- State Key Laboratory of Rice Biology/Chinese National Center for Rice Improvement, China National Rice Research Institute, Hangzhou, China
In rice, the contents of protein and amino acids are the major parameters of nutritional quality. Co-localization of quantitative trait loci (QTLs) for heading date and protein content were reported, but pleiotropism of heading-date genes on protein contents has not been investigated. Here, we reported that rice florigen gene RFT1 plays an important role in controlling amino acid contents of rice grain. Firstly, 73 QTLs for the contents of 17 amino acids in unmilled rice were detected using recombinant inbred lines (RILs) of the indica rice cross Zhenshan 97 (ZS97)/Milyang 46 (MY46). Then, the effect of the largest cluster consisting of 14 QTLs, located in proximity to the rice florigen genes RFT1 and Hd3a, was validated using three populations consisting of near isogenic lines (NILs) that only segregated a region covering the target QTL. The first and second NIL populations were derived from a residual heterozygote identified from the ZS97/MY46 RIL population, consisting of homozygous lines that were only segregated in a 29.9-kb region covering the two florigen genes and a 1.7-kb region for RFT1, respectively. The third NIL population was segregated for the RFT1ZS97 transgene in the background of japonica rice cultivar Zhonghua 11. In all the three NIL populations, RFT1 was shown to have a strong effect on the contents of most amino acids, with the ZS97 allele always having the reducing effects. By comparing QTLs for amino acid contents detected in the ZS97/MY46 RIL population and genes/QTLs previously identified for heading date difference between ZS97 and MY46, possible pleiotropism on amino acid contents was also shown for other key heading-date genes including Hd1, Ghd7, and OsGI.
Introduction
Rice sustains about half of the world's population, providing a source of energy and protein. Protein content (PC) of the rice grain is influenced by both genotype and growing environment. The PC values in the un-milled (brown) and milled rice of a large collection of rice cultivars were found to range as 5.6−11.2 and 6.0−15.7%, respectively, with a high correlation coefficient of 0.96 between the two measurements (Chen et al., 2006). In addition to the quantity of total protein, contribution of different amino acids is also an important factor determining the nutritional value of rice grain (Wang et al., 2008). An understanding of the genetic basis underlying the variation of the total grain protein content and the contribution of individual amino acids has the potential to facilitate the breeding of rice cultivars having nutritionally superior grain.
A number of attempts have been made to identify the genetic architecture of the spectrum of grain amino acids in rice by means of quantitative trait locus (QTL) analysis. Using recombinant inbred lines (RILs) derived from a cross between rice cultivars Zhenshan 97 (ZS97) and Nanyangzhan, 18 QTL clusters for 19 components of the amino acid content (AAC) in milled rice were identified (Wang et al., 2008). In two other RIL populations derived from crosses using ZS97 as the female parent, ZS97/Minghui 63 (Lu et al., 2009) and ZS97/Delong 208 (Zhong et al., 2011), 5 and 29 QTL regions for 10 and 17 components of AAC in milled rice were detected, respectively. Many more studies examined the total protein content without analysis on individual amino acids. A region covering the Wx locus on the short arm of chromosome 6 was frequently found to be associated with PC (Tan et al., 2001; Aluko et al., 2004; Wada et al., 2006; Yu et al., 2009; Kashiwagi and Munakata, 2018), but its influence on AAC was only occasionally observed (Wang et al., 2008; Lu et al., 2009; Zhong et al., 2011). Whether the Wx gene itself or other linked genes are involved in the genetic control of PC and AAC remains to be determined.
In rice, the short arm of chromosome 6 is a region harboring multiple genes that play critical roles in the regulation of heading date (HD), including Hd1, Hd3a, RFT1, and Hd17/Hd3b (Hori et al., 2016). In a number of segregating populations that derived from intra-subspecies or inter-species crosses, negative correlation between HD and PC was observed (Wada et al., 2006; Kwon et al., 2011; Yun et al., 2016), which could be partially ascribed to a QTL region on the short arm of chromosome 6 that affected both HD and PC with opposite allelic directions (Wada et al., 2006; Yun et al., 2016). These results suggest that one or more heading-date genes located in this region may have pleiotropic effects on the contents of proteins and amino acids. In the present study, QTL analysis for 17 components of AAC in unmilled rice was performed using the ZS97/Milyang 46 (MY46) RIL population, followed by the validation of a QTL cluster on the short arm of chromosome 6 using three populations of near isogenic lines (NILs) in either indica or japonica backgrounds. A total of 73 QTLs were detected, and the RFT1 gene was found to have a strong and stable pleiotropic effect on AAC.
Material and Methods
Plant Materials
Four segregating populations of rice (Oryza sativa L.) were used in this study. One was a primary mapping population consisting of 247 RILs developed from a cross between indica rice cultivars ZS97 and MY46. The other three were NIL populations segregating a region involving the RFT1 gene in an isogenic background, all of which have been reported by Zhu et al. (2017). Two of the NIL populations, namely TF6-15 and R1, were derived from a residual heterozygote (RH) of ZS97/MY46. An F10 plant was selected, which was heterozygous in a 29.9-kb region covering the RFT1 and Hd3a loci and homozygous in other regions. The S1 plants were assayed with DNA markers located in the segregating region. Homozygous plants were selfed to produce NILs. The TF6-15 population was established, consisting of 10 lines of ZS97 homozygotes and 10 lines of MY46 homozygotes differing in the 29.9 kb region. New RHs were identified from heterozygous progeny of the F10 plant. An F14 plant that was heterozygous at the RFT1 locus only was selected. The R1 population was constructed, comprising 20 lines of ZS97 homozygotes and 20 lines of MY46 homozygotes differing for the RFT1 gene only. The remaining NIL population consisted of 28 homozygous transgenic lines in the genetic background of japonica rice cultivar Zhonghua 11 (ZH11), of which 14 lines carried the RFT1ZS97 transgene and the others carried no transgene.
Field Experiments
The four populations were planted in the single-cropping rice season at the China National Rice Research Institute in Hangzhou, Zhejiang, China. The RIL population was raised in 2015, 2016, and 2017, and the other three populations were grown in 2017 only. Each line was represented by 12 plants per row, with an inter-plant spacing of 16.7 cm and an inter-row spacing of 26.7 cm. The RILs were grown without replication, while the other populations were represented by two replicates. Plants were managed using standard agricultural practice.
Determination of Amino Acid Content
Grain was bulked from the middle ten plants of each row and dried to a moisture content of ~12%. De-hulling was achieved using THU-35A testing husker (Satake Engineering Co. Ltd., Hiroshima, Japan). The dehusked grain was ground using a Cyclotec 1093 Sample Mill (Tecator, Hoganas, Sweden) and the resulting flour was passed through a 0.42 mm sieve. A 250 mg batch of each flour sample was sealed in a vial containing 10 ml 6.0 M HCl and held at 110°C for 24 h. The resulting hydrolysate was diluted to 50 ml with deionized water and filtered. A 0.2 ml aliquot of the filtrate was transferred to a 2 ml tube and evaporated down to ~0.1 ml by bubbling nitrogen gas. After the addition of 2 ml 20 mM HCl, the solution was passed through a 0.2 μm Acrodisc membrane (Pall Corp., Port Washington, NY, USA). The amino acid content of each sample was acquired using a L8900 amino acid auto analyzer (Hitachi, High-Technology Corporation, Tokyo, Japan). Percentage contributions of each of the 17 amino acids were obtained using Ezchrom Elite software (High-Technology Corporation, Tokyo, Japan).
The amino acids studied were aspartic acid (Asp), threonine (Thr), serine (Ser), glutamic acid (Glu), glycine (Gly), alanine (Ala), cystine (Cys), valine (Val), methionine (Met), isoleucine (Ile), leucine (Leu), tyrosine (Tyr), phenylalanine (Phe), lysine (Lys), histidine (His), arginine (Arg), and proline (Pro). Each sample was analyzed in triplicate. Every analytical batch included a blank (20 mM HCl only) and a GBW (E) 100010 reference sample (Chinese National Research Center for Certified Reference Materials, Beijing, China), except that the reference samples for Asp and Cys were not available. Concentrations in the standard samples of Ile, Leu, Tyr, Phe, Lys, His, Arg, Thr, Ser, Glu, Pro, Gly, Ala, Met, and Val measured based on six replicates were 0.38 ± 0.060, 0.74 ± 0.040, 0.30 ± 0.020, 0.52 ± 0.068, 0.28 ± 0.012, 0.22 ± 0.012, 0.49 ± 0.048, 0.30 ± 0.014, 0.49 ± 0.014, 3.28 ± 0.18, 0.95 ± 0.010, 0.40 ± 0.048, 0.503 ± 0.026, 0.165 ± 0.046, and 0.50 ± 0.024%, respectively, matching well with the Certified Reference Material's values. The day-to-day reproducibility of the assay checked over three days was satisfactory, having the relative standard deviation lower than 5.0% for all the fifteen amino acids.
Quantitative Trait Locus Mapping
Genetic map of the ZS97/MY46 RIL population was previously constructed, consisting of 256 markers and spanning 1,814.7 cM (Wang et al., 2017b). This map was applied for QTL analysis using composite interval mapping (CIM) and multiple interval mapping (MIM) in Windows QTL Cartographer v2.5 (Wang et al., 2011). A candidate QTL was identified with CIM using a threshold of logarithm of odds (LOD) > 2.0 and then evaluated with MIM using the Bayesian Information Criterion c (n) = ln (n). A putative QTL was claimed if it satisfied both criteria. QTLs were designated as proposed by McCouch and CGSNL (2008). Two-way analysis of variance (ANOVA) was conducted to test the differences between the two genotypic groups in each of the three NIL populations, using a general linear model (GLM) of the SAS Program as described by Dai et al. (2008).
Results
Variation of Amino Acid Contents in the ZS97/MY46 Recombinant Inbred Line Population
Variation of the 17 components of AAC in unmilled rice of the ZS97/MY46 RIL population is summarized in Table 1. There was a strong evidence for transgressive segregation in both directions for all the amino acids except His of which the contents of the RILs were all lower than the high-parental value in 2016. It was also shown that differences between the two parental lines varied greatly across the 3 years. Among the contents of the 17 amino acids, ZS97 was found to be the high-value parent for two amino acids only (Phe and His) in 2015, but ZS97 was the high-value parent for seven amino acids (Ser, Glu, Leu, Tyr, Phe, Lys, and His) in 2016 and for 14 amino acids (Asp, Thr, Ser, Glu, Gly, Ala, Val, Ile, Leu, Tyr, Phe, Lys, His, and Arg) in 2017.
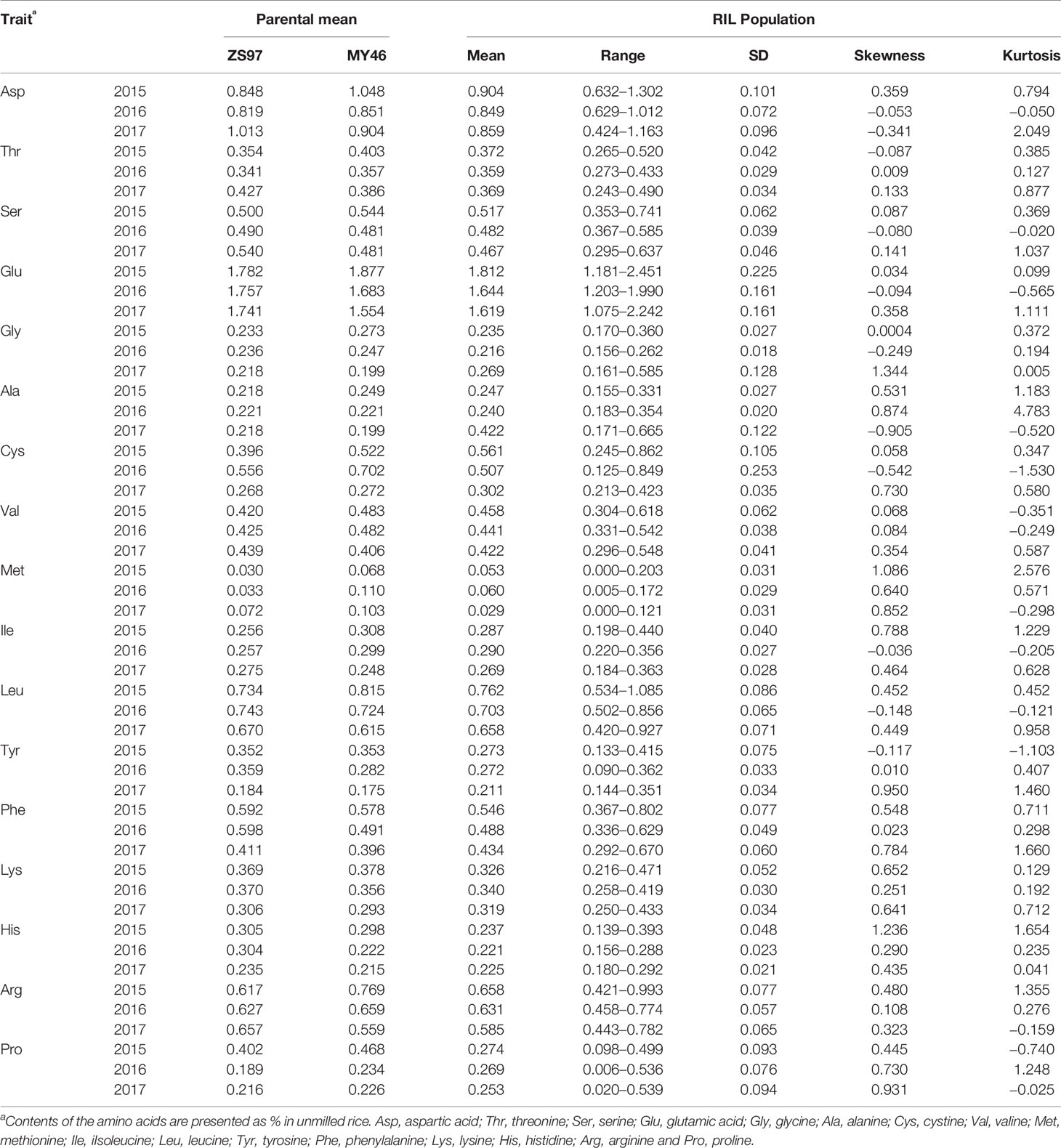
Table 1 Phenotype performance of 17 components of amino acid content in the ZS97/MY46 recombinant inbred line (RIL) population.
Pearson correlation coefficients between the 17 components of AAC were calculated using mean values over 3 years and data of each year. Family error rates were controlled by dividing the P value of 0.05 by 17, thus a threshold of P < 0.003 was used for declaring a significant correlation. It was found that a large majority of the correlations were positively significant. Of the 136 estimates produced from the mean values, 114 were positively significant, 4 were negatively significant, and 18 were non-significant (Table 2). The four negative correlations occurred between Ala and Gly, Cys, Met, and Pro. The 18 non-significant correlations included nine between Ala and others (Ser, Glu, Val, Ile, Leu, Tyr, Phe, Lys, and Arg), 8 between Cys and others (Asp, Thr, Gly, Val, Met, Lys, His, and Pro), and 1 between Tyr and Met. Common occurrence of significantly positive correlations between different AAC components were also observed when data of each year were used (Supplementary Table S1). In 2015, 101 correlations were positively significant and the other 35 were non-significant. In 2016, 122 correlations were positively significant and the other 14 were non-significant. In 2017, 118 correlations were positively significant, three were negatively significant, and 15 were non-significant.
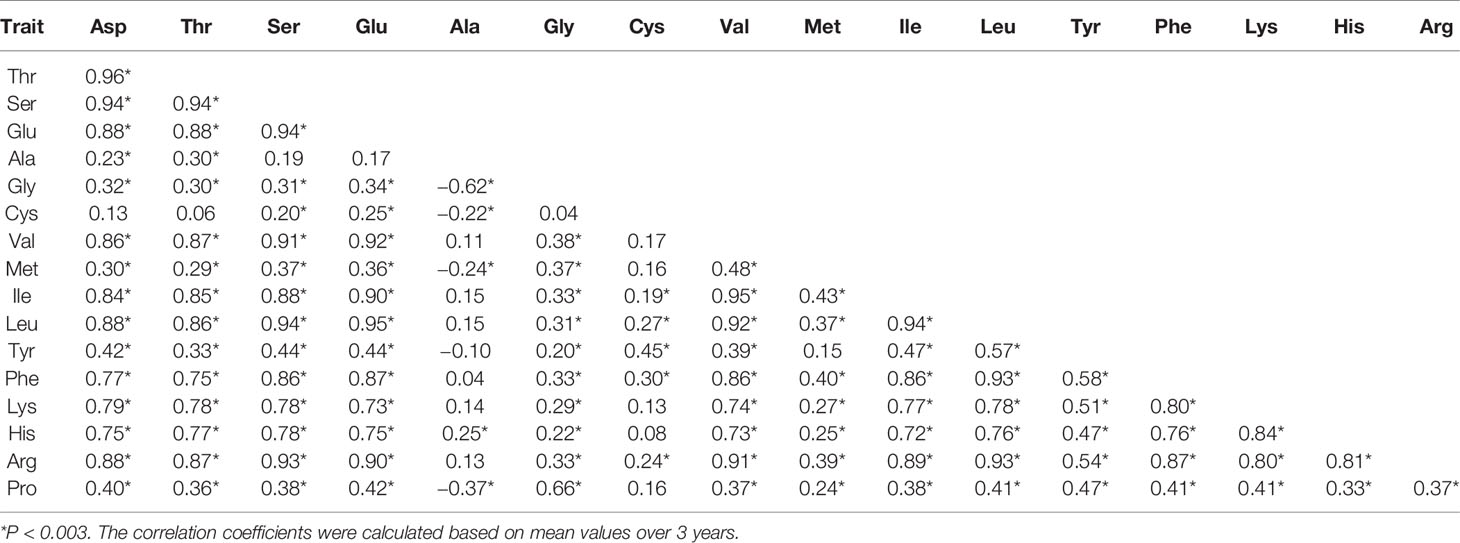
Table 2 Pearson correlation coefficients between 17 components of AAC in the ZS97/MY46 recombinant inbred line (RIL) population.
Quantitative Trait Loci for Amino Acid Contents Detected in the ZS97/MY46 Recombinant Inbred Line Population
A total of 73 QTLs were detected based on 3-year's data of the 17 components of AAC in the ZS97/MY46 RIL population (Supplementary Table S2). Of these QTLs, seven were identified in all the 3 years, eight were found in 2 years, and the others were detected in 1 year only. The number of QTLs detected for each amino acid ranged from two to eight, with the proportion of the variance explained (R2) by a single QTL ranging from 2.2 to 35.9%. These QTL were distributed over all the 12 rice chromosomes except chromosome 5 (Figure 1). Most of them were located in cluster, with chromosomes 1, 6, and 7 harboring the highest number of loci. Of the 15 QTLs detected in 2 or 3 years, 12, 2, and 1 were located in chromosomes 6, 7, and 11, respectively. Except qThr11, allelic directions of these QTLs all remained consistent across different years.

Figure 1 Chromosomal locations of quantitative trait loci for 17 components of amino acid contents detected in the recombinant inbred line population of Zhenshan 97/Milyang 46.
Fourteen QTLs were located in the RM190–RM6917 region on the short arm of chromosome 6, forming the largest cluster in terms of QTL number. Included were seven QTLs detected in three years (qAsp6, qSer6, qGly6, qLeu6, qPhe6, qHis6, and qArg6), five QTLs detected in 2 years (qThr6, qGlu6, qVal6, qMet6, and qTyr6), and two QTLs detected in 1 year (qLys6 and qPro6.1). Enhancing alleles of these QTLs were all derived from the male parent MY46, with qGly6 having the highest R2 of 33.9%. Two other QTLs (qAla6 and qPro6.2) were detected in nearby intervals RM253–RM276 and RZ667–RM19784, respectively, of which the enhancing alleles were both derived from the female parent ZS97. Altogether, 16 QTLs were detected on chromosome 6.
The second largest cluster consisting of nine QTLs was located in the RM3325–RM3859 region on the short arm of chromosome 7. Included were two QTLs detected in 2 years (qAsp7 and qArg7) and seven QTLs detected in 1 year (qThr7, qSer7, qGly7.1, qAla7, qVal7, qTyr7.1, and qPhe7). Enhancing alleles of these QTLs were all derived from ZS97, with qAsp7 having the highest R2 of 19.8%. Five other QTLs were clustered in the RZ471–RZ395 region on the long arm of this chromosome. The enhancing alleles were derived from ZS97 at qTyr7.2 and qPro7.1, and from MY46 at qGly7.2, qCys7, and qPro7.2. The five QTLs had high R2 ranging from 10.7 to 35.9%. Altogether, 14 QTLs were detected on chromosome 7.
The third largest cluster consisting eight QTLs (qSer1, qVal1, qIle1, qLeu1, qTyr1, qLys1.1, qHis1, and qPro1.1) was located in the RM283–RM3746 region on the short arm of chromosome 1. Enhancing alleles of these QTLs were all derived from ZS97, with qLeu1 having the highest R2 of 15.0%. Three other QTLs (qAsp1, qGly1, and qPro1.2) were located in the pericentromeric region of chromosome 1. They had high R2 ranging from 17.0 to 32.8%, and the enhancing alleles were all derived from ZS97. Two sparsely-distributed QTLs (qPro1.3 and qLys1.2) were located in lower regions of the long arm. Altogether, 13 QTLs were detected on chromosome 1.
Among the remaining 30 QTLs, two single QTL were located on chromosomes 8 and 10, respectively, and the others were distributed on chromosomes 2, 3, 4, 9, 11, and 12 with 2–6 QTLs per chromosome. The six QTLs on chromosome 2 were all located in the lower region of the long arm; the two QTLs on chromosome 3 were tightly linked; the six QTLs on chromosome 4 involved two pairs of tightly-linked QTLs with two nearby QTLs; the five QTLs on chromosome 9 may be viewed as one cluster and two single QTL; the five QTLs on chromosome 11 was separated into two clusters; and the four QTLs on chromosome 12 included one cluster and one single QTL.
Effect of RFT1 on Amino Acid Contents Detected Between NILZS97 and NILMY46
As described above, the largest QTL cluster for the 17 components of AAC detected in the ZS97/MY46 RIL population was located in the RM190–RM6917 region on the short arm of chromosome 6. This region covered the two florigen genes of rice, RFT1 and Hd3a (Hori et al., 2016), suggesting a possible involvement of RFT1 and/or Hd3a in controlling AAC of rice grain. This assumption was firstly tested using the NIL population TF6-15 segregating a 29.9-kb interval covering both RFT1 and Hd3a. Significant differences (P < 0.05) between the 10 homozygous lines of NILZS97 and 10 homozygous lines of NILMY46 were detected on 15 of the 17 components of AAC (Table 3). The R2 for individual components ranged from 16.7 to 61.2%. The enhancing alleles were all derived from MY46, which is in agreement with the effects detected in the ZS97/MY46 RIL population.
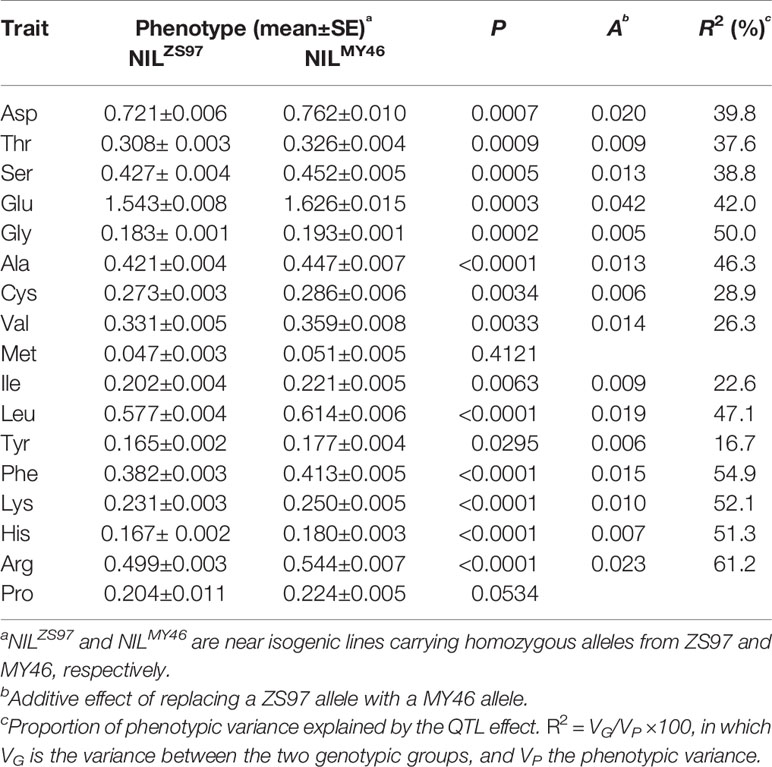
Table 3 Effects of the RFT1-Hd3a region on 17 components of amino acid content (AAC) detected in the TF6-15 population.
Then, QTL analysis was performed using the NIL population R1 that was homozygous at the Hd3a locus but segregated for the RFT1 gene. Significant differences (P < 0.05) between the 20 homozygous lines of NILZS97 and 20 homozygous lines of NILMY46 were detected on 15 of the 17 components of AAC (Table 4). The R2 for individual components ranged from 9.5 to 63.2%. Again, the enhancing alleles were all derived from MY46. It is also noted that the two components showing no significant difference between NILZS97 and NILMY46 were commonly found to be Met and Pro in the TF6-15 and R1 populations. These results indicate that the RFT1 gene has a strong and stable effect on most components of AAC in unmilled rice.
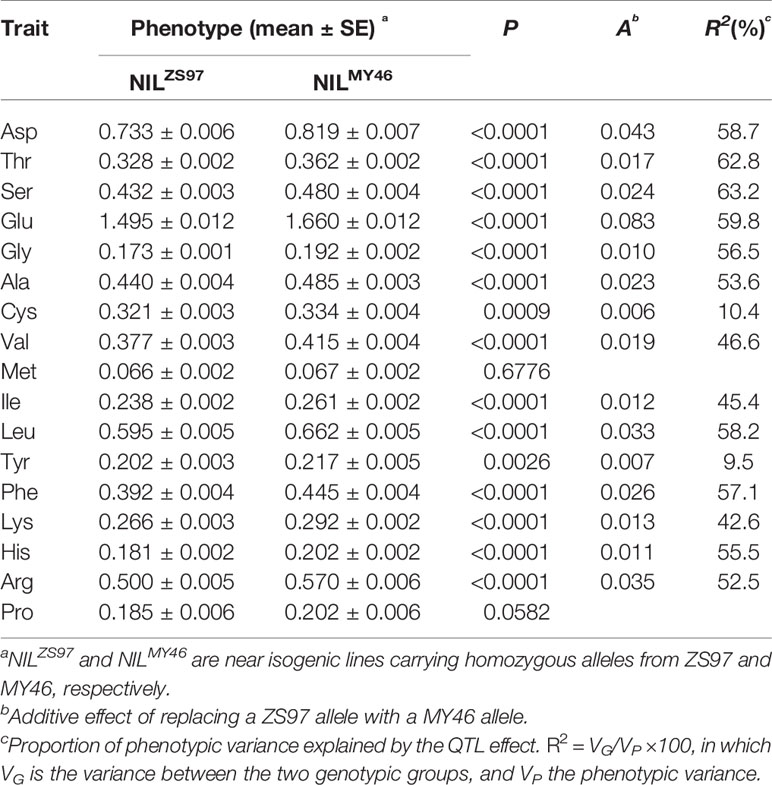
Table 4 Effects of the RFT1 gene on 17 components of amino acid content (AAC) detected in the R1 population.
Effect of the RFT1ZS97 Transgene on Amino Acid Contents in a Japonica Rice Background
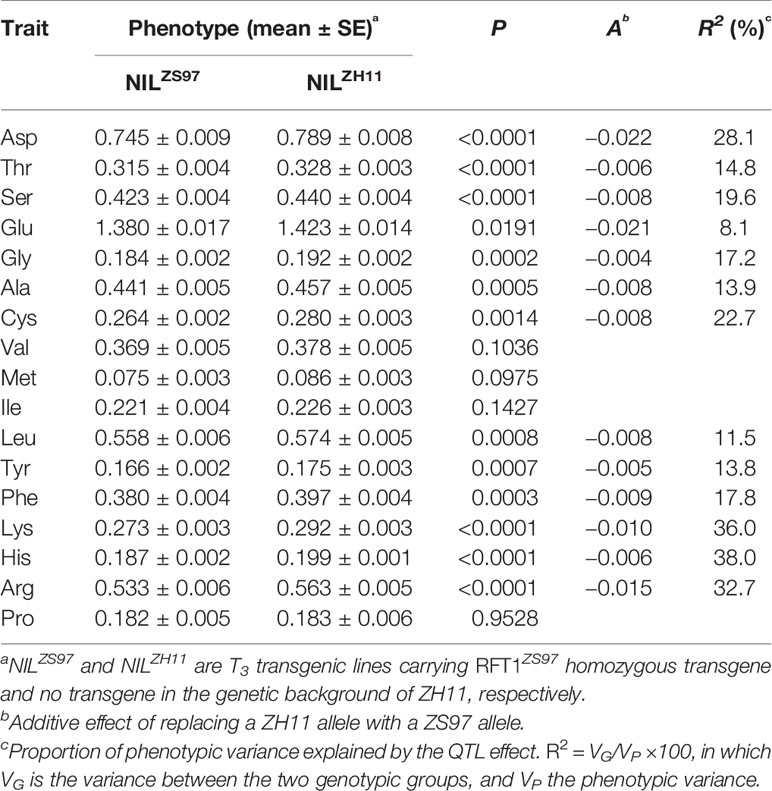
The effect of RFT1 on AAC in unmilled rice was further tested using a transgenic population segregating the RFT1ZS97 transgene in the genetic background of japonica cultivar ZH11. Significant differences (P < 0.05) between the 14 lines of NILZS97 carrying homozygous transgenes and 14 lines of NILZH11 carrying no transgene were detected on 13 of the 17 components of AAC, with R2 ranging from 8.1 to 38.0% (Table 5). Integration of the RFT1ZS97 transgene into the genome of ZH11 reduced the contents of the amino acids. In addition, the two components showing no significant difference between NILZS97 and NILMY46 in the TF6-15 and R1 populations, Met and Pro, were included in the four components having no significant difference between NILZS97 and NILZH11 in the transgenic population. These results indicate that the effects of RFT1 on AAC of unmilled rice are consistent in the genetic background of different subspecies of Asian cultivated rice.
Discussion
Heading date, grain yield, and grain quality are three basic traits influencing the commercial utilization of a rice cultivar. The regional and seasonal adaptation is mostly determined by heading date, the productivity is measured by grain yield, and whether the product can meet the demand of end-users is mainly characterized by grain quality. A number of key genes for flowering regulation in rice have been found to play important roles in the genetic control of yield traits, including Ghd7 (Xue et al., 2008; Weng et al., 2014), DTH8/Ghd8 (Wei et al., 2010; Yan et al., 2011), Hd1 (Zhang et al., 2012; Zhang et al., 2015; Ye et al., 2018), Ghd7.1 (Yan et al., 2013), and RFT1 (Zhu et al., 2017). On the other hand, no study has been reported for the pleiotropic effects of heading-date genes on grain quality in rice. Among traits in the four primary categories of rice grain quality (i.e., milling, appearance, eating and cooking, and nutritional qualities), PC and AAC are the major parameters of nutritional quality (Wang et al., 2008; Wang et al., 2017a). In the present study, a total of 73 QTLs for AAC of unmilled rice were detected using the ZS97/MY46 RIL population, and the largest QTL cluster was validated to be responsible by the RFT1 gene on the short arm of chromosome 6. It is also evident that the effect of RFT1 is consistent across different genetic backgrounds. In accordance with the common occurrence of significantly positive correlations between different components of AAC (Table 2; Supplementary Table S1), most of the QTLs were located in cluster and different QTL in a given region usually had the same allelic direction (Figure 1; Supplementary Table S2).
RFT1 protein is the florigen for promoting the flowering of rice under long-day (LD) conditions (Tsuji et al., 2011). As compared to the ZS97 allele of RFT1, the MY46 and ZH11 alleles were shown to promote heading in rice populations grown under natural LD conditions in Hangzhou (Zhu et al., 2017). Replacing a ZS97 allele by a MY46 allele in the R1 population promoted flowering by 11.63 to 15.61 days over 3 years; and replacing a ZS97 allele by a ZH11 allele promoted flowering by 3.91 and 6.12 days in the transgenic population in 2 years. In the present study, replacement of the ZS97 allele of RFT1 with the MY46 and ZH11 alleles resulted in increasing the contents of most amino acids (Supplementary Table S2; Tables 3–5). Obviously, the effects of RFT1 on HD and AAC have opposite allelic directions, which is in accordance with the opposite allelic directions between QTLs for HD and PC located on the short arm of chromosome 6 (Wada et al., 2006; Yun et al., 2016).
Two other QTLs for AAC (qAla6 and qPro6.2) were detected on the short of chromosome 6 in the ZS97/MY46 RIL population. They were located in the intervals RM253–RM276 and RZ667–RM19784 that are closer to the centromere region than is RFT1. The alleles for increasing AAC were both derived from ZS97 (Supplementary Table S2). At the Hd1 locus that is tightly linked to RM19784, the functional Hd1ZS97 allele acted to decrease HD as compared to the non-functional Hd1MY46 allele in the ZS97 background (Zhang et al., 2012). These results suggest that the Hd1 gene also have pleiotropic effects on HD and AAC with opposite directions.
The second and third largest QTL clusters detected in the ZS97/MY RIL population were located in the RM3325–RM3859 and RM283–RM3746 regions on the short art of chromosomes 7 and 1, covering the heading-date genes Ghd7 (Xue et al., 2008) and OsGI (Hayama et al., 2003), respectively. For all the QTLs included in the two clusters, the alleles for increasing AAC were derived from ZS97 (Supplementary Table S2). In previous studies using the same rice cross, the ZS97 alleles in the two regions were found to decrease HD (Zhang et al., 2011; Zhang et al., 2016). These results suggest that the pleiotropic effects of major heading-date genes on AAC with opposite directions could be a common occurrence.
Similar to previous results reported by other groups (Wang et al., 2008; Lu et al., 2009; Zhong et al., 2011), our study found that it is common that a QTL region affected most components of AAC. However, it is unlikely that a gene can affect the biosynthesis of most amino acids in rice. Given that all the major QTL regions affecting AAC detected in this study were located in approximate to genes/QTLs controlling flowering time, these regions would have large effects on all traits which were influenced by heading date. It is possible that the influence of these regions on most components of AAC could be caused by indirect effects of heading date genes rather than by the direct control of these genes on the biosynthesis of amino acids. It is possible that a heading-date gene is involved in controlling nutrient transportation and accumulation in rice, either by direct involvement in the regulating network or by environmental influences on the nutrition uptake and transport due to heading date variation.
Utilization of the pleiotropic effects of heading-date genes on AAC and PC could help to meet the diverse requirements of protein for human consumption. High contents of protein and amino acids are favorable for enhancing the nutritional value, but unfavorable for eating quality (Martin and Fitzgerald, 2002; Kwon et al., 2011; Yun et al., 2016) and undesirable for some uses such as wine-making (Yoshida et al., 2002) and certain types of diet (Zhang et al., 2008). Alleles for promoting HD and increasing AAC could be selected for developing rice varieties with high nutritional values; and alleles for delaying heading and reducing AAC may be applied for developing high-yielding varieties with good eating quality. In this regard, more efforts are needed to establish a better understanding on the pleiotropism of heading-date genes on multiple traits for grain quality.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
J-YZ and P-SH conceived and designed the experiments. L-HX, Y-JZ, S-QT, X-JW, Z-HS, and G-AJ performed the experiments. L-HX, Y-JZ and J-YZ analysed the data. L-HX, Y-JZ and J-YZ wrote the manuscript.
Funding
This work was funded by the National Key R&D Program of China (Grant number 2017YFD0100300), and the National Natural Science Foundation of China (Grant number 31571637).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mr. Hongfa Chen for rice material preparation and Mrs. Lingli Sun for preparation of amino acid content analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00013/full#supplementary-material
References
Aluko, G., Martinez, C., Tohme, J., Castano, C., Bergman, C., Oard, J. H. (2004). QTL mapping of grain quality traits from the interspecific cross Oryza sativa × O. glaberrima. Theor. Appl. Genet. 109, 630−639. doi: 10.1007/s00122-004-1668-y
Chen, N., Luo, Y. K., Xie, L. H., Zhu, Z. W., Duan, B. W., Zhang, L. P. (2006). Protein content and its correlation with other quality parameters of rice in China. Acta Agron. Sinica. 32, 1193−1196. (in Chinese with English abstract). doi: 10.3321/j.issn:0496-3490.2006.08.015
Dai, W. M., Zhang, K. Q., Wu, J. R., Wang, L., Duan, B. W., Zheng, K. L., et al. (2008). Validating a segment on the short arm of chromosome 6 responsible for genetic variation in the hull silicon content and yield traits of rice. Euphytica 160, 317−324. doi: 10.1007/s10681-007-9501-8
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Hori, K., Matsubara, K., Yano, M. (2016). Genetic control of flowering time in rice: integration of Mendelian genetics and genomics. Theor. Appl. Genet. 129, 2241–2252. doi: 10.1007/s00122-016-2773-4
Kashiwagi, T., Munakata, J. (2018). Identification and characteristics of quantitative trait locus for grain protein content, TGP12, in rice (Oryza sativa L.). Euphytica 214, 165. doi: 10.1007/s10681-018-2249-5
Kwon, S. W., Cho, Y. C., Lee, J. H., Suh, J. P., Kim, J. J., Kim, M. K., et al. (2011). Identification of quantitative trait loci associated with rice eating quality traits using a population of recombinant inbred lines derived from a cross between two temperate japonica cultivars. Mol. Cells 31, 427−445. doi: 10.1007/s10059-011-0289-y
Lu, K., Li, L., Zheng, X., Zhang, Z., Mou, T., Hu, Z. (2009). Genetic dissection of amino acid content in rice grain. J. Sci. Food Agric. 89, 2377−2382. doi: 10.1002/jsfa.3731
Martin, M., Fitzgerald, M. A. (2002). Proteins in rice influence cooking properties. J. Cereal Sci. 36, 285−294. doi: 10.1006/jcrs.2001.0465
McCouch, S. R., CGSNL (Committee on Gene Symbolization, Nomenclature and Linkage, Rice Genetics Cooperative) (2008). Gene nomenclature system for rice. Rice 1, 72−84. doi: 10.1007/s12284-008-9004-9
Tan, Y. F., Sun, M., Xing, Y. Z., Hua, Z. P., Sun, X. L., Zhang, Q. F., et al. (2001). Mapping quantitative trait loci for milling quality, protein content and color characteristics of rice using a recombinant inbred line population derived from an elite rice hybrid. Theor. Appl. Genet. 103, 1037−1045. doi: 10.1007/s001220100665
Tsuji, H., Taoka, K. I., Shimamoto, K. (2011). Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14, 45−52. doi: 10.1016/j.pbi.2010.08.016
Wada, T., Uchimura, Y., Ogata, T., Tsubone, M., Matsue, Y. (2006). Mapping of QTLs for physicochemical properties in japonica rice. Breed. Sci. 56, 253−260. doi: 10.1270/jsbbs.56.253
Wang, L. Q., Zhong, M., Li, X. H., Yuan, D. J., Xu, Y. B., Liu, H. F., et al. (2008). The QTL controlling amino acid content in grains of rice (Oryza sativa) are co-localized with the regions involved in the amino acid metabolism pathway. Mol. Breed. 21, 127−137. doi: 10.1007/s11032-007-9141-7
Wang, S., Basten, C., Zeng, Z. (2011). Windows QTL Cartographer 2.5 (Raleigh, NC: North Carolina State University).
Wang, X., Pang, Y., Zhang, J., Wu, Z., Chen, K., Ali, J., et al. (2017a). Genome-wide and gene-based association mapping for rice eating and cooking characteristics and protein content. Sci. Rep. 7, 17203. doi: 10.1038/s41598-017-17347-5
Wang, Z., Chen, J. Y., Zhu, Y. J., Fan, Y. Y., Zhuang, J. Y. (2017b). Validation of qGS10, a quantitative trait locus for grain size on the long arm of chromosome 10 in rice (Oryza sativa L.). J. Integr. Agr. 16, 16–26. doi: 10.1016/S2095-3119(16)61410-7
Wei, X., Xu, J., Guo, H., Jiang, L., Chen, S., Yu, C., et al. (2010). DTH8 Suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153, 1747–1758. doi: 10.1104/pp.110.156943
Weng, X., Wang, L., Wang, J., Hu, Y., Du, H., Xu, C., et al. (2014). Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 164, 735–747. doi: 10.1104/pp.113.231308
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yan, W. H., Wang, P., Chen, H. X., Zhou, H. J., Li, Q. P., Wang, C. R., et al. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330. doi: 10.1093/mp/ssq070
Yan, W., Liu, H., Zhou, X., Li, Q., Zhang, J., Lu, L., et al. (2013). Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 23, 969–971. doi: 10.1038/cr.2013.43
Ye, J., Niu, X., Yang, Y., Wang, S., Xu, Q., Yuan, X., et al. (2018). Divergent Hd1, Ghd7, and DTH7 alleles control heading date and yield potential of japonica rice in Northeast China. Front. Plant Sci. 9, 35. doi: 10.3389/fpls.2018.00035
Yoshida, S., Ikegami, M., Kuze, J., Sawada, K., Hashimoto, Z., Ishii, T., et al. (2002). QTL analysis for plant and grain characters of sake-brewing rice using a double haploid population. Breed. Sci. 52, 309–317. doi: 10.1270/jsbbs.52.309
Yu, Y. H., Li, G., Fan, Y. Y., Zhang, K. Q., Min, J., Zhu, Z. W., et al. (2009). Genetic relationship between grain yield and the contents of protein and fat in a recombinant inbred population of rice. J. Cereal Sci. 50, 121−125. doi: 10.1016/j.jcs.2009.03.008
Yun, Y. T., Chung, C. T., Lee, Y. J., Na, H. J., Lee, J. C., Lee, S. G., et al. (2016). QTL Mapping of grain quality traits using introgression lines carrying Oryza rufipogon chromosome segments in japonica rice. Rice 9, 62. doi: 10.1186/s12284-016-0135-0
Zhang, W., Bi, J., Chen, L., Zheng, L., Ji, S., Xia, Y., et al. (2008). QTL mapping for crude protein and protein fraction contents in rice (Oryza sativa L.). J. Cereal Sci. 48, 539–547. doi: 10.1016/j.jcs.2007.11.010
Zhang, Z. H., Guo, L., Zhu, Y. J., Fan, Y. Y., Zhuang, J. Y. (2011). Mapping of quantitative trait loci for heading date and plant height in two populations of indica rice. Sci. Agric. Sin. 44, 3069−3077. (in Chinese with English abstract). doi: 10.3864/j.issn.0578-1752.2011.15.001
Zhang, Z. H., Wang, K., Guo, L., Zhu, Y. J., Fan, Y. Y., Cheng, S. H., et al. (2012). Pleiotropism of the photoperiod-insensitive allele of Hd1 on heading date, plant height and yield traits in rice. PloS One 7, e52538. doi: 10.1371/journal.pone.0052538
Zhang, J., Zhou, X., Yan, W., Zhang, Z., Lu, L., Han, Z., et al. (2015). Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 208, 1056–1066. doi: 10.1111/nph.13538
Zhang, Z. H., Cao, L. Y., Chen, J. Y., Zhang, Y. X., Zhuang, J. Y., Cheng, S. H. (2016). Effects of Hd2 in the presence of the photoperiod-insensitive functional allele of Hd1 in rice. Biol. Open 5, 1719–1726. doi: 10.1242/bio.021071
Zhong, M., Wang, L. Q., Yuan, D. J., Luo, L. J., Xu, C. G., He, Y. Q. (2011). Identification of QTL affecting protein and amino acid contents in rice. Rice Sci. 18, 187−195. doi: 10.1016/S1672-6308(11)60026-7
Keywords: amino acid content, heading date, near isogenic line, pleiotropic effect, quantitative trait locus, Oryza sativa L.
Citation: Xie L-H, Zhu Y-J, Tang S-Q, Wei X-J, Sheng Z-H, Jiao G-A, Hu P-S and Zhuang J-Y (2020) Pleiotropic Effects of Rice Florigen Gene RFT1 on the Amino Acid Content of Unmilled Rice. Front. Genet. 11:13. doi: 10.3389/fgene.2020.00013
Received: 22 July 2019; Accepted: 06 January 2020;
Published: 31 January 2020.
Edited by:
Balram Marathi, Professor Jayashankar Telangana State Agricultural University, IndiaReviewed by:
Ashok Kumar Singh, Indian Agricultural Research Institute (ICAR), IndiaKazuki Matsubara, Institute of Crop Science (NARO), Japan
Copyright © 2020 Xie, Zhu, Tang, Wei, Sheng, Jiao, Hu and Zhuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei-Song Hu, cXVhbGl0eWhAMTYzLmNvbQ==; Jie-Yun Zhuang, emh1YW5namlleXVuQGNhYXMuY24=
†These authors have contributed equally to this work
 Li-Hong Xie†
Li-Hong Xie† Yu-Jun Zhu
Yu-Jun Zhu Jie-Yun Zhuang
Jie-Yun Zhuang