- 1Key Laboratory of Beef Cattle Genetics and Breeding in Ministry of Agriculture and Rural Agriculture, Branch of Animal Husbandry, Jilin Academy of Agricultural Sciences, Changchun, China
- 2College of Animal Science and Technology, Northwest A&F University, Yangling, China
- 3Computational Biology Laboratory, Department of Agricutural Biotechnology, National Institute for Biotechnology and Genetic Engineering, Faisalabad, Pakistan
- 4Pakistan Institute of Engineering and Applied Sciences, Nilore, Pakistan
Yanbian cattle is inhabitant of North of China, exhibiting many phenotypic features, such as long, dense body hair, and abundant intramuscular fat, designed to combat the extreme cold climate adaption. In the current study, we studied the cold tolerance of nine Yanbian cattle by whole genome resequencing and compared with African tropical cattle, N’Dama, as a control group. Yanbian cattle was aligned to the Bos taurus reference genome (ARS-UCD1.2) yielding an average of 10.8 fold coverage. The positive selective sweep analysis for the cold adaption in Yanbian cattle were analyzed using composite likelihood ratio (CLR) and nucleotide diversity (θπ), resulting in 292 overlapped genes. The strongest selective signal was found on BTA16 with potential mutation in CORT gene, a regulatory gene of primary hormone in the hypothalamic-pituitary-adrenal (HPA) axis, is reported to be associated with the cold stress, representedfour missense mutations (c.269C > T, p.Lys90Ile; c.251A > G, p.Glu84Gly; c.112C > T, p.Pro38Ser; c.86G > A, p.Pro29His). Meanwhile another gene on BTA6, showed significantly higher selective sweep signals for a cold adapted trait for hair follicle and length development, FGF5 (fibroblast growth factor 5) with a missense mutation (c.191C > T, p.Ser64Phe). Moreover, cold adapted Yanbian cattle was statistically compared with the hot adapted N’Dama cattle, a taurine cattle reported to show superior heat tolerance than zebu cattle, making them better adapted to the hot regions of Africa. XP-CLR, Fst, and θπ ratio were used to compare both breeds, yielding 487, 924, and 346 genes respectively. Among the 12 overlapped genes, (CD36) (c.638A > G, p.Lys 213Arg) involved in fat digestion and absorption plays an important role in membrane transport of long-chain fatty acid and its expression could increase in cold exposure. Henceforth, our study provides a novel genetic insights into the cold climate adaptation of Yanbian cattle and identified three candidate genes (CORT, FGF5, and CD36), which can add to an understanding of the cold climate adaptation of Yanbian cattle.
Introduction
Cold climate adaptation is a general term used to describe the physiological functions associated with cold adaptation. Studies show that cold exposure will lead to increase the blood pressure (De Lorenzo et al., 1999). During cold stress, the body tries to save energy by cheap methods (such as standing hair) and by changes posture to reduce surface area (Cannon and Nedergaard, 2004). To protect tissues from cold damage, the body adopts different processes, which increases warm blood flow near the surface of the skin (Cannon and Nedergaard, 2011). Those adaptation mechanisms, as well as biological processes, suggest the complex mechanisms of adaptation to cold.
Yanbian cattle is a taurine breed that living in northeast China (Xin et al., 2010) and belongs to the “yellow” class of Chinese cattle (Ji et al., 2014). Unlike majority of Chinese indigenous breeds, Yanbian cattle have had no ancestral to breed with indicine cattle (Xin et al., 2014). They are mainly used as herbivores, especially in the rice fields but are also increasingly used for the beef purpose. The living environment of Yanbian cattle has long, freezing winters with snow-covered grounds half a year and only brief summers. The temperature drops as low as −37°C at the peak of the winter season (Figure S1). Yanbian cattle exhibit unique morphoanatomical adaptations to the cold climate with its long and dense hairs as predicted by Allen’s rule (Allen, 1877), compactly built with short limbs. On the other hand, N’Dama has been known for its heat resistance in the harsh climatic conditions in Africa. The temperature rise as high as 50°C in the summers, while the weather remains hotter rest of the year as well. Thus, it is an ideal taurine to be compared with Yanbian cattle to identify the potential temperature regulating genes and pathways.
Along with constant release of whole-genomic sequence data in domesticated cattle (Daetwyler et al., 2014; Stothard et al., 2015; Kim et al., 2017), continuous expansion of directory to genetic variants (Karim et al., 2011), gradual maturity of selective theory and method (Vitti et al., 2013), the genetic basis of phenotypic diversity can be hunted down at the complete genome level. However, to our knowledge, no information has yet been generated regarding the cold climate adaptation based on whole genomes level in Yanbian cattle.
Yanbian cattle living in cold environments can be an excellent model for the identification of genomic loci explaining cold climate adaptation in cattle. Here, we are starting from the whole genome scan of Yanbian and N’Dama cattle, and reported genes that are positively selected in Yanbian cattle associated with cold stress adaptation. We used composite likelihood ratio (CLR) and θπ statistics to study the diverse nature of Yanbian cattle, which can provide genomic materials for genetic improvement of Yanbian cattle adaptive traits. Furthermore, we also employed three different statistical approaches i.e., XP-CLR, Fst, and θπ ratio, in order to detect selection signatures in Yanbian cattle, compared to N’Dama cattle. The high fat content, marbling, and the superior hide of Yanbian cattle needs to be well preserved and further enhanced. However, the genetic predispositions associated with adaption and enhanced cold tolerant parameters remain uncertain. The current study will help us to enunciate the extreme environmental adaptations and the positive selective sweeps in the Yanbian cattle.
Material and Methods
Library Construction and Sequencing
We generated whole-genome resequencing data for nine Yanbian cattle, three of them have been generated from our previous study (Chen et al., 2018). N’Dama cattle (n=10), a natural inhabitant of temporal climate, was included in the study for the genome comparison, in order to understand the heat and cold tolerance in livestock (Kim et al., 2017). The genomic DNA was extracted from the ear tissues using the standard phenol-chloroform protocol (Sterky et al., 2017). In Supplementary Note 1 we have detailed description about the approaches and tools utilized in the current analysis.
Read Mapping and Single-Nucleotide Polymorphism Calling
The clean reads were aligned to the latest reference genome sequence (GCF_002263795.1) using Burrows-Wheeler Aligner (BWA)-MEM (Li et al., 2009) with default parameters. The Picard tools (version 1.106) were used to generate the quality matrices whereas, the Genome Analysis Toolkit (GATK, version 3.8) was employed for the single-nucleotide polymorphism (SNP) calling for mapping (McKenna et al., 2010). We used “HaplotypeCaller,” “GenotypeGVCFs,” and “SelectVariants” argument of GATK to call the raw SNP. The filtration of the raw SNPs was conducted by using “variant Filtration” with the following parameters: 1) the depth of base quality to ensure variant confidence (QD) < 2.0; 2) the quality of mapping reads (MQ) > 40.0; 3) also, the Phred-scaled P-value calculating with Fisher`s exact test (FS) < 60.0; 4) ReadPosRankSum < −8.0, 5) MQRankSum < −12.5; 6) mean sequence depth (for all individuals) > 1/3× and < 3×; while, 7) SOR > 3.0.
Variant Functional Annotation and Enrichment Analysis
The SNP were annotated by SnpEff tool using ARS-UCD1.2 database and gene set enrichment analyses were carried out with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway using KEGG Orthology-Based Annotation System (KOBAS) tool. To provide a preliminary overview of the genomic excess and test its reliability, we performed different GO and KEGG pathway enrichment analyses with KOBAS using different lists of genes located in chromosomal regions from the different selective analysis methods of Yanbian to N’Dama cattle. The enriched pathways and genes were selected stringently with an adjusted probability (P < 0.05).
Selective Sweep Identification
The genome and nucleotide diversity were calculated by θπ, whereas, allele frequency for the positive selection signals were attained by CLR in Yanbian cattle. To infer the scan in progress, we used a 50 kb window for both statistical parameters. The CLR test uses information from allele frequencies to detect selective scans and relies on determining skews in the allele spectrum to bias rare and frequent alleles (Nielsen et al., 2005). Whereas, θπ analyzes the complete polymorphism data to compile the nature of diversity of the species.
We also performed XP-CLR (Chen et al., 2010), Fst, and θπ ratio to identify the potential areas differentially, selected between Yanbian and N’Dama cattle. XP-CLR is used for detecting selective sweeps that models the multilocus allele frequency differentiation between the two populations (Chen et al., 2010). We used a 50 kb non-overlapping sliding windows with the number of SNPs less than 600, and the correlation level reduced the contribution of SNPs to the XP-CLR results to 0.95. XP-CLR values in the top 0.5% of the empirical distribution (XP-CLR> 29.49) are designated as candidate selection scans, and genes that in those window region are defined as potential candidate genes (Lee et al., 2014). It will give more evidence to the same gene regions using different methods (Qanbari and Simianer, 2014). Fst can change the degree of differentiation between the different populations of one species. If populations are differentiated, the number of genetic differentiation in the selected locus region increase while the difference in genomic region is greater than the neutral conditions (Oleksyk et al., 2010). VCFtools were used to calculate the Fst values of the candidate gene regions in a 50 kb window size at an interval of 20 kb steps (Danecek et al., 2011). Finally, the identified selective sweeps regions were annotated to the reference genome (ARS-UCD1.2) and were further explored following the functional analysis. Lastly, θπ ratio between Yanbian and N’Dama cattle groups was calculated as ln(θπ,Yanbian/θπ,N’Dama), which reflected the loss of nucleotide diversity in Yanbian cattle relative to N’Dama.
Results
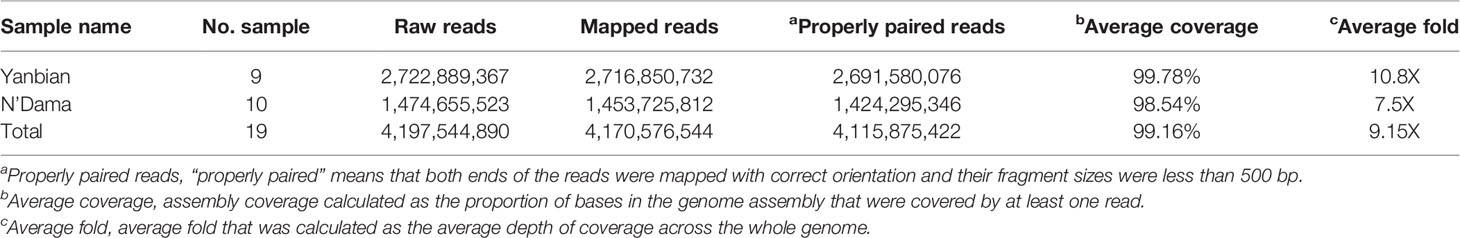
Resequencing of Yanbian Cattle and Single-Nucleotide Polymorphism Call
The re-sequenced Yanbian and the N’Dama cattle were pooled together in a 19 cattle data set, generated to an average of 9.15X coverage. In total, 4.2 billion reads were generated which were aligned against the ARS-UCD1.2 reference genome using BWA MEM algorithm, yielding ~4.1 billion mapping reads, covering 99.16% of the reference sequence across the region (Table 1). After the SNP call, 11,331,903 and 5,720,198 SNPs in Yanbian and N’Dama cattle, respectively were identified. The quality of the SNP call was evaluated with the Ts/Tv ratio (Table 2). The SNPs count of Yanbian cattle was observed to be larger than N’Dama, which might account for the low coverage and low genetic diversity of N’Dama sequencing. At the same time, the average ratios of homozygous versus heterozygous SNPs of Yanbian and N’Dama cattle are 0.487 and 0.848, respectively, representing the more genetic diversity in North China with its elevated heterozygous.
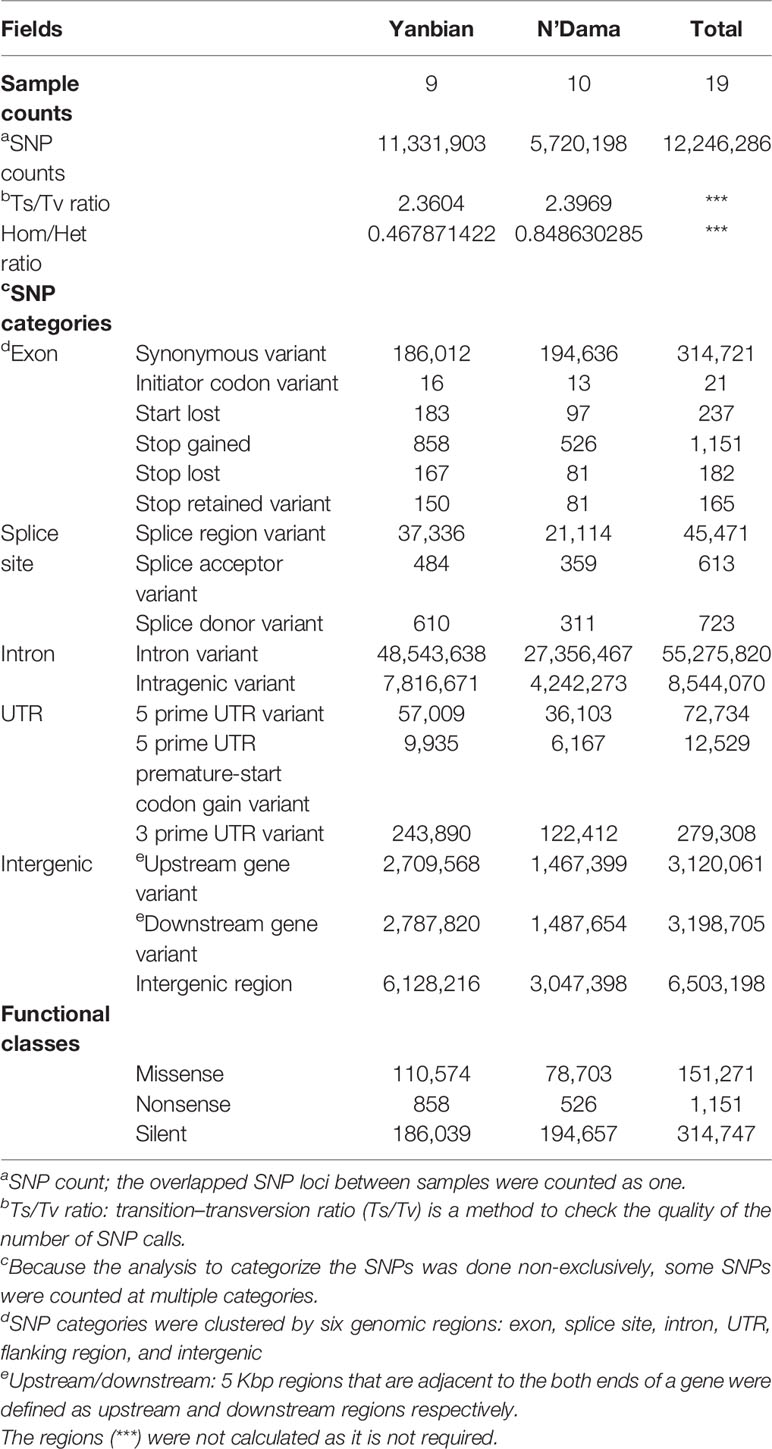
Table 2 Functional annotation of the identified single-nucleotide polymorphisms (SNPs) in Yanbian and N’Dama cattle.
Biological Process and Pathways of Yanbian Cattle Population
CLR and θπ statistics were calculated for the Yanbian cattle alone, to distinguish the positive selection region. Based on CLR test statistic, we obtained 604 putative genes (Table S5) whereas, 680 positively selected genes were detected by θπ test (Table S6). Of these, 292 overlapped genes were detected in both statistics (Figure 1C). GO and KEGG pathways were employed by KOBAS, whereas, only 34 enriched pathways were retained (correct p < 0.05; Table S7; Figure 1D).
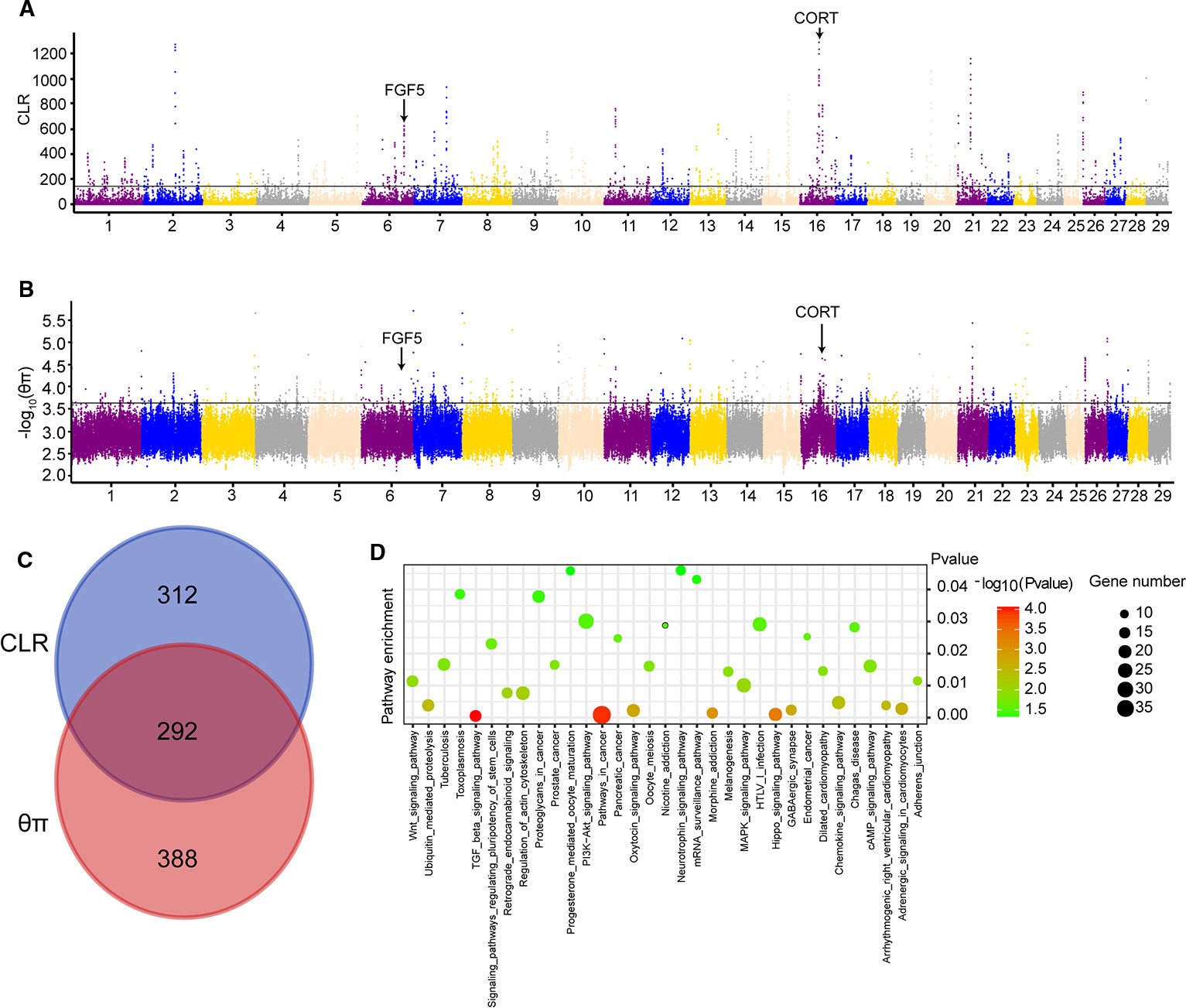
Figure 1 Genome-wide selection scan in Yanbian cattle. (A, B) Manhattan plot of the genome-wide distribution of composite likelihood ratio (CLR) and qp in Yanbian cattle using 50 kb windows size and 20 kb step size, respectively. (C) Venn diagram showing the genes overlap among CLR and qp significant selection region. (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed genes. Advanced bubble chart shows enrichment of differentially expressed genes in signaling pathways. Y-axis label represents pathway, and X-axis label represents rich factor (rich factor = amount of differentially expressed genes enriched in the pathway/amount of all genes in background gene set). Size and color of the bubble represent amount of differentially expressed genes enriched in pathway and enrichment significance, respectively.
Positive Selective Signature in Yanbian Cattle Related to Cold Climate Adaptation
Among 292 candidate genes detected in both CLR and θπ, some positively selected genes were considered to be associated with cold climate adaptation (CORT and FGF5). The strongest selection signal found on BTA16 (16:43127555-43129154) contains the CORT gene (Figures 1A, B), which was reported to be related with the cold stress in mouse, chicken, and humans (Dronjak et al., 2004; Hangalapura et al., 2004). In the cold exposure experiment of rats, CORT in the blood of rats significantly increased after 2 h of cold exposure (Dronjak et al., 2004). Meanwhile, the regulatory mechanism of cold stress and stress was studied in chickens, and it was found that CORT in plasma was considerably different under variable cold stress levels (Hangalapura et al., 2004). To identify the potential causal mutation around CORT locus, we checked all of mutations of Yanbian cattle, and four missense mutations in CORT (c.269C > T, p.Lys90Ile; c.251A > G, p.Glu84Gly; c.112C > T, p.Pro38Ser; c.86G > A, p.Pro29His) were found.
Interestingly, we found a gene, fibroblast growth factor 5 (FGF5) on BTA6 from CLR test (Figure 1A), which has been reported to be related to the development of hair follicles and hair length in cat, dog and human (Drögemüller et al., 2007; Dierks et al., 2013; Higgins et al., 2014). Meanwhile, we also found that FGF5 was located in a significant region on chromosome from θπ test (Figure 1B). Under cold stress conditions, long and dense hairs are very important in order to keep the body warm against the cold environment, a distinctive feature of Yanbian cattle.
Biological Process and Pathway Between Yanbian and N’Dama Cattle
Moreover, XP-CLR test was performed to distinguish the positive selection region between Yanbian and N’Dama cattle. Based on the analysis, we obtained 487 putative genes (Table S1). Whereas, 924 and 346 candidate genes were identified using Fst and θπ ratio between the two populations, respectively. (Tables S2 and S3). In addition, 12 overlapping genes from the three tests were detected and evaluated for the functional studies (Figure 2D). GO and KEGG pathways with KOBAS were used to build on biological modules consisting of clusters of functional terms. After wiping out duplicates, 1,375 genes were retained from XP-CLR, Fst, and θπ ratio for the analysis. KEGG pathway analysis resulted in 98 significantly enriched pathways (correct p < 0.05; Table S4). Vascular smooth muscle contraction, circadian entrainment, hypertrophic cardiomyopathy (HCM), fatty acid metabolism, and fat digestion and absorption were involved as major enrichment pathway, which may play an important role in the cold adaptation of Yanbian cattle.
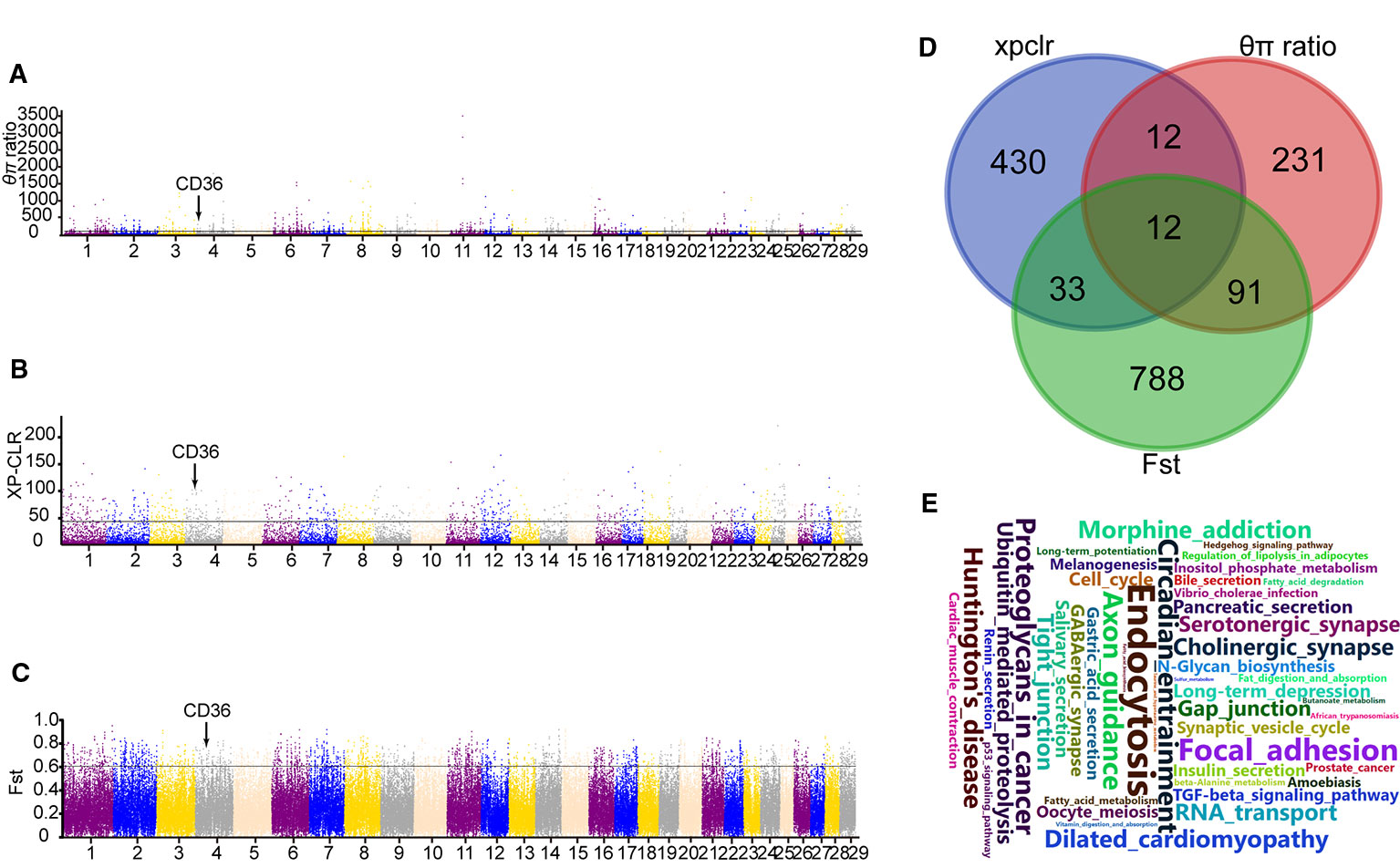
Figure 2 Genome-wide selection scan between Yanbian and N’Dama cattle. (A–C) Manhattan plot of the genome-wide distribution of qp ratio, XP-CLR, and Fst between Yanbian and N’Dama cattle using 50 kb windows size and 20 kb step size, respectively. The threshold corresponding to the top 0.5% of qp ratio, XP-CLR, and Fst are marked with a horizontal line, respectively. (D) Venn diagram showing the genes overlap among qp ratio, XP-CLR, and Fst significant selection region. (E) Word cloud illustrating major enrichment pathway of these genes in qp ratio, XP-CLR, and Fst significant selection region.
Candidate Regions and Genes Under Positive Selection in Yanbian and N’Dama Cattle
Here, we compared the genomes of Yanbian and N’Dama cattle to identify signatures of positive selection within each subspecies following environmental and artificial selection pressures. Twelve genes were overlapped in XPCLR, Fst, and θπ ratio methods including fatty acid synthase (CD36; Figures 2A–C), which plays main role in membrane transport in the heart and adipose tissue. The expression of CD36 is increased in cold exposure, which enhances brown adipose tissue (BAT) uptake of TG-rich lipoprotein (TRL) and of albumin bound FA. Previous studies have shown that CD36 gene in Hanwoo and Yanbian cattle affects the intramuscular fat deposition. Compared with N’Dama, Yanbian cattle have excellent meat quality. Yanbian cattle have been in the cold environment for a long time, and have a lot of fat deposits in their bodies, which is helpful to resist the cold. The divergent mutations of CD36 gene were checked between Yanbian and N’Dama cattle, and one missense mutations (c.638A > G, p.Lys 213Arg) was found.
Discussion
We herein carefully examined the whole genome resequencing of Yanbian and N’Dama cattle. The Ts/Tv ratio indicate the quality of resequencing, which was calculated to be 2.36 and 2.39 in Yanbian and N’Dama cattle respectively, comparable with the previous studies (Tables 1 and 2) (Choi et al., 2013; Choi et al., 2015). As for the heterozygous and homozygous ratio were concerned in the detected SNPs, the higher ratio of Yanbian cattle suggests that it’s population structure maybe normal and have a high heterozygosity rate.
Our study used resequencing data from Yanbian and N’Dama cattle to reveal a detailed genomic information along with candidate genes associated to the cold climate adaptation in Yanbian cattle. Yanbian cattle live in cold north China for a long time. Many biological traits in Yanbian cattle have adapted to the local cold environment, such as long and dense body hairs and increased muscular fat. In order to study the cold-adaptive mechanism of Yanbian cattle, N’Dama cattle was selected, a Bos taurus cattle living in the hot areas of Africa, as the reference. Cold adaptation research indicate that more than one mechanism involved in the biological response to cold stress. And it is a multi-factor complex trait that be affected by different factors at different levels in molecular and mechanical aspects. Consistent with the expectations of biological complexity of cold adaptation, our selection test highlights several different processes that are coherently responsible.
We calculated the CLR and θπ to analyze the positive selection of Yanbian cattle. The strongest selection signal of CLR found on BTA16 (16: 43127555-43129154) contains the CORT gene (Figure 1A), which was reported to be related with the cold stress in mouse, chicken and humans (Dronjak et al., 2004; Hangalapura et al., 2004). Secretion of CORT, a primary hormone in the hypothalamic-pituitary-adrenal (HPA) axis, exhibits a circadian rhythm in many species (Mormède et al., 2007). At the same time, CORT levels are also used as one of the biochemical parameters used to measure the physiological response of animals to stressful environments. Hence, increased cortisol levels in blood are an important stress indicator (Grandin, 1997; Ndlovu et al., 2008). In the cold exposure experiment of rats, CORT in the blood of rats significantly increased after 2 h of cold exposure (Dronjak et al., 2004). Meanwhile, the regulatory mechanism of cold stress and stress was studied in chickens, and it was found that CORT in plasma was considerably different under variable cold stress levels (Hangalapura et al., 2004). At the same time, we found four missense mutations in CORT (c.269C > T, p.Lys90Ile; c.251A > G, p.Glu84Gly; c.112C > T, p.Pro38Ser; c.86G > A, p.Pro29His). Also, CORT is one of the biochemical parameters used to measure the physiological response of animals to stressful environments. Under different cold stress levels, the study on the regulation mechanism of cold stress and stress in chickens found that CORT levels in plasma were significantly different (Hangalapura et al., 2004). Meanwhile, we also identified another gene FGF5 that may influence the hair length and density in Yanbian cattle, which is very important for keeping the body warm against the cold environment. FGF5 has been reported to be related to the development of hair follicles and hair length in cat, dog and human (Drögemüller et al., 2007; Dierks et al., 2013; Higgins et al., 2014). Though, analyzing the selection region of CLR test, FGF5 was found to be on top prominent point on BTA6 (Figure 1A), and also was located in a significant region on BTA6 from θπ test (Figure 1B).
Three methods (XP-CLR, Fst and θπ ratio) were used to analysis the whole genome data and choose the significant signals between Yanbian and N’Dama cattle. Twelve genes (RBFOX1, CD36, GRXCR2, KCNB2, NSG2, ROBO1, NRXN1, LINGO2, GRM5, and AMOTL1) were overlapped among XP-CLR, Fst, and θπ ratio. Among all, CD36 plays an important role in membrane transport of long-chain fatty acid (FA) in the heart, skeletal muscle, and adipose tissue (Glatz et al., 2010). The expression of CD36 is increased in cold exposure, which enhances BAT uptake of TG-rich lipoprotein (TRL) and of albumin bound FA (Bartelt et al., 2011). Previous studies have shown that CD36 gene in Hanwoo and Yanbian cattle affects the intramuscular fat deposition (Jeong et al., 2012). Also, the expression of CD36 has been proven to be positively correlated with obesity in dairy cows (Prodanović et al., 2016). Compared with N’Dama, Yanbian cattle have excellent meat quality. Yanbian cattle have been in the cold environment for a long time, and have a lot of fat deposits in their bodies, which is helpful to resist the cold. There are some pathways [fat digestion and absorption, AMPK signaling pathway, phagosome, extracellular matrix receptors (ECM)-receptor interaction] represented in Yanbian and N’Dama cattle include CD36 genes (Figures 2A–C). Studies have shown that fat digestion and absorption pathway affects the heat production of animals, CD36 gene plays an indispensable role in the heat production (Putri et al., 2015). Also, the expression of CD36 has been reported to be positively correlated with obesity in dairy cows (Prodanović et al., 2016).
As far as we know, Yanbian cattle as beef cattle is rich in muscle fat. The phenomenon of fat abundance in muscle of Yanbian cattle may be one of the mechanisms for resisting cold climate. A missense mutation (c.638A > G, p.Lys 213Arg) was been found between Yanbian and N’Dama cattle, we speculate that this may be one of the causes of Yanbian cattle cold tolerance.
Data Availability Statement
The bioproject number of the sequencing data information about Yanbian cattle is PRJNA565271 in the NCBI Sequence Read Archive. The sequenced N’Dama cattle genomes in this study are publicly available from GenBank with the Bioproject accession number PRJNA312138.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Northwest A&F University following the recommendation of the Regulations for the Administration of Affairs Concerning Experimental Animals of China.
Author Contributions
JS and QH contributed equally towards the construction and execution of this manuscript. YC and YY helped in sample collection, CL revised the manuscript and provided with the valuable suggestion. GZ and YZ contributed in the funding for the research.
Funding
This work was supported by Natural Science Foundation of China (No. 31872317), the Program of National Beef Cattle and Yak Industrial Technology System (No. CARS-37).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00094/full#supplementary-material
References
Allen, J. A. (1877). The influence of physical conditions in the genesis of species. Rad. Rev. 1, 108–140.
Bartelt, A., Bruns, O. T., Reimer, R., Hohenberg, H., Ittrich, H., Peldschus, K., et al. (2011). Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 17, 200–205. doi: 10.1038/nm.2297
Cannon, B., Nedergaard, J. (2004). Brown adipose tissue: function and physiological significance.Physiol. Rev. 84, 277–359. doi: 10.1152/physrev.00015.2003
Cannon, B., Nedergaard, J. (2011). Nonshivering thermogenesis and its adequate measurement in metabolic studies. J. Exp. Biol. 214, 242–253. doi: 10.1242/jeb.050989
Chen, H., Patterson, N., Reich, D. (2010). Population differentiation as a test for selective sweeps.Genome Res. 20, 393–402. doi: 10.1101/gr.100545.109
Chen, N., Cai, Y., Chen, Q., Li, R., Wang, K., Huang, Y., et al. (2018). Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 9, 2337. doi: 10.1038/s41467-018-04737-0
Choi, J.-W., Liao, X., Park, S., Jeon, H.-J., Chung, W.-H., Stothard, P., et al. (2013). Massively parallel sequencing of Chikso (Korean brindle cattle) to discover genome-wide SNPs and In Dels. Mol. Cells 36, 203–211. doi: 10.1007/s10059-013-2347-0
Choi, J., Choi, B., Lee, S., Lee, S., Kim, H., Yu, D., et al. (2015). Whole-genome resequencing analysis of hanwoo and yanbian cattle to identify genome-wide SNPs and signatures of selection. Mol. Cells 38, 466–473. doi: 10.14348/molcells.2015.0019
Daetwyler, H. D., Capitan, A., Pausch, H., Stothard, P., Binsbergen, R. V., Brøndum, R. F., et al. (2014). Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 46, 858–865. doi: 10.1038/ng.3034
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
De Lorenzo, F., Sharma, V., Scully, M., Kakkar, V. V. (1999). Cold adaptation and the seasonal distribution of acute myocardial infarction. Int. J. Med. 92, 747–751. doi: 10.1093/qjmed/92.12.747
Dierks, C., Mömke, S., Philipp, U., Distl, O. (2013). Allelic heterogeneity of FGF 5 mutations causes the long-hair phenotype in dogs. Anim. Genet. 44, 425–431. doi: 10.1111/age.12010
Drögemüller, C., Rüfenacht, S., Wichert, B., Leeb, T. (2007). Mutations within the FGF5 gene are associated with hair length in cats. Anim. Genet. 38, 218–221. doi: 10.1111/j.1365-2052.2007.01590.x
Dronjak, S., Gavrilović, L., Filipović, D., Radojčić, M. B, behavior (2004). Immobilization and cold stress affect sympatho–adrenomedullary system and pituitary–adrenocortical axis of rats exposed to long-term isolation and crowding. Physiol. Behav. 81, 409–415. doi: 10.1016/j.physbeh.2004.01.011
Glatz, J. F., Luiken, J. J., Bonen, A. (2010). Membrane fatty acid transporters as regulators of lipid metabolism: implications for metabolic disease. Physiol. Behav. 90, 367–417. doi: 10.1152/physrev.00003.2009
Grandin, T. (1997). Assessment of stress during handling and transport. J. Anim. Sci.75, 249–257. doi: 10.2527/1997.751249x
Hangalapura, B. N., Nieuwland, M. G. B., Buyse, J., Kemp, B., Parmentier, H. K. (2004). Effect of duration of cold stress on plasma adrenal and thyroid hormone levels and immune responses in chicken lines divergently selected for antibody responses. Poult. Sci. 83, 1644–1649. doi: 10.1093/ps/83.10.1644
Higgins, C. A., Petukhova, L., Harel, S., Ho, Y. Y., Drill, E., Shapiro, L., et al. (2014). FGF5 is a crucial regulator of hair length in humans. PNAS 111, 10648–10653. doi: 10.1073/pnas.1402862111
Jeong, J., Kwon, E., Im, S., Seo, K., Baik, M. (2012). Expression of fat deposition and fat removal genes is associated with intramuscular fat content in longissimus dorsi muscle of Korean cattle steers. J. Anim. Sci. 90, 2044–2053. doi: 10.2527/jas.2011-4753
Ji, S., Yang, R., Lu, C., Qiu, Z., Yan, C., Zhao, Z. (2014). Differential expression of PPARγ, FASN, and ACADM genes in various adipose tissues and longissimus dorsi muscle from yanbian yellow cattle and yan yellow cattle. J. Anim. Sci. 27, 10–18. doi: 10.5713/ajas.2013.13422
Karim, L., Takeda, H., Lin, L., Druet, T., Arias, J. A., Baurain, D., et al. (2011). Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nat. Genet. 43, 405–413. doi: 10.1038/ng.814
Kim, J., Hanotte, O., Mwai, O. A., Dessie, T., Bashir, S., Diallo, B., et al. (2017). The genome landscape of indigenous African cattle. Genome Biol. 18, 34. doi: 10.1186/s13059-017-1153-y
Lee, H.-J., Kim, J., Lee, T., Son, J. K., Yoon, H.-B., Baek, K.-S., et al. (2014). Deciphering the genetic blueprint behind Holstein milk proteins and production.Genome Biol. Evol. 6, 1366–1374. doi: 10.1093/gbe/evu102
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data.Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Mormède, P., Andanson, S., Aupérin, B., Beerda, B., Guémené, D., Malmkvist, J., et al. (2007). Exploration of the hypothalamic–pituitary–adrenal function as a tool to evaluate animal welfare.Physiol. Behav. 92, 317–339. doi: 10.1016/j.physbeh.2006.12.003
Ndlovu, T., Chimonyo, M., Okoh, A. I., Muchenje, V. (2008). A comparison of stress hormone concentrations at slaughter in Nguni, Bonsmara and Angus steers.African J. Agric. Res. 3, 096–100.
Nielsen, R., Williamson, S., Kim, Y., Hubisz, M. J., Clark, A. G., Bustamante, C. (2005). Genomic scans for selective sweeps using SNP data.Genome Res. 15, 1566–1575. doi: 10.1101/gr.4252305
Oleksyk, T. K., Smith, M. W., O'Brien, S. J. (2010). Genome-wide scans for footprints of natural selection. Physiol. Trans. R. Soc. B.365, 185–205. doi: 10.1098/rstb.2009.0219
Prodanović, R., Korićanac, G., Vujanac, I., Djordjević, A., Pantelić, M., Romić, S., et al. (2016). Obesity-driven prepartal hepatic lipid accumulation in dairy cows is associated with increased CD36 and SREBP-1 expression. Res. Vet. Sci. 107, 16–19. doi: 10.1016/j.rvsc.2016.04.007
Putri, M., Syamsunarno, M. R. A., Iso, T., Yamaguchi, A., Hanaoka, H., Sunaga, H., et al. (2015). CD36 is indispensable for thermogenesis under conditions of fasting and cold stress. Biochem. Biophys. Res. Commun. 457, 520–525. doi: 10.1016/j.bbrc.2014.12.124
Qanbari, S., Simianer, H. (2014). Mapping signatures of positive selection in the genome of livestock. Livestock Sci. 166, 133–143. doi: 10.1016/j.livsci.2014.05.003
Sterky, F. H., Trotter, J. H., Lee, S., Recktenwald, C. V., Du, X., Zhou, B., et al. (2017). Carbonic anhydrase-related protein CA10 is an evolutionarily conserved pan-neurexin ligand. PNAS 114, 201621321. doi: 10.1073/pnas.1621321114
Stothard, P., Liao, X., Arantes, A. S., De Pauw, M., Coros, C., Plastow, G., et al. (2015). A large and diverse collection of bovine genome sequences from the Canadian Cattle Genome Project. GigaScience 4, 49–49. doi: 10.1186/s13742-015-0090-5
Vitti, J. J., Grossman, S. R., Sabeti, P. C. (2013). Detecting natural selection in genomic data. Annu. Rev. Genet. 47, 97. doi: 10.1146/annurev-genet-111212-133526
Xin, J., Zhang, L., Li, Z., Liu, X., Jin, H., Yan, C. (2010). Association of polymorphisms in the calpain I gene with meat quality traits in yanbian yellow cattle of china. Asian-Australasian J. Anim. Sci. 24, 9–16. doi: 10.5713/ajas.2011.90407
Keywords: cold adaptability, positive selective, CD36, CORT, FGF5
Citation: Shen J, Hanif Q, Cao Y, Yu Y, Lei C, Zhang G and Zhao Y (2020) Whole Genome Scan and Selection Signatures for Climate Adaption in Yanbian Cattle. Front. Genet. 11:94. doi: 10.3389/fgene.2020.00094
Received: 26 October 2019; Accepted: 28 January 2020;
Published: 25 February 2020.
Edited by:
Fabyano Fonseca Silva, Universidade Federal de Viçosa, BrazilReviewed by:
Lingyang Xu, Chinese Academy of Agricultural Sciences, ChinaMarielle Moura Baena, Universidade Federal de Lavras, Brazil
Copyright © 2020 Shen, Hanif, Cao, Yu, Lei, Zhang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yumin Zhao, eXVtaW56aGFvQDEyNi5jb20=
†These authors have contributed equally to this work
 Jiafei Shen
Jiafei Shen Quratulain Hanif
Quratulain Hanif Yang Cao1
Yang Cao1 Chuzhao Lei
Chuzhao Lei