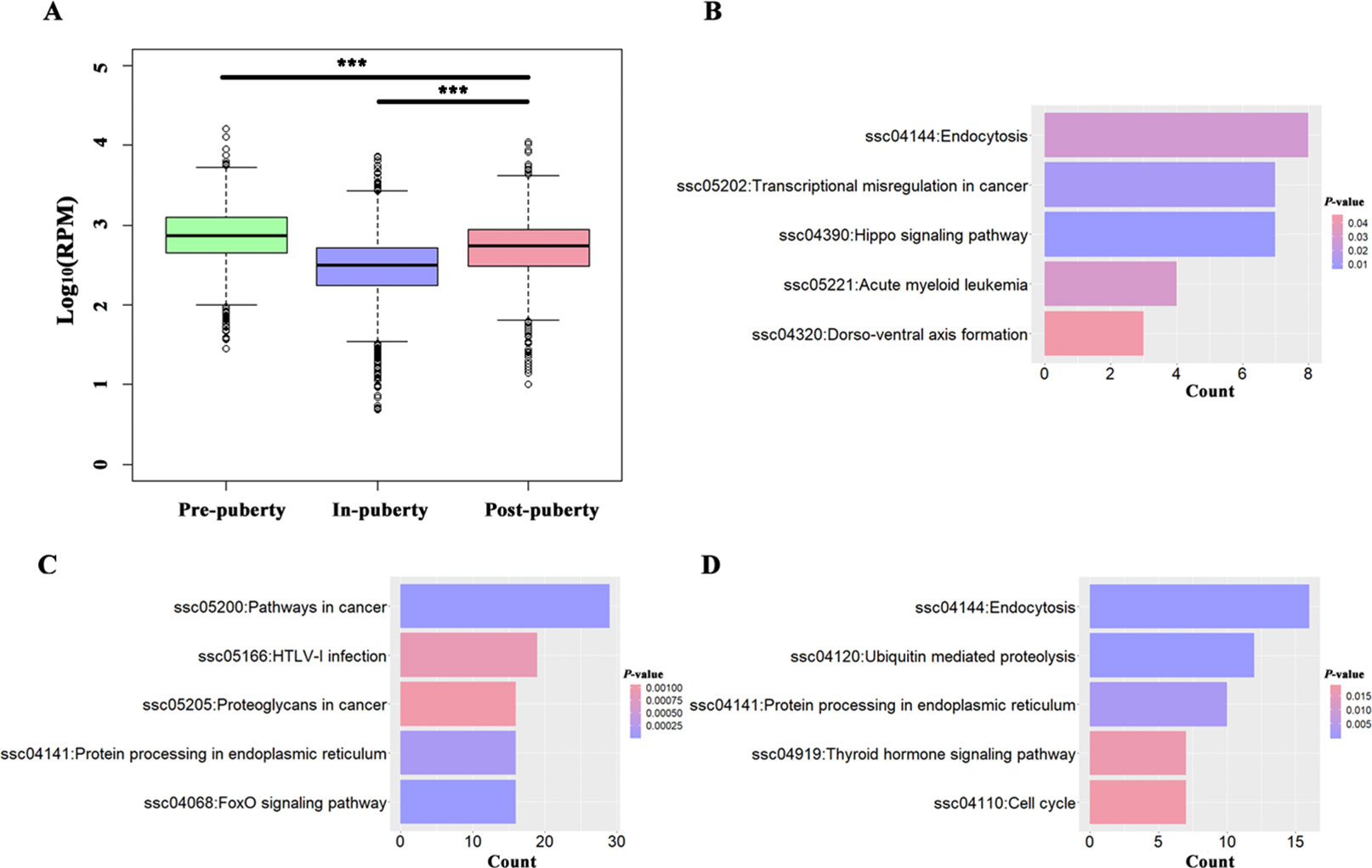
Figure 2 Analysis of potential stage-specific circRNAs in pigs. (A) Boxplots of pre-, in-, and post-puberty stage-specific circRNAs' expression levels; the top 5 KEGG pathways enriched using parental genes of pre- (B), in-, (C) and post-puberty (D) stage-specific circRNAs. *** P < 0.001.
Putative Tissue-Specific circRNAs in Gilts' Pituitary
To explore the specific circRNAs in pituitary tissue, 4990 circRNAs were identified as known circRNAs that overlapped with those in circAtlas 2.0, while another 158 circRNAs were identified as being specifically expressed in pituitary tissue. Furthermore, the latter, hereon the “putative pituitary-specific circRNAs,” were significantly shorter than the known circRNAs (t-test, P = 7.86E-06) (Figure 3A) and these novel circRNAs had significantly higher expression levels than did the known circRNAs during the onset of puberty (t-test, P < 2.20E−16) (Figure 3B). The KEGG enrichment analysis of parental genes of these putative pituitary-specific circRNAs were enriched in “ssc04917: Prolactin signaling pathway,” “ssc04080: Neuroactive ligand-receptor interaction,” and “ssc04728: Dopaminergic synapse” pathways (Figure 3C).
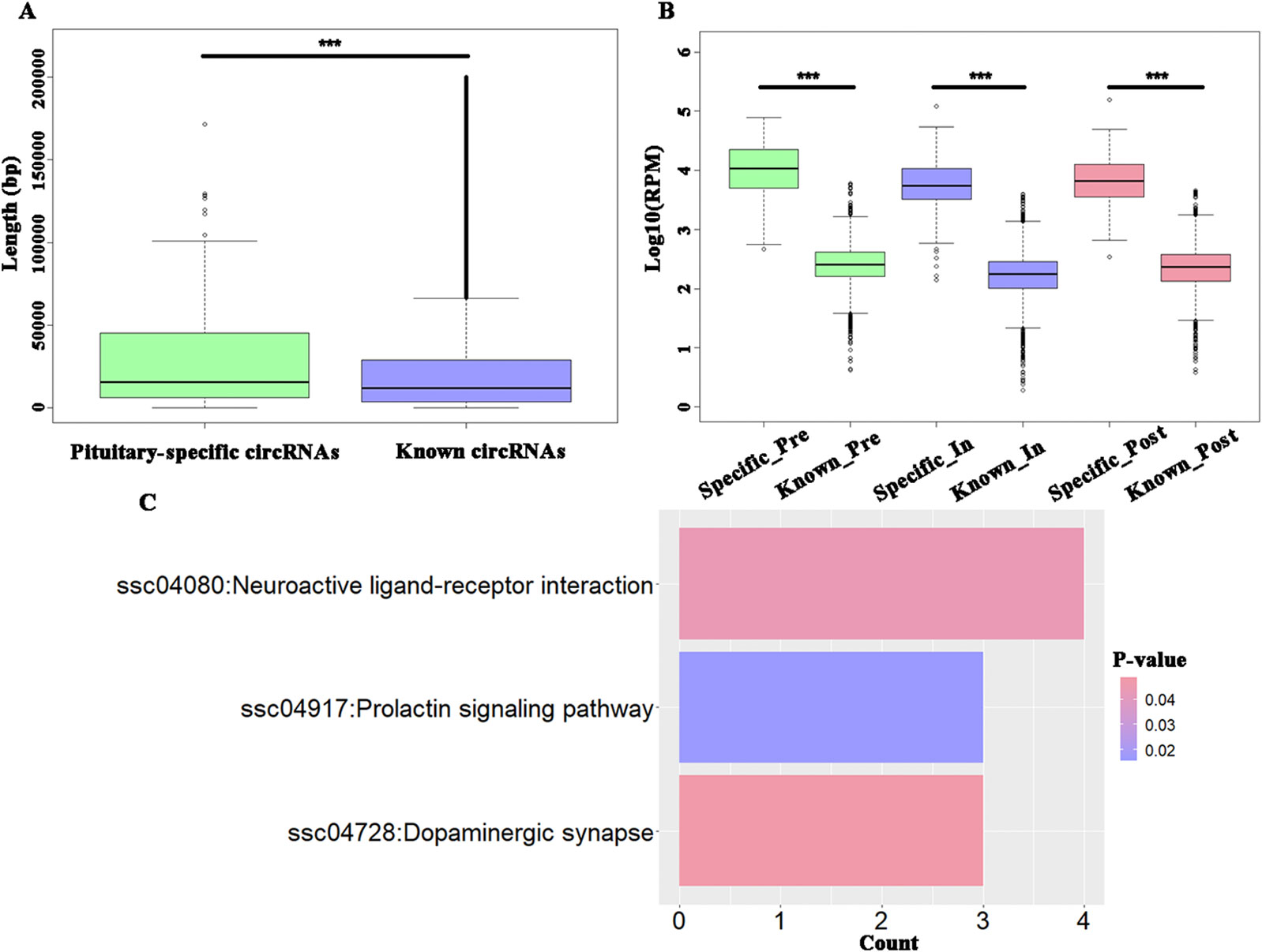
Figure 3 Analysis of potential tissue-specific circRNAs in pigs. (A) Boxplots of potential pituitary-specific and known circRNAs' length; (B) Boxplots of potential pituitary-specific and known circRNAs' expression level; (C) The KEGG pathways enriched using parental genes of potential pituitary-specific circRNAs. *** P < 0.001.
Analysis of Differentially Expressed circRNAs
A total of 14 differentially upregulated circRNAs and three differentially downregulated circRNAs were identified (Table 1). Some of them were derived from different transcripts of the same genes, such as ESR1 and RALGPS1. All differentially regulated circRNAs in the pre- vs. in-puberty groups were both derived from ESR1 (Table 1). Interestingly, one of them, circRNA “Circ 1:14408861|14457143,” was identified here for the first time. Furthermore, the circRNA “Circ 7:121001608|121012600” was downregulated in the in- vs. post-puberty groups yet upregulated in the pre- vs. post-puberty groups.
Validation of circRNAs by qRT-PCR
To validate the accuracy of RNA-seq data, a total of five circRNAs, including four differentially expressed circRNAs—Circ 1:14408861|14457143 (Figure 4A), Circ 9:28120503|28122017 (Figure 4B), Circ 2:88184110|88206327 (Figure 4C), Circ 9:75284452|75290025 (Figure 4D)—and one randomly selected circRNA: Circ 15:74631515|74643464 (Figure 4E) were chosen and validated via qRT-PCR.
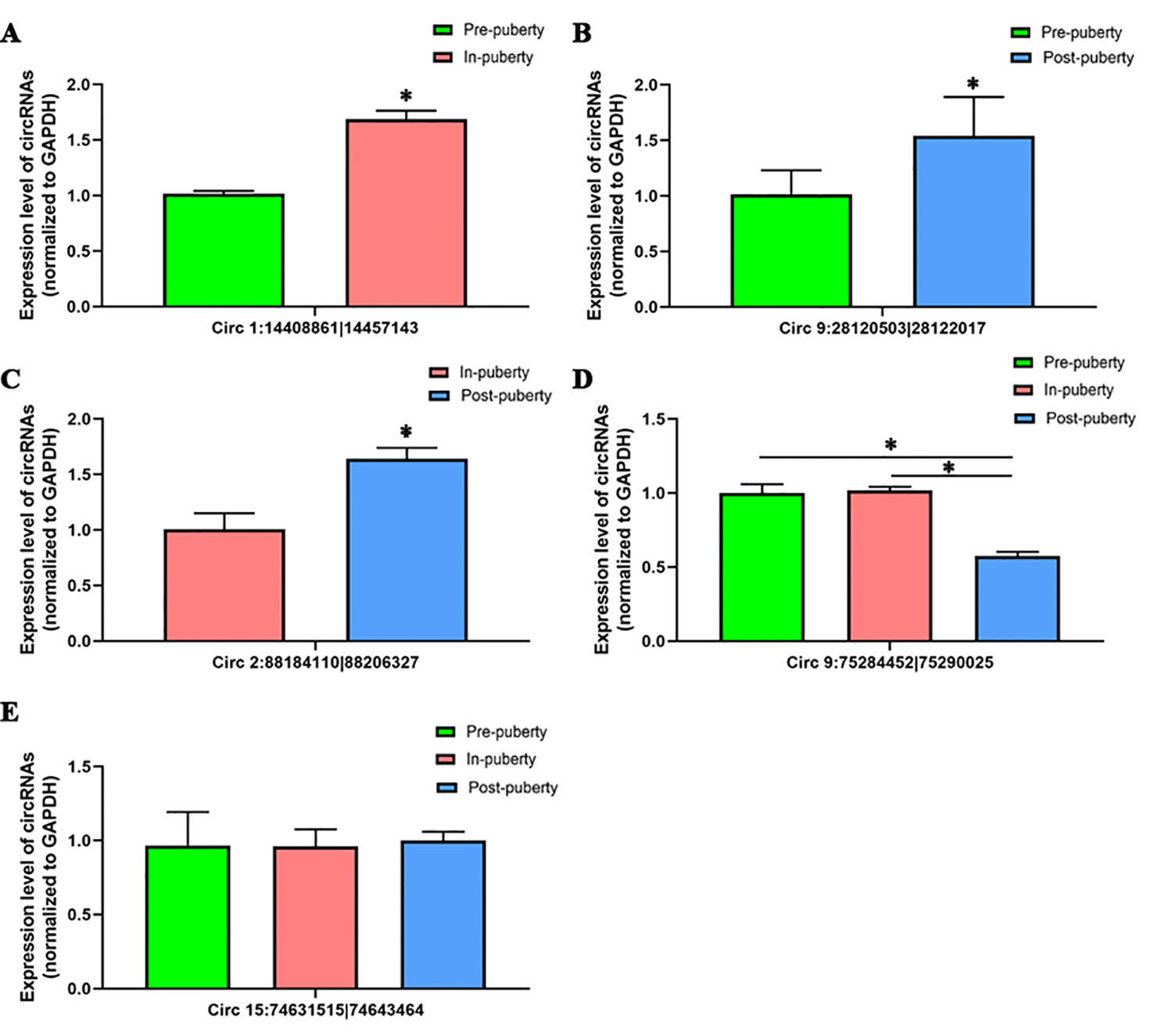
Figure 4 Validation of circRNAs using qRT-PCR. The qRT-PCR results of (A) Circ 1:14408861|14457143, (B) Circ 9:28120503|28122017, (C) Circ 2:88184110|88206327, (D) Circ 9:75284452|75290025, and (E) Circ 15:74631515|74643464 are shown. The green, red, and blue columns represent the pre-, in-, and post-puberty pig groups, respectively. * P < 0.05.
When compared with the RNA-seq data, similar expression trends for the qRT-PCR results of all selected circRNAs were discovered, thus showing that the obtained qRT-PCR results of these above circRNAs were consistent with the RNA-seq data (Figure S1).
CircRNA-Targeted miRNA-Gene Network Prediction
To further explore the putative functions of differentially expressed circRNAs, these circRNAs were conducted to predict the binging sites with miRNA targets (Figure 5). The top five plausible miRNA targets were chosen according to their respective miRanda match score and are listed in Table 1. According to this study, we found that many of differentially expressed circRNAs interact with miRNAs that potentially regulate estrus of pigs. These predicted circRNA-targeted miRNA-gene networks will be the focus of further research.
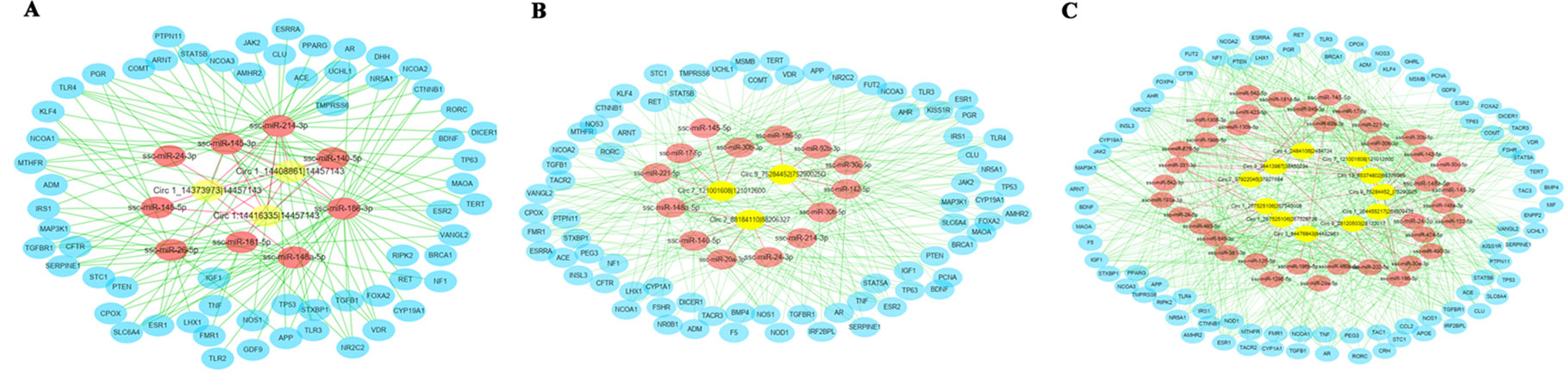
Figure 5 The circRNA-targeted miRNA-gene network prediction results of differentially regulated circRNAs. The network prediction results of differentially regulated circRNAs in (A) the pre- vs. in-puberty group, (B) the in- vs. post-puberty group, and (C) the pre- vs. post-puberty group.
Discussion
As a key physiological process of sexual maturation, the timing of puberty's onset provides a great opportunity for improving the efficiency of gilts' reproduction. In this study, we identified the genome-wide landscape of circRNAs in three important pubertal stages: pre-, in-, and post-puberty. The results showed that the number of circRNAs were dramatically altered among these three stages; the most circRNAs detected from in-puberty pigs, followed by those at post-puberty, with the least number occurring in the stage of pre-puberty. Many genome-wide analyses of circRNAs in mammals have been widely conducted using RNA-seq; these collectively indicate the number of circRNAs can differ between species, as well among different tissues or ontogeny stages. Of the 5148 circRNAs identified in our study, 158 circRNAs were distinguishable as putative pituitary-specific circRNAs that are involved in the prolactin signaling pathway, the neuroactive ligand-receptor interaction, and the dopaminergic synapse. Prolactin secreted by the pituitary was related to the regulation of reproductive function, the immune system, osmotic balance, and angiogenesis (Freeman et al., 2000). The secretion of prolactin is regulated by endocrine neurons between the hypothalamus and pituitary, and its regulation mainly depends on the secretion of dopamine (Bole-Feysot et al., 1998). For the distribution of circRNAs in the genomic regions, previous studies have shown extremely differences between different species and tissues. In pigs, Liang et al. (2017) found that 21.93% of circRNAs in intergenic regions and 68.40% in exon regions through carrying out nine organs. Yan et al. (2018) demonstrated that the found circRNAs consisted of approximately 74.31% exonic, 20.36% intergenic, and 5.33% intronic circRNAs in spleen, and Huang et al. (2018) demonstrated that more than 86% of circRNAs consisted of exons while nearly 10% originated from intronic and intergenic regions in liver. In rats, Yang et al. (2019) found that the circRNAs consisted of approximately 80.18% exonic, 0.15% intergenic, and 19.67% intronic circRNAs in pulmonary. These observations strongly support the view that circRNAs' expression occurs in a specie-specific, tissue-specific, and developmental stage-specific manner.
Importantly, we identified 17 circRNAs that were differentially expressed in the gilts, for which we speculated that some parental genes of differentially regulated circRNAs could influence the fertility and production traits of female mammals, such as ESR1 (Handa and Weiser, 2014), DENND1A (McAllister et al., 2014), RALGPS1 (Cochran et al., 2013), and MAML2 (Whittington et al., 2018). In addition, after identifying the miRNA targets of each differentially regulated circRNA, we found that some candidate miRNAs targeted by several circRNAs are linked to mammalian development of sex differentiation and maturation. For example, miR-145-5p was found likewise up-regulated after sexual maturity in pigs (Li et al., 2016) and miR-214-3p was shown to be involved in the onset of mouse primordial germ and somatic cell sex differentiation (Fernández-Pérez et al., 2018). Those findings coupled to our results suggest that circRNAs probably regulate the onset of puberty.
Interestingly, one of the differentially regulated circRNAs, circRNA “Circ 1:14408861|14457143,” was reported here in pig for the first time, and the top five miRNA targets of this particular circRNA had a predicted interaction with ESR1. ESR1 encodes an estrogen receptor alpha, a nuclear receptor activated by the sex hormone estrogen (Green et al., 1986). Previous studies confirmed that lacking an active ESR1 caused the disruption of normal pituitary tissue development and function. For instance, female mice lacking the estrogen receptor alpha in the pituitary gonadotroph have elevated levels of serum LH and LH beta-subunit gene expression, indicating that lacking estrogen has a negative feedback effect on the gonadotroph, with LH values and estrous cyclicity also found absent in these mice (Singh et al., 2009). Most circRNAs detected in our study are in the circAtlas 2.0 database, whose circRNAs were identified by at least two tools (CIRI2, DCC, find_circ, or CIRCexplorer2) to avoid false positives. Hence, the predictions made in the present study should be reliable.
Our dataset provides fresh insight into the existence of pituitary-derived circRNAs in pigs, yet this study did have a few limitations. Although the rRNA-depleted total RNA-seq analyses have been used to enrich for circRNAs in previous studies (e.g., Memczak et al., 2013; Tan et al., 2017; Sekar et al., 2018), there is no doubt that these sequencing analyses may not have comprehensively captured all occurring circRNAs. Furthermore, these enrichment steps may produce a few false BSJ reads that originated from linear RNAs, which could possibly lead to false detections of circRNAs. To guard against this, we used CIRI algorithms to identify circRNAs in this study, as they are reportedly effective for preventing the false detections of circRNAs that are caused by false BSJ reads (Gao and Zhao, 2018). Finally, the underlying mechanism of these circRNAs during pigs' pubertal onset still requires carefully elucidation and verification.
Conclusions
This investigation identified and described the circRNAs during the onset of puberty in gilts' pituitaries. In all, 5148 circRNAs were found, of which 158 were putative pituitary-specific expressed circRNAs. Because their expression levels were significantly higher than those of the remaining circRNAs during the onset of puberty, this suggested they are involved in regulating the key function of pituitary tissue. Upon further examination, 17 differentially regulated circRNAs were identified and these circRNAs were chosen to construct the posited circRNA-targeted miRNA-gene network. These results suggest circRNAs likely play a critical role in puberty's timing in gilts and thus provide useful information for future investigations of circRNA-mediated puberty at the pituitary level.
Data Availability Statement
The data used in our study has been released in NCBI Sequence Read Archive with the accession number PRJNA576641 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA576641).
Ethics Statement
The animal study was reviewed and approved by The Animal Care and Use Committee of the South China Agricultural University, Guangzhou, China (permit number: SCAU#2013-10).
Author Contributions
ZC, XP, and YK conceived this study, performed the qRT-PCR validations, and wrote the manuscript. ZZ, YZ, XY, XP, YJ, and JL helped the manuscript review and editing. HZ and ZC originally derived the data and helped in the analyses. JL, ZZ, and XY stimulated the idea of the paper and helped in the manuscript. All authors read, edited, and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by the Earmarked Fund for China Agriculture Research System (CARS-35), the National Natural Science Foundation of China (31902131), the Youth Innovative fund of Guangdong Education Department (2018KQNCX019), and Guangdong Sailing Program (2014YT02H042).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00135/full#supplementary-material
Figure S1 | Validation of circRNAs using qRT-PCR (left) and comparison withRNA-seq data (right). * P < 0.05.
Table S1 | The KEGG pathways of pre-, in-, and post-puberty stage-specific circRNAs.
References
Angold, A., Costello, E. J., Erkanli, A., Worthman, C. M. (1999). Pubertal changes in hormone levels and depression in girls. Psychol. Med. 29, 1043–1053. doi: 10.1017/S0033291799008946
Barb, C. R., Hausman, G. J., Rekaya, R. (2012). Sustainable Agriculture Reviews. Ed. Lichtfouse, E. (Dordrecht: Springer Netherlands). doi: 10.1007/978-94-007-5449-2
Blakemore, S.-J., Burnett, S., Dahl, R. E. (2010). The role of puberty in the developing adolescent brain. Hum. Brain Mapp. 31, 926–933. doi: 10.1002/hbm.21052
Bole-Feysot, C., Goffin, V., Edery, M., Binart, N., Kelly, P. A. (1998). Prolactin (PRL) and Its Receptor: Actions, Signal Transduction Pathways and Phenotypes Observed in PRL Receptor Knockout Mice. Endocr. Rev. 19, 225–268. doi: 10.1210/edrv.19.3.0334
Chen, L.-L., Yang, L. (2015). Regulation of circRNA biogenesis. RNA Biol. 12, 381–388. doi: 10.1080/15476286.2015.1020271
Chen, J., Zou, Q., Lv, D., Wei, Y., Raza, M. A., Chen, Y., et al. (2018). Comprehensive transcriptional landscape of porcine cardiac and skeletal muscles reveals differences of aging. Oncotarget. 9. doi: 10.18632/oncotarget.23290
Cochran, S. D., Cole, J. B., Null, D. J., Hansen, P. J. (2013). Discovery of single nucleotide polymorphisms in candidate genes associated with fertility and production traits in Holstein cattle. BMC Genet. 14, 49. doi: 10.1186/1471-2156-14-49
Coe, C. L., Chen, J., Lowe, E. L., Davidson, J. M., Levine, S. (1981). Hormonal and behavioral changes at puberty in the squirrel monkey. Horm. Behav. 15, 36–53. doi: 10.1016/0018-506X(81)90033-7
Fernández-Pérez, D., Brieño-Enríquez, M. A., Isoler-Alcaraz, J., Larriba, E., del Mazo, J. (2018). MicroRNA dynamics at the onset of primordial germ and somatic cell sex differentiation during mouse embryonic gonad development. RNA 24, 287–303. doi: 10.1261/rna.062869.117
Freeman, M. E., Kanyicska, B., Lerant, A., Nagy, G. (2000). Prolactin: Structure, Function, and Regulation of Secretion. Physiol. Rev. 80, 1523–1631. doi: 10.1152/physrev.2000.80.4.1523
Gao, Y., Zhao, F. (2018). Computational Strategies for Exploring Circular RNAs. Trends Genet. 34, 389–400. doi: 10.1016/j.tig.2017.12.016
Gao, Y., Wang, J., Zhao, F. (2015). CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 16, 4. doi: 10.1186/s13059-014-0571-3
Green, S., Walter, P., Greene, G., Krust, A., Goffin, C., Jensen, E., et al. (1986). Cloning of the human oestrogen receptor cDNA. J. Steroid Biochem. 24, 77–83. doi: 10.1016/0022-4731(86)90035-X
Handa, R. J., Weiser, M. J. (2014). Gonadal steroid hormones and the hypothalamo–pituitary–adrenal axis. Front. Neuroendocrinol. 35, 197–220. doi: 10.1016/j.yfrne.2013.11.001
Huang, D. W., Sherman, B. T., Tan, Q., Kir, J., Liu, D., Bryant, D., et al. (2007). DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35, W169–W175. doi: 10.1093/nar/gkm415
Huang, M., Shen, Y., Mao, H., Chen, L., Chen, J., Guo, X., et al. (2018). Circular RNA expression profiles in the porcine liver of two distinct phenotype pig breeds. Asian-Australasian J. Anim. Sci. 31, 812–819. doi: 10.5713/ajas.17.0651
Ji, P., Wu, W., Chen, S., Zheng, Y., Zhou, L., Zhang, J., et al. (2019). Expanded Expression Landscape and Prioritization of Circular RNAs in Mammals. Cell Rep. 26, 3444–3460.e5. doi: 10.1016/j.celrep.2019.02.078
John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C., Marks, D. S. (2004). Human MicroRNA Targets. PloS Biol. 2, e363. doi: 10.1371/journal.pbio.0020363
Leng, N., Dawson, J. A., Thomson, J. A., Ruotti, V., Rissman, A. I., Smits, B. M. G., et al. (2013). EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043. doi: 10.1093/bioinformatics/btt087
Li, H., Durbin, R. (2010). Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Li, Y., Li, J., Fang, C., Shi, L., Tan, J., Xiong, Y., et al. (2016). Genome-wide differential expression of genes and small RNAs in testis of two different porcine breeds and at two different ages. Sci. Rep. 6, 26852. doi: 10.1038/srep26852
Li, A., Huang, W., Zhang, X., Xie, L., Miao, X. (2018). Identification and characterization of CircRNAs of two pig breeds as a new biomarker in metabolism-related diseases. Cell. Physiol. Biochem. 47, 2458–2470. doi: 10.1159/000491619
Liang, G., Yang, Y., Niu, G., Tang, Z., Li, K. (2017). Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 24, 523–535. doi: 10.1093/dnares/dsx022
Luo, L., Yao, Z., Ye, J., Tian, Y., Yang, C., Gao, X., et al. (2017). Identification of differential genomic DNA Methylation in the hypothalamus of pubertal rat using reduced representation Bisulfite sequencing. Reprod. Biol. Endocrinol. 15, 81. doi: 10.1186/s12958-017-0301-2
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10. doi: 10.14806/ej.17.1.200
McAllister, J. M., Modi, B., Miller, B. A., Biegler, J., Bruggeman, R., Legro, R. S., et al. (2014). Overexpression of a DENND1A isoform produces a polycystic ovary syndrome theca phenotype. Proc. Natl. Acad. Sci. 111, E1519–E1527. doi: 10.1073/pnas.1400574111
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495, 333–338. doi: 10.1038/nature11928
Nonneman, D. J., Schneider, J. F., Lents, C. A., Wiedmann, R. T., Vallet, J. L., Rohrer, G. A. (2016). Genome-wide association and identification of candidate genes for age at puberty in swine. BMC Genet. 17, 50. doi: 10.1186/s12863-016-0352-y
Patterson, J. L., Willis, H. J., Kirkwood, R. N., Foxcroft, G. R. (2002). Impact of boar exposure on puberty attainment and breeding outcomes in gilts. Theriogenology. 57, 2015–2025. doi: 10.1016/S0093-691X(02)00674-X
Patterson, J. L., Beltranena, E., Foxcroft, G. R. (2010). The effect of gilt age at first estrus and breeding on third estrus on sow body weight changes and long-term reproductive performance1. J. Anim. Sci. 88, 2500–2513. doi: 10.2527/jas.2008-1756
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., Shang, R., et al. (2015). Circular RNA: A new star of noncoding RNAs. Cancer Lett. 365, 141–148. doi: 10.1016/j.canlet.2015.06.003
R Core Team (2018). R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing.
Rosales Nieto, C. A., Thompson, A. N., Macleay, C. A., Briegel, J. R., Hedger, M. P., Ferguson, M. B., et al. (2014). Relationships among body composition, circulating concentrations of leptin and follistatin, and the onset of puberty and fertility in young female sheep. Anim. Reprod. Sci. 151, 148–156. doi: 10.1016/j.anireprosci.2014.10.008
Saito, H., Sasaki, Y., Koketsu, Y. (2011). Associations between age of gilts at first mating and lifetime performance or culling risk in commercial herds. J. Vet. Med. Sci. 73, 555–559.
Sekar, S., Cuyugan, L., Adkins, J., Geiger, P., Liang, W. S. (2018). Circular RNA expression and regulatory network prediction in posterior cingulate astrocytes in elderly subjects. BMC Genomics 19, 340. doi: 10.1186/s12864-018-4670-5
Singh, S. P., Wolfe, A., Ng, Y., DiVall, S. A., Buggs, C., Levine, J. E., et al. (2009). Impaired Estrogen Feedback and Infertility in Female Mice with Pituitary-Specific Deletion of Estrogen Receptor Alpha (ESR1)1. Biol. Reprod. 81, 488–496. doi: 10.1095/biolreprod.108.075259
Su, G., Morris, J. H., Demchak, B., Bader, G. D. (2014). Biological Network Exploration with Cytoscape 3. Curr. Protoc. Bioinforma. 47, 8.13.1–8.13.24. doi: 10.1002/0471250953.bi0813s47
Szabo, L., Salzman, J. (2016). Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 17, 679–692. doi: 10.1038/nrg.2016.114
Tan, W. L. W., Lim, B. T. S., Anene-Nzelu, C. G. O., Ackers-Johnson, M., Dashi, A., See, K., et al. (2017). A landscape of circular RNA expression in the human heart. Cardiovasc. Res. doi: 10.1093/cvr/cvw250
Whittington, C. M., O'Meally, D., Laird, M. K., Belov, K., Thompson, M. B., McAllan, B. M. (2018). Transcriptomic changes in the pre-implantation uterus highlight histotrophic nutrition of the developing marsupial embryo. Sci. Rep. 8, 2412. doi: 10.1038/s41598-018-20744-z
Witkos, M. T., Koscianska, E., Krzyzosiak, J. W. (2011). Practical Aspects of microRNA Target Prediction. Curr. Mol. Med. 11, 93–109. doi: 10.2174/156652411794859250
Yan, Z., Jiang, T., Wang, P., Huang, X., Yang, Q., Sun, W., et al. (2018). Circular RNA expression profile of spleen in a Clostridium perfringens type C-induced piglet model of necrotizing enteritis. FEBS Open Bio. 8, 1722–1732. doi: 10.1002/2211-5463.12512
Keywords: pituitary, crossbred pig, puberty, circRNAs, miRNA sponges
Citation: Chen Z, Pan X, Kong Y, Jiang Y, Zhong Y, Zhang H, Zhang Z, Yuan X and Li J (2020) Pituitary-Derived Circular RNAs Expression and Regulatory Network Prediction During the Onset of Puberty in Landrace × Yorkshire Crossbred Pigs. Front. Genet. 11:135. doi: 10.3389/fgene.2020.00135
Received: 29 October 2019; Accepted: 04 February 2020;
Published: 28 February 2020.
Edited by:
Shu-Hong Zhao, Huazhong Agricultural University, ChinaReviewed by:
Tang Zhonglin, Chinese Academy of Agricultural Sciences, ChinaXiaodong Zhang, Anhui Agricultural University, China
Copyright © 2020 Chen, Pan, Kong, Jiang, Zhong, Zhang, Zhang, Yuan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolong Yuan, eXhsQHNjYXUuZWR1LmNu; Jiaqi Li, anFsaUBzY2F1LmVkdS5jbg==
 Zitao Chen
Zitao Chen Xiangchun Pan
Xiangchun Pan Yaru Kong
Yaru Kong Yao Jiang
Yao Jiang Hao Zhang
Hao Zhang Zhe Zhang
Zhe Zhang Xiaolong Yuan
Xiaolong Yuan Jiaqi Li
Jiaqi Li